- 1National Biodiversity Future Center (NBFC), Palermo, Italy
- 2Dipartimento di Scienze e Tecnologie Agrarie, Alimentari, Ambientali e Forestali (DAGRI), Università degli Studi di Firenze, Florence, Italy
- 3Istituto di Bioscienze e Biorisorse (IBBR), Consiglio Nazionale delle Ricerche (CNR), Sesto Fiorentino, Italy
Introduction: The molecular and physiological mechanisms activated in plants during drought stress tolerance are regulated by several key genes with both metabolic and regulatory roles. Studies focusing on crop gene expression following plant growth-promoting rhizobacteria (PGPR) inoculation may help understand which bioinoculant is closely related to the induction of abiotic stress responses.
Methods: Here, we performed a meta-analysis following Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines to summarise information regarding plant-PGPR interactions, focusing on the regulation of nine genes involved in plant drought stress response. The literature research yielded 3,338 reports, of which only 41 were included in the meta-analysis based on the chosen inclusion criteria. The meta-analysis was performed on four genes (ACO, APX, ACS and DREB2); the other five genes (ERD15, MYB, MYC, acdS, WRKY) had an insufficient number of eligible articles.
Results: Forest plots obtained through each meta-analysis showed that the overexpression of ACO, APX, ACS and DREB2 genes was not statistically significant. Unlike the other genes, DREB2 showed statistically significant results in both the presence and absence of PGPR. Considering I2>75 %, the results showed a high heterogeneity among the studies included, and the cause for this was examined using subgroup analysis. Moreover, the funnel plot and Egger’s test showed that the analyses were affected by strong publication bias.
Discussion: This study argues that the presence of PGPR may not significantly influence the expression of drought stress response-related crop genes. This finding may be due to high heterogeneity, lack of data on the genes examined, and significant publication bias.
1 Introduction
Current agricultural practices aiming at maximizing targeting maximum yield has serious impacts on natural ecosystems, including the depletion of natural resources, a decline in ecosystem services, increased soil erosion, and a significant losses in biodiversity and soil organic matter (Struik et al., 2014; Watson et al., 2021). Therefore, scientists are focusing on maintaining crop productivity and sustainable agriculture. Accounting for the increasing demand for food and environmentally friendly and economically beneficial practices is necessary to preserve our planet. Climate change has impacted agricultural productivity alongside intensification. In fact, plant health is also affected by abiotic stresses, which limit the overall crop yield by more than 50% every year (Ojuederie et al., 2019). Drought is a significant abiotic stressor strongly impacting crop productivity (Golldack et al., 2014; Anjum et al., 2017; Hussain et al., 2018) and decreases the amount of arable land. The “agroecological practices” developed in the last century, which include livestock manure utilisation, cover crops and intercropping, agroforestry, biological control, and biodiversity conservation, remain the most used practices to realise a more “green” agriculture (Wezel et al., 2014). However, recognizing beneficial soil and plant microorganisms as crucial for enhancing plant nutrition and protection is not a recent development. Indeed, for several decades, the scientific community has deepened the study of complex networks established in the biosphere between plants and their associated microbiota, focusing on the role of microorganisms in plant physiology (Singh et al., 2016). Plants cannot be considered singular entities but are the result of a close relationship established with their microbiota, which can be beneficial, neutral, or pathogenic and is usually controlled by the plant’s genotype and environment (Meier et al., 2022; Wagner, 2022). Most bacteria that initially colonise the rhizosphere or plant surfaces as epiphytes can enter plants and proliferate as endophytes, thereby establishing a mutualistic association. Mutualists are often crucial for plant health and resilience (Vandenkoornhuyse et al., 2015), realising numerous mechanisms that enhance plant growth, such as plant growth-promoting (PGP) microorganisms. The main direct mechanisms involved are i) endogenous phytohormone modulation (auxins, gibberellins, cytokinins, and ethylene), ii) nutrient solubilisation, mainly phosphorus (P) and potassium (K), and iii) nutrient bioavailability (Çakmakçı, 2016) such as biological fixation of atmospheric nitrogen (N2). Among the most well-known indirect mechanisms are the regulation of plant-induced (ISR) and plant-acquired (ASR) systemic resistance (Alori et al., 2017; Enebe and Babalola, 2018), the production of secondary metabolites such as auxins and 1-aminocyclopropane-1-carboxylate (ACC) deaminase, and the general control of pathogenic diseases (Kamle et al., 2020) through the production of antibiotics, lytic enzymes, and siderophores. It is known that PGP microorganisms from the rhizosphere, named PGP rhizobacteria (PGPR), and exploiting their abilities to interact with the host could help plants in harsh environmental conditions. They can be used as bioinoculants in agricultural practices (Singh et al., 2011) to reduce the use of chemical fertilisers. For instance, Andrographis paniculate inoculated with Pseudomonas sp. showed numerous benefits, including enhanced growth, early flowering, and increased production of photosynthetic pigments (Thakur et al., 2023). Moreover, Thakur and Yadav (2023) demonstrated that AB-11, a strain of Streptomyces sp., hold promise as an agent for plant growth promotion, providing an environmentally friendly alternative to chemical fertilizers. The success of their utilisation depends on several factors, such as survival in the soil, ability to interact with the existing microbiota, genetic compatibility with the crop on which they are applied (Cangioli et al., 2021), and environmental factors. Some PGPR are characterised by specific PGP traits, such as heavy metal detoxifying activities, biological control, and abiotic stress tolerance (Egamberdieva and Lugtenberg, 2014), which allow them to be considered a resource for preserving agricultural productivity during adverse conditions (Chandra et al., 2021). It is essential to consider that the plant response to abiotic stresses is a complex phenomenon affecting both plant development and the metabolism and physiology of its microbiota (Omae and Tsuda, 2022). The regulatory mechanisms of plant stress responses include gene expression adjustments aimed at physiological and morphological adaptations (Kooyers, 2015). Several plant genes are involved in abiotic stress responses and drought tolerance (Gong et al., 2020), and many are secondary messengers and transcription factors (TFs) that participate in signalling pathways. A deeper understanding of the extreme complexity of such pathways should be the way to devise new and applicable strategies to improve plant tolerance to drought (Joshi et al., 2016), especially in light of some studies that report that plants inoculated with microbes show different patterns of gene expression compared to non-inoculated plants (Creus et al., 2005; Molina-Favero et al., 2008; Dimkpa C.O.et al., 2009; Lim and Kim, 2013; Sarma and Saikia, 2014; Vurukonda et al., 2016). Thus, studies focusing on plant gene expression in the presence of PGPR can play a significant role in understanding whether PGPR can be used as a resource for drought stress resistance. In this study, we provide a meta-analysis of peer-reviewed studies to verify whether the interaction established between PGPR and host plants may beneficially affect plant resilience to drought stress in a significant and detectable way.
2 Materials and methods
2.1 Protocol used to carry out systematic review and meta-analysis
We performed a systematic review and meta-analysis of the influence of PGPR on plant genes that respond to drought stress following the guidelines of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) statement (Moher et al., 2009). The PRISMA protocol includes four steps for conducting a meta-analysis: i) identification of studies via bibliographic research, ii) screening of obtained studies via title–abstract analysis, iii) full-text screening to identify studies that meet the inclusion and exclusion criteria, and iv) detection of eligible studies that can be included in the meta-analysis (Figure 1) (Moher et al., 2009; Nakagawa et al., 2017).
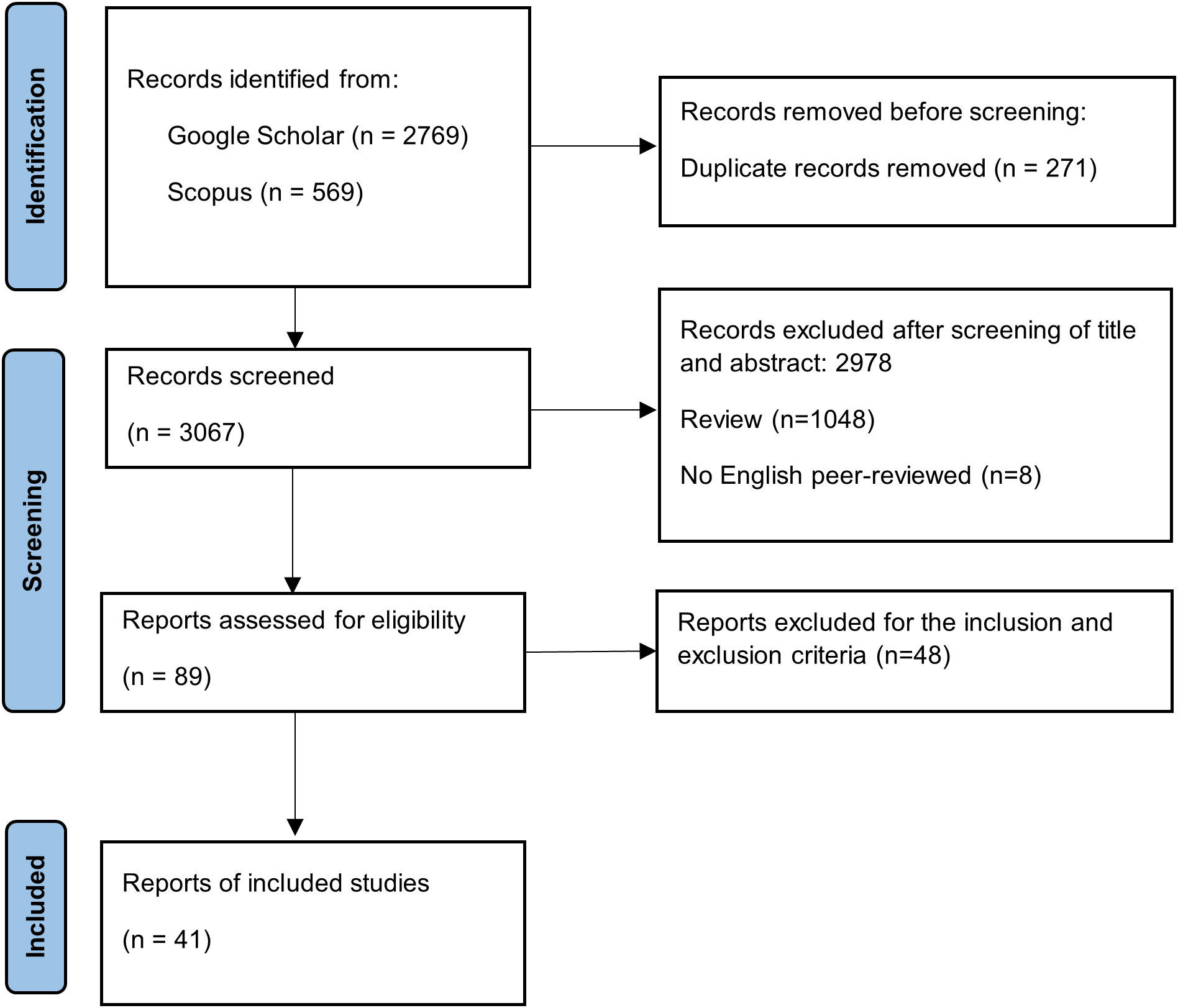
Figure 1 PRISMA workflow diagram to perform the systematic review and the meta-analysis (adapted from flow diagram proposed by Page et al., 2021). PRISMA, Preferred Reporting Items for Systematic Reviews and Meta-Analyses.
2.2 Bibliographic research and search strategy
This study aimed to describe information from different studies on gene expression during water stress with/without the presence of PGPR. Therefore, we mainly focused on the following bacterial genes: 1-aminocyclopropane-1-carboxylate deaminase (acdS) and on plant genes aminocyclopropane-1-carboxylate synthetase (ACS), 1-aminocyclopropane-1-carboxylate (ACO), ascorbate peroxidase (APX), early responsive to dehydration 15 (ERD15), WRKY, myelocytomatosis (MYC), myeloblastosis (MYB), and dehydration-responsive element-binding protein 2 (DREB2). Firstly, a bibliographic search was conducted to collect studies regarding gene expression in crops during drought and in the presence of PGPR. Secondly, bibliographic research was performed to gather data on gene expression in crops during drought in the absence of PGPR. Google Scholar and Scopus were used as search engines. The keywords used for the first bibliographic research on Google Scholar were “gene AND expression AND drought AND stress AND plants AND PGPR”. The word selection used for all the genes in Scopus was ALL (“gene ID” AND “drought stress” AND “PGPR” OR “PGPB”). Only for DREB2 was the bibliographic research regarding DREB2 expression in plants subjected to drought stress and without PGPR yielded positive results. The search terms used were DREB2 AND expression AND drought AND stress AND plant”. All identified studies are listed in the Supplementary Bibliography (Supplementary File 1).
2.3 Study selection
After obtaining articles from Google Scholar and Scopus, a database was created for each analysed gene in order to simplify the preliminary screening of the studies, in particular, the presence of PGPR, ACS (Table S1), APX (Table S2), ERD15 (Table S3), MYB (Table S4), MYC (Table S5), acdS (Table S6), ACO (Table S7), WRKY (Table S8), DREB2 (Table S9), and DREB2 without PGPR (Table S10). The tables report the keywords used to search the studies, the title of the study, the author’s first name, the abstract, the decision of rejection/acceptance, and the reasons for exclusion. Screening was performed by assessing titles and abstracts. After this step, a final screening of the studies based on the inclusion criteria was performed. The inclusion criteria were i) the focus on herbaceous/crop plants, ii) the number of individuals, iii) the presence of data regarding selected gene expression (i.e., log2 fold change (FC)) assessed by quantitative PCR, iv) the presence of dispersion indices, such as standard error (SE) or standard deviation (SD), and v) the presence of controls to compare gene expression between control and treated plants. Unfortunately, some studies did not report SD or SE, which are crucial for calculating the size effect. Therefore, a prognostic method was adopted to allocate an assigned SD (Ma et al., 2008; Weir et al., 2018). Two reviewers identified and examined eligible studies.
2.4 Data extraction
After eligible studies that met the inclusion criteria were identified, data extraction, such as FC, number of individuals/number of replicates, SE, and SD, was performed. These data were indispensable for calculating the effect size. In some studies, FC and SD values were obtained using Web Plot Digitiser (version 4.6), a web tool that allows data acquisition from plots and images (“WebPlotDigitizer - Extract data from plots, images, and maps”, n.d.) (Rohatgi, 2022). As mentioned previously, a prognostic method was adopted to determine the assigned SD. In particular, the SE values were obtained from another distribution consisting of data extracted from Borges et al. (2012); Feng et al. (2019); Le et al. (2012); Nabi et al. (2021), Neves-Borges et al. (2012); Song et al. (2016); Thirumalaikumar et al. (2018), and Wu et al. (2014). Thus, it was possible to predict the missing SE and the SD of our sampling distribution.
2.5 Effect size calculation
It is essential to select an appropriate effect size for a meta-analysis. In this case, continuous outputs were treated; therefore, Cohen’s d was used as the effect size. Cohen’s d estimates the standard mean difference of an effect (Lakens, 2013) between two groups, such as the control and treatment (Lakens, 2013; Nakagawa et al., 2017), and was calculated using Equation 1:
where is the log2 fold change in treated plants, is the log2 fold change in control plants, n1 is the number of treated plants, n2 is the number of control plants, SD1 is the standard deviation of treated plants, and SD2 is the standard deviation of control plants.
The numerator represented the difference between the means of FC ( and FC . The denominator represents the pooled SD. Because the number of samples in some studies included in the meta-analysis was<20, the correction factor, Hedges’s g (Lakens, 2013), was applied, as shown in Equation 2:
where is the effect size calculated before applying the correction factor, n1 is the number of treated plants, and n2 is the number of control plants.
Moreover, the effect size error (SEES) was determined (de Vries et al., 2022), as reported in Equation 3.
where ES is the effect size, n1 is the number of treated plants, and n2 is the number of control plants.
2.6 Data analysis
A minimum of four articles were selected for each meta-analysis. The meta-analysis was performed using the software JASP 0.14 “Meta-analysis module” (JASP Team, 2023). A random-effects model considering the heterogeneity-assorted studies was chosen. The summary of results through forest and funnel plots using JASP was produced, and the heterogeneity of the included studies through forest plots and publication bias via funnel plots were assessed. The inconsistency or heterogeneity of the studies was estimated via an intraclass correlation index (I2), which ranges from 0 to 1 (or 0 to 100%) and indicates how much the variation in effect size is due to the between-variance (τ2) or, more generally, the proportion of variance not attributable to sampling (error) variance (Nakagawa et al., 2017). Publication bias was estimated using Egger’s test. Egger’s statistical test detects funnel plot asymmetry by determining whether the intercept deviates significantly from zero in a regression of standardised effect estimates versus precision (Hayashino et al., 2005). Bias can also be estimated using funnel plots (JASP Team, 2023), which are scatter plots of the treatment effects estimated from individual studies (horizontal axis) against the measure of study precision (vertical axis) (Sterne and Egger, 2001; Sterne et al., 2011).
3 Results
3.1 Study selection and data extraction
The individual bibliographic searches yielded 3,338 total reports through Google Scholar (2,769) and Scopus (569), following the PRISMA protocol, as shown in Figure 1. Among the records found, 271 duplicates were removed before the second screening phase. Then, title and abstract screening reduced records from 3,067 to 89, excluding reviews, non-original research articles, and studies regarding biotic stress. Subsequently, the remaining reports were analysed to assess their suitability for meta-analysis. The screening consisted of meeting the following inclusion criteria: i) studies on herbaceous plants, ii) number of individuals, iii) presence of data on selected gene expression in terms of log2 FC and assessed by quantitative PCR, iv) presence of dispersion indices such as SE or SD, and v) presence of a control to compare gene expression between control and treated plants. Finally, 41 studies were included in this meta-analysis.
After identifying the included studies, two types of databases were set up to organise the data extracted from the studies and calculate the effect size (Table S11 and Table S12). Tables S1–S9 show the organised data obtained from reports concerning studies on gene expression in crop plants inoculated with PGPR under drought conditions. Table S10 shows the data on DREB2 expression in plants subjected to drought but not inoculated with PGPR. Initially, drought stress induced in plants by dehydration (lack of water) and polyethylene glycol (PEG) were considered for meta-analysis. Unfortunately, it was not possible to include acdS, ERD15, WRKY, MYC, and MYB, as there were insufficient eligible articles regarding the expression of these genes in plants inoculated with PGPR and exposed to water shortages.
3.2 Meta-analysis results
Our search strategy yielded 24 studies querying the expression of ACS, ACO, APX, and DREB2 in plants subjected to drought stress and inoculated with PGPR and 17 studies querying DREB2 expression in plants subjected to drought stress without PGPR. Firstly, an independent meta-analysis was performed using the software JASP 0.14 for each of the included genes (ACS, ACO, APX, and DREB2) that met the inclusion criteria. The analysis synthesised different information across studies on gene expression under water stress, with or without PGPR. Table 1 summarises the results obtained from the meta-analysis for each gene, evidencing general pooled effect size, 95% confidence interval (CI), and pooled I2 (%). Then, the effect size for each subgroup, 95% CI, and I2 for each subgroup were obtained. Forest and funnel plots for ACO gene (Figure S1), ACS gene (Figure S2), APX gene (Figure S3), and DREB2 gene (Figure S4) were obtained. Results for ACO, ACS, and APX genes were statistically insignificant since the 95% CI from the pooled estimated effect size crossed zero, and the heterogeneity was high. Therefore, a subgroup analysis based on specific and frequently analysed plant organs (i.e., leaves) was performed. Also, in this case, the results obtained for ACO (Figure S5), ACS (Figure S6), and APX (Figure S7) were statistically insignificant. In contrast, statistically significant results were obtained for DREB2 with and without PGPR inoculation in plants subjected to drought stress (Figures S4 and S8). In these two analyses, the heterogeneity was considered very elevated because it was greater than 75% (Cohen, 1988). An I2 value of 98.52% (Table 1) in DREB2 expression in plants inoculated with PGPR during drought was obtained. The forest plot (Figure S8) and I2 value of 99.248 (Table 1) for DREB2 expression in water-stressed plants and without PGPR showed high heterogeneity. Further subgroup analyses were performed to investigate the cause of this heterogeneity. The first was based on the specific method used for the stress treatment (dehydration) (Figure 2A) and on the plant organs analysed (leaves) (Figure 2B) in the presence of PGPR. The second method was based on the specific method used for stress treatment (dehydration) (Figure 3A) and on the plant organs analysed (leaves) (Figure 3B) but without PGPR.
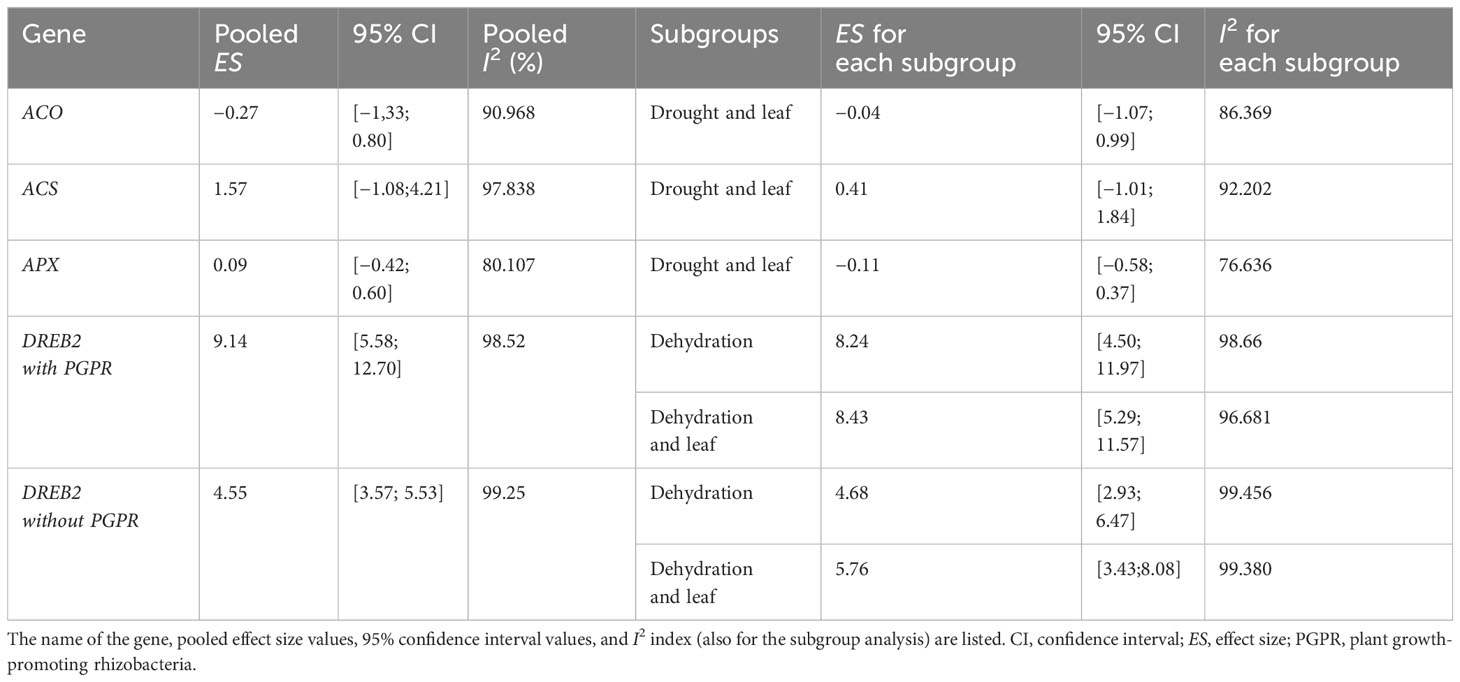
Table 1 Summary of results obtained from the meta-analysis for each gene, namely, 1-aminocyclopropane-1-carboxylate (ACO), 1-aminocyclopropane-1-carboxylate synthetase (ACS), ascorbate peroxidase (APX), and dehydration-responsive element-binding protein 2 (DREB2).
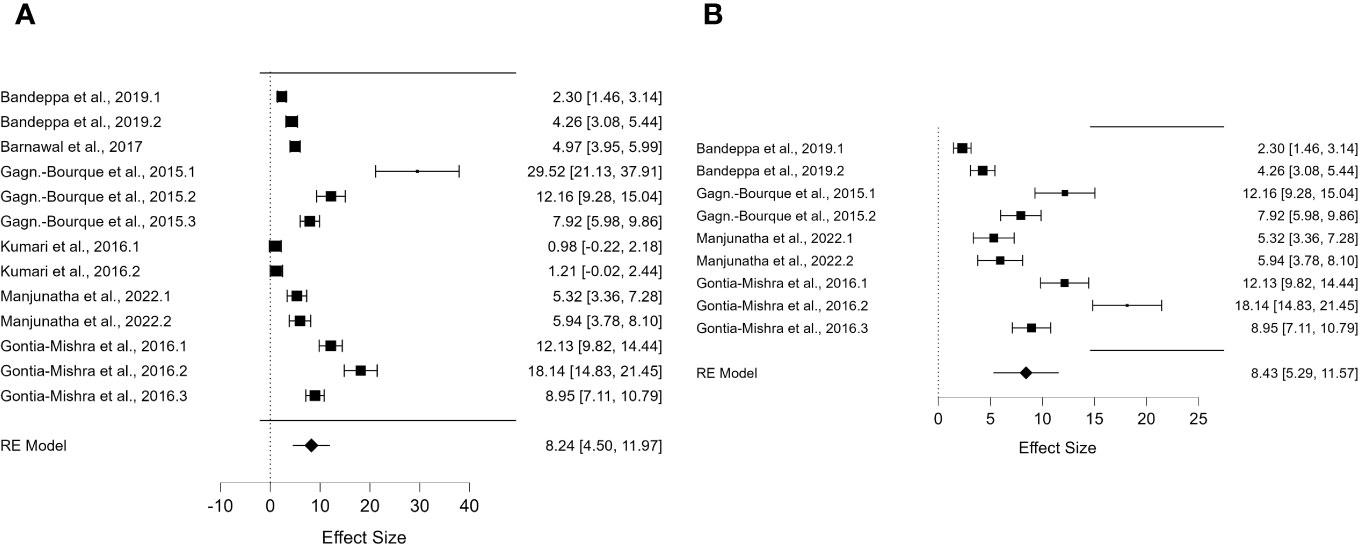
Figure 2 Forest plot of DREB2 expression with PGPR in plants exposed to drought stress. (A) Forest plot of subgroups based on the drought stress treatment selected (dehydration). (B) Forest plot of subgroup analysis based on the drought stress treatment (dehydration) and the stressed plant organ (leaves). PGPR, plant growth-promoting rhizobacteria.
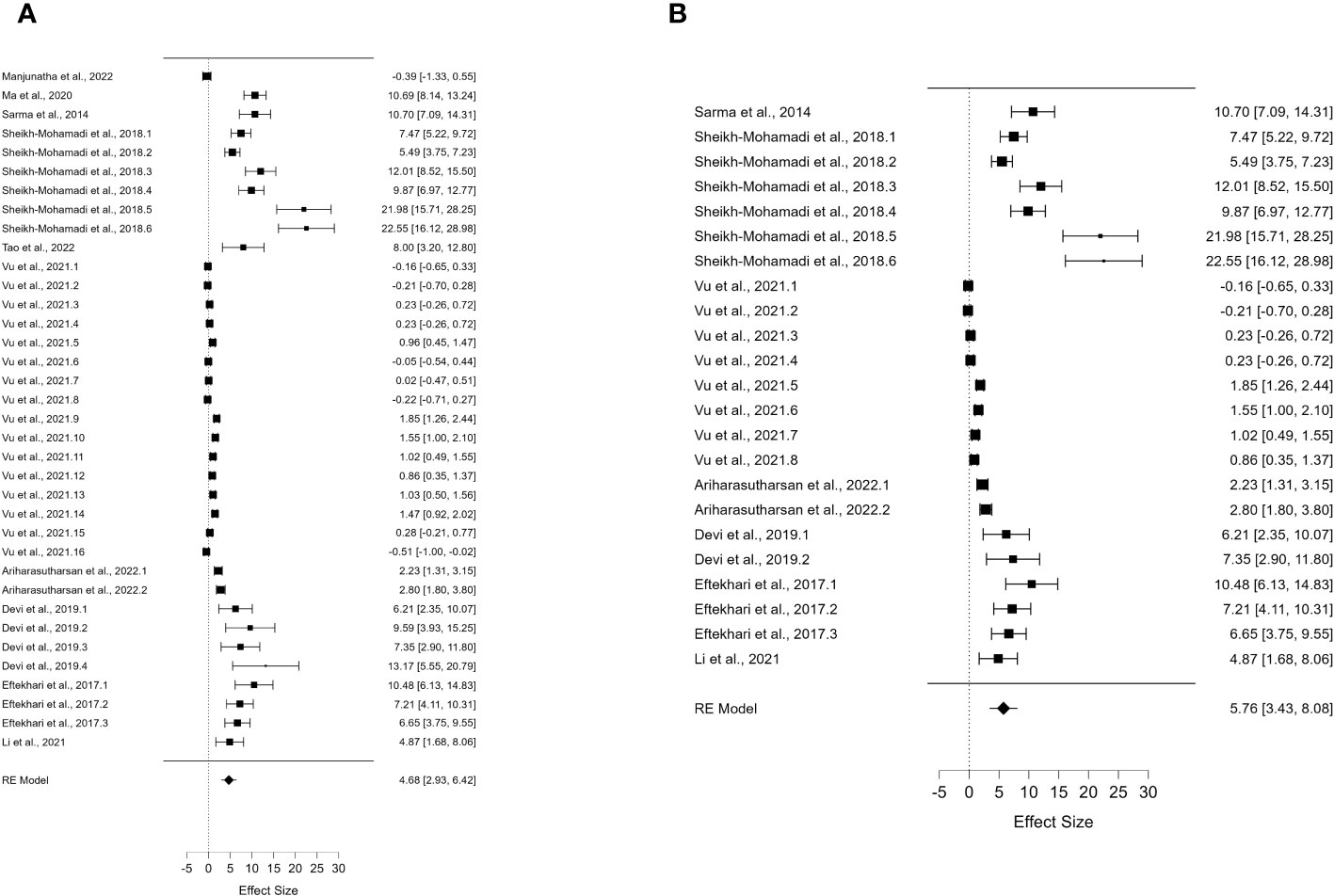
Figure 3 Forest plot of DREB2 expression without PGPR in plants exposed to drought stress. (A) Forest plot of subgroups based on the drought stress treatment selected (dehydration). (B) Forest plot of subgroup analysis based on the drought stress treatment (dehydration) and the stressed plant organ (leaves). PGPR, plant growth-promoting rhizobacteria.
For DREB2, thanks to the sufficient number of papers, it was possible to include further subgroup analyses. To allow the comparison of results and consistent assumptions, the minimum number of articles was considered to be at least four for each subgroup chosen. In particular, for DREB2 with PGPR and DREB2 without PGPR inoculation, the number of studies selected should be the same. In the first case, in the subgroup analysis of DREB2 during dehydration of the entire plant or in different districts and the presence of PGPR, the I2 increased to 98.665% (Figure 2). The subgroup analysis based on dehydration stress (Figure 2A) and drought stress applied to leaves (Figure 2B) in the presence of PGPR showed a decrease in heterogeneity of 96.68% but remained too high for consideration. Therefore, a subgroup analysis for DREB2 expression without PGPR was performed (Figure 3). Both forest plots based on general dehydration (Figure 3A) and dehydration in leaves (Figure 3B) showed a decrease in heterogeneity; in particular, the I2 value reached 98.66% for general dehydration and 96.68% for dehydration applied to leaves. In conclusion, significant heterogeneity was found in each summary effect, which could not be avoided by subgroup analysis. Furthermore, by observing the cumulative ES values, it can be assumed that the expression of the gene encoding DREB2 was positively regulated during drought stress, in both the presence and absence of PGPR (Table 1). Nonetheless, the cumulative effect size values for DREB2 in the presence of PGPR (ES = 9.14; 95% CI 5.58, 12.70) were higher than those of DREB2 in the absence of PGPR inoculation (ES = 5.76; 95% CI 3.43, 8.08). Funnel plots of subgroup analysis of DREB2 expression based on drought stress treatments and in the absence or presence of PGPR inoculation are shown in Figure 4. All funnel plots were asymmetrical along the vertical axis around the meta-analytical pooled effect size estimate. Moreover, almost all the points representing each study were external to the 95% confidence intervals. A significant publication bias in all analyses of DREB2 expression with and without PGPR in plants exposed to drought stress and their subgroups was identified via Egger’s test (p< 0.01). Evident asymmetry, confirmed via Egger’s test, could be interpreted as evidence of publication bias and heterogeneity.
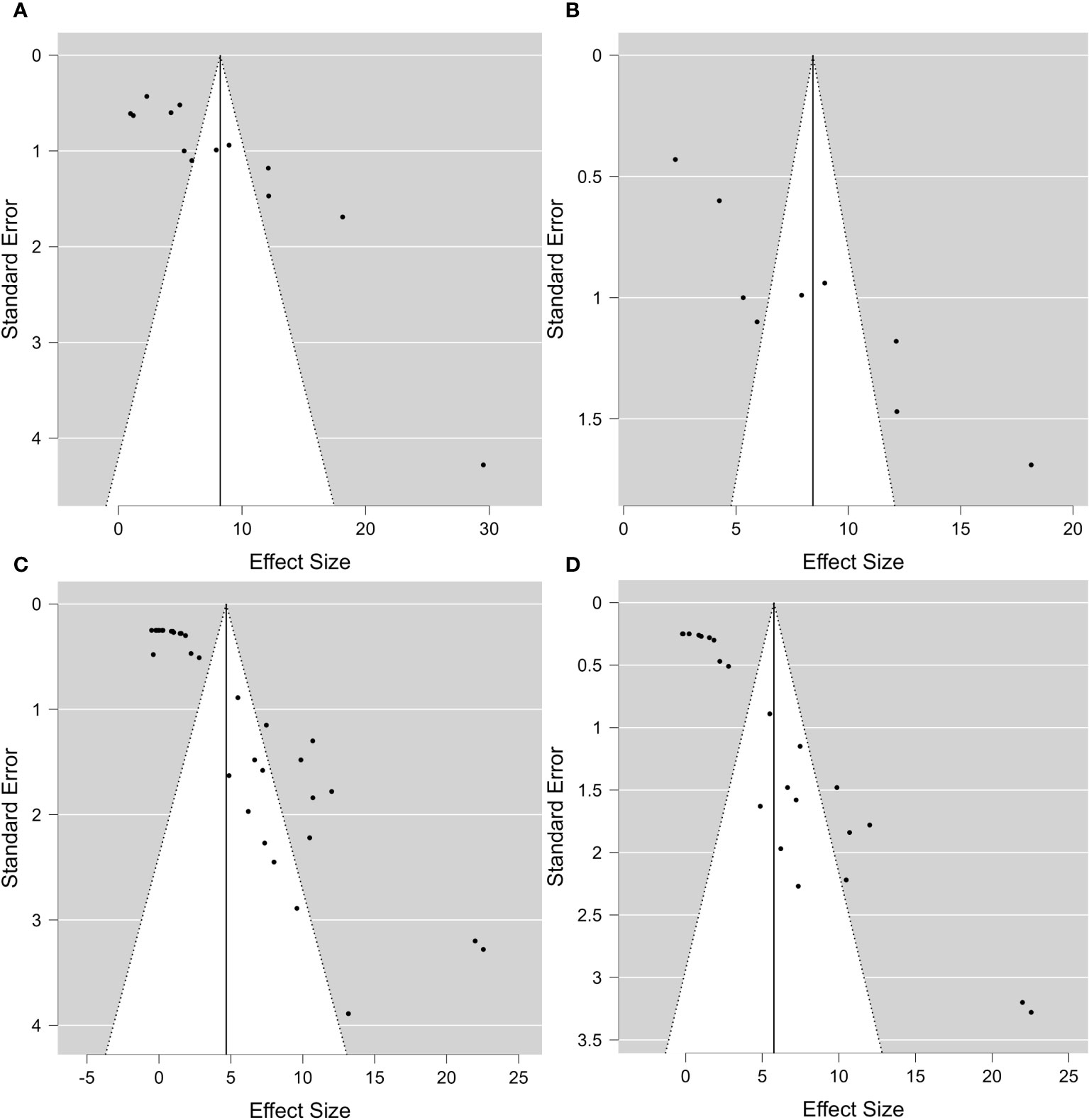
Figure 4 Funnel plot of DREB2 expression with and without PGPR in plants exposed to drought stress. (A) Funnel plot of subgroup analysis based on the drought stress treatment selected (dehydration) and in the presence of PGPR. (B) Funnel plot of subgroup analysis based on drought stress treatment selected and the stressed plant organ (leaves) in the presence of PGPR. (C) Funnel plot of subgroup analysis based on the drought stress treatment selected (dehydration). (D) Funnel plot of subgroup analysis based on the drought stress treatment selected (dehydration) and the stressed plant organ (leaves). Analyses in panels (C, D) are independent of PGPR inoculation. PGPR, plant growth-promoting rhizobacteria.
4 Discussion and conclusion
Climate change has exacerbated the water crisis in the Mediterranean region, affecting agricultural productivity and worsening land overexploitation. Therefore, scientists are challenged to develop new strategies for maintaining sustainable agriculture and coping with the current increase in drought stress. Several recent studies have shown that the association between plants and PGPR is important for resistance and adaptation to abiotic and biotic stresses. Among them, drought stress can be relieved by the presence of positive bacteria at the physio-morphological level, increasing root volume, enhancing nutrient mobilisation and up-taking, and over-regulating antioxidant enzymes, such as ascorbate peroxidase, catalase, superoxide dismutase, ascorbic acid, glutathione, α-tocopherol, and glutathione reductase (Bukhat et al., 2020; Goswami and Deka, 2020; Ahluwalia et al., 2021). Inter-kingdom communication causes the upregulation of drought-response marker genes such as DREB1B-like, ERD15 (Naylor and Coleman-Derr, 2018), and DREB2A (Sarma and Saikia, 2014). The molecular mechanism underlying the plant–PGPR interactions has deepened thanks to advances in molecular techniques, such as the analysis of differential gene expression using cDNA microarrays (van Loon, 2007), qPCR (Ibort et al., 2018), and RNA-seq (Tao et al., 2022). Recent molecular studies have shown that PGPR affects the expression patterns of stress-responsive genes in plants, alleviating the damaging effects of drought stress (Vurukonda et al., 2016; Gupta et al., 2021; Manjunatha et al., 2022).
Saikia et al. (2018) studied Vigna mungo L. and Pisum sativum L. subjected to water stress and inoculated them with a combination of three ACC deaminase-producing rhizobacteria (Ochrobactrum pseudogrignonense RJ12, Pseudomonas sp. RJ15, and Bacillus subtilis RJ46) (Saikia et al., 2018). This study showed that inoculated plants showed a lower amount of ACC than the control due to the upregulation of ACS and the downregulation of ACO genes, consequently decreasing ethylene production (Saikia et al., 2018). Moreover, several studies have argued that the expression of genes encoding MYC, MYB, and WRKY is influenced by plant–microorganism interactions during abiotic stress (Dimkpa C. et al., 2009; Arora et al., 2020; Ha-Tran et al., 2021; Hoque et al., 2023; Rosier et al., 2018; Khan et al., 2020; Oleńska et al., 2020). Kasim et al. (2013) reported that wheat plants inoculated with strains of Bacillus amyloliquefaciens and Azospirillum brasilense and subjected to drought showed an upregulation of APX1 and an increase in the activity of enzymes involved in the ascorbate-glutathione redox cycle, affecting reactive oxygen species (ROS) scavenging (Kasim et al., 2013). Similarly, ERD15 gene expression was upregulated in Arabidopsis thaliana inoculated with Paenibacillus polymyxa strains under water shortage (Timmusk and Wagner, 1999). In addition, the upregulation of DREB2 and dehydrins in Vigna radiata L. inoculated with Pseudomonas aeruginosa GGRJ21 was observed (Sarma and Saikia, 2014).
To further investigate the mechanisms described in these studies and explore the role of PGPR in plant drought stress-responsive genes, a meta-analysis was performed to quantitatively synthesise the data available in the literature. The focus was on the bacterial gene acdS and the plant genes ACO, ACS, APX, WRKY, MYC, MYB, ERD15, and DREB2 to evaluate whether and how they are influenced by the presence of PGPR. These genes are specifically involved in metabolic pathways activated in plant responses to abiotic stress conditions, especially during water shortage. An independent meta-analysis was performed for each gene. Relevant articles were insufficient for acdS, MYC, MYB, WRKY, and ERD15 to proceed with the analysis. The primary obstacle to conducting a meta-analysis for these genes stemmed from the insufficient number of studies providing the log2 fold change, a crucial parameter for our analysis. Additional challenges included the absence of standard deviation (SD) or standard error (SE) values for the relative transcript accumulation of drought-responsive genes. To address this, the prognostic method was employed for DREB2, ACO, ACS, and APX genes. It is essential to underscore that SD and SE are pivotal values in a study, playing a crucial role in determining the effect size in meta-analysis.
Despite the scarcity of data preventing a comprehensive meta-analysis for acdS, MYC, MYB, WRKY, and ERD15 genes, we conducted a meticulous systematic review to gather insights into the expression and roles of these genes in response to drought stress in PGPR-treated plants. Although quantitative synthesis was impossible due to limited available studies, we excluded numerous acdS gene studies for lacking log2 fold change reporting. Many of these studies validated the presence of the ACC deaminase gene (acdS) through PCR in bacterial strains from the rhizosphere of plants like Cyamopsis tetragonoloba and Pennisetum glaucum L., showcasing their potential as PGPR in enhancing plant growth under water stress conditions (Goyal et al., 2022; Murali et al., 2021). It is widely recognized that microorganisms can reduce ethylene levels in the plant through the ACC deaminase enzyme. This hormone plays a crucial role in plant defence responses to abiotic stresses, limiting root and shoot growth (Selvakumar et al., 2012; Goswami and Deka, 2020). For instance, in V. mungo L. and P. sativum L. treated with a consortium of three ACC-deaminase-producing rhizobacteria, there was a reduction in deleterious stress ethylene accumulation during drought stress (Saikia et al., 2018). MYB transcription factors are known to be involved in plant drought responses, impacting development, metabolism, and stress regulation (Baldoni et al., 2015). However, the effect of drought on MYB gene regulation seems to vary based on plant species and organs (Baldoni et al., 2015). From our systematic review, only two articles regarding MYB expression in two different crop species inoculated with PGPR could have been included in the meta-analysis (Cho et al., 2013; Kakar et al., 2016). In Oryza sativa L. subjected to water stress and treated with a consortium of B. amyloliquefaciens–Bacillus methylotrophicus–Brevibacillus laterosporus, OsMYB3R-2 was upregulated, and plants showed improved seedling height and shoot number compared to control plants (Kakar et al., 2016). Conversely, in A. thaliana inoculated with Pseudomonas chlororaphis during water stress, MYB genes were downregulated (Cho et al., 2013). When water was suspended, Arabidopsis plants without inoculation showed a decrease in survival after 14 days of drought stress. The survival rate notably dropped in control plants on days 15 and 16 of the drought treatment. Conversely, plants inoculated with P. chlororaphis O6 did not display such a decline in viability when water was withheld (Cho et al., 2013). Only one study was suitable for meta-analysis regarding MYC gene, which is one of those found for MYB (Cho et al., 2013). Similarly, MYC genes, belonging to the basic helix-loop-helix (bHLH) transcription factor family, exhibited downregulation during water stress (Feng et al., 2023; Cho et al., 2013). WRKY transcription factors play a vital role in regulating plant stress responses. Overexpression of specific WRKY genes has shown increased tolerance to heat, drought, and salinity in various species (Tripathi et al., 2014). The only two WRKY-related studies that met the inclusion criteria for the meta-analysis showed that under drought stress conditions, in tomato inoculated with Streptomyces strains and Bacillus megaterium, WRKY70, SlWRKY75, and SlWRKY45 were downregulated (Abbasi et al., 2020; Morcillo et al., 2021). This significantly impacted the fruit weight of tomato plants subjected to water stress and treated with these strains (Abbasi et al., 2020). ERD15 was initially characterized as a rapidly drought-responsive gene in Arabidopsis; it has recently been reported as a negative regulator of ABA, which can also be induced by salicylic acid (SA), wounds, and pathogenic infections to mediate interactions between abiotic and biotic stress responses (Shao et al., 2014). Unfortunately, for ERD15, we did not find any articles that met the inclusion criteria.
The meta-analysis conducted on ACO, ACS, and APX showed non-significant results, considering that the confidence intervals of the summary effects overlapped by zero (Dong et al., 2017). ACO and ACS participate in the biosynthesis of ethylene, while APX gene codes for the ascorbate peroxide antioxidant enzyme, regulating the ROS defensive mechanism in plant cells in response to abiotic stress (Selvakumar et al., 2012; Nadeem et al., 2019; Goswami and Deka, 2020). From these results, it can be speculated that the lack of statistically significant differences in the expression of these genes, with or without PGPR, was due to the insignificant overall effect size. However, studies included in the meta-analysis for these genes highlighted a significant improvement in plants treated with PGPR during drought stress compared to the control (Kasim et al., 2013; Saikia et al., 2018). Saikia et al. (2018) conducted a study on V. mungo L. and P. sativum L. subjected to water stress and inoculated with a consortium of three ACC deaminase-producing rhizobacteria (O. pseudogrignonense RJ12, Pseudomonas sp. RJ15, and B. subtilis RJ46) (Saikia et al., 2018). This study demonstrated that inoculated plants showed a lower amount of ACC compared to the control due to the upregulation of ACS and the downregulation of ACO gene, consequently decreasing ethylene production (Saikia et al., 2018). Moreover, Kasim et al. (2013) reported that wheat plants inoculated with strains of B. amyloliquefaciens and A. brasilense during drought stress exhibited an upregulation of APX1 and increased activity of enzymes involved in the ascorbate-glutathione redox cycle, impacting ROS scavenging (Kasim et al., 2013). Therefore, we also hypothesized that the statistically insignificant results might be attributed to heterogeneity, even if we could not investigate its sources through subgroup analysis (insufficient number of articles: four for ACO and ACS and five for APX). Variability can arise from mathematical–statistical causes (statistical heterogeneity) and also from the evaluation of plants with different characteristics, treatments, or their assessment (clinical heterogeneity) (Egger et al., 2022). We speculated that the main source of heterogeneity could be attributed to the different induction stress methods used in studies included in the meta-analysis. In fact, the included studies for ACO and ACS gene meta-analysis used different treatments: Saikia et al. (2018); Sandhya and Ali (2018), and Tiwari et al. (2016) employed PEG 6000, while SkZ et al. (2018) induced drought stress by discontinuing water after 14 days of planting. Similarly, studies analysing APX exhibited differences: Gururani et al. (2013) and Tiwari et al. (2016) used PEG to induce drought stress, while Murali et al. (2021) and Singh et al. (2020) induced drought stress by withholding water. The divergence in the stress induction method (using PEG or withholding water) leads to differences in stress precision and practicality, influencing the plants’ responses, and this could be considered a source of heterogeneity in our meta-analysis (Krizek, 1985). DREBs (dehydration-responsive element binding) belong to the ERF family and the ABA-independent signal transduction pathway (Akbudak et al., 2018). Specifically, DREB2A plays a key role in plants’ drought response, primarily functioning in ABA-independent water stress-induced gene expression (Liu et al., 1998). In the first analysis, the expression of DREB2 in the absence of PGPR and stress versus the presence of PGPR under drought stress conditions was highlighted as a statistically significant result. The cumulative ES values in the forest plot clearly indicate DREB2 overexpression. In the second analysis regarding the expression of DREB2 in the absence of PGPR and stress versus the presence of drought stress, the absence of PGPR again highlighted a statistically significant result. Indeed, the cumulative ES values in the forest plot indicate DREB2 overexpression. Subsequently, the results matched. This comparison showed a statistically insignificant overexpression of DREB2 in the presence of PGPR compared to the results obtained for crops not treated with PGPR but still stressed due to drought. Indeed, by analysing the resulting forest plots (Supplementary Figures and Supplementary File 2), an overlap of the cumulative effect size was noticeable. Therefore, our data showed that DREB2 was overexpressed in plants subjected to drought stress, regardless of PGPR inoculation. In the former case, the gene expression level seemed higher, highlighting the slight effect that the bioinoculation of PGPR had on DREB2 expression in plants, but this was not statistically significant. However, studies included in the meta-analysis, which investigated the expression of DREB2 in PGPR-inoculated and drought-stressed plants, demonstrated positive enhancements in terms of dry weight. For instance, Brassica juncea L. inoculated with osmotolerant strains like Bacillus sp. MR D17 and Bacillus cereus NA D17, Glycine max L. Merrill treated with Pseudomonas simiae, and Triticum aestivum inoculated with Dietzia natronolimnaea and B. subtilis LDR2 all showed substantial increases in dry weight in response to drought stress (Barnawal et al., 2017; Bandeppa et al., 2019; Vaishnav and Choudhary, 2019). Despite employing a random effects model and conducting subgroup analysis, we were unable to diminish the high heterogeneity. The sources of heterogeneity in this study were as follows:
a. Treatments used for stressed plants. In some cases, water shortage or dehydration was caused by not administering water for a certain time interval (e.g., Gontia-Mishra et al., 2016; Sheikh-Mohamadi et al., 2018; Vu et al., 2021), while in other cases, PEG was utilised to provide the drought stress (DS) (e.g., Vaishnav and Choudhary, 2019; Zhang et al., 2022). In two out of eight articles included in the meta-analysis for DREB2 in PGPR-inoculated plants (Sarma and Saikia, 2014; Vaishnav and Choudhary, 2019), the authors decided to induce DS with PEG. Therefore, a subgroup analysis based on the water suspension method used to induce DS was performed. Nevertheless, heterogeneity did not diminish sufficiently, allowing us to hypothesise dependence on other variables.
b. Unavailability of a common protocol to stress the plant and measure the relative expression of genes. This reduces the number of data points subjected to comparison.
c. Choice of native and non-native PGPR strains for plant inoculation. Indeed, in some studies, plant non-native PGPR (Gagné-Bourque et al., 2015; Barnawal et al., 2017; Bandeppa et al., 2019) were used. In other works, such as Sarma and Saikia (2014); Vaishnav and Choudhary (2019), and Gontia-Mishra et al. (2016), plant-native PGPR were inoculated. This aspect is relevant because plant-native microbiota originated through a specific selection process that occurred during evolution, overcoming issues in persistence in the plant and rhizosphere with an adaptation process (Banerjee et al., 2017). Thus, the utilisation of non-native PGPR may affect the expression of plant genes.
d. Different plant organs are also subjected to stress. In some cases, the relative expression level of drought stress-related genes is derived from the seedling, whereas in others, it is derived from the root.
e. Although all of the studies considered were conducted under controlled conditions, the biometric parameters used for plant growth evaluation and the soil used for cultivation may differ slightly.
f. The limitations of the sample in this study may have overestimated the treatment effects and contributed to statistical heterogeneity (Zhang et al., 2013). In some cases, there were several hundred replicates, whereas in others, there were three replicates.
g. Gene expression can be induced in different ways during drought stress. Studies did not measure the same time-lapse. DREB2, for instance, is expressed within a range of a few hours; it starts to increase 10 min after stress treatment and reaches its maximum after 10 h of stress (Nakashima and Yamaguchi-Shinozaki, 2006).
Finally, it is crucial to mention the publication bias evident in the asymmetry of the funnel plots for DREB2, ACO, ACS, and APX. Publication bias reflects the tendency to publish studies on the direction of results; namely, it is more probable that studies with statistically significant or positive results will be published and accepted by journals6 than studies showing statistically insignificant or negative results (Song et al., 2013; Olsson and Sundell, 2023), affecting a clear and exhaustive panel of the resulting data. Indeed, it is widely assumed that negative studies that appear to be conducted better than positive ones are much less likely to be accepted for publication (Thornton and Lee, 2000). Some relevant biases concerning scientific literature include time lag, outcome reporting, grey literature, full publication, language, citation, and media-attention biases (Song et al., 2013; Olsson and Sundell, 2023). According to Olsson and Sundell (2023), every type of bias negatively affects the effectiveness of research synthesis concerning biased literature. This can significantly alter the process of effect estimation in a meta-analysis (Thornton and Lee, 2000). In fact, as suggested in the guidelines of the PRISMA statement (Moher et al., 2009), we excluded all non-English-language articles and the “grey literature” such as dissertations, conference proceedings, and congresses. The results reported in the “grey literature” may have limited accessibility, thereby complicating their immediate use with respect to results from peer-reviewed journals (Song et al., 2013). The asymmetry of the funnel plots obtained from our analyses could be due to both the high heterogeneity and publication bias of the examined studies, generating asymmetries due to data irregularities, artefacts, and chances (Nakagawa et al., 2022).
In conclusion, meta-analysis is an enormously powerful and extremely useful tool in many scientific fields and serves as a synopsis of a research question that provides a quantitative assessment of the relationship between two target variables. The purpose is twofold: it can bring out a unique conclusion on a topic and open science to new and more specific research questions. Meta-analyses are also recommended to identify topics for which the available data are insufficient and further studies are required. In light of our findings, plants inoculated with PGPR did not show observable changes in the expression of the defence-related genes acdS, ACS, ACO, APX, and DREB2. These results could depend on the three pivotal challenges faced in this study: lack of data, heterogeneity of studies, and publication bias. However, in the case of any pathogenic attack, rhizobacteria-treated plants showed a strong and quick response compared to the control plants, as confirmed in another meta-analysis (Bukhat et al., 2020). Considering that our results did not show a clear ability of PGPR to influence plant gene expression and promote growth under water stress, further investigation is fundamental. It is important for future work to deepen the understanding of the aspects related to signal sensing, perception, and the metabolic pathways involved in DREB2 expression and the expression of other plant-responsive genes under drought stress. Considering the limitations of the literature, our results emphasise the importance of establishing common and shared protocols and standardising treatments to facilitate reproducibility and statistical analysis.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
RF: Data curation, Formal analysis, Investigation, Writing – original draft. CC: Data curation, Formal analysis, Investigation, Writing – original draft. CV: Funding acquisition, Project administration, Supervision, Writing – review & editing. AC: Writing – review & editing. CG: Writing – review & editing. DP: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by MIUR (Ministero dell’Istruzione e del Merito) within the PRIMA CALL 2020 section 2.
Acknowledgments
We thank all the scientists who produced the published data that were included in this meta-analysis. We would like to thank Catia Boggi (CNR‐IBBR, Italy) for her technical support. The authors RF and DP acknowledge the support of NBFC to Consiglio Nazionale delle Ricerche, funded by the Italian Ministry of University and Research, PNRR, Missione 4, Componente 2, “Dalla ricerca all’impresa”, Investimento 1.4 Project CN00000033.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1282553/full#supplementary-material
References
Abbasi, S., Sadeghi, A., Safaie, N. (2020). Streptomyces alleviate drought stress in tomato plants and modulate the expression of transcription factors ERF1 and WRKY70 genes. Scientia Hortic. 265, 109206. doi: 10.1016/j.scienta.2020.109206
Ahluwalia, O., Singh, P. C., Bhatia, R. (2021). A review on drought stress in plants: Implications, mitigation and the role of plant growth promoting rhizobacteria. Resour. Environ. Sustain. 5, 100032. doi: 10.1016/j.resenv.2021.100032
Akbudak, M. A., Filiz, E., Kontbay, K. (2018). DREB2 (dehydration-responsive element-binding protein 2) type transcription factor in sorghum (Sorghum bicolor): genome-wide identification, characterization and expression profiles under cadmium and salt stresses. 3 Biotech. 8, 426. doi: 10.1007/s13205-018-1454-1
Alori, E., Dare, M. O., Babalola, O. (2017). “Microbial inoculants for soil quality and plant health,” in Sustainable agriculture reviews. Ed. Lichtfouse, E. (Springer Cham, Switzerland), 281–307. doi: 10.1007/978-3-319-48006-0_9
Anjum, S., Ashraf, U., Zohaib, A., Tanveer, M., Naeem, M., Ali, I., et al. (2017). Growth and developmental responses of crop plants under drought stress: A review. Zemdirbyste-Agriculture 104, 267–276. doi: 10.13080/z-a.2017.104.034
Arora, D., Abel, N. B., Liu, C., Van Damme, P., Yperman, K., Eeckhout, D., et al. (2020). Establishment of proximity-dependent biotinylation approaches in different plant model systems. Plant Cell 32, 3388–3407. doi: 10.1105/tpc.20.00235
Baldoni, E., Genga, A., Cominelli, E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16, 15811–15851. doi: 10.3390/ijms160715811
Bandeppa, S., Sangeeta, P., Thakur, J. K., Chandrashekar, N., Umesh, D. K., Aggarwal, C., et al. (2019). Antioxidant, physiological and biochemical responses of drought susceptible and drought tolerant mustard (Brassica juncea L) genotypes to rhizobacterial inoculation under water deficit stress. Plant Physiol. Biochem. 143, 19–28. doi: 10.1016/j.plaphy.2019.08.018
Banerjee, A., Bareh, D. A., Joshi, S. R. (2017). Native microorganisms as potent bioinoculants for plant growth promotion in shifting agriculture (Jhum) systems. J. Soil Sci. Plant Nutr. 17, 127–140. doi: 10.4067/S0718-95162017005000010
Barnawal, D., Bharti, N., Pandey, S. S., Pandey, A., Chanotiya, C. S., Kalra, A. (2017). Plant growth-promoting rhizobacteria enhance wheat salt and drought stress tolerance by altering endogenous phytohormone levels and TaCTR1/TaDREB2 expression. Physiol. Plantarum 161, 502–514. doi: 10.1111/ppl.12614
Borges, A., Tsai, S. M., Caldas, D. G. G. (2012). Validation of reference genes for RT-qPCR normalisation in common bean during biotic and abiotic stresses. Plant Cell Rep. 31, 827–838. doi: 10.1007/s00299-011-1204-x
Bukhat, S., Imran, A., Javaid, S., Shahid, M., Majeed, A., Naqqash, T. (2020). Communication of plants with microbial world: Exploring the regulatory networks for PGPR mediated defense signaling. Microbiol. Res. 238, 126486. doi: 10.1016/j.micres.2020.126486
Çakmakçı, R. (2016). Screening of multi-trait rhizobacteria for improving the growth, enzyme activities, and nutrient uptake of tea (Camellia sinensis). Commun. Soil Sci. Plant Anal. 47, 1680–1690. doi: 10.1080/00103624.2016.1206559
Cangioli, L., Checcucci, A., Mengoni, A., Fagorzi, C. (2021). Legume tasters: symbiotic rhizobia host preference and smart inoculant formulations. Bio. Commun. 66, 47–54. doi: 10.21638/spbu03.2021.106
Chandra, P., Wunnava, A., Verma, P., Chandra, A., Sharma, R. K. (2021). Strategies to mitigate the adverse effect of drought stress on crop plants—influences of soil bacteria: A review. Pedosphere 31, 496–509. doi: 10.1016/S1002-0160(20)60092-3
Cho, S. M., Kang, B. R., Kim, Y. C. (2013). Transcriptome analysis of induced systemic drought tolerance elicited by Pseudomonas chlororaphis O6 in Arabidopsis thaliana. Plant Pathol. J. 29, 209. doi: 10.5423/PPJ.SI.07.2012.0103
Cohen, J. (1988). Statistical power analysis for the behavioral sciences. 2nd ed (New York: Routledge). doi: 10.4324/9780203771587
Creus, C. M., Graziano, M., Casanovas, E. M., Pereyra, M. A., Simontacchi, M., Puntarulo, S., et al. (2005). Nitric oxide is involved in the Azospirillum brasilense-induced lateral root formation in tomato. Planta 221, 297–303. doi: 10.1007/s00425-005-1523-7
de Vries, K., Medawar, E., Korosi, A., Witte, A. V. (2022). The effect of polyphenols on working and episodic memory in non-pathological and pathological aging: A systematic review and meta-analysis. Front. Nutr. 8. doi: 10.3389/fnut.2021.720756
Dimkpa, C. O., Merten, D., Svatos, A., Büchel, G., Kothe, E. (2009). Siderophores mediate reduced and increased uptake of cadmium by Streptomyces tendae F4 and sunflower (Helianthus annuus), respectively. J. Appl. Microbiol. 107, 1687–1696. doi: 10.1111/j.1365-2672.2009.04355.x
Dimkpa, C., Weinand, T., Asch, F. (2009). Plant–rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x
Dong, C., Ma, Y., Wisniewski, M., Cheng, Z.-M. (2017). Meta-analysis of the effect of overexpression of CBF/DREB family genes on drought stress response. Environ. Exp. Bot. 142, 1–14. doi: 10.1016/j.envexpbot.2017.07.014
Egamberdieva, D., Lugtenberg, B. (2014). “Use of plant growth-promoting rhizobacteria to alleviate salinity stress in plants,” in Use of microbes for the alleviation of soil stresses. Ed. Miransari, M. (New York, NY: Springer), 73–96. doi: 10.1007/978-1-4614-9466-9_4
Egger, M., Higgins, J. P. T., Smith, G. D. (2022). Systematic reviews in health research: meta-analysis in context (John Wiley & Sons Ltd, 9600 Garsington Road, Oxford, OX4 2DQ, UK). doi: 10.1002/9781119099369
Enebe, M. C., Babalola, O. O. (2018). The influence of plant growth-promoting rhizobacteria in plant tolerance to abiotic stress: a survival strategy. Appl. Microbiol. Biotechnol. 102, 7821–7835. doi: 10.1007/s00253-018-9214-z
Feng, C., Song, X., Tang, H. (2019). Molecular cloning and expression analysis of GT-2-like genes in strawberry. 3 Biotech. 9, 105. doi: 10.1007/s13205-019-1603-1
Feng, Y., Zeng, S., Yan, J., Li, K., Xu, H. (2023). Genome-wide analysis and expression of MYC family genes in tomato and the functional identification of slmyc1 in response to salt and drought stress. Agronomy 13, 757. doi: 10.3390/agronomy13030757
Gagné-Bourque, F., Mayer, B. F., Charron, J.-B., Vali, H., Bertrand, A., Jabaji, S. (2015). Accelerated Growth Rate and Increased Drought Stress Resilience of the Model Grass Brachypodium distachyon Colonized by Bacillus subtilis B26. PloS One 10, e0130456. doi: 10.1371/journal.pone.0130456
Golldack, D., Li, C., Mohan, H., Probst, N. (2014). Tolerance to drought and salt stress in plants: Unraveling the signaling networks. Front. Plant Sci. 5 (151). doi: 10.3389/fpls.2014.00151
Gong, Z., Xiong, L., Shi, H., Yang, S., Herrera-Estrella, L. R., Xu, G., et al. (2020). Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 63, 635–674. doi: 10.1007/s11427-020-1683-x
Gontia-Mishra, I., Sapre, S., Sharma, A., Tiwari, S. (2016). Amelioration of drought tolerance in wheat by the interaction of plant growth-promoting rhizobacteria. Plant Biol. 18, 992–1000. doi: 10.1111/plb.12505
Goswami, M., Deka, S. (2020). Plant growth-promoting rhizobacteria—alleviators of abiotic stresses in soil: A review. Pedosphere 30, 40–61. doi: 10.1016/S1002-0160(19)60839-8
Goyal, D., Kumar, S., Meena, D., Solanki, S. S., Swaroop, S., Pandey, J. (2022). Selection of ACC deaminase positive, thermohalotolerant and drought tolerance enhancing plant growth-promoting bacteria from rhizospheres of Cyamopsis tetragonoloba grown in arid regions. Lett. Appl. Microbiol. 74, 519–535. doi: 10.1111/lam.13633
Gupta, K., Dubey, N. K., Singh, S. P., Kheni, J. K., Gupta, S., Varshney, A. (2021). “Plant growth-promoting rhizobacteria (PGPR): current and future prospects for crop improvement,” in Current trends in microbial biotechnology for sustainable agriculture. Eds. Yadav, A. N., Singh, J., Singh, C., Yadav, N. (Singapore: Springer), 203–226. doi: 10.1007/978-981-15-6949-4_9
Gururani, M. A., Upadhyaya, C. P., Baskar, V., Venkatesh, J., Nookaraju, A., Park, S. W. (2013). Plant growth-promoting rhizobacteria enhance abiotic stress tolerance in Solanum tuberosum through inducing changes in the expression of ROS-scavenging enzymes and improved photosynthetic performance. J. Plant Growth Regul. 32, 245–258. doi: 10.1007/s00344-012-9292-6
Ha-Tran, D., Nguyen, T., Hung, S.-H., Huang, E., Huang, C.-C. (2021). Roles of plant growth-promoting rhizobacteria (PGPR) in stimulating salinity stress defense in plants: A review. Int. J. Mol. Sci. 22 (6), 3154. doi: 10.3390/ijms22063154
Hayashino, Y., Noguchi, Y., Fukui, T. (2005). Systematic evaluation and comparison of statistical tests for publication bias. J. Epidemiol. 15, 235–243. doi: 10.2188/jea.15.235
Hoque, M., Hannan, A., Imran, S., Paul, N. C., Mondal, M., Sadhin, Md. M.R., et al. (2023). Plant growth-promoting rhizobacteria-mediated adaptive responses of plants under salinity stress. J. Plant Growth Regul. 42, 1307–1326. doi: 10.1007/s00344-022-10633-1
Hussain, H. A., Hussain, S., Khaliq, A., Ashraf, U., Anjum, S. A., Men, S., et al. (2018). Chilling and drought stresses in crop plants: implications, cross talk, and potential management opportunities. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00393
Ibort, P., Molina, S., Ruiz-Lozano, J. M., Aroca, R. (2018). Molecular insights into the involvement of a never ripe receptor in the interaction between two beneficial soil bacteria and tomato plants under well-watered and drought conditions. Mol. Plant Microbe Interact. 31, 633–650. doi: 10.1094/mpmi-12-17-0292-r
JASP Team (2023) JASP (Version 0.14). Available at: https://jasp-stats.org/ (Accessed August 21, 2023).
Joshi, R., Wani, S. H., Singh, B., Bohra, A., Dar, Z. A., Lone, A. A., et al. (2016). Transcription factors and plants response to drought stress: current understanding and future directions. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01029
Kakar, K. U., Ren, X. L., Nawaz, Z., Cui, Z. Q., Li, B., Xie, G. L., et al. (2016). A consortium of rhizobacterial strains and biochemical growth elicitors improve cold and drought stress tolerance in rice (Oryza sativa L.). Plant Biol. 18, 471–483. doi: 10.1111/plb.12427
Kamle, M., Borah, R., Bora, H., Jaiswal, A. K., Singh, R. K., Kumar, P. (2020). “Systemic acquired resistance (SAR) and induced systemic resistance (ISR): role and mechanism of action against phytopathogens,” in Fungal biotechnology and bioengineering, fungal biology. Eds. Hesham, A. E.-L., Upadhyay, R. S., Sharma, G. D., Manoharachary, C., Gupta, V. K. (Springer International Publishing, Cham, Switzerland), 457–470. doi: 10.1007/978-3-030-41870-0_20
Kasim, W. A., Osman, M. E., Omar, M. N., Abd El-Daim, I. A., Bejai, S., Meijer, J. (2013). Control of drought stress in wheat using plant-growth-promoting bacteria. J. Plant Growth Regul. 32, 122–130. doi: 10.1007/s00344-012-9283-7
Khan, N., Bano, A., Ali, S., Babar, M. (2020). Crosstalk amongst phytohormones from planta and PGPR under biotic and abiotic stresses. Plant Growth Regul. 90, 189–203. doi: 10.1007/s10725-020-00571-x
Kooyers, N. J. (2015). The evolution of drought escape and avoidance in natural herbaceous populations. Plant Sci. 234, 155–162. doi: 10.1016/j.plantsci.2015.02.012
Krizek, D. T. (1985). Methods of inducing water stress in plants. HortScience 20, 1028–1038. doi: 10.21273/HORTSCI.20.6.1028
Lakens, D. (2013). Calculating and reporting effect sizes to facilitate cumulative science: a practical primer for t-tests and ANOVAs. Front. Psychol. 4. doi: 10.3389/fpsyg.2013.00863
Le, D. T., Aldrich, D. L., Valliyodan, B., Watanabe, Y., Ha, C. V., Nishiyama, R., et al. (2012). Evaluation of candidate reference genes for normalisation of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PloS One 7, e46487. doi: 10.1371/journal.pone.0046487
Lim, J.-H., Kim, S.-D. (2013). Induction of drought stress resistance by multi-functional PGPR Bacillus licheniformis K11 in pepper. Plant Pathol. J. 29, 201–208. doi: 10.5423/PPJ.SI.02.2013.0021
Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., et al. (1998). Two transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell. 10, 1391–1406. doi: 10.1105/tpc.10.8.1391
Ma, J., Liu, W., Hunter, A., Zhang, W. (2008). Performing meta-analysis with incomplete statistical information in clinical trials. BMC Med. Res. Methodol. 8, 56. doi: 10.1186/1471-2288-8-56
Manjunatha, B. S., Nivetha, N., Krishna, G. K., Elangovan, A., Pushkar, S., Chandrashekar, N., et al. (2022). Plant growth-promoting rhizobacteria Shewanella putrefaciens and Cronobacter dublinensis enhance drought tolerance of pearl millet by modulating hormones and stress-responsive genes. Physiologia Plantarum 174, e13676. doi: 10.1111/ppl.13676
Meier, M. A., Xu, G., Lopez-Guerrero, M. G., Li, G., Smith, C., Sigmon, B. (2022). Association analyses of host genetics, root-colonising microbes, and plant phenotypes under different nitrogen conditions in maise. Elife 11, e75790. doi: 10.7554/eLife.75790
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G., PRISMA Group (2009). Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ 339, b2535. doi: 10.1136/bmj.b2535
Molina-Favero, C., Creus, C. M., Simontacchi, M., Puntarulo, S., Lamattina, L. (2008). Aerobic nitric oxide production by Azospirillum brasilense Sp245 and its influence on root architecture in tomato. Mol. Plant Microbe Interact. 21, 1001–1009. doi: 10.1094/MPMI-21-7-1001
Morcillo, R. J. L., Vílchez, J. I., Zhang, S., Kaushal, R., He, D., Zi, H., et al. (2021). Plant transcriptome reprograming and bacterial extracellular metabolites underlying tomato drought resistance triggered by a beneficial soil bacteria. Metabolites 11, 369. doi: 10.3390/metabo11060369
Murali, M., Singh, S. B., Gowtham, H. G., Shilpa, N., Prasad, M., Aiyaz, M., et al. (2021). Induction of drought tolerance in Pennisetum glaucum by ACC deaminase producing PGPR- Bacillus amyloliquefaciens through Antioxidant defense system. Microbiol. Res. 253, 126891. doi: 10.1016/j.micres.2021.126891
Nabi, R. B. S., Tayade, R., Hussain, A., Adhikari, A., Lee, I.-J., Loake, G. J., et al. (2021). A novel DUF569 gene is a positive regulator of the drought stress response in Arabidopsis. IJMS 22, 5316. doi: 10.3390/ijms22105316
Nadeem, M., Li, J., Yahya, M., Sher, A., Ma, C., Wang, X., et al. (2019). Research progress and perspective on drought stress in legumes: A review. Int. J. Mol. Sci. 20, 2541. doi: 10.3390/ijms20102541
Nakagawa, S., Lagisz, M., Jennions, M. D., Koricheva, J., Noble, D. W. A., Parker, T. H., et al. (2022). Methods for testing publication bias in ecological and evolutionary meta-analyses. Methods Ecol. Evol. 13, 4–21. doi: 10.1111/2041-210X.13724
Nakagawa, S., Noble, D. W. A., Senior, A. M., Lagisz, M. (2017). Meta-evaluation of meta-analysis: ten appraisal questions for biologists. BMC Biol. 15, 18. doi: 10.1186/s12915-017-0357-7
Nakashima, K., Yamaguchi-Shinozaki, K. (2006). Regulons involved in osmotic stress-responsive and cold stress-responsive gene expression in plants. Physiol. Plant 126, 62–71. doi: 10.1111/j.1399-3054.2005.00592.x
Naylor, D., Coleman-Derr, D. (2018). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.02223
Neves-Borges, A. C., Guimarães-Dias, F., Cruz, F., Mesquita, R. O., Nepomuceno, A. L., Romano, E., et al. (2012). Expression pattern of drought stress marker genes in soybean roots under two water deficit systems. Genet. Mol. Biol. 35, 212–221. doi: 10.1590/S1415-47572012000200003
Ojuederie, O., Olanrewaju, O., Babalola, O. (2019). Plant growth promoting rhizobacterial mitigation of drought stress in crop plants: implications for sustainable agriculture. Agronomy 9, 712. doi: 10.3390/agronomy9110712
Oleńska, E., Małek, W., Wójcik, M., Swiecicka, I., Thijs, S., Vangronsveld, J. (2020). Beneficial features of plant growth-promoting rhizobacteria for improving plant growth and health in challenging conditions: A methodical review. Sci. Total Environ. 743, 140682. doi: 10.1016/j.scitotenv.2020.140682
Olsson, T. M., Sundell, K. (2023). Publication bias, time-lag bias, and place-of-publication bias in social intervention research: An exploratory study of 527 Swedish articles published between 1990–2019. PloS One 18, e0281110. doi: 10.1371/journal.pone.0281110
Omae, N., Tsuda, K. (2022). Plant-microbiota interactions in abiotic stress environments. MPMI 35, 511–526. doi: 10.1094/MPMI-11-21-0281-FI
Page, M. J., Moher, D., Bossuyt, P. M., Boutron, I., Hoffmann, T. C., Mulrow, C. D., et al. (2021). PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ 372, n160. doi: 10.1136/bmj.n160
Rohatgi, A. (2022) Web plot digitiser. Available at: http://www.arohatgi.info/WebPlotDigitizer (Accessed August 21, 2023).
Rosier, A., Medeiros, F. H. V., Bais, H. P. (2018). Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 428, 35–55. doi: 10.1007/s11104-018-3679-5
Saikia, J., Sarma, R. K., Dhandia, R., Yadav, A., Bharali, R., Gupta, V. K., et al. (2018). Alleviation of drought stress in pulse crops with ACC deaminase producing rhizobacteria isolated from acidic soil of Northeast India. Sci. Rep. 8, 3560. doi: 10.1038/s41598-018-21921-w
Sandhya, V., Ali, S. Z. (2018). Quantitative mRNA analysis of induced genes in maize inoculated with Acinetobacter baumannii strain MZ30V92. Curr. Biotechnol. 7, 438–452. doi: 10.2174/2211550108666190125114821
Sarma, R. K., Saikia, R. (2014). Alleviation of drought stress in mung bean by strain Pseudomonas aeruginosa GGRJ21. Plant Soil 377, 111–126. doi: 10.1007/s11104-013-1981-9
Selvakumar, G., Panneerselvam, P., Ganeshamurthy, A. N. (2012). “Bacterial mediated alleviation of abiotic stress in crops,” in Bacteria in agrobiology: stress management. Ed. Maheshwari, D. (Berlin, Heidelberg: Springer). doi: 10.1007/978-3-642-23465-1_10
Shao, H. H., Chen, S. D., Zhang, K., Cao, Q. H., Zhou, H., Ma, Q. Q., et al. (2014). Isolation and expression studies of the ERD15 gene involved in drought-stressed responses. Genet. Mol. Res. 13, 10852–10862. doi: 10.4238/2014
Sheikh-Mohamadi, M.-H., Etemadi, N., Arab, M. M., Aalifar, M., Arab, M. (2018). Physiological and Ascorbate -Glutathione pathway-related genes responses under drought and heat stress in crested wheatgrass. Scientia Hortic. 242, 195–206. doi: 10.1016/j.scienta.2018.07.037
Singh, J. S., Abhilash, P. C., Gupta, V. K. (2016). Agriculturally important microbes in sustainable food production. Trends Biotechnol. 34, 773–775. doi: 10.1016/j.tibtech.2016.06.002
Singh, J. S., Pandey, V. C., Singh, D. P. (2011). Efficient soil microorganisms: A new dimension for sustainable agriculture and environmental development. Agric. Ecosyst. Environ. 140, 339–353. doi: 10.1016/j.agee.2011.01.017
Singh, D. P., Singh, V., Gupta, V. K., Shukla, R., Prabha, R., Sarma, B. K., et al. (2020). Microbial inoculation in rice regulates antioxidative reactions and defense related genes to mitigate drought stress. Sci. Rep. 10, 4818. doi: 10.1038/s41598-020-61140-w
SkZ, A., Vardharajula, S., Vurukonda, S. S. K. P. (2018). Transcriptomic profiling of maize (Zea mays L.) seedlings in response to Pseudomonas putida stain FBKV2 inoculation under drought stress. Ann. Microbiol. 68, 331–349. doi: 10.1007/s13213-018-1341-3
Song, F., Hooper, L., Loke, Y. (2013). Publication bias: what is it? How do we measure it? How do we avoid it? Open Access J. Clin. Trials 5, 71–81. doi: 10.2147/OAJCT.S34419
Song, L., Prince, S., Valliyodan, B., Joshi, T., Maldonado dos Santos, J. V., Wang, J., et al. (2016). Genome-wide transcriptome analysis of soybean primary root under varying water-deficit conditions. BMC Genomics 17, 57. doi: 10.1186/s12864-016-2378-y
Sterne, J. A. C., Egger, M. (2001). Funnel plots for detecting bias in meta-analysis. J. Clin. Epidemiol. 54, 1046–1055. doi: 10.1016/S0895-4356(01)00377-8
Sterne, J. A. C., Sutton, A. J., Ioannidis, J. P. A., Terrin, N., Jones, D. R., Lau, J., et al. (2011). Recommendations for examining and interpreting funnel plot asymmetry in meta-analyses of randomised controlled trials. BMJ 343, d4002. doi: 10.1136/bmj.d4002
Struik, P., Kuyper, T., Brussaard, L., Leeuwis, C. (2014). Deconstructing and unpacking scientific controversies in intensification and sustainability: why the tensions in concepts and values? Curr. Opin. Environ. Sustain. 8, 80–88. doi: 10.1016/j.cosust.2014.10.002
Tao, L., Yu, G., Chen, H., Wang, B., Jiang, L., Han, X., et al. (2022). SlDREB2 gene specifically recognising to the universal DRE elements is a transcriptional activator improving drought tolerance in tomato. Sci. Hortic. 295, 110887. doi: 10.1016/j.scienta.2022.110887
Thakur, R., Yadav, S. (2023). Thermotolerant and halotolerant Streptomyces sp. isolated from Ajuga parviflora having biocontrol activity against Pseudomonas syringae and Xanthomonas campestris acts as a sustainable bioadditive in growth promotion of Cicer arietinum. Physiological and Molecular Plant Pathology 127, 102059. doi: 10.1016/j.pmpp.2023.102059
Thakur, R., Srivastava, S., Yadav, S. (2023). Multitrait Pseudomonas sp. isolated from the rhizosphere of Bergenia ciliata acts as a growth-promoting bioinoculant for plants. Front. Sustain. Food Syst. 7, 1097587. doi: 10.3389/fsufs.2023.1097587
Thirumalaikumar, V. P., Devkar, V., Mehterov, N., Ali, S., Ozgur, R., Turkan, I., et al. (2018). NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 16, 354–366. doi: 10.1111/pbi.12776
Thornton, A., Lee, P. (2000). Publication bias in meta-analysis: its causes and consequences. J. Clin. Epidemiol. 53, 207–216. doi: 10.1016/S0895-4356(99)00161-4
Timmusk, S., Wagner, E. G. (1999). The plant-growth-promoting rhizobacterium Paenibacillus polymyxa induces changes in Arabidopsis thaliana gene expression: a possible connection between biotic and abiotic stress responses. Mol. Plant Microbe Interact. 12, 951–959. doi: 10.1094/MPMI.1999.12.11.951
Tiwari, S., Lata, C., Chauhan, P. S., Nautiyal, C. S. (2016). Pseudomonas putida attunes morphophysiological, biochemical and molecular responses in Cicer arietinum L. during drought stress and recovery. Plant Physiol. Biochem. 99, 108–117. doi: 10.1016/j.plaphy.2015.11.001
Tripathi, P., Rabara, R. C., Rushton, P. J. (2014). A systems biology perspective on the role of WRKY transcription factors in drought responses in plants. Planta 239, 255–266. doi: 10.1007/s00425-013-1985-y
Vaishnav, A., Choudhary, D. K. (2019). Regulation of Drought-Responsive Gene Expression in Glycine max L. Merrill is Mediated Through Pseudomonas simiae Strain AU. J. Plant Growth Regul. 38, 333–342. doi: 10.1007/s00344-018-9846-3
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
van Loon, L. C. (2007). Plant responses to plant growth-promoting rhizobacteria. Eur. J. Plant Pathol. 119, 243–254. doi: 10.1007/s10658-007-9165-1
Vu, T. T. H., Le, T. T. C., Pham, T. L. (2021). Growth responses and differential expression of VrDREB2A gene at different growth stages of mungbean (Vigna radiata L. Wilczek) under drought stress. Physiol. Mol. Biol. Plants 27, 2447–2458. doi: 10.1007/s12298-021-01089-w
Vurukonda, S. S. K. P., Vardharajula, S., Shrivastava, M., Sk, Z. A. (2016). Enhancement of drought stress tolerance in crops by plant growth promoting rhizobacteria. Microbiol. Res. 184, 13–24. doi: 10.1016/j.micres.2015.12.003
Wagner, M. R. (2022). How hungry roots get their microbes. eLife 11, e82391. doi: 10.7554/eLife.82391
Watson, S. C. L., Newton, A. C., Ridding, L. E., Evans, P. M., Brand, S., McCracken, M., et al. (2021). Does agricultural intensification cause tipping points in ecosystem services? Landscape Ecol. 36, 3473–3491. doi: 10.1007/s10980-021-01321-8
Weir, C. J., Butcher, I., Assi, V., Lewis, S. C., Murray, G. D., Langhorne, P., et al. (2018). Dealing with missing standard deviation and mean values in meta-analysis of continuous outcomes: a systematic review. BMC Med. Res. Methodol. 18, 25. doi: 10.1186/s12874-018-0483-0
Wezel, A., Casagrande, M., Celette, F., Vian, J.-F., Ferrer, A., Peigné, J. (2014). Agroecological practices for sustainable agriculture. A review. Agron. Sustain. Dev. 34, 1–20. doi: 10.1007/s13593-013-0180-7
Wu, Y., Wei, W., Pang, X., Wang, X., Zhang, H., Dong, B., et al. (2014). Comparative transcriptome profiling of a desert evergreen shrub, Ammopiptanthus mongolicus, in response to drought and cold stresses. BMC Genomics 15, 671. doi: 10.1186/1471-2164-15-671
Zhang, J., Sun, Y., Zhou, Z., Zhang, Y., Yang, Y., Zan, X., et al. (2022). OsSCL30 overexpression reduces the tolerance of rice seedlings to low temperature, drought and salt. Sci. Rep. 12, 8385. doi: 10.1038/s41598-022-12438-4
Keywords: PGPR, plant gene regulation, transcriptional factors regulation, meta-analysis, abiotic stress, crops, drought stress
Citation: Ferrante R, Campagni C, Vettori C, Checcucci A, Garosi C and Paffetti D (2024) Meta-analysis of plant growth-promoting rhizobacteria interaction with host plants: implications for drought stress response gene expression. Front. Plant Sci. 14:1282553. doi: 10.3389/fpls.2023.1282553
Received: 24 August 2023; Accepted: 18 December 2023;
Published: 15 January 2024.
Edited by:
Maurizio Ruzzi, University of Tuscia, ItalyReviewed by:
Saurabh Yadav, Hemwati Nandan Bahuguna Garhwal University, IndiaBaby Tabassum, M. J. P. Rohilkhand University, India
Copyright © 2024 Ferrante, Campagni, Vettori, Checcucci, Garosi and Paffetti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Cristina Vettori, cristina.vettori@cnr.it
†These authors have contributed equally to this work and share first authorship
 Roberta Ferrante1,2†
Roberta Ferrante1,2† Chiara Campagni
Chiara Campagni Cristina Vettori
Cristina Vettori Alice Checcucci
Alice Checcucci Cesare Garosi
Cesare Garosi Donatella Paffetti
Donatella Paffetti