- 1The National Key Facility for Crop Gene Resources and Genetic Improvement (NFCRI) and MOA Key Lab of Soybean Biology, Institute of Crop Science, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Institute of Crop Resources, Heilongjiang Academy of Agricultural Sciences, Harbin, China
The soybean aphid poses a severe threat to soybean quality and yield by sucking phloem sap and transmitting plant viruses. An early-maturing and highly resistant soybean landrace, Fangzheng Moshidou, with markedly reduced aphid colonization has been identified by screening of aphid-resistant soybean accessions. In a population derived from the cross of Fangzheng Moshidou with the susceptible cultivar Beifeng 9, resistance was conferred by a single dominant gene. Three linked markers, Satt114, Satt334, and Sct_033, on chromosome 13 were identified by bulked-segregant analysis. Additional simple-sequence repeat and single-nucleotide polymorphism (SNP) markers were developed for gene mapping. The resistance of Fangzheng Moshidou was fine-mapped to the interval between the SNP markers YCSNP20 and YCSNP80, corresponding to 152.8 kb in the Williams 82 assembly 2 genome. This region was near the reported loci Rag2 and Rag5 but did not overlap the interval containing them. A unique haplotype is described for Fangzheng Moshidou that distinguishes it from soybean accessions PI 587972, PI 594879, and PI 567301B in the interval containing Rag2 and Rag5. These results indicate that Fangzheng Moshidou harbors a novel gene at a tightly linked resistance locus, designated as RagFMD. Fourteen candidate genes were annotated in the fine-mapping region, including seven NBS-LRR genes, which are usually considered resistance genes in plant defense. Most of these candidate genes showed variations distinguishing the resistant and susceptible parents and some genes also showed differences in expression between the two parental lines and at several times after aphid infestation. Isolation of RagFMD would advance the study of molecular mechanisms of soybean aphid resistance and contribute to precise selection of resistant soybeans.
Introduction
Soybean, Glycine max (L.) Merr, is a source of protein and oil worldwide. The yield of soybean is often threatened by soybean aphid (SA, Aphis glycines Matsumura). SAs colonize young leaves and tender stems of plants as adults and nymphs and suck phloem sap with piercing and sucking mouthparts. The chlorophyll of damaged soybean leaves disappears, forming bright yellow irregularly shaped macule that gradually expand and turn brown. Severely damaged plants show curly stems, yellow leaves, short plants, reduced branches and pods, and yield loss (Wu et al., 2004; Ragsdale et al., 2007; Beckendorf et al., 2008; Xiao et al., 2013). The whole plant may die if severely infected with aphids during seedling stage. Soybean aphids reduce soybean quality and can lead to yield losses in excess of 50% in China (Wang et al., 1994; Ragsdale et al., 2007). SAs also carry viruses, and transmit soybean mosaic virus and other plant viruses by secreting honeydew when feeding on soybean plants (Hill et al., 2001; Davis et al., 2005; Gildow et al., 2008; Tilmon et al., 2011; McCarville et al., 2014).
Control of SAs has relied mainly on insecticides (Tilmon et al., 2011). Long-term use of insecticides not only increases production costs, but damages the environment, increases insecticide resistance, and hastens aphid biotype differentiation (Hanson et al., 2017). Introduction of aphid-resistant soybean cultivars is a promising strategy for controlling SAs (Ragsdale et al., 2011; Hodgson et al., 2012). Since the invasion by SA of the United States in 2000, aphid-resistant soybean accessions including Jackson (PI 548657), Dowling (PI 548663) among others have been identified (Hill et al., 2004; Mensah et al., 2005; Mian et al., 2008b). With biological type differentiation of aphid, the resistance of some soybeans has gradually disappeared (Kim et al., 2008; Hill et al., 2010; Alt and Ryan-Mahmutagic, 2013), and new aphid-resistant soybeans are urgently needed. Additional resistant soybeans including K1639, Pioneer 85B97, and others have been reported (Diaz-Montano et al., 2006; Schapaugh et al., 2010; Bhusal et al., 2013, 2014). All of these soybeans can be valuable for soybean breeding programs as sources of aphid resistance.
Some aphid-resistance loci carried by aphid-resistant soybeans have been identified. The resistance genes Rag and Rag1, from respectively Jackson (PI 548657) and Dowling (PI 548663) were mapped to soybean chromosome 7 (Hill et al., 2004; Li et al., 2007; Kim et al., 2010a). Rag2 from PI 243540 to PI 200538 (Kang et al., 2008; Mian et al., 2008a; Hill et al., 2009; Kim et al., 2010b) and Rag5 from PI 567301B (Jun et al., 2012) were mapped to chromosome 13. PI 587972 and PI 594879 were also characterized as carrying one dominant gene located in the Rag2 region (Fox et al., 2014; Hill et al., 2017). Rag3, a single dominant gene from PI 567543C, was mapped to chromosome 16 (Zhang et al., 2010). Two dominant aphid-resistance genes, Rag6 and Rag3c, from wild soybean 85-32 were mapped to chromosomes 8 and 16 (Zhang et al., 2017). Recessive genes rag1c and rag4 carried by PI 567541B, rag1b and rag3 from PI 567598B have been identified (Mensah et al., 2008; Bales et al., 2013). Some quantitative-trait loci (QTLs) and single-nucleotide polymorphisms (SNPs) associated with SA resistance have been identified by QTL mapping and genome-wide association study methods (Jun et al., 2013; Hanson et al., 2018; Lamantia et al., 2019). Among them, Rag1, Rag2, Rag3c, and Rag6 were fine-mapped to a very small interval, although none of them were isolated (Kim et al., 2010a,b; Zhang et al., 2017). These intervals all harbored nucleotide-binding site leucine-rich repeat (NBS-LRR) genes, which are considered resistance genes in plant defense.
Resistance to SA controlled by Rag genes was described to be overcome (Kim et al., 2008; Hill et al., 2010; Alt and Ryan-Mahmutagic, 2013); then, the use of new germplasm or combinations of different resistance genes in one genotype could ensure the durability of host resistance (Michel et al., 2011; Ajayi-Oyetunde et al., 2016). A soybean landrace, Fangzheng Moshidou, from northeast China showed high aphid resistance and early-maturing in our previous study (Liu et al., 2014). The objective of the present study was to characterize the inheritance of SA resistance in this accession, perform fine mapping, and identify candidate resistance genes.
Materials and Methods
Plant Materials
Fangzheng Moshidou (accession ZDD00326 in the National Crop Genebank of China) is a maturity group (MG) 0 soybean landrace collected from Heilongjiang province in China. Beifeng 9 is a MG 0 soybean cultivar that is susceptible to SA. A cross was made between Beifeng 9 and Fangzheng Moshidou to create genetic mapping populations. Eight F1 plants were grown in the field at Heilongjiang Academy of Agricultural Sciences (HAAS), Harbin, China. One hundred and eighty-two F2 seedlings were used for genetic analysis of aphid resistance in the growth chamber and for bulked-segregant analysis (BSA) (Michelmore et al., 1991). Another 130 F2 seeds were planted in the field for developing advanced generation mapping populations by pedigree selection. The F5 generation was grown in Harbin and 650 single plants were used for mapping resistance genes. Eight hundred and fifty-three F5:6 plants were derived for fine mapping from F5 plants that were heterozygous in the initially mapped region. Seeds from identified recombinants were harvested for phenotyping of their progenies in order to further confirm the phenotypes of these recombinants. PI 587972 and PI 594879 carrying Rag2 and PI 567301B carrying Rag5, which were used for controls of published aphid resistance genes in haplotype analysis, were provided by the National Crop Genebank of China.
Soybean Aphid Culture
Soybean aphids were collected from the field at HAAS. The SAs were maintained on a susceptible soybean cultivar Williams 82 in growth chamber at approximately 26/22°C day/night temperatures with 16 h light and 8 h dark daily.
Aphid Resistance Assessment
Soybean aphid resistance assessment was performed in the growth chamber or a net house. Tests in the growth chamber were used mainly to evaluate the aphid resistance of 8 F1 plants, 182 F2 plants, and progenies of recombinants. Soybean plants were grown in 8 × 8 × 8-cm plastic pots with four plants per pot in a 3:1 mixture of fertile soil and perlite. Tests in the net house were used to evaluate aphid resistance in 650 F5 plants and 853 F5:6 plants. Plants were grown in rows with 10-cm spacing within rows and 50 cm between rows using an unreplicated, completely randomized design. Susceptible and resistant parental controls were planted every 10 rows and 30 plants per row. Soybean plants in the growth chamber and net house were each infested with two wingless aphids using a brush at the V1 growth stage (with one set of unfolded trifoliolate leaves) and the number of aphids per plant was recorded 3 weeks after infestation. Resistance evaluation followed Mensah et al. (2008) using a scale of 0–4 where 0 = no SAs; 0.5 = 0–10 SAs; 1.0 = 11–100 SAs; 1.5 = 101–150 SAs; 2.0 = 151–300 SAs; 2.5 = 301–500 SAs, 3.0 = 501–800 SAs; 3.5 = more than 800 SAs, plants growing slowly, and leaves curled slightly and yellow, without black mold; 4 = more than 800 SAs, plants growing slowly, and leaves severely curled and yellow with black mold. In qualitative analyses, plants with phenotypic ratings of 0–1 were assigned as resistant and plants with ratings of 1.5–4 as susceptible. For evaluating whether the resistance of Fangzheng Moshidou was dependent on initial aphid levels, the three initial densities of aphids (2, 10, and 50 aphids/plant) were introduced to incompletely unfolded leaves at the soybean V1 stage with three repeats for each treatment. The number of aphids per plant was recorded 3 weeks post-infestation and resistance rating for each treatment was evaluated.
DNA Extraction and Polymerase Chain Reaction Amplification
Young leaves from each plant were collected in a 2-mL centrifuge tube with steel balls and rapidly frozen in liquid nitrogen. Genomic DNA was extracted by the modified CTAB method (Murray and Thompson, 1980) and a Nanodrop 1000 spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, United States) was used to quantify DNA concentration. All DNA samples were diluted to 20 ng/μL with water for genotype identification. EasyTaq DNA Polymerase (Beijing Transgen Biotech, Beijing, China) was used for polymerase chain reaction (PCR) amplification according to the instructions.
Bulked-Segregant Analysis and Linkage Mapping
A set of 580 SSR markers distributed over the soybean genome were used to screen polymorphic markers in the parental lines, and primer sequences of these SSR markers were retrieved from Soybase.1 BSA was performed on a resistant and a susceptible bulk formed by pooling equal amounts of DNA from 20 resistant to 20 susceptible F2 plants. The bulks and parental lines were genotyped with polymorphic SSR markers. Markers showing heterozygous genotypes in the resistant bulk and homozygous genotypes in the susceptible bulk identical to that of the susceptible parent were assigned as closely linked markers based on the principle of BSA.
Additional polymorphic SSR markers used for linkage mapping were developed based on the results of BSA and predicted positions of markers in the Williams 82 reference genome (see foot note text 2). Initial mapping was performed using F2 and F5 populations and the initial mapping interval was delimited by analysis of the genotypic and phenotypic data. SNP markers were also developed using whole-genome resequencing data of parental lines (accession SAMC286906 and SAMC287315 at the National Genomics Data Center2 lying in the initial mapping interval. Flanking sequences of these markers were retrieved from the database of Glycine max Wm82.a2.v1 genomic sequence in Soybase. Primers were designed on an online site3 and their sequences are listed in Supplementary Table 1. Genotyping was performed by PCR amplification, followed by polyacrylamide gel electrophoresis (for SSR markers) or sequencing (for SNP markers).
Aphid Infestation and RNA Extraction
Plants of the parental lines were grown in a growth chamber (70–80% relative humidity, 25/22°C, 16/8 h light/dark) for infestation with SAs. Each seedling was infested with 15 aphids at the V2 growth stage (two sets of unfolded trifoliolate leaves). Leaves were collected before infestation (0 h) and in the early stage (8 and 24 h) and later stage (7 days or 168 h) following SA infestation. All SAs were brushed off leaves during sampling. Samples were immediately frozen in liquid nitrogen and stored at –80°C for RNA extraction. Total RNA was extracted from leaf samples of about 20 mg with Trizol reagent (Invitrogen, Carlsbad, CA, United States).
Complementary DNA Synthesis and Quantitative Real-Time Polymerase Chain Reaction Analysis
First-strand complementary DNA (cDNA) was synthesized using 1 μg of total RNA with a Primer Script RT reagent Kit (Takara Bio Inc., Tokyo, Japan) according to the manufacturer’s instructions. Primers used for real-time PCR are listed in Supplementary Table 2. A total of 2 μl of the synthesized first-strand cDNA was amplified by PCR in 20-μl reaction mixtures using SYBR Select Master Mix on a ABI 7300 Real-time PCR system (Applied Biosystems, Thermo Fisher Scientific Corp., Waltham, MA, United States) with the following procedure: 95°C for 5 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s, and 72°C for 31 s. The soybean actin 11 gene was used as the reference gene. Melting-curve analysis was performed to ensure that the PCR products were unique. The values from three independent biological samples were averaged, and relative expression levels of selected genes were calculated by the 2–ΔΔt method (Livak and Schmittgen, 2001). The expression levels of candidate genes in the fine-mapping region were compared between Fangzheng Moshidou and Beifeng 9, and during infestation of SA at four time points: 0 h (before aphid infestation) and 8, 24, and 168 h (7 days) after aphid infestation. Significance of differences between parental lines without aphid infestation and two time points in Fangzheng Moshidou or Beifeng 9 after SA infestation was evaluated by one-way analysis of variance.
Haplotype Analysis
A genome-wide genotyping array containing 1,58,327 SNPs (Beijing Compass Biotechnology Co., Beijing, China) was used to examine the haplotypes of mapping regions in Fangzheng Moshidou, Beifeng 9, PI 587972 (Rag2), PI 594879 (Rag2), and PI 567301B (Rag5) using the Illumina platform (Illumina, San Diego, CA, United States). All genotype data was loaded into Genome Studio 2.0 (Illumina, San Diego, CA, United States) for raw-data normalization, clustering, and genotype calling. Flanking SNPs were collected based on the physical positions 30318939–31329840 on chromosome 13, and SNPs with minor allele frequency < 0.25, missing rate > 50%, and heterozygosity ratio > 50% were removed. Graphic representations of initial mapping region in all accessions were drawn with polymorphic SNPs using FlapJack (Milne et al., 2010).
Results
Evaluation of Fangzheng Moshidou for Aphid Resistance
Fangzheng Moshidou showed high SA resistance in both growth chamber and net house. Two weeks after SA infestation, only a few aphids were observed on Fangzheng Moshidou plants, but many on Beifeng 9 plants (Figure 1). Fangzheng Moshidou also showed a resistance score of 1 in the net house test, whereas Beifeng 9 showed a score of 4. Furthermore, Fangzheng Moshidou showed high resistance after infestation even at initial density of 50 aphids per plant (Supplementary Table 3).

Figure 1. Phenotype of Fangzheng Moshidou and Beifeng 9 two weeks after SA infestation. (A) Resistant response of Fangzheng Moshidou seedlings infested with SAs; (B) Susceptible response of Beifeng 9 seedlings infested with SAs.
Inheritance Pattern of Soybean Aphid Resistance in Fangzheng Moshidou
F1 and F2 plants of a cross between Beifeng 9 and Fangzheng Moshidou, together with the two parental lines, were tested for resistance to SA. The mean number of aphids on 12 plants of Fangzheng Moshidou was 21 after aphid infestation, whereas the mean number of aphids on 12 plants of Beifeng 9 was 358. The mean aphid number on eight F1 plants was 14 and all eight showed resistance to SA, suggesting that the resistance of Fangzheng Moshidou was controlled by a dominant gene or genes. Of the 182 F2 plants, 139 were resistant and 43 susceptible (Table 1). These frequencies fitted a 3:1 segregation (x2 = 0.117, p = 0.73), indicating that the aphid resistance of Fangzheng Moshidou was controlled by a single dominant Rag gene.
Initial Mapping by Bulked-Segregant Analysis and Linkage Analysis
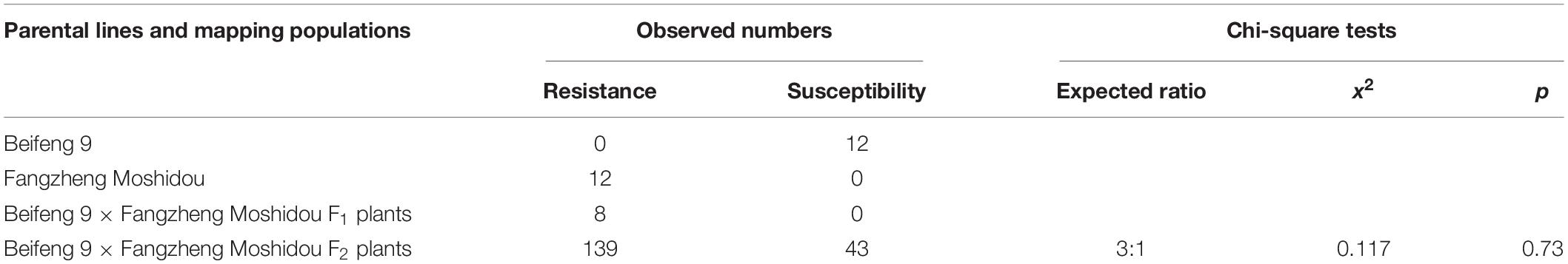
Of the 580 SSR markers distributed over the soybean genome, 180 (31%) were polymorphic between Beifeng 9 and Fangzheng Moshidou. These 180 markers were used to search for linked markers in the resistant and susceptible pools. Three markers (Satt114, Sat_033, and Satt335) located on chromosome 13 were identified as closely linked, all showing heterozygous genotypes in the resistant bulk and homozygous genotypes like those of the susceptible parent Beifeng 9 in the susceptible bulk. The polymorphic SSR markers in other regions were all heterozygous in both bulks. These three markers were then used to genotype all F2 plants and the putative gene conferring SA resistance was assigned to the interval between Satt114 and Sct_033. A set of SSR markers located in this mapping region were then tested for polymorphism between the two parental lines, yielding eight polymorphic markers: BARCSOYSSR_13_1114, 13_1124, 13_1131, 13_1133, 13_1138, 13_1156, 13_1168, and 13_1184. A set of 650 lines of the F5 population derived from the same cross were genotyped with these markers. These genotypic data together with the phenotypic data of the F5 plants and the progenies of recombinants localized the resistance gene between BARCSOYSSR_13_1133 and 13_1184 (Figure 2).
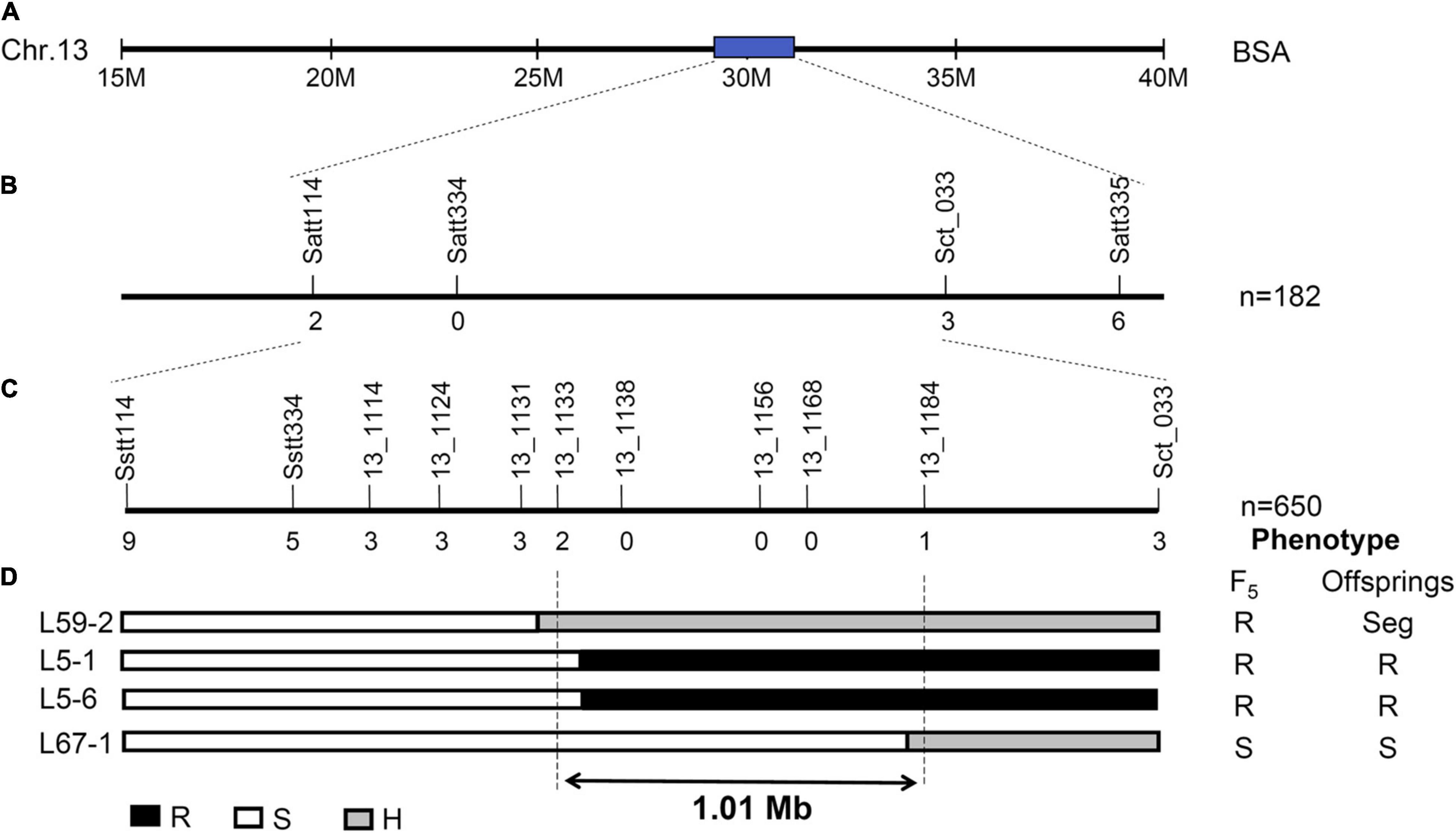
Figure 2. Initial mapping of SA resistance gene from Fangzheng Moshidou. (A) Chromosomal location of resistance gene identified by BSA. (B,C) Initial mapping of the resistance gene using F2 (B) and F5 (C) populations. Vertical lines indicate polymorphic SSR markers. Names of markers are shown above the line and the number of recombinants between gene and each marker is shown below the line. (D) The genotype and phenotype of recombinants used for restricting the mapping region. Black, white, and gray bars represent homozygous segments from Fangzheng Moshidou, homozygous segments from Beifeng 9, and heterozygous segments from the two parental lines, respectively. “Seg” of the phenotype represents segregation of resistance in the offsprings.
Haplotype Analysis Indicated That the Resistance of Fangzheng Moshidou Was Conferred by a Novel Gene
According to their physical locations, the dominant genes Rag2 and Rag5 on chromosome 13 were located in the primary interval identified in Fangzheng Moshidou, and the recessive gene rag4 is far from this region. The haplotype of Fangzheng Moshidou proved distinct from those of the resistant lines PI 587972 (Rag2), PI 594879 (Rag2), and PI 567301B (Rag5). No other genotypes tested, including the susceptible Beifeng 9, carried this haplotype. PI 587972 and PI 594879 carrying the same resistance gene (Rag2) shared the same haplotype (Figure 3). The aphid-resistance gene in Fangzheng Moshidou was accordingly assigned as a new gene and designated as Resistance to Aphids glycines in Fangzheng Moshidou (RagFMD).
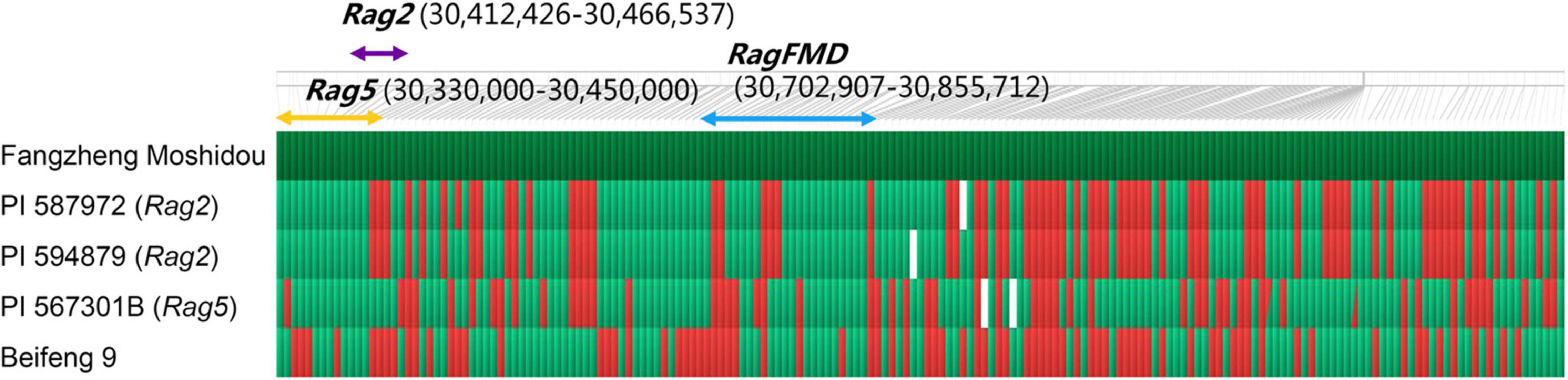
Figure 3. Haplotype analysis of the intial mapping region in different soybean genotypes. The SNPs of Fangzheng Moshidou, PI 587972 (Rag2), PI 594879 (Rag2), PI 567301B (Rag5), and Beifeng 9 were genotyped by a genome-wide genotyping array containing 158,327 SNPs and haplotypes of initial mapping region were carried out by FlapJack software. The reference line Fangzheng Moshidou is shown in dark green. Green, red, and white squares represent same, different, and missing genotypes from the reference, respectively. Purple, yellow, and blue arrows indicate mapping intervals of Rag2, Rag5, and RagFMD, respectively.
Fine Mapping of RagFMD to a 152.8 kb Region
To further map the position of RagFMD locus, 853 F5:6 lines derived from F5 individuals heterozygous in the initially mapped region were genotyped using flanking markers BARCSOYSSR_13_1133, and 13_1184 and recombinants were identified. The additional SSR markers BARCSOYSSR_13_1141, 13_1147, 13_1151, 13_1155, and 13_1177 were used to genotype these recombinants and the mapping interval was narrowed to the region between markers BARCSOYSSR_13_1141 and 13_1155 (Figure 4). This region did not coincide with the mapping regions of other Rag genes including the nearby Rag2 and Rag5 (Kim et al., 2010b; Jun et al., 2012; Lee et al., 2017), further supporting its assignment as a new aphid-resistance locus. Four SNP markers (YCSNP16, YCSNP20, YCSNP80, and YCSNP90) between BARCSOYSSR_13_1141 and 13_1155 were then developed based on the resequencing data of the two parental lines. The combination of the genotypic and phenotypic data of recombinants and their progenies finally localized RagFMD to the region between markers YCSNP20 and YCSNP80, an interval of 152.8 kb (Figure 4).
Figure 4. Fine mapping of RagFMD locus. (A,B) Fine mapping of RagFMD locus using F5:6 populations. Vertical lines indicate polymorphic markers. Names of markers are shown above the line and the number of recombinants between gene and each marker is shown below the line. (C) The genotype and phenotype of recombinants used for restricting the fine mapping region. Black, white, and gray bars represent homozygous segments from Fangzheng Moshidou, homozygous segments from Beifeng 9, and heterozygous segments from the parental lines, respectively. (D) Annotated gene models within 152.8 kb region based on the Williams 82 genome assembly (Wm82.a2.v1).
Candidate Gene Identification
Fourteen genes were annotated in the 152.8-kb interval (30,702,907–30,855,712 bp) of the RagFMD locus based on the Glycine max Wm82.a2.v1 genomic sequence (Table 2). Seven (Glyma.13G194100, Glyma.13G194500, Glyma.13G194600, Glyma.13G194700, Glyma.13G194800, Glyma.13G194900, and Glyma.13G195100) encoded resistance (R) genes with NBS-LRR domains. The other seven encoded other proteins including sigma factor binding protein 1, chaperone DnaJ-domain superfamily protein, ribosome recycling factor, mitochondrial inner membrane protease subunit 1 (IMP1), and albumin I chain b. Of these 14 genes, 13 harbored variations between the two parental lines according to the resequencing data and six harbored non-synonymous variations (Table 2). Eleven genes harbored variations and seven harbored non-synonymous variations between the SA-resistant parent Fangzheng Moshidou and the reference soybean Williams 82, an SA-susceptible soybean like Beifeng 9.
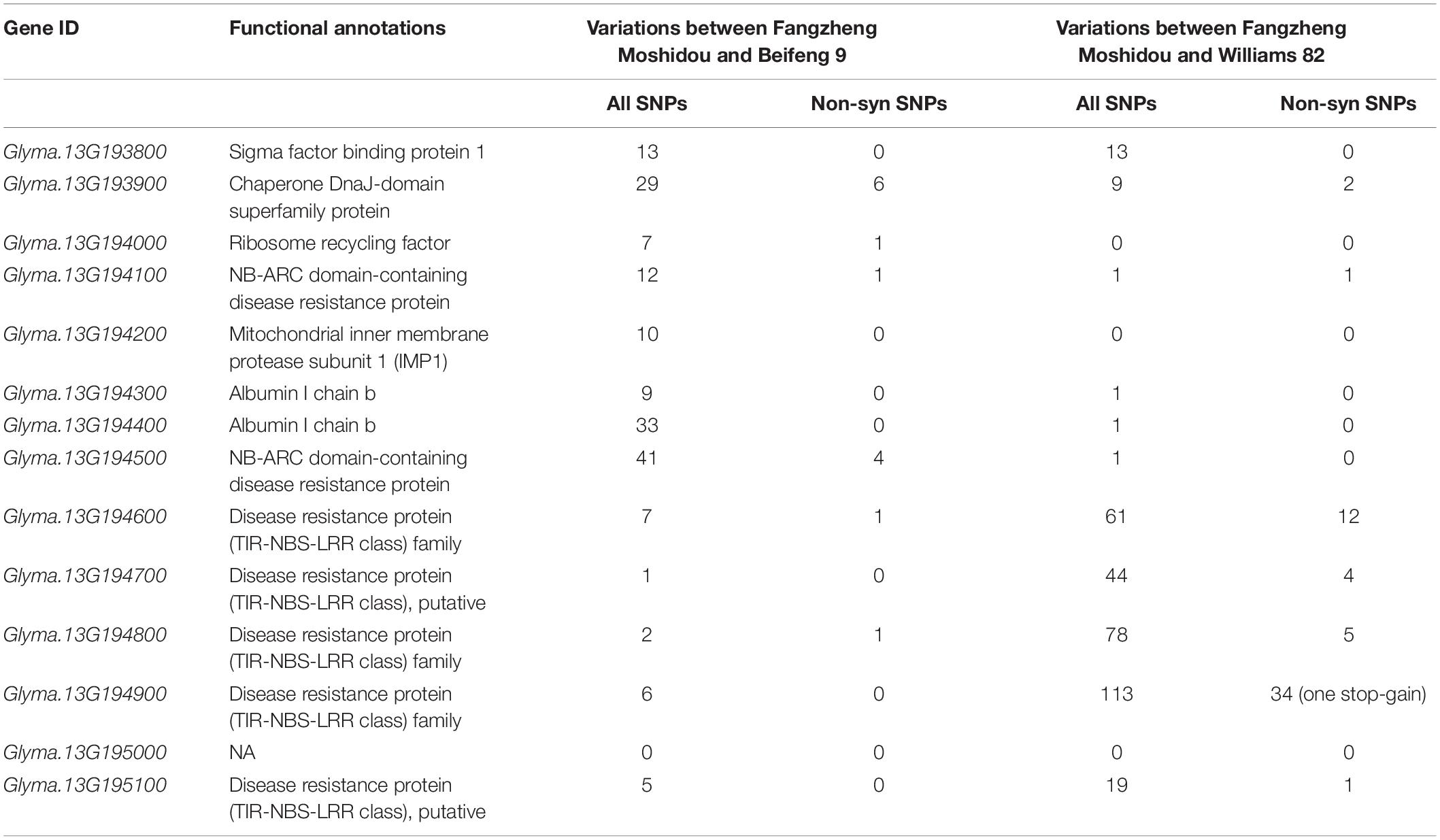
Table 2. Functional annotations and sequence variations of candidate genes in the interval of RagFMD locus.
Expression of 5 genes (Glyma.13g193900, Glyma.13g194300, Glyma.13g194400, Glyma.13g194900, and Glyma.13g195000) was extremely low in both parental lines both before and after aphid infestation. Among the other 9 genes, four (Glyma.13G193800, Glyma.13G194100, Glyma.13G194500 and Glyma.13G195100) showed significant expression differences between the two parental lines before aphid infestation (Figure 5). With aphid infestation, the expression levels of Glyma.13G193800, Glyma.13G194000, and Glyma.13G194100 in Beifeng 9 were significantly changed. Among them, the expression of Glyma.13G194000 was up-regulated at 8 h post-infestation and those of Glyma.13G193800 and Glyma.13G194100 was up-regulated at 7 days post-infestation. However, in Fangzheng Moshidou, the expressions of Glyma.13G194000 and Glyma.13G194800 were significantly up-regulated at 8 h post-infestation and that of Glyma.13G193800 was up-regulated at 7 days post-infestation. In particular, Glyma.13G193800 showed significant expression differences both before and after aphid infestation. The differentially expressed genes were assigned as candidate genes for further investigation.
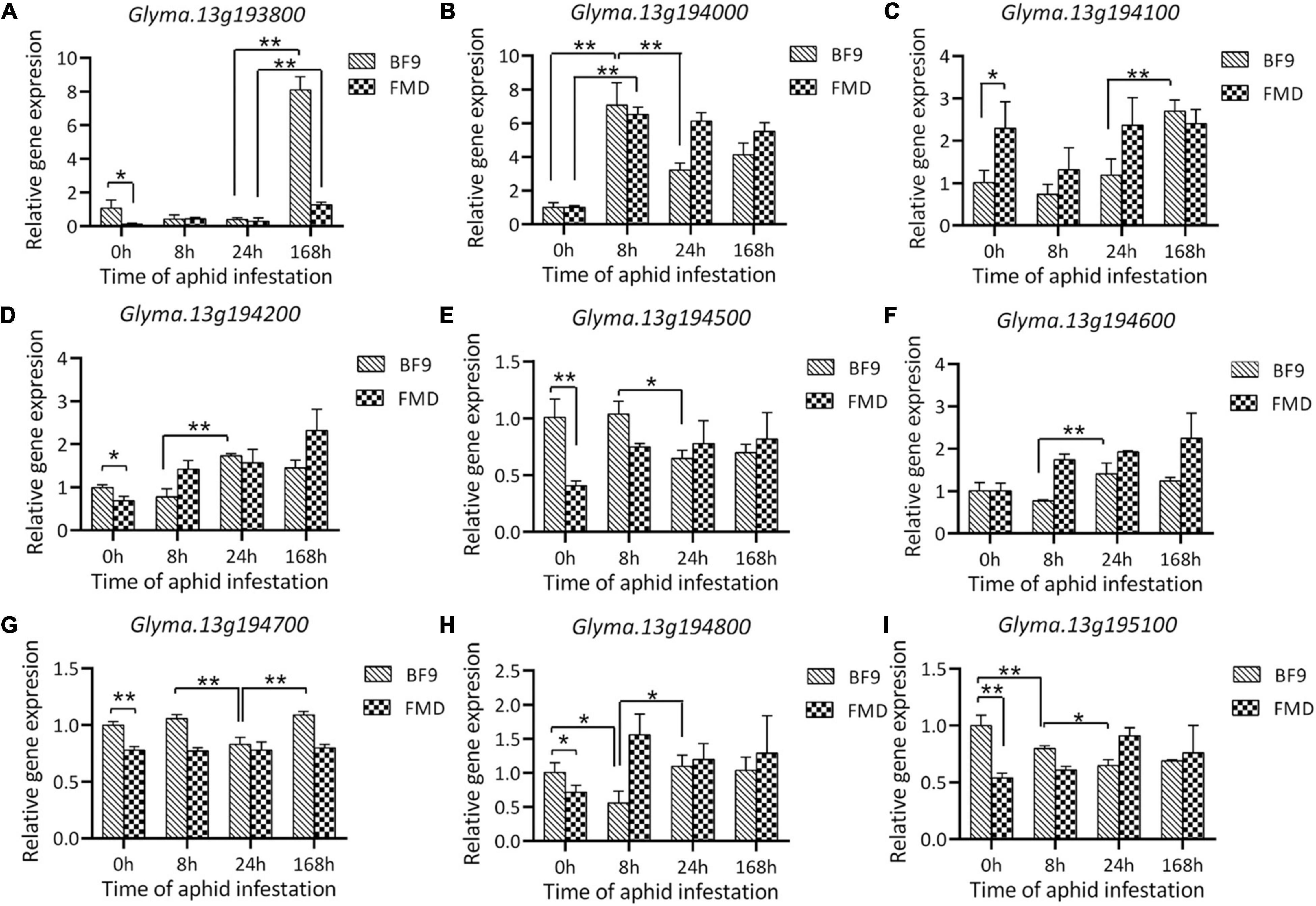
Figure 5. Relative expression analysis of the nine candidate genes (A–I) in Beifeng 9 and Fangzheng Moshidou after SA infestation. Fifteen aphids were introduced to soybean plants and the relative expressions of candidate genes were determined at 0, 8, 24, and 168 h (7 days) after infestation. FMD, Fangzheng Moshidou; BF9, Beifeng 9. Bar represents a standard deviation of three biological replicates. Significance of differences was evaluated by one-way analysis of variance. The * and ** represent significant difference (P < 0.05) and extremely significant difference (P < 0.01) between two labeled parts including either parental lines or time points in BF9 or FMD after SA infestation.
Discussion
In the present study, the aphid resistance of a Chinese soybean landrace, Fangzheng Moshidou, was characterized. A previous report (Diaz-Montano et al., 2006) suggested that the resistance of some soybean accessions is density-dependent, given that they were susceptible when infested with a high aphid density but resistant at lower densities. The SA resistance of Fangzheng Moshidou even at high aphid density suggests its application in soybean breeding. Because it shows early maturity (in MG 0), whereas most resistance sources reported previously (Fox et al., 2014; Hill et al., 2017; Natukunda et al., 2019) have longer growth durations, Fangzheng Moshidou is a promising resource for developing early-maturing SA-resistant soybean cultivars.
Soybean aphid resistance genes have been identified on chromosomes 7, 8, 13, and 16 of soybean (Hanson et al., 2018). Among them, Rag2, Rag5, and rag4 were located on chromosome 13. The gene rag4 is a recessive gene distantly linked to SSR markers BARCSOYSSR_13_0518 and 13_0572 (Mensah et al., 2008). Rag2 from PI 243540 was first mapped on chromosome 13, and the aphid-resistance gene carried by PI 200538 was then mapped to the same physical position and proposed to be allelic to Rag2, which was fine-mapped to the 54-kb interval between 30,412,426 and 30,466,537 bp on chromosome 13 (Kang et al., 2008; Mian et al., 2008a; Hill et al., 2009; Kim et al., 2010b). Rag5 was mapped to a 120-kb interval (Gm13:30.33–30.35 Mb) on chromosome 13, which coincides with the fine mapping region of Rag2, but they were from different soybean accessions and of different (antixenosis and antibiosis) resistance types (Jun et al., 2012; Lee et al., 2017). In the present study, RagFMD carried by Fangzheng Moshidou was also mapped to chromosome 13. Although the initial mapping region overlaps the interval containing Rag2 and Rag5, the haplotype of this region in Fangzheng Moshidou differed sharply from those of the accessions carrying Rag2 and Rag5. Moreover, RagFMD was fine-mapped to the 152.8 kb interval 30,702,907–30,855,712 on chromosome 13 in the Williams 82 reference genome sequence, a region differing from the fine-mapping region of Rag2 and Rag5. For this reason, we propose that RagFMD is a novel aphid resistance gene and can be used to extend resistance durability by pyramiding with other resistance gene.
Nucleotide-binding site leucine-rich repeat proteins function in plant resistance to insects. Some NBS-LRR protein-encoding genes conferring resistance to aphids have been isolated. The Mi-1 gene in tomato (Kaloshian et al., 1995; Vos et al., 1998) and the Vat gene in melon (Boissot et al., 2010, 2016; Dogimont et al., 2014) all have CC-NB-LRR structures and confer resistance to potato aphid (Meloidogyne incognita) and cotton aphid (Aphis gossypii), respectively. The planthopper, like SA, is an insect with piercing–sucking mouth parts, and can kill the rice plant and severely reduce rice yields. Several planthopper-resistance genes including BPH9 (Zhao et al., 2016), BPH14 (Du et al., 2009; Hu et al., 2017), BPH18 (Ji et al., 2016), and BPH26 (Tamura et al., 2014) have been isolated and shown to encode NBS-LRR-type proteins. CC and NB domains of BPH14 may act in rice planthopper resistance by activating the salicylic acid signaling pathway and defense-gene expression (Hu et al., 2017). In soybean, many NBS-LRR type genes have been proposed as candidate genes of Rag loci, although none have been isolated. Of 13 genes in the 115-kb interval of Rag1, one of two NBS-LRR genes was proposed as a candidate gene (Kim et al., 2010a). The only NBS-LRR gene (Glyma13g26000) among the 7 genes in the interval of Rag2 was considered the most likely candidate gene (Kim et al., 2010b). Rag3, Rag3b, Rag3c, and rag3 are all located at the same physical position and an NBS-LRR gene cluster was present in this interval (Zhang et al., 2017). For the candidate genes of RagFMD in our study, 7 genes belong to the NBS-LRR gene family and four showed non-synonymous variations between parental lines. The expression of three NBS-LRR genes was induced after aphid infestation in the aphid-resistant soybean Fangzheng Moshidou. These results further suggest that an NBS-LRR gene mediates aphid resistance in soybean.
Among the 14 annotated genes in the fine-mapped interval of RagFMD, 2 genes (Glyma.13g194300 and Glyma.13g194400) encode albumin I chain b family proteins, which are orthologous to the toxic protein PA1b (Pea albumin 1, subunit b). PA1b is a small and compact 37-amino-acid protein isolated from pea (Pisum sativum) seeds. PA1b confers insecticidal activity against cereal weevils and aphid species, making it a promising bioinsecticide (Gressent et al., 2011; Rahioui et al., 2014). The survival rate of both cotton aphid and pea aphid decreased in response to feeding with the protein (Gressent et al., 2007). Whether these 2 soybean genes function in SA resistance awaits further study.
Conclusion
A novel SA resistance gene was identified in the early-maturing soybean landrace Fangzheng Moshidou and fine-mapped to a 152.8 kb interval on chromosome 13. This region harbors seven NBS-LRR domain-containing proteins and two toxic-protein PA1b-like proteins. Most of these candidate genes showed variations between the resistant and susceptible parental lines and some were induced after aphid infestation. Isolating this gene by map-based cloning will shed light on the molecular mechanism of SA resistance and facilitate marker-assisted selection in soybean breeding programs.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
YG, GL, and L-JQ conceived and designed the experiments. JY, YG, GL, JT, XW, YD, YS, DS, and JS performed the experiments. YG, JY, and GL analyzed the data. YG, JY, GL, and L-JQ wrote and revised the manuscript. All authors read and approved the final version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32172003), the National Key R&D Program of China (2016YFD0100304), and the Agricultural Science and Technology Innovation Program (ASTIP) of Chinese Academy of Agricultural Sciences.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.899212/full#supplementary-material
Footnotes
References
Ajayi-Oyetunde, O. O., Diers, B. W., Lagos-Kutz, D., Hill, C. B., Hartman, G. L., Reuter-Carlson, U., et al. (2016). Differential reactions of soybean isolines with combinations of aphid resistance genes Rag1, Rag2, and Rag3 to four soybean aphid biotypes. J. Econ. Entomol. 109, 1431–1437. doi: 10.1093/jee/tow033
Alt, J., and Ryan-Mahmutagic, M. (2013). Soybean aphid biotype 4 identified. Crop Sci. 53, 1491–1495. doi: 10.2135/cropsci2012.11.0672
Bales, C., Zhang, G., Liu, M., Mensah, C., Gu, C., Song, Q., et al. (2013). Mapping soybean aphid resistance genes in PI 567598B. Theor. Appl. Genet. 126, 2081–2091. doi: 10.1007/s00122-013-2120-y
Beckendorf, E. A., Catangui, M. A., and Riedell, W. E. (2008). Soybean aphid feeding injury and soybean yield, yield components, and seed composition. Agron. J. 100, 237–246. doi: 10.2134/agrojnl2007.0207
Bhusal, S. J., Jiang, G. L., Hesler, L. S., and Orf, J. H. (2014). Soybean aphid resistance in soybean germplasm accessions of maturity group I. Crop. Sci. 54, 2093–2098. doi: 10.2135/cropsci2014.03.0205
Bhusal, S. J., Jiang, G. L., Tilmon, K. J., and Hesler, L. S. (2013). Identification of soybean aphid resistance in early maturing genotypes of soybean. Crop Sci. 53, 491–499. doi: 10.2135/cropsci2012.06.0397
Boissot, N., Schoeny, A., and Vanlerberghe-Masutti, F. (2016). Vat, an amazing gene conferring resistance to aphids and viruses they carry, from molecular structure to field effects. Front. Plant Sci. 7:1420. doi: 10.3389/fpls.2016.01420
Boissot, N., Thomas, S., Sauvion, N., Marchal, C., Pavis, C., and Dogimont, C. (2010). Mapping and validation of QTLs for resistance to aphids and whiteflies in melon. Theor. Appl. Genet. 121, 9–20. doi: 10.1007/s00122-010-1287-8
Davis, J. A., Radcliffe, E. B., and Ragsdale, D. W. (2005). Soybean aphid, Aphis glycines Matsumura, a new vector of potato virus Y in potato. Am. J. Potato Res. 82, 197–201. doi: 10.1007/BF02853585
Diaz-Montano, J., Reese, J. C., Schapaugh, W. T., and Campbell, L. R. (2006). Characterization of antibiosis and antixenosis to the soybean aphid (Hemiptera: Aphididae) in several soybean genotypes. J. Econ. Entomol. 99, 1884–1889. doi: 10.1603/0022-0493-99.5.1884
Dogimont, C., Chovelon, V., Pauquet, J., Boualem, A., and Bendahmane, A. (2014). The Vat locus encodes for a CC-NBS-LRR protein that confers resistance to Aphis gossypii infestation and A. gossypii-mediated virus resistance. Plant J. 80, 993–1004. doi: 10.1111/tpj.12690
Du, B., Zhang, W., Liu, B., Hu, J., Wei, Z., Shi, Z., et al. (2009). Identification and characterization of Bph14, a gene conferring resistance to brown planthopper in rice. Proc. Natl. Acad. Sci. USA 106, 22163–22168. doi: 10.1073/pnas.0912139106
Fox, C. M., Kim, K. S., Cregan, P. B., Hill, C. B., Hartman, G. L., and Diers, B. W. (2014). Inheritance of soybean aphid resistance in 21 soybean plant introductions. Theor. Appl. Genet. 127, 43–50. doi: 10.1007/s00122-013-2199-1
Gildow, F. E., Shah, D. A., Sackett, W. M., Butzler, T., Nault, B., and Fleischer, S. J. (2008). Transmission efficiency of cucumber mosaic virus by aphids associated with virus epidemics in snap bean. Phytopathology 98, 1233–1241. doi: 10.1094/PHYTO-98-11-1233
Gressent, F., Duport, G., Rahioui, I., Pauchet, Y., Bolland, P., Specty, O., et al. (2007). Biological activity and binding site characteristics of the PA1b Entomotoxin on insects from different orders. J. Insect. Sci. 7, 1–10. doi: 10.1673/031.007.1201
Gressent, F., Silva, P. D., Eyraud, V., Karaki, L., and Royer, C. (2011). Pea Albumin 1 subunit b (PA1b), a promising bioinsecticide of plant origin. Toxins 3, 1502–1517. doi: 10.3390/toxins3121502
Hanson, A. A., Lorenz, A. J., Hesler, L. S., Bhusal, S. J., Bansal, R., Michel, A. P., et al. (2018). Genome-wide association mapping of host-plant resistance to soybean aphid. Plant Genome. 11, 1–12. doi: 10.3835/plantgenome2018.02.0011
Hanson, A. A., Menger-Anderson, J., Silverstein, C., Potter, B. D., MacRae, I. V., Hodgson, E. W., et al. (2017). Evidence for soybean aphid (Hemiptera: Aphididae) resistance to pyrethroid insecticides in the upper midwestern United States. J. Econ. Entomol. 110, 2235–2246. doi: 10.1093/jee/tox235
Hill, C. B., Crull, L., Herman, T. K., Voegtlin, D. J., and Hartman, G. L. (2010). A new soybean aphid (Hemiptera: Aphididae) biotype identified. J. Econ. Entomol. 103, 509–515. doi: 10.1603/ec09179
Hill, C. B., Curtis, B., Yan, Li, Glen, L., and Hartman, G. L. (2004). Resistance to the soybean aphid in soybean germplasm. Crop Sci. 44, 98–106. doi: 10.2135/cropsci2004.0098
Hill, C. B., Kim, K. S., Crull, L., Diers, B. W., and Hartman, G. L. (2009). Inheritance of resistance to the soybean aphid in soybean PI 200538. Crop Sci. 49, 1193–1200. doi: 10.2135/cropsci2008.09.0561
Hill, C. B., Shiao, D., Fox, C. M., and Hartman, G. L. (2017). Characterization and genetics of multiple soybean aphid biotype resistance in five soybean plant introductions. Theor. Appl. Genet. 130, 1335–1348. doi: 10.1007/s00122-017-2891-7
Hill, J. H., Alleman, R., Hogg, D. B., and Grau, C. R. (2001). First report of transmission of soybean mosaic virus and alfalfa mosaic virus by Aphis glycines in the new world. Plant Dis. 85:561. doi: 10.1094/PDIS.2001.85.5.561C
Hodgson, E. W., McCornack, B. P., Tilmon, K., and Knodel, J. J. (2012). Management recommendations for soybean aphid (Hemiptera: Aphididae) in the United States. J. Integr. Pest Manag. 3, E1–E10. doi: 10.1603/IPM11019
Hu, L., Wu, Y., Wu, D., Rao, W., Guo, J., Ma, Y., et al. (2017). The coiled-coil and nucleotide binding domains of BROWN PLANTHOPPER RESISTANCE14 function in signaling and resistance against planthopper in rice. Plant Cell 29, 3157–3185. doi: 10.1105/tpc.17.00263
Ji, H., Kim, S. R., Kim, Y. H., Suh, J. P., Park, H. M., Sreenivasulu, N., et al. (2016). Map-based cloning and characterization of the BPH18 gene from wild rice conferring resistance to brown planthopper (BPH) insect pest. Sci. Rep. 6, 1–14. doi: 10.1038/srep34376
Jun, T. H., Mian, M. A. R., and Michel, A. P. (2013). Genetic mapping of three quantitative trait loci for soybean aphid resistance in PI 567324. Heredity 111, 16–22. doi: 10.1038/hdy.2013.10
Jun, T., Mian, M. A. R., and Michel, A. P. (2012). Genetic mapping revealed two loci for soybean aphid resistance in PI 567301B. Theor. Appl. Genet. 124, 13–22. doi: 10.1007/s00122-011-1682-9
Kaloshian, I., Lange, W. H., and Williamson, V. M. (1995). An aphid-resistance locus is tightly linked to the nematode-resistance gene, Mi, in tomato. Proc. Natl. Acad. Sci. USA 92, 622–625. doi: 10.1073/pnas.92.2.622
Kang, S. T., Mian, M. A. R., and Hammond, R. B. (2008). Soybean aphid resistance in PI 243540 is controlled by a single dominant gene. Crop Sci. 48, 1744–1748. doi: 10.2135/cropsci2007.12.0672
Kim, K. S., Bellendir, S., Hudson, K. A., Hill, C. B., Hartman, G. L., Hyten, D. L., et al. (2010a). Fine mapping the soybean aphid resistance gene Rag1 in soybean. Theor. Appl. Genet. 120, 1063–1071. doi: 10.1007/s00122-009-1234-8
Kim, K. S., Hill, C. B., Hartman, G. L., Hyten, D. L., Hudson, M. E., and Diers, B. W. (2010b). Fine mapping of the soybean aphid-resistance gene Rag2 in soybean PI 200538. Theor. Appl. Genet. 121, 599–610. doi: 10.1007/s00122-010-1333-6
Kim, K. S., Hill, C. B., Hartman, G. L., Mian, M. A. R., and Diers, B. W. (2008). Discovery of soybean aphid biotypes. Crop Sci. 48, 923–928. doi: 10.2135/cropsci2007.08.0447
Lamantia, J. M., Mian, M. A. R., and Redinbaugh, M. G. (2019). Genetic mapping of soybean aphid biotype 3 and 4 resistance in PI 606390A. Mol. Breed. 39:53. doi: 10.1007/s11032-019-0956-9
Lee, S., Cassone, B. J., Wijeratne, A., Jun, T. H., Michel, A. P., and Mian, M. A. R. (2017). Transcriptomic dynamics in soybean near-isogenic lines differing in alleles for an aphid resistance gene, following infestation by soybean aphid biotype 2. BMC Genom. 18:1–12. doi: 10.1186/s12864-017-3829-9
Li, Y., Hill, C. B., Carlson, S. R., Diers, B. W., and Hartman, G. L. (2007). Soybean aphid resistance genes in the soybean cultivars Dowling and Jackson map to linkage group M. Mol. Breed. 19, 25–34. doi: 10.1007/s11032-006-9039-9
Liu, G., Xing, H., Diao, Y., Yang, X., Sun, D., Wang, Q., et al. (2014). Identification of resistance to soybean aphids in early germplasm. Crop Sci. 54, 2707–2712. doi: 10.2135/cropsci2013.11.0735
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–△△CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
McCarville, M. T., Soh, D. H., Tylka, G. L., and O’Neal, M. E. (2014). Above ground feeding by soybean aphid, Aphis glycines, affects soybean cyst nematode, Heterodera glycines, reproduction belowground. PLoS One 9:e86415. doi: 10.1371/journal.pone.0086415
Mensah, C., Difonzo, C., and Wang, D. (2008). Inheritance of soybean aphid resistance in PI 567541B and PI 567598B. Crop Sci. 48, 1759–1763. doi: 10.2135/cropsci2007.09.0535
Mensah, C., DiFonzo, C., Nelson, R. L., and Wang, D. (2005). Resistance to soybean aphid in early maturing soybean germplasm. Crop Sci. 45, 2228–2233. doi: 10.2135/cropsci2004.0680
Mian, M. A. R., Hammond, R. B., and Martin, S. K. S. (2008b). New plant introductions with resistance to the soybean aphid. Crop Sci. 48, 1055–1061. doi: 10.2135/cropsci2007.06.0357
Mian, M. A. R., Kang, S. T., Beil, S. E., and Hammond, R. B. (2008a). Genetic linkage mapping of the soybean aphid resistance gene in PI 243540. Theor. Appl. Genet. 117, 955–962. doi: 10.1007/s00122-008-0835-y
Michel, A. P., Omprakah, M., and Mian, R. M. (2011). Evolution of soybean aphid biotypes: understanding and managing virulence to host-plant resistance in Soybean - Molecular Aspects of Breeding. London: In Tech. 355–372.
Michelmore, R. W., Paran, I., and Kesseli, R. V. (1991). Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc. Natl. Acad. Sci. USA 88, 9828–9832. doi: 10.1073/pnas.88.21.9828
Milne, I., Shaw, P., Stephen, G., Baye, R. M., Cardle, L., Thomas, W. T., et al. (2010). Flapjack-graphical genotype visualization. Bioinformatics 26, 3133–3134. doi: 10.1093/bioinformatics/btq580
Murray, M. G., and Thompson, W. F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4325. doi: 10.1093/nar/8.19.4321
Natukunda, M. I., Parmley, K. A., Hohenstein, J. D., Assefa, T., Zhang, J., MacIntosh, G. C., et al. (2019). Identification and genetic characterization of soybean accessions exhibiting antibiosis and antixenosis resistance to Aphis glycines (Hemiptera: Aphididae). J. Econ. Entomol. 112, 1428–1438. doi: 10.1093/jee/toz017
Ragsdale, D. W., Landis, D. A., Brodeur, J., Heimpel, G. E., and Desneux, N. (2011). Ecology and management of the soybean aphid in North America. Annu. Rev. Entomol. 56, 375–399. doi: 10.1146/annurev-ento-120709-144755
Ragsdale, D. W., McCornack, B. P., Venette, R. C., Potter, B. D., MacRae, I. V., Hodgson, E. W., et al. (2007). Economic threshold for soybean aphid (Hemiptera: Aphididae). J. Econ. Entomol. 100, 1258–1267. doi: 10.1603/0022-0493
Rahioui, I., Eyraud, V., Karaki, L., Sasse, F., Carre-Pierrat, C., Qin, A., et al. (2014). Host range of the potential biopesticide Pea Albumin 1b (PA1b) is limited to insects. Toxicon 89, 67–76. doi: 10.1016/j.toxicon.2014.07.004
Schapaugh, W. T., Todd, T., Reese, J., Diaz-Montano, J., Meng, J., and Smith, C. M. (2010). Registration of K1639-2 soybean germplasm resistant to soybean cyst nematode and soybean aphid. J. Plant Regist. 4, 67–69. doi: 10.3198/jpr2009.07.0361crg
Tamura, Y., Hattori, M., Yoshioka, H., Yoshioka, M., Takahashi, A., Wu, J., et al. (2014). Map-based cloning and characterization of a brown planthopper resistance gene BPH26 from Oryza sativa L. ssp. indica cultivar ADR52. Sci. Rep. 4:5872. doi: 10.1038/srep05872
Tilmon, K. J., Hodgson, E. W., O’Neal, M. E., and Ragsdale, D. W. (2011). Biology of the soybean aphid, Aphis glycines (Hemiptera: Aphididae) in the United States. J. Integr. Pest Manag. 2, A1–A7. doi: 10.1603/IPM10016
Vos, P., Simons, G., Jesse, T., Wijbrandi, J., Heinen, L., Hogers, R., et al. (1998). The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nat. Biotechnol. 16, 1365–1369. doi: 10.1038/4350
Wang, X. B., Fang, Y. H., Lin, S. Z., Zhang, L. R., and Wang, H. D. (1994). A study on the damage and economic threshold of the soybean aphid at the seedling stage. Plant Prot. 20, 12–13.
Wu, Z., Schenk-Hamlin, D., Zhan, W., Ragsdale, D. W., and Heimpel, G. E. (2004). The soybean aphid in China: a historical review. Ann. Entomol. Soc. Am. 97, 209–218.
Xiao, L., Hu, Y., Wang, B., and Wu, T. (2013). Genetic mapping of a novel gene for soybean aphid resistance in soybean (Glycine max [L.] Merr.) line P203 from China. Theor. Appl. Genet. 126, 2279–2287. doi: 10.1007/s00122-013-2134-5
Zhang, G., Gu, C., and Wang, D. (2010). A novel locus for soybean aphid resistance. Theor. Appl. Genet. 120, 1183–1191. doi: 10.1007/s00122-009-1245-5
Zhang, S., Zhang, Z., Wen, Z., Gu, C., An, Y. C., Bales, C., et al. (2017). Fine mapping of the soybean aphid-resistance genes Rag6 and Rag3c from Glycine soja 85-32. Theor. Appl. Genet. 130, 2601–2615. doi: 10.1007/s00122-017-2979-0
Keywords: soybean aphid, resistance gene, fine mapping, NBS-LRR, gene expression
Citation: Yang J, Liu G, Tang J, Wang X, Diao Y, Su Y, Sun D, Shang J, Guo Y and Qiu L-J (2022) Fine Mapping and Characterization of an Aphid-Resistance Gene in the Soybean Landrace Fangzheng Moshidou. Front. Plant Sci. 13:899212. doi: 10.3389/fpls.2022.899212
Received: 18 March 2022; Accepted: 10 May 2022;
Published: 15 June 2022.
Edited by:
Ali M. Missaoui, University of Georgia, United StatesReviewed by:
Maria Luisa GomezGuillamon, Spanish National Research Council (CSIC), SpainYong Liu, Hunan Academy of Agricultural Sciences (CAAS), China
Copyright © 2022 Yang, Liu, Tang, Wang, Diao, Su, Sun, Shang, Guo and Qiu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li-Juan Qiu, qiulijuan@caas.cn; Yong Guo, guoyong@caas.cn
†These authors have contributed equally to this work and share first authorship
 Jing Yang1†
Jing Yang1† Xiujun Wang
Xiujun Wang Yong Guo
Yong Guo Li-Juan Qiu
Li-Juan Qiu