First Isolation and Molecular Characterization of blaCTX-M-121-Producing Escherichia coli O157:H7 From Cattle in Xinjiang, China
- College of Veterinary Medicine, Xinjiang Agricultural University, Urumqi, China
The bovine Escherichia coli O157:H7 is a major foodborne pathogen causing severe bloody diarrhea, hemorrhagic colitis, and hemolytic uremic syndrome in humans. Cattle are recognized major reservoir and source of E. coli O157:H7. We investigated the antibiotic resistance, molecular profiles, and intrinsic relationship between 21 isolates of E. coli O157:H7 from cattle farms and slaughtering houses in Xinjiang. Using pulsed-field gel electrophoresis (PFGE) molecular typing, two types of PFGE were revealed through cluster analysis, including clusters I and II, with 66 and 100% similarity of PFGE spectra between 21 isolates. We also detected that 18 isolates (86%) carried at least one virulence gene, 16 isolates (76%) carried the eae gene, and 7 (33%) carried the stx1 + stx2 + eae + hly + tccp genes. Eighteen isolates were susceptible to antibiotics. Three isolates were resistant to antibiotics, and two were multidrug resistant. One of the two multidrug-resistant isolates detectably carried the blaCTX−M−121 gene. This is the first finding of the blaCTX−M−121 gene detected in E. coli O157:H7 isolated from cattle in Xinjiang. The blaCTX−M−121 gene is transferable between the bacterial strains via plasmid transmission. The results indicated that E. coli O157:H7 may have undergone clonal propagation in cattle population and cross-regional transmission in Xinjiang, China.
Introduction
Escherichia coli O157:H7 is a major foodborne pathogen that causes severe bloody diarrhea, hemorrhagic colitis (HC), and hemolytic uremic syndrome (HUS) in humans (1). E. coli O157:H7 was first recognized as a pathogen contributing to an outbreak of HC associated with hamburger consumption in 1982 (2). Since then, E. coli O157:H7 outbreaks have been reported in the United States, Canada, Japan, and China (3–6). E. coli O157:H7 has been reportedly detected in healthy cattle worldwide (7). The infected, asymptomatic cattle irregularly excrete E. coli O157:H7, resulting in contaminating food and water in the environment, as well as infecting humans and other animals (8). Cattle are recognized major reservoir and source of E. coli O157:H7.
Pathogenic virulence of E. coli O157:H7 is attributable to genes coding for Shiga toxin (Stx), the intestinal cell shedding site [locus of entericyte effacement (LEE)] virulence island, and the large plasmid pO157 (9). Stx, comprising Stx1 and Stx2, is able to induce cell necrosis and tissue lesions, and Stx2 is more potent than Stx1 (10, 11). The LEE region encodes a type III secretion system, and the secreted proteins E. coli secreted proteins (Esp) and translocated intimin receptor (Tir). Both Esp and Tir are required for intimate attachment and A/E lesion formation (12). The LEE region also encodes intimin, an outer membrane protein adhesin (Eae) that mediates the intimate attachment of bacteria to the host epithelial cell surface (13). In addition, Tir cytoskeleton-coupling protein (TccP) stimulates actin polymerization during the formation of A/E lesion (14). The large plasmid pO157 carries genes coding for type II secretion systems, such as hemolysin (Hly) and ToxB. All these virulence factors of E. coli O157:H7 reportedly regulate the adhesion of pathogenic bacteria to intestinal epithelial cells, causing the shedding of intestinal cells. These virulence genes have been used to identify bacterial strains isolated from various sources in epidemiological studies (6, 15, 16).
Antimicrobials have been the mainstay for the prevention and treatment of bacterial diseases in animals. However, their use is getting limited due to rising antibiotic resistance, which has become a serious problem worldwide, especially in developing countries where the quality, distribution, and use of antibiotics in human and veterinary medicine is not strictly regulated (15, 17). Extended-spectrum cephalosporins (ESCs), especially the third- and fourth-generation cephalosporins, are classified by the World Health Organization (WHO) to treat infections of multidrug-resistant Gram-negative bacteria (18). However, acquisition of genes encoding extended spectrum β-lactamases (ESBLs), especially CTX-M enzymes, by E. coli plays an important role in the resistance to ESCs (19). The genes encoding these enzymes, i.e., blaCTX−M genes, are usually located on transferable plasmids, which also carry resistance genes for other types of antimicrobials (i.e., fluoroquinolones, aminoglycosides). These plasmids mediate the spread of drug resistance between bacteria via conjugation (20). E. coli O157:H7 isolates collected from humans and animals have shown resistance to a variety of antibiotics; therefore, the emergence of multidrug resistant (MDR) E. coli O157:H7 has become a public health issue (21, 22).
The sustainability of cattle industry and food safety depend upon the effective prevention and control of bovine pathogenic microorganisms. Xinjiang is one of largest cattle-raising regions in China. To further assess the potential public health impact of E. coli O157:H7 in Xinjiang, we investigated the pathogenicity and antibiotic resistance of isolates collected from farms and slaughterhouses. We examined the intrinsic relationship among different isolates and assessed the potential dissemination of MDR profiles in vitro.
Materials and Methods
Sample Collection
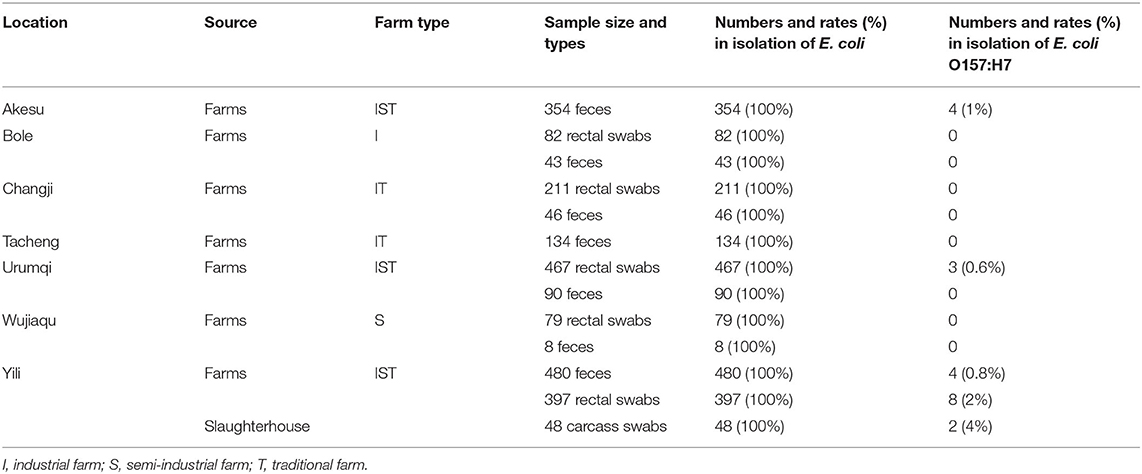
Total samples (n = 2,439) included 1,155 fresh feces, 1,236 rectal swabs, and 48 carcass swabs that were collected from 18 beef cattle and dairy farms (industrial, semi-industrial, and traditional farms, with a herd size of 200–8,000 cattle) and one slaughterhouse in the region of Akesu, Bole, Changji, Tacheng, Urumqi, Wujiaqu, and Yili in Xinjiang, China between October 2012 and March 2017. Samples were collected from Xinjiang brown cattle, Holstein cattle, Simmental cattle, and Angus cattle (1–7 years old, 400–800 kg body weight). Approximately 25 g of fecal samples were collected from each animal by rectal palpation or during defecation using disposable sleeve gloves and then placed in sterile Whirl-Pak bags (Nasco, Fort Atkinson, WI, USA). Rectal swabs were collected when rectal palpation is not applicable or no bowel movement is observed. Sterile cotton swabs (length, 150 mm; Copan Italia SpA) were used to collect mucus samples from the rectal anal junction. Sterile cotton swabs were also used to swab ~10 cm2 of carcass surface. All samples were transported in icebox to the laboratory and stored at 4°C until processed within 2 h.
Escherichia coli O157:H7 Isolation
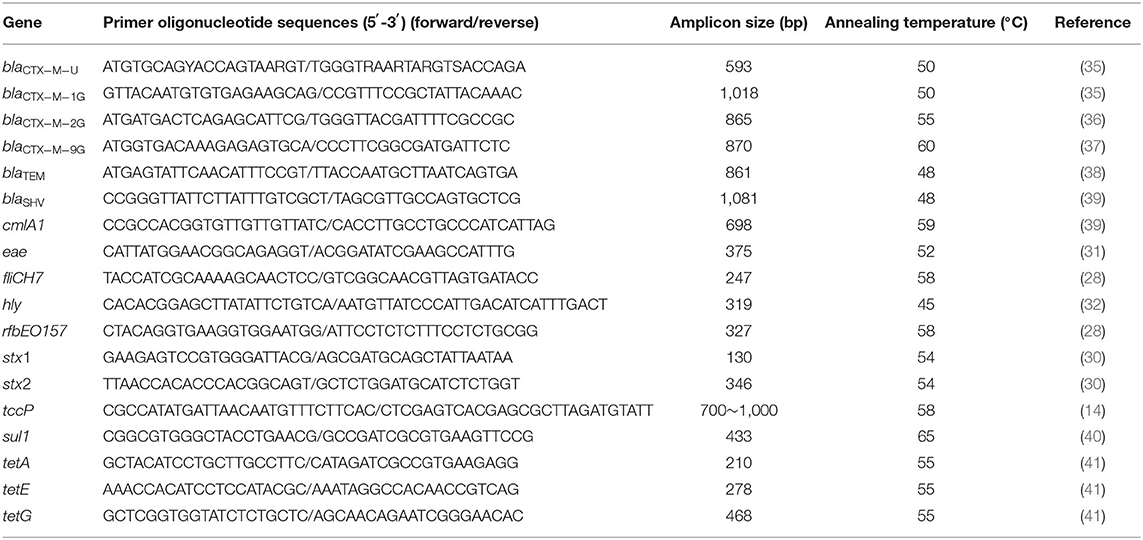
Selective enrichment was carried out according to the method reported by Mersha et al. (23) with minor modifications. One gram of each feces was aseptically added to 9 ml of modified tryptone soya broth containing 20 mg/L novobiocin (mTSB + n) (Hopebio, Qingdao, China) and incubated at 37°C for 16 h. To all the swab samples, 90 ml of mTSB + n was added and homogenized using a vortex mixer. After incubation for 16 h, all the samples were processed for immunomagnetic separation (IMS) using anti-E. coli O157 Dynabeads (Dynal, Invitrogen, USA) as follows. One microliters of the enriched broth culture was put in a sterile screw cupped Eppendorf tubes to which 20 μl of anti-O157:H7 immunomagnetic beads was added, followed by shaking at ambient temperature for 30 min. The tubes were then kept inside the manual magnetic particle concentrator. The beads were washed thrice using 300 μl phosphate-buffered saline (PBS) buffer for each wash. Finally, 100 μl of PBS was added in each tube and mixed gently (24). Fifty microliters of the mixture was streaked onto Sorbitol MacConkey agar containing 0.05 mg/L cefixime and 2.5 mg/L potassium tellurite (CT-SMAC) (Hopebio, Qingdao, China) and incubated at 37°C for 20–24 h to develop colonies. Pale-colored colonies were purified by repeated streak plating until a uniform colony morphology was obtained (25). One or more of the colonies were individually selected as presumptive E. coli O157 per sample. E. coli CICC 21530 (O157:H7, stx1 + stx2 + eae + hly + tccp) (26, 27) and ATCC 25922 strains were used as positive and negative controls, respectively. Two genes (rfbEO157 and fliCH7) were used to identify E. coli O157:H7 (28). Pink colonies (suspected the general E. coli) were purified by restreaking on McConkey agar and confirmed by PCR method as described by Teichmann et al. (29) (Table 1). The PCR amplicons (10 μl) were subjected to electrophoresis on a 1.2% agarose gel in 1× Tris–acetate–EDTA (TAE) buffer at 115 V for 30 min and stained with SYBR Green (Fermentas, Germany). The positive isolates were each inoculated in separate TSB and incubated overnight at 37°C, from which glycerol stocks were made and then stored at −80°C for further analysis.
Analysis of Virulence Genes
Genomic DNA contents were extracted from 21 E. coli O157:H7 isolates as confirmed by PCR serotyping. In brief, 3–5 colonies were individually suspended in 200 μl of sterile distilled water. Bacterial suspensions were then heated at 95°C for 10 min centrifugation at 13,400 × g for 10 min to obtain the supernatant containing the template DNA and were transferred into 1.5-ml Eppendorf tubes without nuclease and stored at −20°C.
A multiplex PCR procedure was used to detect the stx1 and stx2 genes (30), and a single PCR procedure was used to detect the eae (31), hly (32), and tccp (14) genes. The primers, conditions, and references cited are listed in Table 1. E. coli CICC 21530 was used as a positive control for all the five virulence genes, while ATCC 25922 was used as a negative control. Amplification of the targeted gene was carried out using EX Taq (TaKaRa, Dalian, China) with the following PCR program: 94°C for 4 min, 30 cycles of denaturation at 94°C for 30 s, annealing at 54°C for 30 s, and extension at 72°C for 30 s, with a final extension at 72°C for 10 min. The annealing temperature was adjusted according to the primer Tm value (Table 1).
Antimicrobial Susceptibility Tests
Antibiotic susceptibility was tested using the Kirby–Bauer disk diffusion technique. Antibiotic disks of 6 mm in diameter obtained from OXOID, UK, containing ampicillin (AMP, 10 μg/disk), piperacillin (PIP, 100 μg/disk), cefotaxime (CTX, 30 μg/disk), ceftazidime (CAZ, 30 μg/disk), cefepime (FEP, 30 μg/disk), aztreonam (ATM, 30 μg/disk), ampicillin-sulbactam (SAM, 10/10 μg/disk), piperacillin-tazobactam (TZP, 100/10 μg/disk), amoxicillin-clavulanic acid (AMC, 20/10 μg/disk), gentamicin (GEN, 10 μg/disk), amikacin (AMI, 30 μg/disk), streptomycin (STR, 10 μg/disk), cotrimoxazole (SXT, 25 μg/disk), chloramphenicol (CHL, 30 μg/disk), levofloxacin (LEV, 5 μg/disk), ciprofloxacin (CIP, 5 μg/disk), tetracycline (TET, 30 μg/disk), and polymyxin B (PB, 300 U/disk) (20). E. coli ATCC25922, purchased from China Center of Industrial Culture Collection (CICC), was used as a quality control strain in the susceptibility tests. The ESBL-producing isolates were determined by double-disk synergy tests according to CLSI (33). Isolates shown to be resistant to at least three different classes of antimicrobial agents were determined to be multidrug resistant (MDR) (34).
Detection of Antibiotic Resistance Genes
The following resistance determinants were investigated by PCR: blaCTX−M [the CTX-M-type genes were detected using universal primers blaCTX−M−U (35), and the entire CTX-M-type genes were amplified using the primers blaCTX−M−1G (35), blaCTX−M−2G (36), or blaCTX−M−9G (37)], blaTEM (38), and blaSHV (39), which encode β-lactamases, chloramphenicol efflux pumps [cmlA1(39)], sulfonamide resistance gene [sul1 (40)], and the tetA (41), tetE (41), and tetG (41) tetracycline efflux pumps. blaTEM and blaSHV genes were amplified by double PCR; tetA, tetE, and tetG genes were amplified by triplex PCR, while other resistant genes were amplified by single PCR. Primers used for the different genes are listed in (Table 1). The PCR products were sent to Sangon Biotech Co., Ltd. (Shanghai, China) for sequence determination. The DNA sequences and deduced amino acid sequences were compared with sequences reported in GenBank to confirm the subtypes of the β-lactamase gene.
Conjugation Experiments and Plasmid Analysis
Sodium azide-resistant E. coli J53 was used as a recipient and conjugated to a blaCTX-M-producing isolate by filtration. Transconjugants were selected on Mac Conkey agar containing cefotaxime or ceftazidime (4 μg/ml) and sodium azide (200 μg/ml). ESBL and antibiotic susceptibility was also tested in selected transconjugants, and the presence of bla genes was determined using PCR as described above. The resistance plasmids carried by transconjugants were typed using PCR-based replicon typing (42).
Epidemiological Typing
All the 21 E. coli O157:H7 isolates were characterized by pulsed field gel electrophoresis (PFGE) using the CHEF-MAP-PER System (Bio-Rad Laboratories, Hercules, CA, USA) as described by Gautom (43). Briefly, chromosomal DNA of E. coli O157:H7 isolates was isolated, and the inserts were digested with XbaI (TaKaRa Dalian, China) for 16 h at 37°C. The electrophoresis was performed at 6.0 V/cm for 18.5 h with an angle of 120 at 14°C. The pulse time was increased from 0.5 to 60 s. The Salmonella serotype Braenderup H9812 (ATCC BAA-664) was chosen as the molecular weight marker. Gels were then stained in ethidium bromide (1.0 mg/L). Isolates were considered to belong to the same PFGE cluster when the similarity index was >80% (44).
Results
Isolation and Presence of Virulence Genes
To investigate the virulence and antibiotic resistance of E. coli O157:H7, we collected 2,439 samples from farms and slaughterhouses in Xinjiang regions (Table 2). We successfully isolated E. coli clones from all the feces (100%), rectal swabs (100%), and carcass swabs (100%). Studying these E. coli isolates, we detected that 21 isolates were the E. coli O157:H7 strain (19 isolates collected from cattle farms and 2 isolates obtained from one slaughterhouse). As shown in Table 2, the isolation rates of E. coli O157:H7 in feces, rectal swabs, and carcass swabs were 0.7% (8/1155), 1% (11/1236), and 4% (2/48), respectively.
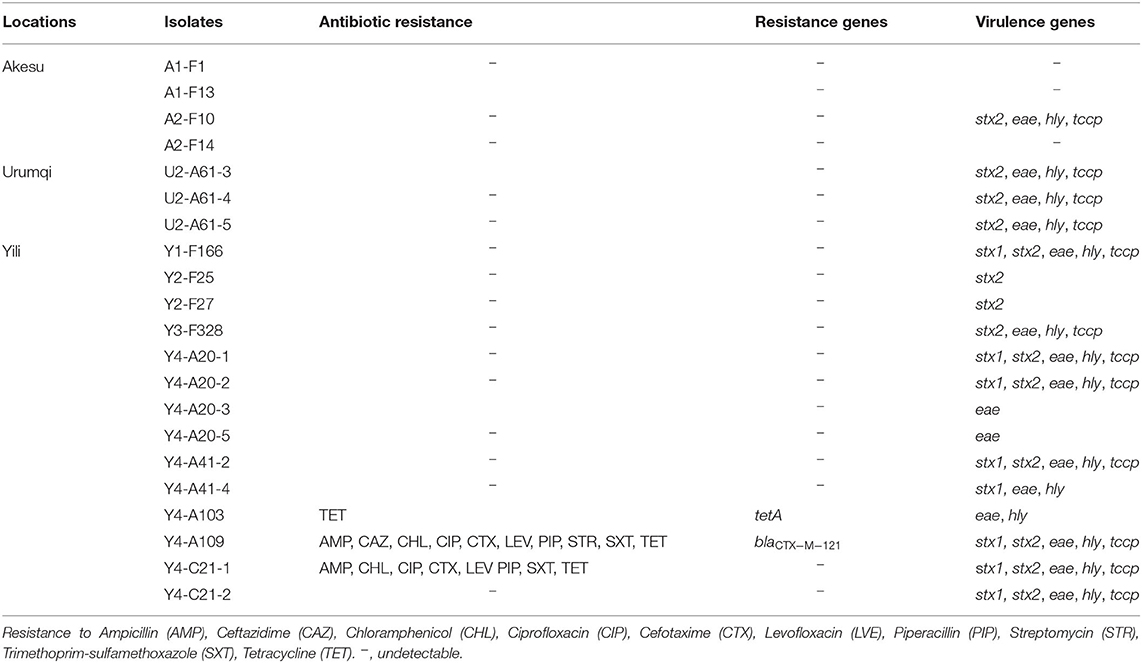
Of the 21 E. coli O157:H7 isolates, 18 (86%) carried at least one virulence gene and 3 (14%) did not carry any (Table 3). Using PCR technique, we detected that seven (33%) possessed only stx2, seven (33%) isolates were positive for stx1 and stx2, and only one (5%) isolate had just stx1 gene. The eae gene and hly gene were detected in 16 (76%) and 14 (67%) E. coli O157:H7 isolates, respectively. Tccp in combination with hly and eae was found in 12 (57%) isolates. In total, six diverse virulence profiles were determined, including stx1/stx2/eae/hly/tccp (seven isolates), stx2/eae/hly/tccp (five isolates), stx2 (two isolates), eae (two isolates), stx1/eae/hly (one isolate), and eae/hly (one isolate) (Table 3).
Antibiotic Resistance Spectrum and Distribution of Antibiotic Resistance Genes
Studying the resistance of isolated E. coli O157:H7 to antibiotics, we detected that one isolate (Y4-A103) was resistant to tetracycline and carried the tetA gene, which encodes a tetracycline efflux pump. Y4-A109 and Y4-C21-1 were MDR isolates with the resistant patterns: AMP/CAZ/CHL/CIP/CTX/LEV/PIP/SXT/TET (Y4-A109) and AMP/CHL/CIP/CTX/LEV/PIP/SXT/TET (Y4-C21-1). In particular, the Y4-A109 was an ESBL-producing isolate carrying the blaCTX−M−121 gene (Table 3). Although both Y4-A109 and Y4-C21-1 isolates were resistant to chloramphenicol and sulfonamides, the cmlAI and sulI genes were not detectable in these isolates, indicating other genes involved in the resistance to chloramphenicol and sulfonamides. In addition, those two MDR isolates (Y4-A109 and Y4-C21-1) simultaneously harbored five virulence genes (stx1/stx2/eae/hly/tccp).
Transferability of blaCTX-M Genes and Plasmid Replicon Typing
Studying transferability, we detected that the blaCTX−M−121 gene of the E. coli O157:H7 Y4-A109 isolate was transferable to the recipient strain azide-resistant E. coli J53 by conjugation at a frequency of approximately 10−6 per donor cell after coincubation of bacteria. We also determined that the resistance of Y4-A109 to ampicillin, cefotaxime, ceftazidime, cotrimoxazole, and tetracycline was also transferable to the recipient. However, plasmid replicon carrying these resistance genes in Y4-A109 remained to be determined.
Epidemiological Typing
Overall, the genetic relatedness ranged from 66 to 100% among the 21 isolates (Figure 1). Furthermore, the studied isolates shared ≤80% genetic similarity to the reference strain 21530. Seventeen of the 21 isolates were grouped into two clusters using >80% similarity of the Dice coefficient. Isolates Y4-A20-1 and Y4-A41-2 (cluster II) were simultaneously isolated from different cattle at Yili in 2015 but shared identical pattern of PFGE, virulence genes, and antibiotic susceptibility (Figure 1). This suggests that potential pathogen transmission might occur from animals to animals within the farm. In addition, three drug-resistant isolates were all identified from Yili. However, they were genetically distantly related (<71% similarity of the Dice coefficient). Noticeably, the PFGE profiles of two isolates from the slaughterhouse were identical to those from the farms. However, Y4-C21-2 carried stx1 gene, which was absent from its identical farm isolate, whereas Y4-C21-1 appeared to be resistant to eight drugs tested, which were not observed in its identical counterparts (Figure 1).
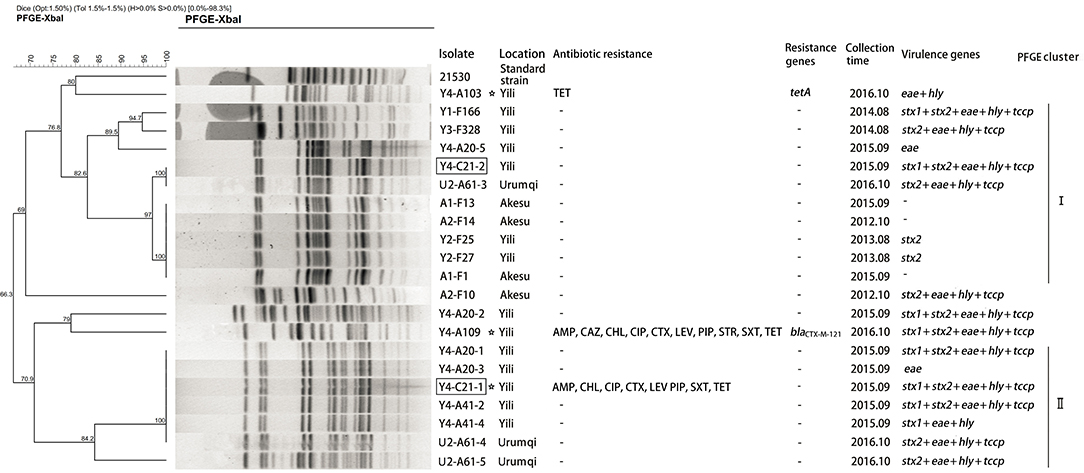
Figure 1. Dendrogram of Xbal pulsed-field gel electrophoresis profiles of E. coli O157:H7 isolates. The box indicates that the strains isolated from slaughterhouse. The asterisk represents the resistant strains.
Discussion
In this communication, we reported, for the first time, that the blaCTX−M−121 gene was detected in E. coli O157:H7 isolated from cattle in Xinjiang. The blaCTX−M−121 gene belongs to the blaCTX−M−9 group. Rao et al. (45) reported the blaCTX−M−121 gene detected in two E. coli isolates collected from farm ducks in China. Zhou et al. (46) identified the blaCTX−M−121 gene in one E. coli isolated from healthy people in Guangdong Province. Jin (47) reported the blaCTX−M−121 gene in chicken E. coli isolated from Guangdong Province. The cephalosporins are used to treat infectious disease such as bovine respiratory infection and mastitis, which may promote production and dissemination of β-lactamase genes (48).
Besides the transferable blaCTX−M−121 gene between the bacterial strains via plasmid transmission, we also detected a wide spectrum of virulence genes, including the stx1, stx2, eae, hly, and tccp genes in E. coli O157:H7 isolates, which were consistent with the virulence gene types of E. coli O157:H7 from bovine in Jiangsu Province (49). We also detected three isolates of E. coli O157:H7 lacking any of these virulence genes, which was similar to the bovine E. coli O157:H7 isolates reported by Akomoneh et al. (50). E. coli O157:H7 isolates possessing only eae or stx2 gene were similar to the isolates obtained from cattle in USA and milk in Nigeria, respectively (51, 52). The attendance of the eae gene in O157:H7 STEC (Shiga toxin-producing E. coli) isolates resulted in the formation of a highly virulent subpathotype, Enterohemorrhage E. coli (EHEC) (53), which was observed in two MDR isolates in the present survey.
In addition, the tet resistance gene has been increasingly detected in bovine O157 and non-O157 STEC isolates worldwide (54–56). Our studies also revealed, for the first time, the presence of the tetA gene detected in bovine E. coli O157:H7 in Xinjiang. Horizontal gene transfer plays a key role in bacterial evolution and transmission of antibiotic resistance genes (57). Resistance traits located in genetic mobile elements, such as plasmids, transposons, and integrons, can be transferred to different strains or bacterial species (58, 59). It is conceivable that virulence gene and drug-resistance gene are carried by the same genetic element; cotransfer may occur under the selection of antibiotics to result in stable virulence clones, thereby leading to production of drug-resistant pathogenic bacteria and persistent bacterial infection in humans and food animals.
We found that the isolates obtained in the same geographical location at the same time had similar PFGE patterns and vice versa, indicating that clonal propagation in cattle population and cross-regional transmission. E. coli O157:H7 with identical PFGE pattern (100% similarity) carry different virulence genes and different drug resistance phenotypes, suggesting that the virulence and drug resistance carried by E. coli O157:H7 may be acquired or lost during the evolution and transfer of the same cluster of strains. The β-lactam-resistant E. coli O157:H7 may give β-lactam resistance to other pathogenic enterobacteria via plasmid-mediated conjugation, thereby posing potential challenges in the management of their associated infectious disease in cattle (60).
E. coli O157:H7 was prevalent in 2–15% population of cattle and other animals in China (47). Our results revealed that the overall isolation rate at ~0.9% (21 of 2,439 samples) of E. coli O157:H7 and the blaCTX−M gene detected in 1 of 21 isolates indicated that the transmission of the blaCTX−M gene in E. coli O157:H7 population was at an early stage in Xinjiang. Thus, it is important to not only continuously monitor but also identify methods to intervene in the transmission of blaCTX−M genotypes to E. coli and other bacterial strains, thereby minimizing potential dissemination of β-lactam resistance from the cattle production to their surrounding environment.
Data Availability Statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Ethics Statement
The animal study was reviewed and approved by the Animal Care and Use Committee of Xinjiang Agricultural University. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author Contributions
ZS and PT conceived and designed the experiments. LZ, MZ, DW, and KM performed the experiments. YZ and YL analyzed the date. PT, LX, and JX contributed to the writing of the manuscript. All authors read and approved the article.
Funding
The project was funded by the National Natural Science Foundation of China (31960695 and 31560485), Natural Science Foundation of Xinjiang Province (2019D01A51), Prior Period Project of Xinjiang Agricultural University (XJAU201703), and by Postdoctoral Science Foundation of Xinjiang Agricultural University and Xinjiang Uygur Autonomous Region High-Level Talent Introduction.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The PFGE analysis was supported by State Key Laboratory for Infectious Disease Prevention and Control National Institute for Communicable Disease Control and Prevention, China CDC for her support. We also thank professor Hwa-Chain R. Wang for his instructive suggestions in manuscript preparation. This manuscript has been released as a pre-print at (61).
References
1. Wang LK, Qu X, Li Z, Cao X, Wang Z, Li Y, et al. Use of bacteriophages to control Escherichia coli O157:H7 in domestic ruminants, meat products, and fruits and vegetables. Foodborne Pathog. Dis. (2017) 14:483–93. doi: 10.1089/fpd.2016.2266
2. Riley LW, Remis RS, Helgerson SD, McGee HB, Wells JG, Davis BR, et al. Hemorrhagic colitis associated with a rare Escherichia coli serotype. N Engl J Med. (1983) 308:681–5. doi: 10.1056/NEJM198303243081203
3. Bosllevac JM, Koohmaraie M. Prevalence and characterization of non-O157 Shiga toxin-producing Escherichia coli isolates from commercial ground beef in the United States. Appl Environ Microbiol. (2011) 77:2103–12. doi: 10.1128/AEM.02833-10
4. Orr P, Lorencz B, Brown R, Kielly Tan B, Holton D, et al. An outbreak of diarrhea due to verotoxin-producing Escherichia coli in the Canadian Northwest Territories. Scand J Infect Dis. (1994) 26:675–84. doi: 10.3109/00365549409008635
5. Ostroff SM, Kobayashi JM, Lewis JH. Infections with Escherichia coli O157:H7 in Washington State. The first year of statewide disease surveillance. JAMA. (1989) 262:355–9. doi: 10.1001/jama.262.3.355
6. Zhang J, Xia S, Shen G, Chen Z, Huang P, Fu B, et al. A study on acute renal failure after an outbreak of diarrhea in Suixian county, Henan province. Zhonghua Liu Xing Bing Xue Za Zhi. (2002) 23:105–7. doi: 10.3760/j.issn:0254-6450.2002.02.008
7. Munns KD, Selinger LB, Stanford K, Guan L, Callaway TR, McAllister TA. Perspectives on super-shedding of Escherichia coli O157:H7 by cattle. Foodborne Pathog Dis. (2015) 12:89–103. doi: 10.1089/fpd.2014.1829
8. Menge C. The role of Escherichia coli Shiga toxins in STEC colonization of cattle. Toxins (Basel). (2020) 12:607. doi: 10.3390/toxins12090607
9. Fu SS, Bai XN, Fan RY, Xu YM, Xu XB, Xiong YW. Molecular characteristics of human-derived non-O157 Shiga toxin-producing Escherichia coli strains isolated in five regions of China. Chin J Microbiol Immunol. (2017) 37:213–8. doi: 10.3760/cma.j.issn.0254-5101.2017.03.009
10. Tarr PI, Gordon CA, Chandler WL. Shiga-toxin-producing Escherichia coli and haemolytic uraemic syndrome. Lancet. (2005) 365:1073–86. doi: 10.1016/S0140-6736(05)74232-X
11. Schüller S, Frankel G, Phillips AD. Interaction of Shiga toxin from Escherichia coli with human intestinal epithelial cell lines and explants: Stx2 induces epithelial damage in organ culture. Cell Microbiol. (2004) 6:289–301. doi: 10.1046/j.1462-5822.2004.00370.x
12. Ghaem-Maghami M, Simmons CP, Daniell S, Pizza M, Lewis D, Frankel G, et al. Intimin-specific immune responses prevent bacterial colonization by the attaching-effacing pathogen Citrobacter rodentium. Infect Immun. (2001) 69:5597–605. doi: 10.1128/IAI.69.9.5597-5605.2001
13. Jores J, Rumer L, Wieler LH. Impact of the locus of enterocyte effacement pathogenicity island on the evolution of pathogenic Escherichia coli. Int J Med Microb. (2004) 294:103–13. doi: 10.1016/j.ijmm.2004.06.024
14. Ji XW, Liao YL, Zhu YF, Wang HG, Gu L, Gu J, et al. Multilocus sequence typing and virulence factors analysis of Escherichia coli O157 strains in China. J Microbiol. (2010) 48:849–55. doi: 10.1007/s12275-010-0132-8
15. Adamu MS, Ugochukwu ICI, Idoko SI, Kwabugge YA, Abubakar NS, Ameh JA. Virulent gene profile and antibiotic susceptibility pattern of Shiga toxin-producing Escherichia coli (STEC) from cattle and camels in Maiduguri, North-Eastern Nigeria. Trop Anim Health Prod. (2018) 50:1327–41. doi: 10.1007/s11250-018-1565-z
16. Stevens MP, Roe AJ, Vlisidou I, Van Diemen PM, La Ragione RM, Best A, et al. Mutation of toxB and truncated version of the efa-1 gene in Escherichia coli O157: H7 influences the expression and secretion of locus of enterocyte effacement-encoded proteins but not intestinal colonization in calves or sheep. Infect Immun. (2004) 72:5402–11. doi: 10.1128/IAI.72.9.5402-5411.2004
17. Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. (2010) 10:597–602. doi: 10.1016/S1473-3099(10)70143-2
18. Collignon P, Powers JH, Chiller TM, Aidara-Kane A, Aarestrup FM. World Health Organization ranking of antimicrobials according to their importance in human medicine: a critical step for developing risk management strategies for the use of antimicrobials in food production animals. Clin Infect Dis. (2009) 49:132–41. doi: 10.1086/599374
19. Liebana E, Carattoli A, Coque TM, Hasman H, Magiorakos AP, Mevius D, et al. Public health risks of enterobacterial isolates producing extended-spectrum β-lactamases or AmpC β-lactamases in food and food-producing animals: an EU perspective of epidemiology, analytical methods, risk factors, and control options. Clin Infect Dis. (2013) 56:1030–7. doi: 10.1093/cid/cis1043
20. Ma JY, Liu JH, Lv LC, Zong ZY, Sun Y, Zheng HQ, et al. Characterization of extended-spectrum β-lactamase genes found among Escherichia coli isolates from duck and environmental samples obtained on a duck farm. Appl Environ Microbiol. (2012) 78:3668–73. doi: 10.1128/AEM.07507-11
21. Fontcuberta M, Planell R, Torrents A, Sabaté S, Gonzalez R, Ramoneda M, et al. Characterization of Shiga toxin-producing Escherichia coli O157 isolates from bovine carcasses. J Food Prot. (2016) 79:1418–23. doi: 10.4315/0362-028X.JFP-15-508
22. Mir RA Kudva IT. Antibiotic-resistant Shiga toxin-producing Escherichia coli: an overview of prevalence and intervention strategies. Zoonoses Public Health. (2019) 66:1–13. doi: 10.1111/zph.12533
23. Mersha G, Asrat D, Zewde BM, Kyule M. Occurrence of Escherichia coli O157:H7 in faeces, skin and carcasses from sheep and goats in Ethiopia. Lett Appl Microbiol. (2010) 50:71–6. doi: 10.1111/j.1472-765X.2009.02757.x
24. NPH (National Public Health Service for Wales). Detection of Escherichia coli O157 by Automated Immunomagnetic Separation. Standard Method. Issued by Standards Unit, Evaluations and standards laboratory with the regional food, Water and Environmental Coordinators Forum. Wales: SOPs from the Health Protection Agency (2006). p. 1–15.
25. Wang Y, Ametaj BN, Ambrose DJ, Gänzle MG. Characterisation of the bacterial microbiota of the vagina of dairy cows and isolation of pediocin-producing Pediococcus acidilactici. BMC Microbiol. (2013) 13:19. doi: 10.1186/1471-2180-13-19
26. Yu LL, Ji SS, Yu JL, FU WJ, Zhang L, Li JL, et al. Effects of salt stress on the survival and virulence genes expression of Escherichia coli O157:H7. Food Science. (2020) 41:95–101. doi: 10.7506/spkx1002-6630-20190613-149
27. Zhang T, Su ZQ, Xin LN, Wang YM, Zhou YP, Liu YY, et al. Isolation and identification of E. coli O157:H7 strain from cattle and detection of its virulence genes. Chin J Zoonoses. (2015) 31:1136–41. doi: 10.3969/j.issn.1002-2694.2015.12.010
28. Wang G, Clifford GC, Frank GR. Detection in Escherichia coli of the genes encoding the major virulence factors, the genes defining the O157:H7 Serotype, and components of the type 2 shiga toxin family by multiplex PCR. J Clin Microbiol. (2002) 40:3613–9. doi: 10.1128/JCM.40.10.3613-3619.2002
29. Teichmann A, Agra HN, Nunes LD, Rocha MP, Renner JDP, Possuelo LG, et al. Antibiotic resistance and detection of the sul2 gene in urinary isolates of Escherichia coli in patients from Brazil. J Infect Dev Ctries. (2014) 8:39–43. doi: 10.3855/jidc.3380
30. Pollard DR, Johnson WM, Lior H, Tyler SD, Rozee KR. Rapid and specific detection of verotoxin genes in Escherichia coli by the polymerase chain reaction. J Clin Microbiol. (1990) 28:540–5. doi: 10.1128/JCM.28.3.540-545.1990
31. Bai JF, Shi XR, Nagaraja TG. A multiplex PCR procedure for the detection of six major virulence genes in Escherichia coli O157:H7. J Microbiol Methods. (2010) 82:85–9. doi: 10.1016/j.mimet.2010.05.003
32. Akashi S, Joh K, Tsuji A, Ito H, Hoshi H, Hayakawa T, et al. A severe outbreak of haemorrhagic colitis and haemolytic uraemic syndrome associated with Escherichia coli O157:H7 in Japan. Eur J Pediatr. (1994) 153:650–5. doi: 10.1007/s004310050206
33. Clinical Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 29th edn. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2019).
34. Magiorakos AP, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
35. Pagani L, Dell'Amico E, Migliavacca R, D'Andrea MM, Giacobone E, Amicosante G, et al. Multiple CTX-M-type extended-spectrum β-lactamases in nosocomial isolates of enterobacteriaceae from a hospital in northern Italy. J Clin Microbiol. (2003) 41:4264–9. doi: 10.1128/JCM.41.9.4264-4269.2003
36. Saladin M, Cao VT, Lambert T, Donay JL, Herrmann JL, Ould-Hocine Z, et al. Diversity of CTX-M beta-lactamases and their promoter regions from Enterobacteriaceae isolated in three Parisian hospitals. FEMS Microbiol Lett. (2002) 209:161–8. doi: 10.1111/j.1574-6968.2002.tb11126.x
37. Eckert C, Gautier V, Saladin-Allard M, Hidri N, Verdet C, Ould-Hocine Z, et al. Dissemination of CTX-M-type beta-lactamases among clinical isolates of Enterobacteriaceae in Paris, France. Antimicrob Agents Chemother. (2004) 48:1249–55. doi: 10.1128/AAC.48.4.1249-1255.2004
38. Lin CF, Hsu SK, Chen CH, Huang JR, Lo HH. Genotypic detection and molecular epidemiology of extended-spectrum β-lactamase-producing Escherichia coli and Klebsiella pneumoniae in a regional hospital in central Taiwan. J Med Microbiol. (2010) 59:665–71. doi: 10.1099/jmm.0.015818-0
39. Keyes K, Hudson C, Maurer JJ, Thayer S, White DG, Lee MD. Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob Agents Chemother. (2000) 44:421–4. doi: 10.1128/AAC.44.2.421-424.2000
40. Kerrn MB, Klemmensen T, Frimodt-Møller N, Espersen F. Susceptibility of Danish Escherichia coli strains isolated from urinary tract infections and bacter-aemia, and distribution of sul genes conferring sulphona-mide resistance. J Antimicrob Chemother. (2002) 50:513–6. doi: 10.1093/jac/dkf164
41. Ng LK, Martin I, Alfa M, Mulvey M. Multiplex PCR for the detection of tetracycline resistant genes. Mol Cell Probes. (2001) 15:209–15. doi: 10.1006/mcpr.2001.0363
42. Carattoli A, Bertini A, Villa L, Falbo V, Hopkins KL, Threlfall EJ. Identification of plasmids by PCR-based replicon typing. J Microbiol Methods. (2005) 63:219–28. doi: 10.1016/j.mimet.2005.03.018
43. Gautom RK. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. (1997) 35:2977–80. doi: 10.1128/JCM.35.11.2977-2980.1997
44. Szijártó V, Pal T, Nagy G, Nagy E, Ghazawi A, al-Haj M, et al. The rapidly emerging ESBL-producing Escherichia coli O25-ST131 clone carries LPS core synthesis genes of the K-12 type. FEMS Microbiol Lett. (2012) 332:131–6. doi: 10.1111/j.1574-6968.2012.02585.x
45. Rao LL, Lv LC, Zeng ZL, Chen S, He DD, Chen XJ, et al. Increasing prevalence of extended-spectrum cephalosporin-resistant Escherichia coli in food animals and the diversity of CTX-M genotypes during 2003-2012. Vet Microbiol. (2014) 172:534–41. doi: 10.1016/j.vetmic.2014.06.013
46. Zhou Y, Wu XW, Zhang J, Tao X, Deng ZA, Hu YS. High prevalence of CTX-M beta-lactamases in Enterobacteriaceae from healthy individuals in Guangzhou, China. Microb Drug Resist. (2015) 21:398–403. doi: 10.1089/mdr.2014.0201
47. Jin C. Prevalence of Antibiotic Resistance β-Lactamas among Escherichia coli Isolated from Pigs and Chickens at Slaughter. Master Thesis, South China Agricultural University (2016) [in Chinese].
48. Mir RA, Weppelmann TA, Teng L, Kirpich A, Elzo MA, Driver JD, et al. Colonization dynamics of cefotaxime resistant bacteria in beef cattle raised without cephalosporin antibiotics. Front Microbiol. (2018) 9:500. doi: 10.3389/fmicb.2018.00500
49. Ye Q, Zhang XH, He KW, Fei RM, Zhao PD, Luan XT. Isolation of Escherichia coli O157:H7 from cattle and detection of its virulence genes. Chin J Vet Sci. (2012) 32:1148–53. doi: 10.16303/j.cnki.1005-4545.2012.08.007
50. Akomoneh EA, Esemu SN, Kfusi AJ, Ndip RN, Ndip LM. Prevalence and virulence gene profiles of Escherichia coli O157 from cattle slaughtered in Buea, Cameroon. PLoS One. (2020) 15:e0235583. doi: 10.1371/journal.pone.0235583
51. Yang H, Carlson B, Geornaras I, Woerner D, Sofos J, Belk K. Draft genome sequence of Shiga toxin-negative Escherichia coli O157:H7 strain C1-057, isolated from feedlot cattle. Genome Announc. (2016) 4:e00049–16. doi: 10.1128/genomeA.00049-16
52. Ivbade A, Ojo OE, Dipeolu MA. Shiga toxin-producing Escherichia coli O157:H7 in milk and milk products in Ogun State, Nigeria. Vet Ital. (2014) 50:185–91. doi: 10.12834/VetIt.129.2187.1
53. Jajarmi M, Badouei MA, Fooladi AAI, Ghanbarpour R, Ahmadi A. Pathogenic potential of Shiga toxin-producing Escherichia coli strains of caprine origin: virulence genes, Shiga toxin subtypes, phylogenetic background and clonal relatedness. BMC Vet Res. (2018) 14:97. doi: 10.1186/s12917-018-1407-2
54. Aslam M, Stanford K, McAllister TA. Characterization of antimicrobial resistance and seasonal prevalence of Escherichia coli O157:H7 recovered from commercial feedlots in Alberta, Canada. Lett Appl Microbiol. (2010) 50:320–6. doi: 10.1111/j.1472-765X.2010.02798.x
55. Hu B, Kou ZQ, Shao CC, Yin HY, Liu ZD, Xu XH, et al. Characteristics and drug resistance of non-O157 Shiga toxin-producing E. coli in animal feces, from Shandong Province. Zhonghua Yu Fang Yi Xue Za Zhi. (2018) 52:271–6. doi: 10.3760/cma.j.issn.0253-9624.2018.03.010
56. Kennedy CA, Fanning S, Karczmarczyk M, Byrne B, Monaghan Á, Bolton D, et al. Characterizing the multidrug resistance of non-O157 Shiga toxin-producing Escherichia coli isolates from cattle farms and abattoirs. Microb Drug Resist. (2017) 23:781–90. doi: 10.1089/mdr.2016.0082
57. Liu Y, Lu T, Zhang LG, Zhao ML, Sun DD, Sun SC, et al. Isolation and identification of Escherichia coli O157:H7 from cattle and analysis of resistant gene. J Northeast Agric Univ. (2014) 45:83–8. doi: 10.19720/j.cnki.issn.1005-9369.2014.11.013
58. Barlow M. What antimicrobial resistance has taught us about horizontal gene transfer. Methods Mol Biol. (2009) 532:397–411. doi: 10.1007/978-1-60327-853-9_23
59. Dionisio F, Zilhão R, Gama JA. Interactions between plasmids and other mobile genetic elements affect their transmission and persistence. Plasmid. (2019) 102:29–36. doi: 10.1016/j.plasmid.2019.01.003
60. Kawahara R, Seto K, Taguchi M, Nakajima C, Kumeda Y, Suzuki Y. Characterization of third-generation-cephalosporin-resistant Shiga Toxin-producing strains of Escherichia coli O157:H7 in Japan. J Clin Microbiol. (2015) 53:3035–8. doi: 10.1128/JCM.01263-15
Keywords: E. coli O157:H7, virulence genes, antibiotic resistance, PFGE, bovine
Citation: Su Z, Tong P, Zhang L, Zhang M, Wang D, Ma K, Zhang Y, Liu Y, Xia L and Xie J (2021) First Isolation and Molecular Characterization of blaCTX-M-121-Producing Escherichia coli O157:H7 From Cattle in Xinjiang, China. Front. Vet. Sci. 8:574801. doi: 10.3389/fvets.2021.574801
Received: 25 June 2020; Accepted: 12 April 2021;
Published: 25 May 2021.
Edited by:
Dongyan Niu, University of Calgary, CanadaReviewed by:
Gerardo M. Nava, Universidad Autónoma de Querétaro, MexicoJames L. Bono, United States Department of Agriculture, United States
Xiaonan Zhao, Shandong Academy of Agricultural Sciences, China
Copyright © 2021 Su, Tong, Zhang, Zhang, Wang, Ma, Zhang, Liu, Xia and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lining Xia, xln750530@163.com; Jinxin Xie, xiejinxin198683@163.com
†These authors have contributed equally to this work
 Zhanqiang Su†
Zhanqiang Su†  Panpan Tong
Panpan Tong