- 1State Key Laboratory of Genetic Engineering, Collaborative Innovation Center for Genetics and Development, Department of Biochemistry, Institute of Plant Biology, School of Life Sciences, Fudan University, Shanghai, China
- 2Ministry of Education Key Laboratory for Biodiversity Science and Ecological Engineering, Institute of Biodiversity Science, School of Life Sciences, Fudan University, Shanghai, China
- 3Institut de Biologie Moléculaire des Plantes (IBMP), CNRS, Université de Strasbourg, Strasbourg, France
As sessile organisms, plants are constantly exposed to changing environments frequently under diverse stresses. Invasion by pathogens, including virus, bacterial and fungal infections, can severely impede plant growth and development, causing important yield loss and thus challenging food/feed security worldwide. During evolution, plants have adapted complex systems, including coordinated global gene expression networks, to defend against pathogen attacks. In recent years, growing evidences indicate that pathogen infections can trigger local and global epigenetic changes that reprogram the transcription of plant defense genes, which in turn helps plants to fight against pathogens. Here, we summarize up plant defense pathways and epigenetic mechanisms and we review in depth current knowledge’s about histone modifications and chromatin-remodeling factors found in the epigenetic regulation of plant response to biotic stresses. It is anticipated that epigenetic mechanisms may be explorable in the design of tools to generate stress-resistant plant varieties.
Plant pathways against pathogens
Naturally growing plants are inevitably exposed to attacks by a variety of harmful pathogens. Invasions by viruses and bacteria rely on the natural openings or damaged tissues of plants, while other pathogens such as oomycetes and fungi can directly penetrate the surface of host plants. Resistance usually manifests itself as a multi-layered defense, which includes the barrier formed by cell wall as well as by the waxy and cuticle layers on the surface of plant organs, which naturally prevents the colonization of pathogens. Importantly plants also have evolved divers defense-responsive mechanisms that are induced upon pathogen attack, such as physical reinforcements of cell walls by generating callose and lignin, and phytoanticipins and antimicrobial metabolites/proteins productions (Gill et al., 2015). Meanwhile the activation of defense responses may have a detrimental effect on plant growth and thus needs to be tightly regulated to strict necessities.
Plants have developed complex sensory mechanisms to identify pathogen invasions (Jones and Dangl, 2006). As a primary step of defense response, plants widely use receptor kinases and receptor-like proteins located on the plasma membrane, namely pattern-recognition receptors (PRRs), which sensitively recognize pathogen-associated molecular patterns (PAMPs) / microbe-associated molecular patterns (MAMPs; e.g., flg22) or damage-associated molecular patterns (DAMPs; e.g., endogenous peptide 1; Boutrot and Zipfel, 2017). Such recognition triggers a range of downstream defense mechanisms that lead to the activation of pattern-triggered immunity (PTI; Bigeard et al., 2015). PTI involves physiological phenomena with distinctive characteristics, such as stomatal closure and callose deposition to limit pathogen penetration, production of reactive oxygen species (ROS) and nitric oxide (NO), limitation of nutrient transfer from the cytoplasmic matrix to apoplast, and biosynthesis of antimicrobial metabolites and defense phytohormones to fight against pathogen invasion and propagation.
Meanwhile, some well-adapted pathogens have evolved to produce specific effectors and deliver them into host plant cells (Nguyen et al., 2021). Many of these pathogenic bacteria complete this physiological process through the Type III secretion system. Such effectors prevent the host from recognizing PAMPs/MAMPs and deregulate immune signaling and expression, leading to effector-triggered susceptibility (ETS; Feng and Zhou, 2012; Schreiber et al., 2021; Hannan Parker et al., 2022).
During evolution, plants have evolved specific R (resistance) genes encoding proteins that can detect the activity of immune-inhibitory effectors, and pathogens co-evolve new effectors to evade or counteract the plant detection. Plant R genes typically encode intracellular receptors with nucleotide-binding leucine-rich repeat (NB-LRR) domains, organized with central NB and C-terminal LRRs. These NB-LRR-containing R proteins (NLRs) can be further classified into Toll/Interleukin1 receptor-like (TIR) or coiled-coil (CC) types according to their N-terminal sequences. NLRs can interact directly or indirectly with pathogen effectors to induce defense responses (Spoel and Dong, 2012; Nguyen et al., 2021). One of the R gene-mediated resistance responses is known as oxidative burst that rapidly produce ROS, which may have a direct antimicrobial effect and/or acts as a signal to activate other defense responses. The other well-known defense response is called hypersensitive response (HR) that is associated with local cell death to limit the pathogen propagation. R gene-mediated resistance is thought to be related to the activation of phytohormone salicylic acid (SA)-dependent signaling pathways, in which most R proteins participate in ROS production and HR development, leading to effector-triggered immunity (ETI; Nguyen et al., 2021).
Although the extent and duration of the immune response triggered by different pathogen effectors vary greatly during PTI and ETI, plasma membrane-localized immune receptors activate similar downstream molecular events, such as activation of mitogen-activated protein kinase (MAPK), Ca2+ burst (Koster et al., 2022), oxidative burst, ion influx, and increased biosynthesis of plant defense hormones, suggesting that the defensive signals initiated on the plasma membrane converge in the downstream pathway (Iakovidis et al., 2022). PTI and ETI pathway-induced signaling share multiple proteins, such as transcription factors CALMODULIN-BINDING PROTEIN 60 g (CBP60g) and SYSTEMIC ACQUIRED RESISTANCE DEFICIENT 1 (SARD1), in which the two proteins bind directly to the promoter of the SA-synthase genes and activate their expression, which in turn leads to the activation of immune-related genes, such as PATHOGENESIS-RELATED GENE 1 (PR1) and PR2 (Peng et al., 2018).
While effective defense against biotrophic pathogens that feed on living host tissues primarily relies on the activation by SA-dependent pathway and R gene-mediated resistance, necrotrophic pathogens that kill host tissue and feed on the remains need to be counteracted by a different set of defense responses activated by jasmonic acid (JA) and ethylene (ET) signaling pathways. As a result, the SA, JA, and ET signal pathways have an extensive interaction. SA and JA have mutual inhibitory effects on the expression of many defense genes. It is widely believed that this antagonism between SA and JA enables plants to be more effective against different types of pathogens, with SA-mediated signaling pathways involved in the defense against biotrophic pathogens, while JA-mediated signaling participates in defense against necrotrophic pathogens (Ghorbel et al., 2021; Peng et al., 2021).
Treatments on genetically susceptible plants by inducing molecules (such as JA) can improve plant innate immunity levels and enhance plant defenses against upcoming pathogens and pests (Luna et al., 2016). This so-called induced resistance (IR) is often considered as an adaptive component of plant immune system as well as a form of phenotypic plasticity (Mauch-Mani et al., 2017; De Kesel et al., 2021). IR is the sum of direct and primed defense responses. Priming refers to an enhanced capacity where the postchallenged/posttreated state mounts a faster and/or stronger defense response to the upcoming attack. A typical example of IR is systemic acquired resistance (SAR) where upon local pathogen attack plants develop strong resistance in distal tissues to against subsequent attacks (Gao et al., 2021). SAR is activated by signal substances produced at the site of infection. Many endogenous metabolites have been shown to be potential elicitors for SAR, including methyl salicylate (MeSA) and free radicals (such as NO and ROS; Gao et al., 2021). These small signaling molecules can be transported to distal tissues through phloem, resulting in hormone SA accumulation and PR proteins secretion to induce SAR. Thus, over the course of weeks to months, the rest of host plant can gain a strong defense response and broad-spectrum disease resistance through SAR against secondary infections (Fu and Dong, 2013). Arabidopsis (Arabidopsis thaliana) NON-EXPRESSER OF PR-GENES 1 (NPR1) protein was identified as the main regulator of SAR (Luna et al., 2012). In general, proper transcriptional programming/reprogramming is widely believed to be critical for priming and plant defense against multiple pathogens (Tsuda and Somssich, 2015; Birkenbihl et al., 2017; Mauch-Mani et al., 2017).
Epigenetic regulation in plant response to pathogens
Epigenetics refers to heritable changes in gene expression that do not involve alterations of DNA sequences. Epigenetic regulation relies on changing chromatin structures. The basic unit of chromatin is nucleosome, which is composed by approximately 146 bp of DNA wrapped around a histone octomer, comprising two molecules of each of histones H2A, H2B, H3 and H4 (Luger et al., 1997). Nucleosomes are joined by variable-length internucleosomal DNA and the linker histone H1, forming a “beads-on-a-string” structure under electron microscopy observation (Luger et al., 2012; Rutowicz et al., 2019). Further compaction results in distinct higher-order structures. Less condensed or open chromatin regions referred to as euchromatin are rich in transcriptionally active genes, and more condensed or closed chromatin regions referred to as heterochromatin are majorly transcriptional inert and often associated with repeat sequences and transposable elements (Vergara and Gutierrez, 2017).
Histone tails protruding from the nucleosome core are subjects to different types of post-translational modifications (PTMs), such as methylation (me), acetylation (ac), ubiquitination (ub) and others (Rando, 2012). PTMs may alter histone-histone or histone-DNA interactions that subsequently affect transcription processes, or alternatively PTMs may serve in recruiting other factors participating in transcription (Krajewski, 2022). In plants, it is generally known that acetylation of histones H3 and H4 (H3ac and H4ac), trimethylation on lysine 4 and lysine 36 of H3 (H3K4me3 and H3K36me3) and monoubiquitination of H2B (H2Bub1) are associated with transcriptional activation, whereas H3K9me2, H3K27me3 and H2Aub1 are associated with silencing/repression of transcription (Alvarez et al., 2010). Transcriptional reprogramming within the chromatin context constitutes a central part of plant defense mechanisms (Tsuda and Somssich, 2015; Birkenbihl et al., 2017).
Treatment of plants with SA or the synthetic SA-analogue benzothiadiazole (BTH), which triggers an intensified response of defense genes to secondary stress, can induce H3/H4ac and H3K4me2/3 levels at chromatin regions within promoters of genes encoding multiple SA pathway-related WRKY transcription factors. These modifications were blocked in priming-deficient plants carrying npr1 mutations. BTH did not directly activate the expression of these defense genes, which still remained inactive, but these changes contributed to a faster and stronger expression after subsequent infection with Pseudomonas syringae pv maculicola (Jaskiewicz et al., 2011). In the same line of histone modifications involved in priming, SAR in Arabidopsis has been associated with activation-related histone PTMs and the priming of SA-dependent defense genes (Conrath et al., 2015). As a synthetic priming activator, non-protein amino acid β-aminobutyric acid (BABA) protects Arabidopsis from oomycete pathogen Peronospora parasitica infection by activating plant innate defense mechanisms, such as callose deposition and HR. BABA can still offer a complete protective effect against Peronospora parasitica even in plants lacking of SA, JA and ET signaling pathways. Similar to BTH, BABA treatment does not directly induce mRNA accumulation of the relevant defense genes, but can enhance the induction of corresponding genes after stimulation, resulting in stronger resistance (Zimmerli et al., 2000). BABA can also enhance the plant resistance to hemibiotrophic bacteria Pseudomonas syringae pv. tomato (Pst) DC3000 and necrotrophic bacteria Pectobacterium carotovorum ssp. carotovorum (Pcc) via priming mechanism (Ton et al., 2005; Po-Wen et al., 2013). After BABA treatment, the increased responsiveness of PTI-responsive genes to necrotrophic bacteria or relevant MAMP is mediated by histone H3K9ac/K14ac and histone H3K4me2. Correspondingly, PTI-mediated callose deposition and immunity to Pcc are also enhanced in BABA-treated Arabidopsis (Po-Wen et al., 2013). The treatment of BABA and 2,6-dichloroisonicotinic acid (INA, the functional synthetic analog of SA) in common bean (Phaseolus vulagris) induced plant resistance to Pseudomonas syringae pv. phaseolicola infection, and changes in H3K4me3 and H3K36me3 markers occur in the promoter-exon regions of defense-related genes (Martinez-Aguilar et al., 2016). In addition, Arabidopsis plants exposed to repetitive heat, cold, or salt stress are more resistant to virulent bacteria than Arabidopsis grown in a more stable environment. Plant resistance is increased by the priming of Arabidopsis PTI-responsive genes and PTI-mediated callose deposition (Singh et al., 2014a).
Histone modifications are dynamically regulated by “writers” that are responsible for the establishment of modifications (such as acetyltransferases, methyltransferases, ubiquitin-ligases, etc.) and by “erasers” that are responsible for the removal of modifications (such as deacetylases, demethylases, deubiquitinases, etc.). In addition, histone modifications are specifically recognized by “readers,” which can help to assemble appropriate transcriptional machineries and/or chromatin-remodeling factors or complexes (CRCs). These catalytic factors/complexes utilize the energy from ATP hydrolysis to drive nucleosome sliding along DNA, to alter nucleosome occupancy and positioning, and/or to mediate nucleosomes removal and replacement by variant histones. Variant histones differ from canonical histones at primary amino acid sequence levels and display significant diversity in genome-wide chromatin distribution and PTMs, thus playing critical roles in regulating genome functions. Frequently, chromatin-remodeling factors work together with histone chaperones, which act in avoiding nonspecific interactions between histone and DNA molecules. More information about these above-mentioned histone/chromatin modifiers and their functions in plant growth and development can be found in several recent review articles (Han et al., 2015; Mehta and Jeffrey, 2015; Kang et al., 2020; Borg et al., 2021; Hu and Du, 2022; Shang and He, 2022). Here below, we focus our review on their implication specifically in plant response against pathogens (Figure 1).
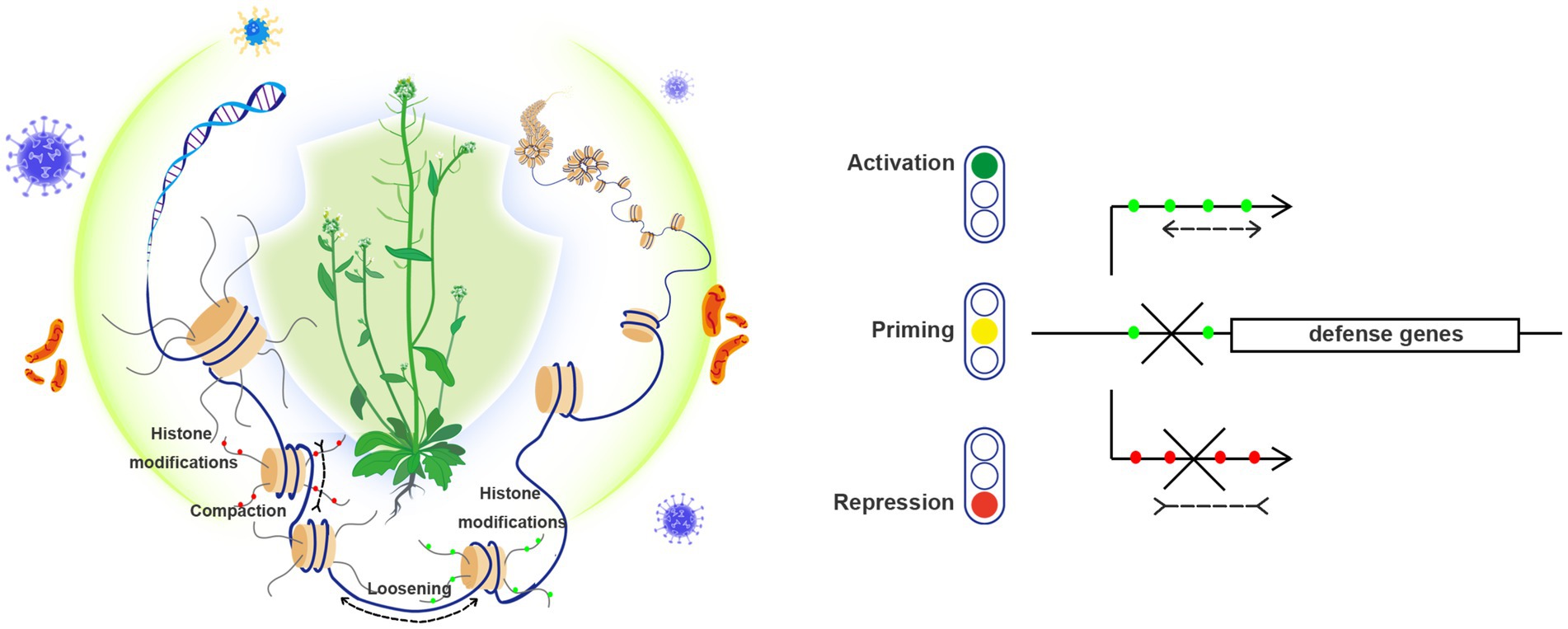
Figure 1. Both histone modification and chromatin remodeling are implicated in plant resistance to pathogens. (Left) Plant nucleosomes are widely subjected to various histone modifications, including repressive histone methylation (such as H3K27me3, labeled as red dots) and permissive histone methylation and acetylation (such as H3K4me3 and H3K14ac, labeled as green dots). Chromatin remodeling includes nucleosome sliding (compaction or loosening of neighboring nucleosomes) and histone variant exchanges. These epigenetic changes coordinate to shape local chromatin, which act as important mechanisms underlying transcriptional reprogramming, and effectively contribute to plant resistance (illustrated by an Arabidopsis plant under protection) to diverse pathogens (virus, bacteria and fungi). (Right) Different transcriptional states of defense genes are illustrated by: gene activation (green light) that is often associated with permissive histone modifications (green dots) and loosened chromatin structure, priming state (yellow light) where permissive histone modifications can also be found but transcription is poised yet uninitiated, and gene repression (red light) that is generally accompanied with repressive histone modifications (red dots) and compacted chromatin structure.
Histone modification regulators involved in plant response to pathogens
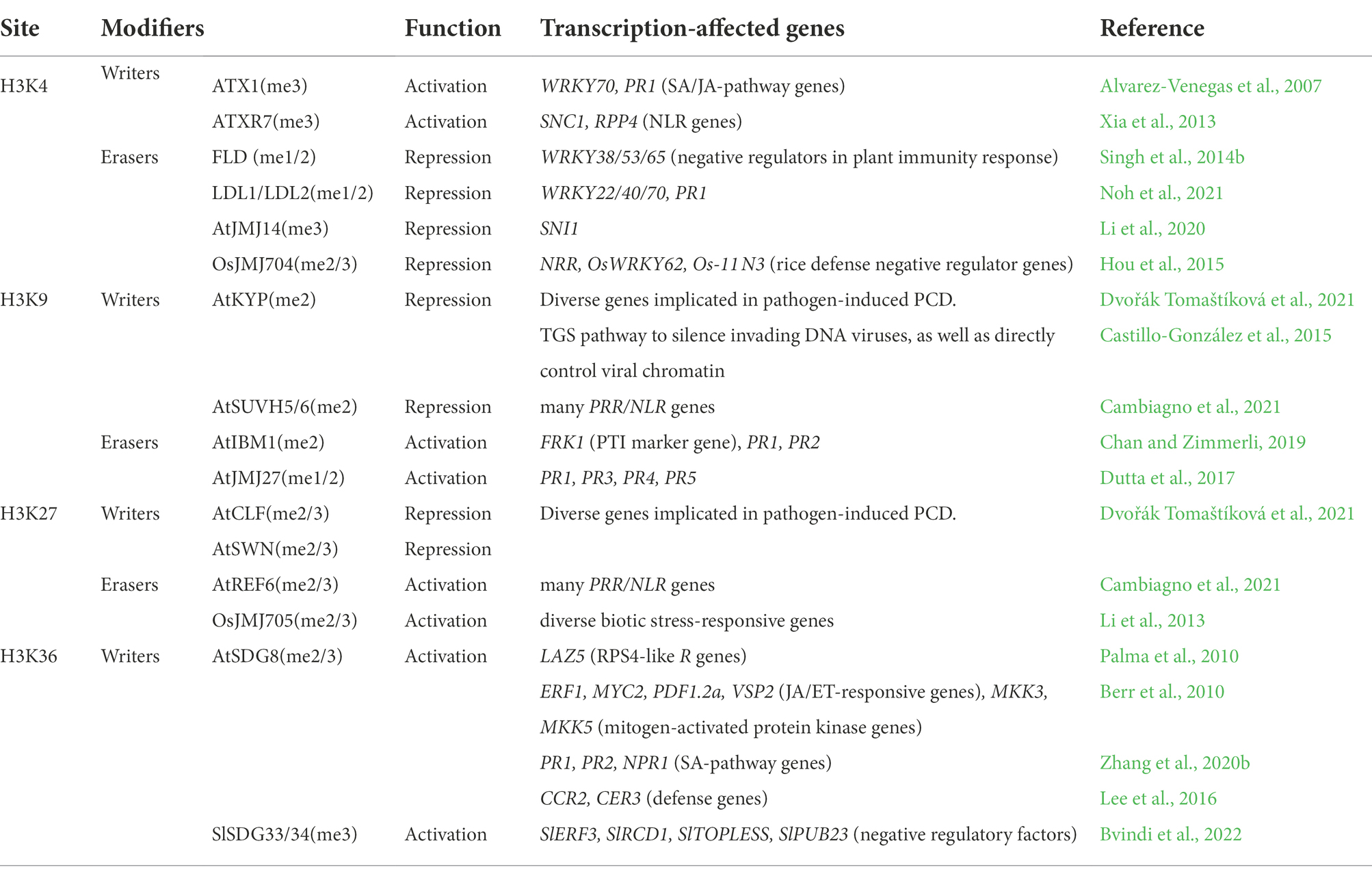
Histone lysine methyltransferases and demethylases
Typically, all known plant histone lysine methyltransferases (HKMTs) contain an evolutionarily conserved SET [named after Su(var)3–9, Enhancer-of-zeste, and Trithorax] domain. Their specificities to different lysine residues (e.g., H3K4, H3K9, H3K27 and H3K36) are determined by distinct SET domains as well as surrounding sequences. Plant histone lysine demethylases (HKDMs) comprise two families: lysine-specific demethylase (LSD1) homologues and Jumonji-C (JmjC) domain-containing proteins (JMJs). So far, a number of HKMTs and HKDMs were reported as involved in plant responses to pathogens (Table 1).
The transcription of WRKY70, a transcriptional factor gene acting at the crosstalk of both SA and JA signaling pathways in Arabidopsis, relies on H3K4me3 deposited by ARABIDOPSIS HOMOLOG OF TRITHORAX 1 (ATX1, Alvarez-Venegas et al., 2007). ATX1 also participates in the transcriptional initiation of the defense response gene PR1, keeping it under an ‘actively’ modified chromatin state to respond rapidly in transcription when needed. Correspondingly, atx1 mutant plants display reduced PR1 gene expression and reduced resistance to the hemibiotrophic bacteria Pst DC3000 (Alvarez-Venegas et al., 2007). Although the mechanism of ATX1 targeting WRKY70 and PR1 remains largely unclear, there have been good advances in the targeting mechanism of its homologue ATX-RELATED 7 (ATXR7). ATXR7 binds to the plant-specific protein MODIFIER OF SNC1 9 (MOS9) to modulate via H3K4 methylation the transcription of SUPPRESSOR OF NPR1-1, CONSTITUTIVE 1 (SNC1) and RECOGNITION OF PERONOSPORA PARASITICA 4 (RPP4), which encode plant NLR proteins acting as intracellular sensors to detect pathogen effectors and trigger immune response, playing thus an important role in Arabidopsis resistance to oomycete pathogen Hyaloperonospora arabidopsidis Emwa1 (Xia et al., 2013). Arabidopsis FLOWERING LOCUS D (FLD) encoding an LSD1-family HKDM involved in removing H3K4me1/2 was reported as necessary for response to SAR signals, guiding systematic accumulation of SA and enhancement of disease resistance (Singh et al., 2013, 2014b). LSD1-LIKE 1 (LDL1) and LDL2 involved in removing H3K4me1/2 were also reported to play roles in plant immunity to infections by several Pst strains. Compared to wild-type plants, H3K4me1 strongly accumulates at the promoter regions of many defense-related genes in the ldl1 ldl2 double mutant, indicating that these two HKDMs are involved in controlling the transcription of a group of defense-related genes (Noh et al., 2021). In addition, the HKDM JMJ14 plays a role in local and systemic Arabidopsis plant immune responses by removing pathogen-induced H3K4me3 enrichment and regulating defense genes involved in hormone (such as SA)-mediated defense pathways (Li et al., 2020). In rice, transcription of 15 JmjC genes was found induced after plant infection by the bacterial blight disease pathogen Xanthomonas oryzae pv. oryzae (Xoo). Among them, JMJ704 inhibits the transcription of negative defense regulator genes, such as NRR, OsWRKY62 and Os-11 N3, by reducing H3K4me2/3 levels, and thus positively regulates rice defense responses (Hou et al., 2015). Nonetheless, the specificity of these different HKDMs activities and the potential redundancy of their roles in plant immunity still remain largely obscure and await future investigations.
H3K36 methylation is also involved in the transcriptional regulation of R genes. In Arabidopsis, SET-DOMAIN GROUP 8 (SDG8) is the major HKMT catalyzing H3K36me2/3 (Xu et al., 2008), and SDG8 is necessary for H3K36me3 deposition at LAZARUS 5 (LAZ5) that encodes a RPS4-like R protein (Palma et al., 2010). In addition, SDG8 plays a vital role in plant defense against fungal pathogens by modulating a range of genes in JA and/or ET signaling pathways. Loss-of-function of SDG8 results in mutant plants exhibiting decreased resistance to the fungal pathogens Alternaria brassicicola and Botrytis cinerea (Berr et al., 2010) as well as to the bacteria pathogen Pst DC3000 (Zhang et al., 2020b). Consistently, several defense genes in wild-type plants infected with fungal pathogens or methyl JA treatment showed dynamic changes in SDG8-dependent histone H3K36 methylation. It was proposed that SDG8-mediated histone H3K36 methylation may act as a memory mechanism for a range of defense genes including those involved in JA/ethylene signaling pathway, helping them to achieve rapid transcription upon fungal infection or JA treatment (Berr et al., 2010). Meanwhile, SDG8 can also physically interact with the C-terminal domain of RNA polymerase II (RNAPII), linking H3K36me3 deposition with RNAPII loading, for defense gene transcriptional induction by SA (Zhang et al., 2020b). Lastly, SDG8 was also reported to regulate CAROTENOID ISOMERASE 2 (CCR2) and ECERIFERUM 3 (CER3), which encode proteins to catalyze the biosynthesis of carotenoids and be involved in the biosynthesis of cuticular wax, respectively, and loss of SDG8 resulted in the reduction of lipid accumulation and loss of cuticle integrity leading to impeded plant immunity (Lee et al., 2016). In tomato (Solanum lycopersicum, Sl), the SDG8-orthologues SlSDG33 and SlSDG34 were required for pathogen- and stress-induced enrichment of H3K36me3 and H3K4me3 at target genes, but their loss increased plant resistance to Botrytis cinerea (Bvindi et al., 2022). It was proposed that mutations in SlSDG33 and SlSDG34 may disrupt permissive transcriptional context promoting expression of negative regulatory factors to allow improvement of plant tolerance without growth trade-offs.
In contrast to the above activation histone methylations, H3K27me2/3 marks the silenced or suppressed state of gene transcription. Arabidopsis LIKE HETEROCHROMATIN PROTEIN 1 (LHP1), which binds to H3K27me3 through its chromodomain and is required for the spreading of H3K27me3, was found to be implicated in the repression of the MYC2 branch of JA/ET pathway of plant immunity, and its loss-of-function leads to reduced SA content and resistance to Pst DC3000 (Ramirez-Prado et al., 2019). Arabidopsis rapidly reprograms transcriptome after programmed cell death (PCD) triggered in response to immunity- and disease-promoting pathogen effectors, and changes in gene expression are associated with H3K27me3. Polycomb Repressive Complex 2 (PRC2) is responsible for H3K27me2/3 deposition and it is SET-domain-containing components CURLY LEAF (CLF) and SWINGER (SWN) have partial non-redundant functions in HR progression after Pst DC3000 infection (Dvořák Tomaštíková et al., 2021). The HKDM RELATIVE OF EARLY FLOWERING 6 (REF6)/JMJ12 involved in removing H3K27me2/3 was shown to contribute to the establishment of specific chromatin states in promoting priming of many genes including PRR and NLR genes that encore immune receptors in Arabidopsis (Cambiagno et al., 2021). In rice, JMJ705 specifically removes H3K27me2/3 and its overexpression results in the preferential activation of biotic stress response genes marked by H3K27me3, which enhances rice resistance to bacterial blight disease pathogen Xoo (Li et al., 2013). Notably, JMJ704 and JMJ705 show a different target specificity (H3K4me2/3 vs. H3K27me2/3), although they are jointly involved in the positive regulation of rice defense mechanisms against Xoo (Hou et al., 2015).
In addition to H3K27me3, H3K9me2 deposited by the HKMT SU(VAR)3–9 HOMOLOG 4 (SUVH4)/ KRYPTONITE (KYP), along with its homologs SUVH5/6, is also involved in repressing the basal expression of PRR/NLR genes (Cambiagno et al., 2021). While PRR/NLR genes determine the plant capability to resist pathogen infections, their excessive activation is detrimental to normal plant growth and development, pointing to a strict modulation role of H3K9 methylation in fine-tuning gene expression in plant immunity. In line with this, the HKDM INCREASE IN BONSAI METHYLATION 1 (IBM1)/JMJ25 and its homologue JMJ27 had been shown as required in removing H3K9me2 to ensure a permissive chromatin context for the correct induction of defense genes (including PR1/2) for Arabidopsis resistance to Pst DC3000 (Dutta et al., 2017; Chan and Zimmerli, 2019).
It is worth noting that epigenetic modifications can also occur in pathogens and contribute to plant-pathogen interactions. It has been reported that in innate immunity against geminivirus SUVH4/KYP binds viral chromatin and controls its methylation to combat virus infection (Castillo-González et al., 2015). The geminivirus-encoded TrAP protein interacts with the catalytic domain of SUVH4/KYP and inhibits its HKTM activity, thus counteracting plant defense, forming an epigenetic regulatory node between the host antiviral defense and virus counter defense (Castillo-González et al., 2015). Soybean (Glycine max) root rot pathogen Phytophthora sojae evades the soybean R-gene Rps1b by its PRC2 PsSu(z)12-mediated transcriptional polymorphism in the effector gene Avr1b without any sequence variation. The PsSu(z)12 mutation leads to the loss of H3K27me3 in the Avr1b region, increased Avr1b expression, and thus the loss of ability to evade the immune recognition by Rps1b and the loss of virulence in soybeans (Wang et al., 2020a). Fungal pathogens secrete effectors that are highly expressed in planta but inhibited ex planta. It was found that H3K27me3 provides significant local transcriptional repression ex planta and is replaced by H3K27ac when highly expressed in planta, enabling a strict regulation of effector genes that contribute to host infection (Zhang et al., 2021).
Histone acetyltransferases and deacetylases
Histone acetylation is catalyzed by histone acetyltransferases (HATs), and removed by histone deacetylases (HDACs; Hollender and Liu, 2008; Chen et al., 2020; Kumar et al., 2021). Based on sequence similarity and domain organization, HATs can be divided into four classes: GENERAL CONTROL NONDEREPRESSIBLE 5 (GCN5)-RELATED ACETYLTRANSFERASE (GNAT, also named HAG), MOZ-YBF2/SAS3-SAS2/TIP60 (MYST, also named HAM), cAMP-RESPONSIVE ELEMENT BINDING PROTEIN (CREB)-BINDING PROTEIN (CBP, also named HAC), and TATA-BINDING PROTEIN ASSOCIATED FACTOR 1 (TAFII250, also named HAF). Plant HDACs can be divided into three classes: RPD3-like, sirtuin-like, and plant-specific Histone Deacetylase 2 (HD2). While RPD3-like HDACs require zinc as a cofactor for deacetylase activity, Sirtuin-like HDACs remove acetyl groups in an NAD+-dependent manner. Both HAT-mediated histone acetylation in transcriptional activation and HDAC-mediated histone deacetylation in transcriptional repression are involved in plant responses to pathogens (Table 2).
Exposure of plants to sub-lethal levels of salt, cold or heat can enhance their resistance to infection by pathogens. In such cross-priming, PTI-responsive genes are enriched by histone acetylation associated with their transcriptional activation. Compared to wild-type Arabidopsis plants, mutants of HISTONE ACETYLTRANSFERASE 1 (HAC1), which encodes a HAT belonging to the HAC/CBP class, are impaired in priming PTI responses and fail to exhibit enhanced resistance to bacteria (Singh et al., 2014a). In the SA signaling pathway, HAC1/5 forms together with NPR1, a coactivator complex that is recruited through the transcription factor TGA, to activate PRs transcription via histone acetylation-mediated epigenetic reprogramming (Jin et al., 2018). HAG1/GCN5 has been considered as a master modulator of plant-environment interaction by inhibiting SA accumulation and SA-mediated immunity (Kim et al., 2020). Being a component of the SPT-ADA-GCN5 ACETYLTRANSFERASE (SAGA) transcriptional co-activator complex, Arabidopsis HAG1/GCN5 acetylates multiple sites of histone H3 (Wu et al., 2021a). While hag1/gcn5 mutation led to a decrease of H3K14ac at the 5′ end of HAG1/GCN5 down-regulated targets, it also led to an increase of H3K14ac at the 3′ end of HAG1/GCN5 up-regulated targets, revealing a dual role of HAG1/GCN5 in establishing acetylation status of its target genes. Moreover, HAG1/GCN5 also affected H3K9ac and H3K27ac levels (Wu et al., 2021a).
Pathogen effectors may inhibit the activation of plant defense genes by interfering with the function of HAT complex during infection. Indeed, the Phytophthora sojae effector PsAvh23 binds the ADA2 subunit of SAGA complex and interrupts the interaction between ADA2 and GCN5, thus leading to the dysregulation of defense-related genes and increased soybean plant susceptibility to the pathogen infection (Kong et al., 2017). PsGCN5-silenced mutant strains show impaired virulence likely due to their higher sensitivity to hydrogen peroxide (H2O2), a major oxidative product induced in host plant during early immune response, and to their incapability to inhibit ROS production in soybean plants (Zhao et al., 2015). Catalase plays a vital role in protecting cells from oxidative damage by catalyzing H2O2 degradation into water and oxygen. In the filamentous fungus Neurospora crassa, NcGCN5 acts as a positive regulator of CATALASE-3 transcription via H3 acetylation and its deprivation leads to the failure of fungus to effectively resist H2O2 accumulation (Qi et al., 2018). It becomes evident that GCN5 functions as a major HAT in plants and in pathogens, both can be detrimental for their interactions.
Besides HATs, HDACs also play essential roles in plant-pathogen interactions. Both Arabidopsis HISTONE DEACETYLASE 6 (HDA6) and HDA19, which belong to the RPD3-like class of HDACs, play key regulatory roles in plant responses to pathogens. HDA19 expression is induced by wound as well as by the Alteraria brassicicola and Pst pathogens (Zhou et al., 2005). Loss of HDA19 increases SA accumulation and the expression of related genes as well as PR genes, resulting in enhanced resistance to Pst (Kim et al., 2008). HDA19 binds directly to PR1/2 promoter and then deacetylates histones, ensuring low basal expression levels of PR1/2 genes under unchallenged conditions as well as avoiding an overstimulated expression of these genes during the defense response to pathogen attack (Choi et al., 2012). Similarly, HDA6 binds and represses the expression of pathogen-responsive genes (including PR1/2, etc.) under both normal growth conditions and pathogen infections (Wang et al., 2017). HDA6 inhibits SA biosynthesis by directly controlling the expression of CBP60g and SARD1 (Wu et al., 2021b). Remarkably, the opposing activities of HDA6 and GCN5 also maintain acetylation homeostasis of TOPLESS (TPL), a non-histone protein, which represses the master transcriptional factor MYC2 involved in plant immunity (An et al., 2022).
The geminivirus-encoded protein V2 protein interacts with Nicotiana benthamiana NbHDA6, and competes the NbMET1-NbHDA6 interaction, thus interfering the recruitment of NbMET1 by NbHDA6 to downregulate the TGS-triggered viral DNA genome methylation, which eventually increases host susceptibility to geminivirus infection (Wang et al., 2018). UvSec117, a key effector secreted by Ustilaginoidea virens, targets the rice RPD3-like class HDAC OsHDA701 to the nucleus to reduce H3K9ac and interfer with the activation of H3K9ac-marked defense gene transcription, which eventually promotes pathogen infection (Chen et al., 2022). Overexpression of BcRPD3 in Botrytis cinerea led to a significant decrease in the levels of histones H3 and H4 acetylation at virulence genes, resulting in a slight delay in fungus vegetative growth and infectious structures formation and significantly weakened virulence (Zhang et al., 2020a).
In addition to the RPD3-like HDACs, the Sirtuin-like HDAC AtSRT2 also has a negative regulatory role in Arabidopsis basal resistance against Pst DC3000, associated with the repression of SA pathway-related genes such as PHYTOALEXIN DEFICIENT 4 (PAD4) and PR1 (Wang et al., 2010). The HD2-class HDAC AtHD2B was identified as targeted by the MAPK-type kinase MPK3 to re-localize from nucleolus to nucleoplasm to repress the transcription of flg22-regulated genes and biotic stress-responsive genes (Latrasse et al., 2017). More recently, another HD2-class HDAC AtHD2C was identified as a positive regulator of plant resistance to Cauliflower mosaic virus (CaMV) infection (Li et al., 2021). AtHD2C is involved in silencing viral genome by decreasing histone acetylation on viral minichromosome. As counteraction, the CaMV P6 protein physically interacts with AtHD2C and interferes with the interaction between AtHD2C and HDA6, leading to HDAC dysfunction and thus promoting CaMV infection. Correspondingly, the susceptibility to CaMV infection is increased in the hd2c mutants (Li et al., 2021). The wheat (Triticum aestivum) HD2-class HDAC TaHDT701 can stabilize the interaction between TaHDA6 and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 15 (TaHOS15), forming a larger HDAC complex involved in plant resistance to wheat powdery mildew (Zhi et al., 2020). Notably, Arabidopsis HOS15 homolog physically interacts with HDA9, another RPD3-like HDAC in Arabidopsis, and function together as the negative regulators of one-third of diverse NLR genes in Arabidopsis likely through altering the local H3K9 acetylation (Yang et al., 2020). Whether or not an Arabidopsis HD2 member also participates together with HOS15-HDA9 to form a larger HDAC complex in plant immunity still remains unclear yet.
Lastly, it is worthy to note that enzymatic inhibitors of HDACs also play essential functions in plant-pathogen interactions. The cyclic tetrapeptide HC-toxin (HCT) from the filamentous fungus Cochliobolus (formerly Helminthosporium) carbonum inhibits enzymatic activities of numerous HDACs in vitro (Brosch et al., 1995). HCT treatments of maize (Zea mays L.) led to hyperacetylation of histones H3 and H4 as well as non-histone proteins including the TPL-homolog RAMOSA1 ENHANCER LOCUS 2 (REL2; Ransom and Walton, 1997; Walley et al., 2018). It is likely that HCT promotes the establishment of pathogenic compatibility between Cochliobolus carbonum and maize by interfering with the reversible acetylation of histones and/or non-histone transcription regulators. It is known that pathogen-induced SA signaling promotes biosynthesis of nitric oxide (NO), and that NO can physically inhibit enzymatic activities of various HDACs. A more recent study in Arabidopsis demonstrated that treatment with S-nitrosoglutathione (GSNO), the physiological NO donor, increased global levels of all examined acetylation markers of histones H3 and H4 (Mengel et al., 2017). Genome-wide H3K9/14 ac-profiles revealed that NO-regulated histone acetylation occurs at many genes involved in plant defense response, suggesting that NO-mediated inhibition of HDAC activities favors hyperacetylation and transcription of these specific genes (Mengel et al., 2017). Sodium valproate (SV) can enhance acetylation of many histone H3 lysines (H3K9ac, H3K14ac and H3K56ac) and can act as a fungicide to make tomato plants more resistant to Botrytis cinerea infection (Xu et al., 2022). SV treatment altered the expression levels of several genes associated with Botrytis cinerea virulence as well as tomato immune response, indicating that changes of acetylation level within chromatin of both plants and pathogens are involved in their interplay.
Other histone modifiers
Arabidopsis RING E3 ligase HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2 mediate histone H2B monoubiquitination (H2Bub1). HUB1-deficient Arabidopsis mutants exhibited higher sensitivity to Botrytis cinerea and Alternaria brassicicola, while HUB1 overexpression showed resistance to Botrytis cinerea (Dhawan et al., 2009). HUB1/HUB2 mediate H2Bub1 directly at the R-gene SNC1 to induce its expression, which is associated with autoimmune phenotypes and enhanced disease resistance (Zou et al., 2014). The HUB1/HUB2 homologues in tomato, SlHUB1/SlHUB2, have similar H2B monoubiquitination E3 ligase activity in vitro, and their silencing leads to increased plant sensitivity to Botrytis cinerea (Zhang et al., 2015). Correspondingly, gene expression in the SA-mediated signaling pathway was significantly upregulated, while gene expression in the JA/ET-mediated signaling pathway was significantly reduced, in SlHUB1/HUB2-silenced plants. Thus, SlHUB1/SlHUB2 likely play a role in defense response against Botrytis cinerea by adjusting the balance between SA and JA/ET-mediated signaling pathways (Zhang et al., 2015).
The lysine 2-hydroxyisobutyrylation (Khib) is a more recently discovered histone modification, and is widely distributed as an active histone marker associated with gene transcriptional activation (Dai et al., 2014; Huang et al., 2018). In rice, histone Khib is involved in regulating the expression of R genes. The ascomycete Ustilaginoidea virens can regulate Khib in rice where most Khib sites in histone H3 were downregulated during infection (Chen et al., 2021). The rice RPD3-like HDAC HDA705 is involved in removing Khib, negatively regulating plant resistance to a wide range of pathogens including Ustilaginoidea virens, while knockingout of HDA705 enhances disease resistance to these pathogens (Chen et al., 2021).
Chromatin-remodeling factors involved in plant immunity
Genome-wide transcriptional analysis reveals that SAR is involved in the processes of transcriptional reprogramming (Thibaud-Nissen et al., 2006; Wang et al., 2006), and chromatin remodeling activity is known to be extensively involved in the corresponding processes (van den Burg and Takken, 2009; Ramirez-Prado et al., 2018). All chromatin-remodeling factors show homology to the yeast SNF2 ATPase protein and contain a common Snf2 domain. They can modulate the structural and dynamic properties of chromatin, thereby influence a wide range of nuclear processes (Flaus et al., 2006). Chromatin-remodeling factors can be divided into four subfamilies based on the catalytic Snf2 domain and other accessory domains: Switch/Sucrose on-fermentable (SWI/SNF), Inositol auxotrophy 80 (INO80), Imitation switch (ISWI), and Chromodomain helicase DNA-binding (CHD; Clapier et al., 2017). These ATPases usually perform remodeling functions individually or in the form of multi-subunit CRCs (Clapier et al., 2017; Hannan Parker et al., 2022), and members from all the four subfamilies were found involved in plant immunity (Table 3).
SWI/SNF subfamily
In Arabidopsis, BRAHMA (BRM) and SPLAYED (SYD) are highly conserved SWI/SNF ATPases, which exhibit similar as well as distinct roles in regulating plant developmental processes (Bezhani et al., 2007). SYD can be directly recruited to the promoter region of target genes to regulate the expression of multiple defense genes downstream of JA and ET signaling pathways. SYD is involved in defense against Botrytis cinerera, which is associated with the activation of JA-pathway defense gene PLANT DEFENSIN1.2a (PDF1.2a; Walley et al., 2008). In addition, together with other proteins, SYD co-represses SNC1 transcription at the chromatin level, and this regulatory role is important for fine-tuning the expression of NLR-encoding genes to prevent adverse autoimmunity (Johnson et al., 2015). In contrast, the syd mutant barely shows modified resistance to Pst. This mild effects of syd may be partly explained by its redundancy with BRM in SA-mediated signaling, because both SYD and BRM have been shown to act on shared as well as unique target genes (Bezhani et al., 2007; Wu et al., 2012). Yet, an overlapping or specific role of BRM in plant immunity still remains to be elucidated.
NLR-mediated immunity is precisely controlled to avoid adverse autoimmunity. SWI/SNF-ASSOCIATED PROTEINS 73 (SWP73A) is a subunit of the SWI/SNF CRC (Jerzmanowski, 2007; Hopfner et al., 2012). SWP73A can directly repress the expression of multiple NLRs by binding to their promoters, or inhibit the expression of gene encoding the key RNA splicing regulatory protein CELL DIVISION CYCLE 5 (CDC5), thereby affecting the alternative splicing of NLRs. Upon the Pst infection, bacteria-induced small RNAs silence SWP73A to activate the correct induction and alternative splicing of NLR genes involved in the ETI pathway. Thus, SWP73A acts as a negative regulator of gene expression, and inhibits the endogenous immune response of plants to avoid autoimmunity in the absence of pathogens (Huang et al., 2021).
INO80 subfamily
The INO80 subfamily comprises two highly conserved groups: INO80 and SWI2/ SNF2-related 1 (SWR1). Both INO80 and SWR1 ATPases act as scaffold and core catalytic subunit for assembling protein complexes with other accessary proteins, the INO80-Complex (INO80-C) and SWR1-C, respectively. INO80-C and SWR1-C comprise their specific subunits, such as ACTIN-RELATED PROTEIN 5 (ARP5) and ARP6, respectively (Wang et al., 2019). Meanwhile, they also share several common subunits including RUVB-LIKE 1 (RVB1), which belongs to a large family of proteins called AAA+ class of chaperone-like ATPases (Neuwald et al., 1999). RVB1 in plants, also known as RESISTANCE TO PSEUDOMONAS SYRINGAE PV MACULICOLA INTERACTOR 1 (RIN1), negatively regulates the expression of many R-genes. RVB1/RIN1 interacts with CC-type NLR protein RESISTANCE TO PSEUDOMONAS SYRINGAE PV MACULICOLA (RPM1) and TIR-type NLR protein RPP5 to participate in plant defense pathways. Reduction of RVB1/RIN1 mRNA level results in the measurable increase of two R-dependent responses without constitutively activating defense responses (Holt 3rd et al., 2002).
SWR1-C can mediate the replacement of histone H2A by the variant histone H2A.Z and is required for repression of SA-dependent defense genes (March-Díaz et al., 2008). All components of Arabidopsis SWR1-C have been identified (Luo et al., 2020), and mutations in the genes encoding catalytic core subunit PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 (PIE1) and subunits SWR1-C SUBUNIT 6 (SWC6) and ARP6 impair H2A.Z replacement, leading to the abnormal expression of a large number of genes that overlaps with the consequence of endogenous H2A.Z gene dysfunction in plants. Many of the misregulated genes are involved in SA-dependent immune pathways. Functional knockout of these genes leads to constitutive activation of SA-mediated defense responses, and these mutants exhibit abnormal SA biosynthesis, overexpression of many SA-responsive genes, and spontaneous cell death under normal conditions, thus altering plant resistance to both biotrophic and necrotrophic pathogens (Berriri et al., 2016). Genome-wide expression analyses further revealed that these SWR1-C subunits have complex and specific functions in defense gene regulation, and PIE1 may play an important role in the regulation of crosstalk between different defense signaling pathways (Berriri et al., 2016).
SWR1-C also interacts with histone chaperone NUCLEOSOME ASSEMBLY PROTEIN-RELATED PROTEIN (NRP) in the H2A.Z regulation. NRP is a histone chaperone exhibiting higher affinities for H2A/H2B than H3/H4, while it can also effectively bind H2A.Z in vivo. Under normal growth conditions, NRP proteins cause a decrease in H2A.Z-containing nucleosomes in Arabidopsis chromatin and thereby counteract SWR1-C activity in the regulation of gene expression (Wang et al., 2020b). Pathogen infection tests have revealed that aging of plant tissues favors necrotrophs, while juvenile plants generally favor biotrophic pathogens (Glazebrook, 2005; Barna et al., 2012). Tissue damage caused by Pst was found in nrp mutant plants at early aging (Barna et al., 2018). The nrp mutants are more susceptible to infection by Sclerotinia sclerotiorum and Pst, while NRP overexpression increases resistance to these pathogens (Barna et al., 2018).
Plants preferentially activate ETI signaling at relatively low temperatures (10–23°C), while switching to PTI signaling at moderately elevated temperatures (23–32°C; Cheng et al., 2013). Elevated temperature suppresses plant immunity (Wang et al., 2009; Alcazar and Parker, 2011). SWR1-C-mediated H2A.Z-containing nucleosome dynamics have been shown to underlie temperature responses in Arabidopsis, which enrich in the genes that respond to environmental and developmental stimuli, such as those involved in immune and temperature responses, and play important roles in controlling gene expression (Kumar and Wigge, 2010; Coleman-Derr and Zilberman, 2012). The arp6 and h2a.z mutants phenocopy plants grown at elevated temperatures, and exhibit enhanced PTI and yet reduced ETI responses (Cheng et al., 2013). Thus, SWR1-C and H2A.Z may participate in the plant resistance to different pathogens in a manner influenced by temperature fluctuation.
In Arabidopsis, another ATPase of INO80 subfamily, CHROMATIN REMODELING 19 (CHR19), has a conserved ATP-dependent nucleosome sliding activity. Loss-of-function of CHR19 causes substantial changes in genome-wide nucleosome positioning and occupancy (Kang et al., 2022). CHR19 is involved in the repression of SA/JA stress-responsive genes under normal growth conditions, coordinating plant growth balance between development and stress response, and contributing to the improvement of plant resistance to fungal pathogens. Moreover, chr19 mutant displayed higher susceptibility to the JA pathway-defended necrotrophic fungal pathogen Botrytis cinerea (Kang et al., 2022).
ISWI and CHD subfamilies
Arabidopsis ISWI proteins are encoded by two functionally redundant genes, CHR11 and CHR17 (Knizewski et al., 2008; Li et al., 2012; Han et al., 2015). CHR11 and CHR17 are required for evenly spaced nucleosome pattern in gene bodies, and the primary role of ISWI is to slide nucleosomes, but not to affect nucleosome structures and density (Racki and Narlikar, 2008; Li et al., 2014). They are negative regulators of plant disease resistance. In the absence of pathogen infection, loss-of-function of CHR11 and CHR17 leads to the upregulation of a large set of defense response genes. The chr11 single mutant shows enhanced resistance against Pst DC3000 (Liu et al., 2021). The mutation of PAD4, which impairs SA accumulation (Jirage et al., 1999), specifically rescued the upregulation of both defense gene expression and resistance to Pst DC3000 in the chr11 chr17 double mutant (Liu et al., 2021).
Arabidopsis CHD1 is encoded by CHR5, which promotes the expression of seed maturation genes and participates in the regulation of plant growth and development (Zou et al., 2017). CHR5 is required for the upregulation of the intracellular immune receptor gene SNC1 likely via the regulation of nucleosome occupancy in the promoter region. Intriguingly, however, CHR5 acts as a positive regulator of the SNC1-independent plant immune pathway against Pst, implying its role in regulation of other defense genes (Zou et al., 2017).
Conclusion and perspectives
Studies in the past 20 years have greatly advanced our understanding of how diverse histone modifications and chromatin remodeling affect plant defenses against pathogens. Accumulating evidences have revealed that both plants and pathogens rely on epigenetic machineries for their proper development and metabolism. These machineries and associated mechanisms are collectively used to establish or maintain specific transcriptional states, thereby effectively activating or repressing gene transcription, or establishing priming states and/or immune memory. Meanwhile, in addition to the characterization of conserved chromatin modifiers, the study of various environmental factors as well as synthetic chemicals and drugs and pathogenic metabolites that directly or indirectly participate in the establishment, maintenance or change of epigenetic states greatly helps to understand the molecular mechanisms underlying plant responses against pathogens.
The regulation of plant immunity at the chromatin level is located most likely at the end step of signaling pathways, which ultimately affects the plant transcriptome, proteome and metabolome. Albeit the identification of a large number of chromatin modifiers involved in plant defense against pathogens, most of the current studies are limited in the analysis of individual modifiers, and their focus is often confined within the transcriptional regulation of several star defense genes. Although these analyses have successfully revealed many regulatory mechanisms in well-known pathways and uncovered the convergence or interplay of different mechanisms on some target genes, they often ignore the spatiotemporal and dynamic nature of pathogenic infection as well as the complexity of plant-pathogen interplays.
Future studies may also be needed to better integrate histone modifications and chromatin remodeling together with other epigenetic regulations such as DNA methylation and small RNAs. High-throughput profiling data and single-cell sequencing techniques would provide a better spatiotemporal resolution for studying plant-microbial interactions to unravel genome-wide epigenetic changes and complex network regulation underlying this important dynamic process. Such knowledge will be helpful for better understanding the initiation of defense gene transcription and plant immune activation as well as the molecular mechanisms that balance plant stress resistance with normal plant growth and development. The molecular and system-level knowledge will be essential for designing crops that enhance immune responses for improving plant defenses while avoiding or minimizing undesirable side effects, such as reduced plant growth and yield.
Author contributions
YZ and W-HS conceived the idea. HK, TF, JW and YZ wrote the first version. W-HS revised and finalized the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant NSFC 32070201 and 31671341) to YZ, and China Postdoctoral Science Foundation (No. 2022 M710782) to HK. W-HS received supports from the Centre National de la Recherche Scientifique (Laboratoire Interna-tional Associé Plant Epigenetic Research) and the Agence National de la Recherche (ANR-19-CE20-0018).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRC, chromatin-remodeling complex; DAMP, damage-associated molecular pattern; ETI, effector-triggered immunity; ETS, effector-triggered susceptibility; HR, hypersensitive response; IR, induced resistance; JA, jasmonic acid; MAMP, microbial-associated molecular patterns; NB-LRR, nucleotide-binding leucine-rich repeat; NLR, NB-LRR-containing R protein; PAMP, pathogen-associated molecular patterns; PRRs, pattern recognition receptors; PTI, pattern-triggered immunity; PTMs, post-translational modifications; ROS, reactive oxygen species; SA, salicylic acid; SAR, systemic acquired resistance.
References
Alcazar, R., and Parker, J. E. (2011). The impact of temperature on balancing immune responsiveness and growth in Arabidopsis. Trends Plant Sci. 16, 666–675. doi: 10.1016/j.tplants.2011.09.001
Alvarez, M. E., Nota, F., and Cambiagno, D. A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11, 563–576. doi: 10.1111/j.1364-3703.2010.00621.x
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
An, C., Deng, L., Zhai, H., You, Y., Wu, F., Zhai, Q., et al. (2022). Regulation of jasmonate signaling by reversible acetylation of TOPLESS in Arabidopsis. Mol. Plant 15, 1329–1346. doi: 10.1016/j.molp.2022.06.014
Barna, B., Fodor, J., Harrach, B. D., Pogany, M., and Kiraly, Z. (2012). The Janus face of reactive oxygen species in resistance and susceptibility of plants to necrotrophic and biotrophic pathogens. Plant Physiol. Biochem. 59, 37–43. doi: 10.1016/j.plaphy.2012.01.014
Barna, B., Gemes, K., Domoki, M., Bernula, D., Ferenc, G., Balint, B., et al. (2018). Arabidopsis NAP-related proteins (NRPs) contribute to the coordination of plant growth, developmental rate, and age-related pathogen resistance under short days. Plant Sci. 267, 124–134. doi: 10.1016/j.plantsci.2017.11.006
Berr, A., McCallum, E. J., Alioua, A., Heintz, D., Heitz, T., and Shen, W. H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414. doi: 10.1104/pp.110.161497
Berriri, S., Gangappa, S. N., and Kumar, S. V. (2016). SWR1 chromatin-remodeling complex subunits and H2A.Z have non-overlapping functions in immunity and gene regulation in Arabidopsis. Mol. Plant 9, 1051–1065. doi: 10.1016/j.molp.2016.04.003
Bezhani, S., Winter, C., Hershman, S., Wagner, J. D., Kennedy, J. F., Kwon, C. S., et al. (2007). Unique, shared, and redundant roles for the Arabidopsis SWI/SNF chromatin remodeling ATPases BRAHMA and SPLAYED. Plant Cell 19, 403–416. doi: 10.1105/tpc.106.048272
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Birkenbihl, R. P., Liu, S., and Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.2017.04.004
Borg, M., Jiang, D., and Berger, F. (2021). Histone variants take center stage in shaping the epigenome. Curr. Opin. Plant Biol. 61:101991. doi: 10.1016/j.pbi.2020.101991
Boutrot, F., and Zipfel, C. (2017). Function, discovery, and exploitation of plant pattern recognition receptors for broad-Spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. doi: 10.1146/annurev-phyto-080614-120106
Brosch, G., Ransom, R., Lechner, T., Walton, J. D., and Loidl, P. (1995). Inhibition of maize histone deacetylases by HC toxin, the host-selective toxin of Cochliobolus carbonum. Plant Cell 7, 1941–1950. doi: 10.1105/tpc.7.11.1941
Bvindi, C., Lee, S., Tang, L., Mickelbart, M. V., Li, Y., and Mengiste, T. (2022). Improved pathogen and stress tolerance in tomato mutants of SET domain histone 3 lysine methyltransferases. New Phytol. 235, 1957–1976. doi: 10.1111/nph.18277
Cambiagno, D. A., Torres, J. R., and Alvarez, M. E. (2021). Convergent epigenetic mechanisms avoid constitutive expression of immune receptor gene subsets. Front. Plant Sci. 12:703667. doi: 10.3389/fpls.2021.703667
Castillo-González, C., Liu, X., Huang, C., Zhao, C., Ma, Z., Hu, T., et al. (2015). Geminivirus-encoded TrAP suppressor inhibits the histone methyltransferase SUVH4/KYP to counter host defense. elife 4:e06671. doi: 10.7554/eLife.06671
Chan, C., and Zimmerli, L. (2019). The histone Demethylase IBM1 positively regulates Arabidopsis immunity by control of defense gene expression. Front. Plant Sci. 10:1587. doi: 10.3389/fpls.2019.01587
Chen, X., Ding, A. B., and Zhong, X. (2020). Functions and mechanisms of plant histone deacetylases. Sci. China Life Sci. 63, 206–216. doi: 10.1007/s11427-019-1587-x
Chen, X., Duan, Y., Qiao, F., Liu, H., Huang, J., Luo, C., et al. (2022). A secreted fungal effector suppresses rice immunity through host histone hypoacetylation. New Phytol. 235, 1977–1994. doi: 10.1111/nph.18265
Chen, X., Xu, Q., Duan, Y., Liu, H., Chen, X., Huang, J., et al. (2021). Ustilaginoidea virens modulates lysine 2-hydroxyisobutyrylation in rice flowers during infection. J. Integr. Plant Biol. 63, 1801–1814. doi: 10.1111/jipb.13149
Cheng, C., Gao, X., Feng, B., Sheen, J., Shan, L., and He, P. (2013). Plant immune response to pathogens differs with changing temperatures. Nat. Commun. 4:2530. doi: 10.1038/ncomms3530
Choi, S. M., Song, H. R., Han, S. K., Han, M., Kim, C. Y., Park, J., et al. (2012). HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 71, 135–146. doi: 10.1111/j.1365-313X.2012.04977.x
Clapier, C. R., Iwasa, J., Cairns, B. R., and Peterson, C. L. (2017). Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18, 407–422. doi: 10.1038/nrm.2017.26
Coleman-Derr, D., and Zilberman, D. (2012). Deposition of histone variant H2A.Z within gene bodies regulates responsive genes. PLoS Genet. 8:e1002988. doi: 10.1371/journal.pgen.1002988
Conrath, U., Beckers, G. J., Langenbach, C. J., and Jaskiewicz, M. R. (2015). Priming for enhanced defense. Annu. Rev. Phytopathol. 53, 97–119. doi: 10.1146/annurev-phyto-080614-120132
Dai, L., Peng, C., Montellier, E., Lu, Z., Chen, Y., Ishii, H., et al. (2014). Lysine 2-hydroxyisobutyrylation is a widely distributed active histone mark. Nat. Chem. Biol. 10, 365–370. doi: 10.1038/nchembio.1497
De Kesel, J., Conrath, U., Flors, V., Luna, E., Mageroy, M. H., Mauch-Mani, B., et al. (2021). The induced resistance lexicon: Do's and Don'ts. Trends Plant Sci. 26, 685–691. doi: 10.1016/j.tplants.2021.01.001
Dhawan, R., Luo, H., Foerster, A. M., Abuqamar, S., Du, H. N., Briggs, S. D., et al. (2009). HISTONE MONOUBIQUITINATION1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019. doi: 10.1105/tpc.108.062364
Dutta, A., Choudhary, P., Caruana, J., and Raina, R. (2017). JMJ27, an Arabidopsis H3K9 histone demethylase, modulates defense against Pseudomonas syringae and flowering time. Plant J. 91, 1015–1028. doi: 10.1111/tpj.13623
Dvořák Tomaštíková, E., Hafren, A., Trejo-Arellano, M. S., Rasmussen, S. R., Sato, H., Santos-Gonzalez, J., et al. (2021). Polycomb repressive complex 2 and KRYPTONITE regulate pathogen-induced programmed cell death in Arabidopsis. Plant Physiol. 185, 2003–2021. doi: 10.1093/plphys/kiab035
Feng, F., and Zhou, J. M. (2012). Plant-bacterial pathogen interactions mediated by type III effectors. Curr. Opin. Plant Biol. 15, 469–476. doi: 10.1016/j.pbi.2012.03.004
Flaus, A., Martin, D. M., Barton, G. J., and Owen-Hughes, T. (2006). Identification of multiple distinct Snf 2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. doi: 10.1093/nar/gkl295
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Gao, H., Guo, M., Song, J., Ma, Y., and Xu, Z. (2021). Signals in systemic acquired resistance of plants against microbial pathogens. Mol. Biol. Rep. 48, 3747–3759. doi: 10.1007/s11033-021-06344-7
Ghorbel, M., Brini, F., Sharma, A., and Landi, M. (2021). Role of jasmonic acid in plants: the molecular point of view. Plant Cell Rep. 40, 1471–1494. doi: 10.1007/s00299-021-02687-4
Gill, U. S., Lee, S., and Mysore, K. S. (2015). Host versus nonhost resistance: distinct wars with similar arsenals. Phytopathology 105, 580–587. doi: 10.1094/PHYTO-11-14-0298-RVW
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Han, S. K., Wu, M. F., Cui, S., and Wagner, D. (2015). Roles and activities of chromatin remodeling ATPases in plants. Plant J. 83, 62–77. doi: 10.1111/tpj.12877
Hannan Parker, A., Wilkinson, S. W., and Ton, J. (2022). Epigenetics: a catalyst of plant immunity against pathogens. New Phytol. 233, 66–83. doi: 10.1111/nph.17699
Hollender, C., and Liu, Z. (2008). Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 50, 875–885. doi: 10.1111/j.1744-7909.2008.00704.x
Holt, B. F. 3rd, Boyes, D. C., Ellerstrom, M., Siefers, N., Wiig, A., Kauffman, S., et al. (2002). An evolutionarily conserved mediator of plant disease resistance gene function is required for normal Arabidopsis development. Dev. Cell 2, 807–817. doi: 10.1016/s1534-5807(02)00174-0
Hopfner, K. P., Gerhold, C. B., Lakomek, K., and Wollmann, P. (2012). Swi 2/Snf 2 remodelers: hybrid views on hybrid molecular machines. Curr. Opin. Struct. Biol. 22, 225–233. doi: 10.1016/j.sbi.2012.02.007
Hou, Y., Wang, L., Wang, L., Liu, L., Li, L., Sun, L., et al. (2015). JMJ704 positively regulates rice defense response against Xanthomonas oryzae pv. Oryzae infection via reducing H3K4me2/3 associated with negative disease resistance regulators. BMC Plant Biol. 15:286. doi: 10.1186/s12870-015-0674-3
Hu, H., and Du, J. (2022). Structure and mechanism of histone methylation dynamics in Arabidopsis. Curr. Opin. Plant Biol. 67:102211. doi: 10.1016/j.pbi.2022.102211
Huang, H., Luo, Z., Qi, S., Huang, J., Xu, P., Wang, X., et al. (2018). Landscape of the regulatory elements for lysine 2-hydroxyisobutyrylation pathway. Cell Res. 28, 111–125. doi: 10.1038/cr.2017.149
Huang, C. Y., Rangel, D. S., Qin, X., Bui, C., Li, R., Jia, Z., et al. (2021). The chromatin-remodeling protein BAF60/SWP73A regulates the plant immune receptor NLRs. Cell Host Microbe 29:e424. doi: 10.1016/j.chom.2021.01.005
Iakovidis, M., Chung, E. H., Saile, S. C., Sauberzweig, E., and El Kasmi, F. (2022). The emerging frontier of plant Immunity’s Core hubs. FEBS J. doi: 10.1111/febs.16549
Jaskiewicz, M., Conrath, U., and Peterhansel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Jerzmanowski, A. (2007). SWI/SNF chromatin remodeling and linker histones in plants. Biochim. Biophys. Acta 1769, 330–345. doi: 10.1016/j.bbaexp.2006.12.003
Jin, H., Choi, S. M., Kang, M. J., Yun, S. H., Kwon, D. J., Noh, Y. S., et al. (2018). Salicylic acid-induced transcriptional reprogramming by the HAC-NPR1-TGA histone acetyltransferase complex in Arabidopsis. Nucleic Acids Res. 46, 11712–11725. doi: 10.1093/nar/gky847
Jirage, D., Tootle, T. L., Reuber, T. L., Frost, L. N., Feys, B. J., Parker, J. E., et al. (1999). Arabidopsis thaliana PAD4 encodes a lipase-like gene that is important for salicylic acid signaling. Proc. Natl. Acad. Sci. U. S. A. 96, 13583–13588. doi: 10.1073/pnas.96.23.13583
Johnson, K. C., Xia, S., Feng, X., and Li, X. (2015). The chromatin remodeler SPLAYED negatively regulates SNC1-mediated immunity. Plant Cell Physiol. 56, 1616–1623. doi: 10.1093/pcp/pcv087
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kang, H., Liu, Y., Fan, T., Ma, J., Wu, D., Heitz, T., et al. (2022). Arabidopsis CHROMATIN REMODELING 19 acts as a transcriptional repressor and contributes to plant pathogen resistance. Plant Cell 34, 1100–1116. doi: 10.1093/plcell/koab318
Kang, H., Wu, D., Fan, T., and Zhu, Y. (2020). Activities of chromatin remodeling factors and histone chaperones and their effects in root apical meristem development. Int. J. Mol. Sci. 21:771. doi: 10.3390/ijms21030771
Kim, K. C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Kim, S., Piquerez, S. J. M., Ramirez-Prado, J. S., Mastorakis, E., Veluchamy, A., Latrasse, D., et al. (2020). GCN5 modulates salicylic acid homeostasis by regulating H3K14ac levels at the 5′ and 3′ ends of its target genes. Nucleic Acids Res. 48, 5953–5966. doi: 10.1093/nar/gkaa369
Knizewski, L., Ginalski, K., and Jerzmanowski, A. (2008). Snf 2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13, 557–565. doi: 10.1016/j.tplants.2008.08.004
Kong, L., Qiu, X., Kang, J., Wang, Y., Chen, H., Huang, J., et al. (2017). A Phytophthora effector manipulates host histone acetylation and reprograms defense gene expression to promote infection. Curr. Biol. 27, 981–991. doi: 10.1016/j.cub.2017.02.044
Koster, P., DeFalco, T. A., and Zipfel, C. (2022). Ca (2+) signals in plant immunity. EMBO J. 41:e110741. doi: 10.15252/embj.2022110741
Krajewski, W. A. (2022). Histone modifications, Internucleosome dynamics, and DNA stresses: how they cooperate to "functionalize" nucleosomes. Front. Genet. 13:873398. doi: 10.3389/fgene.2022.873398
Kumar, V., Thakur, J. K., and Prasad, M. (2021). Histone acetylation dynamics regulating plant development and stress responses. Cell. Mol. Life Sci. 78, 4467–4486. doi: 10.1007/s00018-021-03794-x
Kumar, S. V., and Wigge, P. A. (2010). H2A.Z-containing nucleosomes mediate the thermosensory response in Arabidopsis. Cells 140, 136–147. doi: 10.1016/j.cell.2009.11.006
Latrasse, D., Jegu, T., Li, H., de Zelicourt, A., Raynaud, C., Legras, S., et al. (2017). MAPK-triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Biol. 18:131. doi: 10.1186/s13059-017-1261-8
Lee, S., Fu, F., Xu, S., Lee, S. Y., Yun, D. J., and Mengiste, T. (2016). Global regulation of plant immunity by histone lysine methyl Transferases. Plant Cell 28, 1640–1661. doi: 10.1105/tpc.16.00012
Li, T., Chen, X., Zhong, X., Zhao, Y., Liu, X., Zhou, S., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Li, D., Liu, R., Singh, D., Yuan, X., Kachroo, P., and Raina, R. (2020). JMJ14 encoded H3K4 demethylase modulates immune responses by regulating defence gene expression and pipecolic acid levels. New Phytol. 225, 2108–2121. doi: 10.1111/nph.16270
Li, G., Liu, S., Wang, J., He, J., Huang, H., Zhang, Y., et al. (2014). ISWI proteins participate in the genome-wide nucleosome distribution in Arabidopsis. Plant J. 78, 706–714. doi: 10.1111/tpj.12499
Li, S., Lyu, S., Liu, Y., Luo, M., Shi, S., and Deng, S. (2021). Cauliflower mosaic virus P6 dysfunctions histone Deacetylase HD2C to promote virus infection. Cells 10:278. doi: 10.3390/cells10092278
Li, G., Zhang, J., Li, J., Yang, Z., Huang, H., and Xu, L. (2012). Imitation switch chromatin remodeling factors and their interacting RINGLET proteins act together in controlling the plant vegetative phase in Arabidopsis. Plant J. 72, 261–270. doi: 10.1111/j.1365-313X.2012.05074.x
Liu, H., Li, J., Xu, Y., Hua, J., and Zou, B. (2021). ISWI chromatin remodeling factors repress PAD4-mediated plant immune responses in Arabidopsis. Biochem. Biophys. Res. Commun. 583, 63–70. doi: 10.1016/j.bbrc.2021.10.058
Luger, K., Dechassa, M. L., and Tremethick, D. J. (2012). New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat. Rev. Mol. Cell Biol. 13, 436–447. doi: 10.1038/nrm3382
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 a resolution. Nature 389, 251–260. doi: 10.1038/38444
Luna, E., Beardon, E., Ravnskov, S., Scholes, J., and Ton, J. (2016). Optimizing chemically induced resistance in tomato against Botrytis cinerea. Plant Dis. 100, 704–710. doi: 10.1094/PDIS-03-15-0347-RE
Luna, E., Bruce, T. J., Roberts, M. R., Flors, V., and Ton, J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.187468
Luo, Y. X., Hou, X. M., Zhang, C. J., Tan, L. M., Shao, C. R., Lin, R. N., et al. (2020). A plant-specific SWR1 chromatin-remodeling complex couples histone H2A.Z deposition with nucleosome sliding. EMBO J. 39:e102008. doi: 10.15252/embj.2019102008
March-Díaz, R., Garcia-Dominguez, M., Lozano-Juste, J., Leon, J., Florencio, F. J., and Reyes, J. C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53, 475–487. doi: 10.1111/j.1365-313X.2007.03361.x
Martinez-Aguilar, K., Ramirez-Carrasco, G., Hernandez-Chavez, J. L., Barraza, A., and Alvarez-Venegas, R. (2016). Use of BABA and INA as activators of a primed state in the common bean (Phaseolus vulgaris L.). Front. Plant Sci. 7:653. doi: 10.3389/fpls.2016.00653
Mauch-Mani, B., Baccelli, I., Luna, E., and Flors, V. (2017). Defense priming: An adaptive part of induced resistance. Annu. Rev. Plant Biol. 68, 485–512. doi: 10.1146/annurev-arplant-042916-041132
Mehta, S., and Jeffrey, K. L. (2015). Beyond receptors and signaling: epigenetic factors in the regulation of innate immunity. Immunol. Cell Biol. 93, 233–244. doi: 10.1038/icb.2014.101
Mengel, A., Ageeva, A., Georgii, E., Bernhardt, J., Wu, K., Durner, J., et al. (2017). Nitric oxide modulates histone acetylation at stress genes by inhibition of histone Deacetylases. Plant Physiol. 173, 1434–1452. doi: 10.1104/pp.16.01734
Neuwald, A. F., Aravind, L., Spouge, J. L., and Koonin, E. V. (1999). AAA+: a class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 9, 27–43.
Nguyen, Q. M., Iswanto, A. B. B., Son, G. H., and Kim, S. H. (2021). Recent advances in effector-triggered immunity in plants: new pieces in the puzzle create a different paradigm. Int. J. Mol. Sci. 22:22. doi: 10.3390/ijms22094709
Noh, S. W., Seo, R. R., Park, H. J., and Jung, H. W. (2021). Two Arabidopsis homologs of human lysine-specific Demethylase function in epigenetic regulation of plant defense responses. Front. Plant Sci. 12:688003. doi: 10.3389/fpls.2021.688003
Palma, K., Thorgrimsen, S., Malinovsky, F. G., Fiil, B. K., Nielsen, H. B., Brodersen, P., et al. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog. 6:e1001137. doi: 10.1371/journal.ppat.1001137
Peng, Y., van Wersch, R., and Zhang, Y. (2018). Convergent and divergent signaling in PAMP-triggered immunity and effector-triggered immunity. Mol. Plant-Microbe Interact. 31, 403–409. doi: 10.1094/MPMI-06-17-0145-CR
Peng, Y., Yang, J., Li, X., and Zhang, Y. (2021). Salicylic acid: biosynthesis and signaling. Annu. Rev. Plant Biol. 72, 761–791. doi: 10.1146/annurev-arplant-081320-092855
Po-Wen, C., Singh, P., and Zimmerli, L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14, 58–70. doi: 10.1111/j.1364-3703.2012.00827.x
Qi, S., He, L., Zhang, Q., Dong, Q., Wang, Y., Yang, Q., et al. (2018). Cross-pathway control gene CPC1/GCN4 coordinates with histone acetyltransferase GCN5 to regulate catalase-3 expression under oxidative stress in Neurospora crassa. Free Radic. Biol. Med. 117, 218–227. doi: 10.1016/j.freeradbiomed.2018.02.003
Racki, L. R., and Narlikar, G. J. (2008). ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr. Opin. Genet. Dev. 18, 137–144. doi: 10.1016/j.gde.2008.01.007
Ramirez-Prado, J. S., Abulfaraj, A. A., Rayapuram, N., Benhamed, M., and Hirt, H. (2018). Plant immunity: from signaling to epigenetic control of defense. Trends Plant Sci. 23, 833–844. doi: 10.1016/j.tplants.2018.06.004
Ramirez-Prado, J. S., Latrasse, D., Rodriguez-Granados, N. Y., Huang, Y., Manza-Mianza, D., Brik-Chaouche, R., et al. (2019). The Polycomb protein LHP1 regulates Arabidopsis thaliana stress responses through the repression of the MYC2-dependent branch of immunity. Plant J. 100, 1118–1131. doi: 10.1111/tpj.14502
Rando, O. J. (2012). Combinatorial complexity in chromatin structure and function: revisiting the histone code. Curr. Opin. Genet. Dev. 22, 148–155. doi: 10.1016/j.gde.2012.02.013
Ransom, R. F., and Walton, J. D. (1997). Histone Hyperacetylation in maize in response to treatment with HC-toxin or infection by the filamentous fungus Cochliobolus carbonum. Plant Physiol. 115, 1021–1027. doi: 10.1104/pp.115.3.1021
Rutowicz, K., Lirski, M., Mermaz, B., Teano, G., Schubert, J., Mestiri, I., et al. (2019). Linker histones are fine-scale chromatin architects modulating developmental decisions in Arabidopsis. Genome Biol. 20:157. doi: 10.1186/s13059-019-1767-3
Schreiber, K. J., Chau-Ly, I. J., and Lewis, J. D. (2021). What the wild things do: mechanisms of plant host manipulation by bacterial type III-secreted effector proteins. Microorganisms 9:1029. doi: 10.3390/microorganisms9051029
Shang, J. Y., and He, X. J. (2022). Chromatin-remodeling complexes: conserved and plant-specific subunits in Arabidopsis. J. Integr. Plant Biol. 64, 499–515. doi: 10.1111/jipb.13208
Singh, V., Roy, S., Giri, M. K., Chaturvedi, R., Chowdhury, Z., Shah, J., et al. (2013). Arabidopsis thaliana FLOWERING LOCUS D is required for systemic acquired resistance. Mol. Plant-Microbe Interact. 26, 1079–1088. doi: 10.1094/MPMI-04-13-0096-R
Singh, V., Roy, S., Singh, D., and Nandi, A. K. (2014b). Arabidopsis flowering locus D influences systemic-acquired-resistance- induced expression and histone modifications of WRKY genes. J. Biosci. 39, 119–126. doi: 10.1007/s12038-013-9407-7
Singh, P., Yekondi, S., Chen, P. W., Tsai, C. H., Yu, C. W., Wu, K., et al. (2014a). Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-dependent manner. Plant Cell 26, 2676–2688. doi: 10.1105/tpc.114.123356
Spoel, S. H., and Dong, X. (2012). How do plants achieve immunity? Defence without specialized immune cells. Nat. Rev. Immunol. 12, 89–100. doi: 10.1038/nri3141
Thibaud-Nissen, F., Wu, H., Richmond, T., Redman, J. C., Johnson, C., Green, R., et al. (2006). Development of Arabidopsis whole-genome microarrays and their application to the discovery of binding sites for the TGA2 transcription factor in salicylic acid-treated plants. Plant J. 47, 152–162. doi: 10.1111/j.1365-313X.2006.02770.x
Ton, J., Jakab, G., Toquin, V., Flors, V., Iavicoli, A., Maeder, M. N., et al. (2005). Dissecting the beta-aminobutyric acid-induced priming phenomenon in Arabidopsis. Plant Cell 17, 987–999. doi: 10.1105/tpc.104.029728
Tsuda, K., and Somssich, I. E. (2015). Transcriptional networks in plant immunity. New Phytol. 206, 932–947. doi: 10.1111/nph.13286
van den Burg, H. A., and Takken, F. L. (2009). Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 14, 286–294. doi: 10.1016/j.tplants.2009.02.003
Vergara, Z., and Gutierrez, C. (2017). Emerging roles of chromatin in the maintenance of genome organization and function in plants. Genome Biol. 18:96. doi: 10.1186/s13059-017-1236-9
Walley, J. W., Rowe, H. C., Xiao, Y., Chehab, E. W., Kliebenstein, D. J., Wagner, D., et al. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog. 4:e1000237. doi: 10.1371/journal.ppat.1000237
Walley, J. W., Shen, Z., McReynolds, M. R., Schmelz, E. A., and Briggs, S. P. (2018). Fungal-induced protein hyperacetylation in maize identified by acetylome profiling. Proc. Natl. Acad. Sci. U. S. A. 115, 210–215. doi: 10.1073/pnas.1717519115
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
Wang, Y., Bao, Z., Zhu, Y., and Hua, J. (2009). Analysis of temperature modulation of plant defense against biotrophic microbes. Mol. Plant-Microbe Interact. 22, 498–506. doi: 10.1094/MPMI-22-5-0498
Wang, L., Chen, H., Li, J., Shu, H., Zhang, X., Wang, Y., et al. (2020a). Effector gene silencing mediated by histone methylation underpins host adaptation in an oomycete plant pathogen. Nucleic Acids Res. 48, 1790–1799. doi: 10.1093/nar/gkz1160
Wang, J., Gao, S., Peng, X., Wu, K., and Yang, S. (2019). Roles of the INO80 and SWR1 chromatin remodeling complexes in plants. Int. J. Mol. Sci. 20:4591. doi: 10.3390/ijms20184591
Wang, C., Gao, F., Wu, J., Dai, J., Wei, C., and Li, Y. (2010). Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 51, 1291–1299. doi: 10.1093/pcp/pcq087
Wang, Y., Hu, Q., Wu, Z., Wang, H., Han, S., Jin, Y., et al. (2017). HISTONE DEACETYLASE 6 represses pathogen defence responses in Arabidopsis thaliana. Plant Cell Environ. 40, 2972–2986. doi: 10.1111/pce.13047
Wang, B., Yang, X., Wang, Y., Xie, Y., and Zhou, X. (2018). Tomato yellow Leaf curl virus V2 interacts with host histone Deacetylase 6 to suppress methylation-mediated transcriptional gene silencing in plants. J. Virol. 92:18. doi: 10.1128/JVI.00036-18
Wang, Y., Zhong, Z., Zhang, Y., Xu, L., Feng, S., Rayatpisheh, S., et al. (2020b). NAP1-RELATED PROTEIN1 and 2 negatively regulate H2A.Z abundance in chromatin in Arabidopsis. Nat. Commun. 11:2887. doi: 10.1038/s41467-020-16691-x
Wu, Z., He, L., Jin, Y., Chen, J., Shi, H., Wang, Y., et al. (2021b). HISTONE DEACETYLASE 6 suppresses salicylic acid biosynthesis to repress autoimmunity. Plant Physiol. 187, 2592–2607. doi: 10.1093/plphys/kiab408
Wu, C. J., Liu, Z. Z., Wei, L., Zhou, J. X., Cai, X. W., Su, Y. N., et al. (2021a). Three functionally redundant plant-specific paralogs are core subunits of the SAGA histone acetyltransferase complex in Arabidopsis. Mol. Plant 14, 1071–1087. doi: 10.1016/j.molp.2021.03.014
Wu, M. F., Sang, Y., Bezhani, S., Yamaguchi, N., Han, S. K., Li, Z., et al. (2012). SWI2/SNF2 chromatin remodeling ATPases overcome polycomb repression and control floral organ identity with the LEAFY and SEPALLATA3 transcription factors. Proc. Natl. Acad. Sci. U. S. A. 109, 3576–3581. doi: 10.1073/pnas.1113409109
Xia, S., Cheng, Y. T., Huang, S., Win, J., Soards, A., Jinn, T. L., et al. (2013). Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol. 162, 1694–1705. doi: 10.1104/pp.113.214551
Xu, Y., Wang, Y., Wang, L., Liang, W., and Yang, Q. (2022). Sodium valproate is effective against Botrytis cinerea infection of tomato by enhancing histone H3 acetylation-directed gene transcription and triggering tomato fruit immune response. Phytopathology 112, 1264–1272. doi: 10.1094/PHYTO-11-21-0483-R
Xu, L., Zhao, Z., Dong, A., Soubigou-Taconnat, L., Renou, J. P., Steinmetz, A., et al. (2008). Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28, 1348–1360. doi: 10.1128/MCB.01607-07
Yang, L., Chen, X., Wang, Z., Sun, Q., Hong, A., Zhang, A., et al. (2020). HOS15 and HDA9 negatively regulate immunity through histone deacetylation of intracellular immune receptor NLR genes in Arabidopsis. New Phytol. 226, 507–522. doi: 10.1111/nph.16380
Zhang, W., Huang, J., and Cook, D. E. (2021). Histone modification dynamics at H3K27 are associated with altered transcription of in planta induced genes in Magnaporthe oryzae. PLoS Genet. 17:e1009376. doi: 10.1371/journal.pgen.1009376
Zhang, Y., Li, D., Zhang, H., Hong, Y., Huang, L., Liu, S., et al. (2015). Tomato histone H2B monoubiquitination enzymes SlHUB1 and SlHUB2 contribute to disease resistance against Botrytis cinerea through modulating the balance between SA-and JA/ET-mediated signaling pathways. BMC Plant Biol. 15:252. doi: 10.1186/s12870-015-0614-2
Zhang, X., Menard, R., Li, Y., Coruzzi, G. M., Heitz, T., Shen, W. H., et al. (2020b). Arabidopsis SDG8 potentiates the sustainable transcriptional induction of the Pathogenesis-related genes PR1 and PR2 during plant defense response. Front. Plant Sci. 11:277. doi: 10.3389/fpls.2020.00277
Zhang, N., Yang, Z., Zhang, Z., and Liang, W. (2020a). BcRPD3-mediated histone Deacetylation is involved in growth and pathogenicity of Botrytis cinerea. Front. Microbiol. 11:1832. doi: 10.3389/fmicb.2020.01832
Zhao, W., Wang, T., Liu, S., Chen, Q., and Qi, R. (2015). The histone acetyltransferase PsGcn5 mediates oxidative stress responses and is required for full virulence of Phytophthora sojae. Microb. Pathog. 87, 51–58. doi: 10.1016/j.micpath.2015.07.015
Zhi, P., Kong, L., Liu, J., Zhang, X., Wang, X., Li, H., et al. (2020). Histone Deacetylase TaHDT701 functions in TaHDA6-TaHOS15 complex to regulate wheat defense responses to Blumeria graminis f. sp. tritici. Int. J. Mol. Sci. 21:640. doi: 10.3390/ijms21072640
Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17, 1196–1204. doi: 10.1105/tpc.104.028514
Zimmerli, L., Jakab, G., Metraux, J. P., and Mauch-Mani, B. (2000). Potentiation of pathogen-specific defense mechanisms in Arabidopsis by beta-aminobutyric acid. Proc. Natl. Acad. Sci. U. S. A. 97, 12920–12925. doi: 10.1073/pnas.230416897
Zou, B., Sun, Q., Zhang, W., Ding, Y., Yang, D. L., Shi, Z., et al. (2017). The Arabidopsis chromatin-remodeling factor CHR5 regulates plant immune responses and nucleosome occupancy. Plant Cell Physiol. 58, 2202–2216. doi: 10.1093/pcp/pcx155
Keywords: epigenetics, histone modification, chromatin remodeling, plant defense, pathogen infection
Citation: Kang H, Fan T, Wu J, Zhu Y and Shen W-H (2022) Histone modification and chromatin remodeling in plant response to pathogens. Front. Plant Sci. 13:986940. doi: 10.3389/fpls.2022.986940
Edited by:
Mahmoud Yaish, Sultan Qaboos University, OmanReviewed by:
Namisha Sharma, Institute of Life Sciences, IndiaDaisuke Miki, Chinese Academy of Sciences (CAS), China
Copyright © 2022 Kang, Fan, Wu, Zhu and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yan Zhu, zhu_yan@fudan.edu.cn; Wen-Hui Shen, wen-hui.shen@ibmp-cnrs.unistra.fr
†ORCID: Yan Zhu, https://orcid.org/0000-0001-7206-2612
Wen-Hui Shen, https://orcid.org/0000-0001-7988-6382
‡These authors have contributed equally to this work and share first authorship
 Huijia Kang1,2,3‡
Huijia Kang1,2,3‡ Tianyi Fan
Tianyi Fan Yan Zhu
Yan Zhu Wen-Hui Shen
Wen-Hui Shen