Metallopeptidades 2 and 9 genes epigenetically modulate equine endometrial fibrosis
- 1CIISA - Center for Interdisciplinary Research in Animal Health, Faculty of Veterinary Medicine, University of Lisbon, Lisbon, Portugal
- 2Associate Laboratory for Animal and Veterinary Sciences (AL4AnimalS), Lisbon, Portugal
- 3Unidade Estratégica de Investigação e Serviços de Biotecnologia e Recursos Genéticos (UEISBR), Instituto Nacional de Investigação Agrária e Veterinária, I. P. (INIAV), Vairão, Portugal
- 4Embriovet, Muge, Portugal
- 5Polytechnic of Coimbra, Coimbra Agriculture School, Coimbra, Portugal
- 6Faculty of Medicine and Biomedical Sciences (FMCB), University of Algarve, Faro, Portugal
- 7Algarve Biomedical Center Research Institute (ABC-RI), Faro, Portugal
- 8Institute of Animal Reproduction and Food Research PAS, Olsztyn, Poland
Endometrium type I (COL1) and III (COL3) collagen accumulation, periglandular fibrosis and mare infertility characterize endometrosis. Metalloproteinase-2 (MMP-2), MMP-9 and tissue inhibitors of metalloproteinases (TIMP-1 and TIMP-2) are involved in collagen turnover. Since epigenetic changes may control fibroproliferative diseases, we hypothesized that epigenetic mechanisms could modulate equine endometrosis. Epigenetic changes can be reversed and therefore extremely promising for therapeutic use. Methylation pattern analysis of a particular gene zone is used to detect epigenetic changes. DNA methylation commonly mediates gene repression. Thus, this study aimed to evaluate if the transcription of some genes involved in equine endometrosis was altered with endometrial fibrosis, and if the observed changes were epigenetically modulated, through DNA methylation analysis. Endometrial biopsies collected from cyclic mares were histologically classified (Kenney and Doig category I, n = 6; category IIA, n = 6; category IIB, n = 6 and category III, n = 6). Transcription of COL1A1, COL1A2, COL3A1, MMP2, MMP9, TIMP1, and TIMP2 genes and DNA methylation pattern by pyrosequencing of COL1A1, MMP2, MMP9, TIMP1 genes were evaluated. Both MMP2 and MMP9 transcripts decreased with fibrosis, when compared with healthy endometrium (category I) (P < 0.05). TIMP1 transcripts were higher in category III, when compared to category I endometrium (P < 0.05). No differences were found for COL1A1, COL1A2, COL3A1 and TIMP2 transcripts between endometrial categories. There were higher methylation levels of (i) COL1A1 in category IIB (P < 0.05) and III (P < 0.01), when compared to category I; (ii) MMP2 in category III, when compared to category I (P < 0.001) and IIA (P < 0.05); and (iii) MMP9 in category III, when compared to category I and IIA (P < 0.05). No differences in TIMP1 methylation levels were observed between endometrial categories. The hypermethylation of MMP2 and MMP9, but not of COL1A1 genes, occurred simultaneously with a decrease in their mRNA levels, with endometrial fibrosis, suggesting that this hypermethylation is responsible for repressing their transcription. Our results show that endometrosis is epigenetically modulated by anti-fibrotic genes (MMP2 and MMP9) inhibition, rather than fibrotic genes activation and therefore, might be promising targets for therapeutic use.
Introduction
Equine endometrosis is a multifactorial disease considered to be one of the most important causes of equine infertility, especially in older mares (1) and has an economic impact on the horse breeding history (2). Endometrosis is characterized by periglandular fibrosis of the equine endometrium (3) which compromises the integrity and function of the endometrial glands required for embryonic preimplantation and placental development (4). However, this condition is still a puzzle regarding its pathogenesis and treatment. Periglandular arrangement of myofibroblasts, associated with the deposition of extracellular matrix (ECM), such as collagen (COL), is a cardinal feature of endometrosis in mares (5). Bochsler and Slauson (6) stated that the deposition of collagen types I (COL1) and III (COL3) occurs in fibrotic processes, which promotes the development of cicatricial tissues with major tensile strength. In a healthy endometrium, the first collagen to be synthesized is COL3, which in turn is gradually replaced by COL1 following the development of fibrotic lesions (7, 8). Collagen type I is usually predominant in Kenney's category III endometrium (severe fibrosis) (7), and COL3 in category I healthy endometrium (9). Matrix metalloproteinases (MMPs) are a family of extracellular endopeptidases (10) that are important factors in the process of fibrosis. Data concerning MMP expression in equine endometrial fibrosis are limited but MMP-2 and MMP-9 (now called 72 kDa gelatinase and 92 kDa gelatinase, in the horse), seem to be involved in this process (11–14). MMP-2 and MMP-9 are gelatinases that denature collagens (gelatins) and other ECM substrates (15, 16). The endogenous inhibitors of MMPs are tissue inhibitors of metalloproteinases (TIMPs) and neutralize the activity of MMPs. Among the four types of TIMPs, TIMP-1 is a specific inhibitor for MMP-9 (15) while TIMP-2 regulates MMP-2 activity (17). A key feature of fibrosis is the disbalance between MMPs and TIMPs resulting the loss of the homeostasis between fibrolysis and fibrogenesis (18).
Novel findings implicate a role for epigenetic modifications contributing to the progression of fibrosis by alteration of gene expression profiles (19–22). Epigenetic modifications, heritable changes in the genome that do not alter the DNA sequence, influence, or regulate gene expression (23). Epigenetic changes, unlike genetic alterations, can be reversed (24) as thus extremely promising for therapeutic use. In mammals, the most studied epigenetic events are DNA methylation and histone modifications, such as methylation, acetylation, ubiquitination, and phosphorylation (25, 26). DNA methylation constitutes a major epigenetic modification of the genome and is essential for cellular reprogramming, tissue differentiation, and normal development related to many biological processes including gene expression regulation. DNA methylation is known to occur at the 5′ of cytosine in CpG dinucleotides which are found mostly in so-called CpG islands present in promoters (27–29) and is catalyzed by DNA methyltransferases (DNMTs) such as DNMT1, DNMT3a, and DNMT3b (30). These CpG islands are enriched in promotor regions close to transcriptional start sites and their methylation might prevent the transcription of the respective gene (31, 32). Hypermethylation of a promoter has long been well recognized as an efficient means of repressing transcription (33). The majority of CpG sites outside of CpG islands are methylated, suggesting a role in the global maintenance of the genome, while the majority of CpG islands in gene promoters are unmethylated, which allows active gene transcription (34, 35). Transcriptional factors bind to the unmethylated promotor region of a gene to allow its transcription. But, if that region becomes hypermethylated this binding does not occur, and this transcription is not activated. However, most recently, it was demonstrated that intragenic DNA methylation could also affect the gene expression (36). In fact, differential methylation within the gene body plays a role in several gene regulation processes (37). One way to evaluate epigenetics mechanisms is through DNA methylation by DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B). However, this only reflects the level of these enzymes, which in turn may indicate the level of global methylation. Another way to assess the DNA methylation pattern is by bisulfite pyrosequencing. This method is very accurate and is commonly used for quantitative analysis of DNA methylation at single nucleotide level, and in a particular region of the gene (CpG islands), providing more detailed information.
As such, we proposed to evaluate the transcriptomic pattern of some of the most relevant genes involved in mare endometrosis (COL1A1, COL1A2, COL3A1, MMP2, MMP9, TIMP1, and TIMP2) and secondly, perform epigenetic studies of the genes that have shown alterations (through DNA methylation analysis) to determine whether there is an epigenetic regulation of endometrial fibrosis in mares.
Materials and methods
Animals
During the breeding season, Lusitano cyclic mares (n = 24; 6 per Kenney and Doig's category; 3 in luteal phase and 3 in follicular phase per category) were used for endometrial biopsy procurement. Mare's internal genitalia were examined by transrectal ultrasonography (Sonovet 600). Endometrial biopsies were randomly obtained from cyclic mares (May to July), with a biopsy alligator jaw forceps (ref. 141965; Kruuse), complying to welfare mandates as a clinical procedure, and with owner's consent. The age of mares ranged from 3 ± 0 years within category I; from 3 to10 years (5.17 ± 1.38) in category IIA, from 6 to 14 years (9.50 ± 1.26) in category IIB, and 11 to 23 years (18.5 ± 2.72) in category III.
The tissue was divided into small pieces with a scalpel and then immersed in RNA later for qPCR or in 4% formaldehyde solution for histopathological evaluation. Formaldehyde-fixed endometrium was paraffin embedded and hematoxylin (05-06014E; Bio-Optica) and eosin (HT1103128; Sigma-Aldrich) stained sections were examined under a light microscope (Leica DM500; Leica Microsystems, Mannheim, Germany). Endometrial biopsies were graded based on the extent of inflammation and /or fibrosis, following Kenney and Doig's classification (38). They were assigned to category I (n = 6) when the endometrium was healthy or with slight or sparse inflammation or fibrosis; to category IIA (n = 6) when there was mild and scattered inflammation and fibrosis; to category IIB (n = 6), when moderate inflammation or fibrosis were present; or to category III (n = 6), characterized by severe irreversible fibrosis and/or inflammation.
Real time PCR
Endometrial biopsies, from different Kenney and Doig's categories, were used for the evaluation of COL1A1, COL1A2, COL3A1, MMP2, MMP9, TIMP1, and TIMP2 transcripts, after RNA isolation, cDNA synthesis and qPCR studies, performed as described (39). Briefly, total RNA was extracted using TRI Reagent (Ref T9424; Sigma Life Science), including a DNA-digestion step with an RNase-free DNase (Ref. 79254, RNase-Free DNase Set, Qiagen, Germany), according to manufacturer's instructions. Quantification and quality of RNA was carried out with a Nanodrop system (ND; Fisher Scientific, Spain) and by agarose gel electrophoresis, respectively. The cDNA was obtained from total RNA (1μg), using M-MLV Reverse transcriptase (Ref. M1705; Promega) and oligo (dT) 15 primer (Ref. C101; Promega). Specific primers were designed, as well as the reference gene (Supplementary Table 1), using the Internet-based program Primer-3 (40) and Primer Premier software (Premier Biosoft Interpairs). Mitochondrial ribosomal protein L32 (MRPL32) was chosen as the most stable internal control gene (41) among four validated reference genes, as described (39). Using Power SYBER Green PCR Master Mix (Ref. 4367659; Applied Biosystems) and a StepOne-Plus TM Real-Time PCR System (Applied Biosystems), qPCR studies of target and reference genes were performed simultaneously. Zhao and Fernald (42) method was used to analyze the relative mRNA data.
DNA preparation
DNA extraction was performed using the kit Quick-DNA 96 Plus Kit (Zymo Research®). Briefly the biopsy sample (25 mg) was diluted with 95 μL of DEPC water and 95 μL Buffer Solid Tissue as indicated in the Kit. The samples were then macerated using the TissueLyser (QIAGEN), 5 cycles of 25 mHz for 30 s. The protocol was performed as instructed by the Kit. Quantification and quality of DNA was carried out with a Nanodrop system (ND; Fisher Scientific, Spain). 500 μL of each sample (24 in total; 6 per Kenney and Doig's category) were sent to an external lab for DNA bisulfite pyrosequencing methylation analysis.
Promoter annotations
CpG islands and CG percentage were predicted for COL1A1, MMP2, MMP9 and TIMP1 equine gene sequences with MethPrimer software (43). Within these, hotspot regions with the highest percentage of CpGs were identified and used in our analysis (Supplementary Table 2). Among the four genes (COL1A1, MMP2, MMP9, and TIMP1) studied, three were annotated in this study to possess CpG islands within the promoter region (COL1A1, MMP2 and TIMP1). Only MMP9 did not have CpG islands in the first 1,000 bp and therefore the studied region was exon 8, by homology with human studies, which have demonstrated regulation of MMP9 gene transcription in this region (44).
DNA bisulfite pyrosequencing analysis (CpG Islands)
The bisulfite modified DNA sample was then 10-fold diluted and 1 μL of diluted DNA was used in PCR reactions with 3 μL 10xPCR buffer, 200 μL/L of dNTPs, 6 pmol forward primer, 6 pmol reverse primer, and 3 mmol/L MgCl2, 0.75 U Qiagen HotStar Taq polymerase (Qiagen Inc., Valencia, CA., 205203) in 30 μL total volume adjusted using double distilled H20, as necessary. The PCR cycling condition was as follows: 95 °C 15 min; 45 x (95 °C 30 s; 51 °C 30 s: 72 °C 30 s); 72°C 10 min; 4 °C∞. The PSQ96HS system was used according to standard procedures for the Pyrosequencing TM analysis. DNA methylation pattern of COL1A1, MMP2, MMP9, and TIMP1 was analyzed by bisulfite pyrosequencing in an external lab (IMIBA, Malaga, Spain). Quantitative sodium bisulfite pyrosequencing was performed, as previously described (45), for the genes that showed differences in the transcriptomic analysis between endometrial categories, and comprised: COL1A, MMP2, MMP9 and TIMP1. In brief, targeted assays were designed using the PyroMark Assay Design Software 1.0 (Qiagen). Forward, reverse, and sequencing primers were used for PCR and pyrosequencing (Supplementary Table 2). The % of methylation was calculated as a mean of the CpG sites that passed quality control. Samples were considered for the study where at least 80% of the CpG sites passed quality control.
In silico analysis
The in-silico analysis was performed in the genes where methylation was correlated to transcription: MMP2 and MMP9 genes. CpG islands sequences were analyzed using two different programs: TRANSFAC® database (46) and Alibaba 2.1 (47) in search for possible binding transcription factors of the regulatory region of the genes (Figure 6).
Statistical analysis
Normal distribution of the data was evaluated by Shapiro-Wilk test. Kruskal-Wallis analysis followed by Dunn's multiple comparison test were performed to compare COL1A1 and MMP9 transcripts and COL1A1, TIMP1, and TIMP2 methylation between endometrial categories. One-way analysis of variance (ANOVA) followed by post-hoc Tukey multiple comparison test were used to analyse COL1A2, COL3A1 and TIMP2 mRNA and MMP9 methylation, between endometrial categories. Unpaired t-test was used to compare MMP2 and TIMP1 mRNA and MMP2 methylation, between endometrial categories. Pearson correlation test was performed to analyse transcription and methylation of MMP2 and MMP9 genes. GraphPAD PRISM (Version 8.1.0, 253, San Diego, CA, USA) was used. Significance was considered when P < 0.05. Data are presented as mean ± SEM.
Results
MMP2 and MMP9 expression is downregulated during fibrosis progression
Firstly, we interrogated if expression levels of the COL genes and some of their regulators (MMPs and TIMPs) were altered in the different endometrial categories. Although no statistical differences in mRNA levels were observed in COL1A1, COL1A2 and COL3A1 between different stages of endometrosis (Figure 1), a striking decrease in mRNA levels was observed for the MMP2 and MMP9 metallopeptidase genes (P < 0.05) between endometrial categories (Figure 2). As for the metallopeptidase tissue inhibitors studied, TIMP1 transcripts were higher in category III when compared to category I endometrium (P < 0.05) whereas no differences were found for TIMP2 transcripts (Figure 3). Mares were then grouped by phase of estrous cycle for each category and no differences were found between luteal and follicular phase (data not shown).
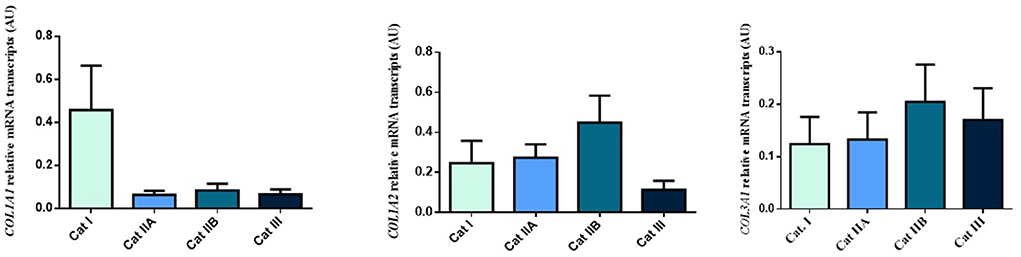
Figure 1. Relative COL1A1, COL1A2 and COL3A1 mRNA transcripts in equine endometrium graded as Kenney and Doig's categories I, IIA, IIB and III. Bars represent mean ± SEM.
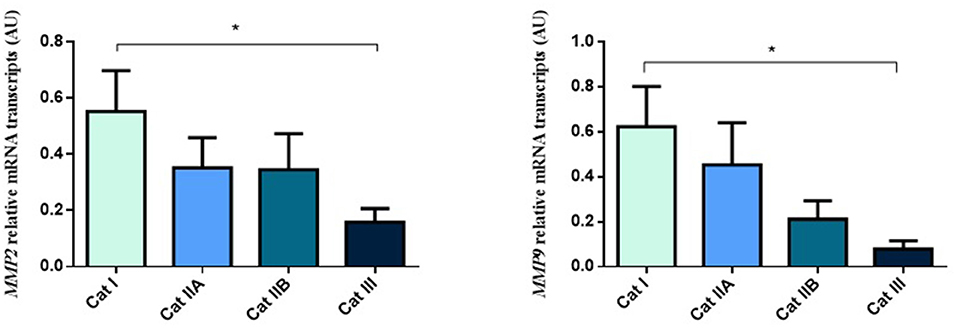
Figure 2. Relative MMP2 and MMP9 transcripts in equine endometrium graded as Kenney and Doig's categories I, IIA, IIB and III. Bars represent mean ± SEM. The asterisk indicates significant differences between endometrial categories (*P < 0.05).
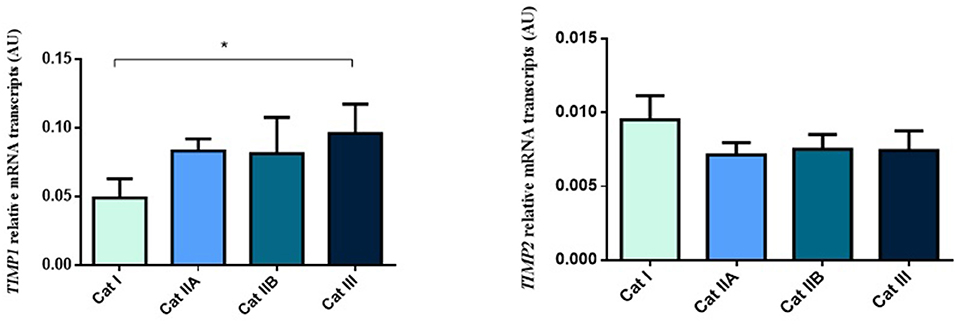
Figure 3. Relative TIMP1 and TIMP2 transcripts in equine endometrium graded as Kenney and Doig's categories I, IIA, IIB and III. Bars represent mean ± SEM. The asterisk indicates significant differences between endometrial categories (*P < 0.05).
DNA methylation plays a role in endometrial fibrosis regulation
Next, we questioned if the observed alterations in mRNA levels of the studied genes were associated with epigenetic mechanisms. As such, we performed DNA methylation analysis and observed higher methylation levels of COL1A1 in category IIB (P < 0.05) and III (P < 0.01), when compared to category I endometrium (Figure 4A). In addition, higher levels of methylation were observed for MMP2 in category III when compared to category I (P < 0.001) and IIA (P < 0.05) (Figure 4B) and for MMP9 gene in category III with respect to category I and IIA endometrium (P < 0.05) (Figure 4C). No difference in methylation levels between endometrium categories were observed for TIMP1 (data not shown). There were no methylation differences between luteal and follicular phase for each endometrial category.
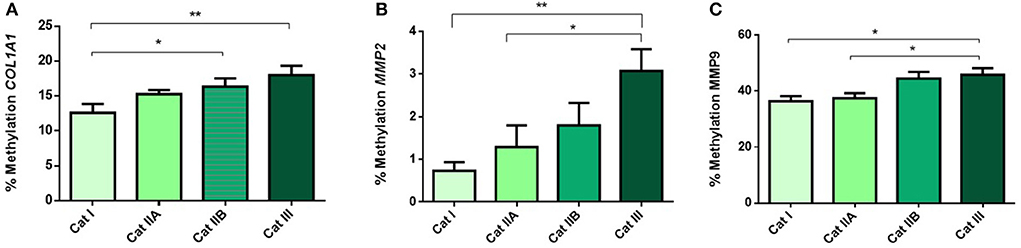
Figure 4. Methylation of DNA (%) of (A) COL1A1, (B) MMP2 and (C) MMP9 in equine endometrium graded as Kenney and Doig's categories I, IIA, IIB and III. Bars represent mean±SEM. The asterisks indicate significant differences between endometrial categories (*P < 0.05, **P < 0.01).
Methylation of MMP2 and MMP9 is negatively correlated with its transcription
We than analyzed the correlation between the observed DNA methylation events and potential alterations in gene expression. The higher methylation levels observed in MMP2 and MMP9 were strongly correlated with the decreased transcription levels of both genes (r = −0.967, P < 0.05 and r = −0.956, P < 0.05, respectively) upon endometrial fibrosis progression (Figure 5).
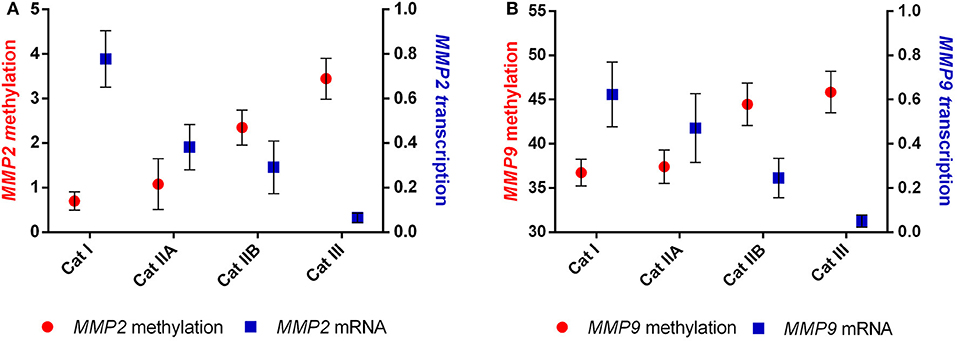
Figure 5. Correlation between methylation and transcription of (A) MMP2 and (B) MMP9 in equine endometrial categories graded as Kenney and Doig's classification.
Sp-1 and Ap-2α transcription factors identified as predicted binding factors in regulatory regions of MMP2 and MMP9
Several transcription factors from each software program were detected and the overlapping factors from the two programs were identified. The predicted transcription factors for MMP2 and MMP9 were Sp-1 and Ap-2α (Figure 6).
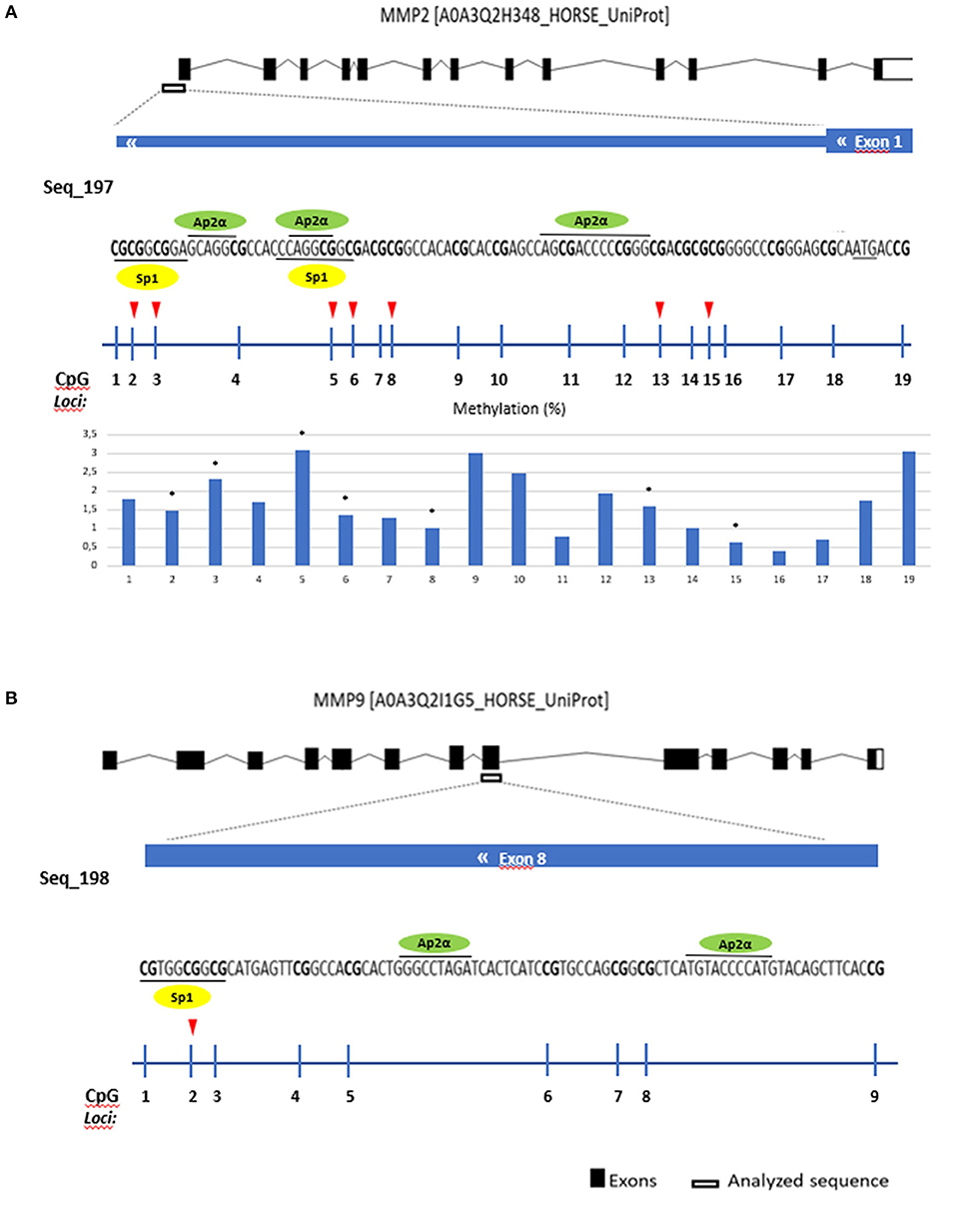
Figure 6. Gene analyzed sequence, binding transcription factors and methylation (%) in equine endometrial categories graded as Kenney and Doig's classification per CpG position in (A) MMP2 and (B) MMP9. Sp1 - proximal specificity protein 1, AP2α- Activator protein 2. Red arrows and asterisks indicate significant differences between CpG positions (*P < 0.05).
Then we analyzed which positions within the CpG island were accountable for the observed alterations. For COL1A1 the CpG positions that showed more alterations in methylation levels between endometrial categories were sites 2, 6, and 8 (Figure 7); for MMP2 site 2, 3, 5, 6, 8, 13, and 15 (Figure 6A); and MMP9 only site 2 (Figure 7B).
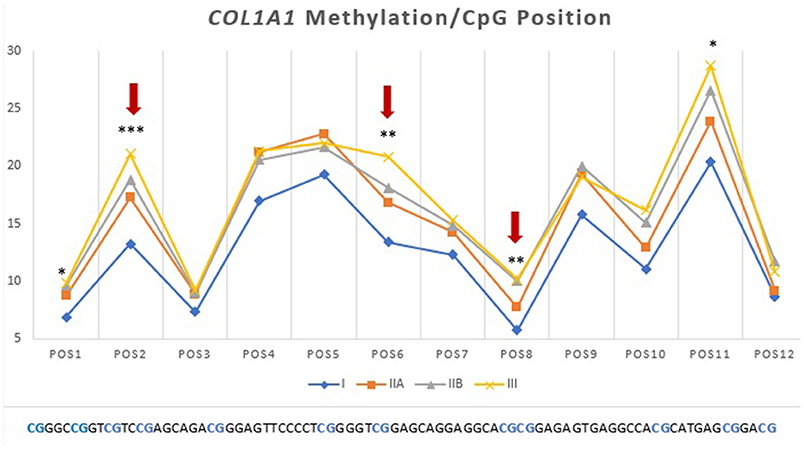
Figure 7. Methylation (%) of COL1A1 by CpG position in equine endometrial categories graded as Kenney and Doig's classification. Red arrows and asterisks indicate significant differences between CpG positions (*P < 0.05, **P < 0.01, ***P < 0.001).
Discussion
Novel findings implicate a role for epigenetic modifications contributing to the progression of fibrosis by alteration of gene expression profiles. Furthermore, accumulating evidence suggests that epigenetic alterations are central in maintaining the myofibroblast phenotype (20). Several studies demonstrated that the hypermethylation of gene promotors of antifibrotic mediators plays important roles in pathologic fibroblast activation and that inhibition of DNMTs prevents fibrosis in many fibrotic diseases (48–55). Previously, we reported an increase in DNMT3B mRNA levels with equine endometrial fibrosis (category III) when compared with healthy endometrium (category I), indicating there was hypermethylation with the advance stage of endometrosis (56). However, more detailed information is needed to understand which genes were involved and if hypermethylation was occurring in the gene promoter region and therefore could be regulating its transcription.
Our first proposed goal to evaluate transcriptomic of some of the genes involved in mare endometrial fibrosis, showed a decrease in MMP2 and MMP9 mRNA transcripts and an increase in TIMP1 mRNA transcripts with endometrial fibrosis. Although the COL1A1 mRNA transcripts did not show statistical differences we decided to include it in the DNA methylation study, along with the genes where an alteration was observed, to understand if they were epigenetically regulated.
The main constituents of fibrotic lesions are interstitial collagens, such as COL1 and COL3, and excessive deposition of these durable fibers can result in disruption of proper tissue structure and function (17). Activated fibroblasts are the central mediators in the pathogenesis of fibrosis and they differentiate into myofibroblasts, which are characterized by the increased secretion of collagen and other components of the extracellular matrix (57, 58). In the present study, no differences were found for COL1A1, COL1A2 or COL3A1 transcripts between different endometrial categories. In a study in equine endometrium (59) neither the estrous cycle phase, nor the age of mares had any effect on COL1 mRNA levels. Another study in jennies showed no differences in COL1A2 and COL3A1 transcripts between endometrial categories (60). However, some in vitro studies with mare cultured endometrial fibroblasts showed increased COL1A1 and COL3A1 mRNA transcripts after TGF-β1induced fibrosis (61) together with high COL1 and COL3 protein levels. Additionally, a study in equine tendon fibroblasts also demonstrated an increase in COL1A1 and COL1A2 gene expression after stimulation with TGF-β (62). This divergence could be due to differences in signaling and regulatory pathways in tissues examined ex vivo or cultured in vitro. Nevertheless, data from other study, with the same mares (unpublished data), have shown elevated COL1 and COL3 protein concentrations in endometrial tissue with fibrosis, regardless of unaltered COL1 and COL3 gene transcription. This suggests that accumulation of collagen in the endometrial tissue might be due to lack of degradation, rather than increased collagen production. Other explanation could be that the mRNA signal for COL1 and COL3 production might have occurred before the biopsy was performed. It is known that mRNAs are less stable than proteins with a maximum half-life of approximately 7 h, compared to 46 h for proteins (63). Thus, protein abundance mainly depends on a dynamic balance amongst transcription, mRNA processing and damage, translation, modification, and destruction of the resulting proteins (63). DNA methylation of the CpG islands in the promotor zone of the COL1A1 gene increased with endometrial fibrosis. However, hypermethylation of COL1A1 gene with fibrosis, did not result in its altered transcription. It is important to consider that collagen synthesis is precisely controlled at multiple levels, including via post-transcriptional and post-translational mechanisms that are still being discovered (64).
One class of molecules that is thought to be important in the maintenance of the ECM and processes of tissue repair is the class of matrix metalloproteinases (MMPs). The MMPs have been considered to play an important role in the extracellular matrix turnover (15) and a balance between activation and inhibition of MMPs is crucial for maintaining tissue homeostasis (14). Dysregulated expression of various MMPs is associated with many pathological processes, such as fibrosis, weakening of ECM or tissue destruction, e.g., in cancer metastasis (65, 66). However, data concerning MMP expression in equine endometrium during endometrial fibrosis is limited. In our study both MMP2 and MMP9 gene expression decreased with endometrial fibrosis, suggesting that the reduced transcription may result in diminished degradation of collagen and its accumulation in the endometrium. Other studies also demonstrated lower MMP2 gene transcription in fibrotic endometrium but MMP9 transcription was higher (67). In another study with endometrial fibroblasts in mares (14), MMP9 gene transcription increased after TGF-β1 stimulation. Also, a study by Centeno and collaborators (12) showed that MMP2 transcription was upregulated in endometrial fibrosis. On the other hand, a study in mice with induced liver fibrosis reported increased MMP2 mRNA and decreased MMP9 mRNA (68). Many studies in other animals and humans have shown a decrease in MMP2 (69, 70) and MMP9 gene expression in several fibrotic diseases (71) while others reported the opposite (72–74). These inconsistent results regarding MMP2 and MMP9 expression might be partially explained by the fact that MMPs demonstrate tissue-dependent and disease-specific expression and function (75). Moreover, current knowledge about MMP regulation is largely based on cell culture systems, raising a major question as to whether identical mechanisms apply to MMP expression in the whole organism as well (76).
Our results showed increased methylation in MMP2 and MMP9 in category III when compared to category I and IIA. Our study agrees with many others in that hypermethylation of antifibrotic gene promotors is involved in fibrosis development (48–54). Furthermore, we observed that hypermethylation of MMP2 and MMP9 genes occurred concomitantly with a decrease in their transcription levels as fibrosis increased, showing an epigenetic regulation as suggested in Figure 8. Other studies also observed that silencing of MMP genes is likely mediated by epigenetic alterations (68).
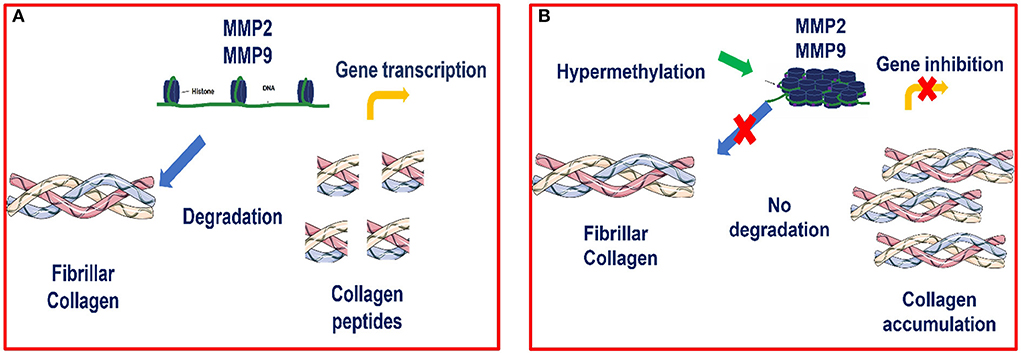
Figure 8. (A) Transcription of MMP2 and MMP9 genes is possible when there is no hypermethylation of these genes, as chromatin is accessible and thus allows transcriptional factors to bind to the promoter region and activate the transcription process (B) Epigenetic modulation of MMP2 and MMP9 in equine endometrial fibrosis. When hypermethylation occurs, the chromatin becomes condensed and the transcriptional factors are not able to bind to this site and therefore, transcription is inhibited.
Tissue inhibitors of metalloproteinases (TIMPs) are the major endogenous regulators of MMP activities in the tissue microenvironment and have the capacity to modify cellular activities and to modulate matrix turnover (77). TIMP-1 and TIMP-2 proteins bind to and inhibit activated collagenases, subsequently protecting newly synthesized collagen from immediate degradation by MMPs (18). In our study TIMP1 mRNA transcripts raised with fibrosis when compared to healthy endometrium whereas no differences were observed for TIMP2 mRNA. Our data agrees with a study in human lung fibrosis, where TIMP1 mRNA was markedly increased in response to lung injury, whereas there was no change in TIMP2 mRNA levels (77). Also, studies by Heymans in human cardiac disease (78) and Wang in rat lungs (79) reported upregulated TIMP1 gene expression during fibrosis. Since in our study, there were no alterations in DNA methylation pattern between endometrial categories for the TIMP1 gene, this might suggest that its inhibition does not occur, continuing to be expressed and increased with fibrosis, thus contributing to collagen accumulation in mare endometrium.
Some of the studied CpG island positions showed more differences in methylation between categories than others. Transcriptional factors (TF) play an important role in gene transcription as they can regulate transcription by binding to the activator or promoter regions of DNA and control gene expression through various mechanisms (80). The in silico analysis performed (81, 82) in the analyzed regions of the MMP2 and MMP9 genes, detected Sp-1 and Ap-2α as possible binding transcription factors for MMP2 and MMP9. These putative binding regions encompass CpG sites that showed significant methylation differences across the four studied endometrial categories. Specificity protein 1 (Sp-1) binding sites are often located close to binding sites for activator proteins (Ap-1/Ap-2) (83), as it was observed in our analysis. Transcription factor Ap-2, an important TF for the expression of many genes (84) has a role in the transcriptional regulation of MMP2 in humans (85, 86). In this study, the binding region for Sp-1 in MMP9 comprised loci 2, where the differences in methylation between endometrial categories occurred. Therefore, we speculate that these factors may play a role in the complex mechanism of endometrial fibrosis regulation and should be further investigated.
Overall, it seems like a different approach might be needed to address fibrosis treatment, as up to date, no effective therapy exists for equine endometrosis. “Epi-drugs” that target active myofibroblasts in fibrotic disorders are a promising direction in the treatment of a myriad of diseases. Nevertheless, we are far from a comprehensive understanding of how epigenetic modulators influence each other and myofibroblast behavior (23). If epigenetic mechanisms are involved in mare endometrial fibrosis development, as suggested by our results, then therapeutic agents that can reverse these epigenetic changes may represent a new and promising approach, for a condition that still has no available treatment.
Conclusion
In this study we have showed that equine endometrial fibrosis seems to be epigenetically modulated. Furthermore, that modulation seems to occur through the inhibition of antifibrotic genes like MMP2 and MMP9, rather than fibrotic genes (COL1 and TIMP1) and therefore might be promising targets for therapeutic use. Nevertheless, further studies are required to understand in depth this mechanism and possible role of other genes involved in mare endometrosis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal study was reviewed and approved by Ethic Committee for Research and Teaching Comissão de Ética para a Investigação e Ensino-CEIE (Ethic Committee No: 013/2022), Faculty of Veterinary Medicine, University of Lisbon, Lisbon, Portugal. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
Conceptualization: JA-M, GF-D, MR, and DS. Methodology: JA-M, CF, and MB. Software and writing—original draft preparation: JA-M. Validation: GF-D, JP, PC-B, and DS. Formal analysis: JA-M and CF. Investigation: JA-M, JP, and MR. Resources: JP and MB. Data curation: JA-M, JP, and MR. Writing - review and editing: JA-M, JP, PC-B, MR, AS, and GF-D. Visualization and funding acquisition: GF-D and DS. Supervision: GF-D and MR. Project administration: GF-D. All authors contributed to the article and approved the submitted version.
Funding
UIDB/00276/2020; PTDC/CVT-REP/4202/2014; LA/0059/2020/ AL4AnimalS funded by Fundação para a Ciência e Tecnologia (FCT; Portugal), OPUS19 nr 2020/37/B/NZ9/03355 funded by the National Science Center, (NSC; Poland), and bilateral Polish-Portugal project under NAWA and FCT agreement (2019–2020).
Acknowledgments
The authors are grateful to Fundação Alter-Real and Eng. Francisco Beja for the administrative and organization support of sample collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.970003/full#supplementary-material
References
1. Buczkowska J, Kozdrowski R, Nowak M, Raś A, Mrowiec J. Endometrosis-significance for horse reproduction, pathogenesis, diagnosis, and proposed therapeutic methods. Pol J Vet Sci. (2014) 17:547–54. doi: 10.2478/pjvs-2014-0083
2. Schöniger S, Schoon HA. The healthy and diseased equine endometrium: a review of morphological features and molecular analyses. Animals. (2020) 10:625. doi: 10.3390/ani10040625
3. Kenney RM. Cyclic and pathologic changes of the mare endometrium as detected by biopsy, with a note on early embryonic death. J Am Vet Med Assoc. (1978) 172:241–62.
4. Gray CA, Bartol FF, Tarleton BJ, Wiley AA, Johnson GA, Bazer FW, et al. Developmental biology of uterine glands. Biol Reprod. (2001) 65:1311–23. doi: 10.1095/biolreprod65.5.1311
5. Walter I, Handler J, Miller I, Aurich C. Matrix metalloproteinase 2 (MMP-2) and tissue transglutaminase (TG 2) are expressed in periglandular fibrosis in horse mares with endometrosis. Histol Histopathol. (2005) 20:1105–13. doi: 10.14670/HH-20.1105
6. Bochsler PN, Slauson DO. “Inflammation and repair of tissue”. In: Slauson DO, Cooper BJ, editors. Mechanisms of Disease: A Textbook of Comparative General Pathology, 3rd Edition. St. Louis, MO: Mosby (2002). p. 140–245.
7. Masseno APB. Caracterização histoquímica do colágeno e expressão de MMP2, MMP9 e TIMP-1 nas endometrites crónicas das éguas (Dissertação mestrado) – Faculdade de Medicina Veterinária e Zootecnia, Universidade Estadual Paulista, Botucatu, Brazil (2009).
8. Costa LD. Histochemical and immunohistochemical characterization of fibrotic changes in mares endometrosis (Master's Dissertation). Faculty of Veterinary Medicine and Animal Science- University Estadual Paulista “Júlio de Mesquita Filho”, Botucatu, Brazil (2015).
9. Lunelli D, Cirio SM, Leite SC, Camargo CE, Kozicki LE. Collagen types in relation to expression of estradiol and progesterone receptors in equine endometrial fibrosis. Adv Biosci Biotechnol. (2013) 4:599–605. doi: 10.4236/abb.2013.44078
10. Ra HJ, Parks WC. Control of matrix metalloproteinase catalytic activity. Matrix Biol. (2007) 26:587–96. doi: 10.1016/j.matbio.2007.07.001
11. Aresu L, Benali S, Giannuzzi D, Mantovani R, Castagnaro M, Falomo ME. The role of inflammation and matrix metalloproteinases in equine endometriosis. J Vet Sci. (2012) 13:171–7. doi: 10.4142/jvs.2012.13.2.171
12. Centeno LAM, Bastos HBA, Bueno VLC, Trentin JM, Fiorenza MF, FialaRechsteiner S, et al. Gene expression of Mmp-1, Mmp-2 And TNF-α in the endometrium of mares with different degrees of fibrosis. J Equine Vet Sci. (2018) 66:143–4. doi: 10.1016/j.jevs.2018.05.182
13. Crociati M, Capomaccio S, Mandara MT, Stradaioli G, Sylla L, Monaci M, et al. Different expression of Defensin-B gene in the endometrium of mares of different age during the breeding season. BMC Vet Res. (2019) 15:465. doi: 10.1186/s12917-019-2215-z
14. Szóstek-Mioduchowska A, Słowińska M, Pacewicz J, Skarzynski DJ, Okuda K. Matrix metallopeptidase expression and modulation by transforming growth factor-β1 in equine endometrosis. Sci Rep. (2020) 10:1119. doi: 10.1038/s41598-020-58109-0
15. Vandooren J, Van den Steen PE, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit Rev Biochem Mol Biol. (2013) 48:222–72. doi: 10.3109/10409238.2013.770819
16. Djuric T, Zivkovic M. Overview of MMP biology and gene associations in human diseases. In: Travascio F, editor. The Role of Matrix Metalloproteinase in Human Body Pathologies. London: IntechOpen (2017). doi: 10.5772/intechopen.70265
17. Giannandrea M, Parks WC. Diverse functions of matrix metalloproteinases during fibrosis. Dis Model Mech. (2014) 7:193–203. doi: 10.1242/dmm.012062
18. Hemmann S, Graf J, Roderfeld M, Roeb E. Expression of MMPs and TIMPs in liver fibrosis - a systematic review with special emphasis on anti-fibrotic strategies. J Hepatol. (2007) 46:955–75. doi: 10.1016/j.jhep.2007.02.003
19. Neary R, Watson C, Baugh JA. Epigenetics and the overhealing wound: the role of DNA methylation in fibrosis. Fibrogenesis Tissue Repair. (2015) 8:18. doi: 10.1186/s13069-015-0035-8
20. Bergmann C, Distler JH. Epigenetic factors as drivers of fibrosis in systemic sclerosis. Epigenomics. (2017) 9:463–77. doi: 10.2217/epi-2016-0150
21. Avci E, Sarvari P, Savai R, Seeger W, Pullamsetti SS. Epigenetic mechanisms in parenchymal lung diseases: bystanders or therapeutic targets? Int J Mol Sci. (2022) 23:546. doi: 10.3390/ijms23010546
22. Yang D, Xu P, Su H, Zhong W, Xu J, Su Z, et al. The histone methyltransferase DOT1L is a new epigenetic regulator of pulmonary fibrosis. Cell Death Dis. (2022) 13:60. doi: 10.1038/s41419-021-04365-5
23. Duong TE. Hagood JS. Epigenetic regulation of myofibroblast phenotypes in fibrosis. Curr Pathobiol Rep. (2018) 6:79–96. doi: 10.1007/s40139-018-0155-0
24. Simmons D. Epigenetic influence and disease. Nat Educ. (2008) 1:6. Available online at: http://www.nature.com/scitable/topicpage/epigenetic-influences-anddisease-895
25. Das PM. Singal, R. DNA methylation and cancer. J Clin Oncol. (2004) 22:4632–42. doi: 10.1200/JCO.2004.07.151
26. Jenuwein T, Allis CD. Translating the histone code. Science. (2001) 293:1074–80. doi: 10.1126/science.1063127
27. Newell-Price J, Clark AJ, King P. DNA methylation and silencing of gene expression. Trends Endocrinol Metab. (2000) 11:142–8. doi: 10.1016/S1043-2760(00)00248-4
28. Curradi M, Izzo A, Badaracco G, Landsberger N. Molecular mechanisms of gene silencing mediated by DNA methylation. Mol Cell Biol. (2002) 22:3157–73. doi: 10.1128/MCB.22.9.3157-3173.2002
29. Ehrlich M, Lacey M. DNA methylation and differentiation: silencing, upregulation and modulation of gene expression. Epigenomics. (2013) 5:553–68. doi: 10.2217/epi.13.43
30. Robertson KD. DNA methylation and chromatin - unraveling the tangled web. Oncogene. (2002) 21:5361–79. doi: 10.1038/sj.onc.1205609
31. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. (2012) 13:484–92. doi: 10.1038/nrg3230
32. Klein K, Gay S. Epigenetics in rheumatoid arthritis. Curr Opin Rheumatol. (2015) 27:76–82. doi: 10.1097/BOR.0000000000000128
33. Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev. (2005) 15:490–5. doi: 10.1016/j.gde.2005.08.002
34. Herman JG, Baylin SB. Gene silencing in cancer in association with promoter hypermethylation. N Engl J Med. (2003) 349:2042–54. doi: 10.1056/NEJMra023075
35. Weber M, Hellmann I, Stadler MB, Ramos L, Paabo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. (2007) 39:457–66. doi: 10.1038/ng1990
36. Singer M, Kosti I, Pachter L, Mandel-Gutfreund Y. A diverse epigenetic landscape at human exons with implication for expression. Nucleic Acids Res. (2015) 43:3498–508. doi: 10.1093/nar/gkv153
37. Kulis M, Queirós AC, Beekman R, Martín-Subero JI. Intragenic DNA methylation in transcriptional regulation, normal differentiation and cancer. Biochim Biophys Acta. (2013) 1829:1161–74. doi: 10.1016/j.bbagrm.2013.08.001
38. Kenney RM, Doig PA. Equine endometrial biopsy. In: Morrow D.A, editor. Current Therapy in Theriogenology. Philadelphia: W.B. Saunders (1986). p. 723–9.
39. Rebordão MR, Amaral A, Lukasik K, Szóstek-Mioduchowska A, Pinto-Bravo P, Galvão A, et al. Constituents of neutrophil extracellular traps induce in vitro collagen formation in mareendometrium. Theriogenology. (2018) 113:8–18. doi: 10.1016/j.theriogenology.2018.02.001
40. Untergasser A, Cutcutache I, Koressaar T, Ye J, Faircloth BC, Remm M, et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. (2012) 40:e115. doi: 10.1093/nar/gks596
41. Dheda K, Huggett JF, Bustin SA, Johnson MA, Rook G, Zumla A. Validation of housekeeping genes for normalizing RNA expression in real-time PCR. Biotechniques. (2004) 37:112–9. doi: 10.2144/04371RR03
42. Zhao S, Fernald RD. Comprehensive algorithm for quantitative real-time polymerase chain reaction. J Comput Biol. (2005) 12:1047–64. doi: 10.1089/cmb.2005.12.1047
43. Li LC, Dahiya R. MethPrimer: designing primers for methylation PCRs. Bioinformatics. (2002) 18:1427–31. doi: 10.1093/bioinformatics/18.11.1427
44. Falzone L, Salemi R, Travali S, Scalisi A, McCubrey JA, Candido S, et al. MMP-9 overexpression is associated with intragenic hypermethylation of MMP9 gene in melanoma. Aging. (2016) 8:933–44. doi: 10.18632/aging.100951
45. Castelo-Branco P, Choufani S, Mack S, Gallagher D, Zhang C, Lipman T, et al. Methylation of the TERT promoter and risk stratification of childhood brain tumours: an integrative genomic and molecular study. Lancet Oncol. (2013) 14:534–42. doi: 10.1016/S1470-2045(13)70110-4
46. Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. (2006) 34:D108-10. doi: 10.1093/nar/gkj143
47. Grabe N. AliBaba2: context specific identification of transcription factor binding sites. In Silico Biol. (2002) 2:S1–15.
48. Wang Y, Fan PS, Kahaleh B. Association between enhanced type I collagen expression and epigenetic repression of the FLI1 gene in scleroderma fibroblasts. Arthritis Rheum. (2006) 54:2271–9. doi: 10.1002/art.21948
49. Bechtel W, McGoohan S, Zeisberg EM, Müller GA, Kalbacher H, Salant DJ, et al. Methylation determines fibroblast activation and fibrogenesis in the kidney. Nat Med. (2010) 16:544–50. doi: 10.1038/nm.2135
50. Huang SK, Fisher AS, Scruggs AM, White ES, Hogaboam CM, Richardson BC, et al. Hypermethylation of PTGER2 confers prostaglandin E2 resistance in fibrotic fibroblasts from humans and mice. Am J Pathol. (2010) 177:2245–55. doi: 10.2353/ajpath.2010.100446
51. Dees C, Schlottmann I, Funke R, Distler A, Palumbo-Zerr K, Zerr P, et al. The Wnt antagonists DKK1 and SFRP1 are downregulated by promoter hypermethylation in systemic sclerosis. Ann Rheum Dis. (2014) 73:1232–9. doi: 10.1136/annrheumdis-2012-203194
52. Noda S, Asano Y, Nishimura S, Taniguchi T, Fujiu K, Manabe I, et al. Simultaneous downregulation of KLF5 and Fli1 is a key feature underlying systemic sclerosis. Nat Commun. (2014) 5:5797. doi: 10.1038/ncomms6797
53. Watson CJ, Collier P, Tea I, Neary R, Watson JA, Robinson C, et al. Hypoxia-induced epigenetic modifications are associated with cardiac tissue fibrosis and the development of a myofibroblast-like phenotype. Hum Mol Genet. (2014) 23:2176–88. doi: 10.1093/hmg/ddt614
54. Altorok N, Kahaleh B. Epigenetics and systemic sclerosis. Semin Immunopathol. (2015) 37:453–62. doi: 10.1007/s00281-015-0504-6
55. Dees C, Pötter S, Zhang Y, Bergmann C, Zhou X, Luber M, et al. TGF-β-induced epigenetic deregulation of SOCS3 facilitates STAT3 signaling to promote fibrosis. J Clin Invest. (2020) 130:2347–63. doi: 10.1172/JCI122462
56. Alpoim-Moreira J, Fernandes C, Rebordão MR, Amaral A, Pinto-Bravo P, Bliebernicht M, et al. Collagens and DNA methyltransferases in mare endometrosis. Reprod Domest Anim. (2019) 46:52–52. doi: 10.1111/rda.13515
57. Abraham DJ, Eckes B, Rajkumar V, Krieg T. New developments in fibroblast and myofibroblast biology: implications for fibrosis and scleroderma. Curr Rheumatol Rep. (2007) 9:136–43. doi: 10.1007/s11926-007-0008-z
58. Wynn TA, Ramalingam TR. Mechanisms of fibrosis: therapeutic translation for fibrotic disease. Nat Med. (2012) 18:1028–40. doi: 10.1038/nm.2807
59. Bravo PNAMP. Involvement of Hormones, Cytokines and Angiogenic Factors on Mare Oviduct Physiological Function and Fibrosis (Tese de Doutoramento). Universidade de Lisboa, Faculdade de Medicina Veterinária, Lisboa, Portugal (2018).
60. Miró J, Gutiérrez-Reinoso M, da Silva JA, Fernandes C, Rebordão MR, Alexandre-Pires G, et al. Collagen and eosinophils in jenny's endometrium: do they differ with endometrial classification? Front Vet Sci. (2020) 7:631. doi: 10.3389/fvets.2020.00631
61. Szóstek-Mioduchowska AZ, Lukasik K, Skarzynski DJ, Okuda K. Effect of transforming growth factor -β1 on α-smooth muscle actin and collagen expression in equine endometrial fibroblasts. Theriogenology. (2019) 124:9–17. doi: 10.1016/j.theriogenology.2018.10.005
62. Gumbs JA. TGF-β Mediated Synthesis of Degradation-Resistant Collagen (I) Homotrimer in Musculoskeletal Cells (PhD thesis). University of Liverpool, Liverpool, United Kingdom (2019).
63. Vogel C, Marcotte EM. Insights into the regulation of protein abundance from proteomic and transcriptomic analyses. Nat Rev Genet. (2012) 13:227–32. doi: 10.1038/nrg3185
64. Roche P., Czubryt MP. Transcriptional control of collagen I gene expression. Cardiovasc Hematol Disord Drug Targets. (2014) 14:107–20. doi: 10.2174/1871529X14666140505122510
65. Di Nezza LA, Misajon A, Zhang J, Jobling T, Quinn MA, Ostör AG, et al. Presence of active gelatinases in endometrial carcinoma and correlation of matrix metalloproteinase expression with increasing tumor grade and invasion. Cancer. (2002) 94:1466–75. doi: 10.1002/cncr.10355
66. Amălinei C, Căruntu ID, Giuşcă SE, Bălan RA. Matrix metalloproteinases involvement in pathologic conditions. Rom J Morphol Embryol. (2010) 51:215–28.
67. Falomo ME, Ferroni L, Tocco I, Gardin C, Zavan B. Immunomodulatory role of adipose-derived stem cells on equine endometriosis. Biomed Res Int. (2015) 2015:141485. doi: 10.1155/2015/141485
68. Qin L, Han YP. Epigenetic repression of matrix metalloproteinases in myofibroblastic hepatic stellate cells through histone deacetylases 4: implication in tissue fibrosis. Am J Pathol. (2010) 177:1915–28. doi: 10.2353/ajpath.2010.100011
69. Sugihara T, Koda M, Matono T, Maeda K, Yamamoto S, Ueki M, et al. Extracellular matrix metabolism-related gene expression in bile duct-ligated rats. Mol Med Rep. (2009) 2:345–51. doi: 10.3892/mmr_00000106
70. Kendziorski JA, Belcher SM. Strain-specific induction of endometrial periglandular fibrosis in mice exposed during adulthood to the endocrine disrupting chemical bisphenol A. Reprod Toxicol. (2015) 58:119–30. doi: 10.1016/j.reprotox.2015.08.001
71. Bailey JR, Bland PW, Tarlton JF, Peters I, Moorghen M, Sylvester PA, et al. IL-13 promotes collagen accumulation in Crohn's disease fibrosis by down-regulation of fibroblast MMP synthesis: a role for innate lymphoid cells? PLoS ONE. (2012) 7:e52332. doi: 10.1371/journal.pone.0052332
72. Kim WU, Min SY, Cho ML, Hong KH, Shin YJ, Cho CS. Elevated matrix metalloproteinase-9 in patients with systemic sclerosis. Arthritis Res Ther. (2005) 7:R71–9. doi: 10.1186/ar1454
73. Andersen GN, Nilsson K, Pourazar J, Hackett TL, Kazzam E, Blomberg A, et al. Bronchoalveolar matrix metalloproteinase 9 relates to restrictive lung function impairment in systemic sclerosis. Respir Med. (2007) 101:2199–206. doi: 10.1016/j.rmed.2007.04.019
74. Dancer RC, Wood AM, Thickett DR. Metalloproteinases in idiopathic pulmonary fibrosis. Eur Respir J. (2011) 38:1461–7. doi: 10.1183/09031936.00024711
75. Leong E, Bezuhly M. Marshall JS. Distinct metalloproteinase expression and functions in systemic sclerosis and fibrosis: what we know and the potential for intervention. Front Physiol. (2021) 12:727451. doi: 10.3389/fphys.2021.727451
76. Yan C, Boyd DD. Regulation of matrix metalloproteinase gene expression. J Cell Physiol. (2007) 211:19–26. doi: 10.1002/jcp.20948
77. Madtes DK, Elston AL, Kaback LA, Clark JG. Selective induction of tissue inhibitor of metalloproteinase-1 in bleomycin-induced pulmonary fibrosis. Am J Respir Cell Mol Biol. (2001) 24:599–607. doi: 10.1165/ajrcmb.24.5.4192
78. Heymans S, Schroen B, Vermeersch P, Milting H, Gao F, Kassner A, et al. Increased cardiac expression of tissue inhibitor of metalloproteinase-1 and tissue inhibitor of metalloproteinase-2 is related to cardiac fibrosis and dysfunction in the chronic pressure-overloaded human heart. Circulation. (2005) 112:1136–44. doi: 10.1161/CIRCULATIONAHA.104.516963
79. Wang BL, Tu YY, Fu JF, Zhong YX, Fu GQ, Tian XX, et al. Unbalanced MMP/TIMP-1 expression during the development of experimental pulmonary fibrosis with acute paraquat poisoning. Mol Med Rep. (2011) 4:243–8. doi: 10.3892/mmr.2011.425
80. Gill G. Regulation of the initiation of eukaryotic transcription. Essays Biochem. (2001) 37:33–43. doi: 10.1042/bse0370033
81. Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM, et al. Detection of known transcription regulatory elements using species-tailored searches. Bioinformatics. (2002) 18:333–4. doi: 10.1093/bioinformatics/18.2.333
82. Farré D, Roset R, Huerta M, Adsuara JE, Roselló L, Albà MM, et al. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res. (2003) 31:3651–3. doi: 10.1093/nar/gkg605
83. Sato T, Furukawa K. Transcriptional regulation of the human beta-1, 4-galactosyltransferase V gene in cancer cells: essential role of transcription factor Sp1. J Biol Chem. (2004) 279:39574–83. doi: 10.1074/jbc.M405805200
84. Wang G, Zeng Y, Chen S, Li D, Li W, Zhou Y, et al. Localization of TFPI-2 in the nucleus modulates MMP-2 gene expression in breast cancer cells. Sci Rep. (2017) 7:13575. doi: 10.1038/s41598-017-14148-8
85. Bergman MR, Cheng S, Honbo N, Piacentini L, Karliner JS, Lovett DH. A functional activating protein 1 (AP-1) site regulates matrix metalloproteinase 2 (MMP-2) transcription by cardiac cells through interactions with JunB-Fra1 and JunB-FosB heterodimers. Biochem J. (2003) 369:485–96. doi: 10.1042/bj20020707
Keywords: endometrial fibrosis, mare, collagen, epigenetics, DNMT, MMP, endometrosis
Citation: Alpoim-Moreira J, Fernandes C, Pimenta J, Bliebernicht M, Rebordão MR, Castelo-Branco P, Szóstek-Mioduchowska A, Skarzynski DJ and Ferreira-Dias G (2022) Metallopeptidades 2 and 9 genes epigenetically modulate equine endometrial fibrosis. Front. Vet. Sci. 9:970003. doi: 10.3389/fvets.2022.970003
Received: 15 June 2022; Accepted: 22 July 2022;
Published: 12 August 2022.
Edited by:
Emilio A. Martinez, University of Murcia, SpainReviewed by:
Manuel Alvarez Rodriguez, Instituto Nacional de Investigación y Tecnología Agroalimentaria (INIA), SpainGuillermo Ramis Vidal, University of Murcia, Spain
Copyright © 2022 Alpoim-Moreira, Fernandes, Pimenta, Bliebernicht, Rebordão, Castelo-Branco, Szóstek-Mioduchowska, Skarzynski and Ferreira-Dias. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Graça Ferreira-Dias, gmlfdias@fmv.ulisboa.pt
 Joana Alpoim-Moreira
Joana Alpoim-Moreira Carina Fernandes
Carina Fernandes Jorge Pimenta
Jorge Pimenta Miguel Bliebernicht4
Miguel Bliebernicht4  Maria Rosa Rebordão
Maria Rosa Rebordão Anna Szóstek-Mioduchowska
Anna Szóstek-Mioduchowska Dariusz J. Skarzynski
Dariusz J. Skarzynski Graça Ferreira-Dias
Graça Ferreira-Dias