- 1Shandong Provincial Key Laboratory of Plant Stress Research, College of Life Science, Shandong Normal University, Jinan, China
- 2Dongying Institute, Shandong Normal University, Dongying, China
RING zinc finger proteins have a conserved RING domain, mainly function as E3 ubiquitin ligases, and play important roles in plant growth, development, and the responses to abiotic stresses such as drought, salt, temperature, reactive oxygen species, and harmful metals. RING zinc finger proteins act in abiotic stress responses mainly by modifying and degrading stress-related proteins. Here, we review the latest progress in research on RING zinc finger proteins, including their structural characteristics, classification, subcellular localization, and physiological functions, with an emphasis on abiotic stress tolerance. Under abiotic stress, RING zinc finger proteins on the plasma membrane may function as sensors or abscisic acid (ABA) receptors in abiotic stress signaling. Some RING zinc finger proteins accumulate in the nucleus may act like transcription factors to regulate the expression of downstream abiotic stress marker genes through direct or indirect ways. Most RING zinc finger proteins usually accumulate in the cytoplasm or nucleus and act as E3 ubiquitin ligases in the abiotic stress response through ABA, mitogen-activated protein kinase (MAPK), and ethylene signaling pathways. We also highlight areas where further research on RING zinc finger proteins in plants is needed.
Introduction
Plants face a variety of abiotic stresses, such as drought, high salinity, cold, and heat stress, during their growth and development (Li et al., 2019). Under these adverse conditions, plants suffer various degrees of damage. For example, under high-salt stress, the imbalance of Na+/K+ inside and outside the cell restricts plant growth (Assaha et al., 2017) and the stress-induced accumulation of ROS causes oxidative stress (Apel and Hirt, 2004). Plants have evolved a variety of physiological and biochemical defense mechanisms to deal with abiotic stressors (Hasanuzzaman et al., 2013), such as antioxidant defenses (Yang and Guo, 2018), programmed cell death (Locato and De Gara, 2018), stomatal movement (Qiao et al., 2016), and autophagy (Su et al., 2020). Functional proteins and transcription factors such as MYBs (Li C. et al., 2015), bHLHs (Sun et al., 2018), WRKYs (Chen et al., 2012), and zinc finger proteins (Han et al., 2020b,2021) are involved in these defense mechanisms.
Zinc finger proteins maintain a finger-like spatial configuration by binding to zinc ions through amino acids in the peptide chain, and are widely distributed in eukaryotes (Han et al., 2020a). RING zinc finger proteins have been identified in many plants, including Arabidopsis thaliana (Linden et al., 2021; Liu et al., 2021; Van Nguyen et al., 2021), rice (Oryza sativa) (Kim et al., 2020; Kim and Jang, 2021; Zhu et al., 2021), maize (Zea mays) (Gao et al., 2012; Xia et al., 2012; Yang et al., 2018), pepper (Capsicum annuum) (Liu et al., 2007; Joo et al., 2019a,2020), wheat (Triticum aestivum) (Agarwal and Khurana, 2018; Liu et al., 2018; Wang et al., 2020), potato (Solanum tuberosum) (Guo et al., 2003; Ni et al., 2010; Qi et al., 2020), wild tomato (Solanum pimpinellifolium) (Yang et al., 2016), wild Chinese grape (Vitis pseudoreticulata) (Yu et al., 2013; Wang et al., 2017), soybean (Glycine max) (Du et al., 2010; Biniek et al., 2017; Zhao et al., 2020), Brassica rapa (Jung et al., 2013), tobacco (Nicotiana benthamiana) (Wu et al., 2014), and cassava (Manihot esculenta Crantz) (Dos Reis et al., 2012). RING zinc finger proteins have a characteristic RING zinc finger domain. They bind to specific gene sequences, interact with a variety of proteins, participate in signal transduction and gene expression, and mediate growth, development, and environmental adaptation (An et al., 2017c; Sun et al., 2019; Han et al., 2020b). RING zinc finger proteins regulate photomorphogenesis (Von Arnim and Deng, 1993; Hardtke et al., 2002; Yang et al., 2015), floral organ size (Disch et al., 2006), rosette leaf shape (Wang et al., 2018), root development (Karlowski and Hirsch, 2003; Fleury et al., 2007; Liu et al., 2007), flowering time (Lee et al., 2001; Chen and Ni, 2006), fruit development (Wu et al., 2014), and symbiotic nodulation (Cai et al., 2018). In addition, RING zinc finger proteins play an important role in abiotic stress tolerance (Lu, 2017), especially in drought, temperature, salt, and ROS stresses (Kim et al., 2019).
Here, we focus on the role of RING zinc finger proteins in plant responses to abiotic stress and provide suggestions for future research.
Structure and Domain Organization of Ring Zinc Finger Proteins
The RING finger domain of RING zinc finger proteins is the site of protein–protein interaction and is necessary for its E3 ligase activity. RING zinc finger proteins contain a conserved cysteine-rich ring finger domain (RING finger domain), which consists of 40–60 residues arranged thus: Cys-X2-Cys-X9–39-Cys-X1–3-His-X2–3-Cys/His-X2-Cys-X4–48-Cys-X2-Cys (Stone et al., 2005). The RING finger domain contains eight spatially conserved Cys and His residues that act as metal ligands to chelate two zinc ions, and form a staggered secondary structure as a platform for the ubiquitin conjugating enzyme E2.
Classification of Ring Zinc Finger Proteins
RING zinc finger proteins are mainly divided into the following 13 types: RING-H2, RING-HC (RING-HCa and RING-HCb), RING-v, RING-C2, RING-D, RING-S/T, RING-G, RING-mh2, RING-mhc, C2SHC4, C3GC3S, C2HC5, and C3HCHC2 (Sun et al., 2019). RING zinc finger proteins have been isolated and identified in many plants. For instance, 508 RING zinc finger domains have been predicted in Arabidopsis thaliana and classified into seven categories (Jiménez-López et al., 2018). In Brassica rapa, 731 RING domains were identified from 715 predicted RING zinc finger protein genes and classified into eight categories (Alam et al., 2017). In tomato (Solanum lycopersicum), 474 RING domains were identified from 469 predicted RING zinc finger proteins and classified into seven categories (Yang et al., 2019). In rice, 425 RING zinc finger protein genes were identified and classified into four categories (Lim et al., 2010). In apple (Malus domestica), 688 RING domains were identified and divided into nine types (Li Y. et al., 2011). In Brassica oleracea, 756 RING domains were identified from 734 predicted RING zinc finger proteins and divided into eight types (Yang and Lu, 2018). In Ostreococcus tauri, the smallest photosynthetic eukaryote yet discovered, only 65 RING finger domains were identified from 65 predicted RING zinc finger proteins and divided into eight RING domain types (Gao et al., 2016). The specific structure domain characteristics and number were showed in Supplementary Table 1. According to the above reports, RING-HC (C3HC4) and RING-H2 (C3H2C3) types are the most abundant. The main difference between RING-HC and RING-H2 types is that the fifth Cys/His residue in the common sequence is Cys or His, respectively (Stone et al., 2005). It can be speculated that RING-HC and RING-H2 types of RING zinc finger proteins play important roles in plant growth and development and adversity stress.
Subcellular Localization of Ring Zinc Finger Proteins
Plant RING zinc finger proteins are located in the plasma membrane, cytoplasm, and nucleus. RING zinc finger proteins located in the plasma membrane with a characteristic transmembrane domain include ABA INSENSITIVE RING PROTEIN 1 (AtAIRP1) (Ryu et al., 2010), RING-H2 FINGER A2A (RHA2a) (Li H. et al., 2011), and ARABIDOPSIS TÓXICOS EN LEVADURA 78 (AtATL78) (Kim S. J. and Kim W. T., 2013) in Arabidopsis; RING DOMAIN-CONTAINING PROTEINS 1 (OsRDCP1) (Bae et al., 2011), SALT-INSENSITIVE RING H2-TYPE 14 (OsSIRH2-14) (Park et al., 2019), and RING FINGER PROTEIN V6 (OsRFPv6) (Kim et al., 2021) in rice; VpRH2 in grape (Wang et al., 2017); LjCZF1 in Lotus japonicus (Cai et al., 2018); and StRFP2 in potato (Qi et al., 2020). Some of the transmembrane RING zinc finger proteins (such as AtATL78, RHA2a, OsRFPv6, and OsRDCP1) involved in abiotic stress are shown in Figure 1.
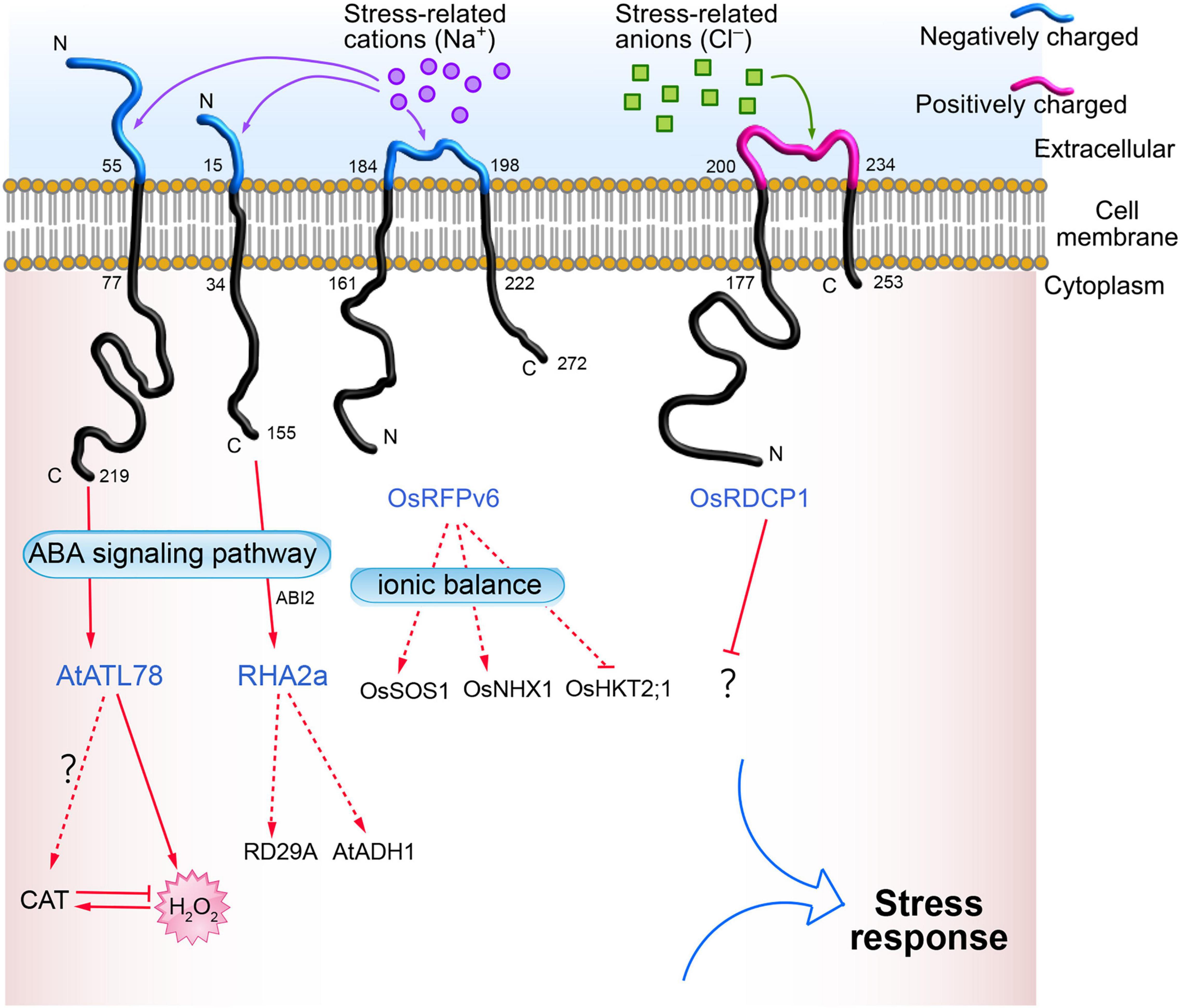
Figure 1. Bioinformatics analysis of transmembrane domains of RING zinc finger proteins located on the plasma membrane. The online tools TMHMM Server v. 2.0 (http://www.cbs.dtu.dk/services/TMHMM/) and Expasy ProtParam (https://web.expasy.org/protparam/) were used to for the analysis.
RING zinc finger proteins located in the cytoplasm include AtAIRP4 (Yang et al., 2016) and ERAD-mediating RING finger protein (EMR) (Park et al., 2018a) in Arabidopsis; OsSIRH2-14 and OsSIRP1 (Hwang et al., 2016) in rice; ABA SENSITIVE RING FINGER E3 LIGASE 1 (CaASRF1) (Joo et al., 2019a) and DROUGHT SENSITIVE RING FINGER PROTEIN 1 (CaDSR1) (Lim et al., 2018) in pepper; and ZmXerico2 in maize (Gao et al., 2012). RING zinc finger proteins located in the nucleus include AL TOLERANCE RING FINGER 1 (AtATRF1) (Qin et al., 2017) and HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 1 (AtHOS1) (Kim J.-H. et al., 2017) in Arabidopsis; SALT-, ABA- AND DROUGHT-INDUCED RING FINGER PROTEIN 1 (OsSADR1) (Park et al., 2018c) in rice; and ADIP1 INTERACTING RING FINGER PROTEIN 1 (CaAIRF1), CaASRF1, and CaDSR1 (Lim et al., 2017) in pepper.
In addition to the cell membrane, cytoplasm, and nucleus, a few RING zinc finger proteins localize in other subcellular organelles, such as the endoplasmic reticulum, Golgi apparatus, and cytoskeleton. The rice RING-HC zinc finger protein HEAT AND COLD INDUCED 1 (OsHCI1) is mainly distributed near the cytoskeleton (Lim et al., 2013). The rice RING-H2 zinc finger protein OsSIRH2-14 is not only located in the cell membrane and cytoplasm, but also found in the Golgi apparatus (Park et al., 2019). The MICROTUBULE-ASSOCIATED RING FINGER PROTEIN 1 (OsMAR1), a rice RING-H2 zinc finger protein, is found in microtubules (Park et al., 2018b), and the wild tomato RING-H2 zinc finger protein SpRing is distributed in the endoplasmic reticulum (Yang et al., 2016). This shows that RING zinc finger proteins function in numerous biological processes in plants.
Roles of Ring Zinc Finger Proteins in the Ubiquitin Proteasome Pathway
Ensuring the normal synthesis and degradation of proteins is critical for maintaining growth and development (Marshall and Vierstra, 2019). Proteins are mainly degraded through autophagy and the UPS (Blasiak et al., 2019). Under abiotic stress, large amounts of non-functional and unfolded proteins accumulate in plant cells. The UPS identifies these aberrant proteins and removes them from the cell, which alters the plant proteome and can improve survival under stress conditions (Stone, 2019).
The ubiquitin proteasome pathway has many components, including the 76-amino acid Ub, UBA, UBC, UBL, DUBs, the 26S ubiquitin proteasome, and the target substrate proteins modified by ubiquitination (Liu W. et al., 2020). UPS-mediated protein degradation consists of two stages. First, Ub attaches to the substrate protein, then the ubiquitinated substrate is degraded by the 26S proteasome (Linden and Callis, 2020). The transfer of Ub requires the participation of E1, E2, and E3 enzymes. First, the Cys on the active site of E1 is covalently bound to the terminal Gly of the Ub molecule through a high-energy thioester bond, consuming ATP to activate the Ub. Then the Ub is transferred to the Cys residue of E2 to form an E2-Ub thioester complex, and finally E3 transfers the Ub from E2 to the substrate protein.
As the last enzyme in the first stage of the ubiquitin proteasome pathway, E3 plays a specific role in the recognition of target substrates and then degrades the target protein or changes the activity of the target protein (Swatek et al., 2019). Most RING zinc finger proteins have E3 ubiquitin ligase activity (Joazeiro and Weissman, 2000). E3 ubiquitin ligases are the most diverse component of the UPS, as most plant genomes encode hundreds of E3 ubiquitin ligases, but have many fewer genes encoding other UPS components.
Roles of Ring Zinc Finger Proteins in Abiotic Stress Responses
Abiotic stress factors include drought, low temperature, high temperature, flooding, saline-alkali soils/water, ROS, and harmful metals. These factors lead to accumulation of misfolded or dysfunctional proteins, which negatively affect the physiological and biochemical processes of plant cells, resulting in reduced growth and production. RING zinc finger proteins respond to a variety of abiotic stresses through different pathways in plants, such as via expression of stress-responsive genes, the ABA pathway, the ubiquitination pathway, ROS and Ca2+ signaling (Figure 2).
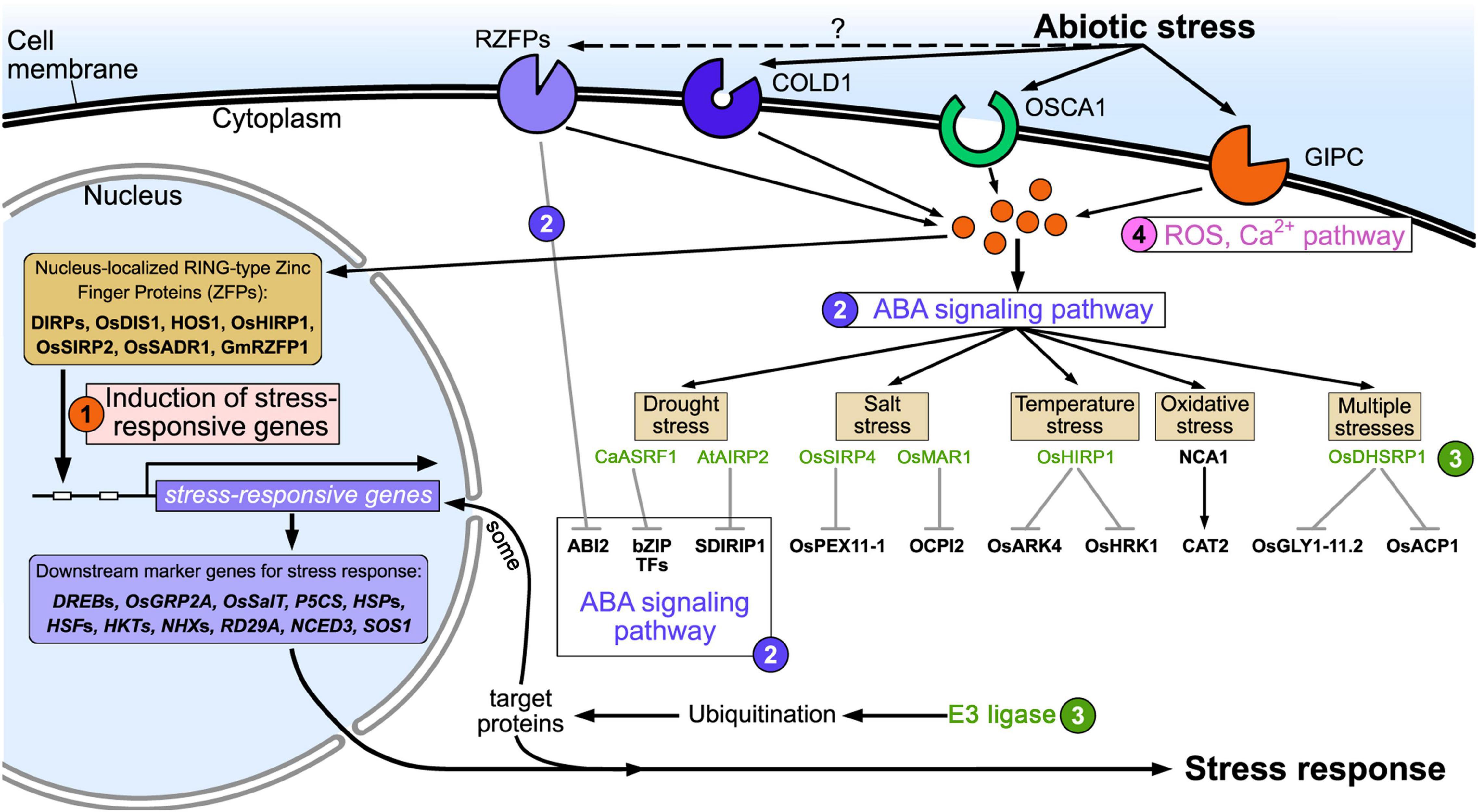
Figure 2. Schematic of RING zinc finger protein regulation of plant adaptation to abiotic stress. ➀ Expression of stress-responsive genes; ➁ ABA pathway; ➂ ubiquitination pathway; ➃ Ca2+ and ROS signaling pathway.
Roles in Drought Stress
Drought stress dramatically affects the survival and distribution of plants (Sharma et al., 2018). RING zinc finger proteins respond to drought tolerance by regulating ABA biosynthesis and signal transduction. As a type of isoprenoid phytohormone, the early steps of ABA biosynthesis occur in plastids, and the key enzymes involved in ABA biosynthesis are ZEP, NCED, short chain dehydrogenase/reductase, and ABA aldolase (Vaičiukynė et al., 2019). The three core components of the ABA signaling pathway include ABA receptors such as PYR1, PYL, and RCAR family, PP2Cs (negative regulators), and SnRK2s (positive regulators). In the presence of ABA, the formation of a PYR1/PYL/RCAR-PP2Cs complex results in the inhibition of PP2Cs activity, which in turn activates SnRK2s. The activated SnRK2s then phosphorylate downstream substrate proteins, such as bZIP transcription factors, thereby promoting the transcription of ABA responsive genes (Sah et al., 2016).
Roles in Abscisic Acid Biosynthesis
RING-H2-type zinc finger proteins in Arabidopsis greatly enhance drought tolerance by increasing ABA synthesis. AtXERICO in Arabidopsis encodes a RING-H2-type zinc finger protein. Upon drought stress, AtNECD3 expression, a key gene in ABA biosynthesis, was rapidly increased in AtXERICO-overexpressing Arabidopsis plants compared with the wild type (Ko et al., 2006). When AtXERICO was introduced into rice, the drought tolerance of transgenic rice was significantly improved compared to the wild type, and the transpiration and water consumption was lower than that of the wild type. Furthermore, under drought stress, the content of ABA and the expression of four ABA-biosynthesis or ABA-responsive genes, OsNCED, OsABA3, OsABI5, and OsLEA3-1, was significantly increased in AtXERICO-overexpressing rice compared with the wild type (Zeng et al., 2015). Maize ZmXerico1 and ZmXerico2 (Brugière et al., 2017) are homologs of AtXERICO. Unlike AtXERICO, ZmXerico genes appear to improve ABA signaling by regulating the stability of ABA 8’-hydroxylase, and ultimately increase the content of ABA in plants. Pepper CaDSR1 (Lim et al., 2018) is a RING-H2-type zinc finger protein with E3 ubiquitin ligase activity. CaDSR1 promotes drought tolerance by degrading the bZIP family protein CaDILZ1, which negatively functions in ABA accumulation.
Roles in the Abscisic Acid Signal Transduction Pathway
RING zinc finger proteins that function as E3 ubiquitin ligases participate in plant drought tolerance via regulating ABA signal transduction. For example, SALT-AND DROUGHT-INDUCED RING FINGER1 (SDIR1) encodes a RING-H2-type E3 ubiquitin ligase and SDIR1-overexpressing Arabidopsis lines show ABA hypersensitivity and ABA-related phenotypes during germination, enhanced ABA-induced stomatal closure, and drought tolerance (Zhang et al., 2007), while the sdir1 loss-of-function mutant exhibits ABA-insensitive phenotypes. Furthermore, complementation with bZIP family genes, such as ABI5, ABF3, and ABF4, can rescue the ABA-insensitive phenotype of the sdir1 mutant, whereas SDIR1 overexpression cannot rescue the ABA-insensitive phenotype of an abi5 loss-of-function mutant. This suggests that SDIR1 acts upstream of some bZIP family transcription factors and may function in the ABA signaling pathway. SDIR1 orthologs in other plant species were found to have similar functions, such as DELAYED SEED GERMINATION 1 (OsDSG1) (Park et al., 2010) and SALT-AND DROUGHT-INDUCED RING FINGER 1 (OsSDIR1) (Gao et al., 2011) in rice, ZmRFP1 in maize (Xia et al., 2012), and ABI3-INTERACTING PROTEIN 2 (AtAIP2) (Zhang et al., 2005) in Arabidopsis. Their target substrates (such as OsABI3 and AtABI3) also share high sequence similarity.
The RING-HC-type zinc finger protein AtAIRP2 (Cho et al., 2011) has E3 ubiquitin ligase activity. AtAIRP2 is strongly induced by drought and ABA. Compared with wild-type Arabidopsis, the atairp2 loss-of-function mutant showed reduced sensitivity to ABA and reduced tolerance to drought stress, whereas AtAIRP2-overexpression lines showed ABA sensitivity and enhanced drought tolerance. Accordingly, the expression of ABA-induced drought-response genes (such as RD20, RD26, RD29A, and RD29B) in the atairp2 loss-of-function lines was much lower than that in the AtAIRP2-overexpression lines. Interestingly, ectopic overexpression of AtAIRP2 in the atairp2 mutant rescues the ABA-insensitive and drought-sensitive phenotypes of the mutants. Yeast two-hybrid assays showed that AtAIRP2 interacts with SDIRIP1 (Oh et al., 2017), which is a negative factor in ABA signal transduction. SDIRIP1 selectively regulates the expression of the downstream bZIP transcription factor ABI5, rather than ABF3 or ABF4. Further results suggest that SDIRIP1 acts downstream of AtAIRP2, and AtAIRP2 may regulate the Arabidopsis response to ABA and drought stress through ubiquitination of SDIRIP1.
AtAIRP4 (Yang et al., 2016) shows high sequence similarity with AtAIRP2 and also plays a positive role in drought stress. When treated with ABA, the expression of ABI2 and ABF3 increased significantly in Arabidopsis lines overexpressing AtAIRP2, while there was no significant change in these genes in AtAIRP4-overexpression lines. These differences suggest that AtAIRP2 and AtAIRP4 may have different targets and play combined roles in drought stress responses.
The RING-H2 zinc finger proteins RHA2a and RHA2b (Li H. et al., 2011) in Arabidopsis also positively regulate ABA signal transduction and drought stress response. Genetic analyses indicated that RHA2a and RHA2b act as E3 ubiquitin ligases to ubiquitinate downstream substrates of ABI2, thus positively regulating ABA signal transduction. The rha2a rha2b double loss-of-function mutant is less sensitive to ABA and more susceptible to drought stress than the single loss-of-function mutants, suggesting that RHA2a functions redundantly with RHA2b in ABA-dependent stress responses.
The pepper RING-HC zinc finger protein CaAIRF1 (Lim et al., 2017) is a RING-type E3 ligase, and positively regulates ABA signaling and drought stress response. CaAIRF1 ubiquitinates and degrades protein phosphatase 1 (CaADIP1), which negatively regulates ABA signaling. Pepper CaASRF1 (Joo et al., 2020) improves the drought tolerance of plants and positively regulates ABA signaling by degrading downstream substrates. As a C3H2C3-type RING E3 ligase, CaASRF1 degrades, via the ubiquitination pathway, the subgroup D bZIP transcription factor CaAIBZ1 and the subgroup A bZIP transcription factor Capsicum annuum ASRF1 target bZIP transcription factor 1 (CaATBZ1) (Joo et al., 2019b), which negatively regulate ABA signaling and drought stress response, respectively. Furthermore, a yeast two-hybrid screening for CaATBZ1 identified another RING-type E3 ubiquitin ligase regulator Capsicum annuum ATBZ1 Interacting RING finger protein 1 (CaATIR1) (Joo et al., 2020), which mediates CaATBZ1 degradation through the 26S proteasome system.
RING zinc finger proteins are involved in ABA-mediated drought stress tolerance by affecting amino acid transport. AtAIRP3 (Kim J. H. and Kim W. T., 2013) is a RING-HC-type zinc finger protein with E3 ubiquitin ligase activity, and its expression was up-regulated by drought and ABA. The atairp3 loss-of-function mutant exhibited impaired ABA-mediated seed germination, impaired stomatal closure, and increased sensitivity to drought. AtAIRP3 is the same gene as LOG2 (Pratelli et al., 2012), which is involved in amino acid export from the plasma membrane. Two downstream target proteins of AtAIRP3/LOG2, GDU1 and RD21, were identified in a yeast two-hybrid assay. GDU1 is considered to be a subunit of the amino acid output complex on the plasma membrane, and AtAIRP3/LOG2 ubiquitinated GDU1 to stabilize the localization of GDU1 on the plasma membrane. RD21 (Koizumi et al., 1993) is a drought-induced cysteine protease, which may function in re-mobilizing intracellular macromolecules under stress. AtAIRP3/LOG2 ubiquitinated RD21 to prevent excessive degradation of cellular proteins under drought stress. These results suggest that AtAIRP3/LOG2 is not only involved in the ABA-mediated drought stress response, but also functions in amino acid output in Arabidopsis.
RING zinc finger proteins are involved in ABA-mediated drought stress via other mechanisms. The Arabidopsis RING-H2-type zinc finger protein AtAIRP1 (Ryu et al., 2010) has E3 ubiquitin ligase activity. AtAIRP1-overexpression lines showed ABA sensitivity and enhanced drought tolerance, whereas the loss-of-function mutants show the opposite phenotypes. The interacting protein of AtAIRP1 is AtKPNB1 (Oh et al., 2020), which is a core component of nuclear transport. AtKPNB1 acts as a negative factor in the ABA-mediated drought stress response. AtAIRP1 regulates the ABA-mediated drought response in Arabidopsis through ubiquitination of AtKPNB1. The wheat RING-HC zinc finger protein TaDIS1 (Liu et al., 2018; Lv et al., 2020) localizes in the Golgi apparatus and has E3 ubiquitin ligase activity. TaDIS1-overexpression plants have increased sensitivity to ABA. TaSTP was screened by yeast two-hybrid assays and interacts with TaDIS1 and positively regulates drought tolerance of wheat. OsDIS1 negatively regulated drought tolerance of rice by promoting TaSTP degradation through the 26S proteasome. ABA induced the expression of the RING zinc finger proteins TaRZF70 (Kam et al., 2007), AdZFP1 (Yang et al., 2008), and BdRHP1 (Zeng et al., 2019), which are involved in drought stress. However, their specific molecular mechanisms need further study.
Roles in Other Signaling Pathways in Drought Stress
In addition to the ABA signaling pathway, RING zinc finger proteins are also involved in drought tolerance through the MAPK signaling pathway. RGLG1 and RGLG2, as E3 ubiquitin ligases, ubiquitinate and degrade MAPKKK18, a mitogen-activated protein kinase, to decrease drought tolerance (Yu et al., 2021).
RING zinc finger proteins also function in drought tolerance by regulating ROS content. AtATL78, located in the plasma membrane, is a RING-H2-type E3 ubiquitin ligase in Arabidopsis (Kim S. J. and Kim W. T., 2013). atatl78 loss-of-function mutants showed decreased tolerance to drought, while AtATL78 overexpression produced the opposite phenotype. Studies found that loss of AtATL78 led to a decrease of CAT and H2O2. AtATL78 may promote ROS-mediated ABA signaling to improve drought tolerance (Suh et al., 2016). The potato RING zinc finger StRFP2 (Qi et al., 2020) is an E3 ubiquitin ligase, which enhanced drought tolerance by increasing the free proline content in potato and CAT activity in StRFP2-overexpressing N. benthamiana lines.
RING zinc finger proteins can also improve drought tolerance by regulating the content of osmoregulatory substances. AtRZF1 (Ju et al., 2013) is a RING-H2-type zinc finger protein with E3 ubiquitin ligase activity. The proline content was higher in the Arabidopsis atrzf1 loss-of-function mutant than that in the wild type and AtRZF1-overexpression plants. In subsequent experiments, atrzf1 mutants were used as materials for T-DNA tag insertion. After screening, three double mutants, atrzf1 pcp17, (Shin et al., 2019) atrzf1 pca21 (Park et al., 2017), and atrzf1 pca22 (Kim A.-R. et al., 2017), were found to have reduced proline content compared with the atrzf1 single mutant under drought conditions. These three mutations inhibited the drought-insensitive phenotype of the atrzf1 mutant by reducing the proline content, suggesting that AtRZF1 may function through the proline pathway.
RING zinc finger proteins also improve drought tolerance by mediating the ubiquitination of drought stress-related proteins. DREB2A-INTERACTING PROTEIN1 (DRIP1) and DRIP2 (Qin et al., 2008) are both RING-HC-type zinc finger proteins with E3 ubiquitin ligase activity in Arabidopsis. Drought-related gene expression was significantly enhanced in the drip1 drip2 double loss-of-function mutant under drought stress. In vitro ubiquitination experiments showed that DRIP1 and DRIP2 mediate the ubiquitination of DREB2A, thereby negatively regulating plant tolerance to drought stress. The RING-HC zinc finger protein OsDIS1 (Ning et al., 2011a,b) functions as an E3 ubiquitin ligase to negatively regulate drought tolerance in rice by promoting OsNek6 and OsSKIPa degradation. RGLG2 (Cheng et al., 2012) translocates from the plasma membrane to the nucleus and interacts with AtERF53 under drought stress. RGLG2 plays a negative role in resisting drought stress by degrading AtERF53 through the ubiquitination pathway. The N. benthamiana RING-H2 zinc finger protein NtRHF1 (Xia et al., 2013) is an ortholog of AtSDIR1. AtUAP1 (Min et al., 2021) interacted with AtRZF1. Ubiquitin-Associated Protein 1 (AtUAP1) contains a Ub-associated motif, and AtUAP1-overexpressing Arabidopsis plants were sensitive to drought stress, but the atrzf1 mutation rescued this phenotype. This suggests that AtRZF1 may also participate in the Arabidopsis response to drought stress through AtUAP1 ubiquitination.
RING zinc finger proteins can also improve drought tolerance through the endoplasmic reticulum stress pathway. AtRMA1 in Arabidopsis (Matsuda et al., 2001) and CaRma1H1 in pepper (Lee et al., 2009) are both RING-HC-type zinc finger proteins with E3 ubiquitin ligase activity. AtRma1 and CaRma1H1 are both located on the endoplasmic reticulum, both are induced by abiotic stress, and both inhibit the transport of the aquaporin PIP2;1 from the endoplasmic reticulum to the plasma membrane through ubiquitination. However, there is also an interesting difference between AtRma1 and CaRma1H1; CaRma1H1 overexpression in Arabidopsis is much more stable than AtRma1 overexpression in Arabidopsis, which explains why Arabidopsis CaRma1H1-overexpression lines have enhanced drought tolerance and Arabidopsis AtRMA1-overexpression lines show no obvious phenotypic changes compared to wild-type Arabidopsis. Overexpression of OsRDCP1 (Bae et al., 2011), a homolog of CaRma1H1, in rice also enhanced drought tolerance. Furthermore, overexpression of CaRma1H1 in cultivated tomato (Solanum lycopersicum) also enhanced drought tolerance (Seo et al., 2012), and the expression of downstream endoplasmic reticulum stress marker genes (such as LeCNX1, LeBIP1, and LePDIL1) was significantly up-regulated in the transgenic tomato, suggesting that CaRma1H1 overexpression may enhance drought tolerance not only by regulating aquaporins but also by modulating endoplasmic reticulum stress.
Roles in Temperature Stress
Extreme temperatures (too low or too high) affect plant metabolic activities, cell membrane fluidity, membrane lipid composition, photosynthesis, and respiration, thus affecting plant growth and development (Ding et al., 2020). RING zinc finger proteins are involved in the response to temperature stress.
Roles in Low Temperature Stress
RING zinc finger proteins are involved in the response to low temperature by ubiquitinating low temperature-induced transcription factors. The RING-HC zinc finger protein HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENES 1 (HOS1) (Dong et al., 2006) with E3 ubiquitin ligase activity in Arabidopsis is a key negative regulator of plant tolerance to low temperature. HOS1 interacted with ICE1 to mediate ICE1 ubiquitination and degradation. Therefore, HOS1 overexpression inhibited the expression of CBF genes and decreased tolerance to low temperature in Arabidopsis.
RING zinc finger proteins can improve the low temperature tolerance of plants by increasing the content of osmoregulation substances. In rice, the RING-H2 zinc finger protein COLD-INDUCIBLE (OsCOIN) (Liu et al., 2007) is induced by low temperature. OsCOIN overexpression in transgenic rice not only significantly improved cold tolerance, but also increased OsP5CS expression and increased the proline content, which plays an important role in cell osmotic balance.
Roles in High Temperature Stress
In addition to being responsive to low temperatures, RING zinc finger proteins are also involved in the response to high temperature.
RING zinc finger proteins participate in high temperature tolerance by activating DNA repair systems. Heat tolerance and DNA repair capacity were significantly reduced in the HOS1-deficient Arabidopsis mutants, in which heat-induced expression of DNA repair system genes, such as the DNA helicase RECQ2, was significantly reduced (Han et al., 2020c). Furthermore, HOS1 is heat stable in a heat shock factor Hsp90-dependent manner. The heat-responsive HSP90-HOS1-RECQ2 module helps maintain genome integrity under high temperature stress, demonstrating a significant relationship between DNA repair and plant heat tolerance.
RING zinc finger proteins can ubiquitinate high-temperature-tolerance-related proteins by trafficking between different subcellular organelles under heat stress. The rice RING-HC zinc finger protein HEAT AND COLD INDUCED 1 (OsHCI1) (Lim et al., 2013) is induced by high temperature. OsHCI1 mainly localizes to the Golgi apparatus and moved rapidly and widely in the cytoskeleton. At high temperature, OsHCI1 accumulates in the nucleus. Heat shock-induced accumulation of OsHCI1 mediates nucleocytoplasmic trafficking of nuclear substrate proteins through monoubiquitination, thereby acting as an inactivation mechanism to improve heat tolerance. Furthermore, the sorghum (Sorghum bicolor) ortholog of OsHCI1, SbHCI1 (Lim et al., 2020), is also a RING E3 ubiquitin ligase, and SbHCI1 predominantly localizes to the cytosol, but it moves to the Golgi under heat stress. Ectopic overexpression of SbHCI1 in Arabidopsis improved ABA sensitivity and heat stress tolerance. Consistent with OsHCI1, SbHCI1 also acts as a positive regulator of the heat stress response.
RING zinc finger proteins improve high temperature tolerance by ubiquitinating high-temperature-tolerance-related proteins. Rice HEAT-INDUCED RING FINGER PROTEIN 1 (OsHIRP1) (Kim et al., 2019) is a RING-HC zinc finger protein with E3 ligase activity, which positively regulates the response to high temperature stress. Two OsHIRP1-interacting proteins, OsARK4 and OsHRK1, were screened and identified by yeast two-hybrid and BiFC assays. In vitro ubiquitination assays revealed that OsHIRP1 directly ubiquitinates OsARK4 and OsHRK1. In addition, under heat stress, heat stress-inducible genes, such as HsfA3, HSP17.3, HSP18.2, and HSP20, were up-regulated in OsHIRP1-overexpressing Arabidopsis lines compared to the control.
RING zinc finger proteins are involved in high temperature tolerance through the ABA signaling pathway. The rice RING-HC zinc finger protein OsRZFP34 (Hsu et al., 2014) is an E3 ubiquitin ligase, and its expression is induced by high temperature and ABA treatment. The relative stomatal opening was increased in rice and Arabidopsis plants overexpressing OsRZFP34 compared with their wild types. The analysis of water loss and leaf temperature showed that the evaporation rate and cooling effects of OsRZFP34 overexpression in rice and Arabidopsis were higher than those of their wild types, and OsRZFP34 expression had protective effects on stomatal opening, leaf cooling, and tolerance to high temperature stress. The rice RING-H2 zinc finger protein HEAT TOLERANCE AT SEEDLING STAGE (OsHTAS) (Liu et al., 2016) is an E3 ubiquitin ligase. OsHTAS regulates H2O2 accumulation in leaves, alters the stomatal pore size of leaves, and promotes ABA biosynthesis. OsHTAS enhances heat tolerance by regulating stomatal closure induced by H2O2, and participates in drought and salt tolerance pathways through the ABA signaling pathway.
RING zinc finger proteins are also involved in high temperature tolerance through the ethylene signaling pathway. Ethylene, a gaseous phytohormone, plays a crucial role in plant adaptive responses to temperature changes. As an E3 ubiquitin ligase, SALT-AND DROUGHT-INDUCED RING FINGER1 (SDIR1) (Hao et al., 2021) positively regulates the ethylene response. SDIR1 mediates temperature-induced EBF1/EBF2 degradation and EIN3 accumulation to fine tune the ethylene response to environmental temperature changes, enabling plants to adapt to fluctuating environmental conditions.
Roles in Salt Stress
Long-term exposure to high salinity results in a high osmotic potential and ion toxicity within the plant, which inhibits growth and development (Acosta-Motos et al., 2017). The expression of genes encoding RING zinc finger proteins was up-regulated under high salt conditions (Dos Reis et al., 2012). Many RING zinc finger proteins have a potential role in salt stress tolerance.
RING zinc finger proteins improve salt stress tolerance by regulating ion homeostasis and ROS scavenging. Arabidopsis SALT TOLERANCE RING FINGER1 (AtSTRF1) (Tian et al., 2015) is a RING-H2-type zinc finger protein. AtSTRF1 has E3 ubiquitin ligase activity in vitro, and STRF1-overexpressing Arabidopsis lines exhibit enhanced tolerance to salt and osmotic stresses, reduced ROS accumulation under salt stress, and increased expression of AtRbohD, which encodes a NADPH oxidase involved in H2O2 production. STRF1 is a ubiquitin ligase related to membrane transport, which helps plants respond to salt stress by monitoring intracellular membrane transport and ROS production. The rice RING-H2-type zinc finger protein OsSIRH2-14 (Park et al., 2019) has E3 ubiquitin ligase activity and functions in salt tolerance by modifying salt stress-related proteins such as OsHKT2. OsSIRH2-14 is expressed in NaCl-treated root and stem tissues. Compared with wild-type plants, OsSIRH2-14-overexpressing rice plants show obvious salt tolerance, and the accumulation of Na+ in the aboveground and root tissues is significantly reduced. In vitro pull-down experiments and BiFC showed that OsSIRH2-14 interacted with salt stress-related proteins, including OsHKT2;1, OsSalT, and OsPRF2. OsSIRH2-14 may positively regulate the salt stress response by regulating the stability of salt-related proteins. Rice RING FINGER PROTEIN V6 (OsRFPv6) (Kim et al., 2021) is a RING-HC-type zinc finger protein with E3 ubiquitin ligase activity that positively regulates salt tolerance in rice. OsRFPv6 was highly expressed under saline conditions. Under salt stress, OsRFPv6-overexpression plants were much less sensitive to salt stress compared with wild-type rice, and OsRFPv6-overexpression lines had higher levels of proline, soluble sugars, and chlorophyll, and lower H2O2 accumulation than wild-type rice. The expression of the Na+ transporter genes OsHKT2;1 and OsCNGC1 was significantly lower in the roots and shoots of the OsRFPv6-overexpression plants than in the wild-type plants under salt stress. However, OsHKT1;5, which encodes a transporter involved in phloem Na+ exclusion to prevent leaf Na+ accumulation, was significantly up-regulated in roots and shoots of the OsRFPv6-overexpression plants. Furthermore, expression of the genes encoding the Na+/H+ antiporter OsNHX1, located in the vacuolar membrane, the Na+/H+ antiporter OsSOS1, located in the plasma membrane, and the K+ transporter OsHAK7 was significantly up-regulated in the roots and shoots of the OsRFPv6-overexpression plants. These results suggest that OsRFPv6 positively regulates salt stress responses by regulating the expression of Na+-related transporter genes. Rice OsSIRP1 (Hwang et al., 2016) is a RING-HC-type zinc finger protein with E3 ubiquitin ligase activity. OsSIRP1-overexpressing Arabidopsis lines had attenuated salt tolerance during seed germination and root growth. Consistent with OsSIRP1, OsSIRP4-overexpression plants also exhibited hypersensitivity to salt responses (Kim and Jang, 2021). Yeast two-hybrid and BiFC analyses showed that OsSIRP4 regulates the degradation of its substrate PEROXISOMAL BIOGENESIS FACTOR 11-1 (OsPEX11-1) through the 26S proteasome system. OsPEX11 is involved in salt stress tolerance by regulating the expression of cation transporter and antioxidant defense genes, and can significantly improve the salt tolerance of rice (Cui et al., 2016). In OsSIRP4-overexpressing plants, the enzymatic activities of CAT, SOD, and APX were significantly reduced, and the accumulation of soluble sugars and proline also decreased, but the H2O2 content was significantly elevated. Furthermore, qRT–PCR showed that the expression of Na+/K+ homeostasis-related genes AtSOS1, AtAKT1, AtNHX1, and AtHKT decreased in the transgenic plants. These results indicate that OsSIRP4 regulates OsPEX11-1 accumulation through the ubiquitination pathway, and OsPEX11-1 most likely regulates its downstream genes (such as AtSOS1, AtAKT1, AtNHX1, and AtHKT) in response to salt stress. The sweet potato (Ipomoea batatas) RING-H2-type zinc finger protein IbATL38 (Du et al., 2021) is an E3 ubiquitin ligase. IbATL38 expression was strongly induced by NaCl and ABA. Subcellular localization showed that IbATL38 localized in the nucleus and plasma membrane. Arabidopsis lines overexpressing IbATL38 exhibited enhanced salt tolerance and reduced H2O2 content. Under salt stress, the upregulation of ROS scavenging system genes (such as AtSOD, AtPOD, and AtAPX) in IbATL38-overexpressing Arabidopsis lines was much higher than that in wild-type Arabidopsis. Therefore, the ROS content in the overexpression lines was lower than that in wild-type Arabidopsis under salt stress. These results suggest that IbATL38 may improve plant tolerance to salt stress by promoting ROS scavenging.
RING zinc finger proteins improve plant salt tolerance via the ABA signaling pathway. The Arabidopsis RING-H2-type RING zinc finger protein SDIR1 (Zhang et al., 2015) is an E3 ligase. SDIR1 expression is up-regulated under salt stress. SDIR1 overexpression leads to increased ABA sensitivity and phenotypic changes related to ABA signaling pathways, such as increased salt sensitivity during germination, ABA-induced stomatal closure, and enhanced drought tolerance. SDIR1 targets SDIRIP1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Wild tomato SpRing (Qi et al., 2016) encodes a RING-H2-type zinc finger protein with E3 ubiquitin ligase activity. SpRing is expressed in all tissues of wild tomato and its expression is regulated by salt stress. Virus-induced gene silencing lines showed increased sensitivity to salt stress, while SpRing-overexpressing transgenic Arabidopsis showed enhanced salt tolerance during seed germination and early seedling development. Under salt stress, the Na+ content in the shoots of SpRing-silenced tomato plants was much higher than that in the control group. Further, expression of the ABA-independent gene SlRD29A and the ABA-dependent gene SlNCED was decreased in SpRing-silenced tomato plants, which was consistent with the increased expression of AtRD29A and AtNCED3 in SpRing-overexpressing Arabidopsis plants. These results indicate that SpRing is involved in ion homeostasis under salt stress and may play a positive regulatory role in both ABA-dependent and ABA-independent pathways.
RING zinc finger proteins improve plant salt tolerance by ubiquitinating salt tolerance-related proteins. Rice plants overexpressing the RING-H2-type zinc finger gene OsSIRP3 (Park et al., 2018d) showed hypersensitive phenotypes to salt stress including reduced seed germination rate and root growth. OsSIRP3 interacted with the salt-induced proteins OsMADS70 and OsABC1P11 in a yeast two-hybrid assay. Therefore, OsSIRP3 may negatively regulate salt stress responses by regulating these two proteins. However, unlike OsSIRP1, OsSIRP3, and OsSIRP4, the RING-HC zinc finger protein OsSIRP2 (Chapagain et al., 2018) showed a positive regulatory effect on the salt stress response. Heterologous overexpression of OsSIRP2 in Arabidopsis plants resulted in enhanced tolerance to salt and osmotic stresses. Rice TRANSKETOLASE1 (OsTKL1) was identified as an OsSIRP2-interacting partner by yeast two-hybrid assay. Transketolases play different roles in plant growth, development, and physiology (Bernacchia et al., 1995). This finding indicates that OsSIRP2 may positively regulate the plant response to salt and osmotic stresses by regulating OsTKL1. The rice RING-H2-type zinc finger protein OsMAR1 (Park et al., 2018b) with E3 ligase activity is highly expressed under high salinity. Compared with the wild type, OsMAR1-overexpressing Arabidopsis lines showed slow root growth and exhibited hypersensitivity under high salinity. Yeast two-hybrid and BiFC assays indicated that OsMAR1 interacts with rice CHYMOTRYPSIN INHIBITOR 2 (OsCPI2) and degrades OsCPI2 through the ubiquitination pathway. While heterologous overexpression of OsCPI2 leads to enhanced growth and enhanced salinity and osmotic stress tolerance in transgenic Arabidopsis (Tiwari et al., 2015), OsMAR1 may negatively regulate the salt stress response by regulating OsCPI2 in rice.
Roles in Oxidative Stress
Under oxidative stress, the metabolic homeostasis of active oxygen is destroyed, resulting in cellular ROS accumulation (Nadarajah, 2020). High concentrations of superoxide radicals, hydroxyl radicals, and H2O2 oxidatively damage biological macromolecules, such as protein and DNA, and affect plant growth and development (Pardo-Hernández et al., 2020). Some RING zinc finger proteins are also involved in oxidative stress responses in plants.
Soybean GmRZFP1 (Wu et al., 2010) is a RING-HC-type zinc finger protein involved in oxidative stress in plants. Under oxidative stress, compared with wild-type soybean, GmRZFP1-overexpressing soybean lines had stronger tolerance to oxidative stress. Arabidopsis AtOHRP1 (Li et al., 2013) is also a HC-type RING zinc finger protein, and AtOHRP expression is significantly induced by H2O2. Analysis of changes of the H2O2 content and antioxidant enzyme activity in plants after oxidative stress revealed that the atohrp mutant accumulated more H2O2, and the activities of key enzymes in the active oxygen scavenging system, such as SOD, CAT, and APX, decreased significantly compared with the wild type. This indicated that AtOHRP1 has an important physiological function in oxidative stress. Arabidopsis AtNCA1 (Li J. et al., 2015), a RING-type zinc finger protein, interacts with and activates CAT, and maintains the proper CAT folding to ensure maximum CAT enzyme activity under oxidative stress, to maintain ROS homeostasis, and to enhance the tolerance to oxidative stress. Arabidopsis lines with mutated NCA1 are hypersensitive to multiple abiotic stresses.
Apple MdMIEL1 (An et al., 2017b) is a RING zinc finger protein with E3 ubiquitin ligase activity. MdMIEL1 expression was significantly induced by H2O2. MdMIEL1-overexpressing Arabidopsis and apple calli exhibited higher H2O2 sensitivity compared to the wild-type controls. DAB and NBT tissue staining showed that MdMIEL1-overexpressing plants exhibited more ROS accumulation than wild-type plants without H2O2 treatment, suggesting that MdMIEL1 may be involved in ROS homeostasis. Yeast two-hybrid assay identified MdMYB1 as the interactor of MdMIEL1 (An et al., 2017a). MdMYB1 regulates anthocyanin biosynthesis in apple, and anthocyanins are closely associated with various abiotic stresses such as oxidative stress. MdMIEL1 negatively regulates anthocyanin accumulation by degrading MdMYB1 through the ubiquitination pathway. MdMIEL1 negatively regulates oxidative stress tolerance by regulating ROS homeostasis and anthocyanin biosynthesis. Further studies showed that MdMIEL1 also regulates lateral root initiation by increasing auxin accumulation in MdMIEL1-overexpressing Arabidopsis roots (An et al., 2017b).
Roles in Harmful Metal Stress
Harmful metals such as cadmium and aluminum can substantially hinder plant growth and development. Heavy metal cadmium accumulation in plants seriously affects the absorption and transportation of nutrients and water, increases oxidative damage, perturbs metabolism, and ultimately inhibits plant growth and development (Haider et al., 2021). Some 50% of the world’s potential arable land is acidic soil (Chauhan et al., 2021), and light metal aluminum ions have become the main factor limiting plant growth in acidic soil by inhibiting root growth, increasing oxidative stress, inhibiting photosynthesis, and interfering with the absorption and transportation of water and nutrients (Sade et al., 2016). Traditional methods such as fertilizer application and physical modification of soil composition to help plants cope with cadmium and aluminum stress have shown limited success.
RING zinc finger proteins enhance harmful metal tolerance by reducing the accumulation of metals and alleviating oxidative stress. The soybean RBR type zinc finger protein GmARI1 (Biniek et al., 2017) enhances plant tolerance to aluminum stress. GmARI1 is an E3 ubiquitin ligase. qRT-PCR showed that GmARI1 expression in soybean roots was strongly induced by aluminum, and GmARI1-overexpressing Arabidopsis plants exhibited significantly enhanced aluminum stress tolerance compared with wild-type Arabidopsis. Glyma02g15070 was the top gene co-expressed with GmARI1, and the Glyma02g15070 ortholog in Arabidopsis, AT1G49670, is involved in oxidative stress tolerance (Rounds and Larsen, 2008). SlRING1 (Ahammed et al., 2020) is a RING E3 ligase that positively regulates cadmium tolerance in cultivated tomato. When cadmium stress was encountered, compared with the wild type, SlRING1-overexpressing tomato lines not only had an increased chlorophyll content, net photosynthetic rate, and maximal photochemical efficiency of photosystem II, but also decreased ROS levels and relative electrolyte leakage. Furthermore, the improvement of cadmium tolerance in SlRING1-overexpression plants was also suggested to be associated with the increased transcript levels of CAT, DHAR, MDHAR, GSH, and PCS genes, which are involved in antioxidant and detoxification systems. More importantly, SlRING1-overexpression plants also had decreased cadmium concentrations in shoots and roots compared to wild-type tomato.
RING zinc finger proteins can temporarily ameliorate the DNA damage caused by harmful metal stress through the ubiquitination pathway. The Arabidopsis RING-RH-type zinc finger protein AL TOLERANCE RING FINGER 1 (AtATRF1) (Qin et al., 2017) also enhances plant tolerance to aluminum stress. Like GmARI1, AtATRF1 also encodes an E3 ubiquitin ligase, and its expression is increased by aluminum treatment. AtATRF1-overexpressing Arabidopsis lines showed enhanced tolerance to aluminum compared with wild-type plants. Curiously, the expression of nine genes related to aluminum stress tolerance showed no significant differences between wild-type plants and AtATRF1-overexpressing lines. However, compared with wild-type plants, the expression of AtATR (a gene that monitors DNA integrity and controls cell cycle progression) (Rounds and Larsen, 2008) was lower in AtATRF1-overexpressing plants.
Mechanisms of Ring Zinc Finger Proteins in Abiotic Stress
When plants face abiotic stresses such as drought, high salt, high or low temperature, and heavy metals, these external stress signals are first sensed by the receptors or sensors on the cell’s plasma membrane, and are transmitted to the cell through ROS or Ca2+ signals (Choi et al., 2017; Marcec et al., 2019; Chen and Yang, 2020). Therefore, abiotic stress receptors on the plasma membrane play a crucial role in the plant’s response to stress. However, our knowledge of abiotic stress receptors on plant cell plasma membranes is limited, and only a few receptors have been reported, such as low temperature receptors (COLD1) (Ma et al., 2015), drought stress receptors (OSCA1) (Yuan et al., 2014), and salt stress receptors (GIPC) (Jiang et al., 2019; Figure 2). Numerous studies have found that RING zinc finger proteins are significantly induced by drought (Joo et al., 2020; Yu et al., 2021), high salt (Liu Y. et al., 2020; Kim and Jang, 2021), high or low temperature (Han et al., 2020c; Lim et al., 2020), and heavy metals (Qin et al., 2017; Ahammed et al., 2020). This indicates that RING zinc finger proteins participate in the response to abiotic stresses either directly or indirectly. Interestingly, subcellular localization found that some RING zinc finger proteins accumulate on the cell plasma membrane and are transmembrane proteins (Cheng et al., 2012; Suh et al., 2016). For example, AtATL78 is involved in cold and drought stress (Suh et al., 2016), RHA2a (Li H. et al., 2011) and OsRDCP1 (Bae et al., 2011) are involved in drought stress, and OsRFPv6 is involved in salt stress (Kim et al., 2021). Bioinformatics analysis found that these transmembrane proteins have an extracellular domain, transmembrane domain, and intracellular domain. The extracellular domains of some RING zinc finger proteins are negatively charged (such as AtATL78, RHA2a, and OsRFPv6), and some are positively charged (such as OsRDCP1), as shown in Figure 1. These charged extracellular domains may react with harmful stress-related ions such as Na+ or Cl– in the response to the external abiotic stress, and thus act as sensors in stress perception. The intracellular domains transmit these abiotic stress signals to the cytoplasm. For example, AtATL78 may promote ROS signal-mediated ABA signaling to improve plant stress tolerance (Suh et al., 2016); the extracellular domain of OsRFPv6 may be involved in salt stress (Kim et al., 2021); and RHA2a mediates drought stress directly through ubiquitinating ABI2 in the ABA receptor signaling pathway. These cases indicate that RING zinc finger proteins on the plasma membrane may function as sensors or ABA receptors in abiotic stress signaling, which needs further study. The amino acid sequence of RING zinc finger proteins expressed on the extracellular plasma membrane is shown in Supplementary Table 2.
In addition, some RING zinc finger proteins accumulate in the nucleus (such as DIRPs, OsDIS1, HOS1, and OsHIRP1). Under abiotic stress, these RING zinc finger proteins may act like transcription factors to regulate the expression of downstream abiotic stress marker genes (such as DREBs, HSPs, HKTs, and RD29A) through direct or indirect ways (Qin et al., 2008; Ning et al., 2011a; Kim et al., 2019; Han et al., 2020c).
RING zinc finger proteins usually accumulate in the cytoplasm or nucleus and act as E3 ubiquitin ligases (such as CaASRF1 and AtAIRP2) in the abiotic stress response through ABA, MAPK, and ethylene signaling pathways. RING zinc finger proteins play a key role in ABA signaling especially under drought conditions. They have positive or negative roles in stress tolerance through ubiquitination and degradation of ABA receptor and ABA synthesis-related proteins (such as bZIP transcription factors and SDIRIP1) (Li J. et al., 2015; Oh et al., 2017; Joo et al., 2019a). In addition, a few RING zinc finger proteins, such as NCA1 (Li J. et al., 2015), interact with and activate target proteins involved in stress tolerance (such as CAT2) to maintain correct folding and functionality of the target protein.
The possible mechanisms of RING zinc finger proteins in plant abiotic stress tolerance are shown in Figure 2, and the expression patterns, resistance manners, and references of RING zinc finger proteins related to abiotic stress are summarized in Supplementary Table 3.
Summary and Future Prospects
The growth and development of plants and their ability to adapt to various stresses are mainly achieved by protein modifications and altering metabolic pathways. Studying the expression and function of RING zinc finger proteins is of great importance to improve plant growth, development, and abiotic stress tolerance.
Protein ubiquitination is one of the most important post-translational modifications of plant proteins. Most RING zinc finger proteins function as E3 ligases in different cellular processes via degrading or modifying their substrate proteins through the 26S proteasome. Ubiquitin E3 ligase determines the specific substrate proteins. However, how E3 ligases interact with substrate proteins, whether they recognize substrate proteins with the same characteristics, and whether they can modify substrate proteins through ubiquitin or polyubiquitin remains to be further determined. The ubiquitin pathway involves many types of RING finger proteins, and each type of RING finger domain may correspond to a variety of E2 ubiquitin binding enzymes. Therefore, further biochemical analysis is needed to combine E2 ubiquitin binding enzymes from different plants with E3 enzymes to further understand how E2-E3 enzymes interact.
Further analysis of the mechanisms of action of RING zinc finger proteins is needed to answer the above questions. New biotechnological tools will also accelerate the research on RING zinc finger proteins. For example, bioinformatics analysis combined with functional genomics and proteomics can be used to infer the corresponding DNA sequences from amino acid sequences of unknown RING zinc finger proteins. The function of unknown RING zinc finger proteins could be identified by gene knockout, gene silencing, or gene overexpression approaches. Additionally, single cell sequencing, spatial transcriptomics, and metabolomics can be used to gain further insight into their structures and functions. Furthermore, the structure and function of RING zinc finger proteins can be artificially changed via clustered regularly interspaced short palindromic repeats (CRISPR)/CRISPR-associated protein 9 (Cas9) genome editing.
Although RING zinc finger proteins have been studied in many species, the current research has mainly focused on Arabidopsis and rice, while little is known about these proteins in other species, especially in stress-resilient plants. Therefore, the function of RING zinc finger proteins still needs to be discovered and studied in halophytes, xerophytes, and heavy metal-tolerant plants.
In addition, the RING zinc finger proteins that are associated with the plasma membrane warrant further study. Plants sometimes have cross adaptation to multiple stresses, and one stress may have multiple effects. For example, salt stress can lead to osmotic stress and ionic stress. Identifying receptors of abiotic stress signals in plants is a hotspot of research and is difficult to study. Studying the characteristics of extracellular domains of transmembrane RING zinc finger proteins could help to clarify their functions in stress signaling. While some transmembrane RING zinc finger proteins (such as AtATL78, RHA2a, and OsRDCP1) with a charged extracellular domain have only been reported to be involved in drought stress, they may also participate in salt and/or heavy metal stresses. Therefore, molecular biological techniques, such as CRISPR-Cas9, for domain modification could help to determine how plasma membrane-localized RING zinc finger proteins perceive abiotic stress signals.
Another important finding is that some RING-type zinc finger proteins undergo nucleocytoplasmic shuttle under stress like CCCH zinc finger proteins (Han et al., 2021). For example, OsHIRP1 has an NLS sequence and should theoretically be located in the nucleus, but at room temperature, 100% of rice protoplasts showed cytoplasmic and nuclear localization. However, under heat treatment (38°C), 11.1% of rice protoplasts showed only nuclear localization, and 88.9% of rice protoplasts still showed cytoplasmic and nuclear localization. Interestingly, under severe heat stress (45°C), 95.8% of rice protoplasts showed nuclear localization, and only 4.2% showed cytoplasmic and nuclear localization (Kim et al., 2019). The mechanism and significance of nucleocytoplasmic shuttle of these RING zinc finger proteins need to be further studied.
Overall, RING zinc finger proteins have great potential in plant stress tolerance. Genetic engineering of RING zinc finger proteins may provide a useful approach to accelerate crop stress tolerance breeding and thus promote sustainable agriculture.
Author Contributions
GH and ZQ wrote this manuscript. YL, ZY, CW, YZ, and LL participated in the writing and modification of this manuscript. GH and BW conceptualized the idea. All authors read and approved the final manuscript.
Funding
This work was supported by National Natural Science Research Foundation of China (Project Nos. 32000209 and 31770288), Natural Science Research Foundation of Shandong Province (Project No. ZR2020QC031), and China Postdoctoral Science Foundation (Project No. 2020M672114).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.877011/full#supplementary-material
Abbreviations
ABA, abscisic acid; MAPK, mitogen-activated protein kinase; ROS, reactive oxygen species; UPS, ubiquitin proteasome system; Ub, ubiquitin molecules; UBA, ubiquitin activating enzyme E1; UBC, ubiquitin conjugating enzyme E2; UBL, ubiquitin ligase E3; DUBs, deubiquitinating enzymes; Cys, cysteine residue; Gly, glycine; ZEP, zeaxanthin epoxidase; NCED, 9-cis-epoxycarotenoid dioxygenase; PYR1, ACTIN RESISTANCE 1; PYL, PYR1-LIKE; RCAR, REGULATORY COMPONENTS OF ABA RECEPTORS; PP2Cs, type 2C protein phosphatases; SnRK2s, SNF1-related protein kinase 2s; bZIP, basic region/leucine zipper; H2O2, hydrogen peroxide; CAT, catalase; SOD, superoxide dismutase; APX, ascorbate peroxidase; POD, guaiacol peroxidase; CBF, C-repeat binding factor; Hsp90, A1/heat shock protein 90; BiFC, biomolecular fluorescence complementation; NADPH, nicotinamide adenine dinucleotide phosphate; RBR, Ring-Between-Ring fingers; qRT-PCR, quantitative real-time polymerase chain reaction; DHAR, dehydroethrin reductase; MDHAR, monodehydroascorbate reductase; GSH, glutathione; PCS, phytochelatin synthase; GST, glutathione S-transferase; COLD1, CHILLING-TOLERANCE DIVERGENCE 1; OSCA1, reduced hyperosmolality-induced [Ca2+]i increase 1; GIPC, glycosyl inositol phosphorylceramide; DAB, 3,3 ′ -diaminobenzidine; NBT, nitro blue tetrazolium.
References
Acosta-Motos, J. R., Ortuño, M. F., Bernal-Vicente, A., Diaz-Vivancos, P., Sanchez-Blanco, M. J., and Hernandez, J. A. (2017). Plant responses to salt stress: adaptive mechanisms. Agronomy 7:18. doi: 10.3390/agronomy7010018
Agarwal, P., and Khurana, P. (2018). Characterization of a novel zinc finger transcription factor (TaZnF) from wheat conferring heat stress tolerance in Arabidopsis. Cell Stress Chaperones 23, 253–267. doi: 10.1007/s12192-017-0838-1
Ahammed, G. J., Li, C. X., Li, X., Liu, A., Chen, S., and Zhou, J. (2020). Overexpression of tomato RING E3 ubiquitin ligase gene SlRING1 confers cadmium tolerance by attenuating cadmium accumulation and oxidative stress. Physiol. Plant. 173, 449–459. doi: 10.1111/ppl.13294
Alam, I., Yang, Y.-Q., Wang, Y., Zhu, M.-L., Wang, H.-B., Chalhoub, B., et al. (2017). Genome-wide identification, evolution and expression analysis of RING finger protein genes in Brassica rapa. Sci. Rep. 7:40690. doi: 10.1038/srep40690
An, J.-P., Liu, X., Song, L., You, C., Wang, X., and Hao, Y. (2017c). Functional characterization of the Apple RING E3 ligase MdMIEL1 in transgenic Arabidopsis. Hortic. Plant J. 3, 53–59. doi: 10.1016/j.hpj.2017.01.001
An, J.-P., Liu, X., Song, L.-Q., You, C.-X., Wang, X.-F., and Hao, Y.-J. (2017b). Apple RING finger E3 ubiquitin ligase MdMIEL1 negatively regulates salt and oxidative stresses tolerance. Journal of Plant Biology 60, 137–145. doi: 10.1007/s12374-016-0457-x
An, J.-P., Liu, X., Li, H.-H., You, C.-X., Wang, X.-F., and Hao, Y.-J. (2017a). Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant Cell Physiol. 58, 1953–1962. doi: 10.1093/pcp/pcx129
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Assaha, D. V., Ueda, A., Saneoka, H., Al-Yahyai, R., and Yaish, M. W. (2017). The role of Na+ and K+ transporters in salt stress adaptation in glycophytes. Front. Physiol. 8:509. doi: 10.3389/fphys.2017.00509
Bae, H., Kim, S. K., Cho, S. K., Kang, B. G., and Kim, W. T. (2011). Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci. 180, 775–782. doi: 10.1016/j.plantsci.2011.02.008
Bernacchia, G., Schwall, G., Lottspeich, F., Salamini, F., and Bartels, D. (1995). The transketolase gene family of the resurrection plant Craterostigma plantagineum: differential expression during the rehydration phase. EMBO J. 14, 610–618. doi: 10.1002/j.1460-2075.1995.tb07037.x
Biniek, C., Heyno, E., Kruk, J., Sparla, F., Trost, P., and Krieger-Liszkay, A. (2017). Role of the NAD (P) H quinone oxidoreductase NQR and the cytochrome b AIR12 in controlling superoxide generation at the plasma membrane. Planta 245, 807–817. doi: 10.1007/s00425-016-2643-y
Blasiak, J., Pawlowska, E., Szczepanska, J., and Kaarniranta, K. (2019). Interplay between autophagy and the ubiquitin-proteasome system and its role in the pathogenesis of age-related macular degeneration. Int. J. Mol. Sci. 20:210. doi: 10.3390/ijms20010210
Brugière, N., Zhang, W., Xu, Q., Scolaro, E. J., Lu, C., Kahsay, R. Y., et al. (2017). Overexpression of RING domain E3 ligase ZmXerico1 confers drought tolerance through regulation of ABA homeostasis. Plant Physiol. 175, 1350–1369. doi: 10.1104/pp.17.01072
Cai, K., Yin, J., Chao, H., Ren, Y., Jin, L., Cao, Y., et al. (2018). A C3HC4-type RING finger protein regulates rhizobial infection and nodule organogenesis in Lotus japonicus. J. Integr. Plant Biol. 60, 878–896. doi: 10.1111/jipb.12703
Chapagain, S., Park, Y. C., Kim, J. H., and Jang, C. S. (2018). Oryza sativa salt-induced RING E3 ligase 2 (OsSIRP2) acts as a positive regulator of transketolase in plant response to salinity and osmotic stress. Planta 247, 925–939. doi: 10.1007/s00425-017-2838-x
Chauhan, D. K., Yadav, V., Vaculík, M., Gassmann, W., Pike, S., Arif, N., et al. (2021). Aluminum toxicity and aluminum stress-induced physiological tolerance responses in higher plants. Crit. Rev. Biotechnol. 41, 715–730. doi: 10.1080/07388551.2021.1874282
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, M., and Ni, M. (2006). RFI2, a RING-domain zinc finger protein, negatively regulates CONSTANS expression and photoperiodic flowering. Plant J. 46, 823–833. doi: 10.1111/j.1365-313X.2006.02740.x
Chen, Q., and Yang, G. (2020). Signal function studies of ROS, especially RBOH-dependent ROS, in plant growth, development and environmental stress. J. Plant Growth Regul. 39, 157–171. doi: 10.1007/s00344-019-09971-4
Cheng, M.-C., Hsieh, E.-J., Chen, J.-H., Chen, H.-Y., and Lin, T.-P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158, 363–375. doi: 10.1104/pp.111.189738
Cho, S. K., Ryu, M. Y., Seo, D. H., Kang, B. G., and Kim, W. T. (2011). The Arabidopsis RING E3 ubiquitin ligase AtAIRP2 plays combinatory roles with AtAIRP1 in abscisic acid-mediated drought stress responses. Plant Physiol. 157, 2240–2257. doi: 10.1104/pp.111.185595
Choi, W. G., Miller, G., Wallace, I., Harper, J., Mittler, R., and Gilroy, S. (2017). Orchestrating Rapid Long-Distance Signaling in Plants with Ca2+, ROS and Electrical Signals. Hoboken, NJ: Wiley Online Library.
Cui, P., Liu, H., Islam, F., Li, L., Farooq, M. A., Ruan, S., et al. (2016). OsPEX11, a peroxisomal biogenesis factor 11, contributes to salt stress tolerance in Oryza sativa. Front. Plant Sci. 7:1357. doi: 10.3389/fpls.2016.01357
Ding, Y., Shi, Y., and Yang, S. (2020). Molecular regulation of plant responses to environmental temperatures. Mol. Plant 13, 544–564. doi: 10.1016/j.molp.2020.02.004
Disch, S., Anastasiou, E., Sharma, V. K., Laux, T., Fletcher, J. C., and Lenhard, M. (2006). The E3 ubiquitin ligase BIG BROTHER controls Arabidopsis organ size in a dosage-dependent manner. Curr. Biol. 16, 272–279. doi: 10.1016/j.cub.2005.12.026
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Dos Reis, S. P., Tavares, L. D. S. C., Costa, C. D. N. M., Santa Brígida, A. B., and De Souza, C. R. B. (2012). Molecular cloning and characterization of a novel RING zinc-finger protein gene up-regulated under in vitro salt stress in cassava. Mol. Biol. Rep. 39, 6513–6519. doi: 10.1007/s11033-012-1479-1
Du, B., Nie, N., Sun, S., Hu, Y., Bai, Y., He, S., et al. (2021). A novel sweetpotato RING-H2 type E3 ubiquitin ligase gene IbATL38 enhances salt tolerance in transgenic Arabidopsis. Plant Sci. 304:110802. doi: 10.1016/j.plantsci.2020.110802
Du, Q.-L., Cui, W.-Z., Zhang, C.-H., and Yu, D.-Y. (2010). GmRFP1 encodes a previously unknown RING-type E3 ubiquitin ligase in Soybean (Glycine max). Mol. Biol. Rep. 37:685. doi: 10.1007/s11033-009-9535-1
Fleury, D., Himanen, K., Cnops, G., Nelissen, H., Boccardi, T. M., Maere, S., et al. (2007). The Arabidopsis thaliana homolog of yeast BRE1 has a function in cell cycle regulation during early leaf and root growth. Plant Cell 19, 417–432. doi: 10.1105/tpc.106.041319
Gao, T., Wu, Y., Zhang, Y., Liu, L., Ning, Y., Wang, D., et al. (2011). OsSDIR1 overexpression greatly improves drought tolerance in transgenic rice. Plant Mol. Biol. 76, 145–156. doi: 10.1007/s11103-011-9775-z
Gao, Y., Li, H., Deng, D., Chen, S., Jiang, W., and Chen, J. (2012). Characterization and expression analysis of the maize RING-H2 finger protein gene ZmXERICO responsive to plant hormones and abiotic stresses. Acta Physiol. Plant. 34, 1529–1535. doi: 10.1007/s11738-012-0951-9
Gao, Y., Li, M.-Y., Zhao, J., Zhang, Y.-C., Xie, Q.-J., and Chen, D.-H. (2016). Genome-wide analysis of RING finger proteins in the smallest free-living photosynthetic eukaryote Ostreococus tauri. Mar. Genom. 26, 51–61. doi: 10.1016/j.margen.2015.12.008
Guo, D., Spetz, C., Saarma, M., and Valkonen, J. P. (2003). Two potato proteins, including a novel RING finger protein (HIP1), interact with the potyviral multifunctional protein HCpro. Mol. Plant Microbe Interact. 16, 405–410. doi: 10.1094/MPMI.2003.16.5.405
Haider, F. U., Liqun, C., Coulter, J. A., Cheema, S. A., Wu, J., Zhang, R., et al. (2021). Cadmium toxicity in plants: Impacts and remediation strategies. Ecotoxicol. Environ. Saf. 211:111887. doi: 10.1016/j.ecoenv.2020.111887
Han, G., Qiao, Z., Li, Y., Wang, C., and Wang, B. (2021). The roles of CCCH zinc-finger proteins in plant abiotic stress tolerance. Int. J. Mol. Sci. 22:8327. doi: 10.3390/ijms22158327
Han, G., Wei, X., Dong, X., Wang, C., Sui, N., Guo, J., et al. (2020b). Arabidopsis ZINC FINGER PROTEIN1 acts downstream of GL2 to repress root hair initiation and elongation by directly suppressing bHLH genes. Plant Cell 32, 206–225. doi: 10.1105/tpc.19.00226
Han, G., Lu, C., Guo, J., Qiao, Z., Sui, N., Qiu, N., et al. (2020a). C2H2 zinc finger proteins: master regulators of abiotic stress responses in plants. Front. Plant Sci. 11:115. doi: 10.3389/fpls.2020.00115
Han, S.-H., Park, Y.-J., and Park, C.-M. (2020c). HOS1 activates DNA repair systems to enhance plant thermotolerance. Nat. Plants 6, 1439–1446. doi: 10.1038/s41477-020-00809-6
Hao, D., Jin, L., Wen, X., Yu, F., Xie, Q., and Guo, H. (2021). The RING E3 ligase SDIR1 destabilizes EBF1/EBF2 and modulates the ethylene response to ambient temperature fluctuations in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 118:e2024592118. doi: 10.1073/pnas.2024592118
Hardtke, C. S., Okamoto, H., Stoop-Myer, C., and Deng, X. W. (2002). Biochemical evidence for ubiquitin ligase activity of the Arabidopsis COP1 interacting protein 8 (CIP8). Plant J. 30, 385–394. doi: 10.1046/j.1365-313x.2002.01298.x
Hasanuzzaman, M., Nahar, K., Alam, M., Roychowdhury, R., and Fujita, M. (2013). Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 14, 9643–9684. doi: 10.3390/ijms14059643
Hsu, K.-H., Liu, C.-C., Wu, S.-J., Kuo, Y.-Y., Lu, C.-A., Wu, C.-R., et al. (2014). Expression of a gene encoding a rice RING zinc-finger protein, OsRZFP34, enhances stomata opening. Plant Mol. Biol. 86, 125–137. doi: 10.1007/s11103-014-0217-6
Hwang, S. G., Kim, J. J., Lim, S. D., Park, Y. C., Moon, J. C., and Jang, C. S. (2016). Molecular dissection of Oryza sativa salt-induced RING Finger Protein 1 (OsSIRP1): possible involvement in the sensitivity response to salinity stress. Physiol. Plant. 158, 168–179. doi: 10.1111/ppl.12459
Jiang, Z., Zhou, X., Tao, M., Yuan, F., Liu, L., Wu, F., et al. (2019). Plant cell-surface GIPC sphingolipids sense salt to trigger Ca2+ influx. Nature 572, 341–346. doi: 10.1038/s41586-019-1449-z
Jiménez-López, D., Muñóz-Belman, F., González-Prieto, J. M., Aguilar-Hernández, V., and Guzmán, P. (2018). Repertoire of plant RING E3 ubiquitin ligases revisited: New groups counting gene families and single genes. PLoS One 13:e0203442. doi: 10.1371/journal.pone.0203442
Joazeiro, C. A., and Weissman, A. M. (2000). RING finger proteins: mediators of ubiquitin ligase activity. Cell 102, 549–552. doi: 10.1016/s0092-8674(00)00077-5
Joo, H., Lim, C. W., and Lee, S. C. (2019a). A pepper RING-type E3 ligase, CaASRF1, plays a positive role in drought tolerance via modulation of CaAIBZ1 stability. Plant J. 98, 5–18. doi: 10.1111/tpj.14191
Joo, H., Lim, C. W., and Lee, S. C. (2019b). Roles of pepper bZIP transcription factor CaATBZ1 and its interacting partner RING-type E3 ligase CaASRF1 in modulation of ABA signalling and drought tolerance. Plant J. 100, 399–410. doi: 10.1111/tpj.14451
Joo, H., Lim, C. W., and Lee, S. C. (2020). The pepper RING-type E3 ligase, CaATIR1, positively regulates abscisic acid signalling and drought response by modulating the stability of CaATBZ1. Plant Cell Environ. 43, 1911–1924. doi: 10.1111/pce.13789
Ju, H.-W., Min, J.-H., Chung, M.-S., and Kim, C. S. (2013). The atrzf1 mutation of the novel RING-type E3 ubiquitin ligase increases proline contents and enhances drought tolerance in Arabidopsis. Plant Sci. 203, 1–7. doi: 10.1016/j.plantsci.2012.12.007
Jung, Y.-J., Lee, I.-H., Nou, I.-S., Lee, K.-D., Rashotte, A. M., and Kang, K.-K. (2013). BrRZFP1 a Brassica rapa C3HC4-type RING zinc finger protein involved in cold, salt and dehydration stress. Plant Biol. 15, 274–283. doi: 10.1111/j.1438-8677.2012.00631.x
Kam, J., Gresshoff, P., Shorter, R., and Xue, G.-P. (2007). Expression analysis of RING zinc finger genes from Triticum aestivum and identification of TaRZF70 that contains four RING-H2 domains and differentially responds to water deficit between leaf and root. Plant Sci. 173, 650–659. doi: 10.1016/j.plantsci.2007.09.001
Karlowski, W. M., and Hirsch, A. M. (2003). The over-expression of an alfalfa RING-H2 gene induces pleiotropic effects on plant growth and development. Plant Mol. Biol. 52, 121–133. doi: 10.1023/a:1023916701669
Kim, A.-R., Min, J.-H., Lee, K.-H., and Kim, C. S. (2017). PCA22 acts as a suppressor of atrzf1 to mediate proline accumulation in response to abiotic stress in Arabidopsis. J. Exp. Bot. 68, 1797–1809. doi: 10.1093/jxb/erx069
Kim, J. H., and Jang, C. S. (2021). E3 ligase, the Oryza sativa salt-induced RING finger protein 4 (OsSIRP4), negatively regulates salt stress responses via degradation of the OsPEX11-1 protein. Plant Mol. Biol. 105, 231–245. doi: 10.1007/s11103-020-01084-x
Kim, S. J., and Kim, W. T. (2013). Suppression of Arabidopsis RING E3 ubiquitin ligase AtATL78 increases tolerance to cold stress and decreases tolerance to drought stress. FEBS Lett. 587, 2584–2590. doi: 10.1016/j.febslet.2013.06.038
Kim, J. H., and Kim, W. T. (2013). The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress responses. Plant Physiol. 162, 1733–1749. doi: 10.1104/pp.113.220103
Kim, J. H., Lim, S. D., and Jang, C. S. (2019). Oryza sativa heat-induced RING finger protein 1 (OsHIRP1) positively regulates plant response to heat stress. Plant molecular biology 99, 545–559. doi: 10.1007/s11103-019-00835-9
Kim, J. H., Lim, S. D., and Jang, C. S. (2020). Oryza sativa drought-, heat-, and salt-induced RING finger protein 1 (OsDHSRP1) negatively regulates abiotic stress-responsive gene expression. Plant Mol. Biol. 103, 235–252. doi: 10.1007/s11103-020-00989-x
Kim, J. H., Lim, S. D., and Jang, C. S. (2021). Oryza sativa, C4HC3-type really interesting new gene (RING), OsRFPv6, is a positive regulator in response to salt stress by regulating Na+ absorption. Physiol. Plant. 173, 883–895. doi: 10.1111/ppl.13481
Kim, J.-H., Lee, H.-J., and Park, C.-M. (2017). HOS1 acts as a key modulator of hypocotyl photomorphogenesis. Plant Signal. Behav. 12:e1315497. doi: 10.1080/15592324.2017.1315497
Ko, J. H., Yang, S. H., and Han, K. H. (2006). Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. Plant J. 47, 343–355. doi: 10.1111/j.1365-313X.2006.02782.x
Koizumi, M., Yamaguchi-Shinozaki, K., Tsuji, H., and Shinozaki, K. (1993). Structure and expression of two genes that encode distinct drought-inducible cysteine proteinases in Arabidopsis thaliana. Gene 129, 175–182. doi: 10.1016/0378-1119(93)90266-6
Lee, H. K., Cho, S. K., Son, O., Xu, Z., Hwang, I., and Kim, W. T. (2009). Drought stress-induced Rma1H1, a RING membrane-anchor E3 ubiquitin ligase homolog, regulates aquaporin levels via ubiquitination in transgenic Arabidopsis plants. Plant Cell 21, 622–641. doi: 10.1105/tpc.108.061994
Lee, H., Xiong, L., Gong, Z., Ishitani, M., Stevenson, B., and Zhu, J.-K. (2001). The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo–cytoplasmic partitioning. Genes Dev. 15, 912–924. doi: 10.1101/gad.866801
Li, C., Ng, C. K.-Y., and Fan, L.-M. (2015). MYB transcription factors, active players in abiotic stress signaling. Environ. Exp. Bot. 114, 80–91. doi: 10.1016/j.envexpbot.2014.06.014
Li, H., Jiang, H., Bu, Q., Zhao, Q., Sun, J., Xie, Q., et al. (2011). The Arabidopsis RING finger E3 ligase RHA2b acts additively with RHA2a in regulating abscisic acid signaling and drought response. Plant Physiol. 156, 550–563. doi: 10.1104/pp.111.176214
Li, J., Han, G., Sun, C., and Sui, N. (2019). Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal Behav. 14:1613131. doi: 10.1080/15592324.2019.1613131
Li, J., Liu, J., Wang, G., Cha, J.-Y., Li, G., Chen, S., et al. (2015). A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27, 908–925. doi: 10.1105/tpc.114.135095
Li, J., Yang, F., Liu, Z., Yang, Y., and Li, X. (2013). First exploration on function of Arabidopsis AtOHRP1 in response to oxidative stress. J. Agric. Sci. Technol. (Beij.) 15, 93–99.
Li, Y., Wu, B., Yu, Y., Yang, G., Wu, C., and Zheng, C. (2011). Genome-wide analysis of the RING finger gene family in apple. Mol. Genet. Genom. 286:81. doi: 10.1007/s00438-011-0625-0
Lim, C. W., Baek, W., and Lee, S. C. (2017). The pepper RING-type E3 ligase CaAIRF1 regulates ABA and drought signaling via CaADIP1 protein phosphatase degradation. Plant Physiol. 173, 2323–2339. doi: 10.1104/pp.16.01817
Lim, C. W., Baek, W., and Lee, S. C. (2018). Roles of pepper bZIP protein Ca DILZ 1 and its interacting partner RING-type E3 ligase Ca DSR 1 in modulation of drought tolerance. Plant J. 96, 452–467. doi: 10.1111/tpj.14046
Lim, S. D., Cho, H. Y., Park, Y. C., Ham, D. J., Lee, J. K., and Jang, C. S. (2013). The rice RING finger E3 ligase, OsHCI1, drives nuclear export of multiple substrate proteins and its heterogeneous overexpression enhances acquired thermotolerance. J. Exp. Bot. 64, 2899–2914. doi: 10.1093/jxb/ert143
Lim, S. D., Oh, D. G., Park, Y. C., and Jang, C. S. (2020). Molecular characterization of a RING E3 ligase SbHCI1 in sorghum under heat and abscisic acid stress. Planta 252:89. doi: 10.1007/s00425-020-03469-0
Lim, S. D., Yim, W. C., Moon, J.-C., Kim, D. S., Lee, B.-M., and Jang, C. S. (2010). A gene family encoding RING finger proteins in rice: their expansion, expression diversity, and co-expressed genes. Plant Mol. Biol. 72, 369–380. doi: 10.1007/s11103-009-9576-9
Linden, K. J., and Callis, J. (2020). The ubiquitin system affects agronomic plant traits. J. Biol. Chem. 295, 13940–13955. doi: 10.1074/jbc.REV120.011303
Linden, K. J., Hsia, M. M., Chen, Y.-T., and Callis, J. (2021). The Arabidopsis thaliana E3 ubiquitin ligase BRIZ functions in abscisic acid response. Front. Plant Sci. 12:641849. doi: 10.3389/fpls.2021.641849
Liu, B., Jiang, Y., Tang, H., Tong, S., Lou, S., Shao, C., et al. (2021). The ubiquitin E3 ligase SR1 modulates the submergence response by degrading phosphorylated WRKY33 in Arabidopsis. Plant Cell 33, 1771–1789. doi: 10.1093/plcell/koab062
Liu, J., Zhang, C., Wei, C., Liu, X., Wang, M., Yu, F., et al. (2016). The RING finger ubiquitin E3 ligase OsHTAS enhances heat tolerance by promoting H2O2-induced stomatal closure in rice. Plant Physiol. 170, 429–443. doi: 10.1104/pp.15.00879
Liu, K., Wang, L., Xu, Y., Chen, N., Ma, Q., Li, F., et al. (2007). Overexpression of OsCOIN, a putative cold inducible zinc finger protein, increased tolerance to chilling, salt and drought, and enhanced proline level in rice. Planta 226, 1007–1016. doi: 10.1007/s00425-007-0548-5
Liu, W., Tang, X., Qi, X., Fu, X., Ghimire, S., Ma, R., et al. (2020). The ubiquitin conjugating enzyme: an important ubiquitin transfer platform in ubiquitin-proteasome system. Int. J. Mol. Sci. 21:2894. doi: 10.3390/ijms21082894
Liu, Y., Li, L., Zhang, L., Lv, Q., Zhao, Y., and Li, X. (2018). Isolation and identification of wheat gene TaDIS1 encoding a RING finger domain protein, which negatively regulates drought stress tolerance in transgenic Arabidopsis. Plant Sci. 275, 49–59. doi: 10.1016/j.plantsci.2018.07.017
Liu, Y., Pei, L., Xiao, S., Peng, L., Liu, Z., Li, X., et al. (2020). AtPPRT1 negatively regulates salt stress response in Arabidopsis seedlings. Plant Signal. Behav. 15, 1732103. doi: 10.1080/15592324.2020.1732103
Locato, V., and De Gara, L. (2018). “Programmed cell death in plants: an overview,” in Plant Programmed Cell Death, eds L. De Gara and V. Locato (New York, NY: Humana Press), 1–8. doi: 10.1007/978-1-4939-7668-3_1
Lu, Y. (2017). Research Progress of RING-type Zinc Finger Protein in Plants. Anhui Agric. Sci. Bull. 23:8. doi: 10.1016/j.plantsci.2013.11.004
Lv, Q., Zhang, L., Zan, T., Li, L., and Li, X. (2020). Wheat RING E3 ubiquitin ligase TaDIS1 degrade TaSTP via the 26S proteasome pathway. Plant Sci. 296:110494. doi: 10.1016/j.plantsci.2020.110494
Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X., Zeng, D., et al. (2015). COLD1 confers chilling tolerance in rice. Cell 160, 1209–1221. doi: 10.1016/j.cell.2015.01.046
Marcec, M. J., Gilroy, S., Poovaiah, B., and Tanaka, K. (2019). Mutual interplay of Ca2+ and ROS signaling in plant immune response. Plant Sci. 283, 343–354. doi: 10.1016/j.plantsci.2019.03.004
Marshall, R. S., and Vierstra, R. D. (2019). Dynamic regulation of the 26S proteasome: from synthesis to degradation. Front. Mol. Biosci. 6:40. doi: 10.3389/fmolb.2019.00040
Matsuda, N., Suzuki, T., Tanaka, K., and Nakano, A. (2001). Rma1, a novel type of RING finger protein conserved from Arabidopsis to human, is a membrane-bound ubiquitin ligase. J. Cell Sci. 114, 1949–1957. doi: 10.1242/jcs.114.10.1949
Min, J.-H., Park, C.-R., Chung, J.-S., and Kim, C. S. (2021). Arabidopsis thaliana Ubiquitin-Associated Protein 1 (AtUAP1) interacts with Arabidopsis thaliana Ring Zinc Finger 1 (AtRZF1) to negatively regulate dehydration response. Plant Cell Physiol. 62, 1044–1057. doi: 10.1093/pcp/pcab082
Nadarajah, K. K. (2020). ROS homeostasis in abiotic stress tolerance in plants. Int. J. Mol. Sci. 21:5208. doi: 10.3390/ijms21155208
Ni, X., Tian, Z., Liu, J., Song, B., and Xie, C. (2010). Cloning and molecular characterization of the potato RING finger protein gene StRFP1 and its function in potato broad-spectrum resistance against Phytophthora infestans. J. Plant Physiol. 167, 488–496. doi: 10.1016/j.jplph.2009.10.019
Ning, Y., Jantasuriyarat, C., Zhao, Q., Zhang, H., Chen, S., Liu, J., et al. (2011a). The SINA E3 ligase OsDIS1 negatively regulates drought response in rice. Plant Physiol. 157, 242–255. doi: 10.1104/pp.111.180893
Ning, Y., Xie, Q., and Wang, G.-L. (2011b). OsDIS1-mediated stress response pathway in rice. Plant Signal. Behav. 6, 1684–1686. doi: 10.4161/psb.6.11.17916
Oh, T. R., Kim, J. H., Cho, S. K., Ryu, M. Y., Yang, S. W., and Kim, W. T. (2017). AtAIRP2 E3 ligase affects ABA and high-salinity responses by stimulating its ATP1/SDIRIP1 substrate turnover. Plant Physiol. 174, 2515–2531. doi: 10.1104/pp.17.00467
Oh, T. R., Yu, S. G., Yang, H. W., Kim, J. H., and Kim, W. T. (2020). AtKPNB1, an Arabidopsis importin-β protein, is downstream of the RING E3 ubiquitin ligase AtAIRP1 in the ABA-mediated drought stress response. Planta 252:93. doi: 10.1007/s00425-020-03500-4
Pardo-Hernández, M., López-Delacalle, M., and Rivero, R. M. (2020). ROS and NO regulation by melatonin under abiotic stress in plants. Antioxidants (Basel) 9:1078. doi: 10.3390/antiox9111078
Park, G.-G., Park, J.-J., Yoon, J., Yu, S.-N., and An, G. (2010). A RING finger E3 ligase gene, Oryza sativa Delayed Seed Germination 1 (OsDSG1), controls seed germination and stress responses in rice. Plant Mol. Biol. 74, 467–478. doi: 10.1007/s11103-010-9687-3
Park, J. H., Kang, C. H., Nawkar, G. M., Lee, E. S., Paeng, S. K., Chae, H. B., et al. (2018a). EMR, a cytosolic-abundant ring finger E3 ligase, mediates ER-associated protein degradation in Arabidopsis. New Phytol. 220, 163–177. doi: 10.1111/nph.15279
Park, Y. C., Chapagain, S., and Jang, C. S. (2018c). A negative regulator in response to salinity in rice: Oryza sativa Salt-, ABA-and drought-induced RING Finger Protein 1 (OsSADR1). Plant Cell Physiol. 59, 575–589. doi: 10.1093/pcp/pcy009
Park, Y. C., Chapagain, S., and Jang, C. S. (2018b). The microtubule-associated RING finger protein 1 (OsMAR1) acts as a negative regulator for salt-stress response through the regulation of OCPI2 (O. sativa chymotrypsin protease inhibitor 2). Planta 247, 875–886. doi: 10.1007/s00425-017-2834-1
Park, Y. C., Moon, J.-C., Chapagain, S., Oh, D. G., Kim, J. J., and Jang, C. S. (2018d). Role of salt-induced RING finger protein 3 (OsSIRP3), a negative regulator of salinity stress response by modulating the level of its target proteins. Environ. Exp. Bot. 155, 21–30.
Park, S.-H., Chung, M.-S., Lee, S., Lee, K.-H., and Kim, C. S. (2017). Loss of Ribosomal Protein L24A (RPL24A) suppresses proline accumulation of Arabidopsis thaliana ring zinc finger 1 (atrzf1) mutant in response to osmotic stress. Biochem. Biophys. Res. Commun. 494, 499–503. doi: 10.1016/j.bbrc.2017.10.092
Park, Y. C., Lim, S. D., Moon, J. C., and Jang, C. S. (2019). A rice really interesting new gene H 2-type E 3 ligase, OsSIRH2-14, enhances salinity tolerance via ubiquitin/26 S proteasome-mediated degradation of salt-related proteins. Plant Cell Environ. 42, 3061–3076. doi: 10.1111/pce.13619
Pratelli, R., Guerra, D. D., Yu, S., Wogulis, M., Kraft, E., Frommer, W. B., et al. (2012). The ubiquitin E3 ligase LOSS OF GDU2 is required for GLUTAMINE DUMPER1-induced amino acid secretion in Arabidopsis. Plant Physiol. 158, 1628–1642. doi: 10.1104/pp.111.191965
Qi, S., Lin, Q., Zhu, H., Gao, F., Zhang, W., and Hua, X. (2016). The RING finger E3 ligase SpRing is a positive regulator of salt stress signaling in salt-tolerant wild tomato species. Plant Cell Physiol. 57, 528–539. doi: 10.1093/pcp/pcw006
Qi, X., Tang, X., Liu, W., Fu, X., Luo, H., Ghimire, S., et al. (2020). A potato RING-finger protein gene StRFP2 is involved in drought tolerance. Plant Physiol. Biochem. 146, 438–446. doi: 10.1016/j.plaphy.2019.11.042
Qiao, Z., Li, C.-L., and Zhang, W. (2016). WRKY1 regulates stomatal movement in drought-stressed Arabidopsis thaliana. Plant Mol. Biol. 91, 53–65. doi: 10.1007/s11103-016-0441-3
Qin, F., Sakuma, Y., Tran, L.-S. P., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress–responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Qin, X., Huang, S., Liu, Y., Bian, M., Shi, W., Zuo, Z., et al. (2017). Overexpression of A RING finger ubiquitin ligase gene AtATRF1 enhances aluminium tolerance in Arabidopsis thaliana. J. Plant Biol. 60, 66–74. doi: 10.1007/s12374-016-0903-9
Rounds, M. A., and Larsen, P. B. (2008). Aluminum-dependent root-growth inhibition in Arabidopsis results from AtATR-regulated cell-cycle arrest. Curr. Biol. 18, 1495–1500. doi: 10.1016/j.cub.2008.08.050
Ryu, M. Y., Cho, S. K., and Kim, W. T. (2010). The Arabidopsis C3H2C3-type RING E3 ubiquitin ligase AtAIRP1 is a positive regulator of an abscisic acid-dependent response to drought stress. Plant Physiol. 154, 1983–1997. doi: 10.1104/pp.110.164749
Sade, H., Meriga, B., Surapu, V., Gadi, J., Sunita, M., Suravajhala, P., et al. (2016). Toxicity and tolerance of aluminum in plants: tailoring plants to suit to acid soils. Biometals 29, 187–210. doi: 10.1007/s10534-016-9910-z
Sah, S. K., Reddy, K. R., and Li, J. (2016). Abscisic acid and abiotic stress tolerance in crop plants. Front. Plant Sci. 7:571. doi: 10.3389/fpls.2016.00571
Seo, Y. S., Choi, J. Y., Kim, S. J., Kim, E. Y., Shin, J. S., and Kim, W. T. (2012). Constitutive expression of CaRma1H1, a hot pepper ER-localized RING E3 ubiquitin ligase, increases tolerance to drought and salt stresses in transgenic tomato plants. Plant Cell Rep. 31, 1659–1665. doi: 10.1007/s00299-012-1278-0
Sharma, R., Singh, G., Bhattacharya, S., and Singh, A. (2018). Comparative transcriptome meta-analysis of Arabidopsis thaliana under drought and cold stress. PLoS One 13:e0203266. doi: 10.1371/journal.pone.0203266
Shin, D.-J., Min, J.-H., Van Nguyen, T., Kim, Y.-M., and Kim, C. S. (2019). Loss of Arabidopsis Halotolerance 2-like (AHL), a 3′-phosphoadenosine-5′-phosphate phosphatase, suppresses insensitive response of Arabidopsis thaliana ring zinc finger 1 (atrzf1) mutant to abiotic stress. Plant Mol. Biol. 99, 363–377. doi: 10.1007/s11103-019-00822-0
Stone, S. L. (2019). Role of the ubiquitin proteasome system in plant response to abiotic stress. Int. Rev. Cell Mol. Biol. 343, 65–110. doi: 10.1016/bs.ircmb.2018.05.012
Stone, S. L., Hauksdóttir, H., Troy, A., Herschleb, J., Kraft, E., and Callis, J. (2005). Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 137, 13–30. doi: 10.1104/pp.104.052423
Su, T., Yang, M., Wang, P., Zhao, Y., and Ma, C. (2020). Interplay between the ubiquitin proteasome system and ubiquitin-mediated autophagy in plants. Cells 9:2219. doi: 10.3390/cells9102219
Suh, J. Y., Kim, S. J., Oh, T. R., Cho, S. K., Yang, S. W., and Kim, W. T. (2016). Arabidopsis Tóxicos en Levadura 78 (AtATL78) mediates ABA-dependent ROS signaling in response to drought stress. Biochem. Biophys. Res. communications 469, 8–14. doi: 10.1016/j.bbrc.2015.11.061
Sun, J., Sun, Y., Ahmed, R. I., Ren, A., and Xie, M. (2019). Research progress on plant RING-finger proteins. Genes 10:973. doi: 10.3390/genes10120973
Sun, X., Wang, Y., and Sui, N. (2018). Transcriptional regulation of bHLH during plant response to stress. Biochem. Biophys. Res. Commun. 503, 397–401. doi: 10.1016/j.bbrc.2018.07.123
Swatek, K. N., Usher, J. L., Kueck, A. F., Gladkova, C., Mevissen, T. E., Pruneda, J. N., et al. (2019). Insights into ubiquitin chain architecture using Ub-clipping. Nature 572, 533–537. doi: 10.1038/s41586-019-1482-y
Tian, M., Lou, L., Liu, L., Yu, F., Zhao, Q., Zhang, H., et al. (2015). The RING finger E3 ligase STRF1 is involved in membrane trafficking and modulates salt-stress response in Arabidopsis thaliana. Plant J. 82, 81–92. doi: 10.1111/tpj.12797
Tiwari, L. D., Mittal, D., Mishra, R. C., and Grover, A. (2015). Constitutive over-expression of rice chymotrypsin protease inhibitor gene OCPI2 results in enhanced growth, salinity and osmotic stress tolerance of the transgenic Arabidopsis plants. Plant Physiol. Biochem. 92, 48–55. doi: 10.1016/j.plaphy.2015.03.012
Vaičiukynė, M., Žiauka, J., Žūkienė, R., Vertelkaitė, L., and Kuusienė, S. (2019). Abscisic acid promotes root system development in birch tissue culture: a comparison to aspen culture and conventional rooting-related growth regulators. Physiol. Plant. 165, 114–122. doi: 10.1111/ppl.12860
Van Nguyen, T., Park, C.-R., Lee, K.-H., Lee, S., and Kim, C. S. (2021). BES1/BZR1 Homolog 3 cooperates with E3 ligase AtRZF1 to regulate osmotic stress and brassinosteroid responses in Arabidopsis. J. Exp. Bot. 72, 636–653. doi: 10.1093/jxb/eraa458
Von Arnim, A., and Deng, X.-W. (1993). Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domain. J. Biol. Chem. 268, 19626–19631. doi: 10.1016/s0021-9258(19)36562-7
Wang, J., Wang, R., Mao, X., Zhang, J., Liu, Y., Xie, Q., et al. (2020). RING finger ubiquitin E3 ligase gene TaSDIR1-4A contributes to determination of grain size in common wheat. J. Exp. Bot. 71, 5377–5388. doi: 10.1093/jxb/eraa271
Wang, L., Xie, X., Yao, W., Wang, J., Ma, F., Wang, C., et al. (2017). RING-H2-type E3 gene VpRH2 from Vitis pseudoreticulata improves resistance to powdery mildew by interacting with VpGRP2A. J. Exp. Bot. 68, 1669–1687. doi: 10.1093/jxb/erx033
Wang, X., Chen, E., Ge, X., Gong, Q., Butt, H., Zhang, C., et al. (2018). Overexpressed BRH1, a RING finger gene, alters rosette leaf shape in Arabidopsis thaliana. Sci. China Life Sci. 61, 79–87. doi: 10.1007/s11427-017-9133-8
Wu, W., Cheng, Z., Liu, M., Yang, X., and Qiu, D. (2014). C3HC4-type RING finger protein Nb ZFP1 is involved in growth and fruit development in Nicotiana benthamiana. PLoS One 9:e99352. doi: 10.1371/journal.pone.0099352
Wu, X.-C., Cao, X.-Y., Chen, M., Zhang, X.-K., Liu, Y.-N., Xu, Z.-S., et al. (2010). Isolation and expression pattern assay of a C3HC4-type RING Zinc Finger Protein Gene GmRZFP1 in Glycine max (L.). J. Plant Genet. Resour. 11, 343–348.
Xia, Z., Liu, Q., Wu, J., and Ding, J. (2012). ZmRFP1, the putative ortholog of SDIR1, encodes a RING-H2 E3 ubiquitin ligase and responds to drought stress in an ABA-dependent manner in maize. Gene 495, 146–153. doi: 10.1016/j.gene.2011.12.028
Xia, Z., Su, X., Liu, J., and Wang, M. (2013). The RING-H2 finger gene 1 (RHF1) encodes an E3 ubiquitin ligase and participates in drought stress response in Nicotiana tabacum. Genetica 141, 11–21. doi: 10.1007/s10709-013-9702-0
Yang, L., Liu, Q., Liu, Z., Yang, H., Wang, J., Li, X., et al. (2016). Arabidopsis C3HC4-RING finger E3 ubiquitin ligase AtAIRP4 positively regulates stress-responsive abscisic acid signaling. J. Integr. Plant Biol. 58, 67–80. doi: 10.1111/jipb.12364
Yang, L., Miao, M., Lyu, H., Cao, X., Li, J., Li, Y., et al. (2019). Genome-wide identification, evolution, and expression analysis of RING finger gene family in Solanum lycopersicum. Int. J. Mol. Sci. 20:4864. doi: 10.3390/ijms20194864
Yang, L., Wu, L., Chang, W., Li, Z., Miao, M., Li, Y., et al. (2018). Overexpression of the maize E3 ubiquitin ligase gene ZmAIRP4 enhances drought stress tolerance in Arabidopsis. Plant Physiol. Biochem. 123, 34–42. doi: 10.1016/j.plaphy.2017.11.017
Yang, X., Sun, C., Hu, Y., and Lin, Z. (2008). Molecular cloning and characterization of a gene encoding RING zinc finger ankyrin protein from drought-tolerant Artemisia desertorum. J. Biosci. 33, 103–112. doi: 10.1007/s12038-008-0026-7
Yang, Y., and Guo, Y. (2018). Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol. 217, 523–539. doi: 10.1111/nph.14920
Yang, Y., Fu, D., Zhu, C., He, Y., Zhang, H., Liu, T., et al. (2015). The RING-finger ubiquitin ligase HAF1 mediates heading date 1 degradation during photoperiodic flowering in rice. Plant Cell 27, 2455–2468. doi: 10.1105/tpc.15.00320
Yang, Y.-Q., and Lu, Y.-H. (2018). Genome-wide survey, characterization, and expression analysis of RING finger protein genes in Brassica oleracea and their syntenic comparison to Brassica rapa and Arabidopsis thaliana. Genome 61, 685–697. doi: 10.1139/gen-2018-0046
Yu, J., Kang, L., Li, Y., Wu, C., Zheng, C., Liu, P., et al. (2021). RING finger protein RGLG1 and RGLG2 negatively modulate MAPKKK18 mediated drought stress tolerance in Arabidopsis. J. Integr. Plant Biol. 63, 484–493. doi: 10.1111/jipb.13019
Yu, Y., Xu, W., Wang, J., Wang, L., Yao, W., Yang, Y., et al. (2013). The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 200, 834–846. doi: 10.1111/nph.12418
Yuan, F., Yang, H., Xue, Y., Kong, D., Ye, R., Li, C., et al. (2014). OSCA1 mediates osmotic-stress-evoked Ca2+ increases vital for osmosensing in Arabidopsis. Nature 514:367. doi: 10.1038/nature13593
Zeng, D., Cai, Z., and Liu, Y. (2019). Overexpression of BdRHP1 improves drought tolerance and reduces yield loss in rice. Biologia Plant. 63, 371–379. doi: 10.32615/bp.2019.043
Zeng, D.-E., Hou, P., Xiao, F., and Liu, Y. (2015). Overexpression of Arabidopsis XERICO gene confers enhanced drought and salt stress tolerance in rice (Oryza sativa L.). J. Plant Biochem. Biotechnol. 24, 56–64. doi: 10.1007/s13562-013-0236-4
Zhang, H., Cui, F., Wu, Y., Lou, L., Liu, L., Tian, M., et al. (2015). The RING finger ubiquitin E3 ligase SDIR1 targets SDIR1-INTERACTING PROTEIN1 for degradation to modulate the salt stress response and ABA signaling in Arabidopsis. Plant Cell 27, 214–227. doi: 10.1105/tpc.114.134163
Zhang, X., Garreton, V., and Chua, N.-H. (2005). The AIP2 E3 ligase acts as a novel negative regulator of ABA signaling by promoting ABI3 degradation. Genes Dev. 19, 1532–1543. doi: 10.1101/gad.1318705
Zhang, Y., Yang, C., Li, Y., Zheng, N., Chen, H., Zhao, Q., et al. (2007). SDIR1 is a RING finger E3 ligase that positively regulates stress-responsive abscisic acid signaling in Arabidopsis. Plant Cell 19, 1912–1929. doi: 10.1105/tpc.106.048488
Zhao, J.-Y., Lu, Z.-W., Sun, Y., Fang, Z.-W., Chen, J., Zhou, Y.-B., et al. (2020). The Ankyrin-repeat gene GmANK114 confers drought and salt tolerance in Arabidopsis and Soybean. Front. Plant Sci. 11:1666. doi: 10.3389/fpls.2020.584167
Keywords: RING zinc finger proteins, plant, abiotic stress, E3 ubiquitin ligase, regulation mechanism
Citation: Han G, Qiao Z, Li Y, Yang Z, Wang C, Zhang Y, Liu L and Wang B (2022) RING Zinc Finger Proteins in Plant Abiotic Stress Tolerance. Front. Plant Sci. 13:877011. doi: 10.3389/fpls.2022.877011
Received: 16 February 2022; Accepted: 22 March 2022;
Published: 14 April 2022.
Edited by:
Oscar Vicente, Universitat Politècnica de València, SpainReviewed by:
Wen-Cheng Liu, Henan University, ChinaEwa Joanna Hanus-Fajerska, University of Agriculture in Kraków, Poland
Copyright © 2022 Han, Qiao, Li, Yang, Wang, Zhang, Liu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoliang Han, gl_han@sdnu.edu.cn; Baoshan Wang, bswang@sdnu.edu.cn
†These authors have contributed equally to this work
 Guoliang Han
Guoliang Han Ziqi Qiao
Ziqi Qiao Yuxia Li
Yuxia Li Zongran Yang1
Zongran Yang1 Chengfeng Wang
Chengfeng Wang Baoshan Wang
Baoshan Wang