Molecular Characterization Analysis of Prevalent Infectious Bronchitis Virus and Pathogenicity Assessment of Recombination Strain in China
- 1Heyuan Branch, Guangdong Provincial Laboratory of Lingnan Modern Agricultural Science and Technology, Guangdong Provincial Key Lab of Agro-Animal Genomics and Molecular Breeding, Key Laboratory of Chicken Genetics, Breeding and Reproduction, Ministry of Agriculture, College of Animal Science, South China Agricultural University, Guangzhou, China
- 2Wen's Group Academy, Wen's Foodstuffs Group Co., Ltd., Xinxing, China
- 3Guangdong Engineering Research Center for Vector Vaccine of Animal Virus, Guangzhou, China
- 4South China Collaborative Innovation Center for Poultry Disease Control and Product Safety, Guangzhou, China
- 5Key Laboratory of Animal Health Aquaculture and Environmental Control, College of Animal Science, South China Agricultural University, Guangzhou, China
Avian coronavirus infectious bronchitis virus (IBV) is a respiratory pathogen of chickens, resulting in severe economic losses in the poultry industry. This study aimed to monitor and isolate the molecular identity of IBV in broiler flocks with respiratory symptoms in eight provinces of China. In total, 910 samples (oropharyngeal and cloacal mixed swabs) from broiler flocks showed IBV positive rates of 17.6% (160/910) using PCR assay. Phylogenetic analysis of the complete S1 genes of 160 IBV isolates was performed and revealed that QX-type (GI-19), TW-type (GI-7), 4/91-type (GI-13), HN08-type (GI-22),TC07-2-type (GVI-1), and LDT3-type (GI-28) exhibited IBV positive rates of 58.15, 25, 8.12, 1.86, 5.62, and 1.25%. In addition, recombination analyses revealed that the four newly IBV isolates presented different recombination patterns. The CK/CH/JS/YC10-3 isolate likely originated from recombination events between strain YX10 (QX-type) and strain TW2575-98 (TW-type), the pathogenicity of which was assessed, comparing it with strain GZ14 (TW-type) and strain CK/CH/GD/JR07-7 (QX-type). The complete S1 gene data from these isolates indicate that IBV has consistently evolved through genetic recombination or mutation, more likely changing the viral pathogenicity and leading to larger outbreaks in chick populations, in China.
Introduction
Infectious bronchitis (IB) is a highly contagious and widespread viral disease of economic significance, affecting severely the global poultry industry (1). Infectious bronchitis virus (IBV), a member of the γ-Coronavirus genus in the Coronaviridae family, is one of the etiologic agents of upper respiratory disease in chickens of all ages and varieties, as well as it causes urogenital illness, which is characterized by gout or nephritis (2, 3). IBV is prone to genetic evolution through the gradual accumulation of mutations and drastic recombination events. Currently, lots of new IBV variants have been continuously emerging, while vaccines have been created and inoculated worldwide. In China, based on S1 gene phylogenetic analysis, IBV is mainly divided into sever genotypes, including QX-type (GI-19), TW-type (GI-7), 4/91-type (GI-13), Mass-type (GI-1), HN08-type (GI-22), TC07-2-type (GVI-1), and LDT3-type (GI-28) (4–6). QX-type strains, with changes in geographical distribution and tissue tropism (7), have been epidemic and predominant in poultry flocks since their initial emergence in 1996 (8, 9). Notably, TW-type and 4/91-type have been identified more frequently in recent years (10). The following are susceptible factors for the continuous emergence of new IBVs: (a) in the process of viral replication, its genomes are prone to insertion, deletion, and substitution; (b) natural recombination occurs randomly in various viral RNAs; (c) live attenuated or inactivated commercial IBV vaccines show little or no cross-protection against different genotype strains and even carry the risk of recombination between vaccine strains and wild type strains.
In this brief report, we analyzed the molecular characterization of 160 IBV isolates from eight provinces in China in 2020–2021. It provides detailed evidence of the severe prevalence of IBV in China, and they also provide a reminder that more scientific vaccine strategies are needed, including strain selection and cross-protection assessment to provide broad protection against different IBVs.
Materials and Methods
Ethics Statement
The use of animals in this study was approved by the South China Agricultural University Committee for Animal Experiments [approval ID: SYXK (Guangdong) 2019-0136]. All study procedures and animal care activities were conducted following the national and institutional guidelines for the care and use of laboratory animals.
The Prevalence of IBV in Chick Flock
Between Sept 2020 and July 2021, several chick flocks, having developed upper respiratory symptoms, including coughing, wheezing, rales, and nasal discharge, were suspected of being infected with IBV. A total of 910 oropharyngeal and cloacal mixed swab samples were collected from the suspected chick flocks from eight provinces in mainland China (Fujian, Guangdong, Guangxi, Yunnan, Hunan, Hubei, Jiangsu, and Zhejiang). The swab specimens were stored in 500 μL of 1 mM phosphate-buffered saline (PBS) containing 200 U/mL of penicillin and 200 μg/mL of streptomycin, and vortexed for 20 min to prepare the virus solution for testing. The total RNA was extracted by using the RaPure Viral RNA/DNA Kit (Magen, Guangdong, China). The viral RNA was measured by quantitative reverse transcription PCR (qRT-PCR) according to the following protocol (10) using the HiScript® II One Step qRT-PCR Probe Kit (Vazyme, Nanjing, China), with the primer pairs designed to target the gene RNA nucleotide sequence (Accession: AF093793.1). The reaction conditions were as follows: 50°C for 5 min, 95°C for 30 s, and 45 cycles of 95°C for 5 s and 60°C for 34 s on the Applied Biosystems™ 7500 (Thermofisher, Beijing, China). The positive judgment value for this assay is between 15 and 35 cycle thresholds (CT) per 2 μL RNA template. The sequences of primers and probes used were as follows: IBV-F (5′-CCTCTAAGGGCTTTTGAG-3′), IBV-R (5′-GTCACTGTCTATTGTATGTC-3′), and IBV-P (5′-FAMCACCACCAGAACCTGTCACCTC BHQ1-3′).
IBV Isolation and S1 Gene Sequencing
Special pathogen free (SPF) embryonated eggs were purchased from Guangdong DHN Poultry and Egg Products Co. Ltd. (Yunfu, Guangdong, China). After the positive swab samples were vortexed shortly and centrifuged at 750 × g for 5 min at 4°C, the supernatants (0.2 mL) were collected and serially diluted 10-fold from 10−1 to 10−3 in PBS, which inoculate into 9-day-old SPF chicken embryonated eggs respectively with 0.2 mL by the allantoic cavity route. Infectious allantoic fluid was harvested from the embryos at 2–3 days post-inoculation and the viral RNA was determined as previously described. Next, specific primer pairs targeting the complete IBV S1 gene were designed for reverse transcription polymerase chain reaction (RT-PCR). The thermal cycling (Bio-Rad, UK) parameters were as follows: 94°C for 1 min, and 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 1.5 min. The complete S1 gene, ~1,640 bp, was amplified using the PrimeScript™ RT-PCR Kit (Takara, Japan), and its products were separated on agarose gel and purified using a QIAquick Gel Extraction Kit (Qiagen, USA). The purified PCR products were cloned into a pMD™19-T vector (Takara, Japan) and then sequenced by BGI Genomics Co., Ltd. The sequences of the primers used were as follows: S1-F (5′-AACTGAACAAAAGACCGACT-3′), S1-R (5′-CAAAACCTGCCATAACTAACA-3′) (11).
Complete S1 Gene Phylogenetic and Recombination Analysis
The sequencing was performed using DNAStar Lasergene software (DNAStar, Madison, WI, USA). Additional publicly available complete S1 gene sequences, obtained from GenBank, were aligned by the MEGA (version 7.0) ClustalW algorithm and the Test Neighbor-Joining Tree for designing the IBV S1 phylogenetic tree. A total of 1,000 bootstraps were calculated. Finally, the tree was visualized using Interactive Tree Of Life (iTOL) web-based tool (http://itol.embl.de/). All of the reference complete IBV S1 sequences representing all currently recognized types and subtypes used in the present study are listed in the Supplementary Material. Furthermore, the potential recombination events assessed by the Recombination Detection Program 4.0 (RDP 4.0, version 4.96) with strong P-values (<10–20), were further investigated by similarity plot and bootscan analyses implemented in Simplot 3.5.1.
IBV Isolates Pathogenicity Test
A total of 30 ten-day-old SPF chickens were purchased from Guangdong DHN Poultry and Egg Products Co. Ltd. (Yunfu, Guangdong, China) and housed in the isolators and randomly divided into four groups. The GZ14 strain (TW-type) and strain CK/CH/GD/JR07-7 (QX-type) was isolated and saved in the laboratory. Subsequently, we inoculated the chicks intranasally with 106 EID50 per 0.2 mL of GZ14 strains, CK/CH/GD/JR07-7 strains and CK/CH/JS/YC10-3 isolates respectively and used the same volume of sterile allantoic fluid for the control group. Postmortem was carried out 7 dpi, and the kidney and air sac lesions of the chickens were scored as previously described (12–14) (Tables 1, 2).
Statistical Analysis
The gross lesion scores of kidney and air arc were analyzed using the one-way ANOVA followed by Tukey's multiple comparison tests. A p-value of <0.05 was considered a statistically significant difference (*), p < 0.01 was considered a moderately significant difference (**), and p < 0.001 was considered a highly significant difference (***).
Results
The Prevalence of IBV
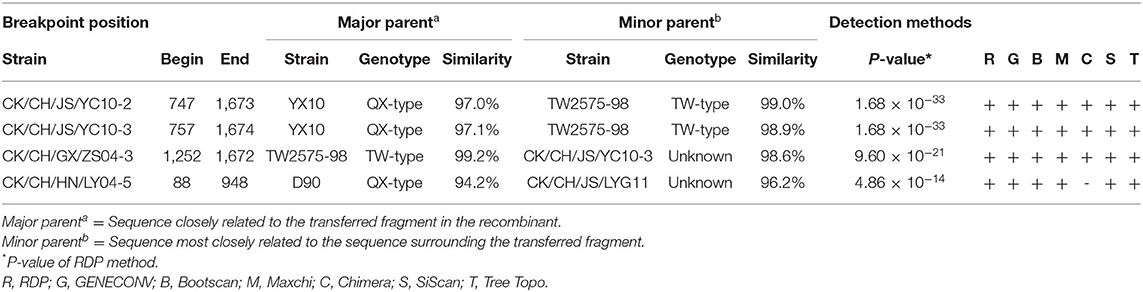
Previous investigations have suggested that IBV will induce respiratory system and kidney disease in chickens (15, 16). In the present study, we collected swabs from chick flocks with suspected respiratory virus infections from eight provinces and analyzed them using RT-qPCR. In the examined flocks showing respiratory symptoms, 17.6% (160 out of 910) were infected with IBV. The most abundant strains were detected in the province of Hebei (Figure 1A). A total of 160 IBV strains were isolated and comparative analysis of the S1 gene revealed that QX-type, TW-type, 4/91-type, HN08-type, TC07-2-type, and LDT3-type exhibited IBV positive rates of 58.15, 25, 8.12, 1.86, 5.62, and 1.25% (Table 3), respectively. Among them, the QX-type was isolated in 73.3% of chicks infected in Fujian, 78.9% in Guangxi, 92.5% in Guangdong, 50% in Yunnan, 58.6% in Hunan, 63.2% in Jiangsu, and 57.1% in Zhejiang. In addition, TW-type threatened 63.6% of infected chicks in the province of Hubei, and Mass-type was not discovered (Figure 1B).
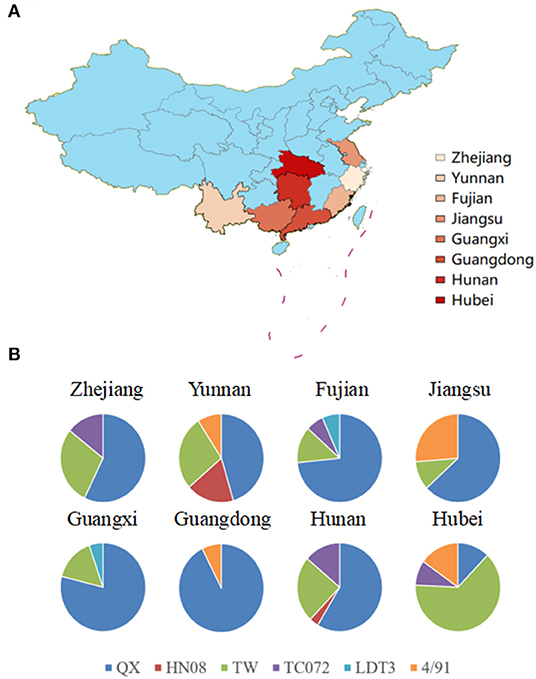
Figure 1. (A) Map of China with IBV strain prevalence in eight provinces in China where samples were collected. The map was created using ArcGis and the darker the color, the higher the level of positive rates of IBV specimens. (B) Different IBV strain prevalence rates in eight provinces. The graphs show the distribution of positive specimens for genotypes of IBV (QX in blue, HN08 in red, TW in green, TC072 in purple, LDT3 in purple, and 4/91 in orange).
The Genetic Evolution and Recombination of IBV
The nucleotide sequence of the complete S1 gene of isolates was aligned to relevant sequences that represent different IBV genotypes available in GenBank using MEGA 7.0. According to neighbor-joining tree phylogenetic analysis (Figure 2).
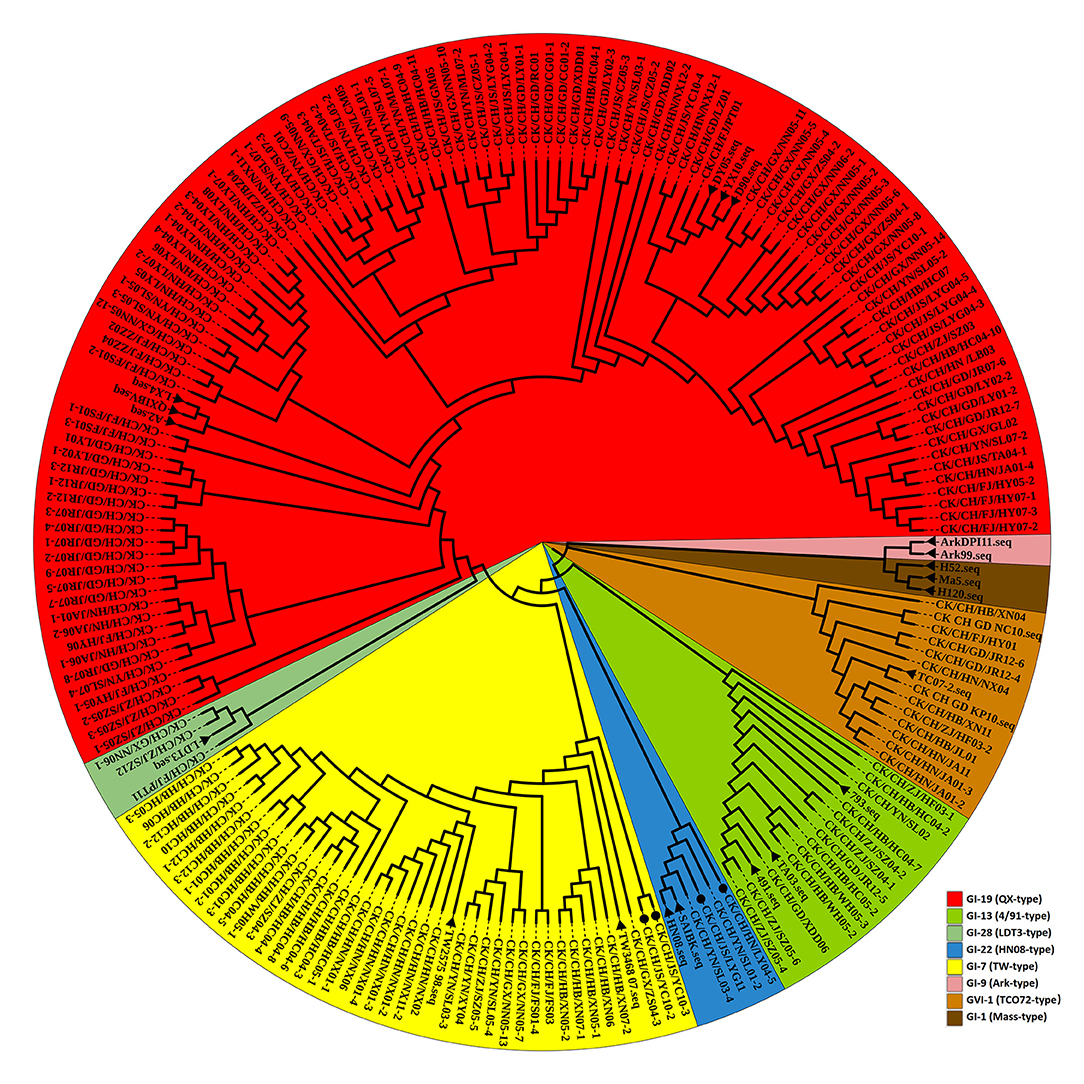
Figure 2. Phylogenetic analysis of the IBV S1 gene. Circle tree: IBV isolates from Sept 2020 to July 2021 in China can be divided into six clusters. The reference strains are marked by a black solid triangle (▴). Newly recombinant strains are indicated with black solid circles (•). Other strains without a sign were excluded from this study.
To reveal possible recombination events, the complete S1 genes of 20 reference strains were used as putative parent strains and recombination detection was conducted with RDP 4.0. Using at least five detection methods, the results suggest that four isolates' S1 genes are possibly recombinant strains, with p-values of ≤ 1 × 10−6 (Table 4). As shown in Figure 3, strain CK/CH/JS/YC10-2 appears to be recombinant with strain YX10 as the major parent and TW2575-98 as the minor parent, with recombination breakpoints mapping to positions 747 (beginning breakpoint) and 1673 (ending breakpoint). Strain CK/CH/JS/YC10-3 appears to be recombinant with strain YX10 as the major parent and TW2575-98 as the minor parent, with recombination breakpoints mapping to positions 757 (beginning breakpoint) and 1674 (ending breakpoint). Strain CK/CH/GX/ZS04-3 appears to be recombinant with strain TW2575-98 as the major parent and CK/CH/JS/YC10-3 as the minor parent, with recombination breakpoints mapping to positions 1252 (beginning breakpoint) and 1672 (ending breakpoint). Strain CK/CH/HN/LY04-5 appears to be recombinant with strain D90 as the major parent and CK/CH/JS/LYG11 as the minor parent, with recombination breakpoints mapping to positions 88 (beginning breakpoint) and 948 (ending breakpoint).
Figure 3. SimPlot analysis of recombination events of strains CK/CH/JS/YC10-2, CK/CH/JS/YC10-3, CK/CH/GX/ZS04-3, and CK/CH/HN/LY04-5. Strain CK/CH/JS/YC10-2 appears to be recombinant with strain YX10 as the major parent and TW2575-98 as the minor parent. Strain CK/CH/JS/YC10-3 appears to be recombinant with strain YX10 as the major parent and TW2575-98 as the minor parent. Strain CK/CH/GX/ZS04-3 appears to be recombinant with strain TW2575-98 as the major parent and CK/CH/JS/YC10-3 as the minor parent. Strain CK/CH/HN/LY04-5 appears to be recombinant with strain D90 as the major parent and CK/CH/JS/LYG11 as the minor parent.
IBV Isolate Pathogenicity Test
Based on the viral recombination results, the CK/CH/JS/YC10-3 isolate, which was formed by the major parent strain YX10 (QX-type) and the minor parent strain TW2575-98 (TW-type), was used for the exposure tests.
Postmortem, all of the birds exposed to strain GZ14 showed obvious lesions in the kidneys, in which they were mainly preformed nephritis, as well as different degrees of air sac turbidness. In contrast, the birds with strain CK/CH/GD/JR07-7 or the CK/CH/JS/YC10-3 isolate generally showed swollen kidneys and obviously thickened air sacs covered with yellow white caseous exudate. No clinical symptoms were observed in the chickens of the control group (Figure 4A). In addition, gross lesions in the air sacs and kidneys were scored and recorded (Figure 4B).
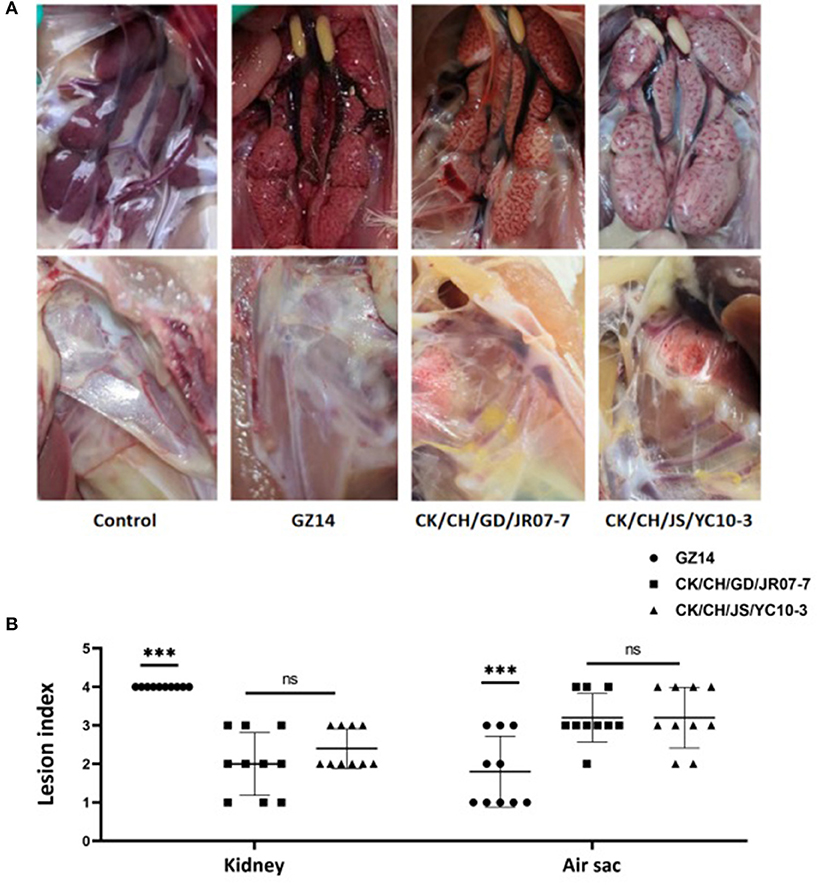
Figure 4. (A) Gross lesions from experimentally exposed chicks. Control group: No gross lesions in the kidneys and air sacs are clean, thin and transparent; GZ14: Nephritis, air sac turbidness; CK/CH/JS/YC10-3: Swollen kidney with urate, yellow white caseous exudate in air sacs. (B) Gross lesion scores of kidney and air sac by exposing with strain GZ14, CK/CH/GC/JR07-7, and CK/CH/JS/YC10-3. Each lesion's score independently reflects each chick's gross lesions. Black solid circles indicate the GZ14 group, black solid circles the CK/CH/GC/JR07-7 group, and black solid triangle indicate the CK/CH/JS/YC10-3 group. Using one-way ANOVA followed by Tukey's multiple comparison test, strain GZ14 and CK/CH/JS/YC10-3 exhibited a highly significant difference (***) in kidneys and air sacs with p < 0.001. Strain CK/CH/GC/JR07-7 and CK/CH/JS/YC10-3 exhibited non-significant (ns) with p > 0.05.
Discussion
Attention has been paid to poultry respiratory system diseases worldwide in recent years. High morbidity rate and reduced egg and meat production are caused by respiratory tract infections, which are of paramount importance in the poultry industry. The potential pathogens induce high mortality rates, growth retardation, and high cost of medications, all of which decrease net income (17, 18). The etiology of respiratory infections is an ongoing exploration, with the majority of the studies have focused on chronic infections and various pathogen co-infections, including infectious bronchitis virus (IBV), Newcastle disease virus (NDV), low pathogenic avian influenza virus (LPAIV) H9N2, Escherichia coli, Staphylococcus aureus, Mycoplasma gallisepticum (MG), and Mycoplasma synoviae (MS); these pathogens can cause respiratory diseases independently, or together (19).
In this research, we focused on the impact of IBV infection in chickens in China. Throughout eight provinces, the situation of IBV prevalence is still serious, even though the chickens have been immunized with commercial live attenuated or inactivated IBV vaccine. IBV strains were successfully detected and isolated from Oropharyngeal and cloacal swabs of chick flocks with target symptoms. Based on phylogenetic analysis, the current prevalent strains can be distributed into six clusters, revealing a high genetic variety and potential pathogenicity complexity among IBV strains emerging in China. Specifically, QX-type strains have played a predominant role and TW-type strains are increasingly emerging. In addition, recombination strains were also identified. Furthermore, pathogenic assessment of selected strains was also performed by challenging 10-day-old SPF chickens. Our research result revealed that recombinant CK/CH/JS/YC10-3 isolate, formed by a major parent QX-type strain and a minor parent TW-type strain, presented lesion of air sacs covered with yellow white caseous exudate and obvious air sac thickening which similar to CK/CH/GC/JR07-7 strain (QX-type). However, GZ14 strain (TW-type) showed its damage mainly with acute nephritis.
IBV, a single molecule RNA virus, can mutate frequently for complex mechanisms, leading to extensive antigenic variation (20). Recombination events are the important factors of generating new strains, and sometimes, random mutations also involve this procedure (21). The genetic alterations found in the S1 gene will urge the corresponding S1 protein to mutate in IBV variants (22). Eventually, it might contribute to differences in viral replication, pathogenicity, antigenicity, immunogenicity, and tissue tropism (23). In this study, analysis of the complete S1 gene of these isolates definitively proved that IBV is continuously undergoing these processes, and simultaneity its biological characteristics has changed probably. In productive activities, chick flocks in China were received already the IBV H120, 4/91, or QX vaccine in 1-day-old. However, it also happened high positive rate of wild type virus infection. The caveat is that the recombination strain hardly shows weaker virulence than the classical strain, which is a significant challenge for the effectiveness of the vaccine. It is difficult for the commercial vaccine to make a difference in cross-protection against various IBVs.
It is particularly notable here that controlling IB in China remains a challenging task due to the co-circulation of numerous IBV genotypes and the increasing isolation of novel strains (24). Therefore, it is necessary to continuously monitor the different genotypes or lineages of IBV and make effort to decrease IBVs co-circulation so that the risks of IBV vaccination failure can be avoided and new effective vaccines can be developed. Nowadays, multiple pathogen co-infections causing respiratory diseases are common in production. Even though we did not detect whether all of the specimens carry other pathogens in this study, it is worth considering that LPAIV or MG possibly plays a role. Therefore, the synergy between IBV to other respiratory etiologic agents should be explored as soon as possible, as it is of considerable significance for prevention. This present pathogenic testing of strain GZ14, CK/CH/GC/JR07-7, and CK/CH/JS/YC10-3 laid the foundation for further research.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Materials, further inquiries can be directed to the corresponding author.
Ethics Statement
The animal study was reviewed and approved by South China Agricultural University.
Author Contributions
Conceptualization, project administration, and funding acquisition: QX. Methodology: HF and ZWu. Software, writing—original draft preparation, and formal analysis: HF. Validation: ZWu, ZXu, and JL. Investigation: ZWu and ZXu. Resources: ZWu. Data curation: HF and ZXi. Writing—review and editing: ZWu and ZWa. Visualization: XZ. All authors have read and agreed to the published version of the manuscript.
Conflict of Interest
ZWu, HF, Zxu, ZWa, and JQ were employed by Wen's Group Academy, Wen's Foodstuffs Group Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
This study was supported by the Key Research and Development Program of Guangdong Province (2020B020222001), the Guangdong Basic and Applied Basic Research Foundation (2019A1515012006), Key-Area Research and Development Program of Guangdong Province (2019B020218004), the National Natural Science Foundation of China (Grant Nos. 31972659 and 31902252), the Special Project of National Modern Agricultural Industrial Technology System (CARS-41), and the Construction of Modern Agricultural Science and Technology Innovation Alliance in Guangdong Province (2020KJ128).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2022.842179/full#supplementary-material
Abbreviations
IBV, infectious bronchitis virus; IB, infectious bronchitis; +ss, single-stranded and positive-sense; kb, kilobases; S, spike glycoprotein; E, envelope protein; M, membrane glycoprotein; N, nucleocapsid protein; PBS, phosphate-buffered saline; RT-qPCR, quantitative reverse transcription polymerase chain reaction; CT, cycle thresholds; SPF, special pathogen free; RT-PCR, reverse transcription polymerase chain reaction; RDP, the recombination detection program; MRs, mortality rates; NDV, newcastle disease virus; LPAIV, low pathogenic avian influenza virus; MG, mycoplasma gallisepticum; MS, mycoplasma synoviae.
References
1. Cavanagh D. Coronavirus avian infectious bronchitis virus. Vet Res. (2007) 2:281–97. doi: 10.1051/vetres:2006055
2. Feng J, Hu Y, Ma Z, Yu Q, Zhao J, Liu X, et al. Virulent avian infectious bronchitis virus, People's Republic of China. Emerg Infect Dis. (2012) 12:1994–2001. doi: 10.3201/eid1812.120552
3. Xu P, Shi Y, Liu P, Yang Y, Zhou C, Li G, et al. 16S rRNA gene sequencing reveals an altered composition of the gut microbiota in chickens infected with a nephropathogenic infectious bronchitis virus. Sci Rep. (2020) 1:3556. doi: 10.1038/s41598-020-60564-8
4. Feng K, Wang F, Xue Y, Zhou Q, Chen F, Bi Y, et al. Epidemiology and characterization of avian infectious bronchitis virus strains circulating in southern China during the period from 2013-2015. Sci Rep. (2017) 1:6576. doi: 10.1038/s41598-017-06987-2
5. Huang M, Zou C, Liu Y, Han Z, Xue C, Cao Y. A novel low virulent respiratory infectious bronchitis virus originating from the recombination of QX, TW and 4/91 genotype strains in China. Vet Microbiol. (2020) 2020:108579. doi: 10.1016/j.vetmic.2020.108579
6. Li L, Xue C, Chen F, Qin J, Xie Q, Bi Y, et al. Isolation and genetic analysis revealed no predominant new strains of avian infectious bronchitis virus circulating in South China during 2004-2008. Vet Microbiol. (2010) 2–4:145–54. doi: 10.1016/j.vetmic.2009.11.022
7. Bande F, Arshad SS, Bejo MH, Moeini H, Omar AR. Progress and challenges toward the development of vaccines against avian infectious bronchitis. J Immunol Res. (2015) 2015:424860. doi: 10.1155/2015/424860
8. Liu S, Kong X. A new genotype of nephropathogenic infectious bronchitis virus circulating in vaccinated and non-vaccinated flocks in China. Avian Pathol. (2004) 3:321–7. doi: 10.1080/0307945042000220697
9. Xu L, Han Z, Jiang L, Sun J, Zhao Y, Liu S. Genetic diversity of avian infectious bronchitis virus in China in recent years. Infect Genet Evol. (2018) 82–94. doi: 10.1016/j.meegid.2018.09.018
10. Chen L, Xiang B, Hong Y, Li Q, Du H, Lin Q, et al. Phylogenetic analysis of infectious bronchitis virus circulating in southern China in 2016-2017 and evaluation of an attenuated strain as a vaccine candidate. Arch Virol. (2021) 1:73–81. doi: 10.1007/s00705-020-04851-9
11. Lian J, Wang Z, Xu Z, Chen T, Shao G, Zhang X, et al. Distribution and molecular characterization of avian infectious bronchitis virus in southern China. Poult Sci. (2021) 7:101169. doi: 10.1016/j.psj.2021.101169
12. Awad F, Forrester A, Baylis M, Lemiere S, Ganapathy K, Hussien HA, et al. Protection conferred by live infectious bronchitis vaccine viruses against variant Middle East IS/885/00-like and IS/1494/06-like isolates in commercial broiler chicks. Vet Rec Open. (2015) 2:e000111. doi: 10.1136/vetreco-2014-000111
13. Chu J, Zhang Q, Zhang T, Han E, Zhao P, Khan A, et al. Chlamydia psittaci infection increases mortality of avian influenza virus H9N2 by suppressing host immune response. Sci Rep. (2016). 2016:29421. doi: 10.1038/srep29421
14. Yoder HW Jr, Hopkins SR. Efficacy of experimental inactivated mycoplasma gallisepticum oil-emulsion bacterin in egg-layer chickens. Avian Dis. (1985) 2:322–34. doi: 10.2307/1590492
15. Domanska-Blicharz K, Lisowska A, Pikula A, Sajewicz-Krukowska J. Specific detection of GII-1 lineage of infectious bronchitis virus. Lett Appl Microbiol. (2017) 2:141–6. doi: 10.1111/lam.12753
16. Winterfield RW, Albassam MA. Nephropathogenicity of infectious bronchitis virus. Poult Sci. (1984) 12:2358–63. doi: 10.3382/ps.0632358
17. Malik YS, Patnayak DP, Goyal SM. Detection of three avian respiratory viruses by single-tube multiplex reverse transcription-polymerase chain reaction assay. J Vet Diagn Invest. (2004) 3:244–8. doi: 10.1177/104063870401600314
18. Roussan DA, Haddad R, Khawaldeh G. Molecular survey of avian respiratory pathogens in commercial broiler chicken flocks with respiratory diseases in Jordan. Poult Sci. (2008) 3:444–8. doi: 10.3382/ps.2007-00415
19. Haghighat-Jahromi M, Asasi K, Nili H, Dadras H, Shooshtari AH. Coinfection of avian influenza virus (H9N2 subtype) with infectious bronchitis live vaccine. Arch Virol. (2008) 4:651–5. doi: 10.1007/s00705-008-0033-x
20. Bochkov YA, Tosi G, Massi P, Drygin VV. Phylogenetic analysis of partial S1 and N gene sequences of infectious bronchitis virus isolates from Italy revealed genetic diversity and recombination. Virus Genes. (2007) 1:65–71. doi: 10.1007/s11262-006-0037-0
21. Lee CW, Jackwood MW. Evidence of genetic diversity generated by recombination among avian coronavirus IBV. Arch Virol. (2000) 10:2135–48. doi: 10.1007/s007050070044
22. Ren M, Sheng J, Ma T, Xu L, Han Z, Li H, et al. Molecular and biological characteristics of the infectious bronchitis virus TC07-2/GVI-1 lineage isolated in China. Infect Genet Evol. (2019) 75:103942. doi: 10.1016/j.meegid.2019.103942
23. Liu S, Zhang X, Gong L, Yan B, Li C, Han Z, et al. Altered pathogenicity, immunogenicity, tissue tropism and 3'-7kb region sequence of an avian infectious bronchitis coronavirus strain after serial passage in embryos. Vaccine. (2009) 27:4630–40. doi: 10.1016/j.vaccine.2009.05.072
24. Naguib MM, El-Kady MF, Luschow D, Hassan KE, Arafa AS, El-Zanaty A, et al. New real time and conventional RT-PCRs for updated molecular diagnosis of infectious bronchitis virus infection (IBV) in chickens in Egypt associated with frequent co-infections with avian influenza and Newcastle Disease viruses. J Virol Methods. (2017) 245:19–27. doi: 10.1016/j.jviromet.2017.02.018
Keywords: infectious bronchitis virus, S1 gene, recombination, mutation, pathogenicity
Citation: Wu Z, Fang H, Xu Z, Lian J, Xie Z, Wang Z, Qin J, Huang B, Feng K, Zhang X, Lin W, Li H, Chen W and Xie Q (2022) Molecular Characterization Analysis of Prevalent Infectious Bronchitis Virus and Pathogenicity Assessment of Recombination Strain in China. Front. Vet. Sci. 9:842179. doi: 10.3389/fvets.2022.842179
Received: 23 December 2021; Accepted: 17 June 2022;
Published: 22 July 2022.
Edited by:
Min Yue, Zhejiang University, ChinaReviewed by:
Ahmed Ragab Elbestawy, Damanhour University, EgyptMin Liao, Zhejiang University, China
Copyright © 2022 Wu, Fang, Xu, Lian, Xie, Wang, Qin, Huang, Feng, Zhang, Lin, Li, Chen and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingmei Xie, qmx@scau.edu.cn
 Zhiqiang Wu
Zhiqiang Wu Huanxin Fang2
Huanxin Fang2  Wencheng Lin
Wencheng Lin Qingmei Xie
Qingmei Xie