Evaluation of Safety and Protective Efficacy of a waaJ and spiC Double Deletion Korean Epidemic Strain of Salmonella enterica Serovar Gallinarum
- Department of Veterinary Infectious Diseases and Avian Diseases, College of Veterinary Medicine and Center for Poultry Diseases Control, Jeonbuk National University, Iksan, South Korea
With an aim to develop a highly attenuated and strongly immunogenic distinguishable vaccine candidate, a waaJ (a gene involved in the synthesis of lipopolysaccharide) and spiC (a virulence gene) double deletion Korean epidemic strain of S. enterica ser. Gallinarum (SG005) was constructed. Our results showed that the growth and biochemical characteristics were not altered by this double deletion. The double deletion strain contained dual markers. One was a bacteriological marker (rough phenotype) and the other was a serological marker helping distinguish infected chickens from vaccinated chickens. The double deletion strain showed good genetic stability and reduced resistance to environmental stresses in vitro; furthermore, it was extremely safe and highly avirulent in broilers. Single intramuscular or oral immunization of 7-day-old broilers with the double deletion strain could stimulate the body to produce antibody levels similar to the conventional vaccine strain SG9R. In addition, against a lethal wild-type challenge, it conferred effective protection that was comparable to that seen in the group vaccinated with SG9R. In conclusion, this double deletion strain may be an effective vaccine candidate for controlling S. enterica ser. Gallinarum infection in broilers.
Introduction
Salmonella enterica serovar Gallinarum (S. enterica ser. Gallinarum) causes fowl typhoid (FT), an acute septicemic disease that occurs in chickens of all ages. Chicken infections are characterized by severe hepatosplenomegaly accompanied with bronze liver, anemia, and septicemia. The mortality and morbidity of FT can be as high as 80% (1). FT outbreaks have important economic significance in many areas, including Africa, Asia, the Middle East, and Central and South America (2). In Korea, S. enterica ser. Gallinarum was first officially reported during an FT outbreak in 1992 and is now found nationwide. FT has become one of the most prevalent bacterial diseases.
Many strategies have been used to prevent S. enterica ser. Gallinarum infection, including the establishment of strict biosecurity measures and the use of antibiotics and vaccines. The high costs of biosecurity measures limit their application in many developing countries, and long-term use of antibiotics can result in the emergence of multidrug-resistant strains. Vaccination seems to be the most effective strategy to control FT (1, 3). Until now, three major types of vaccines, namely inactivated vaccine, subunit vaccine, and live attenuated vaccine, have been developed to control Salmonella infections in the poultry industry. Inactivated and subunit vaccines can induce strong antibodies to eradicate extracellular bacteria; however, it is difficult for these antibodies to eliminate intracellular Salmonella; this goal can be achieved with the live attenuated strains of Salmonella (4). The live attenuated SG9R has been used as a commercial vaccine to control FT for nearly 60 years. However, some drawbacks, such as residual virulence in newly hatched chickens, insufficient protection, and vertical transmission, have been reported with its use (5, 6). Therefore, the development of a safe and effective S. enterica ser. Gallinarum vaccine remains a current research hotspot. Thus far, a number of deletion strains have been developed, and their protection efficiencies against FT have been reported (1, 7–14). However, none of the S. enterica ser. Gallinarum deletion strains have been found to have the feature to distinguish infected chickens from vaccinated chickens.
To construct a live attenuated S. enterica ser. Gallinarum vaccine with distinguishable capability, we firstly chose the waaJ gene. This gene codes for the lipopolysaccharide (LPS) 1,2-glucosyltransferase, which is involved in the synthesis of LPS (15). Deletion of the waaJ gene changes smooth LPS to rough LPS, which cannot react with antibodies against the O antigen. The spiC gene encodes an effector protein that is secreted by the Salmonella pathogenicity island-2 (SPI2) type III secretion system (T3SS), which plays important roles in Salmonella infections (16). Therefore, spiC was selected as the second target gene to be deleted.
In this study, we constructed a waaJ and spiC double deletion Korean epidemic strain of S. enterica ser. Gallinarum and evaluated its safety and protection efficacy for use as a live attenuated distinguishable vaccine candidate for the prevention of FT.
Materials and Methods
Bacterial Strains, Plasmids, Media, and Growth Conditions
A wild-type Korean epidemic virulent strain A17-DW-005 (SG005) of S. enterica ser. Gallinarum with resistance toward ciprofloxacin (CIP), streptomycin (STR), gentamicin (GEN), nalidixic acid (NAL), sulfisoxazole (FIS), and colistin (COL) was originally isolated from the liver of a 10-day-old broiler, which came from a broiler farm in Jeonbuk province in 2017. This strain was classified as type 1 by pulsed-field gel electrophoresis analysis (PFGE). Since type 1 has the highest proportion of all types, this strain was judged to be an epidemic strain. The suicide plasmid pRE112 and its host Escherichia coli (E. coli) χ7,213 were kept in our laboratory. pBluescript II SK (+) and pET-30 b (+) stored in our laboratory were used as intermediate vectors in this study. E. coli and S. enterica ser. Gallinarum were grown at 37°C in Luria–Bertani (LB) broth or on LB agar. When required, antibiotics were added to culture media at the following concentrations: ampicillin (Amp) at 100 μg/ml, kanamycin (Km) at 50 μg/ml, chloramphenicol at 25 μg/ml, and DL-α,ε-diaminopimelic acid (DAP) at 50 μg/ml. LB agar containing 5% sucrose was used for sacB gene-based counterselection in allelic exchange experiments. A rough attenuated S. enterica ser. Gallinarum live vaccine SG9R (Intervet International, Boxmeer, The Netherlands) was used as control vaccine in this study.
Chickens
Ross broiler chickens were obtained from the YangJi Company (Cheonan, Choongnam, South Korea). Serum samples collected from all chickens in the present study were tested for Salmonella specific antibodies using a commercial ELISA kit (Biochek, cat# CK117, Crabethstraatt, Netherland). Moreover, the antibodies level of Avian influenza and Newcastle disease were detected by hemagglutination-inhibition (HI) test virus. The test results were all negative. 10% of the chickens were randomly selected and euthanized. The chickens were dissected to check whether there were any lesions in the internal organs. Liver and fecal samples were aseptically collected for routine Salmonella and avian pathogenic Escherichia coli isolation and culture, and the follow-up Salmonella infection experiment was performed after the complete negative was confirmed. The chickens were used after adaptation for a week. To ensure the best environmental conditions, the conditions of the isolator (temperature, humidity, and ventilation) were continuously monitored. The chickens were taken care of and handled by a well-trained staff.
Ethics Statement
All experimental and animal management procedures were undertaken in accordance with the requirements of the Animal Care and Ethics Committee of Jeonbuk National University. The animal facility at Jeonbuk National University is fully accredited by the National Association of Laboratory Animal Care (approval number: JBNU 2020-0162).
Construction of Deletion Strains of S. enterica Ser. Gallinarum by Allelic Exchange
Construction of deletion strains was performed by the allelic exchange method using the suicide vector pRE112 as described previously (Supplementary Figure 1) (17). The 1,000-bp upstream fragment of the waaJ gene was amplified from the genomic DNA of S. enterica ser. Gallinarum using a pair of primers (W1 and W2) by PCR (Supplementary Table 1). The PCR product was cloned into the XbaI and BamHI sites of the pBluescript II SK(+) vector, resulting in pSK-waaJ-up. The 1,000-bp downstream fragment of the waaJ gene was then PCR-amplified using a pair of primers (W3 and W4) and cloned into the BamHI and KpnI sites of pSK-waaJ-up to obtain pSKΔwaaJ. The 2,000-bp fragment, which included the upstream and downstream fragments of the waaJ gene from the XbaI- and KpnI-digested plasmid pSKΔwaaJ, was ligated into plasmid pRE112 to yield the suicide plasmid pREΔwaaJ. The transfer of recombinant suicide plasmids into SG005 was accomplished by conjugation using E. coli χ7213 (pREΔwaaJ) as the plasmid donor. Strains containing single-crossover plasmid insertions (SG005waaJ::pREΔwaaJ) were isolated on plates containing chloramphenicol. The first homologous recombination was confirmed with the primers W5 and W6. A loss of the suicide vector after the second recombination between homologous regions (i.e., allelic exchange) was selected for by using the sacB-based sucrose sensitivity counterselection system. The second homologous recombination was confirmed with the primers W6 and W7. Primers W8 and W9 were used to confirm whether the full waaJ gene was removed from the genome. Primers W10 and W11 were used to confirm whether the suicide plasmid was removed from the genome (Supplementary Figure 2). This deletion strain was designated as SG005ΔwaaJ. Using the same method, deletion of spiC was introduced into SG005, and this deletion strain was designated as SG005ΔspiC. Notably, in the construction of the suicide plasmid pREΔspiC, pET-30 b (+) was used as an intermediate vector. Similarly, the deletion of spiC was introduced into SG005ΔwaaJ, and the double deletion strain was designated as SG005ΔwaaJΔspiC.
The complemented strains were constructed previously described with some modifications (18–20). The gene waaJ was PCR-amplified from the wild type strain SG005 using the primers pBR322-waaJ-F and pBR322-waaJ-R (Supplementary Table 1), which were designed based on the coding sequence of waaJ gene. After amplification, the DNA fragment was digested by BamHI and SalI and ligated to the plasmid pBR322. The resulting vector pBR322-waaJ was introduced into SG005ΔwaaJ by electroporation to produce the complemented strain SG005ΔwaaJ (pBR322-waaJ). Electroporation on the electroporator (Bio-Rad Laboratories, Hercules, CA, USA) was performed using optimized parameters (2.5 kV, 200 Ω, 25 μF, 5 ms). Similarly, the complemented strains SG005ΔspiC (pBR322-spiC) and SG005ΔwaaJΔspiC (pBR322-waaJ-spiC) were constructed. It should be pointed out that when constructing SG005ΔwaaJΔspiC (pBR322-waaJ-spiC), the waaJ gene and spiC gene were inserted into the plasmid pBR322 in tandem.
Analysis of in vitro Growth and Biochemical Characteristics
For the in vitro growth analysis of SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC, a single colony of each strain was inoculated into 5 ml of LB broth and cultured at 37°C with shaking at 200 rpm for approximately 15 h. Subsequently, the optical density (OD590) of each strain was measured by a Genesys 10S UV-Vis spectrophotometer (Thermo Fisher Scientific, Waltham, USA), and cultures were diluted in 20 ml of LB broth; then, the OD590 was determined to achieve an initial concentration (OD590 = 0.03) at the starting time point (0 h). The cultures were incubated at 37°C with shaking at 200 rpm, and the OD590 was determined at 1, 2, 3, 4, 5, 6, 7, and 8 h. Each strain was tested in triplicate in three independent experiments. The biochemical properties of the strains were tested according to the API20E handbook (Biomerieux, SA, France).
Phenotype Identification
O9 antigen is one of the antigens produced on S. enterica ser. Gallinarum. S. enterica ser. Gallinarum with smooth phenotype have O9 antigen and therefore can be agglutinated by O9 antibody. Briefly, colonies from LB agar plates were mixed with 20 μl of phosphate-buffered saline (PBS) and 20 μl of the O9 factor rabbit antiserum (SSI® SALMONELLA ANTISERA, Denmark) on a glass slide. Mixtures were rotated for 1 min and observed for agglutination. The acriflavine agglutination test was performed to detect the rough phenotype characteristic of the deletion strains (21). Briefly, colonies of a 24-h culture prepared from the deletion strains on LB agar plates were mixed with 20 μl of 0.2% acriflavine on a glass slide. Mixtures were rotated for 1 min and observed for agglutination.
Auto-Aggregation Assay
Auto-aggregation refers to the phenomenon that bacteria aggregate into clusters by themselves during the cultivation process. The stronger is the hydrophobicity of the cell surface, the higher is the auto-aggregation ability of the cell. The auto-aggregation assay was performed based on a previously described method with some modifications (22). Briefly, SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC were statically grown in 5 ml of LB broth at 37°C for 24 h in 15-ml conical tubes. The upper 0.5 ml was carefully removed to measure its OD590 (recorded as OD590 prevortex). Then, the remaining culture in the tube was mixed by vortexing to resuspend the aggregated cells, and 0.5 ml of the suspension was removed, and its OD590 was measured (recorded as OD590 postvortex). The “percent aggregation” was calculated using the formula: 100% × (OD590 postvortex – OD590 prevortex)/OD590 postvortex.
Stability Analysis
To determine the stability of the deletion strains, the liquid culture of each strain was continuously passaged 60 times every 12 h. Every 10 generations, DNA was extracted for PCR identification.
Environmental Safety Evaluation
The sensitivity of SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC to ultraviolet (UV) light and various disinfection agents was evaluated as described with some modifications (23, 24). Bacteria were grown to OD590 = 1.0 at 37°C with shaking at 200 rpm and were diluted in PBS to 109 colony forming units (CFU)/ml. One milliliter of suspension was then transferred to a sterile Petri dish and exposed to UV radiation (Sankyo denki UV G30TB: 30 W, distance: 45 cm) for 1 min. For testing oxidative stress and alkali and acid tolerance, each bacterial suspension and the stressor were mixed in equal volume and incubated at room temperature (RT, 25°C): 100-μM hydrogen peroxide (H2O2) for 5 min, 12.5-mM sodium hydroxide (NaOH) for 5 min, and 50-mM citric acid for 20 min. Finally, the number of viable bacteria was determined by plating serial dilutions onto LB agar plates. The experiment was performed three times.
Assessment of LD50
To determine the 50% lethal dose (LD50), SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC were grown for 12 h at 37°C in LB broth. One milliliter of broth from each grown bacterium was inoculated into 100 ml of LB broth. After the inoculated strains were grown to OD590 = 1.0, the bacterial cells were recovered by centrifugation for 10 min at 2,400 g at RT. The harvested cells were suspended in PBS to a concentration of 1011-1012 CFU/ml. The suspended strains were serially diluted by 10-fold. The 7-day-old chickens were intramuscularly (IM) injected or orally inoculated with each dose of these strains (101-1011 CFU). Six chickens were used for each dose. Feed and water were not provided to the tested chickens for 4 h before and until 1 h after the treatment. The treated chickens were observed twice a day for 2 weeks after administration. Clinical scores were determined and recorded using a system as we have previously described (25). Briefly, disease severity was scored as follows: 0, normal; 1, depression and ruffled feathers; 2, depression, ruffled feathers, and respiratory distress; 3, abovementioned clinical signs plus anorexia, emaciation, and green-yellowish diarrhea; and 4, death. When the birds showed a clinical score of 3 (humane endpoint), they were euthanized immediately by cervical dislocation performed by trained veterinarians. LD50 was calculated by the method of Reed and Muench (26).
Organ Colonization and Persistence
Bacterial persistence and clearance were analyzed by counting the number of the bacteria per gram of liver or spleen. Seven-day-old chickens were randomly divided into four groups (n = 12 for each group): SG005ΔwaaJ group, SG005ΔspiC group, SG005ΔwaaJΔspiC group, and PBS control group. The first three groups were injected intramuscularly with 1 × 108 CFU of SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC, respectively. The last group was inoculated with an equal volume of PBS in the same way. Three chickens from each group were euthanized at 2, 6, 10, and 14 days post-inoculation (dpi), and liver and spleen samples were aseptically removed. The samples were weighed and suspended in 1 ml of PBS and then individually homogenized. The homogenates were diluted serially and subsequently plated on MacConkey agar (Difco Laboratories, Detroit, MI, USA) at 37°C. After overnight cultivation, the bacterial number was counted.
Vaccination Experiment
Sixty 7-day-old chickens were divided into five groups (n = 12 for each group) to evaluate the protective efficacy of the deletion strains. The group 1 (PBS negative control) and group 2 (PBS positive control) were intramuscularly inoculated with 500 μl of PBS (pH 7.2). The group 3 and group 4 were intramuscularly inoculated with 108 CFU of SG9R and SG005ΔwaaJΔspiC, respectively. The group 5 was orally inoculated with 108 CFU of SG005ΔwaaJΔspiC. After 4 weeks, except for the group 1, chickens in other groups were challenged intramuscularly with 108 CFU of virulent SG005. Deaths and clinical signs were recorded daily for 2 weeks. At the end of this period, all surviving chickens were euthanized. Clinical scores were determined and recorded following the method previously described (25).
Antibody Production Assay
A laboratory-made indirect enzyme-linked immunosorbent assay (ELISA) using heat-killed whole S. enterica ser. Gallinarum bacteria as the coating antigen was applied to quantify IgG in the serum. Briefly, bacterial antigen (106 CFU/well) was coated in a 96-well plate and incubated overnight at 4°C. After blocking, the sera samples collected on days 14, 21, and 28 were diluted (1:400) with 2% bovine serum albumin (BSA) and added to the wells. The wells were incubated with a 1:8,000 dilution of affinity-purified, peroxidase-labeled goat anti-chicken IgG (H+L) (KPL Inc., MD, USA) for 1 h. OD450 was measured with a microplate reader VICTORTM X4 (PerkinElmer Inc., Waltham, MA, USA) after the reaction was stopped with 4.5 N H2SO4. The OD450 average value of negative serum samples plus the standard deviation (SD) of three times was used as the cut-off value (27).
Capability of DIVA (Differentiation of Infected and Vaccinated Animals) Based Serodiagnosis
The serum plate agglutination (SPA) test was used to assess the DIVA capability of SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC. Seven-day-old chickens were intramuscularly inoculated with each deletion strain. The sera were prepared on 14, 21, and 28 dpi by collecting blood from wing veins. The blood was allowed to coagulate for 1 h at RT, centrifuged for 10 min at 600 g, and then, the clean phase of serum was collected. Serum samples (30 μl) were mixed with an equal volume of S. enterica ser. Pullorum/Gallinarum standard antigen (Green Cross Veterinary Products, Yongin, South Korea), and the agglutination reaction was observed.
Statistical Analysis
Statistical analysis was performed using SPSS version 21.0 (SPSS Inc., Chicago, IL, USA). The data were analyzed by one-way analysis of variance (ANOVA). Differences were considered statistically significant at: *P < 0.05, **P < 0.01, ***P < 0.001.
Results
Constructions of S. enterica Ser. Gallinarum Deletion Strains
The deletion of the waaJ gene in SG005 was confirmed by PCR using the primers W6 and W7 with the expected amplicon sizes of 2,490 and 1,476 bp for the wild type and deletion type of the waaJ gene, respectively. The deletion of the spiC gene in SG005 was confirmed by PCR using the primers S6 and S7 with the expected amplicon sizes of 2,003 and 1,619 bp for the wild type and deletion type of the spiC gene, respectively (Supplementary Figure 3). In the three deletion strains, the corresponding target gene and suicide plasmid could not be detected by PCR (Supplementary Figure 4). The sequencing results also showed that the deletion strains SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC were constructed successfully (Supplementary Text 1).
Growth and Biochemical Characteristics
Growth curve analysis revealed no significant differences between the wild type and each deletion strain when cultured in LB broth at 37°C (Figure 1A). Results of biochemical tests including 2-nitrophenyl-β D-galactopyranoside, L-arginine, L-lysine, L-ornithine, trisodium citrate, sodium thiosulfate, urea, L-tryptophane, L-tryptophane, sodium pyruvate, gelatin, D-glucose, D-mannitol, inositol, D-sorbitol, L-rhamnose, D-sucrose, D-melibiose, amygdalin, and L-arabinose were the same between wild type and each deletion strain, suggesting that deletions in these two genes did not alter the biochemical characteristics of SG005 (Supplementary Table 2).
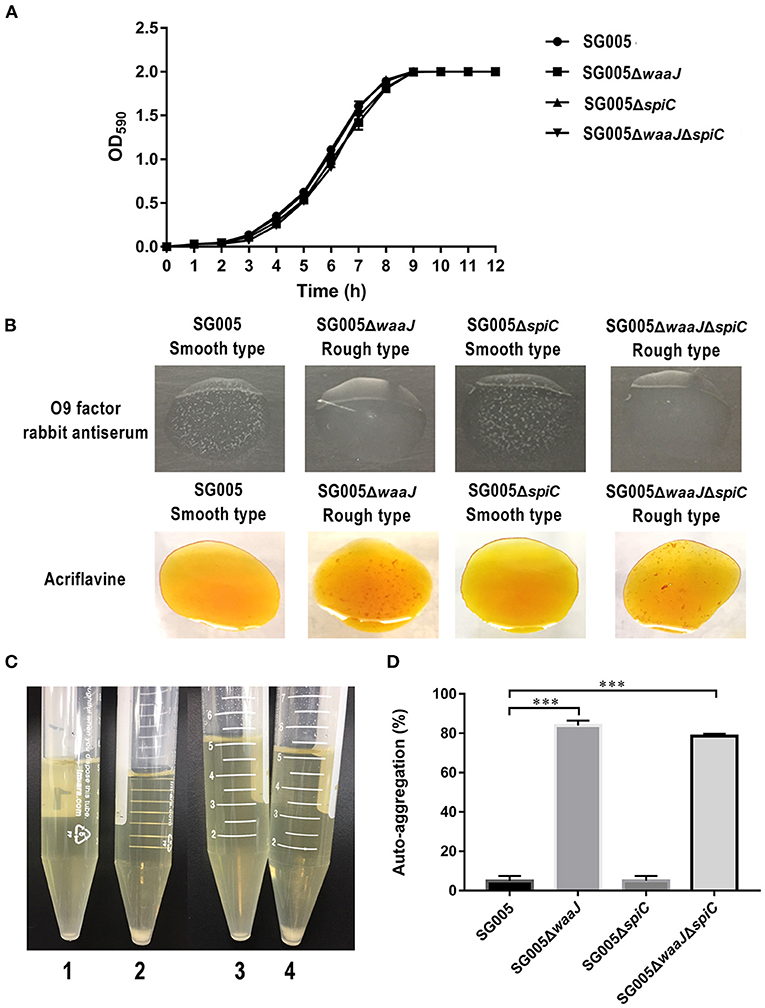
Figure 1. Growth curves, phenotypic characterization, and auto-aggregation assay. (A) Growth curves of wild-type SG005 and the deletion strains. SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC showed a similar growth curve in LB broth. (B) Phenotype identification. Agglutination was examined with O9 factor rabbit antiserum and acriflavine. Pictures were taken within 3 min. (C) Visual auto-aggregation of SG005 (1), SG005ΔwaaJ (2), SG005ΔspiC (3), and SG005ΔwaaJΔspiC (4). (D) Auto-aggregation (%). Bacterial cultures were statically grown in LB broth for 24 h at 37°C. Statistical analysis was done using one-way ANOVA. ***P < 0.001. Each experiment was repeated three times.
Phenotype Identification
SG005 and SG005ΔspiC were agglutinated with O9 factor rabbit antiserum but not with acriflavine, indicating that they had a smooth phenotype. However, SG005ΔwaaJ and SG005ΔwaaJΔspiC were not agglutinated with O9 factor rabbit antiserum but were agglutinated with acriflavine, indicating that they had a rough phenotype (Figure 1B). The complemented strains SG005ΔwaaJ (pBR322-waaJ), SG005ΔspiC (pBR322-spiC) and SG005ΔwaaJΔspiC (pBR322-waaJ-spiC) were all agglutinated with O9 factor rabbit antiserum but not with acriflavine. Therefore, complementary analysis proved that the deletion of the waaJ gene was the direct cause of the bacteria changing from smooth to rough.
Auto-Aggregation Assay
Auto-aggregation was tested in LB broth (Figure 1C), and the aggregation percent was calculated using OD590 measurements from these cultures. Both SG005 and SG005ΔspiC showed ~5% aggregation, whereas SG005ΔwaaJ and SG005ΔwaaJΔspiC demonstrated 84 and 79% aggregation, respectively (Figure 1D).
Stability
The deletion Strains were serially passaged 60 times in LB medium, and the absence of waaJ and spiC genes was confirmed by PCR, indicating that each deletion strain had good genetic stability (Figure 2).
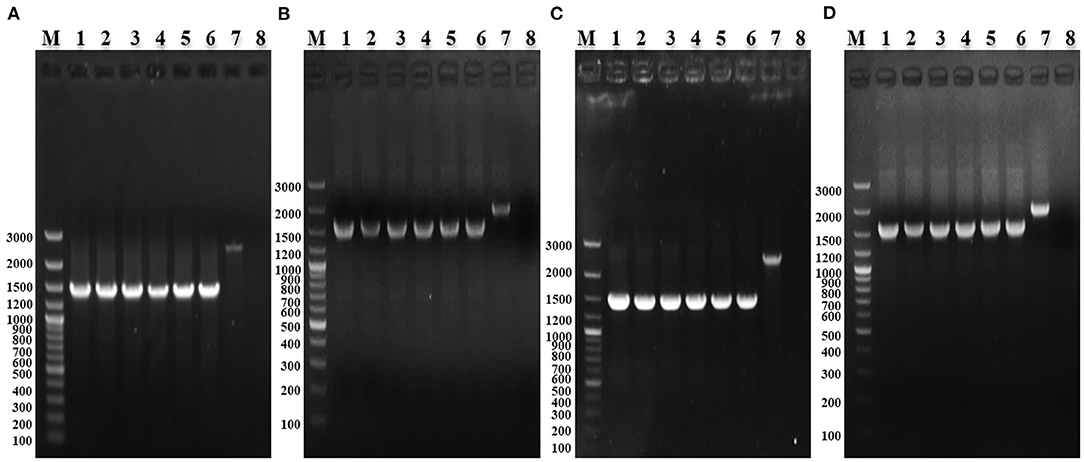
Figure 2. PCR identification of the stability of S. enterica ser. Gallinarum deletion strains. (A) PCR identification of SG005ΔwaaJ with W6/W7. M: GeneRuler 100 bp Plus DNA Ladder (Thermo Fisher Scientific, Waltham, MA, USA); 1–6: PCR-amplified products from the 10th, 20th, 30th, 40th, 50th, and 60th passages of SG005ΔwaaJ; 7: Wild-type SG005; and 8: Negative control. (B) PCR identification of SG005ΔspiC with S6/S7. M: GeneRuler 100 bp Plus DNA Ladder; 1–6: PCR-amplified products from the 10th, 20th, 30th, 40th, 50th, and 60th passages of SG005ΔspiC; 7: Wild-type SG005; and 8: Negative control. (C) PCR identification of SG005ΔwaaJΔspiC with W6/W7. M: GeneRuler 100 bp Plus DNA Ladder; 1–6: PCR-amplified products from the 10th, 20th, 30th, 40th, 50th, and 60th passages of SG005ΔwaaJΔspiC; 7: Wild-type SG005; and 8: Negative control. (D) PCR identification of SG005ΔwaaJΔspiC with S6/S7. M: GeneRuler 100 bp Plus DNA Ladder; 1–6: PCR-amplified products from the 10th, 20th, 30th, 40th, 50th, and 60th passages of SG005ΔwaaJΔspiC; 7: Wild-type SG005; and 8: Negative control.
Bacterial Resistance to Environmental Stress
After UV irradiation, the survival rates for SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC were 48, 37, 45, and 37%, respectively (Figure 3A). A significant difference (P < 0.001) was seen between ΔwaaJ strains (SG005ΔwaaJ and SG005ΔwaaJΔspiC) and non-ΔwaaJ strains (SG005 and SG005ΔspiC), indicating that the deletion of the waaJ gene increased the sensitivity of the strain to UV. In terms of oxidative stress, SG005ΔwaaJ and SG005ΔwaaJΔspiC with survival rates of 52 and 50% showed reduced resistance compared to SG005 and SG005ΔspiC with survival rates of 59 and 60% (P < 0.001) in H2O2 (Figure 3B). For alkali endurance, the survival rates for SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC were 68, 51, 67, and 49%, respectively. A significant difference was noted between ΔwaaJ strains and non-ΔwaaJ strains (P < 0.001; Figure 3C). For acid endurance, the survival rates for SG005, SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC were 34, 31, 35, and 31%, respectively. Statistical analysis revealed a difference between ΔwaaJ strains and non-ΔwaaJ strains (P < 0.05; Figure 3D). However, deletion of spiC gene had no effect on bacterial resistance to these environmental stresses.
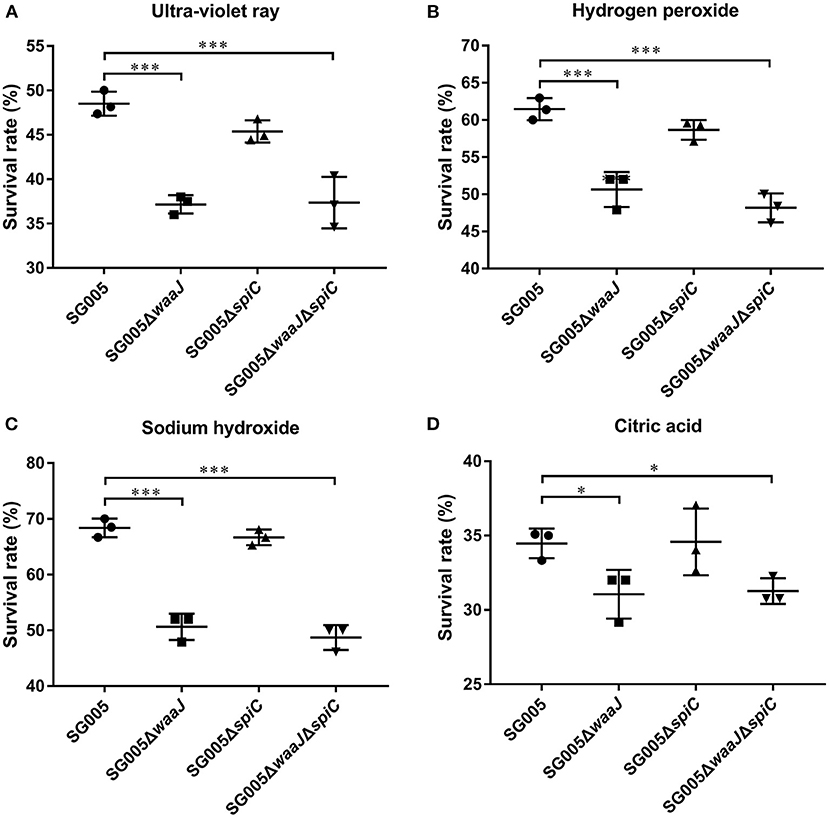
Figure 3. Survival rates of SG005 and deletion strains in different environmental stresses. (A) UV irradiation for 1 min. (B) Hydrogen peroxide (100 μM for 5 min). (C) Sodium hydroxide (12.5 mM for 5 min). (D) Citric acid (50 mM for 20 min). Statistical analysis was done using one-way ANOVA. *P < 0.05, **P < 0.01, ***P < 0.001. Each experiment was repeated three times.
Determination of LD50
To confirm the safety of the deletion strains, the virulence of wild and deletion strains was evaluated in 7-day-old chickens. The IM LD50 of SG005ΔwaaJ and SG005ΔspiC was about 108-fold higher than that of the wild strain. The IM LD50 of SG005ΔwaaJΔspiC was about 1010-fold higher than that of the wild strain. The oral LD50 of the three deletion strains SG005ΔwaaJ, SG005ΔspiC, and SG005ΔwaaJΔspiC was about 105-fold higher than that of the wild strain SG005 (Table 1).
Organ Colonization and Persistence
The results of bacteria persistence and clearance in organs were shown in Figure 4A. SG005ΔwaaJΔspiC colonized and persisted in the liver and spleen for at least 10 days. Within these 10 days, the number of SG005ΔwaaJΔspiC gradually decreased. At 14 dpi, SG005ΔwaaJΔspiC could not be isolated from the liver and spleen. However, SG005ΔwaaJ and SG005ΔspiC could still be isolated, indicating that the double deletion strain could be cleared more quickly than the single deletion strains.
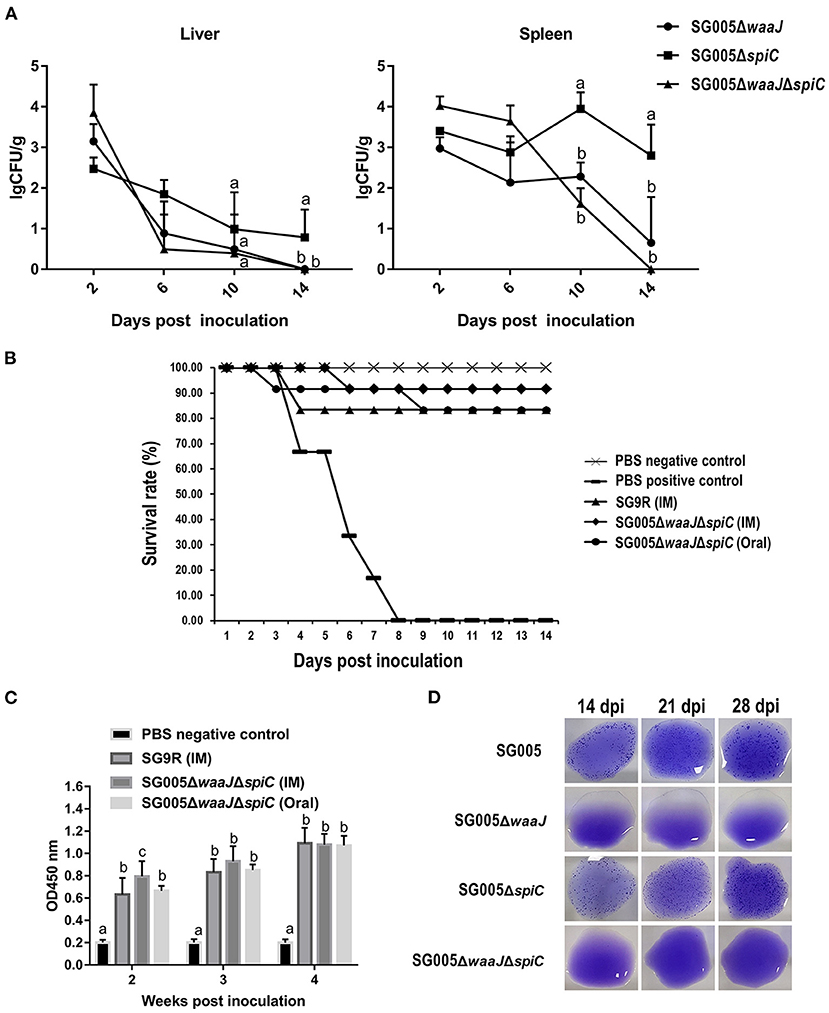
Figure 4. Organ colonization, protective efficacy, humoral immune response, and DIVA capability analysis. (A) Colonization and persistence of the deletion strains in the liver and spleen after inoculation of 7-day-old chickens with 108 CFU bacterial cells. Values were represented as lgCFU/g sample. All broilers inoculated with SG005 died within 3 days, and the number of bacteria isolated from the liver and spleen was about 8 lgCFU/g. (B) Protective ability of the deletion strains against the virulent SG005 challenge. At 14 dpi, chickens from each group were challenged intramuscularly with 1 × 108 CFU wild-type SG005. Protection was expressed by percentage survival after challenge. SG9R was used as a vaccine control. (C) Levels of humoral immune responses in vaccinated vs. control chickens. Serum IgG antibody responses against the deletion strains were measured by indirect ELISA. OD450, optical density at 450 nm. The OD450 average value of negative serum samples plus the standard deviation (SD) of three times was used as the cut-off value, which was found to be 0.281. (D) Capability of DIVA based on a serum plate agglutination test. Sera collected on 14, 21, and 28 dpi were tested for reactivity to the S. enterica ser. Pullorum/Gallinarum standard antigen. Statistical analysis was done using one-way ANOVA. Different letters mean significant differences between groups (P < 0.05).
Protective Efficacy Against Virulent S. enterica Ser. Gallinarum Challenge
Chickens that were intramuscularly inoculated with 108 CFU of SG9R and SG005ΔwaaJΔspiC showed 83 and 92% protection against S. enterica ser. Gallinarum challenge, respectively. Chickens that were orally inoculated with 108 CFU of SG005ΔwaaJΔspiC showed 83% protection. All chickens died in the PBS-positive control group (Figure 4B). Depression, anorexia, and diarrhea were observed in the PBS-positive control group, whereas no clinical signs were observed in the SG005ΔwaaJΔspiC group.
Humoral Immune Response Generated by the Deletion Strains
Statistical analysis showed a significant difference in the humoral immune response in chickens immunized with the deletion strains compared with the PBS control group. Serum IgG levels at each post-immunization time point were significantly higher in the immunized groups than in the PBS control group. The IgG level of the SG005ΔwaaJΔspiC group immunized by intramuscular injection oral administration was not significantly different from that of the SG9R group at 3 and 4 weeks after inoculation (P > 0.05; Figure 4C).
Differentiation of Serum Antibodies to S. enterica Ser. Gallinarum
Antibodies to S. enterica ser. Gallinarum were detected by the SPA test using the SG005 antigen. Sera collected from SG005ΔwaaJ- or SG005ΔwaaJΔspiC-immunized chickens at different time intervals were identified as SPA-negative (Figure 4D). However, sera collected from SG005- or SG005ΔspiC-immunized chickens were identified as SPA-positive. Overall, SG005ΔwaaJ and SG005ΔwaaJΔspiC showed evident distinguishable capability.
Discussion
Many shortcomings of SG9R vaccine limit its application. The residual virulence of SG9R caused liver and spleen lesions in day-old broilers and brown layers, and a single point mutation of the waaJ gene can make SG9R lose DIVA capability. Moreover, SG9R has the potential to revert to a more virulent phenotype in the field (28).
Our goal was to develop a highly attenuated and strongly immunogenic distinguishable vaccine. The reasons we decided to delete waaJ were as follows: Firstly, the protein encoded by the waaJ gene is involved in the synthesis of LPS. LPS is a major virulence factor of S. enterica ser. Typhimurium and is composed of lipid A, core oligosaccharide (C-OS), and O-antigen polysaccharide (O-PS). Partial deletion of LPS will directly reduce bacterial virulence (29). Secondly, deletion of the waaJ (rfaJ) gene can be used as a DIVA marker in current Salmonella serosurveillance programmes based on the detection of antibodies against LPS of Salmonella (30). Thirdly, the deletion of waaJ gene does not affect the protective capacities of the strain. It was reported that applying deletions in the waaJ gene in S. enterica ser. Typhimurium strain 112910a allows differentiation of infected and vaccinated pigs in an LPS based ELISA without reducing the strain's protective capacities in mice (30). Therefore, the waaJ gene is a good choice for constructing a distinguishable vaccine. Reportedly, the virulence of a spiC deletion strain of S. enterica ser. Pullorum was significantly reduced, and this deletion strain could induce high levels of circulating antibody and protective immunity in chickens (31). It has also been reported that a spiC deletion strain of S. enterica ser. Typhimurium was unable to survive within macrophages and had greatly reduced virulence in mice (32). Moreover, a S. enterica ser. Pullorum vaccine candidate S06004ΔspiCΔwaaL (a gene also related to LPS synthesis) with DIVA capability has been reported. Therefore, the virulence gene spiC was selected as the second gene to be deleted.
Here, there was another point that needed to be explained, that is why we did not choose waaL gene, but waaJ gene. Compared with the waaL gene, the reasons for choosing to delete the waaJ gene were as follows: Firstly, waaL gene codes a membrane enzyme implicated in ligating the O-antigen to the lipid A-core oligosaccharide. The waaJ deletion strain deleted more LPS fragment than the waaL deletion strain. The knockout part of the waaJ deletion strain contained the knockout part of the waaL deletion strain (33). Therefore, we speculated that compared with the waaL gene, deleting the waaJ gene may reduce the virulence more significantly and the DIVA effect in waaJ deletion strain may be more obvious than that of waaL deletion strain. Secondly, through whole-genome sequencing, it was found that there was a point mutation in the waaJ gene of the commercial vaccine strain, which prevented the protein encoded by waaJ from being synthesized. LPS defect caused by a point mutation in the waaJ gene may be one of the major mechanisms of vaccine strain attenuation (28). The commercial vaccine has been used for decades, and its immunogenicity is widely recognized by the poultry industry. Therefore, this reminded us that deleting the waaJ gene may be a good choice when developing new vaccines.
The growth and biochemical properties were not altered by deletions of waaJ or spiC or both genes from S. enterica ser. Gallinarum, suggesting that both genes may not be related to the bacterial metabolic pathway (Figure 1A and Supplementary Table 2). S. enterica ser. Gallinarum with smooth phenotype has O9 antigen and can therefore be agglutinated by O9 antibody. Our result showed that SG005 and SG005ΔspiC were agglutinated with O9 factor rabbit antiserum but SG005ΔwaaJ and SG005ΔwaaJΔspiC were not, indicating that the waaJ deletion strain did not contain O9 antigen. We performed the acriflavine agglutination test, which is used to detect the rough phenotype of bacteria (21). Our results were consistent with our expectations. SG005ΔwaaJ and SG005ΔwaaJΔspiC were agglutinated with acriflavine, whereas SG005 and SG00ΔspiC were not, indicating that deletion of the waaJ gene directly caused the LPS to change from smooth to rough (Figure 1B). In addition, both SG005ΔwaaJ and SG005ΔwaaJΔspiC showed evident auto-agglutination (Figures 1C,D). This is because the increased hydrophobicity of the rough Salmonella surface makes the cells aggregate and settle. In brief, the deletion of waaJ and spiC genes had no effect on the growth and biochemical characteristics of the bacteria, and the deletion of the waaJ gene not only changed the phenotype of the bacteria from a smooth type to a rough type but also made the bacteria exhibit obvious auto-aggregation.
The sensitivity of strains to different environmental stresses is an important indicator for testing the environmental safety of vaccines. As an important component of LPS, the O antigen structure may prevent antibacterial agents, such as disinfectants, from reaching the target sites in the cell wall to some extent, thereby improving its environmental resistance. The destruction of the LPS structure could reportedly reduce the resistance of bacteria to environmental stresses (24). In our study, the waaJ deletion strains (SG005ΔwaaJ and SG005ΔwaaJΔspiC) showed reduced resistance to UV, oxidative stress, and alkalis and acids, suggesting that the integrity of LPS is instrumental in the resistance of bacteria to environmental stresses (Figure 3). Similar results have been reported for S. enterica ser. Pullorum, S. enterica ser. Enteritidis, and E. coli (21, 24, 34). These results indicated that the deletion of the waaJ gene significantly increased the sensitivity of bacteria to environmental stress.
The deletion of the waaJ gene was reported to not only confer the DIVA capability to the smooth type strains but also reduce the virulence. The deletion of the waaJ gene consequently obstructed the expression of a part of the outer core and the O antigen of LPS. LPS is an important virulence factor, and partial deletion of LPS will directly reduce bacterial virulence. It has been reported that S. enterica ser. Typhimurium deletion strain 11310 (waaJ44) was significantly attenuated (about 105-fold) in mice (29). S. enterica ser. Enteritidis deletion strain SEM1C3ΔwaaJ had a LD50 at least 100 times higher than that of the parental strain in chickens (35). The spiC gene is closely related to the virulence of bacteria. It has been reported that the LD50 of S. enterica ser. Pullorum S06004ΔspiC was 200 times higher than that of the parent strain (23). Furthermore, the LD50 of S. enterica ser. Enteritidis C50041ΔspiC was reportedly 900 times higher than that of the wild-type strain (16). The LD50 of S. enterica ser. TyphimuriumΔspiC was 105 times higher than that of the parent strain in mice (16). Our research results were similar to these research results. Our result showed that the oral LD50 of both SG005ΔwaaJ and SG005ΔspiC was about 105-fold higher than that of the wild strain SG005 (Table 1). In addition, our results showed that when the 7-day-old broilers were administered intramuscularly, SG005ΔwaaJΔspiC was the least virulent among the three deletion strains, followed by SG005ΔspiC and finally SG005ΔwaaJ (Table 1). In addition, it should be noted that SG005ΔwaaJΔspiC was highly avirulent (IM LD50 increased at least by 1010-fold) because it did not cause mortality even in response to a high-dose challenge (1011 CFU). Furthermore, our results showed that the oral LD50 of SG005ΔwaaJΔspiC (>1011 CFU) was higher than that of SG9R (5 × 1010 CFU) (36). Taken together, these results indicated that SG005ΔwaaJΔspiC was extremely safe for broilers.
After intramuscular inoculation with SG005ΔwaaJΔspiC, the chickens did not show any of the clinical signs, and the weight of the inoculated chickens did not differ from that of the negative control group. This suggested that SG005ΔwaaJΔspiC did not cause any obvious damage to the chickens and was quite safe. In the colonization experiment, our results showed that SG005ΔwaaJΔspiC could exist in the chicken for at least 10 days and could not be detected on 14 dpi (Figure 4A). Some research reports have pointed out that the temporary residence of bacteria in the internal organs of chickens was essential for inducing protective immunity (37). In addition, SG005ΔwaaJΔspiC was cleared faster from the liver and spleen than the two single deletion strains SG005ΔwaaJ and SG005ΔspiC (Figure 4A). In previous experiments using chicken embryos to evaluate the virulence of S. enterica ser. Gallinarum, we have reported that the higher was the virulence of the strain, the stronger was its invasiveness into the liver. In this colonization experiment, since SG005ΔwaaJΔspiC had the lowest virulence had the weakest invasiveness to chickens, it was eliminated first. According to the above results, it can be interpreted that the inoculation of SG005ΔwaaJΔspiC did not cause any clinical signs and had no effect on body weight and that it could colonize in the chicken for at least 10 days.
Generally, the stronger is the virulence of the bacteria, the higher is the immunogenicity. Therefore, the construction of live attenuated Salmonella vaccine needs to balance attenuation and immunogenicity. According to the results of the LD50 assessment, the virulence reduction of SG005ΔwaaJΔspiC was verified, and herein, the protective efficacy was also verified. SG005ΔwaaJΔspiC provided protection similar to SG9R (92 vs. 83%, respectively) by intramuscular injection (Figure 4B). More importantly, through oral immunization, the protection of SG005ΔwaaJΔspiC could reach 83%, indicating that this double strain can be developed as an oral vaccine. In addition, severe clinical signs of FT, such as ruffled feathers, anorexia, somnolence, and greenish diarrhea, were observed in the PBS group 2 (positive control), whereas no obvious clinical signs were observed in the SG005ΔwaaJΔspiC group. For the vaccine strain SG9R, two times of immunization were required, and subcutaneous injection and intramuscular injection were the main ways of vaccination (6). However, for SG005ΔwaaJΔspiC, only a single intramuscular injection or oral immunization can achieve an immune effect no less than that of the vaccine strain SG9R. Taken together, these results indicated that attenuated SG005ΔwaaJΔspiC could confer effective protection against acute systemic FT infection.
Live attenuated Salmonella vaccine can stimulate the body to produce humoral immunity, which is essential for preventing Salmonella infection. In this study, we used indirect ELISA to check the serum IgG antibody to assess the level of specific humoral immune response induced by different deletion strains. The results showed that the antibody level of the SG005ΔwaaJΔspiC group immunized by intramuscular injection was not significantly different from that of the SG9R group (P > 0.05; Figure 4C). This indicated that SG005ΔwaaJΔspiC could provide antibody levels comparable to SG9R. A previous similar study reported that S. enterica ser. Pullorum S06004ΔspiCΔwaaL (waaL, also a gene involved in the synthesis of LPS) led to a significant antibody increase in broilers (21). In brief, SG005ΔwaaJΔspiC induced a strong humoral immune response. The disadvantage of this study was that we did not evaluate the cellular immune responses induced by the deletion strains. The cellular immunity induced by attenuated Salmonella is very important in the evaluation of vaccines, because Salmonella is an intracellular pathogen. Unfortunately, we did not test this indicator. Since this indicator can only be tested in animal experiments, we plan to test the cellular immune response through cell proliferation experiments in the next animal experiment.
It has been described that some LPS deficient mutants (waaJ, waaL, rfaH, rfbH, rfbG, et al.) were used in Salmonella DIVA vaccine research (22, 29, 30, 34, 38). It was reported that immunization of piglets with the ΔwaaJ or ΔwaaL mutants of S. enterica ser. Typhimurium resulted in the induction of a serological response lacking detectable antibodies against LPS, which allowed a clear differentiation between serums from pigs immunized with the ΔwaaJ or ΔwaaL strains and serums from pigs infected with the wild type strain (30). There was also a report showing that vaccination of swine with the rfaH (a gene encoding the RfaH antiterminator that prevents premature termination of long mRNA transcripts) mutant conferred protection against challenge with virulent S. enterica ser. Typhimurium but did not interfere with herd level monitoring for Salmonella spp (38). RfbH and rfbG (both genes are related to LPS biosynthesis) mutants of S. enterica ser. Enteritidis and S. enterica ser. Pullorum S06004ΔspiCΔwaaL were also reported to have DIVA features (22, 34). Our results showed that the antibodies produced by SG005ΔwaaJ and SG005ΔwaaJΔspiC did not react with S. enterica ser. Pullorum/Gallinarum standard antigen, indicating that these two deletion strains have good DIVA capability (Figure 4D). In addition, the limitations of the DIVA need to be pointed out in the present study. For example, when chickens are infected with a low level of wild strains in the field, the resulting serums may not agglutinate with the standard antigen, because the sensitivity of plate agglutination test is lower than that of ELISA (39). Immunization of animals with these deletion strains will not interfere with the Salmonella monitoring program. The vaccine with DIVA capability has the characteristic of distinguishing between infected and immunized animals, and its application to the prevention and control of livestock and poultry diseases is of great significance.
Additionally, although we have verified the deletion strains by PCR and sequencing, to perfect verification of our candidate, we will conduct of characteristics study using southern blot, whole-genome sequencing, immunological study, and field trials.
In this study, we lacked novelty in the selection of target genes, because waaJ deletion strains and spiC deletion strains have been reported on many other Salmonella species (16, 21, 23, 29, 30, 35). However, the waaJ and spiC double-gene deletion strain has not been reported in S. enterica ser. Gallinarum. Another important aspect was that we used a Korean epidemic strain, which was selected through PFGE analysis and antimicrobial sensitivity testing from many clinical isolates. Finally, we want to re-explain the purpose of our experiment. We hope to use the advantages of the commercial vaccine strain (removing the waaJ gene to produce DIVA effects and at the same time reduce virulence) and make up for the disadvantages (removing spiC to further eliminate residual virulence) to develop a new vaccine based on a Korean epidemic strain of S. enterica ser. Gallinarum.
Conclusion
In summary, we have demonstrated that the rough attenuated S. enterica ser. Gallinarum vaccine candidate SG005ΔwaaJΔspiC had attributes required in a potent vaccine, such as high attenuation, good immunogenicity, and distinguishable capability. Therefore, this double deletion strain—SG005ΔwaaJΔspiC—may be an effective vaccine for controlling S. enterica ser. Gallinarum infection in broilers.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by Animal Care and Ethics Committee of Jeonbuk National University.
Author Contributions
J-FZ, MK, and H-KJ contributed to conception and design of experiments. S-YC, KS, Y-JL, and J-YP contributed to acquisition, analysis, and interpretation of data. J-FZ, MK, S-YC, BW, and H-KJ drafted and/or revised the article. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Agriculture, Food and Rural Affairs Convergence Technologies Program for Educating Creative Global Leader (716002-7, 320005-4) funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We have not received for open access publication fees.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.756123/full#supplementary-material
References
1. Revolledo L. Vaccines and vaccination against fowl typhoid and pullorum disease: an overview and approaches in developing countries. J Appl Poult Res. (2018) 27:279–91. doi: 10.3382/japr/pfx066
2. Barrow PA, Freitas Neto OC. Pullorum disease and fowl typhoid–new thoughts on old diseases: a review. Avian Pathol. (2011) 40:1–13. doi: 10.1080/03079457.2010.542575
3. Mastroeni P, Chabalgoity JA, Dunstan SJ, Maskell DJ, Dougan G. Salmonella: immune responses and vaccines. Veterin J. (2001) 161:132–64. doi: 10.1053/tvjl.2000.0502
4. Desin TS, Koster W, Potter AA. Salmonella vaccines in poultry: past, present and future. Exp Rev Vaccines. (2013) 12:87-96. doi: 10.1586/erv.12.138
5. Bouzoubaa K, Nagaraja KV, Kabbaj FZ, Newman JA, Pomeroy BS. Feasibility of using proteins from Salmonella Gallinarum vs. 9R live vaccine for the prevention of fowl typhoid in chickens. Avian Dis. (1989) 33:385–91. doi: 10.2307/1591094
6. Silva EN, Snoeyenbos GH, Weinack OM, Smyser CF. Studies on the use of 9R strain of Salmonella-Gallinarum as a vaccine in chickens. Avian Dis. (1981) 25:38–52. doi: 10.2307/1589825
7. Griffin HG, Barrow PA. Construction of an aroA mutant of Salmonella serotype Gallinarum: its effectiveness in immunization against experimental fowl typhoid. Vaccine. (1993) 11:457–62. doi: 10.1016/0264-410X(93)90288-9
8. Penha Filho RA, de Paiva JB, da Silva MD, de Almeida AM, Berchieri A Jr. Control of Salmonella Enteritidis and Salmonella Gallinarum in birds by using live vaccine candidate containing attenuated Salmonella Gallinarum mutant strain. Vaccine. (2010) 28:2853–9. doi: 10.1016/j.vaccine.2010.01.058
9. Rosu V, Chadfield MS, Santona A, Christensen JP, Thomsen LE, Rubino S, et al. Effects of crp deletion in Salmonella enterica serotype Gallinarum. Acta Vet Scand. (2007) 49:14. doi: 10.1186/1751-0147-49-14
10. Cho Y, Park YM, Barate AK, Park SY, Park HJ, Lee MR, et al. The role of rpoS, hmp, and ssrAB in Salmonella enterica Gallinarum and evaluation of a triple-deletion mutant as a live vaccine candidate in Lohmann layer chickens. J Vet Sci. (2015) 16:187–94. doi: 10.4142/jvs.2015.16.2.187
11. Laniewski P, Mitra A, Karaca K, Khan A, Prasad R, Curtiss R 3rd, et al. Evaluation of protective efficacy of live attenuated Salmonella enterica serovar Gallinarum vaccine strains against fowl typhoid in chickens. Clin Vaccine Immunol. (2014) 21:1267–76. doi: 10.1128/CVI.00310-14
12. Matsuda K, Chaudhari AA, Lee JH. Evaluation of safety and protection efficacy on cpxR and lon deleted mutant of Salmonella Gallinarum as a live vaccine candidate for fowl typhoid. Vaccine. (2011) 29:668–74. doi: 10.1016/j.vaccine.2010.11.039
13. Zhang-Barber L, Turner AK, Dougan G, Barrow PA. Protection of chickens against experimental fowl typhoid using a nuoG mutant of Salmonella serotype Gallinarum. Vaccine. (1998) 16:899–903. doi: 10.1016/S0264-410X(97)00300-9
14. Shah DH, Shringi S, Desai AR, Heo EJ, Park JH, Chae JS. Effect of metC mutation on Salmonella Gallinarum virulence and invasiveness in 1-day-old White Leghorn chickens. Vet Microbiol. (2007) 119:352–7. doi: 10.1016/j.vetmic.2006.09.002
15. Kwon H-J, Cho S-H. Pathogenicity of SG 9R, a rough vaccine strain against fowl typhoid. Vaccine. (2011) 29:1311–8. doi: 10.1016/j.vaccine.2010.11.067
16. Wang Y, Cai Y, Zhang J, Liu D, Gong X, Pan Z, et al. Controversy surrounding the function of SpiC protein in Salmonella: an overview. Front Microbiol. (2019) 10:1784. doi: 10.3389/fmicb.2019.01784
17. Zhao Z, Xue Y, Wu B, Tang X, Hu R, Xu Y, et al. Subcutaneous vaccination with attenuated Salmonella enterica serovar Choleraesuis C500 expressing recombinant filamentous hemagglutinin and pertactin antigens protects mice against fatal infections with both S. enterica serovar Choleraesuis and Bordetella bronchiseptica. Infect Immun. (2008) 76:2157–63. doi: 10.1128/IAI.01495-07
18. Yin J, Xia J, Tao M, Xu L, Li Q, Geng S, et al. Construction and characterization of a cigR deletion mutant of Salmonella enterica serovar Pullorum. Avian Pathol. (2016) 45:569–75. doi: 10.1080/03079457.2016.1187708
19. Zhang X, He L, Zhang C, Yu C, Yang Y, Jia Y, et al. The impact of sseK2 deletion on Salmonella enterica serovar typhimurium virulence in vivo and in vitro. BMC Microbiol. (2019) 19:182. doi: 10.1186/s12866-019-1543-2
20. Xiong K, Chen Z, Xiang G, Wang J, Rao X, Hu F, et al. Deletion of yncD gene in Salmonella enterica ssp. enterica serovar Typhi leads to attenuation in mouse model. FEMS Microbiol Lett. (2012) 328:70–7. doi: 10.1111/j.1574-6968.2011.02481.x
21. Guo R, Jiao Y, Li Z, Zhu S, Fei X, Geng S, et al. Safety, protective immunity, and DIVA capability of a rough mutant Salmonella Pullorum vaccine candidate in broilers. Front Microbiol. (2017) 8:547. doi: 10.3389/fmicb.2017.00547
22. Jiao Y, Guo R, Tang P, Kang X, Yin J, Wu K, et al. Signature-tagged mutagenesis screening revealed a novel smooth-to-rough transition determinant of Salmonella enterica serovar Enteritidis. BMC Microbiol. (2017) 17:48. doi: 10.1186/s12866-017-0951-4
23. Leyman B, Boyen F, Van Parys A, Verbrugghe E, Haesebrouck F, Pasmans F. Tackling the issue of environmental survival of live Salmonella Typhimurium vaccines: deletion of the lon gene. Res Vet Sci. (2012) 93:1168–72. doi: 10.1016/j.rvsc.2012.05.008
24. Han Y, Han X, Wang S, Meng Q, Zhang Y, Ding C, et al. The waaL gene is involved in lipopolysaccharide synthesis and plays a role on the bacterial pathogenesis of avian pathogenic Escherichia coli. Vet Microbiol. (2014) 172:486–91. doi: 10.1016/j.vetmic.2014.05.029
25. Zhang JF, Wei B, Cha SY, Shang K, Jang HK, Kang M. The use of embryonic chicken eggs as an alternative model to evaluate the virulence of Salmonella enterica serovar Gallinarum. PLoS ONE. (2020) 15:e0238630. doi: 10.1371/journal.pone.0238630
26. Reed LJ, Muench H. A simple method of estimating fifty per cent endpoints12. Am J Epidemiol. (1938) 27:493–7. doi: 10.1093/oxfordjournals.aje.a118408
27. Chen R, Shang H, Niu X, Huang J, Miao Y, Sha Z, et al. Establishment and evaluation of an indirect ELISA for detection of antibodies to goat Klebsiella pneumonia. BMC Vet Res. (2021) 17:107. doi: 10.1186/s12917-021-02820-1
28. Van Immerseel F, Studholme DJ, Eeckhaut V, Heyndrickx M, Dewulf J, Dewaele I, et al. Salmonella Gallinarum field isolates from laying hens are related to the vaccine strain SG9R. Vaccine. (2013) 31:4940–5. doi: 10.1016/j.vaccine.2013.08.033
29. Kong Q, Yang J, Liu Q, Alamuri P, Roland KL, Curtiss R 3rd. Effect of deletion of genes involved in lipopolysaccharide core and O-antigen synthesis on virulence and immunogenicity of Salmonella enterica serovar typhimurium. Infect Immun. (2011) 79:4227–39. doi: 10.1128/IAI.05398-11
30. Leyman B, Boyen F, Van Parys A, Verbrugghe E, Haesebrouck F, Pasmans F. Salmonella Typhimurium LPS mutations for use in vaccines allowing differentiation of infected and vaccinated pigs. Vaccine. (2011) 29:3679–85. doi: 10.1016/j.vaccine.2011.03.004
31. Geng S, Jiao X, Barrow P, Pan Z, Chen X. Virulence determinants of Salmonella Gallinarum biovar Pullorum identified by PCR signature-tagged mutagenesis and the spiC mutant as a candidate live attenuated vaccine. Vet Microbiol. (2014) 168:388–94. doi: 10.1016/j.vetmic.2013.11.024
32. Uchiya K, Barbieri MA, Funato K, Shah AH, Stahl PD, Groisman EA. A Salmonella virulence protein that inhibits cellular trafficking. EMBO J. (1999) 18:3924–33. doi: 10.1093/emboj/18.14.3924
33. Liu Q, Liu Q, Zhao X, Liu T, Yi J, Liang K, et al. Immunogenicity and cross-protective efficacy induced by outer membrane proteins from Salmonella Typhimurium mutants with truncated LPS in mice. Int J Mol Sci. (2016) 17:416. doi: 10.3390/ijms17030416
34. Jiao Y, Xia Z, Zhou X, Guo Y, Guo R, Kang X, et al. Signature-tagged mutagenesis screening revealed the role of lipopolysaccharide biosynthesis gene rfbH in smooth-to-rough transition in Salmonella Enteritidis. Microbiol Res. (2018) 212-213:75–9. doi: 10.1016/j.micres.2018.05.001
35. Chang J, Pang E, He H, Kwang J. Identification of novel attenuated Salmonella Enteritidis mutants. FEMS Immunol Med Microbiol. (2008) 53:26–34. doi: 10.1111/j.1574-695X.2008.00394.x
36. Park SI, Jeong JH, Choy HE, Rhee JH, Na HS, Lee TH, et al. Immune response induced by ppGpp-defective Salmonella enterica serovar Gallinarum in chickens. J Microbiol. (2010) 48:674–81. doi: 10.1007/s12275-010-0179-6
37. Cheng Z, Yin J, Kang X, Geng S, Hu M, Pan Z, et al. Safety and protective efficacy of a spiC and crp deletion mutant of Salmonella gallinarum as a live attenuated vaccine for fowl typhoid. Res Vet Sci. (2016) 107:50–4. doi: 10.1016/j.rvsc.2016.05.007
38. Bearson BL, Bearson SM, Kich JD, Lee IS. An rfaH mutant of Salmonella enterica Serovar Typhimurium is attenuated in swine and reduces intestinal colonization, fecal shedding, and disease severity due to virulent Salmonella Typhimurium. Front Vet Sci. (2014) 1:9. doi: 10.3389/fvets.2014.00009
Keywords: DIVA, high attenuation, waaJ, S. enterica ser. Gallinarum, double deletion, spiC
Citation: Zhang J-F, Shang K, Wei B, Lee Y-J, Park J-Y, Jang H-K, Cha S-Y and Kang M (2021) Evaluation of Safety and Protective Efficacy of a waaJ and spiC Double Deletion Korean Epidemic Strain of Salmonella enterica Serovar Gallinarum. Front. Vet. Sci. 8:756123. doi: 10.3389/fvets.2021.756123
Received: 10 August 2021; Accepted: 11 October 2021;
Published: 16 November 2021.
Edited by:
Constantinos S. Kyriakis, Auburn University, United StatesReviewed by:
Ahmed Ragab Elbestawy, Damanhour University, EgyptRaul G. Barletta, University of Nebraska-Lincoln, United States
Copyright © 2021 Zhang, Shang, Wei, Lee, Park, Jang, Cha and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Se-Yeoun Cha, seyeouncha@gmail.com; Min Kang, vet.minkang@gmail.com
†These authors have contributed equally to this work
 Jun-Feng Zhang
Jun-Feng Zhang Ke Shang
Ke Shang Bai Wei
Bai Wei Yea-Jin Lee
Yea-Jin Lee  Hyung-Kwan Jang
Hyung-Kwan Jang Se-Yeoun Cha
Se-Yeoun Cha Min Kang
Min Kang