- 1Technology Center, China Tobacco Hunan Industrial Co., Ltd., Changsha, China
- 2Key Laboratory for Tobacco Gene Resources, Tobacco Research Institute, Chinese Academy of Agricultural Sciences, Qingdao, China
- 3Kunming Branch of Yunnan Provincial Tobacco Company, Kunming, China
- 4Yuxizhongyan Tobacco Seed Co., Ltd., Yuxi, China
- 5Hunan Tobacco Research Institute, Changsha, China
The MYB members play important roles in development, metabolism, and stress tolerance in plants. In the current study, a total of 246 tobacco R2R3-MYB transcription factors were identified and systemically analyzed from the latest genome annotation. The newly identified tobacco members were divided into 33 subgroups together with the Arabidopsis members. Furthermore, 44 NtMYB gene pairs were identified to arise from duplication events, which might lead to the expansion of tobacco MYB genes. The expression patterns were revealed by transcriptomic analysis. Notably, the results from phylogenetic analysis, synthetic analysis, and expression analysis were integrated to predict the potential functions of these members. Particularly, NtMYB102 was found to act as the homolog of AtMYB70 and significantly induced by drought and salt treatments. The further assays revealed that NtMYB102 had transcriptional activities, and the overexpression of the encoding gene enhanced the drought and salt stress tolerance in transgenic tobacco. The results of this study may be relevant for future functional analyses of the MYB genes in tobacco.
Introduction
Transcription factor is the important regulator that controls the expression of target genes, and they play important roles in plant development, metabolism, and stress response (Dubos et al., 2010; Katiyar et al., 2012; Coleto et al., 2021). The MYB transcription factor family is the one of largest transcription factor families in plants (Martin and Paz-Ares, 1997; Stracke et al., 2001). Generally, each MYB member contains a highly conserved DNA binding domain, composed of 50–52 amino acids, which binds the DNA in the form of a helix-turn-helix structure (Moyano et al., 1996). According to the repeat number of MYB domains, the MYB family members could be divided into four groups, including 4R-MYB, R1R2R3-MYB, 1R-MYB, and R2R3-MYB, and the R2R3-MYB members are the most common subgroup in plants (Stracke et al., 2001; Dubos et al., 2010). Interestingly, a conjecture is that R2R3-MYB proteins have likely evolved from R1R2R3-MYB precursors by the loss of the R1 repeat. On the contrary, the R1R2R3-MYB might evolve from R2R3-MYB by acquiring an R1 repeat. (Stracke et al., 2001; Jiang et al., 2004; Dubos et al., 2010). Since the first plant MYB gene was cloned from maize (Paz-Ares et al., 1987), a variety of MYB transcription factors in plants have been identified and analyzed (De Vos et al., 2006; Zou et al., 2013; Stracke et al., 2014; Qiu et al., 2019; Chen et al., 2021; Naik et al., 2021; Qin et al., 2021).
In plants, the R2R3-MYB subfamily members have been found to participate in various biological processes including plant development, secondary metabolism, and biotic/abiotic stress responses (Stracke et al., 2001; Castillejo et al., 2020; Cao et al., 2021, Ding et al., 2021). In Arabidopsis, 126 R2R3-MYB transcription factors have been divided into 25 subgroups, and a number of them had been reported to play an important role in primary and secondary metabolism (Allan et al., 2008; Dubos et al., 2010; Roy, 2016). In subgroup 7, as the typical MYB transcription factors, AtMYB11/PFG2, AtMYB12/PFG1, and AtMYB111/PFG3 were reported to be involved in the accumulation of specific flavonol derivatives in leaves, stems, inflorescences, siliques, and roots (Moyano et al., 1996; Stracke et al., 2007). In subgroup 6, the AtMYB75/PAP1 and AtMYB90/PAP2 transcription factors regulate the biosynthesis of anthocyanins in vegetative tissues (Quattrocchio et al., 1999; Appelhagen et al., 2011). Overexpression of AtMYB113 or AtMYB114 leads to a significant increase in anthocyanins production (Gonzalez et al., 2008). In addition, multiple R2R3-MYB genes have been reported to be involved in the synthesis of Arabidopsis secondary cell walls (Zhao and Dixon, 2011). In subgroup 3, AtMYB58 and AtMYB63 affect the SND1-mediated (secondary wall-associated NAC domain protein 1) transcription network regulating secondary wall formation (Zhou et al., 2009). In subgroup 16, AtMYB83 and AtMYB46 are both direct targets of the SND1 transcription factor and play the redundant role in the transcriptional regulatory cascade, which allows plants to form secondary walls by regulating fibers and blood vessels (Mccarthy et al., 2009).
Some R2R3-MYB genes have also been reported to participate in the growth and development of plants (Stracke et al., 2001; Li et al., 2020; Pucker et al., 2020). AtMYB77 of subgroup 22 was reported to control lateral root growth and development (Shin et al., 2007). In subgroup 14, AtMYB68 specifically regulates root growth, influencing the whole plant development under harsh conditions (Feng et al., 2004). AtMYB59 was reported to regulate the development of roots by regulating the cell cycle of the root tip (Mu et al., 2009). In flower development, AtMYB33 and AtMYB65, which belong to subgroup 18, facilitate both anther and pollen development, and the pollen fails to maintain vitality in myb33/myb65 (Millar and Gubler, 2005). AtMYB120 is studied to be a pollen-specific factor, which controls the pollination of plants and the differentiation and development of pollen (Liang et al., 2013). In subgroup 21, AtMYB105/LOF2 and AtMYB117/LOF1 control the separation of the lateral stem of the plant, and AtMYB91/AS1 participates in the regulation of leaf patterning (Byrne et al., 2000; Lee et al., 2009). In subgroup 20, AtMYB2 is reported to regulate leaf senescence, and in subgroup 22, AtMYB44/MYBR1 was involved in the regulatory network of leaf senescence and ABA signaling (Guo and Gan, 2011; Jaradat et al., 2013).
Unlike animals, plants grow in a complex environment and face multiple biotic/abiotic stresses from the environment; some R2R3-MYB transcription factors have been confirmed to respond to the stresses in plants (Stracke et al., 2001; Dubos et al., 2010). For instance, AtMYB60 and AtMYB96 of subgroup 1 are reported to improve the plant drought resistance by regulating the closure of plant stomata (Seo et al., 2009; Oh et al., 2011). Similarly, in subgroup 22, AtMYB70, AtMYB73, and AtMYB74 also regulate stomatal closure to improve plants’ anti-stress ability (Jung et al., 2008). In addition, several R2R3-MYB members are also involved in cold, salinity, and wounding stresses. In subgroup 2, AtMYB2 is reported to increase plant salt resistance through ABA signaling, while AtMYB15 is involved in the regulation of cold tolerance; myb15 increased plants’ tolerance to cold stress whereas its overexpression reduced plants’ cold tolerance (Abe et al., 2003; Agarwal et al., 2006). In defense response, AtMYB102 keeps plants from being damaged by herbivore Pieris rapae. AtMYB41 is reported to be involved in the negative regulation of short-term transcriptional response to osmotic pressure, and AtMYB72 is essential for Arabidopsis to fight against various fungal and bacterial diseases (De Vos et al., 2006; Cominelli et al., 2008; Van Der Ent et al., 2008; Segarra et al., 2009). Besides, in subgroup 22, overexpression of AtMYB44 could promote Botrytis infection, and AtMYB44 positively regulates the disease resistance of Pseudomonas syringae in Arabidopsis through the salicylic acid signaling pathway (Jung et al., 2008).
Tobacco is not only a considerable economically valuable crop but also a well-studied model organism. However, extreme environment and diseases have always been the potential threat to tobacco yield and quality. Therefore, the study for systematic analysis of the MYB transcription factor family in tobacco is of great significance for the research of tobacco secondary metabolism, growth development, and resistance to biotic/abiotic stresses. With the publication of multiple plant genome sequences, a new insight combining bioinformatics and molecular biology began to identify and analyze the R2R3-MYB transcription factor family and their functions. So far, the function of the R2R3-MYB protein has been discovered and verified in a large number of plants, such as Arabidopsis (Dubos et al., 2010), tomato (Solanum lycopersicum) (Li et al., 2016), potato (Solanum tuberosum) (Li et al., 2019), Chinese pear (Pyrus bretschneideri Rehd) (Cao et al., 2016), maize (Zea mays) (Du et al., 2012), wheat (Triticum aestivum) (Wei et al., 2020), and rice (Oryza sativa) (Katiyar et al., 2012). However, very limited research is available on tobacco R2R3-MYB members. Here, we identified 246 R2R3-MYB members from the tobacco genome sequences using comparative genomic and molecular biology methods, then inferred the function of R2R3-MYB protein from phylogenetic trees, collinearity, and expression patterns. This study will provide a solid foundation for further functional studies on the function of R2R3-MYB members in tobacco growth, metabolism, and biotic/abiotic stresses.
Materials and methods
Identification and classification analysis of tobacco R2R3-MYB members
Version 4.5 of the genome sequence annotations of tobacco (Nicotiana tabacum L.) were downloaded from the SGN database (https://solgenomics.net/). The previously reported AtMYB full-length protein sequences (Stracke et al., 2001; Dubos et al., 2010) were obtained from The Arabidopsis Information Resource (TAIR, https://www.arabidopsis.org/) and used as queries to perform the BLASTP search against the annotated tobacco protein databases with an E-value cutoff of 0.01. Furthermore, with the HMM profile (PF00249), the HMM search was performed against the annotated tobacco protein databases under the E-value cutoff of 0.001. The candidate sequences from the two above-described approaches were integrated, and redundant entries were removed manually. The putative MYB protein sequences were analyzed using both Pfam (https://pfam.xfam.org/) with an E-value cutoff of 1.0 and SMART (http://smart.embl.de/) with an E-value cutoff of 1.0 to detect the presence and number of the MYB domain (Letunic et al., 2020; Mistry et al., 2021).
The full-length protein sequences of Arabidopsis R2R3-MYB members and newly identified tobacco R2R3-MYB members were subjected to the performance of multiple sequence alignment using MAFFT v5.3 under the default settings (Katoh and Standley, 2013). The alignment of R2R3-MYB domain was visualized using TBtools (Chen et al., 2020). Subsequently, the neighbor-joining phylogenetic analysis was conducted using MAGE X based on the alignment of full-length protein sequences with a bootstrap method of 1000 replicates, substitution with the Poisson model, and pairwise deletion (Kumar et al., 2018). The phylogenetic tree was displayed using FigTree v1.3.
Exon–intron structural analysis and identification of conserved motifs
The genomic sequences and coding sequences of AtMYB and NtMYB genes were submitted to the Gene Structure Display Server (Hu et al., 2015) to visualize their exon–intron structure. Further, the MEME Suite 5.1.1 tool (Bailey et al., 2015) was hired to explore the conserved motifs of the AtMYB and NtMYB proteins with following parameters: maximum number of motifs: 10; the optimum width of each motif: between 6 and 100 residues.
Chromosomal localization and duplication event analysis
According to the location information provided by the Solanaceae database, the Perl program was adopted to display the NtMYB genes’ chromosomal location. The tandem gene events were displayed on the chromosomal map according to the previous definition (Li et al., 2018). Afterward, the TBtools’ Circos program (Chen et al., 2020) was recruited to analyze the synteny relationship of the orthologous genes from tobacco and five other species (including Arabidopsis, tomato, potato, maize, and rice).
Promoter analysis of tobacco R2R3-MYB genes
The sequences 2000 bp upstream of the R2R3-MYB genes in tobacco were extracted from the genome sequence database. The obtained sequences were subjected to PlantCARE platform analysis to further search for the putative cis-elements in their promoter regions (Lescot et al., 2002).
Expression patterns analysis
The reported RNA-seq data of tobacco tissues (Edwards et al., 2017) were downloaded from the GEO database (accession number: GSE95717). The processed expression data of the R2R3-MYB genes were extracted (Supplementary Table S1) and transformed with log2 to normalize the raw data using TBtools (Chen et al., 2020).
The RNA-Seq data of the tobacco senescent leaf were produced in our previous work (Li and Guo, 2018) and were uploaded at the NCBI Short Read Archive (SRA) under the accession number SRP102153. The plants were grown with regular practices and topping was done 60 days after transplanting. The middle leaves were collected at nine different time points and used for transcriptomic analysis. The expression data of the R2R3-MYB genes were retrieved (Supplementary Table S2) and normalized by comparing with 5 DAT (days after topping) and log2 fold change transformation. All the normalized RNA-seq data were used to illustrate Heatmap using the pHeatmap R package under the default parameters.
Tobacco plant preparation and stress treatments
Cultivated tobacco K326 was used to analyze the expression pattern of NtMYB in this study. The roots, stems, flowers, upper leaves, middle leaves, and lower leaves were collected and frozen in liquid nitrogen when cultivated tobacco K326 grew to the budding stage; the samples were stored in a refrigerator at -80°C for later use.
For the drought stress treatment assays, T3 transgenic and wild-type (K326) tobacco seeds were sown into the soil and treated after about 6 weeks as with previous conditions (Li et al., 2022). The leaves of the seedlings were detached and air-dried, then the leaf weight was recorded at 180 min.
For the salt stress treatment assays, the tobacco seedlings were germinated in a solid MS medium after disinfection, then they were transferred to a liquid MS medium and adapted for 5 days. Hereafter, some of the seedlings were transferred to a 150 mmol/l NaCl liquid MS medium or absorbent filter paper for salt and drought treatment, respectively. Samples were taken at 1, 3, and 6 h after treatment and frozen in liquid nitrogen for later use. Three biological replicates were performed for each sample.
RNA Extraction and qTR-PCR
Total RNAs from each sample were extracted using the Ultrapure RNA Kit (cwbiotech, Beijing, China), then the first-strand complementary DNA (cDNA) was synthesized using the Evo M-MLV Mix Kit with gDNA Clean for qPCR (Accurate Biotechnology, Changsha, China). Quantitative real-time PCR (qRT-PCR) reactions were performed in a Roche LightCycler 480 Real-Time PCR instrument with SYBR® Green Premix Pro Taq HS qPCR Kit (Accurate Biotechnology, Changsha, China). The tobacco ribosomal protein gene L25 (GenBank No. L18908) was used as control (Li et al., 2021). All experimental data were obtained through three technical repetitions and three biological replicates; the relative expression level was calculated by the 2-△△CT method (Livak and Schmittgen, 2001). The details of the primers are provided in Supplementary Table S3.
Subcellular localization
The CDS of the NtMYB102 gene was amplified from the cDNA of tobacco root using Phanta® Max Master Mix (Vazyme, Nanjing, China) and ligated to the pEasy-Blunt vector for later use; then the sequence of the NtMYB102 encoding gene without stop codon was inserted into the PYG57 vector, which was started by the CaMV-35S promoter and contained the GFP fragment (Sun et al., 2021). The construct was transformed into an Agrobacterium competent cell GV3101, and transiently expressed in the leaves of Nicotiana benthamiana. Simultaneously, the empty vector injected leaves as a control. Three days after the injection, the leaves were soaked in DAPI staining solution to determine the location of the nucleus, as previously reported (Li et al., 2019). Fluorescence signals were captured using a Confocal Microscope (TCS-SP8 Leica, Wetzlar, Germany).
Transcriptional activation assay
The CDS of NtMYB102 was amplified and inserted into the EcoR I site of the pBridge vector, using an Infusion HD Cloning Kit (Takara, Shiga, Japan) to be fused with a GAL4 DNA binding domain. The construct and the control vector were introduced into the yeast strain AH109 separately, followed by growing yeasts on SD/-Trp, and SD/-Trp supplemented with 5-bromo-4-chloro-3-indolyl-a-d-galactopyranoside (X-a-Gal) for 4 days at 30°C. The transcriptional activation activities were evaluated based on the growth status of different transformants.
Tobacco transgenic plant and root length analysis
The coding sequence of the NtMYB102 gene was amplified from the cDNA and inserted into the pCHF3 vector, which was driven by the CaMV-35S promoter, to complete the construction of the overexpression vector. The pCHF3 plasmid containing the NtMYB102 gene was transformed using the Agrobacterium-mediated method (Buschmann, 2016). To obtain T1 generation homozygous lines, the seeds of serval T0 independent lines were selected on 50 mg/l kanamycin MS medium with no separation. The homozygous T1 lines and wild-type (K326) seeds were sterilized and grown in a vertical MS medium for 14 days, then transferred to an MS medium with 0 or 100 mM NaCl, respectively, to observe changes in root length. The significant difference analysis was calculated using SPSS v18.0 with the t-test.
Results
Identification of R2R3-MYB members in tobacco
To identify the R2R3-MYB members in tobacco, the BLASTP and HMMER searches were performed using the previous R2R3-MYB protein sequences from Arabidopsis as the query. Eventually, a total of 246 MYB members were obtained from tobacco genome sequences. Among them, 145 NtMYB genes distribute unevenly on the tobacco chromosomes, while the others map on the scaffolds. To distinguish the newly identified genes, theses R2R3-MYB genes were named according to physical order on the chromosome and scaffolds. The detailed information could be explored in Supplementary Table S4.
Multiple sequence alignment and phylogenetic analysis
In plants, the DNA binding domain with two adjacent MYB repeats is conserved in R2R3-MYB members (Jiang et al., 2004). To explore the features of the R2R3-MYB members of tobacco, the newly identified R2R3-MYB domain performed multiple sequence alignments. As the result, the tobacco R2 and R3 MYB repeats hold the conserved amino acid residues. Notably, the tobacco R2R3-MYB domain features were found to be highly similar with Arabidopsis members (Supplementary Figure 1A). In the R2 MYB repeat, three Tryptophan (W) residues were found to be conserved, while only two Tryptophan (W) residues were found to be conserved in the tobacco R3 MYB repeat; the first Tryptophan (W) was replaced by Phenylalanine (F) or others (Supplementary Figure 1B).
As a result, all of the R2R3-MYB members were divided into 33 subgroups, among them, the S1 to S25 subgroups were consistent with the previous reports (Dubos et al., 2010); the others were named from S26 to S33, which contained some tobacco and Arabidopsis R2R3-MYB members (Figure 1). Results showed that most of the subgroups contained R2R3-MYB members from those two species, indicating that the expansion of R2R3-MYB members may appear before the divergence of tobacco and Arabidopsis. Interestingly, several subgroups contained much more R2R3-MYB members from tobacco than Arabidopsis, such as S1, S2, and S14, implying that the duplication events might occur in those subgroups. Notably, it was found that S17, S29, S31, and S32 only harbored R2R3-MYB members from tobacco and S12 only contained members from Arabidopsis.
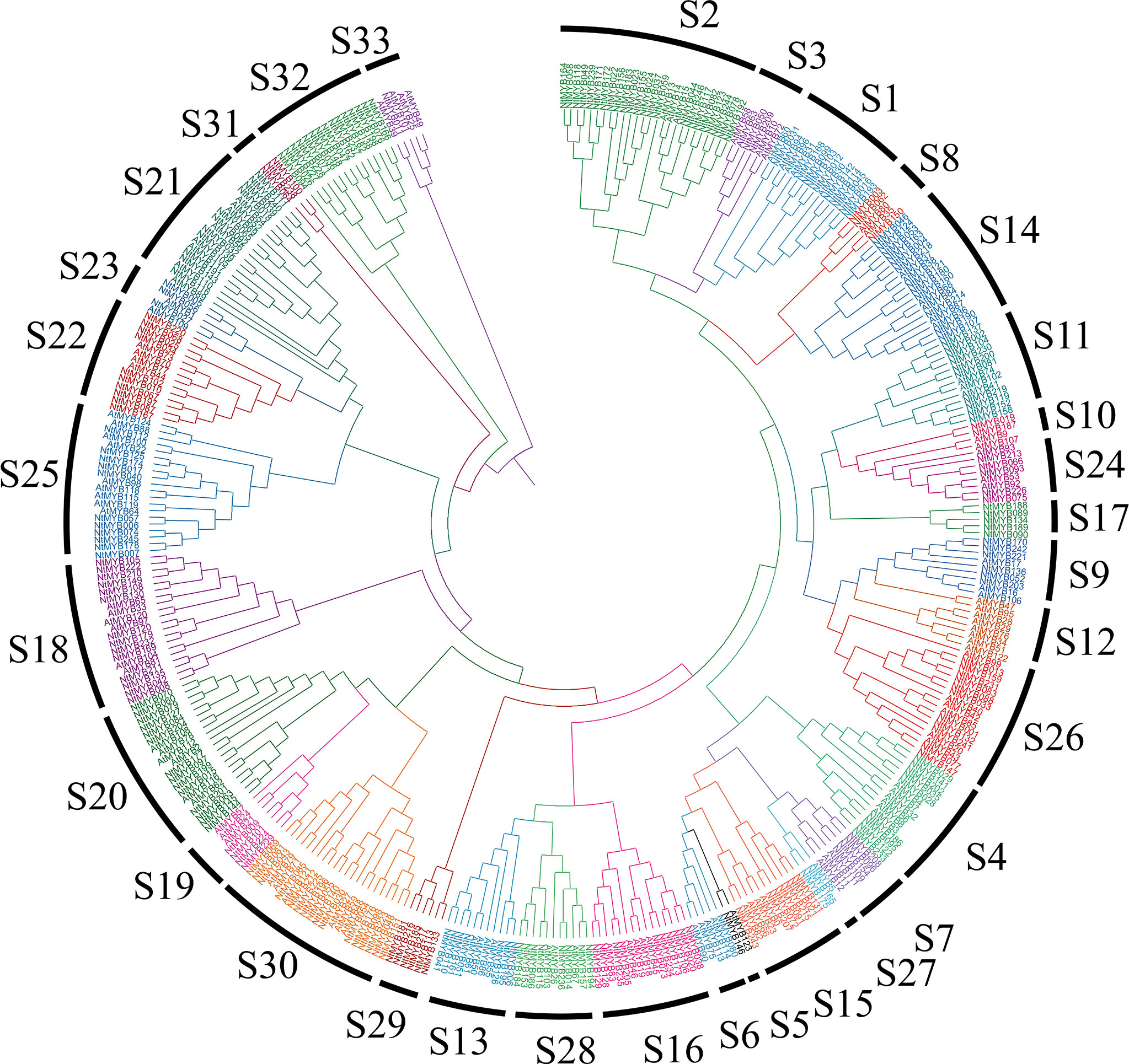
Figure 1 Phylogenetic analysis of tobacco R2R3-MYB family members. The tobacco R2R3-MYB members together with their Arabidopsis homologs were classified into 33 subgroups.
Gene structure and conserved,motif analysis
The gene structure could provide the clues of a gene family evolution history. As a result, the intron number of studied genes was found to range from 0 to 11, and NtMYB genes shared the similar gene structures with Arabidopsis MYB genes in the same group (Supplementary Figure S2). Interestingly, most (82.1%) of the coding sequences of NtMYB proteins were interrupted by one or two introns, whereas 15 (6.1%) NtMYB genes did not hold any introns. Furthermore, more than eight introns were found in NtMYB112, AtMYB88, and AtMYB124, which were all clustered into S25.
In addition, the R2R3-MYB protein sequences of tobacco and Arabidopsis were submitted to the MEME tool to analyze the conserved motifs. As a result, a total of 10 motifs were identified, namely, motif 1–10 (Supplementary Figures S2, S3). Among them, motifs 2, 3, and 4 together constitute the R2R3-MYB domain, which could be found in all studied MYB members. Consistent with the results of gene structure analysis, the R2R3-MYB proteins in the same group usually have similar types and orders of motifs. In addition, several motifs were found to be unique to certain subgroups. For instance, motif 8 was only found in S31, while motif 9 was unique to S20, implying that these unique motifs might undertake different functions. The similarities in characteristic motifs in each group may reflect functional similarities and should be conducive to determining specific functions for each R2R3-MYB gene.
Syntenic analysis of the tobacco R2R3-MYB gene
Syntenic analysis is important in genome sequence comparison, which reveals the genomic evolution of different species, while the syntenic pairs are predicted as orthologs and might share similar functions (Wang et al., 2021). As a result, collinearity pairs of the R2R3-MYB member were found in tobacco and five other species (Figure 2A). The collinear pairings between 81 of the NtMYB genes with MYB members in Arabidopsis were identified, followed by 151 NtMYB genes pairing with tomato, 125 NtMYB genes with potato, and 23 and 10 NtMYB genes with MYB genes from rice and maize, respectively. Notably, more R2R3-MYB collinearity pairs were found between tobacco and dicotyledonous species than monocotyledonous species. Furthermore, a total of six tobacco MYB genes were identified to form collinear pairs with MYB genes from all of the other species, indicating that these MYB genes may have existed before the divergence of these species (Figure 2B). Interestingly, 29 R2R3-MYB collinear pairs were predicted between tobacco and three dicotyledonous plants, but not found in the tested monocotyledonous plants, suggesting that these 29 pairs might arise after the divergence of dicotyledonous and monocotyledonous plants. The details of the tobacco MYB syntenic pairs can be found in Supplementary Table S5.
Chromosomal distribution and duplication events
The chromosomal location information of 246 NtMYB genes was obtained from the SGN database and visualized using the R (Figure 3A). As a result, Chromosome 04 harbored the most R2R3-MYB genes, while Chromosomes 01, 07, 11, 16, and 18 were found to hold only three R2R3-MYB genes. According to the previous definition of tandem gene events (Li et al., 2018), a total of seven clusters (NtMYB022/023, NtMYB023/024, NtMYB089/090, NtMYB118/119, NtMYB155/156, NtMYB171/172, NtMYB188/189) were identified, the first four of which were located on chromosomes and the others were found on scaffolds. Furthermore, NtMYB022/023 and NtMYB171/172 were found to arise from the tandem duplication events (Figure 3A).
The gene segmental duplication event served as the important access for plants to acquire new genes and gene family expansion. As results, a total of 64 tobacco R2R3-MYB genes were identified to form 42 segmental duplication pairs (Figure 3B and Supplementary Table S6). Notably, these results suggested that about 44% of the NtMYB genes may be generated by duplication events, which played the major role in the expansion of the MYB gene family in tobacco.
Promoter element analysis
In a previous study, the MYB genes had been reported to be involved in various developmental and stress responses. Considering these clues, the cis-element analysis was investigated to explore the probability of these NtMYB genes in developmental and stress responses. To study the expression regulation of the NtMYB genes, the promoter region of 246 R2R3-MYB genes was analyzed using the PlantCARE Online toolbox. Generally, it was found that a lot of cis-elements involved in various developmental and stress responses were present in these tobacco R2R3-MYB gene promoters. Furthermore, 13 cis-elements were selected from the PlantCARE database for visualization (Supplementary Figure S4). As a results, a total of 203 (82.5%) NtMYB gene promoters contained ABRE (abscisic acid responsive cis-element), suggesting that those R2R3-MYB genes may function in the abscisic acid signal pathway. Besides, 203 (82.5%) NtMYB gene promoters harbored ERE (ethylene responsive cis-element), while 129 (52.4%) gene promoters possessed a salicylic acid responsive cis-element (TCA-element). In addition, both CGTCA-motif and TGACG-motif are related to MeJA responsive, and 184 and 185 gene promoters were detected to possess these two kinds of cis-element, respectively. Further, a total of 98 (39.8%) gene promoters were detected to hold the CAT-box, which was related to the development of the plant meristem. Notably, the MYB family members were reported to be induced by stress treatments; the stress response-related cis-elements of tobacco MYB gene promoters were also analyzed consequently. The stress-responsive cis-elements including MBS (MYB-binding site), TC-rich repeats, HSE (heat stress-responsive element), LTR (low-temperature-responsive element), WUN-motif (wound-responsive element), and ARE (anaerobic induction element) were found to be abundant in the promoter regions of many NtMYB gene promoters. Interestingly, a total of 151 NtMYB gene promoters were predicted to hold W-box cis-elements, which act as the binding sites of the WRKY transcription factor, implying these NtMYB genes might be regulated by a certain WRKY transcription factor. Overall, the promoters of NtMYB genes possess abundant cis-elements, suggesting that the expression of these NtMYB genes might be regulated by multiple factors.
Expression analysis of the NtMYB genes from RNA-seq
To explore the expression pattern of the newly identified NtMYB genes, their RNA-seq data (GSE95717) was analyzed and visualized by R packages. As a result, the expression levels of 246 NtMYB genes in root, shoot, and shoot apex tissues were investigated (Figure 4). The results showed that several genes were expressed abundantly in the tested tissues, including NtMYB181, NtMYB216, and NtMYB067; while NtMYB176, NtMYB048, NtMYB026, NtMYB151, NtMYB227, NtMYB054, and NtMYB098 were detected to be highly expressed in the shoot and root. In addition, a total of 51 genes such as NtMYB033, NtMYB088, and NtMYB187 were only detected in the root, whereas 15 genes like NtMYB168, NtMYB154, and NtMYB096 were found to be expressed in all tested tissues except the root. It was worth noting that 61 (24.8%) NtMYB genes were not detected in these tested tissues, implying those genes might function in other tissues.
In addition, another RNA-seq data (SRP102153) was also used to analyze the expression pattern of NtMYB genes in different developmental stages of tobacco leaves. As a result, a total of eight leaf growth stages were sequenced, and the expression of 57 genes increased as tobacco leaves grew (Figure 5). In particular, NtMYB008 and NtMYB178 was only expressed in the later stages of leaf growth, suggesting they might have a certain relationship with leaf senescence. AtMYB2 was reported to contribute to the regulation of whole plant senescence (Guo and Gan, 2011), so its homologous genes were also concerned during the senescence of tobacco leaves. Combined with evolutionary analysis, it was found that genes homologous to AtMYB2 were expressed at a high level during leaf senescence, such as the NtMYB060, NtMYB079, NtMYB127, NtMYB084, and NtMYB177. This finding proved the reliability of evolutionary analysis on the one hand and implied the potential function of these genes in regulating leaf senescence.
The validation of expression patterns by qRT-PCR
To enrich the expression profile of the NtMYB genes of RNA-seq data, qRT-PCR was hired to analyze the expression changes of several representative genes. As a result, NtMYB096, NtMYB109, and NtMYB124 were observed to highly be expressed in the root, NtMYB053 showed abundant transcripts in the stem, and NtMYB130 was found to be expressed globally in all tested tissues (Figure 6A), which were consistent with the RNA-seq data. In addition, NtMYB060 and NtMYB079 were detected to be upregulated during leaf senescence; on the contrary, NtMYB210 had down-regulated expression, implying those genes might participate in leaf senescence of tobacco. Notably, qRT-PCR added the floral tissue to enrich the expression profile of tobacco R2R3-MYB genes; the results showed that the selected genes were expressed in varying degrees in flowers. Especially, NtMYB105, NtMYB108, and NtMYB149 were highly expressed in flowers, suggesting they might play the key role in the flower development of tobacco.
Furthermore, a number of R2R3-MYB transcription factors were reported to respond to abiotic stresses in Arabidopsis; representative NtMYB genes were selected to test whether they could respond to abiotic stresses, including salt and drought stress. As a result (Figures 6B, C), NtMYB078 was significantly induced by salt and drought treatments, whereas NtMYB108 and NtMYB177 were repressed by salt treatment. The expression of several NtMYB genes continued to increase under drought treatment, such as NtMYB123, NtMYB201, and NtMYB210. Interestingly, the expression of NtMYB081 in response to salt stress was time-specific, reached the peak at 1 h of salt treatment, and then dropped sharply. Notably, NtMYB102 was found to respond significantly to both drought and salt treatments, whereas NtMYB010 was induced by drought treatment but nearly kept the original expression under salt stress.
Subcellular localization analysis
To explore the potential function of the NtMYB genes, the subcellular localization of the one of the salt-responsive genes, NtMYB102, was analyzed (Supplementary Figure S5). The full-length coding sequence of NtMYB102 without the stop codon was fused to the GFP reporter gene sequence, which was driven by the CaMV35S promoter. The Agrobacterium cultures with the NtMYB102-GFP fusion construct and the 35S::GFP control were transiently expressed in the leaves of N. benthamiana, respectively. As shown by confocal microscopy, the signal of GFP protein was found to distribute throughout the whole cell, whereas the fluorescence signal of the NtMYB102-GFP fusion protein was specifically confined within the nucleus, which was confirmed by staining with DAPI.
The function of NtMYB102 in plant drought and salt tolerance
As the transcriptional activation assay result, all the yeast cells grew well on a SD/-Trp medium, while on the SD/-Trp medium supplemented with X-α-Gal, the yeast cells harboring the NtMYB102 grew well and displayed a blue color; the yeast cells containing the pBridge empty vector were not in blue color (Figure 7A). These results showed that NtMYB102 has transactivation activities. Furthermore, the function of NtMYB102 gene was further examined through genetic experiments. Considering that this gene could be induced by drought and salt stresses, the overexpression lines and wild-type seedlings were firstly treated by drought stress. After the drought treatment, overexpression lines and wild-type seedlings displayed leaf wilting phenotypes, whereas the wild-type seedlings were much more extreme than those overexpression lines (Figure 7B and Supplementary Figure S6) and the survival rates of the OE-1 lines, OE-4 lines, and OE-5 lines were significantly higher than those of the wild-type seedlings (Figure 7C). Besides, the overexpression lines displayed higher water content than wild-type during dehydration (Figure 7D).
Furthermore, the salt tolerance of wild-type and NtMYB102 overexpressing tobacco was examined via root elongation assay (Figure 8). As a result, no significant difference in root length between wild-type and the NtMYB102 overexpressing plants was found under normal conditions. However, on 100 mM NaCl plates, longer roots of the independent overexpression lines were observed compared to the wild type. Hence, the overexpression of NtMYB102 gene could improve the drought and salt tolerance in transgenic tobacco.
Discussion
The MYB transcription factors play important roles in plant development, metabolism, and responding to biotic and abiotic stress (Stracke et al., 2007; Dubos et al., 2010). In this study, the newly identified R2R3-MYB members were studied through a series of analyses. In addition, the MYB genes homologous between Arabidopsis and tobacco were studied to investigate their potential functions.
A total of 246 R2R3-MYB members were identified from tobacco, and divided into 33 subgroups together with Arabidopsis R2R3-MYB members (Figure 1). The syntenic analysis could visualize the location of the homologous or orthologous genes and the presence of collinear R2R3-MYB genes in different species may have conserved functions, which gives an insight into the functions of the R2R3-MYB genes (Li et al., 2019). In the current study, we identified the collinear pairs of the R2R3-MYB genes in five studied species. A total of six tobacco R2R3-MYB genes were identified to form collinear pairs with genes from all the other species (Figure 2), whereas those six collinear R2R3-MYB genes were distributed in different subgroups (Figure 1), suggesting these R2R3-MYB genes may have existed before the divergence of these species. Gene duplication has played a very important role in the expansion of gene families (Kent et al., 2003; Cannon et al., 2004). In the current study, a total of 44 duplication events were identified in the 68 NtMYB genes, most (42) of which involved segmental duplication, and several (2) of which involved tandem duplication. This discovery implied that duplication events might play an important role in the evolution of the tobacco R2R3-MYB gene family.
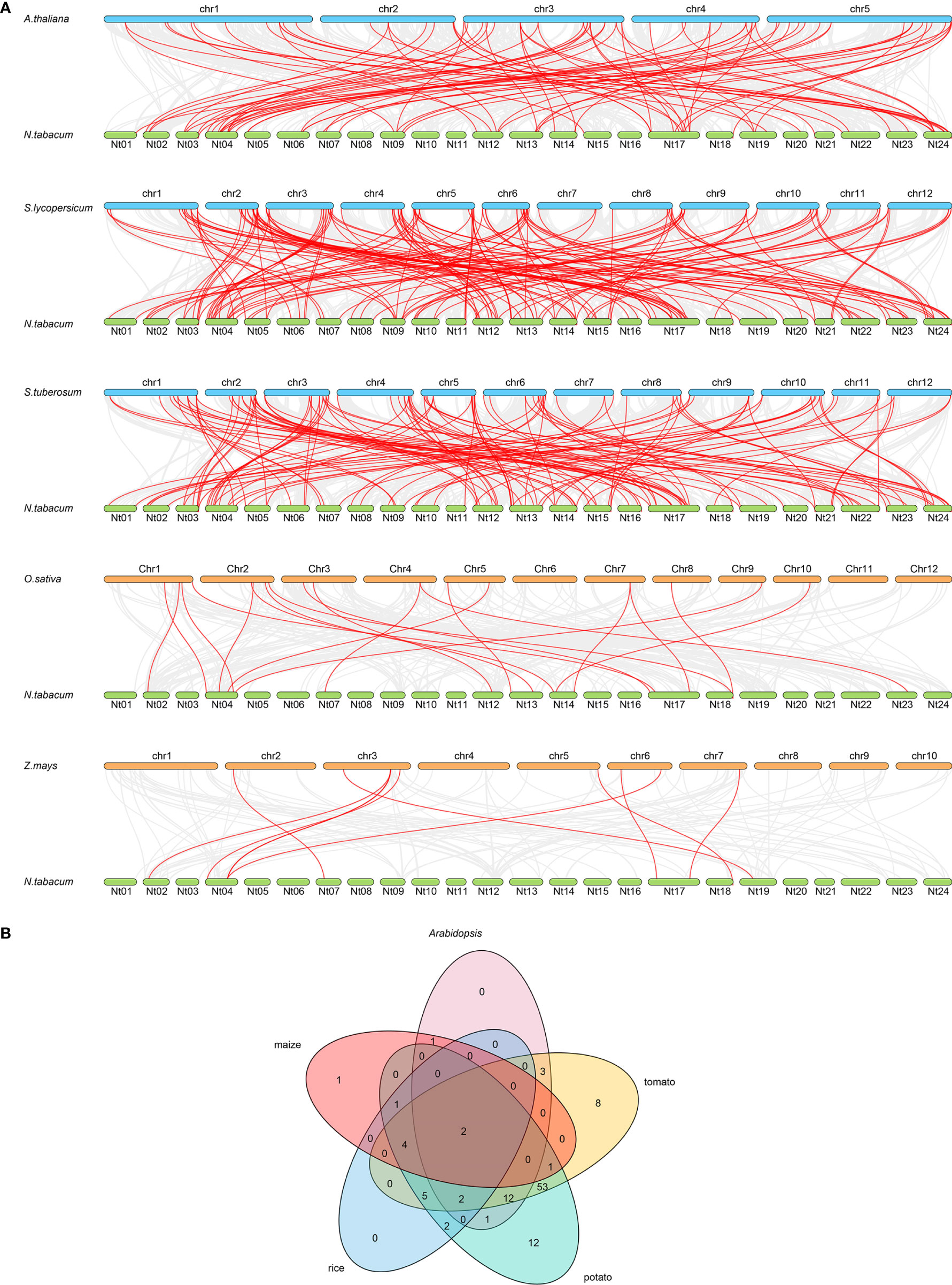
Figure 2 The synteny analysis of R2R3-MYB genes between tobacco and five representative species. (A) The gray line in the background represents the collinear blocks between tobacco and five representative species, while the collinear R2R3-MYB gene pairs are highlighted in red color. (B) The R2R3-MYB genes form the syntenic pairs between tobacco and all the other selected species, which is visualized by the Venn plot.
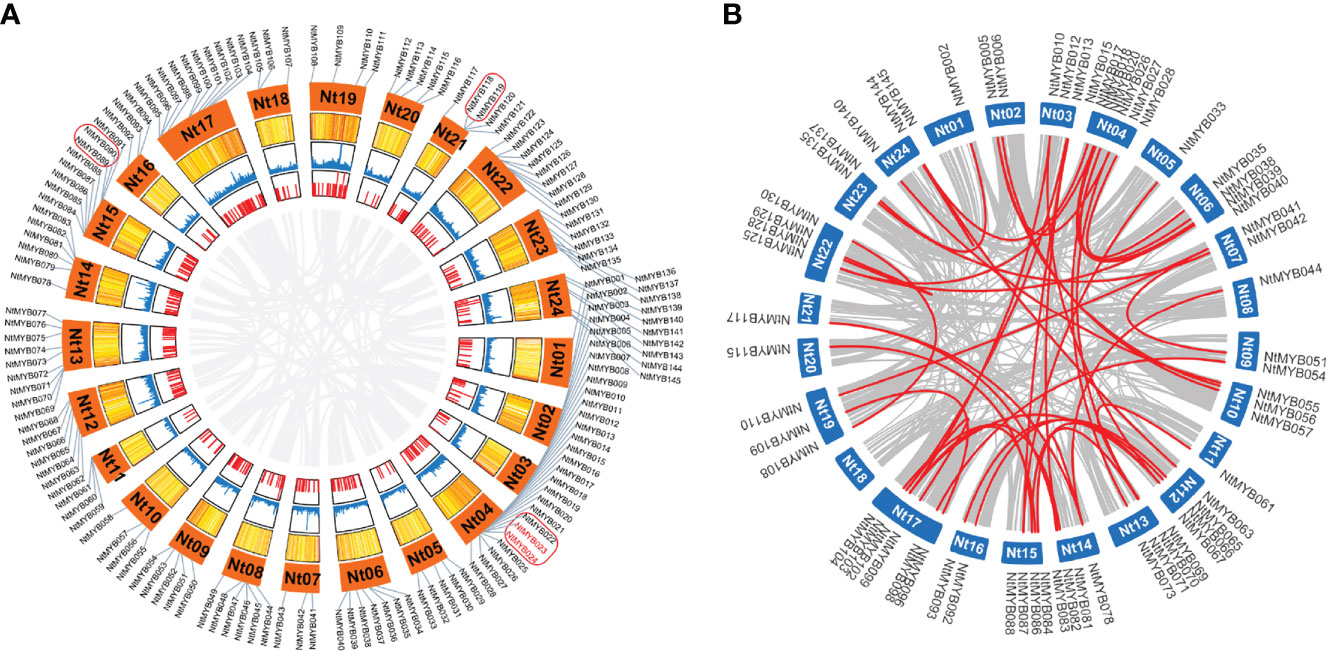
Figure 3 The chromosomal distribution and duplication events. (A) The distribution of tobacco R2R3-MYB genes on chromosomes. A total of 145 R2R3-MYB genes anchored on tobacco chromosomes. The red box suggests the gene cluster, while the tandem duplication gene pair is colored red. (B) The tobacco R2R3-MYB genes segmental duplication events and inter-chromosomal relationships. The 42 segmental duplication pairs of NtMYB genes are predicted by MCScanX and linked by the red lines, respectively. In addition, the gray lines stand for all putative sefmental duplication pairs in the tobacco genome sequences.
The R2R3-MYB transcription factors have been reported to be related to plant development (Stracke et al., 2001; Pucker et al., 2020). In subgroup 14, AtMYB37, which functions in axillary meristem development (Keller et al., 2006), clustered together with NtMYB053, and their coding genes were investigated to form the collinear pair (Figures 1, 2). Interestingly, NtMYB053 was highly expressed in the stem (Figure 4), suggesting that it might be involved in tobacco stem development, while NtMYB124 clustered together with AtMYB68 (Figure 1), which specifically regulates root growth (Feng et al., 2004), and the expression profiling showed NtMYB124 have high expression at the root (Figure 4), suggesting that it might be involved in the root development. In subgroup 18, AtMYB33 and AtMYB65 redundantly facilitate anther development (Millar and Gubler, 2005); NtMYB105, NtMYB222, NtMYB210, NtMYB149, NtMYB108, and NtMYB130 were clustered together with AtMYB33 and AtMYB65, and their coding genes were highly expressed in flowers (Figures 1, 4), suggesting they may be involved in the development of floral organs. These results suggested functional conservation between homologous R2R3-MYB members from Arabidopsis and tobacco.
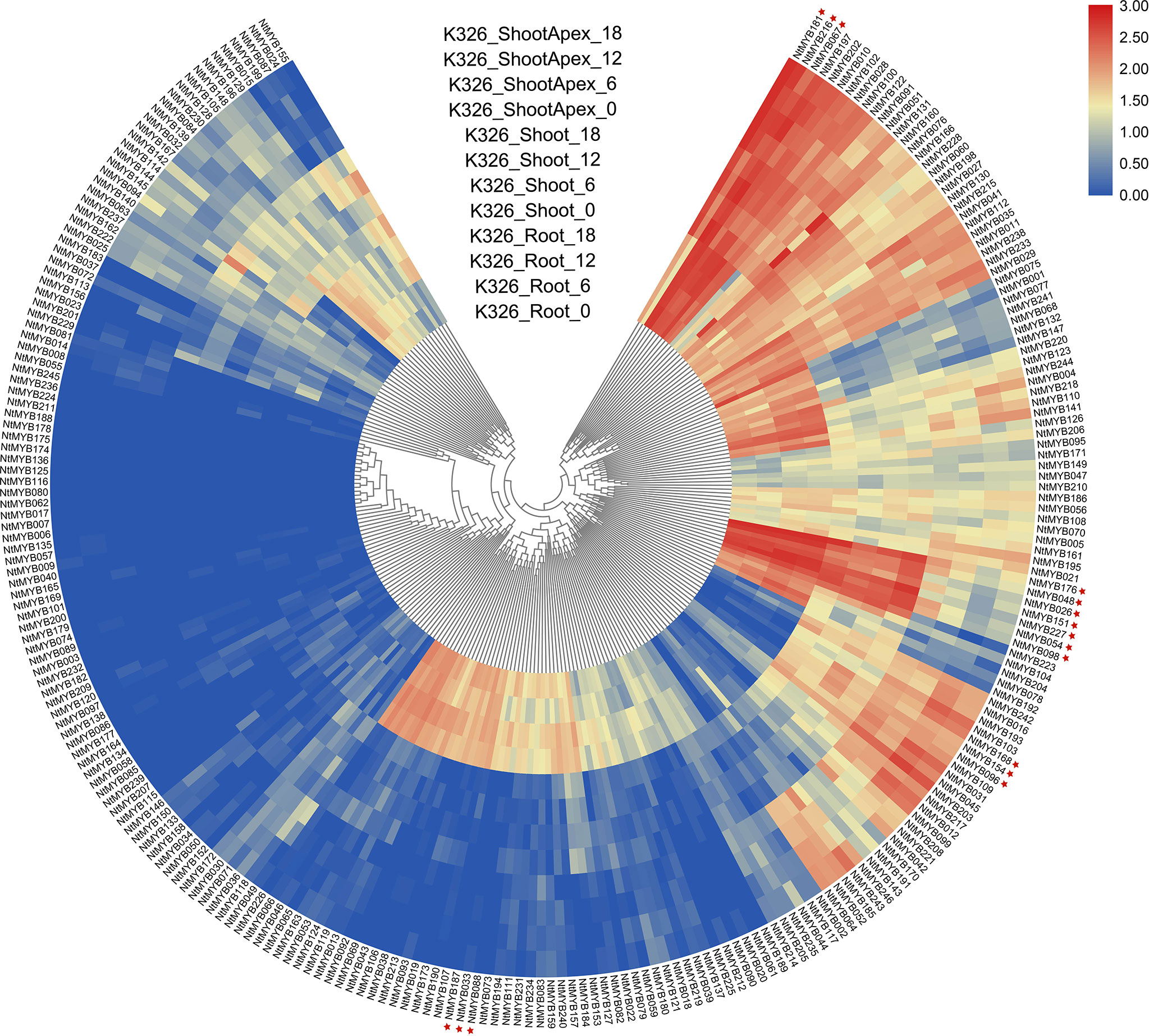
Figure 4 Expression profiles of the NtMYB genes in three different tissues (root, shoot, and shoot apex). The heatmap was constructed based on the transcriptome data of NtMYB genes and visualized by R.
Besides, several MYB members have been reported to control the synthesis of the anthocyanins, proanthocyanidins, flavonols, and flavonoids in plants (Mehrtens et al., 2005; Cao et al., 2021). Notably, AtMYB11/PFG2, AtMYB12/PFG1, and AtMYB111/PFG3 from subgroup 7 were characterized as specific flavonol regulators in Arabidopsis; AtMYB12 controls flavonol biosynthesis mainly in the root, while AtMYB111, primarily in cotyledons (Stracke et al., 2007). Evolutionary analysis showed that NtMYB096 and NtMYB109 were clustered together with these PFG members (Figure 1). Meanwhile, their coding genes were predicted to form five collinear gene pairs with AtMYB11, AtMYB12, and AtMYB111(Figure 2). Further, the transcriptome and qRT-PCR data showed that NtMYB96 and NtMYB109 were highly expressed in roots (Figures 4, 6), hinting that they might control the flavonol biosynthesis in tobacco roots. Notably, the members from subgroup 4 were identified to act as repressors of the monolignol pathway (Liu et al., 2015). NtMYB028, NtMYB029, and NtMYBB035 were found to fall into this subgroup, indicating these members may confer lignin synthesis in tobacco.
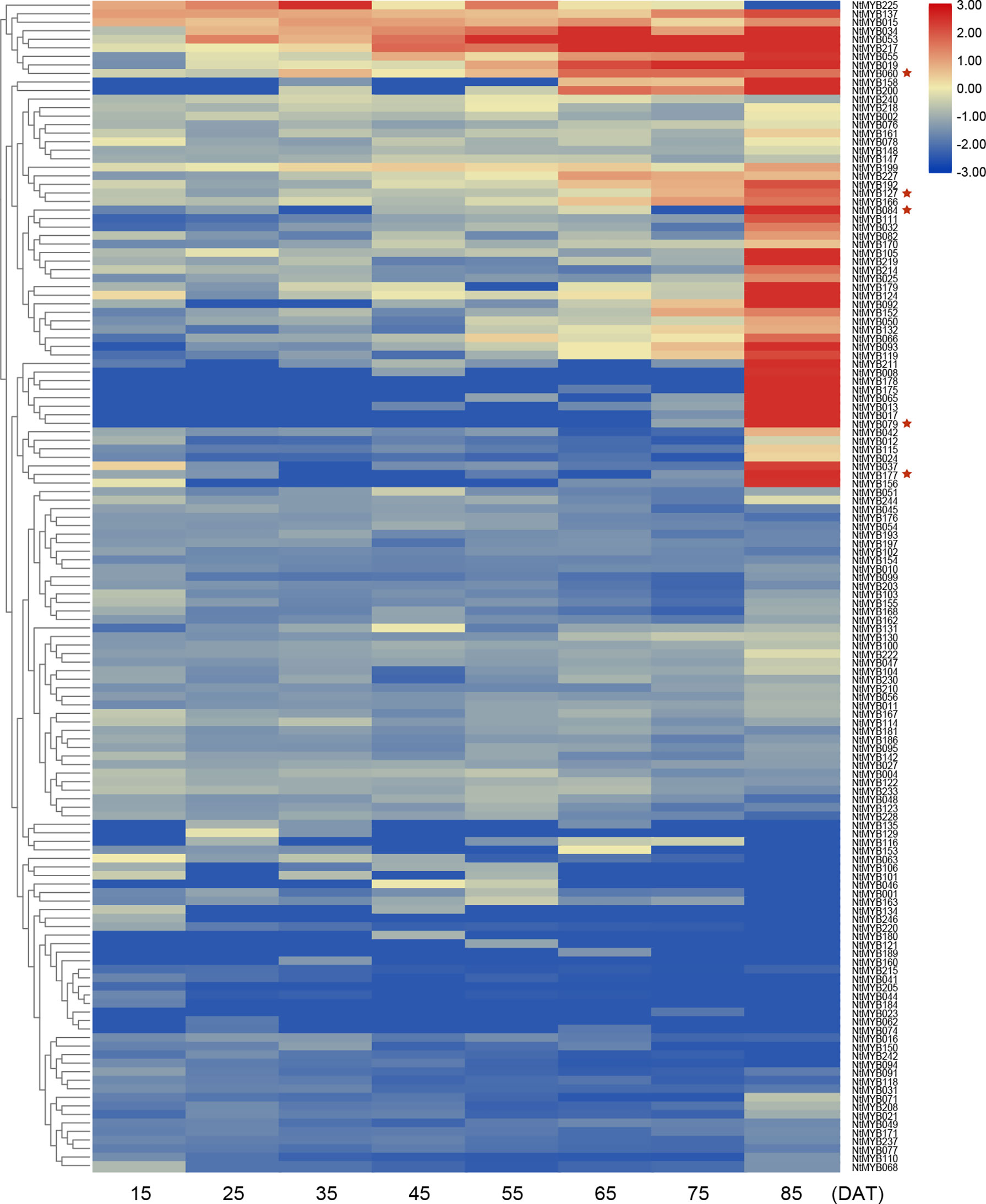
Figure 5 Expression profiles of the NtMYB genes of middle tobacco leaves in different periods. The example of middle leaf harvested from plants from 15 to 85 DAT (days after topping). The heatmap was constructed based on normalized RNA-seq data of NtMYB genes and visualized by R. Red indicates high expression, and blue indicates no detected expression. H indicates Honghuadajingyuan; M, middle leaves. * indicates the genes mentioned in the text.
Furthermore, in subgroup 20, AtMYB2 and AtMYB108 were up-regulated during leaf senescence and participated in the network regulating leaf senescence (Guo and Gan, 2011; Chou et al., 2018). In this study, NtMYB060, NtMYB079, NtMYB084, NtMYB127, and NtMYB177 were found to be clustered together with AtMYB2 and AtMYB108 (Figure 1). The transcriptome data showed that these tobacco homologous genes were detected to be up-regulated during tobacco leaf senescence (Figure 5), suggesting that they might also participate in the regulation of leaf senescence of tobacco. Interestingly, although NtMYB070 and NtMYB084 were predicted to arise from segmental duplication events (Figure 3B), NtMYB070 had lowly expressed during the senescence of the leaves (Figure 5), indicating these two duplicated genes might undergo subfunctionalization.
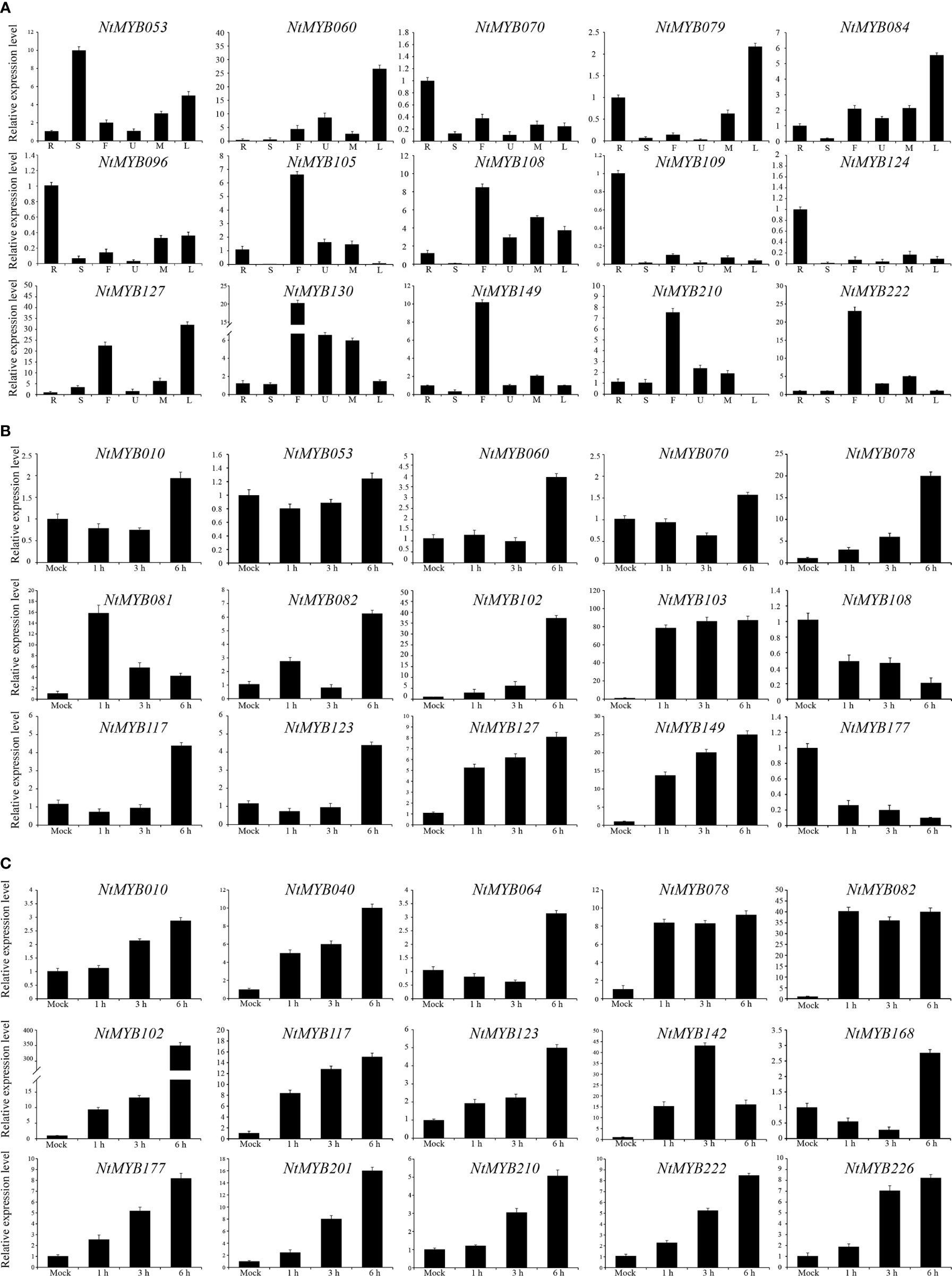
Figure 6 The qRT-PCR analysis of representative NtMYB genes. (A) To verify the tissue specificity expression of the representative NtMYB genes, the expression level of each NtMYB gene was calculated relative to the root. (B) The expression level of representative NtMYB genes under salt stress treatments. (C) The expression level of representative NtMYB genes under drought stress treatments.
Many R2R3-MYB family members were found to confer tolerance to abiotic and biotic stresses in plants. In subgroup 1, NtMYB103 was clustered with AtMYB30, and they were detected to form a collinear gene pair (Figures 1, 2). AtMYB30 had been reported to be involved in abiotic stress responses (Marino et al., 2013). Interestingly, its tobacco homologous gene, NtMYB103, was detected to be induced by salt treatment (Supplementary Figure S4), implying that NtMYB103 might be involved in abiotic stress responses of tobacco. In subgroup 2, overexpression of AtMYB15 improves drought and salt tolerance in Arabidopsis (Agarwal et al., 2006). The collinearity analysis showed that NtMYB078, NtMYB082, NtMYB117, and NtMYB123 in the same subgroup were investigated to be forming the collinear gene pair with AtMYB15 respectively (Figure 3B). Similarly, those NtMYB genes were induced by multiple abiotic stress treatments (Figures 6B, C), implying that the R2R3-MYB members in this subgroup might also confer stress tolerance in tobacco.
In subgroup 22, NtMYB102 clustered together with AtMYB44, AtMYB70, AtMYB73, and AtMYB77 (Figure 1); those Arabidopsis members were reported to function in regulating stomatal closure and abiotic stress responses (Jung et al., 2008). Interestingly, NtMYB102 were found to form the collinear gene pairs with those Arabidopsis members (Figure 2) and the promoter analyses revealed that the NtMYB102 promoter region contains many ABRE cis-elements (Supplementary Figure 4), suggesting that it might be involved in ABA signalling and stress response. Notably, NtMYB102 had high transactivation activities in yeast and the NtMYB102-GFP fusion protein was in the nucleus (Figure 7A and Supplementary Figure S5). Furthermore, NtMYB102 was significantly induced by drought and salt stresses. In addition, the overexpression analyses further demonstrated that NtMYB102 can confer drought and salt tolerances in transgenic tobacco plant (Figures 7, 8). Those clues indicated that NtMYB102 acts as a transcriptional activator to regulate gene expression in response to stresses.
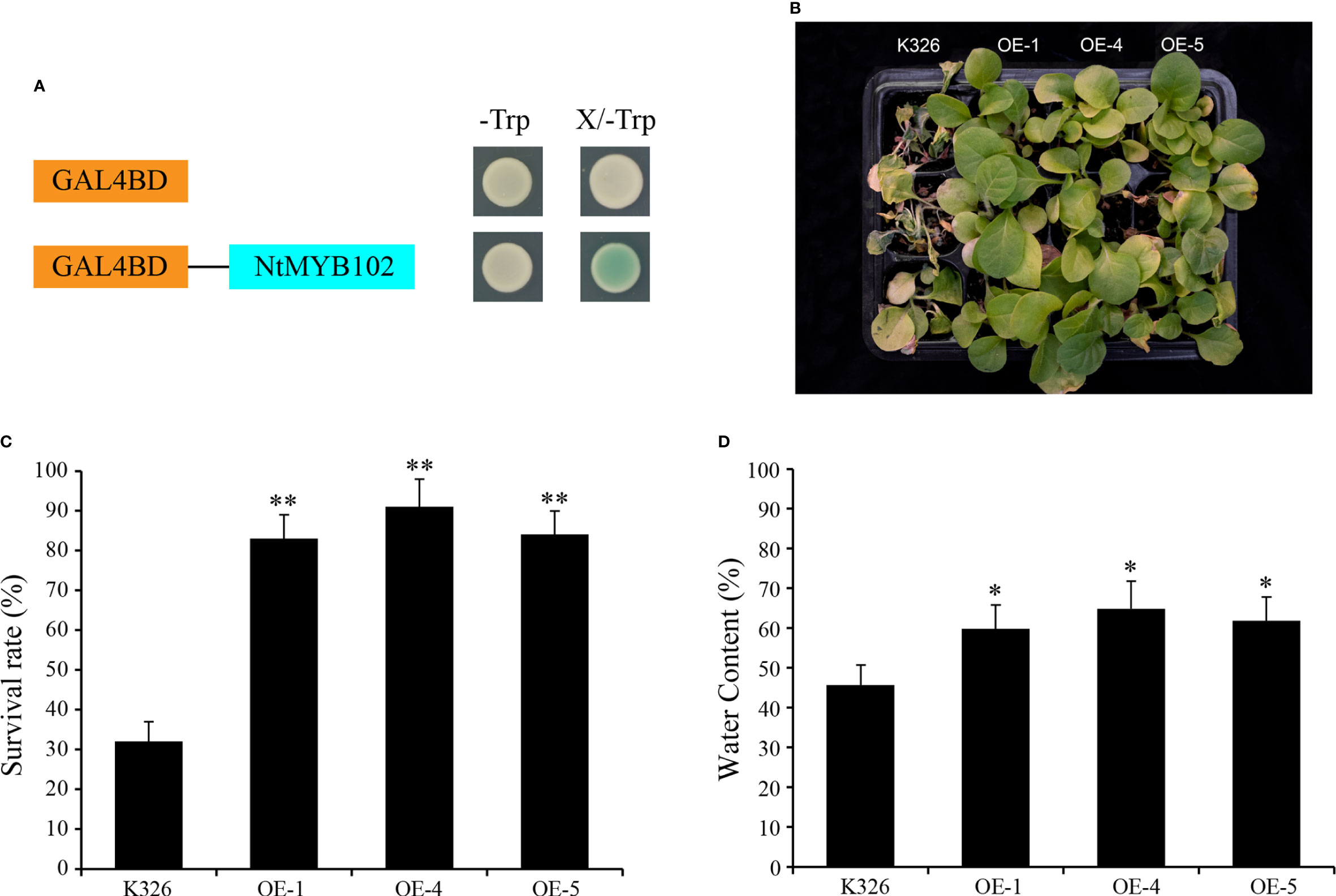
Figure 7 The function of NtMYB102 in drought tolerance. (A) Transactivation analysis of NtMYB102 in yeast. (B) Phenotypes of overexpression lines and wild-type tobacco under drought stress. The overexpression lines displayed higher drought tolerance after 2 weeks of drought stress and three days of recovery. (C) The survival rates of the overexpression lines and wild-type tobacco. (D) The water content of detached leaves of overexpression lines and wild-type tobacco, which was calculated by comparing the weight of the leaves before/after the treatments. The values represent means ± SD. *p < 0.05, **p < 0.01 (t-tests).
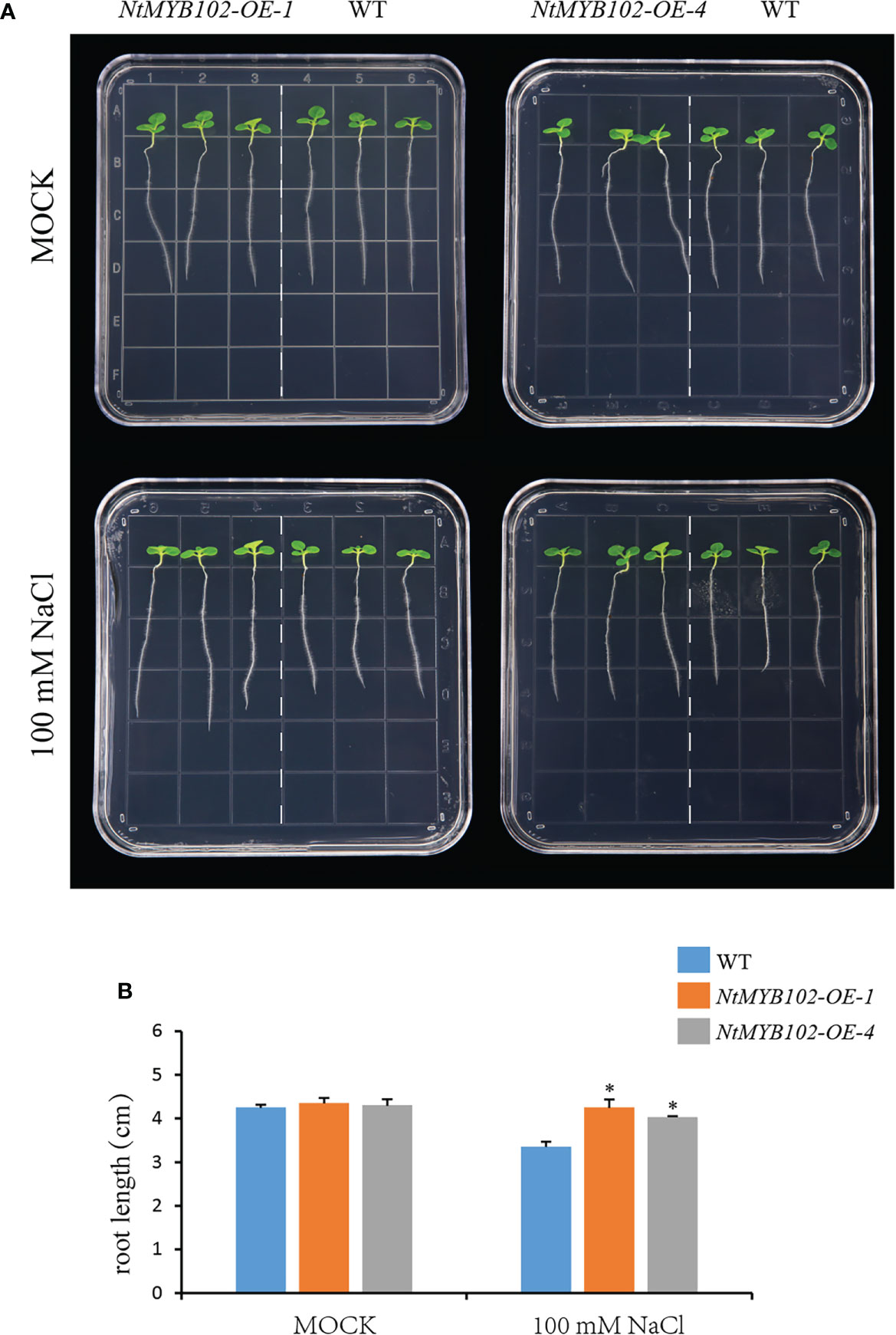
Figure 8 The function of NtMYB102 in plant salt tolerance. (A) Root growth of wild-type and NtMYB102 overexpression lines under 0 or 100 mM NaCl treatment. (B) The quantification of primary root length on medium and the data were retrieved from more than 15 plants per genotype with three biological replicates. WT, wild-type. Values represent means ± SD. *p < 0.05 (t-tests).
Conclusions
The systematic analysis of the tobacco genome sequences in this study was carried out to identify and characterize the R2R3-MYB genes; the phylogeny and expression profiling analysis implied that the tobacco R2R3-MYB gene family might be involved in various biological processes. The R2R3-MYB members homologous between Arabidopsis and tobacco were found to play conserved roles in regulating plant development and stress responses. Notably, NtMYB102 was found to be a nucleus-localized transcription factor with transactivation, and the coding gene was induced by salt treatments. Furthermore, the overexpression of NtMYB102 in tobacco significantly enhanced the drought and salt stress tolerance of the transgenic plants.
Data availability statement
Datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
XL, CG, and ZL conducted the research and drafted the manuscript. The other authors assisted in data collection and analysis. WP and LW conceived the research and drafted the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was financially supported by the China Tobacco Genome Project [110202001029(JY-12)]; The China Tobacco Hunan Industrial Co., Ltd. Technology Project (KY2022YC0010); The Key Science and Technology Program of Yunnan Provincial Tobacco Corporation (2021530000242033); The Key Science and Technology Program of Hunan Provincial Tobacco Corporation (19-23Aa01).
Conflict of interest
XL, GW, LC, JG and WP were employed by China Tobacco Hunan Industrial Co., Ltd. CG was employed by Yunnan Provincial Tobacco Company. GW was employed by Yuxizhongyan Tobacco Seed Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.998606/full#supplementary-material
Supplementary Sequences 1, 2 | The CDS and protein sequences of all NtMYB members.
Supplementary Figure 1 | The R2 and R3 MYB repeats features of MYB members from Arabidopsis (A) and tobacco (B). The asterisks indicate the conserved Tryptophan (W) residues in the MYB repeats.
Supplementary Figure 2 | Conserved motifs and exon-intron structure organizations of NtMYB members;
Supplementary Figure 3 | Detailed sequence information for each motif in NtMYB proteins;
Supplementary Figure 4 | Regulatory elements in the promoter regions of tobacco R2R3-MYB genes. The color represents the number of cis-elements contained in the promoter sequence.
Supplementary Figure 5 | Subcellular localization of NtMYB102. The location of the nucleus was determined by 4,6-diamidino-2-phenylindole (DAPI) staining.
Supplementary Figure 6 | The expression level of NtMYB102 gene in wild-type and two overexpression lines, the expression level of each genotype was calculated relative to the wild-type.
Supplementary Figure 7 | The expression pattern of the housekeeping gene L25 with the reported RNA-Seq data. (A) The expression pattern of the L25 gene in tested tissues. (B) The expression pattern of the L25 gene in tobacco middle leaves under different periods. The numbers represent the normalized data of gene expression.
References
Abe, H., Urao, T., Ito, T., Seki, M., Shinozaki, K., Yamaguchi-Shinozaki, K. (2003). Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) function as transcriptional activators in abscisic acid signaling. Plant Cell 15, 63–78. doi: 10.1105/tpc.006130
Agarwal, M., Hao, Y., Kapoor, A., Dong, C.-H., Fujii, H., Zheng, X., et al. (2006). A R2R3 type MYB transcription factor is involved in the cold regulation of CBF genes and in acquired freezing tolerance. J. Biol. Chem. 281, 37636–37645. doi: 10.1074/jbc.M605895200
Allan, A. C., Hellens, R. P., Laing, W. A. (2008). MYB transcription factors that colour our fruit. Trends Plant Sci. 13, 99–102. doi: 10.1016/j.tplants.2007.11.012
Appelhagen, I., Jahns, O., Bartelniewoehner, L., Sagasser, M., Weisshaar, B., Stracke, R. (2011). Leucoanthocyanidin dioxygenase in Arabidopsis thaliana: characterization of mutant alleles and regulation by MYB–BHLH–TTG1 transcription factor complexes. Gene 484, 61–68. doi: 10.1016/j.gene.2011.05.031
Bailey, T. L., Johnson, J., Grant, C. E., Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43, W39–W49. doi: 10.1093/nar/gkv416
Buschmann, H. (2016). Plant cell division analyzed by transient agrobacterium-mediated transformation of tobacco BY-2 cells Methods Mol Biol. 1370, 17–25. doi: 10.1007/978-1-4939-3142-2_2
Byrne, M. E., Barley, R., Curtis, M., Arroyo, J. M., Dunham, M., Hudson, A., et al. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408, 967–971. doi: 10.1038/35050091
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4, 1–21. doi: 10.1186/1471-2229-4-10
Cao, Y., Han, Y., Li, D., Lin, Y., Cai, Y. (2016). MYB transcription factors in chinese pear (Pyrus bretschneideri rehd.): Genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.00577
Cao, Y., Jia, H., Xing, M., Jin, R., Grierson, D., Gao, Z., et al. (2021). Genome-wide analysis of MYB gene family in Chinese bayberry (Morella rubra) and identification of members regulating flavonoid biosynthesis. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.691384
Castillejo, C., Waurich, V., Wagner, H., Ramos, R., Oiza, N., Muñoz, P., et al. (2020). Allelic variation of MYB10 is the major force controlling natural variation in skin and flesh color in strawberry (Fragaria spp.) fruit. Plant Cell 32, 3723–3749. doi: 10.1105/tpc.20.00474
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Wang, P., Gu, M., Lin, X., Hou, B., Zheng, Y., et al. (2021). R2R3-MYB transcription factor family in tea plant (Camellia sinensis): Genome-wide characterization, phylogeny, chromosome location, structure and expression patterns. Genomics 113, 1565–1578. doi: 10.1016/j.ygeno.2021.03.033
Chou, M. L., Liao, W. Y., Wei, W. C., Li, A. Y. S., Chu, C. Y., Wu, C. L., et al. (2018). The direct involvement of dark-induced Tic55 protein in chlorophyll catabolism and its indirect role in the MYB108-NAC signaling pathway during leaf senescence in Arabidopsis thaliana. Int. J. Mol. Sci. 19, 1854–1877. doi: 10.3390/ijms19071854
Coleto, I., Bejarano, I., Marín-Peña, A. J., Medina, J., Rioja, C., Burow, M., et al. (2021). Arabidopsis thaliana transcription factors MYB28 and MYB29 shape ammonium stress responses by regulating fe homeostasis. New Phytol. 229, 1021–1035. doi: 10.1111/nph.16918
Cominelli, E., Sala, T., Calvi, D., Gusmaroli, G., Tonelli, C. (2008). Over-expression of the Arabidopsis AtMYB41 gene alters cell expansion and leaf surface permeability. Plant J. 53, 53–64. doi: 10.1111/j.1365-313X.2007.03310.x
De Vos, M., Denekamp, M., Dicke, M., Vuylsteke, M., Van Loon, L., Smeekens, S. C., et al. (2006). The Arabidopsis thaliana transcription factor AtMYB102 functions in defense against the insect herbivore Pieris rapae. Plant Signal Behav. 1, 305–311. doi: 10.4161/psb.1.6.3512
Ding, A., Tang, X., Yang, D., Wang, M., Ren, A., Xu, Z., et al. (2021). ERF4 and MYB52 transcription factors play antagonistic roles in regulating homogalacturonan de-methylesterification in Arabidopsis seed coat mucilage. Plant Cell 33 (2), 381–403. doi: 10.1093/plcell/koaa031
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Du, H., Feng, B.-R., Yang, S.-S., Huang, Y.-B., Tang, Y.-X. (2012). The R2R3-MYB transcription factor gene family in maize. PloS One 7, e37463. doi: 10.1371/journal.pone.0037463
Edwards, K., Fernandez-Pozo, N., Drake-Stowe, K., Humphry, M., Evans, A., Bombarely, A., et al. (2017). A reference genome for Nicotiana tabacum enables map-based cloning of homeologous loci implicated in nitrogen utilization efficiency. BMC Genomics 18, 1–14. doi: 10.1186/s12864-017-3791-6
Feng, C., Andreasson, E., Maslak, A., Mock, H. P., Mattsson, O., Mundy, J. (2004). Arabidopsis MYB68 in development and responses to environmental cues. Plant Sci. 167, 1099–1107. doi: 10.1016/j.plantsci.2004.06.014
Gonzalez, A., Zhao, M., Leavitt, J. M., Lloyd, A. M. (2008). Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/MYB transcriptional complex in Arabidopsis seedlings. Plant J. 53, 814–827. doi: 10.1111/j.1365-313X.2007.03373.x
Guo, Y., Gan, S. (2011). AtMYB2 regulates whole plant senescence by inhibiting cytokinin-mediated branching at late stages of development in Arabidopsis. Plant Physiol. 156, 1612–1619. doi: 10.1104/pp.111.177022
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Jaradat, M. R., Feurtado, J. A., Huang, D., Lu, Y., Cutler, A. J. (2013). Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 13, 1–19. doi: 10.1186/1471-2229-13-192
Jiang, C., Gu, J., Chopra, S., Gu, X., Peterson, T. (2004). Ordered origin of the typical two-and three-repeat MYB genes. Gene 326, 13–22. doi: 10.1016/j.gene.2003.09.049
Jung, C., Seo, J. S., Han, S. W., Koo, Y. J., Kim, C. H., Song, S. I., et al. (2008). Overexpression of AtMYB44 enhances stomatal closure to confer abiotic stress tolerance in transgenic Arabidopsis. Plant Physiol. 146, 623–635. doi: 10.1104/pp.107.110981
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13, 1–19. doi: 10.1186/1471-2164-13-544
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Keller, T., Abbott, J., Moritz, T., Doerner, P. (2006). Arabidopsis REGULATOR OF AXILLARY MERISTEMS1 controls a leaf axil stem cell niche and modulates vegetative development. Plant Cell 18, 598–611. doi: 10.1105/tpc.105.038588
Kent, W. J., Baertsch, R., Hinrichs, A., Miller, W., Haussler, D. (2003). Evolution’s cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl. Acad. Sci. 100, 11484–11489. doi: 10.1073/pnas.1932072100
Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, D. K., Geisler, M., Springer, P. S. (2009). LATERAL ORGAN FUSION1 and LATERAL ORGAN FUSION2 function in lateral organ separation and axillary meristem formation in Arabidopsis. Development 136, 2423–2432. doi: 10.1242/dev.031971
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van De Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Khedkar, S., Bork, P. (2020). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49 (D1), D458–D460. doi: 10.1093/nar/gkaa937
Li, Z., Peng, R., Tian, Y., Han, H., Xu, J., Yao, Q. (2016). Genome-wide identification and analysis of the MYB transcription factor superfamily in Solanum lycopersicum. Plant Cell Physiol. 57, 1657–1677. doi: 10.1093/pcp/pcw091
Li, X., Ahmad, S., Guo, C., Yu, J., Cao, S., Gao, X., et al. (2018). Identification and characterization of LRR-RLK family genes in potato reveal their involvement in peptide signaling of cell fate decisions and biotic/abiotic stress responses. Cells 7, 120. doi: 10.3390/cells7090120
Li, W., Guo, Y. (2018). RNA-Seq Analysis of the Transcriptome of Leaf Senescence in Tobacco. Methods Mol Biol 1744, 331–337. doi: 10.1007/978-1-4939-7672-0_27
Li, W., Zhang, H., Li, X., Zhang, F., Liu, C., Du, Y., et al. (2018). Intergrative metabolomic and transcriptomic analyses unveil nutrient remobilization events in leaf senescence of tobacco. Sci. Rep. 7, 1. doi: 10.1038/s41598-017-11615-0
Li, X., Guo, C., Ahmad, S., Wang, Q., Yu, J., Liu, C., et al. (2019). Systematic analysis of MYB family genes in potato and their multiple roles in development and stress responses. Biomolecules 9, 317. doi: 10.3390/biom9080317
Li, J., Liu, H., Yang, C., Wang, J., Yan, G., Si, P., et al. (2020). Genome-wide identification of MYB genes and expression analysis under different biotic and abiotic stresses in Helianthus annuus l. Int. J. Biol. Macromol 143, 111924. doi: 10.1016/j.ijbiomac.2021.08.072
Li, Z., Chao, J., Li, X., Li, G., Song, D., Guo, Y., et al. (2021). Systematic analysis of the bZIP family in tobacco and functional characterization of NtbZIP62 involvement in salt stress. Agronomy 11, 148. doi: 10.3390/agronomy11010148
Li, X., Wang, Q., Guo, C., Sun, J., Li, Z., Wang, Y., et al. (2022). NtNAC053, a novel NAC transcription factor, confers drought and salt tolerances in tobacco. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.817106
Liang, Y., Tan, Z.-M., Zhu, L., Niu, Q.-K., Zhou, J.-J., Li, M., et al. (2013). MYB97, MYB101 and MYB120 function as male factors that control pollen tube-synergid interaction in Arabidopsis thaliana fertilization. PloS Genet. 9, e1003933. doi: 10.1371/journal.pgen.1003933
Liu, J., Osbourn, A., Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2– ΔΔCT method. methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marino, D., Froidure, S., Canonne, J., Khaled, S. B., Khafif, M., Pouzet, C., et al. (2013). Arabidopsis ubiquitin ligase MIEL1 mediates degradation of the transcription factor MYB30 weakening plant defence. Nat. Commun. 4, 1–9. doi: 10.1038/ncomms2479
Martin, C., Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. doi: 10.1016/s0168-9525(96)10049-4
Mccarthy, R. L., Zhong, R., Ye, Z.-H. (2009). MYB83 is a direct target of SND1 and acts redundantly with MYB46 in the regulation of secondary cell wall biosynthesis in Arabidopsis. Plant Cell Physiol. 50, 1950–1964. doi: 10.1093/pcp/pcp139
Mehrtens, F., Kranz, H., Bednarek, P., Weisshaar, B. (2005). The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 138, 1083–1096. doi: 10.1104/pp.104.058032
Millar, A. A., Gubler, F. (2005). The Arabidopsis GAMYB-like genes, MYB33 and MYB65, are microRNA-regulated genes that redundantly facilitate anther development. Plant Cell 17, 705–721. doi: 10.1105/tpc.104.027920
Mistry, J., Chuguransky, S., Williams, L., Qureshi, M., Salazar, G. A., Sonnhammer, E. L., et al. (2021). Pfam: The protein families database in 2021. Nucleic Acids Res. 49, D412–D419. doi: 10.1093/nar/gkaa913
Moyano, E., Martínez-Garcia, J. F., Martin, C. (1996). Apparent redundancy in MYB gene function provides gearing for the control of flavonoid biosynthesis in antirrhinum flowers. Plant Cell 8, 1519–1532. doi: 10.1105/tpc.8.9.1519
Mu, R.-L., Cao, Y.-R., Liu, Y.-F., Lei, G., Zou, H.-F., Liao, Y., et al. (2009). An R2R3-type transcription factor gene AtMYB59 regulates root growth and cell cycle progression in Arabidopsis. Cell Res. 19, 1291–1304. doi: 10.1038/cr.2009.83
Naik, J., Rajput, R., Pucker, B., Stracke, R., Pandey, A. (2021). The R2R3-MYB transcription factor MtMYB134 orchestrates flavonol biosynthesis in Medicago truncatula. Plant Mol. Biol. 106, 157–172. doi: 10.1007/s11103-021-01135-x
Oh, J. E., Kwon, Y., Kim, J. H., Noh, H., Hong, S.-W., Lee, H. (2011). A dual role for MYB60 in stomatal regulation and root growth of Arabidopsis thaliana under drought stress. Plant Mol. Biol. 77, 91–103. doi: 10.1007/s11103-011-9796-7
Paz-Ares, J., Ghosal, D., Wienand, U., Peterson, P. A., Saedler, H. (1987). The regulatory c1 locus of zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J.1 6 (12), 3553–3558. doi: 10.1002/j.1460-2075.1987.tb02684.x
Pucker, B., Pandey, A., Weisshaar, B., Stracke, R. (2020). The R2R3-MYB gene family in banana (Musa acuminata): Genome-wide identification, classification and expression patterns. PloS One 15, e0239275. doi: 10.1371/journal.pone.0239275
Qin, W., Xie, L., Li, Y., Liu, H., Fu, X., Chen, T., et al. (2021). An R2R3-MYB transcription factor positively regulates the glandular secretory trichome initiation in Artemisia annua l. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.657156
Qiu, Z., Yan, S., Xia, B., Jiang, J., Yu, B., Lei, J., et al. (2019). The eggplant transcription factor MYB44 enhances resistance to bacterial wilt by activating the expression of spermidine synthase. J. Exp. Bot. 70 (19), 5343–5354. doi: 10.1093/jxb/erz259
Quattrocchio, F., Wing, J., van der Woude, K., Souer, E., De Vetten, N., Mol, J., et al. (1999). Molecular analysis of the anthocyanin2 gene of petunia and its role in the evolution of flower color. Plant Cell 11, 1433–1444. doi: 10.1105/tpc.11.8.1433
Roy, S. (2016). Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal Behav. 11, e1117723. doi: 10.1080/15592324.2015.1117723
Segarra, G., van der Ent, S., Trillas, I., Pieterse, C. (2009). MYB72, a node of convergence in induced systemic resistance triggered by a fungal and a bacterial beneficial microbe. Plant Biol. 11, 90–96. doi: 10.1111/j.1438-8677.2008.00162.x
Seo, P. J., Xiang, F., Qiao, M., Park, J.-Y., Lee, Y. N., Kim, S.-G., et al. (2009). The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 151, 275–289. doi: 10.1104/pp.109.144220
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453. doi: 10.1105/tpc.107.050963
Stracke, R., Holtgräwe, D., Schneider, J., Pucker, B., Sörensen, T. R., Weisshaar, B. (2014). Genome-wide identification and characterisation of R2R3-MYB genes in sugar beet (Beta vulgaris). BMC Plant Biol. 14, 1–17. doi: 10.1186/s12870-014-0249-8
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Stracke, R., Werber, M., Weisshaar, B. (2001). The R2R3-MYB gene family in arabidopsis thaliana. Curr. Opin. Plant Biol. 4 (5), 447–456. doi: 10.1016/s1369-5266(00)00199-0
Sun, J., Xie, M., Li, X., Li, Z., Wang, Q., Ding, A., et al. (2021). Systematic investigations of the ZF-HD gene family in tobacco reveal their multiple roles in abiotic stresses. Agronomy 11, 406. doi: 10.3390/agronomy11030406
Van Der Ent, S., Verhagen, B. W., Van Doorn, R., Bakker, D., Verlaan, M. G., Pel, M. J., et al. (2008). MYB72 is required in early signaling steps of rhizobacteria-induced systemic resistance in Arabidopsis. Plant Physiol. 146, 1293–1304. doi: 10.1104/pp.107.113829
Wang, Q., Guo, C., Li, Z., Sun, J., Wang, D., Xu, L., et al. (2021). Identification and analysis of bZIP family genes in potato and their potential roles in stress responses. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.637343
Wei, Q., Chen, R., Wei, X., Liu, Y., Zhao, S., Yin, X., et al. (2020). Genome-wide identification of R2R3-MYB family in wheat and functional characteristics of the abiotic stress responsive gene TaMYB344. BMC Genomics 21, 1–16. doi: 10.1186/s12864-020-07175-9
Zhao, Q., Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhou, J., Lee, C., Zhong, R., Ye, Z.-H. (2009). MYB58 and MYB63 are transcriptional activators of the lignin biosynthetic pathway during secondary cell wall formation in Arabidopsis. Plant Cell 21, 248–266. doi: 10.1105/tpc.108.063321
Keywords: MYB transcription factors, tobacco, metabolism, stress tolerance, NtMYB102
Citation: Li X, Guo C, Li Z, Wang G, Yang J, Chen L, Hu Z, Sun J, Gao J, Yang A, Pu W and Wen L (2022) Deciphering the roles of tobacco MYB transcription factors in environmental stress tolerance. Front. Plant Sci. 13:998606. doi: 10.3389/fpls.2022.998606
Received: 20 July 2022; Accepted: 05 September 2022;
Published: 24 October 2022.
Edited by:
Yunpeng Cao, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Gaofei Jiang, Nanjing Agricultural University, ChinaQiang Zhao, Qingdao Agricultural University, China
Copyright © 2022 Li, Guo, Li, Wang, Yang, Chen, Hu, Sun, Gao, Yang, Pu and Wen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxuan Pu, puwx0605@hngytobacco.com; Liuying Wen, wenliuying@caas.cn
†These authors have contributed equally to this work
 Xiaoxu Li
Xiaoxu Li Cun Guo
Cun Guo Zhiyuan Li
Zhiyuan Li Guoping Wang1,4
Guoping Wang1,4 Zhengrong Hu
Zhengrong Hu Jinghao Sun
Jinghao Sun Aiguo Yang
Aiguo Yang Liuying Wen
Liuying Wen