Comparison of differentially expressed genes in longissimus dorsi muscle of Diannan small ears, Wujin and landrace pigs using RNA-seq
- 1Faculty of Animal Science and Technology, Yunnan Agricultural University, Kunming, China
- 2College of Biology and Agriculture, Zunyi Normal University, Zunyi, China
Introduction: Pig growth is an important economic trait that involves the co-regulation of multiple genes and related signaling pathways. High-throughput sequencing has become a powerful technology for establishing the transcriptome profiles and can be used to screen genome-wide differentially expressed genes (DEGs). In order to elucidate the molecular mechanism underlying muscle growth, this study adopted RNA sequencing (RNA-seq) to identify and compare DEGs at the genetic level in the longissimus dorsi muscle (LDM) between two indigenous Chinese pig breeds (Diannan small ears [DSE] pig and Wujin pig [WJ]) and one introduced pig breed (Landrace pig [LP]).
Methods: Animals under study were from two Chinese indigenous pig breeds (DSE pig, n = 3; WJ pig, n = 3) and one introduced pig breed (LP, n = 3) were used for RNA sequencing (RNA-seq) to identify and compare the expression levels of DEGs in the LDM. Then, functional annotation, Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and Protein–Protein Interaction (PPI) network analysis were performed on these DEGs. Then, functional annotation, Gene Ontology (GO) enrichment analysis, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, and Protein-Protein Interaction (PPI) network analysis were performed on these DEGs.
Results: The results revealed that for the DSE, WJ, and LP libraries, more than 66, 65, and 71 million clean reads were generated by transcriptome sequencing, respectively. A total of 11,213 genes were identified in the LDM tissue of these pig breeds, of which 7,127 were co-expressed in the muscle tissue of the three samples. In total, 441 and 339 DEGs were identified between DSE vs. WJ and LP vs. DSE in the study, with 254, 193 up-regulated genes and 187, 193 down-regulated genes in DSE compared to WJ and LP. GO analysis and KEGG signaling pathway analysis showed that DEGs are significantly related to contractile fiber, sarcolemma, and dystrophin-associated glycoprotein complex, myofibril, sarcolemma, and myosin II complex, Glycolysis/Gluconeogenesis, Propanoate metabolism, and Pyruvate metabolism, etc. In combination with functional annotation of DEGs, key genes such as ENO3 and JUN were identified by PPI network analysis.
Discussion: In conclusion, the present study revealed key genes including DES, FLNC, PSMD1, PSMD6, PSME4, PSMB4, RPL11, RPL13A, ROS23, RPS29, MYH1, MYL9, MYL12B, TPM1, TPM4, ENO3, PGK1, PKM2, GPI, and the unannotated new gene ENSSSCG00000020769 and related signaling pathways that influence the difference in muscle growth and could provide a theoretical basis for improving pig muscle growth traits in the future.
1 Introduction
Pork accounts for 35% of global meat consumption. Demand for pork in China continues to increase as the country develops socially and economically. Therefore, it is imperative to improve the pork production and quality of domestic pig breeders. The genetic improvement of swine mainly focuses on the production and reproductive traits that have economic value, focusing in production traits (1). In order to meet the demand for pork, the growth rate is a crucial assessment indicator in pig breeding. Growth in pigs is a complex trait regulated by multiple genes and their regulatory networks. Studying the development and growth properties of skeletal muscle and combining it with modern molecular breeding techniques is an effective way to increase the efficiency of pig breeding.
Pig breeding resources are found all over the world (2). During the process of natural selection, Chinese indigenous pig breeds have exhibited numerous outstanding characteristics and significant phenotypic variations (3). Diannan small ears (DSE) pig is a small Chinese indigenous breed with slow growth and low adult weight (4). They are grown in the southern areas of Mengla, Ruili, and Yingjiang in Yunnan Province, which have a subtropical climate (5). DSE has numerous distinct phenotypic traits and are of superior commercial value (6, 7). DSE pork is abundant in nutrients and has a unique taste, and it has high stress resistance and adaptability to harsh environmental conditions (8). At the same time, DSE is also an ideal animal model for medical research. All these factors make DSE a valuable genetic resource. In addition, the research of genetic diversity and selection signatures within DSE shown it has lower genetic diversity than other breeds, indicating the importance of strengthening conservation strategies for DSE (9). At present, there are limited studies on the genetic improvement of muscle growth traits in DSE. It was therefore necessary to screen for key genes involved in pig muscle growth and development.
Wujin pig (WJ) is another excellent local breed found in the plateau regions of Yunnan, Sichuan, and Guizhou in China. It has valuable characteristics such as strong disease resistance, high fecundity, good mothering, strong adaptability, and high-quality pork products (10). As a typical fatty pig breed, WJ has become a famous high-quality ham product both domestically and internationally due to its delicious meat, abundant nutrition, elevated amino acid content, heavy collagen, and delicate flavor (11). WJ is broadly studied in China. It has strong fatty acid transport capacity and fat deposition rate in the body, and the expression of related genes involved in muscle cell differentiation and muscle growth and development is down-regulated compared with Landrace pig (LP) (12, 13). Western commercial pig breeds have consistently been selected for rapid muscle growth and development (14). LP is a typical lean pig breed originated in Denmark, with stable production performance and genetic stability, and is famous for its fast growth rate, strong feed utilization, and elevated lean meat rate (15). In the study conducted by Fang et al. (16), it was found that the rapid growth of LP may be attributed to the concentration of gene expression in muscle tissue on biological functions associated with muscle contraction.
Phenotypic differences are primarily determined by genetic differences (17). The growth rate and potential of skeletal muscle depends largely on the development of embryonic muscle fibers, with muscle fiber proliferation suspended between 85 and 95 days of gestation and not significantly altered by external conditions (18). Therefore, the disparity in the number of muscle fibers among various pig breeds may be permanent (19). This study used RNA-seq method to construct a transcriptome map of the Longissimus dorsi muscle (LDM) of pigs with different growth phenotypes. Comparison of differentially expressed genes (DEGs) in muscle growth between DSE, Chinese indigenous pig breed WJ, and introduced pig breed LP, and the screening of key candidate genes will provide a theoretical basis for future improvement of pig muscle growth traits.
2 Materials and methods
2.1 Animal and sample preparation
Animals under study were from two Chinese indigenous pig breeds (DSE pig, n = 3; WJ pig, n = 3) and one introduced pig breed (LP, n = 3). They were housed in similar environmental and nutritional conditions. All pigs were offered the same type of feed, and they were involved in the trial from around 30 kg of body weight (BW) until around 100 ± 2 kg BW. The animals were slaughtered according to the standard protocol of the Yunnan Agricultural University, and the LDM samples were collected after the slaughter. Samples were stored immediately in liquid nitrogen and later were stored at −80°C refrigerator for subsequent total RNA extraction. The research proposal and the relevant experimental procedures were approved by the institutional Animal Care and Use Committee of Yunnan Agricultural University (Case Number: 20210915).
2.2 Total RNA isolation from LDM samples
Total RNA was extracted from LDM tissues using the TRIzol Reagent kit (Invitrogen, San Diego, CA). The concentration and purity of the total RNA were checked using the NanoDrop 2000 Biophotometer and their completeness was verified by agarose gel electrophoresis.
2.3 Library preparation and RNA sequencing
A pool of equimolar ratios of RNAs from three individuals was established. The pooled RNA samples were purified with an RNeasy Micro Kit (Cat. Qiagen-74004; venlo, The Netherlands) for the preparation of complementary DNA (cDNA) library (approximately 3 μg of total RNA). Poly (A) messenger RNA (mRNA) isolation, first and second strand cDNA synthesis, fragment and adaptor ligation, and cDNA library preparation were performed sequentially with the TruSeq RNA Sample Prep Kit (Cat. RS-122-2002; Illumina, San Diego, CA). After that, each sample product was loaded onto the flow cell channels of the HighSeq 2000 platform (Illumina) for double-end 150 bp sequencing. The insert size of the double-ended library is 380 bp, and three biological replicas were included in each group.
2.4 Mapping and alignment of sequence reads
The raw reads obtained from RNA-seq were processed using CLC Genomics Workbench 4.8 (Qiagen), which trimmed the adapter sequences and removed low-quality reads. Pair-sequenced read files from each lane were separately mapped to the Sus scrofa reference genome (10.2) using Top Hat11, and two mismatches were allowed for 150-bp reads in each alignment.
2.5 Differential gene expression analysis using RNA-seq
The fragments per kilobase of transcript per million fragments mapped (FPKM) for all genes were calculated using Cuff software (version 2.1.2). Gene expression differences were evaluated using the Fisher exact test after normalizing the total number of mapped reads using the top-quartile normalization method. DEGs between the breeds were identified using the statistical significance criteria: | log2 fold change | > 1 and false discovery rate (FDR) < 0.01.
2.6 Functional annotation of genes with differential expression
Gene Ontology (GO) enrichment analysis of DEGs were performed using the GOseq software to determine their primary biological functions, and conducted Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis on DEGs. The GO terms and KEGG pathway enrichments with an adjusted p-value (Benjamini) less than 0.05 were considered significant.
3 Results
3.1 Summary of RNA-seq data
After removing the adaptor sequence and low-quality reads, each sample yielded clean reads ranging from 6,589,139 bp to 7,171,030 bp. Approximately 98% (ranging from 96% to 99%) of the clean reads were successfully mapped to the Sus scrofa reference genome (Supplementary Table S1). By calculating FPKM values as a function of gene expression levels, we observed that 11,213 genes are expressed in LDM tissue. Among these genes, 7,127 had the same expression across the three groups (Figure 1). The FPKM values of the expressed genes were mostly distributed between 0 ~ 0.1 and 0.3 ~ 15 (Supplementary Figure S1). The heat map of all co-expressed genes showed that the three biological repeats in each group were first clustered together, but the transcriptome patterns of DSE and WJ were different from those of LP (Supplementary Figure S2).
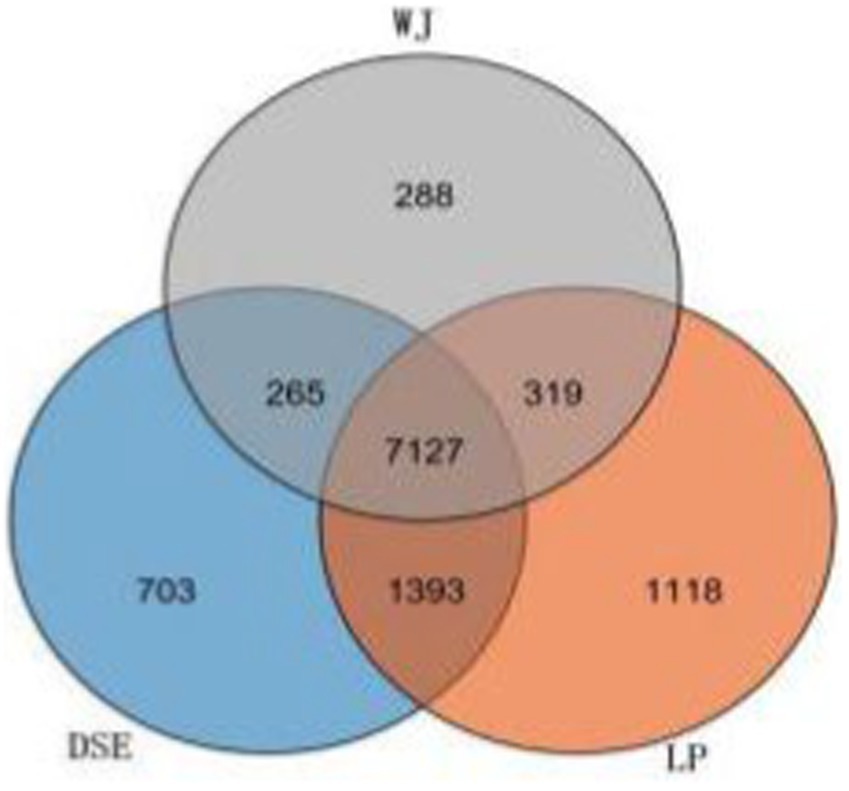
Figure 1. Venn diagrams of the number of genes expressed in each group. DSE, Diannan small ears pig; LP, Landrace pig; WJ, Wujin pig.
3.2 Analysis of differentially expressed genes
Using |log2 (fold change)| > 1 and FDR < 0.01 between groups as strict criteria, 441 DEGs were screened out when comparing DSE and WJ (Supplementary Table S2 and Supplementary Figure S3A), including 254 up-regulated and 187 down-regulated DEGs (Supplementary Table S2 and Supplementary Figure S3A). In the comparison of LP and DSE (Supplementary Table S3 and Supplementary Figure S3B), 339 DEGs were detected, including 146 up-regulated and 193 down-regulated DEGs; in the comparison of WJ and LP (Supplementary Table S4 and Supplementary Figure S3C), 695 DEGs were detected, including 296 up-regulated and 399 down-regulated DEGs. The number of DEGs in DSE vs. LP and DSE vs. WJ is higher than in WJ vs. LP, indicating that DSE is different from WJ and LP and has a unique pattern of gene expression.
About 167 overlapping DEGs were found between WJ vs. LP and LP vs. DSE, indicating that LP has similar differences in prenatal muscles with WJ and DSE (Figure 2). A total of 189 DEGs were identified in the WJ pig LDM, while 86 DEGs were identified in DSE pigs. There are also 24 overlapping DEGs in three groups. In this experiment, 62 DEGs unique to the DSE were focused on. These DEGs are different from WJ and LP, which is helpful for additional studies of DSE.
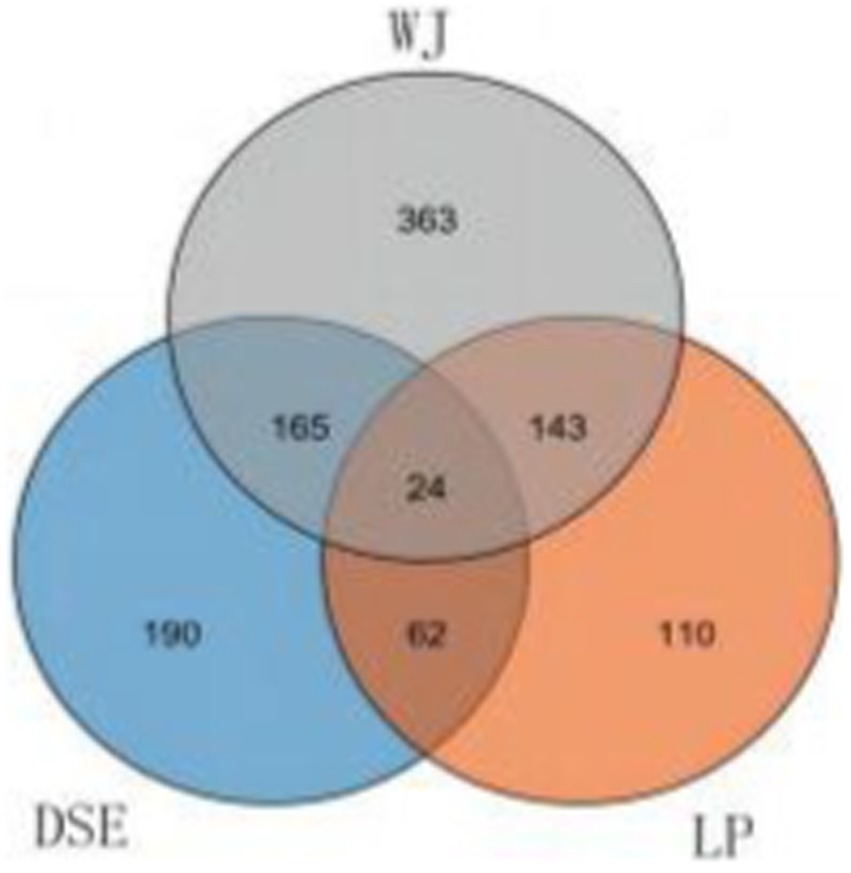
Figure 2. Venn diagrams of differentially expressed genes in the comparisons of LP vs. DSE, DSE vs. WJ, and WJ vs. LP. DSE, Diannan small ears pig; LP, Landrace pig; WJ, Wujin pig.
3.3 Go functional annotation of the differentially expressed genes
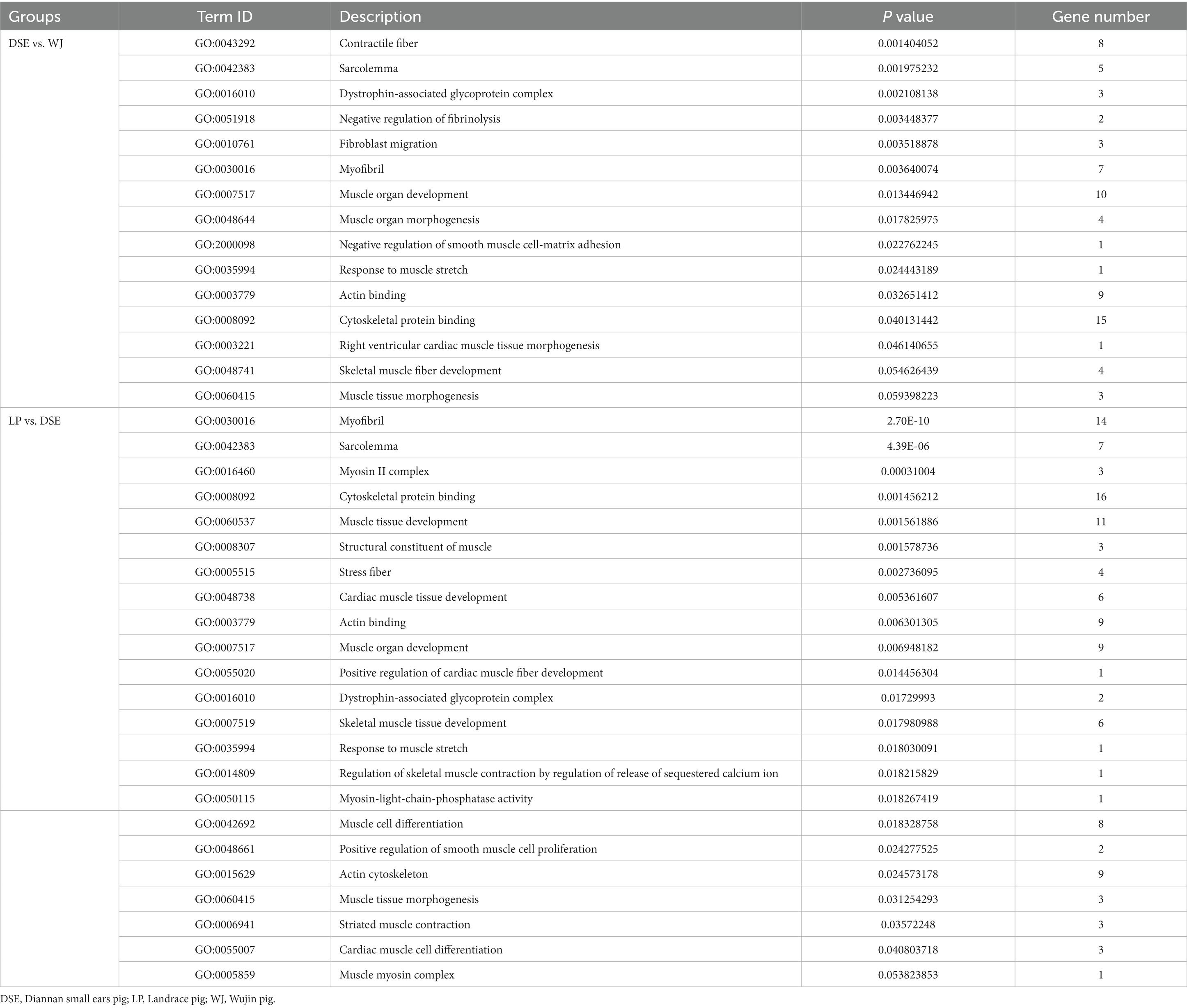
Of the 441 DEGs between DSE and WJ, 361 were annotated. A total of 330 terms (Supplementary Table S5) were significantly enriched in the three categories of cellular components, biological processes, and molecular functions (p < 0.05). Among them, the top 20 terms (the 20 terms with the lowest p value) consist of 12 cellular components, 4 biological process terms, and 4 molecular function terms (Figure 3A). There are 15 terms (Table 1) related to muscle growth and development, including contractile fiber (GO:0043292), sarcolemma (GO:0042383), and dystrophin-associated glycoprotein complex (GO:0016010), etc.
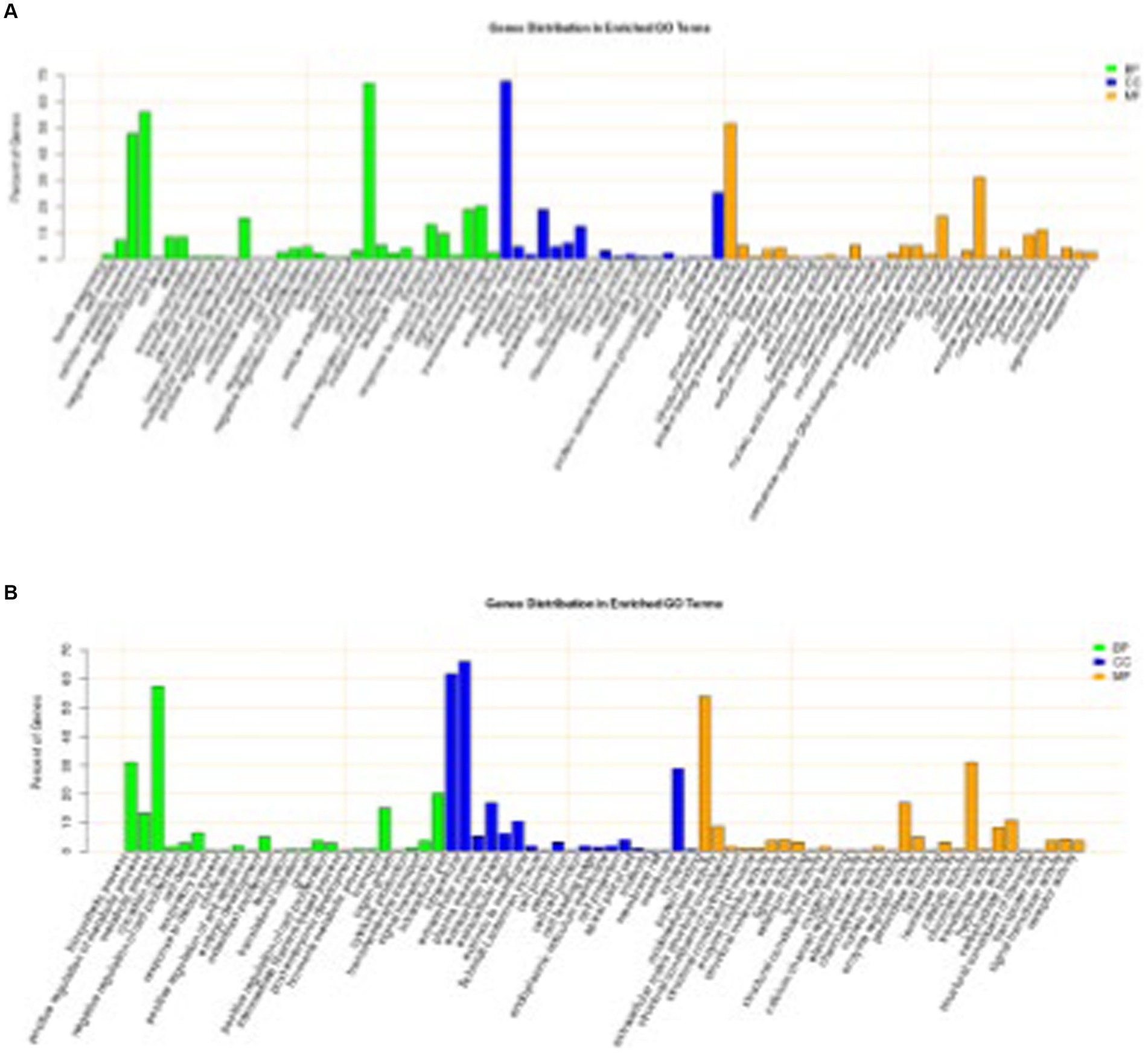
Figure 3. Significantly enriched GO for differentially expressed genes. (A) DSE vs. WJ, (B) LP vs. DSE. DSE, Diannan small ears pig; LP, Landrace pig; WJ, Wujin pig.
Of the 339 DEGs between LP and DSE, 278 were annotated. A total of 496 terms (Supplementary Table S6) were significantly enriched in the three categories of cellular components, biological processes, and molecular functions (p < 0.05). Among them, the top 20 terms consist of 9 cellular components, 9 biological processes, and 2 molecular functions (Figure 3B). There are 23 terms (Table 1) related to muscle growth and development, including myofibril (GO:0030016), sarcolemma (GO:0042383), and myosin II complex (GO:0016460), etc.
3.4 KEGG functional annotation of the differentially expressed genes
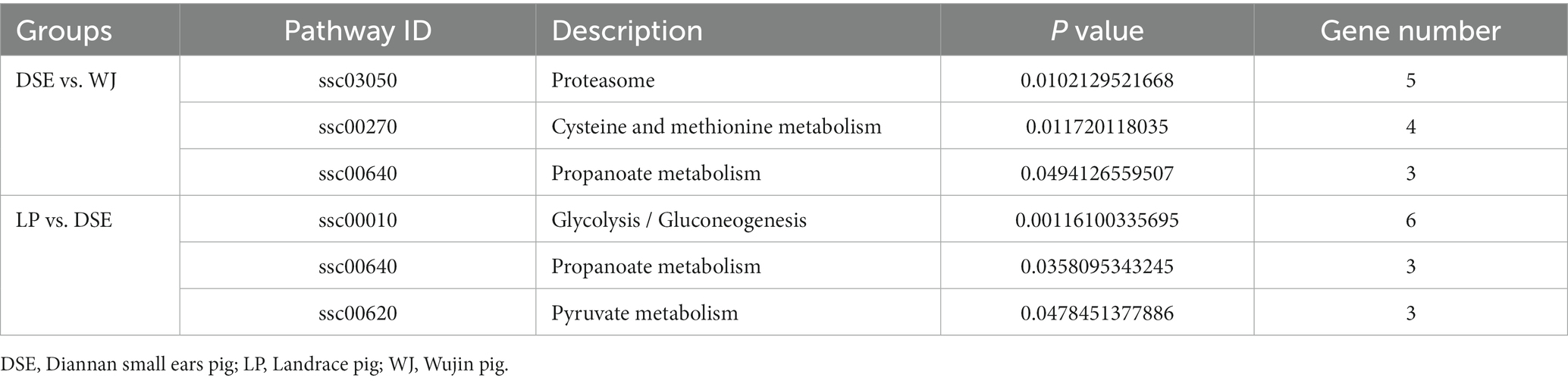
A total of 441 DEGs between DSE and WJ were integrated into the KEGG pathway database. Among these, 13 pathways (p < 0.05) were found to be significantly enriched (Supplementary Table S7 and Figure 4A). Muscle growth and development involve three pathways (Table 2), including the Proteasome pathway, Cysteine and Methionine metabolism, and Propanoate metabolism. 339 DEGs between LP and DSE were integrated into the KEGG pathway database. A total of 10 pathways (p < 0.05) were significantly enriched (Supplementary Table S8 and Figure 4B). Muscle growth and development involve three pathways (Table 2), including Glycolysis/Gluconeogenesis, Propanoate metabolism, and Pyruvate metabolism.
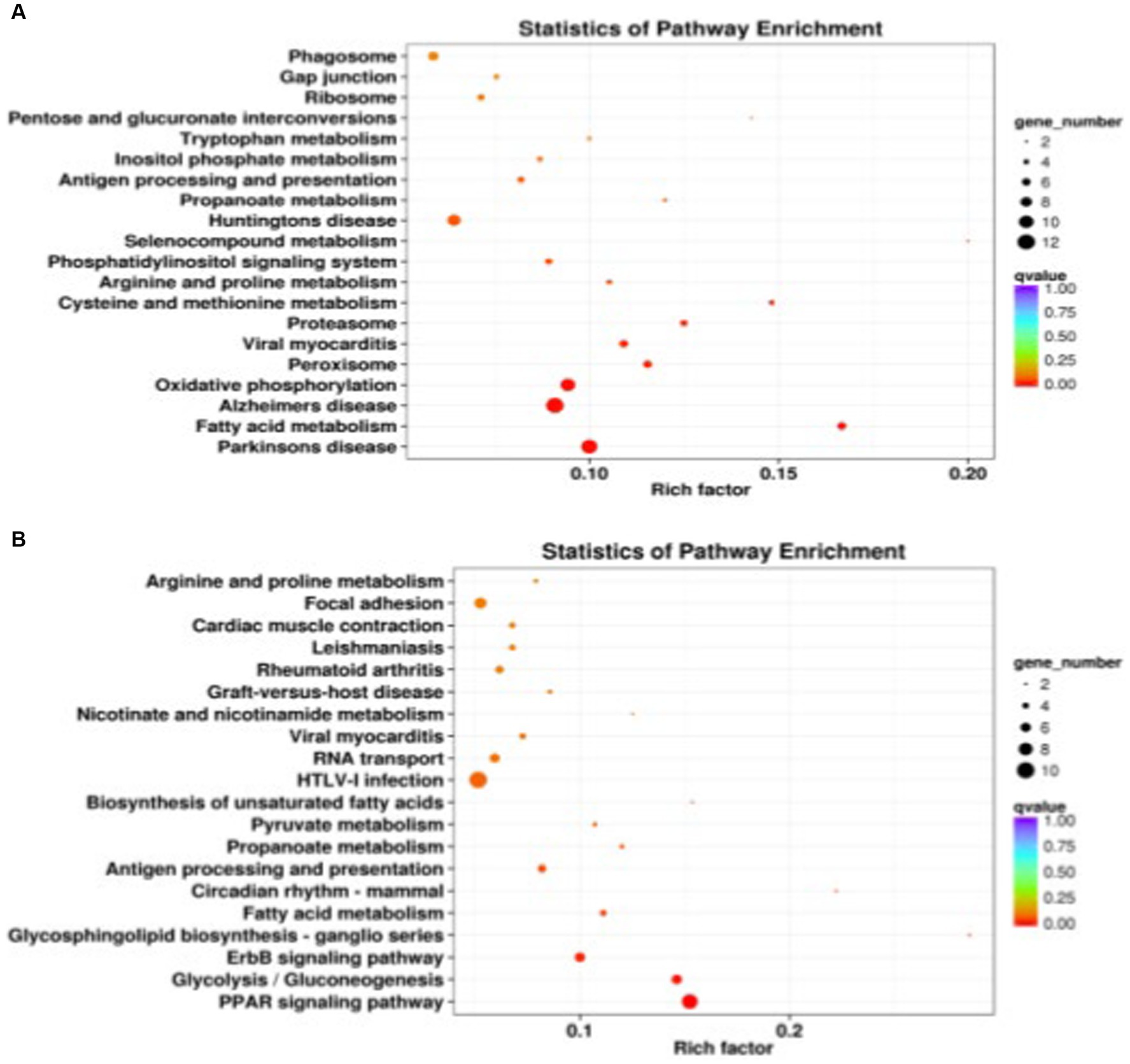
Figure 4. Significantly enriched KEGG for differentially expressed genes. (A) DSE vs. WJ, (B) LP vs. DSE. DSE, Diannan small ears pig; LP, Landrace pig; WJ, Wujin pig.
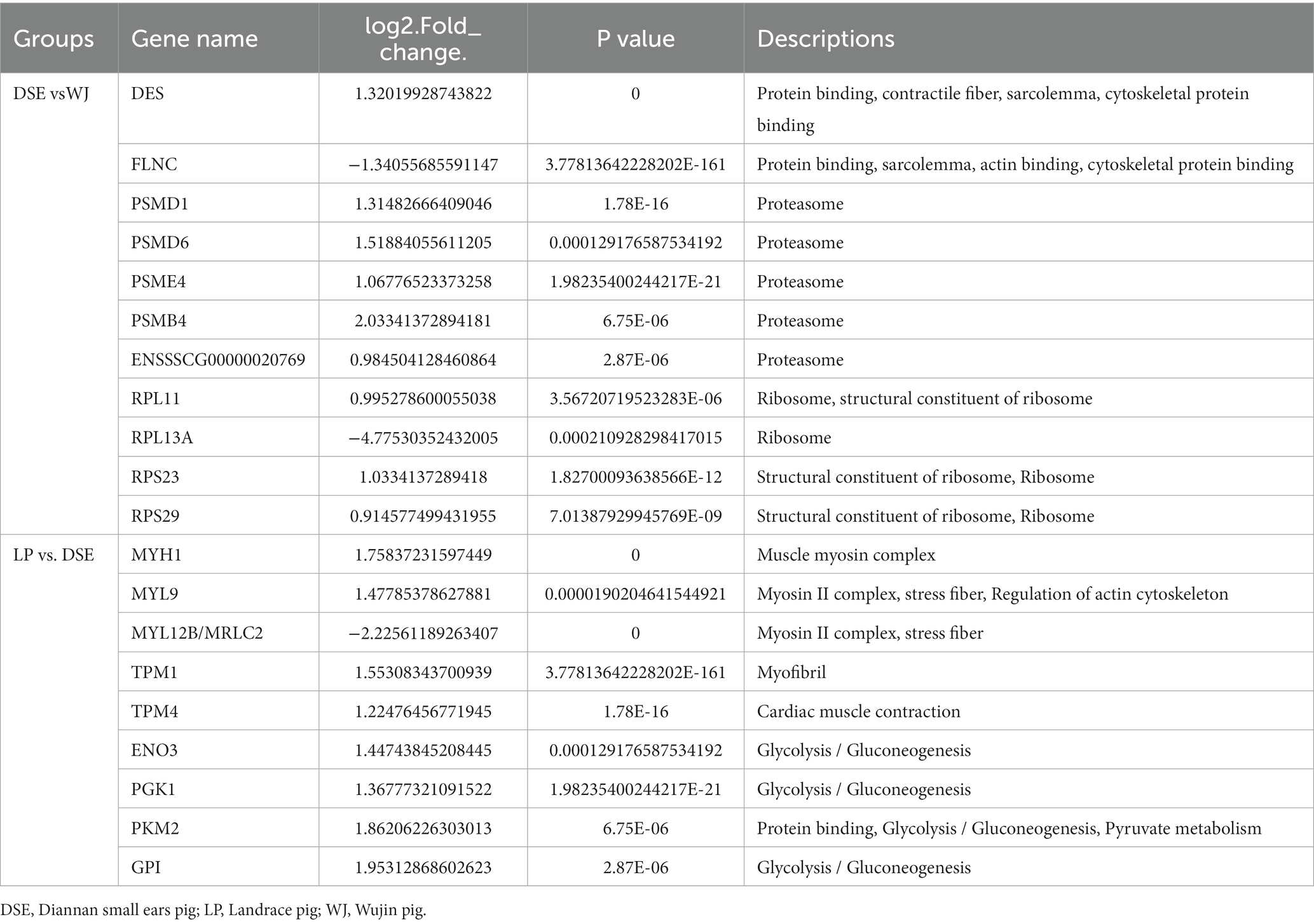
There are 20 genes significantly enriched in Table 3 that are involved in muscle growth and development. Among them, six genes (DES, FLNC, RPL11, MYH1, MYL9, and TPM1) were highly enriched in GO terms and significantly enriched in the KEGG pathway. In addition, PSMD1, PAMD6, PSME4, PSMB4, ENSSSCG000020769, RPL13A, RPS23, RPS29, TPM4, ENO3, PGK1, GPI, and PKM2 were found to be enriched only in the KEGG pathway, while MYL12B was enriched only in the GO term.
3.5 PPI network construction and hub gene identification
Protein–Protein Interaction (PPI) network analysis was performed on the key genes among the screened DEGs to gain a better understanding of their interactions. In the PPI network of DSE vs. WJ, a total of 51 nodes and 61 edges were established (Figure 5A). Additionally, in the PPI network of LP vs. DSE, a total of 50 nodes and 65 edges were established (Figure 5B). In Figure 5A, we found four distinct clusters in the network, consisting of proteins encoded by DEGs. In Figure 5B, most of the proteins in the network interacted with multiple different partners. However, we found that the proteins enolase 3 (ENO3) and Jun proto-oncogene, AP-1 transcription factor subunit (JUN) have the highest degree of connectivity (eight edges), which may enable them to play a role as “hub” proteins, acting as controllers within biochemical pathways.
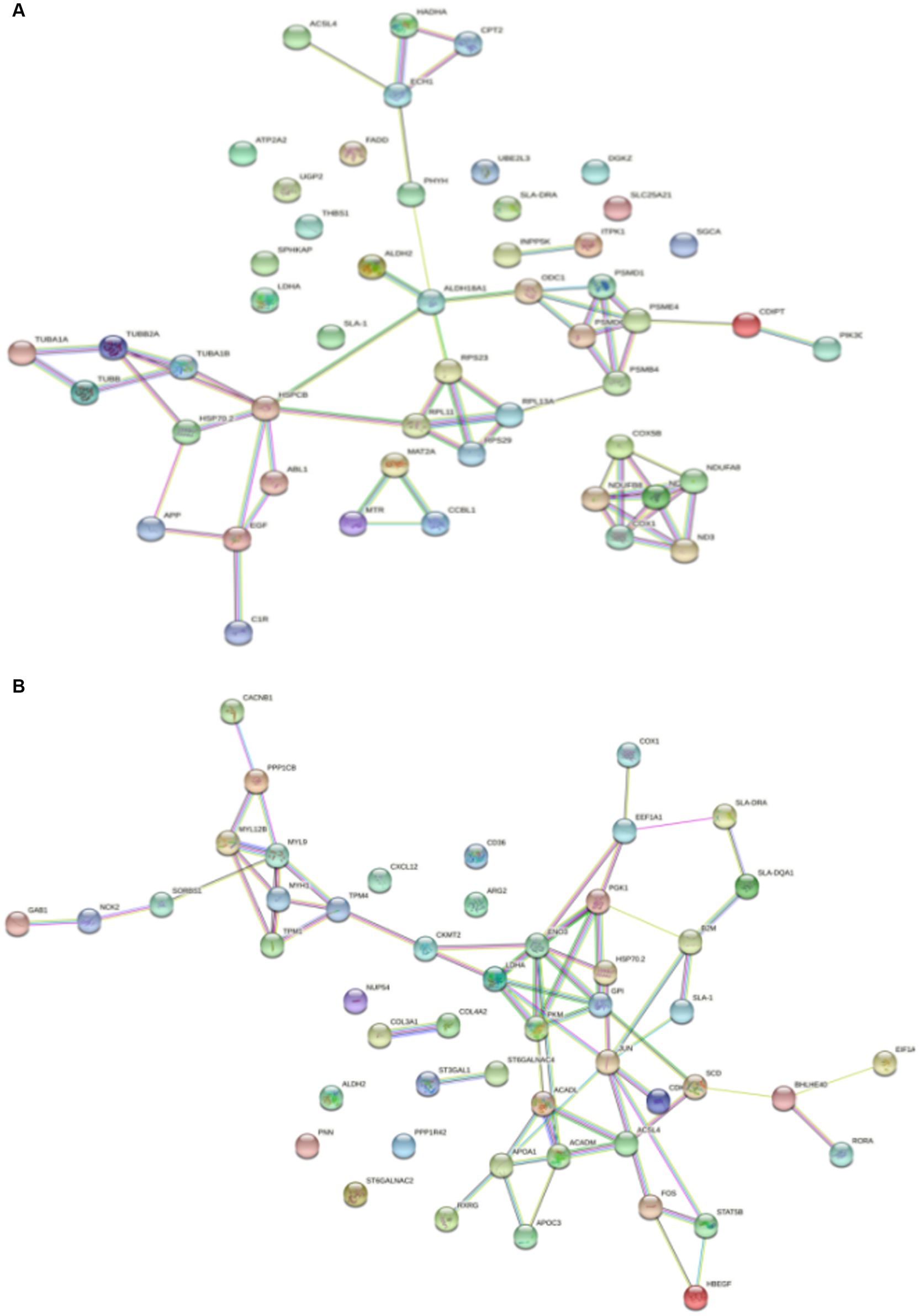
Figure 5. Significantly enriched PPI for differentially expressed genes. (A) DSE vs. WJ, (B) LP vs. DSE. DSE, Diannan small ears pig; LP, Landrace pig; WJ, Wujin pig.
4 Discussion
RNA-seq provides a quantitative and open system for large-scale analysis of transcription results (20). In the animal husbandry industry, meat quality are critical economic characteristics (21). Identifying key candidate genes involved in muscle growth is effective in studying molecular genetic regulatory mechanisms for muscle growth and development. This promotes muscle growth and provides people with a high-quality source of meat. Muscle is an essential part of all meat products. Growth performance and product quality are considered to be the basic conditions for modern breeding (22). Muscle growth is regulated by the homeostatic balance of muscle protein biosynthesis and degradation (23, 24). This is the result of a combination of a distinctive genotype, nutrient status, age, and muscle type in muscle tissue (25). In the study, 11,213 genes were detected in the longest muscles. Moreover, by analyzing the GO and KEGG signaling pathways of selected DEGs, significant functional genes and signaling pathways associated with phenotypic differences between DSE, WJ, and LP were found. A large proportion of previously screened DEGs are involved in metabolic regulation and muscle development. A total of 199 DEGs were classified as associated with muscle growth and development at the time of GO term analysis, representing approximately 25.5% of the total number of DEGs. Among them, 20 DEGs have been identified as key regulators of myocyte differentiation and muscle development. These genes are thought to affect postnatal muscle growth rates, muscle fiber diameter and density, and fiber type.
Intermediate filaments are the main components of the cytoskeleton, and desmin and filament proteins play important roles in skeletal muscle (26). The Desmin (DES) gene encodes desmin, which is a tissue-specific type III intermediate filament expressed in skeletal muscle cells, cardiomyocytes, and smooth muscle cells (27). DES has been found to be related to striated muscle disease (28). DES connects myofibrillar structures such as sarcomeres, nuclei, mitochondria, and lysosomes around the Z-disc. This connection is essential for maintaining the lateral organization and alignment of sarcomere structures during myofibril regeneration (29). Studies have shown that DES can effectively serve as a marker for muscle fiber regeneration (30). Filamins (FLNs) are large dimeric actin-binding proteins that regulate actin cytoskeleton remodeling (31). FLNs are primarily expressed in skeletal muscle and cardiac muscle, and they are located in various structures such as the Z-disc, myotendinous junctions, myotomes, and intercalated discs (32). They are involved in the organization and stabilization of the actin cytoskeleton, as well as in anchoring adhesion sites to transmembrane proteins, which play a role in cell-matrix and cell–cell interactions (33). Fujita et al. (34) found that Filamin C (FLNC) plays an indispensable role in maintaining the structural integrity of cardiac and skeletal muscles, which supports their resistance to stress. In addition, studies have also found that mutations in the FLNC gene can cause hypertrophic cardiomyopathy (35). The results of this study indicate that the expression levels of the DES gene are significantly higher in DSE than in WJ, while the expression levels of the FLNC gene are lower in DSE than in WJ. This result may be due to the significant difference in adult body weight between the two local pig breeds.
The formation of muscle fibers, or myogenesis, is believed to be a complex process involving the proliferation of myoblasts and the increase of structural proteins. This process is initiated by mesenchymal stem cells that originate from mesoblasts (36, 37). Myosin is the structural component of muscle (38). The myoglobin II molecule is an essential component of both muscle cells and non-muscle cells. They consist of two Myosin heavy chain (MyHC) and two Myosin light chain (MyLC) molecules (39, 40). The formation of muscle fibers is precisely regulated at the molecular level by the genes of muscle cells. Musclin heavy chain 1 (MyH1), also known as MyHC II.X, a type of muscle fiber found in fast-twitch muscles. Type II.X belongs to the transition type of oxidation in yeast-type, while the proportion of oxidized muscle fibers is positively related to meat quality (41). In addition, the MyH1 gene is significantly associated with muscle contraction and muscle development (42). Muscle growth inhibition in muscle differentiation can help slow down and inhibit the expression of rapidly condensed heavy chain genes, as well as have some effects on DNA repair (43). Although MyLC is not in the myosin protein family, it plays an important role in myogenase catalysis supramolecular complex formation (44). Among them, MyL9 is a key link for activating muscle global activity to provide power for cells (45). MYL12B regulates the activity of myosin light chain ATP enzyme through phosphorylation (46). Tropomyosin is an essential regulatory protein during muscle contraction. It mainly regulates the contraction of horizontal and smooth muscles (47). TPM1 and TPM4 genes are members of this family (48). In the study, mRNA expression levels of the genes MYH1, MYL9, TPM1, and TPM4 were lower in DSE than in LP. In turn, the expression of the MYL12B gene is higher in DSE than in LP. Protein is a manifestation of life functions (49). The proteasome is responsible for the degradation of most proteins in the cell through the ubiquitin-proteasome system (UPS) of the protein-protease system (50). The protease complex is composed of one 20S core particles (CP) and two 19S regulatory particles (RP). In the study, the mRNA expression levels of the genes PSMD1, PSMD6, PSME4, and PSMB4 were higher in DSE than in WJ. A new gene, ENSSSCG000020769, was found to be involved in proteasome synthesis. Its expression level is higher in DSE than in WJ.
It is well accepted that total protein synthesis in muscle is related to both the ribosome content of the tissue and the activity per ribosome (51). The increase in protein synthesis may not be solely attributable to an increase in ribosomal and mRNA production in muscle. However, the high ribosome concentration may be associated with the rapid rate of protein synthesis (52, 53). The ribosome is a venue for protein synthesis and is involved in cell proliferation, withering and differentiation. Ribosomal protein (RP) is the main component, named ribosomal protein large (RPL) and ribosomal protein Small (RPS) according to the source size sub-tadlies (54). In the study, mRNA expression levels of the genes RPL11, RPS23, and RPS29 were higher in DSE than in WJ. The results show that protein synthesis and metabolism related genes in WJ are down-regulated compared to LP, in particular the ribosomal pathway and several ribosomal protein genes.
Muscle fiber is considered an essential factor influencing numerous biochemical processes and thereby meat quality (55). Intensive selection for lean growth in modern pig breeds has resulted in a shift in muscle metabolism towards more glycolytic and less oxidative fiber types. A high proportion of type II fiber adversely affects meat quality, while oxidative-type fiber generally contains higher intramuscular fat (55, 56). Enolase (ENO) is a kind of glycoling enzyme, participating in the regulation of the energy metabolic pathway of glycolysis and gluconeogenesis (57). Energy metabolism is thought to play an important role in skeletal muscle growth and development, directly affecting muscle fiber type and meat quality. ENO3 is positioned in muscle tissue, expressed in proliferative muscle cells and differentiated muscle pipes (58). Lactate dehydrogenase (LDH) is the hallmark enzyme of glycolysis (59). LDHA is abundantly expressed in skeletal muscle and is mainly responsible for catalyzing lactate and oxidizing NADH to generate NAD+ (60). The reaction catalyzed by phosphoglycerate kinase (PGK) is the first energy-producing reaction in glucose breakdown and is an important metabolic enzyme in the glycolysis pathway (61). Mutations in the PGK1 gene can lead to reduced glycerol kinase activity and interfere with normal glucose disassembly pathways and energy production (62). Glucose can be utilized intracellularly through conversion to hexokinase 2 (HK-2), phosphofructokinase (PFKM), and pyruvate kinase M (PKM). Pyruvate kinase muscle isoenzyme 2 (PKM2) is an isoenzyme of the glycolytic enzyme pyruvate kinase (63). PKM2 and its alternatively spliced mRNA (PKM1) are both produced by the PKM gene (64). PKM1 is mainly expressed in tissues with elevated energy demands, such as muscle and brain (65), while PKM2 is expressed at varying levels in most cell types (66). But PKM2 reduces pyruvate kinase activity and promotes the pentose phosphate pathway (PPP), thus preventing oxidative stress (67). Glucose-6-phosphate isomerase (GPI) is a gene-encoded product that plays an important regulatory role in cell growth, differentiation, and apoptosis (68). In this study, the expression levels of ENO3, PGK1, PKM2 and GPI genes were lower in DSE than in LP.
The WJ has a high body fat content, especially in terms of intramuscular fat, which is higher than other pig breeds (11). DSE is known for its superior meat quality and higher fat deposits compared to Western pig breeds (8). In a previous report by this experimental team, it was shown that the increasing mechanism of intramuscular fat (IMF) content deposition in fatty pigs may be due to higher lipogenesis and fatty acid transport capacity and lower lipolysis capacity (13). Stearoyl-CoA desaturase (SCD) is a regulatory enzyme produced by fat (69) that produces monounsaturated fatty acids (MUFA) that contribute to cell growth, survival, differentiation, metabolic regulation, and signaling (70). Long-chain ACYL-CoA synthase 4 (ACSL4) is an important catalytic enzyme for the preparation of polyunsaturated fatty acids (PUFA). It is present in anabolic steroid tissues, plays an important role in the anabolism and catabolism of fatty acids, and is important for growth and developmental processes, also showing a regulatory role (71, 72). Acyl-CoA dehydrogenase (ACAD) is a mitochia lutease family involved in the metabolism of fatty acids and branches (73). Acyl-CoA dehydrogenase medium chain gene (ACADM) and Acyl-COA dehydrogenase long chain gene (ACADL) can catalyze the mitochondrial oxidation of straight-chain fatty acid mitochondria. In this study, the top 20 GO terms included terms related to triglyceride metabolism processes, cholesterol metabolism processes, negative regulators of fatty acid biosynthesis, and fatty acid biosynthesis processes. In the KEGG functional annotation we found pathways involved in IMF synthesis in the body, like the PPAR signaling pathway, fatty acid metabolism pathway, propionate metabolism pathway, pyruvate metabolism pathway, and biosynthetic unsaturated fatty acid pathway. Based on the PPI analysis, the ACASL4, ACADM and ACADL genes were identified as the three key nodes with higher expression levels in DSE compared to LP. Considering the strong fat deposition ability of Chinese native pigs, this result could explain the slower growth rate of DSE compared to other exotic pig breeds.
5 Conclusion
In summary, this study identified 11,213 genes in the LDM tissue in pigs. By comparing three different pig breeds (WJ, LP, and DSE), we found 62 unique DEGs. Among them, the genes DES, FLNC, PSMD1, PSMD6, PSME4, PSMB4, RPL11, RPL13A, ROS23, RPS29, MYH1, MYL9, MYL12B, TPM1, ENO3, PGK1, PKM2, GPI, and the unannotated new gene ENSSSCG201020769 were identified as key genes that regulate muscle growth and development. These genes determine the pig’s postnatal growth rate, muscle fiber diameter and density, and fiber type. The research results shown here, provide a theoretical basis for improving pig muscle growth traits in the future.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession numbers PRJNA1050857 and SAMN38755984-SAMN38755986.
Ethics statement
The animal study was approved by Animal Care and Use Committee of Yunnan Agricultural University (Case Number: 20230906). The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
QL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. MH: Conceptualization, Investigation, Validation, Writing – original draft, Writing – review & editing. JZ: Investigation, Software, Writing – review & editing. LY: Investigation, Software, Writing – review & editing. WC: Formal analysis, Investigation, Writing – review & editing. YX: Investigation, Writing – review & editing. ZS: Data curation, Funding acquisition, Methodology, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the National Natural Science Foundation of China (32360808, 31760645, 31260592 and 31060331), Key Science and Technology Projects of Yunnan (202202AE090032) and Prefecture-School Cooperation Project of Yunnan (2021533416000035).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1296208/full#supplementary-material
References
1. Kanis, E , De Greef, KH , Hiemstra, A , and van Arendonk, JA . Breeding for societally important traits in pigs. J Anim Sci. (2005) 83:948–57. doi: 10.2527/2005.834948x
2. Huang, Y , Zhou, L , Zhang, J , Liu, X , Zhang, Y , Cai, L, et al. A large-scale comparison of meat quality and intramuscular fatty acid composition among three Chinese indigenous pig breeds. Meat Sci. (2020) 168:108182. doi: 10.1016/j.meatsci.2020.108182
3. Wang, H , Wang, X , Yan, D , Sun, H , Chen, Q , Li, M, et al. Genome-wide association study identifying genetic variants associated with carcass backfat thickness, lean percentage and fat percentage in a four-way crossbred pig population using SLAF-seq technology. BMC Genomics. (2022) 23:594. doi: 10.1186/s12864-022-08827-8
4. Liu, M , Lan, Q , Yang, L , Deng, Q , Wei, T , Zhao, H, et al. Genome-wide association analysis identifies genomic regions and candidate genes for growth and fatness traits in Diannan small-ear (DSE) pigs. Animals (Basel). (2023) 13:1571. doi: 10.3390/ani13091571
5. Yonggang, L . A novel porcine gene, MAPKAPK3, is differentially expressed in the pituitary gland from mini-type Diannan small-ear pigs and large-type Diannan small-ear pigs. Mol Biol Rep. (2010) 37:3345–9. doi: 10.1007/s11033-009-9921-8
6. Lan, Q , Lian, Y , Peng, P , Yang, L , Zhao, H , Huang, P, et al. Association of gut microbiota and SCFAs with finishing weight of Diannan small ear pigs. Front Microbiol. (2023) 14:1117965. doi: 10.3389/fmicb.2023.1117965
7. Lü, MD , Han, XM , Ma, YF , Irwin, DM , Gao, Y , Deng, JK, et al. Genetic variations associated with six-white-point coat pigmentation in Diannan small-ear pigs. Sci Rep. (2016) 6:27534. doi: 10.1038/srep27534
8. Wang, Z , Li, Q , Chamba, Y , Zhang, B , Shang, P , Zhang, H, et al. Identification of genes related to growth and lipid deposition from transcriptome profiles of pig muscle tissue. PLoS One. (2015) 10:e0141138. doi: 10.1371/journal.pone.0141138
9. Wu, F , Sun, H , Lu, S , Gou, X , Yan, D , Xu, Z, et al. Genetic diversity and selection signatures within Diannan small-ear pigs revealed by next-generation sequencing. Front Genet. (2020) 11:733. doi: 10.3389/fgene.2020.00733
10. Zhao, SM , Liu, YG , Pan, HB , Zhang, X , Ge, CR , Jian, JJ, et al. Generation and analysis of expressed sequence tags (ESTs) from muscle full-length cDNA library of Wujin pig. J Integr Agric. (2014) 13:378–86. doi: 10.1016/S2095-3119(13)60414-1
11. Yang, H , Ma, C , Qiao, F , Song, Y , and Du, M . Lipolysis in intramuscular lipids during processing of traditional Xuanwei ham. Meat Sci. (2005) 71:670–5. doi: 10.1016/j.meatsci.2005.05.019
12. Hong, X , Huang, Y , Li, WZ , Yang, MH , Ge, CR , Zhang, X, et al. Muscle biological characteristics of differentially expressed genes in Wujin and landrace pigs. J Integr Agric. (2014) 13:2236–42. doi: 10.1016/S2095-3119(13)60605-X
13. Zhao, S M , Ren, LJ , Chen, L , Zhang, X , Cheng, ML , Li, WZ, et al. Differential expression of lipid metabolism related genes in porcine muscle tissue leading to different intramuscular fat deposition. Lipids (2009) 44:1029–1037. doi: 10.1007/s11745-009-3356-9
14. Cagnazzo, M , te Pas, MF , Priem, J , de Wit, AA , Pool, MH , Davoli, R, et al. Comparison of prenatal muscle tissue expression profiles of two pig breeds differing in muscle characteristics. J Anim Sci. (2006) 84:1–10. doi: 10.2527/2006.8411
15. Gjerlaug-Enger, E , Kongsro, J , Odegård, J , Aass, L , and Vangen, O . Genetic parameters between slaughter pig efficiency and growth rate of different body tissues estimated by computed tomography in live boars of landrace and Duroc. Animal. (2012) 6:9–18. doi: 10.1017/S1751731111001455
16. Fang, C , Guo, F , Zhao, X , Zhang, Z , Lu, J , Pan, H, et al. Biological mechanisms of growth performance and meat quality in porcine muscle tissue. Anim Biotechnol. (2022) 33:1246–54. doi: 10.1080/10495398.2021.1886939
17. Wang, JY , Lan, J , Zhao, J , Chen, L , and Liu, Y . A novel porcine gene, POT1, differentially expressed in the longissimus muscle tissues from Wujin and large white pigs. Cytokine. (2012) 59:22–6. doi: 10.1016/j.cyto.2012.03.028
18. Brown, LD . Endocrine regulation of fetal skeletal muscle growth: impact on future metabolic health. J Endocrinol. (2014) 221:R13–29. doi: 10.1530/JOE-13-0567
19. Wigmore, PM , and Stickland, NC . Muscle development in large and small pig fetuses. J Anat. (1983) 137:235–45.
20. Van den Berge, K , Hembach, KM , Soneson, C , Tiberi, S , Clement, L , Love, MI, et al. RNA sequencing data: hitchhiker's guide to expression analysis. Annu Rev Biomed Data Sci. (2019) 2:139–73. doi: 10.1146/annurev-biodatasci-072018-021255
21. Murray, AC . The evaluation of muscle quality In: SM Jones , editor. Quality and grading of carcasses of meat animals. Boca Raton, FL: CRC Press (2020). 83–107.
22. Ren, L , Liu, A , Wang, Q , Wang, H , Dong, D , and Liu, L . Transcriptome analysis of embryonic muscle development in Chengkou Mountain chicken. BMC Genomics. (2021) 22:431. doi: 10.1186/s12864-021-07740-w
23. Schiaffino, S , Dyar, KA , Ciciliot, S , Blaauw, B , and Sandri, M . Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. (2013) 280:4294–314. doi: 10.1111/febs.12253
24. Tan, KT , Ang, SJ , and Tsai, SY . Sarcopenia: tilting the balance of protein homeostasis. Proteomics. (2020) 20:e1800411. doi: 10.1002/pmic.201800411
25. Liu, Y , Li, F , Kong, X , Tan, B , Li, Y , Duan, Y, et al. Signaling pathways related to protein synthesis and amino acid concentration in pig skeletal muscles depend on the dietary protein level, genotype and developmental stages. PLoS One. (2015) 10:e0138277. doi: 10.1371/journal.pone.0138277
26. Gruenbaum, Y , and Aebi, U . Intermediate filaments: a dynamic network that controls cell mechanics. F1000Prime Reports. (2014) 6:54. doi: 10.12703/P6-54
27. Hol, EM , and Capetanaki, Y . Type III intermediate filaments Desmin, glial fibrillary acidic protein (GFAP), vimentin, and Peripherin. Cold Spring Harb Perspect Biol. (2017) 9:a021642. doi: 10.1101/cshperspect.a021642
28. Maggi, L , Mavroidis, M , Psarras, S , Capetanaki, Y , and Lattanzi, G . Skeletal and cardiac muscle disorders caused by mutations in genes encoding intermediate filament proteins. Int J Mol Sci. (2021) 22:4256. doi: 10.3390/ijms22084256
29. Bornemann, A , and Schmalbruch, H . Desmin and vimentin in regenerating muscles. Muscle Nerve. (1992) 15:14–20. doi: 10.1002/mus.880150104
30. Soglia, F , Bordini, M , Mazzoni, M , Zappaterra, M , Di Nunzio, M , Clavenzani, P, et al. The evolution of vimentin and desmin in pectoralis major muscles of broiler chickens supports their essential role in muscle regeneration. Front Physiol. (2022) 13:970034. doi: 10.3389/fphys.2022.970034
31. Rosa, JP , Raslova, H , and Bryckaert, M . Filamin a: key actor in platelet biology. Blood. (2019) 134:1279–88. doi: 10.1182/blood.2019000014
32. Nakamura, F , Stossel, TP , and Hartwig, JH . The filamins: organizers of cell structure and function. Cell Adhes Migr. (2011) 5:160–9. doi: 10.4161/cam.5.2.14401
33. van der Ven, PF , Obermann, WM , Lemke, B , Gautel, M , Weber, K , and Fürst, DO . Characterization of muscle filamin isoforms suggests a possible role of gamma-filamin/ABP-L in sarcomeric Z-disc formation. Cell Motil Cytoskeleton. (2000) 45:149–62. doi: 10.1002/(SICI)1097-0169(200002)45:2<149::AID-CM6>3.0.CO;2-G
34. Fujita, M , Mitsuhashi, H , Isogai, S , Nakata, T , Kawakami, A , Nonaka, I, et al. Filamin C plays an essential role in the maintenance of the structural integrity of cardiac and skeletal muscles, revealed by the medaka mutant zacro. Dev Biol. (2012) 361:79–89. doi: 10.1016/j.ydbio.2011.10.008
35. Qin, X , Li, P , Qu, HQ , Liu, Y , Xia, Y , Chen, S, et al. FLNC and MYLK2 gene mutations in a Chinese family with different phenotypes of cardiomyopathy. Int Heart J. (2021) 62:127–34. doi: 10.1536/ihj.20-351
36. Zhou, J , Zhang, Y , Wu, J , Qiao, M , Xu, Z , Peng, X, et al. Proteomic and lipidomic analyses reveal saturated fatty acids, phosphatidylinositol, phosphatidylserine, and associated proteins contributing to intramuscular fat deposition. J Proteome. (2021) 241:104235. doi: 10.1016/j.jprot.2021.104235
37. Pittenger, MF , Discher, DE , Péault, BM , Phinney, DG , Hare, JM , and Caplan, AI . Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen Med. (2019) 4:22. doi: 10.1038/s41536-019-0083-6
38. Warburg, O . Annual review of biochemistry. Prefatory chapter. Annu Rev Biochem. (1964) 33:1–15. doi: 10.1146/annurev.bi.33.070164.000245
39. Brooke, MH , and Kaiser, KK . Muscle fiber types: how many and what kind? Arch Neurol. (1970) 23:369–79. doi: 10.1001/archneur.1970.00480280083010
40. Punkt, K . Fibre types in skeletal muscles. Adv Anat Embryol Cell Biol. (2002) 162:1–109. doi: 10.1007/978-3-642-59399-4
41. Schiaffino, S , and Reggiani, C . Fiber types in mammalian skeletal muscles. Physiol Rev. (2011) 91:1447–531. doi: 10.1152/physrev.00031.2010
42. Ghosh, M , Sodhi, SS , Sharma, N , Mongre, RK , Kim, N , Singh, AK, et al. An integrated in silico approach for functional and structural impact of non-synonymous SNPs in the MYH1 gene in Jeju native pigs. BMC Genet. (2016) 17:35. doi: 10.1186/s12863-016-0341-1
43. Joulia, D , Bernardi, H , Garandel, V , Rabenoelina, F , Vernus, B , and Cabello, G . Mechanisms involved in the inhibition of myoblast proliferation and differentiation by myostatin. Exp Cell Res. (2003) 286:263–75. doi: 10.1016/s0014-4827(03)00074-0
44. Coletti, C , Acosta, GF , Keslacy, S , and Coletti, D . Exercise-mediated reinnervation of skeletal muscle in elderly people: an update. Eur J Transl Myol. (2022) 32:10416. doi: 10.4081/ejtm.2022.10416
45. Lv, M , Luo, L , and Chen, X . The landscape of prognostic and immunological role of myosin light chain 9 (MYL9) in human tumors. Immun Inflamm Dis. (2022) 10:241–54. doi: 10.1002/iid3.557
46. Park, I , Han, C , Jin, S , Lee, B , Choi, H , Kwon, JT, et al. Myosin regulatory light chains are required to maintain the stability of myosin II and cellular integrity. Biochem J. (2011) 434:171–80. doi: 10.1042/BJ20101473
47. Zot, AS , and Potter, JD . Structural aspects of troponin-tropomyosin regulation of skeletal muscle contraction. Annu Rev Biophys Biophys Chem. (1987) 16:535–59. doi: 10.1146/annurev.bb.16.060187.002535
48. Marchenko, M , Nefedova, V , Artemova, N , Kleymenov, S , Levitsky, D , and Matyushenko, A . Structural and functional peculiarities of cytoplasmic tropomyosin isoforms, the products of TPM1 and TPM4 genes. Int J Mol Sci. (2021) 22:5141. doi: 10.3390/ijms22105141
49. Kessel, A , and Ben-Tal, N . Introduction to proteins: structure, function, and motion. Boca Raton, FL: CRC Press (2018).
50. Khalil, R . Ubiquitin-proteasome pathway and muscle atrophy. Adv Exp Med Biol. (2018) 1088:235–48. doi: 10.1007/978-981-13-1435-3_10
51. Lin, CS , and Hsu, CW . Differentially transcribed genes in skeletal muscle of Duroc and Taoyuan pigs. J Anim Sci. (2005) 83:2075–86. doi: 10.2527/2005.8392075x
52. Nirenberg, M , and Leder, P . RNA CODEWORDS and protein synthesis. The effect of trinucleotides upon the binding of SRNA to ribosomes. Science. (1964) 145:1399–407. doi: 10.1126/science.145.3639.1399
53. Brostrom, MA , and Brostrom, CO . Calcium dynamics and endoplasmic reticular function in the regulation of protein synthesis: implications for cell growth and adaptability. Cell Calcium. (2003) 34:345–63. doi: 10.1016/s0143-4160(03)00127-1
54. Hummel, M , Dobrenel, T , Cordewener, JJ , Davanture, M , Meyer, C , Smeekens, SJ, et al. Proteomic LC-MS analysis of Arabidopsis cytosolic ribosomes: identification of ribosomal protein paralogs and re-annotation of the ribosomal protein genes. J Proteome. (2015) 128:436–49. doi: 10.1016/j.jprot.2015.07.004
55. Lee, SH , Joo, ST , and Ryu, YC . Skeletal muscle fiber type and myofibrillar proteins in relation to meat quality. Meat Sci. (2010) 86:166–70. doi: 10.1016/j.meatsci.2010.04.040
56. Guo, J , Shan, T , Wu, T , Zhu, LN , Ren, Y , An, S, et al. Comparisons of different muscle metabolic enzymes and muscle fiber types in Jinhua and landrace pigs. J Anim Sci. (2011) 89:185–91. doi: 10.2527/jas.2010-2983
57. Díaz-Ramos, A , Roig-Borrellas, A , García-Melero, A , and López-Alemany, R . α-Enolase, a multifunctional protein: its role on pathophysiological situations. J Biomed Biotechnol. (2012) 2012:156795. doi: 10.1155/2012/156795
58. Didiasova, M , Schaefer, L , and Wygrecka, M . When place matters: shuttling of Enolase-1 across cellular compartments. Front Cell Dev Biol. (2019) 7:61. doi: 10.3389/fcell.2019.00061
59. Everse, J , and Kaplan, NO . Lactate dehydrogenases: structure and function. Adv Enzymol Relat Areas Mol Biol. (1973) 37:61–133. doi: 10.1002/9780470122822.ch2
60. Qiu, H , Xu, X , Fan, B , Rothschild, MF , Martin, Y , and Liu, B . Investigation of LDHA and COPB1 as candidate genes for muscle development in the MYOD1 region of pig chromosome 2. Mol Biol Rep. (2010) 37:629–36. doi: 10.1007/s11033-009-9882-y
61. Rojas-Pirela, M , Andrade-Alviárez, D , Rojas, V , Kemmerling, U , Cáceres, AJ , Michels, PA, et al. Phosphoglycerate kinase: structural aspects and functions, with special emphasis on the enzyme from Kinetoplastea. Open Biol. (2020) 10:200302. doi: 10.1098/rsob.200302
62. Qian, X , Li, X , Cai, Q , Zhang, C , Yu, Q , Jiang, Y, et al. Phosphoglycerate kinase 1 phosphorylates Beclin1 to induce autophagy. Mol Cell. (2017) 65:917–931.e6. doi: 10.1016/j.molcel.2017.01.027
63. Magadum, A , Singh, N , Kurian, AA , Munir, I , Mehmood, T , Brown, K, et al. Pkm2 regulates cardiomyocyte cell cycle and promotes cardiac regeneration. Circulation. (2020) 141:1249–65. doi: 10.1161/CIRCULATIONAHA.119.043067
64. Wong, N , De Melo, J , and Tang, D . PKM2, a central point of regulation in cancer metabolism. Int J Biochem Cell Biol. (2013) 2013:242513. doi: 10.1155/2013/242513
65. Carbonell, J , Felíu, JE , Marco, R , and Sols, A . Pyruvate kinase. Classes of regulatory isoenzymes in mammalian tissues. Eur J Biochem. (1973) 37:148–56. doi: 10.1111/j.1432-1033.1973.tb02969.x
66. Israelsen, WJ , Dayton, TL , Davidson, SM , Fiske, BP , Hosios, AM , Bellinger, G, et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cells. (2013) 155:397–409. doi: 10.1016/j.cell.2013.09.025
67. Luo, W , and Semenza, GL . Pyruvate kinase M2 regulates glucose metabolism by functioning as a coactivator for hypoxia-inducible factor 1 in cancer cells. Oncotarget. (2011) 2:551–6. doi: 10.18632/oncotarget.299
68. Achari, A , Marshall, SE , Muirhead, H , Palmieri, RH , and Noltmann, EA . Glucose-6-phosphate isomerase. Philos Trans R Soc Lond B Biol Sci. (1981) 293:145–57. doi: 10.1098/rstb.1981.0068
69. Ntambi, JM , Miyazaki, M , and Dobrzyn, A . Regulation of stearoyl-CoA desaturase expression. Lipids. (2004) 39:1061–5. doi: 10.1007/s11745-004-1331-2
70. Kikuchi, K , and Tsukamoto, H . Stearoyl-CoA desaturase and tumorigenesis. Chem Biol Interact. (2020) 316:108917. doi: 10.1016/j.cbi.2019.108917
71. Leng, X , and Jiang, H . Effects of arachidonic acid and its major prostaglandin derivatives on bovine myoblast proliferation, differentiation, and fusion. Domest Anim Endocrinol. (2019) 67:28–36. doi: 10.1016/j.domaniend.2018.12.006
72. Orlando, UD , Castillo, AF , Dattilo, MA , Solano, AR , Maloberti, PM , and Podesta, EJ . Acyl-CoA synthetase-4, a new regulator of mTOR and a potential therapeutic target for enhanced estrogen receptor function in receptor-positive and -negative breast cancer. Oncotarget. (2015) 6:42632–50. doi: 10.18632/oncotarget.5822
Keywords: transcriptome sequencing (RNA-seq), longissimus dorsi muscle, differentially expressed genes, Diannan small ears pig, Wujin pig, landrace pig
Citation: Li Q, Hao M, Zhu J, Yi L, Cheng W, Xie Y and Zhao S (2024) Comparison of differentially expressed genes in longissimus dorsi muscle of Diannan small ears, Wujin and landrace pigs using RNA-seq. Front. Vet. Sci. 10:1296208. doi: 10.3389/fvets.2023.1296208
Edited by:
Ning Song, Anhui Agricultural University, ChinaReviewed by:
Bárbara Silva-Vignato, University of São Paulo, BrazilHui Li, Guangxi University, China
Lian Huang, Southwest Minzu University, China
Copyright © 2024 Li, Hao, Zhu, Yi, Cheng, Xie and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sumei Zhao, zhaosm2009@126.com
 Qiuyan Li
Qiuyan Li Meilin Hao
Meilin Hao Junhong Zhu1
Junhong Zhu1  Lanlan Yi
Lanlan Yi Wenjie Cheng
Wenjie Cheng