- 1University Institute of Biotechnology, Chandigarh University, Mohali, India
- 2Department of Botany, Gargi College, New Delhi, India
- 3Department of Botany, Institute of Science, Banaras Hindu University, Varanasi, Uttar Pradesh, India
- 4Department of Botany, Ramjas College, New Delhi, India
The process of plant immune response is orchestrated by intracellular signaling molecules. Since plants are devoid of a humoral system, they develop extensive mechanism of pathogen recognition, signal perception, and intricate cell signaling for their protection from biotic and abiotic stresses. The pathogenic attack induces calcium ion accumulation in the plant cells, resulting in calcium signatures that regulate the synthesis of proteins of defense system. These calcium signatures induct different calcium dependent proteins such as calmodulins (CaMs), calcineurin B-like proteins (CBLs), calcium-dependent protein kinases (CDPKs) and other signaling molecules to orchestrate the complex defense signaling. Using advanced biotechnological tools, the role of Ca2+ signaling during plant-microbe interactions and the role of CaM/CMLs and CDPKs in plant defense mechanism has been revealed to some extent. The Emerging perspectives on calcium signaling in plant-microbe interactions suggest that this complex interplay could be harnessed to improve plant resistance against pathogenic microbes. We present here an overview of current understanding in calcium signatures during plant-microbe interaction so as to imbibe a future direction of research.
Introduction
Calcium ions (Ca2+) play an important role in various plant functions, like providing structural support, nutrition, and inducing stress responses. Besides these, Ca2+ functions as a secondary messenger in cell to cell signaling, and disruptions in its levels occur during biotic or abiotic stress responses (Ghosh et al., 2022). Under different stress conditions, there are spikes in Ca2+ concentration within the cell cytosol, known as “calcium signatures”. These Ca2+ spikes can be sensed by different calcium influx and efflux proteins, which maintain cytosolic Ca2+ homeostasis (Yadav et al., 2022). The calcium influx proteins facilitate the entry of calcium ions into the cell, initiating calcium signaling through the channels that allow Ca2+ to enter the cytosol. Conversely, calcium efflux proteins work to remove excess calcium ions from the cytosol, regulating the duration and magnitude of calcium signals and maintaining cellular calcium homeostasis. These proteins are critical components of calcium signaling pathways in plants, orchestrating various physiological processes encompassing growth, development, and the ability to mount responses against biotic and abiotic stresses (Ranty et al., 2016; Raina et al., 2021a; Verma et al., 2022).
The dynamic control of intracellular calcium level relies on a delicate balance between extracellular Ca2+ entry and intracellular Ca2+ release from various subcellular compartments including mitochondria, endoplasmic reticulum and vacuoles (Resentini et al., 2021). The influx of extracellular Ca2+ is mediated by various calcium channel, such as plasma membrane localized calcium channels and the voltage-dependent calcium channels. These channels play crucial roles in regulating cellular Ca2+ levels and are essential for various physiological processes in plants (Figure 1, right panel). Additionally, the mobilization of Ca2+ from intracellular storage is actively facilitated by various signaling molecules such as inositol triphosphate (InsP3), cyclic adenosine diphosphate ribose (cADPR) and nicotinic acid adenine dinucleotide phosphate (NAADP) (Galione and Churchill, 2002). InsP3 is involved in the activation of Ca2+ release from the endoplasmic reticulum, while cADPR and NAADP participate in Ca2+ release from acidic organelles such as lysosomes and endosomes. In the InsP3 pathway, signaling molecules like hormones or environmental cues activate phospholipase C, which enzymatically hydrolyses phosphatidylinositol 4,5-biphosphate (PIP2) to produce diacylglycerol and InsP3. The InsP3 subsequently interacts with InsP3 receptors located on the endoplasmic reticulum, inducing the discharge of Ca2+ into the cytosol (Kudla et al., 2018). The overexpression of a phospholipase C gene resulted in elevated intracellular Ca2+ levels and increased resistance against Pseudomonas syringae (Meng et al., 2016). On the other hand, cADPR and NAADP promote the release of Ca2+ from acidic compartments such as vacuoles and lysosomes by activating ryanodine receptors (RyRs) and two-pore channels (TPCs), respectively (Berridge et al., 2000; Li et al., 2015). For example, the intracellular bacterial pathogen Legionella pneumophila can hijack host cellular machinery to promote the mobilization of Ca2+ from intracellular reservoirs, leading to increased replication within host cells (Swanson and Isberg, 1995; Machner and Isberg, 2006).
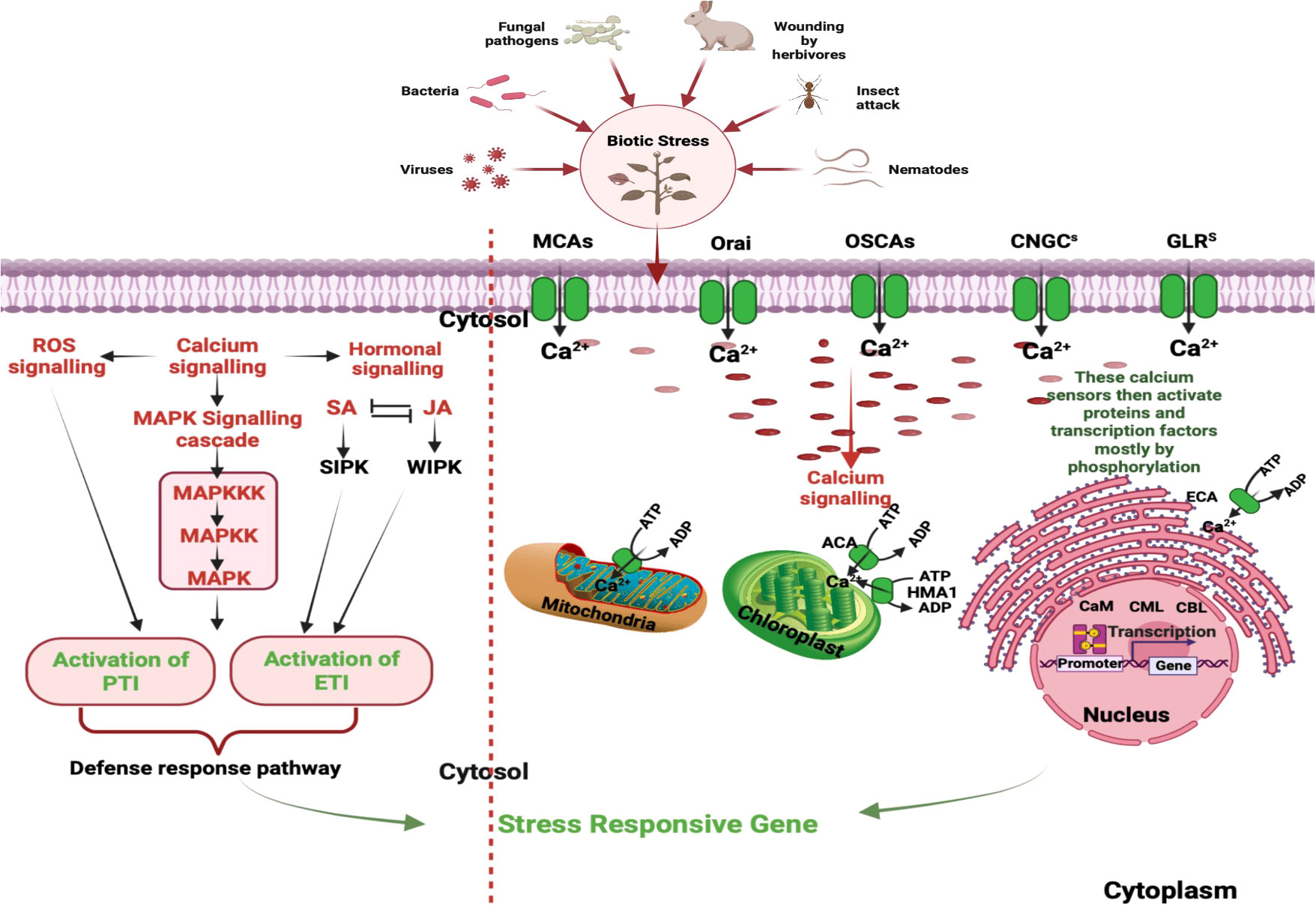
Figure 1 Stages in Ca2+ Signaling Pathways During Plant Interactions with Biotic stressors: This figure underscores the intricate network of calcium signaling components within plant cells. Calcium influx is mediated by diverse channels, including CNGCs, GLRs, TPCs, MCAs, and OSCAs. Meanwhile, calcium efflux is facilitated through systems like ACAs, ECAs, HMA1, MCUC, and CAX. Decoding is executed by specific protein families, encompassing CDPKs), calcineurin B-like protein kinases (CIPKs), calmodulin (CaM), and CaM-like proteins (CMLs). These processes play a pivotal role in perceiving and responding to a myriad of signals during plant interactions with both pathogenic and mutualistic organisms. Initial steps involve distinct cytosolic calcium spikes, referred to as calcium signatures, which encode the primary layer of specificity. Subsequent decoding of these calcium transients contributes to the second layer of specificity, ultimately culminating in the activation of target proteins within the defense cascade. Additionally, phytohormones such SA and JA, along with induced protein kinases and reactive oxygen species (ROS), contribute to the signaling cascade, culminating in intricate and coordinated plant defense reactions. [CNGCs: Cyclic Nucleotide-Gated Channels; GLRs: Ionotropic Glutamate Receptors; TPCs: Two-Pore Channel 1; MCAs: Mechanosensitive Protein Channels; CDPKs, Calcium-Dependent Protein Kinases; CBLs, Calcineurin B-Like Proteins; ACAs, Autoinhibited Ca2+-ATPases; ECAs, ER-Type Ca2+-ATPases; HMA1, P1-ATPase Heavy Metal Transporter 1; MCUC, Mitochondrial Calcium Uniporter Complex; CAX, Ca2+-Exchangers; SA, Salicylic Acid; JA, Jasmonic Acid; ROS, Reactive Oxygen Species.]
Furthermore, the calcium signaling pathway is intricately integrated with other signaling modules, hormonal signaling, and the production of reactive oxygen species (ROS) in plants. The CDPKs target different defense-responsive proteins and bring about resistance response (Singh et al., 2022). Additionally, calcium transport and storage in plants involve calcium-binding proteins, and transporters/pumps, that help chelate and buffer cytosolic Ca2+. Calcium flow occurs through the cell wall, vacuoles, apoplast and different cellular organelles (Mohanta et al., 2018). Figure 1, left panel represents a schematic diagram of calcium signaling components within plant cells.
During stress, changes in the water potential or ion concentration in the plant’s environment can activate signaling pathways that result in the influx of extracellular Ca2+ into the cytoplasm of plant cells (McAinsh and Pittman, 2009). This influx of calcium ions can trigger downstream signaling events that help the plant to adapt to the stressful conditions. For example, calcium influx can activate various CDPKs and CBPs, which can regulate gene expression and various cellular processes such as stomatal closure, ion transport, and osmotic adjustment (Kudla et al., 2010). The transport of Ca2+ across membranes is an essential aspect of calcium signaling in plants. Several classes of membrane transporters have been identified that play critical roles in regulating the intracellular calcium concentration [Ca2+]i. These include Ca2+-ATPases, Ca2+/H+ antiporters, Ca2+ channels, and Ca2+ permeable ion channels. Ca2+-ATPases are involved in active transport of Ca2+ out of the cytosol, contributing to the maintenance of low [Ca2+]i levels (Kudla et al., 2018). The Ca2+/H+ antiporters mediate the exchange of cytosolic Ca2+ and protons across the membranes, thus regulating the pH and Ca2+ homeostasis in different cellular compartments (White and Broadley, 2003). The Ca2+ channels facilitate the influx of extracellular Ca2+ into the cytosol and are activated by various stimuli, including voltage, mechanical stress, and ligands (Kudla et al., 2018). Calcium transporters have been investigated for their plausible role in plant responses to pathogenic stress and environmental cues. For instance, in Arabidopsis, the overexpression of a vacuolar Ca2+ATPase led to an increase in calcium accumulation in vacuoles and thereby increasing the tolerance to NaCl induced salt stress (Yamaguchi et al., 2013). In another study, the silencing of a plasma membrane Ca2+-ATPase reduced calcium transport and increased sensitivity to oxidative stress in Nicotiana tabacum (Crouzet et al., 2013). The compiled data on calcium transporters and their associated protein sensors have been meticulously presented in Table 1.
One of the most well-known calcium sensors in plants is the calmodulin (CaM) protein. CaM binds to calcium ions in a calcium-dependent manner, leading to a conformational change that allows it to interact with downstream effectors. CDPKs are a group of calcium-dependent protein kinases involved in several physiological processes, including stress response, development and hormone signaling (Cheng et al., 2002). CBL-interacting protein kinases (CIPKs) are another group of calcium sensors that interact with calcium-binding proteins called CBLs to regulate downstream signaling events (Kudla et al., 2010). The binding of CBLs to CIPKs is calcium-dependent, and the activation of CIPKs is induced by this interaction. Downstream targets, such as ion channels, transporters, transcription factors and enzymes, are phosphorylated by CIPKs as a consequence (Evans et al., 2016).
Factors like greenhouse conditions, extreme temperatures, drought stress, and chelation can hinder calcium uptake, leading to symptoms like black spots, bushy morphology, and stunted growth (Bose et al., 2011). Therefore, a critical study of the process of calcium transport, storage, and signaling is crucial for developing stress tolerant-high yielding crops. In this review, we aim to provide a comprehensive overview of calcium signaling molecules involved in different kind of plant-microbe interactions. We explore the emerging perspectives and discuss the future directions and challenges for harnessing calcium signatures to generate climate smart crops.
Calcium signaling in plant responses to microbial pathogens
Calcium signaling is an essential factor of plant-microbe interactions, especially in response to microbial pathogens. When pathogens interact with host plants, they may trigger an influx of calcium ions (Ca2+) into host cells, which activates various defense responses. Calcium signaling pathways can also play a role in mediating communication and coordination between microbes, as quorum sensing systems can involve calcium signaling (DeFalco et al., 2023).
When plants encounter pathogens, an early signaling event is an increase in cytosolic Ca2+ concentration. The immune system of plant comprises two distinct pathways; Pathogen-Associated Molecular Patterns (PAMP)-Triggered Immunity (PTI) and Effector-Triggered Immunity (ETI). These pathways have different Ca2+ signatures. The PAMPs are recognized by Pattern Recognition Receptors (PRRs) triggering PTI, which induces a rapid and brief influx of Ca2+ from extracellular sources. This event activates downstream signaling pathways resulting in the expression of defense-related genes, ROS production and callose deposition. ETI is initiated when plants perceive specific pathogen effectors that disrupt host defenses. This recognition results in sustained elevations of Ca2+ and the activation of downstream signaling pathways, transcription factor activation, mitogen activated protein kinase cascade and hypersensitive response (Tian et al., 2020).
Plant-microbe interactions are initiated through the recognition of microbial-associated molecular patterns (MAMPs) or damage associated molecular patterns by plant pattern recognition receptors (PRRs) (Zipfel, 2014). Notably, the recognition of bacterial flagellin or flg22 minimal epitope involves the flagellin-sensitive 2 (FLS2) pattern recognition receptor, whereas LysM-receptor kinase 5 (LYK5) receptor complex and chitin elicitor receptor kinase 1 (CERK1) are crucial for sensing fungal chitin (Sun et al., 2013; Cao et al., 2014). Furthermore, the plant endogenous peptide 1 receptor (PEPR1) and PEPR2 function as Damage Associated DAMP receptors, perceiving host-derived DAMPs that are released during pathogen or insect attacks (Krol et al., 2010). Rhizobia and mycorrhizal fungi communicate their presence to host plant roots using specialized small molecules known as Nod factors and Myc factors, respectively, which are recognized by specific PRRs (Yuan et al., 2017).
Upon the detection of microbial-associated molecular patterns or damage associated molecular patterns by PRRs, there is a swift and temporary surge in cytosolic Ca2+ concentration. PRRs can directly activate calcium-permeable channels. For example, upon activation, FLS2 interacts with Arabidopsis-autoinhibted Ca2+ ATPase 8 (ACA8) and ATPase10, which acts as Ca2+ pumps situated in plasma membrane (Ma et al., 2013). Similarly, during fungal-plant interactions, CERK1 forms an association with Annexin1, which acts as a calcium-permeable channel (Wang et al., 2016). Notably, the presence of 8-mer chitin induces the accumulation of Annexin 1 (ANN1) protein, suggesting a potential supplementary role in chitin-triggered innate defense signaling. Additionally, PRRs- induces calcium influx may indirectly activate other signaling molecules, such as reactive oxygen species (ROS) and cyclic nucleotides, which in turn contribute to the overall defense response (Yuan et al., 2017) (Table 2; Figure 2). Moreover, calcium signaling pathways in plants can also be influenced by factors like abscisic acid (ABA), which induces stomatal closure during pathogen attacks and abiotic stress, thereby limiting pathogen entry and spread. Understanding the intricate role of calcium signaling in plant responses to microbial pathogens holds significant implications for developing strategies to enhance plant resistance and improve crop protection (Upadhyay, 2022).
Table 2 The involvement of various calcium signaling molecules in response to biotic stress, their role in stress response, and downstream effectors in plant-microbe interactions.
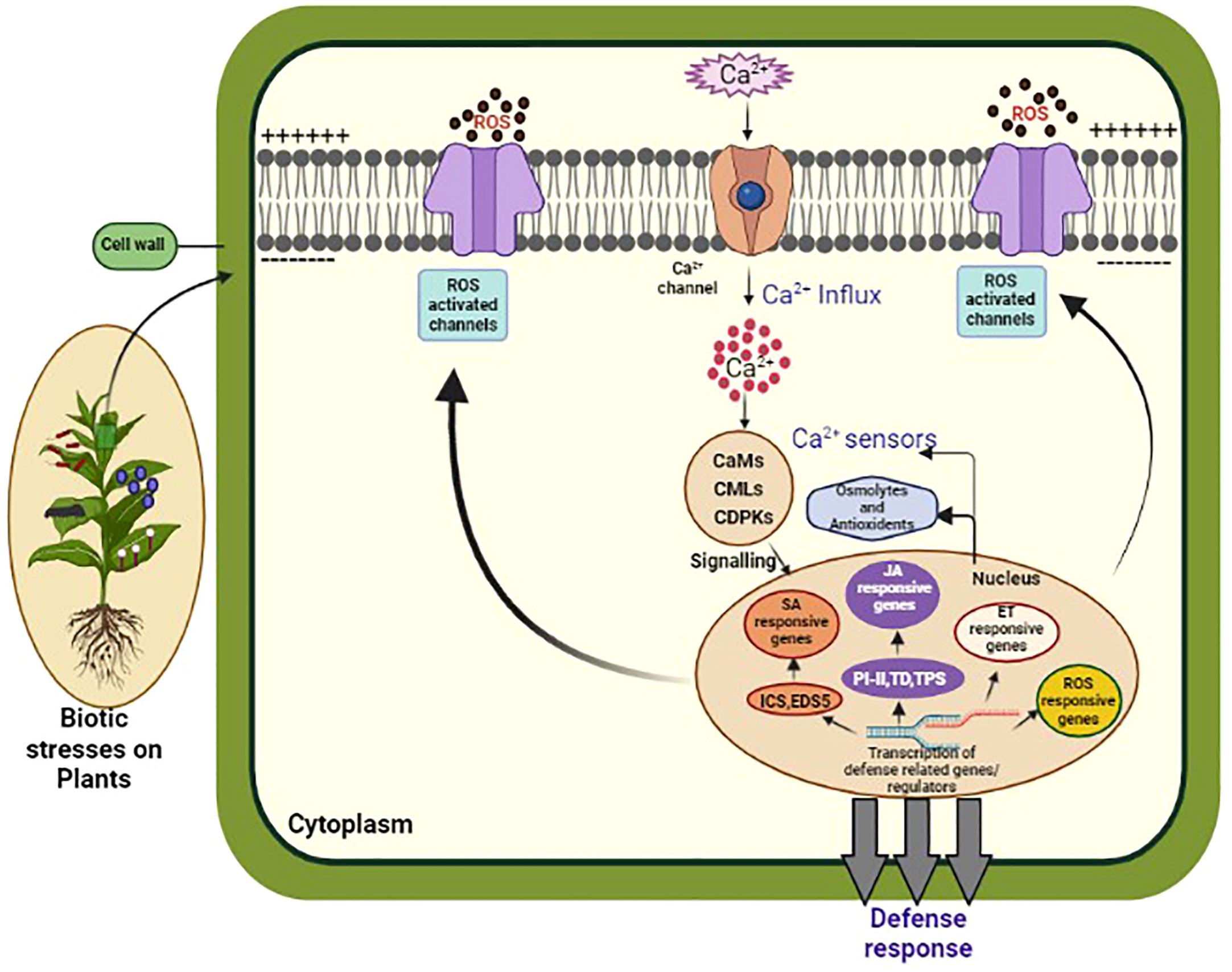
Figure 2 This diagram illustrates the Ca2+ signaling pathway in plants. Calcium ions (Ca2+) are important signaling molecules involved in various plant processes, including biotic and abiotic stress responses, growth, and development. In response to a stimulus, Ca2+ channels on the plasma membrane open, allowing Ca2+ to enter the cytosol. This leads to the activation of Ca2+ binding proteins, such as calmodulins (CaMs), and Ca2+ dependent protein kinases (CDPKs), which in turn activate downstream signaling pathways. These pathways can trigger gene expression changes, ion channel regulation, and activation of enzymes involved in secondary messenger production. The Ca2+ signaling pathway is tightly regulated by Ca2+ ATPases, which remove Ca2+ from the cytosol, and Ca2+ binding proteins, which buffer the Ca2+ concentration to prevent excessive Ca2+ accumulation and toxicity. Understanding the intricacies of Ca2+ signaling in plants is crucial for developing strategies to enhance plant growth, improve stress tolerance, and combat diseases.
Calcium channels
Several channel proteins facilitate the exchange of calcium ions in plants, including cyclic nucleotide-gated channels (CNGCs), two pore channels 1 (TPC1), glutamate receptors (GLRs) and annexins. In Arabidopsis, for example, around 150 cation transporters exist, and 20 of these are CNGC-class calcium transporters that sense intracellular levels of cyclic nucleotide monophosphates (cNMPs), such as cAMP and cGMP, to regulate Ca2+ levels and transduce different signaling events (Xu et al., 2022).
Several CNGC channels in Arabidopsis have been identified to play significant roles in the immune response against pathogens. CNGC2 and CNGC4 were found to trigger ROS generation and ETI response upon recognition of flg22, a bacterial PAMP (Eichstadt et al., 2021). Additionally, CNGC2/defense, no death1 (DND1) regulates intracellular nitric oxide levels and defense responses, controlling HR-mediated cell death (Zhao et al., 2021). CNGC11 and CNGC12 participate in caspase-dependent programmed cell death (PCD) in response to pathogenesis, while CNGC20 has a significant role in plant immune response by interacting with CNGC19 BOTRYTIS INDUCED KINASE 1 (BIK1) (Ren et al., 2021). Interestingly, the cpr22 mutant contains a chimeric CNGC channel (AtCNGC11/12) resulting from a deletion between AtCNGC11 and AtCNGC12, which leads to constitutive expression of PR genes22, and thus provides resistance to virulent Hyaloperonospora parasitica. The cpr22 mutant is semi-dominant, and homozygosity for the mutant allele is lethal (Clough et al., 2000; Moeder et al., 2011). Moreover, the Ca2+ dependent cell-death observed in the cpr22 mutant suggests the involvement of calcium ions in this process (Urquhart et al., 2007).
Plant GLR-type Ca2+ receptors are a common feature in plants and are structurally similar to ionotropic glutamate receptors found in animals. Arabidopsis contains 20 GLR-type Ca2+ transporters, with most of them performing developmental functions. While most studies on glutamate receptor-like proteins have focused on their role in ion homeostasis and neurotransmission in animals, recent research has highlighted their potential involvement in immune signaling and plant stress adaptations (Ghosh et al., 2022). Responsiveness to various abiotic stresses like cold, drought and salt, as well as biotic stress like infection with the fungal pathogen Phytophthora sojae has been demonstrated by GLRs in soybean (Zhang et al., 2021). An immune response has been observed in Arabidopsis, upon topical application of glutamate, as evidenced by the activation of various PTI-responsive gees, such as BAK1, BKK1. BIK1, PBL1 and CERK1, as well as chitin receptor LYSIN-MOTIF RECEPTOR-LIKE KINASE 5 (LYK5) and genes involved in the biosynthesis of plant hormone salicylic acid (SID2) (Bhar et al., 2023). The results suggest that GLRs have a previously unappreciated role in plant immune signaling and could open new avenues for research into the molecular mechanisms underlying plant stress responses.
Annexins are a conserved family of cytoplasmic proteins that interact with phospholipids in the cell membrane in a Ca2+-dependent or -independent manner. They play essential roles in regulating various cellular processes in plants, including responses to abiotic and biotic stresses as well as developmental processes (Gerke and Moss, 2002). The unique feature of annexins is their annexin core domain, a Ca2+ and membrane-binding module, which allows Ca2+ bound annexins to associate with membranes containing negatively charged phospholipids. This interaction between annexins and phospholipids is critical for their function in cellular processes.
In addition to the annexin core domain, each annexin contains a highly variable region known as the N-terminal interaction domain. This region serves as a binding site for cytoplasmic protein ligands that can be directed to membranes through the annexin-core-mediated phospholipid interaction. Due to their ability to interact with membranes and modulate calcium-dependent responses, annexins are versatile regulators involved in a wide range of cellular activities, contributing to the overall functioning and adaptation of plants in their environment. Mechanosensitive channels are a class of ion channels found in various organisms, including plants, animals, bacteria and fungi (Martinac, 1993). These channels are known for their ability to respond to mechanical forces and changes in membrane tension. One significant aspect of their function is their role in mediating calcium signaling in response to mechanical stimuli. When a cell experiences mechanical stress or changes in membrane tension, mechanosensitive channels are activated, allowing the influx of ions, including calcium (Ca2+), into the cell. This increase in intracellular calcium concentration serves as a crucial second messenger, initiating a cascade of signaling events that regulate various cellular processes. In plants, mechanosensitive channels have been extensively studied, particularly in Arabidopsis (Hamilton et al., 2015). They include mechanosensitive-like channels (MSLs), Mid1-complementing activity channels (MCAs), and two-pore potassium (TPK) families. The mechanosensitive-like channels (MSLs) located on mitochondria, chloroplasts, and the plasma membrane (PM) possess transmembrane domains that form a pore, allowing the influx of calcium ions upon mechanical stimuli. The MSLs lacking voltage sensor regions respond solely to mechanical cues, highlighting their specialized role in mechanotransduction. Mid1-complementing activity channels (MCAs) are formed by subunits containing a single transmembrane domain. These subunits come together to form a pore at the cell membrane. When activated by mechanical forces, MCAs facilitate calcium entry, contributing to calcium signaling (Shigematsu et al., 2014). The two-pore potassium (TPK) channels, as the name suggests, contain two K+ channel pore domains. They exhibit unique structural features and are involved in both potassium and calcium transport. TPKs can respond to mechanical stress and osmotic changes, modulating calcium signaling and ion fluxes in response to these stimuli. The activation of mechanosensitive channels and subsequent calcium signaling play vital roles in diverse physiological processes. For instance, in plants, they are involved in touch and gravity sensing, osmoregulation, and responses to environmental cues such as wind or touch (Becker et al., 2004).
OSCAs are a family of conserved channels found in eukaryotes. These channels have nine transmembranous domains, and a pore domain is located between the eighth and ninth domains. In Arabidopsis, there are 15 OSCA genes encoded in the genome. Two of these genes, OSCA1.1 and OSCA1.2, have been identified as being involved in the transport of calcium ions (Yuan et al., 2014). On the other hand, Piezo channels are composed of 2000 to 4000 amino acid residues and are predicted to have 20 to 40 transmembranous domains (Hamilton et al., 2015). Both OSCAs and Piezo channels are mechanosensitive, meaning they respond to mechanical forces.
From calcium sensors to signaling pathways: deciphering the intricate language of calcium signatures
Conformational changes induced by calcium play a crucial role in relying of Ca2+ signals to downstream signaling components, such as CBPs, in response to biotic and abiotic stress involving different kind of calcium receptors. On the other hand, CBLs relay Ca2+ signals to CIPKs, which regulate various physiological processes in plants. While CaM/CMLs and CBLs are crucial for Ca2+ signaling, within the same molecule, CPKs contain both a kinase domain and a CaM like calcium sensor domain. Therefore, they can directly decode Ca2+ signaling into phosphorylation events, providing a more efficient and rapid response to Ca2+ signals (Seybold et al., 2014; Verma et al., 2022).
Calmodulin (CaM) and CaM-like proteins (CML)
Plants have various forms of Calmodulins, also termed CaM-like proteins (CMLs), contain EF-hand motifs, which are common calcium-binding structural domains found in calcium sensor proteins. In CMLs, the EF-hand motifs are typically present in pairs, and each motif consists of approximately 30 amino acids, forming a helix-loop-helix structure. The functional significance of these EF-hand motifs in CMLs lies in their calcium-binding properties and their role as calcium sensors in plant cells (Mohanta et al., 2015; Kaur and Upadhyay, 2022). The Calmodulin and CaM-like protein family members may have functional redundancy, individual CaM/CML gene expression deregulation or loss of CML function in mutated plants which can impact pathogenic defense responses. The Calmodulin and CaM-like protein family members play a crucial role in mitigating different abiotic stresses too (Raina et al., 2021b). Gain-and loss-of-function genetic approaches have further strengthened the role of CMLs in plant immunity. Several studies have explored the calcium sensors like CaMs and CMLs for their role in plant defense responses. The overexpression of soybean CMLs in tobacco plants has shown improved resistance against various pathogens, including bacteria, viruses, and fungi. The overexpression of CMLs in Transgenic tobacco plants with increased expression of GmCaM4 or GmCaM5 demonstrate spontaneous necrotic lesions and constitutive expression of systemic acquired resistance (SAR)-associated genes, which occurs independently of salicylic acid (SA) production (Heo et al., 1999). Additionally, these genetically modified plants display heightened resistance against a diverse range of pathogens, including Phytophthora parasitica var. nicotianae, Pseudomonas syringae pv. tabaci, and Tobacco mosaic virus. These findings indicate that specific CaM isoforms are involved in an SA-independent signaling pathway that triggers disease resistance in the transgenic tobacco plants.
Plants possess a diverse range of CaM-like proteins (CMLs) that are distinct from typical CaMs. In Arabidopsis thaliana alone, there are 50 members of the CML family, each of which may have a specific role in plant physiology due to their varying expression patterns in response to developmental stages, tissues, and environmental stimuli. Recent research has highlighted the critical role of CMLs in plant immunity, where they act as essential Ca2+ sensors involved in defense mechanisms against microbial pathogens and herbivores. Numerous studies have demonstrated the importance of individual CaM/CML gene expression in plant defense responses to various pathogens. Gain-of-function experiments have demonstrated that overexpression of specific CMLs, such as soybean CMLs, can boost plant tolerance to a broad range of pathogens. For example, overexpressing soybean CMLs in tobacco results in increased resistance to various pathogens, while overexpression of a typical CaM does not have the same effect, suggesting that CMLs are selectively activated in response to pathogen invasion.
The reduction of hypersensitive response was demonstrated in tomato when APR134 was silenced (Chiasson et al., 2005). Conversely, the stimulation of HR was observed to an avirulent strain of P. syringae by overexpressing the APR134 ortholog from Arabidopsis CML43. Studies have shown crucial roles in regulating plant defense against various pathogens in Arabidopsis are played by calcium binding proteins such as CML8, CML9, CML24 and CML41. The hypersensitive response is impaired and nitric oxide production is reduced upon recognition pf pathogen-associated molecular patterns when CML24 is knocked out. Plant defense against different strains of Pseudomonas syringae is positively regulated by CML9 and CML8 and CML8 functions mainly through salicylic acid (SA)-dependent pathways. Furthermore, defense against P. syringae is positively regulated by CML41 through the facilitation of plasmodesmata closure in response to bacterial flagellin (Leba et al., 2012; Zhu et al., 2017). Resistance to Spodoptera littoralis is enhanced by CML42, whereas defense against the same insect herbivory is positively influenced by CML37, according to Scholz et al. (2014). These calcium-binding proteins act as calcium sensors, enabling changes in calcium levels to be perceived by plants and translated into various signaling pathways that activate defense responses against pathogens. Overexpression of SCaM-5 in Arabidopsis also enhances resistance to Pseudomonas syringae infection, while overexpression of a typical CaM (SCAM-1) does not (Bouche et al., 2002; Park et al., 2004). SCaM-4 is a calcium-binding protein, and its overexpression triggers molecular and cellular responses that strengthen the plant’s defense mechanisms. This enhanced resistance could potentially offer a valuable strategy for improving soybean crop protection against these harmful fungal diseases (Aldon et al., 2018). Additionally, overexpressing SCaM-4 in soybean stimulates resistance to fungal pathogens Alternaria tenuissima, Phomopsis longicolla and Phytophthora sojae (Rao et al., 2014).
Loss-of-function genetic studies have provided valuable insights into the roles of different CMLs in plant immunity. The reduction in expression of pathogen-induced CaM isoforms, specifically NtCaM1 and NtCaM13, had distinct effects on disease resistance in tobacco (Takabatake et al., 2007). Silencing NtCaM13 increased the plant’s vulnerability to viral, bacterial, and fungal pathogens, while knockdown of NtCaM1 did not. Conversely, when pepper CaM1 was transiently overexpressed, it stimulated the production of reactive oxygen species and NO, resulting in the appearance of HR-like lesions and the activation of defense-related genes in pepper leaves. This led to local resistance against bacterial pathogens. Zhu et al. (2017) demonstrated that silencing of APR134 in tomato suppresses the hypersensitive response (HR) in tomato plants. They also observed that the overexpression of Arabidopsis CML43 ortholog of APR134 stimulates the HR in response to an avirulent strain of P. syringae. Similarly, knockout of CML24 in Arabidopsis reduces nitric oxide production and impairs the HR response after PAMP recognition (Dubreuil-Maurizi et al., 2011). CML8 and CML9 act as positive regulators of plant defense against different strains of P. syringae, with CML9 also contributing to plant immune responses despite its initial identification as a gene involved in plant responses to abiotic stress (Delk et al., 2005; Luan et al., 2009; Reddy et al., 2011). Recent research by Boudsocq and Sheen (2013) has identified plasmodesmal-localized CML41 as a positive regulator of defense against P. syringae. The Ca2+ signaling is also involved in plant responses to herbivores. Studies have shown that Arabidopsis CML42 knockout harbor increased resistance against Spodoptera littoralis, with an upregulation of JA-responsive genes and accumulation of glucosinolates. Furthermore, Zhang et al. (2022a) reported that silencing SlCML55 in tomatoes (Solanum lycopersicum) leads to higher tolerance against the oomycete pathogen Phytophthora capsici, as this CML was found to exert negative control on the activation of PR genes.
Calcium-dependent protein kinases (CDPKs)
CDPKs play a crucial role in plant defense signaling against various biotic and abiotic stresses. Unlike other Ca2+ decoders, CDPKs integrate both Ca2+ sensing and downstream signal propagation capabilities into a single module. In angiosperms, the number of CDPK members is around 30, and they are organized into four distinct groups based on their family architecture. CDPKs or calcium dependent protein kinase are diverse group of serine/threonine protein kinase found in plants. They play crucial roles in regulating various aspects of plant growth development and responses to abiotic and biotic stresses. CDPKs consists of three main structural components: a variable N-terminal, a central kinase domain, and an activation domain. It typically consists of an auto-inhibitory pseudo-substrate linked to a CaM like domain containing four EF- hands and acts as calcium sensor. When calcium ions bind to the EF-hands, a conformation change occurs, relieving the auto-inhibition and enabling kinase activation. This allows the kinase to become active and initiate downstream phosphorylation events, which can lead to various cellular responses such as gene expression, ion fluxes, and protein degradation (Hu et al., 2016; Kaur and Upadhyay, 2022). The role of calcium-dependent protein kinases (CDPKs) in potatoes and their involvement in various biological processes, including hormone signaling, plant growth, and responses to abiotic and biotic stresses have been demonstrated. Fantino et al. (2017) studied StCDPK7 and found high transcript levels in swollen stolons, roots, and mini tubers, with induced expression upon Phytophthora infestans infection in systemic leaves. Surprisingly, StCDPK7 displayed cytosolic/nuclear localization despite a predicted chloroplast transit peptide. The recombinant protein, StCDPK7:6xHis, exhibited Ca2+-dependent kinase activity and could phosphorylate phenylalanine ammonia lyase, an enzyme involved in plant defense response. Another study Mori et al. (2006) provides compelling evidence for the involvement of CDPK5 (also known as CPK6) in tomato (Solanum lycopersicum) in regulating guard cell S-type anion- and Ca2+-permeable channels, which are crucial for stomatal closure. The research demonstrates that CDPK5/CPK6 plays a pivotal role in ABA-mediated signaling pathways, leading to the activation of downstream defense responses in guard cells. This example sheds light on the specific functions of CDPKs in tomato and highlights their significant contribution to enhancing plant defense against pathogens.
The plant immune system is impacted by the presence of CDPKs, as defense gene expression upon bacterial flagellin perception is positively regulated by them. PTI-induced resistance against Pseudomonas syringae is collectively contributed by CDPK4, CDPK5, CDPK6 and CDPK11, while AtRBOHD, the primary reactive oxygen species producing enzymes involved in immunity, can be phosphorylated by CDPK5, CDPK6, CDPK11 and CDPK4 (Boudsocq et al., 2010). The phosphorylation of the HsfB2a transcription factor regulates the induction of the PDF1.2 defense gene after caterpillar Sodoptera littoralis-induced wounding, and this is facilitated by CDPK3 and CDPK13 (Kanchiswamy et al., 2010). Disease resistance against Magnaporthe oryzae in rice is enhanced through the overexpression of full-lenth OsCDPK4, resulting in elevated basal levels of salicylic acid and augmented defense gene induction (Bundo and Coca, 2016).
Plant cell death in response to pathogens is controlled by CDPKs as well. In Nicotiana sp., the programmed cell death response triggered by the perception of the Cladosporium fulvum race-specific Avr4 or Avr9 elicitors requires CDPK2 and CDPK3. In Arabidopsis, the onset of hypersensitive response upon challenge with avirulent P. syringae and subsequent NLR-mediated effector recognition is controlled by CDPK1 and CDPK2 (Romeis et al., 2001). Several constitutively active CDPKs harbor cell death-inducing activity. Cell death-inducing activity is harbored by several constitutively active CDPKs. The activation domains of auto-active CDPKs are absent, and their active state does not rely on calcium ion inputs. Cell death and kinase activity are both required when CDPK5 is expressed in Arabidopsis leaf protoplasts. The cell death-inducing activity of barley CDPK4 or Arabidopsis CDPK5, when transiently expressed, has been observed in tobacco leaves. It appears that the activation of a specific CDPKs alone is not sufficient to trigger PCD, as not all auto-active configurations of tested CDPKs induce PCD (Dubiella et al., 2013).
Calcineurin B-like proteins (CBLs) and CBL-interacting protein kinases (CIPKs)
Calcineurin B-like proteins (CBLs) and CBL-interacting protein kinases (CIPKs) have emerged as a new class of plant calcium sensors that play a crucial role in response to both biotic and abiotic stresses (Thakur and Negi, 2021). Arabidopsis has 10 CBLs and 25 CIPKs, while maize has 12 CBL genes that have been linked to abiotic stress tolerance. However, recent genome-wide analysis of Lagerstroemia indica (crape myrtle) revealed 37 CIPKs, indicating their abundance in plants (Yu et al., 2022).
The role of CBL-CIPKs in biotic stress response is currently receiving increased attention, although their functions in abiotic stress tolerance are well established (Ma et al., 2020; Plasencia et al., 2021; Xiaolin et al., 2022). In rice, the upregulation of OsCIPK14 and OsCIPK15 were observed in response to PAMP treatment, resulting in resistance through the activation of ROS-mediated HR and cell dealth (Kurusu et al., 2010). Conversely, a recent study in wheat demonstrated that CIPK14 negatively regulates resistance against rust fungi, specifically Puccinia striiformis f. sp. tritici (Pst) (He et al., 2023).
In addition to biotic stress response, CIPKs have also been linked to nitrogen uptake and root development. Recent research has revealed the involvement of CmCIPK23, a CIPK from Chrysanthemum, in the regulation of CmTGA1 and activation of nitrogen uptake during root development (Liu et al., 2022). Interestingly, TGA transcription factors, which are essential for NPR1-dependent PR1 activation, may also be linked to pathogenesis, suggesting a potential role for CBLs and CIPKs in plant defense mechanisms that require further exploration. Studies have identified the role of CBLs and CIPKs in stress responses in other plant species as well. In pepper (Capsicum annum L.), nine CaCBLs and twenty-six CaCIPKs were induced under stress conditions. CIPK1 specifically participates in biotic stress responses in C. annum, as shown by the increased sensitivity of cacipk1 mutant lines to the fungal pathogen Phytophthora capsici. Conversely, CaCIPK1-OE lines exhibit heightened defense activity, accompanied by H2O2 accumulation and cell death (Ma et al., 2019). In wheat (Triticum aestivum), TaCBL4 has been shown to be involved in defense responses against Puccinia striiformis f. sp. tritici infection. The upregulation of TaCBL4 following infection and increased susceptibility in the loss-of-TaCBL4 function mutants suggest its role in the defense mechanism. TaCBL4 interacts with TaCIPK5, indicating their joint participation in positive defense mechanisms (Liu et al., 2018). The enhanced tolerance to Pst in wheat can be achieved by overexpressing TaCIPK10, which triggers hypersensitive cell death and accumulation of reactive oxygen species (ROS). TaNH2, a counterpart of Arabidopsis NPR3/4, has been shown to play a crucial role in salicylic acid (SA) signaling and collaborate with TaCIPK10 in the defense response by interacting and phosphorylating it (Liu et al., 2019). In tomato, SlCBL10 interacts with SlCIPK6 to regulate its kinase activity. Overexpressing SlCIPK6 in Nicotiana benthamiana induces ROS production by regulating NbRBOH and the kinase activity of SlCIPK6. The complex of SlCBL10-SlCIPK6 interacts with the downstream protein RBOH, which leads to an increase in ROS, thereby contributing to plant immunity (de la Torre et al., 2013). These findings emphasize the essential role of Ca2+ signaling components in stress responses and suggest their potential application in the development of stress-tolerant crops.
Modulation of calcium signaling by microbial effectors
The Ca2+ ion concentration inside the cell system changes, as soon a microbe is sensed in the external milieu. An elevation in Ca2+ ions has been observed during symbiotic or pathogenic interaction of microbes with plants. The Ca2+ influx and the oxidative burst are the plant’s first and foremost immunologic response to pathogenic elicitors. Increased Ca2+ ions have been known to be associated with the gene expression for suitable defense mechanism (Gilroy et al., 1990). The involvement of calcium signaling during plant-microbe interactions to bring about pathogenic or symbiotic outcome is compiled in Table 3. The effectors are protein molecules of microbial origin, which upon suitable molecular interactions, bring about either a pathogenic or a defense response. These microbial effectors are responsible for debilitating the host plant to bring about an effective pathogenesis. These effectors also aid in host invasion and nutrient acquisition for the pathogen.
The bacterial cells have been known to influence their positive or negative interaction by virtue of Ca2+ modulation. Grant et al. (2000) measured the cytosolic Ca2+ concentration in leaves of Arabidopsis plants, infected with Pseudomonas syringae using aequorin-mediated bioluminescence. The Ca2+ signal peaked after ~10 min and a second, Ca2+ signal was observed after 1.5–2 h, which was much stronger and robust. They studied avrRpm/RPM1 gene to gene interaction during early stages of hypersensitive response (HR) and observed a constitutively enhanced Ca2+ in cytosol. Since the Ca2+ signals are very localized and are influenced by other cellular and sub-cellular factors, they pioneered to use a whole plant system to explore calcium connections during plant-pathogen interaction. Botrytis–induced kinase 1 (BIK1) is transcribed in plants upon infection by Botrytis and defends plants from biotrophic and necrotrophic pathogens by regulating salicylic acid (SA) synthesis in plant cell (Veronese et al., 2006). It plays a role in regulation of Ca2+ influxes, triggered by membrane protein Flagellin 22 (flg22). It is also known to increase ROS generation by modulating the calcium concentration owing to site specific phosphorylation of NADPH oxidase RbohD (Li et al., 2014). Thor and Peiter (2014) used fluorescent genetically encoded Ca2+ indicators (GECIs) to investigate the oscillatory nature of flg22‐induced cytosolic Ca2+ signals at single cell level in the guard cells of the leaves of Arabidopsis thaliana. They observed that the flg22 could elicit oscillations in single cell and accordingly, all the oscillations occurring in all the cells contribute to the total oscillation of the whole plant. The pathogenic and mutualistic fungi too establish their relation with plants by altering Ca2+ efflux or influx. The cytoplasmic free calcium ([Ca2+]cyt) transiently increased to about 1 μM in transgenic Parsley expressing apoaequorin, upon treatment with Pep-13, an oligopeptide elicitor obtained from Phytophthora sojae. This peaked concentration of ([Ca2+]cyt), later returned to normal level of about 300 nM. This biphasic variation in calcium concentration was observed to be associated with oxidative burst and phytoalexin production (Blume et al., 2000). The transgenic cell suspension of Nicotiana plumbaginifolia expressing apoaequorin proteins have been studied for changes in ([Ca2+]cyt), when treated with cryptogein and oligogalacturonides, the elicitors of defense mechanism in plants. The elicitor treated cells showed Ca2+ influx and released calcium from internal stores (Lecourieux et al., 2002). The endophyte, Piriformospora indica is known to mitigate biotic and abiotic stresses, when co-cultured with plant roots. The subcellular levels of Ca2+ increased in roots of germinating seedlings of Arabidopsis, when treated with cell wall extract of P. indica, thereby establishing calcium connection between successful mutualism and stress mitigation (Vadassery et al., 2009). The PRRs and downstream signaling components such as receptor-like cytoplasmic kinases (RLCKs) are required for PTI (Ranf et al., 2014). The Ca2+-dependent protein kinase CPK28 degrades the excess BIK1 protein to maintain its optimal concentration. The cpk28 knockout mutants accumulate an increased level of BIK1 protein while CPK28 over-expressing Arabidopsis plants have below optimal levels of BIK1 protein and are immune compromised. Therefore, Monaghan et al. (2015) concluded that CPK28 is a negative regulator of BIK1 and has a role in PAMP-induced Ca2+ burst.
The role of endophytic microbes in calcium signaling
The endophytic microbes, usually fungal or bacterial, inhabit the internal tissues of plants but without causing disease. The endophytes reside in the seeds and along with the process of germination, they promote healthy growth of the plant. The endophytes are present in nearly all the plants. In some cases, they may have originated from soil, which upon successful interaction in the rhizosphere, colonize the plants internally to the benefit of the host. Likewise, the endophytes become an integral component of plants. These endophytes are beneficial to plants in multiparous ways like they enhance nutrients uptake by plant roots; strengthen the defense mechanism of plants; alleviate stress induced damages; modulate plant development and keep a check on the weed growth. Wais et al. (2000) injected oregon-dextran dye in the cells of root hairs by iontophoresis to demonstrate Ca2+ spiking in plant mutants lacking early symbiotic responses to bacteria. All mutant lines failed to form nodules. Five complementation groups were studied, and two mutants dmi1 and dmi2 were almost blocked in calcium spiking. Therefore, they concluded that DMI1 and DMI2 were involved upstream of the Ca2+ spikes during rhizobia symbiosis. Two years later, Wais et al. (2002) studied spatiotemporal Ca2+ spiking for analysis of the regulation of nod gene expression during S. meliloti-M. truncatula interaction. They differentiated the strains on the basis of their ability to produce Nod factor, which is attributed to the Ca2+ oscillations induced by them. Thus, two strains of S. meliloti (Rm1021 and Rm, 2011) had different kinetics of Ca2+ spiking. The strain Rm1021 elicited a robust Ca2+ spiking after an initial lag phase of 10–15 min, while no Ca2+ spiking was detected in the strain Rm2011. They also observed in their study that the calcium spiking triggered by bacteria is no different than the calcium spiking observed in response to purified nodulation factors. Therefore, it was concluded that the calcium spiking is a nod gene-dependent host response. The availability of genetically encoded Ca2+ probes facilitated the study of Ca2+ concentrations at the cell and organ level during symbiotic interaction of plants with microbes. Kosuta et al. (2008) compared and discriminated two types of symbiosis by virtue of Ca2+ oscillations induced by them. The two symbiotic pathways required both DMI1 and DMI2 for functional symbiosis and for Ca2+ oscillations. The results also indicate that Ca2+ oscillations are an essential and necessary process for rhizobial or mycorrhizal colonization of plant roots. It is notable that Ca2+ oscillations induced by symbiotic fungus differ in its amplitude and periodicity from the Ca2+ oscillations induced by Nod-factors. Sieberer et al. (2009) carried their research on Medicago plants to evaluate Ca2+ responses in nucleus of mutant lines defective for NFP, DMI1, and DMI2. These three genes function upstream of Ca2+ spiking in Nodulation factor (NF) mediated signal transduction. The defective mutant of DMI3, which encodes for CCaMK was also studied. When treated with NF elicitors, no change in Ca2+ concentration in nucleus of nfp, dmi1, and dmi2 mutants was observed. However, the dmi3 mutant showed an immediate surge in nuclear calcium spiking. Using a nucleoplasmin-tagged cameleon (NupYC2.1) coupled with mathematical modeling and time lapse imaging, they concluded that the initial spiking in Ca2+ originates in the vicinity of both sides of the nuclear envelope.
Calcium signaling in beneficial plant-microbe interactions
The beneficial symbiotic and mutualistic associations of plants and microbes are also governed by Ca2+ signals. In rhizobia-legume symbiosis, Ca2+ signals are necessary for recognition and infection process for root nodule formation (Oldroyd, 2013). The infection process starts with the secretion of bacterial nodulation factors (NFs), which are recognized by plant receptors, resulting in a transitionary surge in cellular Ca2+ in plant cells (Gage, 2004). The Ca2+ signal transcriptionally activates the genes involved in the formation and differentiation of the nodules (Mitra et al., 2004).
Garcia et al., 2016 concluded that Ca2+ signaling is required for successful establishment of arbuscular mycorrhizal (AM) symbiosis as they investigated the mutualistic associations of plant roots and fungi of the Glomeromycota phylum. The hyphae enter the root cells and forms arbuscules, wherein nutrient exchange between the plant and the fungus takes place. The Ca2+ signals regulate this nutrient exchange and is involved in the establishment of the plant-fungus interface.
In mycorrhizal symbiosis, calcium signaling is essential for the recognition and coordination of the mutualistic association between plant roots and mycorrhizal fungi to establish a successful association. Flavonoid compounds released from plant roots signal compatible fungal species, while chitin oligosaccharides secreted by the fungal hyphae stimulate calcium signaling in plant cells. Calcium signaling activates downstream signaling pathways that lead to the formation of a symbiotic interface, where the exchange of nutrients between the plant and the fungus can occur. The nuclei in plants respond to rhizobacteria and AM fungi through the induction of a Ca2+ spiking response. This spiking in Ca2+ are recognized by the plant-specific CCaMK that contains a kinase domain and three Ca2+-binding EF hands. The activation of CCaMK is determined by free Ca2+ or activated calmodulin, while variations in the Ca2+ signatures determine whether the Ca2+ information is decoded into symbiosis or nodule formation. The autophosphorylation site of CCaMK is responsible for nodule formation, indicating that it acts as a regulatory switch.
During the establishment of a symbiotic relationship between plants and mycorrhizal fungi, transient changes in cytosolic calcium (Ca2+) occur, which indicate that host cells perceive the signaling molecules diffused by fungi. The Ca2+ signal is induced by these diffusible molecules and there appears a biphasic Ca2+ trace, which is characterized by a sudden surge in (Ca2+) cyt and a small transitionary increase thereafter, that dissipates in about 30 minutes. This Ca2+ response is essential for the formation of a polarized cytoplasmic assembly called the perpetration apparatus (PPA), which is required for successful colonization of the plant root by the fungi.
Rhizobial symbiosis is a type of plant-microbe interaction, where legumes establish a mutually beneficial relationship with soil bacteria for fixation of atmospheric nitrogen. Calcium signaling is crucial for this symbiosis as it regulates several key events such as the early stages of root hair deformation, the formation of infection pegs and threads, and the development of nodules. Studies have shown that rhizobia release Nod factors, which trigger the activation of host plant calcium channels, resulting in a rapid influx of Ca2+ into the cytosol. This surge in cytoplasmic calcium concentration ([Ca2+]cyt) leads to downstream events that promote root hair deformation and initiate infection thread formation (Kosuta et al., 2008). Furthermore, during the later stages of symbiosis, calcium signaling is also involved in nodule development. Research has demonstrated that the activity of the calcium-dependent protein kinase (CDPK) is necessary for the differentiation and proper functioning of nodule cells (Singh et al., 2014). In addition, the process of symbiotic nitrogen fixation is also driven by calcium signaling as it regulates the synthesis of key protein leghaemoglobin (Wang et al., 2016). Calcium-based fertilizers have shown great potential in enhancing symbiotic interactions in plants. The addition of calcium to soil has been reported to enhance nodulation and nitrogen fixation in leguminous crops (Santachiara et al., 2019). The topical application of calcium also enhances the growth of arbuscular mycorrhizal fungi (AMF) and improves the development of AMF structures in roots (Harrison, 2005). This is because calcium ions play a crucial role in the establishment of symbiotic interactions by regulating the signaling pathways involved in the recognition and response of plants to symbiotic microorganisms (Bapaume and Reinhardt, 2012). Studies have shown that calcium-based fertilizers can significantly increase the colonization of plant roots by AMF. For example, Bao et al. (2022) reported that the application of a calcium-based fertilizer improved the colonization rate of maize roots by AMF by up to 40%. Similarly, Berruti et al. (2016) observed that the enrichment of soil with calcium increased the spore density of AMF and improved the growth and yield of tomato plants. Furthermore, calcium-based fertilizers have known to enhance the nodulation and nitrogen fixation in legume crops. For instance, Nakei et al., 2022 demonstrated that the application of calcium nitrate increased the nodulation, nitrogenase activity, and yield of soybean plants. Similarly, Garg and Singh (2018) reported that the application of calcium improved the nodulation, nitrogenase activity, and yield of chickpea plants.
Novel techniques for studying calcium signaling in plants and microbes
The change in Ca2+ signals in the cytosol of plant cells could be measured using Ca2+ radioisotopes, Ca2+‐sensitive dyes, and other electrophysiological means (Atkinson et al., 1996; Zimmermann et al., 1997). However, recent advances in the study of calcium signaling in plant-microbe interactions offer a more detailed understanding of the complex molecular mechanisms involved. The promising tools and approaches that are currently being used are as follows:
Live-cell imaging and fluorescent probes
These are powerful tools for studying calcium signaling in real-time. By using fluorescent probes that specifically bind calcium ions, researchers can visualize changes in calcium concentration within live plant cells during interactions with microbes. This technique has been used to monitor calcium changes in plant cells in response to pathogens, hormonal cues, and environmental stressors. Several calcium indicators have been developed, including aequorin, cameleon, and GCaMP, which have different characteristics in terms of sensitivity, kinetics, and spectral properties.
Calcium imaging studies have helped to explore the calcium signaling in diverse plant and microbial systems, including Arabidopsis, rice, tobacco, and yeast. Aequorin, one of the earliest developed calcium indicators, which demonstrated that various stimuli can trigger Ca2+ signals in plant cells, including biotic stimuli such as yeast preparations, fungal cell wall components, and bacterial pathogens. Aequorin based calcium signaling has been studied in Arabidopsis and rice plants (Mithöfer et al., 1999). Ca2+ fluctuations have been linked to ROS-mediated defense signaling pathways, and sustained high Ca2+ concentration responses have been associated with phytoalexin production. Herbivore-associated molecular patterns (HAMPs) such as volicitin or linolenoyl-L-glutamine have been demonstrated to cause changes in cytosolic Ca2+ levels. Studies with lima bean leaves and soybean cell cultures have demonstrated that Ca2+ signaling is activated by Spodoptera littoralis larvae bites and certain components present in their regurgitate, like linolenoyl-L-Glutamine and volicitin (Maffei et al., 2004). These Ca2+ transients are associated with early membrane depolarization and/or hyperpolarization, which strengthens plant defense through systemin, ROS synthesis, and jasmonic acid signaling. Therefore, the regulatory function of Ca2+ signaling is highlighted in plant defense mechanisms, where different stimuli can elicit distinct Ca2+ variations, exhibited by their form, amplitude, frequency, duration, spatial localization, and the involvement of calcium ion pool, which are all dependent on the type of stimulus perceived by plant cell. Studies on guard cells further confirmed this concept, where the manipulation of Ca2+ oscillations can control the stomatal opening and closure for a long term.
The GECIs have been instrumental in studying the real‐time kinetics of Ca2+ surge in plant tissues, as a consequence of pathogenic infection or elicitor treatment. Aequorin (AEQ) is a luminescent calcium-sensitive protein, isolated from the bioluminescent crystal jelly, Aequorea victoria and is the pioneering GECI used to study Ca2+ signaling in plants. The AEQ luminesces upon binding with Ca2+ ions and thereby serves as a reporter for any change in Ca2+ ions concentration, brought about by touch, cold stress or pathogenic stress (Knight et al., 1991). Miyawaki et al. (1997) pioneered to construct a Ca2+ dependent fluorescent indicators that are genetically encoded and can be targeted for specific sub-cellular locations. The GECI cameleon, an engineered protein created for studying Ca2+ movement in live cells has been used to study calcium signaling in plants. The Ca2+ signature concept was developed through research on Medicago truncatula plants expressing the Ca2+ probe cameleon YC2.1. A link between Ca2+ oscillations induced by bacterial Nod factor and corresponding activation of selected nodulation marker genes by modulating Ca2+ homeostasis has been demonstrated pharmacologically. For instance, the ENOD11 nodulin gene required about 30 consecutive Ca2+ spikes for induction. Wang et al. (2019), used the fluorescent calcium sensor Cameleon 3.60 to study the dynamics of calcium signaling during P. indica and Arabidopsis roots interaction. They found that calcium signaling was critical for the colonization of roots by P. indica, and that calcium oscillations occurred in a specific temporal pattern during the interaction. Yet another GECI, GCaMP, has been studied for calcium signaling in yeast (Wu et al., 2022). Tang et al. (2017), used the genetically-encoded calcium sensor GCaMP6s to study calcium dynamics during the interaction between the pathogenic bacterium Ralstonia solanacearum and tomato plants. They found that calcium signaling activated defense responses in tomato plants and that the bacterial effector protein RipAC disrupted calcium signaling to suppress plant defense responses. Calcium imaging has also been used to study calcium signaling in response to various biotic and abiotic stresses in plants. For instance, calcium imaging has been used to study calcium signaling in response to pathogen infection in Arabidopsis (Mithöfer et al., 1999) and rice (Zhang et al., 2015). Calcium imaging has also been used to study calcium signaling during drought and salt stress in Arabidopsis (Bartels and Sunkar, 2005; Du et al., 2018). By providing real-time visualization of calcium dynamics, live-cell imaging and fluorescent probes can provide significant insights to explore the molecular mechanisms involved in plant-microbe interactions.
Genome editing
The CRISPR Cas9 techniques have allowed researchers to directly and objectively monitor the complex interactions between stress signaling and response in plants. As a result, many new connections have been uncovered and many lacunae have been bridged in the complex mechanism of stress tolerance by plants. Crop improvement has been accelerated over the past three decades through genome editing methods that modulate gene activity. The Ca2+/CaM-binding proteins have been targeted to generate stress-tolerant crop lines by generating knockout mutation of CML 24 (Ma et al., 2008). While the specific stress-responsiveness of individual Ca2+/CaM-binding partners is unclear, CDPK/CIPK regulations have been linked to several abiotic and biotic factors, likely due to their upstream position in the signaling cascade (Pan et al., 2018). Additionally, CAMTAs regulate downstream genes of signaling cascade to mitigate the deleterious effects of various stressors (Doherty et al., 2009; Rahman et al., 2016). CRISPR-Cas9 was used to develop disease-resistant wheat varieties. They identified TaCIPK14, a wheat CIPK homologue gene, and found that TaCIPK14 knockout mutants have higher tolerance to wheat stripe rust caused by Puccinia striiformis f. sp. tritici (Pst). Furthermore, the researchers generated Tacipk14 mutant plants via CRISPR/Cas9 technology that showed higher resistance to Pst without any significant difference in crop yield in comparison to control plants. These results demonstrate the potential of using CRISPR/Cas9 technology to develop disease-resistant wheat varieties by targeting the genes involved in various calcium signatures (He et al., 2023).
Virus-induced gene silencing (VIGS) using Barley stripe mosaic virus (BSMV) has been employed to study the function of various genes, including those encoding Cyclic nucleotide-gated channels (CNGCs) during plant-pathogen interactions. Kang et al. (2019) used BSMV-VIGS to identify TaCNGC14 and TaCNGC16 gene interactions during stipe-rust infection. This BSMV-VIGS has been used to investigate the role of CNGCs in alleviation of biotic and abiotic stresses (Guo et al., 2018; Tian et al., 2019). The significance of CNGCs in plant defense against pathogens has been studied using the gene silencing approach. For instance, Saand et al. (2015) employed BSMV-VIGS to silence CaM2 and CaM6 genes, which implicate SlCNGC17 and SlCNGC18 genes for resistance to tomato against P. aphanidermatum, a soil-borne fungal pathogen that causes root rot in tomato. These studies demonstrate the versatility of the BSMV-VIGS approach in studying the functional aspects of CNGCs for their potential role in pathogenic defense. Therefore, identifying a master switch for Ca2+ signaling is a high priority for the scientific community to address the challenges posed by biotic stressors.
Multi omics approaches
The use of innovative “omics” techniques, combined with remarkable advancements in modern molecular biology, has empowered scientists to uncover the intricate interactions between stress signaling and response in plants. Genome-wide studies have helped to identify genes encoding Ca2+ signaling elements, which have been extensively studied in plants. The diversity of Ca2+ transport elements in plants reflects their evolutionary origins, functional diversification, and roles in stress response. In order to fulfil the intricate and myriad functions of the Ca2+ transport system, different types of Ca2+ transport elements are present in plants (Taneja and Upadhyay, 2021). In silico investigations of genes involved in Ca2+ signaling has identified various Ca2+ transporters in Arabidopsis and rice. Comparative transcriptomic analysis has further revealed the involvement of Ca2+ transporters, particularly CNGCs, in mitigation of biotic and abiotic stresses. For example, CNGCs have been implicated in clubroot resistance, sunflower genotype resistance to Verticillium dahliae, and rice responses to bacterial pathogens and drought stress (Guo et al., 2017). Also, CNGC genes were implicated in response to Pseudomonas fuscovaginae and Xanthomonas oryzae pv. oryzae (Xoo), in rice as well as in drought stress in tobacco (Nawaz et al., 2014). Similarly, transcriptomic profiling of Botryosphaeria dothidea-infected apple leaves revealed differential expression of MdCNGC genes in response to the pathogen (Zhang et al., 2018). These studies indicate the importance of CNGC genes in plant defense mechanisms against biotic and abiotic stresses and suggest their potential as targets for crop improvement. The transcriptomic profiling has shown that Ca2+ signaling elements play crucial roles in regulating the resistance to pathogens in plants. For instance, a study by Tian et al. (2020) used transcriptomic analysis to identify genes that were upregulated in tomato plants infected with the pathogenic bacterium Ralstonia solanacearum. The research found that numerous genes involved in calcium signaling were associated with plant defense responses. Previous studies have typically investigated bacteria and host plants separately; however, meta-transcriptomic analysis, provides a more comprehensive information on the simultaneous transcriptional state of a variety of microorganisms. Samaras et al. (2021) performed transcriptomic and metabolomic analysis to investigate changes in cucumber roots after inoculation with Bacillus subtilis MBI600, a biocontrol agent. They found that an upregulation of expression of genes involved in plant growth promotion, stress response, and defense-related pathways were upregulated in the treated plants. The metabolomic analysis revealed changes in the levels of metabolites involved in primary and secondary metabolism, including amino acids, organic acids, and phenolic compounds. These results suggest that the Bacillus subtilis MBI600-mediated growth promotion and enhanced defense response in cucumber plants may be attributed to the activation of signaling pathways and the biosynthesis of defense-related metabolites. Proteomics studies have also been utilized for identification of proteins involved in calcium signaling pathways in plant-microbe interactions. For instance, a proteomic study discovered calcium-binding proteins and ion transporters as vital components of calcium signaling in the interactions between soybean roots and Bradyrhizobium japonicum (Wang et al., 2016).
Future directions and challenges
The intricate involvement of calcium signaling in plant-microbe interactions presents opportunities for biotechnological applications aimed at enhancing plant growth and improving crop yields. Calcium-based fertilizers have been proposed as potential tools for promoting symbiotic interactions between plants and beneficial microbes, such as endophytes, mycorrhizal fungi and rhizobia. Additionally, the manipulation of calcium signaling pathways in plants through genetic engineering or chemical treatments could be utilized to confer resistance against microbial pathogens or improve stress tolerance.
Despite the potential benefits of harnessing calcium signaling in plant-microbe interactions, there are several challenges that must be overcome. One major hurdle is the complex nature of calcium signaling pathways, which involves a large number of components and regulatory mechanisms. The identification and characterization of key components and their interactions will require further research and development of new techniques for studying calcium signaling dynamics in vivo. Another challenge is the specificity of calcium signaling responses, which must be finely tuned to ensure appropriate responses to different microbes and environmental conditions. Understanding the mechanisms of specificity and cross-talk between different signaling pathways will be essential for developing strategies for manipulating calcium signaling in plants.
New research avenues are emerging to provide deep insights into the intricacies of calcium signaling in plant-microbe interactions. For example, advances in imaging techniques and biosensors are enabling the visualization and measurement of calcium signals in real-time, providing a detailed understanding of the spatiotemporal dynamics of calcium connections in plants and microbes. Furthermore, the identification and characterization of novel components of calcium signaling, such as ion channels and transporters, will help to further unravel the complex mechanisms of calcium signaling in plant-microbe interactions. The omics approaches, such as transcriptomics and proteomics, may provide a system-level understanding of calcium signatures in plants and microbes.
Conclusion
This review highlights the crucial role of calcium signaling in plant-microbe interactions. Calcium signaling pathways in plants involve the release of calcium ions, transporters, and sensors, which play a critical role in plant responses to microbial pathogens and beneficial microbes. The modulation of calcium signaling by microbial effectors, particularly in pathogenic interactions, has been extensively studied. Additionally, calcium signaling has been identified as a crucial mediator in plant-microbe symbiosis, particularly in the establishment of arbuscular mycorrhizal and rhizobia interactions. Since a large number of Ca2+ binding and Ca2+ decoding proteins are involved in the process of Ca2+ dependent signaling, mutation studies for identification of genes responsible for Ca2+ surge is recommended as a promising method to unravel this complexity. The study of calcium signaling mechanisms in plant-microbe interactions has significant implications for biotechnological applications, particularly in the development of calcium-based fertilizers to enhance symbiotic interactions. However, there are still key challenges to overcome in understanding the intricacies of calcium signaling in plant-microbe interactions, particularly in the development of novel techniques for studying calcium signaling in plants and microbes. The precise manipulation of Ca2+ signals may be harnessed to obtain climate smart crops with enhanced stress tolerance. The generation and dissipation of Ca2+ could be controlled using reverse genetic approaches. Overall, the unravelling of calcium signaling mechanisms in plant-microbe interactions provides exciting research avenues for future investigations.
Author contributions
NN and AR conceived and conceptualized the idea. NN, AR, PN, RP, GP, BC, and DK contributed to writing, editing, reviewing and finalizing the article. All authors contributed to the article and approved the submitted version.
Acknowledgments
The authors acknowledge Prof. Vishnu Bhat, Department of Botany, University of Delhi for infrastructural support and scientific consultations.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Aldon, D., Mbengue, M., Mazars, C., Galaud, J. P. (2018). Calcium signalling in plant biotic interactions. Int. J. Mol. Sci. 19 (3), 665. doi: 10.3390/ijms19030665
Atkinson, M. M., Midland, S. L., Sims, J. J., Keen, N. T. (1996). Syringolide 1 triggers Ca2+ influx, K+ efflux, and extracellular alkalization in soybean cells carrying the disease-resistance gene Rpg4. Plant Physiol. 112 (1), 297–302. doi: 10.1104/pp.112.1.297\
Bai, F., Stratmann, J. W., Matton, D. P. (2022). A complete MAPK cascade, a calmodulin, and a protein phosphatase act downstream of CRK receptor kinases and regulate Arabidopsis innate immunity. BioRxiv, 2022–2003. doi: 10.1101/2022.03.27.486008
Bao, Z., Shi, C., Tu, W., Li, L., Li, Q. (2022). Recent developments in modification of biochar and its application in soil pollution control and ecoegulation. Environ. Pollution. 313, 120184. doi: 10.1016/j.envpol.2022.120184
Bapaume, L., Reinhardt, D. (2012). How membranes shape plant symbioses: signaling and transport in nodulation and arbuscular mycorrhiza. Front. Plant Sci. 3. doi: 10.3389/fpls.2012.00223
Bartels, D., Sunkar, R. (2005). Drought and salt tolerance in plants. ritical Rev. Plant Sci. 24 (1), 23–58. doi: 10.1080/07352680590910410
Batistic, O., Kudla, J. (2004). Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219, 915–924. doi: 10.1007/s00425-004-1333-3
Becker, D., Geiger, D., Dunkel, M., Roller, A., Bertl, A., Latz, A., et al. (2004). AtTPK4, an Arabidopsis tandem-pore K+ channel, poised to control the pollen membrane voltage in a pH-and Ca2+-dependent manner. Proc. Natl. Acad. Sci. 101 (44), 15621–15626. doi: 10.1073/pnas.040150210
Berridge, M. J., Lipp, P., Bootman, M. D. (2000). The versatility and universality of calcium signalling. Nat. Rev. Mol. Cell Biol. 1 (1), 11–21. doi: 10.1038/35036035
Berruti, A., Lumini, E., Balestrini, R., Bianciotto, V. (2016). Arbuscular mycorrhizal fungi as natural biofertilizers: let's benefit from past successes. Front. Microbiol. 6. doi: 10.3389/fmicb.2015.01559
Bhar, A., Chakraborty, A., Roy, A. (2023). The captivating role of calcium in plant-microbe interaction. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1138252
Bi, G. Z., Su, M., Li, N., Liang, Y., Dang, S., Xu, J. C., et al. (2021). The ZAR1 resistosome is a calcium-permeable channel triggering plant immune signaling. Cells. 184 (13), 3528–3541. doi: 10.1016/j.cell.2021.05.003
Blume, B., Nürnberger, T., Nass, N., Scheel, D. (2000). Receptor-mediated increase in cytoplasmic free calcium required for activation of pathogen defense in parsley. Plant Cell. 12 (8), 1425–1440. doi: 10.1105/tpc.12.8.1425
Bohm, H., Albert, I., Oome, S., Raaymakers, T. M., Van den Ackerveken, G., Nürnberger, T. (2014). A conserved peptide pattern from a widespread microbial virulence factor triggers pattern-induced immunity in Arabidopsis. PLoS Pathog. 10 (11), e1004491. doi: 10.1371/journal.ppat.1004491
Bose, J., Pottosin, I. I., Shabala, S. S., Palmgren, M. G., Shabala, S. (2011). Calcium efflux systems in stress signaling and adaptation in plants. Front. Plant Science. 2. doi: 10.3389/fpls.2011.00085
Bouche, N., Scharlat, A., Snedden, W., Bouchez, D., Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. J. Biol. Chem. 277, 21851–21861. doi: 10.1074/jbc.M200268200
Boudsocq, M., Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant science. 18 (1), 30–40. doi: 10.1016/j.tplants.2012.08.008
Boudsocq, M., Willmann, M. R., McCormack, M., Lee, H., Shan, L., He, P., et al. (2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature. 464 (7287), 418–422. doi: 10.1038/nature08794
Boursiac, Y., Lee, S. M., ROmanowsky, S., Blank, R., Sladek, C., Chung, W. S., et al. (2010). Disruption of the vacuolar calcium-ATPases in Arabidopsis results in the activation of a salicylic acid-dependent programmed cell death pathway. Plant Physiol. 154, 1158–1171. doi: 10.1104/pp.110.159038
Bundo, M., Coca, M. (2016). Enhancing blast disease resistance by overexpression of the calcium-dependent protein kinase Os CPK 4 in rice. Plant Biotechnol. J. 14 (6), 1357–1367. doi: 10.1111/pbi.12500
Cao, Y., Liang, Y., Tanaka, K., Nguyen, C. T., Jedrzejczak, R. P., Joachimiak, A., et al. (2014). The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin-induced complex with related kinase CERK1. Elife. 3, e03766. doi: 10.7554/eLife.03766
Chen, G., Cui, J., Wang, L., Zhu, Y., Lu, Z., Jin, B. (2017). Genome-wide identification of circular RNAs in Arabidopsis thaliana. Front. Plant Sci. 8 1678, 1–12. doi: 10.3389/fpls.2017.01678
Cheng, S. H., Willmann, M. R., Chen, H. C., Sheen, J. (2002). Calcium signaling through protein kinases. The Arabidopsis calcium-dependent protein kinase gene family. Plant Physiol. 129 (2), 469–485. doi: 10.1104/pp.005645
Chiasson, D., Ekengren, S. K., Martin, G. B., Dobney, S. L., Snedden, W. A. (2005). Calmodulin-like Proteins from Arabidopsis and Tomato are Involved in Host Defense Against Pseudomonas syringae pv. tomato. Plant Mol. Biol. 58, 887–897. doi: 10.1007/s11103-005-8395-x
Chung, W. S., Lee, S. H., Kim, J. C., Do Heo, W., Kim, M. C., Park, C. Y., et al. (2000). Identification of a calmodulin-regulated soybean Ca2+-ATPase (SCA1) that is located in the plasma membrane. Plant Cell. 12 (8), 1393–1407. doi: 10.1105/tpc.12.8.1393
Clough, S. J., Fengler, K. A., Yu, I. C., Lippok, B., Smith, R. K., Jr., Bent, A. F. (2000). The Arabidopsis dnd1 “defense, no death” gene encodes a mutated cyclic nucleotide-gated ion channel. Proc. Natl. Acad. Sci. 97 (16), 9323–9328. doi: 10.1073/pnas.150005697
Crouzet, J., Roland, J., Peeters, E., Trombik, T., Ducos, E., Nader, J., et al. (2013). NtPDR1, a plasma membrane ABC transporter from Nicotiana tabacum, is involved in diterpene transport. Plant Mol. Biol. 82, 181–192. doi: 10.1007/s11103-013-0053-0
Cutler, S. R., Rodriguez, P. L., Finkelstein, R. R., Abrams, S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679. doi: 10.1146/annurev-arplant-042809-112122
DeFalco, T. A., Moeder, W., Yoshioka, K. (2023). Ca2+ signalling in plant biotic interactions. Front. Plant Science. 14. doi: 10.3389/fpls.2023.1137001
de la Torre, F., Gutiérrez-Beltrán, E., Pareja-Jaime, Y., Chakravarthy, S., Martin, G. B., del Pozo, O. (2013). The tomato calcium sensor Cbl10 and its interacting protein kinase Cipk6 define a signaling pathway in plant immunity. Plant Cell. 25 (7), 2748–2764. doi: 10.1105/tpc.113.113530
Delk, N. A., Johnson, K. A., Chowdhury, N. I., Braam, J. (2005). CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 139 (1), 240–253. doi: 10.1104/pp.105.062612
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell. 21 (3), 972–984. doi: 10.1046/j.1365-313X.1999.00375.x
Du, Y. T., Zhao, M. J., Wang, C. T., Gao, Y., Wang, Y. X., Liu, Y. W., et al. (2018). Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 18 (1), 1–18. doi: 10.1186/s12870-018-1551-7
Dubiella, U., Seybold, H., Durian, G., KOmander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. USA. 110 (21), 8744–8749. doi: 10.1073/pnas.1221294110
Dubreuil-Maurizi, C., Vitecek, J., Marty, L., Branciard, L., Frettinger, P., Wendehenne, D., et al. (2011). Glutathione deficiency of the Arabidopsis mutant pad2-1 affects oxidative stress-related events, defense gene expression, and the hypersensitive response. Plant Physiol. 157 (4), 2000–2012. doi: 10.1104/pp.111.182667
Eichstadt, B., Lederer, S., Trempel, F., Jiang, X., Guerra, T., Waadt, R., et al. (2021). Plant immune memory in systemic tissue does not involve changes in rapid calcium signaling. Front. Plant Science. 12, 3020. doi: 10.3389/fpls.2021.798230
Evangelisti, E., Rey, T., Schornack, S. (2014). Cross-interference of plant development and plant–microbe interactions. Curr. Opin. Plant Biol. 20, 118–126. doi: 10.1016/j.pbi.2014.05.014
Evans, M. J., Choi, W. G., Gilroy, S., Morris, R. J. (2016). A ROS-assisted calcium wave dependent on the AtRBOHD NADPH oxidase and TPC1 cation channel propagates the systemic response to salt stress. Plant Physiol. 171 (3), 1771–1784. doi: 10.1104/pp.16.00215
Fantino, E., Segretin, M. E., Santin, F., Mirkin, F. G., Ulloa, R. M. (2017). Analysis of the potato calcium-dependent protein kinase family and characterization of StCDPK7, a member induced upon infection with Phytophthora infestans. Plant Cell Rep. 36, 1137–1157. doi: 10.1007/s00299-017-2144-x
Forde, B. G., Roberts, M. R. (2014). Glutamate receptor-like channels in plants: a role as amino acid sensors in plant defence? F1000Prime Rep. 6, 37. doi: 10.12703/P6-37
Gage, D. J. (2004). Infection and invasion of roots by symbiotic, nitrogen-fixing rhizobia during nodulation of temperate legumes. Microbiol. Mol. Biol. Rev. 68 (2), 280–300. doi: 10.1128/mmbr.68.2.280-300.2004
Galione, A., Churchill, G. C. (2002). Interactions between calcium release pathways: multiple messengers and multiple stores. Cell Calcium. 32 (5-6), 343–354. doi: 10.1016/S0143416002001902
Garcia, K., Doidy, J., Zimmermann, S. D., Wipf, D., Courty, P. E. (2016). Take a trip through the plant and fungal transportome of mycorrhiza. Trends Plant Sci. 21 (11), 937–950. doi: 10.1016/j.tplants.2016.07.010
Garg, N., Singh, S. (2018). Mycorrhizal inoculations and silicon fortifications improve rhizobial symbiosis, antioxidant defense, trehalose turnover in pigeon pea genotypes under cadmium and zinc stress. Plant Growth Regulation. 86 (1), 105–119. doi: 10.1007/s10725-018-0414-4
Gelli, A., Blumwald, E. (1997). Hyperpolarization-activated Ca2+-permeable channels in the plasma membrane of tomato cells. J. Membr. Biol. 155, 35–45. doi: 10.1007/s002329900156
Gerke, V., Moss, S. E. (2002). Annexins: from structure to function. Physiol. Rev. 82 (2), 331–371. doi: 10.1152/physrev.00030.2001
Ghosh, S., Bheri, M., Bisht, D., Pandey, G. K. (2022). Calcium signaling and transport machinery: Potential for development of stress tolerance in plants. Curr. Plant Biol. 29, 100235. doi: 10.1016/j.cpb.2022.100235
Gilroy, S., Read, N., Trewavas, A. J. (1990). Elevation of cytoplasmic calcium by caged calcium or caged inositol trisphosphate initiates stomatal closure. Nature. 346 (6286), 769–771. doi: 10.1038/346769a0
Gonçalves, M. F., Nunes, R. B., Tilleman, L., Van de Peer, Y., Deforce, D., Van Nieuwerburgh, F., et al. (2019). Dual RNA sequencing of Vitis vinifera during Lasiodiplodia theobromae infection unveils host–pathogen interactions. Int. J. Mol. Sci. 20 (23), 6083. doi: 10.3390/ijms20236083
Grant, M., Brown, I., Adams, S., Knight, M., Ainslie, A., Mansfield, J. (2000). The RPM1 plant disease resistance gene facilitates a rapid and sustained increase in cytosolic calcium that is necessary for the oxidative burst and hypersensitive cell death. Plant J. 23 (4), 441–450. doi: 10.1046/j.1365-313x.2000.00804.x
Guo, J., Islam, M. A., Lin, H., Ji, C., Duan, Y., Liu, P., et al. (2018). Genome-wide identification of cyclic nucleotide-gated ion channel gene family in wheat and functional analyses of TaCNGC14 and TaCNGC16. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00018
Guo, S., Zuo, Y., Zhang, Y., Wu, C., Su, W., Jin, W., et al. (2017). Large-scale transcriptome comparison of sunflower genes responsive to Verticillium dahliae. BMC Genomics 18, 1–13. doi: 10.1186/s12864-016-3386-7
Hamilton, E. S., Schlegel, A. M., Haswell, E. S. (2015). United in diversity: mechanosensitive ion channels in plants. Annu. Rev. Plant Biol. 66, 113–137. doi: 10.1146/annurev-arplant-043014-114700
Harrison, M. J. (2005). Signaling in the arbuscular mycorrhizal symbiosis. Annu. Rev. Microbiol. 59, 19–42. doi: 10.1146/annurev.micro.58.030603.123749
He, F., Wang, C., Sun, H., Tian, S., Zhao, G., Liu, C., et al. (2023). Simultaneous editing of three homoeologues of TaCIPK14 confers broad-spectrum resistance to stripe rust in wheat. Plant Biotechnol. J. 21 (2), 354–368. doi: 10.1111/pbi.13956
Heo, W. D., Lee, S. H., Kim, M. C., Kim, J. C., Chung, W. S., Chun, H. J., et al. (1999). Involvement of specific calmodulin isoforms in salicylic acid-independent activation of plant disease resistance responses. Proc. Natl. Acad. Sci. 96 (2), 766–771. doi: 10.1073/pnas.96.2.766
Hu, Z., Lv, X., Xia, X., Zhou, J., Shi, K., Yu, J., et al. (2016). Genome-wide identification and expression analysis of calcium-dependent protein kinase in tomato. Front. Plant science. 7. doi: 10.3389/fpls.2016.00469
Jiang, Z., Watanabe, C. K., Miyagi, A., Kawai-Yamada, M., Terashima, I., Noguchi, K. (2019). Mitochondrial AOX supports redox balance of photosynthetic electron transport, primary metabolite balance, and growth in Arabidopsis thaliana under high light. Int. J. Mol. Sci. 20 (12), 3067. doi: 10.3390/ijms20123067
Jiang, S., Zhang, D., Wang, L., Pan, J., Liu, Y., Kong, X., et al. (2013). A maize calcium-dependent protein kinase gene, ZmCPK4, positively regulated abscisic acid signalling and enhanced drought stress tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 71, 112–120. doi: 10.1016/j.plaphy.2013.07.004
Jin, F., Liu, J., Wu, E., Yang, P., Gao, J., Gao, X., et al. (2021). Leaf transcriptome analysis of broomcorn millet uncovers key genes and pathways in response to sporisorium destruens. Int. J. Mol. Sci. 22 (17), 9542. doi: 10.3390/ijms22179542
Kamal, H., Minhas, F. U. A. A., Tripathi, D., Abbasi, W. A., Hamza, M., Mustafa, R., et al. (2019). βC1, pathogenicity determinant encoded by Cotton leaf curl Multan betasatellite, interacts with calmodulin-like protein 11 (Gh-CML11) in Gossypium hirsutum. PLoS One 14 (12), e0225876. doi: 10.1371/journal.pone.0225876
Kanchiswamy, C. N., Takahashi, H., Quadro, S., Maffei, M. E., Bossi, S., Bertea, C., et al. (2010). Regulation of Arabidopsis defense responses against Spodoptera littoralis by CPK-mediated calcium signaling. BMC Plant Biol. 10, 1–10. doi: 10.1186/1471-2229-10-97
Kang, G., Wu, Y., Li, G., Wang, P., Han, Q., Wang, Y., et al. (2019). Proteomics combined with BSMV-VIGS methods identified some N deficiency-responsive protein species and ABA role in wheat seedling. Plant Soil. 444, 177–191. doi: 10.1007/s11104-019-04260-1
Kaur, A., Upadhyay, S. K. (2022). “EF-hand domain-containing proteins: diversity and role in plants,” in Cation transporters in plants (London: Academic Press), 185–203. doi: 10.1016/B978-0-323-85790-1.00010-5
Kiep, V., Vadassery, J., Lattke, J., Maaß, J. P., Boland, W., Peiter, E., et al. (2015). Systemic cytosolic Ca2+ elevation is activated upon wounding and herbivory in Arabidopsis. New Phytol. 207 (4), 996–1004. doi: 10.1111/nph.13493
Knight, M. R., Campbell, A. K., Smith, S. M., Trewavas, A. J. (1991). Transgenic plant aequorin reports the effects of touch and cold-shock and elicitors on cytoplasmic calcium. Nature. 352 (6335), 524–526. doi: 10.1038/352524a0
Kosuta, S., Hazledine, S., Sun, J., Miwa, H., Morris, R. J., Downie, J. A., et al. (2008). Differential and chaotic calcium signatures in the symbiosis signaling pathway of legumes. Proc. Natl. Acad. Sci. 105 (28), 9823–9828. doi: 10.1073/pnas.0803499105
Krol, E., Mentzel, T., Chinchilla, D., Boller, T., Felix, G., Kemmerling, B., et al. (2010). Perception of the Arabidopsis danger signal peptide 1 involves the pattern recognition receptor AtPEPR1 and its close homologue AtPEPR2. J. Biol. Chem. 285 (18), 13471–13479. doi: 10.1074/jbc.M109.097394
Kudla, J., Batistic, O., Hashimoto, K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell. 22 (3), 541–563. doi: 10.1105/tpc.109.072686
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytologist. 218 (2), 414–431. doi: 10.1111/nph.14966
Kurusu, T., Hamada, J., Nokajima, H., Kitagawa, Y., Kiyoduka, M., Takahashi, A., et al. (2010). Regulation of microbe-associated molecular pattern-induced hypersensitive cell death, phytoalexin production, and defense gene expression by calcineurin B-like protein-interacting protein kinases, OsCIPK14/15, in rice cultured cells. Plant Physiol. 153 (2), 678–692. doi: 10.1104/pp.109.151852
Kurusu, T., Yagala, T., Miyao, A., Hirochika, H., Kuchitsu, K. (2005). Identification of a putative voltage-gated Ca2+ channel as a key regulator of elicitor-induced hypersensitive cell death and mitogen-activated protein kinase activation in rice. Plant J. 42, 798–809. doi: 10.1111/j.1365-313X.2005.02415.x
Laxalt, A. M., Munnik, T. (2002). Phospholipid signalling in plant defence. Curr. Opin. Plant Biol. 5 (4), 332–338. doi: 10.1016/S1369-5266(02)00268-6
Leba, L. J., Cheval, C., Ortiz-Martín, I., Ranty, B., Beuzón, C. R., Galaud, J. P., et al. (2012). CML9, an Arabidopsis calmodulin-like protein, contributes to plant innate immunity through a flagellin-dependent signalling pathway. Plant J. 71 (6), 976–989. doi: 10.1111/j.1365-313X.2012.05045.x
Lecourieux, D., Mazars, C., Pauly, N., Ranjeva, R., Pugin, A. (2002). Analysis and effects of cytosolic free calcium increases in response to elicitors in Nicotiana plumbaginifolia cells. Plant Cell. 14 (10), 2627–2641. doi: 10.1105/tpc.005579
Li, Z., Cao, L., Fan, M., Xu, Q. (2015). Direct pulp capping with calcium hydroxide or mineral trioxide aggregate: a meta-analysis. J. Endodontics. 41 (9), 1412–1417. doi: 10.1016/j.joen.2015.04.012
Li, L., Li, M., Yu, L., Zhou, Z., Liang, X., Liu, Z., et al. (2014). The FLS2-associated kinase BIK1 directly phosphorylates the NADPH oxidase RbohD to control plant immunity. Cell Host Microbe 15 (3), 329–338. doi: 10.1016/j.chom.2014.02.009
Li, G., Liu, S., Wu, L., Wang, X., Cuan, R., Zheng, Y., et al. (2022). Characterization and functional analysis of a new Calcium/Calmodulin-dependent Protein Kinase (CaMK1) in the citrus pathogenic fungus Penicillium italicum. J. Fungi 8 (7), 667. doi: 10.3390/jof8070667
Liao, W., Huang, G., Yu, J., Zhang, M., Shi, X. (2011). Nitric oxide and hydrogen peroxide are involved in indole-3-butyric acid-induced adventitious root development in marigold. J. Hortic. Sci. Biotechnol. 86 (2), 159–165. doi: 10.1080/14620316.2011.11512742
Liu, P., Duan, Y., Liu, C., Xue, Q., Guo, J., Qi, T., et al. (2018). The calcium sensor TaCBL4 and its interacting protein TaCIPK5 are required for wheat resistance to stripe rust fungus. J. Exp. botany. 69 (18), 4443–4457. doi: 10.1093/jxb/ery227
Liu, B., Fan, H., Sun, C., Yuan, M., Geng, X., Ding, X., et al. (2022). New insights into the role of chrysanthemum calcineurin B–like interacting protein kinase CmCIPK23 in nitrate signaling in Arabidopsis roots. Sci. Rep. 12 (1), 1018. doi: 10.1038/s41598-021-04758-8
Liu, P., Guo, J., Zhang, R., Zhao, J., Liu, C., Qi, T., et al. (2019). TaCIPK10 interacts with and phosphorylates TaNH2 to activate wheat defense responses to stripe rust. Plant Biotechnol. J. 17 (5), 956–968. doi: 10.1111/pbi.13031
Lu, L., Rong, W., Zhou, R., Huo, N., Zhang, Z. (2019). TaCML36, a wheat calmodulin-like protein, positively participates in an immune response to Rhizoctonia cerealis. Crop J. 7 (5), 608–618. doi: 10.1016/j.cj.2019.02.001
Luan, S. (2009). The CBL–CIPK network in plant calcium signalling. Trends Plant Sci. 14 (1), 37–42. doi: 10.1016/j.tplants.2008.10.005
Luan, S., Lan, W., Lee, S. C. (2009). Potassium nutrition, sodium toxicity, and calcium signaling: connections through the CBL–CIPK network. Curr. Opin. Plant Biol. 12 (3), 339–346. doi: 10.1016/j.pbi.2009.05.003
Ma, Y., Berkowitz, G. A. (2012). “Biotic stress signalling: calcium-mediated pathogen defence programmes,” in Plant stress physiology (Wallingford UK: CABI), 291–309. doi: 10.1079/9781845939953.0291
Ma, X., Gai, W. X., Qiao, Y. M., Ali, M., Wei, A. M., Luo, D. X., et al. (2019). Identification of CBL and CIPK gene families and functional characterization of CaCIPK1 under Phytophthora capsici in pepper (Capsicum annuum L.). BMC Genomics 20, 1–18. doi: 10.1186/s12864-019-6125-z
Ma, S. C., Lapin, D., Liu, L., Sun, Y., Song, W., Zhang, X. X., et al. (2020). Direct pathogen-induced assembly of an NLR immune receptor complex to form a holoenzyme. Science. 370, 1184. doi: 10.1126/science.abe3069
Ma, W., Qi, Z., Smigel, A., Walker, R. K., Verma, R., Berkowitz, G. A. (2009). Ca2+, cAMP, and transduction of non-self perception during plant immune responses. Proc. Natl. Acad. Sci. 106 (49), 20995–21000. doi: 10.1073/pnas.0905831106
Ma, W., Smigel, A., Tsai, Y. C., Braam, J., Berkowitz, G. A. (2008). Innate immunity signaling: cytosolic Ca2+ elevation is linked to downstream nitric oxide generation through the action of calmodulin or a calmodulin-like protein. Plant Physiol. 148 (2), 818–828. doi: 10.1104/pp.108.125104
Ma, Y., Zhao, Y., Walker, R. K., Berkowitz, G. A. (2013). Molecular steps in the immune signaling pathway evoked by plant elicitor peptides: Ca2+-dependent protein kinases, nitric oxide, and reactive oxygen species are downstream from the early Ca2+ signal. Plant Physiol. 163 (3), 1459–1471. doi: 10.1104/pp.113.226068
Machner, M. P., Isberg, R. R. (2006). Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev. Cell. 11 (1), 47–56. doi: 10.1016/j.devcel.2006.05.013
Maffei, M., Bossi, S., Spiteller, D., Mithoüfer, A., Boland, W. (2004). Effects of feeding Spodoptera littoralis on lima bean leaves. I. Membrane potentials, intracellular calcium variations, oral secretions, and regurgitate components. Plant Physiol. 134 (4), 1752–1762. doi: 10.1104/pp.103.034165
Manohar, M., Shigaki, T., Mei, H., Park, S., Marshall, J., Aguilar, J., et al. (2011). Characterization of arabidopsis ca2+/H+ exchanger CAX3. Biochemistry. 50 (28), 6189–6195. doi: 10.1021/bi2003839
Martin, R., Qi, T., Zhang, H., Liu, F., King, M., Toth, C., et al. (2020). Structure of the activated ROQ1 resistosome directly recognizing the pathogen effector XopQ. Science 370 (6521), eabd9993. doi: 10.1126/science.abd9993
Martinac, B. (1993). “Mechanosensitive ion channels: biophysics and physiology,” in Thermodynamics of membrane receptors and channels (Florida, USA: CRC Press), 327–351.
Martínez-Medina, A., Appels, F. V., van Wees, S. C. (2017). Impact of salicylic acid-and jasmonic acid-regulated defences on root colonization by Trichoderma harzianum T-78. Plant Signaling Behav. 12 (8), e1345404. doi: 10.1080/15592324.2017.1345404
McAinsh, M. R., Pittman, J. K. (2009). Shaping the calcium signature. New Phytologist. 181 (2), 275–294. doi: 10.1111/j.1469-8137.2008.02682.x
Meng, X., Chai, A., Chen, L., Shi, Y., Xie, X., Ma, Z., et al. (2016). Rapid detection and quantification of viable Pseudomonas syringae pv. lachrymans cells in contaminated cucumber seeds using propidium monoazide and a real-time PCR assay. Can. J. Plant Pathology. 38 (3), 296–306. doi: 10.1080/07060661.2016.1216897
Meng, H., Sun, M., Jiang, Z., Liu, Y., Sun, Y., Liu, D., et al. (2021). Comparative transcriptome analysis reveals resistant and susceptible genes in tobacco cultivars in response to infection by Phytophthora nicotianae. Sci. Rep. 11 (1), 1–13. doi: 10.1038/s41598-020-80280-7
Mishra, G. P., Aski, M. S., Bosamia, T., Chaurasia, S., Mishra, D. C., Bhati, J., et al. (2022). Insights into the host-pathogen interaction pathways through RNA-Seq analysis of Lens culinaris Medik. in response to Rhizoctonia bataticola infection. Genes 13 (1), 90. doi: 10.3390/genes13010090
Mithöfer, A., Schulze, B., Boland, W. (1999). Biotic and heavy metal stress response in plants: evidence for common signals. FEBS Lett. 454, 173–177. doi: 10.1016/j.febslet.2004.04.011
Mitra, R. M., Gleason, C. A., Edwards, A., Hadfield, J., Downie, J. A., Oldroyd, G. E., et al. (2004). A Ca2+/calmodulin-dependent protein kinase required for symbiotic nodule development: gene identification by transcript-based cloning. Proc. Natl. Acad. Sci. 101 (13), 4701–4705. doi: 10.1073/pnas.0400595101
Miyawaki, A., Llopis, J., Heim, R., McCaffery, J. M., Adams, J. A., Ikura, M., et al. (1997). Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature. 388 (6645), 882–887. doi: 10.1038/42264
Moeder, W., Urquhart, W., Ung, H., Yoshioka, K. (2011). The role of cyclic nucleotide-gated ion channels in plant immunity. Mol. Plant 4 (3), 442–452. doi: 10.1093/mp/ssr018
Mohanta, T. K., Bashir, T., Hashem, A., Abd_Allah, E. F., Khan, A. L., Al-Harrasi, A. S. (2018). Early events in plant abiotic stress signaling: interplay between calcium, reactive oxygen species and phytohormones. J. Plant Growth Regulation. 37, 1033–1049. doi: 10.1007/s00344-018-9833-8
Mohanta, T. K., Mohanta, N., Mohanta, Y. K., Parida, P., Bae, H. (2015). Genome-wide identification of Calcineurin B-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biol. 15 (1), 1–15. doi: 10.1186/s12870-015-0543-0
Monaghan, J., Matschi, S., Romeis, T., Zipfel, C. (2015). The calcium-dependent protein kinase CPK28 negatively regulates the BIK1-mediated PAMP-induced calcium burst. Plant Signaling behavior. 10 (5), e1018497. doi: 10.1080/15592324.2015.1018497
Mori, I. C., Murata, Y., Yang, Y., Munemasa, S., Wang, Y. F., Andreoli, S., et al. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion-and Ca2+-permeable channels and stomatal closure. PLoS Biol. 4 (10), e327. doi: 10.1371/journal.pbio.0040327
Nakei, M. D., Venkataramana, P. B., Ndakidemi, P. A. (2022). Soybean-nodulating rhizobia: ecology, characterization, diversity, and growth promoting functions. Front. Sustain. Food Syst. 6. doi: 10.3389/fsufs.2022.824444
Nawaz, Z., Kakar, K. U., Saand, M. A., Shu, Q. Y. (2014). Cyclic nucleotide-gated ion channel gene family in rice, identification, characterization and experimental analysis of expression response to plant hormones, biotic and abiotic stresses. BMC Genomics 15, 853. doi: 10.1186/1471-2164-15-853
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11 (4), 252–263. doi: 10.1038/nrmicro2990
Pan, W., Shen, J., Zheng, Z., Yan, X., Shou, J., Wang, W., et al. (2018). Overexpression of the Tibetan Plateau annual wild barley (Hordeum spontaneum) HsCIPKs enhances rice tolerance to heavy metal toxicities and other abiotic stresses. Rice. 11 (1), 1–13. doi: 10.1186/s12284-018-0242-1
Pandey, G. K., Cheong, Y. H., Kim, K. N., Grant, J. J., Li, L., Hung, W., et al. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16 (7), 1912–1924. doi: 10.1105/tpc.021311
Pandey, G. K., Cheong, Y. H., Kim, B. G., Grant, J. J., Li, L., Luan, S. (2007). CIPK9: a calcium sensor-interacting protein kinase required for low-potassium tolerance in Arabidopsis. Cell Res. 17 (5), 411–421. doi: 10.1038/cr.2007.39
Park, H. C., Kim, M. L., Kang, Y. H., Jeon, J. M., Yoo, J. H., Kim, M. C., et al. (2004). Pathogen-and NaCl-induced expression of the SCaM-4 promoter is mediated in part by a GT-1 box that interacts with a GT-1-like transcription factor. Plant Physiol. 135 (4), 2150–2161. doi: 10.1104/pp.104.041442
Plasencia, F. A., Estrada, Y., Flores, F. B., Ortíz-Atienza, A., Lozano, R., Egea, I. (2021). The Ca2+ sensor Calcineurin B–like protein 10 in plants: emerging new crucial roles for plant abiotic stress tolerance. Front. Plant Science. 11. doi: 10.3389/fpls.2020.599944
Qi, Z., Stephens, N. R., Spalding, E. P. (2006). Calcium entry mediated by GLR3.3, an Arabidopsis glutamate receptor with a broad agonist profile. Plant Physiol. 142, 963–971. doi: 10.1104/pp.106.088989
Rahman, H., Xu, Y. P., Zhang, X. R., Cai, X. Z. (2016). Brassica napus genome possesses extraordinary high number of CAMTA genes and CAMTA3 contributes to PAMP triggered immunity and resistance to Sclerotinia sclerotiorum. Front. Plant Science. 7. doi: 10.3389/fpls.2016.00581
Raina, M., Kisku, A. V., Joon, S., Kumar, S., Kumar, D. (2021b). “Calmodulin and Calmodulin-like Ca2+ binding proteins as molecular players of abiotic stress response in plants,” in Calcium transport element in plants. Ed. Upadhyay, S. K. (Singapore: Academic Press, Springer Nature), ISBN: ISBN- 9780128217924. doi: 10.1016/B978-0-12-821792-4.00001-1
Raina, M., Kumar, A., Yadav, N., Kumari, S., Yusuf, M. A., Mustafiz, A., et al. (2021a). StCaM2, a Calcium Binding Protein, alleviates negative effects of salinity and drought stress in Tobacco. Plant Mol. Biol. 106 (1-2), 85–108. doi: 10.1007/s11103-021-01131-1
Ranf, S., Eschen-Lippold, L., Fröhlich, K., Westphal, L., Scheel, D., Lee, J. (2014). Microbe-associated molecular pattern-induced calcium signaling requires the receptor-like cytoplasmic kinases, PBL1 and BIK1. BMC Plant Biol. 14 (1), 1–15. doi: 10.1186/s12870-014-0374-4
Ranf, S., Wunnenberg, P., Lee, J., Becker, D., Dunkel, M., Hedrich, R., et al. (2008). Loss of the vacuolar cation channel, AtTPC1, does not impair Ca2+ signals induced by abiotic and biotic stresses. Plant J. 53, 287–299. doi: 10.1111/j.1365-313X.2007.03342.x
Ranty, B., Aldon, D., Cotelle, V., Galaud, J. P., Thuleau, P., Mazars, C. (2016). Calcium sensors as key hubs in plant responses to biotic and abiotic stresses. Front. Plant Science. 7. doi: 10.3389/fpls.2016.00327
Rao, S. S., El-Habbak, M. H., Havens, W. M., Singh, A., Zheng, D., Vaughn, L., et al. (2014). Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 15, 145–160. doi: 10.1111/mpp.12075
Reddy, A. S. N., Ali, G. S., Celesnik, H., Day, I. S. (2011). Coping with stresses: roles of calcium- and calcium/calmodulin-regulated gene expression. Plant Cell. 23, 2010–2032. doi: 10.1105/tpc.111.084988
Ren, H., Zhao, X., Li, W., Hussain, J., Qi, G., Liu, S. (2021). Calcium signaling in plant programmed cell death. Cells. 10 (5), 1089. doi: 10.3390/cells10051089
Resentini, F., Ruberti, C., Grenzi, M., Bonza, M. C., Costa, A. (2021). The signatures of organellar calcium. Plant Physiol. 187 (4), 1985–2004. doi: 10.1093/plphys/kiab189
Romeis, T., Ludwig, A. A., Martin, R., Jones, J. D. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. EMBO J. 20 (20), 5556–5567. doi: 10.1093/emboj/20.20.5556
Saand, M. A., Xu, Y. P., Li, W., Wang, J. P., Cai, X. Z. (2015). Cyclic nucleotide gated channel gene family in tomato: genome-wide identification and functional analyses in disease resistance. Front. Plant Science. 6. doi: 10.3389/fpls.2015.00303
Samaras, A., Nikolaidis, M., Antequera-Gómez, M. L., Cámara-Almirón, J., Romero, D., Moschakis, T., et al. (2021). Whole genome sequencing and root colonization studies reveal novel insights in the biocontrol potential and growth promotion by Bacillus subtilis MBI 600 on cucumber. Front. Microbiol. 11. doi: 10.3389/fmicb.2020.600393
Santachiara, G., Salvagiotti, F., Rotundo, J. L. (2019). Nutritional and environmental effects on biological nitrogen fixation in soybean: A meta-analysis. Field Crops Res. 240, 106–115. doi: 10.1016/j.fcr.2019.05.006
Scholz, S. S., Vadassery, J., Heyer, M., Reichelt, M., Bender, K. W., Snedden, W. A., et al. (2014). Mutation of the Arabidopsis calmodulin-like protein CML37 deregulates the jasmonate pathway and enhances susceptibility to herbivory. Mol. Plant 7 (12), 1712–1726. doi: 10.1093/mp/ssu102
Schuurink, R. C., Shartzer, S. F., Fath, A., Jones, R. L. (1998). Characterization of a calmodulin-binding transporter from the plasma membrane of barley aleurone. Proc. Natl. Acad. Sci. U. S. A. 95, 1944–1949. doi: 10.1073/pnas.95.4.1944
Seybold, H., Trempel, F., Ranf, S., Scheel, D., Romeis, T., Lee, J. (2014). Ca2+ signalling in plant immune response: from pattern recognition receptors to Ca2+ decoding mechanisms. New Phytologist. 204 (4), 782–790. doi: 10.1111/nph.13031
Shigematsu, H., Iida, K., Nakano, M., Chaudhuri, P., Iida, H., Nagayama, K. (2014). Structural characterization of the mechanosensitive channel candidate MCA2 from Arabidopsis thaliana. PLoS One 9 (1), e87724. doi: 10.1371/journal.pone.0087724
Shu, B., Jue, D., Zhang, F., Zhang, D., Liu, C., Wu, Q., et al. (2020). Genome-wide identification and expression analysis of the citrus calcium-dependent protein kinase (CDPK) genes in response to arbuscular mycorrhizal fungi colonization and drought. Biotechnol. Biotechnol. Equip. 34 (1), 1304–1314. doi: 10.1080/13102818.2020.1837011
Sieberer, B. J., Chabaud, M., Timmers, A. C., Monin, A., Fournier, J., Barker, D. G. (2009). A nuclear-targeted cameleon demonstrates intranuclear Ca2+ spiking in Medicago truncatula root hairs in response to rhizobial nodulation factors. Plant Physiol. 151 (3), 1197–1206. doi: 10.1104/pp.109.142851
Singh, R., Bartok, A., Paillard, M., Tyburski, A., Elliott, M., Hajnóczky, G. (2022). Uncontrolled mitochondrial calcium uptake underlies the pathogenesis of neurodegeneration in MICU1-deficient mice and patients. Sci. Advances. 8 (11), eabj4716. doi: 10.1126/sciadv.abj4716
Singh, A., Kanwar, P., Yadav, A. K., Mishra, M., Jha, S. K., Baranwal, V., et al. (2014). Genome-wide expressional and functional analysis of calcium transport elements during abiotic stress and development in rice. FEBS J. 281 (3), 894–915. doi: 10.1111/febs.12656
Sun, Y., Li, L., Macho, A. P., Han, Z., Hu, Z., Zipfel, C., et al. (2013). Structural basis for flg22-induced activation of the Arabidopsis FLS2-BAK1 immune complex. Science. 342 (6158), 624–628. doi: 10.1126/science.1243825
Sun, L., Qin, J., Wu, X., Zhang, J., Zhang, J. (2022a). TOUCH 3 and CALMODULIN 1/4/6 cooperate with calcium-dependent protein kinases to trigger calcium-dependent activation of CAM-BINDING PROTEIN 60-LIKE G and regulate fungal resistance in plants. Plant Cell 34 (10), 4088–4104. doi: 10.1093/plcell/koac209
Sun, X., Xie, F., Chen, Y., Guo, Z., Dong, L., Qin, L., et al. (2022b). Glutamine synthetase gene PpGS1. 1 negatively regulates the powdery mildew resistance in Kentucky bluegrass. Horticulture Res. 9, uhac196. doi: 10.1093/hr/uhac196
Swanson, M. S., Isberg, R. R. (1995). Association of Legionella pneumophila with the macrophage endoplasmic reticulum. Infection immunity. 63 (9), 3609–3620. doi: 10.1128/iai.63.9.3609-3620.1995
Takabatake, R., Karita, E., Seo, S., Mitsuhara, I., Kuchitsu, K., Ohashi, Y. (2007). Pathogen-induced calmodulin isoforms in basal resistance against bacterial and fungal pathogens in tobacco. Plant Cell Physiol. 48 (3), 414–423. doi: 10.1093/pcp/pcm011
Taneja, M., Upadhyay, S. K. (2021). “An introduction to the calcium transport elements in plants,” in Calcium transport elements in plants. Ed. Upadhyay, S. K. (Academic Press), 1–18. doi: 10.1016/C2019-0-04172-6
Tang, M., Mao, D., Xu, L., Li, D., Song, S., Chen, C. (2017). The effector AvrRxo1 phosphorylates NAD in planta. PLoS Pathogens. 13 (8), e1006442. doi: 10.1371/journal.ppat.1006442
Tang, M., Zhang, H., Wan, Y., Deng, Z., Qin, X., Li, R., et al. (2022). Transcriptome Analysis in Response to Infection of Xanthomonas oryzae pv. oryzicola Strains with Different Pathogenicity. Int. J. Mol. Sci. 24 (1), 14. doi: 10.3390/ijms24010014
Tapken, D., Anschutz, U., Liu, L. H., Huelsken, T., Seebohm, G., Becker, D., et al. (2013). A plant homolog of animal glutamate receptors is an ion channel gated by multiple hydrophobic amino acids. Sci. Signal. 6, ra47. doi: 10.1126/scisignal.2003762
Thakur, P., Negi, N. P. (2021). “CBL-CIPK: The ca+ Signals during abiotic stress response,” in Crop improvement: Biotechnological advances (Boca Raton and London: CRC Press), 63–74.
Thor, K., Peiter, E. (2014). Cytosolic calcium signals elicited by the pathogen-associated molecular pattern flg22 in stomatal guard cells are of an oscillatory nature. New Phytologist. 204 (4), 873–881. doi: 10.1111/nph.13064
Tian, W., Hou, C., Ren, Z., Wang, C., Zhao, F., Dahlbeck, D., et al. (2019). A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572 (7767), 131–135. doi: 10.1038/s41586-019-1413-y
Tian, W., Wang, C., Gao, Q., Li, L., Luan, S. (2020). Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 6 (7), 750–759. doi: 10.1038/s41477-020-0667-6
Upadhyay, S. K. (2022). Calcium channels, OST1 and stomatal defence: Current status and beyond. Cells. 12 (1), 127. doi: 10.3390/cells12010127
Urquhart, W., Gunawardena, A. H., Moeder, W., Ali, R., Berkowitz, G. A., Yoshioka, K. (2007). The chimeric cyclic nucleotide-gated ion channel ATCNGC11/12 constitutively induces programmed cell death in a Ca2+ dependent manner. Plant Mol. Biol. 65, 747–761. doi: 10.1007/s11103-007-9239-7
Vadassery, J., Ranf, S., Drzewiecki, C., Mithöfer, A., Mazars, C., Scheel, D., et al. (2009). A cell wall extract from the endophytic fungus Piriformospora indica promotes growth of Arabidopsis seedlings and induces intracellular calcium elevation in roots. Plant J. 59 (2), 193–206. doi: 10.1111/j.1365-313X.2009.03867.x
Verma, S., Negi, N. P., Narwal, P., Kumari, P., Kisku, A. V., Gahlot, P., et al. (2022). Calcium signaling in coordinating plant development, circadian oscillations and environmental stress responses in plants. Environ. Exp. Botany. 201, 104935. doi: 10.1016/j.envexpbot.2022.104935
Veronese, P., Nakagami, H., Bluhm, B., AbuQamar, S., Chen, X., Salmeron, J., et al. (2006). The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 18 (1), 257–273. doi: 10.1105/tpc.105.035576
Wais, R. J., Galera, C., Oldroyd, G., Catoira, R., Penmetsa, R. V., Cook, D., et al. (2000). Genetic analysis of calcium spiking responses in nodulation mutants of Medicago truncatula. Proc. Natl. Acad. Sci. 97 (24), 13407–13412. doi: 10.1073/pnas.230439797
Wais, R. J., Keating, D. H., Long, S. R. (2002). Structure-function analysis of nod factor-induced root hair calcium spiking in Rhizobium-legume symbiosis. Plant Physiol. 129 (1), 211–224. doi: 10.1104/pp.010690
Wan, P., Toudeshki, A., Tan, H., Ehsani, R. (2018). A methodology for fresh tomato maturity detection using computer vision. Comput. Electron. Agric. 146, 43–50. doi: 10.1016/j.compag.2018.01.011
Wang, X., Cai, X., Xu, C., Wang, Q., Dai, S. (2016). Drought-responsive mechanisms in plant leaves revealed by proteomics. Int. J. Mol. Sci. 17 (10), 1706. doi: 10.3390/ijms17101706
Wang, Z., Ma, L. Y., Cao, J., Li, Y. L., Ding, L. N., Zhu, K. M., et al. (2019). Recent advances in mechanisms of plant defense to Sclerotinia sclerotiorum. Front. Plant Science. 10. doi: 10.3389/fpls.2019.01314
Wang, C., Mao, X., Zhao, D., Yu, H., Duo, H., Sun, E., et al. (2022). Transcriptomic analysis reveals that cell wall-and hypersensitive response (HR)-related genes are involved in the responses of apple to Valsa Mali. Plant Biotechnol. Rep. 16 (5), 539–551. doi: 10.1007/s11816-022-00763-z
White, P. J., Broadley, M. R. (2003). Calcium in plants. Ann. botany. 92 (4), 487–511. doi: 10.1093/aob/mcg164
Wu, S. Y., Shen, Y., Shkolnikov, I., Campbell, R. E. (2022). Fluorescent indicators for biological imaging of monatomic ions. Front. Cell Dev. Biol. 10. doi: 10.3389/fcell.2022.885440
Xiang, Y., Huang, Y., Xiong, L. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144 (3), 1416–1428. doi: 10.1104/pp.107.101295
Xiaolin, Z., Baoqiang, W., Xian, W., Xiaohong, W. (2022). Identification of the CIPK-CBL family gene and functional characterization of CqCIPK14 gene under drought stress in quinoa. BMC Genomics 23 (1), 1–18. doi: 10.1186/s12864-022-08683-6
Xiong, J. S., Zhu, H. Y., Bai, Y. B., Liu, H., Cheng, Z. M. (2018). RNA sequencing-based transcriptome analysis of mature strawberry fruit infected by necrotrophic fungal pathogen Botrytis cinerea. Physiol. Mol. Plant Pathol. 104, 77–85. doi: 10.1016/j.pmpp.2018.08.005
Xu, G., Moeder, W., Yoshioka, K., Shan, L. (2022). A tale of many families: calcium channels in plant immunity. Plant Cell. 34 (5), 1551–1567. doi: 10.1093/plcell/koac033
Yadav, M., Pandey, J., Chakraborty, A., Hassan, M. I., Kundu, J. K., Roy, A., et al. (2022). A comprehensive analysis of calmodulin-like proteins of glycine max indicates their role in calcium signaling and plant defense against insect attack. Front. Plant Science. 13. doi: 10.3389/fpls.2022.817950
Yamaguchi, T., Hamamoto, S., Uozumi, N. (2013). Sodium transport system in plant cells. Front. Plant science. 4. doi: 10.3389/fpls.2013.00410
Yu, B., Liu, N., Tang, S., Qin, T., Huang, J. (2022). Roles of glutamate receptor-like channels (GLRs) in plant growth and response to environmental stimuli. Plants. 11 (24), 3450. doi: 10.3390/plants11243450
Yu, H., Xiao, A., Dong, R., Fan, Y., Zhang, X., Liu, C., et al. (2018). Suppression of innate immunity mediated by the CDPK-Rboh complex is required for rhizobial colonization in Medicago truncatula nodules. New Phytol. 220 (2), 425–434. doi: 10.1111/nph.15410
Yuan, P., Jauregui, E., Du, L., Tanaka, K., Poovaiah, B. W. (2017). Calcium signatures and signaling events orchestrate plant–microbe interactions. Curr. Opin. Plant Biol. 38, 173–183. doi: 10.1016/j.pbi.2017.06.003
Yuan, P., Tanaka, K., Poovaiah, B. W. (2022). Calcium/calmodulin-mediated defense signaling: What is looming on the horizon for AtSR1/CAMTA3-mediated signaling in plant immunity. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.795353
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature. 514 (7522), 367–371. doi: 10.1038/nature13593
Zhang, C., Cheng, Q., Wang, H., Gao, H., Fang, X., Chen, X., et al. (2021). GmBTB/POZ promotes the ubiquitination and degradation of LHP1 to regulate the response of soybean to Phytophthora sojae. Commun. Biol. 4 (1), 372. doi: 10.1038/s42003-021-01907-7
Zhang, W., Dong, C., Zhang, Y., Zhu, J., Dai, H., Bai, S. (2018). An apple cyclic nucleotide-gated ion channel gene highly responsive to Botryosphaeria dothidea infection enhances the susceptibility of Nicotiana benthamiana to bacterial and fungal pathogens. Plant Science. 269, 94–105. doi: 10.1016/j.plantsci.2018.01.009
Zhang, L. I., Poo, M. M. (2001). Electrical activity and development of neural circuits. Nat. Neurosci. 4 (Suppl 11), 1207–1214. doi: 10.1038/nn753
Zhang, W., Wang, Z., Dan, Z., Zhang, L., Xu, M., Yang, G., et al. (2022b). Transcriptome Analysis of Fusarium Root-Rot-Resistant and-Susceptible Alfalfa (Medicago sativa L.) Plants during Plant–Pathogen Interactions. Genes 13 (5), 788. doi: 10.3390/genes13050788
Zhang, Y., Wang, Y., Taylor, J. L., Jiang, Z., Zhang, S., Mei, F., et al. (2015). Aequorin-based luminescence imaging reveals differential calcium signalling responses to salt and reactive oxygen species in rice roots. J. Exp. botany. 66 (9), 2535–2545. doi: 10.1093/jxb/erv043
Zhang, J., Zou, A., Wen, Y., Wei, X., Liu, C., Lv, X., et al. (2022a). SlCML55, a novel Solanum lycopersicum calmodulin-like gene, negatively regulates plant immunity to Phytophthora pathogens. Scientia Horticulturae. 299, 111049. doi: 10.1016/j.scienta.2022.111049
Zhao, C., Tang, Y., Wang, J., Zeng, Y., Sun, H., Zheng, Z., et al. (2021). A mis-regulated cyclic nucleotide-gated channel mediates cytosolic calcium elevation and activates immunity in Arabidopsis. New Phytologist. 230 (3), 1078–1094. doi: 10.1111/nph.17218
Zhu, X., Caplan, J., Mamillapalli, P., Czymmek, K., Dinesh-Kumar, S. P. (2010). Function of endoplasmic reticulum calcium ATPase in innate immunity-mediated programmed cell death. EMBO J. 29 (5), 1007–1018. doi: 10.1038/emboj.2009.402
Zhu, X., Robe, E., Jomat, L., Aldon, D., Mazars, C., Galaud, J. P. (2017). CML8, an Arabidopsis calmodulin-like protein, plays a role in Pseudomonas syringae plant immunity. Plant Cell Physiol. 58 (2), 307–319. doi: 10.1080/15592324.2017.1322246
Zhu, K., Yang, L. T., Li, C. X., Lakshmanan, P., Xing, Y. X., Li, Y. R. (2021). A transcriptomic analysis of sugarcane response to Leifsonia xyli subsp. xyli infection. PLoS One 16 (2), e0245613. doi: 10.1371/journal.pone.0245613
Zimmermann, S., Nürnberger, T., Frachisse, J. M., Wirtz, W., Guern, J., Hedrich, R., et al. (1997). Receptor-mediated activation of a plant Ca2+-permeable ion channel involved in pathogen defense. Proc. Natl. Acad. Sci. 94 (6), 2751–2755. doi: 10.1073/pnas.94.6.275
Zipfel, C. (2014). Plant pattern-recognition receptors. Trends Immunol. 35 (7), 345–351. doi: 10.1016/j.mib.2014.10.009
Keywords: Ca2+ signaling, plant microbe interaction, endophytes, biotic stress, omics, CRISPR
Citation: Negi NP, Prakash G, Narwal P, Panwar R, Kumar D, Chaudhry B and Rustagi A (2023) The calcium connection: exploring the intricacies of calcium signaling in plant-microbe interactions. Front. Plant Sci. 14:1248648. doi: 10.3389/fpls.2023.1248648
Received: 27 June 2023; Accepted: 24 August 2023;
Published: 02 October 2023.
Edited by:
Dhandapani Gurusamy, Kongunadu Arts and Science College, IndiaReviewed by:
Prafull Salvi, National Agri-Food Biotechnology Institute, IndiaSantosh Kumar Upadhyay, Panjab University, India
Copyright © 2023 Negi, Prakash, Narwal, Panwar, Kumar, Chaudhry and Rustagi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anjana Rustagi, anjana.rustagi@gargi.du.ac.in
 Neelam Prabha Negi
Neelam Prabha Negi Geeta Prakash
Geeta Prakash Parul Narwal
Parul Narwal Ruby Panwar
Ruby Panwar Deepak Kumar
Deepak Kumar Bharti Chaudhry
Bharti Chaudhry Anjana Rustagi
Anjana Rustagi