- 1Department of Bioscience, Tokyo University of Agriculture, Tokyo, Japan
- 2Department of Life Sciences, Faculty of Life and Environmental Sciences, Shimane University, Matsue, Japan
- 3Department of Plant Sciences, Institute of Agrobiological Science, NARO, Tsukuba, Ibaraki, Japan
- 4Nodai Genome Center, Tokyo University of Agriculture, Tokyo, Japan
- 5RIKEN BioResource Research Center, Tsukuba, Ibaraki, Japan
We have previously reported a wide variation in salt tolerance among Arabidopsis thaliana accessions and identified ACQOS, encoding a nucleotide-binding leucine-rich repeat (NLR) protein, as the causal gene responsible for the disturbance of acquired osmotolerance induced after mild salt stress. ACQOS is conserved among Arabidopsis osmosensitive accessions, including Col-0. In response to osmotic stress, it induces detrimental autoimmunity, resulting in suppression of osmotolerance, but how ACQOS triggers autoimmunity remains unclear. Here, we screened acquired osmotolerance (aot) mutants from EMS-mutagenized Col-0 seeds and isolated the aot19 mutant. In comparison with the wild type (WT), this mutant had acquired osmotolerance and decreased expression levels of pathogenesis-related genes. It had a mutation in a splicing acceptor site in NUCLEOPORIN 85 (NUP85), which encodes a component of the nuclear pore complex. A mutant with a T-DNA insertion in NUP85 acquired osmotolerance similar to aot19. The WT gene complemented the osmotolerant phenotype of aot19. We evaluated the acquired osmotolerance of five nup mutants of outer-ring NUPs and found that nup96, nup107, and aot19/nup85, but not nup43 or nup133, showed acquired osmotolerance. We examined the subcellular localization of the GFP–ACQOS protein and found that its nuclear translocation in response to osmotic stress was suppressed in aot19. We suggest that NUP85 is essential for the nuclear translocation of ACQOS, and the loss-of-function mutation of NUP85 results in acquired osmotolerance by suppressing ACQOS-induced autoimmunity in response to osmotic stress.
Introduction
Osmotic stress caused by drought, salt or cold reduces plant growth. Acquired stress tolerance is defined as the ability of plants to withstand stress following an initial stress exposure (Sung et al., 2003). Acquired osmotolerance after salt stress is widespread among Arabidopsis thaliana accessions (Katori et al., 2010). Pre-exposure of 7-d-old seedlings to 100 mM NaCl for 7 d (acclimation period) leads to acquired osmotolerance to 750 mM sorbitol in some A. thaliana accessions (Ariga et al., 2017). We have identified ACQOS as the gene responsible for acquired osmotolerance. ACQOS is identical to VICTR, which encodes a nucleotide-binding leucine-rich repeat (NLR) protein (Ariga et al., 2017). This protein interacts with PHYTOALEXIN DEFICIENT4 (PAD4) and ENHANCED DISEASE SUSCEPTIBILITY1 (EDS1) in the nucleus to activate the immune response (Kim et al., 2012). ACQOS contributes to bacterial resistance in the absence of osmotic stress, but causes detrimental autoimmunity via EDS1 and PAD4, thereby reducing osmotolerance under osmotic stress (Ariga et al., 2017). The enhanced immune response mediated by EDS1/PAD4 induces programmed cell death (PCD) (Bartsch et al., 2006). Accessions with functional ACQOS alleles (e.g., Col-0) are impaired in acquired osmotolerance, whereas accessions with non-functional alleles (e.g., Bu-5) show acquired osmotolerance (Ariga et al., 2017).
ACQOS induces the immune response via EDS1 and PAD4 (Kim et al., 2012; Ariga et al., 2017). EDS1 and PAD4 localize in the nucleus, while NLRs localize in the cytoplasm and nucleus (Zhu et al., 2010; Alcázar and Parker, 2011; Roth et al., 2017; Zhang et al., 2020). The mechanism of ACQOS translocation to the nucleus is unknown. The nuclear pore complex (NPC) is responsible for the nuclear–cytoplasmic transport of mRNA and proteins (Wiermer et al., 2012). It allows cytoplasmic-to-nuclear transport of proteins larger than 40–60 kDa through importins and exportins (Wang and Brattain, 2007; Saier et al., 2014). The NPC consists of about 30 different nucleoporins (Nups) (Wälde and Kehlenbach, 2010), which form an outer cytoplasmic region and an inner nucleoplasmic region. The symmetrical core region consists of outer ring-, linker-, inner ring-, transmembrane ring-, and central phenylalanine-glycine NUPs (Yang et al., 2017). Abscisic acid (ABA) is important in osmotic stress responses, and its enhanced signaling improves osmotic tolerance (Nakashima et al., 2009). A mutation in an NPC component, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES1 (HOS1) increases transcript levels of cold-responsive genes such as RESPONSIVE TO DESICCATION 29A (RD29A) and COLD-REGULATED 15A (COR15A) under cold stress relative to WT (Ishitani et al., 1998). On the other hand, both hos1 and nup85 mutations suppress their expression under ABA, salt, and osmotic stress (Zhu et al., 2017). NUP85 interacts with the transcription factor MED18, which controls the expression of osmotic stress–responsive genes (Lai et al., 2014; Liao et al., 2016) In both mutants, sensitivity to ABA and mild salt stress is higher than in WT (Zhu et al., 2017). The double mutant bak1 bkk1, a co-receptor of the brassinosteroid (BR) receptor BRI1, BRI1-ASSOCIATED KINASE 1 (BAK1), and its homolog BAK1-LIKE 1 (BKK1), exhibits a salicylic acid-dependent cell death phenotype and constitutive expression of PR genes even without pathogen invasion (He et al., 2007). NUP85 was identified as the causal gene for a suppressor of bak1 bkk1 (sbb1-1) mutant, which suppresses the cell death phenotype of bak1 bkk1 (Du et al., 2016). It suggests that NUP is also involved in plant defense and the regulation of cell death.
Suppressor of npr1-1, constitutive 1 (snc1) is a gain-of-function Arabidopsis mutant carrying a mutation in NLR that constitutively activates resistance responses to pathogens (Li et al., 2001; Zhang et al., 2003). Modifiers of snc (mos) 1, 3, and 6 suppress the autoimmune responses mediated by snc1 (Palma et al., 2005; Zhang and Li, 2005). MOS3 encodes NUP96 and MOS6 encodes importin α3. Nuclear accumulation of SNC1 is inhibited in those mutants, which explains the suppression of the autoimmune responses (Zhu et al., 2010; Roth et al., 2017; Zhang et al., 2020). In addition to ACQOS, mutations in EDS1 and PAD4 (Alcázar and Parker, 2011; Cui et al., 2017), as well as RAR1 and SGT1, whose products facilitate stable NLR protein accumulation and function (Takahashi et al., 2003) result in acquired osmotolerance in the Col-0 background (Ariga et al., 2017). However, the molecules involved in the localization of ACQOS are unknown.
Here, we EMS-mutagenized Col-0 seeds, isolated acquired osmotolerance (aot) mutants. We sequenced 5 genes including ACQOS, EDS1, PAD4, RAR1, and SGT1 known to be involved in acquired osmotolerance in the Col-0 background (Ariga et al., 2017) and characterized one of the candidate mutants that none of these genes was mutated.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana seeds were sown on agar (1.0% w/v) plates containing full-strength Murashige and Skoog (MS) salts with a vitamin mixture (10 mg L−1 myoinositol, 200 µg L−1 glycine, 50 µg L−1 nicotinic acid, 50 µg L−1 pyridoxine hydrochloride, 10 µg L−1 thiamine hydrochloride, pH 5.7) and 1% w/w sucrose. Plates were sealed with surgical tape, the seeds were stratified at 4°C for 4 to 7 d, and transferred to a growth chamber (80 µmol photons m2 s−1; 16/8-h light/dark cycle; 22°C) for germination and growth (Isono et al., 2020).
Seeds of the A. thaliana Be-1 and Col-0 background mutants nup85-2 (SALK_113274), nup43 (SALK_095344C), mos3-2/nup96 (CS69987), nup107 (SALK_057072C), nup133 (SALK_092608C), and mos6 (SALK_119474C) were obtained from the Arabidopsis Biological Resource Center (ABRC, Ohio State University).
Mutagenesis and aot mutant screening
EMS-mutagenesis was performed as described in Isono et al. (2020). Acquired osmotolerance assay: Seedlings (7-d old) were grown on nylon mesh (990 μm) on an MS agar plate supplemented with 100 mM NaCl for 7 d and were then mesh-transferred to a plate supplemented with 750 mM sorbitol for 15 d or 38 d. Though Col-0 WT plants cannot survive in this assay, plants that survive under these conditions were isolated as aot candidates.
Abiotic stress assays
Seedlings (10-d old) were grown on nylon mesh (990 μm) on an MS agar plate supplemented with 600 mM sorbitol for 16 d (osmotic-shock stress) or 225 mM NaCl for 8 d (salt-shock stress). Their chlorophyll content was determined according to Porra et al. (1989).
RNA extraction, RT-PCR and qRT-PCR
Total RNA extraction and RT-PCR or qRT-PCR analysis were performed as described in Isono et al. (2020) (Isono et al., 2020). ACTIN2 was used as the internal standard for qRT-PCR. The primers are listed in Supplementary Table S1.
Genetic mapping of the causative gene of the aot19 phenotype
We crossed the aot19 mutant with Be-1, an accession that does not acquire osmotolerance like Col-0, and selfed the resulting F1 progeny to generate an F2 population. Genomic DNA was prepared from individual F2 plants with the osmotolerant phenotype and used as a PCR template. The simple-sequence-length polymorphism markers for mapping are listed in Supplementary Table S1. PCR conditions were as follows: initial denaturation at 94°C for 2 min; 34 cycles at 94°C for 20 s, 56 to 59°C for 20 s, and 72°C for 20 s; and final extension at 72°C for 2 min. Microsatellites were fractionated in 5% to 7% agarose gel, and recombination frequencies (%) were calculated from the band pattern.
DNA library construction, whole-genome sequencing of the aod19 mutant, and detection of mutations
All procedures were performed as in Tsukimoto et al. (2021). The read data were submitted to the DNA Data Bank of Japan Sequence Read Archive (accession number DRA016084).
Plasmid construction and transformation
For complementation test, the genomic region of At4g32910 (793 bp upstream of the ATG initiation codon to 690 bp downstream of the terminator) derived from WT Col-0 was amplified by PCR with AOT19/NUP85 primers and cloned into the binary vector pGreen0029. The construct was introduced into Agrobacterium tumefaciens GV3101, which was used for plant transformation to aot19 by the floral dip method. Transgenic plants were selected on MS agar plates containing 200 µg ml−1 claforan and 25 µg ml−1 kanamycin. Ten-day-old seedlings (T1 plants) were transferred to soil in pots.
For construction of p35S:mGFP-ACQOS, to avoid GFP dimerization, a A207K substitution (Zacharias et al., 2002), which eliminates the dimerization, was introduced into the sGFP gene of pGH35S-sGFP (Fujita et al., 2009) and the construct obtained was named pGH35S-mGFP. To obtain the coding sequence (CDS) of ACQOS, six exons of At5g46520 were independently amplified and then joined by overlapping-PCR. Using ACQOS_CDS_F and ACQOS_CDS_R primers, the final CDS was amplified without a stop codon and ligated into the EcoRV site of the pGH35S-mGFP vector. All primers are listed in Supplementary Table S1.
Cell death detection by trypan blue staining
Trypan blue was dissolved in a mixture of equal volumes of lactic acid (85% w/w), phenol (TE buffered, pH 7.5–8.0), glycerol, and distilled water in a plastic test tube. Seedlings were immersed in fresh trypan blue solution (0.2% w/v), boiled in a water bath for 1 min, incubated for 3 h at room temperature, and destained in chloral hydrate solution (500 g of chloral hydrate dissolved in 195 ml of distilled water) for 2 d. Destained seedlings were mounted in glycerol solution (70% v/v) for microscopy. Photographs were taken under a microscope (SZ61 or DP22, Olympus, Tokyo) under white light.
Microscopic analysis
Transgenic seedlings (4-day old) grown on an MS agar plate were transferred to a plate supplemented with 600 mM sorbitol for 2 d (osmotic-shock stress). Nuclei in the seedlings were stained in PBS containing 0.5 μg/ml Hoechst 33342 for 20–30 min before fluorescence microscopy. Images were obtained with an all-in-one fluorescence microscope (BZ-X800, Keyence, Osaka, Japan) equipped with an optical sectioning module and a Plan Apochromat 40× objective (NA0.95 and BZ-PA40, respectively, both from Keyence, Tokyo). Green fluorescence was detected with a GFP filter (ex 470/40 nm, em 525/50 nm, dichroic 495 nm; OP-87763, Keyence). Blue fluorescence (Hoechst 33342) was detected with a DAPI filter (ex 360/40 nm, em 460/50 nm, dichroic 400 nm; OP-87762, Keyence).
Results
Isolation of the acquired osmotolerance 19 mutant
We screened 19 500 M2 seedlings from an EMS-mutagenized Col-0 population for acquired osmotolerance to severe osmotic stress (750 mM sorbitol) after mild-salt acclimatization (100 mM NaCl for 7 d) and isolated 13 aot mutant candidates. We sequenced 5 genes known to be involved in acquired osmotolerance in the Col-0 background (Ariga et al., 2017) and detected mutations in ACQOS (7 lines), EDS1 (3 lines), PAD4 (1 line), and RAR1 (1 line), but no mutations in SGT1 (data not shown), but none of these genes was mutated in aot19. WT Col-0 did not acquire osmotolerance, but aot19 did (Figure 1A). Chlorophyll content did not differ significantly between WT and aot19 under normal conditions, but was significantly higher in aot19 after exposure to 750 mM sorbitol (Figure 1A). The fresh weight of aot19 plants tended to be smaller than that of WT plants under normal growth conditions (Figure 1B).
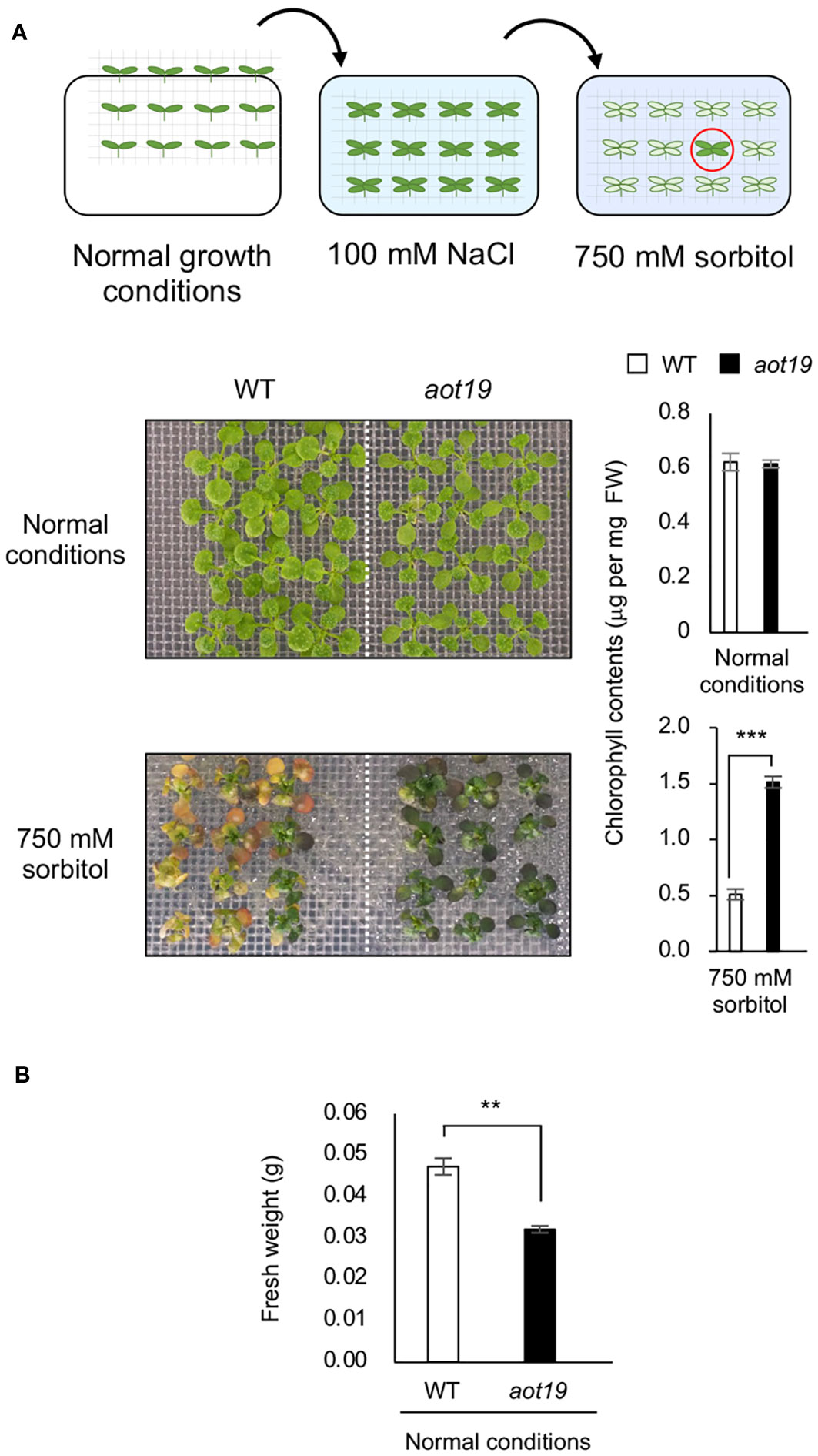
Figure 1 Identification of the acquired osmotolerance (aot) 19 mutant. (A) Flow chart of the acquired osmotolerance assay. A total of 19 500 EMS-mutagenized, salt-acclimatized, 14-d-old seedlings of accession Col-0 were transferred to Murashige and Skoog (MS) agar plates containing 750 mM sorbitol for 15 d, and osmotolerant seedlings (red circle) were selected. Upper photos: 14-day-old wild-type (WT) and aot19 seedlings grown under normal conditions. Lower photos: acquired osmotolerance of the WT and aot19 plants. Right panels: chlorophyll content as an index of acquired osmotolerance. FW, fresh weight. (B) Fresh weight of plants grown as described in A under normal growth conditions. Differences between WT and aot19 were analyzed by Student’s t-test (mean ± SE, n = 3, **P < 0.01, ***P < 0.001).
Characterization of the aot19 mutant
The aot19 plants were directly exposed to osmotic or salt stress without salt acclimatization (Figure 2A). Tolerance to osmo-shock (600 mM sorbitol for 16 d) and salt-shock (225 mM NaCl for 8 d) stresses was higher in aot19 than in WT plants (Figure 2A), suggesting that AOT19 negatively regulates acquired osmotolerance and osmo-shock tolerance. The mRNA levels of the ABA/osmostress marker genes RD29A, COR15A, and KIN1 were comparable between WT and aot19, although they were rather lower in aot19 than in WT at some time point under osmotic stress (Figure 2B), indicating that the osmotolerance of aot19 was not due to an increase in osmotic- and ABA-responsive gene expressions.
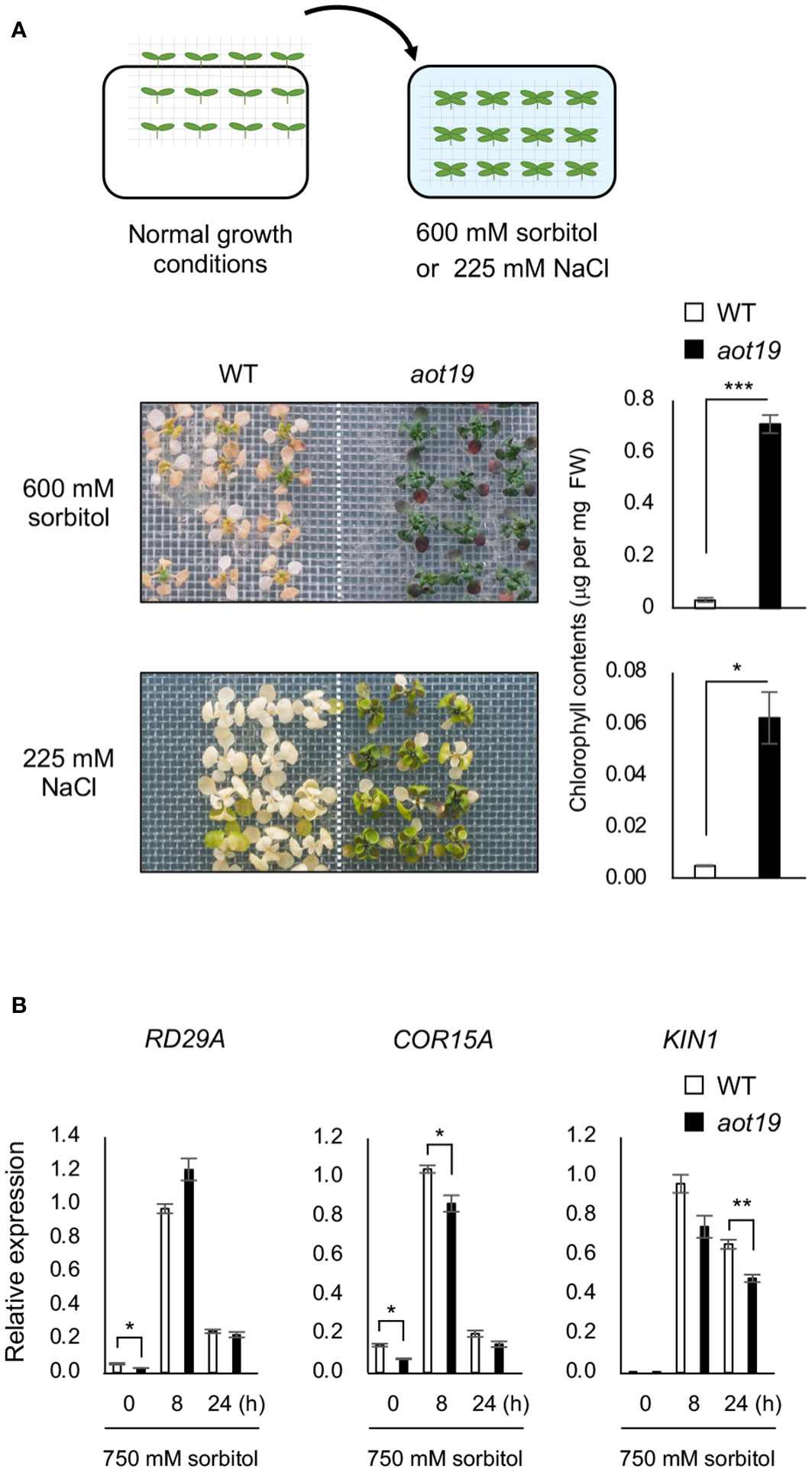
Figure 2 Characterization of the aot19 mutant. (A) Top: Flow chart of the salt- and osmo-shock tolerance assay. Seedlings (14-d old) were transferred to MS agar plates containing 225 mM NaCl for 8 d or 600 mM sorbitol for 16 d. Upper photos: osmo-shock tolerance of the WT and aot19 plants. Lower photos: salt-shock tolerance of the WT and aot19 plants. Right panels: Chlorophyll content of the salt- and osmo-shock tolerances. (B) Expression of osmostress-responsive marker genes in WT and aot19 under normal conditions and acquired osmotic stress (100 mM NaCl for 7 d and subsequent 750 mM sorbitol for 8 or 24 h). Expression levels were determined by quantitative real-time polymerase chain reaction (qRT-PCR) and were normalized to those of Actin2 (mean ± SE, n = 3). Differences between WT and aot19 were analyzed by Student’s t-test (mean ± SE, n = 3, *P < 0.05, **P < 0.01, ***P < 0.001).
Identification of the causal gene in aot19
High-resolution mapping using the F2 progeny from a cross between Be-1 and aot19 plants revealed the locus responsible for the osmotolerance of aot19 on chromosome 4 near position 16 000 k within a 900-kbp region (Figure 3A). Whole-genome sequencing revealed non-synonymous mutations in three genes within this region. We examined the acquired osmotolerance of the T-DNA insertion mutants of these genes that obtained from ABRC and found that only the at4g32910 mutant exhibited the osmotolerance compared with WT plants (Figure 3B; Supplementary Figure S1). In aot19, there is a mutation at the acceptor site of the third intron of the At4g32910 gene, resulting in intron retention (Figure 3A). The intron retention was confirmed by RT-PCR with primers designed for the exons adjacent to or surrounding the mutation (Figure 3C). It resulted in a stop codon within the intron, presumably resulting in a truncated At4g32910 protein (Figure 3C). In a complementation test, the aot19 plants transformed with At4g32910 (with its native promoter) had impaired acquired osmotolerance; chlorophyll content was significantly lower in these lines than in aot19 (Figure 3D). These results suggest that At4g32910 is the causal gene for aot19. AOT19 is identical to NUCLEOPORIN 85 (NUP85) (Figure 3A) and encodes a nuclear pore complex (NPC) component.
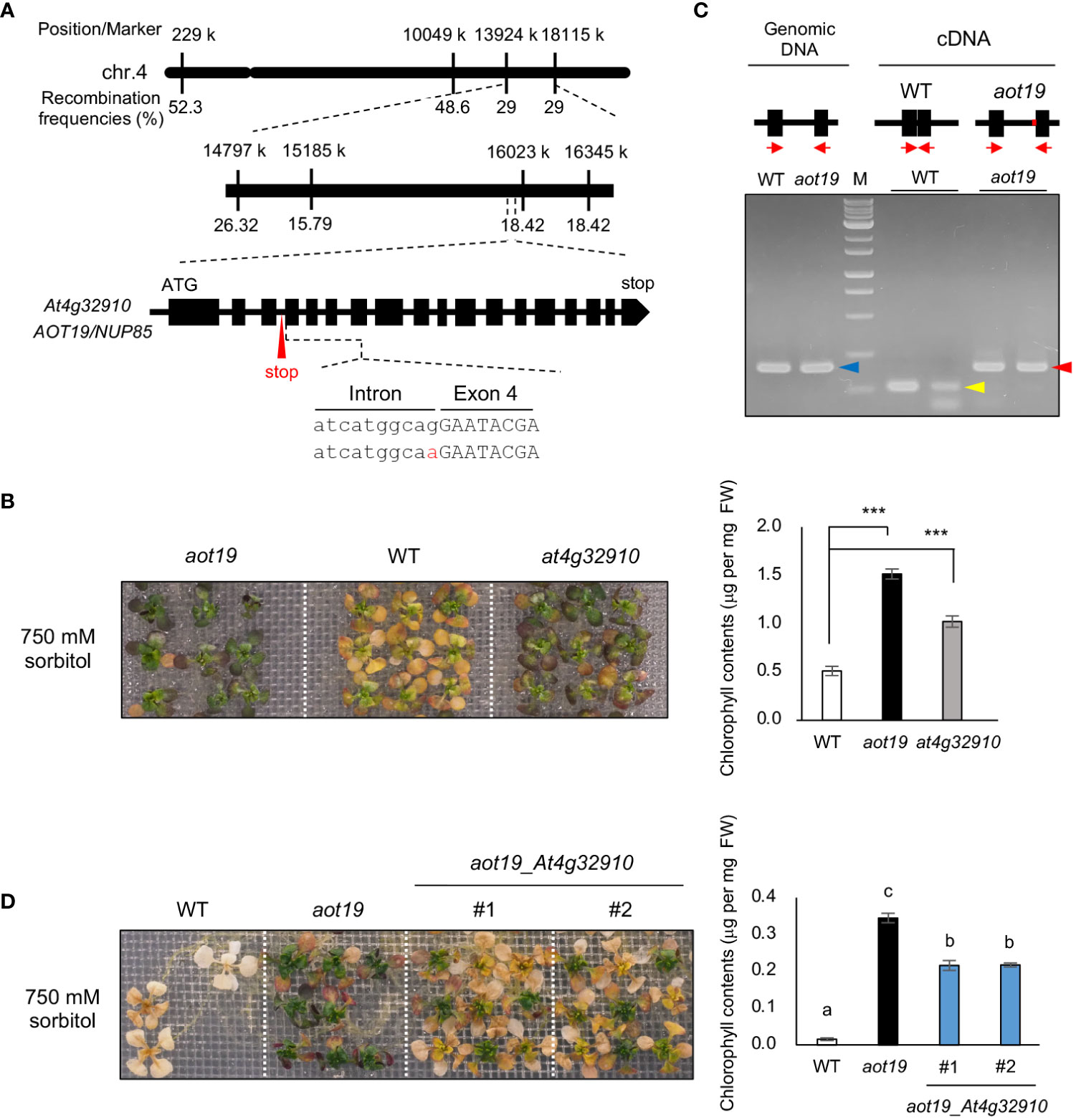
Figure 3 Identification of the causal gene in aot19. (A) High-resolution mapping of the causal locus in aot19 using F2 progeny between aot19 and Be-1. A mutation in aot19 (red) results in intron retention and a stop codon (red arrow). (B) Acquired osmotolerance assay of aot19, WT, and a T-DNA insertion mutant of At4g32910 in WT (at4g32910). The assay was performed as in Figure 1A. Photo: acquired osmotolerance of aot19, WT, and at4g32910 plants. Right panel: chlorophyll contents of the seedlings. Differences between the WT and the mutants were analyzed by Student’s t-test (mean ± SE, n = 3, ***P < 0.001). (C) Detection of intron retention by PCR. Upper panel: The block and line mean exon and intron, respectively. The red on the line means the mutation occurred in aot19. The red arrows indicate the positions of primers. Genomic PCR (blue arrow) and RT-PCR in aot19 (red arrow) and WT (yellow arrow) with primers designed for the exons before and after the mutation (n = 2). (D) Complementation test of aot19 with At4g32910. T2 plants transformed with their native promoter: At4g32910 (aot19_ At4g32910) derived from the WT Col-0 were used. The salt -acclimated plants were mesh-transferred to a plate supplemented with 750 mM sorbitol for 38 d. Right panel: chlorophyll contents of the seedlings. Bars labeled with different letters differed significantly (P < 0.05, one-way ANOVA with post hoc Tukey HSD test, mean ± SE, n = 3).
Analysis of AOT19/NUP85 function for osmotolerance
To investigate whether aot19 suppresses the osmostress-induced immune responses, we analyzed the expression levels of PATHOGENESIS-RELATED (PR) genes. Under osmotic stress (750 mM sorbitol for 3 d), expression of the PR1 and PR2 genes increased in WT, whereas this increase was significantly suppressed in aot19 (Figure 4A). We examined the expression levels of osmotic- and ABA-responsive genes RD29A, COR47, COR15A and ABI5, in addition to immune responsive genes PR1, PR2 and PR5 under mild and severe osmotic stress (Supplementary Figure S2). There were no substantial differences in osmotic- and ABA-responsive gene expressions between WT and aot19 mutants under either osmotic stress, although they were lower in aot19 than in WT at some point. As for the expression levels of PR1 and PR2, little expression was observed in both WT and aot19 mutant under mild osmotic stress. However, under severe osmotic stress, the expression levels of PR1, PR2 and PR5 increased significantly in WT compared with aot19. These results suggest that the mechanism by which aot19 exhibits osmotolerance is not due to an increase in osmotic- and ABA-responsive gene expressions, but rather to suppression of the enhanced immune response under severe osmotic stress.
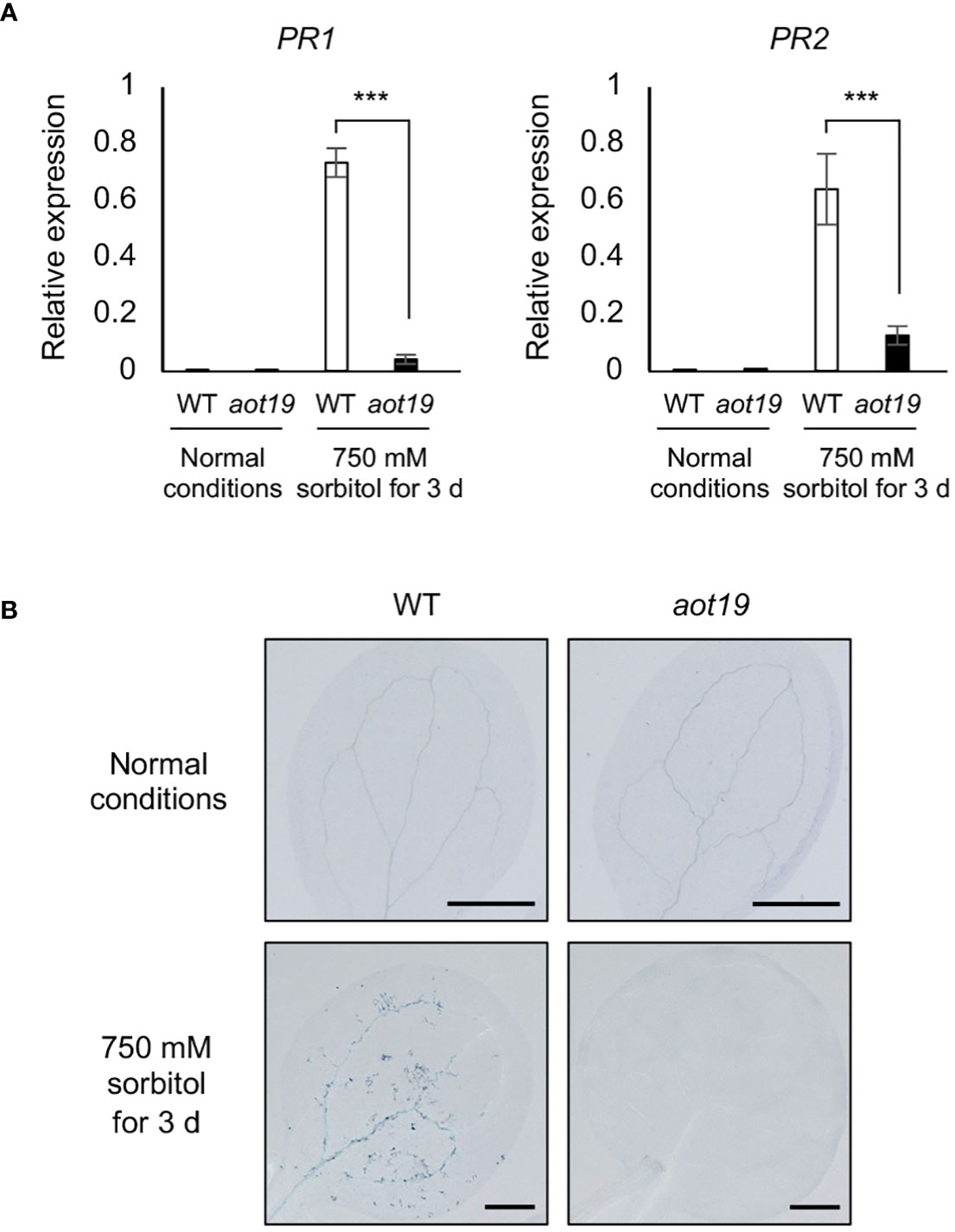
Figure 4 Immune response and programmed cell death in aot19. (A) Expression of pathogenesis-related (PR) 1 and PR2 genes in WT and aot19 plants under normal and acquired osmotolerance conditions (100 mM NaCl for 7 d followed by 750 mM sorbitol for 3 d). Expression levels were determined by qRT-PCR and were normalized to those of Actin2. Differences between WT and aot19 plants were analyzed by Student’s t-test (mean ± SE, n = 3, ***P < 0.001). (B) Trypan blue staining of seedling leaves.
We investigated whether PCD may be suppressed in rosette leaves of aot19 under osmotic stress. Osmotic stress increased trypan blue staining in WT leaves, but aot19 leaves were scarcely stained under either normal or osmotic-stress conditions (Figure 4B), suggesting that AOT19/NUP85 regulates osmostress-induced PCD.
We evaluated the acquired osmotolerance of five nup mutants of outer-ring NUPs and found that nup96, nup107, and aot19/nup85, but not nup43 or nup133, showed acquired osmotolerance (Figure 5; Supplementary Figure S1). These findings suggest that mutations in some NPC components suppress acquired osmotolerance in Col-0.

Figure 5 Acquired osmotolerance of nup mutants. (A) Acquired osmotolerance of Col-0 WT and Col-0-background aot19/nup85, nup43, nup96, nup107, and nup133 mutants. (B) Chlorophyll contents of the seedlings shown in (A) Bars labeled with different letters differed significantly (P < 0.05, one-way ANOVA with post hoc Tukey HSD test, mean ± SE, n = 3).
Subcellular localization of ACQOS in aot19
We hypothesized that ACQOS is unable to translocate from the cytoplasm into the nucleus in aot19. To test this hypothesis, we examined the subcellular localization of a GFP-ACQOS chimera under osmotic stress. In WT, ACQOS translocates from cytoplasm into nucleus in response to osmotic stress, whereas less osmostress-triggered nuclear translocation was not observed in aot19 than in WT (Figure 6). These results suggest that the AOT19/NUP85 mutation in aot19 prevents the nuclear translocation from the cytoplasm of ACQOS under osmotic stress; therefore, the immune response mediated by the nucleus-localized immune-regulators EDS1 and PAD4 is interrupted, resulting in enhanced osmotolerance.
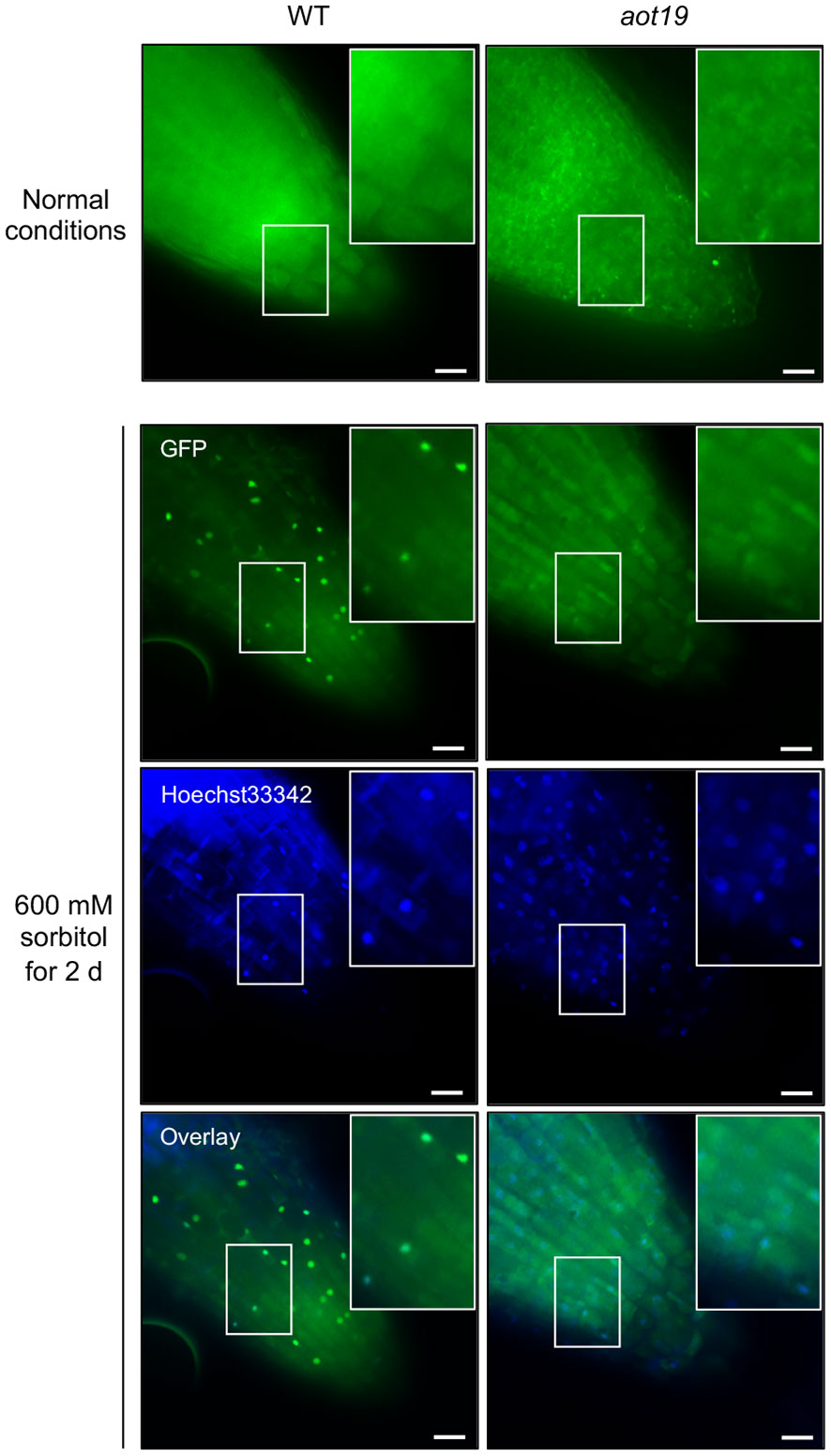
Figure 6 Subcellular localization of mGFP-ACQOS in roots. Seedlings of transgenic Arabidopsis Col-0 plants stably expressing ACQOS fused to the C-terminus of monomeric green fluorescent protein (mGFP) under the control of the promoter of the cauliflower mosaic virus (CaMV) 35S gene (p35S:mGFP-ACQOS). Seedlings were grown under normal conditions or osmotic stress (600 mM sorbitol for 2 d). Single-section images of the GFP channel (top), nucleus channel showing staining with Hoechst 33342 (middle), and their overlay (bottom) are shown in each panel. In each image, a large rectangle shows an enlargement of the area enclosed by the small rectangle. Scale bars, 20 μm.
Discussion
Here we report the A. thaliana aot19 mutant that acquired osmotolerance despite the Col-0 background. This mutant carries the functional ACQOS allele, which encodes an NLR protein and suppresses osmotolerance owing to detrimental autoimmunity via EDS1/PAD4. An AOT19/NUP85 mutation in aot19 inhibited nuclear translocation of ACQOS in response to osmotic stress. We suggest that the aot19 mutant acquires osmotolerance by suppressing the immune response due to the defect in ACQOS nuclear translocation, which is essential for ACQOS function to trigger the immune response via interaction with nuclear-localized EDS1 and PAD4 (Zhu et al., 2010; Alcázar and Parker, 2011; Roth et al., 2017; Zhang et al., 2020).
Nuclear translocation of proteins larger than 40–60 kDa requires an importin (Stewart, 2007; Wang and Brattain, 2007; Beck and Hurt, 2017). Importin MOS6 supports the nuclear translocation of the NLR-protein SNC1 (Palma et al., 2005; Roth et al., 2017; Lüdke et al., 2021). In response to osmotic stress, ACQOS (137 kDa) translocated into the nucleus in WT plants, so a yet-unidentified importin might be involved. It may be useful to examine the osmotolerance of importin knockout mutants or to isolate proteins that interact with ACQOS through the use of co-immunoprecipitation.
In aot19, the AOT19/NUP85 mutation may also affect the transport of other nuclear-localized proteins. The nup85 mutant of Lotus japonicus produces fewer seeds (Saito et al., 2007), as similarly seen in the aot19 mutant. In addition, aot19 mutant tended to have lower fresh weight than the WT plants, suggesting that the AOT19/NUP85 mutation impacts growth and reproduction. However, the impact on growth and reproduction was not severe, perhaps because the NUP85 mutation does not completely inhibit the cytoplasmic-to-nuclear transport of ACQOS and other proteins in aot19. ACQOS was detected in the nucleus in aot19, although less than in WT. The osmotolerance of F1 seedlings from the cross between Col-0 and NIL-Bu-5, which carried non-functional ACQOS alleles from Bu-5 in the genetic background of Col-0, was intermediate between those of Col-0 and NIL-Bu-5 (Ariga et al., 2017). This suggests that ACQOS decreases acquired osmotolerance in a protein level–dependent manner. Therefore, osmotolerance can be improved by partial limitation of ACQOS nuclear translocation.
NUP85 has been identified as the causal gene for a mutant with a decreased expression of an abiotic stress–responsive luciferase reporter (RD29A-LUC) in response to ABA and salt stress (Zhu et al., 2017). The expression of ABA- and salt stress–inducible genes is lower in the nup85 mutant than in WT, resulting in higher sensitivity of the mutant to ABA and mild salt stress of 100 mM NaCl (Zhu et al., 2017). On the other hand, aot19/nup85 was highly tolerant to severe osmotic (600 mM sorbitol) and salt (225 mM NaCl) stresses in this study. The improved osmotolerance of aot19 to severe osmotic stress may depend on whether ACQOS translocates to the nucleus at a particular stress intensity. Under mild osmotic stress, little expression of PR1, PR2 and PR5 was observed in both WT and aot19 mutants, but under severe osmotic stress, the expression levels were significantly increased in WT compared to aot19. Thus, under severe salt and osmotic stress, aot19/nup85 suppresses the excessive immune response by inhibiting the nuclear translocation of ACQOS, resulting in its higher tolerance than that of WT plants.
Of the nup mutants tested, nup96, nup107, and aot19/nup85, but not nup43 or nup133, acquired osmotolerance. These findings suggest that NUP85, NUP96, and NUP107 are important for ACQOS nuclear translocation. These NUPs are components of the outer NPC, and NUP96 is important in the nuclear migration of SNC1. NUP85 and NUP96 affect the expression of auxin, ABA, and osmotic stress–responsive genes (Zhu et al., 2017; Zhang et al., 2020). Despite reports that the NUP85 mutation results in decreased expression of osmotic stress-responsive genes, no substantial differences in expression levels between WT and aot19 mutant were observed. The T-DNA mutant of NUP85 exhibited the osmotolerance compared with WT plants. However, the aot19 mutant was more tolerant to osmotic stress than the T-DNA mutant. In addition, the complementary line aot19_At4g32910 impaired acquired osmotolerance compared to aot19, but more tolerant than WT. This may be because the immature truncated protein from aot19 competes with the correct protein from At4g32910, and as a result, the expressions of osmotic stress-responsive genes are not reduced in aot19. The Arabidopsis genome encodes approximately 150 NLRs, but little is known about their nuclear translocation. These NUPs may be important not only in immune responses but also in various environmental responses via nuclear translocation of NLRs.
Conclusion
We isolated the aot19 mutant, which shows acquired osmotolerance despite its Col-0 background and the presence of functional ACQOS alleles. The causal gene of aot19 is identical to NUP85, which encodes an NPC component. In aot19, the nuclear translocation of ACQOS and detrimental autoimmunity in response to osmotic stress were suppressed. These findings suggest that ACQOS translocates into the nucleus via the NPC, with NUP85, NUP96, and NUP107 playing an important role.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ddbj.nig.ac.jp/, DRA016084.
Author contributions
KM: Conceptualization, Data curation, Formal analysis, Investigation, Writing – original draft. YM: Data curation, Formal analysis, Writing – review & editing. MT: Formal analysis, Writing – original draft. SK: Formal analysis, Writing – original draft. KN: Data curation, Formal analysis, Writing – review & editing. HA: Data curation, Writing – review & editing. KT: Data curation, Writing – review & editing. SI: Resources, Writing – review & editing. IY: Data curation, Writing – review & editing. YS: Data curation, Writing – review & editing. TT: Conceptualization, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by KAKENHI grants from the Ministry of Education, Culture, Sports, Science and Technology of Japan (21H05668, 23H04206, and 23H00334 to TT).
Acknowledgments
We thank Dr. Kazuho Isono for his technical assistance in identifying the mutations in the aot19 mutant.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1304366/full#supplementary-material
Supplementary Figure 1 | T-DNA insertion mutants of five nup mutants. (A) T-DNA insertion site of T-DNA insertion mutants. (B) Expression of NUP85 in WT and nup85 mutant by RT-PCR. The fragments were separated on an agarose gel and stained with ethidium bromide. Arabidopsis ACTIN2 (ACT2) was used as the semiquantitative control.
Supplementary Figure 2 | Expression levels of osmotic-, ABA- and immune-responsive genes in WT and aot19 mutant under mild and severe osmotic stress. Expression of RD29A, COR15A, COR47, ABI5, PR1, PR2 and PR5 genes in WT and aot19 plants under normal, mild (300 mM sorbitol for 5 h and 3 d) and severe (750 mM sorbitol for 5 h and 3 d) osmotic stress conditions. Expression levels were determined by qRT-PCR and were normalized to those of Actin2. Differences between WT and aot19 plants were analyzed by Student’s t-test (mean ± SE, n = 3, *P < 0.05).
References
Alcázar, R., Parker, J. E. (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16, 666–675. doi: 10.1016/j.tplants.2011.09.001
Ariga, H., Katori, T., Tsuchimatsu, T., Hirase, T., Tajima, Y., Parker, J. E., et al. (2017). NLR locus-mediated trade-off between abiotic and biotic stress adaptation in Arabidopsis. Nat. Plants 3, 1–8. doi: 10.1038/nplants.2017.72
Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J. L., Bautor, J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell 18, 1038–1051. doi: 10.1105/tpc.105.039982
Beck, M., Hurt, E. (2017). The nuclear pore complex: understanding its function through structural insight. Nat. Rev. Mol. Cell Biol. 18, 73–89. doi: 10.1038/nrm.2016.147
Cui, H., Lan, X., Lu, S., Zhang, F., Zhang, W. (2017). Bioinformatic prediction and functional characterization of human KIAA0100 gene. J. Pharm. Anal. 7, 10–18. doi: 10.1016/j.jpha.2016.09.003
Du, J., Gao, Y., Zhan, Y., Zhang, S., Wu, Y., Xiao, Y., et al. (2016). Nucleocytoplasmic trafficking is essential for BAK1- and BKK1-mediated cell-death control. Plant J. 85, 520–531. doi: 10.1111/tpj.13125
Fujita, Y., Nakashima, K., Yoshida, T., Katagiri, T., Kidokoro, S., Kanamori, N., et al. (2009). Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132. doi: 10.1093/pcp/pcp147
He, K., Gou, X., Yuan, T., Lin, H., Asami, T., Yoshida, S., et al. (2007). BAK1 and BKK1 regulate brassinosteroid-dependent growth and brassinosteroid-independent cell-death pathways. Curr. Biol. 17, 1109–1115. doi: 10.1016/j.cub.2007.05.036
Ishitani, M., Xiong, L., Lee, H., Stevenson, B., Zhu, J.-K. (1998). HOS1 , a genetic locus involved in cold-responsive gene expression in arabidopsis. Plant Cell 10, 1151–1161. doi: 10.1105/tpc.10.7.1151
Isono, K., Tsukimoto, R., Iuchi, S., Shinozawa, A., Yotsui, I., Sakata, Y., et al. (2020). An ER–golgi tethering factor SLOH4/MIP3 is involved in long-term heat tolerance of arabidopsis. Plant Cell Physiol. 62, 272–279. doi: 10.1093/pcp/pcaa157
Katori, T., Ikeda, A., Iuchi, S., Kobayashi, M., Shinozaki, K., Maehashi, K., et al. (2010). Dissecting the genetic control of natural variation in salt tolerance of Arabidopsis thaliana accessions. J. Exp. Bot. 61, 1125–1138. doi: 10.1093/jxb/erp376
Kim, T.-H., Kunz, H.-H., Bhattacharjee, S., Hauser, F., Park, J., Engineer, C., et al. (2012). Natural variation in small molecule–induced TIR-NB-LRR signaling induces root growth arrest via EDS1- and PAD4-complexed R protein VICTR in arabidopsis. Plant Cell 24, 5177–5192. doi: 10.1105/tpc.112.107235
Lai, Z., Schluttenhofer, C. M., Bhide, K., Shreve, J., Thimmapuram, J., Lee, S. Y., et al. (2014). MED18 interaction with distinct transcription factors regulates multiple plant functions. Nat. Commun. 5, 3064. doi: 10.1038/ncomms4064
Li, X., Clarke, J. D., Zhang, Y., Dong, X. (2001). Activation of an EDS1-mediated R -gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol. Plant-Microbe Interact. 14, 1131–1139. doi: 10.1094/mpmi.2001.14.10.1131
Liao, C.-J., Lai, Z., Lee, S., Yun, D.-J., Mengiste, T. (2016). Arabidopsis HOOKLESS1 regulates responses to pathogens and abscisic acid through interaction with MED18 and acetylation of WRKY33 and ABI5 chromatin. Plant Cell 28, 1662–1681. doi: 10.1105/tpc.16.00105
Lüdke, D., Roth, C., Kamrad, S. A., Messerschmidt, J., Hartken, D., Appel, J., et al. (2021). Functional requirement of the Arabidopsis importin-α nuclear transport receptor family in autoimmunity mediated by the NLR protein SNC1. Plant J. 105, 994–1009. doi: 10.1111/tpj.15082
Nakashima, K., Ito, Y., Yamaguchi-Shinozaki, K. (2009). Transcriptional regulatory networks in response to abiotic stresses in Arabidopsis and grasses. Plant Physiol. 149, 88–95. doi: 10.1104/pp.108.129791
Palma, K., Zhang, Y., Li, X. (2005). An importin α Homolog, MOS6, plays an important role in plant innate immunity. Curr. Biol. 15, 1129–1135. doi: 10.1016/j.cub.2005.05.022
Porra, R. J., Thompson, W. A., Kriedemann, P. E. (1989). Determination ofaccurate extinction coefficients and simultaneous equations for assaying chlorophyllsa and b extracted with four different solvents: verification of the concentrationof chlorophyll standards by atomic absorption spectroscopy. Biochim. Biophys.Acta Bioenerg. 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Roth, C., Lüdke, D., Klenke, M., Quathamer, A., Valerius, O., Braus, G. H., et al. (2017). The truncated NLR protein TIR-NBS13 is a MOS6/IMPORTIN-α3 interaction partner required for plant immunity. Plant J. 92, 808–821. doi: 10.1111/tpj.13717
Saier, M. H., Reddy, V. S., Tamang, D. G., Västermark, Å. (2014). The transporter classification database. Nucleic Acids Res. 42, D251–D258. doi: 10.1093/nar/gkt1097
Saito, K., Yoshikawa, M., Yano, K., Miwa, H., Uchida, H., Asamizu, E., et al. (2007). NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in lotus japonicus. Plant Cell 19, 610–624. doi: 10.1105/tpc.106.046938
Stewart, M. (2007). Molecular mechanism of the nuclear protein import cycle. Nat. Rev. Mol. Cell Biol. 8, 195–208. doi: 10.1038/nrm2114
Sung, D.-Y., Kaplan, F., Lee, K.-J., Guy, C. L. (2003). Acquired tolerance to temperature extremes. Trends Plant Sci. 8, 179–187. doi: 10.1016/s1360-1385(03)00047-5
Takahashi, A., Casais, C., Ichimura, K., Shirasu, K. (2003). HSP90 interacts with RAR1 and SGT1 and is essential for RPS2-mediated disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. 100, 11777–11782. doi: 10.1073/pnas.2033934100
Tsukimoto, R., Isono, K., Kajino, T., Iuchi, S., Shinozawa, A., Yotsui, I., et al (2021). Mitochondrial fission complex is required for long-term heat tolerance of Arabidopsis. Plant Cell Physiol. 63, 296–304. doi: 10.1093/pcp/pcab171
Wälde, S., Kehlenbach, R. H. (2010). The Part and the Whole: functions of nucleoporins in nucleocytoplasmic transport. Trends Cell Biol. 20, 461–469. doi: 10.1016/j.tcb.2010.05.001
Wang, R., Brattain, M. G. (2007). The maximal size of protein to diffuse through the nuclear pore is larger than 60 kDa. FEBS Lett. 581, 3164–3170. doi: 10.1016/j.febslet.2007.05.082
Wiermer, M., Cheng, Y. T., Imkampe, J., Li, M., Wang, D., Lipka, V., et al. (2012). Putative members of the Arabidopsis Nup107-160 nuclear pore sub-complex contribute to pathogen defense. Plant J. 70, 796–808. doi: 10.1111/j.1365-313x.2012.04928.x
Yang, Y., Wang, W., Chu, Z., Zhu, J.-K., Zhang, H. (2017). Roles of nuclear pores and nucleo-cytoplasmic trafficking in plant stress responses. Front. Plant Sci. 08. doi: 10.3389/fpls.2017.00574
Zacharias, D. A., Violin, J. D., Newton, A. C., Tsien, R. Y. (2002). Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science 296, 913–916. doi: 10.1126/science.1068539
Zhang, Y., Goritschnig, S., Dong, X., Li, X. (2003). A Gain-of-Function Mutation in a Plant Disease Resistance Gene Leads to Constitutive Activation of Downstream Signal Transduction Pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15, 2636–2646. doi: 10.1105/tpc.015842
Zhang, Y., Li, X. (2005). A Putative Nucleoporin 96 Is Required for Both Basal Defense and Constitutive Resistance Responses Mediated by suppressor of npr1-1,constitutive 1. Plant Cell 17, 1306–1316. doi: 10.1105/tpc.104.029926
Zhang, A., Wang, S., Kim, J., Yan, J., Yan, X., Pang, Q., et al. (2020). Nuclear pore complex components have temperature-influenced roles in plant growth and immunity. Plant Cell Environ. 43, 1452–1466. doi: 10.1111/pce.13741
Zhu, Y., Qian, W., Hua, J. (2010). Temperature modulates plant defense responses through NB-LRR proteins. PloS Pathog. 6, e1000844. doi: 10.1371/journal.ppat.1000844
Keywords: osmotolerance, nuclear pore complex, NB-LRR transport, immune response, Arabidopsis thaliana accession
Citation: Mori K, Murakoshi Y, Tamura M, Kunitake S, Nishimura K, Ariga H, Tanaka K, Iuchi S, Yotsui I, Sakata Y and Taji T (2024) Mutations in nuclear pore complex promote osmotolerance in Arabidopsis by suppressing the nuclear translocation of ACQOS and its osmotically induced immunity. Front. Plant Sci. 15:1304366. doi: 10.3389/fpls.2024.1304366
Received: 29 September 2023; Accepted: 02 January 2024;
Published: 22 January 2024.
Edited by:
Weiqiang Li, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Zhihua Hua, Ohio University, United StatesEnrique Castano, Centro de Investigación Científica de Yucatán, Mexico
Copyright © 2024 Mori, Murakoshi, Tamura, Kunitake, Nishimura, Ariga, Tanaka, Iuchi, Yotsui, Sakata and Taji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Teruaki Taji, t3teruak@nodai.ac.jp
 Kento Mori1
Kento Mori1 Masashi Tamura
Masashi Tamura Kohji Nishimura
Kohji Nishimura Hirotaka Ariga
Hirotaka Ariga Yoichi Sakata
Yoichi Sakata Teruaki Taji
Teruaki Taji