Molecular characterization of porcine reproductive and respiratory syndrome virus identified in 2021 from Nepal
- 1National Animal Health Research Centre, Nepal Agricultural Research Council, Lalitpur, Nepal
- 2Central Department of Microbiology, Tribhuvan University, Kathmandu, Nepal
- 3College of Animal Husbandry and Veterinary Medicine, Southwest Minzu University, Chengdu, China
- 4Animal and Plant Health Agency, Addlestone, United Kingdom
Porcine reproductive and respiratory syndrome (PRRS), an important viral disease of swine caused by PRRS virus (PRRSV) was first confirmed in Nepal in 2013. Since then, the virus has spread throughout the country and has now become endemic affecting the pig production nationally. However, molecular characterization of circulating strains has not been done in Nepal yet. In the present study, serum samples were collected from outbreak areas of different districts of Nepal and samples positive for PRRSV by ELISA were sent to Animal and Plant Health Agency (APHA), United Kingdom for sequence analysis. Out of 35 samples that were sent to APHA, only one sample was found positive by PCR and subjected to sequence analysis based on ORF5, ORF7 and Nsp2. The results from the phylogenetic analysis demonstrated that the PRRSV strain belongs to PRRSV-2 and lineage 8 strain. The sequences from the Nepalese PRRSV strain revealed a high degree of similarity with the strains isolated from India, China and Vietnam, with the closest genetic relatedness to the Indian isolates from 2020 and 2018. This is the first study on molecular characterization of PRRS virus circulating in Nepal. Further studies on strains circulating in Nepal are very essential to understand the virus diversity, its spread and evolution.
1 Introduction
Porcine reproductive and respiratory syndrome (PRRS) is an important infectious viral disease of swine which has caused huge economic impact in pig production worldwide. The disease is caused by PRRS virus (PRRSV) which is an enveloped, positive sense single-stranded RNA virus within the family Arteriviridae (1). PRRSV genome consists of nine open reading frames (ORFs) and is approximately 15 kb long (2). The nonstructural proteins are encoded by ORF 1a and 1b which are related to replication whereas ORF 2–7 encode structural proteins GP2, GP2b, E, GP3, GP4, GP5, GP5a, M, and N, respectively (3). Nsp2 is the largest PRRSV nonstructural proteins and recognized as a variable gene where frequent mutations, insertions, or deletions are observed. This distinctive feature makes Nsp2 a valuable marker for tracking the genetic changes and evolution of PRRSVs (4). ORF5 encodes the major envelope glycoprotein GP5, which is involved in viral attachment to cells and contains a neutralization epitope. ORF5 exhibits marked genetic variations compared to other genes and is therefore widely used for phylogenetic analyses (5, 6). Nucleocapsid protein (N) encapsidating the viral RNA genome is highly immunogenic in infected animals and is encoded by the ORF7 gene (7).
In the mid to late 1980s, PRRSV was reported for the first time in North America followed by Europe in 1990 (8). The two species of PRRSV, i.e., PRRSV-1 (previously European genotype) and PRRSV-2 (previously North American genotype) cause similar clinical infection to pigs though they have approximately 60% nucleotide identity at the genome level and 50–80% amino acid similarity (9, 10). In addition, within species, the Nsp2 and ORF5 genes vary considerably with sequence differences as high as 20% (10–12). Therefore, ORF5 and Nsp2 are preferred for studying the evolution and molecular epidemiology research on PRRSV (13–15). Based on molecular characterization of ORF5, PRRSV-2 have been further categorized into 9 lineages with several sublineages of each lineage and PRRSV-1 into three subtypes (subtype 1–3) (16).
PRRSV is transmitted through direct contact between infected pigs via ingestion of contaminated feed, inhalation of infected aerosols by coitus and semen of infected pigs (17). Indirect transmission occurs through fomite (18). Clinically the disease is manifested in two forms; reproductive and respiratory form (19). The reproductive signs include birth of still born piglets, abortions, mummified fetuses and weak born pigs. Respiratory signs include pneumonia, reduced feed intake, debilitation, chronic recurring illness and often high mortality.
Clinical cases of PRRS was confirmed for the first time in 2013 when the pig industry was booming in Nepal (20). In the same year, India also reported its first outbreak of PRRSV in pig population of Mizoram state (21). Since then, the virus has spread throughout the country affecting the pig production nationally. However, not much molecular epidemiological studies have been carried out. In addition, there is no active surveillance of PRRS to generate information on detection and distribution of disease or infection in the animal population. This study aimed to investigate the presence of PRRSV and characterize based on ORF5, ORF7 and Nsp2 sequence analysis.
2 Methodology
2.1 Sampling
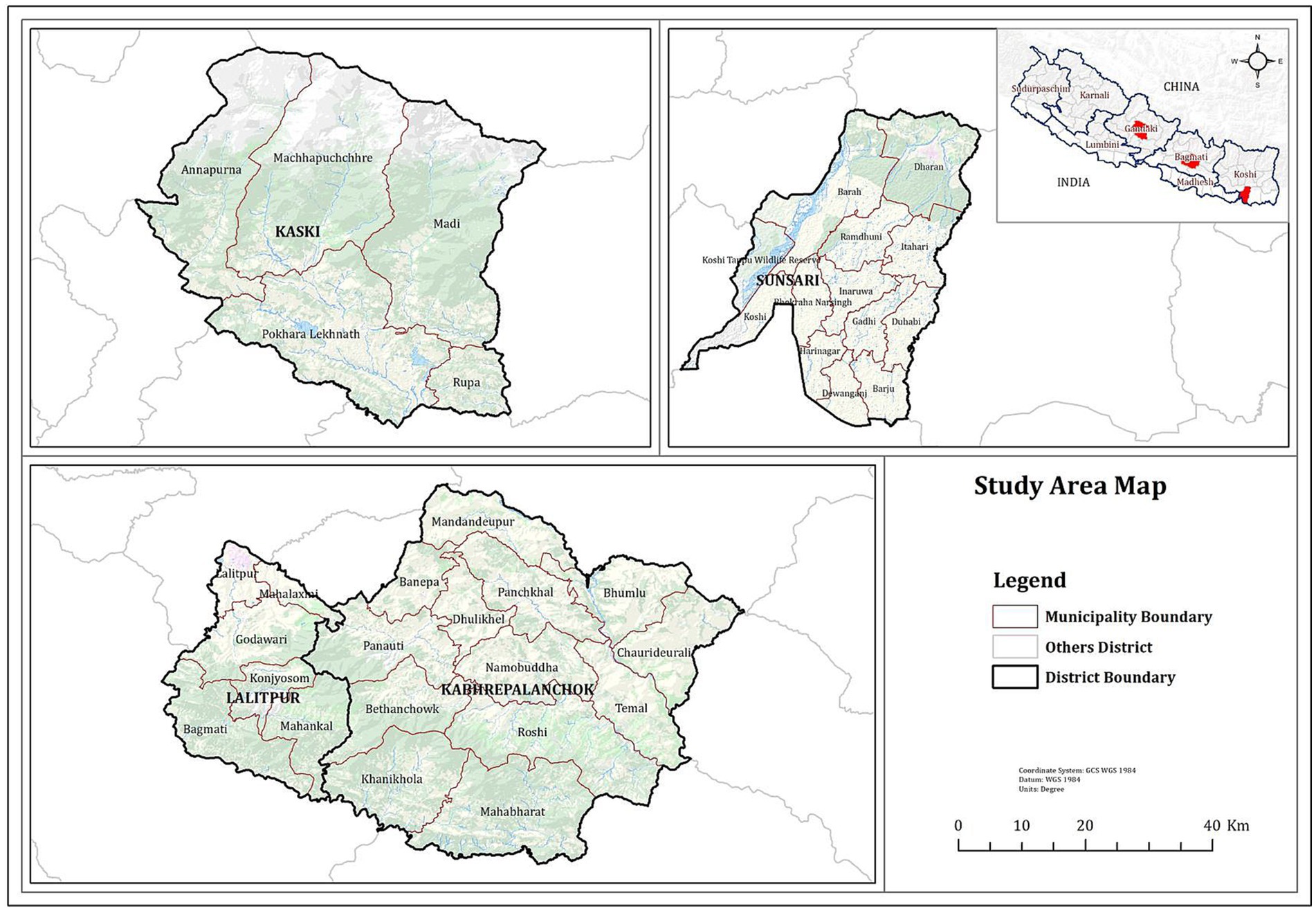
Altogether 180 serum samples were collected from pig farms in four districts, namely Kaski, Lalitpur, Kabhrepalanchok and Sunsari of Nepal (see Figure 1) which had a history of PRRSV outbreaks at different time periods. The farmers’ consent was taken before collecting samples. Animals were controlled and the blood was withdrawn in a humane way using a sterile 3 mL syringe. Samples were collected from the age group of 4 months to 2 years. Serum samples were tested to detect PRRSV antibodies using a commercial ELISA kit (22). Thus, collected serum samples were stored for further studies and only the 35 sera samples which were detected positive by ELISA was sent to the Animal and Plant Health Agency, UK for genome sequence analysis.
2.2 RNA extraction, PCR amplification and sequencing
Nucleic acids from all samples were extracted using the KingFisher™ Flex Purification System (Thermo Fisher Scientific) and the MagMAX™ CORE Nucleic Acid Purification Kit (Applied Biosystems) following the manufacturer’s instructions for low cell content samples using the protocol MagMAX_CORE_Flex_no_heat.bdz.
All samples were tested for the presence of PRRSV RNA by performing a RT-qPCR using the VetMAX™ PRRSV EU & NA 2.0 Kit (Life Technologies) following the manufacturer’s instructions on an Agilent Aria machine.
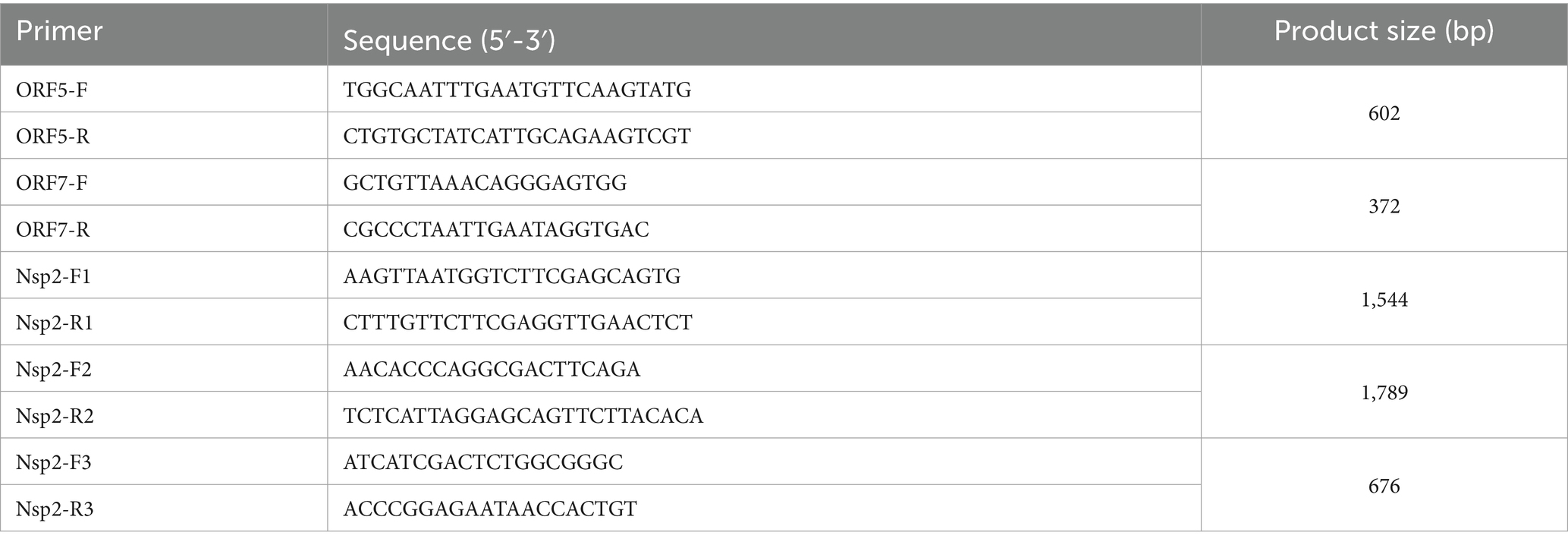
For positive samples sequencing PCRs were carried out for ORF5, ORF7 and Nsp2. In brief, a conventional PCR was performed as a single step RT-PCR reaction using the Invitrogen™ Superscript™ IV One-step RT-PCR kit, with cycling conditions: 50°C for 10 min, 98°C for 2 min, (98°C for 10 s, 61°C for 10 s, 72°C for 30 s) × 40, 72°C for 5 min. The products were visualized on a 1.8% agarose gel using SYBR Safe gel stain (Thermo Fisher Scientific). The primers for ORF5, ORF7 and Nsp2 were listed in Table 1. Sanger sequencing was carried out using the same primers as for amplification, with cycle sequencing using the Big Dye Terminator v3.1 Cycle Sequencing Kit, followed by sequencing by capillary electrophoresis on an AB 3130xl instrument. The sequences generated during this study have been deposited in NCBI (OP037986.1, OP037987.1, and PP227416).
2.3 Phylogenetic and sequence analysis of PRRSV strains
Altogether, 35 sequences of ORF5 and 30 sequences of ORF7 of PRRSV and 25 sequences of Nsp2 were analyzed. The sequences include the sequence generated during this study together with sequences downloaded from NCBI. The sequences used in this study are listed in Tables 2–4. Phylogenetic and molecular analysis were carried out using MegaX software (23) and the phylogenetic trees were generated (see Figures 2–4) (neighbor joining method with 1,000 bootstrap replicates).
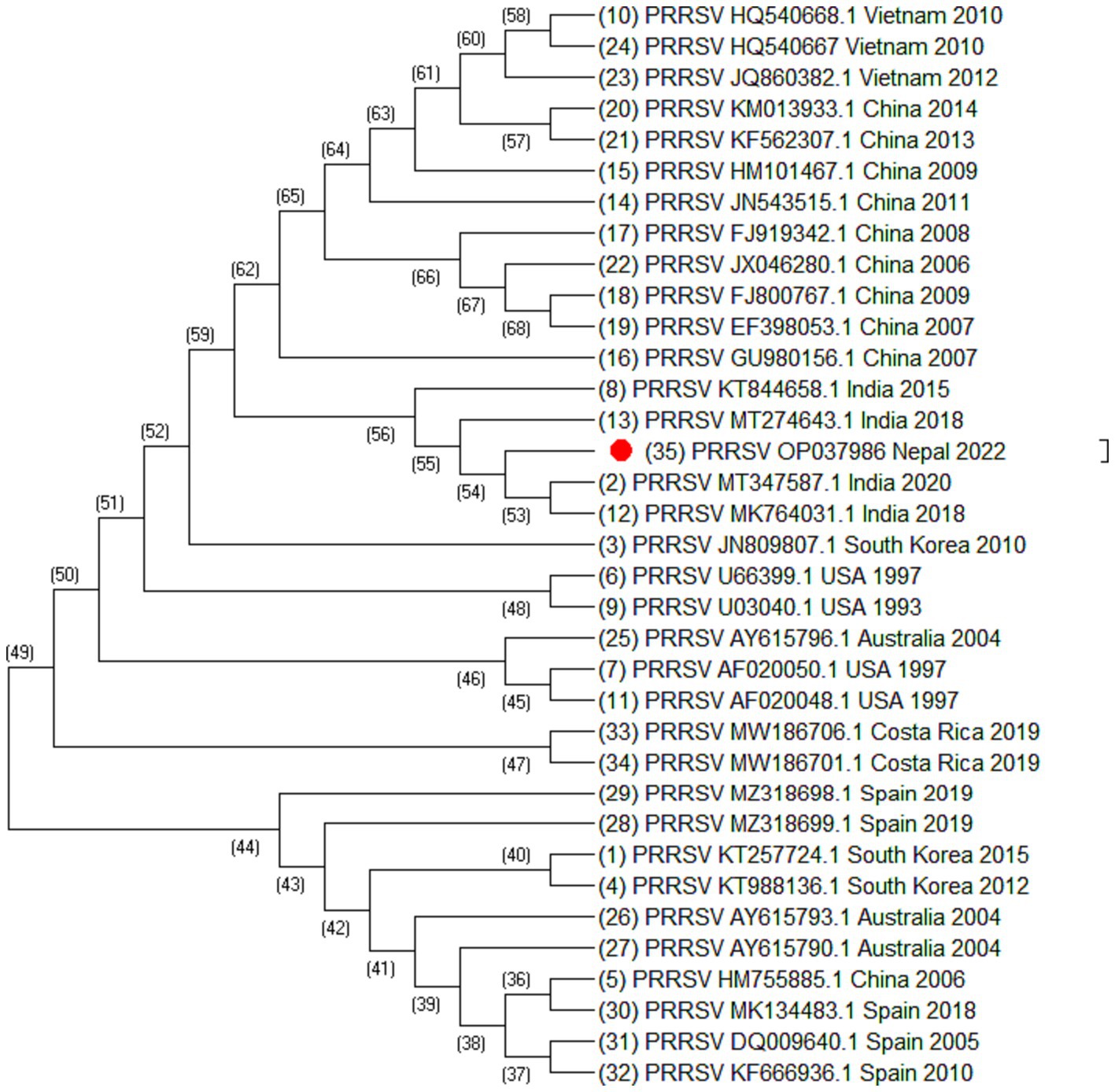
Figure 2. Phylogenetic tree of PRRSV based on ORF5 sequences. Altogether 35 sequences were used for analysis. The labels represent the name of virus followed by NCBI number country of isolation and year of isolation. The strain from this study is highlighted in red.
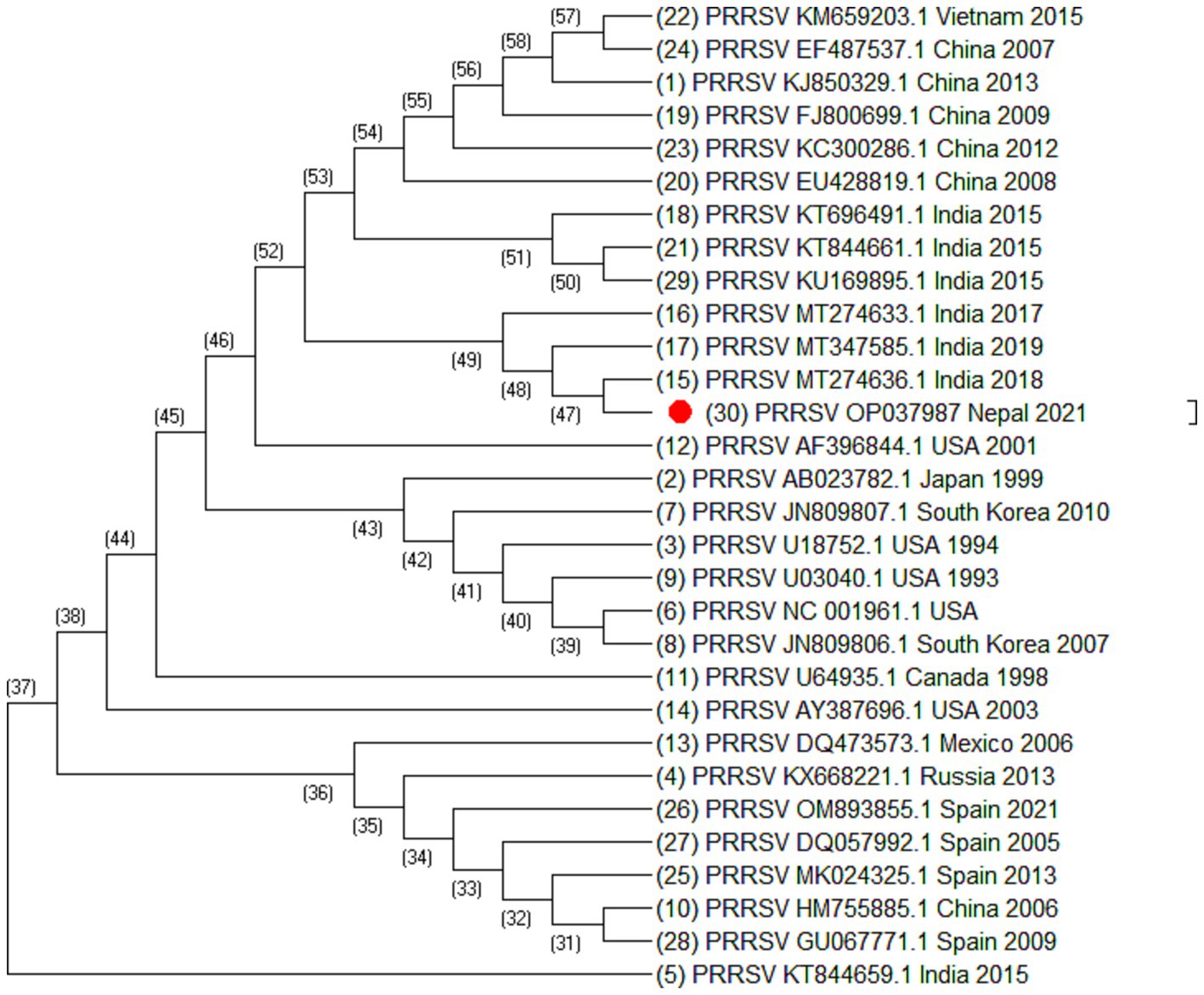
Figure 3. Phylogenetic tree of PRRSV based on ORF7 sequences. Altogether 30 sequences were used for analysis. The labels represent the name of virus followed by NCBI number country of isolation and year of isolation. The strain from this study is highlighted in red.

Figure 4. Phylogenetic tree of PRRSV based on Nsp2 sequences. Altogether 25 sequences were used for analysis. The labels represent the name of virus followed by NCBI number country of isolation and year of isolation. The strain from this study is highlighted in red.
3 Results
Out of 180 serum samples collected from different districts, 37 were found positive for PRRSV antibodies resulting in an overall seroprevalence of 20.5% (22). However ORF5, ORF7 and Nsp2 virus sequence was generated from one PCR positive serum sample. Of 35 ORF5 sequences, it was observed that most of the sequences were similar except the sequences isolated from China (HM755885.1), South Korea (KT257724.1 and KT988136.1), Australia (AY615793.1 and AY615790.1) and Spain (KF666936.1). These sequences were longer at 5′ end by 15 nucleotides. Similarly, of 30 ORF7 sequences, it was also observed that Spain (MK024325.1, OM893855.1, GU067771.1, and DQ057992.1), China (HM755885.1), Russia (KX668221.1), and Vietnam (KM659203.1) were different as compared to the other sequences. The phylogenetic analysis of Nsp2 revealed that the strain isolated from Nepal is clustered into Indian isolate and Chinese isolates. All these sequences were elongated by 12 nucleotides except Vietnam which was elongated by 48 nucleotides at the 3′ end. This indicated that the sequences of PRRSV throughout the world are variable.
Blast analysis of ORF5 sequences of Nepal with that of reference strain showed that the nucleotide sequence identities ranged from 98.34% to 84.31% and the amino acid similarities ranged from 99% to 58.79%. It shares 98.34% to 77% nucleotide identity with Indian isolates, isolate no. MT347587.1, MK764031.1, MT274643.1, and KT844658.1. The sequence identity of ORF5 shares 96.52% to 96.35% with the Chinese isolates. Similarly, the nucleotide sequence identities of ORF7 ranged from 100% to 90.05% and the amino acid similarities ranged from 100% to 60.91%. ORF7 sequence identity with the Indian isolates ranged from 100% to 97.58% and with the Chinese isolates ranged from 97.58% to 97.31%. Blast analysis of Nsp2 sequence revealed that the Nsp2 nucleotide identities ranged from 92.85% to 80.03% and amino acid similarities ranged from 91.59% to 41%.
3.1 Phylogenetic tree
From the phylogenetic tree constructed from 35 sequences of ORF5 (Figure 2), 30 sequences of ORF7 (Figure 3), and 25 sequences of Nsp2 (Figure 4) it was observed that the Nepalese strain of PRRSV belongs to the PRRSV-2 species. It can be observed that the strain identified in Nepal is closely related with the strains isolated from India in 2020 and 2018, and belongs to lineage 8 as defined by Shi et al. (15).
4 Discussion
The first outbreak of PRRSV in Nepal occurred in a national pig farm of Khumaltar in 2013 showing the signs of abortion and stillbirth in sows (20). Since then, the disease is spreading widely all over the country causing a huge impact in the national economy. Field observation in the farm showed the poor biosecurity measures, lack of PRRSV knowledge and awareness among farmers. Furthermore, the practice of keeping both seropositive pigs and the recovered pigs by farmers has led to the persistence of infection in the animal population increasing the risk of spread of disease (22). Despite the severe economic loss caused by this disease, adequate studies on molecular epidemiological has not been conducted. This study has attempted to characterize the PRRSV based on ORF5, ORF7, and Nsp2 from the samples collected during PRRSV outbreak in different districts of the country in 2021.
ORF5/ORF7 sequence analysis have been widely used in PRRS genotyping showing ORF dependent clustering in phylogenetic trees with some possible recombination (24, 25). ORF5 having a mutation rate of approximately 0.5%–1% per year, consists of approximately 600 nucleotides which shows great genetic diversity (24, 26). Even the isolates sharing identical genotypes can exhibit significant variations in their genetic sequences, particularly within the Nsp2 gene and ORF5 region (27). The phylogenetic analysis of Nsp2 revealed that the strain isolated from Nepal is clustered into India and China. Although whole genome sequencing of PRRSV may provide more complete information, this study could not generate complete PRRSV sequence as there was not sufficient viable RNA in the sample. However, sequencing of ORF5, ORF7, and Nsp2 has provided sufficient information to determine phylogenetic inferences. The sequences from a PRRSV strain from Nepal revealed a high degree of similarity with the strains isolated from India, China, and Vietnam, with the closest genetic relatedness to the Indian isolates from 2020 and 2018. From the phylogenetic tree, it can be inferred that the strain identified in Nepal was closely related to the strains isolated from India whereas studies have shown that the Indian isolates resembles to HP-PRRSV of China (21). Likewise, PRRSV isolate of Nepal shares 97.01% nucleotide identity with the Indian strain isolated during the outbreak in Mizoram state, India. Though the analysis of additional samples is required to provide the complete information about all the strains circulating in Nepal and their source, this study has demonstrated for the first time in Nepal the active circulation of a PRRSV-2, lineage 8 strain in domestic pigs based on ORF5, ORF7 and Nsp2 sequence analysis.
PRRSVs are rapidly evolving RNA viruses which have highly affected the pig industry worldwide. Emergence of new highly pathogenic PRRSV strains leads to widespread outbreak of PRRS. In Nepal, the disease could be spreading mainly through occasional movements of pigs from one place to another, via contaminated vehicles, and via semen from infected boars. The porous international border with India also contributes to transmission of the disease where the animals and animal products are transported without any barrier. Studies have indicated that wild boars, being a reservoir of PRRSV, may act as a source of infection for domestic pigs (28, 29). However, the status of PRRSV infection in wild boars of Nepal is yet to be revealed.
Control of PRRSV is not only important but also very challenging due to the high genetic variability of the virus (30). Till now, neither national control measures nor compensation to the farmers have been implemented in Nepal despite the disease-causing major constraints to the pig production and productivity. All-in all-out measures, conducting PRRSV diagnostic testing before the introduction of animals or semen into the farm and strict biosecurity measures seems only the possible means to prevent the introduction of the disease.
PRRSV genotyping is one of the major tools to better understand the virus ecology. The present study demonstrates that a PRRSV strain from Nepal belongs to the PRRSV-2 species (previously North American genotype). Since the disease has a high economic impact, monitoring and genotyping of circulating viruses is an important tool in order to improve diagnostics and increase vaccine efficacy.
5 Conclusion
Here a PRRSV-2 phylogenetic analysis was conducted and described based on ORF5, ORF7 and Nsp2 sequence analysis. A field strain from this study belonged to lineage 8. Further studies are very necessary to elucidate its geographic distribution, dynamic of genotypes and potential vaccine implementation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
This study was approved by Nepal Agricultural Research Council. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
MP: Conceptualization, Investigation, Writing – original draft. MA: Data curation, Writing – review & editing. YL: Writing – review & editing. ZZ: Writing – review & editing. MPA: Project administration, Writing – review & editing. SC: Methodology, Writing – review & editing. J-PF: Data curation, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by Nepal Agricultural Research Council, Nepal. Fund for sample transportation was supported by Southwest Minzu University, China.
Acknowledgments
The authors would like to acknowledge Southwest Minzu University, China for providing fund for sample transportation, Defra project SV3000 for funding the National Reference Laboratory for PRRSV, Animal and Plant Health Agency, UK for sample analysis and Nepal Agricultural Research Council for providing the fund.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Brinton, MA, Gulyaeva, AA, Balasuriya, UBR, Dunowska, M, Faaberg, KS, Goldberg, T, et al. ICTV virus taxonomy profile: Arteriviridae 2021. J Gen Virol. (2021) 102:001632. doi: 10.1099/jgv.0.001632
2. Stadejek, T, Stankevicius, A, Storgaard, T, Oleksiewicz, MB, Belak, S, Drew, TW, et al. Identification of radically different variants of porcine reproductive and respiratory syndrome virus in Eastern Europe: towards a common ancestor for European and American viruses. J Gen Virol. (2002) 83:1861–73. doi: 10.1099/0022-1317-83-8-1861
3. Stadejek, T, Oleksiewicz, MB, Potapchuk, D, and Podgorska, K. Porcine reproductive and respiratory syndrome virus strains of exceptional diversity in Eastern Europe support the definition of new genetic subtypes. J Gen Virol. (2006) 87:1835–41. doi: 10.1099/vir.0.81782-0
4. Saenglub, W, Jantafong, T, Mungkundar, C, Romlamduan, N, Pinitkiatisakul, S, and Lekcharoensuk, P. Genetic signatures of the immune-escaping type 2 porcine reproductive and respiratory syndrome virus in farms with a robust vaccination program. Microb Pathog. (2020) 144:104166. doi: 10.1016/j.micpath.2020.104166
5. Martinez-Lobo, FJ, Diez-Fuertes, F, Simarro, I, Castro, JM, and Prieto, C. Porcine reproductive and respiratory syndrome virus isolates differ in their susceptibility to neutralization. Vaccine. (2011) 29:6928–40. doi: 10.1016/j.vaccine.2011.07.076
6. Li, J, and Murtaugh, MP. Dissociation of porcine reproductive and respiratory syndrome virus neutralization from antibodies specific to major envelope protein surface epitopes. Virology. (2012) 433:367–76. doi: 10.1016/j.virol.2012.08.026
7. Spilman, MS, Welbon, C, Nelson, E, and Dokland, T. Cryo-electron tomography of porcine reproductive and respiratory syndrome virus: organization of the nucleocapsid. J Gen Virol. (2009) 90:527–35. doi: 10.1099/vir.0.007674-0
8. Keffaber, K. Swine reproductive failure of unknown etiology In: The George A. Young swine conference & annual nebraska spf swine conference. Lincoln, NE: University of Nebraska (1990)
9. Wensvoort, G, de Kluyver, EP, Luijtze, EA, den Besten, A, Harris, L, Collins, JE, et al. Antigenic comparison of Lelystad virus and swine infertility and respiratory syndrome (SIRS) virus. J Vet Diagn Invest. (1992) 4:134–8. doi: 10.1177/104063879200400203
10. Nelsen, CJ, Murtaugh, MP, and Faaberg, KS. Porcine reproductive and respiratory syndrome virus comparison: divergent evolution on two continents. J Virol. (1999) 73:270–80. doi: 10.1128/JVI.73.1.270-280.1999
11. Han, J, Liu, G, Wang, Y, and Faaberg, KS. Identification of nonessential regions of the Nsp2 replicase protein of porcine reproductive and respiratory syndrome virus strain VR-2332 for replication in cell culture. J Virol. (2007) 81:9878–90. doi: 10.1128/JVI.00562-07
12. Meng, XJ. Heterogeneity of porcine reproductive and respiratory syndrome virus: implications for current vaccine efficacy and future vaccine development. Vet Microbiol. (2000) 74:309–29. doi: 10.1016/s0378-1135(00)00196-6
13. Cha, SH, Chang, CC, and Yoon, KJ. Instability of the restriction fragment length polymorphism pattern of open reading frame 5 of porcine reproductive and respiratory syndrome virus during sequential pig-to-pig passages. J Clin Microbiol. (2004) 42:4462–7. doi: 10.1128/JCM.42.10.4462-4467.2004
14. Murtaugh, MP, Elam, MR, and Kakach, LT. Comparison of the structural protein coding sequences of the VR-2332 and Lelystad virus strains of the PRRS virus. Arch Virol. (1995) 140:1451–60. doi: 10.1007/BF01322671
15. Shi, M, Lam, TT, Hon, CC, Murtaugh, MP, Davies, PR, Hui, RK, et al. Phylogeny-based evolutionary, demographical, and geographical dissection of North American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. (2010) 84:8700–11. doi: 10.1128/JVI.02551-09
16. Shi, M, Lam, TT, Hon, CC, Hui, RK, Faaberg, KS, Wennblom, T, et al. Molecular epidemiology of PRRSV: a phylogenetic perspective. Virus Res. (2010) 154:7–17. doi: 10.1016/j.virusres.2010.08.014
17. Zimmerman, JJ, Benfield, DA, Dee, SA, Murtaugh, MP, Stadejek, T, Stevenson, GW, et al. eds. Porcine reproductive and respiratory syndrome virus (porcine arterivirus). 10th ed. Iowa: Wiley-Blackwell (2012).
18. Cho, JG, and Dee, SA. Porcine reproductive and respiratory syndrome virus. Theriogenology. (2006) 66:655–62. doi: 10.1016/j.theriogenology.2006.04.024
19. Iowa State University. (2023) Porcine reproductive and respiratory syndrome (PRRS). Available at: https://vetmed.iastate.edu/vdpam/FSVD/swine/index-diseases/porcine-reproductive#:~:text=PRRS%20is%20an%20acronym%20(porcine,in%20pigs%20of%20any%20age
20. Prajapati, M, Thakur, RP, Acharya, MP, Shrestha, SP, Khanal, DR, Shrestha, P, et al. eds. Outbreak of porcine reproductive and respiratory syndrome in Khumaltar farm: a case report In: Pig & pork industry in Nepal: harmonizing production and marketing networks for promotion and commercialization. Kathmandu: Nepal Agricultural Research Council (2014)
21. Rajkhowa, TK, Jagan Mohanarao, G, Gogoi, A, Hauhnar, L, and Isaac, L. Porcine reproductive and respiratory syndrome virus (PRRSV) from the first outbreak of India shows close relationship with the highly pathogenic variant of China. Vet Q. (2015) 35:186–93. doi: 10.1080/01652176.2015.1066043
22. Prajapati, M, Acharya, MP, Yadav, P, and Frossard, JP. Farm characteristics and sero-prevalence of porcine reproductive and respiratory syndrome virus (PRRSV) antibodies in pigs of Nepal. Vet Med Sci. (2023) 9:174–80. doi: 10.1002/vms3.1011
23. Chaithra, G, Ravishankar, C, Sebastian, SR, Rajasekhar, R, Anoopraj, R, Mani, BK, et al. Molecular characterisation of porcine reproductive and respiratory syndrome virus from pigs in Kerala. Virus. (2020) 31:560–5. doi: 10.1007/s13337-020-00634-7
24. Dortmans, J, Buter, GJ, Dijkman, R, Houben, M, and Duinhof, TF. Molecular characterization of type 1 porcine reproductive and respiratory syndrome viruses (PRRSV) isolated in the Netherlands from 2014 to 2016. PLoS One. (2019) 14:e0218481. doi: 10.1371/journal.pone.0218481
25. Music, N, and Gagnon, CA. The role of porcine reproductive and respiratory syndrome (PRRS) virus structural and non-structural proteins in virus pathogenesis. Anim Health Res Rev. (2010) 11:135–63. doi: 10.1017/S1466252310000034
26. Martinez-Bautista, NR, Sciutto-Conde, E, Cervantes-Torres, J, Segura-Velazquez, R, Mercado Garcia, MC, Ramirez-Mendoza, H, et al. Phylogenetic analysis of ORF5 and ORF7 of porcine reproductive and respiratory syndrome (PRRS) virus and the frequency of wild-type PRRS virus in Mexico. Transbound Emerg Dis. (2018) 65:993–1008. doi: 10.1111/tbed.12831
27. Deng, Y, Pan, Y, Wang, D, Zhou, Q, Bi, Y, Chen, F, et al. Complete genome sequence of porcine reproductive and respiratory syndrome virus strain Qy2010 reveals a novel subgroup emerging in China. J Virol. (2012) 86:7719–20. doi: 10.1128/JVI.00977-12
28. Elbers, AR, Dekkers, LJ, and van der Giessen, JW. Sero-surveillance of wild boar in the Netherlands, 1996–1999. Rev Sci Tech. (2000) 19:848–54. doi: 10.20506/rst.19.3.1254
29. Laddomada, A. Incidence and control of CSF in wild boar in Europe. Vet Microbiol. (2000) 73:121–30. doi: 10.1016/s0378-1135(00)00139-5
Keywords: ORF5, ORF7, Nsp2, PRRS, PRRSV
Citation: Prajapati M, Aryal M, Li Y, Zhang Z, Acharya MP, Clive S and Frossard J-P (2024) Molecular characterization of porcine reproductive and respiratory syndrome virus identified in 2021 from Nepal. Front. Vet. Sci. 11:1267571. doi: 10.3389/fvets.2024.1267571
Edited by:
Walid Azab, Free University of Berlin, GermanyReviewed by:
Rui Li, Henan Academy of Agricultural Sciences (HNAAS), ChinaPavulraj Selvaraj, Louisiana State University, United States
Copyright © 2024 Prajapati, Aryal, Li, Zhang, Acharya, Clive and Frossard. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Meera Prajapati, prajapati_m@hotmail.com; Jean-Pierre Frossard, jean-pierre.frossard@apha.gov.uk
 Meera Prajapati
Meera Prajapati Manita Aryal2
Manita Aryal2  Zhidong Zhang
Zhidong Zhang Jean-Pierre Frossard
Jean-Pierre Frossard