Genetic analysis of the equine orthologues for human CYP2D6: unraveling the complexity of the CYP2D family in horses
- 1Pharmgenetix GmbH, Niederalm-Anif, Austria
- 2Privatklinik Maria Hilf GmbH, Klagenfurt, Austria
Introduction: Because of their importance as companion animals or as racehorses, horses can be treated with various drugs. Although it is known that drug withdrawal times can vary for each horse, pharmacogenetics for these animals has not been adequately studied and requires further development. Since CYP2D6 is responsible for the metabolism of 25–30% of drugs in humans, including some used to treat horses, a study of the CYP2D family in horses was conducted to define its genetic structure as well as its expression pattern in the liver.
Methods: Genomic DNA extracted from venous blood and mRNA from fresh liver tissue were amplified and sequenced to analyze the genomic structure, genotype, and expression of the various enzymes that are part of the equine orthologous family for CYP2D6.
Results: Amplification and sequencing of the gDNA of CYP2D50, the major CYP2D6 orthologue identified in previous studies, revealed a novel putative genomic structure for this gene compared with that reported from the EquCab3.0 assembly, including the formation of a hybrid structure similar to what happens in human CYP2D6. At the mRNA level, transcripts from six different members of the equine CYP2D family were detected in horse liver. In addition, genotyping of CYP2D50 and CYP2D82 revealed the presence of several polymorphisms, six of which result in novel, nonsynonymous amino acid changes for each of the two genes.
Discussion: This study aimed to elucidate the pharmacogenetic analysis of the CYP2D family in horses and resulted in the identification of a novel gene structure for CYP2D50, the expression of six different members of the CYP2D family in horse liver, and several novel polymorphisms for CYP2D50 and CYP2D82.
Introduction
Pharmacogenetics has become an increasingly important topic in recent years, driving medicine toward more personalized therapy. Genetic variations can greatly affect an enzyme’s ability to metabolize drugs, and by identifying these functional enzymatic variations, a more efficient therapy can be prescribed (1). The acquisition of pharmacogenetic knowledge is not limited to human genetics but is also echoed in veterinary medicine. Horses are among the companion and sport animals that can be widely treated with drugs such as NSAIDs (2–4), corticosteroids (5, 6), local anesthetics (7, 8), mucolytics (9), sedatives (10), antibiotics (11–13), diuretics (14–16) and PPIs (17, 18). In addition to certain medications that are banned in racehorses, others may be administered as needed, but are subject to control through drug testing. The Federation Equestre Internationale (FEI) provides a List of Detection Times (19) that indicates the approximate length of time that a drug or its metabolite remains in a horse’s system and is therefore detectable. This document also notes that the withdrawal time for a drug should not be decided simply on the basis of the stated detection time, but that individual differences between horses such as size, fitness level, metabolism, and concurrent administration of other drugs should be considered. Genetic variations that impair the functionality of enzymes involved in the metabolism of these controlled drugs can result in positive doping findings even when standard drug withdrawal is followed, leading to regulatory problems for racehorses (20, 21). Considering that the annual economic value of the combined sport horse and breeding industry in Europe is 52.1 billion euros per year (22), pharmacogenetics may become an important turning point in the pharmaceutical treatment of horses.
The cytochrome P450 (CYP450) superfamily of monooxygenase enzymes is well known in humans and is responsible for various reactions involved in life processes, mainly drug metabolism and detoxification of xenobiotics, but also for the synthesis of endogenous molecules such as steroids or vitamin B3 (23). Cytochrome 2D6 (CYP2D6), which is responsible for the metabolism of 25–30% of currently available drugs, including antiarrhythmics, antidepressants, chemotherapeutic agents, and opioids, is one of the most important members of the family in humans, although it accounts for only 2% of the total CYP content in the liver (24, 25). One of the main features of CYP2D6 is that it is highly polymorphic, leading to high interindividual variability in its activity. Accordingly, these differences are reflected in four different phenotypes, termed Ultrarapid (UM), Extensive (EM), Intermediate (IM), or Poor (PM) metabolizers (26). The clinical relevance of these specific phenotypes is high, especially for UM or PM, which can lead to adverse drug reactions or impaired treatment efficacy, depending on the metabolic pathway of the drug (27). These four phenotypes have also been observed in horses (28, 29). Several drugs that are metabolized by CYP2D6 in humans, such as dextromethorphan, lidocaine, codeine, or tramadol, can also be administered to horses for the treatment of crib-biting (30), as cough suppressants (31), or for general analgesia (32, 33). In addition, the risk of accidental exposure to drugs or substances that test positive in racehorses is high (34, 35), even considering that dextromethorphan, its metabolite dextrorphan, and other common human therapeutics that use the same hepatic pathway, such as tramadol and venlafaxine, are frequently present and stable in the environment, with a peak during the cold season (36). However, to date, knowledge of the equine orthologues for CYP2D6 is limited, as is the presence of possible polymorphisms leading to impaired drug metabolism, although some studies indicate metabolic variability in equine CYP2Ds (29, 37). In contrast to humans (where only one member of the CYP2D family is active), but similar to other species such as rodents or rabbits, a total of six CYP2Ds have been identified in horses (38). To date, only two of these genes, CYP2D50 and CYP2D82, have been shown to be responsible for the metabolism of dextromethorphan (28) and codeine (29, 39), respectively. Moreover, similar to humans, CYP2Ds in horses and their polymorphisms can have strong clinical effects on drug metabolism, resulting in adverse effects or lack of efficacy (29). However, little is known about the expression and activity of all other CYP2Ds in horses (38). Given the importance that pharmacogenetic may have in horses, the aim of this study is to analyze the genetic structure and liver expression patterns of CYP2D6 equine orthologues and to define the presence of polymorphisms in these genes that may lead to functional effects.
Materials and methods
Sample collection
Venous whole blood samples were collected in EDTA K Monovettes® (Sarstedt; Wr. Neudorf, Austria) by venipuncture from 71 horses (4 Noriker, 1 Quarter, 1 Icelandic, and 51 Warmblood) at the Tillysburg Pferdeklinik in Sankt Florian (Austria). Fresh liver tissue was also collected immediately after euthanasia from 4 of these horses (1 Noriker, 1 Quarter, and 2 Warmblood). 0,5 cm cubes were cut from the fresh tissue and immediately submerged in RNA Later stabilization reagent (Qiagen, Hilden, Germany). Transport was performed at 4°C.
Genomic DNA (gDNA) isolation from whole blood samples
gDNA was isolated from ~350 mL blood using the EZ1 DNA Blood 350 mL Kit (Qiagen, Hilden, Germany) on the EZ1 Advanced XL platform (Qiagen, Hilden, Germany) according to the manufacturer’s instructions with an elution volume of 50 μL. gDNA was quantified with the QIAxpert (Qiagen, Hilden, Germany) spectrophotometer. Only samples with an A260/A280 between 1.700 and 1.900 were used for downstream analyses.
mRNA isolation from liver tissue and reverse transcription
mRNA was isolated from 60-65 mg of liver tissue stabilized with RNAlater using the QIAamp RNA Blood Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions with an elution volume of 30 μL. The tissue was disrupted using a mortar and pestle and then homogenized using a QIAshredder homogenizer. The mRNA was quantified with the QIAxpert (Qiagen, Hilden, Germany) spectrophotometer. Only samples with an A260/A280 between 1.900 and 2.000 were used for downstream analyses. mRNA was reverse transcribed using the QuantiTect Reverse Transcription Kit (Qiagen, Hilden, Germany). 1 μg RNA was incubated with 1X gDNA wipe-out buffer in an 84 μL reaction for 2 min. at 42°C, followed by 2 min. cooling on ice. 41.5 μL of the incubated mixture were then added to 18 μL + RT and -RT reactions. Both these reactions contained 1X Quantiscript RT buffer and 3 μL RT primer mix. 3 μL Quantiscript reverse transcriptase was added to the +RT mix and 3 μL RNAse-free water was added to the -RT mix. The resulting 60 μL reactions were incubated for 30 min. at 42°C, followed by 3 min. at 95°C and cooling on ice.
PCR and sanger sequencing
PCR reactions (50 μL) were prepared using JumpStart REDAccuTaq LA polymerase (Sigma, St. Louis, Missouri) and consisted of 1X AccuTaq LA Buffer, 0.5 mM dNTPs, 5% DMSO and 0.4 μM forward and reverse primers. The PCR parameters for amplification of transcripts are 96°C for 30 s. followed by 35 PCR cycles with denaturation at 94°C for 15 s., 30 s. annealing and elongation at 68°C for 1 min. 50 s. followed by an additional elongation at 68°C for 30 min. For the amplification of genes, the parameters are 96°C for 30 s. followed by 35 PCR cycles with denaturation at 94°C for 15 s., 30 s. annealing and elongation at 68°C for 4 min. 45 s. followed by an additional elongation at 68°C for 30 min. Primers sequences and specific annealing temperature for the different targets are listed in Supplemental Table S1. The presence of the expected amplicon was verified on a 0.5% agarose gel. The PCR product was purified using the QIAquick PCR purification kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The amplicon was sequenced by Sanger sequencing (Microsynth, Balgach, Switzerland) in the forward and reverse orientations with the following primers for each specific target: CYP2D50–1 transcript (211, 212, 220, 221, 222, 223, 331), CYP2D50–1 gene (220, 221, 222, 223, 228, 265, 266, 267, 254, 331), CYP2D50–2 transcript (220, 221, 223, 228, 253, 331), CYP2D50–2 gene (220, 221, 223, 228, 231, 265, 266, 267, 253, 331), CYP2D82 transcript (395, 396, 397, 398), CYP2D82 gene (395, 396, 397, 398, 507, 508, 509, 510, 511, 512, 513, 514). For sequencing primers sequence, see Supplemental Table S2. For the other CYP2D transcripts, the sequencing was performed using the PCR primers. Sequencing results were analysed using MacVector Software version 18.1.3.
Real-time PCR
10 μL real-time PCR reactions were performed using SYBR Green in white 384-well plates on the Quanstudio 12 K Flex instrument (Thermo Fisher Scientific, United States). Each reaction consisted of 1X SYBR Green (Thermo Fisher Scientific), 250 nM forward and reverse primers and 1:10 diluted cDNA or -RT reaction. Primer sequences for the different targets are listed in Supplemental Table S1. Cycling parameters were as follows: an initial denaturation at 55°C for 2 min. and 95°C for 2 min., followed by 40 cycles at 95°C for 15 s., 55°C for 15 s. and 72°C for 1 min. At the end of the cycles, a melting curve step was added to detect the formation of non-specific products. Gene expression values were determined using the relative standard curve method and actin as the reference gene. GraphPad Prism Software version 9.1.1 was used to plot the values and for the unpaired t-test with Welch’s correction for statistical analysis.
Protein function prediction tools
Sorting Intolerant From Tolerant (SIFT) (40, 41) and PROtein Variation Effect ANalyzer (PROVEAN) (42, 43) were used as protein function prediction tools. For both tools, the wild-type protein sequence was submitted in FASTA format along with a list of amino acid substitutions as described on the website. In SIFT, the analysis parameters were kept as default, and a single SIFT score from 0 to 1 was determined for each of the submitted substitutions (where 0 is considered the most deleterious and 1 as fully tolerated), with all substitutions with a score below 0,05 considered deleterious. For PROVEAN, the threshold score was left at the default value (−2.5) to ensure consistency in sensitivity and specificity, with all amino acid substitutions falling below a score of −2.5 defined as harmful.
Results
Identification of a new genetic structure for CYP2D50 gene
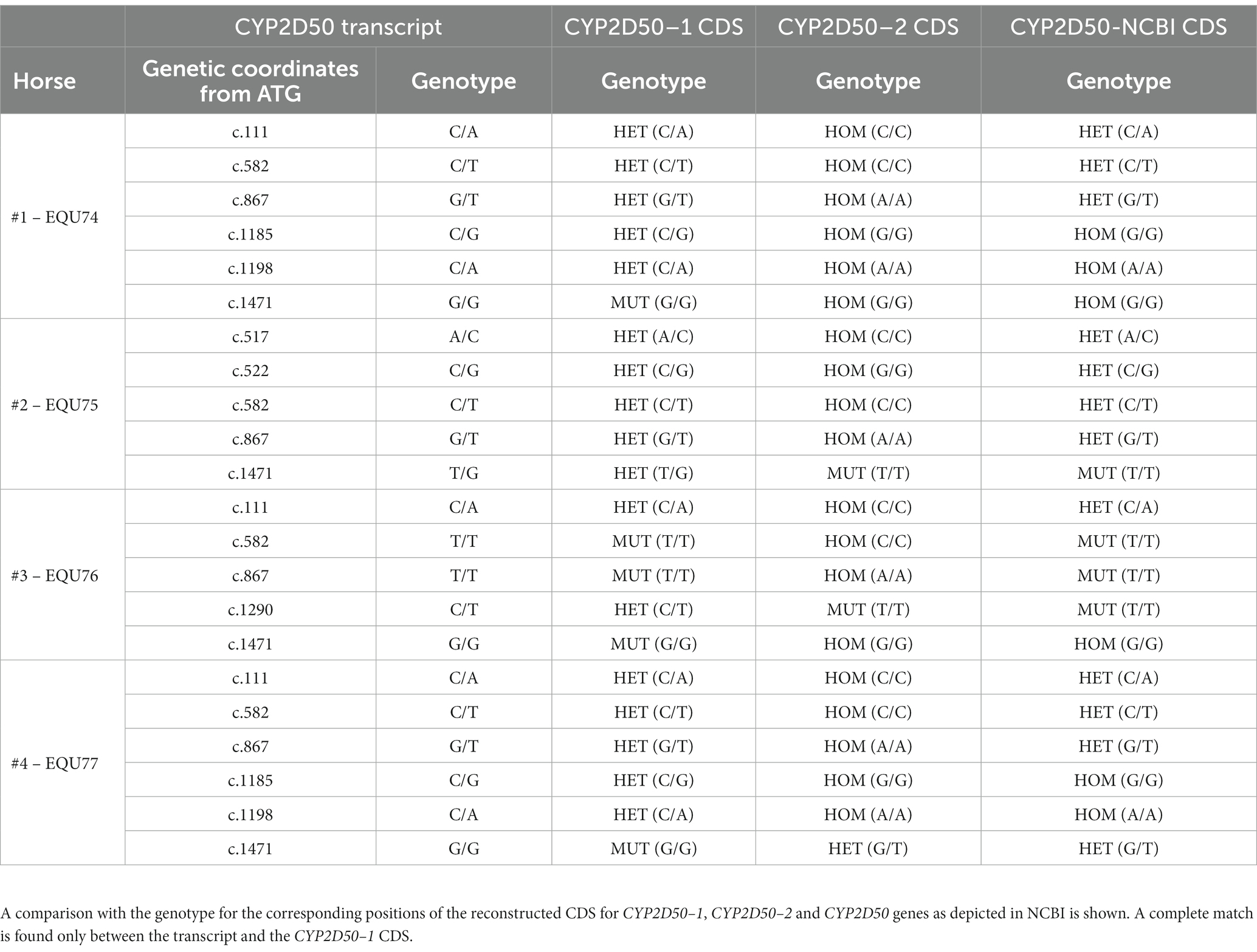
According to the EquCab3.0 assembly, CYP2D50 (NC_009171.3) is represented in NCBI as an approximately 22Kb gene with 9 exons and a 17Kb long intron 7 (Figure 1A). Primer design revealed high sequence similarity between the region containing exons 1 to 7 and the sequence upstream of exon 8 (dashed area in Figure 1A); similarly, this was found also between the region containing exons 8 and 9 and the sequence downstream of exon 7 (dotted area in Figure 1A). This led to a deeper examination of sequence similarity within the gene, which revealed the presence of two putative genetic structures, both containing 9 exons separated by a 13Kb intergenic region (Figure 1B). The two putative genes are hereafter referred to as CYP2D50–1 and CYP2D50–2 and show nearly 95% identity at the coding sequence level (Table 1). Therefore, the structure currently depicted in NCBI would be a fusion between the first 7 exons of putative CYP2D50–1 and the last two exons (8, 9) of putative CYP2D50–2. Exons 8 and 9 between the two putative genes differ by only 3 nucleotides, two in exon 8 and one in exon 9 (Figure 2).
Figure 1. Cartoon of the CYP2D50. (A) Original genetic structure as proposed in the EQUCab3.0 assembly and (B). Two genes structure as proposed in this study. The line represents the genomic sequence, while the numbered boxes represent the exons. The dashed and dotted boxes under (A) indicate the two regions that have a high degree of identity.
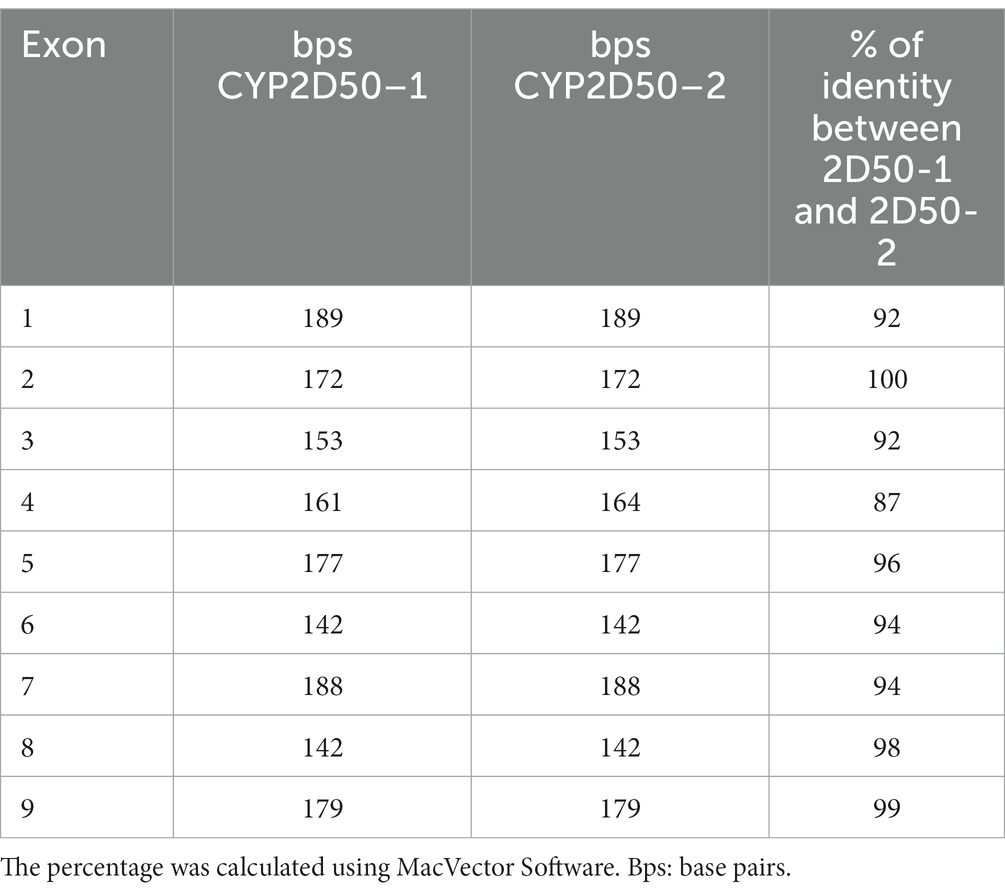
Table 1. Percentage of identity between exons in the two putative genetic structures, CYP2D50–1 and CYP2D50–2, defined by our model.

Figure 2. Alignment and comparison of exon 8 and exon 9 sequence between CYP2D50–1 and CYP2D50–2. Positions are relative to genetic position, starting from ATG, for the two different genetic structures.
We first focused on CYP2D50–1 because its genetic structure contains most of the exons defined for the CYP2D50 gene in NCBI (7 of 9) and an alternative transcript for CYP2D50 that resembles the structure of CYP2D50–1 has already been proposed (XM_023631011.1). A PCR was designed to amplify all possible transcripts produced by CYP2D50–1 and/or by the structure indicated in NCBI in mRNA extracts from fresh horse liver obtained from four different horses. For each of the four horses, the expected coding sequences (CDS) for CYP2D50–1 and the NCBI genetic structure were reconstructed following genes amplification and sequencing from extracted gDNA. Alignment of the sequenced transcript of each horse with the corresponding CDS showed a complete match with the proposed CYP2D50–1 gene and not with the NCBI structure (Table 2). The transcripts were also aligned with the corresponding CYP2D50–2 CDS, and no match was found. A complete list of all mismatches found between the transcript and the CYP2D50–2 CDS for each of the four horses can be found in Supplemental Tables S3–S6. Critical to the analysis of the results was the specific pattern of polymorphisms and resulting genotype identified in the amplified transcripts, particularly in the region of exon 8 and 9, which represents the only CDS difference between CYP2D50–1 and the NCBI genetic structures. The polymorphisms identified in the transcript were completely replicated only in the CYP2D50–1 gene with respect to each of the four sequenced horses (Table 2). A complete match to the corresponding genetic structure was also achieved by designing PCRs specific for CYP2D50–1 and CYP2D50–2 transcripts (data not shown). Since both CYP2D50–1 and CYP2D50–2 transcripts were detected, gene-specific qPCRs were developed to determine their relative expression in horse livers. The results showed that CYP2D50–1 mRNA was expressed 7.5-fold more than CYP2D50–2 (Figure 3). According to the overall results, the following sequences were submitted to the NCBI GenBank database: CYP2D50–1 mRNA, complete CDS (accession numbers OP596321, OP596322, OP596323, OP596324), CYP2D50–2 mRNA, complete CDS (accession number OP715798, requested CYP2D51 as new name for the gene).
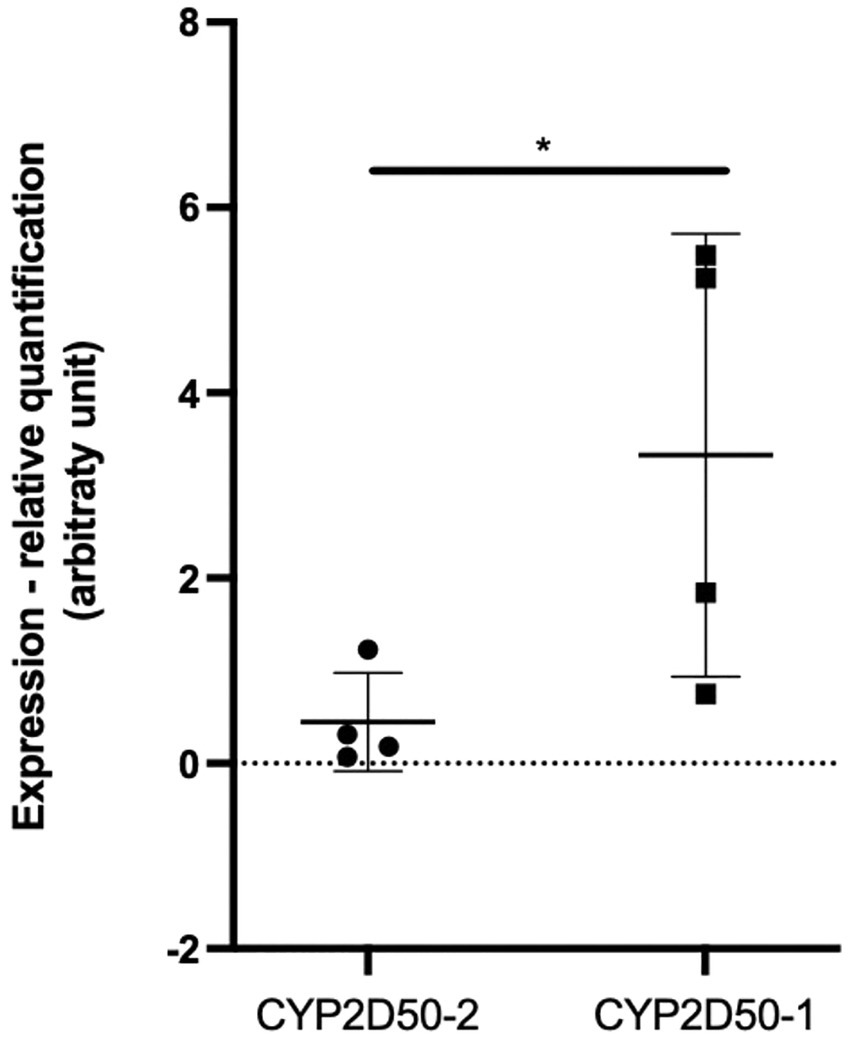
Figure 3. Real-Time PCR quantification of CYP2D50–1 and CYP2D50–2 transcripts on fresh horse liver mRNA extracted from 4 different horses. An asterisk (*) indicates a p value <0.05 after an unpaired t-test with Welch’s correction.
The complexity of the CYP2D genes family
Several genes in the region around the CYP2D50 location on chromosome 28 show a high degree of similarity to each other and to the CYP2D50 gene at the genetic level (Table 3). For some of these genes, expression remains to be confirmed. PCR amplification identified full-length transcripts for LOC100146596 (CYP2D84), LOC100147480 (CYP2D86), and LOC100070990 (CYP2D82) in fresh liver tissue. LOC100070905 was also shown to be expressed, although the PCR used was not-target specific and cloning of the PCR product into vectors was necessary to screen the different amplicons obtained. On the other hand, LOC100056087 (CYP2D89) was shown not to be expressed using a series of primers targeting its specific transcript. LOC100070895 and LOC100070962 (CYP2D88) were also not expressed, although this conclusion was drawn indirectly. Indeed, the primers used to amplify CYP2D50–1 based on the genomic sequence of LOC100070895 and LOC100070962 would also have been able to amplify the transcript of these two genes, if present, resulting in three different sequences. The fact that such PCR always resulted in the specific amplification of only the CYP2D50–1 transcript proves that LOC100070895 and LOC100070962 are not expressed or at least not detectable.
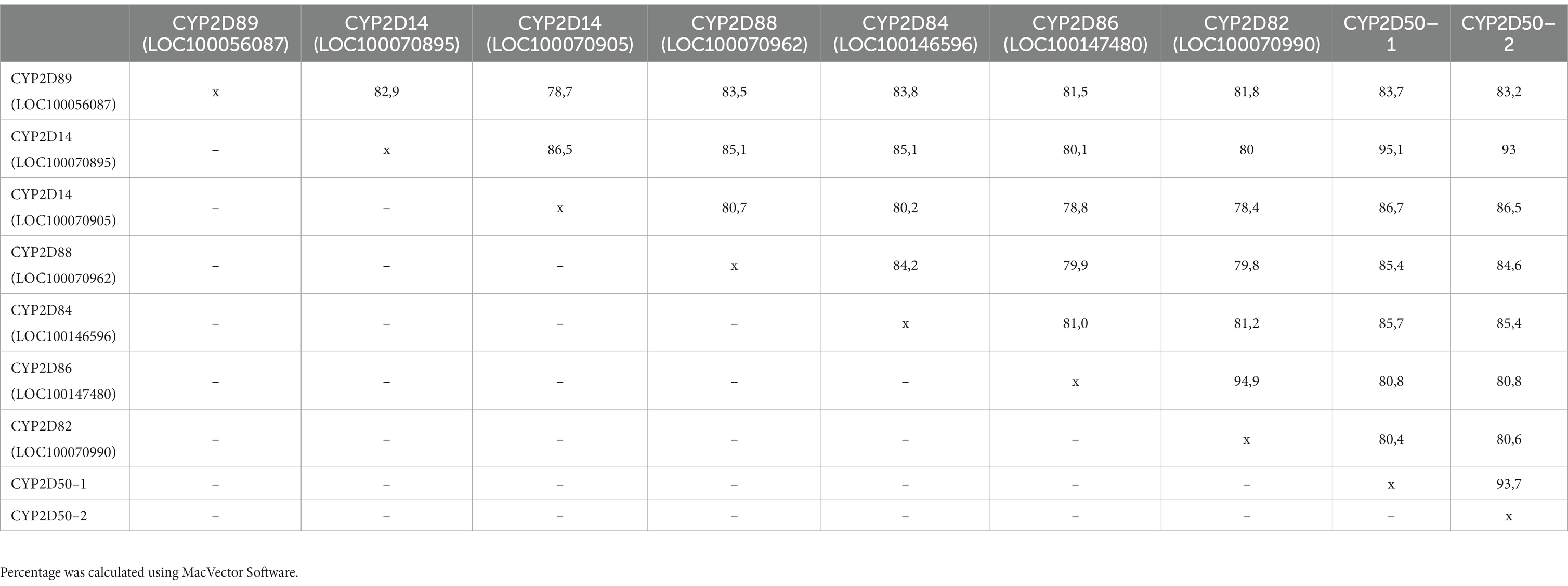
Table 3. Percentage of identity between adjacent loci in the CYP2D50 region at the genetic level (gDNA) from the ATG to the stop codon. 5’ UTR and 3’UTR regions were not considered in the analysis.
Sequencing of CYP2D50–1 and CYP2D82 genes revealed several polymorphisms
Of the members of the CYP2D family in horses, only CYP2D50 and CYP2D82 have been shown to be involved in drug metabolism. To determine the possible SNPs that might affect their function, gDNA corresponding to CYP2D50–1 and CYP2D82 was sequenced from all 72 horses in our cohort.
For the CYP2D50–1 gene, 54 mutations were identified, 23 of which were in exons, including 10 that caused an amino acid change (Supplemental Table S7). Most of these mutations are already associated with an rs number in Ensembl, but 16 of them were observed for the first time. Of the newly identified mutations, 6 cause an amino acid change at positions R31H, G45E, G376A, T486I, L487V, and S489F. All 6 substitutions were tested for functional impact using predictive tools such as SIFT and PROVEAN, and two of them, R31H and G45E, were predicted to be deleterious mutations that severely impair protein function (Table 4).
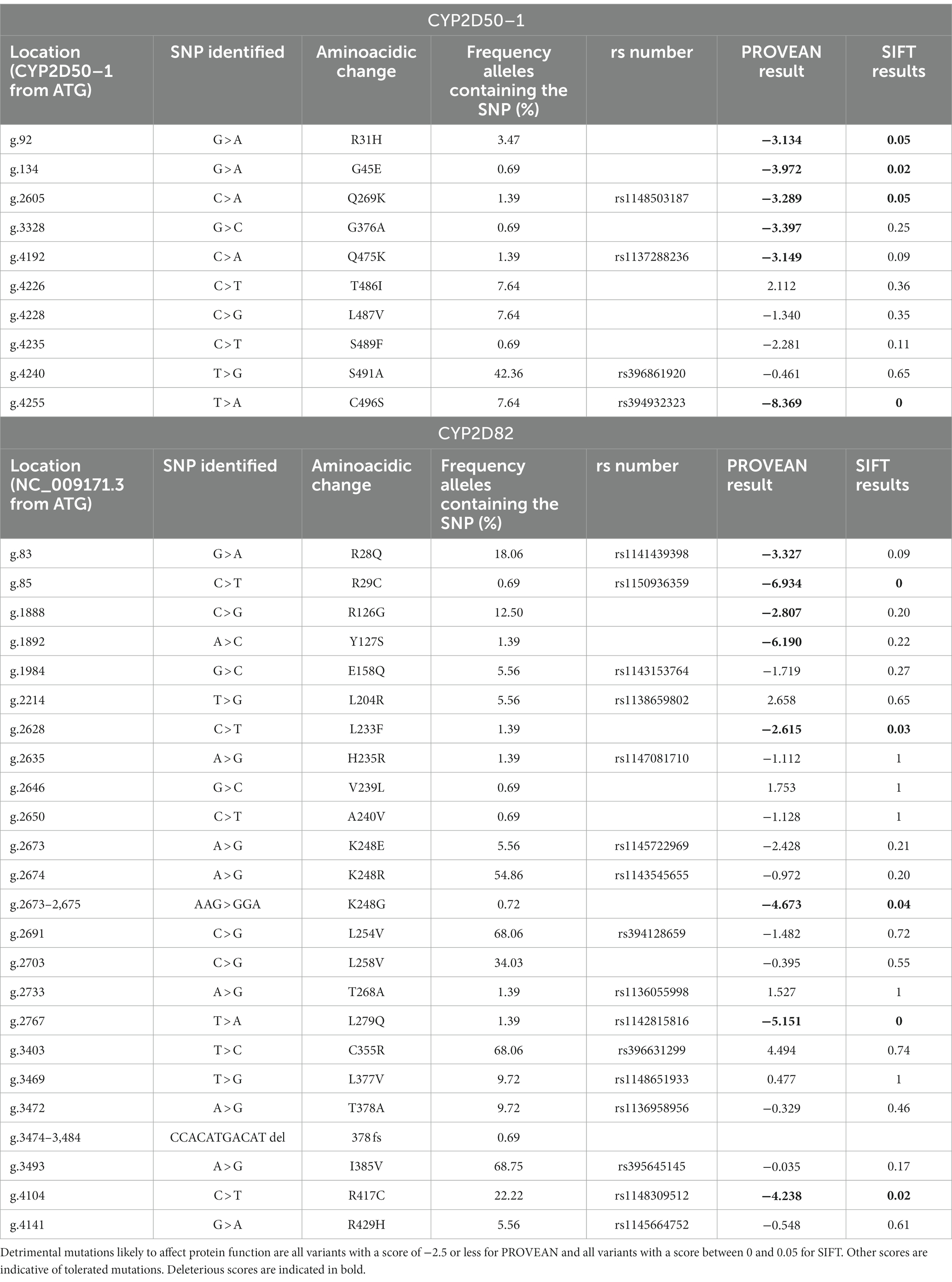
Table 4. List of mutations altering an amino acid found in the CYP2D50–1 and CYP2D82 genes with the resulting score for effects on enzymatic activity derived from PROVEAN and SIFT.
Some of the SNPs identified are very common in the horses tested. 12 mutations were identified in more than 50% of the animals, and 6 of them were frequently detected in homozygosity. One SNP, g.1096 T > C in intron 2, occur in homozygosity in all horses. Although all of these mutations are located in introns or are synonymous variants, an exception is g.4240 T > G, which causes the amino acid mutation S491A and results in complete identity of exon 9 to the sequence of CYP2D50–2.
For the CYP2D82 gene, 88 mutations were identified, 38 of which were in exons, of which 23 caused an amino acid change (Supplemental Table S8). Of the 40 SNPs that were not yet assigned an rs number in Ensembl, 6 cause an amino acid change at positions R126G, Y127S, L233F, V239L, A240V, and L258V. By analysis with SIFT and PROVEAN, only L233F was clearly defined as deleterious, whereas R126G and Y127S appeared as deleterious only with the PROVEAN tool (Table 4). Another newly identified mutation that adversely affects protein function is CCACATGACATdel at position g.3474-3484, resulting in a frameshift (378 fs).
Definition of a CYP2D50–1 hybrid genetic structure
One variation observed in the genetic analysis of CYP2D50 is a CYP2D50–1 hybrid sequence determined by the conversion of exons 8 and 9 with CYP2D50–2. The switching region was located in intron 7 in the area between positions g.3625 and g.3687 on CYP2D50–1, and the hybrid structure was confirmed by the identification of several mismatches in the intron 7 region that perfectly reconstitute the CYP2D50–2 sequence (Figure 4; Table 5). Among the mismatches that convert the CYP2D50–1 sequence to a CYP2D50–2 sequence, nucleotide C in position g.3805 in the CYP250-1 sequence was never found converted to a T, as it should have been for CYP2D50–2. Sequencing of the CYP2D50–2 gene from 9 horses in our cohort revealed a SNP present in homozygosity in all sequenced horses, mutating the T in position g.3864 to a C. Position g.3864 on the CYP2D50–2 gene corresponds to position g.3805 on the CYP2D50–1 gene. The presence of this T > C SNP in the CYP2D50–2 gene explains why the corresponding position on the CYP2D50–1 gene is never found converted in the CYP2D50–1 hybrid structure. The CYP2D50–1 hybrid sequence was identified in 29.17% of the alleles analyzed.

Figure 4. Comparison between the genomic sequence of CYP2D50–1 and CYP2D50–2 in parts of intron 7, exon 8, and exon 9. The sequence of intron 8 sequence is not shown because it is identical between the two genes. Positions are relative to the genetic position, starting with ATG for the two different genetic structures.
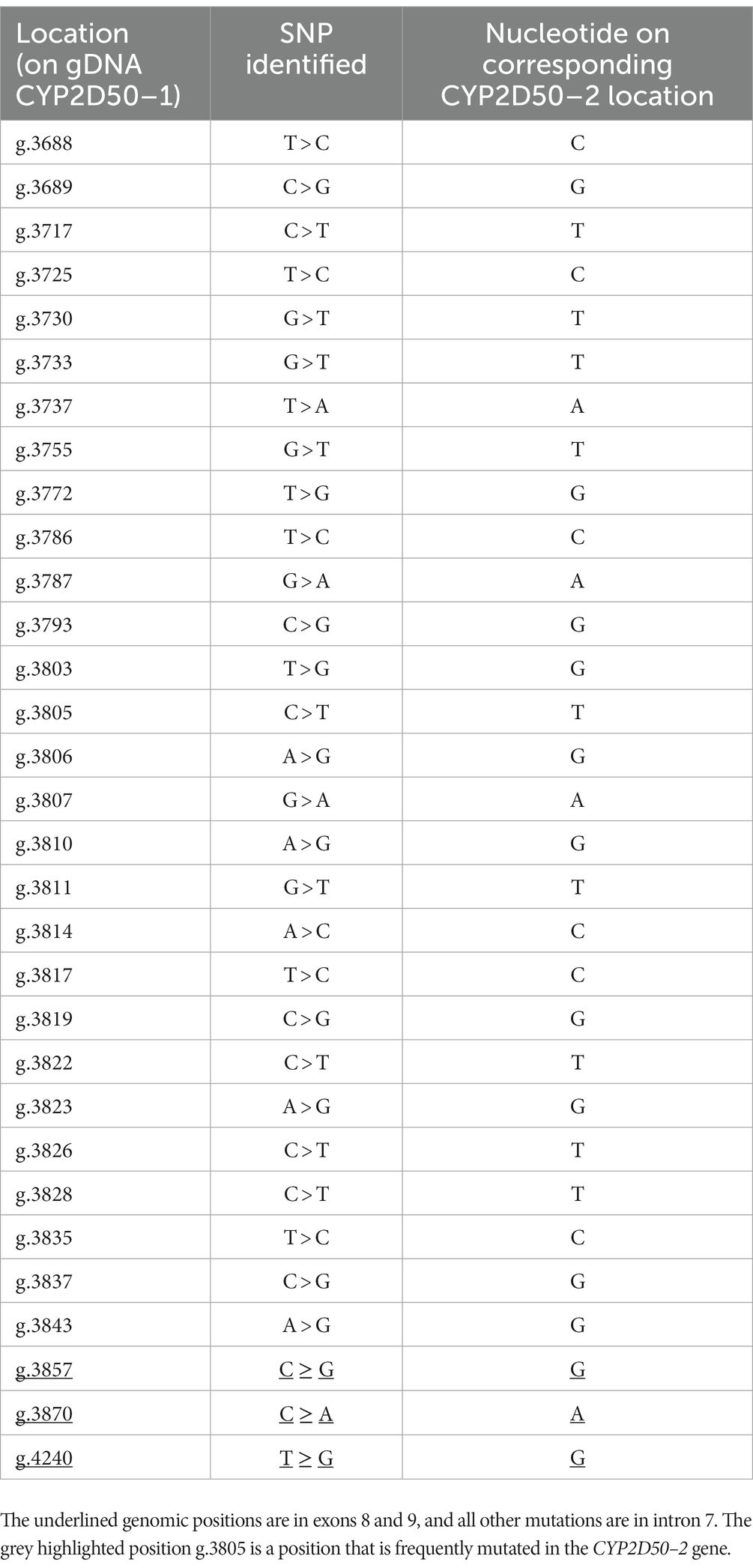
Table 5. List of all SNPs found in the CYP2D50–1 gene between intron 7 and exon 9 converting the sequence to a perfect match with the CYP2D50–2 gene, proving a gene conversion event.
Discussion
Although equine pharmacogenetics may play an increasingly important role in safer therapeutic treatment and appropriate drug withdrawal (20), the involvement of equine metabolic enzymes has not yet been extensively studied. In this view, we analyzed the genetics of the equine orthologue family for CYP2D6. Our initial focus was on CYP2D50, which had a higher level of complexity than previously known. Two very similar genes were identified in the genetic structure previously assigned to the single CYP2D50 gene in NCBI. These two genes, designated as CYP2D50–1 and CYP2D50–2, are both expressed in liver tissue from 4 different horses, although CYP2D50–1 is 7.5-fold more expressed than CYP2D50–2. Overall, the data suggest that the genetic structure of CYP2D50 defined in the latest NCBI EquCab3.0 assembly is likely a fusion of two different genes, CYP2D50–1 and CYP2D50–2, and the former is the most abundant of the two transcripts in horse liver. CYP2D50 is the first gene thought to be the orthologous gene of CYP2D6 in the horse. The CYP2D6 gene locus in humans contains three genes, CYP2D6, CYP2D7, and CYP2D8, the latter two of which are considered pseudogenes and have no functional relevance. However, they are very similar to CYP2D6 at the sequence level and this should always be taken into account in genetic analysis and assay design based on primer and/or probe binding. The two genes CYP2D50–1 and CYP2D50–2 may resemble the gene locus structure in humans, with one being expressed and enzymatically functional and the second being a pseudogene. From the literature, CYP2D50–1 is most likely a functional gene (28), whereas further studies are needed for CYP2D50–2. For specific PCR design, sequence identity between these two genes must be considered, as well as identity with the other 7 genes previously identified in horses that belong to the CYP2D family, 4 of which have been shown to be expressed at the mRNA level in horse liver. For one of them, CYP2D82, expression and function have already been defined (29, 39), whereas for the other 3 (CYP2D84, CYP2D86, and CYP2D14-LOC100070905) no evidence for functionality is available and their expression in other tissues is also unknown. Considering that CYP2D50 and CYP2D82 are involved in the metabolism of two different substrates that are metabolized by CYP2D6 in humans, a study of the target specificity or redundancy of metabolic pathways for the different enzymes would be necessary to identify genes relevant to specific pharmacological treatments if other CYP2D genes are functionally active in horse. Understanding gene locus structure, expression patterns, and enzyme functionality is an essential step toward deeper pharmacogenetic analysis.
An in-depth study of genetic variations was developed only for the two genes for which a function has already been defined, namely CYP2D50–1 and CYP2D82. Several new polymorphisms have been discovered for both genes, some of which determine an amino acid change. In a study with the in-silico prediction tools SIFT and PROVEAN, only 2 of these mutations (R31H and G45E) in CYP2D50–1 and one (L233F) in CYP2D82 were clearly identified as deleterious. These mutations were found in heterozygosity in 5 horses for R31H, in 1 horse for G45E, and in 2 horses for L233F, and if the predictions are correct, it would not be surprising to find metabolic impairment for some drugs in these animals. However, the effect of polymorphisms on the function of an enzyme could only be specifically analyzed if a functional assay was developed, and unfortunately this was not possible in the current study.
Interestingly, analysis of CYP2D50–1 also revealed the possibility of a genetic conversion, a well-known phenomenon that has been described in detail for CYP2D6 in humans (44). An exon 9 conversion and various hybrid structures can be formed between CYP2D6 and CYP2D7, and the results of the genetic analyzes in this study indicate that they are formed similarly between CYP2D50–1 and CYP2D50–2 in the horse genome. Considering that all human CYP2D6::CYP2D7 hybrids are known to be nonfunctional, we might expect the same to be true for CYP2D50–1 and CYP2D50–2 hybrids, although this assumption could only be confirmed experimentally with a functional assay. Since multiplication and deletion of alleles are two other known phenomena for CYP2D6 in humans and can similarly occur in horses (45), the development of specific copy number variation (CNV) assays would be required to overcome some limitations (such as the use of complex algorithms, signal normalization, high background noise, and high error rate in regions of low SNP density (46)) and complement assays based only on SNP to correctly define the equine genotype.
The road to pharmacogenetic analysis in horses is still long because of the lack of knowledge about these genes, their function, and the effects of polymorphisms on them. This study allowed us to gain new insights into the genetic structure of CYP2D50 by identifying two new genes (CYP2D50–1 and CYP2D50–2) and describing the presence of hybrid structures similar to those observed in humans between CYP2D6 and CYP2D7. Extensive sequencing also allowed the identification of novel polymorphisms in CYP250-1 and CYP2D82 also associated with amino acid changes, whose impact would only be defined with a dedicated functional test. In addition, we have begun to unravel the complex structure of the CYP2D family in the horse, highlighting the high sequence similarity between these genes and the associated difficulties for the design of specific PCRs.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
Ethical approval was not required for the studies involving animals in accordance with the local legislation and institutional requirements because venous blood from horses was collected for clinical (rather than scientific) purposes at a veterinary hospital, and the excess sample was donated to PharmGenetix. Liver tissue from horses in the study was obtained from animals that were not euthanized for scientific purposes and donated to PharmGenetix by a veterinary clinic. Written informed consent was not obtained from the owners for the participation of their animals in this study because venous blood from horses was collected for clinical (rather than scientific) purposes at a veterinary hospital, and the excess sample was donated to PharmGenetix. Liver tissue from horses in the study was obtained from animals that were not euthanized for scientific purposes and donated to PharmGenetix by a veterinary clinic.
Author contributions
GS, SV, and CN participated in the study conceptualization and experimental design. GS and SV drafted the manuscript. GS performed the experiments. GS, SV, MP, and CN participated in data analysis and/or interpretation. All authors contributed to the article and approved the submitted version.
Funding
PharmGenetix acknowledges the Österreichische Forschungsförderungsgesellschaft GmbH (FFG) for support via the PGx-Next-Generation Analytics Part 2 grant (FO0999891633/42175800).
Acknowledgments
PharmGenetix thanks Mahringer and the Pferdeklinik Tillysburg for providing the equine specimens used in the study.
Conflict of interest
GS, CN and SV are employees of PharmGenetix GmbH. MP is employed by Privatklinik Maria Hilf GmbH.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1188633/full#supplementary-material
References
1. Oates, JT, and Lopez, D. Pharmacogenetics: an important part of drug development with a focus on its application. Int J Biomed Investig. (2018) 1:1–16. doi: 10.31531/2581-4745.1000111
2. Clark, JO, and Clark, TP. Analgesia. Vet Clin North Am Equine Pract. (1999) 15:705–23. doi: 10.1016/S0749-0739(17)30140-2
3. Goodrich, LR, and Nixon, AJ. Medical treatment of osteoarthritis in the horse - a review. Vet J. (2006) 171:51–69. doi: 10.1016/j.tvjl.2004.07.008
4. Ziegler, AL, and Blikslager, AT. Sparing the gut: COX-2 inhibitors herald a new era for treatment of horses with surgical colic. Equine Vet Educ. (2020) 32:611–6. doi: 10.1111/eve.13189
5. Couetil, LL, Cardwell, JM, Gerber, V, Lavoie, JP, Leguillette, R, and Richard, EA. Inflammatory airway disease of horses--revised consensus statement. J Vet Intern Med. (2016) 30:503–15. doi: 10.1111/jvim.13824
6. Mainguy-Seers, S, and Lavoie, JP. Glucocorticoid treatment in horses with asthma: a narrative review. J Vet Intern Med. (2021) 35:2045–57. doi: 10.1111/jvim.16189
7. Haga, HA, Lykkjen, S, Revold, T, and Ranheim, B. Effect of intratesticular injection of lidocaine on cardiovascular responses to castration in isoflurane-anesthetized stallions. Am J Vet Res. (2006) 67:403–8. doi: 10.2460/ajvr.67.3.403
8. Portier, KG, Jaillardon, L, Leece, EA, and Walsh, CM. Castration of horses under total intravenous anaesthesia: analgesic effects of lidocaine. Vet Anaesth Analg. (2009) 36:173–9. doi: 10.1111/j.1467-2995.2008.00445.x
9. Matsuda, Y, Hobo, S, and Naito, H. Transferability of cephalothin to the alveolar cavity in thoroughbreds. J Vet Med Sci. (1999) 61:209–12. doi: 10.1292/jvms.61.209
10. Valverde, A. Alpha-2 agonists as pain therapy in horses. Vet Clin North Am Equine Pract. (2010) 26:515–32. doi: 10.1016/j.cveq.2010.07.003
11. Giguere, S, Burton, AJ, Berghaus, LJ, and Haspel, AD. Comparative pharmacokinetics of minocycline in foals and adult horses. J Vet Pharmacol Ther. (2017) 40:335–41. doi: 10.1111/jvp.12366
12. Patel, T, Magdesian, KG, Estell, KE, Edman, JM, and Knych, HK. Pharmacokinetics of chloramphenicol base in horses and comparison to compounded formulations. J Vet Pharmacol Ther. (2019) 42:609–16. doi: 10.1111/jvp.12777
13. Chapuis, RJJ, Smith, JS, Uehlinger, FD, Meachem, M, Johnson, R, and Dowling, PM. Pharmacokinetics and pharmacodynamics of doxycycline in a Streptococcusequi subsp. zooepidemicus infection model in horses. J Vet Pharmacol Ther. (2021) 44:766–75. doi: 10.1111/jvp.12982
14. Baggot, JD. The pharmacological basis of cardiac drug selection for use in horses. Equine Vet J Suppl. (1995) 27:97–100. doi: 10.1111/j.2042-3306.1995.tb04995.x
15. Ballester, MR, Roig, E, Gich, I, Puntes, M, Delgadillo, J, Santos, B, et al. Randomized, open-label, blinded-endpoint, crossover, single-dose study to compare the pharmacodynamics of torasemide-PR 10 mg, torasemide-IR 10 mg, and furosemide-IR 40 mg, in patients with chronic heart failure. Drug Des Devel Ther. (2015) 9:4291–302. doi: 10.2147/DDDT.S86300
16. Agne, GF, Jung, SW, Wooldridge, AA, Duran, SH, Ravis, W, and Toribio, R. Pharmacokinetic and pharmacodynamic properties of orally administered torsemide in healthy horses. J Vet Intern Med. (2018) 32:1428–35. doi: 10.1111/jvim.15213
17. Andrews, FM, Sifferman, RL, Bernard, W, Hughes, FE, Holste, JE, Daurio, CP, et al. Efficacy of omeprazole paste in the treatment and prevention of gastric ulcers in horses. Equine Vet J Suppl. (1999) 31:81–6. doi: 10.1111/j.2042-3306.1999.tb05176.x
18. Videla, R, Sommardahl, CS, Elliott, SB, Vasili, A, and Andrews, FM. Effects of intravenously administered esomeprazole sodium on gastric juice pH in adult female horses. J Vet Intern Med. (2011) 25:558–62. doi: 10.1111/j.1939-1676.2011.0716.x
19. Internationale F-FE. (2022). FEI list of detection times 2022 [updated 2022. Available at: https://inside.fei.org/system/files/FEI%20Detection%20Times%202022.pdf.
20. Tobin, T, Dirikolu, L, Brewer, K, and Hughes, CG. A clinician's guide to factors affecting withdrawal times for equine therapeutic medications. Vet J. (2013) 198:313–21. doi: 10.1016/j.tvjl.2013.07.002
21. Waller, P, Lomnicka, I, Lucas, C, Johnson, S, and Dirikolu, L. The medication violations in racehorses at Louisiana racetracks from 2016 to 2020. Vet Med Sci. (2022) 8:553–60. doi: 10.1002/vms3.724
22. Internationale F-FE. (2021). Protecting the EU equine industry and equine health adn welfare 2021 Available at: https://inside.fei.org/system/files/PROTECTING%20THE%20EU%20EQUINE%20INDUSTRY.pdf.
23. Nebert, DW, and Russell, DW. Clinical importance of the cytochromes P450. Lancet. (2002) 360:1155–62. doi: 10.1016/S0140-6736(02)11203-7
24. Ingelman-Sundberg, M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics J. (2005) 5:6–13. doi: 10.1038/sj.tpj.6500285
25. Ingelman-Sundberg, M, Sim, SC, Gomez, A, and Rodriguez-Antona, C. Influence of cytochrome P450 polymorphisms on drug therapies: pharmacogenetic, pharmacoepigenetic and clinical aspects. Pharmacol Ther. (2007) 116:496–526. doi: 10.1016/j.pharmthera.2007.09.004
26. Zanger, UM, and Schwab, M. Cytochrome P450 enzymes in drug metabolism: regulation of gene expression, enzyme activities, and impact of genetic variation. Pharmacol Ther. (2013) 138:103–41. doi: 10.1016/j.pharmthera.2012.12.007
27. Kirchheiner, J, Nickchen, K, Bauer, M, Wong, ML, Licinio, J, Roots, I, et al. Pharmacogenetics of antidepressants and antipsychotics: the contribution of allelic variations to the phenotype of drug response. Mol Psychiatry. (2004) 9:442–73. doi: 10.1038/sj.mp.4001494
28. Corado, CR, McKemie, DS, and Knych, HK. Dextromethorphan and debrisoquine metabolism and polymorphism of the gene for cytochrome P450 isozyme 2D50 in thoroughbreds. Am J Vet Res. (2016) 77:1029–35. doi: 10.2460/ajvr.77.9.1029
29. Gretler, SR, Finno, CJ, Kass, PH, and Knych, HK. Functional phenotyping of the CYP2D6 probe drug codeine in the horse. BMC Vet Res. (2021) 17:77. doi: 10.1186/s12917-021-02788-y
30. Rendon, RA, Shuster, L, and Dodman, NH. The effect of the NMDA receptor blocker, dextromethorphan, on cribbing in horses. Pharmacol Biochem Behav. (2001) 68:49–51. doi: 10.1016/S0091-3057(00)00437-8
31. Westermann, CM, Laan, TT, van Nieuwstadt, RA, Bull, S, and Fink-Gremmels, J. Effects of antitussive agents administered before bronchoalveolar lavage in horses. Am J Vet Res. (2005) 66:1420–4. doi: 10.2460/ajvr.2005.66.1420
32. Cox, S, Villarino, N, and Doherty, T. Determination of oral tramadol pharmacokinetics in horses. Res Vet Sci. (2010) 89:236–41. doi: 10.1016/j.rvsc.2010.02.011
33. Knych, HK, Stucker, K, Gretler, SR, Kass, PH, and McKemie, DS. Pharmacokinetics, adverse effects and effects on thermal nociception following administration of three doses of codeine to horses. BMC Vet Res. (2022) 18:196. doi: 10.1186/s12917-022-03299-0
34. Camargo FCH, C, Lehner, AF, and Tobin, T. Trace level benzoylecgonine identifications in post-race urines: probable sources and the regulatory significance of such identifications. AAEP Proceedings. (2006) 52:331–6.
35. Camargo FCL, AF, Karpiesiuk, W, Stirling, K, Kavanagh, PV, Brennan, N, Dowling, M, et al. Rewiew of environmental morphine identifications: worldwide occurrences and responses of authoriites. AAEP Proceedings. (2005) 61:58–64.
36. Campos-Manas, MC, Ferrer, I, Thurman, EM, Sanchez Perez, JA, and Aguera, A. Identification of opioids in surface and wastewaters by LC/QTOF-MS using retrospective data analysis. Sci Total Environ. (2019) 664:874–84. doi: 10.1016/j.scitotenv.2019.01.389
37. Corado, CR, McKemie, DS, Young, A, and Knych, HK. Evidence for polymorphism in the cytochrome P450 2D50 gene in horses. J Vet Pharmacol Ther. (2016) 39:245–54. doi: 10.1111/jvp.12269
38. Yasukochi, Y, and Satta, Y. Evolution of the CYP2D gene cluster in humans and four non-human primates. Genes Genet Syst. (2011) 86:109–16. doi: 10.1266/ggs.86.109
39. Knych, HK, Baden, RW, Gretler, SR, and McKemie, DS. Characterization of the in vitro CYP450 mediated metabolism of the polymorphic CYP2D6 probe drug codeine in horses. Biochem Pharmacol. (2019) 168:184–92. doi: 10.1016/j.bcp.2019.07.005
40. Ng, PC, and Henikoff, S. Predicting deleterious amino acid substitutions. Genome Res. (2001) 11:863–74. doi: 10.1101/gr.176601
41. Kumar, P, Henikoff, S, and Ng, PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. (2009) 4:1073–81. doi: 10.1038/nprot.2009.86
42. Choi, Y, and Chan, AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. (2015) 31:2745–7. doi: 10.1093/bioinformatics/btv195
43. Choi, Y, Sims, GE, Murphy, S, Miller, JR, and Chan, AP. Predicting the functional effect of amino acid substitutions and indels. PLoS One. (2012) 7:e46688. doi: 10.1371/journal.pone.0046688
44. Gaedigk, A, Bradford, LD, Alander, SW, and Leeder, JS. CYP2D6*36 gene arrangements within the cyp2d6 locus: association of CYP2D6*36 with poor metabolizer status. Drug Metab Dispos. (2006) 34:563–9. doi: 10.1124/dmd.105.008292
45. Ghosh, S, Qu, Z, Das, PJ, Fang, E, Juras, R, Cothran, EG, et al. Copy number variation in the horse genome. PLoS Genet. (2014) 10:e1004712. doi: 10.1371/journal.pgen.1004712
Keywords: horse pharmacogenetics, CYP2D6 equine orthologues, CYP2D50, CYP2D82, drug metabolism
Citation: Scantamburlo G, Nofziger C, Paulmichl M and Vanoni S (2023) Genetic analysis of the equine orthologues for human CYP2D6: unraveling the complexity of the CYP2D family in horses. Front. Vet. Sci. 10:1188633. doi: 10.3389/fvets.2023.1188633
Edited by:
Stephen M. Reed, Rood and Riddle Equine Hospital, United StatesReviewed by:
Heather Knych, University of California, Davis, United StatesMaria Cristina Cozzi, University of Milan, Italy
Copyright © 2023 Scantamburlo, Nofziger, Paulmichl and Vanoni. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Giada Scantamburlo, giada.scantamburlo@pharmgenetix.com
 Giada Scantamburlo1*
Giada Scantamburlo1*  Simone Vanoni
Simone Vanoni