- Department of Horticulture, College of Plant Science, Jilin University, Changchun, China
Ultraviolet-B (UV-B) promotes anthocyanin accumulation and improves fruit quality in plants. To explore the underlying network of MYB transcription factors that regulates UV-B-induced anthocyanin biosynthesis in blueberry (Vaccinium corymbosum), we analyzed the response of MYB transcription factor genes to UV-B treatment. Transcriptome sequencing analysis revealed that VcMYBA2 and VcMYB114 expression were upregulated and were positively correlated with the expression of anthocyanin structural genes under UV-B radiation according to weighted gene co-expression network analysis (WGCNA) data. The VcUVR8-VcCOP1-VcHY5 pathway perceives UV-B signals and promotes the expression of anthocyanin structural genes by upregulating VcMYBA2 and VcMYB114 or by regulating the VcBBXs-VcMYB pathway, ultimately promoting anthocyanin accumulation. By contrast, VcMYB4a and VcUSP1 were downregulated under UV-B treatment, and VcMYB4a expression was negatively correlated with that of anthocyanin biosynthesis genes in response to UV-B. Analysis of VcMYB4a-overexpressing and wild-type blueberry calli exposed to UV-B radiation revealed that VcMYB4a represses UV-B-induced anthocyanin accumulation. Yeast one-hybrid and dual luciferase assays showed that the universal stress protein VcUSP1 directly bound to the promoter of VcMYB4a. These results suggest that the VcUSP1-VcMYB4a pathway negatively regulates UV-B-induced anthocyanin biosynthesis and provide insight into UV-B-induced anthocyanin biosynthesis.
Introduction
Ultraviolet-B (UV-B) is a component of sunlight that affects the plant defense system, leading to increased accumulation of secondary metabolites, especially anthocyanin (Nguyen et al., 2017; Song et al., 2022). The UV-B-induced anthocyanin biosynthesis pathway is well characterized in many plants (Liu et al., 2018; Ding et al., 2021). In Arabidopsis (Arabidopsis thaliana), the UV-B photoreceptor UV RESISTANT LOCUS 8 (UVR8) interacts with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) to activate UV-B signaling and initiate UV-B adaptation pathways in response to UV-B radiation (Oravecz et al., 2006; Rizzini et al., 2011). COP1 is a critical regulator of plant responses to UV-B that promotes flavonoid accumulation. COP1 activates ELONGATED HYPOCOTYL 5 (HY5, a bZIP transcription factor) and coordinates HY5-dependent and -independent pathways, ultimately resulting in UV-B tolerance (Oravecz et al., 2006; Bhatia et al., 2021). HY5 is a central transcription factor that promotes anthocyanin accumulation by regulating the expression of MYB transcription factors and anthocyanin biosynthesis genes (Stracke et al., 2010; Shin et al., 2013; An et al., 2017). B-box (BBX) proteins interact with HY5 or directly bind to the promoter sequences of MYBs or anthocyanin biosynthesis genes to regulate anthocyanin biosynthesis (Bai et al., 2019; Fang et al., 2019). Thus, UVR8, COP1, HY5, BBXs, MYBs, and anthocyanin biosynthesis genes form a UV-B-induced anthocyanin biosynthesis network and MYBs play an important role in this network.
MYB proteins are a superfamily of transcription factors that are responsive to one or multiple stress treatments and regulate the accumulation of secondary metabolites (Hartmann et al., 2005; Chen et al., 2006). In the R2R3-MYB gene family, subgroup 6 members promote anthocyanin biosynthesis and subgroup 4 members inhibit anthocyanin biosynthesis. For example, in the Vaccinium genus, VbMYBA, VmMYBA1, VmMYBA2, and VcMYBA, which belong to subgroup 6, activate anthocyanin biosynthesis, and, in strawberry (Fragaria × ananassa), FaMYB1, a member of subgroup 4, suppresses anthocyanin accumulation (Aharoni et al., 2001; Plunkett et al., 2018; Karppinen et al., 2021; Zhang et al., 2021). UV-B also affects the expression of MYB transcription factor genes from subgroups 4 and 6. In Arabidopsis, AtMYB75 from subgroup 6 is upregulated and AtMYB4 from subgroup 4 is downregulated upon exposure to UV-B light (Jin et al., 2000; Tohge et al., 2011). In apple (Malus domestica), MdMYB10 of subgroup 6 is upregulated throughout fruit development and influences anthocyanin production under UV radiation (Henry-Kirk et al., 2018). However, how MYB pathway genes function under UV-B radiation conditions merits further research.
Rich in anthocyanin compounds, blueberries (Vaccinium corymbosum) are often referred to as a “superfood” (Ribera et al., 2010; Norberto et al., 2013). The anthocyanin biosynthesis pathway in blueberry has been elucidated, and several structural genes associated with this pathway have been characterized (Zhang et al., 2017; Song et al., 2022). VcMYBA of subgroup 6 from blueberry has also been characterized; this transcription factor transactivates the VcDFR promoter to activate anthocyanin production (Plunkett et al., 2018). UV-B promotes anthocyanin accumulation in blueberry fruits and upregulates the expression of VcBBX, VcMYB21, VcR2R3MYB, and structural genes involved in anthocyanin biosynthesis (Nguyen et al., 2017). However, the UV-B-induced network underlying anthocyanin biosynthesis is unclear.
In this study, we identified MYB transcription factors that function in the response of blueberry to UV-B radiation based on transcriptome deep sequencing (RNA-seq) data and elucidated the network of transcription factors that positively regulates UV-B-induced anthocyanin biosynthesis according to weighted gene co-expression network analysis (WGCNA) data. In addition, analysis of blueberry calli overexpressing VcMYB4a and yeast one-hybrid and dual luciferase assays suggested the existence of a UV-B-induced anthocyanin biosynthesis network that is negatively regulated by VcMYB4a. Our findings broaden our understanding of UV-B-induced anthocyanin biosynthesis in blueberry.
Materials and methods
Plant materials and UV-B treatment
Wild-type (WT) and VcMYB4a-overexpressing blueberry calli (OE-1, OE-2, and OE-3 lines) were generated in our laboratory (Zhang et al., 2020; Yang et al., 2022).
All the calli were exposed to UV-B lamps (TL20/01; 311-nm Philips, Netherlands). WT calli were harvested to measure anthocyanin contents and WT and OE-1 calli were used to measure mRNA expression levels by reverse-transcription quantitative PCR (RT-qPCR) after 0, 6, 12, 24, and 48 h of UV-B radiation. The anthocyanin contents of WT and VcMYB4a-overexpressing calli treated with UV-B for 4 days were measured, using untreated calli as a control.
Identification of transcription factor genes
RNA-seq data from calli treated for 0, 1, 3, 6, 12, and 24 h with UV-B radiation were downloaded from the BioProject database in the NCBI repository (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA831018). The data were analyzed on the MKCloud online platform (https://international.biocloud.net). Using 0 h as a control, differentially expressed transcription factor genes from calli treated with 1, 3, 6, 12, and 24 h of UV-B radiation were identified according to the criteria of the absolute value of log2 (fold change) ≥ 1 and a false discovery rate (FDR) < 0.01 by the DEGSeq2 R package. Gene expression levels were quantified as fragments per kilobase of transcript per million fragments mapped (FPKM) values. The transcription factors were annotated using Protein family (Pfam) and Swiss-Prot databases (Apweiler et al., 2004; Finn et al., 2014).
Cluster analysis of VcMYB transcription factors
The differentially expressed VcMYB transcription factors were obtained by deleting the redundant sequences. A phylogenetic tree was constructed from 52 VcMYBs of blueberry and 142 AtMYBs of Arabidopsis using the neighbor-joining method with the MEGA X program (Kumar et al., 2018). The phylogenetic tree was divided into different subgroups based on MYB transcription factors from Arabidopsis (Kranz et al., 1998; Dubos et al., 2010). The VcMYBs were named based on cluster and functional annotations from Swiss-Prot databases.
Correlation analysis between VcMYB4a and genes involved in the UV-B-induced anthocyanin pathway
The differentially expressed genes from the WGCNA kMEblue module were selected to identify genes involved in the UV-B-induced anthocyanin pathway (Song et al., 2022). Pearson’s correlation coefficients (r) between the expression levels of differentially expressed genes were calculated using SPSS 19.0 software. Heatmap of UV-B-induced anthocyanin biosynthesis pathway genes and transcription factor genes were drawn using log10 (FPKM) values with TBtools software (v1.098761) (Chen et al., 2020).
Reverse-transcription quantitative PCR
RT-qPCR analysis was performed for WT and VcMYB4a-overexpressing (OE-1) calli after 0, 6, 12, 24, and 48 h of UV-B radiation using an ABI 7900HT real-time PCR system. The relative expression levels of VcPAL1, VcPAL3, Vc4CL2, VcCHS1, VcDFR, VcCHI3, VcF3H-1, VcF3H-2, and VcUFGT genes were calculated using the 2–ΔΔCt method and VcGAPDH was used as the reference gene. Primer sequences are shown in Table S1.
Promoter analysis of VcMYB4a gene
The upstream sequences of VcMYB4a were cloned using a Genome Walking Kit (TaKaRa, Japan). The cis-elements of the VcMYB4a upstream sequence was analyzed for the presence of cis-acting regulatory sequences in the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
Yeast one-hybrid assays
The promoter fragment (1,718 bp) of VcMYB4a was inserted into the pHIS2 vector as bait (BD Biosciences, Shanghai, China). The VcUSP1 (GenBank accession no. OP957062) sequence was recombined into the pGADT7 vector as prey (Clontech). The primer sequences are shown in Table S1. Yeast strain AY187 was co-transformed with different combinations of vectors and selected on SD medium lacking tryptophan and leucine. A series of 5-µL aliquots of 10× diluted yeast co-transformants were dropped onto SD medium lacking tryptophan, leucine, and histidine (SD/−Trp/−Leu/−His) and containing 10, 20, 30, 40, 50, and 75 mM 3-amino-1,2,4-triazole (3-AT), which was used to suppress the background histidine leakiness of the pHIS2 vectors. The co-transformed yeast was grown at 30°C for 3 days. Yeast cells harboring pHIS2-VcMYB4a/pGAD53m and pHIS2-p53/pGAD53m were used as negative and positive controls, respectively.
LUC complementation imaging assay
The promoter fragment of VcMYB4a and CDS of VcUSP1 were inserted into the pGreenII 0800-miRNA and Pcambia1302 vectors to construct the pVcMYB4a-LUC and GFP-VcUSP1 vectors, respectively (Table S1). Recombining vectors were co-injected into Nicotiana benthamiana leaves and luminescence was detected 48 h after infiltration with a live imaging apparatus. The ratio of Luc to Ren activity was determined using the Dual-Glo® Luciferase Assay System (Promega). As a negative control, Pcambia1302 (GFP) and pVcMYB4a-LUC vectors were co-injected into the N. benthamiana leaves.
Anthocyanin content measurement
The anthocyanin content of the samples was measured as described by Yang et al. (2022). Each sample (0.5 g) was soaked in 3 mL methanol with 1% HCl and incubated for 16 h in darkness at 4°C. After centrifugation, the supernatant was diluted with an equal amount of water and the absorbance of each diluted extract at 530 and 650 nm was measured with a spectrophotometer (UV-1600, Shimadzu). Total anthocyanin content was quantified using the equation A530-0.25×A650 (Rabino and Mancinelli, 1986).
Statistical analysis
In the figures, each error bar represents the standard deviation (SD) of the mean of three independent biological replicates, and three technical replicates were performed for each biological replicate. Significant differences were detected using Tukey’s test, implemented in SPSS 19.0 software.
Results
UV-B radiation induces anthocyanin accumulation
To investigate the function of UV-B radiation on anthocyanin accumulation, blueberry calli were exposed to UV-B radiation for 0, 6, 12, 24, and 48 h. The proportion of callus that was red gradually increased during UV-B treatments (Figure 1A). Furthermore, the anthocyanin content significantly increased 2.9-, 4.2-, 5.1-, and 9.0-fold after 6, 12, 24, and 48 h of UV-B treatment relative to the 0 h treatment, respectively (Figure 1B). Thus, UV-B radiation promotes anthocyanin accumulation in blueberry calli.
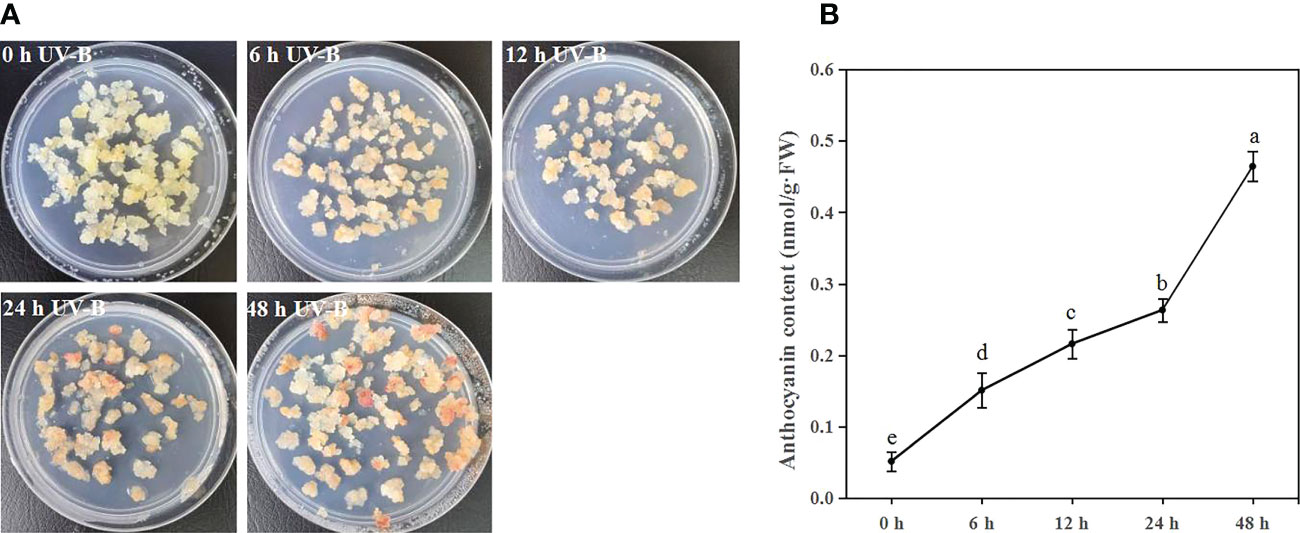
Figure 1 UV-B radiation induces anthocyanin accumulation in blueberry calli. The phenotype (A) and anthocyanin contents (B) of blueberry calli exposed to UV-B radiation for 0, 6, 12, 24, and 48 h. Error bars indicate ± SD of the mean of three independent biological replicates, each with three technical replicates; different letters indicate significant differences (p < 0.05) among samples by Tukey’s test.
UV-B radiation regulates the expression of transcription factor genes
An analysis of Pfam and Swiss-Prot functional annotations revealed 370 transcription factor genes that were differentially expressed in response to UV-B radiation. The most highly enriched differentially expressed genes encoded MYB family members (23.77%), followed by AP2 (17.48%), WRKY (13.21%), bZIP (8.81%), and bHLH (6.92%) transcription factors (Figure S1; Table S2). After analyzing the sequences of VcMYB transcription factor genes and deleting the redundant VcMYB transcription factors, 52 VcMYB transcription factor genes were identified, namely 28 R2R3-MYB, 22 1R-MYB, 1 3R-MYB, and 1 3R/4R-MYB transcription factor genes (Table S3).
To predict the potential functions of the VcMYBs, phylogenetic analysis of the deduced protein sequences of 52 VcMYB and 142 AtMYB genes were performed (Figure 2). Of the 52 VcMYB genes, 28 VcMYBs from the R2R3-MYB subfamily belonging to 15 subgroups were upregulated and/or downregulated in response to UV-B radiation. VcMYB114 and VcMYBA2 were clustered in subgroup 6, and VcMYB4a was clustered in subgroup 4. Most genes from subgroup 6 promote anthocyanin accumulation, whereas most genes from subgroup 4 inhibit anthocyanin biosynthesis (Aharoni et al., 2001; Plunkett et al., 2018). VcMYB114 and VcMYBA2 expression increased and that of VcMYB4a decreased during UV-B treatment.
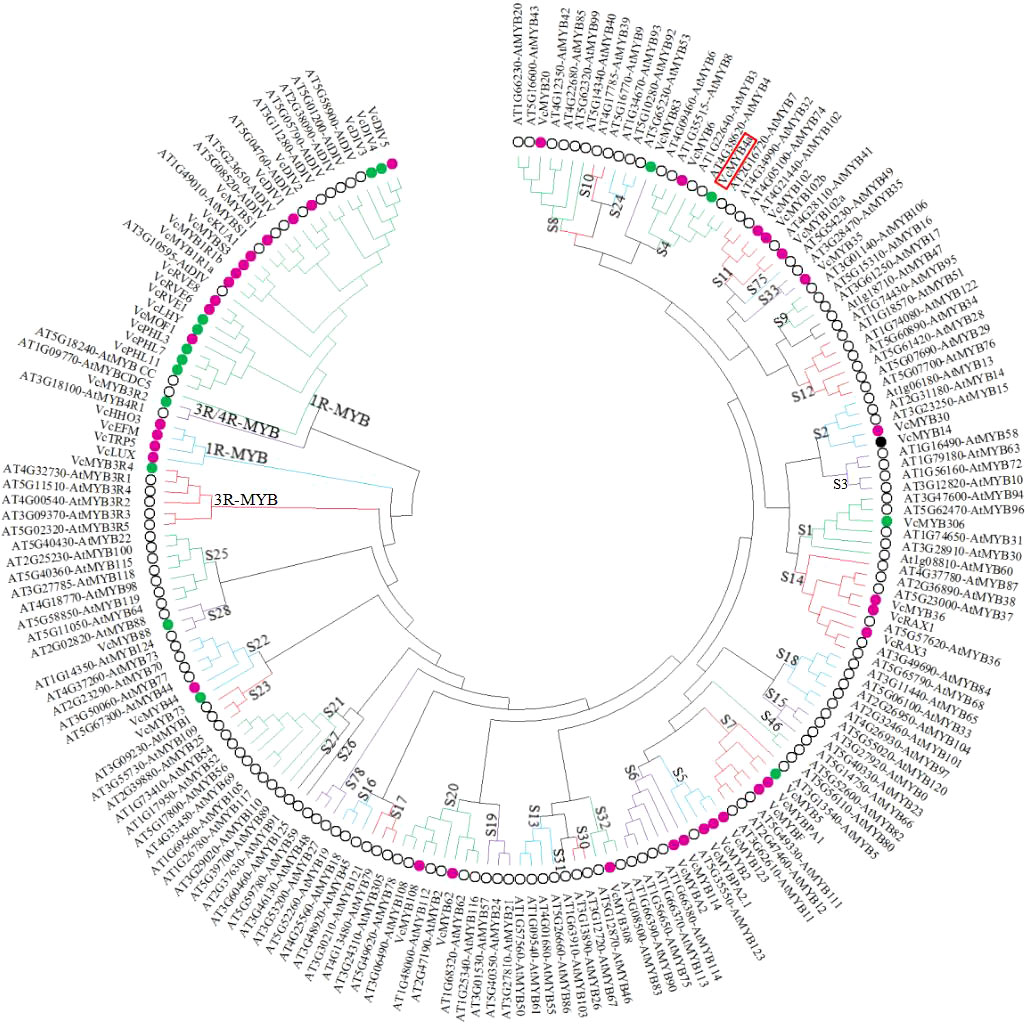
Figure 2 The 52 differentially expressed VcMYB transcription factor genes from blueberry and 142 MYB transcription factor genes from Arabidopsis thaliana were clustered into different subgroups using MEGA X with the neighbor-joining method. Magenta, green, and black dots represent upregulated, downregulated, and both up- and downregulated VcMYBs under UV-B radiation, respectively. Black circle represents MYB transcription factors from Arabidopsis. Red box indicates the position of VcMYB4a.
VcMYB4a represses UV-B-induced anthocyanin accumulation
To explore the role of VcMYB4a in the plant response to UV-B radiation, we treated three VcMYB4a-overexpressing lines (OE-1, OE-2, and OE-3) and wild-type (WT) calli with UV-B for 4 days and measured anthocyanin contents (Figure 3). In the absence of UV-B treatment, the anthocyanin content of VcMYB4a-overexpressing calli was not significantly different from that of WT calli (Figure 3A). Under UV-B treatment, anthocyanin rapidly accumulated in WT calli compared with VcMYB4a-overexpressing calli (Figures 3B, C). Thus, VcMYB4a was a repressor of UV-B-induced anthocyanin biosynthesis.
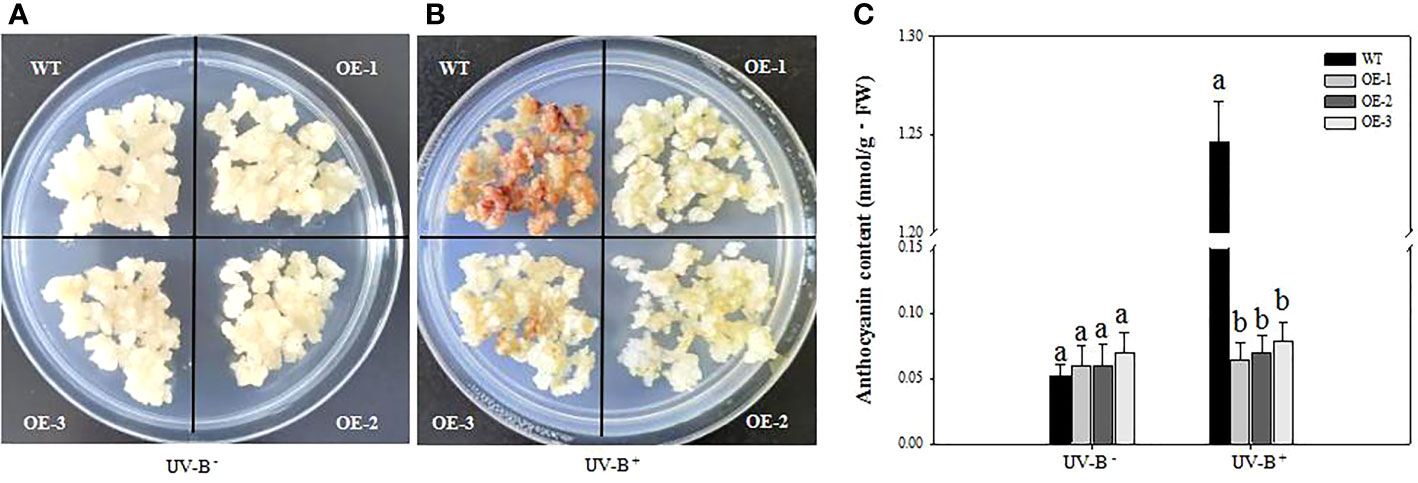
Figure 3 Anthocyanin contents of wild-type (WT) and VcMYB4a-overexpressing (OE-1, OE-2, and OE-3) blueberry calli after 4 days of exposure to UV-B radiation. Untreated calli (A) and calli after 4 days of UV-B treatment are shown (B). (C) Anthocyanin contents of WT and VcMYB4a-overexpressing calli treated or not with UV-B radiation for 4 days. Error bars indicate SD of the mean of three independent biological replicates, each with three technical replicates; different letters indicate significant differences (p < 0.05) among samples by Tukey’s test.
VcMYB4a expression is negatively correlated with that of genes involved in UV-B-induced anthocyanin accumulation
To reveal the role of VcMYB4a in the regulatory network underlying anthocyanin biosynthesis under UV-B radiation, we searched for genes that were co-expressed with VcMYB4a and involved in this network using the WGCNA package in R and calculated their Pearson’s correlation coefficients (r) (Table 1). The expression level of VcMYB4a was negatively correlated with that of VcUVR8, VcCOP1, and most genes involved in the anthocyanin pathway, including VcPAL1, VcPAL3, Vc4CL2, VcCHS1, VcCHI3, VcDFR, VcF3H-1, VcF3H-2, and VcUFGT. Both VcMYB114 and VcMYBA2 expression was positively correlated with the expression of VcCOP1, VcHY5, VcBBX32, and VcBBX30 as well as that of the anthocyanin biosynthesis genes VcPAL1, VcPAL3, VcC4H, Vc4CL2, VcCHS1, VcF3H-2, and VcUFGT. However, VcMYB114 expression was negatively correlated with VcBBX21 expression. In a heatmap, VcMYB4a and VcBBX21 were clustered together and were downregulated by UV-B radiation, whereas the other genes were upregulated by this treatment (Figure 4A; Table S4). These results indicate that VcUVR8, VcCOP1, VcHY5, VcBBXs, VcMYBs, and anthocyanin biosynthesis genes form a UV-B-induced anthocyanin biosynthesis network and that VcMYB4a is negative regulator of this network (Figure 4B).
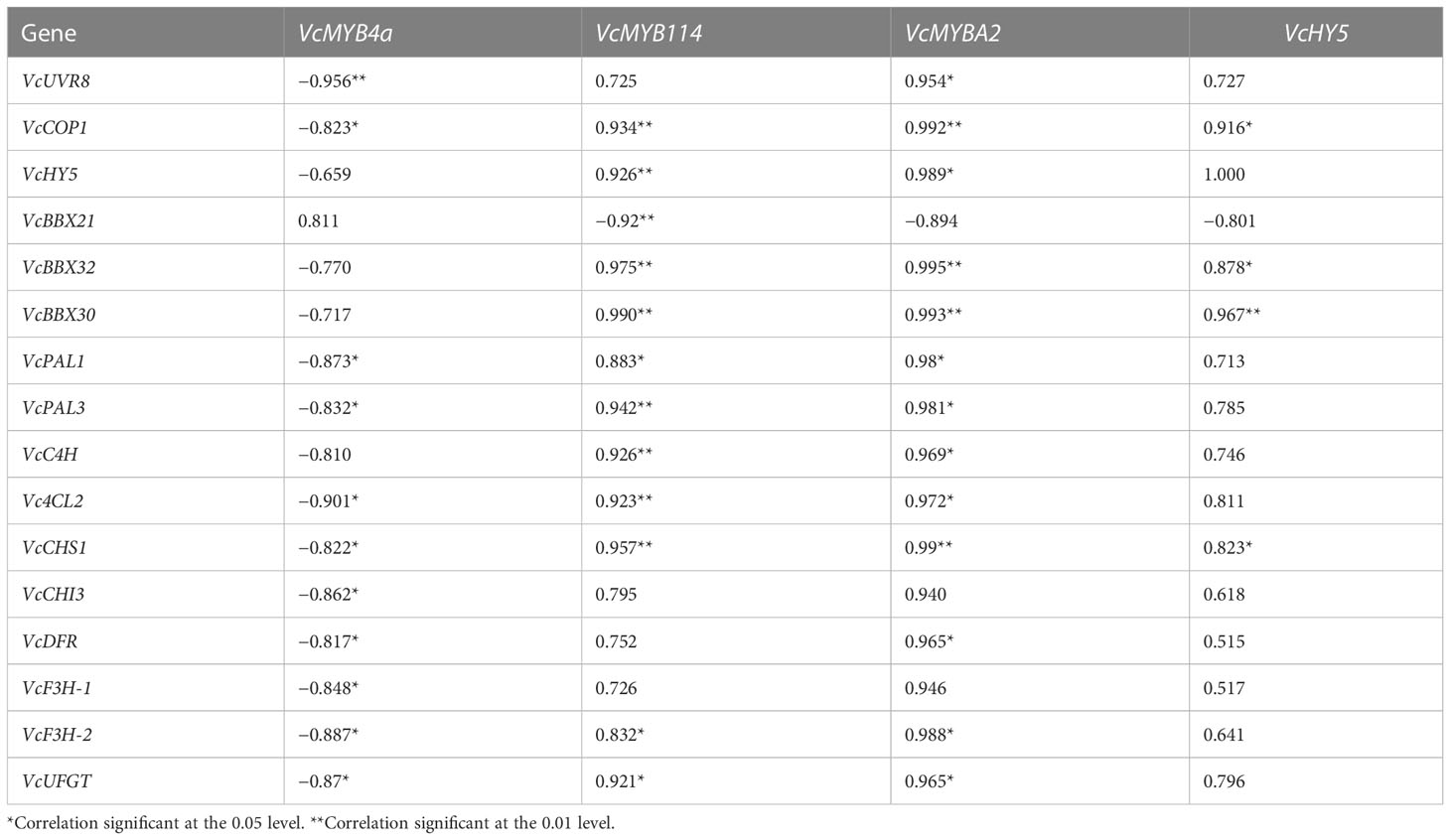
Table 1 Pearson’s correlation coefficients (r) between the expression levels of genes from the UV-B-induced anthocyanin biosynthesis pathway.
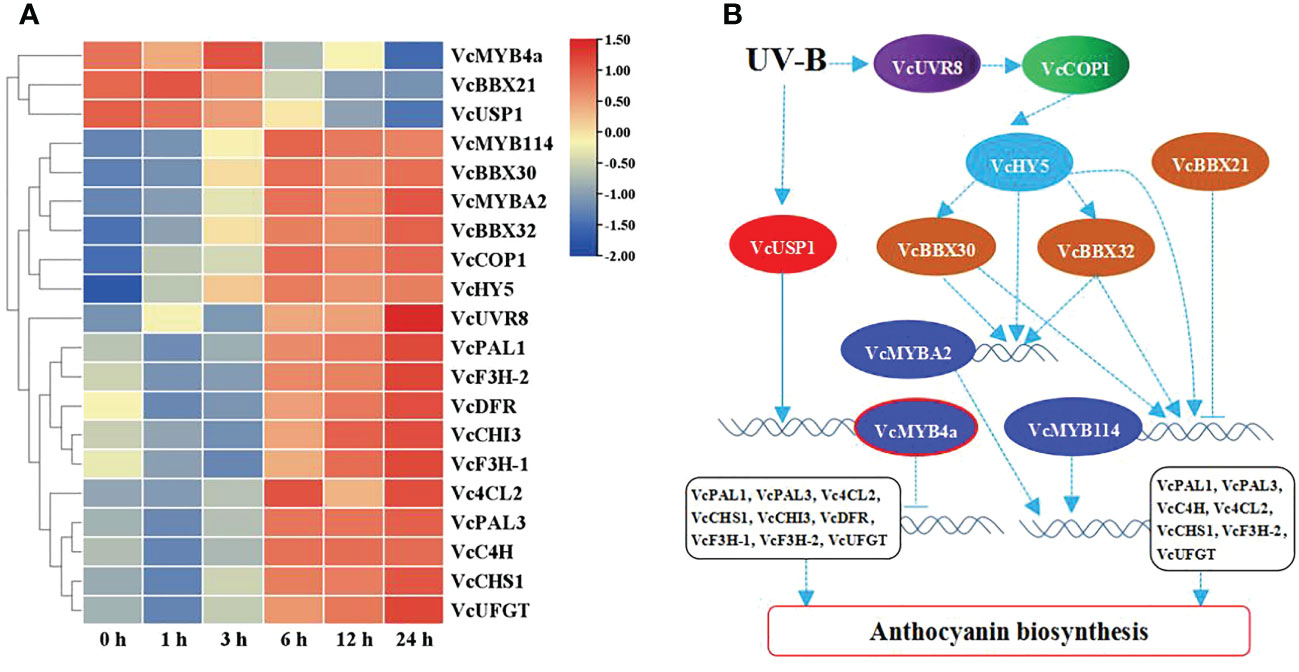
Figure 4 UV-B-induced anthocyanin biosynthesis pathway. (A) Heatmap representation of hierarchical clustering analysis of UV-B-induced anthocyanin biosynthesis pathway genes and transcription factor genes. Blue, low expression; red, high expression, based on log10(FPKM). (B) Proposed model of UV-B-induced anthocyanin biosynthesis pathways. Solid line indicates known signaling pathway; dotted line indicates putative signaling pathway. Arrows indicate activation; short lines indicate inhibition.
VcMYB4a represses the expression of anthocyanin biosynthesis genes in transgenic blueberry calli
To identify potential downstream target genes of VcMYB4a, we compared the transcript levels of VcPAL1, VcPAL3, Vc4CL2, VcCHS1, VcCHI3, VcDFR, VcF3H-1, VcF3H-2, and VcUFGT between VcMYB4a-overexpressing calli (OE-1) and WT calli during UV-B treatment by RT-qPCR analysis (Figure 5). Overexpressing VcMYB4a in blueberry calli resulted in the downregulation of the anthocyanin biosynthesis genes VcPAL3, VcDFR, VcF3H-2, and VcUFGT. The transcript levels of VcPAL1, VcPAL3, VcCHS1, VcDFR, VcF3H-1, VcF3H-2, and VcUFGT genes rapidly increased and peaked after 24 h of UV-B radiation, and then decreased in both OE-1 and WT calli. For the same UV-B radiation time, the expression levels of VcCHS1, VcDFR, VcCHI3, and VcF3H-1 genes in WT calli were higher than in OE-1 calli. These results suggest that VcMYB4a inhibits anthocyanin biosynthesis by downregulating anthocyanin biosynthesis genes.
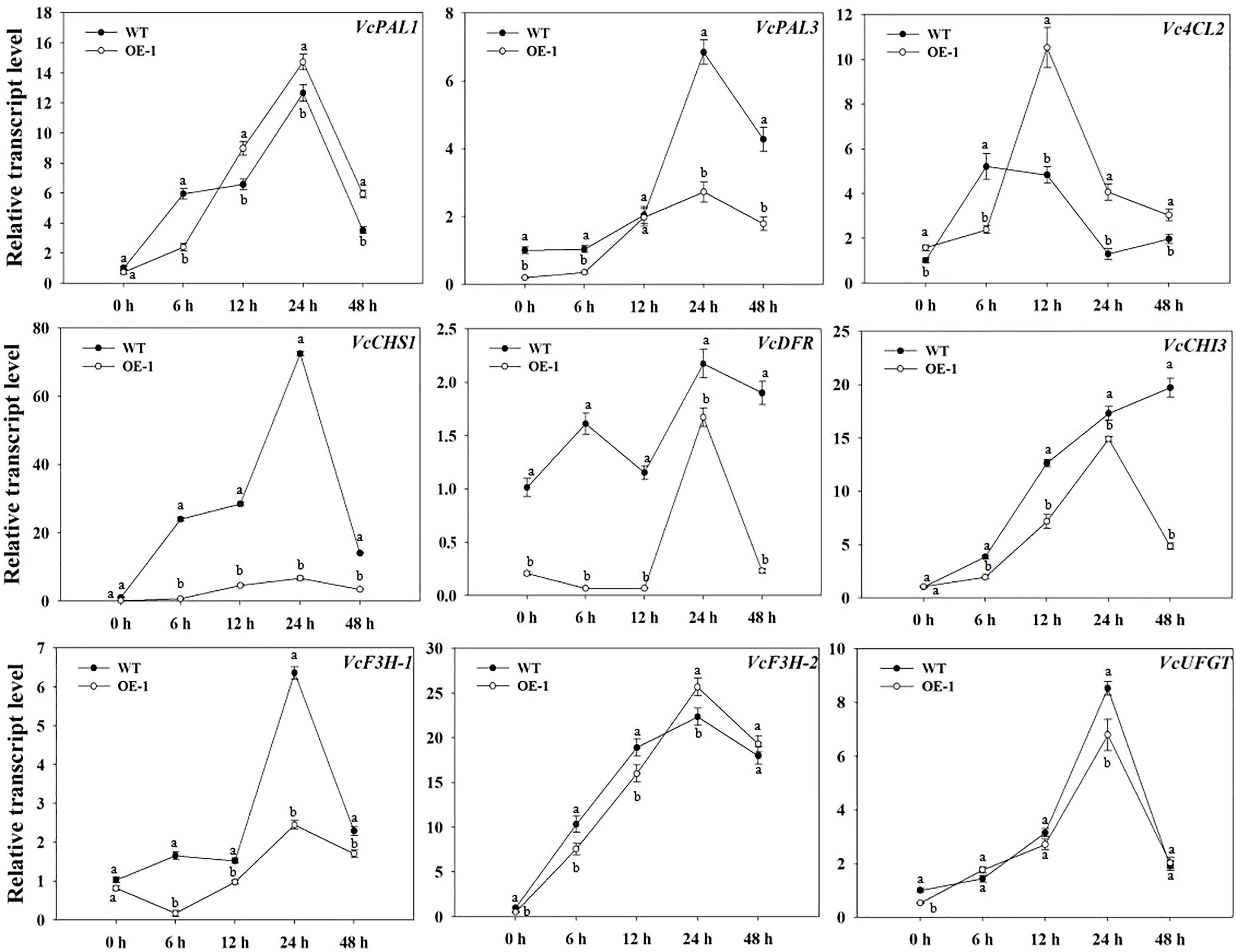
Figure 5 Expression of anthocyanin biosynthesis genes was repressed in the VcMYB4a-overexpressing blueberry calli (OE-1) compared with that in wild-type (WT) calli under UV-B treatment. Error bars indicate ± SD of the mean of three independent biological replicates, each with three technical replicates; different letters indicate significant differences (p < 0.05) among samples by Tukey’s test.
The upstream sequence of VcMYB4a is regulated by VcUSP1
To elucidate the mechanism by which VcMYB4a suppresses anthocyanin metabolism under UV-B radiation, we isolated the promoter sequence of VcMYB4a from blueberry using a previously reported genome-walking protocol and predicted cis-elements using the PlantCARE database (Tables S5; S6). The main elements included light responsive elements, defense-related elements (drought and low-temperature), and phytohormone-related elements (particularly, those associated with methyl jasmonate, abscisic acid, and gibberellin). We cloned the 1,718-bp sequence upstream of the coding region of VcMYB4a and inserted it into the pHIS2 vector. We then performed a yeast one-hybrid assay to identify proteins that interact with the VcMYB4a promoter sequence from a yeast library. The universal stress protein VcUSP1 directly bound to the promoter region of VcMYB4a (Figure 6A). To validate the result of the yeast one-hybrid assay, we performed luciferase complementation imaging assays in N. benthamiana leaves. Co-expression of GFP-VcUSP1 with pVcMYB4a-LUC led to a clear increase in LUC signal and Luc/Ren ratio compared with leaves co-expressing the control vectors (Figure 6B; 6C). These data confirmed that VcUSP1 directly bound to the promoter region of VcMYB4a to regulate its expression.
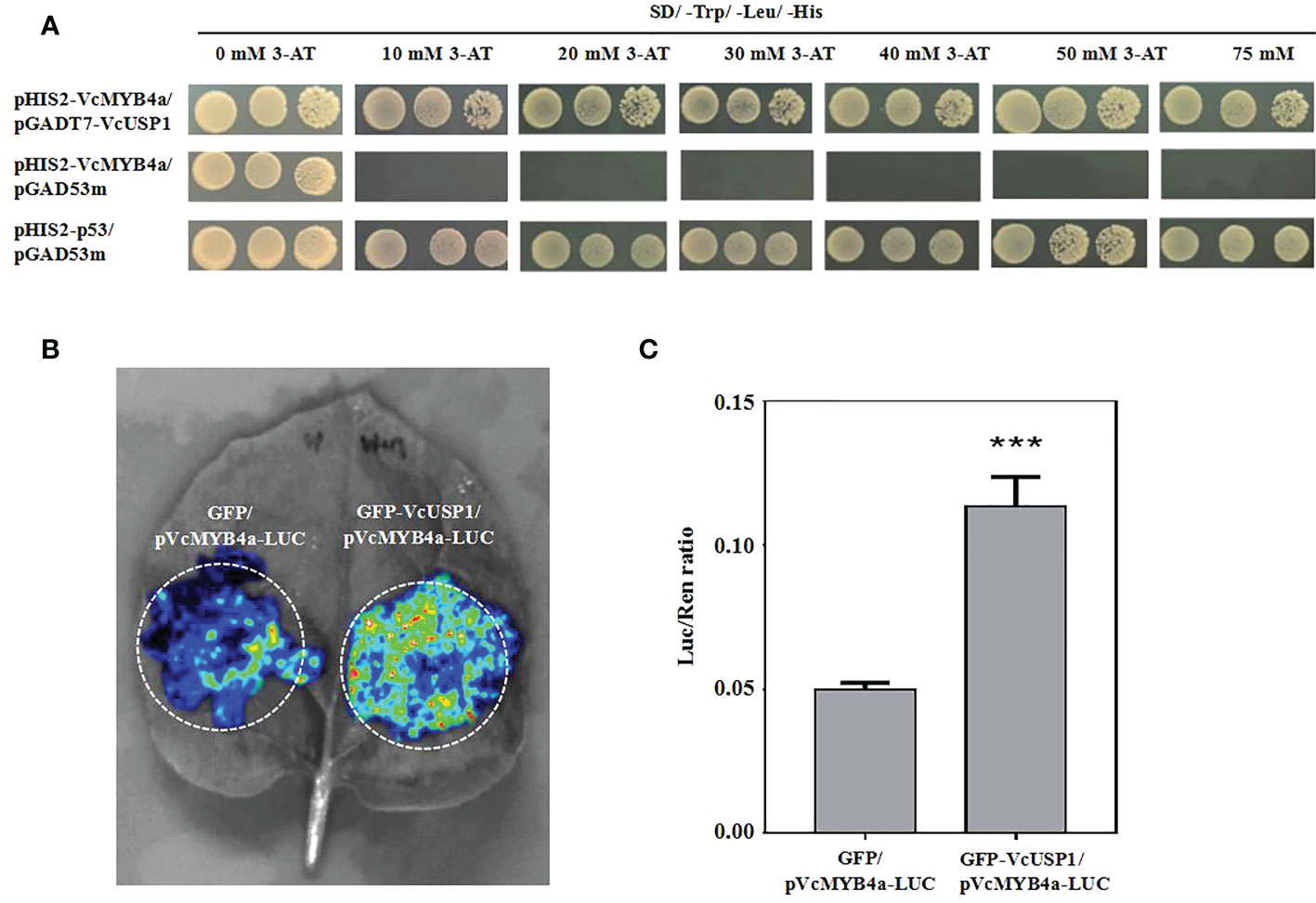
Figure 6 The VcUSP1 protein binds to upstream sequences of VcMYB4a. (A) Interaction of VcUSP1 with the VcMYB4a promoter in a yeast one-hybrid assay. pHIS2-VcMYB4a/pGAD53m was used as a negative control and pHIS2-p53/pGAD53m as a positive control. A series of 5-µL aliquots of 10 × diluted yeast cultures was grown on SD medium lacking tryptophan, leucine, and histidine (SD/–Trp/–Leu/–His) and with different concentrations of 3-amino-1,2,4-triazole (3-AT) for 3 days at 30°C. (B) Interaction of VcUSP1 with the VcMYB4a promoter using dual luciferase complementation imaging assays in Nicotiana benthamiana leaves. GFP/pVcMYB4a-LUC was used as a negative control. (C) The ratio of Luc-to-Ren activity was measured for the dual luciferase assay. Error bars indicate the SD of three biological replicates. The asterisks indicate statistically significant differences by Tukey’s test (***Correlation significant at the 0.001 level).
Discussion
UV-B radiation promotes anthocyanin accumulation via the UVR8-COP1-HY5 pathway (Rizzini et al., 2011; Bhatia et al., 2021). The HY5 transcription factor directly binds to upstream sequences of MYB transcription factor genes to promote their expression (Stracke et al., 2010; Shin et al., 2013; An et al., 2017). BBX proteins directly interact with HY5 to enhance its binding to the promoters of MYBs or regulates the expression of MYBs to promote anthocyanin accumulation. However, B-box protein also suppresses anthocyanin accumulation by interacting with HY5 to synergistically inhibit the expression of MYBs (Bai et al., 2014; An et al., 2019; Fang et al., 2019). In the current study, we established that VcUVR8, VcCOP1, and VcHY5 are upregulated under UV-B treatment, and identified a positive correlation between VcHY5 and VcCOP1 expression (Figure 4A; Table 1). Furthermore, VcHY5 expression was found to be positively correlated with VcBBX30, VcBBX32, VcMYB114, and VcMYBA2 expression. These results suggest that VcUVR8 perceives UV-B radiation and then upregulates VcCOP1 and VcHY5. VcHY5 then combines with VcBBX32 and VcBBX30 to promote VcMYB114 and VcMYBA2 expression. Therefore, VcHY5, VcBBX32, VcBBX30, VcMYB114, and VcMYBA2 form a positive regulatory network of anthocyanin biosynthesis under UV-B treatment (Figure 4B).
Transcription factors play essential roles in plant responses to abiotic stress (Chen et al., 2010; Seo and Park, 2010). UV-B radiation regulates the expression of most transcription factor genes, including MYB, bHLH, WRKY, NAC, and bZIP family genes (Ma et al., 2020; Ding et al., 2021). The current results support this finding. To date, 229 non-redundant MYB sequences have been identified in the blueberry genome, including 72 that respond to drought treatment in leaves and 69 in roots (Wang et al., 2021). In this study, 52 non-redundant MYB transcription factor genes from the R2R3-MYB, 1R-MYB, 3R-MYB, and 3R/4R-MYB subfamilies were found to function in the response of blueberry to UV-B radiation. Fifteen subgroups of R2R3-MYB subfamily members are involved in the response to UV-B radiation, as they were upregulated or downregulated by this treatment (Figure 2). These results improve our understanding of the roles of blueberry MYB TFs in response to abiotic stress.
R2R3-MYB transcription factors from subgroup 6 control anthocyanin biosynthesis by regulating the expression of structural genes involved in this process. For example, AtMYB113, AtMYB114, AtMYB75, and AtMYB90 promote anthocyanin biosynthesis by upregulating the expression of late anthocyanin structural genes in Arabidopsis (Teng et al., 2005; Gonzalez et al., 2008). Blueberry (Vaccinium section Cyanococcus) expressing MYBA of subgroup 6 activated anthocyanin production in N. benthamiana, and MYBA transactivated the DFR promoter from blueberry and other species (Plunkett et al., 2018). In the current study, we identified two VcMYB genes of subgroup 6, VcMYB114 and VcMYBA2, whose expression was positively correlated with that of structural genes involved in anthocyanin biosynthesis that were upregulated under UV-B radiation (Table 1; Figure 4). Thus, VcMYB114 and VcMYBA2 function as activators in the UV-B-induced anthocyanin biosynthesis network by promoting downstream gene expression.
VcMYB4a, belonging to subgroup 4, inhibits lignin biosynthesis and negatively regulates plant responses to salt, drought, and temperature stress (Zhang et al., 2020; Yang et al., 2022). Our study showed that the expression levels of VcMYB4a were negatively correlated with those of anthocyanin biosynthesis genes. Thus, VcMYB4a likely encodes an inhibitor of the anthocyanin biosynthesis pathway. Yeast one-hybrid and dual luciferase assays showed that VcUSP1 binds to the promoter region of VcMYB4a and directly regulates its expression (Figure 6). USP is a ubiquitous protein that plays an important role in abiotic stress tolerance in plants (Jung et al., 2015). Some USP proteins enhance osmotic stress, salinity, drought, cold, freezing, or heat tolerance in plants (Loukehaich et al., 2012; Jung et al., 2015; Udawat et al., 2016; Guo et al., 2020). However, a recent study showed that AtUSP17, a multiple stress-inducible gene, encodes a universal stress protein in Arabidopsis that negatively regulates salt tolerance by modulating multiple signaling pathways (Bhuria et al., 2022). In this study, VcUSP1 was found to be downregulated under UV-B treatment (Figure 4A). These findings suggest that VcUSP1 and VcMYB4a form a negative regulatory network and that UV-B radiation downregulates the expression of VcMYB4a by repressing VcUSP1 expression.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
CZ conceived and designed the project. YS, BM, QG, LZ, XZ, ZM and HY participated in the experiments and analyzed the data. YS and BM drafted the manuscript. CZ modified the manuscript. All authors read and approved the final manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (grant number 31700260).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1125382/full#supplementary-material
Supplementary Figure 1 | Various transcription factor families were induced by UV-B radiation in blueberry callus. Transcriptome deep sequencing (RNA-seq) data were obtained for blueberry callus treated with UV-B radiation for 0, 1, 3, 6, 12, and 24 h. MYB, MYB proteins; bHLH, basic helix-loop helix; WRKY, WRKY proteins; NAC, NAC domain–containing protein; bZIP, basic region/leucine zipper; GATA, GATA zinc finger; AUX/IAA, auxin-responsive protein; HSF, heat stress transcription factor; Auxin, auxin response factor; DOF, Dof zinc finger protein; AP2, AP2 domain–containing protein.
References
Aharoni, A., Vos, C. H. R. D., Wein, M., Sun, Z., Greco, R., Kroon, A., et al. (2001). The strawberry FaMYB1 transcription factor suppresses anthocyanin and flavonol accumulation in transgenic tobacco. Plant J. 28, 319–332. doi: 10.1046/j.1365-313X.2001.01154.x
An, J. P., Qu, F. J., Yao, J. F., Wang, X. N., You, C. X., Wang, X. F., et al. (2017). The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 4, 17023. doi: 10.1038/hortres.2017.23
An, J. P., Wang, X. F., Zhang, X. W., Bi, S. Q., You, C. X., Hao, Y. J. (2019). MdBBX22 regulates UV-b-induced anthocyanin biosynthesis through regulating the function of MdHY5 and is targeted by MdBT2 for 26s proteasome-mediated degradation. Plant Biotechnol. J. 17, 2231–2233. doi: 10.1111/pbi.13196
Apweiler, R., Bairoch, A., Wu, C. H., Barker, W. C., Boeckmann, B., Ferro, S., et al. (2004). UniProt: the universal protein knowledgebase. Nucleic Acids Res. 32, D115–D119. doi: 10.1093/nar/gkh131
Bai, S., Saito, T., Honda, C., Hatsuyama, Y., Ito, A., Moriguchi, T. (2014). An apple b-box protein, MdCOL11, is involved in UV-b- and temperature-induced anthocyanin biosynthesis. Planta 240, 1051–1062. doi: 10.1007/s00425-014-2129-8
Bai, S., Tao, R., Tang, Y., Yin, L., Ma, Y., Ni., J., et al. (2019). BBX16, a b-box protein, positively regulates light-induced anthocyanin accumulation by activating MYB10 in red pear. Plant Biotechnol. J. 17, 1985–1997. doi: 10.1111/pbi.13114
Bhatia, C., Gaddam, S. R., Pandey, A., Trivedi, P. K. (2021). COP1 mediates light-dependent regulation of flavonol biosynthesis through HY5 in Arabidopsis. Plant Sci. 303, 110760. doi: 10.1016/j.plantsci.2020.110760
Bhuria, M., Goel, P., Kumar, S., Singh, A. K. (2022). AtUSP17 negatively regulates salt stress tolerance through modulation of multiple signaling pathways in Arabidopsis. Physiol. Plantarum 174, e13635. doi: 10.1111/ppl.13635
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, H., Lai, Z., Shi, J., Xiao, Y., Chen, Z., Xu, X. (2010). Roles of arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors in plant responses to abscisic acid and abiotic stress. BMC Plant Biol. 10, 281. doi: 10.1186/1471-2229-10-281
Chen, Y., Yang, X., He, K., Liu, M., Li, J., Gao, Z., et al. (2006). The MYB transcription factor superfamily of arabidopsis: expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 60, 107–124. doi: 10.1007/s11103-005-2910-y
Ding, R., Che, X., Shen, Z., Zhang, Y. (2021). Metabolome and transcriptome profiling provide insights into green apple peel reveals light- and UV-b-responsive pathway in anthocyanins accumulation. BMC Plant Biol. 21, 351. doi: 10.1186/s12870-021-03121-3
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Fang, H., Dong, Y., Yue, X., Hu, J., Jiang, S., Xu, H., et al. (2019). The b-box zinc finger protein MdBBX20 integrates anthocyanin accumulation in response to ultraviolet radiation and low temperature. Plant Cell Environ. 42, 2090–2104. doi: 10.1111/pce.13552
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Guo, L., Zhou, C., Lu, S., Guo, Z. (2020). A universal stress protein from Medicago falcata (MfUSP1) confers multiple stress tolerance by regulating antioxidant defense and proline accumulation. Environ. Exp. Bot. 178, 104168. doi: 10.1016/j.envexpbot.2020.104168
Hartmann, U., Sagasser, M., Mehrtens, F., Stracke, R., Weisshaar, B. (2005). Differential combinatorial interactions of cis-acting elements recognized by R2R3-MYB, BZIP, and BHLH facotors control light-responsive and tissue-specific activation of phenylpropanoid biosynthesis genes. Plant Mol. Biol. 57, 155–171. doi: 10.1007/s11103-004-6910-0
Henry-Kirk, R. A., Plunkett, B., Hall, M., McGhie, T., Alla, A. C., Wargent, J. J., et al. (2018). Solar UV light regulates flavonoid metabolism in apple (Malus × domestica). Plant Cell Enviro. 41, 675–688. doi: 10.1111/pce.13125
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jung, Y. J., Melencion, S. M. B., Lee, E. S., Park, J. H., Alinapon, C. V., Oh, H. T., et al. (2015). Universal stress protein exhibits a redox-dependent chaperone function in Arabidopsis and enhances plant tolerance to heat shock and oxidative stress. Front. Plant Sci. 6. doi: 10.3389/fpls.2015.01141
Karppinen, K., Lafferty, D. J., Albert, N. W., Mikkola, N., McGhie, T., Allan, A. C., et al. (2021). MYBA and MYBPA transcription factors co-regulate anthocyanin biosynthesis in blue-coloured berries. New Phytol. 232, 1350–1367. doi: 10.1111/nph.17669
Kranz, H. D., Denekamp, M., Greco, R., Jin, H., Leyva, A., Meissner, R. C., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. doi: 10.1046/j.1365-313x.1998.00278.x
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Liu, L., Li, Y., She, G., Zhang, X., Jordan, B., Chen, Q., et al. (2018). Metabolite profiling and transcriptomic analyses reveal an essential role of UVR8-mediated signal transduction pathway in regulating flavonoid biosynthesis in tea plants (Camellia sinensis) in response to shading. BMC Plant Biol. 18, 233. doi: 10.1186/s12870-018-1440-0
Loukehaich, R., Wang, T., Ouyang, B., Ziaf, K., Li, H., Zhang, J., et al. (2012). SpUSP, an annexin-interacting universal stress protein, enhances drought tolerance in tomato. J. Exp. Bot. 63, 5593–5606. doi: 10.1093/jxb/ers220
Ma, T., Gao, H., Zhang, D., Shi, Y., Zhang, T., Shen, X., et al. (2020). Transcriptome analyses revealed the ultraviolet b irradiation and phytohormone gibberellins coordinately promoted the accumulation of artemisinin in Artemisia annua l. Chin. Med. 15, 67. doi: 10.1186/s13020-020-00344-8
Nguyen, C. T. T., Lim, S., Lee, J. G., Lee, E. J. (2017). VcBBX, VcMYB21, VcR2R3 MYB transcription factors are involved in UV-b induced anthocyanin biosynthesis in the peel of harvested blueberry fruit. J. Agric. Food Chem. 65, 2066–2073. doi: 10.1021/acs.jafc.6b05253
Norberto, S., Silva, S., Meireles, M., Faria, A., Pintado, M., Calhau, C. (2013). Blueberry anthocyanins in health promotion: a metabolic overview. J. Funct. Foods 5, 1518–1528. doi: 10.1016/j.jffff.2013.08.015
Oravecz, A., Baumann, A., Máté, Z., Brzezinska, A., Molinier, J., Oakeley, E., et al. (2006). CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-b response in. Arabidopsis. Plant Cell 18, 1975–1990. doi: 10.1105/tpc.105.040097
Plunkett, B. J., Espley, R. V., Dare, A. P., Warren, B. A. W., Grierson, E. R. P., Cordiner, S., et al. (2018). MYBA from blueberry (Vaccinium section cyanococcus) is a subgroup 6 type R2R3MYB transcription factor that activates anthocyanin production. Front. Plant Sci. 9, 1300. doi: 10.3389/fpls.2018.01300
Rabino, I., Mancinelli, A. L. (1986). Light, temperature, and anthocyanin production. Plant Physiol. 81, 922–924. doi: 10.1104/pp.81.3.922
Ribera, A. E., Reyes-Díaz, M., Alberdi, M., Zuniga, G. E., Mora, M. L. (2010). Antioxidant compounds in skin and pulp of fruits change among genotypes and maturity stages in highbush blueberry (Vaccinium corymbosum l.) grown in southern Chile. J. Soil Sci. Plant Nutr. 10, 509–536. doi: 10.4067/S0718-95162010000200010
Rizzini, L., Favory, J., Cloix, C., Faggionato, D., O’Hara, A., Kaiserli, E., et al. (2011). Perception of UV-b by the arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
Seo, P. J., Park, C. (2010). A membrane-bound NAC transcription factor as an integrator of biotic and abiotic stress signals. Plant Signal. Behav. 5, 481–483. doi: 10.4161/psb.11083
Shin, D. H., Choi, M. G., Kim, K., Bang, G., Cho, M., Choi, S. B., et al. (2013). HY5 regulates anthocyanin biosynthesis by inducing the transcriptional activation of the MYB75/PAP1 transcription factor in Arabidopsis. FEBS Lett. 587, 1543–1547. doi: 10.1016/j.febslet/j.febslet.2013.03.037
Song, Y., Ma, B., Guo, Q., Zhou, L., Lv, C., Liu, X., et al. (2022). UV-B induces the expression of flavonoid biosynthetic pathways in blueberry (Vaccinium cormbosum) calli. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1079087
Stracke, R., Favory, J. J., Gruber, H., Bartelniewoehner, L., Bartels, S., Binkert, M., et al. (2010). The Arabidopsis bZIP transcription factor HY5 regulates expression of the PFG1/MYB12 gene in response to light and ultraviolet-b radiation. Plant Cell Environ. 33, 88. doi: 10.1111/j.1365-3040.2009.02016.x
Teng, S., Keurentjes, J., Bentsink, L., Koornneef, M., Smeekens, S. (2005). Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiol. 139, 1840–1852. doi: 10.1104/pp.105.066688
Tohge, T., Kusano, M., Fukushima, A., Saito, K., Fernie, A. R. (2011). Transcriptional and metabolic programs following exposure of plants to UV-b irradiation. Plant Signal Behav. 6, 1987–1992. doi: 10.4161/psb.6.12.18240
Udawat, P., Jha, R. K., Sinha, D., Mishra, A., Jha, B. (2016). Overexpression of a cytosolic abiotic stress responsive universal stress protein (SbUSP) mitigates salt and osmotic stress in transgenic tobacco plants. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00518
Wang, A., Liang, K., Yang, S., Cao, Y., Wang, L., Zhang, M., et al. (2021). Genome-wide analysis of MYB transcription factors of Vaccinium corymbosum and their positive responses to drought stress. BMC Genomics 22, 565. doi: 10.1186/s12864-021-07850-5
Yang, B., Li, Y., Song, Y., Wang, X., Guo, Q., Zhou, L., et al. (2022). The R2R3-MYB transcription factor VcMYB4a inhibits lignin biosynthesis in blueberry (Vaccinium corymbosum). Tree Genet. Genomes 18, 27. doi: 10.1007/s11295-022-01560-z
Zhang, C., Guo, Q., Liu, Y., Liu, H., Wang, F., Jia, C. (2017). Molecular cloning and functional analysis of a flavanone 3-hydroxylase gene from blueberry. J. Hortic. Sci. Biotech. 92, 57–64. doi: 10.1080/14620316.2016.1224604
Zhang, C., Liu, H., Zhang, X., Guo, Q., Bian, S., Wang, L., et al. (2020). VcMYB4a, an R2R3-MYB transcription factor from Vaccinium corymbosun, negatively regulates salt, drought, and temperature stress. Gene 757, 144935. doi: 10.1016/j.gene.2020.144935
Keywords: blueberry, MYB transcription factors, UV-B radiation, anthocyanin, universal stress protein
Citation: Song Y, Ma B, Guo Q, Zhou L, Zhou X, Ming Z, You H and Zhang C (2023) MYB pathways that regulate UV-B-induced anthocyanin biosynthesis in blueberry (Vaccinium corymbosum). Front. Plant Sci. 14:1125382. doi: 10.3389/fpls.2023.1125382
Received: 16 December 2022; Accepted: 16 January 2023;
Published: 30 January 2023.
Edited by:
Dong Meng, Cornell University, United StatesReviewed by:
Yamei Shen, Zhejiang Agriculture and Forestry University, ChinaYuan-Yuan Li, Shandong Agricultural University, China
Copyright © 2023 Song, Ma, Guo, Zhou, Zhou, Ming, You and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyu Zhang, cy_zhang@jlu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Yan Song
Yan Song Bin Ma
Bin Ma Qingxun Guo
Qingxun Guo Lianxia Zhou
Lianxia Zhou Xintong Zhou
Xintong Zhou Ziqing Ming
Ziqing Ming Honglin You
Honglin You Chunyu Zhang
Chunyu Zhang