Recent Progress in the Study of Peroxiredoxin in the Harmful Algal Bloom Species Chattonella marina
Abstract
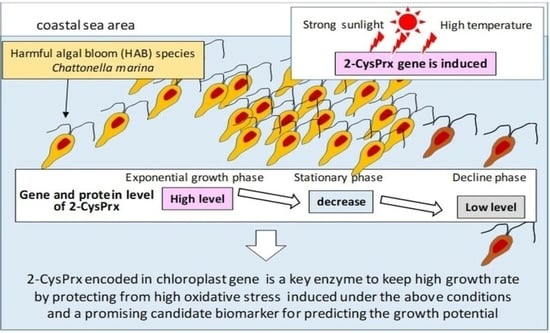
:1. Introduction
2. Temporal Changes of Levels of 2-CysPRX Enzymes During Algal Growth
3. Structure of the 2-CysPrx Gene in Chattonella marina
4. Induction of Gene Expression for the Production of Antioxidant Enzymes by Light, Thermal, and Oxidative Stress under Laboratory Conditions
5. Effect of 2-CysPrx Protein Levels on Survival and Production of Lipid Peroxide under High Irradiance
6. Survey of Prx Family of Genes Expressed in Chattonella marina by RNA-seq Analysis and Possible Application for Evaluation of Cell Division Activity
7. Ecological Significance of Circadian Rhythms in HAB Species
8. Possibility of Relationship between Antioxidant Activity and Toxicity to Fish
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kim, K.; Kim, I.H.; Lee, K.Y.; Rhee, S.G.; Stadtman, E.R. The isolation and purification of a specific “protector” protein which inhibits enzyme inactivation by a thiol/Fe(III)/O2 mixed-function oxidation system. J. Biol. Chem. 1988, 263, 4704–4711. [Google Scholar] [CrossRef]
- Rhee, S.G.; Kil, I.S. Multiple functions and regulation of mammalian peroxiredoxins. Annu. Rev. Biochem. 2017, 86, 749–775. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Miyake, C.; Dietz, K.J.; Tomizawa, K.; Murata, N.; Yokota, A. Thioredoxin peroxidase in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 1999, 477, 269–273. [Google Scholar] [CrossRef] [Green Version]
- Dietz, K.J. Peroxiredoxins in plants and cyanobacteria. Antioxid. Redox Signal. 2011, 15, 1129–1159. [Google Scholar] [CrossRef] [Green Version]
- Fujii, J.; Ikeda, Y. Advances in our understanding of peroxiredoxin, a multifunctional, mammalian redox protein. Redox Rep. 2002, 7, 123–130. [Google Scholar] [CrossRef] [PubMed]
- Nott, A.; Jung, H.S.; Koussevitzky, S.; Chory, J. Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 2006, 57, 739–759. [Google Scholar] [CrossRef] [PubMed]
- Dayer, R.; Fischert, B.B.; Eggen, R.I.L.; Lemaire, S.D. The peroxiredoxin and glutathione peroxidase families in Chlamydomonas reinhardtii. Genetics 2008, 179, 41–57. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomanek, K. Proteomic responses to environmentally induced oxidative stress. J. Exp. Biol. 2015, 218, 1867–1879. [Google Scholar] [CrossRef] [Green Version]
- Dietz, K.J. Thiol-based peroxidases and ascorbate peroxidases: Why plants rely on multiple peroxidase systems in the photosynthesizing chloroplast? Mol. Cells 2016, 39, 20–25. [Google Scholar] [PubMed] [Green Version]
- Okaichi, T.; Yanagi, T. Sustainable development in the Seto Inland Sea, Japan. Terra Sci. Publ. Co. (Terrapub) 1997, 251–304. [Google Scholar]
- Onitsuka, G.; Aoki, K.; Matsuyama, Y.; Kimoto, K.; Matsuo, H.; Kitadai, Y.; Nishi, H.; Tahara, Y.; Sakurada, K. Short-term dynamics of a Chattonella antiqua bloom in the Yatsushiro Sea, Japan, in summer 2010: Characteristics of its appearance in the southern area. Bull. Jpn. Soc. Fish. Oceanogr. 2011, 75, 143–153. [Google Scholar]
- Hallegraeff, G.M.; Munday, B.L.; Baden, D.G.; Whitney, P.L. Chattonella marina Raphidophyte bloom associated with mortality of cultured bluefin tuna (Thunnus maccoyii) in South Australia. In Harmful Algae; Reguera, B., Blanco, J., Fernandez, M.L., Wyatt, T., Eds.; Xunta de Galicia, IOC of UNESCO, Grafisant: Santiago de Compostela, Spain, 1998; pp. 93–96. [Google Scholar]
- Elbraechter, M. Exotic flagellates of coastal North Sea waters. Helgoländer Wiss. Meeresunters 1999, 52, 235–242. [Google Scholar] [CrossRef] [Green Version]
- Amano, K.; Watanabe, M.; Kohata, K.; Harada, S. Conditions necessary for Chattonella antiqua red tide outbreaks. Limnol. Oceanogr. 1998, 43, 117–128. [Google Scholar] [CrossRef]
- Marshall, J.A.; Hallegraeff, G.M. Comparative ecophysiology of the harmful alga Chattonella marina (Raphidophyceae) from South Australian and Japanese waters. J. Plankton Res. 1999, 21, 1809–1822. [Google Scholar] [CrossRef] [Green Version]
- Katano, T.; Yoshida, M.; Yamaguchi, S.; Yoshino, K.; Hamada, T.; Koriyama, M.; Hayami, Y. Effect of nutrient concentration and salinity on diel vertical migration of Chattonella marina (Raphidophyceae). Mar. Biol. Res. 2014, 10, 1007–1018. [Google Scholar] [CrossRef]
- Satta, C.T.; Padedda, B.M.; Sechi, N.; Pulina, S.; Loria, A.; Lugliè, A. Multiannual Chattonella subsalsa Biecheler (Raphidophyceae) blooms in a Mediterranean lagoon (Santa Giusta Lagoon, Sardinia Island, Italy). Harmful Algae 2017, 67, 61–73. [Google Scholar] [CrossRef]
- Giddings, S.N.; MacCready, P.; Hickey, B.M.; Banas, N.S.; Davis, K.A.; Siedlecki, S.A.; Trainer, V.L.; Kudela, R.M.; Pelland, N.A.; Connolly, T.P. Hindcasts of potential harmful algal bloom transport pathways on the Pacific Northwest coast. J. Geophys. Res. 2014, 4, 2439–2461. [Google Scholar] [CrossRef]
- Pinto, L.; Mateus, M.; Silva, A. Modeling the transport pathways of harmful algal blooms in the Iberian coast. Harmful Algae 2016, 53, 8–16. [Google Scholar] [CrossRef]
- Yamaguchi, M.; Imai, I.; Honjo, T. Effects of temperature, salinity and irradiance on the growth rates of the noxious red tide flagellates Chattonella antiqua and C. marina (Raphidophyceae). Nippon Suisan Gakkaishi 1991, 57, 1277–1284. [Google Scholar] [CrossRef]
- Qiu, X.; Shimasaki, Y.; Tsuyama, M.; Yamada, T.; Kuwahara, R.; Kawaguchi, M.; Honda, M.; Gunjikake, H.; Tasmin, R.; Shimizu, M.; et al. Growth phase dependent variation of photosynthetic activity and cellular protein expression profile in harmful raphidophyte Chattonella antiqua. Biosci. Biotechnol. Biochem. 2013, 77, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portune, K.J.; Cary, S.C.; Warner, M.E. Antioxidant enzyme response and reactive oxygen species production in marine raphydophytes. J. Phycol. 2010, 46, 1161–1171. [Google Scholar] [CrossRef]
- Miao, Y.L.D.; Wang, P.; Wang, X.; Chen, J.; Miao, C.; Song, C. An Arabidopsis glutathione peroxidase functions as both a redox transducer and a scavenger in abscisic acid and drought stress responses. Plant Cell 2006, 18, 2749–2766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machida, T.; Ishibashi, A.; Kirino, A.; Sato, J.; Kawasaki, S.; Niimura, Y.; Honjoh, K.; Miyamoto, T. Chloroplast NADPH-dependent thioredoxin reductase from Chlorella vulgaris alleviates environmental stresses in yeast together with 2-Cys Peroxiredoxin. PLoS ONE 2012, 7, 45988. [Google Scholar] [CrossRef] [Green Version]
- Ribeiro, C.W.; Korbes, A.P.; Garighan, J.A.; Jardim-Messeder, D.; Carvalho, F.E.L.; Sousa, R.H.V.; Caverzan, A.; Teixeira, F.K.; Silveira, J.A.G.; Margis-Pinheiro, M. Rice peroxisomal ascorbate peroxidase knockdown affects ROS signaling and triggers early leaf senescence. Plant Sci. 2017, 263, 55–65. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Nakamura, A.; Shikayama, M.; Kawano, I.; Ishimatsu, A.; Muramatsu, T. Generation of reactive oxygen species by raphidophycean phytoplankton. Biosci. Biotechnol. Biochem. 1997, 61, 1658–1662. [Google Scholar] [CrossRef] [PubMed]
- Oda, T.; Moritomi, J.; Kawano, I.; Hamaguchi, S.; Ishimatsu, A.; Muramatsu, T. Catalase- and superoxide dismutase-induced morphological changes and growth inhibition in the red tide phytoplankton Chattonella marina. Biosci. Biotechnol. Biochem. 1995, 59, 2044–2048. [Google Scholar] [CrossRef]
- Johnson, K.J.; Fantone, J.C.; Kaplan, P.A. In vivo damage of rat lungs by oxygen metabolites. J. Clin. Investig. 1981, 67, 983–993. [Google Scholar] [CrossRef] [Green Version]
- Halliwell, B.; Gutteridge, J.M. Oxygen toxicity, oxygen radicals, transition metals and disease. Biochem. J. 1984, 219, 1–14. [Google Scholar] [CrossRef]
- Yamamoto, S. Enzymatic lipid-peroxidation-reactions of mammalian lipoxygenases. Free Radic. Biol. Med. 1991, 10, 149–159. [Google Scholar] [CrossRef]
- Yin, H.; Xu, L.; Porter, N.A. Free radical lipid peroxidation: Mechanisms and analysis. Chem. Rev. 2011, 111, 5944–5972. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, F.M.; Argüelles, S. Lipid peroxidation: Production, metabolism, and signaling mechanisms of malondialdehyde and 4-hydroxy-2-nonenal. Oxid. Med. Cell Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef] [PubMed]
- Chan, L.L.; Hodgkiss, I.J.; Wan, J.M.F.; Lum, J.H.K.; Mak, A.S.C.; Sit, W.H.; Lo, S.C.L. Proteomic study of a model causative agent of harmful algal blooms, Prorocentrum triestinum II: The use of differentially expressed protein profiles under different growth phases and growth conditions for bloom prediction. Proteomics 2004, 4, 3214–3226. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sui, Z.; Chang, L.; Kang, K.; Ma, J.; Kong, F.; Zhou, W.; Wang, J.; Guo, L.; Geng, H.; et al. Transcriptome de novo assembly sequencing and analysis of the toxic dinoflagellate Alexandrium catenella using the Illumina platform. Gene 2014, 537, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Kimura, K.; Okuda, S.; Nakayama, K.; Shikata, T.; Takahashi, F.; Yamaguchi, H.; Skamoto, S.; Yamaguchi, M.; Tomaru, Y. RNA Sequencing revealed numerous polyketide synthase genes in the harmful dinoflagellate Karenia mikimotoi. PLoS ONE 2015, 10, 0142731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shikata, T.; Takahashi, F.; Nishide, H.; Shigenobu, S.; Kamei, Y.; Sakamoto, S.; Yuasa, K.; Nishiyama, Y.; Yamasaki, Y.; Uchiyama, I. RNA-Seq Analysis reveals genes related to photoreception, nutrient uptake, and toxicity in a noxious red-tide raphidophyte Chattonella antiqua. Front. Microbiol. 2019, 10, 1764. [Google Scholar] [CrossRef] [Green Version]
- Hano, T.; Tomaru, Y. Metabolomics-based approach to explore growth phase-dependent markers lit in cultured diatom Chaetoceros tenuissimus. J. Chromatogr. B 2019, 1128, 121779. [Google Scholar] [CrossRef]
- Vingiani, G.M.; Štālberga, D.; De Luca, P.; Ianora, A.; De Luca, D.; Lauritano, C. De novo transcriptome of the non-saxitoxin producing Alexandrium tamutum reveals new insights on harmful dinoflagellates. Mar. Drugs 2020, 18, 386. [Google Scholar] [CrossRef] [PubMed]
- Chae, H.Z.; Kim, H.J.; Kang, S.W.; Rhee, S.G. Characterization of three isoforms of mammalian peroxiredoxin that reduce peroxides in the presence of thioredoxin. Diabetes Res. Clin. Pract. 1999, 45, 101–112. [Google Scholar] [CrossRef]
- Qiu, X.; Mukai, K.; Shimasaki, Y.; Tsuyama, M.; Matsubara, T.; Nakajima, Y.; Honjo, T.; Oshima, Y. Potential maximum quantum yield of photosystem II reflects the growth rate of Chattonella marina in field bloom samples. J. Fac. Agric. Kyushu Univ. 2016, 61, 331–335. [Google Scholar]
- Qiu, X.; Mukai, K.; Shimasaki, Y.; Tsuyama, M.; Matsubara, T.; Nakashima, T.; Ichinose, H.; Nakazima, Y.; Honjo, T.; Oshima, Y. Variations in the expression of photosynthesis–related proteins in field Chattonella marina cells during a harmful algal bloom. J. Fac. Agric. Kyushu Univ. 2017, 62, 373–380. [Google Scholar]
- Dietz, K.J.; Horling, F.; Konig, J.; Baier, M. The function of the chloroplast 2-cysteine peroxiredoxin in peroxide detoxification and its regulation. J. Exp. Bot. 2002, 53, 1321–1329. [Google Scholar] [PubMed]
- Baier, M.; Dietz, K.J. Primary structure and expression of plant homologues of animal and fungal thioredoxin-dependent peroxide reductases and bacterial alkyl hydroperoxide reductases. Plant Mol. Biol. 1996, 31, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Baier, M.; Dietz, K.J. Protective function of chloroplast 2-cysteine peroxiredoxin in photosynthesis. Evidence from transgenic Arabidopsis. Plant Physiol. 1999, 119, 1407–1414. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horling, F.; Baier, M.; Dietz, K.J. Redox-regulation of the expression of the peroxide-detoxifying chloroplast 2-Cys peroxiredoxin in the liverwort Riccia fluitans. Planta 2001, 214, 304–313. [Google Scholar] [CrossRef] [PubMed]
- Klughammer, B.; Baier, M.; Dietz, K.J. Inactivation by gene disruption of 2-cysteine-peroxiredoxin in Synechocystis sp. PCC 6803 leads to increased stress sensitivity. Physiol. Plant. 1998, 104, 699–706. [Google Scholar] [CrossRef]
- Sandrini, G.; Piel, T.; Xu, T.S.; White, E.; Qin, H.J.; Slot, P.C.; Huisman, J.; Visser, P.M. Sensitivity to hydrogen peroxide of the bloom-forming cyanobacterium Microcystis PCC 7806 depends on nutrient availability. Harmful Algae 2020, 99, 101916. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Watanabe, M.; Nakayasu, Y.; Kohata, K. Production of superoxide anion and hydrogen peroxide associated with cell growth of Chattonella antiqua. Aquat. Microb. Ecol. 2004, 35, 57–64. [Google Scholar] [CrossRef]
- Yuasa, K.; Shikata, T.; Ichikawa, T.; Tamura, Y.; Nishiyama, Y. Nutrient deficiency stimulates the production of superoxide in the noxious red-tide-forming raphidophyte Chattonella antiqua. Harmful Algae 2020, 99, 101938. [Google Scholar] [CrossRef] [PubMed]
- Zachary, Z.A.; Poole, L.B.; Karplus, P.A. Peroxiredoxin evolution and the regulation of hydrogen peroxide signaling. Science 2003, 300, 650–653. [Google Scholar]
- Mukai, K.; Teramoto, A.; Qiu, X.; Shimasaki, Y.; Kato-Unoki, Y.; Lee, J.M.; Mizoguchi, M.; Khanam, M.R.M.; Satone, H.; Tatsuke, T.; et al. Gene structure and cDNA sequence of 2-Cys peroxiredoxin in the harmful algal bloom species Chattonella marina and its gene transcription under different light intensities. Eur. J. Phycol. 2018, 53, 29–38. [Google Scholar] [CrossRef]
- Cattolico, R.A.; Jacobs, M.A.; Zhou, Y.; Chang, J.; Duplessis, M.; Lybrand, T.; Mckay, J.; Ong, H.C.; Sims, E.; Rocap, G. Chloroplast genome sequencing analysis of Heterosigma akashiwo CCMP452 (West Atlantic) and NIES293 (West Pacific) strains. BMC Genomics 2008, 9, 211. [Google Scholar] [CrossRef] [Green Version]
- Börner, T.; Aleynikova, A.Y.; Zubo, Y.O.; Kusnetsov, V.V. Chloroplast RNA polymerases: Role in chloroplast biogenesis. Biochim. Biophys. Acta Bioenerg. 2015, 9, 761–769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, C.P. An inspection of the domain between putative TATA box and translation start site in 79 plant genes. Nucleic Acids Res. 1987, 15, 6643–6653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Magee, A.M.; Kavanagh, T.A. Plastid genes transcribed by the nucleus-encoded plastid RNA polymerase show increased transcript accumulation in transgenic plants expressing a chloroplast-localized phage T7 RNA polymerase. J. Exp. Bot. 2002, 53, 2341–2349. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.C.; Purton, S. The transcriptional apparatus of algal plastids. Eur. J. Phycol. 2002, 37, 301–311. [Google Scholar] [CrossRef]
- Mukai, K.; Shimasaki, Y.; Qiu, X.; Kato-Unoki, Y.; Chen, K.; Khanam, M.R.M.; Oshima, Y. Effects of light and hydrogen peroxide on gene expression of newly identified antioxidant enzymes in the harmful algal bloom species Chattonella marina. Eur. J. Phycol. 2019, 54, 393–403. [Google Scholar] [CrossRef]
- Mukai, K.; Shimasaki, Y.; Qiu, X.; Kato-Unoki, Y.; Chen, K.; Takai, Y.; Khanam, M.R.M.; Chairil, A.E.; Oshima, Y. Gene expression stability of candidate reference genes under different culture conditions for quantitative PCR in the raphidophyte Chattonella marina. Phycologia 2020. [Google Scholar] [CrossRef]
- Qiu, X.; Chen, C.; Shimasaki, Y.; Mukai, K.; Teramoto, A.; Wu, M.; Oshima, Y. Time-series responses in photosynthetic activity, 2-cysteine peroxiredoxin gene expression, and proteomics of Chattonella marina var. antiqua under different oxidative stress. Harmful Algae 2020, 94, 101808. [Google Scholar] [CrossRef]
- Qiu, X.; Wu, M.; Mukai, K.; Shimasaki, Y.; Oshima, Y. Effects of Elevated irradiance, temperature, and rapid shifts of salinity on the chlorophyll a fluorescence (OJIP) transient of Chattonella marina var. antiqua. J. Fac. Agric. Kyushu Univ. 2019, 64, 293–300. [Google Scholar]
- Frugoli, J.A.; Zhong, H.H.; Nuccio, M.L.; McCourt, P.; McPeek, M.A.; Thomas, T.L.; McClung, C.R. Catalase is encoded by a multigene family in Arabidopsis thaliana (L.) Heynh. Plant Physiol. 1996, 112, 327–336. [Google Scholar] [CrossRef] [Green Version]
- Kliebenstein, D.J.; Monde, R.A.; Last, R.L. Superoxide dismutase in Arabidopsis: An eclectic enzyme family with disparate regulation and protein localization. Plant Physiol. 1998, 118, 637–650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Milla, M.A.R.; Maurer, A.; Huete, A.R.; Gustafson, J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003, 36, 602–615. [Google Scholar] [CrossRef]
- Najami, N.; Janda, T.; Barriah, W.; Kayam, G.; Tal, M.; Guy, M.; Volokita, M. Ascorbate peroxidase gene family in tomato: Its identification and characterization. Mol. Genet. Genomics 2008, 279, 171–182. [Google Scholar] [CrossRef] [PubMed]
- Nuruzzaman, M.; Gupta, M.; Zhang, C.; Wang, L.; Xie, W.; Xiong, L.; Zhang, Q.; Lian, X. Sequence and expression analysis of the thioredoxin protein gene family in rice. Mol. Genet. Genomics 2008, 280, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Kunert, K.J. Lipid peroxidation in higher plants. Plant Physiol. 1986, 82, 700–702. [Google Scholar] [CrossRef] [PubMed]
- Hurng, W.P.; Kao, C.H. Lipid peroxidation and antioxidative enzymes in senescing tobacco leaves during post-flooding. Plant Sci. 1994, 96, 41–44. [Google Scholar] [CrossRef]
- He, Y.Y.; Klisch, M.; Häder, D.P. Adaptation of cyanobacteria to UV-B stress correlated with oxidative stress and oxidative damage. Photochem. Photobiol. 2002, 76, 188–196. [Google Scholar] [CrossRef]
- Jakhar, S.; Mukherjee, D. Chloroplast pigments, proteins, lipid peroxidation and activities of antioxidative enzymes during maturation and senescence of leaves and reproductive organs of Cajanus cajan L. Physiol. Mol. Biol. Plants 2014, 20, 171–180. [Google Scholar] [CrossRef] [Green Version]
- Oda, T.; Nakamura, A.; Okamoto, T.; Ishimatsu, A.; Muramatsu, T. Lectin-induced enhancement of superoxide anion production by red tide phytoplankton. Mar. Biol. 1998, 131, 383–390. [Google Scholar] [CrossRef]
- Wolfe-Simon, F.; Grzebyk, D.; Schofield, O.; Falkowski, P.G. The role and evolution of superoxide dismutases in algae. J. Phycol. 2005, 41, 453–465. [Google Scholar] [CrossRef]
- Fahnenstich, H.; Scarpeci, T.E.; Valle, E.M.; Flügge, U.; Maurino, V.G. Generation of hydrogen peroxide in chloroplasts of Arabidopsis overexpressing glycolate oxidase as an inducible system to study oxidative stress. Plant Physiol. 2008, 148, 719–729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pospíšil, P. Production of reactive oxygen species by photosystem II as a response to light and temperature stress. Front Plant Sci. 2016, 7, 1950. [Google Scholar] [CrossRef] [PubMed]
- Demmig-Adams, B.; Adams, W.W. Photoprotection and other responses of plants to high light stress. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 599–626. [Google Scholar] [CrossRef]
- Rintamäki, E.; Salo, R.; Lehtonen, E.; Aro, E.M. Regulation of D1-protein degradation during photoinhibition of photosystem II in vivo: Phosphorylation of the D1 protein in various plant groups. Planta 1995, 195, 379–386. [Google Scholar] [CrossRef]
- Shimada, M.; Kawamoto, S.; Nakatsuka, Y.; Watanabe, M. Localization of superoxide anion in the red tide alga Chattonella antiqua. J. Histochem. Cytochem. 1993, 41, 507–511. [Google Scholar] [CrossRef] [PubMed]
- Walling, C. Oxidases and Related Redox Systems; King, T.E., Mason, H.S., Morrison, M., Eds.; Pergamon Press: Oxford, UK, 1982; pp. 85–97. [Google Scholar]
- Cho, E.J.; Yokozawa, T.; Rhyu, D.Y.; Kim, H.Y.; Shibahara, N. The inhibitory effects of 12 medicinal plants and their component compounds on lipid peroxidation. Am. J. Chin. Med. 2003, 31, 907–917. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Zhang, Z.; Shen, Y.; Duan, X.; Jiang, Y. Effect of tea polyphenols on lipid peroxidation and antioxidant activity of litchi (Litchi chinensis Sonn.) fruit during cold storage. Molecules 2014, 19, 16837–16850. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Havaux, M.; Eymery, F.; Porfirov, S.; Rey, P.; Dörmannb, P. Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 2005, 17, 3451–3469. [Google Scholar] [CrossRef] [Green Version]
- Karimi, E.; Jaafar, H.Z.E.; Ghasemzadeh, A.; Ibrahim, M.H. Light intensity effects on production and antioxidant activity of flavonoids and phenolic compounds in leaves, stems and roots of three varieties of Labisia pumila Benth. Aust. J. Crop Sci. 2013, 7, 1016–1023. [Google Scholar]
- Rhee, S.G.; Kang, S.W.; Chang, T.S.; Jeong, W.; Kim, K. Peroxiredoxin, a novel family of peroxidase. IUBMB Life 2001, 52, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Benger, G.; Govindjee, G. The mechanism of photosynthetic water oxidation. Photosynth. Res. 1985, 6, 33–55. [Google Scholar] [CrossRef] [PubMed]
- Armbruster, U.; Galvis, V.C.; Kunz, H.H.; Strand, D.D. The regulation of the chloroplast proton motive force plays a key role for photosynthesis in fluctuating light. Curr. Opin. Plan. Biol. 2017, 37, 56–62. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Vonck, J.; Mills, D.J.; Meier, T.; Kühlbrandt, W. Structure, mechanism, and regulation of the chloroplast ATP synthase. Science 2018, 11, 360. [Google Scholar]
- Koizumi, Y.; Uchida, T.; Honjo, T. Diurnal vertical migration of Gymnodinium mikimotoi during a red tide in Hoketsu Bay, Japan. J. Plankton Res. 1996, 18, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Watanabe, M.; Kohata, K.; Kimura, T. Diel vertical migration and nocturnal uptake of nutrients by Chattonella antiqua under stable stratification. Limnol. Oceanogr. 1991, 36, 593–602. [Google Scholar] [CrossRef]
- Tilney, C.L.; Hoadley, K.D.; Warner, M.E. Comparing the diel vertical migration of Karlodinium veneficum (dinophyceae) and Chattonella subsalsa (Raphidophyceae): PSII photochemistry, circadian control, and carbon assimilation. J. Photochem. Photobiol. B 2015, 143, 107–119. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kohata, K.; Kimura, T.; Takamatsu, T.; Yamaguchi, S.; Ioriya, T. Generation of a Chattonella antiqua bloom by imposing a shallow nutricline in a mesocosm. Limnol. Oceanogr. 1995, 40, 1447–1460. [Google Scholar] [CrossRef]
- Warner, M.E.; Madden, M.L. The impact of shifts to elevated irradiance on the growth and photochemical activity of the harmful algae Chattonella subsalsa and Prorocentrum minimum from Delaware. Harmful Algae 2007, 6, 332–342. [Google Scholar] [CrossRef]
- Kimura, T.; Watanabe, M.; Kohata, K.; Sudo, R. Phosphate metabolism during diel vertical migration in the raphidophycean alga, Chattonella antiqua. J. Appl. Phycol. 1999, 11, 301–311. [Google Scholar] [CrossRef]
- Wang, Y.; Bouchard, J.N.; Coyne, K.J. Expression of novel nitrate reductase genes in the harmful alga, Chattonella subsalsa. Sci. Rep. 2018, 8, 13417. [Google Scholar] [CrossRef]
- Nemoto, Y.; Kuroiwa, T.; Furuya, M. Photocontrol of nuclear DNA replication in Chattonella antiqua (Raphidophyceae). Plant Cell Physiol. 1987, 28, 1043–1049. [Google Scholar]
- Kim, D.; Watanabe, M.; Nakayasu, Y.; Kohata, K. Changes in O2− and H2O2 production by Chattonella antiqua during diel vertical migration under nutrient stratification. Aquat. Microb. Ecol. 2005, 39, 183–191. [Google Scholar] [CrossRef]
- Shikata, T.; Matsunaga, S.; Nishide, H.; Sakamoto, S.; Onistuka, G.; Yamaguchi, M. Diurnal vertical migration rhythms and their photoresponse in four phytoflagellates causing harmful algal blooms. Limnol. Oceanogr. 2015, 60, 1251–1264. [Google Scholar] [CrossRef]
- Noordally, Z.B.; Millar, A.J. Clocks in algae. Biochemistry 2015, 54, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.S.; Green, E.W.; Zhao, Y.W.; van Ooijen, G.; Olmedo, M.; Qin, X.M.; Xu, Y.; Pan, M.; Valekunja, U.K.; Feeney, K.A.; et al. Peroxiredoxins are conserved markers of circadian rhythms. Nature 2012, 485, 459–464. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, X.C.; Mukai, K.; Shimasaki, Y.; Wu, M.; Chen, C.; Lu, Y.J.; Ichinose, H.; Nakashima, T.; Kato-Unoki, Y.; Oshima, Y. Diurnal variations in expression of photosynthesis-related proteins in the harmful Raphidophyceae Chattonella marina var. antiqua. J. Exp. Mar. Biol. Ecol. 2020, 527, 151361. [Google Scholar] [CrossRef]
- Imai, I.; Yamaguchi, M. Life cycle, physiology, ecology and red tide occurrences of the fish-killing raphidophyte Chattonella. Harmful Algae 2012, 14, 46–70. [Google Scholar] [CrossRef]
- Ishimatsu, A.; Maruta, H.; Oda, T.; Ozaki, M. A comparison of physiological responses in yellowtail to fatal environmental hypoxia and exposure to Chattonella marina. Fish. Sci. 1997, 63, 557–562. [Google Scholar] [CrossRef] [Green Version]
- Woo, S.P.S.; Liu, W.; Au, D.W.T.; Anderson, D.M.; Wu, R.S.S. Antioxidant responses and lipid peroxidation in gills and erythrocytes of fish (Rhabdosarga sarba) upon exposure to Chattonella marina and hydrogen peroxide: Implications on the cause of fish kills. J. Exp. Mar. Biol. Ecol. 2006, 336, 230–241. [Google Scholar] [CrossRef]
- Shen, M.; Xu, J.; Tsang, T.Y.; Au, D.W.T. Toxicity comparison between Chattonella marina and Karenia brevis using marine medaka (Oryzias melastigma): Evidence against the suspected ichthyotoxins of Chattonella marina. Chemosphere 2010, 80, 585–591. [Google Scholar] [CrossRef]
- García-Mendoza, E.; Cáceres-Martínez, J.; Rivas, D.; Fimbres-Martinez, M.; SánchezBravo, Y.; Vásquez-Yeomans, R.; Medina-Elizalde, J. Mass mortality of cultivated northern bluefin tuna Thunnus thynnus orientalis associated with Chattonella species in Baja California, Mexico. Front. Mar. Sci. 2018, 5, 454. [Google Scholar] [CrossRef]
- Ishimatsu, A.; Sameshima, M.; Tamura, A.; Oda, T. Histological analysis of the mechanisms of Chattonella-induced hypoxemia in yellowtail. Fish. Sci. 1996, 62, 50–58. [Google Scholar] [CrossRef] [Green Version]
- Marshall, J.A.; Ross, T.; Pyecroft, S.; Hallegraeff, G. Superoxide production by marine microalgae—II. Towards understanding ecological consequences and possible functions. Mar. Biol. 2005, 147, 541–549. [Google Scholar] [CrossRef]
- Yuasa, K.; Shikata, T.; Kitatsuji, S.; Yamasaki, Y.; Nishiyama, Y. Extracellular secretion of superoxide is regulated by photosynthetic electron transport in the noxious red-tide-forming raphidophyte Chattonella antiqua. J. Photochem. Photobiol. B Biol. 2020, 205, 111839. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Nakashima, T.; Matsuyama, Y.; Niwano, Y.; Yamaguchi, K.; Oda, T. Presence of the distinct systems responsible for superoxide anion and hydrogen peroxide generation in red tide phytoplankton Chattonella marina and Chattonella ovata. J. Plankton Res. 2007, 29, 241–247. [Google Scholar] [CrossRef] [Green Version]
- Marshall, J.A.; Nichols, P.D.; Hamilton, B.; Lewis, R.J.; Hallegraeff, G.M. Ichthyotoxicity of Chattonella marina (Raphidophyceae) to damselfish (Acanthochromis polycanthus): The synergistic role of reactive oxygen species and free fatty acids. Harmful Algae 2003, 2, 273–281. [Google Scholar] [CrossRef]
- Cho, K.; Sakamoto, J.; Noda, T.; Nishiguchi, T.; Ueno, M.; Yamasaki, Y.; Yagi, M.; Kim, D.; Oda, T. Comparative studies on the fish-killing activities of Chattonella marina isolated in 1985 and Chattonella antiqua isolated in 2010, and their possible toxic factors. Biosci. Biotechnol. Biochem. 2016, 80, 811–817. [Google Scholar] [CrossRef] [Green Version]
- Dorantes-Aranda, J.J.; Seger, A.; Mardones, J.I.; Nichols, P.D.; Hallegraeff, G.M. Progress in understanding algal bloom-mediated fish kills: The role of superoxide radicals, phycotoxins and fatty acids. PLoS ONE 2015, 10, e0133549. [Google Scholar] [CrossRef]
- Dorantes-Aranda, J.J.; Nichols, P.D.; Waite, T.D.; Hallegraeff, G.M. Strain variability in fatty acid composition of Chattonella marina (Raphidophyceae) and its relation to differing ichthyotoxicity toward rainbow trout gill cells. J. Phycol. 2013, 49, 427–438. [Google Scholar] [CrossRef]
- Shimasaki, Y.; Tsuyama, M.; Tasmin, R.; Qiu, X.; Shimizu, M.; Sato, Y.; Yamasaki, Y.; Kato-Unoki, Y.; Nukata, A.; Nakashima, T.; Ichinose, H.; et al. Thiobencarb herbicide reduces growth, photosynthetic activity, and amount of Rieske iron-sulfur protein in the diatom Thalassiosira pseudonana. J. Biochem. Mol. Toxicol. 2013, 27, 437–444. [Google Scholar] [CrossRef]
- Glibert, P.M.; Anderson, D.M.; Gentien, P.; Graneli, E.; Sellner, K.G. The global, complex phenomena of harmful algae blooms. Oceanography 2005, 18, 136–147. [Google Scholar] [CrossRef]








| Chattonella marina var. antiqua | Chattonella marina var. marina | ||
|---|---|---|---|
| (NIES-1) | (NIES-3) | ||
| Light intensity vs. | Fv/Fm ratio | −0.860 ** | −0.936 ** |
| O2− | 0.018 | 0.122 | |
| H2O2 | 0.472 ** | 0.540 ** | |
| CAT | −0.188 | 0.388 * | |
| LPO | 0.390 * | 0.448 ** |
| Contig (Number of Amino Acids) | Database a | Classification b | Retrieved Gene (Organism Name) | Accession Number of Retrieved Gene | Query Cover | E Value | Identity |
|---|---|---|---|---|---|---|---|
| 659c3g1i2 (195) | S | 2-CysPrx | 2-cysteine peroxiredoxin, chloroplastic (Chattonella marina var. antiqua) | A0A2Z5VKM8.1 | 100% | 5 × 10−144 | 100% |
| 5745c0g1i1 (179) | S | PrxQ | peroxiredoxin q (Nannochloropsis gaditana CCMP526) | XP_005855889.1 | 93% | 9 × 10−69 | 61% |
| HP | PrxQ | peroxiredoxin Q, chloroplastic (Erythranthe guttata) | XP_012851266.1 | 84% | 5 × 10−52 | 55% | |
| 6598c0g1i1 (221) | S | Prx4 | peroxiredoxin-4, partial (Globisporangium splendens) | KAF1329794.1 | 89% | 1 × 10−93 | 65% |
| HP | 2-CysPrxBAS1 | 2-Cys peroxiredoxin BAS1,chloroplastic (Oryza brachyantha) | XP_006648704.1 | 96% | 2 × 10−74 | 51% | |
| 6926c0g1i1 (200) | S | Prx2 | peroxiredoxin-2 (Pythium insidiosum) | GAY02440.1 | 98% | 3 × 10−96 | 68% |
| HP | Prx1 | peroxiredoxin-1-like (Rhodamnia argentea) | XP_030535131.1 | 97% | 9 × 10−79 | 61% | |
| 14890c0g1i1 (143) | S | Prx (unclassified) | peroxiredoxin (Thraustotheca clavata) | OQS00377.1 | 87% | 1 × 10−31 | 44% |
| HP | PrxIIB | peroxiredoxin-2B (Brassica napus) | XP_013676688.1 | 92% | 9 × 10−39 | 52% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shimasaki, Y.; Mukai, K.; Takai, Y.; Qiu, X.; Oshima, Y. Recent Progress in the Study of Peroxiredoxin in the Harmful Algal Bloom Species Chattonella marina. Antioxidants 2021, 10, 162. https://doi.org/10.3390/antiox10020162
Shimasaki Y, Mukai K, Takai Y, Qiu X, Oshima Y. Recent Progress in the Study of Peroxiredoxin in the Harmful Algal Bloom Species Chattonella marina. Antioxidants. 2021; 10(2):162. https://doi.org/10.3390/antiox10020162
Chicago/Turabian StyleShimasaki, Yohei, Koki Mukai, Yuki Takai, Xuchun Qiu, and Yuji Oshima. 2021. "Recent Progress in the Study of Peroxiredoxin in the Harmful Algal Bloom Species Chattonella marina" Antioxidants 10, no. 2: 162. https://doi.org/10.3390/antiox10020162