- 1Division of Plant Pathology, Indian Agriculture Research Institute, New Delhi, India
- 2Division of Plant Pathology, Directorate of Rapeseed-Mustard Research, Bharatpur, India
Oilseed brassica has become instrumental in securing global food and nutritional security. B. juncea, colloquially known as Indian mustard, is cultivated across tropics and subtropics including Indian subcontinent. The production of Indian mustard is severely hampered by fungal pathogens which necessitates human interventions. Chemicals are often resorted to as they are quick and effective, but due to their economic and ecological unsustainability, there is a need to explore their alternatives. The B. juncea-fungal pathosystem is quite diverse as it covers broad-host range necrotrophs (Sclerotinia sclerotiorum), narrow-host range necrotrophs (Alternaria brassicae and A. brassicicola) and biotrophic oomycetes (Albugo candida and Hyaloperonospora brassica). Plants ward off fungal pathogens through two-step resistance mechanism; PTI which involves recognition of elicitors and ETI where the resistance gene (R gene) interacts with the fungal effectors. The hormonal signalling is also found to play a vital role in defense as the JA/ET pathway is initiated at the time of necrotroph infection and SA pathway is induced when the biotrophs attack plants. The review discuss the prevalence of fungal pathogens of Indian mustard and the studies conducted on effectoromics. It covers both pathogenicity conferring genes and host-specific toxins (HSTs) that can be used for a variety of purposes such as identifying cognate R genes, understanding pathogenicity and virulence mechanisms, and establishing the phylogeny of fungal pathogens. It further encompasses the studies on identifying resistant sources and characterisation of R genes/quantitative trait loci and defense-related genes identified in Brassicaceae and unrelated species which, upon introgression or overexpression, confer resistance. Finally, the studies conducted on developing resistant transgenics in Brassicaceae have been covered in which chitinase and glucanase genes are mostly used. The knowledge gained from this review can further be used for imparting resistance against major fungal pathogens.
Introduction
The Brassicaceae family showcases tremendous diversity with approximately 3709-member species and 338 genera (Warwick et al., 2006).This group offers great economic significance with a range of utilities such as edible and industrial oil, condiments, and vegetables (Nesi et al., 2008). To establish evolutionary relationship between the members of Brassicaceae, Nagaharu (1935) gave the ‘triangle of U’ model, in which he proposed the three amphidiploid species, Brassica napus (AACC), B. juncea (AABB), and B. carinata (BBCC), evolved through the interspecific hybridization in nature between diploid species B. rapa (AA), B. nigra (BB), and B. oleracea (CC). B. rapa and B. napus are important in temperate countries, while B. juncea dominates the subtropics such as the Indian subcontinent (OECD, 2016). B. juncea (L.) Czern & Coss (AABB, 2n = 4x = 36) traces its origin to Brassica rapa (AA, 2n = 20) and Brassica nigra (BB, 2n = 16). Axelsson et al. (2000) proved that both the parental genomes in B. juncea were conserved and have not undergone any change since polyploidization. A similar study was also carried out by Parkin and Lydiate (1997) in B. napus to establish the intact nature of parental genomes in it. Yang et al. (2016) sequenced 954.90 Mb genome of B. juncea and found that, the A subgenomes in B. napus and B. juncea have independent origins. Globally, Brassica or rapeseed-mustard is grown over 36.59 Mha with production and productivity of 72.37 Mt, and 1980 kg/ha, respectively. India’s global acreage and production share stood at 19.8% and 9.8% (USDA, 2020).
The B. juncea production is hampered by several biotic and abiotic stresses. Out of which, fungal diseases are a serious concern. The crop is vulnerable to fungal pathogens owing to the genetic uniformity between all prevalent cultivars. Alternaria leaf blight, White rust, Sclerotinia stem rot, and Downy mildew are the major fungal diseases of B. juncea (Singh et al., 2021; Meena et al., 2022). Though, chemicals are a good option for quick and effective control of diseases, they are unsustainable from both economic and ecological perspectives. For developing a robust disease management plan, the focus must shift from pathogen management to host management and there should be substantial efforts to alter crop ecology in the host’s favour (He et al., 2016). The specific host-pathogen interaction must be kept in mind for deciding the RAER (Resistance, Avoidance, Elimination, and Remediation) strategy (Xie et al., 1984). Genetic resistance is considered best to manage plant diseases as it is compatible with all other disease management strategies (Saharan, 1992; Saharan et al, 2005; Ren et al., 2016). The holy grail of resistance development in Brassica is identifying the pathogen virulence factors (effectors and toxins) and the complementary host R genes. In addition to these, the role of defense-related genes must be established as they are thought to contribute in broad-spectrum non-host resistance (NHR).
Plants have a two-step resistance mechanism against invading pathogens. The first one is the pattern triggered immunity (PTI), which is also known as basal resistance (Jones and Dangl, 2006). The pathogen-associated molecular patterns (PAMPs) (exogenous elicitors) are recognised by the pattern recognition receptors (PRRs), while wall-associated kinases (WAKs) detect the damage-associated molecular patterns (DAMPs) (endogenous elicitors) (Heil and Land, 2014). A plethora of defense responses is activated by PTI, which covers the influx of extracellular Ca2+ into the cytosol (Ranf et al., 2011), followed by the activation of MAP kinases (Zhang et al., 2007), reactive oxygen species (ROS) (Scheler et al., 2013), and other hormonal signaling molecules, such as salicylic acid, jasmonic acid, ethylene, and cytokinin (Jin et al., 2008, Nakano et al., 2013, Zhang et al., 2018; Huang et al., 2020).
This basal response is overcome by pathogen effectors, which results in effector triggered susceptibility. The plant fights back by recognising these effectors with intracellular receptors (R genes), thus activating the effector triggered immunity (ETI) (Peng et al., 2018; Alhoraibi et al., 2019). The host-pathogen interaction depends upon the trophic strategy of the pathogen. Biotrophs generally rely on the subtle manipulation of the host defenses for evading detection and effectors are the major weapons for this purpose. The plant’s response is characterised by the hypersensitive response (HR), which is a trusted ally against these pathogens as it limits the food source. The Effector-R gene interaction leads to ETI in the case of biotrophs. On the other hand, necrotrophs have a vast array of disease agents such as toxins, cell death inducing proteins (CDIPs), secondary metabolites, and CWDEs (cell wall degrading enzymes). Hypersensitive response against necrotrophs aids the pathogen instead and it is avoided by plants. The effector-intracellular receptor interaction is characterised by ETS for Necrotrophs (Laluk and Mengiste, 2010; Ghozlan et al., 2020; Liao et al., 2022). The difference in defense response has also been observed in terms of hormonal signaling pathways. The rise in salicylic acid (SA) levels increases resistance against biotrophs but it makes the plant vulnerable to necrotrophs (Caarls et al., 2015). The jasmonic acid (JA) pathway is antagonistic to SA and initiated at the time of attack by a necrotroph (Kravchuk et al., 2011; Niu et al., 2011; Weller et al., 2012) Figure 1 (Figure 2).
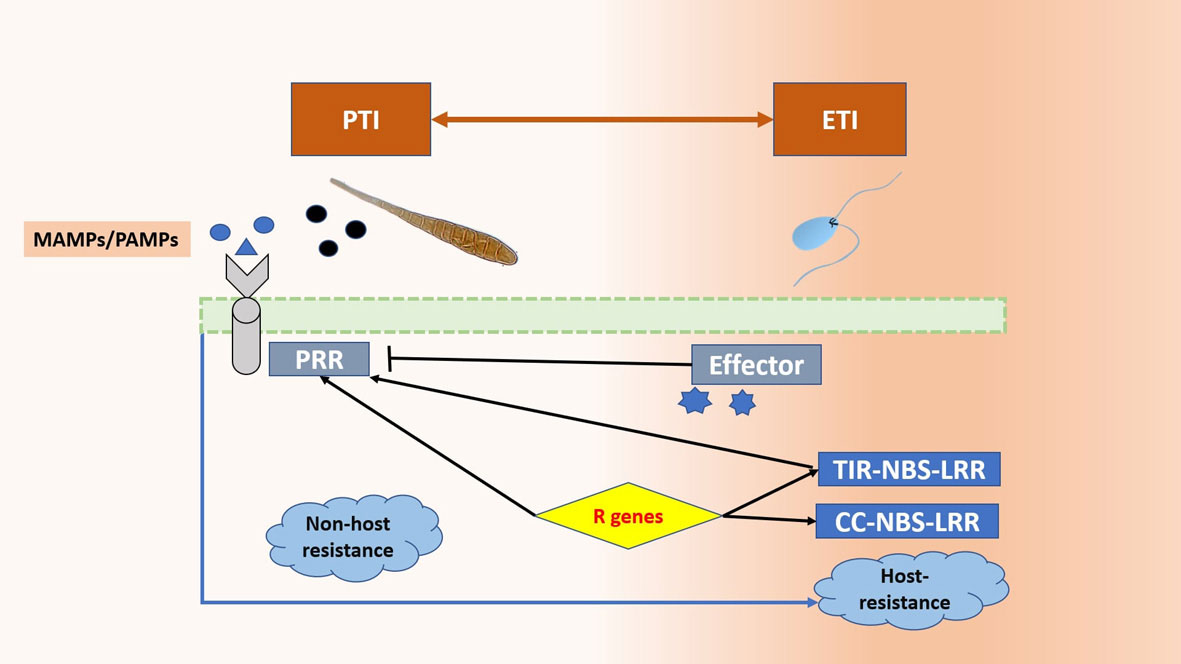
Figure 1 Two-step resistance mechanism in plants. The PAMPs/MAMPs are recognised by the PRRs. This leads to PTI (Pattern Triggered Immunity). To overcome this, the pathogen employs effectors and leads to ETS (Effector Triggered Susceptibility) and the plant responds by initiating ETI (Effector triggered immunity) in which the intracellular receptor recognises the effectors and leads to resistance.
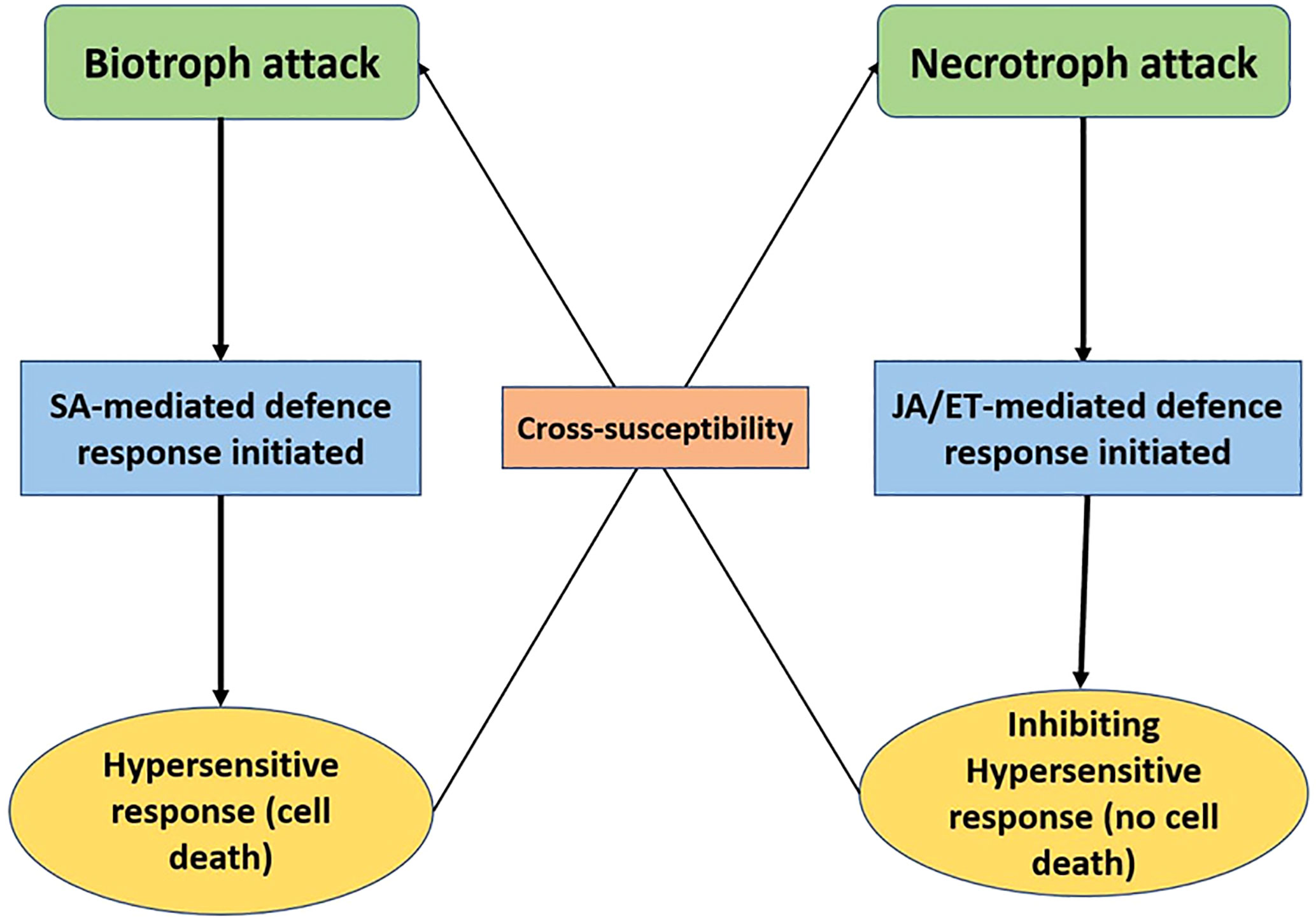
Figure 2 Schematic diagram of differential hormonal signalling defence response given at the time of attack by biotrophs and necrotrophs, where the SA pathway is initiated in the former and JA/ET in the latter.
A robust disease management strategy requires an understanding of the pathogenesis mechanisms which will enable identification of candidate effectors and disease resistance targets (Kim et al., 2016; Van de Wouw and Idnurm, 2019). The principle is based on the fact that resistant varieties will be insensitive to pathogen effectors. This functional assay can help to avoid tedious field trials and infection assays and further act as markers for fastening resistance breeding programs. Effector-based functional assays can be utilised for accelerating R gene cloning and finding out redundancy. It can help us to bring more specificity in breeding programs as the effector-based distinction of resistant sources is better than isolate-based (Du and Vleeshouwers, 2014). A Pathogen-Host Interactions database (PHI-base) can also be set up for analysis of phenotypic and biological data on virulence, pathogenicity, and effector proteins (Urban et al., 2017). Next step is the identification and utilisation of the resistant sources in cultivated and wild Brassica, which has been aided by the release of reference genome data of cultivated brassica species (Wang et al., 2011; Chalhoub et al., 2014; Liu et al., 2014; Parkin et al., 2014; Yang et al., 2016). Other economically viable and ecologically sustainable solutions such as healthy crop husbandry and biological control can be employed depending on their suitability in each pathosystem.
The present review aims to encompass the studies conducted on the 4 major fungal diseases of rapeseed-mustard with special emphasis on effectoromics of pathogens (Table 1), the sources of resistance genes/QTLs and defense-related genes identified across the Brassicaceae family (Table 2) and the transgenics developed for disease resistance (Table 3). Thus, this knowledge can be harnessed to improve Brassica against devastating fungal diseases such as Alternaria leaf blight (Figures 3A, B), Sclerotinia stem rot (Figures 3C, D), white rust (Figures 3E, F), and downy mildew (Figures 3G, H).
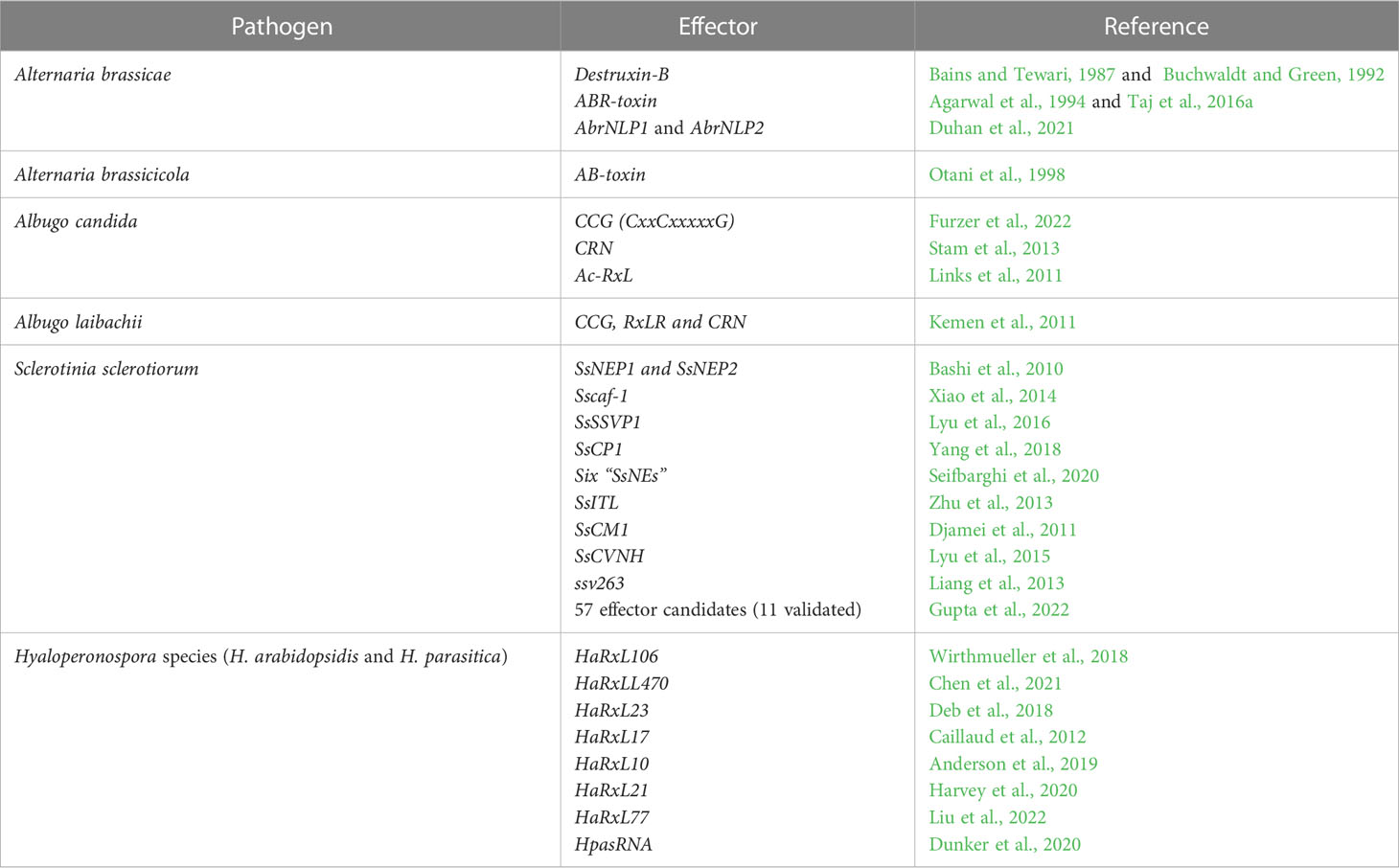
Table 1 The effectors identified and characterized in major fungal pathogens of the Brassicaceae family.
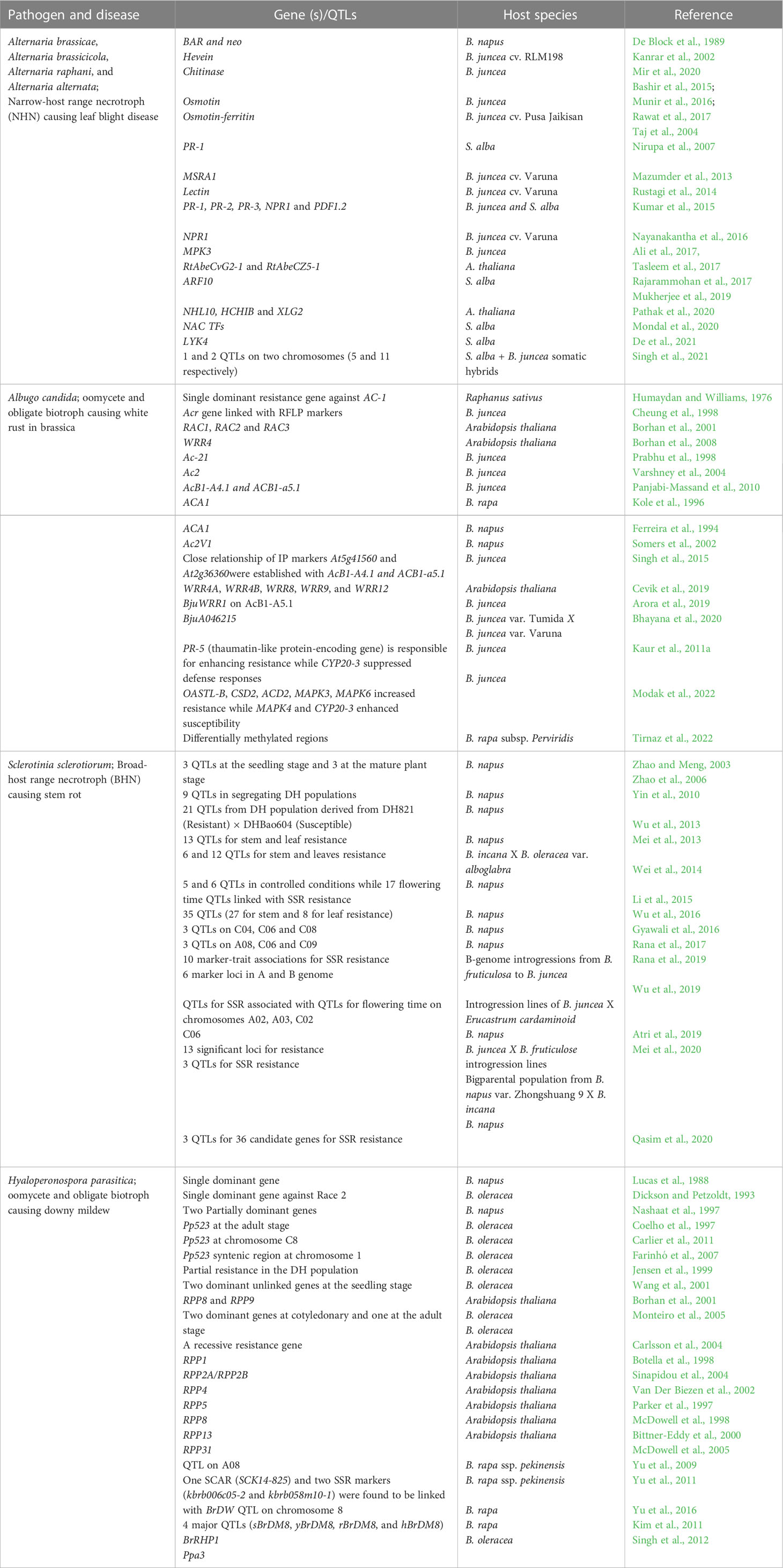
Table 2 The R gene(s)/QTLs and the defense-related genes identified in the Brassicaceae family against major fungal pathogens.
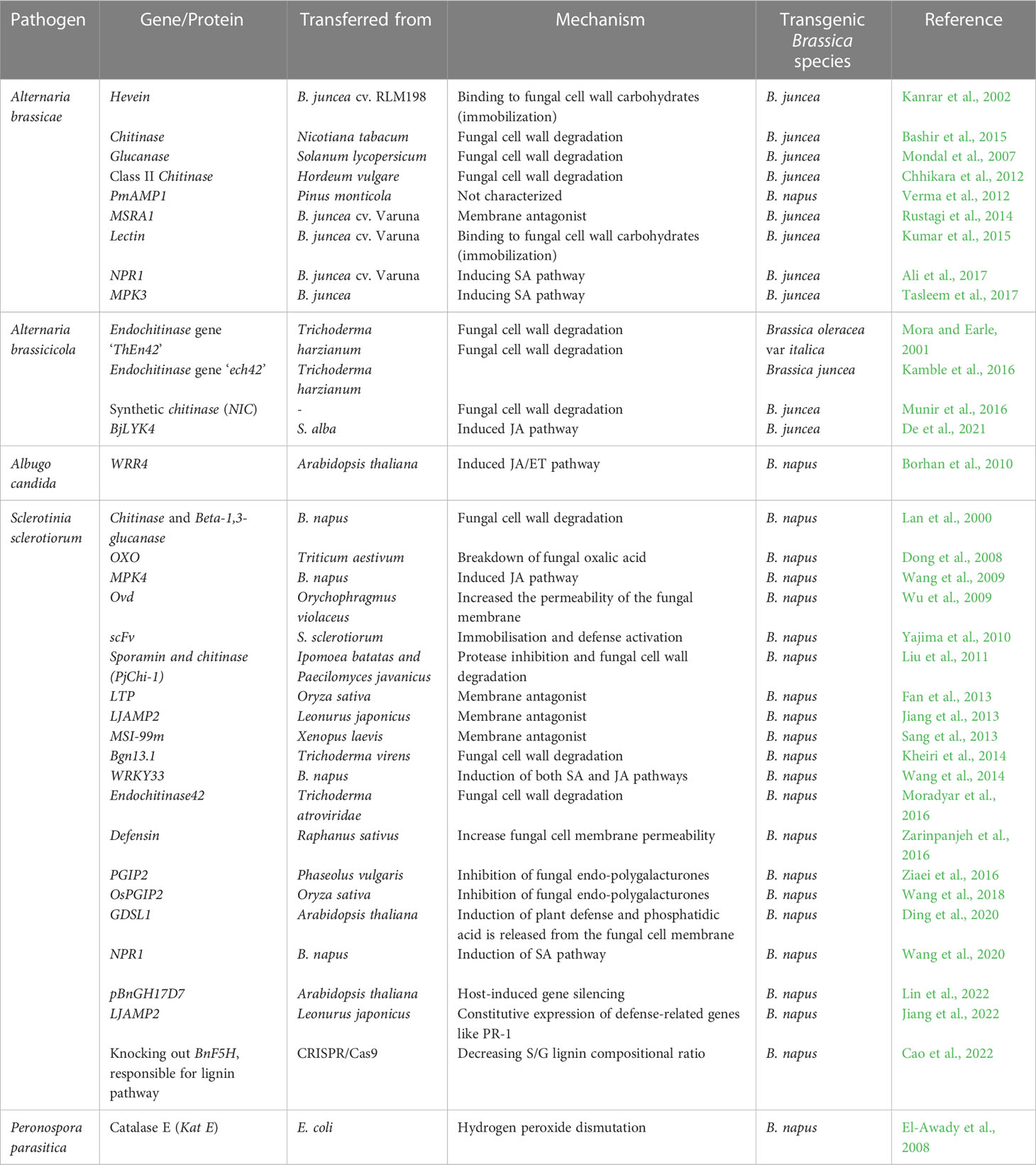
Table 3 The genes transferred from other species to Brassica members for conferring resistance against major fungal pathogens.

Figure 3 (A) Alternaria leaf blight symptons on Indian mustard (B. juncea); (B) Conidia of Alternaria brassicae; (C) Sclerotinia stem rot symptoms; (D) Sclerotinia sclerotiorum mycelium and sclerotia on PDA; (E) White rust on Indian mustard leaf (inset: hypertrophy); (F) Germinating oospores of Albugo candida; (G) Downy mildew on Indian mustard; (H) Sporangia and sporangiospores of Hyaloperonospora brassicae.
Alternaria leaf blight
Alternaria leaf blight or leaf spot (ALB) is a devastating disease caused by members of the genus Alternaria, which is a narrow-host range necrotroph (NHN). Nees von Esenbeck (1816) illustrated the Alternaria genus for the first time. The pathogen is cosmopolitan and found across the globe including India (Kadian and Saharan, 1983; Saharan et al, 2016a), Europe (Gladders, 1987), and Canada (Berkenkamp and Kirkham, 1989). The Alternaria group contains four species that infect Brassica (Meena et al., 2022). Quarantine testing of exotic rapeseed-mustard germplasm from as many as 20 countries, reported that A. brassicicola was more prevalent in seeds as compared to A. brassicae. Though A. raphani is mainly seen in radish, it may also attack other rapeseed-mustard members (Akhtar et al., 2017). On the other hand, A. alternata has a relatively broader host range but is weak as compared to its other counterparts (Verma and Saharan, 1994). Heavy losses ranging from 10% to 70% have been reported due to ALB Saharan et al, 2016a The pathogen severely infects all the above-ground parts of a plant such as pods/siliques, leaves and stems. The leaves give a characteristic target-board symptom caused due to formation of an interrupted necrotic zone (Meena et al., 2016; Jyoti et al., 2021). Grey spots are the characteristic feature of plants infected by A. brassicae while A. brassicicola causes black velvety sooty spots (Figures 3A, B). The hypocotyl of the plant has symptoms of sharp-edged lesions of dark brown colour produced by A. raphani (Meena et al., 2010). The environment is an important determinant for disease development as landing, adhesion, penetration, and host colonisation are all dependent on it (Macioszek et al, 2018). In temperate countries, the pathogen survives on plant debris (Humpherson-Jones, 1991), but for sub-tropical or tropical conditions like Indian subcontinent, the survival takes place on unconventional off-season crops, vegetable brassica, and the alternative hosts (Verma and Saharan, 1994; Mehta et al., 2002). The soil-borne and airborne conidia are responsible for secondary spread of the disease (Fatima et al., 2019).
CAZymes (Carbohydrate-active enzymes) are secreted by Alternaria for degrading and breaking the host cell wall. When checked at 2 and 4 dpi, 14 Polysaccharide lyases (PLs) were found to be upregulated but polygalacturonases, which cause degradation of pectin by hydrolysing the α-1,4 glycosidic bonds, were found to be upregulated only at 4 dpi (Rajarammohan, 2022). Secondary metabolites/toxins play a huge role in the pathogenicity of Alternaria. Around 70 toxins have been reported so far, and out of those 20 are HSTs (Host-specific toxins) (Nishimura and Kohmoto, 1983; Wolpert et al., 2002; Keswani et al., 2016; Taj et al., 2016b). The HSTs interact with the dominant or specific host-susceptibility gene and cause disease and toxicity symptoms. Thus, they are also considered effectors (Friesen et al., 2008). Suzuki et al. (1970) isolated Destruxin from Metarrhizium anisopliae. A. brassicae produces Destruxin-B, which is an important HST giving peculiar symptoms of necrosis and chlorosis in the host-specific plant (Meena and Samal, 2019). Another HST produced by A. brassicae is the ABR-toxin which induces water-soaked lesions followed by chlorosis (Agarwal et al., 1994). AB-toxin is a 35 kDa HST produced by A. brassicicola. (Otani et al., 1998) The pathogen spores recognise a host-derived oligosaccharide and initiate the production of AB-toxin (Oka et al., 2005). Both ABR and AB-toxin have similar host range and host-specificity, with molecular weight being the only recognisable differential (Meena and Samal, 2019). Most of the HST gene clusters are located on dispensable or supernumerary chromosomes (Akagi et al., 2009).
Rajarammohan et al. (2019a) prepared a contiguous genomic assembly for A. brassicae by Nanopore MinION sequencing. Two isolates of A. alternata; PN1 and PN2 were also sequenced by Rajarammohan et al. (2019b). Potential effector repertoire was predicted for six Alternaria species out of which most of them were found to be overlapping and common. It was found that the proportion of effectors is more in the dispensable chromosomes (2.39%) as compared to the core chromosomes (1.69%) (Rajarammohan et al., 2019b). Nep1-like proteins (NLPs) are an important class of necrotrophic effectors that induce necrosis and ethylene production. Duhan et al. (2021) identified and characterised two NLPs of A. brassicae; AbrNLP1 and AbrNLP2. Both the NLPs happened to be secretory in nature but differed in their localization in plants. The transcript level of both genes in the initial stages of infection was found to be upregulated, thus hinting at their role in pathogenesis. PTI and necrosis were induced by AbrNLP2 in both host and non-host species, but the necrosis-inducing ability was absent in AbrNLP1. Most of the necrotrophic species of the Alternaria genus had two copies of the NLP gene but some endophytes possessed only one. Necrotrophs have a relatively less expanded effector repertoire as compared to their biotrophic counterparts (Dong et al., 2012).
Some Brassica species such as B. napus, B. juncea, and B. carinata have been reported to possess inherent resistance. In this regard, B. rapa has been reported to be more susceptible as compared to its counterparts (Chadar et al., 2016). The resistance was also reported in the wild relatives such as Camelina sativa, Capsella bursa-pastoris, Eruca sativa, Diplotaxis catholica, D. erucoides, Hemicrambe fruticulose (Singh et al., 2021a). When A. brassicae was inoculated on B. rapa, 3-butenyl and 4-pentenyl isothiocyanates were found to be released along with sulphides and sesquiterpenes. The catabolism of glucosinolates is established by the release of isothiocyanate and is an important feature of host resistance (Doughty et al., 1996). Camalexin is an indole-derived phytoalexin synthesised by the MAPK6 signalling cascade and is responsible for imparting resistance to Camelina sativa. Gaur et al. (2018) performed molecular docking to find out the interaction of MKK9 with MKK1, MKK4, MKK5, MAPK3 and MAPK6. Different genes were also identified that play a role in camalexin synthesis. Yadav et al. (2020) did a histopathological, transcriptional, and biochemical study on B. juncea, Sinapis alba and C. sativa. They identified necrosis to occur early in B. juncea (1dpi) as compared to Sinapis alba (2dpi) and C. sativa (3dpi). An enhanced catalase and hydrogen peroxide activity was observed in Sinapis alba and C. sativa. Two pathogenesis-related genes, PR-3 and PR-12 were expressed only in the two wild relatives, hinting towards their role in Alternaria resistance. On the other hand, SA-induced genes were found to be highly expressed in B. juncea, which explains its antagonistic activity towards necrotrophs. Thus, both camalexin and jasmonic acid are vital for Alternaria resistance. Taj et al. (2011) reported that both MAPK3 and LOX interact to start the biosynthesis of JA. One more phytoalexin, sinalexin (sinalbin) produced by Sinapis alba was found to induce resistance against ALB.
B. oleracea and B. napus were transformed in a genotype-independent manner using selectable marker genes such as bar or neo. Agrobacterium strains having bar and neo genes infected the hypocotyl explants. Cytokinin concentration, water potential, and RH were reduced to avoid vitrification. 30% efficiency was observed in obtaining the transformed shoots. The copy number of chimeric genes varied from one to three in transformants and was confirmed by Southern blotting and genetic analysis (De Block et al., 1989). Mora and Earle (2001) developed ALB resistance in broccoli by transferring an endochitinase gene from Trichoderma in an Agrobacterium-mediated transfer. Kanamycin resistance followed by PCR and southern blotting was done to confirm the presence of genes in transformants. T0 Plants (Primary transformants) were found to have as much as 37 times more endochitinase activity than the control. In selfed progeny, it went up to 200 times. On the other hand, when inoculated with S. sclerotiorum no substantial difference was observed from the control. Hevein is a chitin-binding lectin having an antifungal activity which was analysed in transformed Indian mustard (B. juncea cv. RLM-198) against Alternaria brassicae. A cDNA encoding hevein was transferred and expressed resistance attributes such as small necrotic lesions, longer latent and incubation period, reduced disease intensity and delayed senescence (Kanrar et al., 2002). Glucan is an important component of the fungal cell wall and is hydrolysed by PR-protein glucanase. Mondal et al. (2007) used the CaMV 35S promoter to express the class I basic glucanase gene. A stable integration was confirmed by northern and southern hybridisation. Transgenics inhibited Alternaria brassicae hyphal growth by 15-54% and heavily delayed the disease incidence as compared to untransformed. Thus, the experiment showcased the efficiency of heterologous PR genes having the ability to impart ALB resistance. Taj et al. (2004) reported that osmotin provides resistance against the ALB disease by influencing the cell cycle and cell death pathways. B. juncea calli was taken as a model for investigating this phenomenon and P53 and caspase-like proteins were found to be overexpressed and CDC and cyclin B were suppressed. Finally, they concluded that osmotin is not able to provide resistance but delays the onset of symptoms. B. juncea cv. Pusa Jaikisan was transformed by Agrobacterium strain GV2260 containing binary vector p35SGUSINT and optimum conditions for transformation were identified based on transient GUS expression. The pre-culture period, age of the explant, bacterial density and silver nitrate were optimised for transformation. PCR and southern and western blotting were done to confirm the presence of transformants in T0 and T1 (Nirupa et al., 2007). Chhikara et al. (2012) developed resistance by coexpressing type I ribosome-inactivating protein and barley antifungal gene class II chitinase through Agrobacterium-mediated transfer. Transgenic plants showed a Mendelian inheritance pattern (3:1) and 44% protection was observed in them. Also, a reduction in lesion size, number and expansion, and delay of onset of symptoms was observed in transformed lines.
PmAMP1 is an antimicrobial peptide isolated from Pinus monticola and was able to provide protection to oilseed-brassica against many fungal genera such as Leptosphaeria maculans, A. brassicae and S. sclerotiorum. Higher resistance was observed in planta when the cDNA encoding gene was transferred to the B. napus genome. Even the in vitro extracted proteins were observed to have antifungal activity (Verma et al., 2012). Mazumder et al. (2013) observed a peculiar difference in disease response given by the host (B. juncea) and non-hosts (Sinapis alba and Arabidopsis thaliana) against A. brassicicola, where the former activates the SA pathway and JA/ABA is induced in the latter ones. This explains that the pathogen modulates the plant defense response in favour of biotrophic mode in host species whereas it is unable to do so in non-hosts as they can counter it by the JA pathway. Broad spectrum resistance against fungal necrotrophs is imparted by cationic antimicrobial peptides (CAPs). Many synthetic compounds have been derived from these proteins. MsrA1 is a chimeric protein formed by the combination of melittin CAPs and cecropin A. Five transgenic lines were checked for resistance against ALB and SSR. Up to 71.5% and 85% protection were observed for SSR and ALB, respectively in transgenic B. juncea (Rustagi et al., 2014). A chitinase gene from Streptomyces griseus was used to transform cotyledons and hypocotyls. Hypocotyls were reported to be more responsive, and 2 mg/L BAP and 0.2 mg/L were the best hormonal combination for callus transformation. Transgenic callus was confirmed by PCR (Bashir et al., 2015). Kumar et al. (2015) transformed B. juncea using chickpea lectin. Lectin is a vital secondary metabolite present everywhere and binds reversibly and specifically to carbohydrates, providing resistance against biotic stresses such as ALB and abiotic stresses such as drought and salinity. As much as 60% protection was observed in the transformants and higher proline content and augmented water retention capacity was also seen. CaMV 35S promoter was used in an Agrobacterium tumefaciens-mediated (EHA101) transfer having synthetic chitinase gene (NIC) in pEKB/NIC. Considerable resistance was seen against A. brassicicola and, the presence of chitinase gene in transformants was validated by PCR and southern hybridisation (Munir et al., 2016). Another endochitinase gene ‘ech42’ was transferred from Trichoderma virens to B. juncea and up to 73% protection was seen against both A. brassicae and A. brassicicola. The presence of the transgene was confirmed by a fluorimetric zymogram (Kamble et al., 2016).
Nayanakantha et al. (2016) comparatively analysed the expression of defense-related genes in Sinapis alba and B. juncea and reported all 5 genes, viz. PR-1, PR-2, PR-3, NPR-1 and Plant Defensin 1.2 (PDF1.2) to be highly expressed in S. alba as compared to Indian mustard; when inoculated with A. brassicae. PDF1.2 is a jasmonic acid-induced gene and was shown to have higher transcript levels in A. thaliana against the necrotrophs. PR-1 on the other hand was induced by the SA pathway. But, in this case, both SA and JA-induced genes were expressed earlier and more in Sinapis alba than in B. juncea. This confirmed that the response varied from Indian mustard to A. thaliana. An expression analysis experiment was conducted to characterise B. juncea class IV chitinase against JA, SA, wounding and Alternaria infection. A chitinase promoter of size 2.5 kb was isolated and in-silico analysis was done further. Finally, it was fused with the GUS gene and introduced into A. thaliana and, it reportedly showed higher activity after wounding and JA treatment but lower in SA treatment. Organ specificity was analysed based on GUS activity in seeds, siliques, leaves and meristematic cells (Rawat et al., 2017). Ali et al. (2017) isolated and characterised SA-receptor gene BJNPR1 to play an important role in conferring broad-spectrum resistance against ALB and powdery mildew. No phenotypic abnormalities were observed, and the gene was found to be constitutively expressed. The gene was upregulated on fungal attack or SA treatment, but similar trends were not observed upon ABA and JA treatment. Studies on MAPK3, MAPK4 and MAPK6 in A. thaliana have indicated that, they are involved in conferring resistance against multiple biotic stresses. Tasleem et al. (2017) checked the expression of these genes in transgenic B. juncea (over expressed MAPK3). MAPK3 and MAPK6 were expressed in the early stages of Alternaria infection whereas MAPK4 was activated in the later part. This showed the SA pathway to be an important determinant of ALB resistance and MAPK3 interacts with it in a positive manner. Arabidopsis can be used for mapping the QTLs for ALB resistance and identifying paralogues in Brassica. In this regard, Rajarammohan et al. (2017) developed three biparental mapping populations with the help of two susceptible lines (Gre-0 and Zdr-1) and three resistant lines (CIBC-5, Ei-2 and Cvi-0). Six QTLs were identified, out of which five were population-specific and one was common to all accessions. 50% of the variation was conferred by two QTLs which had a larger effect, and resistance was confirmed to be quantitative in nature as both population-specific and common QTLs were found. ABA-auxin cross talk is less studied as compared to JA-SA.
Mukherjee et al. (2019) reported an enhanced expression of the auxin-responsive factor ARF10 along with, the expression of ARF16 and ARF17. When ARF10 was expressed in transgenic Brassica juncea, it led to ABA sensitivity and increased resistance against A. brassicicola. Many ABA-responsive genes like ABI3, ABI4 and ABI5 were also induced without having any profound effect on auxin-biosynthesis. The ARF10 interacted with the promoter of ABI5 and conferred ABA sensitivity, finally culminating in a defense response. HCHIB and NHL10 were identified as major defense-related genes against ALB in Arabidopsis. Some genes such as WRKY, CZF1, MP, AXR3, IAA1, IAA19, ARF6, and XLG2 modulated the JA, SA, and ET pathways. XLG2 showed a more elevated response against Alternaria brassicicola. Similar genes were also found and characterised in the B. rapa genome by using Arabidopsis as a model plant system (Pathak et al., 2020). Mir et al. (2020) evaluated C. sativa and B. juncea for chitinase genes and reported 79 and 47 of them respectively. The expression of these chitinase gene was confirmed by qRT-PCR which was found relatively more in C. sativa confirming its comparative tolerance. NAC TFs are a specific class of transcription factors which impart resistance against multiple abiotic and biotic factors. The NAC TFs were comparatively analysed in S. alba (resistant) and B. juncea (susceptible). Out of thirteen selected NAC TFs, six were found to be highly expressed in treated and tolerant B. juncea and S. alba. The NACs were instrumental in resistance against both wounding (abiotic stress) and Alternaria inoculation (Mondal et al., 2020). Plant defense response is activated by the Lysin motif receptor-like kinases (LYKs) by recognising chitin and De et al. (2021) reported it to be strongly induced in S. alba as compared to B. juncea. Though B. juncea had the LYK4 domain, it lacked many key protein kinases and was found to be inactive. ALB resistance was gained by overexpressing the BjLYK4 gene and many JA-induced genes and chitin-responding WRKY transcription factors were also highly expressed upon pathogen attack. Singh et al. (2021b) introgressed QTLs from S. alba to the backcross progenies of S. alba and B. juncea somatic hybrids which were more stable. The quantitative nature of resistance was confirmed by differential response seen in the backcross population ranging from highly resistant to susceptible. One and two QTLs were detected by ICIM-ADD mapping in chromosomes 5 and 11 respectively. 5.51-10.87% variation was observed for disease resistance in the backcross population. Before this, the QTL introgression for Alternaria leaf blight has never taken place from a brassica family member to cultivated rapeseed-mustard.
White rust
Albugo candida is an obligate biotroph (Kaur et al., 2011b) belonging to the class oomycetes, causing the dreaded white rust (WR) disease in all the major oilseed brassica (OSB) growing countries such as India (Chowdhary, 1944; Kolte, 1985), Pakistan (Perwaiz et al., 1969) Canada (Petrie, 1973), Germany (Klemm, 1938), Japan (Hirata, 1954), South Korea (Choi et al., 2007), China (Zhang et al., 1984), New Zealand (Hammett, 1969), United Kingdom (Berkeley, 1848), and USA (Walker, 1957). Petrie and Vanterpool (1974) reported that systemic infection of white rust can lead to a 60% loss in seed yield. In Indian conditions, 60% yield losses are reported due to the combined effect of leaf and inflorescence infection (Lakra and Saharan, 1989). Bal and Kumar (2014) reported average 36.88% yield loss due to white rust disease. As white rust is an obligate biotroph, signs are more pronounced as compared to symptoms. The white blister-like pustules starting from the underside of foliage and spreading to the foliar parts are characteristic symptoms of the disease (Figures 3E, F). Further, the systemic infection leads to the formation of the staghead phase (SP), which is a combined effect of hypertrophy and hyperplasia (Verma and Petrie, 1980; Kolte, 1985) (Figure 3E). Sporangiophore produces basipetal chains of sporangia between the epidermis and mesophyll layer of host tissue. Direct or indirect germination by zoospores takes place after the release of sporangia (Xiao et al., 2022). Finally, intercellular hyphae penetrate the host cell and sporangia and oospores are established as resting structures (Meena et al., 2014a). A. candida has a wide host range and can cause infection in around 63 genera and 241 host species (Gupta and Saharan, 2002; Choi et al., 2007; Mishra et al., 2009; Saharan et al., 2014, Pound G.S. and Williams, 1963 isolated pathogen from various host species and identified 6 races of A. candida. Singh et al. (2021) collected and characterised 13 isolates of A. candida and 1 isolate of Wilsonia bliti based on their morphological features and the highly conserved COX2 and ITS data.
Albugo candida has a relatively smaller genome (45.3 Mb) and less abundance of pathogenicity-related proteins encoding genes like RxLR effector, Elicitins and CRINKLER-like genes as compared to other biotrophic oomycetes like Hyaloperonospora arabidopsidis (99Mb). Still, 26 Ac-RxLs were identified in the pathogen effector repertoire and despite possessing a degenerate RxLR-dEER motif, it was able to cause death in host cells (Links et al., 2011). An improved reference genome was prepared by Furzer et al. (2022) which led to a 175% expansion in the candidate repertoire of CCG class effectors. Albuginales have pathogenetically evolved separately from the Peronosporales, which are characterised by having large families of CRN and RxLR effectors (Haas et al., 2009; Baxter et al., 2010). 13 similar proteins to CRN were identified in the secretome of Albugo candida. (Stam et al., 2013). Their presence and expansion indicate a common CRN present before acquiring pathogenicity and their possible role in parasitising the host. Castel et al. (2021) found out that the col-0 (Columbia) accession of Arabidopsis thaliana has three TIR-NLR encoded genes at White Rust Resistance 4 locus (WRR4). The CCG effectors of A. candida races 2, 4, 7 and 9 (from B. juncea, Capsella bursa-pastoris, B. rapa and B. oleracea) were recognised by the col-0 alleles of WRR4A and WRR4B. On further analysis, it was concluded that four CCG effectors were recognised by WRR4B and eight of them interacted with WRR4A. Thus, WRR4 paralog-based broad-spectrum resistance for multiple A. candida races was established. This resistance was found to be broken by an isolate AcEx1. A WRR4A allele; WRR4AOy-0 in Arabidopsis accession was identified and found to confer resistance against the AcEx1, but it substantially reduced the recognition of WRR4A-recognised effectors (Castel et al., 2021). Races 2 (non-host) and 7 (host) were intercrossed by Adhikari et al. (2003) and a ratio of three avirulent to one virulent was obtained upon inoculating F2 population of B. rapa. This evidently proves a resistance-gene mediated recognition of the Ac2V effector and may hint towards the genetic basis of non-host resistance. The mechanism of NHR in Brassica species is vital for resistance breeding as new effector repertoire can evolve due to genetic exchanges between races of A. candida (McMullan et al., 2015). The recognition of effectors by the non-host intracellular receptors may indicate a common ancestral host species for the pathogen (Lee et al., 2014; Cevik et al., 2019).
The white rust resistance gene for race 1 of Albugo candida (AC1) in R. sativus was studied along with six other characters. The pink pigmentation (pi) in plants was found to be closely linked with the AC1 gene (Humaydan and Williams, 1976). A restriction fragment length polymorphism marker (RFLP) was identified in close association with the white rust resistance gene (Acr) in B. juncea (Cheung et al., 1998). Panjabi-Massand et al. (2010) selected two east European lines, Heera and Donskaja-IV and tagged two independent loci governing white rust resistance. They acquired two doubled haploid populations by crossing Heera with Varuna and TM4 with Donskaja-IV. After crossing, a single major locus was identified in both cases, AcB1-A4.1 was mapped on linkage group A4 in Heera while in Donskaja-IV, the linkage group was AcB1-A5.1 on A5. Synteny between Arabidopsis and B. juncea formed the basis for developing closely flanked markers and introgression of the resistant loci is easier with these markers. Singh et al. (2015) checked the genotype non-specific intron polymorphic (IP) markers At5g41560 and At2g36360 from Arabidopsis and concluded their proximity with loci responsible for white rust resistance AcB1-A4.1 and AcB1-A5.1, respectively in B. juncea. Two accessions were selected from A. thaliana, Ksk-1 and Ksk-2 and three white rust resistance genes (RAC1, RAC2 and RAC3) belonging to class TIR-NB-LRR were mapped and RAC1 was cloned (Borhan et al., 2001). Another R gene, WRR4 belonging to class TIR-NB-LRR was characterised by Borhan et al. (2008). This gene was identified as the basis of broad-spectrum resistance conferred against four races of A. candida. Borhan et al. (2010) suppressed the enhanced disease susceptibility-1 (eds-1) gene through RNA interference, and they transferred the WRR4 gene and expressed it in the susceptible lines of B. napus and B. juncea to get resistance against race 2 and 7 of A. candida.
Prabhu et al. (1998) crossed resistant and susceptible varieties of B. juncea to obtain the doubled-haploid population derived from F1. They identified two markers WR2 and WR3 which determined the presence or absence of the white rust resistance gene, AC21. A white rust resistance locus, AC2(t) was mapped with the help of the RAPD marker by Varshney et al. (2004) and they also used bulk segregant analysis and AFLP to develop a more tightly linked marker. Two cultivars of B. napus were crossed (Major and Stellar) and a backcross population (BCP2), F1-derived doubled haploid and F2 population were analysed to identify a single dominant locus for white rust resistance, ACA1 on linkage group 9 (Ferreira et al., 1994). A similar experiment was carried out on B. rapa by Kole et al. (1996) in which they crossed a resistant (Per) and susceptible (R-500) cultivar. They mapped the ACA1 locus on linkage group 4 with 144 RFLP loci segregating in the F3 generation. The ACA1 locus was 13.3 cM apart from the leaf pubescence locus (PUB1), so both PUB1 and RFLP markers can be used for the introgression of ACA1 (Kole et al., 1996). For transferring the canola quality, an interspecific cross has been made between B. napus and B. juncea. This has facilitated the introgression of a white rust resistance gene from B. napus to B. juncea. A BC3F2 population of B. juncea was used to identify DNA markers for the white rust resistance trait. Eight B. napus-derived AFLP markers along with white rust resistance gene (AC2V1) were identified (Somers et al., 2002). R-gene-mediated non-host resistance (NHR) can be instrumental in providing durable resistance to crop varieties so that they can be transferred from source to the susceptible plants. The adult plants of Arabidopsis were found to be resistant to white rust disease. 593 inbred lines developed from the Arabidopsis MAGIC population were screened and two susceptible transgressive segregants were identified. One of four genes was speculated to be the basis of resistance against the AC2V isolate of A. candida. An additional gene was identified for race 9 infecting the B. oleracea crop (Cevik et al., 2019).
Arora et al. (2019) identified and functionally characterised a CC-NB-LRR protein-encoding gene, BjuWRR1. This constitutively expressed gene was responsible for conferring broad-spectrum complete resistance against white rust in Donskaja-IV. BjuWRR1 located on AcB1-A5.1 was introgressed in Varuna, Rohini, Pusa Jaikisan and Pusa Bold. The developed NILs were found to be resistant against all the six isolates (Pantnagar, Meerut, Bharatpur, Samastipur, Hisar and Alwar) tested. The allele-specific molecular markers can be used for transferring the locus into new lines and developing hybrids. A cross was performed between B. juncea var. Tumida and B. juncea var. Varuna and the F1DH population were used to map a new R-gene locus for isolate ACB1 which is located on LGA6 of B. juncea var. Tumida. The candidate gene, BjuA046215 is a CC-NB-LRR and its alleles in susceptible varieties produce a truncated LRR-domain protein. Both BjuWRR1 and BjuA046215 belonged to the CNL-D group of R genes, and they were phylogenetically similar (Bhayana et al., 2020). DNA methylation as a determinant of epigenetic resistance plays an important role in plant immunity. Its role in Brassica has not been characterised yet. Tirnaz et al. (2022) evaluated B. rapa subsp. perviridis for modification in its whole genome DNA methylation against white rust. “Misugi” (susceptible) and “Nanane” (resistant) were analysed for differentially methylated regions (DMR) and 233 and 275 DMRs were identified respectively. DMRs were located within genes in “Mishugi”, while they were found to be either upstream or downstream in “Nanane”. This study points towards a potential role of DNA methylation in white rust resistance.
Kaur et al. (2011a) carried out a comprehensive proteomic analysis to find the role of defense compounds and genes in imparting white rust resistance. A total of nineteen proteins showcased a reproducible difference in resistant (CBJ001) and susceptible (RH819) cultivars upon inoculation with A. candida. Q-TOF MS/MS was used to confirm the identity of proteins and out of these, five were only reported in the resistant cultivar. PR-5, which encoded a thaumatin-like protein, earlier not associated with defense response was found to play role in white rust resistance. CYP20-3 is an isoform of peptidyl-prolyl cis/trans isomerase (PPIase) and was only identified in the susceptible cultivar, establishing its role in susceptibility. Expression analysis of defense-related genes and MAPKs cascade was checked in wild and transgenic Varuna (overexpressing MAPK3). The transgenic Varuna was found to be relatively more tolerant as confirmed by disease indexing. MAPK6 was found to mimic the MAPK3 pathway and suppress the expression of MAPK4 in transgenic Varuna. WRKY29 and WRKY33 were also reported to be highly expressed in the transformed one. Similarly, transcripts of OASTL-B, ACD2 and CSD2 also increased, and the CYP20-3 had a reverse trend as its transcript accumulated more in non-transgenic Varuna. In-silico studies were done to study protein-protein interaction, secondary & tertiary structures and finally predict putative phosphorylation sites (Modak et al., 2022).
Sclerotinia stem rot
Sclerotinia stem rot (SSR), caused by Sclerotinia sclerotiorum, is a serious production-impeding factor for rapeseed-mustard. The pathogen attacks a plethora of host species and is thus characterised as a broad-host range necrotroph (BHN). The disease resistance is difficult to develop as host specialisation is rare and around 600 plant species are reportedly infected by it (Liang and Rollins, 2018). Though it is rare, host specialisation does occur and was conclusively proven by Kull et al. (2004). There has been a striking preference for dicots against monocots which can be ascribed to dicots possessing a peculiar form of glycosylinositol phosphorylceramide (Lenarčič et al., 2017) and monocots producing germins against the pathogen (Davidson et al., 2009). Libert (1837) described it for the first time and named it Peziza sclerotiorum. Later, Purdy (1979) established the name S. sclerotiorum (Lib.) de Bary. Cool and moist weather conditions have proven to be conducive for SSR epiphytotics causing 100% losses (Ghasolia et al., 2004, McDonald and Boland, 2004, Shukla, 2005; Singh et al., 2008). It has been established as one of the major yield-limiting factors of B. napus in India, China, Europe, Australia, and Canada (Rana et al., 2017). It was considered a minor disease in India 20 years back, but irrigation facilities and monocropping have made it emerge as one of the major yield-limiting factors in rapeseed-mustard (Sharma et al., 2015). The disease is characterised by white, fluffy mycelia seen on siliquae, stem and leaves (Christias and Lockwood, 1973). Though Sclerotinia infects all parts of the plant, the stem is most severely affected which leads to lodging and girdling causing yield reduction (Melouk et al., 1989; Uloth et al., 2016) (Figures 3C, D). Sclerotinia showcases both myceliogenic (soil-borne infection) and carpogenic (air-borne infection) germination which leads to symptoms on stem and siliquae, respectively (Singh et al., 2021).
Being a necrotroph; although recent studies point towards a short initial biotrophic phase, the pathogen is thought to employ brute force for damaging the host, but it has some sophisticated mechanisms up its sleeve (Saharan et al., 2017). Sharma et al. (2018) conducted virulence and phylogenetic analysis for 65 isolates of S. sclerotiorum utilising morphological features, SSR profiling, and ITS sequencing, and finally established three evolutionary lineages. Oxalic acid was proven to be an important pathogenicity determinant with the help of UV-induced mutations (Godoy et al., 1990; Dutton and Evans, 1996). Later studies have concluded that, rather than directly affecting host physiology, pH manipulation is the main job of oxalic acid (Liang et al., 2015; Xu et al., 2015; Jingtao et al., 2018). Several putative effectors have also been identified in S. sclerotiorum. Cell death is vital for a necrotroph to obtain nutrients and in this regard, two NLPs; SsNEP1 and SsNEP2 were the first cell death-inducing effectors to be identified in tobacco leaves and shown by Agrobacterium tumefaciens-mediated expression (Bashi et al., 2010). Xiao et al. (2014) concluded that a gene named, Sscaf-1 led to the development of sclerotia and compound appressoria formation. Its protein contains a Calcium ion-binding EF-hand motif. Disruption of Sscaf-1 was done by the transfer DNA insertion and, further knockdown and gene complementation established it as the cause of changes observed in Sunf-MT6. SsSSVP1 is a small cysteine-rich protein that has the capacity to induce cell death and a reduction in virulence was observed upon silencing the gene. It is a potent plant energy metabolism manipulator as it interacts with and disturbs the subcellular localisation of QCR8 (Lyu et al., 2016). A cerato-platanin protein (CP), SsCP1 was characterised, and an accumulation of transcripts occurred during initial stages of infection. It facilitated infection by interacting with PR1 in apoplast. In transgenic plants possessing SsCP1 gene, an increased level of SA was observed along with broad-spectrum resistance to pathogens such as Botrytis cinerea, Alternaria brassicicola and Golovinomyces orontii (Yang et al., 2018). Six novel effectors; SsNEs were discovered by Seifbarghi et al. (2020). A reduction but not nullification in virulence was observed in SsNE2 when cysteine was substituted by alanine.
Besides inducing necrosis, the S. sclerotiorum effectors also target the host defense response. One such effector, SsITL was found to be upregulated at 1.5-3 hpi and on further analysis, it was seen that upon inoculating the silenced transformant the defense-related genes such as PR1 and PDF 1.2 produced their highest transcript level at 3 hpi which was 9 hours before the response observed for wild strain. It also inhibited the jasmonic acid/ethylene (JA/ET) pathway which is crucial for resistance against necrotrophs (Zhu et al., 2013). A further study by Tang et al. (2020) reported that SsITL interacts with the Calcium-sensing receptor (CAS) located in the chloroplast. This interaction inhibited the Calcium ion signalling and finally, the salicylic acid (SA) pathway was inhibited in the early stages of infection. This interaction is essential for virulence as SsITLs, which lost their ability to interact with CAS, were unable to infect the host. SsCM1, a putative effector shares structural similarities to Cmu1, an Ustilago maydis effector which also targets the SA accumulation (Djamei et al., 2011; Kabbage et al., 2013). SsCVNH is an effector that contains CVNH carbohydrate-binding domain and was conclusively found to be upregulated during infection and played an important role in virulence, growth, and formation of sclerotia. The SsCVNH is thought to either protect the fungal cell wall or evade PTI response of the host like other fungal effectors having LysM domain (Lyu et al., 2015; Sánchez-Vallet et al., 2020). The ssv263 is an identified orthologue of B. cinerea protein and putatively reported to inflict symptoms in B. napus, though the mechanism remains unclear (Liang et al., 2013). Gupta et al. (2022) did a whole genome-sequencing of a highly virulent isolate of S. sclerotiorum, “ESR-01” which yielded 57 candidate effectors, out of which 30 were reported to be novel. Expression profiling of the isolate validated 11 of the effector candidates. Metabolisation of the host defense compounds is an effective strategy employed by S. sclerotiorum and it also plays a significant role in determining its host range. Similarly, cross-kingdom RNAi transfer from pathogen to host is also seen as a potential area of research in the upcoming future (Derbyshire et al., 2022).
Map-maker QTL was used to detect six QTLs for S. sclerotiorum resistance in B. napus. Out of these, three were identified at the seedling stage while three were at the adult plant stage. Additive epistatic interactions were also reported and they, along with single locus QTL were responsible for SSR resistance (Zhao and Meng, 2003). Nine QTLs were identified at A2, A3, A5, C2, C4, C6 and C9 for SSR resistance in B. napus from two segregating DH lines (HUA population) developed by crossing Chinese and European spring lines (Zhao et al., 2006). Advanced molecular tools were used to map the QTLs or putative R genes for SSR resistance. B. juncea was crossed with B. fruticulose to produce fertile introgression lines with B-genome chromosomes being terminally introgressed. Microsatellite markers were used for genotyping these resistant lines. Association mapping was done to identify ten significant marker-trait associations (Rana et al., 2017). 96 sets of B. juncea–Erucastrum cardaminoid introgression lines were developed by Rana et al. (2019). Genotyping was done by both transferrable microsatellite markers and sequencing to confirm marker-trait associations. SSR markers were used to identify the association between resistance and six marker loci in A and B genomes. Based on GWAS analysis, SNP markers were characterised to be linked to SSR resistance in B03, A06 and A03 chromosomes.
A plethora of resistance mechanisms was identified by annotation studies such as production of anti-fungal metabolites, hypersensitive response and signal transduction. The LRR-RLK genes were found to be associated with total five SNPs on A03 chromosome and genetic factors for both PTI and ETI were found on A03. Three R genes encoding TIR-NBS-LRR was identified, but till now no cloning has been done for any R gene against SSR (Wu et al., 2009; Wu et al., 2013). The susceptible B. oleracea var. alboglabra was crossed with resistant wild B. oleracea (B. incana) to give a biparental population exhibiting 6 and 12 QTLs for stem and leaf resistance, respectively. Two QTLs were reportedly identified on C09 for both leaf and stem resistance. With the help of blasting to B. rapa as a reference genome, it was found that chromosome C09 harbours the candidate R gene. About 30 genes were identified at 2.7 Mb genomic region of A09 as being involved in defense response and resistance-related functions. The putative genes were characterised as CC-NS-LRR (Mei et al., 2013). Yin et al. (2010) identified a total of 21 QTLs on A3, A4, C1, C2 and C7 for SSR by evaluating the DH population obtained by crossing B. napus cultivars DHBao604 (susceptible) and DH821 (resistant). Putative QTLs have been identified in B. napus for SSR resistance. Three candidate QTLs involved in SSR resistance have been identified with the help of GWAS on C04, C06, and C08 (Wu et al., 2016) and A08, C06, and C09 (Gyawali et al., 2016). Few more QTLs on chromosomes A02, A03, C02 and C06 in B. napus were identified for SSR resistance by the SNP-array genotyping. The flowering time QTLs were also located in these regions harbouring SSR resistance QTLs (Wu et al., 2019). Introgression lines were prepared from a cross between B. juncea and B. fruticulosa and were analysed for resistance. A total of 13 loci were found to be significant and the annotation experiment provided 20 candidate genes belonging to defense families such as Chitinase, Malectin/receptor-like protein kinase defensin-like (DEFL), desulfoglucosinolate sulfotransferase protein and lipoxygenase (Atri et al., 2019). Mei et al. (2020) pyramided three SSR resistance QTLs located on C01, C09-1, and C09-2 chromosomes by developing BC1F8 population of cross between B. napus var. Zhongshuang 9 and B. incana.
Wu et al. (2013) identified a candidate R gene BnaC.IGMT5.a in B. napus based on the differential expression pattern shown by two parental lines. One out of thirteen identified QTLs for SSR was found to be located on C06. Chromosomes A9 and C6 were found to be harbouring the 8-leaf and 27-stem resistance QTLs. This hints towards some of the genotypes sharing common leaf and stem resistance regions (Li et al., 2015). Seventeen QTLs based on the DH population derived from a cross between European winter and Chinese semi-winter for the flowering time trait were found to be in close association with the SSR QTLs on C02 and LG A02 and five and six QTLs were identified in controlled and field conditions, respectively. Common resistance genes were found on both chromosomes A and C pointing towards common chromosome ancestry and SSR resistance being specific at the subpopulation level (Wei et al., 2014). Qasim et al. (2020) used SNP markers to identify 17 QTLs for SSR resistance over three different seasons. No common QTL was identified across all three seasons but three QTLs, SRA9a, SRC2a and SRC3a appeared in two seasons. Stem width was identified as having weak relationship with the resistance trait. Flowering time shared a very strong negative correlation with SSR resistance as the early maturing varieties were found to be more susceptible as compared to the late maturing ones.
Lan et al. (2000) transformed a good-yielding B. napus variety H165 by constitutively expressing the chitinase and beta-1,3-glucanase genes transferred through an Agrobacterium-mediated transfer of expression vector pBLGC. Oxalic acid is an important determinant for pathogenicity and oxalate oxidase (OXO) can oxidise this compound into CO2 and H2O2. A wheat OXO gene was constitutively expressed in transgenic rape, and it imparted a considerable amount of disease resistance against SSR which accounted for as high as a 90.2% reduction (Dong et al., 2008). MPK4 is known to enhance the jasmonic acid-activated defense response which is evident from an experiment conducted by Wang et al. (2009) in which they developed transgenic B. napus by overexpressing BnMPK4 gene that yielded disease resistance against broad-host range necrotrophs such as S. sclerotiorum and Botrytis cinerea. B. napus was transformed by Agrobacterium-mediated transfer of a plant defensin gene Ovd, that was cloned from Orychophragmus violaceus. The RT-PCR analysis confirmed that the expression of Ovd was much lower in antisense and non-transgenic plants as compared to the sense lines. 20% reduction in lesion size was reported and the gene was confirmed to confer a strong defense against the SSR disease (Wu et al., 2009). Yajima et al. (2010) transferred a recombinant pathogen-specific antibody (scFv) to B. napus lines and reported enhanced tolerance. Brassica napus var. ZS758 was transformed by a binary vector harbouring sporamin and chitinase PjChi-1 gene. The transformed plants showcased increased resistance against Plutella xylostella and S. sclerotiorum (Liu et al., 2011). Anti-fungal and anit-bacterial activities have been reported due to lipid transfer protein (LTP) and, the Agrobacterium-mediated transfer of this protein into B. napus enhanced the peroxidase (POD) and superoxide dismutase (SOD) activities along with providing tolerance to the transgenic against SSR (Fan et al., 2013). Jiang et al. (2013) employed Agrobacterium tumefaciens to transfer a non-specific lipid transfer protein-like antimicrobial protein gene (LJAMP2) from Leonurus japonicus into B. napus genome. It was uniformly transcribed in all transgenics as confirmed by the RT-PCR analysis.
The defense response was initiated in transgenics with an increase in H2O2 and PR-1 gene but PDF 1.2 remained comparatively inactive. Tolerance to SSR was seen in B. napus transformed with the MSI-99m gene which belongs to the magainins class of antimicrobial peptides and is responsible for broad-spectrum resistance against bacteria and fungi (Sang et al., 2013). Kheiri et al. (2014) transferred a β-1,3-glucanase (bgn13.1) gene cloned in a pUC19 cloning vector from Trichoderma virens-10. The transgenic rape showed a high quantum of resistance. Various studies have repeatedly emphasised the role of WRKY transcription factors in plant defense. Transformed plants that overexpressed the BnWRKY33 gene (WRKY gene isolated from B. napus) enhanced the accumulation of H2O2 along with increased transcription of PR-1 and PDF 1.2 genes (Wang et al., 2014). Moradyar et al. (2016) concluded that along with SP-DDE synthetic promoter, the SP-DDE controlled expression of the chimeric chitinase gene was also responsible for inhibiting fungal growth and development. The chimeric Chit42 gene from Trichoderma atroviridae and a defensin gene from Raphanus sativus along with a Serratia marcescens C-terminal fused Chitin binding domain, were transferred and coexpressed in B. napus. The results shown were positive as a heterologous source can be used to transfer and induce resistance for SSR (Zarinpanjeh et al., 2016). Ziaei et al. (2016) developed SSR resistance by transferring the Chit42 gene along with the chitin-binding domain and polygalacturonase-inhibiting protein 2 (PGIP2) of Phaseolus vulgaris. Another PGIP protein was transferred to confer SSR resistance in rape. The Oryza sativa gene (OsPGIP2) was constitutively expressed in transgenic plants as confirmed by RT-PCR (Wang et al., 2018). An extracellular GDSL lipase gene, GDSL1 was identified in Arabidopsis thaliana. Loss of AtGDSL1 amounted to increased susceptibility while overexpression of the gene was responsible for enhanced SA, JA, and ROS accumulation (Ding et al., 2020). An NPR1, BnNPR1 gene was cloned from B. napus, which on overexpression resulted in increased resistance. This gene negatively regulated the JA pathway but had a positive modulation of SA (Wang et al., 2020).
Yield penalty is a serious issue caused by the ectopic expression of defense-related genes, so an S. sclerotiorum-induced promoter needs to be developed. In this line, Lin et al. (2022) developed pBnGH17D7 which overcame this issue. It was also used for host-induced gene silencing control of the disease. Foliar application of Verticillium dahliae Aspf2-like protein induced many defense-related compounds in B. rapa and possessed a great ability to increase the SSR resistance (Jiang et al., 2022). In angiosperms, guaiacyl monolignol is converted to syringyl monolignol with the help of Ferulate-5-hydroxylase. Resistance and biomass recalcitrance are reportedly affected by the monolignol ratio. CRISPR/Cas9-mediated knocking of the F5H gene decreased the S/G ratio thereby enhancing the defense response (Cao et al., 2022).
Downy mildew
Downy mildew (DM) is caused by a biotrophic oomycete, Hyaloperonospora brassicae (formerly Hyaloperonospora parasitica). Butler (1918) reported the disease, and it is found to be well spread in the major Brassica-growing regions of the world such as Canada, Australia, Europe, China and India (Saharan, 1992). All above-ground parts are affected by this seed-borne disease and the seedling stage is the most susceptible stage of all. Severe yield reduction has been reported from the adult plant stage on both floral and non-floral parts (Thines and Kummer, 2013; Meena et al., 2014b). Yellow or yellow-brown chlorotic lesions are visible on the upper surface of leaves, while the under surface has a prominent white powdery growth (Van De Wouw et al., 2016). Kaur et al. (2011b) reported that the severity of A. candida increased when co-infected with H. brassicae (Figures 3G, H). Multiple reports of the co-existence of A. candida and H. brassicae have been done in B. juncea in the Indian subcontinent, thus the development of resistant cultivars for both pathogens is essential to prevent yield losses (Saharan et al., 2016b; Mehta et al., 2018; Inturrisi et al., 2021).
HaRxL106 is an effector secreted by Hyaloperonospora arabidopsis which interacts with the Arabidopsis RADICAL-INDUCED CELL DEATH1 (RCD1). It inhibits the transcription of both defense-related and salicylic acid-triggered genes. Along with that, Mut9-like kinases (MLKs) also play a significant role in modulating the SA levels as SA-induced defense genes were highly expressed in mlk1,3,4 triple mutants (Wirthmueller et al., 2018). Another effector from the RxLR super family; HaRxLL470 interacted with HY5 in the host which was responsible for photomorphogenesis regulation. It is a vital protein for the activation of defense-related genes and the effector binds with the DNA of bZIP transcription factor in HY5 gene (Chen et al., 2021). Deb et al. (2018) characterised two effectors, PsAvh73 and HaRxL23 from Phytophthora sojae and H. Arabidopsis, respectively. These genes were found to be expressed very early during infection and suppressed PTI in Tobacco and ETI in Soybean. When the effector gene was constitutively expressed in Arabidopsis, it was found to impart resistance to these effectors. JA and SA signalling pathways are important for disease resistance in plants. An effector, HaRxL10 attacks JA signalling by targeting the JAZ3 (transcriptional repressor) and finally attenuates the SA signalling which is detrimental for oomycetes. It functionally resembles the bacterial toxin coronatine that mimics the jasmonic acid signalling via TTSS. This further signifies the vulnerability of the plant defense system due to SA-JA crosstalk (Anderson et al., 2019).
Caillaud et al. (2012) observed the sub-cellular compartments of host mesophyll during haustorial growth. Huge changes occurred in the tonoplast which is located close to the extra-haustorial membrane. HaRxL17 was in close association with tonoplast in un-infected cells and was found to be localised near the extra-haustorial membrane in case of infected ones. This establishes it as a potent effector which enhanced disease susceptibility. HaRxL21 interacted with Topless protein (TPL) which is an Arabidopsis transcriptional corepressor. The interaction occurs at the C-terminus EAR motif and was proven to be essential for virulence as it mimics the recruitment of TPL to transcriptional repression sites (Harvey et al., 2020). The plant is well connected in a cell-to-cell manner via plasmodesmata, but it is not beneficial for it to keep them open in infected tissue as isolation of infected cells is a prerequisite for avoiding their transmission to healthy cells. Thus, the pathogen targets this isolation mechanism and keeps the plasmodesmata open in the host tissue. HaRxL77 was reported to suppress the flg22-induced ROS response and promote hypermobility through manipulation of plasmodesmatal permeability. The study opens a new avenue that should be looked at, for plant-pathogen interaction (Liu et al., 2022). Dunker et al. (2020) reported an sRNA (HpasRNA), which utilises the Argonaute (AGO)/RNA-induced silencing complex of the host for causing disease. The transgenic Arabidopsis inhibiting HpasRNAs and atago1 mutants were used to determine their combined role in virulence. Though the role of sRNA is evidently proven in virulence, the cross-kingdom RNAi (ck-RNAi) mechanism must be explored.
Fourteen isolates of Peronospora parasitica were tested on the B. napus cultivar Cresor and it was found resistant to all. This resistance was shown to be conferred by a single dominant gene. When two homothallic isolates which were avirulent on the cultivar were combined, it led to virulence (Lucas et al., 1988). Dickson and Petzoldt (1993) reported that seedling stage resistance in broccoli for downy mildew is independent of the mature-plant stage. As the resistance varied between different stages, the selection must be done at the mature stage rather than the former. An analysis was done to check the response of the downy mildew attack on B. napus and two accession lines were deemed to be resistant viz., RES-26 and RES-02. The resistance for isolate P003 was governed by two independent dominant genes in RES-02 and one partially dominant gene in RES-26. Another isolate R1 was inoculated on RES-02 and the resistance pattern was found to be incompletely dominant. The genes for resistance against P003 and R1 were either identical or closely linked (Nashaat et al., 1997). Coelho et al. (1997) identified a single dominant R gene (Pp523) that is responsible for imparting resistance to broccoli plants. This locus Pp523 was situated on C8 and was the first gene to be identified to confer resistance to adult plants against P. parasitica. The flower colour character was found to be on C3, and the newly prepared map was denser than the earlier ones. This information can be used to perform map-based cloning of Pp523 (Carlier et al., 2011). The syntenic region was located at the top arm of chromosome 1 in A. thaliana (Farinhó et al., 2007).
Jensen et al. (1999) evaluated 20 DH broccoli lines and found br8 and br9 lines to allow fewer conidia production by 50-70% as compared to susceptible ones. The br9 line was somewhat more uniform in resistance response than br8, in which a sort of isolate-specificity was found. 31.8% and 45.8% variations were observed in conidia production and sporulation score, respectively. Recurrent selection for partial resistance in the early generation is an effective method for selecting resistant lines against downy mildew. A resistant DH line (from Everest) was crossed to the susceptible (from Marathon). All F1 plants were found to be resistant and a 9:7 resistant to susceptible ratio was observed in the F2 generation. This confirmed the gene’s dominant nature, which can be incorporated in F1 hybrids and commercially released (Wang et al., 2001). An NBS-LRR gene, RPP8 was identified and cloned by McDowell et al. (1998) and they did a comparative study of alleles in susceptible (Col-0) and resistant (Ler-0) accessions. In Ler-0, the RPP8 haplotype had a functional gene along with non-functional RPHA8. On the other hand, the rpp8 locus had a single chimeric gene in Col-0 accession. McDowell et al. (2005) mapped an adult resistance gene, RPP31 on chromosome no. 5 of A. thaliana. A SA degrading transgene, NahG and mutation leading to loss of function in defense-related genes such as PAD4, NPR1, PBS3 and RAR1 were able to suppress the adult-plant resistance. Three white rust resistance-conferring loci (RAC1, RAC2 and RAC3) were mapped and identified using two Arabidopsis accessions (ksk-1 and ksk-2). The two P. parasitica resistance genes RPP8 and RPP9 were found to be closely attached to RAC3 and RAC1(Borhan et al., 2001). The RPP5 gene in A. thaliana is responsible for downy mildew resistance and, it was positionally cloned by Parker et al. (1997). It encodes a protein with an NBS-LRR that resembles N and L6 proteins coded by R genes in plants. RPP5 produces a single transcript against the N and L6 which uses alternative splicing to produce truncated proteins. The terminal segment of gene looks like the cytoplasmic domain of Drosophila Toll and mammalian interleukin1-transmembrane receptors (TIR). 52 germplasm accessions of B. oleracea and its family members were evaluated for DM resistance. A recessive gene was involved in resistance which goes against the earlier studies in favour of the dominant R gene. This may be because the isolate of P. parasitica was collected from the B. napus field (Carlsson et al., 2004).
Four tightly linked genes were identified on the RPP1 region of chromosome no. 3 in Wassilewskija accession of A. thaliana. Out of these four, three (RPP1-WSSA, RPP1-WSSB, RPP1-WSSC) were found to encode NBS-LRR. Previously, the resistance was thought to be conferred by the RPP1 gene, but all three genes acted as complex loci and were responsible for resistance against multiple pathogen races (Botella et al., 1998). The RPP-13 locus in Arabidopsis had a variation in its LRR domain which was instrumental in resistance against multiple races of P. parasitica, previously thought to be conferred by different R genes. RPP13-Nd was responsible for resistance against 5 isolates of the pathogen and RPP13-Rld recognised some other specific isolates (Bittner-Eddy et al., 2000). An orthologue of RPP5; RPP4 was identified in Col-0 accession and found to provide resistance against Emoy2, Emwa1 and Noco2 races of P. parasitica. It required the action of 12 defense compounds such as Sa, EDS1, PAD4, DTH9, PBS2, PBS3, SID1 and SID2. Its expression at the cotyledonary stage was inhibited by mutations in rps5-1, ndr1 and npr1 but, no such effect was seen at adult plant stage (Van Der Biezen et al., 2002). RPP2A/RPP2B is a specific R gene which recognises P. parasitica isolate Cala2, mutational analysis and map-based cloning were done to study effector-receptor interaction. When this R gene was transferred into a new plant variety, it conferred complete resistance (Sinapidou et al., 2004). Yu et al. (2009) used MQM and interval mapping to construct an improved genetic map for Brassica rapa ssp. pekinensis. This pointed towards a major QTL on A08 and a major gene that imparted seedling resistance.
Yu et al. (2011) identified two microsatellite simple sequence repeat markers (kbrb058m10-1 and kbrb006c05-2) and one sequence-characterised amplified region marker (SCK14-825) being closely linked to seedling resistance QTL (BrDW) located on chromosome A8 in B. rapa ssp. pekinensis. Extending their work, four major QTLs, sBrDM8, yBrDM8, rBrDM8 and hBrDM8 were mapped by Yu et al. (2016) for seedling, young plant, rosette and heading stages, respectively on chromosome A08. Two minor QTLs, hBrDM6 on A06 and rBrDM6 on A04 were also found to be active at the heading and rosette stage. Downy mildew resistance was imparted by a single locus, dominant gene Ppa3 in cauliflower plants. 13 polymorphic markers were identified between two parental lines ‘Pusa Himjyoti’ and ‘BR-2’, out of which six were RAPD and seven were ISSR. Finally, a linkage map was constructed based on 120 F2 plants (Singh et al., 2012). Two inbred lines, RS1 and SS1 were selected from Chinese cabbage for downy mildew resistance and susceptibility, respectively. Evaluation for resistance was done in F1, F2 and BC1F1 populations and BrRHP1 was identified as a dominant single locus. Two molecular markers, BrPERK15A and BrPERK15B were developed along with an RAPD marker that is closely attached to BrRHP1 (Kim et al., 2011). Monteiro et al, 2005 checked the inheritance pattern for DM resistance at the cotyledonary and adult plant stage in B. oleracea var. tronchuda. For the cotyledonary stage, the F2 of the cross between resistant and susceptible varieties segregated in the ratio of 15:1, indicating that the trait is controlled by two duplicate dominant genes. The F2 adult plant stage segregated in the ratio 3:1, signifying a single dominant gene. B. napus was transformed by the Agrobacterium mediation and a bacterial catalase gene katE was introduced in the host chloroplast. The transformed and untransformed plants were checked for both downy mildew and powdery mildew resistance. The growth of fungi was seriously hampered in the transgenics and enzymes such as catalase, polyphenol oxidase and peroxidases were constitutively expressed, thus providing resistance to both diseases (El-Awady et al., 2008). Liu et al. (2021) evaluated forty members of Chinese cabbage CC-NBS-LRR. The phylogenetic relationship of CC-NBS-LRR was analysed in A. thaliana, B. rapa and Oryza sativa. They were also classified based on their conserved domains and role of BrCC-NBS-LRR was established in pathogenesis-related defense. Finally, expression profiling was done on both short-duration and long-duration basis.
Conclusion
Effectoromics is an emerging concept which can be used to identify the pathogen and establish a phylogenetic relationship between members of the same species or across a larger group. It involves a high-throughput functional genomic way for examining plant genomes to find new R genes in cultivated brassica and its relatives. The cognate R gene interacting with the effector of the pathogen once identified can be introgressed in the high-yielding susceptible varieties. The R genes mostly belong to NBS-LRR class of protein-encoding genes and are responsible for narrow-spectrum resistance which is overcome by more virulent races. The introgression is always not without challenges, as sometimes the R genes are closely linked with other non-desirable agronomic traits or in some instances, R genes for biotrophs can behave as pathogen targets (S genes) for necrotrophs. The narrow spectrum nature of this resistance can be overcome by overexpressing the defense-related genes which form the basis of broad-spectrum resistance and relies on hormonal signalling pathways and strengthening of host tissues rather than targeting or interacting with the pathogenicity factors (effectors). There have been many studies conducted on modulating the expression of these genes that leads to resistance. Transgenics go beyond the conventional search of resistance sources in the host family. Many genes that are transferred from across the species, genera, and in some cases cross-kingdoms as well have been able to confer resistance. Thus, for developing a robust disease management strategy, identifying essential genes conferring the pathogenicity and the role of HSTs in each host-pathogen system for identifying the R gene sources, is a must. A better analysis of reaction of transgenics with its biotic and abiotic environment is also essential to check its compatibility with other control measures such as biocontrol. This review focuses on three major concepts: fungal effectors, R genes/QTLs and defense-related genes and transgenics so that in the future better resistance breeding modules can be developed.
Author contributions
PR wrote the draft. PKR and LP edited the manuscript and provided critical review. All authors contributed to the article and approved the submitted version.
Acknowledgments
PR acknowledges ICAR-Indian Agriculture Research Institute for scholarship support and other facilities.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adhikari, T. B., Liu, J. Q., Mathur, S., Wu, C. X., Rimmer, S. R. (2003). Genetic and molecular analyses in crosses of race 2 and race 7 of Albugo candida. Phytopathology 93 (8), 959–965. doi: 10.1094/PHYTO.2003.93.8.959
Agarwal, A., Garg, G. K., Singh, U. S., Mishra, D. P. (1994). Detection and role of chlorotic toxin and phytohormones in the pathogenesis of alternaria blight in Brassica napus. Curr. Sci. (Bangalore) 66 (6), 442–443.
Akagi, Y., Akamatsu, H., Otani, H., Kodama, M. (2009). Horizontal chromosome transfer, a mechanism for the evolution and differentiation of a plant-pathogenic fungus. Eukaryotic Cell 8 (11), 1732–1738. doi: 10.1128/EC.00135-09
Akhtar, J., Singh, B., Kumar, A. K. P., Maurya, A. K., Dubey, S. C. (2017). Interception of pathogens during quarantine processing: an effort towards safe import of oilseed and vegetable brassicas germplasm in India. J. Oilseed Brassica 81 (2), 120–130.
Alhoraibi, H., Bigeard, J., Rayapuram, N., Colcombet, J., Hirt, H. (2019). Plant immunity: the MTI-ETI model and beyond. Curr. Issues Mol. Biol. 30 (1), 39–58. doi: 10.21775/cimb.030.039
Ali, S., Mir, Z. A., Tyagi, A., Mehari, H., Meena, R. P., Bhat, J. A., et al. (2017). Overexpression of NPR1 in Brassica juncea confers broad spectrum resistance to fungal pathogens. Front. Plant Sci. 8, 1693. doi: 10.3389/fpls.2017.01693
Anderson, R., Deb, D., Withers, J., He, S. Y., McDowell, J. (2019). An oomycete RXLR effector triggers antagonistic plant hormone crosstalk to suppress host immunity. bioRxiv 561605, 1–40. doi: 10.1101/561605
Arora, H., Padmaja, K. L., Paritosh, K., Mukhi, N., Tewari, A. K., Mukhopadhyay, A., et al. (2019). BjuWRR1, a CC-NB-LRR gene identified in Brassica juncea, confers resistance to white rust caused by Albugo candida. Theor. Appl. Genet. 132 (8), 2223–2236. doi: 10.1007/s00122-019-03350-z
Atri, C., Akhatar, J., Gupta, M., Gupta, N., Goyal, A., Rana, K., et al. (2019). Molecular and genetic analysis of defensive responses of Brassica juncea–B. fruticulosa introgression lines to Sclerotinia infection. Sci. Rep. 9 (1), 1–12. doi: 10.1038/s41598-019-53444-3
Axelsson, T., Bowman, C. M., Sharpe, A. G., Lydiate, D. J., Lagercrantz, U. (2000). Amphidiploid brassica juncea contains conserved progenitor genomes. Genome 43, 679–688. doi: 10.1139/gen-43-4-679
Bains, P., Tewari, J. P. (1987). Purification, chemical characterization and host-specificity of the toxin produced by Alternaria brassicae. Physiol. Mol. Plant Pathol. 30 (2), 259–271. doi: 10.1016/0885-5765(87)90039-7
Bal, R., Kumar, A. (2014). Studies on the epidemiology of white rust and alternaria leaf blight and their effect on the yield of Indian mustard. Afr. J. Agric. Res. 9 (2), 302–306. doi: 10.5897/AJAR2013.7352
Bashi, Z. D., Hegedus, D. D., Buchwaldt, L., Rimmer, S. R., Borhan, M. H. (2010). Expression and regulation of Sclerotinia sclerotiorum necrosis and ethylene-inducing peptides (NEPs). Mol. Plant Pathol. 11 (1), 43–53. doi: 10.1111/j.1364-3703.2009.00571.x
Bashir, A., Khan, M. S., Haider, A., Khan, I., Ambreen (2015). Agrobacterium mediated transformation of brassica juncea (L.) czern. with chitinase gene conferring resistance against fungal infections. Pak. J. Bot. 47 (1), 211–216.
Baxter, L., Tripathy, S., Ishaque, N., Boot, N., Cabral, A., Kemen, E., et al. (2010). Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science 330 (6010), 1549–1551. doi: 10.1126/science.1195203
Berkenkamp, B., Kirkham, C. (1989). Canola disease survey in NE saskatchewa. Can. Plant Dis. Sur. 69, 62.
Bhayana, L., Paritosh, K., Arora, H., Yadava, S. K., Singh, P., Nandan, D., et al. (2020). A mapped locus on LG A6 of Brassica juncea line tumida conferring resistance to white rust contains a CNL type R gene. Front. Plant Sci. 10, 1690. doi: 10.3389/fpls.2019.01690
Bittner-Eddy, P. D., Crute, I. R., Holub, E. B., Beynon, J. L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21 (2), 177–188. doi: 10.1046/j.1365-313x.2000.00664.x
Borhan, M. H., Brose, E., Beynon, J. L., Holub, E. B. (2001). White rust (Albugo candida) resistance loci on three Arabidopsis chromosomes are closely linked to downy mildew (Peronospora parasitica) resistance loci. Mol. Plant Pathol. 2 (2), 87–95. doi: 10.1046/j.1364-3703.2001.00056.x
Borhan, M. H., Gunn, N., Cooper, A., Gulden, S., Tör, M., Rimmer, S. R., et al. (2008). WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Mol. Plant-Microbe Interact. 21 (6), 757–768. doi: 10.1094/MPMI-21-6-0757
Borhan, M. H., Holub, E. B., Kindrachuk, C., Omidi, M., Bozorgmanesh-Frad, G., Rimmer, S. R. (2010). WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed brassica crops. Mol. Plant Pathol. 11 (2), 283–291. doi: 10.1111/j.1364-3703.2009.00599.x
Botella, M. A., Parker, J. E., Frost, L. N., Bittner-Eddy, P. D., Beynon, J. L., Daniels, M. J., et al. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct peronospora parasitica avirulence determinants. Plant Cell 10 (11), 1847–1860. doi: 10.1105/tpc.10.11.1847
Buchwaldt, L., Green, H. (1992). Phytotoxicity of destruxin b and its possible role in the pathogenesis of Alternaria brassicae. Plant Pathol. 41 (1), 55–63. doi: 10.1111/j.1365-3059.1992.tb02316.x
Caarls, L., Pieterse, C. M., Van Wees, S. C. (2015). How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6, 170. doi: 10.3389/fpls.2015.00170
Caillaud, M. C., Piquerez, S. J., Fabro, G., Steinbrenner, J., Ishaque, N., Beynon, J., et al. (2012). Subcellular localization of the hpa RxLR effector repertoire identifies a tonoplast-associated protein HaRxL17 that confers enhanced plant susceptibility. Plant J. 69 (2), 252–265. doi: 10.1111/j.1365-313X.2011.04787.x
Cao, Y., Yan, X., Ran, S., Ralph, J., Smith, R. A., Chen, X., et al. (2022). Knockout of the lignin pathway gene BnF5H decreases the S/G lignin compositional ratio and improves Sclerotinia sclerotiorum resistance in Brassica napus. Plant Cell Environ. 45 (1), 248–261. doi: 10.1111/pce.14208
Carlier, J. D., Alabaça, C. A., Coelho, P. S., Monteiro, A. A., Leitão, J. M. (2011). The downy mildew resistance locus Pp523 is located on chromosome C8 of Brassica oleracea l. Plant Breed. 131, 170–175. doi: 10.1111/j.1439-0523.2011.01904.x
Carlsson, M., Bothmer, R. V., Merker, A. (2004). Screening and evaluation of resistance to downy mildew (Peronospora parasitica) and clubroot (Plasmodiophora brassicae) in genetic resources of Brassica oleracea. Hereditas 141 (3), 293–300. doi: 10.1111/j.1601-5223.2004.01818.x
Castel, B., Fairhead, S., Furzer, O. J., Redkar, A., Wang, S., Cevik, V., et al. (2021). Evolutionary trade-offs at the Arabidopsis WRR4A resistance locus underpin alternate Albugo candida race recognition specificities. Plant J. 107 (5), 1490–1502. doi: 10.1111/tpj.15396
Cevik, V., Boutrot, F., Apel, W., Robert-Seilaniantz, A., Furzer, O. J., Redkar, A., et al. (2019). Transgressive segregation reveals mechanisms of Arabidopsis immunity to brassica-infecting races of white rust (Albugo candida). Proc. Natl. Acad. Sci. 116 (7), 2767–2773. doi: 10.1073/pnas.1812911116
Chadar, L. K., Singh, R. P., Singh, R. K., Yadav, R. R., Mishra, M. K., Pratap, N., et al. (2016). Studies on alternaria blight of rapeseed-mustard (Brassica juncea l.) caused by Alternaria brassicae (Berk.) sacc. and its integrated management. Plant Arch. 16 (2), 897–901.
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-neolithic brassica napus oilseed genome. Science 345 (6199), 950–953. doi: 10.1111/nph.17280
Chen, S., Ma, T., Song, S., Li, X., Fu, P., Wu, W., et al. (2021). Arabidopsis downy mildew effector HaRxLL470 suppresses plant immunity by attenuating the DNA-binding activity of bZI Ptranscription factor HY5. New Phycologist 230, 1562–1577. doi: 10.1111/nph.17280
Cheung, W. Y., Gugel, R. K., Landry, B. S. (1998). Identification of RFLP markers linked to the white rust resistance gene (Acr) in mustard (Brassica juncea (L.) czern. and coss.). Genome 41 (4), 626–628. doi: 10.1139/g98-043
Chhikara, S., Chaudhury, D., Dhankher, O. P., Jaiwal, P. K. (2012). Combined expression of a barley class II chitinase and type I ribosome inactivating protein in transgenic Brassica juncea provides protection against Alternaria brassicae. Plant Cell Tissue Organ Culture (PCTOC) 108 (1), 83–89. doi: 10.1007/s11240-011-0015-7
Choi, Y. J., Shin, H. D., Hong, S. B., Thines, M. (2007). Morphological and molecular discrimination among Albugo candida materials infecting Capsella bursa-pastoris world-wide. Fungal Divers. 27, 11–34.
Christias, C., Lockwood, J. L. (1973). Conservation of mycelia constituents in four sclerotium forming fungi in nutrient deprived conditions. Phytopath. 63, 602–605. doi: 10.1094/Phyto-63-602
Coelho, P., Leckie, D., Bahcevandziev, K., Valerio, L., Astley, D., Boukema, I. W., et al. (1997). The relationship between cotyledon and adult plant resistance to downy mildew (Peronospora parasitica) in Brassica oleracea. Acta Hortic. 459, 335–342. doi: 10.17660/ActaHortic.1998.459.39
Davidson, R. M., Reeves, P. A., Manosalva, P. M., Leach, J. E. (2009). Germins: a diverse protein family important for crop improvement. Plant Sci. 177, 499–510. doi: 10.1016/j.plantsci.2009.08.012
De, A., Maity, A., Mazumder, M., Mondal, B., Mukherjee, A., Ghosh, S., et al. (2021). Overexpression of LYK4, a lysin motif receptor with non-functional kinase domain, enhances tolerance to Alternaria brassicicola and increases trichome density in Brassica juncea. Plant Sci. 309, 110953. doi: 10.1016/j.plantsci.2021.110953
Deb, D., Anderson, R. G., How-Yew-Kin, T., Tyler, B. M., McDowell, J. M. (2018). Conserved RxLR effectors from oomycetes Hyaloperonospora arabidopsidis and Phytophthora sojae suppress PAMP-and effector-triggered immunity in diverse plants. Mol. Plant-Microbe Interact. 31 (3), 374–385. doi: 10.1094/MPMI-07-17-0169-FI
De Block, M., De Brouwer, D., Tenning, P. (1989). Transformation of Brassica napus and Brassica oleracea using Agrobacterium tumefaciens and the expression of the bar and neo genes in the transgenic plants. Plant Physiol. 91 (2), 694–701. doi: 10.1104/pp.91.2.694
Derbyshire, M. C., Newman, T. E., Khentry, Y., Owolabi Taiwo, A. (2022). The evolutionary and molecular features of the broad-host-range plant pathogen Sclerotinia sclerotiorum. Mol. Plant Pathol. 23 (8), 1075–1090. doi: 10.1111/mpp.13221
Dickson, M. H., Petzoldt, R. (1993). Plant age and isolate source affect expression of downy mildew resistance in broccoli. HortScience 28 (7), 730–731. doi: 10.21273/HORTSCI.28.7.730
Ding, L. N., Li, M., Guo, X. J., Tang, M. Q., Cao, J., Wang, Z., et al. (2020). Arabidopsis GDSL1 overexpression enhances rapeseed Sclerotinia sclerotiorum resistance and the functional identification of its homolog in Brassica napus. Plant Biotechnol. J. 18 (5), 1255–1270. doi: 10.1111/pbi.13289
Djamei, A., Schipper, K., Rabe, F., Ghosh, A., Vincon, V., Kahnt, J., et al. (2011). Metabolic priming by a secreted fungal effector. Nature 478 (7369), 395–398. doi: 10.1038/nature10454
Dong, X., Ji, R., Guo, X., Foster, S. J., Chen, H., Dong, C., et al. (2008). Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228 (2), 331–340. doi: 10.1007/s00425-008-0740-2
Dong, S., Kong, G., Qutob, D., Yu, X., Tang, J., Kang, J., et al. (2012). The NLP toxin family in Phytophthora sojae includes rapidly evolving groups that lack necrosis-inducing activity. Mol. Plant-Microbe Interact. 25 (7), 896–909. doi: 10.1094/MPMI-01-12-0023-R
Doughty, K. J., Blight, M. M., Bock, C. H., Fieldsend, J. K., Pickett, J. A. (1996). Release of alkenyl isothiocyanates and other volatiles from Brassica rapa seedlings during infection by Alternaria brassicae. Phytochemistry 43 (2), 371–374. doi: 10.1016/0031-9422(96)00189-6
Du, J., Vleeshouwers, V. G. (2014). “The do’s and don’ts of effectoromics. plant-pathogen interactions,” in Methods in microbiology, pp257–pp268. doi: 10.1007/978-1-62703-986-4_19
Duhan, D., Gajbhiye, S., Jaswal, R., Singh, R. P., Sharma, T. R., Rajarammohan, S. (2021). Functional characterization of the Nep1-like protein effectors of the necrotrophic pathogen–Alternaria brassicae. Front. Microbiol. 12, 738617. doi: 10.3389/fmicb.2021.738617
Dunker, F., Trutzenberg, A., Rothenpieler, J. S., Kuhn, S., Pröls, R., Schreiber, T., et al. (2020). Oomycete small RNAs bind to the plant RNA-induced silencing complex for virulence. Elife 9, e56096. doi: 10.7554/eLife.56096
Dutton, M. V., Evans, C. S. (1996). Oxalate production by fungi: its role in pathogenicity and ecology in the soil environment. Can. J. Microbiol. 42, 881–895. doi: 10.1139/m96-114
El-Awady, M., Reda, E. A. M., Haggag, W., Sawsan, S. Y., Ahmed, M. (2008). Transgenic canola plants over-expressing bacterial catalase exhibit enhanced resistance to Peronospora parasitica and Erysiphe polygoni. Arab. J. Biotechnol. 11, 71–84.
Fan, Y., Du, K., Gao, Y., Kong, Y., Chu, C., Sokolov, V., et al. (2013). Transformation of LTP gene into Brassica napus to enhance its resistance to Sclerotinia sclerotiorum. Russian J. Genet. 49 (4), 380–387. doi: 10.1134/S1022795413040042
Farinhó, M., Coelho, P., Monteiro, A., Leitão, J. (2007). SCAR and CAPS markers flanking the Brassica oleracea l. Pp523 downy mildew resistance locus demarcate a genomic region syntenic to the top arm end of Arabidopsis thaliana l. chromosome 1. Euphytica 157 (1), 215–221. doi: 10.1007/s10681-007-9414-6
Fatima, U., Bhorali, P., Borah, S., Senthil-Kumar, M. (2019). Perspectives on the utilization of resistance mechanisms from host and nonhost plants for durable protection of brassica crops against alternaria blight. PeerJ 7, e7486. doi: 10.7717/peerj.7486
Ferreira, M. E., Williams, P. H., Osborn, T. C. (1994). RFLP mapping of Brassica napus using doubled haploid lines. Theor. Appl. Genet. 89 (5), 615–621. doi: 10.1007/BF00222456
Friesen, T. L., Faris, J. D., Solomon, P. S., Oliver, R. P. (2008). Host-specific toxins: effectors of necrotrophic pathogenicity. Cell. Microbiol. 10 (7), 1421–1428. doi: 10.1111/j.1462-5822.2008.01153.x
Furzer, O. J., Cevik, V., Fairhead, S. (2022). An improved assembly of the Albugo candida Ac2V genome reveals the expansion of the “CCG” class of effectors. Mol. Plant-Microbe Interact. 35 (1), 39–48. doi: 10.1094/MPMI-04-21-0075-R
Gaur, M., Tiwari, A., Chauhan, R. P., Pandey, D., Kumar, A. (2018). Molecular modeling, docking and protein-protein interaction analysis of MAPK signalling cascade involved in camalexin biosynthesis in Brassica rapa. Bioinformation 14 (4), 145. doi: 10.6026/97320630014145
Ghasolia, R. P., Shivpuri, A., Bhargava, A. K. (2004). Sclerotinia rot of Indian mustard (Brassica juncea) in rajasthan. Indian Phytopathol. 57, 76–79.
Ghozlan, M. H., Eman, E. A., Tokgöz, S., Lakshman, D. K., Mitra, A. (2020). Plant defense against necrotrophic pathogens. Am. J. Plant Sci. 11 (12), 2122–2138. doi: 10.4236/ajps.2020.1112149
Gladders, P. (1987). Current status of diseases and disease control in winter oilseed rape in England and Wales. Bull. SROP 10, 7–10.
Godoy, G., Steadman, J. R., Dickman, M. B., Dam, R. (1990). Use of mutants to demonstrate the role of oxalic acid in pathogenicity of Sclerotinia sclerotiorum on Phaseolus vulgaris. Physiol. Mol. Plant Pathol. 37, 179–191. doi: 10.1016/0885-5765(90)90010-U
Gupta, K., Saharan, G. S. (2002). Identification of pathotype of Albugo candida with stable characteristic symptoms on Indian mustard. J. Mycol Plant Pathol. 32, 46–51.
Gupta, N. C., Yadav, S., Arora, S., Mishra, D. C., Budhlakoti, N., Gaikwad, K., et al. (2022). Draft genome sequencing and secretome profiling of sclerotinia sclerotiorum revealed effector repertoire diversity and allied broad-host range necrotrophy. Sci. Rep. 12 (1), 21855. doi: 10.1038/s41598-022-22028-z
Gyawali, S., Harrington, M., Durkin, J., Horner, K., Parkin, I. A., Hegedus, D. D., et al. (2016). Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol. Breed. 36 (6), 1–13. doi: 10.1007/s11032-016-0496-5
Haas, B. J., Kamoun, S., Zody, M. C., Jiang, R. H., Handsaker, R. E., Cano, L. M., et al. (2009). Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461 (7262), 393–398. doi: 10.1038/nature08358
Harvey, S., Kumari, P., Lapin, D., Griebel, T., Hickman, R., Guo, W., et al. (2020). Downy mildew effector HaRxL21 interacts with the transcriptional repressor TOPLESS to promote pathogen susceptibility. PloS Pathog. 16 (8), e1008835. doi: 10.1371/journal.ppat.1008835
He, D. C., Zhan, J. S., Xie, L. H. (2016). Problems, challenges, and future of plant disease management: from an ecological point of view. J. Integr. Agric. 15 (4), 705–715. doi: 10.1016/S2095-3119(15)61300-4
Heil, M., Land, W. G. (2014). Danger signals - damaged-self recognition across the tree of life. Frontiersin Plant Sci. 5. doi: 10.3389/fpls.2014.00578
Hirata, S. (1954). Studies on the phytohormone in the malformed portion of the diseased plants. i. the relation between the growth rate and the amount of free auxin in the fungous galls and virus infected plants. Ann. Phytopathol. Soc Jpn. 19, 33–38. doi: 10.3186/jjphytopath.19.33
Huang, W., Wang, Y., Li, X., Zhang, Y. (2020). Biosynthesis and regulation of salicylic acid and n-hydroxypipecolic acid in plant immunity. Mol. Plant 13 (1), 31–41. doi: 10.1016/j.molp.2019.12.008
Humaydan, H. S., Williams, P. H. (1976). Inheritance of seven characters in Raphanus sativus l. 1. HortScience 11 (2), 146–147. doi: 10.21273/HORTSCI.11.2.146
Humpherson-Jones, F. M. (1991). The development of weather-related disease forecasts for vegetable crops in the UK. problems and prospects 1. EPPO Bull. 21 (3), 425–429. doi: 10.1111/j.1365-2338.1991.tb01272.x
Inturrisi, F. C., Barbetti, M. J., Tirnaz, S., Patel, D. A., Edwards, D., Batley, J. (2021). Molecular characterization of disease resistance in Brassica juncea–the current status and the way forward. Plant Pathol. 70 (1), 13–34. doi: 10.1111/ppa.13277
Jensen, B. D., Værbak, S., Munk, L., Andersen, S. B. (1999). Characterization and inheritance of partial resistance to downy mildew, Peronospora parasitica, in breeding material of broccoli, Brassica oleracea convar. botrytis var. italica. Plant Breed. 118 (6), 549–554. doi: 10.1046/j.1439-0523.1999.00409.x
Jiang, Y., Fu, X., Wen, M., Wang, F., Tang, Q., Tian, Q., et al. (2013). Overexpression of an nsLTPs-like antimicrobial protein gene (LJAMP2) from motherwort (Leonurus japonicus) enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Physiol. Mol. Plant Pathol. 82, 81–87. doi: 10.1016/j.pmpp.2012.11.001
Jiang, S., Zheng, W., Li, Z., Tan, J., Wu, M., Li, X., et al. (2022). Enhanced resistance to Sclerotinia sclerotiorum in Brassica rapa by activating host immunity through exogenous Verticillium dahliae Aspf2-like protein (VDAL) treatment. Int. J. Mol. Sci. 23 (22), 13958. doi: 10.3390/ijms232213958
Jin, J. B., Jin, Y. H., Lee, J., Miura, K., Yoo, C. Y., Kim, W. Y., et al. (2008). The SUMO E3 ligase, AtSIZ1, regulates flowering by controlling a salicylic acid-mediated floral promotion pathway and through affects on FLC chromatin structure. Plant J. 53, 530–540. doi: 10.1111/j.1365-313X.2007.03359.x
Jingtao, L., Zhang, Y., Zhang, Y., Yu, P., Pan, H., Rollins, J. A. (2018). Introduction of large sequence inserts by CRISPR-Cas9 to create pathogenicity mutants in the multinucleate filamentous pathogen. Sclerotinia sclerotiorum. MBio 9, e00567–e0e618.
Jones, J. D. G., Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jyoti, S. D., Sultana, N., Hassan, L., Robin, A. H. K. (2021). Epidemiology, Genetics and Resistance of Alternaria Blight in Oilseed Brassica. In: Brassica Breeding and Biotechnology. (IntechOpen). Pp 1–174. doi: 10.5772/intechopen.96454
Kabbage, M., Williams, B., Dickman, M. B. (2013). Cell death control: the interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PloS Pathog. 9 (4), e1003287. doi: 10.1371/journal.ppat.1003287
Kadian, A. K., Saharan, G. S. (1983). Symptomatology, host range and assessment of losses due to Alternaria brassicae infection in rapeseed and mustard. Indian J. Mycol. Pl. Pathol. 13, 319–323.
Kamble, S., Mukherjee, P. K., Eapen, S. (2016). Expression of an endochitinase gene from Trichoderma virens confers enhanced tolerance to alternaria blight in transgenic Brassica juncea (L.) czern and coss lines. Physiol. Mol. Biol. Plants 22 (1), 69–76. doi: 10.1007/s12298-016-0340-8
Kanrar, S., Venkateswari, J. C., Kirti, P. B., Chopra, V. L. (2002). Transgenic expression of hevein, the rubber tree lectin, in Indian mustard confers protection against Alternaria brassicae. Plant Sci. 162 (3), 441–448. doi: 10.1016/S0168-9452(01)00588-X
Kaur, P., Jost, R., Sivasithamparam, K., Barbetti, M. J. (2011a). Proteome analysis of the Albugo candida–brassica juncea pathosystem reveals that the timing of the expression of defense-related genes is a crucial determinant of pathogenesis. J. Exp. Bot. 62 (3), 1285–1298. doi: 10.1093/jxb/erq365
Kaur, P., Sivasithamparam, K., Barbetti, M. (2011b). Host range and phylogenetic relationships of Albugo candida from cruciferous hosts in Western Australia, with special reference to Brassica juncea. Plant Dis. 95 (6), 712–718. doi: 10.1094/PDIS-10-10-0765
Kemen, E., Gardiner, A., Schultz-Larsen, T., Kemen, A. C., Balmuth, A. L., Robert-Seilaniantz, A., et al. (2011). Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PloS Biol. 9 (7), e1001094. doi: 10.1371/journal.pbio.1001094
Keswani, C., Bisen, K., Singh, S. P., Sarma, B. K., Singh, H. B. (2016). A proteomic approach to understand the tripartite interactions between plant-Trichoderma-pathogen: investigating the potential for efficient biological control. in. Plant Soil Microbes, (Springer International Publishing Switzerland), pp. 79–93. doi: 10.1007/978-3-319-29573-2_5
Kheiri, H. R., Motallebi, M., Zamani, M. R., Deljo, A. (2014). Beta glucanase (Bgn13. 1) expressed in transgenic Brassica napus confers antifungal activity against Sclerotinia sclerotiorum. J. Crop Prot. 3 (1), 31–42.
Kim, K. T., Jeon, J., Choi, J., Cheong, K., Song, H., Choi, G., et al. (2016). Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00186
Kim, S., Song, Y. H., Lee, J. Y., Choi, S. R., Dhandapani, V., Jang, C. S., et al. (2011). Identification of the BrRHP1 locus that confers resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis) and development of linked molecular markers. Theor. Appl. Genet. 123 (7), 1183–1192. doi: 10.1007/s00122-011-1658-9
Klemm, M. (1938). The most important diseases and pests of colza and rape. Dtsch. Landw. 20, 251–252.
Kole, C., Teutonico, R., Mengistu, A., Williams, P. H., Osborn, T. C. (1996). Molecular mapping of a locus controlling resistance to Albugo candida in Brassica rapa. Phytopathology 86 (4), 367–369. doi: 10.1094/Phyto-86-367
Kolte, S. J. (1985). Diseases of annual edible oilseed crops. volume II: rapeseed mustard and sesame diseases (Boca Raton, Florida: CRC Press, Inc.), 135 p.
Kravchuk, Z., Vicedo, B., Flors, V., et al. (2011). Priming for JA-dependent defenses using hexanoic acid is an effective mechanism to protect Arabidopsis against B. cinerea. J. Plant Physiol. 168 (4), 359–366. doi: 10.1016/j.jplph.2010.07.028
Kull, L. S., Pedersen, W. L., Palmquist, D., Hartman, G. L. (2004). Mycelial compatibility grouping and aggressiveness of Sclerotinia sclerotiorum. Plant Dis. 88, 325–332. doi: 10.1094/PDIS.2004.88.4.325
Kumar, D., Shekhar, S., Bisht, S., Kumar, V., Varma, A., Kumar, M. (2015). Ectopic overexpression of lectin in transgenic Brassica juncea plants exhibit resistance to fungal phytopathogen and showed alleviation to salt and drought stress. J. Bioeng BioMed. Sci. 5 (1), 147. doi: 10.4172/2155-9538.1000147
Lakra, B. S., Saharan, G. S. (1989). Sources of resistance and effective screening techniques in Brassica-albugo system. Indian Phytopathol. 42, 293.
Laluk, K., Mengiste, T. (2010). Necrotroph attacks on plants: wanton destruction or covert extortion? Arabidopsis Book/American Soc. Plant Biologists 8, e0136. doi: 10.1199/tab.0136
Lan, H. Y., Wang, C. H., Zhang, L. H., Liu, G. Z., Wan, L. L., Chen, Z. H., et al. (2000). Studies on transgenic oilseed rape (Brassica napus) plants transformed with beta-1, 3-glucanase and chitinase genes and its resistance to Sclerotinia sclerotiorium. Sheng Wu Gong Cheng Xue Bao 16 (2), 142–146.
Lee, H. A., Kim, S. Y., Oh, S. K., Yeom, S. I., Kim, S. B., Kim, M. S., et al. (2014). Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol. 203 (3), 926–938. doi: 10.1111/nph.12861
Lenarčič, T., Albert, I., Böhm, H., Hodnik, V., Pirc, K., Zavec, A. B., et al. (2017). Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins. Science 358, 1431–1434. doi: 10.1126/science.aan6874
Li, J., Zhao, Z., Hayward, A., Cheng, H., Fu, D. (2015). Integration analysis of quantitative trait loci for resistance to Sclerotinia sclerotiorum in Brassica napus. Euphytica 205 (2), 483–489. doi: 10.1007/s10681-015-1417-0
Liang, X., Liberti, D., Li, M., Kim, Y. T., Hutchens, A., Wilson, R., et al. (2015). Oxaloacetate acetylhydrolase gene mutants of Sclerotinia sclerotiorum do not accumulate oxalic acid, but do produce limited lesions on host plants. Mol. Plant Pathol. 16, 559–571. doi: 10.1111/mpp.12211
Liang, X., Rollins, J. A. (2018). Mechanisms of broad host range necrotrophic pathogenesis in Sclerotinia sclerotiorum. Phytopathology 108 (10), 1128–1140. doi: 10.1094/PHYTO-06-18-0197-RVW
Liang, Y., Yajima, W., Davis, M. R., Kav, N. N. V., Strelkov, S. E. (2013). Disruption of a gene encoding a hypothetical secreted protein from Sclerotinia sclerotiorum reduces its virulence on canola (Brassica napus). Can. J. Plant Pathol. 35, 46–55. doi: 10.1080/07060661.2012.745904
Liao, C. J., Hailemariam, S., Sharon, A., Mengiste, T. (2022). Pathogenic strategies and immune mechanisms to necrotrophs: differences and similarities to biotrophs and hemibiotrophs. Curr. Opin. Plant Biol. 69, 102291. doi: 10.1016/j.pbi.2022.102291
Lin, L., Fan, J., Li, P., Liu, D., Ren, S., Lin, K., et al. (2022). The Sclerotinia sclerotiorum-inducible promoter pBnGH17 D7 in Brassica napus: isolation, characterization, and application in host-induced gene silencing. J. Exp. Bot. 73 (19), 6663–6677. doi: 10.1093/jxb/erac328
Links, M. G., Holub, E., Jiang, R. H. Y., Sharpe, A. G., Hegedus, D., Beynon, E., et al. (2011). De novo sequence assembly of Albugo candida reveals a small genome relative to other biotrophic oomycetes. BMC Genomics 12 (1), 1–12. doi: 10.1186/1471-2164-12-503
Liu, X., Bellandi, A., Johnston, M. G., Faulkner, C. (2022). The Hyaloperanospora arabidopsidis effector HaRxL77 is hypermobile between cells and manipulates host defense. bioRxiv, 1–31. doi: 10.1101/2022.01.24.477405
Liu, H., Guo, X., Naeem, M., Liu, D., Xu, L., Zhang, W., et al. (2011). Transgenic Brassica napus l. lines carrying a two gene construct demonstrate enhanced resistance against Plutella xylostella and Sclerotinia sclerotiorum. Plant Cell Tissue Organ Culture (PCTOC) 106 (1), 143–15.1. doi: 10.1007/s11240-010-9902-6
Liu, Y., Li, D., Yang, N., Zhu, X., Han, K., Gu, R., et al. (2021). Genome-wide identification and analysis of CC-NBS-LRR family in response to downy mildew and black rot in Chinese cabbage. Int. J. Mol. Sci. 22 (8), 1–17. doi: 10.3390/ijms22084266
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5 (1), 1–11. doi: 10.1038/ncomms4930
Lucas, J. A., Crute, I. R. C., Gordon, P. L. (1988). The identification of a gene for race-specific resistance to Peronospora parasitica (downy mildew) in Brassica napus var. oleifera (oilseed rape). Plant Pathol. 37 (4), 538–545. doi: 10.1111/j.1365-3059.1988.tb02112.x
Lyu, X., Shen, C., Fu, Y., Xie, J., Jiang, D., Li, G., et al. (2015). Comparative genomic and transcriptional analyses of the carbohydrate-active enzymes and secretomes of phytopathogenic fungi reveal their significant roles during infection and development. Sci. Rep. 5 (1), 1–16. doi: 10.1038/srep15565
Lyu, X., Shen, C., Fu, Y., Xie, J., Jiang, D., Li, G., et al. (2016). A small secreted virulence-related protein is essential for the necrotrophic interactions of Sclerotinia sclerotiorum with its host plants. PloS Pathog. 12 (2), e1005435. doi: 10.1371/journal.ppat.1005435
Macioszek, V. K., Lawrence, C. B., Kononowicz, A. K. (2018). Infection cycle of Alternaria brassicicola on Brassica oleracea leaves under growth room conditions. Plant Pathol. 67 (5), 1088–1096. doi: 10.1111/ppa.12828
Mazumder, M., Das, S., Saha, U., Chatterjee, M., Bannerjee, K., Basu, D. (2013). Salicylic acid-mediated establishment of the compatibility between Alternaria brassicicola and Brassica juncea is mitigated by abscisic acid in Sinapis alba. Plant physiology and biochemistry. PPB 70, 43–51. doi: 10.1016/j.plaphy.2013.04.025
McDonald, M. R., Boland, G. J. (2004). Forecasting diseases caused by sclerotinia spp. in eastern Canada: fact or fiction? Can. J. Plant Pathol. 26, 480–488. doi: 10.1080/07060660409507168
McDowell, J. M., Dhandaydham, M., Long, T. A., Aarts, M. G., Goff, S., Holub, E. B., et al. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10 (11), 1861–1874. doi: 10.1105/tpc.10.11.1861
McDowell, J. M., Williams, S. G., Funderburg, N. T., Eulgem, T., Dangl, J. L.. (2005). Genetic analysis of developmentally regulated resistance to downy mildew (Hyaloperonospora parasitica) in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 18 (11), 1226–1234. doi: 10.1094/MPMI-18-1226
McMullan, M., Gardiner, A., Bailey, K., Kemen, E., Ward, B. J., Cevik, V., et al. (2015). Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite. eLife 4, e0455. doi: 10.7554/eLife.04550
Meena, P. D., Akhtar, J., Meena, B. R., Mehta, S., Monika, K., Saharan, G. S., et al. (2022). Alternaria-brassica pathosystem: development and perspective. J. Oilseed Brassica 13 (2), 65–89. doi: 10.4454/JPP.V98I3.033
Meena, P. D., Awasthi, R., Chattopadhyay, C., Kolte, S. J., Kumar, A. (2010). Alternaria blight: a chronic disease in rapeseed-mustard. J. Oilseed Brassica 1, 1–11.
Meena, P. D., Jambhulkar, S. J., Gupta, R., Meena, H. S., Singh, D. (2016). Rapid screening technique for alternaria blight resistance in Indian mustard (Brassica juncea l.) using cotyledonary leaf method. J. Plant Pathol. 98 (3), 463–469.
Meena, M., Samal, S. (2019). Alternaria host-specific (HSTs) toxins: an overview of chemical characterization, target sites, regulation and their toxic effects. Toxicol. Rep. 6, 745–758. doi: 10.1016/j.toxrep.2019.06.021
Meena, P. D., Thomas, L., Singh, D. (2014a). Assessment of yield losses in Brassica juncea due to downy mildew (Hyaloperonospora brassicae). J. Oilseed Brassica 1 (1), 73–77.
Meena, P. D., Verma, P. R., Saharan, G. S., Borhan, M. H. (2014b). Historical perspectives of white rust caused by Albugo candida in oilseed brassica. J. Oilseed Brassica 5 (Special), 1–41.
Mehta, N., Saharan, G. S., Meena, P. D. (2018). Expression of disease resistance in brassica-Hyaloperonospora host-patho system- a review. Pl. Dis. Res. 33 (2), 112–141).
Mehta, N., Sangwan, M. S., Srivastava, M. P., Kumar, R. (2002). Survival of alternaria brassicae causing alternaria blight rapeseed-mustard. J. Mycology Plant Pathol. (India) 32 (1), 64–67.
Mei, J., Ding, Y., Lu, K., Wei, D., Liu, Y., Disi, J. O., et al. (2013). Identification of genomic regions involved in resistance against Sclerotinia sclerotiorum from wild Brassica oleracea. Theor. Appl. Genet. 126 (2), 549–556. doi: 10.1007/s00122-012-2000-x
Mei, J., Shao, C., Yang, R., Feng, Y., Gao, Y., Ding, Y., et al. (2020). Introgression and pyramiding of genetic loci from wild Brassica oleracea into B. napus for improving sclerotinia resistance of rapeseed. Theor. Appl. Genet. 133 (4), 1313–1319. doi: 10.1007/s00122-020-03552-w
Melouk, H. A., Akem, C. N., Smith, O. D. (1989). Reaction of peanut genotypes to sclerotinia blight in field plot And 1987. Biol. Cult. Tests Control Plant Dis. 4, 39.
Mir, Z. A., Ali, S., Shivaraj, S. M., Bhat, J. A., Singh, A., Yadav, P., et al. (2020). Genome-wide identification and characterization of chitinase gene family in brassica juncea and camelina sativa in response to alternaria brassicae. Genomics 112 (1), 749–763. doi: 10.1016/j.ygeno.2019.05.011
Mishra, K. K., Kolte, S. J., Nashaat, N. I., Awasthi, R. P. (2009). Pathological and biochemical changes in Brassica juncea (mustard) infected with Albugo candida (white rust). Plant Pathol. 58, 80–86. doi: 10.1111/j.1365-3059.2008.01939.x
Modak, A., Singh, B. R., Dubey, A., Tewari, A. K., Taj, G. (2022). Comparative expression analysis of defense-related genes in both transgenic and nontransgenic Brassica juncea (var.) varuna harbouring overexpressed MAPK3 gene in response to infection by Albugo candida. J. Crop Sci. Biotechnol. 25 (1), 63–72. doi: 10.1007/s12892-021-00113-5
Mondal, K. K., Bhattacharya, R. C., Koundal, K. R., Chatterjee, S. C. (2007). Transgenic Indian mustard (Brassica juncea) expressing tomato glucanase leads to arrested growth of Alternaria brassicae. Plant Cell Rep. 26 (2), 247–252. doi: 10.1007/s00299-006-0241-3
Mondal, B., Mazumder, M., Mukherjee, A., Ghosh, S., De, A., Bose, R., et al. (2020). Association of Alternaria brassicicola induced NAC transcription factors with desiccation and wound responses in Indian mustard. Physiol. Mol. Plant Pathol. 112, 101540. doi: 10.1016/j.pmpp.2020.101540
Monteiro, A. A., Coelho, P. S., Bahcevandziev, K., Valerio, L. (2005). Inheritance of downy mildew resistance at cotyledon and adult-plant stages in ‘Couve algarvia’ (Brassica oleracea var. tronchuda). Euphytica 141, 85–92. doi: 10.1007/s10681-005-5696-8
Mora, A. A., Earle, E. D. (2001). Resistance to Alternaria brassicicola in transgenic broccoli expressing a trichoderma harzianum endochitinase gene. Mol. Breed. 8 (1), 1–9. doi: 10.1023/A:1011913100783
Moradyar, M., Motallebi, M., Zamani, M. R., Aghazadeh, R. (2016). Pathogen-induced expression of chimeric chitinase gene containing synthetic promoter confers antifungal resistance in transgenic canola. In Vitro Cell. Dev. Biology-Plant 52 (2), 119–129. doi: 10.1007/s11627-016-9751-z
Mukherjee, A., Mazumder, M., Jana, J., Srivastava, A. K., Mondal, B., De, A., et al. (2019). Enhancement of ABA sensitivity through conditional expression of the ARF10 gene in Brassica juncea reveals fertile plants with tolerance against Alternaria brassicicola. Mol. Plant-Microbe Interact. 32 (10), 1429–1447. doi: 10.1094/MPMI-05-19-0132-R
Munir, I., Hussan, W., Kazi, M. S. K., Farhatullah Mian, A. A., Aqib, I., et al. (2016). Production of transgenic Brassica juncea with the synthetic chitinase gene (NIC) conferring resistance to Alternaria brassicicola. Pakistan J. Bot. 48 (5), 2063–2070.
Nagaharu, U. (1935). Genome analysis in brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Japanese J. Bot. 7, 389–452.
Nakano, M., Nishihara, M., Yoshioka, H., Takahashi, H., Sawasaki, T., Ohnishi, K., et al. (2013). Suppression of DS1 phosphatidic acid phosphatase confirms resistance to Ralstonia solanacearum in Nicotiana benthamiana. PloS One 8 (9), e75124. doi: 10.1371/journal.pone.0075124
Nashaat, N. I., Heran, A., Mitchell, S. E., Awasthi, R. P. (1997). New genes for resistance to downy mildew (Peronospora parasitica) in oilseed rape (Brassica napus ssp. oleifrea). Plant Pathol. 46, 964–968. doi: 10.1046/j.1365-3059.1997.d01-76.x
Nayanakantha, N. M. C., Rawat, S., Ali, S., Grover, A. (2016). Differential expression of defense-related genes in Sinapis alba and Brassica juncea upon the infection of. Alternaria brassicae. Trop. Agric. Res. 27 (2), 123–136. doi: 10.4038/tar.v27i2.8161
Nees von Esenbeck, C. G. (1816). Das system der pilze und schwämme. Ein versuch. (Legare Street Press). pp. 1–470. doi: 10.11588/diglit.3117
Nesi, N., Delourme, R., Brégeon, M., Falentin, C., Renard, M. (2008). Genetic and molecular approaches to improve nutritional value of Brassica napus l. seed. Comptes rendus biologies 331 (10), 763–771. doi: 10.1016/j.crvi.2008.07.018
Nirupa, N., Prasad, M. N. V., Jami, S. K., Kirti, P. B. (2007). Optimization of agrobacterium-mediated overexpression of osmotin-ferritin genes in Brassica juncea. Trans. Plant J. 1, 384–392.
Nishimura, S., Kohmoto, K. (1983). Host-specific toxins and chemical structures from alternaria species. Annu. Rev. Phytopathol. 21 (1), 87–116. doi: 10.1146/annurev.py.21.090183.000511
Niu, Y., Figueroa, P., Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62 (6), 2143–2154. doi: 10.1093/jxb/erq408
OECD. (2016). “Brassica crops (Brassica species), safety assessment of transgenic organisms in the environment, vol 5,” in OECD consensus documents (Paris: OECD Publishing), pp 151–pp 291.
Oka, K., Akamatsu, H., Kodama, M., Nakajima, H., Kawada, T., Otani, H. (2005). Host-specific AB-toxin production by germinating spores of Alternaria brassicicola is induced by a host-derived oligosaccharide. Physiol. Mol. Plant Pathol. 66 (1-2), 12–19. doi: 10.1016/j.pmpp.2005.03.005
Otani, H., Kohnobe, A., Kodama, M., Kohmoto, K. (1998). Production of a host-specific toxin by germinating spores of Alternaria brassicicola. Physiol. Mol. Plant Pathol. 52 (5), 285–295. doi: 10.1006/pmpp.1998.0147
Panjabi-Massand, P., Yadava, S. K., Sharma, P., Kaur, A., Kumar, A., Arumugam, N., et al. (2010). Molecular mapping reveals two independent loci conferring resistance to Albugo candida in the east European germplasm of oilseed mustard Brassica juncea. Theor. Appl. Genet. 121 (1), 137–145. doi: 10.1007/s00122-010-1297-6
Parker, J. E., Coleman, M. J., Szabò, V., Frost, L. N., Schmidt, R., van der Biezen, E. A., et al. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the toll and interleukin-1 receptors with n and L6. Plant Cell 9 (6), 879–894. doi: 10.1105/tpc.9.6.879
Parkin, I. A., Koh, C., Tang, H., Robinson, S. J., Kagale, S., Clarke, W. E., et al. (2014). Transcriptome and methylome profiling reveals relics of genome dominance in the mesopolyploid brassica oleracea. Genome Biol. 15 (6), 1–18. doi: 10.1186/gb-2014-15-6-r77
Parkin, I. A. P., Lydiate, D. J. (1997). Conserved patterns of chromosome pairing and recombination in Brassica napus crosses. Genome 40 (4), 496–504. doi: 10.1139/g97-066
Pathak, R. K., Baunthiyal, M., Pandey, D., Kumar, A. (2020). Computational analysis of microarray data of Arabidopsis thaliana challenged with Alternaria brassicicola for identification of key genes in brassica. J. Genet. Eng. Biotechnol. 18 (1), 1–20. doi: 10.1186/s43141-020-00032-y
Peng, Y., van Wersch, R., Zhang, Y. (2018). Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 31 (4), 403–409. doi: 10.1094/MPMI-06-17-0145-CR
Perwaiz, M. S., Moghal, S. M., Kamal, M. (1969). Studies on the chemical control of white rust and downy mildew of rape (Sarsoon). West Pak. J. Agric. Res. 7, 71–75.
Petrie, G. A., Vanterpool, T. C. (1974). Fungi associated with hypertophies caused by infection of cruciferae by albugo cruciferarum. Can. Plant Dis. Survey 54 (2), 37–42.
Pound G.S. and Williams, P. H. (1963). Biological races of Albugo candida. Phytopathology 53, 1146–1149.
Prabhu, K., Somers, D. J., Rakow, G., Gugel, R. K. (1998). Molecular markers linked to white rust resistance in mustard Brassica juncea. Theor. Appl. Genet. 97 (5), 865–870. doi: 10.1007/s001220050966
Purdy, L. H. (1979). Sclerotinia sclerotiorum: history, diseases and symptomatology, host range, geographic distribution and impact. Phytopathol. 69, 875–880. doi: 10.1094/Phyto-69-875
Qasim, M. U., Zhao, Q., Shahid, M., Samad, R. A., Ahmar, S., Wu, J., et al. (2020). Identification of QTLs containing resistance genes for sclerotinia stem rot in Brassica napus using comparative transcriptomic studies. Front. Plant Sci. 11, 776. doi: 10.3389/fpls.2020.00776
Rajarammohan, S. (2022). Transcriptome analysis of the necrotrophic pathogen Alternaria brassicae reveals a biphasic mode of pathogenesis in Brassica juncea. bioRxiv, 1–22. doi: 10.1101/2022.09.12.507536
Rajarammohan, S., Kumar, A., Gupta, V., Pental, D., Pradhan, A. K., Kaur, J. (2017). Genetic architecture of resistance to Alternaria brassicae in Arabidopsis thaliana: QTL mapping reveals two major resistance-conferring loci. Front. Plant Sci. 8, 260. doi: 10.3389/fpls.2017.00260
Rajarammohan, S., Paritosh, K., Pental, D., Kaur, J. (2019b). Comparative genomics of alternaria species provides insights into the pathogenic lifestyle of Alternaria brassicae–a pathogen of the brassicaceae family. BMC Genomics 20 (1), 1–13. doi: 10.1186/s12864-019-6414-6
Rajarammohan, S., Pental, D., Kaur, J. (2019a). Near-complete genome assembly of Alternaria brassicae–a necrotrophic pathogen of brassica crops. Mol. Plant-Microbe Interact. 32 (8), 928–930. doi: 10.1094/MPMI-03-19-0084-A
Rana, K., Atri, C., Akhatar, J., Kaur, R., Goyal, A., Singh, M. P., et al. (2019). Detection of first marker trait associations for resistance against Sclerotinia sclerotiorum in Brassica juncea–erucastrum cardaminoides introgression lines. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01015
Rana, K., Atri, C., Gupta, M., Akhatar, J., Sandhu, P. S., Kumar, N., et al. (2017). Mapping resistance responses to sclerotinia infestation in introgression lines of Brassica juncea carrying genomic segments from wild brassicaceae b. fruticulosa. Sci. Rep. 7 (1), 1–12. doi: 10.1038/s41598-017-05992-9
Ranf, S., Eschen-Lippold, L., Pecher, P., Lee, J., Scheel, D. (2011). Interplay between calcium signalling and early signalling elements during defense responses to microbe-or damage-associated molecular patterns. Plant J. 68 (1), 100–113. doi: 10.1111/j.1365-313X.2011.04671.x
Rawat, S., Ali, S., Mittra, B., Grover, A. (2017). Expression analysis of chitinase upon challenge inoculation to alternaria wounding and defense inducers in B. juncea. Biotechnol. Rep. 13, 72–79. doi: 10.1016/j.btre.2017.01.001
Ren, L., Xu, L., Liu, F., Chen, K., Sun, C., Li, J., et al. (2016). Host range of Plasmodiophora brassicae on cruciferous crops and weeds in China. Plant Dis. 100, 933–939. doi: 10.1094/PDIS-09-15-1082-RE
Rustagi, A., Kumar, D., Shekhar, S., Yusuf, M. A., Misra, S., Sarin, N. B. (2014). Transgenic Brassica juncea plants expressing MsrA1, a synthetic cationic antimicrobial peptide, exhibit resistance to fungal phytopathogens. Mol. Biotechnol. 56 (6), 535–545. doi: 10.1007/s12033-013-9727-8
Saharan, G. S., Mehta, N., Meena, P. D. (2016b). Alternaria blight of crucifers: biology, ecology and management (Singapore: Springer Verlag), 326, ISBN: ISBN 978-981-10-0019-5.
Saharan, G. S., Mehta, N., Meena, P. D., Dayal, P. (2016a). Alternaria diseases of crucifers: biology, ecology and disease management (Singapore: Springer), (pp. 17–51).
Saharan, G. S., Mehta, N., Sangwan, M. S. (2005). “Development of disease resistance in rapeseed-mustard,” in Diseases of oilseed crops (New Delhi: Indus Pub Co), 561–617 pp.
Saharan, G. S., Naresh, M., Meena, P. D. (2017). Downy mildew disease of crucifers: biology, ecology and disease management (Singapore, LVI: Springer Verlag), 357, ISBN: ISBN 978-981-10-7499-8.
Saharan, G. S., Verma, P. R., Meena, P. D., Kumar, A. (2014). White rust of crucifers: biology, ecology and management (Frankfurt, Germany: Springer Verlag), 244.
Sánchez-Vallet, A., Tian, H., Rodriguez-Moreno, L., Valkenburg, D. J., Saleem-Batcha, R., Wawra, S., et al. (2020). A secreted LysM effector protects fungal hyphae through chitin-dependent homodimer polymerization. PloS Pathog. 16 (6), e1008652. doi: 10.1371/journal.ppat.1008652
Sang, X., Jue, D., Yang, L., Bai, X., Chen, M., Yang, Q. (2013). Genetic transformation of Brassica napus with MSI-99m gene increases resistance in transgenic plants to Sclerotinia sclerotiorum. Mol. Plant Breed. 4 (30), 247–253. doi: 10.5376/mpb.2013.04.0030
Scheler, C., Durner, J., Astier, J. (2013). Nitric oxide and reactive oxygen species in plant biotic interactions. Curr. Opin. Plant Biol. 16 (4), 534–539. doi: 10.1016/j.pbi.2013.06.020
Seifbarghi, S., Borhan, M. H., Wei, Y., Ma, L., Coutu, C., Bekkaoui, D., et al. (2020). Receptor-like kinases BAK1 and SOBIR1 are required for necrotizing activity of a novel group of Sclerotinia sclerotiorum necrosis-inducing effectors. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01021
Sharma, P., Meena, P. D., Verma, P. R., Saharan, G. S., Mehta, N., Singh, D., et al. (2015). Sclerotinia sclerotiorum (Lib.) de bary causing sclerotinia rot in oilseed brassicas: a review. J. Oilseed Brassica 6 (Special), 1–44.
Sharma, P., Samkumar, A., Rao, M., Prasad, L., Mishra, D. C., Bhattacharya, R., et al. (2018). Genetic diversity studies based on morphological variability, pathogenicity and molecular phylogeny of the Sclerotinia sclerotiorum population from Indian mustard (Brassica juncea). Front. Microbiol. 9. doi: 10.3389/fmicb.2018.01169
Shukla, A. K. (2005). Estimation of yield losses to Indian mustard (Brassica juncea) due to sclerotinia stem rot. J. Phytol. Res. 18, 267–268.
Sinapidou, E., Williams, K., Nott, L., Bahkt, S., Tör, M., Crute, I., et al. (2004). Two TIR: NB: LRR genes are required to specify resistance to Peronospora parasitica isolate Cala2 in Arabidopsis. Plant J. 38 (6), 898–909. doi: 10.1111/j.1365-313X.2004.02099.x
Singh, M., Avtar, R., Lakra, N., Hooda, E., Singh, V. K., Bishnoi, M., et al. (2021). Genetic and proteomic basis of sclerotinia stem rot resistance in Indian mustard [Brassica juncea (L.) czern & coss.]. Genes 12 (11), 1–20. doi: 10.3390/genes12111784
Singh, K. P., Kumari, P., Rai, P. K. (2021a). Current status of the disease-resistant gene (s)/QTLs, and strategies for improvement in Brassica juncea. Front. Plant Sci. 12, 617405. doi: 10.3389/fpls.2021.617405
Singh, K. P., Kumari, P., Yadava, D. K. (2021b). Introgression and QTL mapping conferring resistance for Alternaria brassicae in the backcross progeny of Sinapis alba+ Brassica juncea somatic hybrids. Plant Cell Rep. 40 (12), 2409–2419. doi: 10.1007/s00299-021-02785-3
Singh, B. K., Nandan, D., Supriya, A., Ram, B., Kumar, A., Singh, T., et al. (2015). Validation of molecular markers for marker-assisted pyramiding of white rust resistance loci in Indian mustard (Brassica juncea l.). Can. J. Plant Sci. 95 (5), 939–945. doi: 10.4141/cjps-2014-215
Singh, S., Sharma, S. R., Kalia, P., Deshmukh, R., Kumar, V., Sharma, P., et al. (2012). Molecular mapping of the downy mildew resistance gene Ppa3 in cauliflower (Brassica oleracea var. botrytis l.). J. Hortic. Sci. Biotechnol. 87 (2), 137–143. doi: 10.1080/14620316.2012.11512844
Singh, R., Singh, D., Barbetti, M., Wade, S. M. S., Singh, H., Banga, S. S., et al. (2008). “Sclerotinia rot tolerance in oilseed brassica”. in Proceedings of the 12th International Rapeseed Congress; March 26-30,2007; (Wuhan, China), 94–97.
Singh, O. W., Singh, N., Kamil, D., Singh, V. K., Devi, T. P., Prasad, L. (2021). Morpho-molecular variability and host reactivity of Albugo candida isolates infecting Brassica juncea genotypes in India. J. Plant Pathol. 103 (1), 139–153. doi: 10.1007/s42161-020-00690-4
Somers, D., Rakow, G., Rimmer, S. (2002). Brassica napus DNA markers linked to white rust resistance in Brassica juncea. Theor. Appl. Genet. 104 (6), 1121–1124. doi: 10.1007/s00122-001-0812-1
Stam, R., Jupe, J., Howden, A. J., Morris, J. A., Boevink, P. C., Hedley, P. E., et al. (2013). Identification and characterisation CRN effectors in Phytophthora capsici shows modularity and functional diversity. PloS One 8 (3), e59517. doi: 10.1371/annotation/90bd45cb-33a7-426f-a928-9ddc351b08cc
Suzuki, A., Taguchi, H., Tamura, S. (1970). Isolation and structure elucidation of three new insecticidal cyclodepsipeptides, destruxins c and d and desmethyldestruxin b, produced by Metarrhizium anisopliae. Agric. Biol. Chem. 34 (5), 813–816. doi: 10.1080/00021369.1970.10859690
Taj, G., Agarwal, P., Grant, M., Kumar, A. (2016a). Co-Expression and in-silico interaction studies for inter-linking the activation of MAPK3 and LOX genes during pathogenesis of Alternaria brassicae in Brassica juncea. J. Oilseed Brassica 1 (1), 13–20.
Taj, G., Gaur, V. S., Kumar, A. (2011). Prediction of downstream interaction of transcription factors with MAPK3 in Arabidopsis thaliana using protein sequence information. Int. J. Bioinf. Res. 3 (1), 167–177. doi: 10.9735/0975-3087.3.1.167-177
Taj, G., Kumar, A., Bansal, K. C., Garg, G. K. (2004). Introgression of osmotin gene for creation of resistance against alternaira blight by perturbation of cell cycle machinery. Indian J. Biotechnol. 3, 291–298.
Taj, G., Meena, P. D., Giri, P., Pandey, D., Kumar, A., Kumar, A. (2016b). Pathogenesis mechanisms employed by alternaria species. J. Oilseed Brassica 1 (1), 213–240.
Tang, L., Yang, G., Ma, M., Liu, X., Li, B., Xie, J., et al. (2020). An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Mol. Plant Pathol. 21 (5), 686–701. doi: 10.1111/mpp.12922
Tasleem, M., Baunthiyal, M., Taj, G. (2017). Induction of MPK3, MPK6 and MPK4 mediated defense signaling in response to alternaria blight in transgenic Brassica juncea. Biosci. Biotechnol. Res. Asia 14 (4), 1469–1474. doi: 10.13005/bbra/2593
Thines, M., Kummer, V. (2013). Diversity and species boundaries in floricolous downy mildews. Mycological Prog. 12, 321–329. doi: 10.1007/s11557-012-0837-7
Tirnaz, S., Miyaji, N., Takuno, S., Bayer, P. E., Shimizu, M., Akter, M. A., et al. (2022). Whole-genome DNA methylation analysis in Brassica rapa subsp. perviridis in response to Albugo candida infection. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.849358
Uloth, M. B., Clode, P. L., You, M. P., Barbetti, M. J. (2016). Attack modes and defense reactions in pathosystems involving Sclerotinia sclerotiorum, Brassica carinata, B. juncea and B. napus. Ann. Bot. 117 (1), 79–95. doi: 10.1093/aob/mcv150
Urban, M., Cuzick, A., Rutherford, K., Irvine, A., Pedro, H., Pant, R., et al. (2017). PHI-base: a new interface and further additions for the multi-species pathogen–host interactions database. Nucleic Acids Res. 45 (D1), D604–D610. doi: 10.1093/nar/gkw1089
USDA. (2020). Foreign agriculture service (United States Department of Agriculture). Available at: https://www.fas.usda.gov/regions/india.
Van Der Biezen, E. A., Freddie, C. T., Kahn, K., Parker, J. E., Jones, J. D. (2002). Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J. 29 (4), 439–451. doi: 10.1046/j.0960-7412.2001.01229.x
Van de Wouw, A. P., Idnurm, A. (2019). Biotechnological potential of engineering pathogen effector proteins for use in plant disease management. Biotechnol. Adv. 37 (6), 107387. doi: 10.1016/j.biotechadv.2019.04.009
Van De Wouw, A. P., Idnurm, A., Davidson, J. A., Sprague, S. J., Khangura, R. K., Ware, A. H., et al. (2016). Fungal diseases of canola in Australia: identification of trends, threats and potential therapies. Australas. Plant Pathol. 45415, 23. doi: 10.1007/s13313-016-0428-1
Varshney, A., Mohapatra, T., Sharma, R. P. (2004). Development and validation of CAPS and AFLP markers for white rust resistance gene in Brassica juncea. Theor. Appl. Genet. 109 (1), 153–159. doi: 10.1007/s00122-004-1607-y
Verma, P. R., Petrie, G. A. (1980). Effect of seed infestation and flower bud inoculation on systemic infection of turnip rape by Albugo candida. Can. J. Plant Sci. 60, 267–271. doi: 10.4141/cjps80-038
Verma, P. R., Saharan, G. S. (1994). Monograph on Alternaria diseases of crucifers. Research BranchTechnical Bulletin 1994-6E. Saskatoon Research Centre Research Branch, Agriculture and Agri-Food Canada 107 Science Place Saskatoon, Saskatchewan, Canada. pp 1–162.
Verma, S. S., Yajima, W. R., Rahman, M. H., Shah, S., Liu, J. J., Ekramoddoullah, A. K., et al. (2012). A cysteine-rich antimicrobial peptide from Pinus monticola (PmAMP1) confers resistance to multiple fungal pathogens in canola (Brassica napus). Plant Mol. Biol. 79 (1), 61–74. doi: 10.1007/s11103-012-9895-0
Wang, Z., Fang, H., Chen, Y., Chen, K., Li, G., Gu, S., et al. (2014). Overexpression of BnWRKY33 in oilseed rape enhances resistance to Sclerotinia sclerotiorum. Mol. Plant Pathol. 15 (7), 677–689. doi: 10.1111/mpp.12123
Wang, M., Farnham, M. W., Thomas, C. E. (2001). Inheritance of true leaf stage downy mildew resistance in broccoli. J. Am. Soc. Hortic. Sci. 126 (6), 727–729. doi: 10.21273/JASHS.126.6.727
Wang, Z., Mao, H., Dong, C., Ji, R., Cai, L., Fu, H., et al. (2009). Overexpression of Brassica napus MPK4 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Mol. Plant-Microbe Interact. 22 (3), 235–244. doi: 10.1094/MPMI-22-3-0235
Wang, Z., Wan, L., Xin, Q., Chen, Y., Zhang, X., Dong, F., et al. (2018). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. J. Exp. Bot. 69 (12), 3141–3155. doi: 10.1093/jxb/ery138
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43 (10), 1035–1039. doi: 10.1038/ng.919
Wang, Z., Zhang, W. H., Ma, L. Y., Li, X., Zhao, F. Y., Tan, X. L. (2020). Overexpression of Brassica napus NPR1 enhances resistance to Sclerotinia sclerotiorum in oilseed rape. Physiol. Mol. Plant Pathol. 110, 101460. doi: 10.1016/j.pmpp.2020.101460
Warwick, S. I., Francis, A., Al-Shehbaz, I. A. (2006). Brassicaceae: species checklist and database on CD-rom. Plant Systematics Evol. 259 (2), 249–258. doi: 10.1007/s00606-006-0422-0
Wei, D., Mei, J., Fu, Y., Disi, J. O., Li, J., Qian, W. (2014). Quantitative trait loci analyses for resistance to Sclerotinia sclerotiorum and flowering time in Brassica napus. Mol. Breed. 34 (4), 1797–1804. doi: 10.1007/s11032-014-0139-7
Weller, D. M., Mavrodi, D. V., Van Pelt, J. A., Pieterse, C. M., van Loon, L. C., Bakker, P. A. (2012). Induced systemic resistance (ISR) in Arabidopsis thaliana against Pseudomonas syringae pv. tomato by 2,4-Diacetylphloroglucinol-Producing Pseudomonas fluoresces. Phytopathology 102, 403–412. doi: 10.1094/PHYTO-08-11-0222
Wirthmueller, L., Asai, S., Rallapalli, G., Sklenar, J., Fabro, G., Kim, D. S., et al. (2018). Arabidopsis downy mildew effector HaRxL106 suppresses plant immunity by binding to RADICAL-INDUCED CELL DEATH1. New Phytol. 220 (1), 232–248. doi: 10.1111/nph.15277
Wolpert, T. J., Dunkle, L. D., Ciuffetti, L. M. (2002). Host-selective toxins and avirulence determinants: what’s in a name? Annu. Rev. Phytopathol. 40 (1), 251–285. doi: 10.1146/annurev.phyto.40.011402.114210
Wu, J., Cai, G., Tu, J., Li, L., Liu, S., Luo, X., et al. (2013). Identification of QTLs for resistance to sclerotinia stem rot and BnaC. IGMT5. a as a candidate gene of the major resistant QTL SRC6 in Brassica napus. PloS One 8 (7), e67740. doi: 10.1371/journal.pone.0067740
Wu, J., Chen, P., Zhao, Q., Cai, G., Hu, Y., Xiang, Y., et al. (2019). Co-Location of QTL for sclerotinia stem rot resistance and flowering time in Brassica napus. Crop J. 7 (2), 227–237. doi: 10.1016/j.cj.2018.12.007
Wu, J., Wu, L. T., Liu, Z. B., Qian, L., Wang, M., Zhou, L., et al. (2009). A plant defensin gene from orychophragmus violaceus can improve Brassica napus’ resistance to Sclerotinia sclerotiorum. Afr. J. Biotechnol. 8 (22), 6101–6109.
Wu, J., Zhao, Q., Yang, Q., Liu, H, Li, Q, Yi, X, et al. (2016). Comparative transcriptomic analysis uncovers the complex genetic network for resistance to Sclerotinia sclerotiorum in Brassica napus. Sci. Rep. 6 (1), 1–16. doi: 10.1038/srep19007
Xiao, Z., Gong, N., Zhou, X., Zhu, L., He, X., Zheng, J., et al. (2022). Developmental characteristics of sporogenous hyphae: a new observation between Brassica juncea var. tumida and Albugo candida. Eur. J. Plant Pathol. 162 (2), 343–355. doi: 10.1007/s10658-021-02406-5
Xiao, X., Xie, J., Cheng, J., Li, G., Yi, X., Jiang, D., et al. (2014). Novel secretory protein ss-Caf1 of the plant-pathogenic fungus Sclerotinia sclerotiorum is required for host penetration and normal sclerotial development. Mol. Plant-Microbe Interact. 27 (1), 40–55. doi: 10.1094/MPMI-05-13-0145-R
Xie, L. H., Lin, J. Y., Xie, L. M., Nai, G. B. (1984). On the bunchy stunt disease of rice IV: the experiments of occurrence, development, and control of rice bunchy stunt. Acta Phytopathologica Sin. 14 (1), 33–38.
Xu, L., Xiang, M., White, D., Chen, W. (2015). pH dependency of sclerotial development and pathogenicity revealed by using genetically defined oxalate-minus mutants of Sclerotinia sclerotiorum. Environ. Microbiol. 17, 2896–2909. doi: 10.1111/1462-2920.12818
Yadav, P., Mir, Z. A., Ali, S., Papolu, P. K., Grover, A. (2020). A combined transcriptional, biochemical, and histopathological study unravels the complexity of alternaria resistance and susceptibility in brassica coenospecies. Fungal Biol. 124 (1), 44–53. doi: 10.1016/j.funbio.2019.11.002
Yajima, W., Verma, S. S., Shah, S., Rahman, M. H., Liang, Y., Kav, N. N. (2010). Expression of anti-sclerotinia scFv in transgenic Brassica napus enhances tolerance against stem rot. New Biotechnol. 27 (6), 816–821. doi: 10.1016/j.nbt.2010.09.010
Yang, J., Liu, D., Wang, X., Ji, C., Cheng, F., Liu, B., et al. (2016). The genome sequence of allopolyploid Brassica juncea and analysis of differential homoeolog gene expression influencing selection. Nat. Genet. 48 (10), 1225–1232. doi: 10.1038/ng.3657
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., et al. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytol. 217 (2), 739–755. doi: 10.1111/nph.14842
Yin, X., Yi, B., Chen, W., Zhang, W., Tu, J., Fernando, W. G. D., et al. (2010). Mapping of QTLs detected in a Brassica napus DH population for resistance to Sclerotinia sclerotiorum in multiple environments. Euphytica 173 (1), 25–35. doi: 10.1007/s10681-009-0095-1
Yu, S., Su, T., Zhi, S., Zhang, F., Wang, W., Zhang, D., et al. (2016). Construction of a sequence-based bin map and mapping of QTLs for downy mildew resistance at four developmental stages in Chinese cabbage (Brassica rapa l. ssp. pekinensis). Mol. Breed. 36 (4), 1–12. doi: 10.1186/s12864-019-5810-2
Yu, S., Zhang, F., Yu, R., Zou, Y., Qi, J., Zhao, X., et al. (2009). Genetic mapping and localization of a major QTL for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Breed. 23 (4), 573–590. doi: 10.1007/s11032-009-9257-z
Yu, S., Zhang, F., Zhao, X., Yu, Y., Zhang, D. (2011). Sequence-characterized amplified region and simple sequence repeat markers for identifying the major quantitative trait locus responsible for seedling resistance to downy mildew in Chinese cabbage (Brassica rapa ssp. pekinensis). Plant Breed. 130 (5), 580–583. doi: 10.1111/j.1439-0523.2011.01874.x
Zarinpanjeh, N., Motallebi, M., Zamani, M. R., Ziaei, M. (2016). Enhanced resistance to Sclerotinia sclerotiorum in Brassica napus by co-expression of defensin and chimeric chitinase genes. J. Appl. Genet. 57 (4), 417–425. doi: 10.1007/s13353-016-0340-y
Zhang, J., Shao, F., Li, Y., Cui, H., Chen, L., Li, H., et al. (2007). A Pseudomonas syringae effector inactivates MAPKs to suppress PAMP-induced immunity in plants. Cell Host Microbe 1 (3), 175–185. doi: 10.1016/j.chom.2007.03.006
Zhang, Z. Y., Wang, Y. X., Liu, Y. L. (1984). Taxonomic studies of the family albuginaceae of china. II. a new species of Albugo on acanthaceae and known species of Albugo on cruciferae. Acta Mycol. Sin. 3, 65–71.
Zhang, W., Zhao, F., Jiang, L., Chen, C., Wu, L., Liu, Z. (2018). Different pathogen defense strategies in arabidopsis: more than pathogen recognition. Cells 7 (12), 252. doi: 10.3390/cells7120252
Zhao, J., Meng, J. (2003). Genetic analysis of loci associated with partial resistance to Sclerotinia sclerotiorum in rapeseed (Brassica napus l.). Theor. Appl. Genet. 106 (4), 759–764. doi: 10.1007/s00122-002-1171-2
Zhao, J., Udall, J. A., Quijada, P. A., Grau, C. R., Meng, J., Osborn, T. C. (2006). Quantitative trait loci for resistance to Sclerotinia sclerotiorum and its association with a homeologous non-reciprocal transposition in Brassica napus l. Theor. Appl. Genet. 112 (3), 509–516. doi: 10.1007/s00122-005-0154-5
Zhu, W., Wei, W., Fu, Y., Cheng, J., Xie, J., Li, G., et al. (2013). A secretory protein of necrotrophic fungus Sclerotinia sclerotiorum that suppresses host resistance. PloS One 8 (1), e53901. doi: 10.1371/journal.pone.0053901
Ziaei, M., Motallebi, M., Zamani, M. R., Panjeh, N. Z. (2016). Co-Expression of chimeric chitinase and a polygalacturonase-inhibiting protein in transgenic canola (Brassica napus) confers enhanced resistance to Sclerotinia sclerotiorum. Biotechnol. Lett. 38 (6), 1021–1032. doi: 10.1007/s10529-016-2058-7
Keywords: B. juncea, fungal pathogens, effectoromics, resistance genes, quantitative trait loci, defense-related genes, transgenics
Citation: Rai P, Prasad L and Rai PK (2023) Fungal effectors versus defense-related genes of B. juncea and the status of resistant transgenics against fungal pathogens. Front. Plant Sci. 14:1139009. doi: 10.3389/fpls.2023.1139009
Received: 07 January 2023; Accepted: 09 May 2023;
Published: 08 June 2023.
Edited by:
Mahesh Rao, Indian Council of Agricultural Research, IndiaReviewed by:
Govind Singh Saharan, Chaudhary Charan Singh Haryana Agricultural University, IndiaGunjan Sharma, Gujarat Biotechnology University, India
Shyam Saran Vaish, Banaras Hindu University, India
Ajay Kumar Thakur, Central Potato Research Institute (ICAR), India
Kartikeya Srivasrava, Banaras Hindu University, India
Copyright © 2023 Rai, Prasad and Rai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pramod Kumar Rai, pramodkrai68@gmail.com
 Prajjwal Rai
Prajjwal Rai Laxman Prasad1
Laxman Prasad1 Pramod Kumar Rai
Pramod Kumar Rai