- 1Department of Neurology, The Fifth Affiliated Hospital of Guangxi Medical University, Nanning, China
- 2Department of Internal Medicine, The Fifth Affiliated Hospital of Guangxi Medical University, Nanning, China
- 3Maternal and Child Health Hospital and Obstetrics and Gynecology Hospital of Guangxi Zhuang Autonomous Region, Guangxi, China
Competing endogenous RNA (ceRNA) and autophagy were related to neurological diseases. But the relationship among ceRNA, autophagy and Schizophrenia (SZ) was not clear. In this study, we obtained gene expression profile of SZ patients (GSE38484, GSE54578, and GSE16930) from Gene Expression Omnibus (GEO) database. Then we screened the autophagy-related differentially expressed lncRNA, miRNA, and mRNA (DElncRNA, DEmiRNA, and DEmRNA) combined with Gene database from The National Center for Biotechnology Information (NCBI). In addition, we performed enrichment analysis. The result showed that biological processes (BPs) mainly were associated with cellular responses to oxygen concentration. The enriched pathways mainly included ErbB, AMPK, mTOR signaling pathway and cell cycle. Furthermore, we constructed autophagy-related ceRNA network based on the TargetScan database. Moreover, we explored the diagnostic efficiency of lncRNA, miRNA and mRNA in ceRNA, through gene set variation analysis (GSVA). The result showed that the diagnostic efficiency was robust, especially miRNA (AUC = 0.884). The miRNA included hsa-miR-423-5p, hsa-miR-4532, hsa-miR-593-3p, hsa-miR-618, hsa-miR-4723-3p, hsa-miR-4640-3p, hsa-miR-296-5p, and hsa-miR-3943. The result of this study may be helpful for deepening the pathophysiology of SZ. In addition, our finding may provide a guideline for the clinical diagnosis of SZ.
Introduction
Schizophrenia is a serious genetic psychiatric disease that usually occurs in late adolescence or early adulthood, and it affected 1.13 million people worldwide in 2017 (1, 2). Lifetime prevalence of the disease is close to 1%, and only 10–15% of patients are able to engage in paid work (3). The main risk factors of the disease include disorders of the dopamine system (4); early brain trauma, especially damage to the frontal and temporal lobes (5); use of illicit drugs (6); and infections during pregnancy caused by various factors (7). The pathogenesis of schizophrenia is unclear, and most studies have shown it to involve interactions between genes and the environment (8). The disease is diagnosed based on positive symptoms, such as hallucinations, delusions, and unusual behavior; or negative symptoms, such as blunted emotional reactions, lack of emotion and lack of language. The presence of two or more symptoms is usually indicative of the disease. First-line treatments against schizophrenia are haloperidol and chlordiazepoxide, but they often show poor efficacy and are associated with high risk of serious adverse reactions (9). Identifying better treatments requires a deeper understanding of the biological basis of schizophrenia.
Autophagy, the process of degrading intracellular components in lysosomes, plays an important role in the central nervous system by contributing to neuronal homeostasis (10). Loss of autophagy can destroy neuronal homeostasis (11), leading to abnormal neuronal activity, which in turn may contribute to various neurological disease (12). In fact, loss of autophagy in animal model can seriously damage social and cognitive functions, which may lead to mood disorders, psychotic-like symptoms and behavioral change (13, 14). Dysregulation of autophagy in neurological diseases may involve altered gene regulation. In particular, it may involve changes in how much microRNAs (miRNAs) repress the translation of target genes, perhaps as a result of changes in the levels of long non-coding RNAs (lncRNAs) (15). According to the competing endogenous RNA (ceRNA) hypothesis, lncRNAs compete with target mRNAs for binding to miRNAs, acting as miRNA “sponges” (16). In support of this hypothesis, altered lncRNA-mediated gene regulation has been implicated in schizophrenia (17), and certain miRNAs are up-regulated in schizophrenia and other neurological diseases (18).
Whether schizophrenia involves altered interactions among lncRNAs, miRNAs, and mRNAs is unclear. Based on comparison of blood samples from schizophrenia patients and healthy controls in public datasets, the present study identified a ceRNA network that may regulate autophagy-related genes in the disease. These insights may help clarify the disease process, guide new drug development, and improve diagnosis.
Materials and Methods
Data Collection and Processing
We downloaded the datasets from the Gene Expression Omnibus (GEO) database, each dataset had been normalized with MAS5 when the authors submitted them into the database as required (http://www.ncbi.nlm.nih.gov/geo/). The whole-blood RNA (mRNAs and lncRNAs) expression profiles of GSE38484 based on GPL6947 platform, taken from 106 patients with schizophrenia and 96 controls (19, 20). Peripheral-blood miRNA expression profiles of GSE54578 based on GPL16016 platform included 15 patients and 15 controls were also downloaded (21). The lncRNAs and mRNAs were distinguished according to the file Homo_sapiens.GRCh38.97.chr.gtf on the Ensembl website (http://asia.ensembl.org) (22, 23). The above two datasets (GSE38484 and GSE54587) were used to construct a potential ceRNA network in schizophrenia. The dataset of GSE16930 based on GPL2879 platform, containing 18 patients and 2 controls (24), was used to validate diagnostic performance and expression of RNA in ceRNA network. If one gene corresponded to multiple probes, the average expression value the these probes was considered to be the expression of the gene. The work flow was shown in Figure 1.
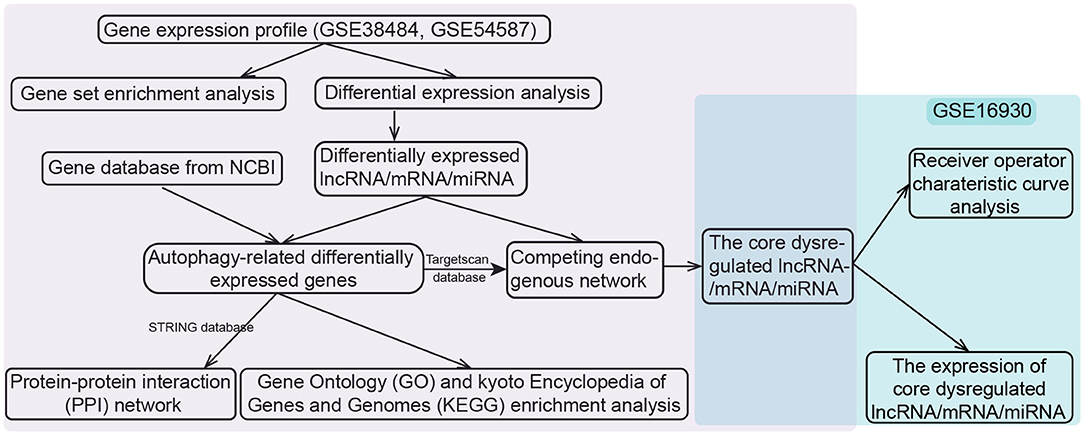
Figure 1. Flowchart of the study design. lncRNA, long noncoding RNA; miRNA, microRNA; NCBI, National Center for Biotechnology Information.
Screening for Autophagy-Related Differentially Expressed RNAs in Schizophrenia
The limma package (25) was used to identify differentially expressed mRNAs, lncRNAs, and miRNAs (DEmRNA, DElncRNA, and DEmiRNA) between patients with schizophrenia and controls. The RNA that was log2|fold change (FC)| >1 and adjusted p <0.05 was considered differentially expressed. The autophagy-related genes were obtained combined the differentially expressed RNAs (DERNAs) and the autophagy-related genes in Gene database (www.ncbi.nlm.nih.gov/gene). The autophagy-related genes were obtained using the autophagy as the search key word in the Gene database.
Functional Enrichment Analysis
Potential interactions among autophagy-related genes were identified using the STRING database (26), and protein-protein interactions (PPIs) network was visualized using Cytoscape (27). In order to further explore the biological functions of autophagy-related genes, gene ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was performed using the clusterProfiler R package (28). For further exploring the differences of biological functions between SZ patients and controls, the gene set variation analysis (GSVA) was performed using GSVA package (29) in R. Gene set enrichment analysis (GSEA) was performed using GSEA2-2.2.4 (Java version) (30). The reference gene set (c5.bp.v6.2.symbols.gmt and c2.cp.kegg.v6.2.symbols.gmt) were obtained from The Molecular Signatures Database (version 6.2) (31). GO and KEGG networks were analyzed and drawn using the ClueGO plug-in (32) in Cytoscape.
Exploration of an Autophagy-Related ceRNA Network in Schizophrenia
To construct a ceRNA regulation network, interactions between DEmRNAs and DEmiRNAs were predicted using the TargetScan database (version: release 7.2) (33). Then the DEmiRNAs in these interaction pairs were used to identify target mRNAs, again based on the TargetScan database. Target mRNAs that we found to be differentially expressed in schizophrenia were considered candidate target mRNAs in the ceRNA network. The co-expression network comprising DElncRNAs, DEmiRNAs and DEmRNAs was visualized using Cytoscape.
Identifying Core Dysregulated DElncRNAs, DEmiRNAs, and DEmRNAs in Schizophrenia
GSVA scores were calculated using an unsupervised, non-parametric algorithm in the GSVA package (29) separately for DElncRNAs, DEmiRNAs, and DEmRNAs. Core genes are also called hub genes, genes that play a vital role in biological processes. In related pathways, the regulation of other genes is often affected by this gene. The ability of the core sets identified based on GSVA score to diagnose schizophrenia was assessed in terms of the area under the receiver operator characteristic curve (AUC) (34).
Statistical Analysis
We screened the differentially expressed genes in the two groups using unpaired t-tests provided by limma package. Unless otherwise stated, we considered p-value < 0.05 to be statistically significant.
Results
DElncRNAs, DEmiRNAs, and DEmRNAs in Schizophrenia
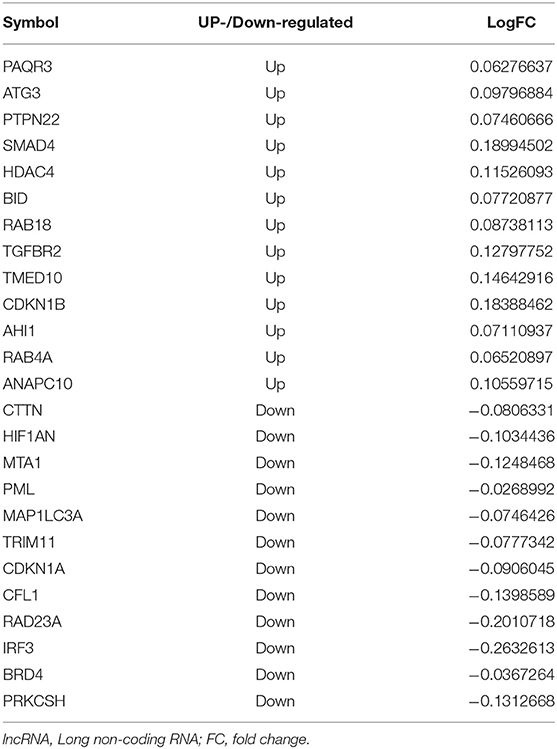
Comparison between patients with schizophrenia and controls revealed 2,400 DElncRNAs (1,130 up-regulated, 1,270 down-regulated), 69 DEmiRNAs (19 up-regulated, 50 down-regulated), and 3,859 DEmRNAs (811 up-regulated, 2,048 down-regulated) (Figure 2A). Of the total set of DEmRNAs, 375 were related to autophagy, of which 176 were up-regulated and 199 down-regulated (Figure 2B). The heatmap suggested that DEmRNAs could distinguish patients from controls to a certain extent (Figure 2C).
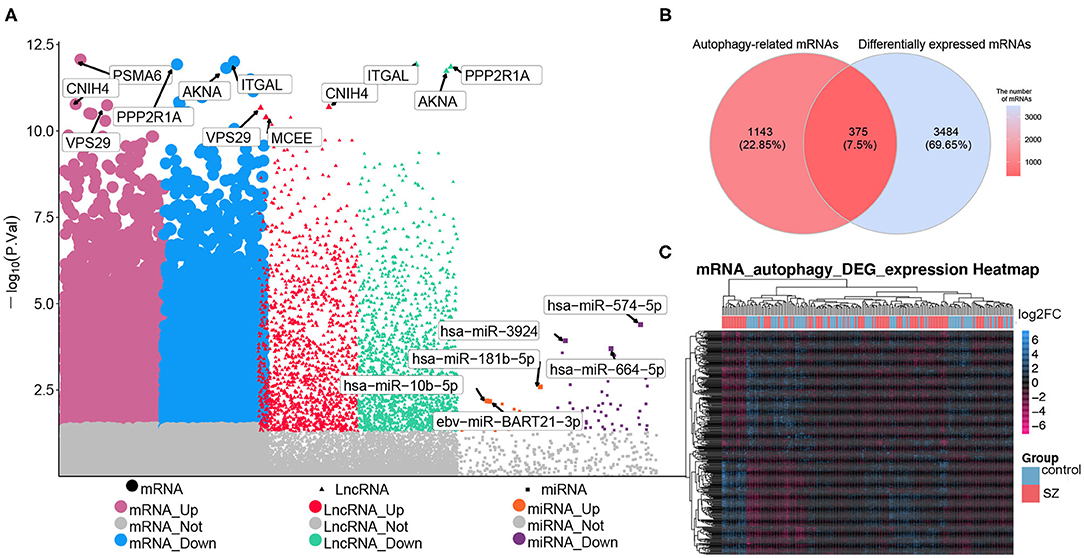
Figure 2. Differential expression analysis and screening of autophagy-related mRNAs. (A) Manhattan diagram showing differentially expressed (DE) lncRNAs, DEmiRNAs and DEmRNAs in schizophrenia (SZ). (B) Genes overlapping between the set of autophagy genes and the set of DEmRNAs. (C) Heatmap showing the expression of autophagy-related DEmRNAs. Yellow means up-regulation, blue means down-regulation.
Biological Functions and Pathways Involving Autophagy-Related DEmRNAs in Schizophrenia
The autophagy-related DEmRNAs encoded a wide range of proteins, based on the STRING database. The analysis identified 161 interaction pairs and 130 nodes in the network when the score was higher than 980 (Supplementary Figure 1). Enrichment analysis showed that autophagy-related DEmRNAs supported cellular responses to oxidative stress, regulation of protein catabolism, apoptosis signaling, as well as biological processes related to cellular responses to oxygen concentration (Figure 3A). They were also closely related to ErbB signaling, AMPK signaling, mTOR signaling and the cell cycle (Figure 3B). The GSEA result showed that there were common GO function and KEGG pathways combined with Figures 3B,C. Two GO functions, “positive regulation of autophagy” and “response to oxygen levels,” were up-regulated in SZ patients compared with controls (Figure 3C). Only one KEGG pathway, “ubiquitin mediated proteolysis,” was upregulated in SZ patients compared to controls (Figure 3D). ClueGO analysis showed that autophagy-related DEmRNAs may also be related to apoptosis and to signaling mediated by mTOR, MAPK, and ErbB (Figure 3E). The results showed that autophagy-related DEmRNAs may be involved in positive regulation of catabolism, apoptosis signal, and regulation of transcription factors (Figure 3F).
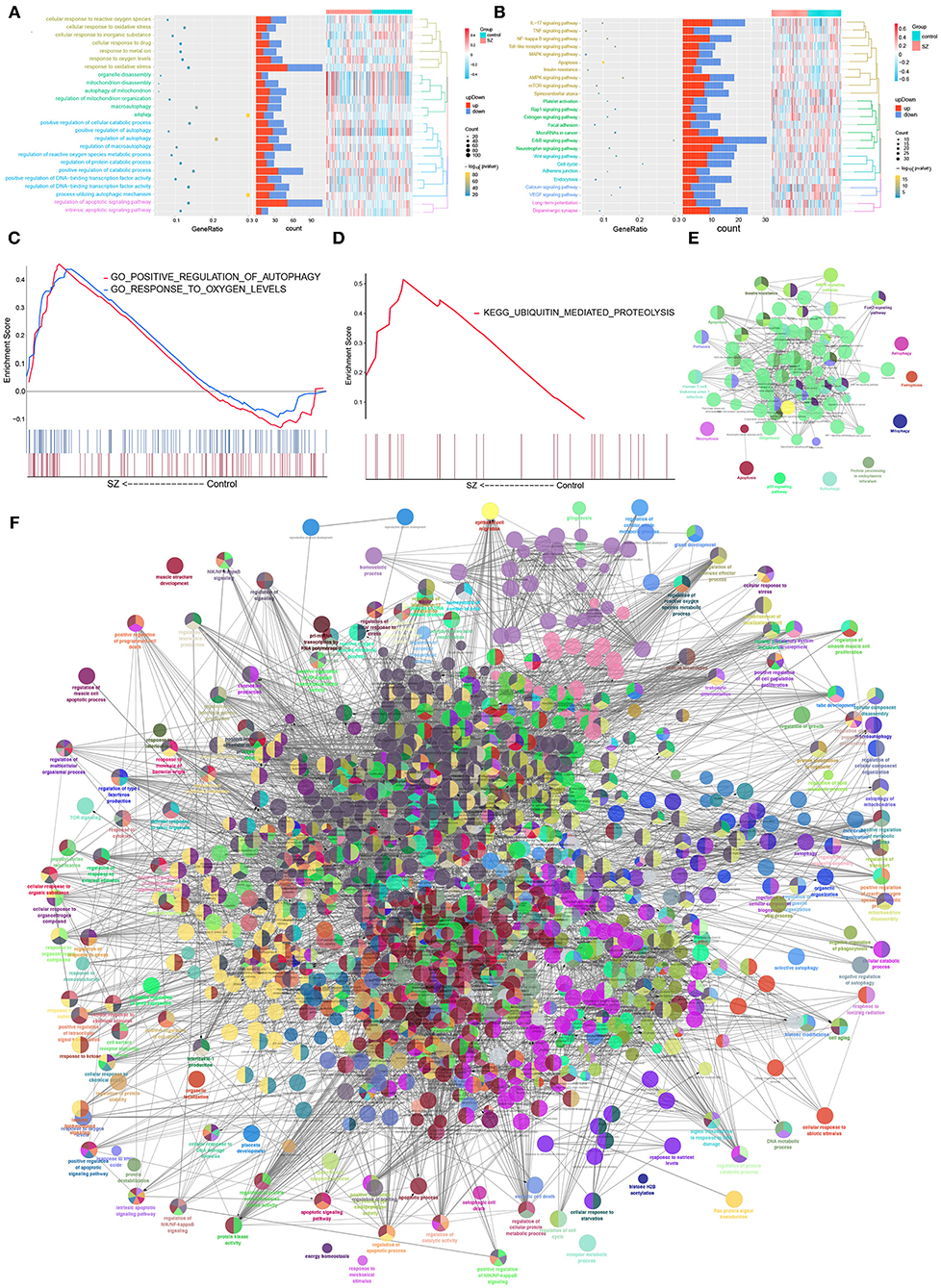
Figure 3. Biological functions and KEGG pathways involving autophagy-related DEmRNAs in schizophrenia (SZ). (A) Biological processes involving autophagy-related DEmRNAs. (B) Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways involved in autophagy-related DEmRNAs. (C) Biological processes involving autophagy-related DEmRNAs. (D) KEGG pathways involved in autophagy-related DEmRNAs. (E) Pathway enrichment analysis of 375 autophagy-related DEmRNAs using ClueGO. (F) Biological enrichment analysis of 375 autophagy-related DEmRNAs using ClueGO. Each node represents a biological process or KEGG pathway. Edges represent connections between the nodes, and the length of each edge reflects the relatedness of two processes.
Involvement of a ceRNA Network in Autophagy-Related DEmRNAs in Schizophrenia
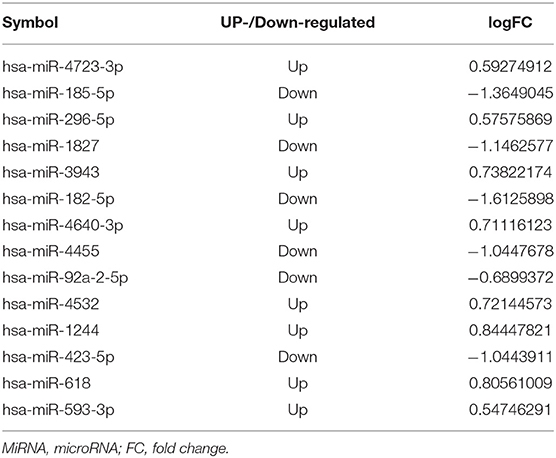
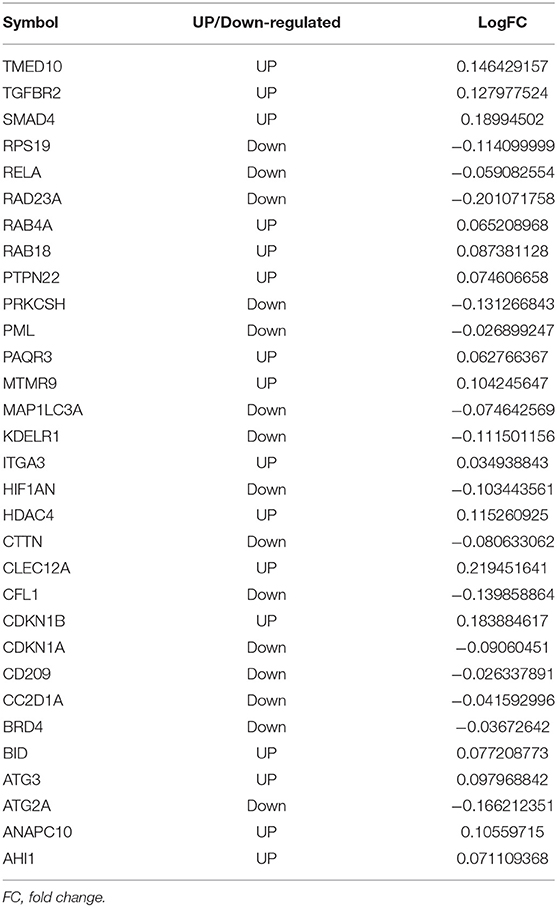
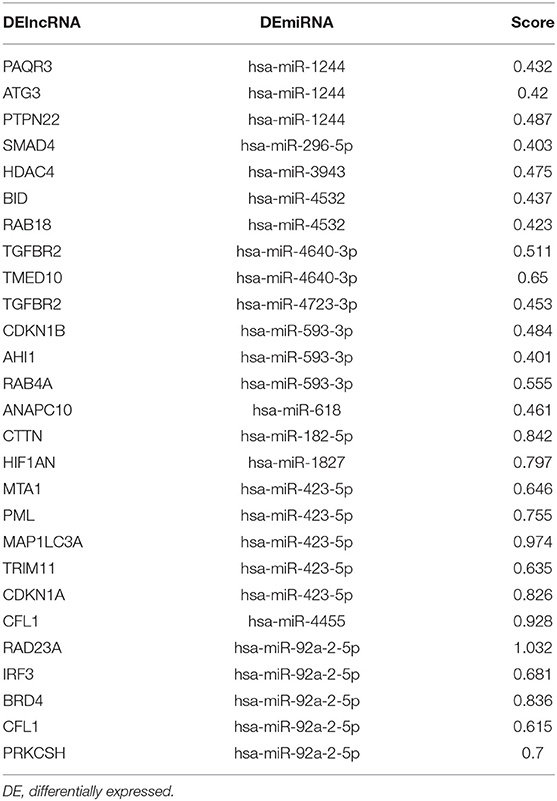
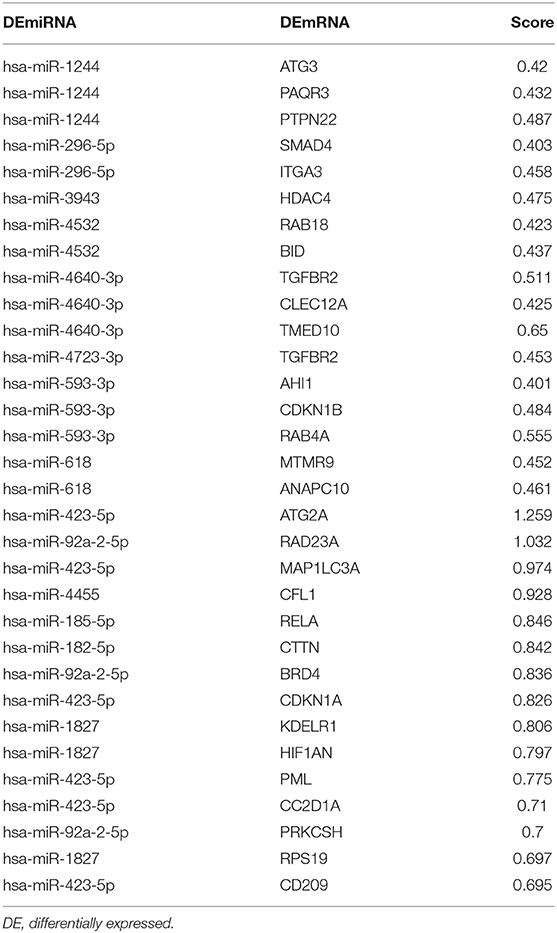
Next, potential interactions among the above genes were explored according to the ceRNA hypothesis. Based on a minimal score of 0.4, we identified 31 autophagy-related DEmRNAs that may interact with 14 DEmiRNAs (Figure 4A, Tables 1–3). In total, there were 25 DElncRNAs, 13 DEmiRNAs and 30 autophagy-related DEmRNAs, with the threshold of score >0.4 (Figure 4B, Table 4). These results, combined with the enrichment analysis, suggest that lncRNAs may regulate the phenotype through ceRNA. We identified 15 DElncRNAs, 8 DEmiRNAs and 11 autophagy-related DEmRNAs and 10 KEGG pathways (Figure 4C, Tables 4, 5). We focused on the nine KEGG pathways previously linked to schizophrenia in the literature: Wnt signaling pathway, adherence junctions, ErbB signaling pathway, spinocerebellar ataxia, apoptosis, MAPK signaling pathway, cell cycle, endocytosis, and focal adhesion (Figure 4D).
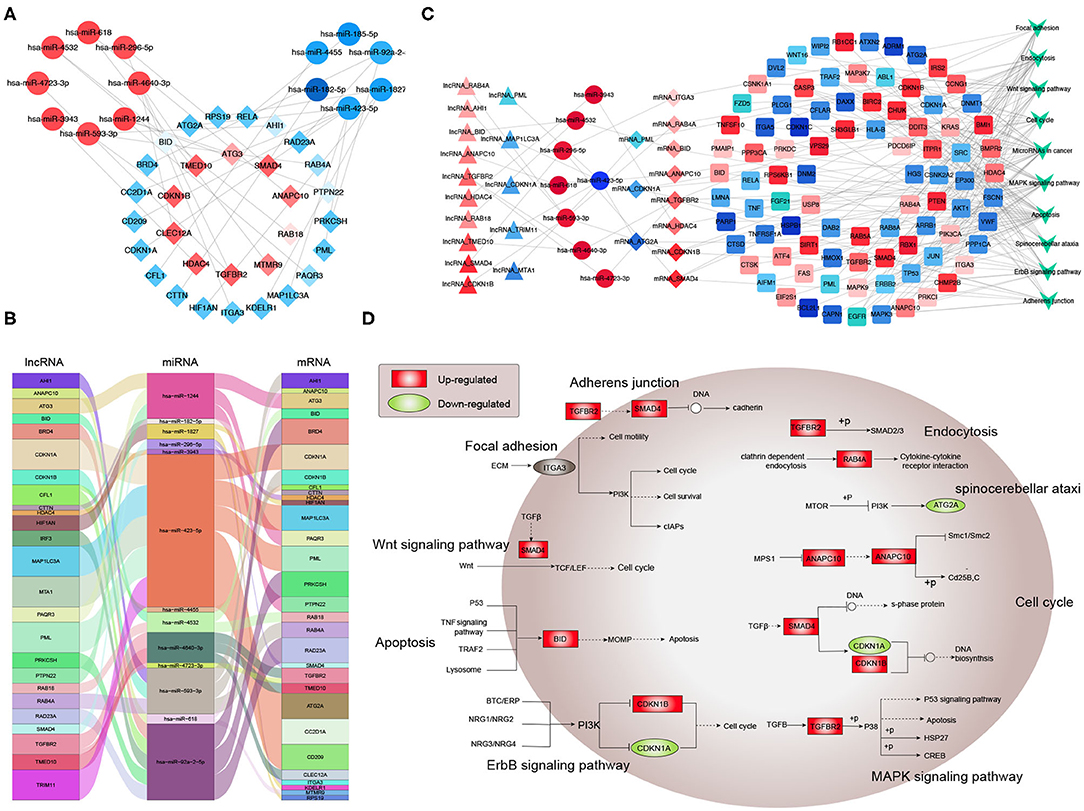
Figure 4. Exploration of a competing endogenous (ce) RNA network in schizophrenia. (A) Network of miRNAs, showing relationships between 14 miRNAs and 31 target genes (mRNAs). Red means up-regulation and blue means down-regulation. Circles indicate miRNAs; diamonds, mRNAs. The darker the color, the greater the absolute value of logFC. (B) Sankey plot, showing relationships among mRNAs, lncRNAs, and miRNAs. (C) Network of lncRNAs, miRNAs, mRNAs, and KEGG pathways. Relationships involving 8 miRNAs, 15 lncRNAs, 11 mRNAs, and 10 KEGG pathways are shown. Triangles represent lncRNAs; circles, miRNAs; diamonds, mRNAs; squares, genes in KEGG pathways; V,KEGG pathway. Blue means down-regulation; red means up-regulation. (D) Nine KEGG pathways in the ceRNA network that have previously been associated with schizophrenia.
Diagnostic Ability of Autophagy-Related Core DElncRNAs, DEmiRNAs and DEmRNAs for Schizophrenia
Most core dysregulated DElncRNAs were up-regulated in schizophrenia compared to controls (Figure 5A). However, the GSVA score based on the core DElncRNAs did not differ significantly between patients and controls (Figure 5B, Table 5). Similarly, the core DElncRNAs showed a poor ability to differentiate patients from controls in the test set (GSE38484, AUC = 0.606) and validation set (GSE16930, AUC = 0.694) (Figure 5C). Although core dysregulated DEmiRNAs did not give a significantly different GSVA score between patients and controls (Figure 5D, Table 6), the score proved to differentiate the two groups well (Figure 5E). This suggests its potential as a diagnostic biomarker. Most core dysregulated DEmRNAs were up-regulated in schizophrenia compared to controls (Figure 5F, Table 7). The GSVA score based on core dysregulated DEmRNAs were significantly higher in patients (p = 0.0087, Figure 5G), and it differentiated patients from controls with good AUCs in the test set (GSE38484, AUC = 0.659) and validation set (GSE16930, AUC = 0.778) (Figure 5H).
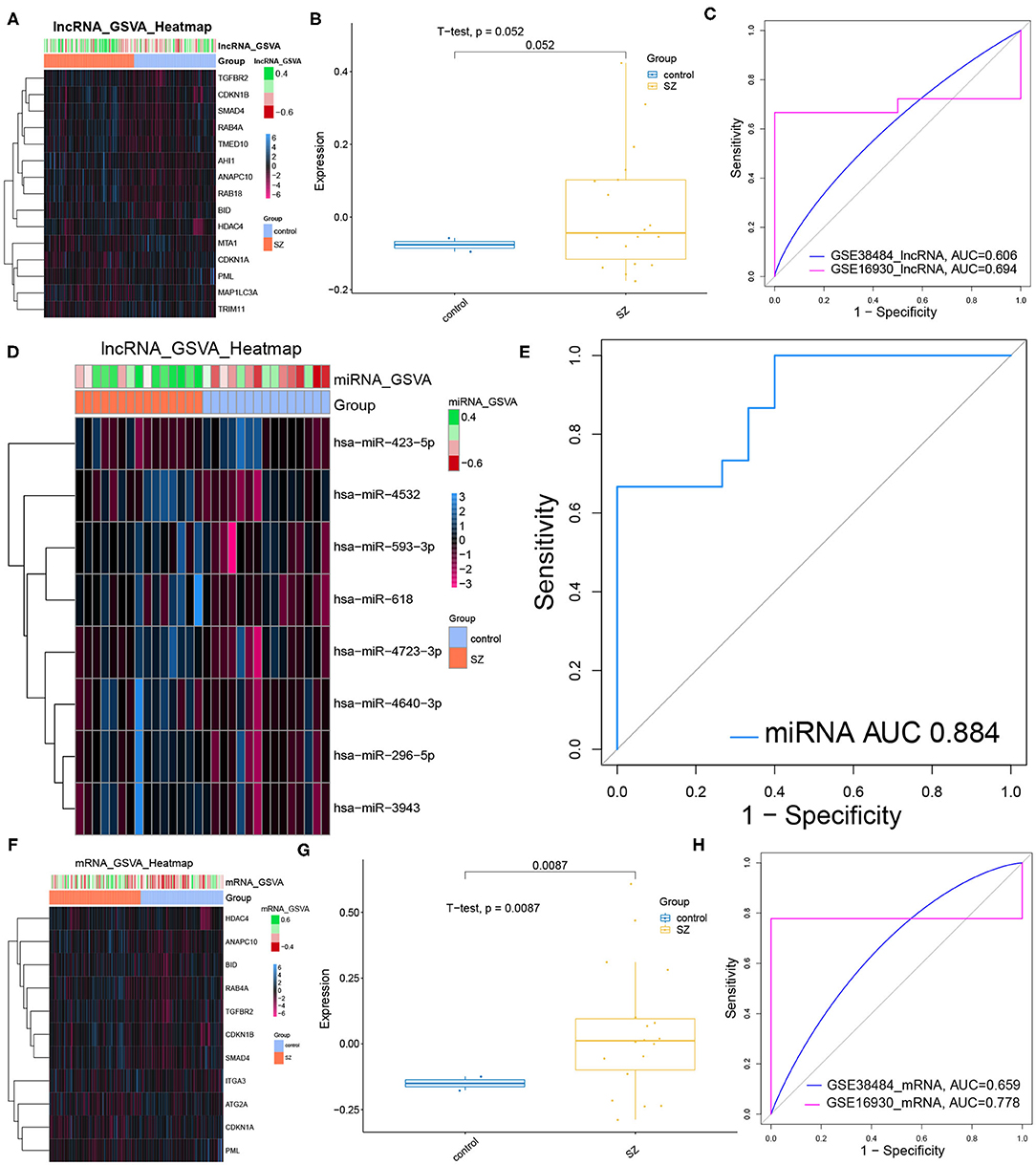
Figure 5. Performance of core dysregulated autophagy-related DElncRNAs, DEmiRNAs and DEmRNAs for diagnosing schizophrenia (SZ). (A) Gene set variation analysis (GSVA) of lncRNA expression. Blue means up-regulation; red means down-regulation. (B) GVSA score for core dysregulated DElncRNAs in the validation set (GSE16930). The horizontal axis shows sample names; the vertical axis, gene expression. Control data are shown in blue, patient data in yellow. (C) Receiver operating characteristic curves assessing how well the GSVA score for core dysregulated DElncRNAs diagnosed schizophrenia in the test set (GSE38484) and validation set (GSE16930). (D) GSVA-miRNA expression heat map. (E) ROC curve analysis for the GSVA score of core dysregulated DEmiRNAs in test set (GSE38484). (F) GSVA-mRNA expression heat map. (G) The expression of the GVSA score of core dysregulated mRNAs in validation set (GSE16930). (H) ROC analysis for the GSVA score of core dysregulated DEmRNAs in test set (GSE38484) and validation set (GSE16930).
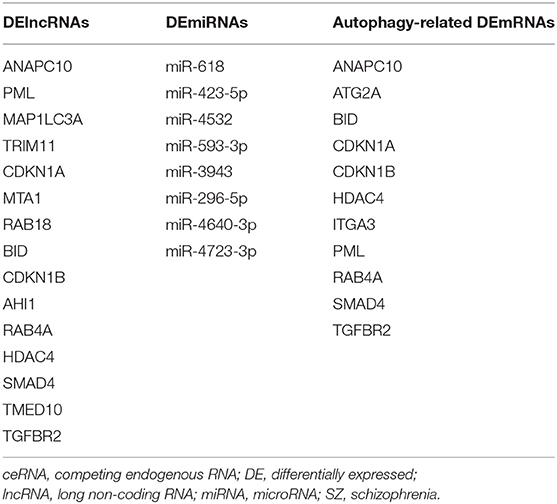
Table 6. Autophagy-related DElncRNAs, DEmiRNAs, and DEmRNAs in the ceRNA network potentially involved in schizophrenia.
Discussion
Schizophrenia is a persistent mental illness that disrupts normal thinking, function and mobility, and it can seriously impact patients and their families. Current anti-schizophrenia drugs can treat only the symptoms of the disease (35). To deepen the understanding of pathology of SZ to guide the diagnosis and treatment, the present study exlpored a ceRNA network that may be related to the disease by altering the regulation of genes involved in autophagy.
At the core of the ceRNA network, we identified 15 lncRNAs, 8 miRNAs, 11 mRNAs, and 10 KEGG pathways. Several of these RNAs have already been associated with schizophrenia. The miRNA137, which maps to chromosome 1p21.3, appears to confer susceptibility to the disease (36), while miR-219 is significantly up-regulated in the dorsolateral pre-frontal cortex of patients (37). The lncRNA MIAT (38), also called Gomafu (39), is down-regulated in schizophrenia, and this lower expression appears to reduce the activity of neurons (40).
Among the 10 core KEGG pathways in our ceRNA network, nine have already been associated with schizophrenia: cell cycle (41), spinocerebellar ataxia (42), apoptosis (43), ErbB signaling (44), focal adhesion (45), endocytosis (46), adhesions junction (47), Wnt signaling (48), and MAPK signaling (49). Mammalian mTOR target mTOR complex 1 (mTORC1) phosphorylates Unc51-like autophagy-activated kinase to block the initiation of autophagy. Both AMPK and oxidative stress can activate the transcription factors EB, FOXO1/3, transcription factor 4, and NF-κB to turn on expression of the autophagy-activated kinase (50).
The results of this study show that based on the exploration of the ceRNA network in schizophrenia, eight core disorders of DEmiRNA (hsa-miR-423-5p, hsa-miR-4532, hsa-miR-593-3P, hsa-miR-618, hsa-miR-4723-3p, hsa-miR-4640-3p, hsa-miR-296-5p, and hsa-miR-3943) may play a role in the diagnosis and treatment of schizophrenia. This article provides some basis for the study of ceRNA in schizophrenia. A previous study showed that hsa-miR-423-5p expressed in brain and were associated with amyotrophic (50). Hsa-miR-296-5p can be used as the prognostic marker for anaplastic glioma, secondary and anterior glioma patient (51). These studies indicated that the miRNA may be used as biomarker for neurological diseases.
In short, this study provides deeper insights into the construction of lncRNA-miRNA-mRNA network involving autophagy-related genes in SZ, and provides new targets for the diagnosis of SZ patients. However, there are some limitations at present. Firstly, due to the small sample size of the lncRNA and mRNA verification sets, and the lack of miRNA verification sets. The expression profiles of lncRNA and mRNA are obtained from the same sample, but miRNA is obtained from a separate data set. The combination of two data sets into a network may lead to selection bias due to batch effect. Secondly, the results of our study only indicate that these ceRNA network may exist in patients with SZ. However, it needs further evidence whether ceRNA exists in SZ patients, with the help of systematic biological experiment in vivo or in vitro. Relevant molecular biology experiments are required to obtain more credible results.
Conclusion
Our results suggest that the ceRNA network is involved in schizophrenia, which may deepen our understanding of the disease and guide the development of new treatments. The GSVA score based on the following eight core dysregulated DEmiRNAs may improve diagnosis of the disease: hsa-miR-423-5p, hsa-miR-4532, hsa-miR-593-3P, hsa-miR-618, hsa-miR-4723-3p, hsa-miR-4640-3p, hsa-miR-296-5p, and hsa-miR-3943.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
RL, QW, YQ, DZ, and CL designed the study and contributed to drafting the manuscript. RL, QW, YQ, YM, LW, HW, RM, DZ, and CL collated data and carried out data analyses. All authors have read and approved the final submitted manuscript.
Funding
This study was supported by the Guangxi Natural Science Foundation (2016GXNSFCA380012), the Project of Qingxiu District of Nanning Scientific Research and Technology Development Plan (2020058), the Scientific Research Project of Guangxi Zhuang Autonomous Region Health Commission (Z20180081 and Z20200201), the High-Level Medical Expert Training Program of Guangxi 139 Plan Funding (G201903049), and the Nanning Excellent Young Scientist Program and the Guangxi Beibu Gulf Economic Zone Major Talent Program (RC20190103).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2021.628361/full#supplementary-material
References
1. Vita A, Barlati S, De Peri L, Deste G, Sacchetti E. Schizophrenia. Lancet. (2016) 388:1280. doi: 10.1016/S0140-6736(16)31674-9
2. Hemager N, Plessen KJ, Thorup A, Christiani C, Ellersgaard D, Spang KS, et al. Assessment of neurocognitive functions in 7-year-old children at familial high risk for schizophrenia or bipolar disorder: the danish high risk and resilience study VIA 7. JAMA Psychiatry. (2018) 75:844–52. doi: 10.1001/jamapsychiatry.2018.1415
3. Dixon L. what it will take to make coordinated specialty care available to anyone experiencing early schizophrenia: getting over the hump. JAMA Psychiatry. (2017) 74:7–8. doi: 10.1001/jamapsychiatry.2016.2665
4. Weinstein JJ, Chohan MO, Slifstein M, Kegeles LS, Moore H, Abi-Dargham A. Pathway-specific dopamine abnormalities in schizophrenia. Biol Psychiatry. (2017) 81:31–42. doi: 10.1016/j.biopsych.2016.03.2104
5. Nicholl J, LaFrance WC Jr. Neuropsychiatric sequelae of traumatic brain injury. Semin Neurol. (2009) 29:247–55. doi: 10.1055/s-0029-1223878
6. van Os J, Bak M, Hanssen M, Bijl RV, de Graaf R, Verdoux H. Cannabis use and psychosis: a longitudinal population-based study. Am J Epidemiol. (2002) 156:319–27. doi: 10.1093/aje/kwf043
7. Svrakic DM, Zorumski CF, Svrakic NM, Zwir I, Cloninger CR. Risk architecture of schizophrenia: the role of epigenetics. Curr Opin Psychiatry. (2013) 26:188–95. doi: 10.1097/YCO.0b013e32835d8329
8. Ng MY, Levinson DF, Faraone SV, Suarez BK, DeLisi LE, Arinami T, et al. Meta-analysis of 32 genome-wide linkage studies of schizophrenia. Mol Psychiatry. (2009) 14:774–85. doi: 10.1038/mp.2008.135
9. Stepnicki P, Kondej M, Kaczor AA. Current concepts and treatments of schizophrenia. Molecules. (2018) 23:2087. doi: 10.3390/molecules23082087
10. Vucicevic L, Misirkic-Marjanovic M, Harhaji-Trajkovic L, Maric N, Trajkovic V. Mechanisms and therapeutic significance of autophagy modulation by antipsychotic drugs. Cell Stress. (2018) 2:282–91. doi: 10.15698/cst2018.11.161
11. Mizushima N. A brief history of autophagy from cell biology to physiology and disease. Nat Cell Biol. (2018) 20:521–7. doi: 10.1038/s41556-018-0092-5
12. Zou D, Chen L, Deng D, Jiang D, Dong F, McSweeney C, et al. DREADD in parvalbumin interneurons of the dentate gyrus modulates anxiety, social interaction and memory extinction. Curr Mol Med. (2016) 16:91–102. doi: 10.2174/1566524016666151222150024
13. Polajnar M, Zerovnik E. Impaired autophagy: a link between neurodegenerative and neuropsychiatric diseases. J Cell Mol Med. (2014) 18:1705–11. doi: 10.1111/jcmm.12349
14. Hsin KY, Matsuoka Y, Asai Y, Kamiyoshi K, Watanabe T, Kawaoka Y, et al. systemsDock: a web server for network pharmacology-based prediction and analysis. Nucleic Acids Res. (2016) 44:W507–13. doi: 10.1093/nar/gkw335
15. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
16. Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. (2011) 146:353–8. doi: 10.1016/j.cell.2011.07.014
17. Tian T, Wei Z, Chang X, Liu Y, Gur RE, Sleiman PMA, et al. The LONG Noncoding RNA landscape in amygdala tissues from schizophrenia patients. EBioMedicine. (2018) 34:171–81. doi: 10.1016/j.ebiom.2018.07.022
18. He K, Guo C, He L, Shi Y. MiRNAs of peripheral blood as the biomarker of schizophrenia. Hereditas. (2018) 155:9. doi: 10.1186/s41065-017-0044-2
19. van Eijk KR, de Jong S, Strengman E, Buizer-Voskamp JE, Kahn RS, Boks MP, et al. Identification of schizophrenia-associated loci by combining DNA methylation and gene expression data from whole blood. Eur J Hum Genet. (2015) 23:1106–10. doi: 10.1038/ejhg.2014.245
20. de Jong S, Boks MP, Fuller TF, Strengman E, Janson E, de Kovel CG, et al. A gene co-expression network in whole blood of schizophrenia patients is independent of antipsychotic-use and enriched for brain-expressed genes. PLoS ONE. (2012) 7:e39498. doi: 10.1371/journal.pone.0039498
21. Zhang F, Xu Y, Shugart YY, Yue W, Qi G, Yuan G, et al. Converging evidence implicates the abnormal microRNA system in schizophrenia. Schizophr Bull. (2015) 41:728–35. doi: 10.1093/schbul/sbu148
22. Qin G, Hu B, Li X, Li R, Meng Y, Wang Y, et al. Identification of key differentially expressed transcription factors in glioblastoma. J Oncol. (2020) 2020:9235101. doi: 10.1155/2020/9235101
23. Liu Z, Mi M, Li X, Zheng X, Wu G, Zhang L. lncRNA OSTN-AS1 may represent a novel immune-related prognostic marker for triple-negative breast cancer based on integrated analysis of a ceRNA network. Front Genet. (2019) 10:850. doi: 10.3389/fgene.2019.00850
24. Lee CH, Liu CM, Wen CC, Chang SM, Hwu HG. Genetic copy number variants in sib pairs both affected with schizophrenia. J Biomed Sci. (2010) 17:2. doi: 10.1186/1423-0127-17-2
25. Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. (2015) 43:e47. doi: 10.1093/nar/gkv007
26. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. (2015) 43:D447–52. doi: 10.1093/nar/gku1003
27. Feng H, Gu ZY, Li Q, Liu QH, Yang XY, Zhang JJ. Identification of significant genes with poor prognosis in ovarian cancer via bioinformatical analysis. J Ovarian Res. (2019) 12:35. doi: 10.1186/s13048-019-0508-2
28. Li MM, Liu XH, Zhao YC, Ma XY, Zhou YC, Zhao YX, et al. Long noncoding RNA KCNQ1OT1 promotes apoptosis in neuroblastoma cells by regulating miR-296-5p/Bax axis. FEBS J. (2020) 287:561–77. doi: 10.1111/febs.15047
29. Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics. (2013) 14:7. doi: 10.1186/1471-2105-14-7
30. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. (2005) 102:15545–50. doi: 10.1073/pnas.0506580102
31. Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. (2015) 1:417–25. doi: 10.1016/j.cels.2015.12.004
32. Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, et al. ClueGO: a Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics. (2009) 25:1091–3. doi: 10.1093/bioinformatics/btp101
33. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. (2015) 4:e05005. doi: 10.7554/eLife.05005
34. Guo Q, Guan GF, Cheng W, Zou CY, Zhu C, Cheng P, et al. Integrated profiling identifies caveolae-associated protein 1 as a prognostic biomarker of malignancy in glioblastoma patients. CNS Neurosci Ther. (2019) 25:343–54. doi: 10.1111/cns.13072
35. Peng S, Li W, Lv L, Zhang Z, Zhan X. BDNF as a biomarker in diagnosis and evaluation of treatment for schizophrenia and depression. Discov Med. (2018) 26:127–36.
36. International Schizophrenia C, Purcell SM, Wray NR, Stone JL, Visscher PM, O'Donovan MC, et al. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature. (2009) 460:748–52. doi: 10.1038/nature08185
37. Darcq E, Warnault V, Phamluong K, Besserer GM, Liu F, Ron D. MicroRNA-30a-5p in the prefrontal cortex controls the transition from moderate to excessive alcohol consumption. Mol Psychiatry. (2015) 20:1219–31. doi: 10.1038/mp.2014.120
38. Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, et al. Genomic analysis of mouse retinal development. PLoS Biol. (2004) 2:E247. doi: 10.1371/journal.pbio.0020247
39. Sone M, Hayashi T, Tarui H, Agata K, Takeichi M, Nakagawa S. The mRNA-like noncoding RNA Gomafu constitutes a novel nuclear domain in a subset of neurons. J Cell Sci. (2007) 120(Pt 15):2498–506. doi: 10.1242/jcs.009357
40. Chung DW, Volk DW, Arion D, Zhang Y, Sampson AR, Lewis DA. Dysregulated ErbB4 splicing in schizophrenia: selective effects on parvalbumin expression. Am J Psychiatry. (2016) 173:60–8. doi: 10.1176/appi.ajp.2015.15020150
41. Okazaki S, Boku S, Otsuka I, Mouri K, Aoyama S, Shiroiwa K, et al. The cell cycle-related genes as biomarkers for schizophrenia. Prog Neuropsychopharmacol Biol Psychiatry. (2016) 70:85–91. doi: 10.1016/j.pnpbp.2016.05.005
42. Trikamji B, Singh P, Mishra S. Spinocerebellar ataxia-10 with paranoid schizophrenia. Ann Indian Acad Neurol. (2015) 18:93–5. doi: 10.4103/0972-2327.144285
43. Beyazyuz M, Kufeciler T, Bulut L, Unsal C, Albayrak Y, Akyol ES, et al. Increased serum levels of apoptosis in deficit syndrome schizophrenia patients: a preliminary study. Neuropsychiatr Dis Treat. (2016) 12:1261–8. doi: 10.2147/NDT.S106993
44. Mostaid MS, Lee TT, Chana G, Sundram S, Shannon Weickert C, Pantelis C, et al. Peripheral transcription of NRG-ErbB pathway genes are upregulated in treatment-resistant schizophrenia. Front Psychiatry. (2017) 8:225. doi: 10.3389/fpsyt.2017.00225
45. Fan Y, Abrahamsen G, Mills R, Calderon CC, Tee JY, Leyton L, et al. Focal adhesion dynamics are altered in schizophrenia. Biol Psychiatry. (2013) 74:418–26. doi: 10.1016/j.biopsych.2013.01.020
46. Wang C, Wang Y, Hu M, Chai Z, Wu Q, Huang R, et al. Synaptotagmin-11 inhibits clathrin-mediated and bulk endocytosis. EMBO Rep. (2016) 17:47–63. doi: 10.15252/embr.201540689
47. Luciano M, Hansell NK, Lahti J, Davies G, Medland SE, Raikkonen K, et al. Whole genome association scan for genetic polymorphisms influencing information processing speed. Biol Psychol. (2011) 86:193–202. doi: 10.1016/j.biopsycho.2010.11.008
48. Hoseth EZ, Krull F, Dieset I, Morch RH, Hope S, Gardsjord ES, et al. Exploring the Wnt signaling pathway in schizophrenia and bipolar disorder. Transl Psychiatry. (2018) 8:55. doi: 10.1038/s41398-018-0102-1
49. Zou D, Qiu Y, Li R, Meng Y, Wu Y. A novel schizophrenia diagnostic model based on statistically significant changes in gene methylation in specific brain regions. Biomed Res Int. (2020) 2020:8047146. doi: 10.1155/2020/8047146
50. Feng Y, Yao Z, Klionsky DJ. How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy. Trends Cell Biol. (2015) 25:354–63. doi: 10.1016/j.tcb.2015.02.002
Keywords: schizophrenia, autophagy, ceRNA, competing endogenous RNA, lncRNA-miRNA-mRNA
Citation: Li R, Wang Q, Qiu Y, Meng Y, Wei L, Wang H, Mo R, Zou D and Liu C (2021) A Potential Autophagy-Related Competing Endogenous RNA Network and Corresponding Diagnostic Efficacy in Schizophrenia. Front. Psychiatry 12:628361. doi: 10.3389/fpsyt.2021.628361
Received: 11 November 2020; Accepted: 02 February 2021;
Published: 23 February 2021.
Edited by:
Antonios Stamatakis, National and Kapodistrian University of Athens, GreeceReviewed by:
Lilach Soreq, University College London, United KingdomTheodora Kalpachidou, Innsbruck Medical University, Austria
Copyright © 2021 Li, Wang, Qiu, Meng, Wei, Wang, Mo, Zou and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Donghua Zou, danvor0922@hotmail.com; Chunbin Liu, liuchunbin@163.com
†These authors have contributed equally to this work
 Rongjie Li1,2†
Rongjie Li1,2† Donghua Zou
Donghua Zou