The Histone Acetyltransferase GCN5 and the Associated Coactivators ADA2: From Evolution of the SAGA Complex to the Biological Roles in Plants
Abstract
:1. The Discovery of Histone Acetyltransferase GCN5 and the Associated SAGA Complex
2. The Plant SAGA Complex
| SAGA Modules | Yeast | Arabidopsis thaliana | Arabidopsis Mutant Phenotype |
|---|---|---|---|
| HATm | GCN5 | GCN5 (AT3G54610, HAG1) | Pleiotropic effects on development and responses to stress [22,23,25,26] |
| ADA2 | ADA2b (AT4G16420, PRZ1) | Pleiotropic effects on development and responses to stress [22,24,27] | |
| ADA2a (AT3G07740) | No developmental abnormalities [28] | ||
| ADA3 | ADA3a (AT2G19390) | Involved in flowering (Vlachonasios, under review) | |
| ADA3b (AT4G29790) | No developmental abnormalities [39] | ||
| SGF29 | SGF29a (AT3G27460) | No developmental abnormalities [29] | |
| SGF29b (AT5G40550) | No developmental abnormalities [29] | ||
| COREm | ADA1 | ADA1a (AT2G24530) | Not available |
| ADA1b (AT4G31440) | Not available | ||
| SPT3 | TAF13 (AT1G026280) | Seed development [40] | |
| SPT7 | HAF1 (AT1G32750, HAC13, TAF1) | Light responses [41] | |
| SPT8 | Not detected | ||
| SPT20 | SPT20 (AT1G72390) | Late flowering [34] | |
| TAF5 | TAF5 (AT5G25150) | Lethal [31] | |
| TAF6 | TAF6a (AT1G04950) | Lethal [32] | |
| TAF6b (AT1G54360) | |||
| TAF9 | TAF9 (AT1G54140) | Not available | |
| TAF10 | TAF10 (AT4G31720) | Involved in osmotic stress [35] | |
| TAF12 | TAF12a (AT3G10070) | ||
| TAF12b (AT1G17440, EER4, CKH1) | Involved in ethylene and cytokinin responses [36,37] | ||
| TRA1m | TRA1 | TRA1a (AT2G17930) | Early flowering [33] |
| TRA1b (AT4G36080) | No developmental abnormalities [33] | ||
| DUBm | SGF73 | Not detected | |
| SGF11 | SGF11 (AT5G58575) | No developmental abnormalities [38] | |
| UBP8 | UBP22 (AT5G10790) | No developmental abnormalities [20] | |
| SUS1 | SUS1 (AT3G27100, ENY2) | No developmental abnormalities [38] |
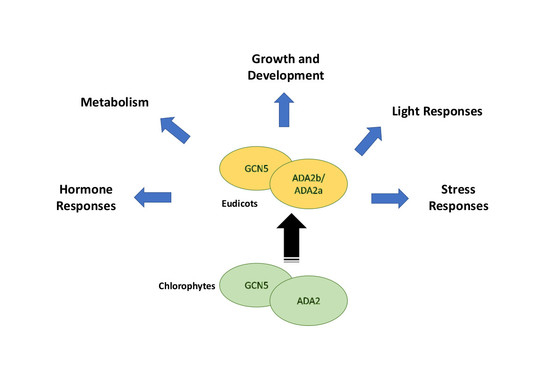
3. Origin of Plant SAGA Complexes
4. The Biological Role of GCN5 and ADA2b in Plants
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Roeder, R.G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem. Sci. 1996, 21, 327–335. [Google Scholar] [CrossRef]
- Li, B.; Carey, M.; Workman, J.L. The role of chromatin during transcription. Cell 2007, 128, 707–719. [Google Scholar] [CrossRef] [Green Version]
- Bannister, A.J.; Kouzarides, T. Regulation of chromatin by histone modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef] [PubMed]
- Strahl, D.B.; Briggs, S.D. The SAGA continues: The rise of cis- and trans-histone crosstalk pathways. Biochim. Biophys. Acta Gene Reg. Mechan. 2020, 194600. [Google Scholar] [CrossRef] [PubMed]
- Berger, S.L.; Pina, B.; Silverman, N.; Marcus, G.A.; Agapite, J.; Reiger, J.L.; Triezenberg, S.J.; Guarente, L. Genetic isolation of ADA2: A potential transcriptional adaptor required for function of certain acidic activation domains. Cell 1992, 70, 251–265. [Google Scholar] [CrossRef]
- Georgakopoulos, T.; Thireos, G. Two distinct yeast transcriptional activators require the function of the GCN5 protein to promote normal levels of transcription. EMBO J. 1992, 11, 4145–4152. [Google Scholar] [CrossRef] [PubMed]
- Brownell, J.E.; Zhou, J.; Ranalli, T.; Kobayashi, R.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Tetrahymena histone Acetyltransferase a: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell 1996, 84, 843–851. [Google Scholar] [CrossRef] [Green Version]
- Kuo, M.H.; Brownell, J.E.; Sobel, R.E.; Ranalli, T.A.; Cook, R.G.; Edmondson, D.G.; Roth, S.Y.; Allis, C.D. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature 1996, 383, 269–272. [Google Scholar] [CrossRef] [PubMed]
- Grant, P.A.; Duggan, L.; Côté, J.; Roberts, S.M.; Brownell, J.E.; Candau, R.; Ohba, R.; Owen-Hughes, T.; Allis, C.D.; Winston, F.; et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997, 11, 1640–1650. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, S.A.; Rao, B.; Garcia, B.A.; Hake, S.B.; Diaz, R.L.; Shabanowitz, J.; Hunt, D.F.; Allis, C.D.; Lieb, J.D.; Strahl, B.D. Identification of histone H3 lysine 36 acetylation as a highly conserved histone modification. J. Biol. Chem. 2007, 282, 7632–7640. [Google Scholar] [CrossRef] [Green Version]
- Lee, K.K.; Workman, J.L. Histone acetyltransferase complexes: One size doesn’t fit all. Nat. Rev. Mol. Cell Biol. 2007, 8, 284–295. [Google Scholar] [CrossRef] [PubMed]
- Baker, S.P.; Grant, P.A. The SAGA continues: Expanding the cellular role of a transcriptional co-activator complex. Oncogene 2007, 26, 5329–5340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Dienemann, C.; Stützer, A.; Urlaub, H.; Cheung, A.C.M.; Cramer, P. Structure of the transcription coactivator SAGA. Nature 2020, 577, 717–720. [Google Scholar] [CrossRef]
- Papai, G.; Frechard, A.; Kolesnikova, O.; Crucifix, C.; Schultz, P.; Ben-Shem, A. Structure of SAGA and mechanism of TBP deposition on gene promoters. Nature 2020, 577, 711–716. [Google Scholar] [CrossRef]
- Helmlinger, D.; Tora, L. Sharing the SAGA. Trends Biochem. Sci. 2017, 42, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Brown, C.E.; Howe, L.; Sousa, K.; Alley, S.C.; Carrozza, M.J.; Tan, S.; Workman, J.L. Recruitment of HAT complexes by direct activator interactions with the ATM-related Tra1 subunit. Science 2001, 292, 2333–2337. [Google Scholar] [CrossRef] [PubMed]
- Lee, T.I.; Causton, H.C.; Holstege, F.C.; Shen, W.C.; Hannett, N.; Jennings, E.G.; Winston, F.; Green, M.R.; Young, R.A. Redundant roles for the TFIID and SAGA complexes in global transcription. Nature 2000, 405, 701–704. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.X.; Hou, X.M.; Zhang, C.J.; Tan, L.M.; Shao, C.R.; Lin, R.N.; Su, Y.N.; Cai, X.W.; Li, L.; Chen, S.; et al. A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J. 2020, 39, e102008. [Google Scholar] [CrossRef] [PubMed]
- Grasser, K.D.; Rubio, V.; Barneche, F. Multifaceted activities of the plant SAGA complex. Biochim. Biophys. Acta (BBA) Gene Regul. Mech. 2020, 194613. [Google Scholar] [CrossRef]
- Nassrallah, A.; Rougée, M.; Bourbousse, C.; Drevensek, S.; Fonseca, S.; Iniesto, E.; Ait-Mohamed, O.; Deton-Cabanillas, A.F.; Zabulon, G.; Ahmed, I.; et al. DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. eLife 2018, 7, e37892. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Dent, S.Y.R. Functions of SAGA in development and disease. Epigenomics 2014, 6, 329–339. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vlachonasios, K.E.; Thomashow, M.F.; Triezenberg, S.J. Disruption mutations of ADA2b and GCN5 transcriptional adaptor genes dramatically affect Arabidopsis growth, development, and gene expression. Plant Cell 2003, 15, 626–638. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, C.; Bergounioux, C.; Domenichini, S.; Delarue, M.; Zhou, D.-X. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J. Biol. Chem. 2003, 278, 28246–28251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sieberer, T.; Hauser, M.-T.; Seifert, G.J.; Lusching, C. PROPORZ1, a putative Arabidopsis transcriptional adaptor protein, mediates auxin and cytokinin signals in the control of cell proliferation. Curr. Biol. 2003, 13, 837–842. [Google Scholar] [CrossRef] [Green Version]
- Vlachonasios, K.E.; Kaldis, A.; Nikoloudi, A.; Tsementzi, D. The role of transcriptional coactivator ADA2b in Arabidopsis abiotic stress responses. Plant Signal. Behav. 2011, 6, 1475–1478. [Google Scholar] [CrossRef] [Green Version]
- Benhamed, M.; Bertrand, C.; Servet, C.; Zhou, D.-X. Arabidopsis GCN5, HD1, and TAF1/HAF2 interact to regulate histone acetylation required for light-responsive gene expression. Plant Cell 2006, 18, 2893–2903. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.; Piquerez, S.J.M.; Ramirez-Prado, J.S.; Mastorakis, E.; Veluchamy, A.; Latrasse, D.; Manza-Mianza, D.; Brik-Chaouche, R.; Huang, Y.; Rodriguez-Granados, N.Y.; et al. GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5’ and 3’ ends of its target genes. Nucleic Acids Res. 2020, 48, 5953–5966. [Google Scholar] [CrossRef]
- Hark, A.T.; Vlachonasios, K.E.; Pavangadkar, K.A.; Rao, S.; Gordon, H.; Adamakis, I.D.; Kaldis, A.; Thomashow, M.F.; Triezenberg, S.J. Two Arabidopsis orthologs of the transcriptional coactivator ADA2 have distinct biological functions. Biochim. Biophys. Acta 2009, 1789, 117–124. [Google Scholar] [CrossRef]
- Kaldis, A.; Tsementzi, D.; Tanriverdi, O.; Vlachonasios, K.E. Arabidopsis thaliana transcriptional co-activators ADA2b and SGF29a are implicated in salt stress responses. Planta 2011, 233, 749–762. [Google Scholar] [CrossRef]
- Mao, Y.; Pavangadkar, K.A.; Thomashow, M.F.; Triezenberg, S.J. Physical and functional interactions of Arabidopsis ADA2 transcriptional coactivator proteins with the acetyltransferase GCN5 and with the cold-induced transcription factor CBF1. Biochim. Biophys. Acta 2006, 1759, 69–79. [Google Scholar] [CrossRef]
- Mastorakis, E. Chromatin Remodeling during Plant-Pathogen Interactions. Ph.D. Thesis, University of Warwick, Coventry, UK, 2017. [Google Scholar]
- Mougiou, N.; Poulios, S.; Kaldis, A.; Vlachonasios, K.E. Arabidopsis thaliana TBP-Associated Factor 5 is essential for plant growth and development. Mol. Breed. 2012, 30, 355–366. [Google Scholar] [CrossRef]
- Lago, C.; Clerici, E.; Dreni, L.; Horlow, C.; Caporali, E.; Colombo, L.; Kater, M.M. The Arabidopsis TFIID factor AtTAF6 controls pollen tube growth. Dev. Biol. 2005, 285, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Tanigawa, Y.; Murakami, T.; Araki, T.; Nagatani, A. Phytochrome-dependent late-flowering accelerates flowering through physical interactions with phytochrome B and constans. Proc. Natl. Acad. Sci. USA 2013, 110, 18017–18022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, X.; Ren, F.; Lu, Y.T. The Arabidopsis mutant stg1 identifies a function for TBP-associated factor 10 in plant osmotic stress adaptation. Plant Cell Physiol. 2006, 47, 1285–1294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robles, L.M.; Wampole, J.S.; Christians, M.J.; Larsen, P.B. Arabidopsis enhanced ethylene response 4 encodes an EIN3-interacting TFIID transcription factor required for proper ethylene response, including ERF1 induction. J. Exp. Bot. 2007, 58, 2627–2639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubo, M.; Furuta, K.; Demura, T.; Fukuda, H.; Liu, Y.G.; Shibata, D.; Kakimoto, T. The CKH1/EER4 gene encoding a TAF12-like protein negatively regulates cytokinin sensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2011, 52, 629–637. [Google Scholar] [CrossRef] [Green Version]
- Pfab, A.; Bruckmann, A.; Nazet, J.; Merkl, R.; Grasser, K.D. The Adaptor Protein ENY2 Is a Component of the Deubiquitination Module of the Arabidopsis SAGA Transcriptional Co-activator Complex but not of the TREX-2 Complex. J. Mol. Biol. 2018, 430, 1479–1494. [Google Scholar] [CrossRef]
- Srivastava, R.; Rai, K.M.; Pandey, B.; Singh, S.P.; Sawant, S.V. Spt-Ada-Gcn5-Acetyltransferase (SAGA) complex in plants: Genome wide identification, evolutionary conservation and functional determination. PLoS ONE 2015, 10, e0134709. [Google Scholar] [CrossRef] [Green Version]
- Lindner, M.; Simonini, S.; Kooiker, M.; Gagliardini, V.; Somssich, M.; Hohenstatt, M.; Simon, R.; Grossniklaus, U.; Kater, M.M. TAF13 interacts with PRC2 members and is essential for Arabidopsis seed development. Dev. Biol. 2013, 379, 28–37. [Google Scholar] [CrossRef] [Green Version]
- Bertrand, C.; Benhamed, M.; Li, Y.F.; Ayadi, M.; Lemonnier, G.; Renou, J.P.; Delarue, M.; Zhou, D.-X. Arabidopsis HAF2 gene encoding TATA-binding protein (TBP)-associated factor TAF1, is required to integrate light signals to regulate gene expression and growth. J. Biol. Chem. 2005, 280, 1465–1473. [Google Scholar] [CrossRef] [Green Version]
- Spedale, G.; Timmers, M.H.T.; Pijnappel, P.W.W.M. ATAC-king the complexity of SAGA during evolution. Genes Dev. 2012, 26, 527–541. [Google Scholar] [CrossRef] [Green Version]
- One Thousand Plant Transcriptomes Initiative; Leebens-Mack, J.H.; Barker, M.S. One thousand plant transcriptomes and the phylogenomics of green plants. Nature 2019, 574, 679–685. [Google Scholar]
- Wang, S.; Li, L.; Li, H. Genomes of early-diverging streptophyte algae shed light on plant terrestrialization. Nat. Plants 2020, 6, 95–106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palenik, B.; Grimwood, J.; Aerts, A. The tiny eukaryote Ostreococcus provides genomic insights into the paradox of plankton speciation. Proc. Natl. Acad. Sci. USA 2007, 104, 7705–7710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirooka, S.; Hirose, Y.; Kanesaki, Y.; Higuchi, S.; Fujiwara, T.; Onuma, R.; Era, A.; Ohbayashi, R.; Uzuka, A.; Nozaki, H.; et al. Acidophilic green algal genome provides insights into adaptation to an acidic environment. Proc. Natl. Acad. Sci. USA 2017, 114, E8304–E8313. [Google Scholar] [CrossRef] [Green Version]
- Worden, A.Z.; Lee, J.H.; Mock, T.; Rouzé, P.; Simmons, M.P.; Aerts, A.L.; Allen, A.E.; Cuvelier, M.L.; Derelle, E.; Everett, M.V.; et al. Green evolution and dynamic adaptations revealed by genomes of the marine picoeukaryotes Micromonas. Science 2009, 324, 268–272. [Google Scholar] [CrossRef] [Green Version]
- Lemieux, C.; Turmel, M.; Otis, C.; Pombert, J.F. A streamlined and predominantly diploid genome in the tiny marine green alga Chloropicon primus. Nat. Commun. 2019, 10, 4061. [Google Scholar] [CrossRef] [Green Version]
- Bowman, J.L.; Kohchi, T.; Yamato, K.T. Insights into land plant evolution garnered from the Marchantia polymorpha genome. Cell 2017, 171, 287–304. [Google Scholar] [CrossRef] [PubMed]
- Rensing, S.A.; Lang, D.; Zimmer, A.D. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 2008, 319, 64–69. [Google Scholar] [CrossRef] [Green Version]
- Antonova, S.V.; Boeren, J.; Timmers, H.T.M.; Snel, B. Epigenetics and transcription regulation during eukaryotic diversification: The saga of TFIID. Genes Dev. 2019, 33, 888–902. [Google Scholar] [CrossRef] [Green Version]
- Szövényi, P.; Perroud, P.F.; Symeonidi, A.; Stevenson, S.; Quatrano, R.S.; Rensing, S.A.; Cuming, A.C.; McDaniel, S.F. De novo assembly and comparative analysis of the Ceratodon purpureus transcriptome. Mol. Ecol. Resour. 2015, 15, 203–215. [Google Scholar] [CrossRef]
- Banks, J.A.; Nishiyama, T.; Hasebe, M. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 2011, 332, 960–963. [Google Scholar] [CrossRef] [Green Version]
- Chamala, S.; Chanderbali, A.S.; Der, J.P. Assembly and validation of the genome of the nonmodel basal angiosperm Amborella. Science 2013, 342, 1516–1517. [Google Scholar] [CrossRef]
- Povilus, R.A.; DaCosta, J.M.; Grassa, C.; Satyaki, P.R.; Moeglein, M.; Jaenisch, J.; Xi, Z.; Mathews, S.; Gehring, M.; Davis, C.C.; et al. Water lily (Nymphaea thermarum) genome reveals variable genomic signatures of ancient vascular cambium losses. Proc. Natl. Acad. Sci. USA 2020, 117, 8649–8656. [Google Scholar] [CrossRef] [PubMed]
- Cohen, R.; Schocken, J.; Kaldis, A.; Vlachonasios, K.E.; Hark, A.T.; McCain, E.R. The histone acetyltransferase GCN5 affects the inflorescence meristem and stamen development in Arabidopsis. Planta 2009, 230, 1207–1221. [Google Scholar] [CrossRef]
- Kornet, N.; Scheres, B. Members of the GCN5 histone acetyltransferase complex regulate PLETHORA-mediated root stem cell niche maintenance and transit amplifying cell proliferation in Arabidopsis. Plant Cell 2009, 21, 1070–1079. [Google Scholar] [CrossRef] [Green Version]
- Anzola, J.M.; Sieberer, T.; Ortbauer, M.; Butt, H.; Korbei, B.; Weinhofer, I.; Müllner, A.E.; Luschnig, C. Putative Arabidopsis transcriptional adaptor protein (PROPORZ1) is required to modulate histone acetylation in response to auxin. Proc. Natl. Acad. Sci. USA 2010, 107, 10308–10313. [Google Scholar] [CrossRef] [Green Version]
- Servet, C.; e Silva, N.C.; Zhou, D.-X. Histone acetyltransferase AtGCN5/HAG1 is a versatile regulator of developmental and inducible gene expression in Arabidopsis. Mol. Plant 2010, 3, 670–677. [Google Scholar] [CrossRef] [PubMed]
- Earley, K.W.; Shook, M.S.; Brower-Toland, B.; Hicks, L.; Pikaard, C.S. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J. 2007, 52, 615–626. [Google Scholar] [CrossRef] [PubMed]
- Long, J.A.; Ohno, C.; Smith, Z.R.; Meyerowitz, E.M. Topless regulates apical embryonic fate in Arabidopsis. Science 2006, 312, 1520–1523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, W.Q.; Li, D.X.; Zhao, F.; Xu, Z.H.; Bai, S.N. One additional histone deacetylase and 2 histone acetyltransferases are involved in cellular patterning of Arabidopsis root epidermis. Plant Signal. Behav. 2016, 11, e1131373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.Y.; Yang, W.; Forner, J.; Lohmann, J.U.; Noh, B.; Noh, Y.S. Epigenetic reprogramming by histone acetyltransferase HAG1/AtGCN5 is required for pluripotency acquisition in Arabidopsis. EMBO J. 2018, 37, e98726. [Google Scholar] [CrossRef] [PubMed]
- Weiste, C.; Dröge-Laser, W. The Arabidopsis transcription factor bZIP11 activates auxin-mediated transcription by recruiting the histone acetylation machinery. Nat. Commun. 2014, 5, 3883. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.Y.; Oh, J.E.; Noh, Y.S.; Noh, B. Epigenetic control of juvenile-to-adult phase transition by the Arabidopsis SAGA-like complex. Plant J. 2015, 83, 537–545. [Google Scholar] [CrossRef] [PubMed]
- Kotak, J.; Saisana, M.; Gegas, V. The histone acetyltransferase GCN5 and the transcriptional coactivator ADA2b affect leaf development and trichome morphogenesis in Arabidopsis. Planta 2018, 248, 613–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, T.; Jia, Q.; Wang, W. GCN5 modulates trichome initiation in Arabidopsis by manipulating histone acetylation of core trichome initiation regulator genes. Plant Cell Rep. 2019, 38, 755–765. [Google Scholar] [CrossRef] [PubMed]
- Poulios, S.; Vlachonasios, K.E. Synergistic action of GCN5 and CLAVATA1 in the regulation of gynoecium development in Arabidopsis thaliana. New Phytol. 2018, 220, 593–608. [Google Scholar] [CrossRef] [Green Version]
- Lenhard, M.; Bohnert, A.; Jürgens, G.; Laux, T. Termination of stem cell maintenance in Arabidopsis floral meristems by interactions between wuschel and agamous. Cell 2001, 105, 805–814. [Google Scholar] [CrossRef] [Green Version]
- Lohmann, J.U.; Hong, R.L.; Hobe, M.; Busch, M.A.; Parcy, F.; Simon, R.; Weigel, D. A molecular link between stem cell regulation and floral patterning in Arabidopsis. Cell 2001, 105, 793–803. [Google Scholar] [CrossRef] [Green Version]
- Sun, B.; Ito, T. Regulation of floral stem cell termination in Arabidopsis. Front. Plant Sci. 2015, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [Green Version]
- Müller, C.J.; Larsson, E.; Spíchal, L.; Sundberg, E. Cytokinin-auxin crosstalk in the gynoecial primordium ensures correct domain patterning. Plant Physiol. 2017, 175, 1144–1157. [Google Scholar] [CrossRef]
- Reyes-Olalde, J.I.; Zúñiga-Mayo, V.M.; Serwatowska, J. The bHLH transcription factor SPATULA enables cytokinin signaling, and both activate auxin biosynthesis and transport genes at the medial domain of the gynoecium. PLoS Genet. 2017, 13, e1006726. [Google Scholar] [CrossRef]
- Alvarez, J.; Smyth, D.R. CRABS CLAW and SPATULA, two Arabidopsis genes that control carpel development in parallel with AGAMOUS. Development 1999, 126, 2377–2386. [Google Scholar]
- Xing, J.; Wang, T.; Liu, Z.; Xu, J.; Yao, Y.; Hu, Z.; Peng, H.; Xin, M.; Yu, F.; Zhou, D.-X.; et al. GCN5-mediated histone acetylation of FRD3 contributes to iron homeostasis in Arabidopsis thaliana. Plant Phys. 2015, 168, 1309–1320. [Google Scholar] [CrossRef] [Green Version]
- Wang, T.; Xing, J.; Liu, Z.; Zheng, M.; Yao, Y.; Hu, Z.; Peng, H.; Xin, M.; Zhou, D.-X.; Ni, Z. Histone acetyltransferase GCN5-mediated regulation of long non-coding RNA At4 contributes to phosphate starvation response in Arabidopsis. J. Exp. Bot. 2019, 70, 6337–6348. [Google Scholar] [CrossRef] [PubMed]
- Stockinger, E.J.; Mao, Y.; Regier, M.K.; Triezenberg, S.J.; Thomashow, M.F. Transcriptional adaptor and histone acetyltransferase proteins in Arabidopsis and their interactions with CBF1, a transcriptional activator involved in cold-regulated gene expression. Nucleic Acids Res. 2001, 29, 1524–1533. [Google Scholar] [CrossRef] [PubMed]
- Pavangadkar, K.; Thomashow, M.F.; Triezenberg, S.J. Histone dynamics and roles of histone acetyltransferases during cold-induced gene regulation in Arabidopsis. Plant Mol. Biol. 2010, 74, 183–200. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.; Jiang, J.; Wu, Q.; Mao, N.; Han, D.; Hu, H.; Yang, C. The transcriptional coactivator ADA2b recruits a structural maintenance protein to double-strand breaks during DNA repair in plants. Plant Physiol. 2018, 176, 2613–2622. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H.; et al. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, X.; Lin, J.; Liu, X.; Wang, Z.; Xin, M.; Yao, Y.; Peng, H.; Zhou, D.X.; Ni, Z.; et al. Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. 2018, 97, 587–602. [Google Scholar] [PubMed]
- Li, S.; Lin, Y.C.; Wang, P. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell 2019, 31, 663–686. [Google Scholar] [CrossRef] [Green Version]
- Poulios, S.; Vlachonasios, K.E. Synergistic action of histone acetyltransferase GCN5 and receptor CLAVATA1 negatively affects ethylene responses in Arabidopsis thaliana. J. Exp. Bot. 2016, 67, 905–918. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Qi, B.; Wang, L.; Zhao, B.; Rode, S.; Riggan, N.D.; Ecker, J.R.; Qiao, H. EIN2-dependent regulation of acetylation of histone H3K14 and non-canonical histone H3K23 in ethylene signalling. Nat. Commun. 2016, 7, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Zhang, F.; Qiao, H. Chromatin Regulation in the Response of Ethylene: Nuclear Events in Ethylene Signaling. Small Methods 2019, 1900288. [Google Scholar] [CrossRef]
- Kong, L.; Qiu, X.; Kang, J. A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 2017, 27, 981–991. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vlachonasios, K.; Poulios, S.; Mougiou, N. The Histone Acetyltransferase GCN5 and the Associated Coactivators ADA2: From Evolution of the SAGA Complex to the Biological Roles in Plants. Plants 2021, 10, 308. https://doi.org/10.3390/plants10020308
Vlachonasios K, Poulios S, Mougiou N. The Histone Acetyltransferase GCN5 and the Associated Coactivators ADA2: From Evolution of the SAGA Complex to the Biological Roles in Plants. Plants. 2021; 10(2):308. https://doi.org/10.3390/plants10020308
Chicago/Turabian StyleVlachonasios, Konstantinos, Stylianos Poulios, and Niki Mougiou. 2021. "The Histone Acetyltransferase GCN5 and the Associated Coactivators ADA2: From Evolution of the SAGA Complex to the Biological Roles in Plants" Plants 10, no. 2: 308. https://doi.org/10.3390/plants10020308