- 1Department of Epidemiology and Biostatistics, College of Public Health, Zhengzhou University, Zhengzhou, China
- 2Department of Occupational Health and Occupational Diseases, College of Public Health, Zhengzhou University, Zhengzhou, China
- 3Internet Medical and System Applications of National Engineering Laboratory, The First Affiliated Hospital of Zhengzhou University, Zhengzhou, China
- 4Gangarosa Department of Environmental Health, Rollins School of Public Health, Emory University, Atlanta, GA, United States
The mitochondrial DNA (mtDNA) copy number is a vital component in maintaining normal mitochondrial function. It is affected by environmental and occupational exposures, as well as polymorphisms in nuclear genes. Nonetheless, the specific roles of polymorphisms in cell-cycle genes and mtDNA copy number are still unknown. This study enrolled a sample of 544 coke oven workers and 238 non-exposed controls so as to assess the effect of exposure of coke oven emissions (COEs) and polymorphisms in cell-cycle genes on the mtDNA copy number. We found that the mtDNA copy number in the exposed group (0.60 ± 0.29) was significantly lower than that in the control group (1.03 ± 0.31) (t =18.931, P < 0.001). The analysis of covariance showed that both the rs1801270 (CA+CC) and the rs1059234 (CT+CC) in p21 gene were associated with lower mtDNA copy number in the exposed group (P = 0.001). Generalized linear models indicated COEs-exposure (β = −0.432, P < 0.001) and rs1059234 (CT+CC) in p21 gene (β = −0.060, P = 0.024) were the factors in mtDNA copy number reduction. In conclusion, this study suggests that the decrease of the mtDNA copy number is associated with COEs-exposure and the rs1059234 (CT+CC) in the p21 gene.
Introduction
Mitochondria are subcellular organelles that sustain life through adenosine triphosphate (ATP) generation. Mitochondria also participate in various vital physiological processes such as calcium homeostasis, generation of free radicals, cell apoptosis, among others (1). Additionally, mitochondria uniquely possess their own genome, known as mitochondrial DNA (mtDNA), that present variable copy numbers in cells based on surroundings the mitochondria inhabit (2). In brief, mtDNA copy number is affected by exogenous and genetic factors.
Coke oven emissions (COEs) are major environmental and occupational hazard factors and contain a variety of toxic substances, including polycyclic aromatic hydrocarbons (PAHs) (3). It is well-established that metabolites of PAHs have carcinogenic and genotoxic potential; long-term exposure can cause the formation of DNA adducts, eventually leading to DNA damage (4). Our previous studies have shown that exposure to PAHs can lead to the decrease in antioxidant capacity and telomere damage (5, 6). If subjected to exogenous stimulation, mtDNA is particularly susceptible to damage due to greater vulnerability to oxidative stress and limited DNA repair capacity (7). To further investigate this process, our lab explored the effects of exposure to PAHs from coke stoves on mtDNA (8).
MtDNA replication is a continuous process which requires coordinated action of several mitochondrial functioning proteins. In addition to nuclear genes encoding functional mitochondrial proteins, some cell-cycle genes influence mtDNA copy number by regulating certain mitochondrial-related genes. The gene p53 contributes to the regulation of mitochondrial transcription factor A (TFAM) and peroxisome proliferator-activated receptor gamma coactivator 1-alpha (PGC-1alpha), which could modulate replication and transcription of mtDNA (9, 10). Moreover, p53 also plays an important direct role in the control of mitochondrial genome stability or mtDNA copy number checkpoint (11). MDM2, a negative regulator of p53, may also control mtDNA copy number through inhibiting p53 expression (12). The gene p21 maintains mtDNA integrity by regulating antioxidant enzymes and thus controlling ROS levels (13). And p21 could modulate mitochondrial biogenesis by controlling the expression of other genes through its transcriptional regulatory function (14). Additionally, several genome-wide association studies have shown that the mtDNA copy number was related to polymorphisms in some nuclear genes (15–17). Our previous study found that the mtDNA copy number was associated with miR-210 rs11246190 in coke oven workers (8). However, the association between mtDNA copy number and polymorphisms in cell-cycle genes has not yet been examined. Therefore, this study aims to further explore the association between polymorphisms in cell-cycle genes and the mtDNA copy number in coke oven workers.
Materials and Methods
Study Subjects
A total of 782 participants were enrolled in the study, comprising 544 in exposed group and 238 in control group. The coke factory workers in Henan, China, exposed to COEs for more than 1 year were included in exposed group. The healthy participants with no history of exposure to occupational PAHs or other toxicants in the same city during the same period were included in control group. And the participants with liver or kidney diseases or other chronic diseases were excluded from the study. Demographic characteristics, occupational exposure level, and blood samples from all subjects were collected by trained interviewers. The study was approved by the Ethics Committee in Zhengzhou University (IRB 00006861, FWA00014064), and written informed consent was obtained from all subjects.
COEs-Exposure Levels Assessment
The air samples were collected at four coking plants. Five representative sampling sites were chosen from each coking plant, including the furnace top, furnace side, hearth, duty room and office. We obtained the time-weighted average (TWA) concentration (mg/m3) and working time (year) of each participant according to the onsite detecting results and detailed occupational exposure history. Cumulative exposure concentration of COEs (mg/m3·year) was used to assess COEs-exposure levels, which is equal to the product of the TWA concentration (mg/m3) and working time (year) in the exposed group. The detailed protocol for measurement of COEs-exposure concentration was described in our previous research (18). The cumulative exposure concentration of COEs in the control group was estimated based on their living environmental concentration and age.
mtDNA Copy Number Detection
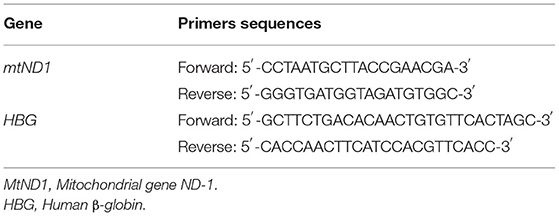
The mtDNA copy number of genomic DNA from whole peripheral blood samples was measured using real-time quantitative polymerase chain reaction (PCR) assay by determining the ratio of the mitochondrial gene ND-1 (mtND1) to the nuclear reference gene human β-globin (HBG) (19). Table 1 shows primer sequences of the two genes. The PCR was conducted using the 2 × SYBR Green Master Mix, and was carried out in the following steps: an initial heating step of 95°C for 5 min was followed by 40 cycles of 10 s at 95°C for denaturation and 30 s at 60°C for annealing/extension. Each sample was run using two parallels. Relative mtDNA copy number was calculated using threshold cycle (Ct) values and the formula of 2−ΔΔCt (ΔCt = CtmtDNA-CtReference, ΔΔCt = ΔCt-ΔCtaveragenormalcontrols).
Genotyping Analysis
Genomic DNA was isolated from whole peripheral blood samples in order to conduct genotyping. This study focused on important functional gene polymorphisms and supplemented susceptible gene polymorphisms, and screened six polymorphic loci in three genes (p53 rs17878362, p53 rs1042522, p53rs1625895, p21 rs1801270, p21rs1059234, and MDM2rs3730485). Polymorphisms in p53 rs17878362 and MDM2 rs3730485 were detected by PCR directly, and other loci were genotyped by the PCR-restriction fragment length polymorphism (PCR-RFLP) method. The details of genotyping, primer sequences, restriction endonucleases, and PCR have been reported in previous research (20–22).
Statistical Analysis
Statistical analysis was conducted by SPSS 27.0 software (SPSS Inc., Chicago, IL, USA). The student t-test was performed to compare the differences between the exposure group and the control group for mtDNA copy number showed a normal distribution. Partial correlations or the analysis of variance (ANOVA) was conducted to explore the association between cumulative exposure dose or individual characteristics and mtDNA copy number. The effects of gene polymorphisms on the mtDNA copy number were examined using the analysis of covariance (ANCOVA), and the Least Significant Difference test was used to conduct the pairwise comparisons. Generalized linear models (GLMs) were applied to analyze the environment, gene, and interaction on mtDNA copy number (23). The genotype distribution in the control group was tested with Hardy-Weinberg equilibrium. Linkage disequilibrium analysis was tested online based on the method described by Shi and He (24). All statistical tests were two-sided, and the level of statistical significance was set at a = 0.05.
Results
Population-Based Data
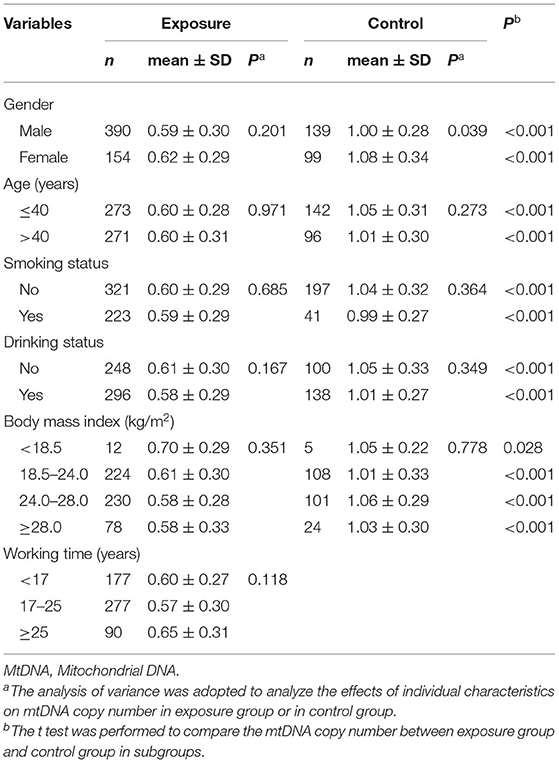
Comparative analysis of cumulative exposure dose and individual characteristics including gender, age, smoking status, drinking status, and body mass index (BMI), between the exposed group and the control group can be seen from our previous research (18). The results showed that the differences between the exposed group and the control group were significant (P < 0.05) in terms of cumulative exposure dose [1.12 (0.34, 2.14) mg/m3·year vs. 0.07 (0.06, 0.09) mg/m3·year], male (71.7 vs. 58.4%), age (40.10 ± 6.30 a vs. 38.39 ± 8.43 a), smoking (41.0 vs. 17.2%), and drinking (54.4 vs. 42.0%).
mtDNA Copy Number and COEs-Exposure
The mtDNA copy number in the exposed group (0.60 ± 0.29) was lower than in the control group (1.03 ± 0.31) (t = 18.931, P < 0.001). Partial correlations showed that the mtDNA copy number was negatively correlated with cumulative exposure dose (r = −0.253, P < 0.001) after controlling for gender, age, smoking status, drinking status and BMI.
mtDNA Copy Number and Individual Characteristics
As reported in our previous research (25), the results were shown in Table 2. The effect of gender on mtDNA copy number was statistically significant in the control group but not in the exposed group. The effect of age, smoking status, drinking status and BMI on mtDNA copy number was not found in either the exposed group or the control group (P > 0.05). And the effect of working time was not significant in exposed group (P = 0.118). The differences of mtDNA copy number between the exposed group and the control group were also observed in subgroups, and the analysis showed that the mtDNA copy number between the exposed group and the control group had significant differences between each subgroup.
mtDNA Copy Number and Polymorphisms in Cell-Cycle Genes
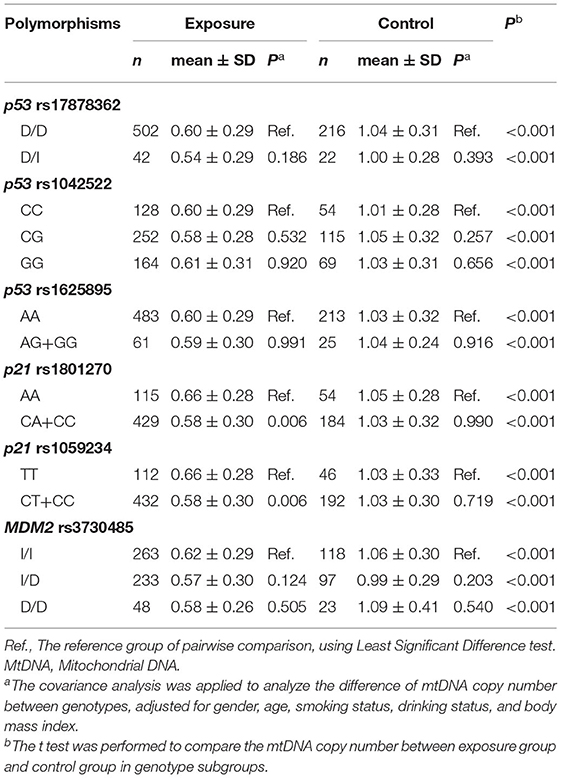
The genotype distribution for the six polymorphisms in cell-cycle genes did not deviate from Hardy-Weinberg equilibrium (P > 0.05). Table 3 presents the differences of mtDNA copy number between genotypes in polymorphisms. The number of homozygous GG genotype group in p53 rs1625895 was 0 in the exposed group and 1 in the control group. Consequently, they were merged with the heterozygous AG genotype group for statistical analysis. Due to the mtDNA copy number in the heterozygous CA genotype group in p21 rs1801270 being similar to that in the wild homozygous genotype CC group, the CA genotype group was merged with CC group. The mtDNA copy number in the CA+CC group in p21 rs1801270 was statistically lower than in the mutation homozygous AA genotype group in the exposed group. For the same reason, the heterozygous CT genotype group in p21 rs1059234 was also merged with the wild homozygous genotype CC group. The mtDNA copy number in the CT+CC group in p21 rs1059234 was statistically lower than in the mutation homozygous TT genotype group in exposure group.
mtDNA Copy Number and the Interaction of COEs-Exposure and the CA+CC Genotypes in p21 rs1059234
Linkage disequilibrium analysis was used to examine the connection between rs1801270 and rs1059234 in p21 gene and showed the D' value was 0.873. This suggests that there is a strong linkage disequilibrium between the two loci. Therefore, we analyzed the effects of the two loci on the mtDNA copy number by focusing on rs1059234 in p21 gene.
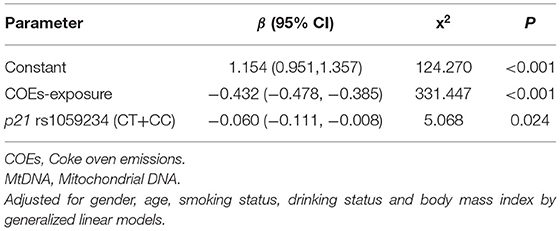
Table 4 summarizes the combined effect of COEs and p21 rs1059234 on mtDNA copy number. The results of the GLMs showed that the combined effect of COEs-exposure and the CT+CC genotypes in p21 rs1059234 on the mtDNA copy number was statistically significant, whereas the interaction of the two factors on the mtDNA copy number was not statistically significant [β (95% CI) = −0.096 (−0.209,0.018), x2 = 2.723, P = 0.099].
Influencing Factors of mtDNA Copy Number
GLMs were also used to screen influencing factors, using the mtDNA copy number as the dependent variable, COEs and rs1059234 in p21 gene as predicators, and gender, age, smoking status, drinking status, and BMI as covariates. The variables kept in the model included COEs-exposure (β = −0.432, P < 0.001) and rs1059234 (CT+CC) (β = −0.060, P = 0.024) (Table 5).
Discussion
MtDNA copy number, as an index of mitochondrial biogenesis, is vital for maintaining normal mitochondrial function and energy production in the body, and it is influenced by both exogenous and genetic factors. In the present research, we demonstrated that COEs exposure, polymorphisms in p21 and their combined effect significantly affected the alteration of mtDNA copy number.
Emerging epidemiological evidence suggests that exogenous factors and occupational exposures may be related to alterations of mtDNA copy number. The gender (26), age (27), smoking (28), drinking and BMI (29) have been reported to be associated with mtDNA copy number. Our results also showed that the mtDNA copy number was different in male vs. female in the control group. However, we did not observe the significant effect of working time on mtDNA copy number. Possible reasons are as follows, the exposure level varies in five representative sampling sites and workers may have different exposure due to job transfer. Since total COEs-exposure levels is a comprehensive indicator of working time and working location, neither one of these can reflect exposure level alone. Moreover, several researches have shown that benzene and black carbon (19) could cause an increased mtDNA copy number and conversely, lead (30), elemental carbon and arsenic (31) could lead to a decreased mtDNA copy number. PAH, a known carcinogen and mutagen, has also been reported to be related with changes in mtDNA copy number, although the relationships are inconsistent. Pavanello, Dioni (32) found that mtDNA copy number in peripheral blood lymphocytes was higher among coke oven workers with exposure to PAHs. However, more studies reported lower mtDNA copy number in PAH-exposed individuals. Pieters, Koppen (33) observed that blood mtDNA copy number was inversely associated with indoor exposure to PAHs in house dust. Ling, Zhang (34) found an association between decreased sperm mtDNA copy number and PAH exposure in a prospective cohort “Male Reproduction Health in Chongqing College Students.” Our previous study showed that mtDNA copy number in the COEs-exposed group was lower than that in the control group (8). PAHs, the main harmful component of COEs, can cause ROS-induced mtDNA damage. Cells challenged with ROS synthesize more copies of mtDNA and increase the number of mitochondria to compensate for the damage. This causes a vicious cycle to develop over time. The increasing abundance of mitochondria causes the overproduction of ROS and further oxidative damage, which may initiate cell senescence or death, eventually resulting in the decreased mtDNA copy number (19).
In addition, the polymorphisms in cell-cycle genes could affect mtDNA copy number. The genes p53 and p21 participate in cell cycle arrest in G1/S transition upon DNA damage and regulate mitochondrial biogenesis. It has been shown that polymorphisms in these cell-cycle genes could affect gene transcription or protein expression, and then modulate gene functions, which consequently may affect mitochondrial biogenesis. Therefore, we screened six polymorphic loci in cell-cycle genes (p53 rs17878362, p53 rs1042522, p53 rs1625895; p21 rs1801270, p21rs1059234, and MDM2rs3730485) to explore the association between polymorphisms in cell-cycle genes and mtDNA copy number in coke oven workers. In this study, we demonstrated that rs1059234 and rs1801270 in p21 have significant effect on the mtDNA copy number. The rs1059234 is a C-to-T transition that occurs 20 nucleotides downstream of the stop codon in 3′-untranslated region (UTR) of p21 gene (35). Studies exhibited that the polymorphisms within 3′-UTR were associated with disease susceptibility (36). Accordingly, we found that the mtDNA copy number in the TT genotype in p21 rs1059234 was statistically higher than that in the mutation homozygous CT+CC genotype in the exposed group. Previous research has shown that the 3′-UTR can affect mRNA degradation by regulating its nucleotide responsive region to interact with mRNA-binding protein, which eventually results in an alteration in protein expression levels (37, 38). It is likely that C-to-T transition in rs1059234 could increase p21 mRNA expression by preventing degradation of mRNA, and thus possess a greater ability to control ROS levels in response to oxidative stress, ultimately reducing the loss of the mtDNA copy number caused by exposure to COEs. Moreover, the rs1801270 is a C-to-A transversion occurring in codon 31 of p21 gene that causes an amino acid substitution from serine to arginine, which is expected to affect the DNA binding zinc finger domain of protein, eventually affecting expression and activity of p21 (39). Our study showed that the rs1801270 was in strong linkage disequilibrium with the rs1059234, which suggests genotypes in the two loci are strongly correlated (35). Thus, the rs1801270 and the rs1059234 may have the same effect on the mtDNA copy number. Similar to the rs1059234, we found that the mutation homozygous AA genotype in rs1801270 had statistically higher mtDNA copy number than the CA+CC genotypes in coke oven workers.
Furthermore, we explored the interactions of COEs-exposure and gene on mtDNA copy number. It has shown that interactions of gene-environment play a determined role in many complex diseases (23). In this study, we found the significant combined effect of COEs-exposure and the CT+CC genotypes in p21 rs1059234 on the mtDNA copy number, though the interaction was not statistically significant. Long-term exposure to PAHs could increase the level of ROS, and CT+CC genotype in p21 rs1059234 also shows a lower ability to control ROS levels, both of which together lead to the gradual accumulation of ROS, which in turn cause mtDNA instability (40). In addition, the associations between polymorphisms in p53 and MDM2 genes and mtDNA copy number were not significant. This could be due to the environment-gene interaction, which may also cause alterations of mtDNA copy number.
In this study, we explored the association between mtDNA copy number and polymorphisms in cell-cycle genes compared to previous researches that mainly focused on the effect of nuclear genes on mtDNA copy number. Moreover, we further explored the combined effect of COEs-exposure and gene polymorphisms on mtDNA copy number in coke oven workers. However, several limitations need to be considered. First, we assessed the external COEs-exposure based on air concentration of COEs and working time, which could not represent the accurate individual exposure level. Second, the research only discovered the effects of the gene and COEs-exposure on mtDNA copy number, further experimental studies may be applied to elucidate the underlying mechanisms.
In conclusions, our study shows that COEs-exposure and the CT+CC genotypes in p21 rs1059234 are the major influencing factors of the mtDNA copy number. The findings can provide strong evidence for researching the mechanism of mtDNA copy number alterations caused by genetic and occupational factors and for screening effective predisposing biomarkers and early prevention of high-risk population.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Ethics Statement
The studies involving human participants were reviewed and approved by Life Sciences of Institutional Review Board of Zhengzhou University Ethics, China. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
YY and XS: conceptualization. JT: formal analysis. WW: funding acquisition. XD: investigation. YW: writing—original draft. BL and LS: writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
The work was supported by the National Science Foundation of China (NSFC81872597 and 81001239) and the HERCULES Exposome Research Centre (P30ES019776).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors express their gratitude to all the individuals who volunteered to participate in this study.
References
1. Bhatti JS, Bhatti GK, Reddy PH. Mitochondrial dysfunction and oxidative stress in metabolic disorders - a step towards mitochondria based therapeutic strategies. Biochim Biophys Acta. (2017) 1863:1066–77. doi: 10.1016/j.bbadis.2016.11.010
2. Guyatt AL, Burrows K, Guthrie PAI, Ring S, McArdle W, Day INM, et al. Cardiometabolic phenotypes and mitochondrial DNA copy number in two cohorts of UK women. Mitochondrion. (2018) 39:9–19. doi: 10.1016/j.mito.2017.08.007
3. Yang K, Jiang X, Cheng S, Chen C, Cao X, Tu B. Effects of coke oven emissions and benzo a pyrene on blood pressure and electrocardiogram in coke oven workers. J Occup Health. (2017) 59:1–7. doi: 10.1539/joh.15-0264-OA
4. Moorthy B, Chu C, Carlin DJ. Polycyclic aromatic hydrocarbons: from metabolism to lung cancer. Toxicol Sci. (2015) 145:5–15. doi: 10.1093/toxsci/kfv040
5. Zhang H, Wang S, Duan X, Feng X, Wang T, Wang P, et al. The interaction effects of coke oven emissions exposure and metabolic enzyme Gene variants on total antioxidant capacity of workers. Environ Toxicol Pharmacol. (2019) 70:103197. doi: 10.1016/j.etap.2019.103197
6. Duan X, Yang Y, Wang S, Feng X, Wang T, Wang P, et al. Dose-related telomere damage associated with the genetic polymorphisms of cGAS/STING signaling pathway in the workers exposed by PAHs. Environ Pollut. (2020) 260:113995. doi: 10.1016/j.envpol.2020.113995
7. Li Z, Zhu M, Du J, Ma H, Jin G, Dai J. Genetic variants in nuclear DNA along with environmental factors modify mitochondrial DNA copy number: a population-based exome-wide association study. BMC Genom. (2018) 19:752. doi: 10.1186/s12864-018-5142-7
8. Duan X, Yang Y, Zhang H, Liu B, Wei W, Wang L, et al. Polycyclic aromatic hydrocarbon exposure, miRNA genetic variations, and associated leukocyte mitochondrial DNA copy number: a cross-sectional study in China. Chemosphere. (2020) 246:125773. doi: 10.1016/j.chemosphere.2019.125773
9. Kamp WM, Wang P-Y, Hwang PM. TP53 mutation, mitochondria and cancer. Curr Opin Genet Dev. (2016) 38:16–22. doi: 10.1016/j.gde.2016.02.007
10. Scarpulla RC, Vega RB, Kelly DP. Transcriptional integration of mitochondrial biogenesis. Trends Endocrinol Metab. (2012) 23:459–66. doi: 10.1016/j.tem.2012.06.006
11. Park J-H, Zhuang J, Li J, Hwang PM. p53 as guardian of the mitochondrial genome. FEBS Lett. (2016) 590:924–34. doi: 10.1002/1873-3468.12061
12. Shangary S, Wang S. Small-molecule inhibitors of the MDM2-p53 protein-protein interaction to reactivate p53 function: a novel approach for cancer therapy. Annu Rev Pharmacol Toxicol. (2009) 49:223–41. doi: 10.1146/annurev.pharmtox.48.113006.094723
13. Yang M, Luna L, Sørbø JG, Alseth I, Johansen RF, Backe PH, et al. Human OXR1 maintains mitochondrial DNA integrity and counteracts hydrogen peroxide-induced oxidative stress by regulating antioxidant pathways involving p21. Free Radical Biol Med. (2014) 77:41–8. doi: 10.1016/j.freeradbiomed.2014.09.003
14. Kim AJ, Jee HJ, Song N, Kim M, Jeong SY, Yun J. p21(WAF1/C1P1) deficiency induces mitochondrial dysfunction in HCT116 colon cancer cells. Biochem Biophys Res Commun. (2013) 430:653–8. doi: 10.1016/j.bbrc.2012.11.096
15. López S, Buil A, Souto JC, Casademont J, Martinez-Perez A, Almasy L, et al. A genome-wide association study in the genetic analysis of idiopathic thrombophilia project suggests sex-specific regulation of mitochondrial DNA levels. Mitochondrion. (2014) 18:34–40. doi: 10.1016/j.mito.2014.09.004
16. Workalemahu T, Enquobahrie DA, Tadesse MG, Hevner K, Gelaye B, Sanchez SE, et al. Genetic variations related to maternal whole blood mitochondrial DNA copy number: a genome-wide and candidate gene study. J Maternal Fetal Neonatal Med. (2017) 30:2433–9. doi: 10.1080/14767058.2016.1252747
17. Lopez S, Buil A, Carlos Souto J, Casademont J, Blangero J, Martinez-Perez A, et al. Sex-specific regulation of mitochondrial DNA levels: genome-wide linkage analysis to identify quantitative trait loci. PLoS ONE. (2012) 7:e0042711. doi: 10.1371/journal.pone.0042711
18. Duan X, Yang Y, Zhang D, Wang S, Feng X, Wang T, et al. Genetic polymorphisms, mRNA expression levels of telomere-binding proteins, and associates with telomere damage in PAHs-Exposure workers. Chemosphere. (2019) 231:442–9. doi: 10.1016/j.chemosphere.2019.05.134
19. Carugno M, Pesatori AC, Dioni L, Hoxha M, Bollati V, Albetti B, et al. Increased mitochondrial DNA copy number in occupations associated with low-dose benzene exposure. Environ Health Perspect. (2012) 120:210–5. doi: 10.1289/ehp.1103979
20. Hu ZB, Ma HX, Lu DR, Qian J, Zhou JN, Chen YJ, et al. Genetic variants in the MDM2 promoter and lung cancer risk in a Chinese population. Int J Cancer. (2006) 118:1275–8. doi: 10.1002/ijc.21463
21. Qiu Y-L, Wang W, Sun P, Liu J, Li J, Chai S-J, et al. Genetic polymorphism in cell cycle control genes and susceptibility of chromosomal damage in vinyl chloride monomer exposed workers. Zhonghua lao dong wei sheng zhi ye bing za zhi. (2007) 25:649–53.
22. Duan X, Yang Y, Wang S, Feng X, Wang T, Wang P, et al. Changes in the expression of genes involved in cell cycle regulation and the relative telomere length in the process of canceration induced by omethoate. Tumor Biology. (2017) 39:1010428317719782. doi: 10.1177/1010428317719782
23. Wang W, Zhang H, Duan X, Feng X, Wang T, Wang P, et al. Telomere length in workers was effected by omethoate exposure, GSTM1 deletion, interaction between smoking and GSTP1 polymorphisms. J Occup Environ Med. (2019) 61:E19–23. doi: 10.1097/JOM.0000000000001503
24. Shi YY, He L. SHEsis, a powerful software platform for analyses of linkage disequilibrium, haplotype construction, and genetic association at polymorphism loci (vol 15, pg 97, 2005). Cell Res. (2006) 16:851. doi: 10.1038/sj.cr.7310101
25. Li X, Duan X, Zhang H, Ding M, Wang Y, Yang Y, et al. Genetic polymorphisms of metabolic enzyme genes associated with leukocyte mitochondrial DNA copy number in PAHs exposure workers. Cancer Rep. (2021) 4:E1361. doi: 10.1002/cnr2.1361
26. Kristensen TN, Loeschcke V, Tan Q, Pertoldi C, Mengel-From J. Sex and age specific reduction in stress resistance and mitochondrial DNA copy number in Drosophila melanogaster. Sci Rep. (2019) 9:12305. doi: 10.1038/s41598-019-48752-7
27. Jang JY, Blum A, Liu J, Finkel T. The role of mitochondria in aging. J Clin Investig. (2018) 128:3662–70. doi: 10.1172/JCI120842
28. Wu S, Li X, Meng S, Fung T, Chan AT, Liang G, et al. Fruit and vegetable consumption, cigarette smoke, and leukocyte mitochondrial DNA copy number. Am J Clin Nutr. (2019) 109:424–32. doi: 10.1093/ajcn/nqy286
29. von Wurmb-Schwark N, Ringleb A, Schwark T, Broese T, Weirich S, Schlaefke D, et al. The effect of chronic alcohol consumption on mitochondrial DNA mutagenesis in human blood. Mutation Res. (2008) 637:73–9. doi: 10.1016/j.mrfmmm.2007.07.003
30. Gong S, Tu Y, Fang Y, Wang T, Zhang G, Xia Z. Association between mitochondrial DNA copy number and lead level in peripheral blood among leadexposed workers. Chin J Environ Occup Med. (2018) 35:189–95. doi: 10.13213/j.cnki.jeom.2018.17670
31. Martínez-Acuña MI, Mejía-Saavedra JJ, Peña LCS, Matousek T, Del Razo LM, Alegría-Torres JA. Mitochondrial DNA copy number in Mexican children co-exposed to inorganic arsenic and fluoride from Zacatecas, Mexico. Toxicol Lett. (2016) 259:S126. doi: 10.1016/j.toxlet.2016.07.324
32. Pavanello S, Dioni L, Hoxha M, Fedeli U, Svach DM, Baccarelli AA. Mitochondrial DNA copy number and exposure to polycyclic aromatic hydrocarbons. Cancer Epidemiol Biomarkers Prev. (2013) 22:1722–9. doi: 10.1158/1055-9965.EPI-13-0118
33. Pieters N, Koppen G, Smeets K, Napierska D, Plusquin M, De Prins S, et al. Decreased mitochondrial DNA content in association with exposure to polycyclic aromatic hydrocarbons in house dust during wintertime: from a population enquiry to cell culture. PLoS ONE. (2013) 8:e0063208. doi: 10.1371/journal.pone.0063208
34. Ling X, Zhang G, Sun L, Wang Z, Zou P, Gao J, et al. Polycyclic aromatic hydrocarbons exposure decreased sperm mitochondrial DNA copy number: a cross-sectional study (MARHCS) in Chongqing, China. Environ Pollut. (2017) 220:680–7. doi: 10.1016/j.envpol.2016.10.026
35. Silva Ribamar Carvalho IN, de Oliveira Reis AH, Cabello PH, Vargas FR. Polymorphisms of CDKN1A gene and risk of retinoblastoma. Carcinogenesis. (2013) 34:2774–7. doi: 10.1093/carcin/bgt308
36. Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. (2010) 10:389–402. doi: 10.1038/nrc2867
37. Li J, Li Z, Kan Q, Sun S, Li Y, Wang S. Association of p21 3' UTR gene polymorphism with cancer risk: evidence from a meta-analysis. Sci Rep. (2015) 5:13189. doi: 10.1038/srep13189
38. Chen FY, Amara FM, Wright JA. Mammalian ribonucleotide reductase R1 mRNA stability under normal and phorbol ester stimulating conditions: involvement of a cis-trans interaction at the 3' untranslated region. EMBO J. (1993) 12:3977–86. doi: 10.1002/j.1460-2075.1993.tb06075.x
39. Cao Q, Wang Y, Song X, Yang W. Association between MDM2 rs2279744, MDM2 rs937283, and p21 rs1801270 polymorphisms and retinoblastoma susceptibility. Medicine. (2018) 97:E13547. doi: 10.1097/MD.0000000000013547
40. Bucher S, Le Guillou D, Allard J, Pinon G, Begriche K, Tête A, et al. Possible involvement of mitochondrial dysfunction and oxidative stress in a cellular model of NAFLD progression induced by benzo[a]pyrene/ethanol coexposure. Oxidative Med Cell Longevity. (2018) 2018:4396403. doi: 10.1155/2018/4396403
Keywords: mitochondrial DNA copy number, coke oven emissions, cell-cycle genes, polymorphisms, generalized linear models (GLM)
Citation: Wang Y, Tan J, Wang W, Duan X, Lappe B, Shi L, Yang Y and Shi X (2022) The Association Between Polymorphisms in Cell-Cycle Genes and Mitochondrial DNA Copy Number in Coke Oven Workers. Front. Public Health 10:904856. doi: 10.3389/fpubh.2022.904856
Received: 30 March 2022; Accepted: 30 May 2022;
Published: 05 July 2022.
Edited by:
Radu Corneliu Duca, Laboratoire National de Santé (LNS), LuxembourgReviewed by:
Rao L. Divi, National Institutes of Health (NIH), United StatesYansen Bai, Guangzhou Medical University, China
Copyright © 2022 Wang, Tan, Wang, Duan, Lappe, Shi, Yang and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongli Yang, ylyang377@zzu.edu.cn; Xuezhong Shi, xzshi@zzu.edu.cn
 Yuping Wang1
Yuping Wang1 Xiaoran Duan
Xiaoran Duan Liuhua Shi
Liuhua Shi Yongli Yang
Yongli Yang Xuezhong Shi
Xuezhong Shi