Effects of Moringa oleifera Leaf Extract on Diabetes-Induced Alterations in Paraoxonase 1 and Catalase in Rats Analyzed through Progress Kinetic and Blind Docking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Preparation of the Extract
2.2. Ethics Statement
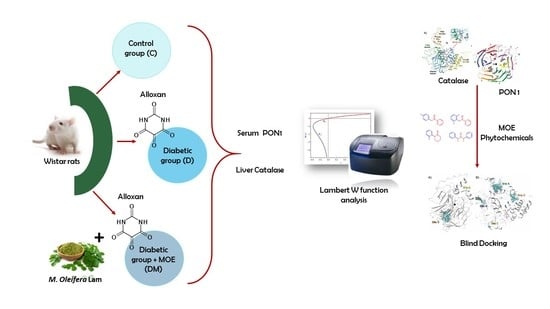
2.3. Diabetic Model and Treatment
2.4. Separation of Serum
2.5. Isolation of Liver Cytosol
2.6. Paraoxonase 1 Activity
2.7. Catalase Activity
2.8. Protein Determination
2.9. Kinetics Data Processing
2.10. Rat Paraoxonase 1 and Catalase Homology Modeling
2.11. Molecular Docking
2.12. Statistical Analysis
3. Results and Discussion
3.1. Effect of M. oleifera Leaves Extract on rPON1 and rCAT Kinetics Parameters
3.2. In-Silico Identification of Potential Interactions between rPON1 and rCAT with Phytochemicals of M. oleifera
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sherwani, S.I.; Khan, H.; Ekhzaimy, A.; Masood, A.; Sakharkar, M.K. Significance of HbA1c Test in Diagnosis and Prognosis of Diabetic Patients. Biomark. Insights 2016, 11, 95–104. [Google Scholar] [CrossRef] [PubMed]
- Milani, E.; Nikfar, S.; Khorasani, R.; Zamani, M.J.; Abdollahi, M. Reduction of diabetes-induced oxidative stress by phosphodiesterase inhibitors in rats. Comp. Biochem. Physiol. Part C Toxicol. Pharmacol. 2005, 140, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Ugochukwu, N.H.; Babady, N.E.; Cobourne, M.; Gasset, S.R. The effect of Gongronema latifolium extracts on serum lipid profile and oxidative stress in hepatocytes of diabetic rats. J. Biosci. 2003, 28, 1–5. [Google Scholar] [CrossRef]
- Cournot, M.; Burillo, E.; Saulnier, P.; Planesse, C.; Gand, E.; Rehman, M.; Ragot, S.; Rondeau, P.; Catan, A.; Gonthier, M.; et al. Circulating Concentrations of Redox Biomarkers Do Not Improve the Prediction of Adverse Cardiovascular Events in Patients with Type 2 Diabetes Mellitus. J. Am. Heart Assoc. 2018, 7, e007397. [Google Scholar] [CrossRef] [Green Version]
- Veskoukis, A.S.; Paschalis, V.; Kyparos, A.; Nikolaidis, M.G. Administration of exercise-conditioned plasma alters muscle catalase kinetics in rat: An argument for in vivo-like Km instead of in vitro-like V max. Redox Boil. 2018, 15, 375–379. [Google Scholar] [CrossRef]
- Sözmen, E.Y.; Sözmen, B.; Delen, Y.; Onat, T. Catalase/superoxide dismutase (SOD) and catalase/paraoxonase (PON) ratios may implicate poor glycemic control. Arch. Med. Res. 2001, 32, 283–287. [Google Scholar] [CrossRef]
- Klejdus, B.; Zehnalek, J.; Adam, V.; Petřek, J.; Kizek, R.; Vacek, J.; Trnková, L.; Rozik, R.; Havel, L.; Kubán, V. Sub-picomole high-performance liquid chromatographic/mass spectrometric determination of glutathione in the maize (Zea mays L.) kernels exposed to cadmium. Anal. Chim. Acta 2004, 520, 117–124. [Google Scholar] [CrossRef]
- Furlong, C.E.; Marsillach, J.; Jarvik, G.; Costa, L.G. Paraoxonases-1, -2 and -3: What are their functions? Chem. Biol. Interact. 2016, 259, 51–62. [Google Scholar] [CrossRef] [Green Version]
- Mackness, M.I.; Mackness, B.; Durrington, P.N. Paraoxonase and coronary heart disease. Atheroscler. Suppl. 2002, 3, 49–55. [Google Scholar] [CrossRef]
- Mackness, M.I.; Durrington, P.N.; Mackness, B. The Role of Paraoxonase 1 Activity in Cardiovascular Disease. Am. J. Cardiovasc. Drugs 2004, 4, 211–217. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, X.; Zhang, Y.; Wang, Y.; Liu, Y.; Sun, R.; Xia, M. Anthocyanin Supplementation Improves HDL-Associated Paraoxonase 1 Activity and Enhances Cholesterol Efflux Capacity in Subjects with Hypercholesterolemia. J. Clin. Endocrinol. Metab. 2014, 99, 561–569. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farid, A.S.; Mido, S.; Linh, B.K.; Hayashi, T.; Horii, Y. An atherogenic lipid profile with low serum paraoxonase-1 activity during nematode infection in rats. Eur. J. Clin. Investig. 2010, 40, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Oda, M.N.; Bielicki, J.K.; Ho, T.T.; Berger, T.; Rubin, E.M.; Forte, T.M. Paraoxonase 1 Overexpression in Mice and Its Effect on High-Density Lipoproteins. Biochem. Biophys. Res. Commun. 2002, 290, 921–927. [Google Scholar] [CrossRef]
- Flekač, M.; Škrha, J.; Zídková, K.; Lacinová, Z.; Hilgertová, J. Paraoxonase 1 gene polymorphisms and enzyme activities in diabetes mellitus. Physiol. Res. 2008, 57, 717–726. [Google Scholar] [PubMed]
- Suvarna, R.; Rao, S.S.; Joshi, C.; Kedage, V.; Muttigi, M.S.; Shetty, J.K.; Prakash, M. Paraoxonase activity in type 2 diabetes mellitus patients with and without complications. J. Clin. Diagn. Res. 2011, 5, 63–65. [Google Scholar]
- Karakaya, P.; Mert, M.; Okuturlar, Y.; Gedikbaşi, A.; Islim, F.; Harmankaya, Ö.; Kumbasar, A. Relation of MPV with serum PON-1 activity and BA-IMT in diabetic patients with respect to obesity and diabetic complications. Diabetes Res. Clin. Pract. 2016, 120, S183. [Google Scholar] [CrossRef]
- Nicholls, P. Classical catalase: Ancient and modern. Arch. Biochem. Biophys. 2012, 525, 95–101. [Google Scholar] [CrossRef]
- Vendemiale, G.; Grattagliano, I.; Altomare, E. An update on the role of free radicals and antioxidant defense in human disease. Int. J. Clin. Lab. Res. 1999, 29, 49. [Google Scholar] [CrossRef]
- Takemoto, K.; Tanaka, M.; Iwata, H.; Nishihara, R.; Ishihara, K.; Wang, D.-H.; Ogino, K.; Taniuchi, K.; Masuoka, N. Low catalase activity in blood is associated with the diabetes caused by alloxan. Clin. Chim. Acta 2009, 407, 43–46. [Google Scholar] [CrossRef]
- Sigfrid, L.; Cunningham, J.; Beeharry, N.; Lortz, S.; Tiedge, M.; Lenzen, S.; Carlsson, C.; Green, I. Cytokines and nitric oxide inhibit the enzyme activity of catalase but not its protein or mRNA expression in insulin-producing cells. J. Mol. Endocrinol. 2003, 31, 509–518. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Janocha, A.J.; Aronica, M.A.; Swaidani, S.; Comhair, S.A.; Xu, W.; Zheng, L.; Kaveti, S.; Kinter, M.; Hazen, S.L.; et al. Nitrotyrosine Proteome Survey in Asthma Identifies Oxidative Mechanism of Catalase Inactivation. J. Immunol. 2006, 176, 5587–5597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rashtbari, S.; Khataee, S.; Iranshahi, M.; Moosavi-Movahedi, A.A.; Hosseinzadeh, G.; Dehghan, G. Experimental investigation and molecular dynamics simulation of the binding of ellagic acid to bovine liver catalase: Activation study and interaction mechanism. Int. J. Boil. Macromol. 2020, 143, 850–861. [Google Scholar] [CrossRef]
- Rashtbari, S.; Dehghan, G.; Yekta, R.; Jouyban, A. Investigation of the binding mechanism and inhibition of bovine liver catalase by quercetin: Multi-spectroscopic and computational study. Bioimpacts Bi 2017, 7, 147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sánchez-Muñoz, M.A.; Valdez-Solana, M.A.; Campos-Almazán, M.I.; Flores-Herrera, O.; Esparza-Perusquía, M.; Olvera-Sanchez, S.; García-Arenas, G.; Avitia-Domínguez, C.; Tellez-Valencia, A.; Sierra-Campos, E. Streptozotocin-Induced Adaptive Modification of Mitochondrial Supercomplexes in Liver of Wistar Rats and the Protective Effect of Moringa oleifera Lam. Biochem. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Ma, G.; Wang, Y.; Zhang, Y. Moringa oleifera leaf flavonoids protect bovine mammary epithelial cells from hydrogen peroxide-induced oxidative stress in vitro. Reprod. Domest. Anim. 2020, 55, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Aekthammarat, D.; Pannangpetch, P.; Tangsucharit, P. Moringa oleifera leaf extract induces vasorelaxation via endothelium-dependent hyperpolarization and calcium channel blockade in mesenteric arterial beds isolated from L-NAME hypertensive rats. Clin. Exp. Hypertens. 2020, 42, 490–501. [Google Scholar] [CrossRef]
- Mohamed, M.A.; Ahmed, M.A.; El Sayed, R.A. Molecular effects of Moringa leaf extract on insulin resistance and reproductive function in hyperinsulinemic male rats. J. Diabetes Metab. Disord. 2019, 18, 487–494. [Google Scholar] [CrossRef]
- Waterman, C.; Rojas-Silva, P.; Tumer, T.B.; Kuhn, P.; Richard, A.J.; Wicks, S.; Stephens, J.M.; Wang, Z.; Mynatt, R.; Cefalu, W.; et al. Isothiocyanate-rich Moringa oleifera extract reduces weight gain, insulin resistance, and hepatic gluconeogenesis in mice. Mol. Nutr. Food Res. 2015, 59, 1013–1024. [Google Scholar] [CrossRef] [Green Version]
- Mbikay, M. Therapeutic Potential of Moringa oleifera Leaves in Chronic Hyperglycemia and Dyslipidemia: A Review. Front. Pharmacol. 2012, 3. [Google Scholar] [CrossRef] [Green Version]
- Vergara-Jimenez, M.; AlMatrafi, M.M.; Fernandez, M.L. Bioactive Components in Moringa Oleifera Leaves Protect against Chronic Disease. Antioxidants 2017, 6, 91. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, J.; Khan, I.; Blundell, R. Moringa oleifera and glycemic control: A review of current evidence and possible mechanisms. Phytother. Res. 2019, 33, 2841–2848. [Google Scholar] [CrossRef] [PubMed]
- Vongsak, B.; Mangmool, S.; Gritsanapan, W. Antioxidant Activity and Induction of mRNA Expressions of Antioxidant Enzymes in HEK-293 Cells of Moringa oleifera Leaf Extract. Planta Medica 2015, 81, 1084–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- AbdElazem, H. Effect of Moringa oleifera on antioxidant enzymes and oxidative stress induced by aluminium exposure in male albino rat testes. Int. J. Cancer Biomed. Res. 2019, 4, 34–41. [Google Scholar] [CrossRef]
- Kesavulu, M.M.; Giri, R.; Rao, B.K.; Apparao, C. Lipid peroxidation and antioxidant enzyme levels in type 2 diabetics with microvascular complications. Diabetes Metab. 2000, 26, 387–392. [Google Scholar]
- Parthiban, A.; Vijayalingam, S.; Shanmugasundaram, K.R.; Mohan, R. Oxidative stress and the development of diabetic complications-antioxidants and lipid peroxidation in erythrocytes and cell membrane. Cell Boil. Int. 1995, 19, 987–994. [Google Scholar] [CrossRef]
- Peuchant, E.; Delmas-Beauvieux, M.-C.; Dubourg, L.; Thomas, M.-J.; Perromat, A.; Aparicio, M.; Clerc, M.; Combe, C. Antioxidant Effects of a Supplemented Very Low Protein Diet in Chronic Renal Failure. Free Radic. Boil. Med. 1997, 22, 313–320. [Google Scholar] [CrossRef]
- Bäuerle, F.; Zotter, A.; Schreiber, G. Direct determination of enzyme kinetic parameters from single reactions using a new progress curve analysis tool. Protein Eng. Des. Sel. 2017, 30, 151–158. [Google Scholar] [CrossRef] [Green Version]
- Goličnik, M.; Bavec, A. Evaluation of the paraoxonase-1 kinetic parameters of the lactonase activity by nonlinear fit of progress curves. J. Enzym. Inhib. Med. Chem. 2020, 35, 261–264. [Google Scholar] [CrossRef] [Green Version]
- Kitchen, D.B.; Decornez, H.; Furr, J.R.; Bajorath, J. Docking and scoring in virtual screening for drug discovery: Methods and applications. Nat. Rev. Drug Discov. 2004, 3, 935–949. [Google Scholar] [CrossRef]
- Pendota, S.; Ndhlala, A.; Aremu, A.; Aderogba, M.; Van Staden, J. Anti-inflammatory, antioxidant and in silico studies of Buddleja salviifolia (L). Lam leaf constituents. S. Afr. J. Bot. 2014, 93, 79–85. [Google Scholar] [CrossRef] [Green Version]
- Rana, S.; Dixit, S.; Mittal, A. In Silico Target Identification and Validation for Antioxidant and Anti-inflammatory Activity of Selective Phytochemicals. Braz. Arch. Boil. Technol. 2019, 62. [Google Scholar] [CrossRef]
- Ighodaro, O.M.; Adeosun, A.M.; Akinloye, O.A. Alloxan-induced diabetes, a common model for evaluating the glycemic-control potential of therapeutic compounds and plants extracts in experimental studies. Medicana (B. Aires) 2017, 53, 365–374. [Google Scholar] [CrossRef] [PubMed]
- Munday, R. Dialuric acid autoxidation: Effects of transition metals on the reaction rate and on the generation of “active oxygen” species. Biochem. Pharmacol. 1988, 37, 409–413. [Google Scholar] [CrossRef]
- Dell, R.B.; Holleran, S.; Ramakrishnan, R. Sample size determination. ILAR J. 2002, 43, 207–213. [Google Scholar] [CrossRef]
- University Boston Sample Size Calculations (IACUC). Available online: https://www.bu.edu/researchsupport/compliance/animal-care/working-with-animals/research/sample-size-calculations-iacuc/#sample-size-calculations-for-a-difference-in-means (accessed on 5 August 2019).
- Aebi, H. Catalase In Vitro. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 1984; Volume 105, pp. 121–126. [Google Scholar]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Boil. Chem. 1951, 193, 265–275. [Google Scholar]
- Nielsen, M.; Lundegaard, C.; Lund, O.; Petersen, T.N. CPHmodels-3.0—Remote homology modeling using structure-guided sequence profiles. Nucleic Acids Res. 2010, 38, W576–W581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwede, T.; Kopp, J.; Guex, N.; Peitsch, M.C. SWISS-MODEL: An automated protein homology-modeling server. Nucleic Acids Res. 2003, 31, 3381–3385. [Google Scholar] [CrossRef] [Green Version]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J.E. The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [Green Version]
- Eswar, N.; Eramian, D.; Webb, B.; Shen, M.-Y.; Sali, A. Protein Structure Modeling with MODELLER. In Advanced Structural Safety Studies; Springer: Berlin, Germany, 2008; Volume 426, pp. 145–159. [Google Scholar]
- Harel, M.; Aharoni, A.; Gaidukov, L.; Brumshtein, B.; Khersonsky, O.; Meged, R.; Dvir, H.; Ravelli, R.B.G.; McCarthy, A.; Toker, L.; et al. Structure and evolution of the serum paraoxonase family of detoxifying and anti-atherosclerotic enzymes. Nat. Struct. Mol. Boil. 2004, 11, 412–419. [Google Scholar] [CrossRef]
- Chen, V.B.; Arendall, W.B.; Headd, J.J.; Keedy, D.A.; Immormino, R.M.; Kapral, G.J.; Murray, L.W.; Richardson, J.S.; Richardson, D.C. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. Sect. D Boil. Crystallogr. 2010, 66, 12–21. [Google Scholar] [CrossRef] [Green Version]
- Benkert, P.; Biasini, M.; Schwede, T. Toward the estimation of the absolute quality of individual protein structure models. Bioinformatics 2011, 27, 343–350. [Google Scholar] [CrossRef] [PubMed]
- DeLano, W.L. The PyMOL Molecular Graphics System; DeLano Scientific: San Carlos, CA, USA, 2002. [Google Scholar]
- Fita, I.; Rossmann, M.G. The NADPH binding site on beef liver catalase. Proc. Natl. Acad. Sci. USA 1985, 82, 1604–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besler, B.H.; Merz, K.M.; Kollman, P.A. Atomic charges derived from semiempirical methods. J. Comput. Chem. 1990, 11, 431–439. [Google Scholar] [CrossRef]
- Forli, S.; Huey, R.; Pique, M.E.; Sanner, M.F.; Goodsell, D.S.; Olson, A.J. Computational protein–ligand docking and virtual drug screening with the AutoDock suite. Nat. Protoc. 2016, 11, 905–919. [Google Scholar] [CrossRef] [Green Version]
- Kim, S.; Thiessen, P.A.; Bolton, E.; Chen, J.; Fu, G.; Gindulyte, A.; Han, L.; He, J.; He, S.; Shoemaker, B.A.; et al. PubChem Substance and Compound databases. Nucleic Acids Res. 2016, 44, D1202–D1213. [Google Scholar] [CrossRef]
- Sanner, M.F. Python: A programming language for software integration and development. J. Mol. Graph. Model. 1999, 17, 57–61. [Google Scholar]
- Trott, O.; Olson, A.J. AutoDock Vina: Improving the Speed and Accuracy of Docking with a New Scoring Function, Efficient Optimization, and Multithreading. J. Comput. Chem. 2010, 31, 455–461. [Google Scholar] [CrossRef] [Green Version]
- O’Boyle, N.M.; Banck, M.; James, C.A.; Morley, C.; Vandermeersch, T.; Hutchison, G.R. Open Babel: An open chemical toolbox. J. Cheminf. 2011, 3, 33. [Google Scholar] [CrossRef] [Green Version]
- Schrödinger Release 2020-1: Maestro, Schrödinger, LLC, New York, NY. 2020. Available online: https://www.schrodinger.com/maestro (accessed on 1 September 2019).
- Zhang, Y.-J.; Gan, R.-Y.; Li, S.; Zhou, Y.; Li, A.-N.; Xu, D.-P.; Li, H.-B. Antioxidant Phytochemicals for the Prevention and Treatment of Chronic Diseases. Molucules 2015, 20, 21138–21156. [Google Scholar] [CrossRef]
- Vargas-Sánchez, K.; Garay-Jaramillo, E.; González-Reyes, R.E. Effects of Moringa oleifera on Glycaemia and Insulin Levels: A Review of Animal and Human Studies. Nutrients 2019, 11, 2907. [Google Scholar] [CrossRef] [Green Version]
- Roskoski, R. Sunitinib: A VEGF and PDGF receptor protein kinase and angiogenesis inhibitor. Biochem. Biophys. Res. Commun. 2007, 356, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Sierra-Campos, E.; A Valdez-Solana, M.; Pérez-Velázquez, J.R.; García-Arenas, G.; Téllez-Valencia, A.; Avitia-Domínguez, C. Moringa oleifera leaves extract regulate the activity of nitric oxide synthases and paraoxonase 1 in diabetic rat. MOJ Biorgan. Org. Chem. 2018, 2, 236–241. [Google Scholar]
- Kıyıcı, A.; Okudan, N.; Gökbel, H.; Belviranlı, M. The Effect of Grape Seed Extracts on Serum Paraoxonase Activities in Streptozotocin-Induced Diabetic Rats. J. Med. Food 2010, 13, 725–728. [Google Scholar] [CrossRef] [PubMed]
- Bosak, A.; Bavec, A.; Konte, T.; Šinko, G.; Kovarik, Z.; Goličnik, M. Interactions of Paraoxonase-1 with Pharmacologically Relevant Carbamates. Molecules 2020, 25, 211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khateeb, J.; Gantman, A.; Kreitenberg, A.J.; Aviram, M.; Fuhrman, B. Paraoxonase 1 (PON1) expression in hepatocytes is upregulated by pomegranate polyphenols: A role for PPAR-γ pathway. Atherosclerosis 2010, 208, 119–125. [Google Scholar] [CrossRef]
- Gong, M.; Garige, M.; Varatharajalu, R.; Marmillot, P.; Gottipatti, C.; Leckey, L.C.; Lakshman, M.R. Quercetin up-regulates paraoxonase 1 gene expression with concomitant protection against LDL oxidation. Biochem. Biophys. Res. Commun. 2009, 379, 1001–1004. [Google Scholar] [CrossRef]
- Mahrooz, A.; Nouri, M.; Rashidi, M.-R. Naringenin is an inhibitor of human serum paraoxonase (PON1): An In Vitro study. J. Clin. Lab. Anal. 2011, 25, 395–401. [Google Scholar] [CrossRef]
- Góth, L. Catalase Deficiency and Type 2 Diabetes. Diabetes Care 2008, 31, e93. [Google Scholar] [CrossRef] [Green Version]
- Vetrano, A.M.; Heck, D.E.; Mariano, T.M.; Mishin, V.; Laskin, D.L.; Laskin, J.D. Characterization of the Oxidase Activity in Mammalian Catalase. J. Boil. Chem. 2005, 280, 35372–35381. [Google Scholar] [CrossRef] [Green Version]
- Moradi, M.; Rahimi, Z.; Amiri, S.; Rahimi, Z.; Vessal, M.; Nasri, H. AT1R A1166C variants in patients with type 2 diabetes mellitus and diabetic nephropathy. J. Nephropathol. 2015, 4, 69. [Google Scholar]
- Panahi, Y.; Yekta, R.; Dehghan, G.; Rashtbari, S.; Baradaran, B.; Jafari, N.J.; Moosavi-Movahedi, A.A. Aspirin in retrieving the inactivated catalase to active form: Displacement of one inhibitor with a protective agent. Int. J. Boil. Macromol. 2019, 122, 306–311. [Google Scholar] [CrossRef]
- Glorieux, C.; Sandoval, J.M.; Fattaccioli, A.; Dejeans, N.; Garbe, J.C.; Dieu, M.; Verrax, J.; Renard, P.; Huang, P.; Calderon, P.B. Chromatin remodeling regulates catalase expression during cancer cells adaptation to chronic oxidative stress. Free Radic. Boil. Med. 2016, 99, 436–450. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.F.; Ahmad, J.; Zhang, H.; Khan, I.; Muhammad, S. Evaluation of phytochemical and medicinal properties of Moringa (Moringa oleifera) as a potential functional food. S. Afr. J. Bot. 2020, 129, 40–46. [Google Scholar] [CrossRef]
- Karaoz, E.; Gultekin, F.; Akdoğan, M.; Oncu, M.; Gökçimen, A. Protective role of melatonin and a combination of vitamin C and vitamin E on lung toxicity induced by chlorpyrifos-ethyl in rats. Exp. Toxicol. Pathol. 2002, 54, 97–108. [Google Scholar] [CrossRef] [PubMed]
- Štefek, M. Natural flavonoids as potential multifunctional agents in prevention of diabetic cataract. Interdiscip. Toxicol. 2011, 4, 69–77. [Google Scholar] [CrossRef] [Green Version]
- Ranilla, L.G.; Kwon, Y.-I.; Apostolidis, E.; Shetty, K. Phenolic compounds, antioxidant activity and in vitro inhibitory potential against key enzymes relevant for hyperglycemia and hypertension of commonly used medicinal plants, herbs and spices in Latin America. Bioresour. Technol. 2010, 101, 4676–4689. [Google Scholar] [CrossRef]
- Lü, J.-M.; Lin, P.H.; Yao, Q.; Chen, C. Chemical and molecular mechanisms of antioxidants: Experimental approaches and model systems. J. Cell. Mol. Med. 2010, 14, 840–860. [Google Scholar] [CrossRef]
- Litvinov, D.; Mahini, H.; Garelnabi, M. Antioxidant and anti-inflammatory role of paraoxonase 1: Implication in arteriosclerosis diseases. N. Am. J. Med Sci. 2012, 4, 523. [Google Scholar]
- Rozenberg, O.; Shih, D.M.; Aviram, M. Paraoxonase 1 (PON1) attenuates macrophage oxidative status: Studies in PON1 transfected cells and in PON1 transgenic mice. Atherosclerosis 2005, 181, 9–18. [Google Scholar] [CrossRef]
- Mohan, Y.; Jesuthankaraj, G.N.; Thangavelu, N.R. Antidiabetic and Antioxidant Properties ofTriticum aestivumin Streptozotocin-Induced Diabetic Rats. Adv. Pharmacol. Sci. 2013, 2013. [Google Scholar] [CrossRef] [Green Version]
- Aju, B.; Rajalakshmi, R.; Mini, S. Corrigendum to “Protective role of Moringa oleifera leaf extract on cardiac antioxidant status and lipid peroxidation in streptozotocin induced diabetic rats” [Heliyon 5, (12), (December 2019), e02935]. Heliyon 2020, 6. [Google Scholar] [CrossRef] [PubMed]
- Omodanisi, E.I.; Aboua, G.Y.; Oguntibeju, O.O. Therapeutic potentials and pharmacological properties of Moringa oleifera Lam in the treatment of diabetes mellitus and related complications. Trop. J. Pharm. Res. 2017, 16, 1737–1746. [Google Scholar] [CrossRef] [Green Version]
- Jaiswal, D.; Rai, P.K.; Mehta, S.; Chatterji, S.; Shukla, S.; Rai, D.K.; Sharma, G.; Sharma, B.; Khair, S.; Watal, G. Role of Moringa oleifera in regulation of diabetes-induced oxidative stress. Asian Pac. J. Trop. Med. 2013, 6, 426–432. [Google Scholar] [CrossRef]
- Blaha-Nelson, D.; Krüger, D.M.; Szeler, K.; Ben-David, M.; Kamerlin, S.C.L. Active Site Hydrophobicity and the Convergent Evolution of Paraoxonase Activity in Structurally Divergent Enzymes: The Case of Serum Paraoxonase 1. J. Am. Chem. Soc. 2017, 139, 1155–1167. [Google Scholar] [CrossRef] [Green Version]
- Murthy, M.R.; Reid, T.J.; Sicignano, A.; Tanaka, N.; Rossmann, M.G. Structure of beef liver catalase. J. Mol. Boil. 1981, 152, 465–499. [Google Scholar] [CrossRef]
- Sierra-Campos, E.; Valdez-Solana, M.A.; Avitia-Domínguez, C.I.; Tellez-Valencia, A.; Meza-Velázquez, J.A.; Aguilera-Ortíz, M.; Escobar-Ramirez, A. Standarization based on chemical markers of Moringa oleifera herbal products using bioautography assay, thin layer chromatography and high performance liquid chromatography-diode array detector. Malays. J. Anal. Sci. 2020, 24, 449–463. [Google Scholar]
- Lin, H.; Zhu, H.; Tan, J.; Wang, H.; Wang, Z.; Li, P.; Zhao, C.; Liu, J. Comparative Analysis of Chemical Constituents of Moringa oleifera Leaves from China and India by Ultra-Performance Liquid Chromatography Coupled with Quadrupole-Time-Of-Flight Mass Spectrometry. Molecules 2019, 24, 942. [Google Scholar] [CrossRef] [Green Version]
- Billecke, S.; Draganov, D.; Counsell, R.; Stetson, P.; Watson, C.; Hsu, C.; La Du, B.N. Human serum paraoxonase (PON1) isozymes Q and R hydrolyze lactones and cyclic carbonate esters. Drug Metab. Dispos. 2000, 28, 1335–1342. [Google Scholar]
- Teiber, J.F.; Draganov, D.I.; La Du, B.N. Lactonase and lactonizing activities of human serum paraoxonase (PON1) and rabbit serum PON3. Biochem. Pharmacol. 2003, 66, 887–896. [Google Scholar] [CrossRef]
- Tavori, H.; Vaya, J.; Aviram, M. Paraoxonase 1 attenuates human plaque atherogenicity: Relevance to the enzyme lactonase activity. In Paraoxonases in Inflammation, Infection, and Toxicology; Springer: Berlin, Germany, 2010; pp. 99–111. [Google Scholar]
- Atrahimovich, D.; Vaya, J.; Tavori, H.; Khateb, S. Glabridin Protects Paraoxonase 1 from Linoleic Acid Hydroperoxide Inhibition via Specific Interaction: A Fluorescence-Quenching Study. J. Agric. Food Chem. 2012, 60, 3679–3685. [Google Scholar] [CrossRef]
- Atrahimovich, D.; Vaya, J.; Khateb, S. The effects and mechanism of flavonoid–rePON1 interactions. Structure–activity relationship study. Bioorganic Med. Chem. 2013, 21, 3348–3355. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.; Zhang, X.; Li, D.; Jin, J. Probing the binding of flavonoids to catalase by molecular spectroscopy. J. Mol. Struct. 2007, 843, 38–44. [Google Scholar] [CrossRef]
- Zapelini, P.H.; Rezin, G.T.; Cardoso, M.R.; Ritter, C.; Klamt, F.; Moreira, J.C.; Streck, E.L.; Dal-Pizzol, F. Antioxidant treatment reverses mitochondrial dysfunction in a sepsis animal model. Mitochondrion 2008, 8, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.-J.; Chen, Y.-C.; Chang, Y.-S. Insight into two antioxidants binding to the catalase NADPH binding site from traditional Chinese medicines. RSC Adv. 2015, 5, 6625–6635. [Google Scholar] [CrossRef]
- Kowalska, K.; Socha, E.; Milnerowicz, H. Review: The role of paraoxonase in cardiovascular diseases. Ann. Clin. Lab. Sci. 2015, 45, 226–233. [Google Scholar] [PubMed]
- Li, R.; Li, J.; Cai, L.; Hu, C.-M.; Zhang, L. Suppression of adjuvant arthritis by hesperidin in rats and its mechanisms. J. Pharm. Pharmacol. 2008, 60, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Shahraki, S.; Delarami, H.S.; Saeidifar, M.; Nejat, R. Catalytic activity and structural changes of catalase in the presence of Levothyroxine and Isoxsuprine hydrochloride. Int. J. Boil. Macromol. 2020, 152, 126–136. [Google Scholar] [CrossRef]
- Krych, J.; Gebicka, L. Catalase is inhibited by flavonoids. Int. J. Boil. Macromol. 2013, 58, 148–153. [Google Scholar] [CrossRef]






| Parameters | Control (C) | Diabetic (D) | Diabetic + MOE (DM) |
|---|---|---|---|
| Km | 0.4146 ± 0.04 * μM | 0.325 ± 0.097 *,† μM | 0.0043 ± 0.0026 μM |
| Vm | 33.15 ± 4.61 U/(min L) | 17.00 ± 0.99 U/(min L) * | 10.32 ± 3.36 U/(min L) *,† |
| Vm/Km | 80.07 ± 2.8 * min−1 | 52.30 ± 9.66 * min−1 | 2400.0 ± 41.74 min−1 |
| Parameters | Control (C) | Diabetic (D) | Diabetic + MOE (DM) |
|---|---|---|---|
| Km | 2.152 ± 0.19 mM | 6.557 ± 0.42 * mM | 60.7 ± 0.03 * mM |
| Km | 652.4 ± 8 U/(min L) | 1074 ± 17 U/(min L) * | 6110 ± 22 U/(min L) * |
| Vm/Km | 303.16 ± 7.8 min−1 | 1637.94 ± 15.63 min−1 * | 1006.58 ± 10.1 min−1 * |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sierra-Campos, E.; Valdez-Solana, M.; Avitia-Domínguez, C.; Campos-Almazán, M.; Flores-Molina, I.; García-Arenas, G.; Téllez-Valencia, A. Effects of Moringa oleifera Leaf Extract on Diabetes-Induced Alterations in Paraoxonase 1 and Catalase in Rats Analyzed through Progress Kinetic and Blind Docking. Antioxidants 2020, 9, 840. https://doi.org/10.3390/antiox9090840
Sierra-Campos E, Valdez-Solana M, Avitia-Domínguez C, Campos-Almazán M, Flores-Molina I, García-Arenas G, Téllez-Valencia A. Effects of Moringa oleifera Leaf Extract on Diabetes-Induced Alterations in Paraoxonase 1 and Catalase in Rats Analyzed through Progress Kinetic and Blind Docking. Antioxidants. 2020; 9(9):840. https://doi.org/10.3390/antiox9090840
Chicago/Turabian StyleSierra-Campos, Erick, Mónica Valdez-Solana, Claudia Avitia-Domínguez, Mara Campos-Almazán, Ismael Flores-Molina, Guadalupe García-Arenas, and Alfredo Téllez-Valencia. 2020. "Effects of Moringa oleifera Leaf Extract on Diabetes-Induced Alterations in Paraoxonase 1 and Catalase in Rats Analyzed through Progress Kinetic and Blind Docking" Antioxidants 9, no. 9: 840. https://doi.org/10.3390/antiox9090840