High Predatory Capacity of a Novel Arthrobotrys oligospora Variety on the Ovine Gastrointestinal Nematode Haemonchus contortus (Rhabditomorpha: Trichostrongylidae)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Gathering of Samples for Isolation (Area of Study)
2.2. Infective Larvae of Haemochus Contortus
2.3. Isolation of Nematophagous Fungi
2.4. Radial Growth
2.5. Conidia Production
2.6. Morphological Identification
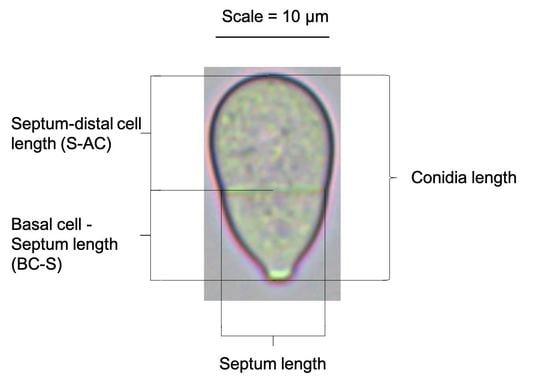
2.7. Conidial Measurements
2.8. Molecular Characterization of the Isolated Fungi
2.8.1. DNA Extraction
2.8.2. Amplification of Ribosomal DNA
2.8.3. Phylogenetic Analyses
2.9. Evaluation of the Predatory Ability
2.10. Statistical Analysis
3. Results
3.1. Isolation of Nematode-Trapping Fungi
3.2. Nematode-Trapping Fungi Growth
3.3. Nematode-Trapping Fungi Asexual Reproduction
3.4. Morphological Characterization of Nematode-Trapping Fungi
3.5. Phylogenetic Analyses of Arthrobotrys Isolates
3.6. High Depredatory Capacity of Nematophagous Native against H. Contortus In Vitro
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Köler, P. The biochemical basis of anthelmintic action and resistance. Int. J. Parasitol. 2001, 31, 336–345. [Google Scholar] [CrossRef]
- Barron, G.L. The Nematode-Destroying Fungi; Canadian Biological Publications Ltd.: Guelph, ON, Canada, 1977; p. 140. [Google Scholar]
- Sagüés, M.F.; Purslow, P.; Fernández, S.; Fusé, L.; Iglesias, L.; Saumell, C. Nematophagous fungi used for the biological control of gastrointestinal nematodes in livestock and administration routes. Rev. Iber. Micol. 2011, 28, 143–147. [Google Scholar] [CrossRef]
- Hernández, J.A.; Vázquez-Ruiz, R.A.; Cazapal-Monteiro, C.F.; Valderrábano, E.; Arroyo, F.L.; Francisco, I.; Miguélez, S.; Sánchez-Andrade, R.; Paz-Silva, A.; Arias, M.S. Isolation of ovicidal fungi from fecal samples of captive animals maintained in a zoological park. J. Fungi. 2017, 3, 29. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Tian, J.; Xiang, M.; Liu, X. How carnivorous fungi use three-celled constricting rings to trap nematodes. Protein Cell 2012, 3, 325–328. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Yang, E.; An, Z.; Liu, X. Evolution of nematode-trapping cells of predatory fungi of the Orbiliaceae based on evidence from rRNA-encoding DNA and multiprotein sequences. Proc. Natl. Acad. Sci. USA 2007, 104, 8379–8384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Braga, F.; De Araújo, J. Nematophagous fungi for biological control of gastrointestinal nematodes in domestic animals. Appl. Microbiol. Biotechnol. 2014, 98, 71–82. [Google Scholar] [CrossRef]
- Ojeda-Robertos, N.F.; Torres-Acosta, J.F.J.; Ayala-Burgos, A.J.; Sandoval-Castro, C.A.; Valero-Coss, R.O.; Mendoza-de-Gives, P. Digestibility of Duddingtonia flagrans chlamydospores in ruminants: In vitro and in vivo studies. BMC Vet. Res. 2009, 5, 46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paz-Silva, A.; Francisco, I.; Valero-Coss, R.O.; Cortiñas, F.J.; Sánchez, J.A.; Francisco, R.; Arias, M.; Suárez, J.L.; López-Arellano, M.E.; Sánchez-Andrade, R.; et al. Ability of the fungus Duddingtonia flagrans to adapt to the cyathostomin egg-output by spreading chlamydospores. Vet. Parasitol. 2011, 179, 277–282. [Google Scholar] [CrossRef] [PubMed]
- Fitz-Aranda, J.A.; Mendoza-de-Gives, P.; Torres-Acosta, J.F.; Liébano-Hernández, E.; López-Arellano, M.E.; Sandoval-Castro, C.A.; Quiroz-Romero, H. Duddingtonia flagrans chlamydospores in nutritional pellets: Effect of storage time and conditions on the trapping ability against Haemonchus contortus larvae. J. Helminthol. 2015, 16, 1–6. [Google Scholar]
- Tavela, A.O.; Araújo, J.V.; Braga, F.R.; Silveira, W.F.; Silva, V.H.D.; Júnior, M.C.; Alcántar Borges, L.; Milani Araujo, J.; dos Anjos Benjamin, L.; Carvalho, G.R.; et al. Coadministration of sodium alginate pellets containing the fungi Duddingtonia flagrans and Monacrosporium thaumasium on cyathostomin infective larvae after passing through the gastrointestinal tract of horses. Res. Vet. Sci. 2013, 94, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Ojeda-Robertos, N.F.; Aguilar-Marcelino, L.; Olmedo-Juárez, A.; Luna-Palomera, C.; Peralta-Torres, J.A.; López-Arellano, M.E.; Mendoza-de-Gives, P. In vitro predatory activity of nematophagous fungi isolated from water buffalo feces and from soil in the Mexican southeastern. Braz. J. Vet. Parasitol. Jaboticabal. 2019, 28, 314–319. [Google Scholar] [CrossRef] [PubMed]
- Mendoza-de-Gives, P.; López-Arellano, M.E.; Aguilar-Marcelino, L.; Olazarán-Henkins, S.; Reyes-Guerrero, D.; Ramírez-Várgas, G.; Vega-Murillo, V.E. The nematophagous fungus Duddingtonia flagrans reduces the gastrointestinal parasitic nematode larvae population in faeces of orally treated calves maintained under tropical conditions—Dose/response assessment. Vet. Parasitol. 2018, 263, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Xue-Mei, N.; Ke-Qin, Z. Arthrobotrys oligospora: A model organism for understanding the interaction between fungi and nematodes. Mycology 2011, 2, 59–78. [Google Scholar] [CrossRef]
- Yang, C.T.; de Ulzurrun, G.V.; Goncalves, A.P.; Lin, H.C.; Chang, C.W.; Huang, T.Y.; Chen, S.A.; Lai, C.K.; Tsai, I.J.; Schroeder, F.C.; et al. Natural diversity in the predatory behavior facilitate the stablishment of a robust model strain for nematode-trapping fungi. Proc. Natl. Acad. Sci. USA 2020, 117, 6762–6770. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Hyde, K.; Jeewon, R.; Cai, L.; Vijaykrishna, D.; Zhang, K.Q. Phylogenetics and evolution of nematode-trapping fungi (Orbiliales) estimated from nuclear and protein coding genes. Mycologia 2005, 97, 1034–1046. [Google Scholar] [CrossRef]
- Wang, B.B.; Liu, W.; Chen, M.Y.; Li, X.; Han, Y.; Xu, Q.; Sun, L.J.; Xie, D.Q.; Cai, K.Z.; Liu, Y.Z.; et al. Isolation and characterization of china isolates of Duddingtonia flagrans, a candidate of the nematophagous fungi for biocontrol of animal parasitic nematodes. J. Parasitol. 2015, 101, 476–484. [Google Scholar] [CrossRef]
- Arias, M.S.; Arroyo, F.L.; Cazapal-Monteiro, C.; Hernández, J.A.; Suárez, J.; Francisco, I.; López-Arellano, M.E.; Sánchez-Andrade, R.; Mendoza de Gives, P.; Paz-Silva, A. Formulating Duddingtonia flagrans in nutritional pellets for the sustainable control of equine strongyles. J. Sci. Technol. Environ. 2015, 5, 1–16. [Google Scholar]
- Cooke, R.C.; Godfrey, B.E.S. A key to the nematode destroying fungi. Trans. Br. Mycol. Soc. 1964, 47, 61–74. [Google Scholar] [CrossRef]
- White, T.J.; Bruns, T.; Lee, S.; Taylor, J. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M.A., Gelfand, D.H., Sninsky, J.J., White, T.J., Eds.; Academic Press: San Diego, CA, USA, 1990; pp. 315–322. [Google Scholar]
- Kimie-Falbo, M.; Thomaz-Soccol, V.; Eloi-Sandini, I.; Aparecida-Vicente, V.; Dobl, D.; Soccol, C.R. Isolation and characterization of the nematophagous fungus Arthrobotrys conoides. Parasitol. Res. 2013, 112, 177–185. [Google Scholar] [CrossRef] [PubMed]
- Maddison, D.R.; Maddison, W.P. MacClade 4.08a: Analysis of Phylogeny and Character Evolution; Sinauer Associates: Sunderland, MA, USA, 2005. [Google Scholar]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [PubMed]
- Posada, D.; Crandall, K.A. Modeltest: Testing the model of DNA substitution. Bioinformatics 1998, 14, 817–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, M.E.; Jaffee, B.A. PCR Primers with Ehhanced Specificity for Nematode-Trapping Fungi (Orbiliales). Environ. Microbiol. 2009, 58, 117–128. [Google Scholar] [CrossRef]
- Raja, H.A.; Miller, A.N.; Pearce, C.J.; Oberlies, N.H. Fungal identification using molecular tools: A primer for the natural products research community. Nat. Prod. 2017, 80, 756–770. [Google Scholar] [CrossRef]
- Yang, J.; Wang, L.; Ji, X.; Feng, Y.; Li, X.; Zou, C.; Xu, J.; Ren, Y.; Mi, Q.; Wu, J.; et al. Genomic and proteomic analyses of the fungus Arthrobotrys oligospora provide insights into nematode-trap formation. PLoS Pathog. 2011, 7, e1002179. [Google Scholar] [CrossRef] [Green Version]
- Schmoll, M.; Esquivel-Naranjo, E.U.; Herrera-Estrella, A. Trichoderma in the light of day–physiology and development. Fungal Genet. Biol. 2010, 47, 909–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santos, C.P.; Charles, T.P.; Rodrigues, M.L.A. Predatory activity of Arthrobotrys oligospora and Duddingtonia flagrans on preparasitic larval stages of cyathostominae under different constant temperatures. Cienc. Rural 2001, 31, 839–842. [Google Scholar] [CrossRef] [Green Version]






| Species | Source | Region | Gene Bank | Reference |
|---|---|---|---|---|
| Dactylellina haptospora | ND | ND | DQ999820 | [6] |
| Dactylellina querci | ND | ND | AY773453 | [6] |
| Arthrobotrys anomala | Soil | China | AY773451 | [6] |
| A. amerospora | ND | ND | EF192178 | [21] |
| A. amerospora | ND | Germany | AF106533 | [21] |
| A. conoides | Soil | Brazil, Paraná | JN191309 | [21] |
| A. conoides | Soil | China | AY773455 | [6] |
| A. eudermata | Soil | China | AY773465 | [6] |
| Duddingtonia flagrans | Jardine soil | Germany, Berlín-Dahlem | AF106520 | [21] |
| D. flagrans | Vineyard soil | USA, California | EF445995 | [21] |
| A. iridis | Soil | China | AY773452 | [6] |
| A. janus | Soil | China | AY773459 | [6] |
| A. javanica | Soil | Indonesia, Java Island | U51947 | [21] |
| A. javanica | Soil | ND | EU977514 | [21] |
| A. musiformis | Soil | China | AY773469 | [6] |
| A. oligospora | Soil | China | AY773462 | [6] |
| A. oligospora | Decaying wood | ND | EU977526 | [21] |
| A. oligospora | Decaying wood | ND | EU977563 | [21] |
| A. oligospora | dung | ND | EU977523 | [21] |
| A. pseudoclavata | Soil | China | AY773446 | [6] |
| A. pyriformis | Soil | China | AY773450 | [6] |
| A. sinensis | Soil | China | AY773445 | [6] |
| A. thaumasia | Decaying wood | ND | EU977535 | [21] |
| A. thaumasia | Cold greenhouse | Germany, Berlín-Dahlem | AF106526 | [21] |
| A. vermicola | Soil | China | AY773454 | [6] |
| A. vermicola | Decaying leaves | ND | EU977508 | [21] |
| A. oligospora var. queretana A6 | Soil underneath a tree | México, Querétaro, El Marqués | MW748504 | This work |
| A. oligospora var. queretana A12 | Compost | México, Querétaro, El Marqués | MW748505 | This work |
| A. oligospora A13 | Ovine feces | México, Querétaro, El Marqués | MW748506 | This work |
| A. oligospora var. queretana R2-1 | Donkey feces | México, Querétaro, Jalpan | MW748501 | This work |
| A. oligospora var. queretana R2-6 | Ovine feces | México, Querétaro, Jalpan | MW748502 | This work |
| A. oligospora var. queretana R2-13 | Bovine feces | México, Querétaro, Jalpan | MW748500 | This work |
| A. oligospora var. queretana R2-14 | Compost | México, Querétaro, Jalpan | MW748503 | This work |
| Strain | Conidia Length (μm) | Septum (μm) | BC-S (μm) | S-DC (μm) | S-DC/BC-S |
|---|---|---|---|---|---|
| A6 (n = 132) | 15.2–24.4 (18.4) | 5.3–11.3 (8.4) | 4.0–11.4 (6.7) | 8.4–15.3 (11.7) | 1.74 |
| A12 (n = 100) | 15.6–21.6 (18.6) | 6.7–10.8 (8.0) | 4.4–9.1 (6.7) | 9.5–14.7 (11.9) | 1.79 |
| A13 (n = 105) | 15.4–23.1 (18.8) | 6.4–11.3 (8.6) | 4.5–9.2 (7.0) | 8.6–14.4 (11.9) | 1.71 |
| R2-1 (n = 96) | 15.0–21.9 (18.5) | 6.5–11.6 (9.0) | 3.8–8.8 (6.7) | 9.1–14.6 (11.9) | 1.78 |
| R2-6 (n = 99) | 13.9–22.1 (17.5) | 6.2–12.1 (8.4) | 3.6–8.9 (6.4) | 7.3–13.9 (11.1) | 1.75 |
| R2-13 (n = 100) | 16.0–30.3 (21.4) | 7.3–14.8 (8.2) | 6.4–15.8 (8.9) | 9.0–15.9 (12.4) | 1.39 |
| R2-14 (n = 100) | 15.1–22.2 (18.2) | 7.0–11.9 (9.0) | 4.1–8.9 (6.4) | 8.4–15.1 (11.8) | 1.85 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arroyo-Balán, F.; Landeros-Jaime, F.; González-Garduño, R.; Cazapal-Monteiro, C.; Arias-Vázquez, M.S.; Aguilar-Tipacamú, G.; Esquivel-Naranjo, E.U.; Mosqueda, J. High Predatory Capacity of a Novel Arthrobotrys oligospora Variety on the Ovine Gastrointestinal Nematode Haemonchus contortus (Rhabditomorpha: Trichostrongylidae). Pathogens 2021, 10, 815. https://doi.org/10.3390/pathogens10070815
Arroyo-Balán F, Landeros-Jaime F, González-Garduño R, Cazapal-Monteiro C, Arias-Vázquez MS, Aguilar-Tipacamú G, Esquivel-Naranjo EU, Mosqueda J. High Predatory Capacity of a Novel Arthrobotrys oligospora Variety on the Ovine Gastrointestinal Nematode Haemonchus contortus (Rhabditomorpha: Trichostrongylidae). Pathogens. 2021; 10(7):815. https://doi.org/10.3390/pathogens10070815
Chicago/Turabian StyleArroyo-Balán, Fabián, Fidel Landeros-Jaime, Roberto González-Garduño, Cristiana Cazapal-Monteiro, Maria Sol Arias-Vázquez, Gabriela Aguilar-Tipacamú, Edgardo Ulises Esquivel-Naranjo, and Juan Mosqueda. 2021. "High Predatory Capacity of a Novel Arthrobotrys oligospora Variety on the Ovine Gastrointestinal Nematode Haemonchus contortus (Rhabditomorpha: Trichostrongylidae)" Pathogens 10, no. 7: 815. https://doi.org/10.3390/pathogens10070815