Lard Injection Matrix for Serial Crystallography
Abstract
:1. Introduction
2. Results
2.1. Preparation of Lard Material for SX Experiment
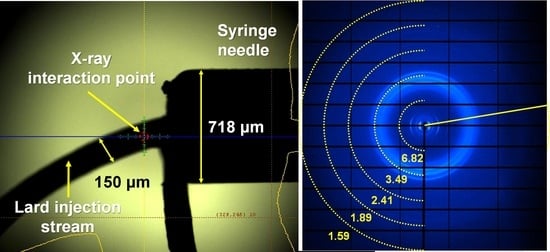
2.2. Application of Lard Injection Matrix for SMX
2.3. Measurement of Background Scattering
3. Discussion
4. Materials and Methods
4.1. Preparation of Lard Injection Matrix
4.2. Crystallization and Mixing of Crystals with the Lard Matrix
4.3. X-ray Data Collection
4.4. Structure Determination
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Schlichting, I. Serial femtosecond crystallography: The first five years. IUCrJ 2015, 2, 246–255. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kim, H.Y.; Park, J.; Kim, S.; Kim, S.; Rah, S.; Lim, J.; Namh, K.H. Focusing X-ray free-electron laser pulses using Kirkpatrick-Baez mirrors at the NCI hutch of the PAL-XFEL. J. Synchrotron Radiat. 2018, 25, 289–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weinert, T.; Olieric, N.; Cheng, R.; Brunle, S.; James, D.; Ozerov, D.; Gashi, D.; Vera, L.; Marsh, M.; Jaeger, K.; et al. Serial millisecond crystallography for routine room-temperature structure determination at synchrotrons. Nat. Commun. 2017, 8, 542. [Google Scholar] [CrossRef] [PubMed]
- Standfuss, J.; Spence, J. Serial crystallography at synchrotrons and X-ray lasers. IUCrJ 2017, 4 Pt 2, 100–101. [Google Scholar] [CrossRef] [Green Version]
- Ebrahim, A.; Moreno-Chicano, T.; Appleby, M.V.; Chaplin, A.K.; Beale, J.H.; Sherrell, D.A.; Duyvesteyn, H.M.E.; Owada, S.; Tono, K.; Sugimoto, H.; et al. Dose-resolved serial synchrotron and XFEL structures of radiation-sensitive metalloproteins. IUCrJ 2019, 6 Pt 4, 543–551. [Google Scholar] [CrossRef]
- Spence, J.C.H. Approaches to time-resolved diffraction using an XFEL. Faraday Discuss. 2014, 171, 429–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin-Garcia, J.M.; Conrad, C.E.; Nelson, G.; Stander, N.; Zatsepin, N.A.; Zook, J.; Zhu, L.; Geiger, J.; Chun, E.; Kissick, D.; et al. Serial millisecond crystallography of membrane and soluble protein microcrystals using synchrotron radiation. IUCrJ 2017, 4 Pt 4, 439–454. [Google Scholar] [CrossRef]
- Martiel, I.; Muller-Werkmeister, H.M.; Cohen, A.E. Strategies for sample delivery for femtosecond crystallography. Acta Crystallogr. Sect. D Struct. Biol. 2019, 75 Pt 2, 160–177. [Google Scholar] [CrossRef] [Green Version]
- DePonte, D.P.; Weierstall, U.; Schmidt, K.; Warner, J.; Starodub, D.; Spence, J.C.H.; Doak, R.B. Gas dynamic virtual nozzle for generation of microscopic droplet streams. J. Phys. D Appl. Phys. 2008, 41, 195505. [Google Scholar] [CrossRef] [Green Version]
- Weierstall, U.; James, D.; Wang, C.; White, T.A.; Wang, D.J.; Liu, W.; Spence, J.C.H.; Doak, R.B.; Nelson, G.; Fromme, P.; et al. Lipidic cubic phase injector facilitates membrane protein serial femtosecond crystallography. Nat. Commun. 2014, 5, 3309. [Google Scholar] [CrossRef]
- Sugahara, M.; Mizohata, E.; Nango, E.; Suzuki, M.; Tanaka, T.; Masudala, T.; Tanaka, R.; Shimamura, T.; Tanaka, Y.; Suno, C.; et al. Grease matrix as a versatile carrier of proteins for serial crystallography. Nat. Methods 2015, 12, 61–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berntsen, P.; Hadian Jazi, M.; Kusel, M.; Martin, A.V.; Ericsson, T.; Call, M.J.; Trenker, R.; Roque, F.G.; Darmanin, C.; Abbey, B. The serial millisecond crystallography instrument at the Australian Synchrotron incorporating the “Lipidico” injector. Rev. Sci. Instrum. 2019, 90, 085110. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Nam, K.H. Sample delivery using viscous media, a syringe and a syringe pump for serial crystallography. J. Synchrotron Radiat. 2019, 26 Pt 5, 1815–1819. [Google Scholar] [CrossRef] [Green Version]
- Stellato, F.; Oberthur, D.; Liang, M.; Bean, R.; Gati, C.; Yefanov, O.; Barty, A.; Burkhardt, A.; Fischer, P.; Galli, L.; et al. Room-temperature macromolecular serial crystallography using synchrotron radiation. IUCrJ 2014, 1 Pt 4, 204–212. [Google Scholar] [CrossRef]
- Nam, K.H. Stable sample delivery in viscous media via a capillary for serial crystallography. J. Appl. Crystallogr. 2020, 53, 45–50. [Google Scholar] [CrossRef]
- Hunter, M.S.; Segelke, B.; Messerschmidt, M.; Williams, G.J.; Zatsepin, N.A.; Barty, A.; Benner, W.H.; Carlson, D.B.; Coleman, M.; Graf, A.; et al. Fixed-target protein serial microcrystallography with an X-ray free electron laser. Sci. Rep. 2014, 4, 6026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roedig, P.; Ginn, H.M.; Pakendorf, T.; Sutton, G.; Harlos, K.; Walter, T.S.; Meyer, J.; Fischer, P.; Duman, R.; Vartiainen, I.; et al. High-speed fixed-target serial virus crystallography. Nat. Methods 2017, 14, 805–810. [Google Scholar] [CrossRef]
- Lee, D.; Baek, S.; Park, J.; Lee, K.; Kim, J.; Lee, S.J.; Chung, W.K.; Lee, J.L.; Cho, Y.; Nam, K.H. Nylon mesh-based sample holder for fixed-target serial femtosecond crystallography. Sci. Rep. 2019, 9, 6971. [Google Scholar] [CrossRef] [Green Version]
- Cohen, A.E.; Soltis, S.M.; Gonzalez, A.; Aguila, L.; Alonso-Mori, R.; Barnes, C.O.; Baxter, E.L.; Brehmer, W.; Brewster, A.S.; Brunger, A.T.; et al. Goniometer-based femtosecond crystallography with X-ray free electron lasers. Proc. Natl. Acad. Sci. USA 2014, 111, 17122–17127. [Google Scholar] [CrossRef] [Green Version]
- Roessler, C.G.; Kuczewski, A.; Stearns, R.; Ellson, R.; Olechno, J.; Orville, A.M.; Allaire, M.; Soares, A.S.; Héroux, A. Acoustic methods for high-throughput protein crystal mounting at next-generation macromolecular crystallographic beamlines. J. Synchrotron Radiat. 2013, 20, 805–808. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.; Park, S.; Lee, K.; Kim, J.; Park, G.; Nam, K.H.; Baek, S.; Chung, W.K.; Lee, J.-L.; Cho, Y.; et al. Application of a high-throughput microcrystal delivery system to serial femtosecond crystallography. J. Appl. Crystallogr. 2020, 53, 477–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monteiro, D.C.F.; Vakili, M.; Harich, J.; Sztucki, M.; Meier, S.M.; Horrell, S.; Josts, I.; Trebbin, M. A microfluidic flow-focusing device for low sample consumption serial synchrotron crystallography experiments in liquid flow. J. Synchrotron Radiat. 2019, 26 Pt 2, 406–412. [Google Scholar] [CrossRef]
- Calvey, G.D.; Katz, A.M.; Pollack, L. Microfluidic Mixing Injector Holder Enables Routine Structural Enzymology Measurements with Mix-and-Inject Serial Crystallography Using X-ray Free Electron Lasers. Anal. Chem. 2019, 91, 7139–7144. [Google Scholar] [CrossRef] [PubMed]
- Nam, K.H. Sample Delivery Media for Serial Crystallography. Int. J. Mol. Sci. 2019, 20, 1094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, K.; Lee, D.; Baek, S.; Park, J.; Lee, S.J.; Park, S.; Chung, W.K.; Lee, J.-L.; Cho, H.-S.; Cho, Y.; et al. Viscous-medium-based crystal support in a sample holder for fixed-target serial femtosecond crystallography. J. Appl. Crystallogr. 2020, 53, 1051–1059. [Google Scholar] [CrossRef]
- Conrad, C.E.; Basu, S.; James, D.; Wang, D.; Schaffer, A.; Roy-Chowdhury, S.; Zatsepin, N.A.; Aquila, A.; Coe, J.; Gati, C.; et al. A novel inert crystal delivery medium for serial femtosecond crystallography. IUCrJ 2015, 2 Pt 4, 421–430. [Google Scholar] [CrossRef]
- Nogly, P.; James, D.; Wang, D.; White, T.A.; Zatsepin, N.; Shilova, A.; Nelson, G.; Liu, H.; Johansson, L.; Heymann, M.; et al. Lipidic cubic phase serial millisecond crystallography using synchrotron radiation. IUCrJ 2015, 2 Pt 2, 168–176. [Google Scholar] [CrossRef]
- Park, J.; Park, S.; Kim, J.; Park, G.; Cho, Y.; Nam, K.H. Polyacrylamide injection matrix for serial femtosecond crystallography. Sci. Rep. 2019, 9, 2525. [Google Scholar] [CrossRef] [Green Version]
- Sugahara, M.; Song, C.Y.; Suzuki, M.; Masuda, T.; Inoue, S.; Nakane, T.; Yumoto, F.; Nango, E.; Tanaka, R.; Tono, K.; et al. Oil-free hyaluronic acid matrix for serial femtosecond crystallography. Sci. Rep. 2016, 6, 24484. [Google Scholar] [CrossRef] [Green Version]
- Sugahara, M.; Nakane, T.; Masuda, T.; Suzuki, M.; Inoue, S.; Song, C.Y.; Tanaka, R.; Nakatsu, T.; Mizohata, E.; Yumoto, F.; et al. Hydroxyethyl cellulose matrix applied to serial crystallography. Sci. Rep. 2017, 7, 703. [Google Scholar] [CrossRef]
- Nam, K.H. Shortening injection matrix for serial crystallography. Sci. Rep. 2020, 10, 107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nam, K.H. Polysaccharide-Based Injection Matrix for Serial Crystallography. Int. J. Mol. Sci. 2020, 21, 3332. [Google Scholar] [CrossRef] [PubMed]
- Sugahara, M.; Motomura, K.; Suzuki, M.; Masuda, T.; Joti, Y.; Numata, K.; Tono, K.; Yabashi, M.; Ishikawa, T. Viscosity-adjustable grease matrices for serial nanocrystallography. Sci. Rep. 2020, 10, 1371. [Google Scholar] [CrossRef] [PubMed]
- Ishchenko, A.; Peng, L.; Zinovev, E.; Vlasov, A.; Lee, S.C.; Kuklin, A.; Mishin, A.; Borshchevskiy, V.; Zhang, Q.; Cherezov, V. Chemically Stable Lipids for Membrane Protein Crystallization. Cryst. Growth Des. 2017, 17, 3502–3511. [Google Scholar] [CrossRef]
- Liu, W.; Ishchenko, A.; Cherezov, V. Preparation of microcrystals in lipidic cubic phase for serial femtosecond crystallography. Nat. Protoc. 2014, 9, 2123–2134. [Google Scholar] [CrossRef] [Green Version]
- Botha, S.; Nass, K.; Barends, T.R.; Kabsch, W.; Latz, B.; Dworkowski, F.; Foucar, L.; Panepucci, E.; Wang, M.; Shoeman, R.L.; et al. Room-temperature serial crystallography at synchrotron X-ray sources using slowly flowing free-standing high-viscosity microstreams. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71 Pt 2, 387–397. [Google Scholar] [CrossRef] [Green Version]
- Kovacsova, G.; Grunbein, M.L.; Kloos, M.; Barends, T.R.M.; Schlesinger, R.; Heberle, J.; Kabsch, W.; Shoeman, R.L.; Doak, R.B.; Schlichting, I. Viscous hydrophilic injection matrices for serial crystallography. IUCrJ 2017, 4, 400–410. [Google Scholar] [CrossRef] [Green Version]
- Santacatalina, J.V.; Garcia-Perez, J.V.; Benedito, E.C.J. Ultrasonic monitoring of lard crystallization during storage. Food Res. Int. 2011, 44, 146–155. [Google Scholar] [CrossRef]
- Devi, A.; Khatkar, B.S. Physicochemical, rheological and functional properties of fats and oils in relation to cookie quality: A review. J. Food Sci. Technol. 2016, 53, 3633–3641. [Google Scholar] [CrossRef] [Green Version]
- Madhaven, B.N. Final Report on the Safety Assessment of Lard Glyceride, Hydrogenated Lard Glyceride, Lard Glycerides, Hydrogenated Lard Glycerides, Lard, and Hydrogenated Lard. Int. J. Toxicol. 2001, 20 (Suppl. 2), 57–64. [Google Scholar]
- Ezekannagha, C.B.; Ude, C.N.; Onukwuli, O.D. Optimization of the methanolysis of lard oil in the production of biodiesel with response surface methodology. Egypt. J. Pet. 2017, 26, 1001–1011. [Google Scholar] [CrossRef]
- Ramli, S.; Talib, R.A.; Rahman, R.A.; Zainuddin, N.; Othman, S.H.; Rashid, N.M. Detection of Lard in Ink Extracted from Printed Food Packaging Using Fourier Transform Infrared Spectroscopy and Multivariate Analysis. J. Spectrosc. 2015, 2015, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Sharma, H.; Giriprasad, R.; Goswami, M. Animal fat-Processing and Its Quality Control. J. Food Process. Technol. 2013, 4, 1000252. [Google Scholar]
- Ahmad Nizar, N.N.; Nazrim Marikkar, J.M.; Hashim, D.M. Differentiation of Lard, Chicken Fat, Beef Fat and Mutton Fat by GCMS and EA-IRMS Techniques. J. Oleo Sci. 2013, 62, 459–464. [Google Scholar] [CrossRef] [PubMed]
- Park, S.Y.; Ha, S.C.; Kim, Y.G. The Protein Crystallography Beamlines at the Pohang Light Source II. Biodesign 2017, 5, 30–34. [Google Scholar]
- Barty, A.; Kirian, R.A.; Maia, F.R.; Hantke, M.; Yoon, C.H.; White, T.A.; Chapman, H. Cheetah: Software for high-throughput reduction and analysis of serial femtosecond X-ray diffraction data. J. Appl. Crystallogr. 2014, 47 Pt 3, 1118–1131. [Google Scholar] [CrossRef] [Green Version]
- White, T.A.; Mariani, V.; Brehm, W.; Yefanov, O.; Barty, A.; Beyerlein, K.R.; Chervinskii, F.; Galli, L.; Gati, C.; Nakane, T.; et al. Recent developments in CrystFEL. J. Appl. Crystallogr. 2016, 49 Pt 2, 680–689. [Google Scholar] [CrossRef] [Green Version]
- Adams, P.D.; Afonine, P.V.; Bunkoczi, G.; Chen, V.B.; Davis, I.W.; Echols, N.; Headd, J.J.; Hung, L.W.; Kapral, G.J.; Grosse-Kunstleve, R.W.; et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2010, 66, 213–221. [Google Scholar] [CrossRef] [Green Version]
- Bae, J.E.; Hwang, K.Y.; Nam, K.H. Structural analysis of substrate recognition by glucose isomerase in Mn2+ binding mode at M2 site in S. rubiginosus. Biochem. Biophys. Res. Commun. 2018, 503, 770–775. [Google Scholar] [CrossRef]
- Emsley, P.; Cowtan, K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. Sect. D Biol. Crystallogr. 2004, 60 Pt 12 Pt 1, 2126–2132. [Google Scholar] [CrossRef] [Green Version]
- Afonine, P.V.; Grosse-Kunstleve, R.W.; Echols, N.; Headd, J.J.; Moriarty, N.W.; Mustyakimov, M.; Terwilliger, T.C.; Urzhumtsev, A.; Zwart, P.H.; Adams, P.D. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D Biol. Crystallogr. 2012, 68 Pt 4, 352–367. [Google Scholar] [CrossRef] [Green Version]
- Williams, C.J.; Headd, J.J.; Moriarty, N.W.; Prisant, M.G.; Videau, L.L.; Deis, L.N.; Verma, V.; Keedy, D.A.; Hintze, B.J.; Chen, V.B.; et al. MolProbity: More and better reference data for improved all-atom structure validation. Protein Sci. 2018, 27, 293–315. [Google Scholar] [CrossRef] [PubMed]




| Data Collection | Lysozyme | Glucose Isomerase |
|---|---|---|
| Energy (eV) | 12,659 | 12,659 |
| Exposure time (ms) | 100 | 100 |
| Space group | P43212 | I222 |
| Cell dimensions (Å) | ||
| a | 79.45 | 94.19 |
| b | 79.45 | 99.92 |
| c | 38.47 | 103.25 |
| Collected images | 60,000 | 110,000 |
| Hits images | 16,136 | 48,506 |
| Indexed pattern | 14,365 | 18,889 |
| Resolution (Å) | 80.0–1.75 (1.81–1.75) | 72.46–1.80 (1.86–1.80) |
| Unique reflections | 12,954 (1256) | 45,422 (4492) |
| Completeness (%) | 100.0 (100.0) | 100.0 (100.0) |
| Redundancy | 385.2 (266.8) | 565.1 (387.8) |
| SNR | 9.84 (3.60) | 5.72 (2.81) |
| CC | 0.9933 (0.8049) | 0.9772 (0.5475) |
| CC* | 0.9983 (0.9444) | 0.9943 (0.8412) |
| Rsplit (%) a | 7.21 (33.84) | 12.90 (39.09) |
| Wilson B factor (Å2) | 33.85 | 30.62 |
| Refinement | ||
| Resolution (Å) | 56.18–1.75 | 71.80–1.80 |
| Rwork | 16.62 | 15.81 |
| Rfree b | 18.65 | 17.66 |
| Rms deviations | ||
| Bond length (Å) | 0.014 | 0.007 |
| Bond angle (°) | 1.937 | 0.975 |
| B factors (Å2) | ||
| Protein | 39.29 | 29.95 |
| Ligands | 42.42 | 22.70 |
| Water | 41.71 | 39.43 |
| Ramachandran (%) | ||
| Preferred | 98.43 | 96.61 |
| Allowed | 1.57 | 3.12 |
| Outliers | 0.00 | 0.26 |
© 2020 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nam, K.H. Lard Injection Matrix for Serial Crystallography. Int. J. Mol. Sci. 2020, 21, 5977. https://doi.org/10.3390/ijms21175977
Nam KH. Lard Injection Matrix for Serial Crystallography. International Journal of Molecular Sciences. 2020; 21(17):5977. https://doi.org/10.3390/ijms21175977
Chicago/Turabian StyleNam, Ki Hyun. 2020. "Lard Injection Matrix for Serial Crystallography" International Journal of Molecular Sciences 21, no. 17: 5977. https://doi.org/10.3390/ijms21175977