- 1Department of Applied Plant Science, College of Agriculture and Life Sciences, Chonnam National University, Gwangju, Republic of Korea
- 2Interdisciplinary Program in IT-Bio Convergence System, Chonnam National University, Gwangju, Republic of Korea
- 3Plant Omics Division, Council of Scientific and Industrial Research-Central Salt and Marine Chemical Research Institute (CSIR-CSMCRI), Bhavnagar, Gujarat, India
The DMP (DUF679 membrane proteins) family is a plant-specific gene family that encodes membrane proteins. The DMP family genes are suggested to be involved in various programmed cell death processes and gamete fusion during double fertilization in Arabidopsis. However, their functional relevance in other crops remains unknown. This study identified 14 genes from the DMP family in soybean (Glycine max) and characterized their physiochemical properties, subcellular location, gene structure, and promoter regions using bioinformatics tools. Additionally, their tissue-specific and stress-responsive expressions were analyzed using publicly available transcriptome data. Phylogenetic analysis of 198 DMPs from monocots and dicots revealed six clades, with clade-I encoding senescence-related AtDMP1/2 orthologues and clade-II including pollen-specific AtDMP8/9 orthologues. The largest clade, clade-III, predominantly included monocot DMPs, while monocot- and dicot-specific DMPs were assembled in clade-IV and clade-VI, respectively. Evolutionary analysis suggests that soybean GmDMPs underwent purifying selection during evolution. Using 68 transcriptome datasets, expression profiling revealed expression in diverse tissues and distinct responses to abiotic and biotic stresses. The genes Glyma.09G237500 and Glyma.18G098300 showed pistil-abundant expression by qPCR, suggesting they could be potential targets for female organ-mediated haploid induction. Furthermore, cis-acting regulatory elements primarily related to stress-, hormone-, and light-induced pathways regulate GmDMPs, which is consistent with their divergent expression and suggests involvement in growth and stress responses. Overall, our study provides a comprehensive report on the soybean GmDMP family and a framework for further biological functional analysis of DMP genes in soybean or other crops.
1 Introduction
The DMPs (DUF679 membrane proteins) are membrane proteins found exclusively in green plants (Viridiplantae) and especially in flowering plants (Cyprys et al., 2019). The members of this uncharacterized plant-specific gene family are predicted to be involved in various physiological processes, particularly senescence and reproduction (Kasaras and Kunze, 2010; Cyprys et al., 2019; Zhu et al., 2021). The DMPs are integral membrane proteins with no sequence similarity to functionally assigned protein domains, channels, transporters, or any other membrane proteins in any kingdom (Kasaras and Kunze, 2010). A total of ten members, located across four chromosomes, were identified in the DMP gene family in Arabidopsis (Kasaras and Kunze, 2010). The Arabidopsis DMP family proteins have four transmembrane spans and amino- and carboxy-termini in the cytosol (Kasaras and Kunze, 2010). Among 10 AtDMPs, AtDMP1 was reported as a senescence-associated gene, as it was upregulated during natural or developmental senescence of siliques, rosettes, and cauline leaves, as well as in dark-induced senescence of attached and detached leaves (Van der Graaff et al., 2006). Additionally, AtDMP1 is highly expressed in dehiscence and abscission zones of siliques (Kasaras and Kunze, 2010). Furthermore, AtDMP1, with dual endoplasmic reticulum (ER) and tonoplast localization, is engaged in membrane fission during the breakdown of these organelles during leaf senescence as well as membrane fusion during root vacuole biogenesis (Kasaras et al., 2012). Like AtDMP1, AtDMP3 and AtDMP4 were upregulated in senescing rosette leaves, cauline leaves, and siliques, indicating overlapping functions during senescence. Additionally, AtDMP3 and AtDMP4 transcripts were detected in roots and flowers, respectively. Three AtDMPs, AtDMP2, AtDMP6, and AtDMP7, were expressed in all organs (Kasaras and Kunze, 2010). AtDMP4 was coregulated with the core of dPCD (developmental-programmed cell death) marker genes, including BIFUNCTIONAL NUCLEASE1 (BFN1), PUTATIVE ASPARTIC PROTEASE A3 (PASPA3), RIBONUCLEASE3 (RNS3), CYSTEINE ENDOPEPDITASE 1 (CEP1), and EXITUS1 (EXI1). They were commonly upregulated in differentiation-induced and age-induced dPCD types (Olvera-Carrillo et al., 2015). Additionally, during Arabidopsis stigma senescence, AtDMP4, along with BFN1, RNS3, EXI1, CEP1, DMP4, and PASPA3 were reported to copromote senescence and dPCD (Gao et al., 2018). Furthermore, a comprehensive genome-wide analysis of four cotton species (Gossypium arboreum, G. raimondii, G. hirsutum, and G. barbadense) identified a total of 58 DMPs. Analysis of the expression patterns of these DMPs unveiled their possible involvement in key biological processes, such as plant senescence, flower reproductive development, and stress response (Zhu et al., 2021).
The sperm-specific AtDMP8 and AtDMP9 are reported to involve gamete fusion with a more significant effect on sperm-egg fusion (Takahashi et al., 2018; Cyprys et al., 2019). Moreover, the detection of a mutation in the haploid inducer-associated locus qhir8 (specifically the ZmDMP gene) in maize and loss-of-function mutations in the Arabidopsis AtDMP8 and AtDMP9 led to the development of an effective haploid induction system in dicots (Zhong et al., 2019; Zhong et al., 2020). Although AtDMP8 and AtDMP9 orthologues of the DMP gene family are utilized to induce maternal haploids in dicots (Zhong et al., 2020; Wang et al., 2022; Zhong et al., 2022a; Zhong et al., 2022b), information about other members in the family and their functional relevance needs to be investigated in crops. Soybean (Glycine max), the most widely grown commercial legume and oilseed crop, is commonly used for human consumption, livestock feed, oil production (Hartman et al., 2011). Soybeans contributed 70.39% of plant-based protein meals and 28.55% of plant-based oils in 2020/2021 globally (Market View Data Base, United Soybean Board 2021, accessed on 26 July 2022) and have been referred to as a ‘Wonder crop’ and the ‘Golden bean’ for their high nutritional content, oil content (18%), high-quality proteins (40%), high productivity, versatile uses, and profitability (Rajendran et al., 2022). Being a major oilseed crop, soybean has gained increasing attention in the genomics area, and the availability of genome sequence information accelerated the characterization of important gene families in soybean (Zhang et al., 2022a). However, studies on soybean DMPs investigating their genome-wide occurrence, phylogenetics, and functions are still lacking.
CRISPR (clustered regularly interspaced short palindromic repeats)/Cas9 technology relies on the precise and efficient introduction of double-stranded DNA breaks by the Cas9 nuclease, guided by a sgRNA. Predicting the cleavage efficacy of sgRNAs in vitro prior to their use in plant genome editing offers time, labor, and cost savings by enabling the selection of efficient sgRNAs, optimizing experimental design, and enhancing the success rate and accuracy of editing experiments (Mehravar et al., 2019; Bente et al., 2020). The integration of preassembled Cas9 enzyme with in vitro-transcribed sgRNA, known as the ribonucleoprotein complex (RNP), has been successfully employed in various plant species such as soybean (Kim et al., 2017; Subburaj et al., 2022), wheat (Liang et al., 2017), Brassica (Murovec et al., 2018; Jeong et al., 2019), maize (Sant’Ana et al., 2020), grapevine (Malnoy et al., 2016), apple (Malnoy et al., 2016), and pepper (Kim et al., 2020). This approach is used to assess the in vitro cleavage efficiency sgRNAs and their suitability for subsequent genome editing experiments.
In this study, we identified a total of 198 DMP genes from monocot and dicot crops and analyzed their phylogenetic relationships. Further, we comprehensively analyzed the physiochemical properties, subcellular location, gene structures, motifs, and promoters of the GmDMP family using bioinformatics tools. In order to understand the age- and tissue-specific expression levels of DMPs, as well as their responses to different stresses, publicly available transcriptome data were processed for analysis. Additionally, we conducted real-time quantitative PCR (qPCR) analysis specifically on floral and vegetative tissues to gain further insights into their expression profiles. Subsequently, based on floral expression pattern, we selected two genes with potential applications in haploid induction and conducted an in vitro cleavage assay to assess the cleavage efficiency of the selected sgRNAs. Our findings consolidate the information available in public databases on the DMP gene family and provide a comparative account of DMPs from soybean that would be useful for subsequent functional characterization. This study also identified potential candidate genes associated with haploid induction, growth, and stress response, which have the potential to accelerate soybean breeding.
2 Materials and methods
2.1 Identification and retrieval of DMP genes from monocot and dicot plants
Protein sequences of Arabidopsis DMPs were retrieved from Phytozome (https://phytozome-next.jgi.doe.gov/) and used as queries to perform blastp searches within the genome sequences of Glycine max (Wm82.a2.v1), acquired from the Soybase database (https://soybase.org/soyseq/); Arachis hypogea (v1), acquired from the Peanutbase database (https://peanutbase.org/); Gossypium hirsutum (v1.1), Helianthus annuus (r1.2), Linum usitatissimum (v1.0), Setaria italica (v2.2), Medicago truncatula (Mt4.0v1), Oryza sativa (v7.0), Solanum lycopersicum (ITAG2.4), Sorghum bicolor (BTx642 v1.1), and Zea mays (RefGen_V4), acquired from the Phytozome database (https://phytozome-next.jgi.doe.gov/); Sesamum indicum, obtained from the Ensembl Plants database (http://plants.ensembl.org/index.html); and Brassica napus obtained from the Brassicaceae Database (BRAD, http://brassicadb.cn/#/). All were accessed in April 2022. Subsequently, retrieved gene sequences were verified for the DMP domain (IPR007770) using the National Center for Biotechnology Information (NCBI) Conserved Domain Database (https://www.ncbi.nlm.nih.gov/cdd) (Marchler-Bauer et al., 2010), Simple Modular Architecture Research Tool (SMART; http://smart.embl.de/) (Letunic and Bork, 2018), and the Pfam tool (http://pfam.xfam.org/) (El-Gebali et al., 2019). All verification databases and tools were accessed in May 2022.
2.2 Protein sequence alignment and phylogenetic analysis
The full-length amino acid sequences of DMP family members from different oilseed crops—including Glycine max, Arachis hypogea, Brassica napus, Gossypium hirsutum, Helianthus annuus, Linum usitatissimum, and Sesamum indicum—were aligned using the ClustalW program with the default parameters, and a phylogenetic tree was constructed using the neighbor-joining (NJ) method, both in the MEGA software suite (version 11.0) (Tamura et al., 2021). The phylogenetic analysis was performed using the p-distance model, pairwise deletion, and 1,000 bootstrap replicates. Then, the iTOL tool (https://itol.embl.de/) was used to visualize the unrooted phylogenetic tree (Letunic and Bork, 2021). The accession numbers of the DMPs used for the phylogenetic analysis are listed in Supplementary Table S1.
2.3 Physiochemical properties and subcellular location prediction
The physicochemical properties, viz., molecular weight (Mw), theoretical isoelectric point (pI), instability index, grand average of hydropathicity (GRAVY), and aliphatic index of soybean DMP proteins were predicted using the ProtParam tool on the ExPASy server (https://web.expasy.org/protparam/) (Gasteiger et al., 2005). The subcellular location of DMP proteins was predicted using DeepLoc 2.0 (https://services.healthtech.dtu.dk/service.php?DeepLoc-2.0) (Thumuluri et al., 2022). The numbers of transmembrane domains in soybean DMP proteins were predicted using the DeepTMHMM tool (https://dtu.biolib.com/DeepTMHMM) (Hallgren et al., 2022).
2.4 Conserved motif and intron/exon structure analysis
The conserved motifs were predicted by the MEME (Multiple Expectation Maximization for Motif Elicitation) tool (http://meme-suite.org/tools/meme), an online program for motif discovery (Bailey et al., 2006). Using MEME suite (Version 5.4.1), the motifs were searched with these parameters: the ‘motif discovery mode’ was set to ‘classic mode’, the ‘site distribution’ to ‘zero or one occurrence per sequence’, the ‘number of motifs’ to 10, the width of motifs was set to between 6 and 50, and the required number of sites for each motif to between 2 and 600. The exon-intron structures of the DMP genes were analyzed using the Gene Structure Display Server (GSDS v2.0, http://gsds.gao-lab.org/) (Hu et al., 2015). TBtools software was employed to visualize the distribution of the motif along with the phylogenetic tree and gene structures (Chen et al., 2020).
2.5 Analysis of promoter regions
The 2000 bp sequences upstream of the start codon for each DMP gene were retrieved from the soybean and Arabidopsis database from Phytozome (https://phytozome-next.jgi.doe.gov/). The sequences were submitted to the CARE (Cis-Acting Regulatory Element) search tool (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) in the PlantCARE database (Lescot et al., 2002) to predict and analyze the cis-acting elements related to plant growth and development, hormones, and stress and light responses in the promotor regions of soybean DMP genes.
2.6 Selection pressure and duplicated gene pairs analysis
The Ka/Ks ratio was used to assess selection history and divergence time. The number of synonymous (Ks) and nonsynonymous (Ka) substitutions of duplicated DMP genes were computed using the Ka/Ks Calculator tool (http://services.cbu.uib.no/tools/kaks). The divergence time (T) was calculated using the formula T=Ks/(2× 6. 1× 10−9) ×10−6 million years ago (Mya) (Kim et al., 2013).
2.7 In silico expression analyzes of soybean DMPs in various tissues and developmental stages
To understand the differential expression patterns of soybean DMPs during plant growth and senescence and determine their responses to different stresses, a total of 68 SRA (Sequence Read Archive) datasets comprised of 12 different treatments were downloaded from the NCBI database (Supplementary Table S2). The raw reads were first analyzed using FastQC (www.bioinformatics.babraham.ac.uk/projects/fastqc), and then low-quality bases (< 30 phred score), low-complexity and short sequences (< 50bp), and adapter sequences were removed using an in-house perl script and TrimGalore v0.6.5 (Krueger, 2015). The remaining high-quality reads were used to further analyze transcript abundance using the Galaxy platform. The high-quality reads from each dataset were mapped to the soybean genome (Gmax_275_v2.0.fa, downloaded on 17.07.2022) using the HiSAT2 aligner with the default parameters (Kim et al., 2015). The mapped reads for each sample were counted (‘wcountedount’) using StringTie v1.2.0 (Pertea et al., 2015), and the abundance of genes and transcripts, in fragments per kilobase of transcript per million mapped reads (FPKM), were computed from those mapped to the Glycine max genome annotation. Furthermore, to compare the normalized read count data (FPKM) across the different tissues and treatments, the Z-score was calculated on a gene-by-gene basis by subtracting the overall mean and then dividing by standard deviation. A heatmap was constructed using Multi Experiment Viewer (MeV) v4.9.0. Additionally, publicly available expression data for 65 anatomical parts and seven developmental stages housed in the Genevestigator database (www.genevestigator.com) (Hruz et al., 2008) were retrieved using the Phytozome IDs for soybean.
2.8 Quantitative real-time PCR analysis
Seeds of the soybean cultivar Williams 82 were germinated and cultivated under controlled conditions in a growth chamber, maintaining a temperature of 25 ± 1°C and a 16 h/8 h light/dark cycle. After 60 days of growth, total RNA was extracted from different tissues of the plants, including leaves, stems, flower buds, sepals, petals, pistils, and pollen. To ensure precise sampling, we collected three newly formed leaves from the top of the plants. Stems were sampled 3 cm below the uppermost shoot apex. To obtain sufficient cDNA for qPCR experiments, floral organs were collected from four plants (25 open flowers per plant). Thus, in total, 100 open flowers were pooled in a single biological sample representing four plants. RNA was extracted from pooled samples with subsequent construction of the corresponding cDNA samples. Therefore, the qPCR analysis involved three technical replicates and pooled single biological replicate from four plants. Pooling samples from several independent plants is a common practice in gene expression studies to increase sample size and statistical power while reducing inter-individual variability (Rego et al., 2019; Moebes et al., 2022). Microscopy (EZ4 HD, Leica, Wetzlar, Germany) was used for observing and sampling the floral organs. All flower organs were collected from 100 open flowers measuring approximately 2 mm in size. Sepals and petals were carefully separated from the flower using a needle. For the pistil, only those free from pollen contamination were collected by cutting them with a needle. Immediately after collection anthers were immersed in distilled water and opened using a needle to obtain pollen. Subsequently, we isolated the pollen by passing it through a 50-um nylon filter (04-0042-2317, Sysmex, Görlitz, Germany) to exclude any other tissues. Plant RNA extraction kits (Takara, Shiga, Japan) were used to extract the RNA, following the manufacturer’s instructions. The quality of RNA was checked by agarose gel electrophoresis, and its quantity was determined using a spectrophotometer (Nano-MD UV-Vis, Scinco, Seoul, Korea). For cDNA synthesis, the RevertAid Reverse Transcriptase (Thermo, Waltham, MA, USA) was used in 20 μL reaction volumes. Real-time quantitative PCR (qPCR) was performed in a Thermal Cycler Dice real-time PCR system (Takara, Shiga, Japan) using TB Green™ Premix Ex Taq™ Master Mix (Takara, Shiga, Japan). The relative expression levels of the target genes were quantified in comparison to leaf tissue using the 2−▵▵CT method. Soybean actin11 (Glyma.18G290800) was used as the reference gene for normalization. The analysis was performed with the sample representing three technical replicates and pooled single biological sample from four plants. Gene-specific primers were designed using Primer3Plus (https://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi), and their details are listed in Supplementary Table S3.
2.9 In vitro cleavage assay
2.9.1 Designing the single-guide RNAs
Cas-Designer, a web-based tool RGENs (http://www.rgenome.net/) (Park et al., 2015), was utilized for designing the sgRNAs against the Glycine max (Wm82.a2.v1) genome with default settings. This process resulted in the prediction of a set of candidate sgRNAs along with their respective cleavage positions, out-of-frame scores, and potential mismatches. However, it is important to note that not all sgRNAs display the same cleavage efficiency (Karmakar et al., 2021). To evaluate cleavage efficiency, two sgRNAs, namely SgGmDMP#1 (5’-GGAGGACCATCTCAAAAGTGAGG-3’) and SgGmDMP#2 (5’-CTCCATATCCTTATCCTTCCCGG-3’), were selected. An in vitro screening method was optimized to assess and identify efficient sgRNA.
2.9.2 In vitro synthesis of SgRNAs
The SgRNAs were transcribed in vitro using the GeneArt™ Precision gRNA Synthesis Kit (Invitrogen, USA) following the manufacturer’s protocol. Briefly, the transcription templates were prepared by PCR assembly of the gRNA-DNA template using synthetic forward and reverse oligonucleotides with the Tracer Fragment + T7 Primer Mix (Supplementary Table S3). The resulting PCR product containing a T7 promoter sequence (5’-TAATACGACTCACTATA-3’) and the sgRNA sequence without the PAM region was purified using the gel-purification kit (GeneAll, Seoul, Korea). The in vitro transcription reaction was conducted with a total volume of 20 μL, consisting of 6 μL of purified gRNA-DNA template, 8 μL of NTP mix (25 mM each NTP), 4 μL of 5X TranscriptAid™ reaction buffer, and 2 μL of TranscriptAid™ enzyme mix. The reaction was incubated at 37°C for 3 h. Following the transcription reaction, the synthesized sgRNAs were treated with DNAase to remove any residual DNA and purified using the gRNA Clean Up Kit (Invitrogen, USA). The concentration of the purified sgRNA was determined using a UV spectrophotometer (Nano-MD UV-Vis, Scinco, Seoul, Korea). The sgRNA samples were then diluted to a final concentration of 1 μM and stored at -80°C for future use.
2.9.3 In vitro Cas9-cleavage assay of PCR products
To generate DNA templates containing the sgRNA target sites, a PCR amplification step was performed using soybean genomic DNA and flaking primers (Supplementary Table S3). The resulting PCR products were gel purified and quantified before stored in aliquots at −20°C. Cas9 cleavage reactions (final volume 15 µl) were assembled by combining: 1x Orange Buffer (O-buffer, Thermo Fisher Scientific), 300 ng sgRNA, 250 ng of recombinant Streptococcus pyogenes Cas9-NLS protein (TrueCut™ Cas9 Protein v2, Invitrogen), and ddH2O to reach a final volume of 15 µL. The mixtures were incubated for 10 mins at 22°C to allow the formation of the ribonucleoprotein (RNP) complex. Subsequently, 250 ng of template DNA was added, and the reactions were incubated for 2 h at 37°C. After incubation, 1.2 µL of RNase A (5 µg) and 1.2 µL of Proteinase K (2 mg/mL) were added to the reaction, followed by incubation at 37°C for 20 min. Finally, the reactions were heat-inactivated at 80°C for 10 min. The products of each reaction were analyzed by electrophoresis on a 2% agarose gel.
3 Results
3.1 Identification of DMP genes in soybean and other crops
The genomes of monocot and dicot plants were mined for DMP genes using the Pfam database-derived HMM profile of the DMP domain (PF05078) as the query. We retrieved 198 putative DMP genes after verification. Among them, we detected 14 DMP genes in soybean, 14 in peanut, 7 in tomato, 14 in foxtail millet, 19 in rice, and 16 in sorghum. The DMP gene names, locus IDs, and other features are shown in Supplementary Table S1. The four monocots, foxtail millet, sorghum, maize, and rice, contained 11–16 DMP homologues, whereas the dicots contained from 5 (M. truncatula) to 14 (G. max) homologues.
3.2 Phylogenetic analysis
To analyze the phylogenetic relationships of the DMP family, an unrooted phylogenetic tree was constructed using the 198 DMP proteins of monocot and dicot crops (Figure 1). The DMP proteins were clustered into seven major clades. Clade-I, with 40 DMPs, encoded senescence-related AtDMP1 and AtDMP2 orthologues, while the pollen-specific AtDMP8 and AtDMP9 orthologous proteins (23 DMPs) were grouped into clade II, which included three soybean DMPs: Glyma.09G237500, Glyma.18G097400, and Glyma.18G098300. Clade-III was the largest clade with 45 DMPs predominated by 28 monocot DMPs, which clustered to form sub-clade-III-2. Two soybean DMPs, Glyma.16G157800 and Glyma.02G075800, formed sub-group III-1 along with orthologues of AtDMP10. Notably, clade-IV, containing nine DMPs, and clade-VI, containing 16, were monocot and dicot specific. Interestingly, group V, comprising AtDMP3 and AtDMP5, did not contain any soybean DMP proteins. It is possible that the region containing these orthologues was deleted during the evolution of the soybean genome. Dicot-predominant clade-VII, with 42 DMPs, harbored AtDMP4, and AtDMP6 orthologues, including three soybean DMPs: Glyma.07G201500, Glyma.13G175000, and Glyma.13G235100.
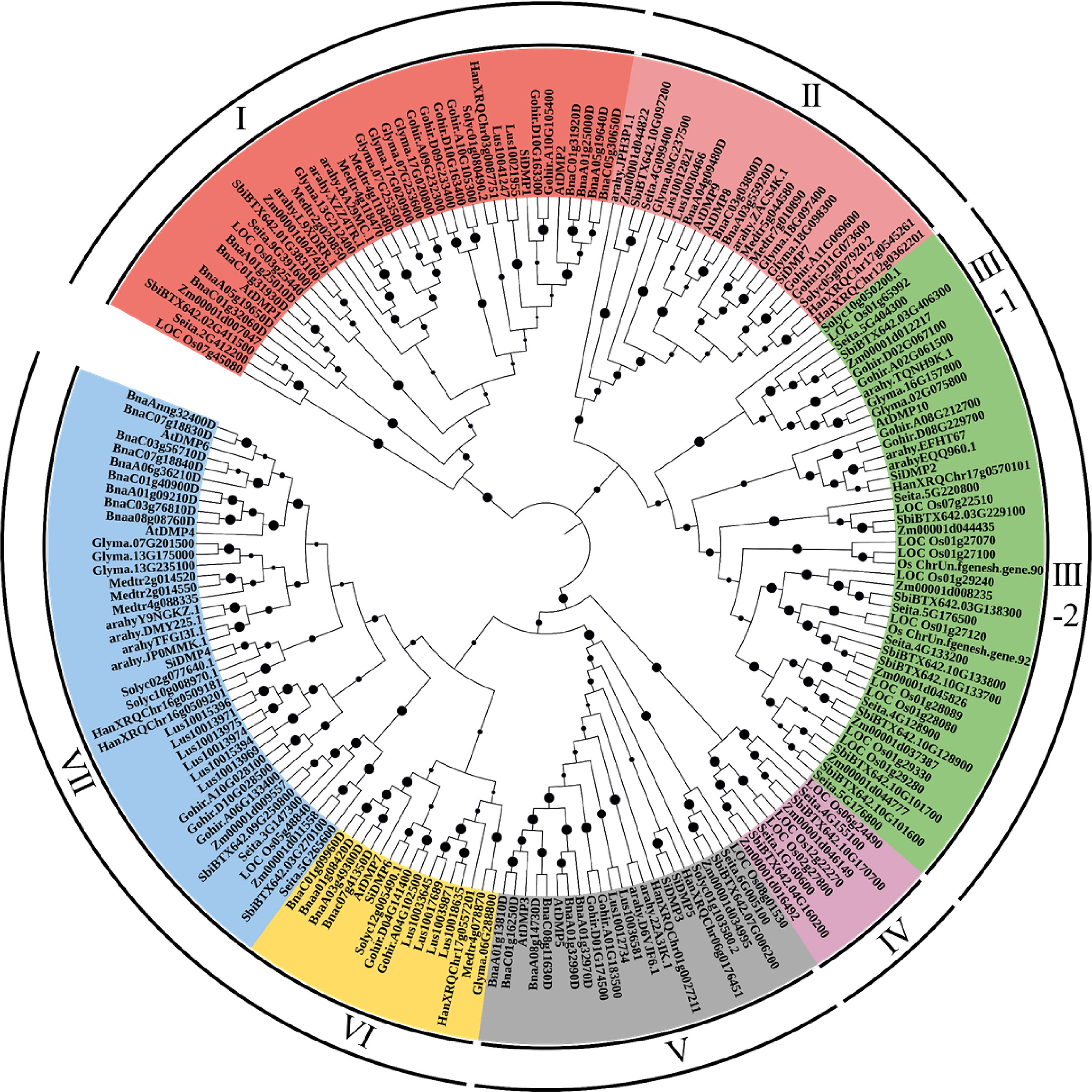
Figure 1 Phylogenetic relationships of the DMP proteins identified from monocot and dicot crops constructed using the N-J method with 1000 bootstraps in MEGAv11. Bootstrap values are shown on the nodes. DMP proteins were grouped into seven clades, which are denoted by color. For accession ID details of the genes, refer to Supplementary Table S1.
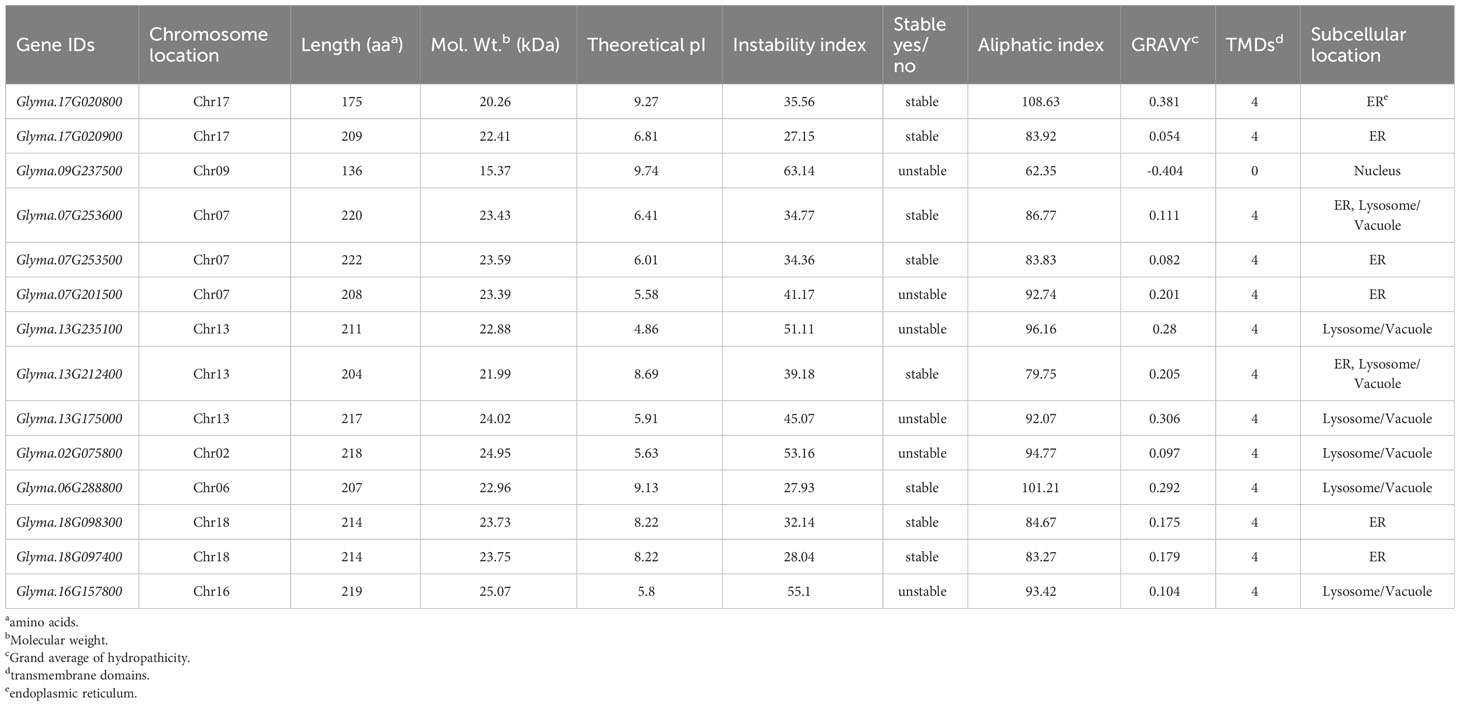
3.3 Physicochemical properties of soybean DMP proteins
The physicochemical properties of soybean DMP proteins were analyzed (Table 1). The GmDMP proteins varied in length, molecular weight, theoretical isoelectric point, aliphatic index, and other properties. The GmDMPs were predicted to encode polypeptides from 136 to 222 amino acids in length, with predicted molecular weights ranging from 15.37 to 25.07 kD. The predicted aliphatic indices ranged from 62.35 to 108.63. The aliphatic index indicates the thermostability and half-life of a protein (Kyte and Doolittle, 1982). The theoretical isoelectric points (pIs) ranged from 4.86 (Glyma.13G235100) to 9.74 (Glyma.09G237500), and the grand average of the hydropathicity values of all GmDMP proteins was positive except Glyma.09G237500, indicating their transmembrane nature. The subcellular localization was predicted using DeepLoc 2.0, and GmDMPs were found to be located on various parts of different cell organelles, including one in the nucleus, six in the ER, five in the lysosome/vacuole and two in ER, lysosome/vacuole both. All GmDMP proteins, except Glyma.09G237500, are predicted to have four transmembrane helix domains.
3.4 Exon-intron structure, motif, and sequence analysis
The exon and intron arrangement among the GmDMP gene family members was analyzed using the GSDS web server, revealing that, except for Glyma.07G201500, none possessed any introns. All gene structures of the DMP family had only one exon without an intron and with a conserved domain. To understand the diversity and similarity of gene structure and motif among the Arabidopsis and soybean proteins, we constructed a separate phylogenetic tree using GSDS (Figure 2). Among the ten motifs identified, five motifs encoded a DMP domain, and all DMP proteins have motifs ranging from 3 to 10 (Figure 2B, Supplementary Table S4). Members of the same group shared a somewhat common motif distribution pattern, suggesting their similar functional relevance and conserved protein architecture. Some motifs were absent in specific groups. For example, motifs 9 and 10 were absent in all the members of the clade I proteins, and motifs 8, 9, and 10 were absent in the members of subclade III-1. The structure and motif conservation within groups support the results of the phylogenetic analysis.
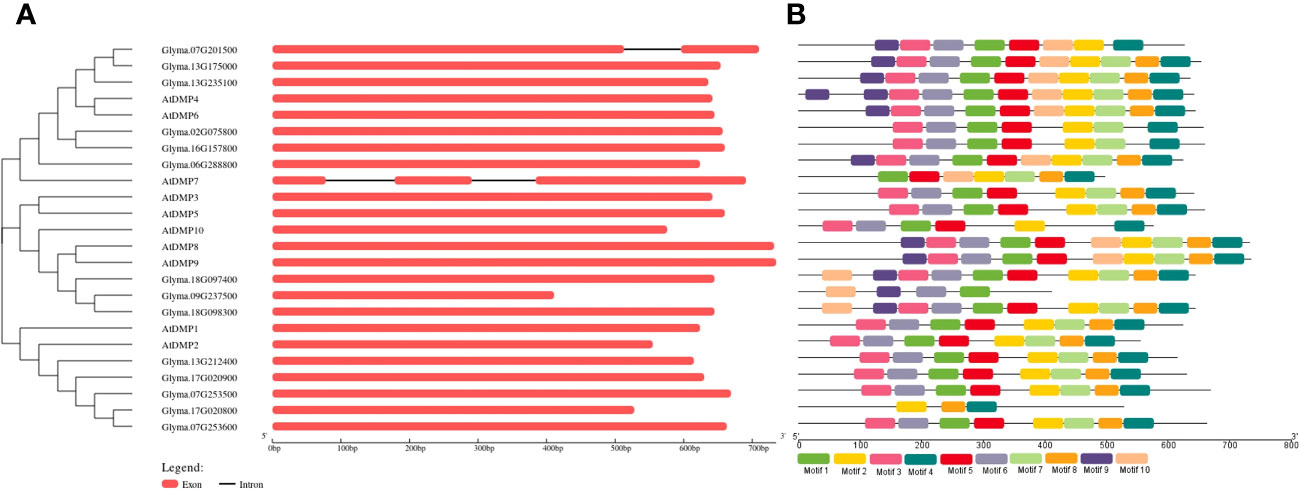
Figure 2 Genomic structure and motif composition comparisons between Arabidopsis and soybean DMP genes. (A) Phylogenetic tree and genomic structures of the genes. Exons and introns are indicated with boxes and black lines, respectively. (B) Motif composition of the DMP genes. Conserved motifs are indicated by colored boxes. For the details of each motif, refer to Supplementary Table S4.
3.5 Ka/Ks selective pressure analysis of soybean DMP gene family
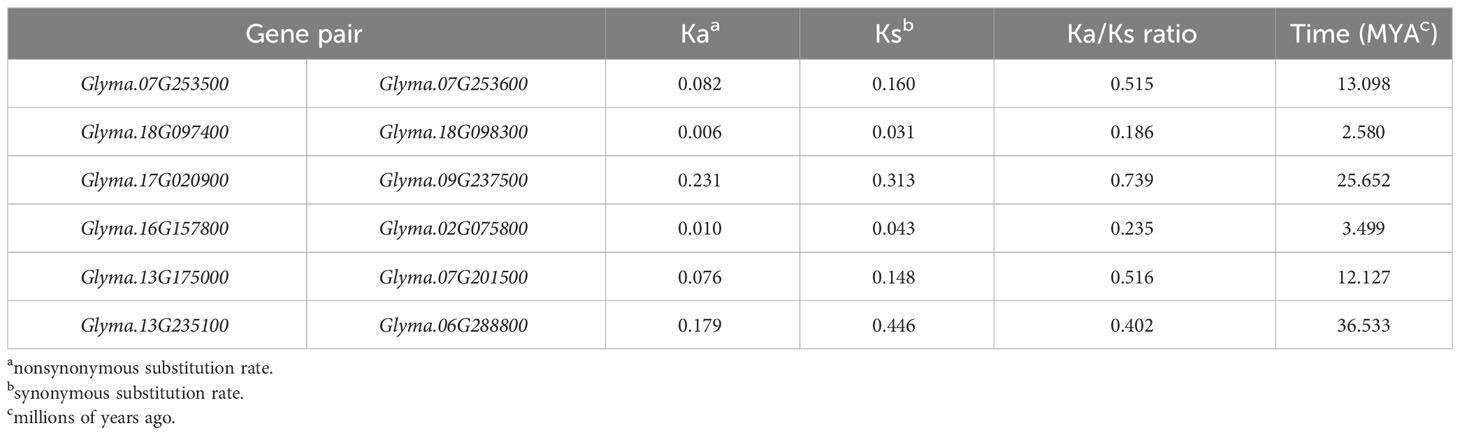
The substitution rates among the duplicated gene pairs were less than 1 (Table 2), signifying a strong purifying selective pressure during the evolution of the soybean DMP gene family. In addition, the approximate time of the duplication events were calculated to determine the extent and nature of selection pressure in GmDMP genes. The five GmDMP pairs were duplicated from 2.580 Mya (Glyma.18G097400 and Glyma.18G098300) to 36.533 Mya (Glyma.13G235100 and Glyma.06G288800) (Table 2). The soybean genome is reported to have experienced two rounds of whole genome duplication (WGD), the first duplication occurred prior to the divergence of legume subfamily Papilionoideae (58–60 Mya) and a recent Glycine-specific WGD occurred approximately 13 Mya (Schmutz et al., 2010). Among the six DMP duplicate gene pairs, three pairs were derived after the first WGD and three after the second WGD, including Glyma.07G253500 and Glyma.07G253600, Glyma.17G020900 and Glyma.09G237500, and Glyma.13G235100 and Glyma.06G288800.
3.6 Cis-element analysis of GmDMP gene promotors
The PlantCARE database-based analysis of the sequences upstream (2 kb) from the start codon of the DMP genes identified 56 CAREs (cis-acting regulatory elements; Supplementary Table S5). These CAREs are grouped into different categories based on their functional relevance, viz., growth and development-, light-, phytohormonal-, and stress-responsive elements (Figure 3; Supplementary Table S5). Binding sites for CAREs associated with stress responsiveness were predominant in most of the promoters; however, promoters of Glyma.06G288800 and Glyma.07G253600 had higher binding sites for light-responsive and phytohormonal-responsive elements (Figure 3B). The highest number of binding sites were detected in the Glyma.13G175000 gene promoter (65 sites) followed by Glyma.13G212400 (60 sites). In comparison, the lowest number of binding sites were identified in the Glyma.02G075800 promoter (25 sites). A higher availability of stress-responsive elements in promoters suggests that their expression is linked to and regulated by stressors. The detected stress-related CAREs included dehydration-responsive (Myeloblastosis-MYB, Myelocytometosis-MYC, and Myb binding site-MBS), low-temperature-responsive (LTR), defense- and stress-responsive (TC-rich repeats), elicitor-mediated activation (AT-rich sequence), anaerobic induction (ARE), fungal elicitor-responsive (W box), and wound-responsive (WRE3) elements.
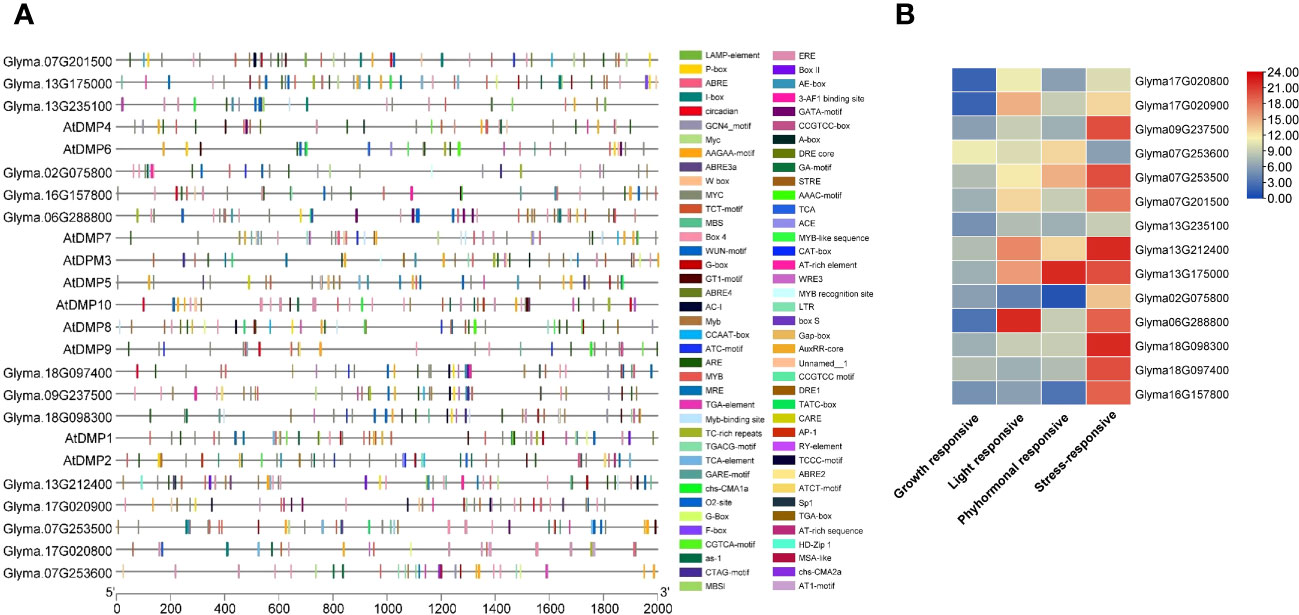
Figure 3 Cis-element analysis of the promoter regions of the DMP family. (A) Variation in different types of cis-acting regulatory elements (CAREs). (B) Graphical representation of CAREs. Different colored shapes represent the different elements. For details of each CARE binding site, refer to Supplementary Table S5.
In the phytohormonal-responsive groups, sites for ABRE (abscisic acid responsiveness element) were observed in all GmDMP genes except Glyma.02G075800 and Glyma.16G157800, while MeJA-responsive motifs, such as CGTCA and TGACG were found in 10 GmDMPs, and gibberellin-responsive motifs—such as P-box, GARE, and TATC-box—were found in 8, 6, and 1 GmDMPs, respectively. The auxin-responsive element TGA and salicylic acid-responsive element TCA appeared in five GmDMPs, and the ethylene-responsive element (ERE) in nine. The gene Glyma.07G253500 contained all six hormone-related CAREs in its promoter, while Glyma.02G075800 contained only salicylic acid-responsive elements. Most GmDMPs possessed at least three hormone-related elements, signifying their involvement in hormone metabolism processes and signal transduction networks regulating growth and developmental processes in soybean (Figure 3B). In addition, light-responsive elements—18 in total, including multiple copies of Box-4 and G-Box—were also found in most GmDMPs. Elements in this category are reported to play significant roles in light regulation and its related activities. Additionally, CAREs involved in growth and development responses—including circadian (circadian control), CCAAT-box (MYBHv1 binding site), MSA-like (cell cycle regulation), RY (seed-specific regulation), and AT-rich (binding site of AT-rich DNA binding protein) elements and the GCN4 motif (involved in endosperm expression)—were identified. These CAREs are associated with the vital processes of flowering, maturity, and seed development. Other CAREs involved in zein metabolism, flavonoid biosynthetic regulation, meristem expression, and secondary xylem development—Opaque2 (O2)-site (nine GmDMPs), Myb binding site-MBSI (Glyma.13G235100), CAT-box (three GmDMPs), and the AAGAA-motif (12 GmDMPs), respectively—were also detected (Supplementary Table S5). These results suggest that the DMP family genes may be involved in development and growth, hormone response processes, and stress responses in soybean.
3.7 Expression profiles in the soybean DMP gene family
3.7.1 Tissue-specific expression patterns
To better understand the function of the soybean GmDMP genes, data from publicly available transcriptome datasets and Genevestigator were used to investigate the expression profiles of DMP genes in various tissues under different stresses. We found that among GmDMP genes, Glyma.13G212400 is highly expressed in all organs, followed by Glyma.13G235100 and Glyma.17G020800. Two GmDMPs, Glyma.18G097400 and Glyma.18G098300, were expressed only in flower tissues, while another two, Glyma.07G253600 and Glyma.07G253500, showed root-specific expression profiles (Figure 4A, Supplementary Table S6). The gene Glyma.06G288800 was only slightly expressed in the pods. In the Genevestigator database, like transcriptome data, Glyma.13G212400 expressed in all organs, and Glyma.18G097400 and Glyma.18G098300 showed anther-specific expression. Seven GmDMPs were mainly expressed in the anther, suggesting the role of these genes in regulating reproductive development (Supplementary Figure S1). To investigate the age-dependent expression of GmDMPs, public expression data repositories for young (20-day-old) and mature (80-day-old) leaf samples were investigated. Most of the GmDMP genes showed increased expression during the mature stage, including the highest increase in Glyma.02G075800, followed by Glyma.09G237500, Glyma.07G253600 and then Glyma.07G253500. At the same time, the expression of two genes, Glyma.07G201500 and Glyma.13G235100, decreased as leaves aged (Figure 4A, Supplementary Table S6).

Figure 4 Expression pattern of soybean DMP genes. (A) Tissue- and age-dependent expression of GmDMP genes. (B) Expression profiles of GmDMP genes in response to abiotic and biotic stresses (SAM: shoot apical meristem, hpi: hours post-inoculation, SCN: soybean cyst nematode). Scale bar represent mean Z-score obtained from normalization of FPKM values of three replicates. Red represents high expression; green indicates low expression. For details of each gene expression, refer to Supplementary Table S6.
3.7.2 Expression under abiotic and biotic stresses
To explore the potential functions of soybean DMP genes in response to abiotic and biotic stressors, we analyzed previously reported Illumina RNA-seq data collected under drought, salt, and heat stress as well as biotic stresses including Fusarium oxysporum, Macrophomina phasiolina, soybean cyst nematode (SCN), and aphid infestation (Supplementary Table S2). The expression levels and patterns of GmDMPs varied considerably (Figure 4B, Supplementary Table S6). Genes Glyma.07G253600, Glyma.09G237500, and Glyma.13G235100 had higher expression under 37 days of drought stress. Under salt stress, Glyma.17G020800, Glyma.02G075800, and Glyma.09G237500 were highly expressed, while six genes—Glyma.07G253500, Glyma.17G020900, Glyma.06G288800, Glyma.07G201500, Glyma.13G175000, and Glyma.13G235100—were found to have higher transcription during heat stress. Looking at biotic stressors, Fusarium infection strongly upregulated the expression of Glyma.07G253600, Glyma.13G212400, Glyma.17G020900, and Glyma.13G175000. Interestingly, the transcripts of Glyma.18G097400 and Glyma.18G098300 showed a significantly increase under Fusarium infection, while their expression remained negligible under all other stress conditions. During Macrophomina infection, four genes—Glyma.07G253500, Glyma.13G212400, Glyma.13G235100, and Glyma.02G075800—were highly upregulated in resistant genotype but not in more susceptible ones, while two genes, Glyma.17G020800 and Glyma.13G175000, were highly down-regulated. Under insect and nematode infestation, DMP genes such as Glyma.07G253500, Glyma.07G253600, Glyma.13G212400, Glyma.02G075800, Glyma.13G175000, and Glyma.13G235100 showed higher transcript abundance. The differential expression patterns of soybean DMP genes illustrated that they play an important role in responses to diverse abiotic and biotic stressors.
3.7.3 qPCR validation
We selected nine genes to investigate tissue-specific expression patterns at the reproductive stage (60-day-old, early flowering-stage plants) of soybean. In our qPCR analysis, we saw similar gene expression patterns to those in the in silico expression analysis (Figure 5), indicating the reliability of the computational analysis. To gain a better understanding of the expression in reproductive organs, we conducted an in-depth analysis of flower parts. Most of the GmDMP genes were much more highly expressed in reproductive tissues than in leaf and stem tissues. Notably, among the three AtDMP8/9 orthologues, Glyma.18G097400 displayed pollen-specific expression, Glyma.18G098300 had its highest expression in the pistil, and Glyma.09G237500 showed high expression in both pollen and pistils. In addition, Glyma.13G212400, Glyma.06G288800, Glyma.07G253600, Glyma.17G020800, and Glyma.13G235100 exhibited their highest expression levels in sepals. Consistent with the in silico expression pattern, Glyma.13G212400 showed high expression in all organs, followed by Glyma.13G235100. Overall, our findings provide valuable insights into the tissue-specific expression patterns of soybean DMP genes and highlight their potential roles in reproductive processes.
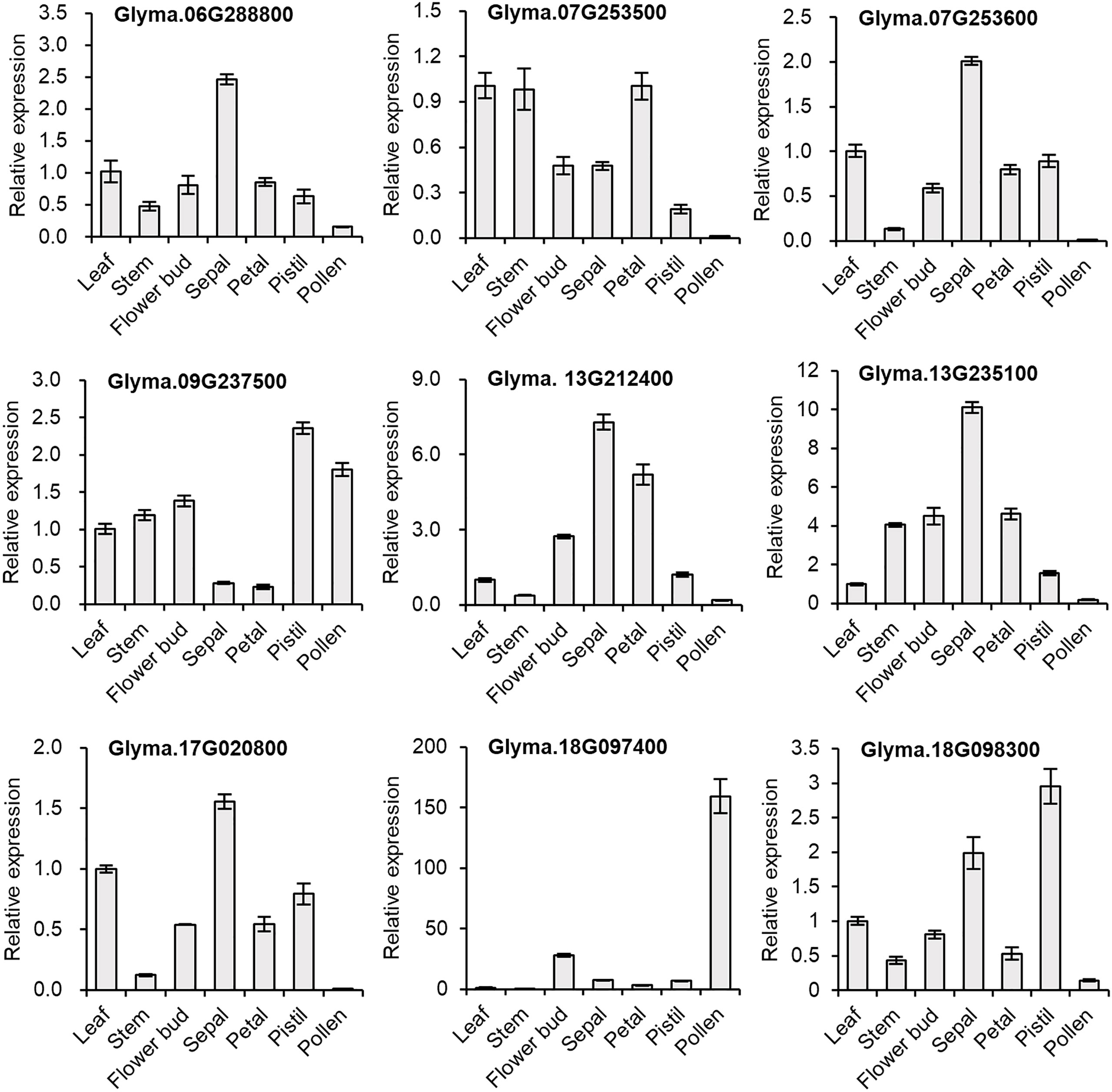
Figure 5 Relative expression levels of selected GmDMP genes in different tissues as quantified by qPCR analysis. Each data point represents the average ± SE from three independent technical replicates, obtained from pooled single biological sample from four plants.
3.8 In vitro cleavage efficiency analysis
As the first step in identifying sgRNAs that could efficiently edit the target genes in soybean, we conducted an in vitro cleavage assay to assess the endonuclease activity of sgRNA candidates We selected AtDMP8/9 orthologues, Glyma.18G097400 (GmDMP8) and Glyma.18G098300 (GmDMP9), which have 97.6% identity and exhibit pollen-specific and pistil-abundant expression patterns, respectively. Through in silico prediction, a set of sgRNAs targeting the conserved sites of these genes was identified, and two sgRNAs were chosen for the in vitro cleavage assay (Supplementary Table S7). The assay demonstrated that both sgRNAs effectively guided Cas9 to cleave the target DNA sequences, resulting in the generation of specific fragment sizes. In the SgDMP#1 assay, 607 and 295 bp fragments were generated, while the SgDMP#2 assay produced fragments of 624 and 278 bp (Figure 6). Notably, based on intensity digestion of target PCR product, SgDMP#1 displayed higher cleavage efficiency compared to SgDMP2.
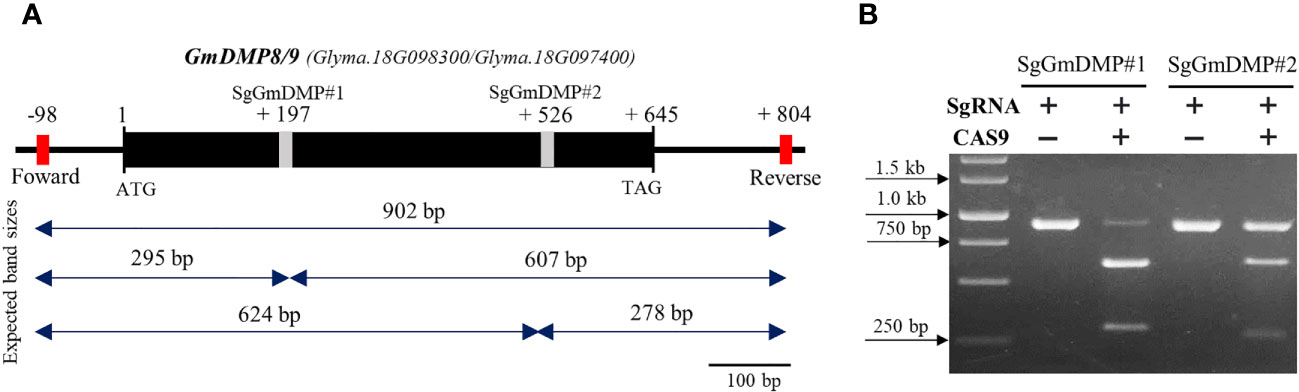
Figure 6 In vitro SgRNA cleavage efficiency analysis. (A) Genomic DNA structure of GmDMP8 and GmDMP9. Grey boxes indicate sgRNA target sites. Expected band sizes after cleavage assay using each sgRNA were indicated by double sided arrow. (B) In vitro cleavage assay to check the efficiency of selected SgRNAs from GmDMP8 and GmDMP9 genes.
4 Discussion
With the availability of high-throughput technologies, genome datasets, and bioanalytical tools, soybean is receiving growing attention in genome-wide gene analyzes, identifying and characterizing multiple gene copies in each gene family. In this study, we mined the soybean genome for DUF679 family genes and characterized them using publicly available transcriptome datasets and various bioinformatics tools. A total of 14 DMP genes were identified in the soybean genome, which was in the range of other dicots and monocots (Kasaras and Kunze, 2010; Cyprys et al., 2019). The lengths of the identified DMP proteins ranged from 136 to 222 amino acids (Table 1). The instability index showed that eight DMP proteins are stable, whereas six are unstable. The majority of the GmDMPs had positive GRAVY index scores, suggesting that they might be membrane proteins, mainly interacting with hydrophobic regions that span membranes, rather than globular proteins. Peptides with fewer charged groups are generally less soluble in water and are disposed to aggregation in and interaction with hydrophobic pockets of larger proteins (Lawrence et al., 2007; Niwa et al., 2009).
In the phylogenetic analysis, seven major clades were formed. The monocot-/dicot-specific sub-clustering patterns corroborate previous studies, suggesting a common ancestor before the divergence of monocot and dicot DMP genes (Cyprys et al., 2019; Zhu et al., 2021). The pollen-specific AtDMP8/9 orthologues (23 DMPs) included three soybean DMPs (Glyma.09G237500, Glyma.18G097400, and Glyma.18G098300). Orthologues of AtDMP8/9 were utilized in a DMP-based haploid inducer system to efficiently induce maternal haploids in maize (Zhong et al., 2019), Medicago truncatula (Wang et al., 2022), Brassica napus (Zhong et al., 2022b), tobacco (Zhang et al., 2022b), and tomato (Zhong et al., 2022a). Our qPCR analysis revealed that among the AtDMP8/9 orthologues, Glyma.18G097400 and Glyma.18G098300 exhibited pollen-specific and pistil-abundant expression patterns, respectively. While, Glyma.09G237500 was highly expressed in reproductive organs, pollen and pistil, at the reproductive stage (Figure 5). This suggests their potential involvement in pollination and fertilization. A recent study demonstrated that the loss of function of the gynoecium-expressed phospholipase AII (pPLAIIγ) created haploid plants in dicotyledonous Arabidopsis (Jang et al., 2023), where the gynoecium-expressed pPLAIIγ induced female organ-mediated haploid induction. This highlights a potential of pistil-expressed Glyma.09G237500 and Glyma.18G098300 in female organ-mediated haploid induction, which could be further explored in future studies. So far, pollen-specific phospholipase A type of gene named MTL/NLD/ZmPLA1 was reported to the functional maternal haploid inducer in monocotyledonous plants such as maize, rice, wheat, and foxtail millet (Gilles et al., 2017; Kelliher et al., 2017; Liu et al., 2017; Yao et al., 2018; Liu et al., 2020; Cheng et al., 2021). Thus, we suggest that Glyma.18G097400 might be the potential target gene of maternal haploid inducer. Female organ-mediated haploid induction lines could be a promising tool as they allow the production of haploid plants without laborious emasculation procedures. Additionally, depending on germ lines, it may be more efficient than male-mediated haploid induction, as the female reproductive organ is the site of fertilization and zygote formation, providing a more direct route to haploid embryo production.
Subsequently, in an in vitro cleavage assay, we observed variations in the digestion efficiency of SgRNAs. SgRNA#1, targeting Glyma.18G097400, and Glyma.18G098300, exhibited higher cleavage efficiency compared to SgRNA#2, showing more cleaved bands from the originally amplified band size (Figure 6). Comparative studies, have found that RNP complexes capable of efficiently cleaving their target sites in vitro consistently produce similar results in protoplast-based screening methods (Kim et al., 2017; Jeong et al., 2019; Kim et al., 2020; Sant’Ana et al., 2020; Subburaj et al., 2022). Notably, the use of this in vitro cleavage protocol offers several advantages over alternative methods. All required components are commercially available or can be easily produced in the laboratory at a low cost (Mehravar et al., 2019; Bente et al., 2020). Additionally, the entire procedure can be completed within a single day. These findings highlight the effectiveness of the rapid, simple, and cost-effective in vitro cleavage protocol in eliminating inefficient candidate SgRNAs and identifying those with optimal performance, thereby increasing the likelihood of successful in vivo functions.
To elucidate the potential regulatory roles of GmDMPs in the development and stress response of soybean, we studied the distribution and frequency of CAREs. The identification of CAREs is currently an intriguing area, allowing the study of complex gene expression by integrating computational, comparative, and functional genomics (Li et al., 2015; Ho and Geisler, 2019; Nawade et al., 2022). The light-responsive elements (LREs) were found to be prevalent in the GmDMP promoters. Most common LREs, Box 4,G-box, and GT1-motif have been demonstrated to be critical for the regulation of light-mediated transcriptional activity (Gangappa et al., 2013; Ezer et al., 2017). The highly conserved G-box motif (CACGTG) binds to the basic helix-loop-helix (bHLH) and basic leucine zipper (bZIP) families of protein motifs (Heim et al., 2003; Carretero-Paulet et al., 2010) and reported to involved in the regulation of chlorophyll biosynthesis in Arabidopsis (Menkens et al., 1995; Kobayashi et al., 2012). The GATA motif (detected in 4 GmDMPs) plays a role in light responsiveness and tissue specificity, and is involved in the light-dependent development of phloem tissues (Trishla et al., 2020). In photosynthetic-responsive gene promoters, both I-box and G-box elements have been shown to be essential for activation in response to phytochrome, cryptochrome, and plastid signals. (Martıínez-Hernández et al., 2002; López-Ochoa et al., 2007). Twelve GmDMP promoters have either I-box or G-box, suggesting their functional relevance as regulators of ribulose 1,5-bisphosphate carboxylase/oxygenase small subunit light-responsive units (Murata et al., 2002). The promoter of Glyma.18G098300 was found to have a site for the sp1 (GGGCGG) element (Figure 3, Supplementary Table S5), which is considered a mammalian promoter element, implicated in the regulation of a wide variety of housekeeping genes and tissue-specific genes (Hagen and Guilfoyle, 2002). Interestingly it was not found in the Arabidopsis or rice genomes (Yamamoto et al., 2007). The AAGAA-motif, activation sequence-1 (as-1; TGACG), O2-site, and GCN4_motif were the most frequently distributed CAREs related to plant growth and development (Figure 3, Supplementary Table S5). The as-1 element was found to be responsible for auxin- or salicylic acid-dependent enhanced expression in leaves (Niggeweg et al., 2000), whereas the AAGAA-motif was associated with secondary xylem development (Ain-Ali et al., 2021).
The promoters of soybean DMP genes have been also found to contain CAREs that are known to modulate gene expression in response to various stresses. Among abiotic stress-responsive CAREs, MYC, and MYB binding sites were present in all promoters. They have been reported to play an important role in drought-inducible expression, indicating that GmDMP expression is associated with abiotic stress (Smita et al., 2015). Glyma.07G201500, Glyma.09G237500, and Glyma.07G253500, which showed higher expression during drought, harbored multiple binding sites for dehydration-responsive elements (Figure 3, Supplementary Table S5). Moreover, the presence of LTR (low temperature responsive) in the Glyma.16G157800 promoter suggests its involvement in cold stress response (Baker et al., 1994; Zhang et al., 2020). The ARE (anaerobic responsive elements) motifs, known as low oxygen and dehydration-induced elements (Dolferus et al., 2001), were found in 10 GmDMPs (Supplementary Table S5). The abscisic acid responsive elements (ABREs) have higher binding sites in most of the GmDMP promoters (Figure 3, Supplementary Table S5). They are regulators of various processes, including stomatal closure and seed and bud dormancy, as well as mediators in plant responses to cold, drought, and salinity stress (Choi et al., 2000; Yoshida et al., 2015). CAREs involved in the methyl-jasmonate responsive elements (TGACG-motif and CGTCA-motif) were also present in 10 GmDMP promoters (Figure 3, Supplementary Table S5). These elements are crucial in TF-mediated gene regulation (Rouster et al., 1997). For example, TGA1, a bZIP TF, was reported to act as a positive regulator of disease resistance by binding at the TGACG-motif and CGTCA-motif of the pathogenesis-related (PR-1) gene promoter in the Arabidopsis (Shearer et al., 2012). Likewise, the TGACG motif from the rice 12-oxophytodienoic acid reductase-1 (OsOPR1) promoter has been shown to play essential roles in defense responses (Sobajima et al., 2007). Moreover, the existence of W-box, WRE3, and the WUN-motif in promoters suggested that GmDMPs might play a vital role in biotic stress responses. The WUN-motif was characterized as wound responsive in the WUN1 gene (Siebertz et al., 1989), while WRKY TFs are known to function in wound response by binding to W-boxes (Eulgem et al., 2000). Moreover, the presence of these CAREs and their elevated expression upon Fusarium infection suggest the possible role of Glyma.18G097400 and Glyma.18G098300 in fungal response (Figure 4B, Supplementary Table S6). Between the two GmDMPs in sub-clade III-1, Glyma.16G157800 and Glyma.02G075800, Glyma.02G075800 expression was 3-fold more elevated in Macrophomina resistant genotype. Notably, these AtDMP10 orthologue harbors wounding and pathogen responsive CAREs (Supplementary Table S5). The W-box (TTGACC) element, which interacts with WRKY transcription factors (TFs) and regulates the expression of defense-related (pathogenesis-related 10, PR-10) genes, has a role in biotic and abiotic stresses, seed dormancy, and senescence (Dhatterwal et al., 2019). Under different biotic and abiotic stresses, the DMP genes exhibited diverse expression patterns and presence of stress responsive CAREs, indicating their potential roles in improving stress resistance and survival in soybean (Figure 7). However, further studies are required to validate the functional relevance of these genes during stress.
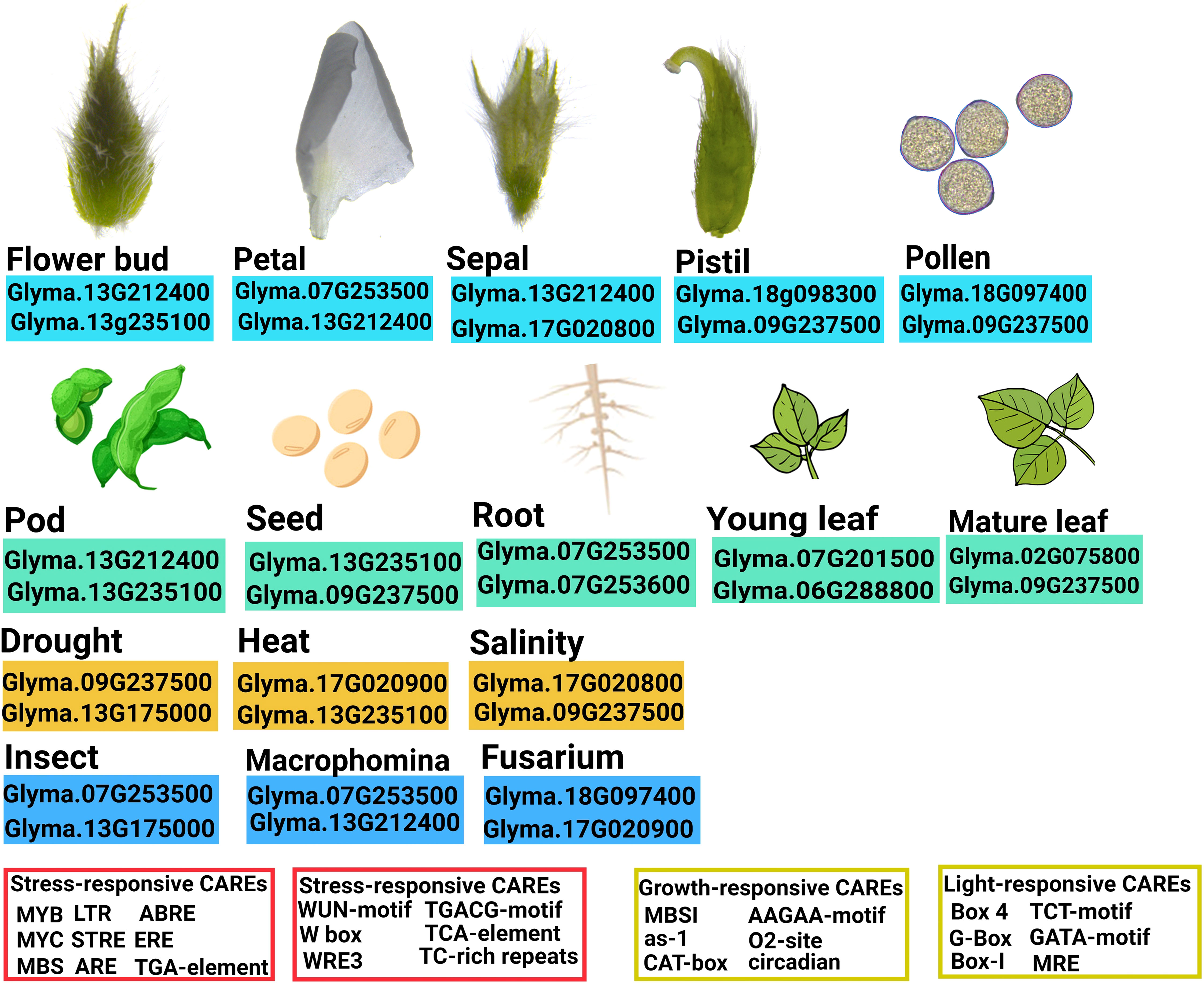
Figure 7 Overview of the expression profiles and key cis-acting regulatory elements of soybean DMP genes.
In summary, DMP genes retrieved from soybean were subjected to bioinformatics analyzes for characterization. In the phylogenetic analyzes, group V, composed of AtDMP3 and AtDMP5, orthologues were absent from the soybean genome. The AtDMP8/9 orthologues Glyma.18G097400 and Glyma.18G098300, showing pollen-specific and pistil-abundant expression, could be potential targets for haploid induction in soybean. The expression profiles for most GmDMPs showed varying expression patterns in response to biotic and abiotic stresses. The presence of binding sites for various regulatory elements in GmDMP promoter sequences is consistent with this divergent expression pattern and implicates their possible involvement in growth and stress responses.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
OL acquired the funds and resources, designed the experiment, and administered and supervised the project. OL, BN, and TB conceptualized the project. OL, BN, JL, and JJ designed the methodology, curated the data, and performed the formal analysis. TB performed the RNA-Seq analysis. BN and TB implemented the software and visualized and validated the data. OL, BN, TB, and JJ wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIT) (2019R1A2C1004140 and RS-2023-00217064); a grant from the New Breeding Technologies Development program (No. PJ01652301), Rural Development Administration, Republic of Korea
Acknowledgments
We extend our sincere gratitude to the researchers from DOE Joint Genome Institute, USA; Nanjing Agricultural University, China; Universidade Federal de Viçosa, Brazil; Shenyang Agricultural University, China; University of North Texas, USA; Yangzhou University, China; Iowa State University, USA; Jawaharlal Nehru Agriculture University, India; and South Dakota State University, USA for their invaluable contributions in submitting the RNA-sequencing datasets to the NCBI public database.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1216082/full#supplementary-material
References
Ain-Ali, Q., Mushtaq, N., Amir, R., Gul, A., Tahir, M., Munir, F. (2021). Genome-wide promoter analysis, homology modeling and protein interaction network of dehydration responsive element binding (DREB) gene family in Solanum tuberosum. PloS One 16, e0261215. doi: 10.1371/journal.pone.0261215
Bailey, T. L., Williams, N., Misleh, C., Li, W. W. (2006). MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, 369–373. doi: 10.1093/nar/gkl198
Baker, S. S., Wilhelm, K. S., Thomashow, M. F. (1994). The 5’-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Bente, H., Mittelsten Scheid, O., Donà, M. (2020). Versatile in vitro assay to recognize Cas9-induced mutations. Plant Direct. 4 (9), e00269. doi: 10.1002/pld3.269
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martínez-García, J. F., Bilbao-Castro, J. R., Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Cheng, Z., Sun, Y., Yang, S., Zhi, H., Yin, T., Ma, X., et al. (2021). Establishing in planta haploid inducer line by edited SiMTL in foxtail millet (Setaria italica). Plant Biotechnol. J. 19, 1089–1091. doi: 10.1111/pbi.13584
Choi, H., Hong, J., Ha, J., Kang, J., Kim, S. Y. (2000). ABFs, a family of ABA-responsive element binding factors. J. Biol. Chem. 275, 1723–1730. doi: 10.1074/jbc.275.3.1723
Cyprys, P., Lindemeier, M., Sprunck, S. (2019). Gamete fusion is facilitated by two sperm cell-expressed DUF679 membrane proteins. Nat. Plants. 5, 253–257. doi: 10.1038/s41477-019-0382-3
Dhatterwal, P., Basu, S., Mehrotra, S., Mehrotra, R. (2019). Genome wide analysis of W-box element in Arabidopsis thaliana reveals TGAC motif with genes down regulated by heat and salinity. Sci. Rep. 9, 1681. doi: 10.1038/s41598-019-38757-7
Dolferus, R., Klok, E. J., Ismond, K., Delessert, C., Wilson, S., Good, A., et al. (2001). Molecular basis of the anaerobic response in plants. IUBMB Life. 51, 79–82. doi: 10.1080/15216540120263
El-Gebali, S., Mistry, J., Bateman, A., Eddy, S. R., Luciani, A., Potter, S. C., et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, 427–432. doi: 10.1093/nar/gky995
Eulgem, T., Rushton, P. J., Robatzek, S., Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Ezer, D., Shepherd, S. J. K., Brestovitsky, A., Dickinson, P., Cortijo, S., Charoensawan, V., et al. (2017). The G-Box transcriptional regulatory code in Arabidopsis. Plant Physiol. 175, 628–640. doi: 10.1104/pp.17.01086
Gangappa, S. N., Maurya, J. P., Yadav, V., Chattopadhyay, S. (2013). The regulation of the Z- and G-Box containing promoters by light signaling components, SPA1 and MYC2, in Arabidopsis. PloS One 8, e62194. doi: 10.1371/journal.pone.0062194
Gao, Z., Daneva, A., Salanenka, Y., Van Durme, M., Huysmans, M., Lin, Z., et al. (2018). KIRA1 and ORESARA1 terminate flower receptivity by promoting cell death in the stigma of Arabidopsis. Nat. Plants. 4, 365–375. doi: 10.1038/s41477-018-0160-7
Gasteiger, E., Hoogland, C., Gattiker, A., Wilkins, M. R., Appel, R. D., Bairoch, A. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook. Ed. Walker, J. M. (Totowa, New Jersey: Springer), 571–607. doi: 10.1385/1-59259-890-0:571
Gilles, L. M., Martinant, J.-P., Rogowsky, P. M., Widiez, T. (2017). Haploid induction in plants. Curr. Biol. 27, 1095–1097. doi: 10.1016/j.cub.2017.07.055
Hagen, G., Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Hallgren, J., Tsirigos, K. D., Pedersen, M. D., Armenteros, J. J. A., Marcatili, P., Nielsen, H., et al. (2022). DeepTMHMM predicts alpha and beta transmembrane proteins using deep neural networks. bioRxiv. doi: 10.1101/2022.04.08.487609
Hartman, G. L., West, E. D., Herman, T. K. (2011). Crops that feed the world 2. Soybean—worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 3, 5–17. doi: 10.1007/s12571-010-0108-x
Heim, M. A., Jakoby, M., Werber, M., Martin, C., Weisshaar, B., Bailey, P. C. (2003). The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 20, 735–747. doi: 10.1093/molbev/msg088
Ho, C. L., Geisler, M. (2019). Genome-wide computational identification of biologically significant cis-regulatory elements and associated transcription factors from rice. Plants 8, 441. doi: 10.3390/plants8110441
Hruz, T., Laule, O., Szabo, G., Wessendorp, F., Bleuler, S., Oertle, L., et al. (2008). Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Adv. Bioinf. 2008, 420747. doi: 10.1155/2008/420747
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jang, J. H., Seo, H. S., Widiez, T., Lee, O. R. (2023). Loss-of-function of gynoecium-expressed phospholipase pPLAIIγ triggers maternal haploid induction in Arabidopsis. New Phytol. 238, 1813–1824. doi: 10.1111/nph.18898
Jeong, S. Y., Ahn, H., Ryu, J., Oh, Y., Sivanandhan, G., Won, K. H., et al. (2019). Generation of early-flowering Chinese cabbage (Brassica rapa spp. pekinensis) through CRISPR/Cas9-mediated genome editing. Plant Biotech. Rep. 13, 491–499. doi: 10.1007/s11816-019-00566-9
Karmakar, S., Behera, D., Baig, M. J., Molla, K. A. (2021). In vitro Cas9 cleavage assay to check guide RNA efficiency. CRISPR-Cas Methods 2, 23–39. doi: 10.1007/978-1-0716-1657-4_3
Kasaras, A., Kunze, R. (2010). Expression, localisation and phylogeny of a novel family of plant-specific membrane proteins. Plant Biol. 12, 140–152. doi: 10.1111/j.1438-8677.2010.00381.x
Kasaras, A., Melzer, M., Kunze, R. (2012). Arabidopsis senescence-associated protein DMP1 is involved in membrane remodeling of the ER and tonoplast. BMC Plant Biol. 12, 54. doi: 10.1186/1471-2229-12-54
Kelliher, T., Starr, D., Richbourg, L., Chintamanani, S., Delzer, B., Nuccio, M. L., et al. (2017). MATRILINEAL, a sperm-specific phospholipase, triggers maize haploid induction. Nature 542, 105–109. doi: 10.1038/nature20827
Kim, H., Choi, J., Won, K. H. (2020). A stable DNA-free screening system for CRISPR/RNPs-mediated gene editing in hot and sweet cultivars of Capsicum annuum. BMC Plant Biol. 20, 1–12. doi: 10.1186/s12870-020-02665-0
Kim, M. Y., Kang, Y. J., Lee, T., Lee, S. H. (2013). Divergence of flowering-related genes in three legume species. Plant Genome. 6, 3. doi: 10.3835/plantgenome2013.03.0008
Kim, H., Kim, S. T., Ryu, J., Kang, B. C., Kim, J. S., Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8 (1), 14406. doi: 10.1038/ncomms14406
Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360. doi: 10.1038/nmeth.3317
Kobayashi, K., Obayashi, T., Masuda, T. (2012). Role of the G-box element in regulation of chlorophyll biosynthesis in Arabidopsis roots. Plant Signal Behav. 7, 922–926. doi: 10.4161/psb.20760
Krueger, F. (2015). Trim Galore!: A wrapper around Cutadapt and FastQC to consistently apply adapter and quality trimming to FastQ files, with extra functionality for RRBS data. Babraham Institute 516.
Kyte, J., Doolittle, R. F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. doi: 10.1016/0022-2836(82)90515-0
Lawrence, M. S., Phillips, K. J., Liu, D. R. (2007). Supercharging proteins can impart unusual resilience. J. Am. Chem. Soc 129, 10110–10112. doi: 10.1021/ja071641y
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Bork, P. (2018). 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46, 493–496. doi: 10.1093/nar/gkx922
Letunic, I., Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, Y., Chen, C. Y., Kaye, A. M., Wasserman, W. W. (2015). The identification of cis-regulatory elements: A review from a machine learning perspective. Biosystems 138, 6–17. doi: 10.1016/j.biosystems.2015.10.002
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8 (1), 14261. doi: 10.1038/ncomms14261
Liu, C., Li, X., Meng, D., Zhong, Y., Chen, C., Dong, X., et al. (2017). A 4-bp insertion at ZmPLA1 encoding a putative phospholipase A generates haploid induction in maize. Mol. Plant 10, 520–522. doi: 10.1016/j.molp.2017.01.011
Liu, C., Zhong, Y., Qi, X., Chen, M., Liu, Z., Chen, C., et al. (2020). Extension of the in vivo haploid induction system from diploid maize to hexaploid wheat. Plant Biotechnol. J. 18, 316–318. doi: 10.1111/pbi.13218
López-Ochoa, L., Acevedo-Hernández, G., Martínez-Hernández, A., Argüello-Astorga, G., Herrera-Estrella, L. (2007). Structural relationships between diverse cis-acting elements are critical for the functional properties of a rbcS minimal light regulatory unit. J. Exp. Bot. 58, 4397–4406. doi: 10.1093/jxb/erm307
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01904
Marchler-Bauer, A., Lu, S., Anderson, J. B., Chitsaz, F., Derbyshire, M. K., DeWeese-Scott, C., et al. (2010). CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229. doi: 10.1093/nar/gkq1189
Martıínez-Hernández, A., López-Ochoa, L., Argüello-Astorga, G., Herrera-Estrella, L. (2002). Functional properties and regulatory complexity of a minimal RBCS light-responsive unit activated by phytochrome, cryptochrome, and plastid signals. Plant Physiol. 128, 1223–1233. doi: 10.1104/pp.010678
Mehravar, M., Shirazi, A., Mehrazar, M. M., Nazari, M. (2019). In vitro pre-validation of gene editing by CRISPR/Cas9 ribonucleoprotein. Avicenna J. @ Med. Biotech. 11 (3), 259.
Menkens, A. E., Schindler, U., Cashmore, A. R. (1995). The G-box: a ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. doi: 10.1016/s0968-0004(00)89118-5
Moebes, M., Kuhlmann, H., Demidov, D., Lermontova, I. (2022). Optimization of quantitative reverse transcription PCR method for analysis of weakly expressed genes in crops based on rapeseed. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.954976
Murata, J., Takase, H., Hiratsuka, K. (2002). Characterization of a novel GT-box binding protein from Arabidopsis. Plant Biotechnol. 19, 103–112. doi: 10.5511/plantbiotechnology.19.103
Murovec, J., Guček, K., Bohanec, B., Avbelj, M., Jerala, R. (2018). DNA-free genome editing of Brassica oleracea and B. rapa protoplasts using CRISPR-Cas9 ribonucleoprotein complexes. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01594
Nawade, B., Kumar, A., Maurya, R., Subramani, R., Yadav, R., Singh, K., et al. (2022). Longer duration of active oil biosynthesis during seed development is crucial for high oil yield—lessons from genome-wide in silico mining and RNA-seq validation in sesame. Plants 11, 2980. doi: 10.3390/plants11212980
Niggeweg, R., Thurow, C., Kegler, C., Gatz, C. (2000). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. doi: 10.1074/jbc.M909267199
Niwa, T., Ying, B. W., Saito, K., Jin, W., Takada, S., Ueda, T., et al. (2009). Bimodal protein solubility distribution revealed by an aggregation analysis of the entire ensemble of Escherichia coli proteins. Proc. Natl. Acad. Sci. U S A. 106, 4201–4206. doi: 10.1073/pnas.0811922106
Olvera-Carrillo, Y., Van Bel, M., Van Hautegem, T., Fendrych, M., Huysmans, M., Simaskova, M., et al. (2015). A conserved core of programmed cell death indicator genes discriminates developmentally and environmentally induced programmed cell death in plants. Plant Physiol. 169, 2684–2699. doi: 10.1104/pp.15.00769
Park, J., Bae, S., Kim, J. S. (2015). Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31 (24), 4014–4016. doi: 10.1093/bioinformatics/btv537
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T. C., Mendell, J. T., Salzberg, S. L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Rajendran, A., Lal, S. K., Raju, D., Ramlal, A. (2022). Associations of direct and indirect selection for pregermination anaerobic stress tolerance in soybean (Glycine max). Plant Breed. 141, 634–643. doi: 10.1111/pbr.13048
Rego, E. C. S., Pinheiro, T. D. M., Antonino, J. D., Alves, G. S. C., Cotta, M. G., Fonseca, F. C. D. A., et al. (2019). Stable reference genes for RT-qPCR analysis of gene expression in the Musa acuminata-Pseudocercospora musae interaction. Sci. Rep. 9, 14592. doi: 10.1038/s41598-019-51040-z
Rouster, J., Leah, R., Mundy, J., Cameron-Mills, V. (1997). Identification of a methyl jasmonate-responsive region in the promoter of a lipoxygenase 1 gene expressed in barley grain. Plant J. 11, 513–523. doi: 10.1046/j.1365-313X.1997.11030513.x
Sant’Ana, R. R. A., Caprestano, C. A., Nodari, R. O., Agapito-Tenfen, S. Z. (2020). PEG-delivered CRISPR-Cas9 ribonucleoproteins system for gene-editing screening of maize protoplasts. Genes 11 (9), 1029. doi: 10.3390/genes11091029
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Shearer, H. L., Cheng, Y. T., Wang, L., Liu, J., Boyle, P., Després, C., et al. (2012). Arabidopsis clade I TGA transcription factors regulate plant defenses in an NPR1-independent fashion. Mol. Plant Microbe Interact. 25, 1459–1468. doi: 10.1094/MPMI-09-11-0256
Siebertz, B., Logemann, J., Willmitzer, L., Schell, J. (1989). Cis-analysis of the wound-inducible promoter wun1 in transgenic tobacco plants and histochemical localization of its expression. Plant Cell. 1, 961–968. doi: 10.1105/tpc.1.10.961
Smita, S., Katiyar, A., Chinnusamy, V., Pandey, D. M., Bansal, K. C. (2015). Transcriptional regulatory network analysis of MYB transcription factor family genes in rice. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01157
Sobajima, H., Tani, T., Chujo, T., Okada, K., Suzuki, K., MORI, S., et al. (2007). Identification of a jasmonic acid-responsive region in the promoter of the rice 12-Oxophytodienoic acid reductase 1 gene OsOPR1. Biosci. Biotechnol. Biochem. 71, 3110–3115. doi: 10.1271/bbb.70532
Subburaj, S., Zanatta, C. B., Nunn, J. A., Hoepers, A. M., Nodari, R. O., Agapito-Tenfen, S. Z. (2022). A DNA-free editing platform for genetic screens in soybean via CRISPR/Cas9 ribonucleoprotein delivery. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.939997
Takahashi, T., Mori, T., Ueda, K., Yamada, L., Nagahara, S., Higashiyama, T., et al. (2018). The male gamete membrane protein DMP9/DAU2 is required for double fertilization in flowering plants. Development 145, dev170076. doi: 10.1242/dev.170076
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Thumuluri, V., Almagro Armenteros, J. J., Johansen, A. R., Nielsen, H., Winther, O. (2022). DeepLoc 2.0: Multi-label subcellular localization prediction using protein language models. Nucleic Acids Res. 5, W228–W234. doi: 10.1093/nar/gkac278
Trishla, V. S., Marriboina, S., Boyidi, P., Kirti, P. B. (2020). GUS-reporter based analysis of the promoter activity of Gossypium hirsutum NAC transcription factor, GhNAC4 that is induced by phytohormones and environmental stresses. Plant Cell Tiss. Organ Cult. 141, 643–654. doi: 10.1007/s11240-020-01825-2
Van der Graaff, E., Schwacke, R., Schneider, A., Desimone, M., Flugge, U. I., Kunze, R. (2006). Transcription analysis of Arabidopsis membrane transporters and hormone pathways during developmental and induced leaf senescence. Plant Physiol. 141, 776–792. doi: 10.1104/pp.106.079293
Wang, N., Xia, X., Jiang, T., Li, L., Zhang, P., Niu, L., et al. (2022). In planta haploid induction by genome editing of DMP in the model legume Medicago truncatula. Plant Biotechnol. J. 20, 22–24. doi: 10.1111/pbi.13740
Yamamoto, Y. Y., Ichida, H., Abe, T., Suzuki, Y., Sugano, S., Obokata, J. (2007). Differentiation of core promoter architecture between plants and mammals revealed by LDSS analysis. Nucleic Acids Res. 35, 6219–6226. doi: 10.1093/nar/gkm685
Yao, L., Zhang, Y., Liu, C., Liu, Y., Wang, Y., Liang, D., et al. (2018). OsMATL mutation induces haploid seed formation in indica rice. Nat. Plants. 4, 530–533. doi: 10.1038/s41477-018-0193-y
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Zhang, M., Liu, S., Wang, Z., Yuan, Y., Zhang, Z., Liang, Q., et al. (2022a). Progress in soybean functional genomics over the past decade. Plant Biotechnol. J. 20, 256. doi: 10.1111/pbi.13682
Zhang, Y., Luo, M., Cheng, L., Lin, Y., Chen, Q., Sun, B., et al. (2020). Identification of the cytosolic glucose-6-phosphate dehydrogenase gene from strawberry involved in cold stress response. Int. J. Mol. Sci. 21, 7322. doi: 10.3390/ijms21197322
Zhang, X., Zhang, L., Zhang, J., Jia, M., Cao, L., Yu, J., et al. (2022b). Haploid induction in allotetraploid tobacco using DMPs mutation. Planta 255, 1–11. doi: 10.1007/s00425-022-03877-4
Zhong, Y., Chen, B., Li, M., Wang, D., Jiao, Y., Qi, X., et al. (2020). A DMP-triggered in vivo maternal haploid induction system in the dicotyledonous Arabidopsis. Nat. Plants. 6, 466–472. doi: 10.1038/s41477-020-0658-7
Zhong, Y., Chen, B., Wang, D., Zhu, X., Li, M., Zhang, J., et al. (2022a). In vivo maternal haploid induction in tomato. Plant Biotechnol. J. 20, 250–252. doi: 10.1111/pbi.13755
Zhong, Y., Liu, C., Qi, X., Jiao, Y., Wang, D., Wang, Y., et al. (2019). Mutation of ZmDMP enhances haploid induction in maize. Nat. Plants. 5, 575–580. doi: 10.1038/s41477-019-0443-7
Zhong, Y., Wang, Y., Chen, B., Liu, J., Wang, D., Li, M., et al. (2022b). Establishment of a dmp based maternal haploid induction system for polyploid Brassica napus and Nicotiana tabacum. J. Integr. Plant Biol. 64, 1281–1294. doi: 10.1111/jipb.13244
Keywords: DUF679, soybean, abiotic stress, genomic analysis, cis-acting elements
Citation: Nawade B, Bosamia TC, Lee JH, Jang JH and Lee OR (2023) Genome-wide characterization of the soybean DOMAIN OF UNKNOWN FUNCTION 679 membrane protein gene family highlights their potential involvement in growth and stress response. Front. Plant Sci. 14:1216082. doi: 10.3389/fpls.2023.1216082
Received: 03 May 2023; Accepted: 22 August 2023;
Published: 08 September 2023.
Edited by:
S. K. Lal, Indian Agricultural Research Institute (ICAR), IndiaReviewed by:
Yuri Shavrukov, Flinders University, AustraliaRafaqat Ali Gill, Lushan Botanical Garden (CAS), China
Arun Jagannath, University of Delhi, India
Copyright © 2023 Nawade, Bosamia, Lee, Jang and Lee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ok Ran Lee, mpizlee@jnu.ac.kr
†These authors have contributed equally to this work
 Bhagwat Nawade
Bhagwat Nawade Tejas C. Bosamia
Tejas C. Bosamia Jae Hyun Lee
Jae Hyun Lee Jin Hoon Jang
Jin Hoon Jang Ok Ran Lee
Ok Ran Lee