Molecular Cloning and Functional Characterization of Catalase in Stress Physiology, Innate Immunity, Testicular Development, Metamorphosis, and Cryopreserved Sperm of Pacific Abalone
Abstract
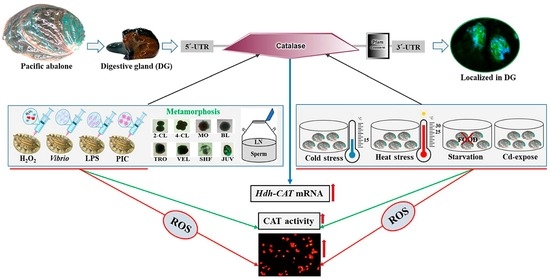
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Experimental Abalone Collection
2.3. Tissue Collection for Gene Cloning
2.4. Tissue Collection of Different Organs of Pacific Abalone
2.5. Fluorescence In Situ Hybridization (FISH) to Localize Hdh-CAT in Digestive Gland Tissue of Pacific Abalone
2.5.1. Tissue Collection for Fluorescence In Situ Hybridization
2.5.2. Riboprobe Synthesis
2.5.3. Preparation of Tissue Sections
2.5.4. Fluorescence In Situ Hybridization (FISH)
2.6. Thermal Stress Experiments
2.6.1. Cold Stress of Pacific Abalone at 15 °C
2.6.2. Heat Stress of Pacific Abalone at 25 °C and 30 °C
2.7. Seasonal Sample Collection
2.8. H2O2 Induction Treatments
2.9. CdCl2 Exposed Treatments
2.10. Tissue Collection of Starved Pacific Abalone
2.11. Immune Challenges of Pacific Abalone to Check the Hdh-CAT Activity and mRNA Abundance
2.12. Tissue Collection of Testicular Developmental Stages
2.13. Sperm Cryopreservation of Pacific Abalone
2.14. Samples Collection of Metamorphosis Stages of Pacific Abalone
2.15. Total RNA Extraction and cDNA Synthesis
2.16. Molecular Cloning and Sequencing of Catalase Gene (Hdh-CAT) in Haliotis discus hannai
2.16.1. Cloning of a Partial Hdh-CAT Sequence
2.16.2. Cloning of the Full-Length Hdh-CAT Sequence
2.17. Sequence Analysis
2.18. Phylogenetic Analysis
2.19. The Three-Dimensional Protein Structure of Hdh-CAT
2.20. Quantitative Real-Time PCR (qRT-PCR) Analysis
2.21. Catalase Activity
2.22. Fluorescent Technique to Detect ROS in DG Tissue Samples of Pacific Abalone
2.23. Statistical Analysis
3. Results
3.1. Cloning and Bioinformatic Analysis of H. discus hannai Catalase (Hdh-CAT) Sequence
3.2. Homology Modeling of Hdh-CAT
3.3. Phylogenetic Analysis
3.4. Tissue Distribution Analysis of Hdh-CAT
3.5. Fluorescence In Situ Hybridization
3.6. Hdh-CAT mRNA Expression in Thermal Stressed Tissue Samples of Pacific Abalone
3.6.1. Hdh-CAT mRNA Expression in Cold Stressed (15 °C) Samples
3.6.2. Hdh-CAT mRNA Expression in Heat Stressed (25 °C) Samples
3.6.3. Hdh-CAT mRNA Expression in Heat Stressed (30 °C) Samples
3.7. Hdh-CAT mRNA Expression in Seasonal Samples
3.8. Hdh-CAT mRNA Expression in H2O2 Induced Tissue Samples of Pacific Abalone
3.9. Hdh-CAT mRNA Expression in CdCl2 Exposed Tissues of Pacific Abalone
3.10. Hdh-CAT mRNA Expression in Starved Tissue Samples of Pacific Abalone
3.11. Hdh-CAT mRNA Expression in Immune Challenged Tissues of Pacific Abalone
3.11.1. Hdh-CAT Expression in Vibrio parahaemolyticus Challenged Samples
3.11.2. Hdh-CAT Expression in Lipopolysaccharides (LPS) Challenged Samples
3.11.3. Hdh-CAT Expression in Poly I:C Challenged Samples
3.12. Hdh-CAT mRNA Expression in Testicular Developmental Stages
3.13. Hdh-CAT mRNA Expression in Cryopreserved Sperm
3.14. Hdh-CAT mRNA Expression in Embryonic and Larval Developmental Stages
3.15. Catalase (CAT) Activity in the Hemolymph
3.16. ROS in DG Tissue Samples of Pacific Abalone
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Vidal-Liñán, L.; Bellas, J.; Campillo, J.A.; Beiras, R. Integrated use of antioxidant enzymes in mussels, Mytilus galloprovincialis, for monitoring pollution in highly productive coastal areas of Galicia (NW Spain). Chemosphere 2010, 78, 265–272. [Google Scholar] [CrossRef] [PubMed]
- Karadag, H.; Fırat, Ö.; Fırat, Ö. Use of oxidative stress biomarkers in Cyprinus carpio L. for the evaluation of water pollution in Ataturk Dam Lake (Adiyaman, Turkey). Bull. Environ. Contam. Toxicol. 2014, 92, 289–293. [Google Scholar] [CrossRef] [Green Version]
- Ekanayake, P.M.; De Zoysa, M.; Kang, H.S.; Wan, Q.; Jee, Y.; Lee, Y.H.; Kim, S.J.; Lee, J. Cloning, characterization and tissue expression of disk abalone (Haliotis discus discus) catalase. Fish Shellfish Immunol. 2008, 24, 267–278. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Tan, K.; Zheng, H. Characterization of a novel catalase gene and its response to low temperature stress in noble scallop Chlamys nobilis with different total carotenoids content. Aquac. Res. 2021, 52, 6179–6188. [Google Scholar] [CrossRef]
- Vetrano, A.M.; Heck, D.E.; Mariano, T.M.; Mishin, V.; Laskin, D.L.; Laskin, J.D. Characterization of the oxidase activity in mammalian catalase. J. Biol. Chem. 2005, 280, 35372–35381. [Google Scholar] [CrossRef] [Green Version]
- Sellaththurai, S.; Priyathilaka, T.T.; Lee, J. Molecular cloning, characterization, and expression level analysis of a marine teleost homolog of catalase from big belly seahorse (Hippocampus abdominalis). Fish Shellfish Immunol. 2019, 89, 647–659. [Google Scholar] [CrossRef]
- Yuan, H.; Jin, C.; Pei, H.; Zhao, L.; Li, X.; Li, J.; Huang, W.; Fan, R.; Liu, W.; Shen, Q.H. The powdery mildew effector CSEP0027 interacts with barley catalase to regulate host immunity. Front. Plant Sci. 2021, 12, 733237. [Google Scholar] [CrossRef]
- Li, C.; Ni, D.; Song, L.; Zhao, J.; Zhang, H.; Li, L. Molecular cloning and characterization of a catalase gene from Zhikong scallop Chlamys farreri. Fish Shellfish Immunol. 2008, 24, 26–34. [Google Scholar] [CrossRef]
- Shaeer, A.; Aslam, M.; Rashid, N. Structural and functional analyses of a novel manganese-catalase from Bacillus subtilis R5. Int. J. Biol. Macromol. 2021, 180, 222–233. [Google Scholar] [CrossRef]
- Galasso, M.; Gambino, S.; Romanelli, M.G.; Donadelli, M.; Scupoli, M.T. Browsing the oldest antioxidant enzyme: Catalase and its multiple regulation in cancer. Free Radic. Biol. Med. 2021, 172, 264–272. [Google Scholar] [CrossRef]
- Chakravarty, D.; Bihani, S.C.; Banerjee, M.; Kalwani, P.; Ballal, A. Unique functional insights into the antioxidant response of the cyanobacterial Mn-catalase (KatB). Free Radic. Biol. Med. 2021, 179, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Wan, F.; Dutta, S.; Welsh, S.; Liu, Z.; Freundt, E.; Baehrecke, E.H.; Lenardo, E. Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. USA 2006, 103, 4952–4957. [Google Scholar] [CrossRef] [Green Version]
- Slimen, I.B.; Najar, T.; Ghram, A.; Dabbebi, H.; Ben Mrad, M.; Abdrabbah, M. Reactive oxygen species, heat stress and oxidative-induced mitochondrial damage. A review. Int. J. Hyperth. 2014, 30, 513–523. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Riquelme, N.; Huerta-Retamal, N.; Gómez-Torres, M.J.; Martínez-Espinosa, R.M. Catalase as a molecular target for male infertility diagnosis and monitoring: An overview. Antioxidants 2020, 9, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, C.; Yue, X.; Lu, X.; Liu, B. The role of catalase in the immune response to oxidative stress and pathogen challenge in the clam Meretrix meretrix. Fish Shellfish Immunol. 2013, 34, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Q.L.; Fujiwara, Y.; Kondo, T. Mechanism of cell death induction by nitroxide and hyperthermia. Free Radic. Biol. Med. 2006, 40, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Kikusato, M.; Toyomizu, M. Crucial role of membrane potential in heat stress-induced overproduction of reactive oxygen species in avian skeletal muscle mitochondria. PLoS ONE 2013, 8, e64412. [Google Scholar] [CrossRef]
- Yu, H.; Deng, W.; Zhang, D.; Gao, Y.; Yang, Z.; Shi, X.; Sun, J.; Zhou, J.; Ji, H. Antioxidant defenses of Onychostoma macrolepis in response to thermal stress: Insight from mRNA expression and activity of superoxide dismutase and catalase. Fish Shellfish Immunol. 2017, 66, 50–61. [Google Scholar] [CrossRef]
- Bai, H.; Ukita, H.; Kawahara, M.; Mitani, T.; Furukawa, E.; Yanagawa, Y.; Yabuuchi, N.; Kim, H.; Takahashi, M. Effect of summer heat stress on gene expression in bovine uterine endometrial tissues. Anim. Sci. J. 2020, 91, e13474. [Google Scholar] [CrossRef]
- Miao, Z.Q.; Tu, Y.Q.; Guo, P.Y.; He, W.; Jing, T.X.; Wang, J.J.; Wei, D.D. Antioxidant enzymes and heat shock protein genes from Liposcelis bostrychophila are involved in stress defense upon heat shock. Insects 2020, 11, 839. [Google Scholar] [CrossRef]
- Zou, D.; Ning, J.; Lu, X.; Wang, X.; Chen, M.; Liu, B.; Fang, J.; Wang, C. Physiological and Transcriptional Responses to Acute and Chronic Thermal Stress in the Ark Shell Scapharca subcrenata. Front. Mar. Sci. 2021, 8, 739662. [Google Scholar] [CrossRef]
- Dar, S.A.; Srivastava, P.P.; Varghese, T.; Nazir, M.I.; Gupta, S.; Krishna, G. Temporal changes in superoxide dismutase, catalase, and heat shock protein 70 gene expression, cortisol and antioxidant enzymes activity of Labeo rohita fingerlings subjected to starvation and refeeding. Gene 2019, 692, 94–101. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.; Wang, A.; Cheng, J.; Chen, L.; Pan, Y.; Li, H.; Zhang, Q.; Zhang, J.; Chu, W.; Zhang, J. Effects of starvation on antioxidant-related signaling molecules, oxidative stress, and autophagy in juvenile Chinese perch skeletal muscle. Mar. Biotechnol. 2020, 22, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.C.; Ma, K.X.; Yang, Y.J.; Shi, C.Y.; Chen, G.W.; Liu, D.Z. Molecular cloning, characterization, expression and enzyme activity of catalase from planarian Dugesia japonica in response to environmental pollutants. Ecotoxicol. Environ. Saf. 2018, 165, 88–95. [Google Scholar] [CrossRef]
- Wang, X.; Li, P.; He, S.; Xing, S.; Cao, Z.; Cao, X.; Liu, B.; Li, Z.H. Effects of tralopyril on histological, biochemical and molecular impacts in Pacific oyster, Crassostrea gigas. Chemosphere 2022, 289, 133157. [Google Scholar] [CrossRef]
- Zhao, Y.Q.; Liu, G.D.; Hou, C.C.; Han, Y.L.; Zhu, J.Q. Effect of cadmium exposure on antioxidant enzyme catalase in different tissues of Acrossocheilus fasciatus. Mol. Cell Toxicol. 2016, 12, 255–263. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, X.; Huang, X.; Gu, L.; Chen, Y.; Yang, Z. Combined effects of cadmium and salinity on juvenile Takifugu obscurus: Cadmium moderates salinity tolerance; salinity decreases the toxicity of cadmium. Sci. Rep. 2016, 6, 30968. [Google Scholar] [CrossRef]
- Zoysa, M.D.; Whang, I.; Nikapitiya, C.; Oh, C.; Choi, C.Y.; Lee, J. Transcriptional analysis of disk abalone (Haliotis discus discus) antioxidant enzymes against marine bacteria and virus challenge. Fish Shellfish Immunol. 2011, 31, 155–160. [Google Scholar] [CrossRef]
- Chainy, G.B.N.; Paital, B.; Dandapat, J. An overview of seasonal changes in oxidative stress and antioxidant defence parameters in some invertebrate and vertebrate species. Scientifica 2016, 2016, 6126570. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, M.R.; Braghirolli, F.M.; Lanés, L.E.K.; Verrastro, L.; Oliveira, G.T. Evaluation of the seasonal variation of parameters of oxidative status of Tropidurus catalanensis Gudynas and Skuk, 1983. S. Am. J. Herpetol. 2021, 19, 12–21. [Google Scholar] [CrossRef]
- Nash, S.; Rahman, M.S. Short-term heat stress impairs testicular functions in the American oyster, Crassostrea virginica: Molecular mechanisms and induction of oxidative stress and apoptosis in spermatogenic cells. Mol. Reprod. Dev. 2019, 86, 1444–1458. [Google Scholar] [CrossRef] [PubMed]
- Mazurais, D.; Covès, D.; Papandroulakis, N.; Ortega, A.; Desbruyeres, E.; Huelvan, C.; Le Gall, M.M.; De La Gándara, F.; Cahu, C.L. Gene expression pattern of digestive and antioxidant enzymes during the larval development of reared Atlantic bluefin tuna (ABFT), Thunnus thynnus L. Aquac. Res. 2015, 46, 2323–2331. [Google Scholar] [CrossRef] [Green Version]
- Zengin, H.; Yılmaz, Ö.; Demir, E.; Gökçe, Z. Antioxidant enzymatic defences during embryogenesis of rainbow trout Oncorhynchus mykiss (Walbaum 1792). Turk. J. Fish. Aquat. Sci. 2015, 15, 443–452. [Google Scholar] [CrossRef]
- Rampon, C.; Volovitch, M.; Joliot, A.; Vriz, S. Hydrogen peroxide and redox regulation of developments. Antioxidants 2018, 7, 159. [Google Scholar] [CrossRef] [Green Version]
- Mirzaei, A.H.; Deldar, H.; Pirsaraei, Z.A.; Shohreh, B. Royal jelly may improve sperm characteristics during preservation of rooster semen: Gene expression of antioxidant enzymes. Reprod. Domest. Anim. 2021, 56, 658–666. [Google Scholar] [CrossRef]
- Gale, S.L.; Burritt, D.J.; Tervit, H.R.; Adams, S.L.; McGowan, L.T. An investigation of oxidative stress and antioxidant biomarkers during Greenshell mussel (Perna canaliculus) oocyte cryopreservation. Theriogenology 2014, 82, 779–789. [Google Scholar] [CrossRef]
- Sharker, M.; Kim, S.C.; Hossen, S.; Kho, K.H. Characterization of insulin-like growth factor binding Protein-5 (IGFBP-5) gene and its potential roles in ontogenesis in the Pacific Abalone, Haliotis discus hannai. Biology 2020, 9, 216. [Google Scholar] [CrossRef]
- Sandamalika, W.G.; Priyathilaka, T.T.; Liyanage, D.S.; Lee, S.; Lim, H.K.; Lee, J. Molecular characterization of kappa class glutathione S-transferase from the disk abalone (Haliotis discus discus) and changes in expression following immune and stress challenges. Fish Shellfish Immunol. 2018, 77, 252–263. [Google Scholar] [CrossRef]
- Elvitigala, D.A.S.; Jayasooriya, R.G.P.T.; Lee, J. First report on the gastropod proapoptotic AIF3 counterpart from disk abalone (Haliotis discus discus) deciphering its transcriptional modulation by induced pathogenic stress. Fish Shellfish Immunol. 2015, 47, 697–705. [Google Scholar] [CrossRef]
- Hossen, S.; Sukhan, Z.P.; Cho, Y.; Kho, K.H. Effects of cryopreservation on gene expression and post thaw sperm quality of Pacific abalone, Haliotis discus hannai. Front. Mar. Sci. 2021, 8, 652390. [Google Scholar] [CrossRef]
- Kim, S.C.; Hossen, S.; Kho, K.H. Cryopreservation of sperm from farmed Pacific abalone, Haliotis discus hannai. Cryobiology 2020, 94, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Kong, N.; Li, H.; Yang, W.; Fu, Q.; Gong, C.; Wang, L.; Song, L. The effects of protein kinase a catalytic subunit on sperm motility regulation in Pacific abalone Haliotis discus hannai. Aquac. Res. 2020, 51, 2525–2534. [Google Scholar] [CrossRef]
- Hossen, S.; Sukhan, Z.P.; Cho, Y.; Lee, W.K.; Kho, K.H. Antioxidant Activity and Oxidative Stress-Oriented Apoptosis Pathway in Saccharides Supplemented Cryopreserved Sperm of Pacific Abalone, Haliotis discus hannai. Antioxidants 2022, 11, 1303. [Google Scholar] [CrossRef] [PubMed]
- Qiao, K.; Fang, C.; Chen, B.; Liu, Z.; Pan, N.; Peng, H.; Hao, H.; Xu, M.; Wu, J.; Liu, S. Molecular characterization, purification, and antioxidant activity of recombinant superoxide dismutase from the Pacific abalone Haliotis discus hannai Ino. World J. Microbiol. Biotechnol. 2020, 36, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sukhan, Z.P.; Hossen, S.; Cho, Y.; Lee, W.K.; Kho, K.H. Hdh-Tektin-4 Regulates Motility of Fresh and Cryopreserved Sperm in Pacific Abalone, Haliotis discus hannai. Front. Cell Dev. Biol. 2022, 10, 870743. [Google Scholar] [CrossRef]
- Silva-Aciares, F.; Moraga, D.; Auffret, M.; Tanguy, A.; Riquelme, C. Transcriptomic and cellular response to bacterial challenge (pathogenic Vibrio parahaemolyticus) in farmed juvenile Haliotis rufescens fed with or without probiotic diet. J. Invertebr. Pathol. 2013, 113, 163–176. [Google Scholar] [CrossRef]
- Sukhan, Z.P.; Cho, Y.; Sharker, M.R.; Hossen, S.; Rha, S.J.; Kho, K.H. Effective accumulative temperature affects gonadal maturation by controlling expression of GnRH, GnRH receptor, serotonin receptor and APGWamide gene in Pacific abalone, Haliotis discus hannai during broodstock conditioning in hatcheries. J. Therm. Biol. 2021, 100, 103037. [Google Scholar] [CrossRef]
- Hossen, S.; Sharker, M.; Cho, Y.; Sukhan, Z.P.; Kho, K.H. Effects of antifreeze protein III on sperm cryopreservation of Pacific abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2021, 22, 3917. [Google Scholar] [CrossRef]
- Sharker, M.R.; Hossen, S.; Nou, I.S.; Kho, K.H. Characterization of insulin-like growth factor binding protein 7 (igfbp7) and its potential involvement in shell formation and metamorphosis of Pacific abalone, Haliotis discus hannai. Int. J. Mol. Sci. 2020, 21, 6529. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Bankoglu, E.E.; Tschopp, O.; Schmitt, J.; Burkard, P.; Jahn, D.; Geier, A.; Stopper, H. Role of PTEN in oxidative stress and DNA damage in the liver of whole-body Pten haplodeficient mice. PLoS ONE 2016, 11, e0166956. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Orbea, A.; Dariush, H.F.; Cajaraville, M.P. Immunolocalization of four antioxidant enzymes in digestive glands of mollusks and crustaceans and fish liver. Histochem. Cell Biol. 2000, 114, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Zarai, Z.; Boulais, N.; Marcorelles, P.; Gobin, E.; Bezzine, S.; Mejdoub, H.; Gargouri, Y. Immunohistochemical localization of hepatopancreatic phospholipase in gastropods mollusc, Littorina littorea and Buccinum undatum digestive cells. Lipids Health Dis. 2011, 10, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gronemeyer, T.; Wiese, S.; Ofman, R.; Bunse, C.; Pawlas, M.; Hayen, H.; Eisenacher, M.; Stephan, C.; Meyer, H.E.; Waterham, H.R.; et al. The proteome of human liver peroxisomes: Identification of five new peroxisomal constituents by a label-free quantitative proteomics survey. PLoS ONE 2013, 8, e57395. [Google Scholar] [CrossRef]
- Marín-García, J. Oxidative stress and cell death in cardiovascular disease. In Post-Genomic Cardiology; Elsevier: Amsterdam, The Netherlands, 2014; pp. 471–498. [Google Scholar]
- Hoegh-Guldberg, O.; Mumby, P.J.; Hooten, A.J.; Steneck, R.S.; Greenfield, P.; Gomez, E.; Harvell, C.D.; Sale, P.F.; Edwards, A.J.; Caldeira, K.; et al. Coral reefs under rapid climate change and ocean acidification. Science 2007, 318, 1737–1742. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abele, D.; Heise, K.; Pörtner, H.O.; Puntarulo, S. Temperature-dependence of mitochondrial function and production of reactive oxygen species in the intertidal mud clam Mya arenaria. J. Exp. Biol. 2002, 205, 1831–1841. [Google Scholar] [CrossRef]
- Soldatov, A.A.; Gostyukhina, O.L.; Golovina, I.V. Antioxidant enzyme complex of tissues of the bivalve Mytilus galloprovincialis Lam. Under normal and oxidative-stress conditions: A review. Appl. Biochem. Microbiol. 2007, 43, 556–562. [Google Scholar] [CrossRef]
- Cheng, C.H.; Guo, Z.X.; Luo, S.W.; Wang, A.L. Effects of high temperature on biochemical parameters, oxidative stress, DNA damage and apoptosis of pufferfish (Takifugu obscurus). Ecotox. Environ. Safe 2018, 150, 190–198. [Google Scholar] [CrossRef]
- Wang, J.; Ren, R.M.; Yao, C.L. Oxidative stress responses of Mytilus galloprovincialis to acute cold and heat during air exposure. J. Molluscan Stud. 2018, 84, 285–292. [Google Scholar] [CrossRef] [Green Version]
- Ding, F.F.; Li, A.; Cong, R.H.; Wang, X.X.; Wang, W.; Que, H.Y.; Zhang, G.; Li, l. The phenotypic and the genetic response to the extreme high temperature provides new insight into thermal tolerance for the pacific oyster Crassostrea gigas. Front. Mar. Sci. 2020, 7, 399. [Google Scholar] [CrossRef]
- Nita, M.; Grzybowski, A. The role of the reactive oxygen species and oxidative stress in the pathomechanism of the age-related ocular diseases and other pathologies of the anterior and posterior eye segments in adults. Oxid. Med. Cell. Longev. 2016, 2016, 3164734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hopwood, M.J.; Rapp, I.; Schlosser, C.; Achterberg, E.P. Hydrogen peroxide in deep waters from the Mediterranean Sea, South Atlantic and South Pacific oceans. Sci. Rep. 2017, 7, 43436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alam, M.R.; Ehiguese, F.O.; Vitale, D.; Martín-Díaz, M.L. Oxidative stress response to hydrogen peroxide exposure of Mytilus galloprovincialis and Ruditapes philippinarum: Reduced embryogenesis success and altered biochemical response of sentinel marine bivalve species. J. Environ. Chem. Ecotoxicol. 2022, 4, 97–105. [Google Scholar] [CrossRef]
- Jo, P.G.; Choi, Y.K.; Choi, C.Y. Cloning and mRNA expression of antioxidant enzymes in the Pacific oyster, Crassostrea gigas in response to cadmium exposure. Comp. Biochem. Physiol. Part-C Toxicol. Pharmacol. 2008, 147, 460–469. [Google Scholar] [CrossRef]
- Qi, Z.; Wang, Q.; Song, S.; Wang, H.; Tan, M. Enhanced cytotoxicity of cadmium by a sulfated polysaccharide from abalone. J. Agric. Food Chem. 2020, 68, 14996–15004. [Google Scholar] [CrossRef]
- Peng, Y.Q.; Wang, M.J.; Chen, H.G.; Chen, J.H.; Gao, H.; Huang, H.H. Immunological responses in haemolymph and histologic changes in the hepatopancreas of Charybdis japonica (A. Milne-Edwards, 1861) (Decapoda: Brachyura: Portunidae) exposed to bisphenol A. J. Crustac. Biol. 2018, 38, 489–496. [Google Scholar] [CrossRef] [Green Version]
- Sakyi, M.E.; Cai, J.; Tang, J.; Abarike, E.D.; Xia, L.; Li, P.; Kuebutornye, F.K.A.; Zou, Z.; Liang, Z.; Jian, J. Effects of starvation and subsequent re-feeding on intestinal microbiota, and metabolic responses in Nile tilapia, Oreochromis niloticus. Aquac. Rep. 2020, 17, 100370. [Google Scholar] [CrossRef]
- Shi, Q.; Xiong, X.; Wen, Z.; Qin, C.; Li, R.; Zhang, Z.; Gong, Q.; Wu, X. Cu/Zn Superoxide Dismutase and Catalase of Yangtze Sturgeon, Acipenser dabryanus: Molecular Cloning, Tissue Distribution and Response to Fasting and Refeeding. Fishes 2022, 7, 35. [Google Scholar] [CrossRef]
- Canesi, L.; Barmo, C.; Fabbri, R.; Ciacci, C.; Vergani, L.; Roch, P.; Gallo, G. Effects of vibrio challenge on digestive gland biomarkers and antioxidant gene expression in Mytilus galloprovincialis. Comp. Biochem. Physio. 2010, 152, 399e406. [Google Scholar] [CrossRef]
- Liu, H.P.; Chen, F.Y.; Gopalakrishnan, S.; Qiao, K.; Bo, J.; Wang, K.J. Antioxidant enzymes from the crab Scylla paramamosain: Gene cloning and gene/protein expression profiles against LPS challenge. Fish Shellfish Immunol. 2010, 28, 862–871. [Google Scholar] [CrossRef]
- Priyathilaka, T.T.; Bathige, S.D.N.K.; Lee, S.; Yang, H.; Jeong, T.; Lee, S.; Lee, J. Structural and functional analysis of three Iκb kinases (IKK) in disk abalone (Haliotis discus discus): Investigating their role in the innate immune responses. Fish Shellfish Immunol. 2020, 103, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Wei, K.; Ding, Y.; Zhang, J. Molecular cloning, mRNA expression and functional characterization of a catalase from Chinese black sleeper (Bostrychus sinensis). Fish Shellfish Immunol. 2020, 103, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Lobo-da-Cunha, A.; Alves, Â.; Oliveira, E.; Guimarães, F.; Calado, G. Endocytosis, lysosomes, calcium storage and other features of digestive-gland cells in cephalaspidean gastropods (Euopisthobranchia). J. Molluscan Stud. 2018, 84, 451–462. [Google Scholar] [CrossRef] [Green Version]
- Figueroa, E.; Farias, J.G.; Lee-Estevez, M.; Valdebenito, I.; Risopatrón, J.; Magnotti, C.; Romero, J.; Watanabe, I.S.; Oliveira, R.P.S. Sperm cryopreservation with supplementation of α-tocopherol and ascorbic acid in freezing media increase sperm function and fertility rate in Atlantic salmon (Salmo salar). Aquaculture 2018, 493, 1–8. [Google Scholar] [CrossRef]
- De Viçose, G.C.; Viera, M.P.; Bilbao, A.; Izquierdo, M.S. Embryonic and larval development of Haliotis tuberculata coccinea Reeve: An indexed micro-photographic sequence. J. Shellfish. Res. 2007, 26, 847–854. [Google Scholar] [CrossRef] [Green Version]
- Kalaimani, N.; Chakravarthy, N.; Shanmugham, R.; Thirunavukkarasu, A.R.; Alavandi, S.V.; Santiago, T.C. Antioxidant status in embryonic, post-hatch and larval stages of Asian seabass (Lates calcarifer). Fish Physiol. Biochem. 2008, 34, 151–158. [Google Scholar] [CrossRef]










Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hossen, S.; Sukhan, Z.P.; Kim, S.C.; Hanif, M.A.; Kong, I.-K.; Kho, K.H. Molecular Cloning and Functional Characterization of Catalase in Stress Physiology, Innate Immunity, Testicular Development, Metamorphosis, and Cryopreserved Sperm of Pacific Abalone. Antioxidants 2023, 12, 109. https://doi.org/10.3390/antiox12010109
Hossen S, Sukhan ZP, Kim SC, Hanif MA, Kong I-K, Kho KH. Molecular Cloning and Functional Characterization of Catalase in Stress Physiology, Innate Immunity, Testicular Development, Metamorphosis, and Cryopreserved Sperm of Pacific Abalone. Antioxidants. 2023; 12(1):109. https://doi.org/10.3390/antiox12010109
Chicago/Turabian StyleHossen, Shaharior, Zahid Parvez Sukhan, Soo Cheol Kim, Md. Abu Hanif, Il-Keun Kong, and Kang Hee Kho. 2023. "Molecular Cloning and Functional Characterization of Catalase in Stress Physiology, Innate Immunity, Testicular Development, Metamorphosis, and Cryopreserved Sperm of Pacific Abalone" Antioxidants 12, no. 1: 109. https://doi.org/10.3390/antiox12010109