The nodD1 Gene of Sinorhizobium fredii HH103 Restores Nodulation Capacity on Bean in a Rhizobium tropici CIAT 899 nodD1/nodD2 Mutant, but the Secondary Symbiotic Regulators nolR, nodD2 or syrM Prevent HH103 to Nodulate with This Legume
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microbiological Techniques
2.2. Identification of Nod Factors and Biological Activity Assays
2.3. Nodulation Assays
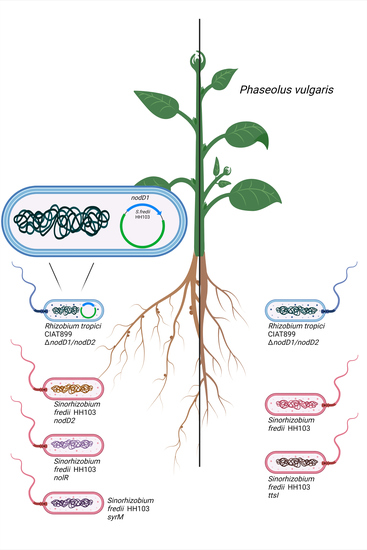
3. Results
3.1. The nodD1 Gene of S. fredii HH103, but Not nodD2, Restores Nodulation Ability in a R. tropici CIAT 899 nodD1 nodD2 Mutant
3.2. Inactivation of the Symbiotic Regulators nodD2, syrM or nolR Extends the Nodulation Range of S. fredii HH103 to Phaseolus vulgaris
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Poole, P.; Ramachandran, V.; Terpolilli, J. Rhizobia: From saprophytes to endosymbionts. Nat. Rev. Microbiol. 2018, 16, 291–303. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef] [Green Version]
- Basile, L.A.; Lepek, V.C. Legume–rhizobium dance: An agricultural tool that could be improved? Microb. Biotechnol. 2021, 14, 1897–1917. [Google Scholar] [CrossRef] [PubMed]
- Goyal, R.K.; Mattoo, A.K.; Schmidt, M.A. Rhizobial–host interactions and symbiotic nitrogen fixation in legume crops toward agriculture sustainability. Front. Microbiol. 2021, 12, 669404. [Google Scholar] [CrossRef]
- Perret, X.; Staehelin, C.; Broughton, W.J. Molecular basis of symbiotic promiscuity. Microbiol. Mol. Biol. Rev. 2000, 64, 180–201. [Google Scholar] [CrossRef] [Green Version]
- Ormeño-Orrillo, E.; Menna, P.; Gonzaga, L.A.; Ollero, F.J.; Nicolas, M.F.; Rodrigues, E.P.; Nakatami, A.S.; Batista, J.S.S.; Chueire, L.M.O.; Souza, R.C.; et al. Genomic basis of broad host range and environmental adaptability of Rhizobium tropici CIAT 899 and Rhizobium sp. PRF 81 which are used in inoculants for common bean (Phaseolus vulgaris L.). BMC Genom. 2012, 13, 735. [Google Scholar] [CrossRef] [Green Version]
- Margaret, I.; Becker, A.; Blom, J.; Bonilla, I.; Goesmann, A.; Göttfert, M.; Lloret, J.; Mittard-Runte, V.; Rückert, C.; Ruiz-Sainz, J.E.; et al. Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 2011, 155, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Hungría, M.; Andrade, D.S.; Chueire, L.M.O.; Probanza, A.; Gutiérrez-Mañero, F.J.; Megías, M. Isolation and characterization of new efficient and competitive bean (Phaseolus vulgaris L.) rhizobia from Brazil. Soil Biol. Biochem. 2000, 21, 1515–1528. [Google Scholar] [CrossRef]
- Del Cerro, P.; Rolla-Santos, A.P.; Gomes, D.F.; Marks, B.B.; Pérez-Montaño, F.; Rodríguez-Carvajal, M.A.; Nakatani, A.S.; Gil-Serrano, A.; Megías, M.; Ollero, F.J.; et al. Regulatory nodD1 and nodD2 genes of Rhizobium tropici strain CIAT 899 and their roles in the early stages of molecular signaling and host-legume nodulation. BMC Genom. 2015, 16, 251. [Google Scholar] [CrossRef] [Green Version]
- Del Cerro, P.; Rolla-Santos, A.P.; Gomes, D.F.; Marks, B.B.; Espuny, M.R.; Rodríguez-Carvajal, M.A.; Soria-Díaz, M.E.; Nakatani, A.S.; Hungría, M.; Ollero, F.J.; et al. Opening the “black box” of nodD3, nodD4 and nodD5 genes of Rhizobium tropici strain CIAT 899. BMC Genom. 2015, 16, 864. [Google Scholar] [CrossRef]
- Hernández-Lucas, I.; Segovia, L.; Martínez-Romero, E.; Pueppke, S.G. Phylogenetic relationships and host range of Rhizobium spp. that nodulates Phaseolus vulgaris L. Appl. Environ. Microbiol. 1995, 61, 2775–2779. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Jurado, S.; Rodríguez-Navarro, D.N.; Kawaharada, Y.; Perea, J.F.; Gil-Serrano, A.; Jin, H.; An, Q.; Rodríguez-Carvajal, M.A.; Andersen, S.U.; Sandal, N.; et al. Sinorhizobium fredii HH103 invades Lotus burttii by crack entry in a Nod factor- and surface polysaccharide-dependent manner. Mol. Plant Microbe Interact. 2016, 29, 925–937. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Baena, F.J.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.A.; Vinardell, J.M. Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 2016, 17, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khokhani, D.; Carrera Carriel, C.; Vayla, S.; Irving, T.B.; Stonoha-Arther, C.; Keller, N.P.; Ané, J.M. Deciphering the chitin code in plant symbiosis, defense, and microbial networks. Annu. Rev. Microbiol. 2021, 75, 583–607. [Google Scholar] [CrossRef] [PubMed]
- Staehelin, C.; Krishnan, H.B. Nodulation outer proteins: Double-edged swords of symbiotic rhizobia. Biochem. J. 2015, 470, 263–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acosta-Jurado, S.; Fuentes-Romero, F.; Ruiz-Sainz, J.E.; Janczarek, M.; Vinardell, J.M. Rhizobial exopolysaccharides: Genetic regulation of their synthesis and relevance in symbiosis with legumes. Int. J. Mol. Sci. 2021, 22, 6233. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Rodríguez-Navarro, D.N.; Kawaharada, Y.; Rodríguez-Carvajal, M.A.; Gil-Serrano, A.; Soria-Díaz, M.E.; Pérez-Montaño, F.; Fernández-Perea, J.; Niu, Y.; Alias-Villegas, C.; et al. Sinorhizobium fredii HH103 nolR and nodD2 mutants gain capacity for infection thread invasion of Lotus japonicus Gifu and Lotus burttii. Environ. Microbiol. 2019, 21, 1718–1739. [Google Scholar] [CrossRef]
- Acosta-Jurado, S.; Alias-Villegas, C.; Navarro-Gómez, P.; Almozara, A.; Rodríguez-Carvajal, M.A.; Medina, C.; Vinardell, J.M. Sinorhizobium fredii HH103 syrM inactivation affects the expression of a large number of genes, impairs nodulation with soybean and extends the host-range to Lotus japonicus. Environ. Microbiol. 2020, 22, 1104–1124. [Google Scholar] [CrossRef]
- López-Baena, F.J.; Vinardell, J.M.; Pérez-Montaño, F.; Crespo-Rivas, J.C.; Bellogín, R.A.; Espuny, M.R.; Ollero, F.J. Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiology 2008, 154, 1825–1836. [Google Scholar] [CrossRef] [Green Version]
- Vinardell, J.M.; Ollero, F.J.; Hidalgo, A.; López-Baena, F.J.; Medina, C.; Ivanov-Vangelov, K.; Parada, M.; Madinabeitia, N.; Espuny, M.R.; Bellogín, R.A.; et al. NolR regulates diverse symbiotic signals of Sinorhizobium fredii HH103. Mol. Plant Microbe Interact. 2004, 17, 676–685. [Google Scholar] [CrossRef] [Green Version]
- Del Cerro, P.; Pérez-Montaño, F.; Gil-Serrano, A.; López-Baena, F.J.; Megías, M.; Hungria, M.; Ollero, F.J. The Rhizobium tropici CIAT 899 NodD2 protein regulates the production of Nod factors under salt stress in a flavonoid independent manner. Sci. Rep. 2017, 7, 46712. [Google Scholar] [CrossRef] [Green Version]
- Morón, B.; Soria-Díaz, M.E.; Ault, J.; Verroios, G.; Noreen, S.; Rodríguez-Navarro, D.N.; Gil-Serrano, A.; Thomas-Oates, J.; Megías, M.; Sousa, C. Low pH changes the profile of nodulation factors produced by Rhizobium tropici CIAT899. Chem. Biol. 2005, 12, 1029–1040. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Estévez, J.; Soria-Díaz, M.E.; Fernández de Cordoba, F.; Morón, B.; Manyani, H.; Gil, A.; Thomas-Oates, J.; van Brussel, A.A.N.; Dardanelli, M.S.; Sousa, C.; et al. Different and new Nod factors produced by Rhizobium tropici CIAT899 following Na+ stress. FEMS Microbiol. Lett. 2009, 293, 220–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guasch-Vidal, B.; Estévez, J.; Dardanelli, M.S.; Soria-Díaz, M.E.; Fernández de Córdoba, F.; Balog, C.I.A.; Manyani, H.; Gil-Serrano, A.; Thomas-Oates, J.; Hensbergen, P.J.; et al. High NaCl concentrations induce the nod genes of Rhizobium tropici CIAT899 in the absence of flavonoid inducers. Mol. Plant Microbe Interact. 2013, 26, 451–460. [Google Scholar] [CrossRef] [Green Version]
- Pérez-Montaño, F.; del Cerro, P.; Jiménez-Guerrero, I.; López-Baena, F.J.; Cubo, M.T.; Hungria, M.; Megías, M.; Ollero, F.J. RNA-seq analysis of the Rhizobium tropici CIAT 899 transcriptome shows similarities in the activation patterns of symbiotic genes in the presence of apigenin and salt. BMC Genom. 2016, 17, 198. [Google Scholar] [CrossRef]
- Del Cerro, P.; Ayala-García, P.; Buzón, P.; Castells-Graells, R.; López-Baena, F.J.; Ollero, F.J.; Pérez-Montaño, F. OnfD, an AraC-Type transcriptional regulator encoded by Rhizobium tropici CIAT 899 and involved in Nod factor synthesis and symbiosis. Appl. Environ. Microbiol. 2020, 86, e01297-20. [Google Scholar] [CrossRef]
- Vinardell, J.M.; López-Baena, F.J.; Hidalgo, A.; Ollero, F.J.; Bellogín, R.; Espuny, M.R.; Temprano, F.; Romero, F.; Krishnan, H.B.; Pueppke, S.G.; et al. The effect of FITA mutations on the symbiotic properties of Sinorhizobium fredii varies in a chromosomal-background-dependent manner. Arch. Microbiol. 2004, 181, 144–154. [Google Scholar] [CrossRef]
- Pérez-Montaño, F.; Jiménez-Guerrero, I.; Acosta-Jurado, S.; Navarro-Gómez, P.; Ollero, F.J.; Ruiz-Sainz, J.E.; López-Baena, F.J.; Vinardell, J.M. A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 2016, 6, 31592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vinardell, J.M.; Acosta-Jurado, S.; Zehner, S.; Göttfert, M.; Becker, A.; Baena, I.; Blom, J.; Crespo-Rivas, J.C.; Goesmann, A.; Jaenicke, S.; et al. The Sinorhizobium fredii HH103 genome: A comparative analysis with S. fredii strains differing in their symbiotic behavior with soybean. Mol. Plant Microbe Interact. 2015, 28, 811–824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Machado, D.; Pueppke, S.G.; Vinardell, J.M.; Ruiz-Sainz, J.E.; Krishnan, H.B. Expression of nodD1 and nodD2 in Sinorhizobium fredii, a nitrogen-fixing symbiont of soybean and other legumes. Mol. Plant-Microbe Interact. 1998, 11, 375–382. [Google Scholar] [CrossRef] [Green Version]
- Jiménez-Guerrero, I.; Acosta-Jurado, S.; Medina, C.; Ollero, F.J.; Alias-Villegas, C.; Vinardell, J.M.; Pérez-Montaño, F.; López-Baena, F.J. The Sinorhizobium fredii HH103 type III secretion system effector NopC blocks nodulation with Lotus japonicus Gifu. J. Exp. Bot. 2020, 71, 6043–6056. [Google Scholar] [CrossRef] [PubMed]
- Vincent, J.M. The modified Fåhraeus slide technique. In A Manual for the Practical Study of Root Nodule Bacteria; Vincent, J.M., Ed.; Blackwell Scientific Publications: Oxford, UK, 1970; pp. 144–145. [Google Scholar]
- Beringer, J.E. R factor transfer in Rhizobium leguminosarum. J. Gen. Microbiol. 1974, 84, 188–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spaink, H.P. Rhizobial lipo-oligosaccharides: Answers and questions. Plant Mol. Biol. 1992, 20, 977–986. [Google Scholar] [CrossRef]
- Sambrook, J.; Fritsch, E.F.; Maniatis, T. Molecular Cloning. A Laboratory Manual, 2nd ed.; Cold Spring Harbor: New York, NY, USA, 1989. [Google Scholar]
- Simon, R. High frequency mobilization of gramnegative bacterial replicons by the in vivo constructed Tn5-mob transposon. Mol. Gen. Genet. 1984, 196, 413–420. [Google Scholar] [CrossRef]
- Farhaeus, G. The infection of clover root hairs by nodule bacteria studied by a simple glass slide technique. J. Gen. Microbiol. 1957, 16, 374–381. [Google Scholar] [CrossRef] [Green Version]
- Buendía-Clavería, A.M.; Ruiz-Sainz, J.E.; Cubo-Sánchez, T.; Pérez Silva, J. Studies of symbiotic plasmids in Rhizobium trifolii and fast-growing bacteria that nodulate soybean. J. Appl. Bacteriol. 1986, 17, 155–160. [Google Scholar] [CrossRef]
- Schlaman, H.R.M.; Philips, D.A.; Kondorosi, E. Genetic organization and transcriptional regulation of rhizobial nodulation genes. In The Rhizobiaceae; Spaink, H.P., Kondorosi, A., Hooykaas, P.J.J., Eds.; Kluwer: Dordrecht, The Netherlands, 1998; pp. 361–386. [Google Scholar]
- Peck, M.C.; Fisher, R.F.; Long, S.R. Diverse flavonoids stimulate NodD1 binding to nod gene promoters in Sinorhizobium meliloti. J. Bacteriol. 2006, 188, 5417–5427. [Google Scholar] [CrossRef] [Green Version]
- Gyorgypal, Z.; Kondorosi, E.; Kondorosi, A. Diverse signal sensitivity of NodD protein homologs from narrow and broad host range rhizobia. Mol. Plant-Microbe Interact. 1991, 4, 356–364. [Google Scholar] [CrossRef]
- Fellay, R.; Hanin, M.; Montorzi, G.; Frey, J.; Freiberg, C.; Golinowski, W.; Staehelin, C.; Broughton, W.J.; Jabbouri, S. nodD2 of Rhizobium sp. NGR234 is involved in the repression of the nodABC operon. Mol. Microbiol. 1998, 27, 1039–1050. [Google Scholar] [CrossRef] [PubMed]
- Barnett, M.J.; Long, S.R. The Sinorhizobium meliloti SyrM regulon: Effects on global gene expression are mediated by syrA and nodD3. J. Bacteriol. 2015, 197, 1792–1806. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Romero, E.; Segovia, L.; Mercante, F.M.; Franco, A.A.; Graham, P.; Pardo, M.A. Rhizobium tropici, a novel species nodulating Phaseolus vulgaris L. beans and Leucaena sp. trees. Int. J. Syst. Bacteriol. 1991, 41, 417–426. [Google Scholar] [CrossRef] [Green Version]
- Madinabeitia, N.; Bellogín, R.A.; Buendía-Clavería, A.M.; Camacho, M.; Cubo, T.; Espuny, M.R.; Gil-Serrano, A.M.; Lyra, M.C.; Moussaid, A.; Ollero, F.J.; et al. Sinorhizobium fredii HH103 has a truncated nolO gene due to a -1 frameshift mutation that is conserved among other geographically distant S. fredii strains. Mol. Plant Microbe Interact. 2002, 15, 150–159. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Jurado, S.; Navarro-Gómez, P.; Murdoch, P.S.; Crespo-Rivas, J.C.; Jie, S.; Cuesta-Berrio, L.; Ruiz-Sainz, J.E.; Rodríguez-Carvajal, M.Á.; Vinardell, J.M. Exopolysaccharide production by Sinorhizobium fredii HH103 is repressed by genistein in a NodD1-dependent manner. PLoS ONE 2016, 11, e0160499. [Google Scholar] [CrossRef]
- Figurski, D.H.; Helinski, D.R. Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 1979, 76, 1648–1652. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fuentes-Romero, F.; Navarro-Gómez, P.; Ayala-García, P.; Moyano-Bravo, I.; López-Baena, F.-J.; Pérez-Montaño, F.; Ollero-Márquez, F.-J.; Acosta-Jurado, S.; Vinardell, J.-M. The nodD1 Gene of Sinorhizobium fredii HH103 Restores Nodulation Capacity on Bean in a Rhizobium tropici CIAT 899 nodD1/nodD2 Mutant, but the Secondary Symbiotic Regulators nolR, nodD2 or syrM Prevent HH103 to Nodulate with This Legume. Microorganisms 2022, 10, 139. https://doi.org/10.3390/microorganisms10010139
Fuentes-Romero F, Navarro-Gómez P, Ayala-García P, Moyano-Bravo I, López-Baena F-J, Pérez-Montaño F, Ollero-Márquez F-J, Acosta-Jurado S, Vinardell J-M. The nodD1 Gene of Sinorhizobium fredii HH103 Restores Nodulation Capacity on Bean in a Rhizobium tropici CIAT 899 nodD1/nodD2 Mutant, but the Secondary Symbiotic Regulators nolR, nodD2 or syrM Prevent HH103 to Nodulate with This Legume. Microorganisms. 2022; 10(1):139. https://doi.org/10.3390/microorganisms10010139
Chicago/Turabian StyleFuentes-Romero, Francisco, Pilar Navarro-Gómez, Paula Ayala-García, Isamar Moyano-Bravo, Francisco-Javier López-Baena, Francisco Pérez-Montaño, Francisco-Javier Ollero-Márquez, Sebastián Acosta-Jurado, and José-María Vinardell. 2022. "The nodD1 Gene of Sinorhizobium fredii HH103 Restores Nodulation Capacity on Bean in a Rhizobium tropici CIAT 899 nodD1/nodD2 Mutant, but the Secondary Symbiotic Regulators nolR, nodD2 or syrM Prevent HH103 to Nodulate with This Legume" Microorganisms 10, no. 1: 139. https://doi.org/10.3390/microorganisms10010139