- 1State Key Laboratory of Tree Genetics and Breeding, Northeast Forestry University, Harbin, China
- 2College of Forestry and Grassland, Jilin Agricultural University, Changchun, China
- 3Jilin Provincial Academy of Forestry Sciences, Changchun, China
- 4Qiqihar Branch of Heilongjiang Academy of Forestry, Qiqihar, China
- 5Liaoning Academy of Forest Science, Shenyang, China
- 6Linjiang Forestry Bureau of Jilin Province, Lijiang, China
Juglans mandshurica is a native tree species in Northeast China. Due to habitat destruction and human disturbance, its population size has sharply decreased. Currently, information on molecular markers of J. mandshurica is limited and cannot meet the needs of germplasm resource evaluation and molecular marker-assisted breeding of J. mandshurica. Based on transcriptomic data from three tissues (leaves, bark, and fruit pericarp), we developed expressed sequence tag-simple sequence repeats (EST-SSRs) for J. mandshurica, and 15 polymorphic EST-SSR primers were initially selected. The average number of alleles (Na), expected heterozygosity (He), and the polymorphic information content (PIC) at different loci were 18.27, 0.670, and 0.797, respectively. Population genetic diversity analysis revealed that the average Na, He, and Shannon information indices (I) for 15 J. mandshurica populations were 6.993, 0.670, and 1.455, respectively. Among them, population Hunchun exhibited the highest genetic diversity (Na = 7.933, He = 0.723, and I = 1.617), while population Heihe exhibited the lowest genetic diversity (Na = 4.200, He = 0.605, and I = 1.158). STRUCTURE analysis, neighbor-joining method cluster analysis, and principal coordinate analysis showed that the 343 individuals of J. mandshurica from 15 populations were clustered into three categories. Category 1 (green) had 147 individuals from eight populations in Qingyuan, Caohekou, Jian, Ningan, Yongji, Baishishan, Helong, and Maoershan; category 2 (blue) had 81 individuals from three populations in Hulin, Boli, and Sanchazi; and category 3 (red) had 115 individuals from four populations in Heihe, Hunchun, Fangzheng, and Liangshui. Analysis of molecular variance (AMOVA) showed that genetic variations among and within individuals accounted for 16.22% and 21.10% of the total genetic variation, respectively, indicating that genetic variations within populations were greater than genetic variations among populations. The average genetic differentiation coefficient (Fst) and gene flow (Nm) between different populations were 0.109 and 4.063, respectively, implying moderate levels of genetic differentiation and gene flow. Based on the genetic diversity characteristics of different populations, we proposed various genetic conservation strategies for J. mandshurica.
Introduction
Juglans mandshurica Maxim., a deciduous tree of the Juglandaceae family, is native to northern and northeastern China and is mainly distributed in the Changbai Mountains and Lesser Khingan Mountains at elevations of 500–1,000 m. It is also distributed in Russia, Japan, and Korea and can withstand low temperatures of −50°C. As an ancient fruit tree species, it is economically valuable (Tian et al., 2009; Zhou et al., 2015). It is composed of a hard, dense, elastic, corrosion-resistant, and easy-to-process elite material for military joins and furniture (Zhu et al., 2011). Its fruits are of high quality with high oil content. Moreover, it is an excellent dried fruit species with a promising future in Northeast China (Pang et al., 2019). J. mandshurica also has high medicinal value. Its green husk, bark, roots, and leaves contain juglone, which exhibits good antitumor (Mallavadhani et al., 2014), antifungal (Sharma et al., 2009), antiviral (Vardhini, 2014), antioxidant (Chobot and Hadacek, 2009), anthelmintic (Islam and Widhalm, 2020), and hypoglycemic activities (Hosseini et al., 2014). It has potential applications in the pharmaceutical, agroforestry, food, and cosmetic industries. Collection and evaluation of plant germplasm resources is a prerequisite for the utilization of wild cultivated genes, which enhances plant genetic improvement and further assists in breeding programs (El-Esawi, 2017; Mohammed et al., 2020). In China, the northeast forest area is the largest natural forest area, with a rich abundance of J. mandshurica and rich variation in the growth, wood, and seed properties of different resources (Song et al., 2017). However, driven by financial gain, J. mandshurica in Northeast China has been illegally and excessively cut, leading to the loss of a large number of good genes of J. mandshurica and the destruction of natural forests. The frequent occurrence of forest fires has also constrained the development and utilization of germplasm resources, leading to a sharp decline in local economic and ecological benefits. Studies on J. mandshurica have been conducted in China from the 1980s to 1990s (Yang et al., 1990); however, the long growth cycle and low survival rate of asexual propagation, such as cutting and grafting, have led to slow progress in research on J. mandshurica. Most studies have focused on medicinal value development (Jin et al., 2015), pharmacological effects (Yao et al., 2017), clinical effects (Park et al., 2012), and toxicity tests (Sun et al., 2007), with a limited number of studies focusing on germplasm collection, conservation, development, and utilization.
In recent years, with advances in molecular biology and high-throughput sequencing technologies, molecular markers, transcriptome sequencing, and genome sequencing have been gradually applied to study the genetic structure, genetic diversity, and relatedness among or within species, which have allowed the exploration of the evolutionary origin of species at the individual and population levels. Molecular marker technologies, such as SRAP (Wang, 2007; Zhang et al., 2020), ISSR (Wang et al., 2011), and SSR (Liu, 2015; Wang et al., 2016; Dang et al., 2021), have been used to study the genetic diversity of J. mandshurica; however, a limited number of studies have used EST-SSRs as markers. We collected germplasm resources for J. mandshurica in the forest area of northeastern China and studied the intraspecific genetic diversity based on transcriptome sequencing and EST-SSR molecular marker technologies. We aimed to: (1) use Illumina HiSeq 2000 to sequence the RNA of J. mandshurica and to develop polymorphic EST-SSR primers using the obtained transcriptome data; (2) analyze the genetic diversity of different J. mandshurica populations in northeastern China using the developed EST-SSR primers; and (3) propose genetic conservation strategies for J. mandshurica in northeastern China.
Materials and Methods
Plant Materials and DNA Extraction
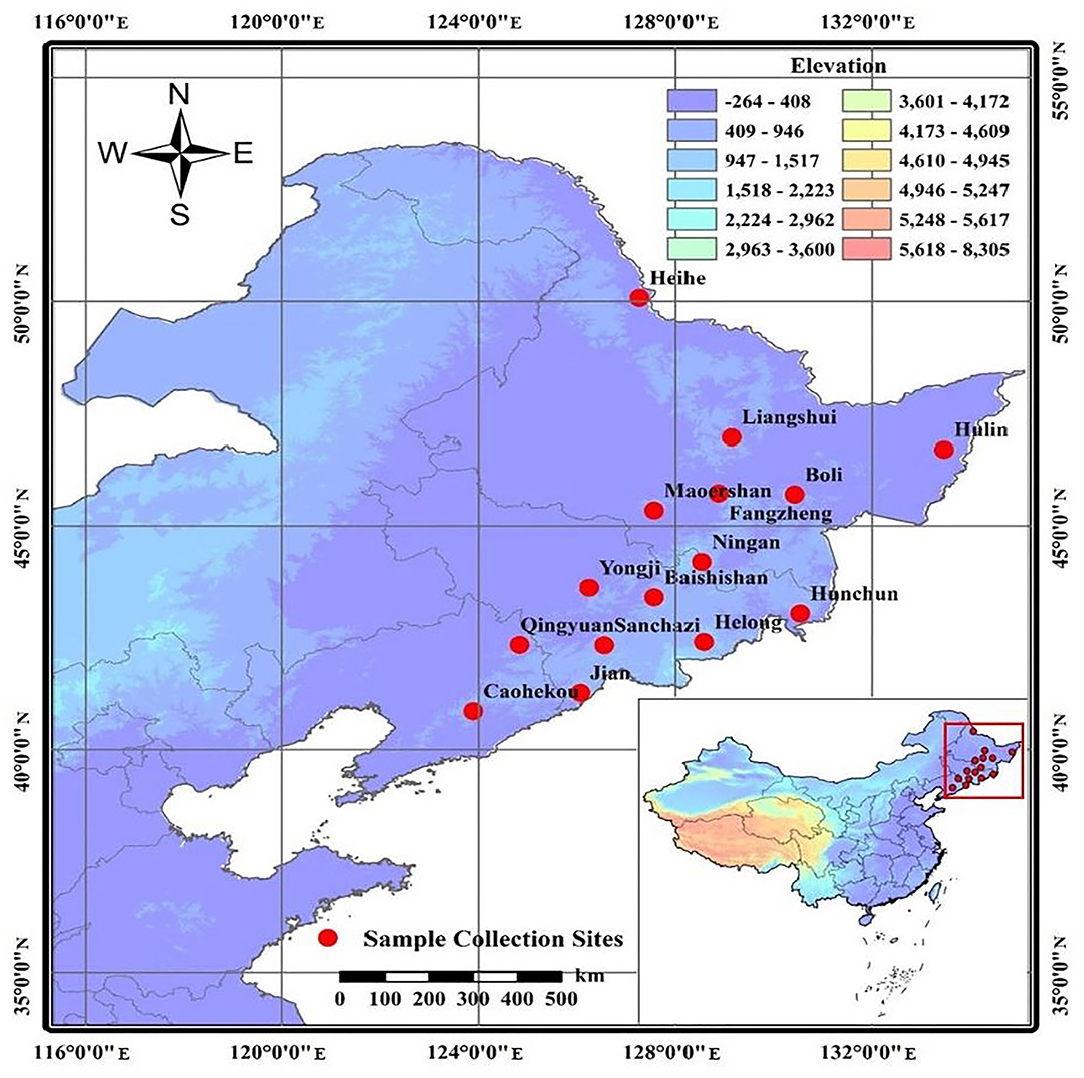
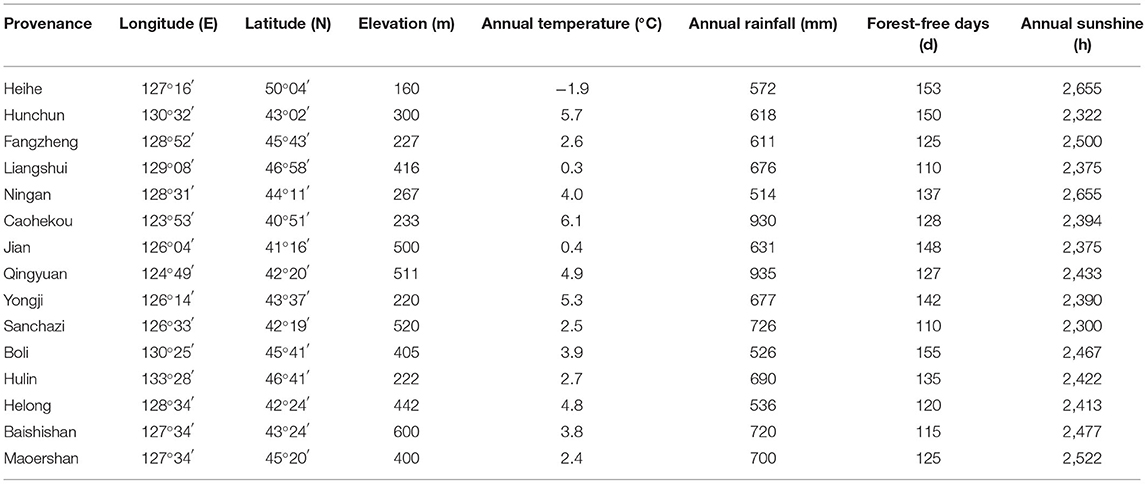
A total of 343 samples from 15 representative J. mandshurica populations were collected from the main distribution areas of Heilongjiang, Jilin, and Liaoning Provinces in Northeast China (Figure 1, Table 1). Sampling was spaced at least 100 m apart. Fresh, pest-free leaves of J. mandshurica were collected and snap-frozen in liquid nitrogen for DNA extraction, which was performed by the CTAB method, as described by Zhao and Woeste (2011), with some modifications. Assessment of DNA quality was performed by 1% agarose gel electrophoresis, while DNA purity and concentration were evaluated by a Micro-Spectrophotometer (KAIAO K5600, Beijing KAIAO Technology Development Co., Ltd., China). Sample DNA solutions were diluted to a working concentration of 35 ng/μL and stored at −20°C.
SSR Primer Development
Three tissue samples (leaves, bark, and fruit pericarp) were collected from a healthy J. mandshurica plant in Harbin, Heilongjiang Province, China. RNA extraction and sequencing were performed by Annaroad Gene Technology (Beijing) Co., Ltd., Beijing, China and Biomarker Technologies Co., Ltd., Beijing, China. Based on transcriptome data, SSRs were detected and localized by the MIcroSAtellite identification tool (MISA) (http://pgrc.ipk-gatersleben.de/misa/) (Patil et al., 2020). Minimum SSR motif repeats were identified by 10 mononucleotides, six dinucleotides, five trinucleotides, tetranucleotide, pentanucleotide, and hexanucleotide motif repeats. Primer 3 version 4.1.0 (https://bioinfo.ut.ee/primer3/) (Preethi et al., 2020) was used to design the primers. Primer length ranged from 18 to 24 bp, GC contents ranged from 40 to 60%, annealing temperatures ranged from 55 to 60°C, and the size of PCR products ranged from 100 to 350 bp.
Primer Screening and PCR Amplification
A total of 240 primer pair sequences were randomly selected and synthesized (Tsingke Biotechnology Co., Ltd., Herbin, China) (Supplementary Table S1), and a universal M13 sequence (5′-TGTAAAACGACGGGCCAGT-3′) labeled with four fluorescent dyes (HEX, ROX, FAM, and TAMRA) was added in front of the 5′ end of the forward primers (Li et al., 2020). To screen the primers, 16 samples were randomly selected from eight populations for PCR amplification. The reaction system (20 μL) contained 10 μL of 2× Super PCR Mix (Beijing Genomics Institution, Beijing, China), 0.2 μL of forward primer (10 μM), 0.8 μL of reverse primer (10 μM), 0.5 μL of M13 primer with a fluorescent label (Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China), 2 μL of DNA, and 2.0 μL of ddH2O. The PCR conditions were as follows: predenaturation at 94°C for 4 min; 30 cycles of denaturation at 94°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 30 s; and a final extension step at 72°C for 10 min. All PCR products were sent to Sangon Biotech (Shanghai) Co., Ltd., Shanghai, China for capillary electrophoresis sequencing. Ultimately, 15 pairs of polymorphic EST-SSRs were selected and characterized (Table 2), and 15 primer pairs were used to analyze the genetic diversity of 343 samples from 15 natural J. mandshurica populations. The different Mantel tests were performed with the R package “vegan” and were based on 1,000 permutations.
Statistical Analysis
For subsequent analyses, the primary microsatellite data obtained by capillary electrophoresis were converted into GenePOP format using the MS-Tools program (Li et al., 2015). The total number of alleles (Na), effective number of alleles (Ne), Shannon information index (I), observed heterozygosity (Ho), expected heterozygosity (He), overall fixation index (Fit), inbreeding coefficient (Fis), genetic differentiation coefficient (Fst), gene flow (Nm), number of private alleles (NPA), and Hardy–Weinberg equilibrium (HWE) were calculated by the GenAlEx 6.502 program (Gultyaeva et al., 2019). PIC calc software was used to calculate the polymorphic information content (PIC) (Nagy et al., 2012), while the CONVERT 1.31 program (Marta et al., 2012) was used to convert the GenePOP data format to STRUCTURE data format. The Bayesian clustering method in STRUCTURE 2.3.4 was used to analyze the genetic structure of the J. mandshurica population (Mohammed et al., 2020), while the K value was tested from 2 to 10, with three replications at each K, 100,000 generations burnin period, and 100,000 Markov Chain Monte Carlo (MCMC) repetitions. The LnP (K) and ΔK plots were generated by the online tool Structure Harvester (http://taylor0.biology.ucla.edu/structureHarvester/) (Ravelombola et al., 2018), and the best K value was selected using the ΔK maximum value principle. Nei's (1983) genetic distances between populations were calculated by PowerMarker 3.25 program (Poudel et al., 2019), and the computed distances were used for the construction of phylogenetic trees by using the neighbor-joining method (NJ). Annotation and visualization of phylogenetic trees were performed using Mega 7.0.26 software and the Interactive Tree of Life online tool (https://itol.embl.de) (Letunic and Bork, 2021). Analysis of molecular variance (AMOVA) and principal coordinate analysis (PCoA) were performed using GeneAlEx 6.502 software (Gultyaeva et al., 2019).
Results
EST-SSR Loci Detection
A total of 63,552 EST-SSR loci were detected based on the transcriptome data of J. mandshurica. Among them, mononucleotide repeat units were the most abundant, accounting for 55.80% of all SSRs, followed by dinucleotides and trinucleotides with 32.14 and 10.43%, respectively (Figure 2A). The lowest number of tandem repeats was 5 (4,292, 6.75%), with 10 tandem repeats accounting for the highest percentage (13,382, 21.06%), followed by 11 (7,384, 11.62%), 6 (6,600, 10.39%), and 12 (5,949, 9.36%) repeats (Figure 2D). In addition, there were 40 sequence types of 63,552 EST-SSRs, among which the A/T mononucleotide motif (34,503, 5.29%) was the most dominant, followed by AG/CT (7,529, 11.85%) and GA/TC (7,294, 11.48%) motifs (Figures 2B,C).
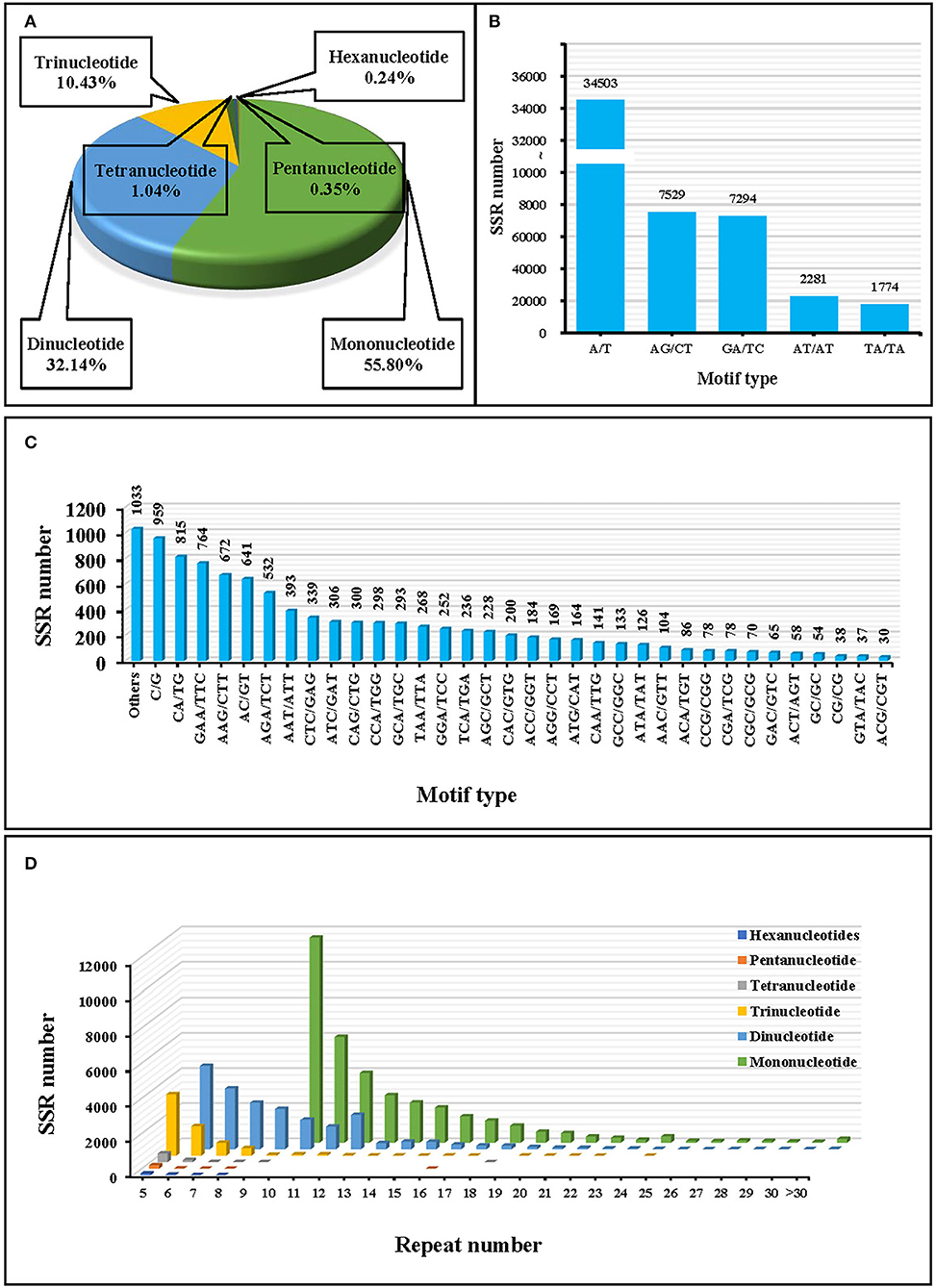
Figure 2. Statistics of simple sequence repeats (SSRs) in J. mandshurica. (A) Statistics of the proportion of different motif types. (B,C) Statistics of the SSR numbers of different motif types. (D) Statistics of the number of repeat types for different motifs.
Polymorphic EST-SSR Primer Development
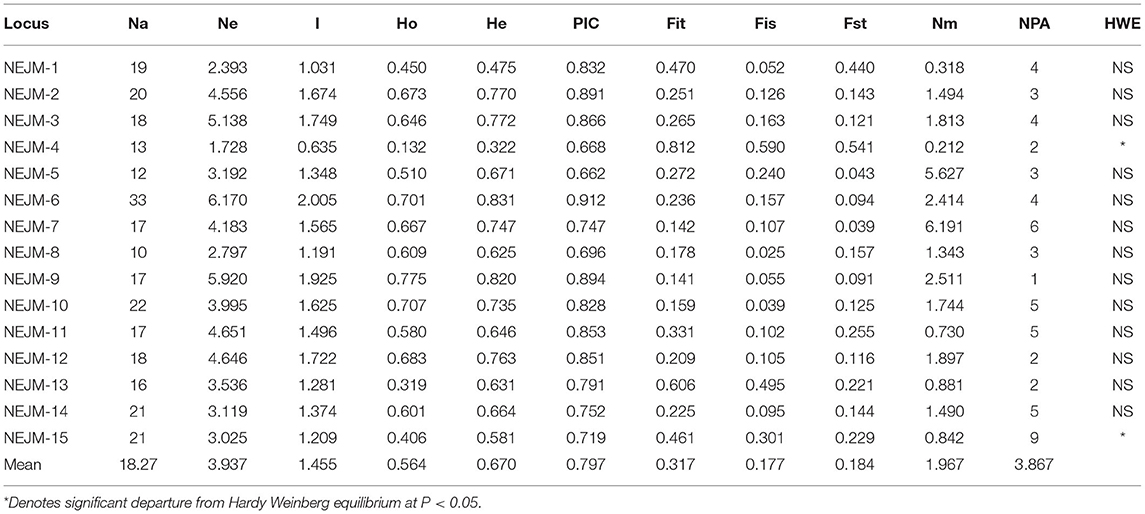
A total of 240 EST-SSRs were randomly selected and synthesized for polymorphism investigation (Table 3). A total of 274 alleles were detected at the 15 EST-SSR loci. The Na per locus varied from 10 (NEJM-8) to 33 (NEJM-6), with a mean of 18.27, whereas the Ne ranged from 1.728 (NEJM-4) to 6.170 (NEJM-6) with a mean of 3.937. The I ranged from 0.635 (NEJM-4) to 2.005 (NEJM-6) with a mean of 1.455. The Ho ranged from 0.132 (NEJM-4) to 0.775 (NEJM-9) with a mean of 0.564; He ranged from 0.322 (NEJM-4) to 0.831 (NEJM-6) with a mean of 0.670. The PIC ranged from 0.662 (NEJM-5) to 0.912 (NEJM-6) with a mean of 0.797. The Fit ranged from 0.141 (NEJM-9) to 0.812 (NEJM-4) with a mean of 0.317; Fis ranged from 0.025 (NEJM-8) to 0.590 (NEJM-4) with a mean of 0.177; Fst ranged from 0.039 (NEJM-7) to 0.541 (NEJM-4) with a mean of 0.184; Nm ranged from 0.212 (NEJM-4) to 6.191 (NEJM-7) with a mean of 1.967; and NPA ranged from 1 (NEJM-9) to 9 (NEJM-15) with a mean of 3.867, accounting for 21.17% of Na.
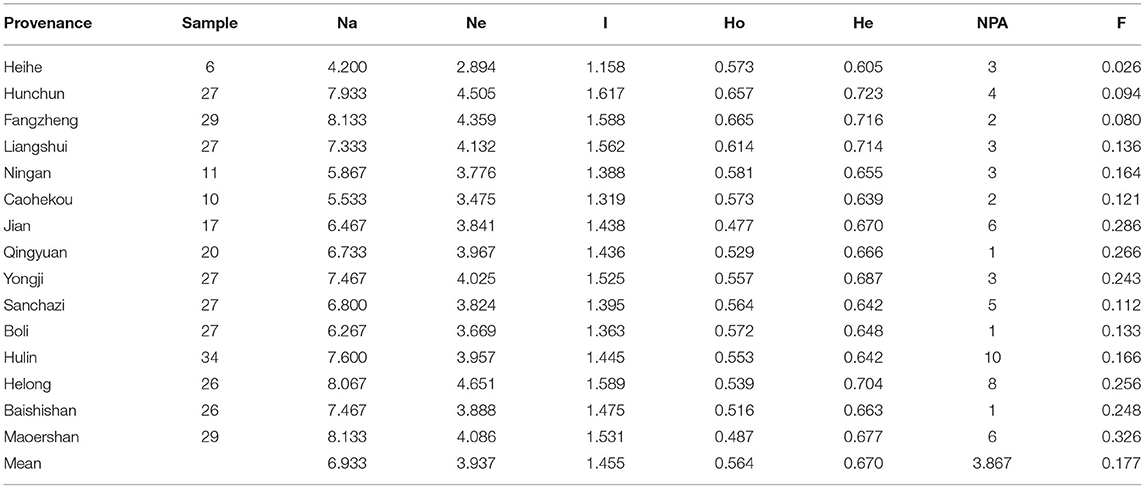
Genetic Diversity of the J. mandshurica Population
Genetic diversity analysis was performed on 343 individuals from 15 populations of J. mandshurica using the primers shown in Table 4. The range of variations in Na among the different populations was from 4.200 (Heihe) to 8.133 (Fangzheng and Maoershan), and the mean value was 6.933. For Ne, the range was from 2.894 (Heihe) to 4.651 (Helong), with an average of 3.937. For I, variations ranged from 1.158 (Heihe) to 1.617 (Hunchun), with a mean value of 1.455. Variations in Ho ranged from 0.477 (Jian) to 0.665 (Fangzheng), with a mean of 0.564, whereas variations in He ranged from 0.605 (Heihe) to 0.723 (Hunchun), with a mean value of 0.670. NPA varied from 1 (Qingyuan, Boli and Baishishan) to 10 (Hulin) with a mean value of 3.867. The fixation index (F) varied from 0.026 (Heihe) to 0.326 (Maoershan) with a mean value of 0.177. In this study, F > 0 indicates the absence of heterozygotes and excess of pure heterozygotes in the J. mandshurica population and the presence of inbreeding. Furthermore, the Hunchun population showed high genetic diversity (Na = 7.933, He = 0.723, and I = 1.617), whereas the Heihe population exhibited a relatively low level of genetic diversity (Na = 4.200, He = 0.605, and I = 1.158).
Genetic Structure of the J. mandshurica Population
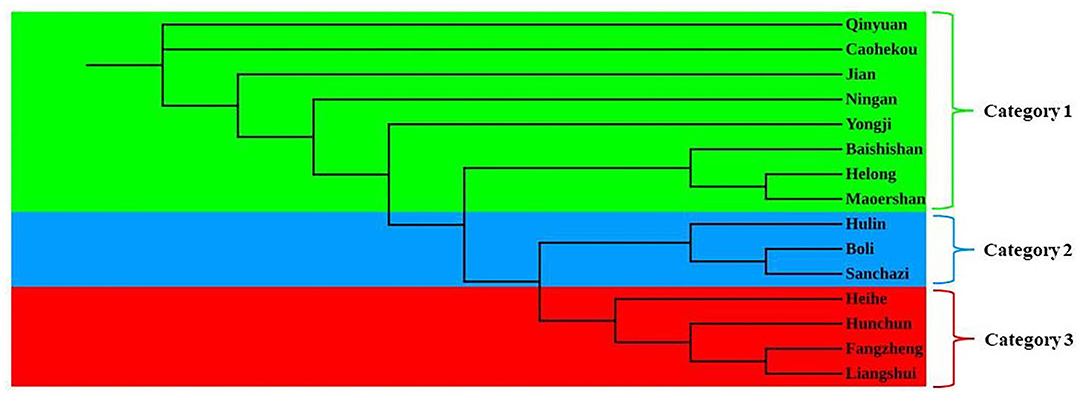
The genetic structure of the J. mandshurica population in Northeast China was analyzed using the STRUCTURE program. The optimal number of clusters (K) was determined by comparing the LnP (D) and ΔK based on the rate of change in LnP (D). The LnP (D) value increased continuously with the number of clusters (K) from 2 to 10 based on estimated posterior probability of genotype data, and the inflection point appears when K = 3 and K = 5 (Figure 3A). Two pronounced peaks of ΔK were observed (Figure 3B). Because the structure analysis cannot clearly classify the test samples into five categories when K = 5, K = 3 was used as the optimal K value. Based on this value, 343 individuals of J. mandshurica were grouped into three genetic categories (category 1 in green, category 2 in blue, and category 3 in yellow) (Figure 3C). Category 1 consisted of 147 individuals from eight populations for Qingyuan, Caohekou, Jian, Ningan, Yongji, Baishishan, Helong, and Maoershan; category 2 consisted of 81 individuals from three populations for Hulin, Boli, and Sanchazi; and category 3 consisted of 115 individuals from four populations for Heihe, Hunchun, Fangzheng, and Liangshui.
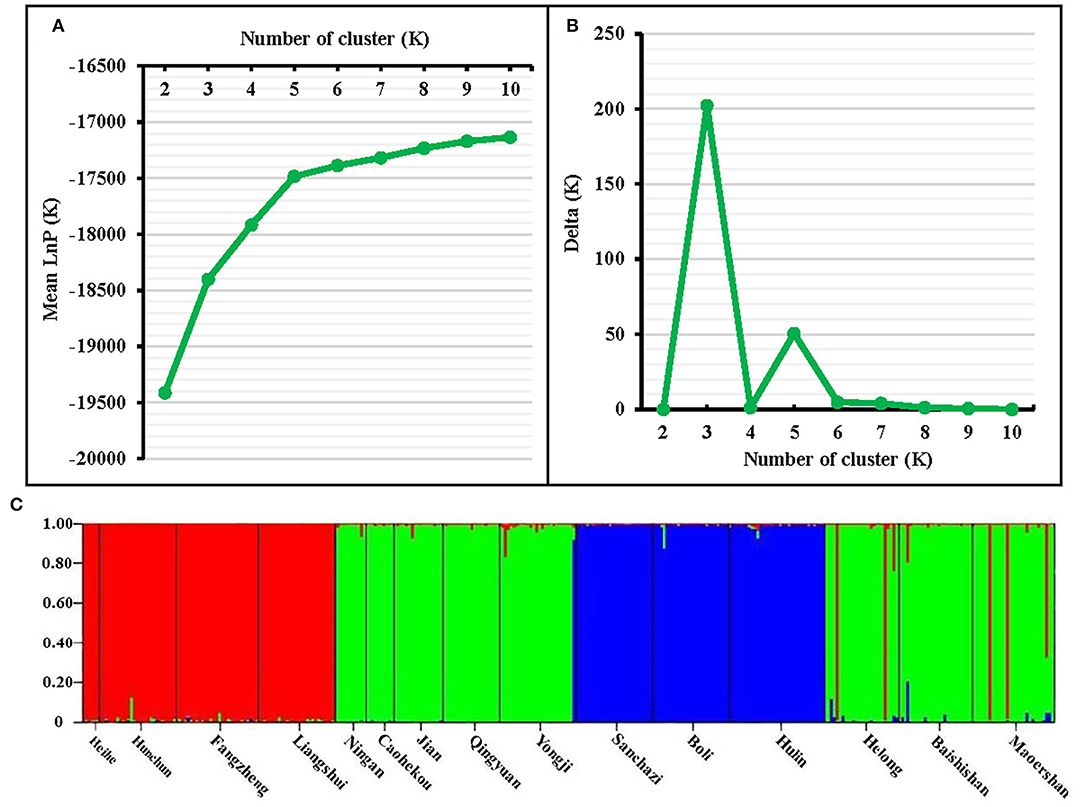
Figure 3. STRUCTURE analysis of the natural population of J. mandshurica based on 15 EST-SSRs. (A) Calculation of population structure using Mean LnP (K). (B) Relations between the optional number of cluster K and Delta K. (C) Genetic structure map of 15 natural populations of J. mandshurica based on STRUCTURE analysis (K = 3).
Principal Coordinates and Evolutionary Tree Analysis of the J. mandshurica Population
To further analyze the genetic relationships among natural populations of J. mandshurica, principal coordinate analysis (PCoA) was performed using Nei's genetic distances of 343 individuals from 15 populations (Figure 4). The PC1 horizontal axis explained 13.90% of the total variations, while the PC2 vertical axis explained 5.86% of the total variations. The two principal axes explained 19.76% of the total genetic variation. The 15 natural populations were divided into three categories, which were generally consistent with the results obtained in the population structure analysis: category 1 included the Qingyuan, Caohekou, Jian, Ningan, Yongji, Baishishan, Helong, and Maoershan populations; category 2 included the Hulin, Boli, and Sanchazi populations; and category 3 included the Heihe, Fangzheng, and Liangshui populations. Nei's genetic distance was used to construct a neighbor-joining dendrogram (Figures 5, 6) in which all samples were clustered into three categories. This was inconsistent with the aforementioned results, and some individuals were assigned to different categories (Figure 6).
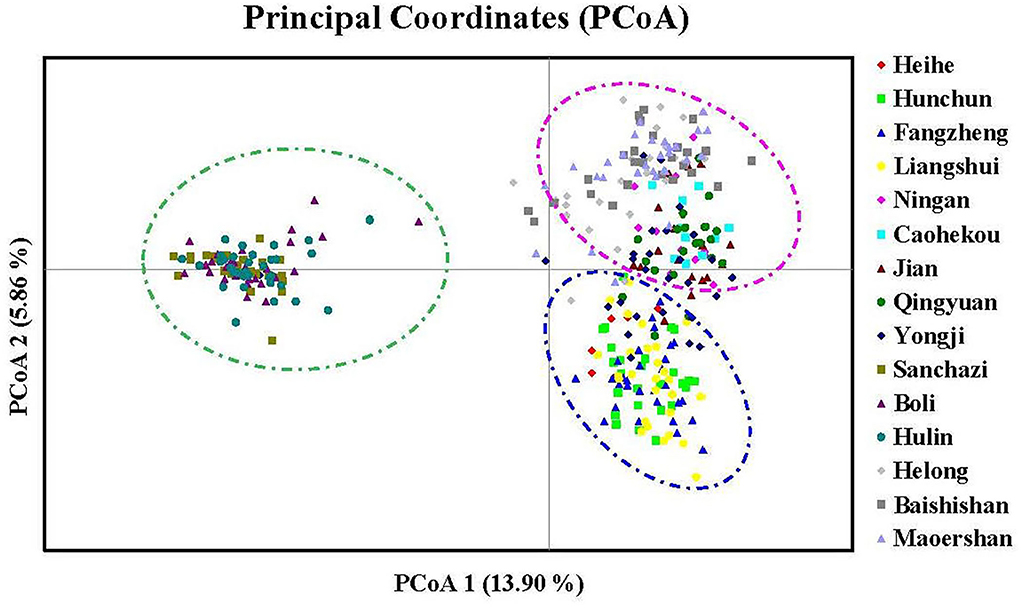
Figure 4. Principal coordinate analysis (PCoA) based on the genetic distance of 343 individuals for J. mandshurica.
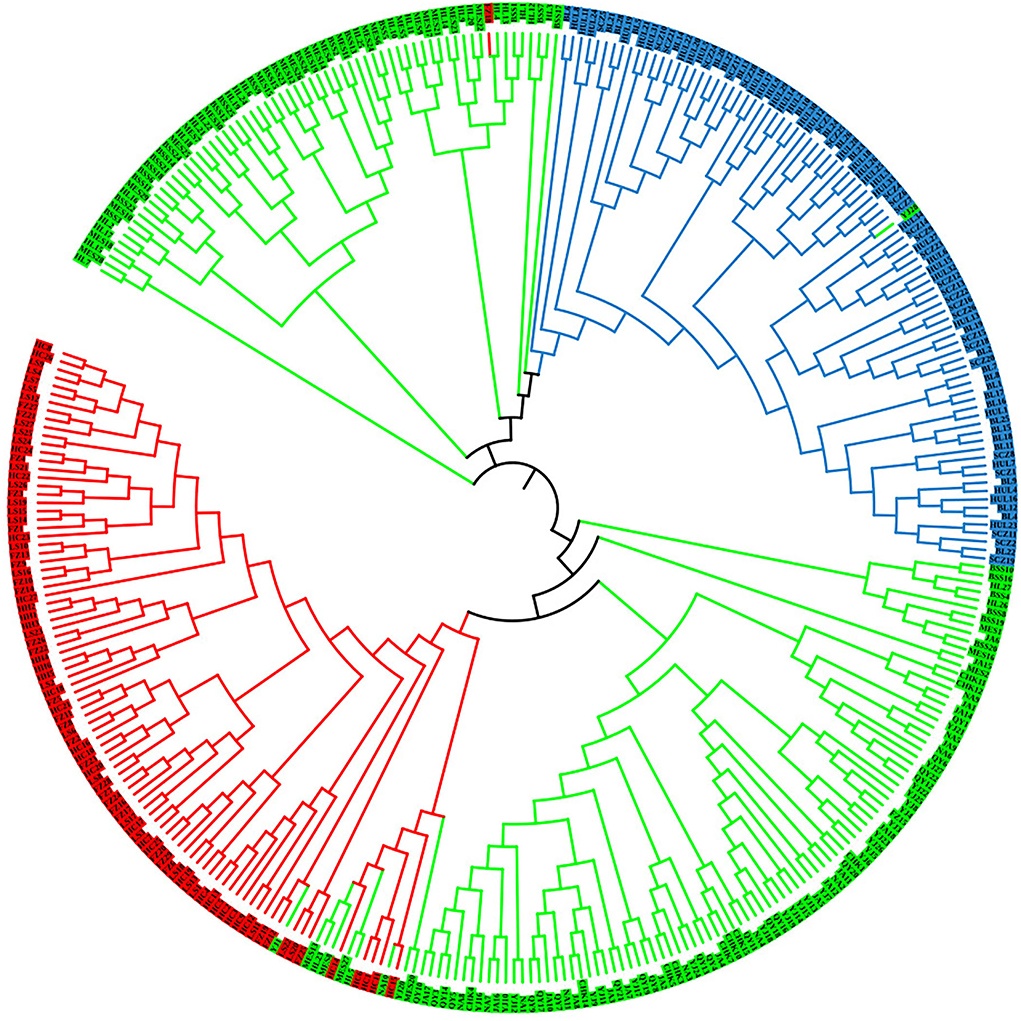
Figure 6. Neighbor-joining tree of 343 J. mandshurica individuals (HH: Heihe; HC: Hunchun; FZ: Fangzheng; LS: Liangshui; NA: Ningan; CHK: Caohekou; JA: Jian; QY: Qingyuan; YJ: Yongji; SCZ: Sanchazi; BL: Boli; HUL: Hulin; HL: Helong; BSS: Baishishan; and MES: Maoershan).
Genetic Differentiation and Gene Flow of the J. mandshurica Population
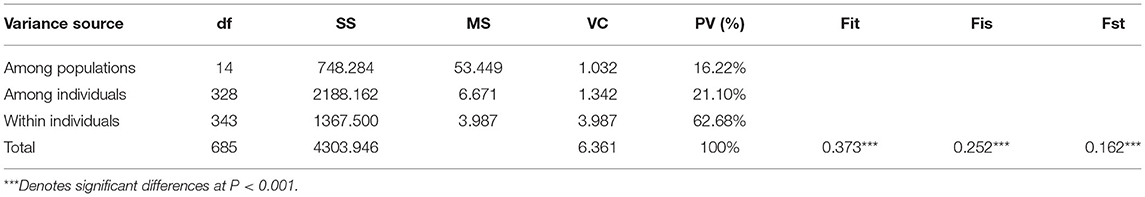
Genetic variation among different populations of J. mandshurica was investigated using the AMOVA tool (Table 5). The obtained results revealed significant genetic differentiation at the 0.01 level among the populations (Fst = 0.162, P < 0.001). Moreover, 21.10 and 62.68% of the genetic variations were attributed to variations among individuals and within individuals, respectively, whereas variations among populations accounted for the total genetic variance of 16.22%, indicating that genetic variations within populations were much higher compared to genetic differentiation among populations. The Fst value was 0.162 (0.15 < Fst < 0.25), indicating a moderate level of genetic differentiation among populations. The Fis value was 0.252 (Fis > 0), indicating the loss of heterozygotes and the presence of inbreeding between populations. The Fst (Figure 7, lower left) and Nm (Figure 7, upper right) values among 15 populations of J. mandshurica were calculated. The data revealed genetic differences and gene flow among different populations of J. mandshurica. The Fst value varied from 0.011 to 0.208 with a mean value of 0.109, and the Nm value varied from 0.950 to 22.422 with a mean value of 4.063 among the different populations. The highest Fst and the lowest Nm values were found between the Sanchazi and Caohekou populations, followed by the Hulin and Caohekou populations. The lowest Fst and the highest Nm values were found between the Sanchazi and Hulin populations.
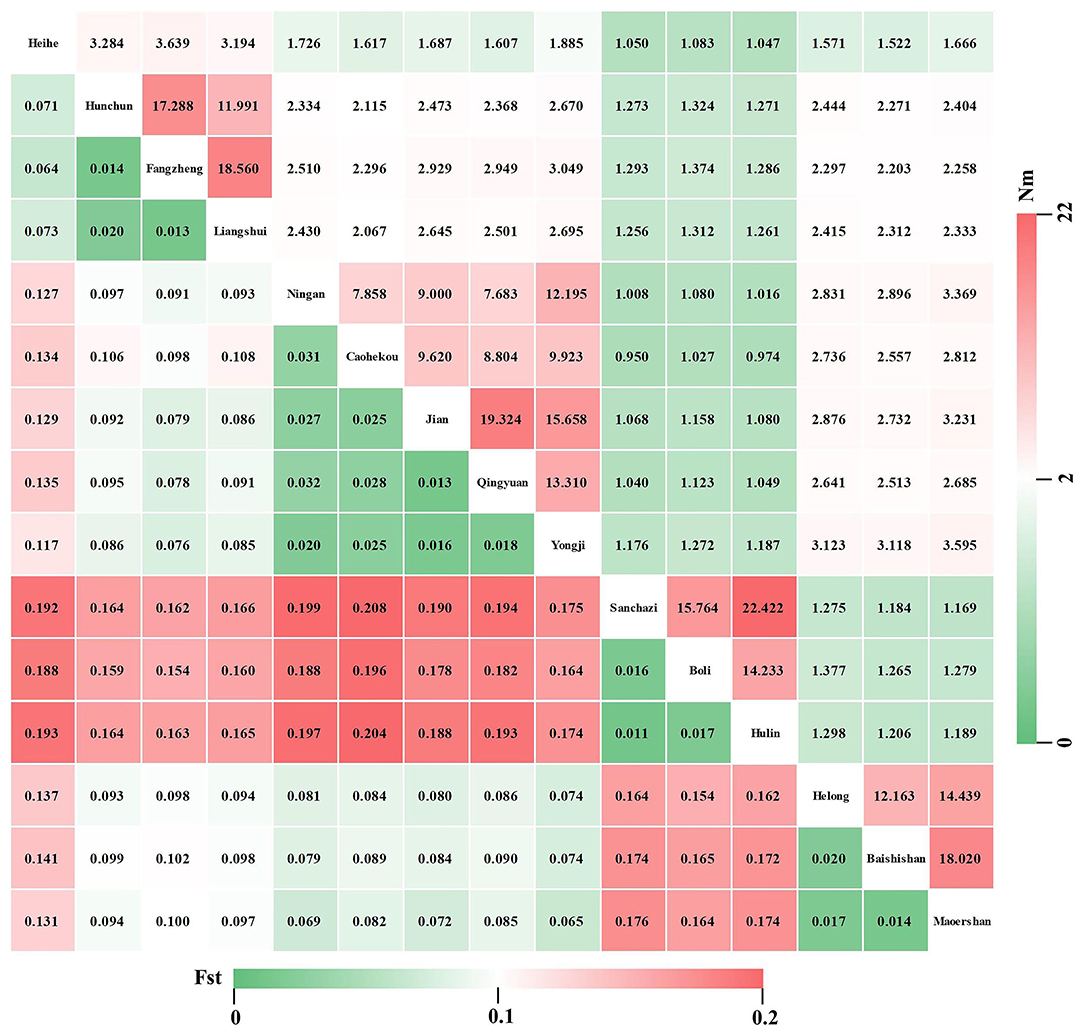
Figure 7. Genetic differentiation coefficient (lower left) and gene flow (upper right) among different populations of J. mandshurica.
Genetic Distance and Genetic Uniformity of the J. mandshurica Population
Nei's genetic distances were used to depict the genetic relationships among the 15 J. mandshurica populations, and a smaller genetic distance and larger genetic uniformity indicate a close genetic relationship. In this study, pairwise population genetic distance ranged from 0.073 to 0.743 (Figure 8, lower left), and the genetic uniformity between populations ranged from 0.156 to 0.958 (Figure 8, upper right). The maximum genetic distance appeared between the Sanchazi and Heihe populations, suggesting that they are not closely related. This was followed by the Heihe and Boli populations, and the minimum genetic distance was observed between the Sanchazi and Hulin populations, suggesting that the two populations are closely related.

Figure 8. Nei's genetic distance (lower left) and genetic uniformity (upper right) among different populations of J. mandshurica.
The Correlations Between the Molecular Traits and Environmental Factors
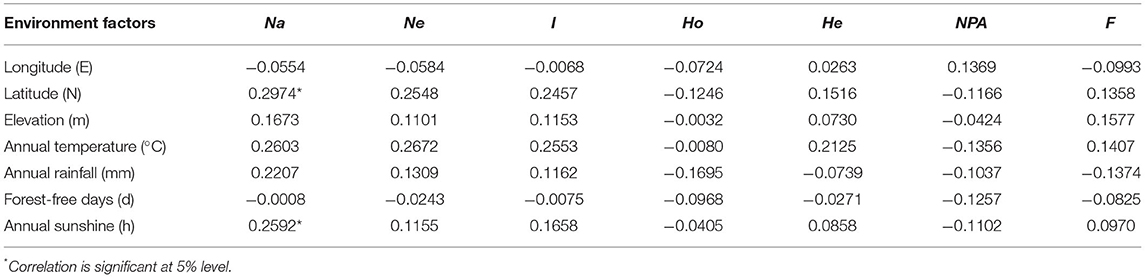
Mantel tests were performed to explore the correlations between molecular traits and environmental factors. The results showed that except for Na, no significant correlation was observed between genetic diversity parameters and environmental factors (Table 6). Na had a significantly positive correlation with latitude (r = 0.2974) and annual sunshine (r = 0.2592), but with weak correlation coefficients. These results indicate that the molecular traits of J. mandshurica populations were less influenced by environmental factors.
Discussion
Due to the challenges associated with asexual reproduction and with most of the J. mandshurica resources basically existing in a wild or semi-wild state, genetic improvement of J. mandshurica is lagging (Zhang et al., 2021). To develop a reasonable, effective breeding strategy and accelerate genetic improvements of J. mandshurica, a comprehensive evaluation of J. mandshurica resources in natural populations is important. We used the EST-SSR molecular marker to analyze the genetic diversity of J. mandshurica populations in Northeast China. Compared to other molecular marker technologies, this molecular marker is characterized by codominance, stable amplification, and good repeatability. It is a commonly used genetic diversity analysis method (Rahemi et al., 2012). Our findings have positive implications for conservation and genetic improvements of J. mandshurica resources in Northeast China.
EST-SSR Primer Development
The EST-SSRs have been extensively studied in the plant, animal, and microbial species. Compared to genomic SSRs, EST-SSRs have many advantages, including low sequencing costs, identification of the genetic relationship between similar species, and analysis of population genetics (Durand et al., 2010). We found 63,552 EST-SSR loci in the transcriptome sequence of J. mandshurica. Then, 240 loci were randomly selected to synthesize primers, and 15 pairs of highly polymorphic EST-SSR primers were finally obtained after screening. In the transcriptome of J. mandshurica, among the six nucleotide repeat types, mononucleotide and dinucleotide repeat motifs were the most abundant, accounting for 55.80 and 32.14%, respectively. Distributions of EST-SSR repeat motifs for J. mandshurica are comparable to those of Ziziphus jujuba (Xiao et al., 2015), Sesamum indicum (Uncu et al., 2015), and Tectona grandis (Yasodha et al., 2018). SSR repeat motifs are associated with the level of species evolution, and the content of short repeat motifs increases with the time of species evolution or variation frequency (Liu et al., 2020). In this study, the proportion of short repeat motifs was relatively high, indicating a high species variation frequency of J. mandshurica. Mononucleotide repeats (A/T) were the most abundant motif class, followed by dinucleotide repeats (AG/CT), while the most common trinucleotide repeats were GAA/TTC and AAG/CTT. Comparable findings have been reported for Vigna angularis (Chen et al., 2015) and J. sigillata (Feng et al., 2018). In most species, dinucleotides and trinucleotides are the most common types of transcriptomic SSR repeat motifs (Kantety et al., 2002; Varshney et al., 2002), which is confirmed by the results of this study.
Initially, we screened 15 EST-SSR loci, and the mean values for Na, He, and PIC for the 15 loci were 18.27, 0.670, and 0.797, respectively. The polymorphism level of the loci for J. mandshurica was higher than that reported by Hu et al., 2016 for the same plant and higher than that of J. regia (Torokeldiev et al., 2019), J. cathayensis (Dang et al., 2015), J. hopeiensis (Hu et al., 2015), and J. sigillata (Feng et al., 2018). PIC is an important parameter that expresses the degree of genetic diversity among plants, and its evaluation is beneficial for the establishment of plant gene pools and accelerating the breeding process (Avval, 2017). The PICs for the EST-SSR loci screened in this study were all >0.5, implying highly polymorphic loci (Botstein et al., 1980). Therefore, the EST-SSR primers developed in this study are highly polymorphic and can be used for genetic diversity evaluation of J. mandshurica breeding resources.
Population Genetic Diversity
Genetic diversity is the product of the long-term evolution of species and is a key factor in population survival and evolution (Barrett and Kidwell, 1998; González et al., 2020). Genetic diversity studies have important implications for forest breeding and the management of endangered tree species (Millar and Westfall, 1992). Heterozygosity is often used to measure the degree of genetic variation and can provide useful information for the protection of endangered species (Schmidt et al., 2021). In this study, the results based on EST-SSR markers showed that the He and Ho of 15 J. mandshurica populations were 0.670 and 0.564, respectively, which was similar to previous research results (Liu, 2015) and higher than those of J. sigillata (Chen et al., 2020) and Amygdalus Mira (Bao et al., 2018), and the level of genetic diversity was relatively high but lower than that of J. regia (Shamlu et al., 2018; Guney et al., 2021). Many factors, including mating systems, evolutionary history, and the level of environmental heterogeneity, influence the level of genetic diversity (Booy et al., 2000). As a tertiary relict plant, J. mandshurica is mostly distributed in the eastern mountains of Northeast China (Tang et al., 2019), and the mutations accumulated from a longer evolutionary history have led to a rich genetic basis for the peccary. Phylogeographical studies have shown that J. mandshurica had two separate refugia during the Quaternary ice age and that there was asymmetric gene flow (Bai et al., 2010). The existing surviving populations may have retained and inherited an overall rich genetic base of ancestors, giving them a high genetic diversity. The results of Wang et al. (2016) on the phylogeography of J. mandshurica also prove this point. Biologically, J. mandshurica is a hermaphroditic heterozygous species (Zhang et al., 2019), which is mainly outcrossing. It can produce new genotypes through sexual recombination. The heterodichogamy flowering mechanism effectively prevents self-pollination in J. mandshurica. These are probably the main reasons why the current population of J. mandshurica still maintains a high level of genetic diversity (Nybom and Bartish, 2000). Additionally, the Ho value was found to be lower than the He value in all populations, indicating heterozygous deletion. Inbreeding, non-random mating, or population structure destruction may lead to the lack of heterozygotes (Liao et al., 2019). Thus, further analysis is warranted.
Genetic diversity analysis of the 15 J. mandshurica populations revealed various differences. The Hunchun population had the highest genetic diversity level (Na = 7.933, He = 0.723, and I = 1.158), while the Heihe population had the lowest (Na = 4.200, He = 0.605, and I = 1.617). Bai et al. (2010) reported that there may be two refugia in the distribution area of J. mandshurica, one of which is in the northeast and is mainly distributed in Changbai Mountain. Changbai Mountain is a scenic tourist site with frequent human activities, which may affect the population of J. mandshurica, leading to plant habitat fragmentation (Aguilar et al., 2008). Fragmented plant habitats inhibit pollen dispersal and gene flow between populations, thereby affecting genetic diversity in the population (Vranckx et al., 2012). The Hunchun population is located near the Northeast refuge, which is not a tourist city. The existence of refuge and less human disturbance may be the reason for the high genetic diversity of the Hunchun population. The Heihe population is located in the northernmost part of China and is distant from other J. mandshurica populations. During field investigations, few J. mandshurica were found in this population, yet J. mandshurica is a tall tree that is suitable for wind-induced cross-pollination. Marked environmental differences and population isolation may destroy the coadaptive allele combination of parents and reduce the adaptability of distant offspring (Hufford et al., 2012). The fixed index (F) for both populations was small but >0, indicating an excess of pure heterozygotes in both populations and a slight inbreeding phenomenon (Wright, 1950).
Environmental factors, such as temperature, elevation, and rainfall, may influence the genetic variation of forests. We used the Mantel test to study the relationship between molecular traits of J. mandshurica populations and environmental factors. However, no clear geographical variation pattern was observed in molecular diversity. This is different from what was found in the studies of other species (Hu et al., 2010; Li et al., 2013; Zhang et al., 2017). This finding indicates that the geographic variation at the gene level in the natural populations of J. mandshurica may be located with some randomness and is less influenced by the environment, the specific reasons for which need to be further investigated.
Population Genetic Structure
The genetic structure reveals the distribution pattern of genetic diversity among and within populations, reflecting the adaptive potential of various species to the environment (Melo et al., 2014). The 15 natural populations of J. mandshurica in this study can be divided into three categories: category 1 denotes the Changbai Mountain area, category 2 denotes the Wanda Mountain area, and category 3 denotes the Lesser Khingan Mountain area. Comparable findings were obtained by neighbor-joining method cluster analysis and principal coordinate analysis. The Sanchazi population, which is located in the Changbai Mountain area, is not included in category 1. The reason may be that the Sanchazi population is located in Jingyu County, Jilin Province, which was affected by the war. Human factors lead to fragmentation of the population's habitat and inhibit gene exchange (Haag et al., 2010). In addition, relatively isolated or dispersed populations may also reduce the success rate of pollination (Ghazoul, 2005), thereby affecting the genetic structure.
Population Genetic Differentiation
To elucidate the genetic variations among J. mandshurica populations, AMOVA was conducted. Genetic variations in J. mandshurica were mainly found at the individual level, accounting for 63% of the total genetic variation. Genetic variations among populations accounted for only 16%, indicating that within-population genetic differentiation is greater than among populations, which is consistent with the findings of Wang (2007), Wang et al. (2011), Dang et al. (2021), and Zhang et al. (2021). This may be attributed to the life history and habitat of J. mandshurica. J. mandshurica is a heterodichogamy deciduous species whose cross-pollination feature results in a high gene flow within the population (Hamrick and Godt, 1996) and a high degree of heterozygosity, thereby improving genetic diversity within the population and reducing the degree of differentiation among populations (Hao et al., 2017). Therefore, when selecting J. mandshurica populations with high genetic diversity, the focus should be on the selection of individuals within the population, which is beneficial for the genetic improvement of J. mandshurica.
The detection of genetic differentiation is a key to promote the genetic improvement of forests (Li et al., 2021). Fst measures the degree of differentiation among populations and is a widely used descriptive statistical indicator in population and evolutionary genetics (Holsinger and Weir, 2009). In this study, the mean Fst for the J. mandshurica population was 0.109, implying a moderate degree of genetic differentiation (Wright, 1950). This is higher than that reported by Wang et al. (2016). Differences may be attributed to varying research materials and different selection pressures of gene fragments where the selected SSRs were located (Hu et al., 2016). Gene flow (Nm) refers to the movement of genes within and among populations, which in the plant kingdom is mainly achieved through the migration or movement of genetic material carriers, such as pollen, seeds, spores, and vegetative bodies. It affects the degree of genetic variation in the population (Zhang et al., 2003). The effects of genetic drift and genetic differentiation can be effectively eliminated when gene flow is >1 (Slatkin, 1985). In this study, the average Nm of the J. mandshurica population was 4.063, which is relatively high when compared to the results of Wang et al. (2011). J. mandshurica is pollinated by the wind; therefore, in this study, J. mandshurica may have achieved gene exchange in different geographical locations through long-distance pollen flow (Victory et al., 2006), thereby eliminating geographic isolation and reducing genetic differentiation among populations. Genetic distance and genetic uniformity are indicators of genetic relationships among populations or individuals, which can be further used to analyze the degree of genetic differentiation among the 15 populations. In this study, the variation range of genetic uniformity among populations was 0.156–0.958, with an average of 0.524, which is lower than those reported by Wang (0.83 and 0.9217 in 2007 and 2011, respectively). The average genetic distance was 0.393, which is higher than that of Zhang (0.0049) (2021). The results show great differences, indicating that genetic relationships among J. mandshurica populations in this study are relatively distantly related. The differences in genetic uniformity and genetic distance among different populations indicate great fluctuations in genetic variations.
Genetic Conservation Strategy
Studies on genetic diversity and genetic structure are prerequisites for formulating species protection measures (Newton et al., 1999; Wuyun et al., 2015). J. mandshurica is a native tree species in Northeast China. It has been listed as a local protected wild plant in Jilin Province, Hebei Province, and Beijing; however, only a nature reserve has been established in Changbai Mountain, and no measures have been undertaken in other distribution areas where it is still the main cut tree species (http://www.iplant.cn/rep/). We show that the J. mandshurica population in Northeast China has high genetic diversity, indicating that the endangerment of J. mandshurica is not the cause of genetic variation but is mainly affected by human disturbance and habitat destruction and degradation (Wang et al., 2011). In this study, the genetic diversity of the Heihe population was relatively low, and the number of individuals was small. Botanical gardens or arboretums can be used for ex situ protection of living J. mandshurica in this area. The genetic diversity of the Hunchun population is relatively high and can be protected by establishing a nature reserve. In addition, the germplasm resources of J. mandshurica should be systematically collected. Given that there are challenges in asexual propagation strategies, such as cuttings, grafting, and tissue cultures of J. mandshurica, the Germplasm Resources Bank of J. mandshurica can be established by collecting seeds. On this basis, combined with whole-genome information of J. mandshurica (Bai et al., 2018; Yan et al., 2021), SNP and other molecular marker technologies should be used to perform molecular marker-assisted breeding and whole-genome selection breeding to accelerate the progress of genetic improvement for J. mandshurica.
Conclusion
We developed 15 EST-SSR primers using the transcriptome data of J. mandshurica, and 15 loci exhibited high levels of genetic polymorphism, which can be used for genetic diversity evaluation of J. mandshurica populations. Genetic diversity analysis was performed on 343 individuals from 15 natural populations of J. mandshurica in Northeast China using 15 EST-SSR primers. The J. mandshurica populations exhibited high levels of genetic diversity and moderate genetic differentiation. Based on STRUCTURE analysis, neighbor-joining method cluster analysis, and principal coordinate analysis, the 15 populations can be divided into three categories: Changbai Mountain area, Wanda Mountain area, and Lesser Khingan Mountain area. With regard to the characteristics of the J. mandshurica population, there is a need to formulate corresponding genetic conservation strategies to maintain the genetic diversity of the J. mandshurica population in Northeast China to the greatest extent possible and to provide a basis for the subsequent development and utilization of important genetic resources of J. mandshurica.
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: https://www.ncbi.nlm.nih.gov/. Transcriptome data of different tissues for Juglans mandshurica PRJNA816294.
Author Contributions
Conceptualization: QZ and XZhao. Methodology: XZhan and XP. Validation: YY. Resources: LX, JF, JW, and YT. Writing—original draft preparation: QZ. Writing—review and editing: XZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Fundamental Research Funds for the Central Universities (No. 2572021AW31) and the Scientific Research Start-up Funds of Jilin Agricultural University (No. 2021002).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.931578/full#supplementary-material
References
Aguilar, R., Quesada, M., Ashworth, L., Herreriasdiego, Y., and Lobo, J. (2008). Genetic consequences of habitat fragmentation in plant populations: susceptible signals in plant traits and methodological approaches. Mol. Ecol. 17, 5177–5188. doi: 10.1111/j.1365-294X.2008.03971.x
Avval, S. E. (2017). Assessing polymorphism information content (PIC) using SSR molecular markers on local species of Citrullus colocynthis. Case study: Iran, Sistan-Balouchestan province. J. Mol. Biol. Res. 7, 42–49. doi: 10.5539/jmbr.v7n1p42
Bai, W. N., Liao, W. J., and Zhang, D. Y. (2010). Nuclear and chloroplast DNA phylogeography reveal two refuge areas with asymmetrical gene flow in a temperate walnut tree from East Asia. New Phytol. 188, 892–901. doi: 10.1111/j.1469-8137.2010.03407.x
Bai, W. N., Yan, P. C., Zhang, B. W., Woeste, K. E., Lin, K., and Zhang, D. Y. (2018). Demographically idiosyncratic responses to climate change and rapid Pleistocene diversification of the walnut genus Juglans (Juglandaceae) revealed by whole-genome sequences. New Phytol. 217, 1726–1736.
Bao, W. Q., Wuyun, T. N., Du, H. Y., Li, T. Z., Liu, H. M., Wang, L., et al. (2018). Genetic diversity and population structure of Amygdalus mira in the Tibet plateau in China based on SSR markers. Sci. Silvae Sin. 54, 30–41. doi: 10.11707/j.1001-7488.20180204
Barrett, B. A., and Kidwell, K. K. (1998). AFLP-based genetic diversity assessment among wheat cultivars from the pacific northwest. Crop Sci. 38, 1261–1271. doi: 10.2135/cropsci1998.0011183X003800050025x
Booy, G., Hendriks, R. J. J., Smulders, M. J. M., Groenendael, J. M. V., and Vosman, B. (2000). Genetic diversity and the survival of populations. Plant Biol. 2, 379–395. doi: 10.1055/s-2000-5958
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314.
Chen, H. L., Liu, L. P., Wang, L. X., Wang, S. H., Somta, P., and Chen, X. Z. (2015). Development and validation of EST-SSR markers from the transcriptome of adzuki bean (Vigna angularis). PLoS ONE 10:e0131939. doi: 10.1371/journal.pone.0131939
Chen, S. Y., Wu, T., Xiao, L. J., Ning, D. L., and Pan, L. (2020). Genetic diversity of Juglans sigillata Dode germplasm in Yunnan Province, China, as revealed by SSRs. Plant Genet. Resour. 18, 417–426. doi: 10.1017/S1479262120000441
Chobot, V., and Hadacek, F. (2009). Milieu-dependent pro-and antioxidant activity of juglone may explain linear and nonlinear effects on seedling development. J. Chem. Ecol. 35, 383–390. doi: 10.1007/s10886-009-9609-5
Dang, M., Liu, Z. X., Chen, X., Zhang, T., Zhou, H. J., Hu, Y. H., et al. (2015). Identification, development, and application of 12 polymorphic EST-SSR markers for an endemic Chinese walnut (Juglans cathayensis L.) using next-generation sequencing technology. Biochem. Syst. Ecol. 60, 74–80. doi: 10.1016/j.bse.2015.04.004
Dang, M., Zhou, H. J., Woeste, K. E., Yue, M., Zhang, Y., Zhao, G. F., et al. (2021). Comparative phylogeography of Juglans regia and J. mandshurica combining organellar and nuclear DNA markers to assess genetic diversity and introgression in regions of sympatry. Trees 35, 1993–2007. doi: 10.1007/s00468-021-02167-y
Durand, J., Bodénès, C., Chancerel, E., Frigerio, J. M., Vendramin, G., Sebastiani, F., et al. (2010). A fast and cost-effective approach to develop and map EST-SSR markers: oak as a case study. BMC Genom. 11:570. doi: 10.1186/1471-2164-11-570
El-Esawi, M. A. (2017). SSR analysis of genetic diversity and structure of the germplasm of faba bean (Vicia faba L.). C. R. Biol. 340, 474–480. doi: 10.1016/j.crvi.2017.09.008
Feng, X. J., Yuan, X. Y., Sun, Y. W., Hu, Y. H., Zulfiqar, S., Ouyang, X. H., et al. (2018). Resources for studies of iron walnut (Juglans sigillata) gene expression, genetic diversity, and evolution. Tree Genet. Genom. 14, 1–15. doi: 10.1007/s11295-018-1263-z
Ghazoul, J. (2005). Pollen and seed dispersal among dispersed plants. Biol. Rev. 80, 413–443. doi: 10.1017/S1464793105006731
González, A. V., Gómez-Silva, V., Ram'irez, M. J., and Font'urbel, F. E. (2020). Meta-analysis of the differential effects of habitat fragmentation and degradation on plant genetic diversity. Conserv. Biol. 34, 711–720. doi: 10.1111/cobi.13422
Gultyaeva, E. I., Kazartsev, I. A., and Shaydayuk, E. L. (2019). Molecular-genetic polymorphism of Puccinia triticina in southern Dagestan relating to the center of the common evolution between agent causing leaf rust and wheat. Russ. J. Genet. 55, 418–425. doi: 10.1134/S1022795419040045
Guney, M., Kafkas, S., Keles, H., Zarifikhosroshahi, M., Gundesli, M. A., Ercisli, S., et al. (2021). Genetic diversity among some walnut (Juglans regia L.) genotypes by SSR markers. Sustainability 13, 6830. doi: 10.3390/su13126830
Haag, T., Santos, A. S., Sana, D. A., Morato, R. G., Cullenjr, L., Crawshawjr, P. G., et al. (2010). The effect of habitat fragmentation on the genetic structure of a top predator: loss of diversity and high differentiation among remnant populations of Atlantic Forest jaguars (Panthera onca). Mol. Ecol. 19, 4906–4921. doi: 10.1111/j.1365-294X.2010.04856.x
Hamrick, J. L., and Godt, M. J. W. (1996). Conservation genetics of endemic plant species. Conserv. Genet. 1996, 281–304. doi: 10.1007/978-1-4757-2504-9_9
Hao, L., Zhang, L., Zhang, G. S., Wang, Y., Han, S. L., and Bai, Y. R. (2017). Genetic diversity and population genetic structure of Salix psammophila. Acta Bot. Boreal-Occident. Sin. 37, 1507–1516. doi: 10.7606/j.issn.1000-4025.2017.08.1507
Holsinger, K. E., and Weir, B. S. (2009). Genetics in geographically structured populations: defining, estimating and interpreting FST. Nat. Rev. Genet. 10, 639–650. doi: 10.1038/nrg2611
Hosseini, S., Huseini, H. F., Larijani, B., Mohammad, K., Najmizadeh, A., Nourijelyani, K., et al. (2014). The hypoglycemic effect of Juglans regia leaves aqueous extract in diabetic patients: a first human trial. DARU, J. Pharm. Sci. 22, 1–5. doi: 10.1186/2008-2231-22-19
Hu, Y. H., Zhao, P., Zhang, Q., Wang, Y., Gao, X. X., Zhang, T., et al. (2015). De novo assembly and characterization of transcriptome using Illumina sequencing and development of twenty five microsatellite markers for an endemic tree Juglans hopeiensis Hu in China. Biochem. Syst. Ecol. 63, 201–211. doi: 10.1016/j.bse.2015.10.011
Hu, Y. P., Wang, L., Xie, X. L., Yang, J., Li, Y., and Zhang, H. G. (2010). Genetic diversity of wild populations of Rheum tanguticum endemic to China as revealed by ISSR analysis. Biochem. Syst. Ecol. 38, 264–274. doi: 10.1016/j.bse.2010.01.006
Hu, Z., Zhang, T., Gao, X. X., Wang, Y., Zhang, Q., Zhou, H. J., et al. (2016). De novo assembly and characterization of the leaf, bud, and fruit transcriptome from the vulnerable tree Juglans mandshurica for the development of 20 new microsatellite markers using Illumina sequencing. Mol. Genet. Genom. 291, 849–862. doi: 10.1007/s00438-015-1147-y
Hufford, K. M., Krauss, S. L., and Veneklaas, E. J. (2012). Inbreeding and outbreeding depression in Stylidium hispidum: implications for mixing seed sources for ecological restoration. Ecol. Evol. 2, 2262–2273. doi: 10.1002/ece3.302
Islam, A. K. M. M., and Widhalm, J. R. (2020). Agricultural uses of juglone: opportunities and challenges. Agronomy 10, 1500. doi: 10.3390/agronomy10101500
Jin, M., Diao, S. B., Zhang, C. H., Cao, S., Sun, J. F., Li, R., et al. (2015). Two new diarylheptanoids isolated from the roots of Juglans mandshurica. Nat. Prod. Res. 29, 1839–1844. doi: 10.1080/14786419.2015.1009063
Kantety, R. V., Rota, M. L., Matthews, D. E., and Sorrells, M. E. (2002). Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol. Biol. 48, 501–510. doi: 10.1023/A:1014875206165
Letunic, I., and Bork, P. (2021). Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi: 10.1093/nar/gkab301
Li, D. Y., Long, C., Pang, X. M., Ning, D. L., Wu, T., Dong, M. L., et al. (2020). The newly developed genomic-SSR markers uncover the genetic characteristics and relationships of olive accessions. PeerJ. 8, e8573. doi: 10.7717/peerj.8573
Li, J., Zhao, M., Wei, S. C., Luo, Z. H., and Wu, H. (2015). Geologic events coupled with Pleistocene climatic oscillations drove genetic variation of Omei treefrog (Rhacophorus omeimontis) in southern China. BMC Evol. Biol. 15:289. doi: 10.1186/s12862-015-0572-1
Li, M., Wang, S. X., Gao, B. J. (2013). Analysis of genetic diversity of chinese pine (Pinus tabulaeformis) natural secondary forest populations and correlation with theirs habitat ecological factors. Acta Ecol. Sin. 33, 3602–3610. doi: 10.5846/stxb201211271680
Li, X., Zhao, M. H., Xu, Y. J., Li, Y., Tigabu, M., and Zhao, X. Y. (2021). Genetic diversity and population differentiation of Pinus koraiensis in China. Horticulturae 7, 104. doi: 10.3390/horticulturae7050104
Liao, R. Y., Luo, Y. X., Yisilam, G., Lu, R. S., Wang, Y. G., and Li, P. (2019). Development and characterization of SSR markers for Sanguinaria canadensis based on genome skimming. Appl. Plant Sci. 7, e11289. doi: 10.1002/aps3.11289
Liu, J. Q., Fang, Y., and Chen, Y. X. (2020). Analysis of SSR distribution characteristics and primer development of Cornus florida transcriptome. Mol. Plant Breed. 18, 6769–6775. doi: 10.13271/j.mpb.018.006769
Liu, J. Z. (2015). Based on the analysis of species distribution model of Juglans mandshurica genetic diversity and virtual screening of flavonoids compounds on Juglans mandshurica (Master). Northeast Forestry University, Harbin.
Mallavadhani, U. V., Prasad, C. V., Shrivastava, S., and Naidu, V. G. M. (2014). Synthesis and anticancer activity of some novel 5, 6-fused hybrids of juglone based 1, 4-naphthoquinones. Eur. J. Med. Chem. 83, 84–91. doi: 10.1016/j.ejmech.2014.06.012
Marta, B., Lenka, P., Petra, B., Katerina, K., and Tomáš, U. (2012). Genetic differences between wild and captive populations of the peregrine falcon (Falco peregrinus) and the saker falcon (Falco cherrug) living in the czech republic. J. Agric. Sci. Technol. 2, 642–651.
Melo, A. T. D. O., Coelho, A. S. G., Pereira, M. F., Blanco, A. J. V., and Franceschinelli, E. V. (2014). High genetic diversity and strong spatial genetic structure in Cabralea canjerana (Vell.) Mart. (Meliaceae): implications to Brazilian Atlantic Forest tree conservation. Nat. Conserv. 12, 129–133. doi: 10.1016/j.ncon.2014.08.001
Millar, C. I., and Westfall, R. D. (1992). Allozyme markers in forest genetic conservation. New For. 6, 347–371. doi: 10.1007/BF00120652
Mohammed, H., Jaiswal, S. K., Mohammed, M., Mbah, G. C., and Dakora, F. D. (2020). Insights into nitrogen fixing traits and population structure analyses in cowpea (Vigna unguiculata L. Walp) accessions grown in Ghana. Physiol. Mol. Biol. Plants 26, 1263–1280. doi: 10.1007/s12298-020-00811-4
Nagy, S., Poczai, P., Cernák, I., Gorji, A. M., Hegedus, G., and Taller, J. (2012). PICcalc: an online program to calculate polymorphic information content for molecular genetic studies. Biochem. Genet. 50, 670–672. doi: 10.1007/s10528-012-9509-1
Newton, A. C., Allnutt, T. R., Gillies, A. C. M., Lowe, A. J., and Ennos, R. A. (1999). Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol. Evol. 14, 140–145. doi: 10.1016/S0169-5347(98)01555-9
Nybom, H., and Bartish, I. V. (2000). Effects of life history traits and sampling strategies on genetic diversity estimates obtained with RAPD markers in plants. Perspect. Plant Ecol. Evol. Syst. 3, 93–114. doi: 10.1078/1433-8319-00006
Pang, H. Y., Li, H. L., Long, Z. Y., and Qi, Y. H. (2019). Study on seed traits and seedling growth characteristics of Juglans mandshurica Maxim. from different provenances. Seed 38, 89–92. doi: 10.16590/j.cnki.1001-4705.2019.09.089
Park, G., Jang, D. S., and Oh, M. S. (2012). Juglans mandshurica leaf extract protects skin fibroblasts from damage by regulating the oxidative defense system. Biochem. Biophys. Res. Commun. 421, 343–348. doi: 10.1016/j.bbrc.2012.04.013
Patil, P. G., Singh, N. V., Parashuram, S., Bohra, A., Mundewadikar, D. M., Sangnure, V. R., et al. (2020). Genome wide identification, characterization and validation of novel miRNA-based SSR markers in pomegranate (Punica granatum L.). Physiol. Mol. Biol. Plants. 26, 683–696. doi: 10.1007/s12298-020-00790-6
Poudel, M. R., Ghimire, S. K., Pandey, M. P., Dhakal, K. H., Thapa, D. B., and Khadka, D. K. (2019). Assessing genetic diversity for drought and heat stress tolerance of Nepalese wheat genotypes by SSR markers. EurAsian J. BioSci. 13, 941.
Preethi, P., Rahman, S., Naganeeswaran, S., Sabana, A. A., Gangaraj, K. P., Jerard, B. A., et al. (2020). Development of EST-SSR markers for genetic diversity analysis in coconut (Cocos nucifera L.). Mol. Biol. Rep. 47, 9385–9397. doi: 10.1007/s11033-020-05981-8
Rahemi, A., Fatahi, R., Ebadi, A., Taghavi, T., Hassani, D., Gradziel, T., et al. (2012). Genetic diversity of some wild almonds and related Prunus species revealed by SSR and EST-SSR molecular markers. Plant Syst. Evol. 298, 173–192. doi: 10.1007/s00606-011-0536-x
Ravelombola, W., Shi, A. N., Weng, Y. J., Mou, B. Q., Motes, D., Clark, K., et al. (2018). Association analysis of salt tolerance in cowpea (Vigna unguiculata (L.) Walp) at germination and seedling stages. Theor. Appl. Genet. 131, 79–91. doi: 10.1007/s00122-017-2987-0
Schmidt, T. L., Jasper, M. E., Weeks, A. R., and Hoffmann, A. A. (2021). Unbiased population heterozygosity estimates from genome-wide sequence data. Methods Ecol. Evol. 12, 1888–1898. doi: 10.1111/2041-210X.13659
Shamlu, F., Rezaei, M., Lawson, S., Ebrahimi, A., Biabani, A., and Khan-Ahmadi, A. (2018). Genetic diversity of superior Persian walnut genotypes in Azadshahr, Iran. Physiol. Mol. Biol. Plants 24, 939–949. doi: 10.1007/s12298-018-0573-9
Sharma, N., Ghosh, P., Sharma, U. K., Sood, S., Sinha, A. K., and Gulati, A. (2009). Microwave-assisted efficient extraction and stability of juglone in different solvents from Juglans regia: quantification of six phenolic constituents by validated RP-HPLC and evaluation of antimicrobial activity. Anal. Lett. 42, 2592–2609. doi: 10.1080/00032710903202055
Slatkin, M. (1985). Gene flow in natural populations. Annu. Rev. Ecol. Syst. 16, 393–430. doi: 10.1146/annurev.es.16.110185.002141
Song, J. X., Li, J., Guo, C., Zhou, Y. C., Wang, H., and Zhang, L. J. (2017). Selection of fruit variation type of Juglans mandshurica germplasm resources in the eastern Liaoning Mountain region. Mol. Plant Breed. 15, 3798–3802. doi: 10.13271/j.mpb.015.003798
Sun, M. L., Wang, Y. M., Song, Z. Q., and Fang, G. Z. (2007). Insecticidal activities and active components of the alcohol extract from green peel of Juglans mandshurica. J. For. Res. 18, 62–64. doi: 10.1007/s11676-007-0011-2
Tang, L. L., Zhang, M., Zhao, X. L., Kang, M. Y., Liu, H. Y., Gao, X. M., et al. (2019). Species distribution and community assembly rules of Juglans mandshurica in North China. Chin. J. Plant Ecol. 43, 753–761. doi: 10.17521/cjpe.2018.0161
Tian, J., Wu, Y., Wang, Y., and Han, F. (2009). Development and prospects of the walnut industry in China. VI Int. Walnut Symp. 861, 31–38. doi: 10.17660/ActaHortic.2010.861.2
Torokeldiev, N., Ziehe, M., Gailing, O., and Finkeldey, R. (2019). Genetic diversity and structure of natural Juglans regia L. populations in the southern Kyrgyz Republic revealed by nuclear SSR and EST-SSR markers. Tree Genet. Genom. 15, 1–12. doi: 10.1007/s11295-018-1311-8
Uncu, A. Ö., Gültekin, V., Allmer, J., Allmer, J., Frary, A., and Doganlar, S. (2015). Genomic simple sequence repeat markers reveal patterns of genetic relatedness and diversity in sesame. Plant Genome 8, 1–12. doi: 10.3835/plantgenome2014.11.0087
Vardhini, S. R. D. (2014). Exploring the antiviral activity of juglone by computational method. J. Recept. Signal Transduct. 34, 456–457. doi: 10.3109/10799893.2014.917325
Varshney, R. K., Thiel, T., Stein, N., Langridge, P., and Graner, A. (2002). In silico analysis on frequency and distribution of microsatellites in ESTs of some cereal species. Cell. Mol. Biol. Lett. 7, 537–546.
Victory, E. R., Glaubitz, J. C., Rhodes, O. E., and Woeste, K. (2006). Genetic homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am. J. Bot. 93, 118–126. doi: 10.3732/ajb.93.1.118
Vranckx, G. U. Y., Jacquemyn, H., Muys, B., and Honnay, O. (2012). Meta-analysis of susceptibility of woody plants to loss of genetic diversity through habitat fragmentation. Conserv. Biol. 26, 228–237. doi: 10.1111/j.1523-1739.2011.01778.x
Wang, D. N., Mu, C. C., Gao, Z., and Feng, F. J. (2011). ISSR analysis of genetic diversity of Juglans mandshurica Maxim populations. Nonwood For. Res. 29, 22–29. doi: 10.14067/j.cnki.1003-8981.2011.02.017
Wang, W. T., Xu, B., Zhang, D. Y., Bai, W. N. (2016). Phylogeography of postglacial range expansion in Juglans mandshurica (Juglandaceae) reveals no evidence of bottleneck, loss of genetic diversity, or isolation by distance in the leading-edge populations. Mol. Phylogenet. Evol. 102, 255–264. doi: 10.1016/j.ympev.2016.06.005
Wang, Y. (2007). The study on the genetic diversity of Juglans mandshurica Maxim in the northeast of China (Master). Northeast Forestry University, Harbin.
Wright, S. (1950). The genetical structure of populations. Annal. Eugen. 15, 323–354. doi: 10.1038/166247a0
Wuyun, T., Amo, H., Xu, J. S., Ma, T., Uematsu, C., and Katayama, H. (2015). Population structure of and conservation strategies for wild Pyrus ussuriensis Maxim. in China. PLoS ONE 100:e0133686. doi: 10.1371/journal.pone.0133686
Xiao, J., Zhao, J., Liu, M. J., Liu, P., Dai, L., and Zhao, Z. H. (2015). Genome-wide characterization of simple sequence repeat (SSR) loci in Chinese jujube and jujube SSR primer transferability. PLoS ONE 10:e0127812. doi: 10.1371/journal.pone.0127812
Yan, F., Xi, R. M., She, R. X., Chen, P. P., Yan, Y. J., Yang, G., et al. (2021). Improved de novo chromosome-level genome assembly of the vulnerable walnut tree Juglans mandshurica reveals gene family evolution and possible genome basis of resistance to lesion nematode. Mol. Ecol. Resour. 21, 2063–2076. doi: 10.1111/1755-0998.13394
Yang, S. W., Liu, G. F., Zhang, S. Y., Wang, H. R., Peng, H. M., Yang, C. P., et al. (1990). A study on the geographic variation and preliminary selection for the best provenances of Juglans mandshurica. J. Northeast For. Univ. 18(S2),72–76.
Yao, G. D., Cheng, Z. Y., Shang, X. Y., Gao, P. Y., Huang, X. X., and Song, S. J. (2017). Coumarins from the bark of Juglans mandshurica exhibited anti-hepatoma activities via inducing apoptosis. J. Asian Nat. Prod. Res. 19, 1134–1142. doi: 10.1080/10286020.2017.1292256
Yasodha, R., Vasudeva, R., Balakrishnan, S., Sakthi, A. R., Abel, N., Binai, N., et al. (2018). Draft genome of a high value tropical timber tree, Teak (Tectona grandis L. f): insights into SSR diversity, phylogeny and conservation. DNA Res. 25, 409–419. doi: 10.1093/dnares/dsy013
Zhang, D. M., Shen, X. H., Zhang, H. X., and Shen, J. M. (2003). Advances in gene flow and paternity analysis of forest population. For. Res. 16, 488–494. doi: 10.13275/j.cnki.lykxyj.2003.04.019
Zhang, H., Hu, D. C., Xie, X. M., Liu, D., Zhang, P., and Wang, Z. L. (2020). Analyses on population structure and genetic diversity of Juglans mandshurica in Queshan provincial nature reserve of Shandong. J. Plant Resour. Environ. 29, 34–42. doi: 10.3969/j.issn.1674-7895.2020.03.05
Zhang, L. J., Guo, C., Qin, B. T., Lu, X. J., Yang, Y. C., Qi, Y. H., et al. (2019). Phonological characteristics of flowering and pollen viability of Juglans mandshurica. J. Northeast For. Univ. 47, 4–8. doi: 10.13759/j.cnki.dlxb.2019.05.002
Zhang, Q. H., Yu, S. H., Pei, X. N., Wang, Q. C., Lu, A. J., Cao, Y., et al. (2021). Within-and between-population variations in seed and seedling traits of Juglans mandshurica. J. For. Res. 2021, 1–12. doi: 10.1007/s11676-021-01381-1
Zhang, S., Gu, J. T., Wang, J. M., Zuo, L. H., and Yang, M. S. (2017). Analysis on introduction trial and genetic diversity of black locust populations. Acta Hortic. Sin. 44, 1609–1618. 10.16420/j.issn.0513-353x.2016-0964
Zhao, P., and Woeste, K. E. (2011). DNA markers identify hybrids between butternut (Juglans cinerea L.) and Japanese walnut (Juglans ailantifolia Carr.). Tree Genet. Genom. 7, 511–533. doi: 10.1007/s11295-010-0352-4
Keywords: Juglans mandshurica, EST-SSR, genetic diversity, genetic structure, genetic differentiation, conservation strategies
Citation: Zhang Q, Zhang X, Yang Y, Xu L, Feng J, Wang J, Tang Y, Pei X and Zhao X (2022) Genetic Diversity of Juglans mandshurica Populations in Northeast China Based on SSR Markers. Front. Plant Sci. 13:931578. doi: 10.3389/fpls.2022.931578
Received: 29 April 2022; Accepted: 03 June 2022;
Published: 30 June 2022.
Edited by:
Daniel Pinero, National Autonomous University of Mexico, MexicoReviewed by:
Xiaoming Pang, Beijing Forestry University, ChinaAsit Ray, Siksha O Anusandhan University, India
Copyright © 2022 Zhang, Zhang, Yang, Xu, Feng, Wang, Tang, Pei and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaona Pei, xiaonapei2020@163.com; Xiyang Zhao, zhaoxyphd@163.com
 Qinhui Zhang1,2
Qinhui Zhang1,2 Xinxin Zhang
Xinxin Zhang Xiaona Pei
Xiaona Pei Xiyang Zhao
Xiyang Zhao