- 1State Key Laboratory of Ecological Pest Control for Fujian and Taiwan Crops, College of Plant Protection, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Agriculture, Fujian Agriculture and Forestry University, Fuzhou, China
- 3College of Horticulture, Fujian Agriculture and Forestry University, Fuzhou, China
- 4State Key Laboratory for Conservation and Utilization of Subtropical Agro-Bioresources, Guangxi Key Lab of Sugarcane Biology, College of Agriculture, Guangxi University, Nanning, China
- 5Fujian Provincial Key Laboratory of Haixia Applied Plant Systems Biology, College of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 6Biotechnology Research Institute, Fujian Provincial Key Laboratory of Genetic Engineering for Agriculture, Fujian Academy of Agricultural Sciences, Fuzhou, China
- 7Center for Crop Biotechnology, College of Agriculture, Anhui Science and Technology University, Fengyang, China
Snf2 family proteins are the crucial subunits of chromatin-remodeling complexes (CRCs), which contributes to the biological processes of transcription, replication, and DNA repair using ATP as energy. Some CRC subunits have been confirmed to be the critical regulators in various aspects of plant growth and development and in epigenetic mechanisms such as histone modification, DNA methylation, and histone variants. However, the functions of Snf2 family genes in rice were poorly investigated. In this study, the relative expression profile of 40 members of Snf2 family in rice was studied at certain developmental stages of seed. Our results revealed that OsCHR741/OsDDM1b (Decrease in DNA methylation 1) was accumulated highly in the early developmental stage of seeds. We further analyzed the OsDDM1b T-DNA insertion loss-of-function of mutant, which exhibited dwarfism, smaller organ size, and shorter and wider grain size than the wild type (Hwayoung, HY), yet no difference in 1,000-grain weight. Consistent with the grain size, the outer parenchyma cell layers of lemma in osddm1b developed more cells with decreased size. OsDDM1b encoded a nucleus, membrane-localized protein and was distributed predominately in young spikelets and seeds, asserting its role in grain size. Meanwhile, the osddm1b was less sensitive to brassinosteroids (BRs) while the endogenous BR levels increased. We detected changes in the expression levels of the BR signaling pathway and feedback-inhibited genes with and without exogenous BR application, and the alterations of expression were also observed in grain size-related genes in the osddm1b. Altogether, our results suggest that OsDDM1b plays a crucial role in grain size via influencing cell proliferation and regulating BR signaling and homeostasis.
Introduction
In eukaryotes, genomic DNA is wrapped around the histone octamer to form a nucleosome which is the subunit of chromatin. A highly organized chromatin structure is essential for gene expression in eukaryotes. Chromatin packages large quantities of genetic information into the nucleus and provides an efficient integrative platform that translates signals to regulate gene responses (Hu et al., 2013; Zhang et al., 2019). The mechanism of epigenetic regulation involves histone modification, histone variants, DNA methylation, and chromatin remodeling (Hu et al., 2013). A recent study reveals that many proteins have been identified to mediate these processes, among which Snf2 family proteins are responsible for chromatin remodeling to regulate gene expression using ATP energy (Cairns, 2005; Hu et al., 2013; Zhang et al., 2019). According to the helicase-like region, the Snf2 family proteins are classified into six groups with two conserved domains (SNF2_N and Helicase_C) (Andrew et al., 2006). Each group could be subdivided into 24 subfamilies, and some of these subfamilies are unique for specific organisms and others are ubiquitous (Andrew et al., 2006; Song et al., 2021).
Snf2 family proteins have been identified to contain 41, 45, and 40 members in Arabidopsis, tomato, and rice, respectively. Many of these proteins have been reported to play the essential roles in regulating plant development and stress responses (Knizewski et al., 2008). For example, the loss-of-function of SPLAYED (SYD) causes shoot apical meristem (SAM) defects in Arabidopsis; SYD interacts with WUSCHEL (WUS), the central regulator in SAM (Kwon et al., 2005); mutations of BRM cause reduced root length and plant size (Farrona et al., 2004), curly leaves (Hurtado et al., 2006), sensitive to abscisic acid (ABA) (Han et al., 2012), and early flowering (Farrona et al., 2011). Indeed, BRM is always recruited and associated with other nuclear proteins, such as RELATIVE OF EARLY FLOWERING 6 (REF6) and FORGETTER1 (FGT1), to modulate gene transcription (Brzezinka et al., 2016; Li et al., 2016). BRM also interacts with TEOSINTE BRANCHED1 CYCLOIDEA AND PCF-CODING GENE (TCP4), ANGUSTIFOLIA3 (AN3), and BREVIPEDICELLUS (BP) to regulate leaf development and inflorescence architecture (Efroni et al., 2013; Vercruyssen et al., 2014) and PHY-INTERACTING FACTOR1 (PIF1) to modulate PROTOCHLOROPHYLLIDE OXIDOREDUCTASE C (PROC) expression for chlorophyll biosynthesis (Zhang et al., 2017). The plants overexpressing AtCHR12 showed growth arrest of primary buds and stem under drought and heat stresses (Mlynarova et al., 2007). Moreover, both DEFECTIVE IN RNA-DIRECTED DNA METHYLATION 1 (DRD1, a DRD1 subfamily member) and DECREASED DNA METHYLATION 1 (DDM1, an Lsh subfamily member) are involved in DNA methylation process (Jeddeloh et al., 1999; Kanno et al., 2004). In addition, the mutant of DRD1 and DDM1 exhibits the delay of leaf senescence (Cho et al., 2016). Furthermore, PHOTOPERIOD-INDEPENDENT EARLY FLOWERING 1 (PIE1, an Swr1 subfamily member) is responsible for H2A.Z deposition to regulate flowering and plant development (Choi et al., 2007), and PICKLE (a Mi-2 subfamily member) regulates the H3K27me3-enriched genes (Zhang et al., 2012) and the plant hormone signaling, such as brassinosteroid (BR), gibberellic acid (GA), and cytokinin (CK) (Kakimoto and Physiology, 2011; Zhang et al., 2014). CHROMATIN REMODELING 4 (CHR4) interacts with transcription factors and affects the expression of critical floral regulators to mediate the flowering response pathways of inflorescence meristem to promote floral identity (Sang et al., 2020).
In addition to Arabidopsis, several Snf2 family proteins have been identified and studied in other plant species. For example, constitutively, SlCHR1 (Solyc01g079690) overexpression causes reduced plant growth (Folta et al., 2016). DRD1 and Snf2 subfamilies are involved in stress responses in tomato plants (Bargsten et al., 2013). In rice (Oryza sativa L.), the loss-of-function of CHR729 (a member of Mi-2 subfamily) results in dwarf, later flowering, less tiller, and narrow leaf by affecting contents of GA3 (Hu et al., 2012; Ma et al., 2015). OsALT1 (alkaline tolerance 1, OsCHR706), an Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance (Guo et al., 2014), and OsBRHIS1 plays a critical role in SA-independent disease resistance by suppressing innate immunity (Li et al., 2015).
OsDDM1a and OsDDM1b, as the homologous genes to Arabidopsis DDM1, are involved in DNA methylation (Higo et al., 2012), which results in high methylation levels at CHG and CG contexts (Tan et al., 2016). The mutation of OsDDM1 decreases histone H3K9me2 and increases the small heterochromatic RNA and long non-coding RNA (Tan et al., 2018). However, the relative expression profile of Snf2 gene family in rice during reproductive development has not been systematically analyzed. Here, we deeply investigated the expression profiles of the rice Snf2 genes during different developmental stages of seed and characterized the phenotype of osddm1b. The mutant displayed dwarfism and shorter and wider grain size and was insensitive to BR treatments. Our results provide an in-depth knowledge for further exploration of the function of Snf2 family proteins in rice, which indicates that OsDDM1b helps to regulate organ size by influencing cell proliferation and involving in BR response regulation.
Materials and Methods
Plant Material and Growth Condition
The OsDDM1b T-DNA insertion mutant (PFG_2B-60109) and wild-type cultivar Hwayoung were obtained from the Crop Biotech Institute, Department of Plant Systems Biotech, Kyung Hee University. The T-DNA information was obtained from SIGnAL database at http://signal.salk.edu/cgi-bin/RiceGE. The plants were grown in the greenhouse at 22–32°C and 80–90% humidity with a 14-h/10-h (light/dark) photoperiod. About 1, 3, 7, 10, and 25 days after fertilization (DAF) of seeds were harvested using micro-dissection needles, frozen in liquid nitrogen immediately, and stored at -80°C for total RNA extraction. A total of three biological replicates of each sample were collected for experimental analysis.
The T-DNA insertion in osddm1b was confirmed by PCR using the primers 2B-60109-Lp and 2B-60109-Rp and the T-DNA-specific primer Rb (2707). The full-length coding sequence of OsDDM1b was amplified from WT cDNA by PCR and cloned into the EcoRI/KpnI sites of the pCAMBIA2300 vector. The derived constructs were introduced into the osddm1b by Agrobacterium tumefaciens-mediated transformation. The relative expression levels of OsDDM1b were detected in 7-day-old seedling leaves of osddm1b and complementary mutants by the primers P1 and P2. The primers sequences are described in Supplementary Table 2 for genotyping identification. The gene structure of OsDDM1b was searched from Gene Structure Display Server.1
RNA Isolation and qRT-PCR Analysis
Total RNAs of all collected samples were extracted using Plant RNeasy Mini kit (Qiagen, Hilden, Germany). About 1 μg RNA was reverse-transcribed using the PrimeScript RT-PCR kit (Takara, Kyoto, Japan) according to the manufacturer’s instruction (Cai et al., 2019). The relative expression level was detected by qRT-PCR using the Bio-Rad qRT-PCR system (Foster City, CA, United States) and SYBR Premix Ex TaqII (TaKaRa Perfect Real Time) (Zhang et al., 2020). The qRT-PCR program was 95°C for 30 s; 40 cycles of annealing at 95°C for 5 s and extension at 60°C for 35 s; and 95°C for 15 s (Zhang et al., 2020; Zhao et al., 2021). The rice OsUBQ5 gene was used as an internal control. To evaluate the relative expression levels of the examined genes, we used the comparative CT method (Su et al., 2017). The gene-specific primers are listed in Supplementary Tables 3–6.
Observation of Pollen and Ovule in Rice
Several anthers were randomly selected before pollination and placed on microscopic slide for dissecting with the micro-dissection needles using a microscope. The pollen was stained with 1% I2-KI solution and observed under the microscope (Ren et al., 2019). For the analysis of the ovule fertility, the spikelets before pollination were fixed in the FAA solution (50% ethanol: acetic acid: formaldehyde = 89:6:5) and using the whole-mount eosin B-staining confocal laser scanning microscopy (WECLSM) to observe the ovule development (Zeng et al., 2007, 2009). After fixation, eosin B staining, and clearing, the ovaries were divided from spikelets and observed using a Leica SP8 CLSM (Leica Microsystems) to screen the ovule (Zhao et al., 2020).
Promoter Fusion and GUS Staining
A 2,559-bp fragment upstream of OsDDM1b ATG codon sequence was amplified by PCR from DNA of rice leaf using the primers listed in Supplementary Table 3. Then, the products were constructed into a pENTER/D-TOPO vector (Invitrogen, CA, United States). After that, the positive clones were recombined with the pGWB533 vector by LR Clonase II enzyme (Invitrogen, CA, United States). The wild-type ZH11 (Zhonghua 11) callus was transformed using the Agrobacterium-mediated transformation using the pOsDDM1b: green fluorescent protein (GUS) recombinant construction. The transgenic plant tissues were incubated in β-glucuronidase (GUS) staining buffer overnight at 37°C (Jefferson et al., 1987) and dehydrated in an ethanol series to remove the chlorophyll (Jiang et al., 2012). The images were viewed under a Leica (M205 FA) microscope.
Subcellular Localization of OsDDM1b
A 2,547-bp segment of OsDDM1b coding sequence was amplified from WT cDNA using the primers listed in Supplementary Table 3. The PCR fragments were cloned into the pENTER/D-TOPO vector (Invitrogen, CA, United States), and pENTER/D-TOPO clones were recombined into the pGWB505 vector using LR Clonase II enzyme (Invitrogen, CA, United States). The 35S:OsDDM1b-GFP recombinant construction and 35S: GFP (vector control) were transformed into Agrobacterium tumefaciens (GV3101) and infiltrated with ER-mCherry marker to tobacco leaves. The fluorescence signals were observed using a confocal microscope (SP8, Leica, Germany), and the excitation wavelength was 488 nm.
Histological Observation and Scanning Electron Microscope
The spikelets were fixed in FAA (50% ethanol, 5% acetic acid, and 3.7% formaldehyde) at 4°C overnight and dehydrated in a series of ethanol for observing the spikelet cell size and number. After fixing with chloroform, the samples were embedded in Paraplast Plus (Sigma). The samples were sliced into the 8-μm-thickness samples using an RM2245 rotary microtome (Leica). Sections were dewaxed in xylene and gradually rehydrated and dehydrated before staining with toluidine blue for light microscopy. For scanning electron microscope (SEM), fresh materials were applied directly to the scanning electron microscope (Ren et al., 2018; Yu et al., 2020). The cell number and cell size in the outer parenchyma layer of the spikelet hulls were measured by ImageJ2 (Yu et al., 2020).
BR Treatment
For the lamina joint test, the seeds were germinated in water at 37°C and grown hydroponically in a nutrient solution containing 0, 0.01, 0.1, and 1 μm epi-BL for 1 week. Then, the second leaf lamina joint was measured. For the coleoptile elongation assay, the seeds were sterilized with 10% hypochlorous acid and germinated under dark conditions in half-strength MS medium supplemented with 0, 0.01, 0.1, and 1 μm epi-BL. Coleoptile length were measured after a week (Liu S. Y. et al., 2015).
Measuring Endogenous BRs
Plants were grown hydroponically in nutrient solution for 10 days. The shoots and roots (equivalent to 1 g of fresh weight) of wild type and osddm1b were harvested and ground with phosphate-buffered saline solution immediately. BR endogenous contents were analyzed using the BR ELISA kit according to the manufacturer’s protocol (SINOBESTBIO, Shanghai, China) by UV spectrophotometer in 450 nm.
Results
The Comparative Expression Level of Snf2 Gene Family During Seed Development
The development of seeds is important to get high yield, and many genes highly regulate the developmental process. To explore the expression level of the Snf2 family genes during seed development, we collected the seeds from 1, 3, 7, 10, and 25 DAF. We checked the gene expression level shown in Figure 1 and Supplementary Figure 1. The results showed that OsCHR712 and OsCHR730 were predominantly expressed in early developmental stages of seeds (S1), and OsCHR703, OsCHR735, and OsCHR741 were preferentially expressed in both S1 and S5 stages (Figure 1A). In addition, OsCHR701, OsCHR707, OsCHR711, OsCHR739, and OsCHR745 were preferentially expressed in both S1 and S4 stages (Figure 1B). There were 23 Snf2 genes that showed similar expression pattern which were predominantly expressed in the S4 stage, such as OsCHR702, OsCHR704, OsCHR705, OsCHR706, OsCHR708, OsCHR709, OsCHR710, OsCHR713, OsCHR715, OsCHR719, OsCHR721, OsCHR722, OsCHR725, OsCHR727, OsCHR729, OsCHR731, OsCHR732, OsCHR736, OsCHR737, OsCHR742 (Figure 1C), OsCHR720, OsCHR726, and OsCHR740 (Supplementary Figure 1A). These results indicated that Snf2, a sizeable family that exhibits specific expression patterns, may have unique functions in different developmental stages of seed in rice.
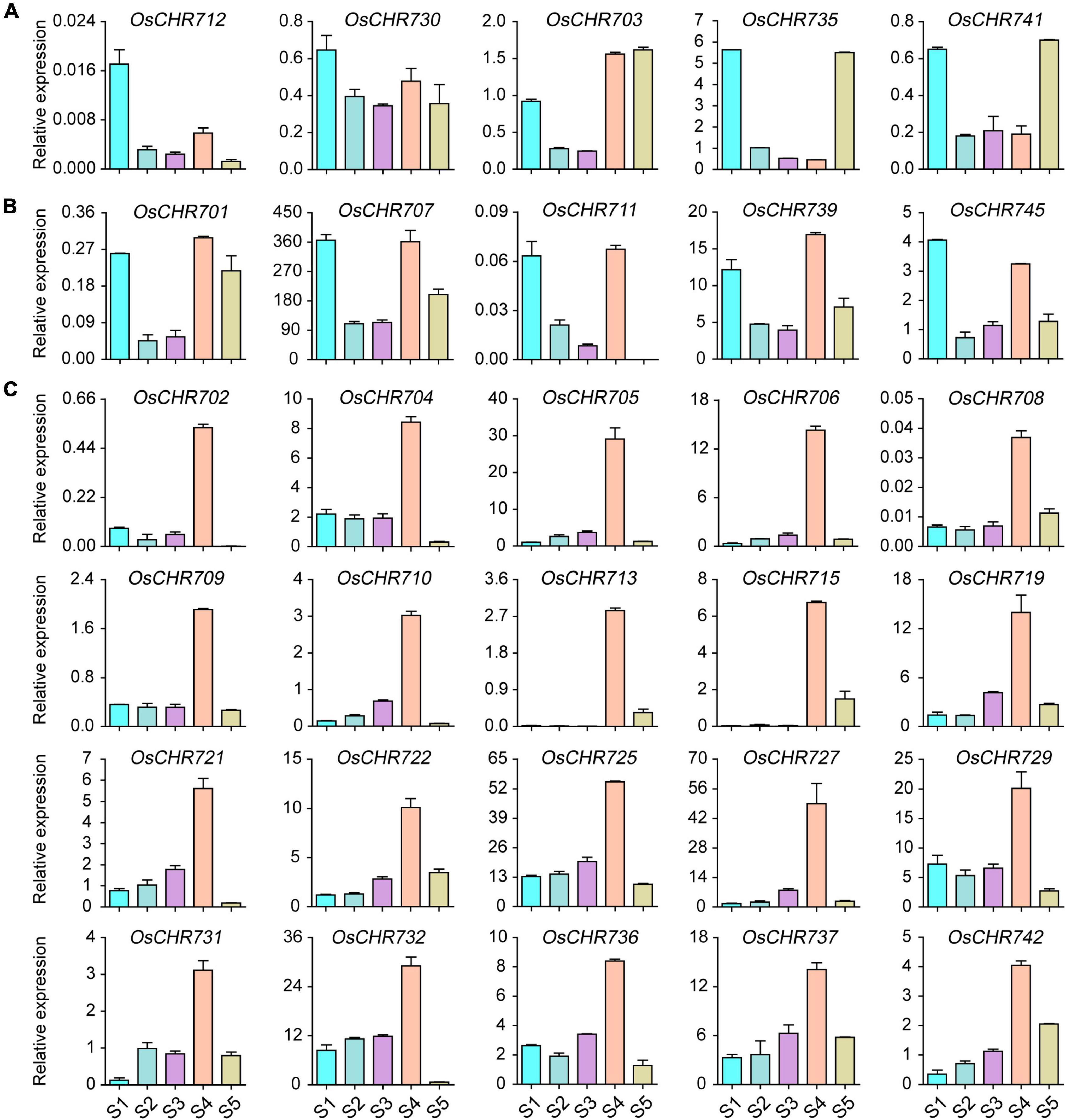
Figure 1. The relative expression level of Snf2 gene family in the wild-type (ZH11) seeds. (A) Preferentially expressed Snf2 genes in the seeds’ early or late developmental stages. (B) Predominantly expressed Snf2 genes in the seeds’ early and mid-term developmental stages. (C) Explicitly expressed Snf2 genes in the seeds mid-term developmental stages. S1, the seeds of 1 DAF; S2, the seeds of 3 DAF; S3, the seeds of 7 DAF; S4, the seeds of 10 DAF; S5, the seeds of 25 DAF.
Characterization of the OsDDM1b Mutant
The previous studies showed that two homologous genes in rice (Oryza sativa var. Nipponbare) offered 60% identity to DDM1 (AT5G66750) (Higo et al., 2012). The two genes, OsDDM1a (LOC_Os09g27060) and OsDDM1b (LOC_Os03g51230), share 93% amino acid identity to each other (Higo et al., 2012; Tan et al., 2016). Here, we identified a T-DNA insertion approximately 0.55 kb downstream of the translation start site ATG in osddm1b (2B-60109.L) from Hwayoung background (Figure 2A), which were identified in the SIGnAL database.3 The homozygous plants for the T-DNA insertion were identified and obtained by PCR (Figure 2H). In addition, the PCR products of homozygous plants were sequenced, and the sequencing results were aligned with the genome sequence (OsDDM1b) and vector sequence (2707), and then, we found the T-DNA insertion site (Supplementary Figure 3).
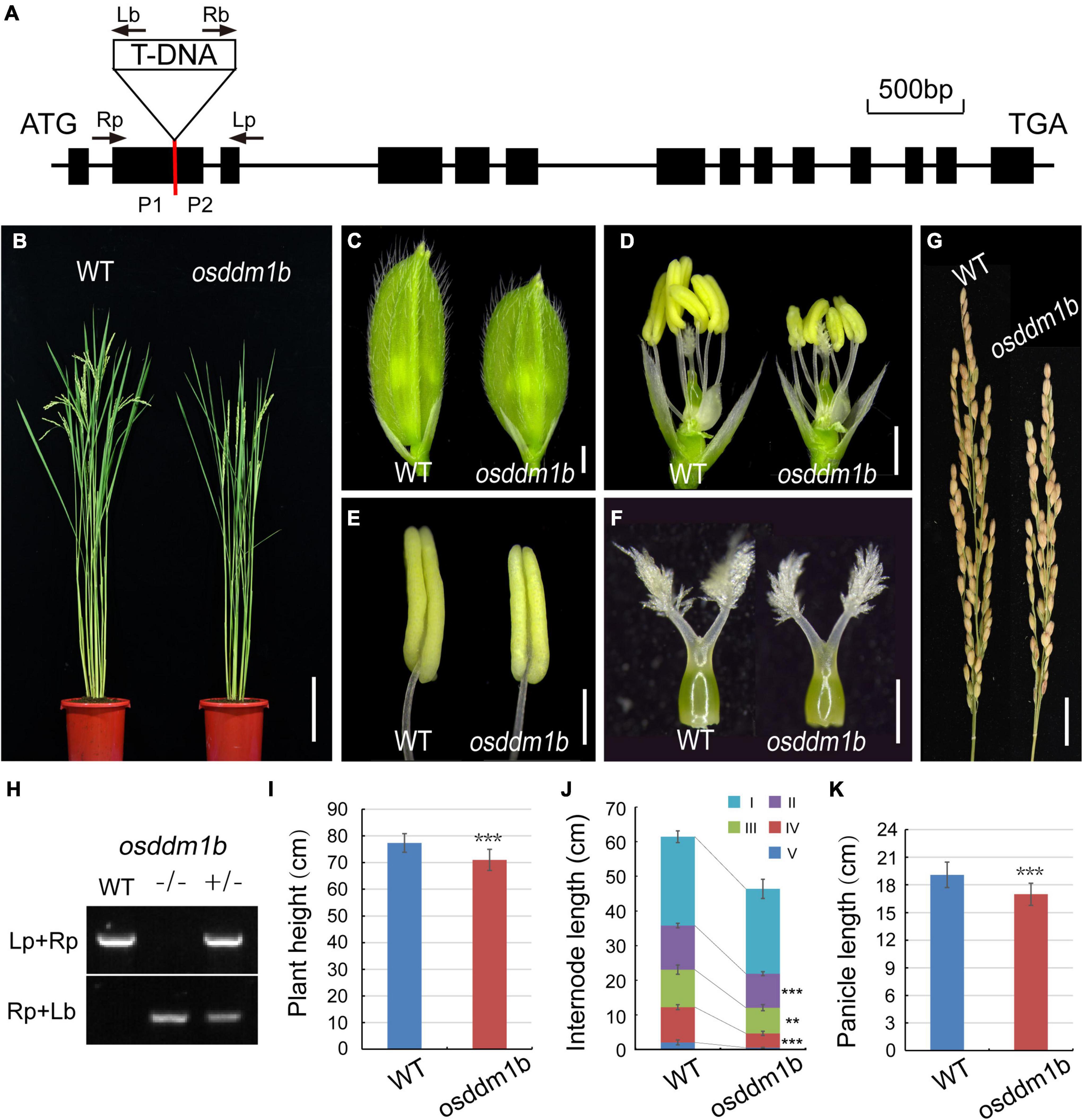
Figure 2. Morphology analysis of the wild type (Hwayoung, HY) and osddm1b. (A) The gene structure of OsDDM1b and the triangle indicate the T-DNA insertion site. The arrows show the primers which were used in the genotyping study, and P1 and P2 are the positions of primers using qRT-PCR. (B) The phenotype of wild type (left, HY) and osddm1b (right) plants at maturity. Bar = 15 cm. (C,D) The spikelets of wild type (HY) and osddm1b. Bar = 1 mm. (E,F) Anthers and pistils of wild type (left) (HY) and osddm1b (right). Bar = 500 μm, 250 μm. (G) Panicles of wild type (HY) and osddm1b. Bar = 2 cm. (H) Genotyping of T-DNA insertion plants. (I–K) The average plant height, internode length, panicles length of wild type (HY), and osddm1b, respectively (n = 20 per sample). Data are given as means ± SD. **p < 0.01, ***p < 0.001 compared with wild type (HY) using Student’s t-test.
In osddm1b, the dwarfism was observed in the seedlings and mature plants (Figures 2B,I and Supplementary Figure 2A and Table 1). The length of first internode was similar to the wild type. However, the second, third, and fourth internodes of osddm1b were shortened, which suggests that abnormal internode elongation leads to dwarf phenotype (Figure 2J and Supplementary Figure 2B). The reproductive organs of the osddm1b, such as spikelets, anthers, and pistils, were much smaller than those of wild type (Figures 2C–F). The panicle morphology in osddm1b exhibited a significantly reduced growth of 11.03% than the wild type at mature stage (Figures 2G,K). Then, we compared male and female gametophyte morphology in the osddm1b and wild-type plants. The staining of pollen with iodine potassium iodide (I2-KI) solution and the observation of ovule with eosin B by confocal showed that the male (Supplementary Figures 2C–D) and female gametophytes (Supplementary Figures 2E,F) have no noticeable difference compared to the wild type. However, the seed setting in osddm1b was decreased to 84.39% (Supplementary Figure 2G).
To further confirm the effect of OsDDM1b on grain size, we examined the grain length, width, thickness, and 1,000-grain weight. The results showed that osddm1b had significantly reduced grain length of 8.15% than the wild type (Figures 3A,D, Supplementary Figure 2I,J), but it increased 17.59% in grain width (Figures 3B,E and Supplementary Figures 2K,M) and 10.67% in grain thickness, respectively (Figures 3C,F and Supplementary Figures 2L,N). Even though there was a change in the grain size, there was no noticeable change in the 1,000-grain weight (Supplementary Figure 2H). To verify the role of OsDDM1b in determining grain size in rice, we performed a genetic complementation test. An overexpression plasmid containing full-length coding sequence was inserted by the transformation into the osddm1b. Then, we got two positive complementary lines. The overexpression of OsDDM1b in osddm1b could partially complement the grain size defection of mutant (Figures 3A–C). The grain size (length, width, and thickness) was significantly different between the complementary lines and osddm1b (Figures 3D–F). Particularly, the grain width in the complementary lines was similar to wild type (Figure 3E). The OsDDM1b expression level was dramatically reduced in 7-day-old young leaves in osddm1b but increased in the complementary lines compared with those of the wild type (Figure 3G). The other defection phenotypes could be partially rescued in the complementation lines. This result indicated that the mutation in OsDDM1b was responsible for the phenotype of grain size exhibited in osddm1b.
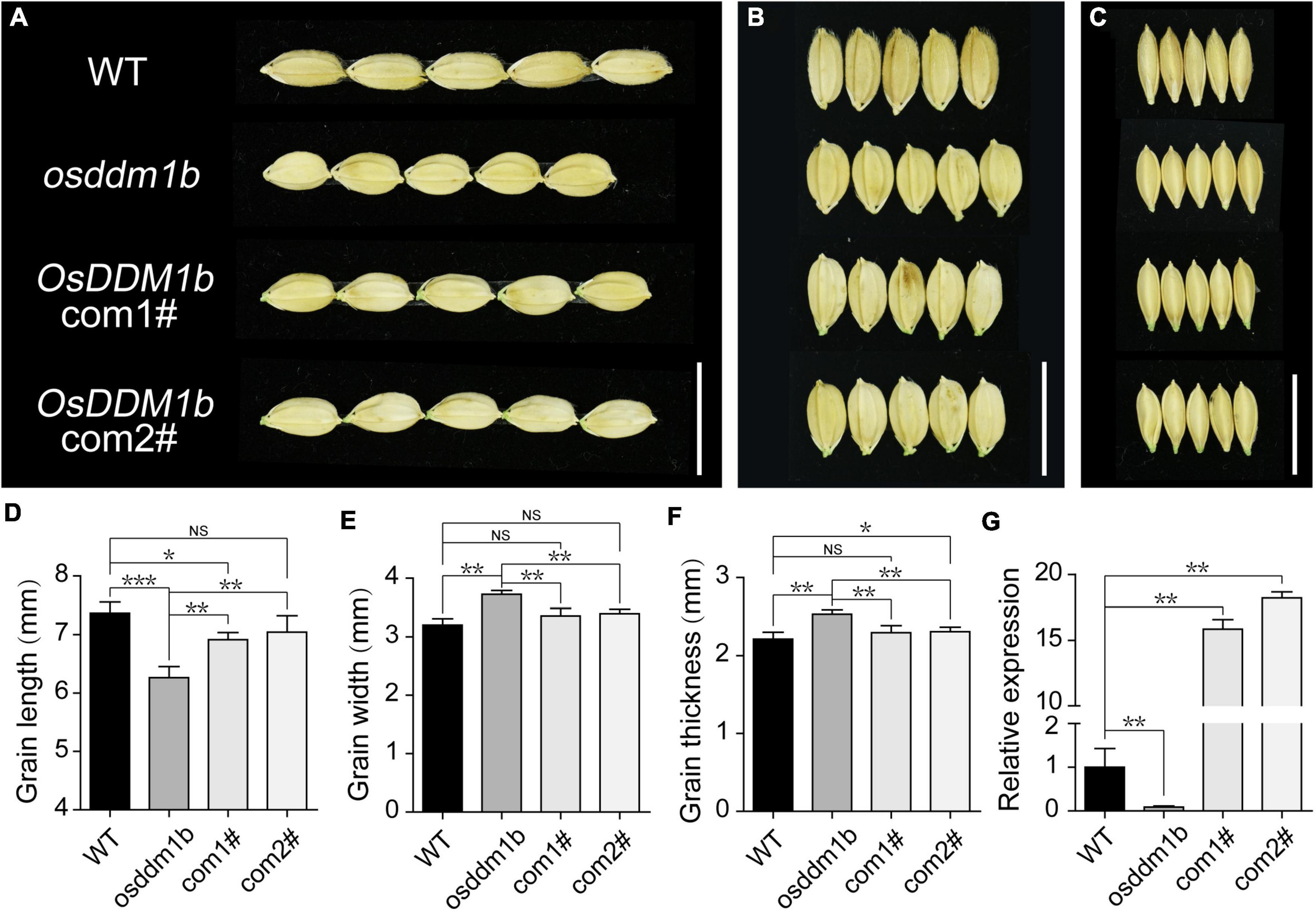
Figure 3. Functional verification of OsDDM1b in grain size. (A–C) Comparation on grain size of osddm1b, complementation mutants with wild type (Hwayoung, HY), respectively. Bar = 1 cm. (D–F) Grain length, width, and thickness of wild type (HY) and osddm1b (n = 60 per sample). (G) The relative expression levels of OsDDM1b in osddm1b, complementation mutants, and wild type (HY) in the leaves of 7-day-old seedlings. Data are given as means ± SD. NSp > 0.05, *p < 0.05, **p < 0.01, ***p < 0.001 compared with wild type (HY) using Student’s t-test. NS: not significant.
OsDDM1b Regulates Grain Size by Affecting Cell Proliferation
The spikelet hullin osddm1b became shorter and more comprehensive compared with wild type before fertilization (Figure 4A). Since the cell division and/or cell expansion are responsible for the alteration in the final spikelet hull size and grain size, we carefully examined the cross-section of the central part of lemma and palea in mature spikelets and compared the epidermal cells of osddm1b and HY by scanning electron microscope. The hull cross-section of the spikelet hullin osddm1b revealed a significant increase of 27.84% in the length of the total cells and 8.15% in cell number of outer parenchyma in the osddm1b. In comparison, the cell width was decreased by 18.07% in the outer parenchyma cells (Figures 4B–F). It also changed the number of rows of specialized cells with a rigid wall in the upper epidermis (Supplementary Figures 4A–B). The data suggested that the cell division was significantly increased in a transverse direction in the osddm1b spikelet hulls. We also investigated the expression of genes involved in the cell cycle, such as CYCA2.2, CYClaZm, E2F2, CYCB2, MCM3, MCM4, and MCM5. We found that these genes were upregulated in young panicles of osddm1b, which suggests that the increased cell number might have resulted from the elevated expression of genes that promote cell proliferation (Supplementary Figure 4C). In addition, we observed the average length and width of longitudinal epidermal cells in outer and inner glumes of wild type and osddm1b by scanning electron microscopy. An apparent 58.43% decrease in cell width was observed in the outer glum, but 10.41% in cell width was increased in the inner glum. Then, there is no significant difference in outer and inner glumes’ cell length in osddm1b (Figures 4G–I). These observations suggest that OsDDM1b might promote latitudinal growth by increasing cell proliferation.
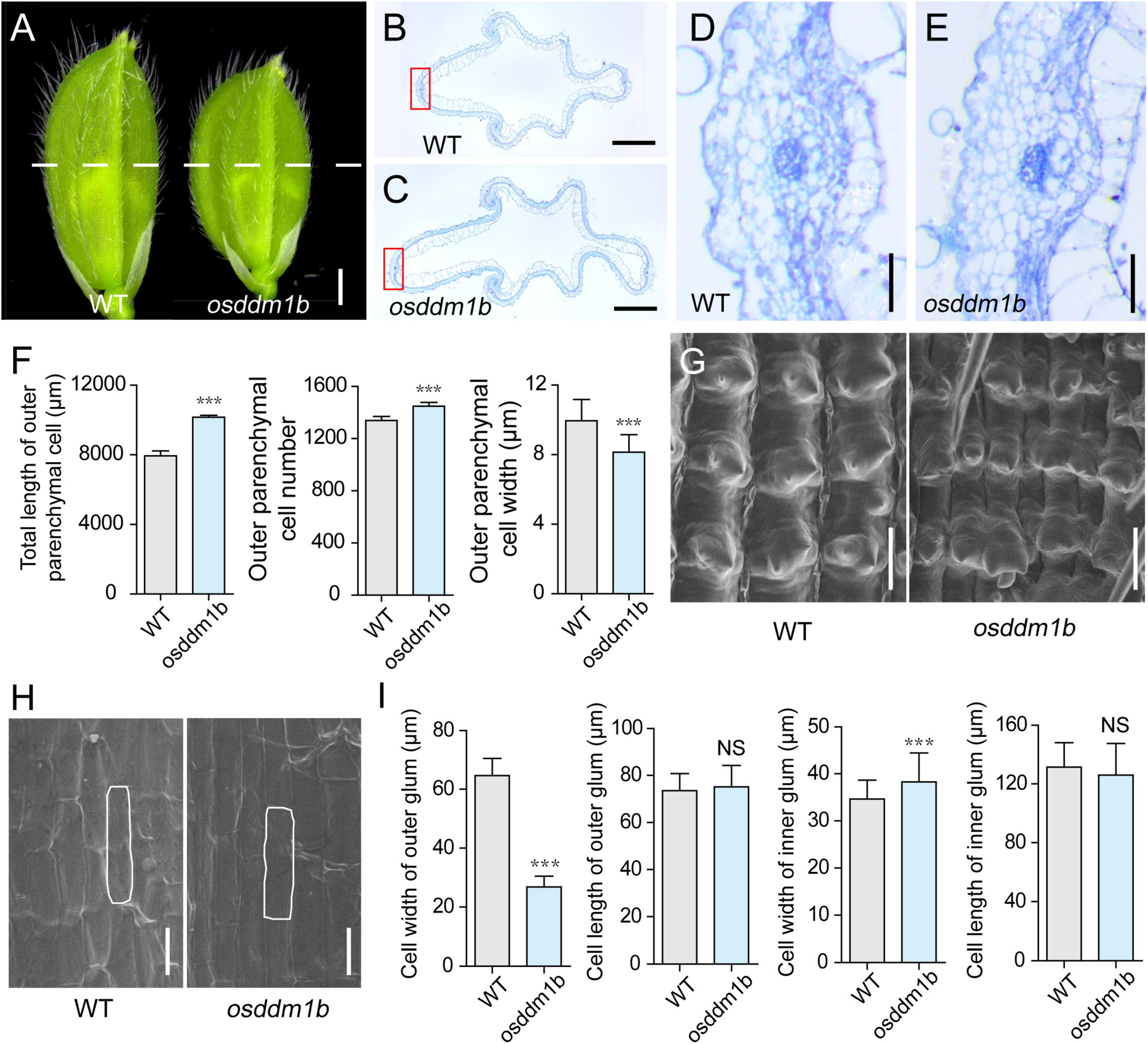
Figure 4. Histological analysis of spikelet hulls. (A) Young spikelet hulls of wild type (Hwayoung, HY) and osddm1b. The white line indicates the position of the cross-section. Bar = 1 mm. (B,C) Cross-sections of spikelet hulls from wild type (HY) and osddm1b, respectively. Bar = 500 μm. (D,E) Magnified view of the cross-section area boxed in panels (B,C), respectively. Bar = 50 μm. (F) Statistical data of the total length, cell number, and length in the outer parenchyma layer (n = 12). (G,H) Scanning electron microscope (SEM) analysis of the outer and inner surfaces of glumes. Bar = 60 μm. (I) Statistical analysis of cell width and length in outer and inner glumes (n = 40, 60, respectively). Data are given as means ± SD. ***p < 0.001 compared with wild type (HY) using Student’s t-test.
OsDDM1b Localization and Expression Pattern
To investigate the subcellular localization of OsDDM1b, we performed a transient transformation assay. When OsDDM1b-green fluorescent protein (GFP) was transformed into tobacco leaves, the signals mainly targeted the nucleus and cell membrane (Figure 5A).
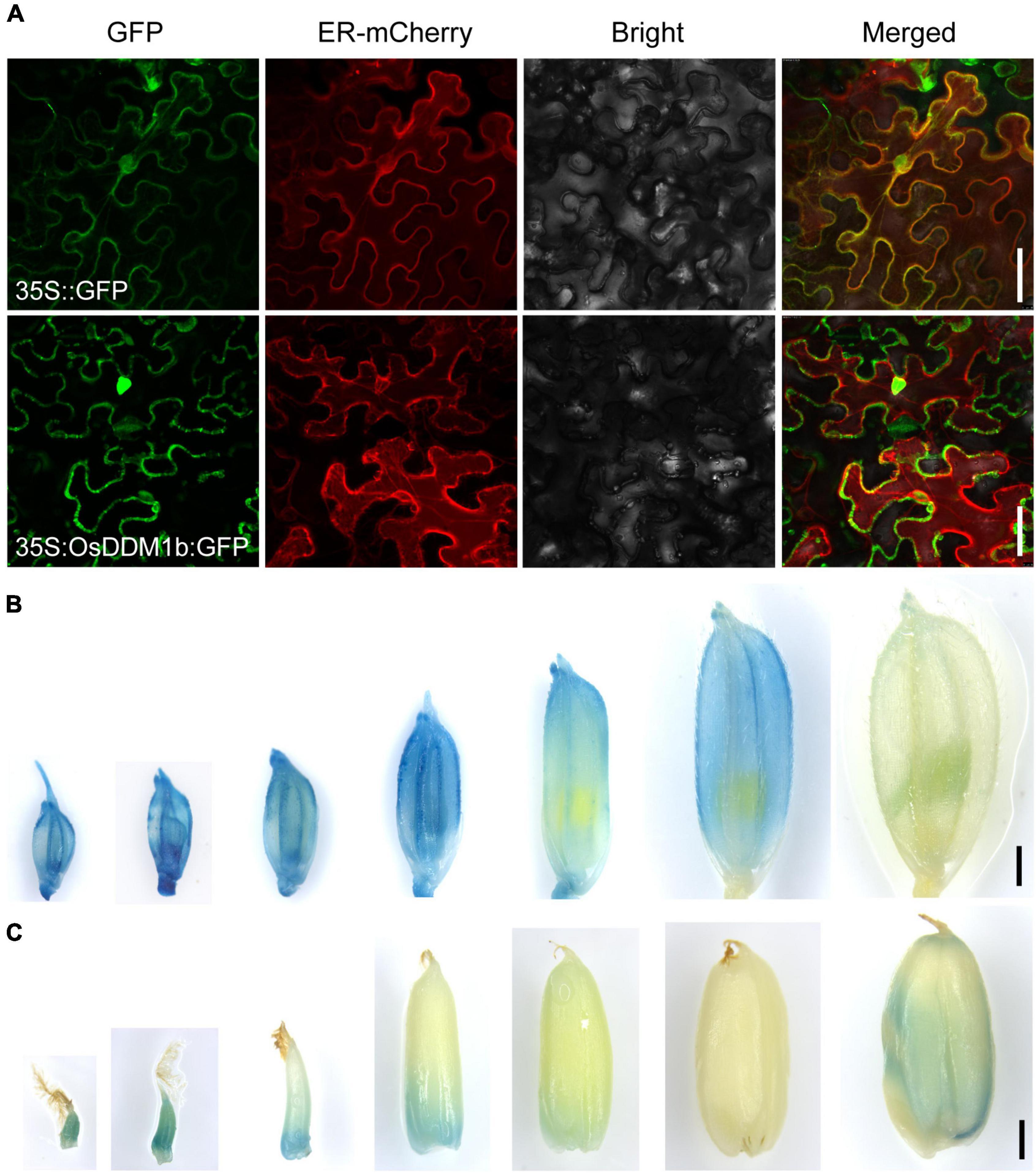
Figure 5. The expression profile of OsDDM1b. (A) Subcellular localization of OsDDM1b protein was shown to target the cytomembrane and nuclear by transient expression of 35:OsDDM1b-GFP in tobacco. The upper row is the 35S:GFP as the control. Bar = 50 μm. (B,C) Expression pattern detected in transgenic plants carrying the pOsDDM1b:GUS vector in ZH11 background. Developing spikelets in turn 1–2, 2–3, 3–4, 4–5, 5–6, 6–7 cm, and mature spikelet (B); The seeds of 5 HAF and 1, 3, 7, 10, 15, and 25 DAF, respectively (C). Bar = 1 mm. HAF, hours after fertilization; DAF, days after fertilization.
To further examine the expression profile of OsDDM1b, a construct was produced in which the OsDDM1b promoter (approximately 2.5 kb upstream of the ATG site) was fused to the β-glucuronidase gene (GUS) to transform wild-type (ZH11) plants. GUS signals were detected in the vegetative organs, such as root tips, young and mature leaf blades, stem, and internodes (Supplementary Figures 5A–D). We also detected at different developmental stages of the reproductive organs, such as spikelets, stamens, pistils, and seeds. GUS expression was visualized during the developmental stage of spikelet hulls except for the mature stage (Figure 5B). The developmental period of the spikelet can provide a reference for the pistil and stamen. Then, we estimated the developmental stage of pistil and stamen according to the length of the spikelet. Filaments of early stamens and the stamens of 4–5 mm and mature spikelets had the GUS signals (Supplementary Figure 5E). It was detected in the basal of pistils during the early stage (≤5 mm spikelet) and stigma of the mature stage (Supplementary Figure 5F). The GUS staining in seeds showed that it was highly expressed in the early stage [such as 5 h after fertilization (HAF), 1 DAF, 3 DAF, and 10 DAF] and mature seeds (Figure 5C), consistent with the results of the qRT-PCR. This expression profile indicated that OsDDM1b functioned in different young tissues, in line with the results presented above that OsDDM1b regulated cell division and proliferation rate.
The OsDDM1b Mutant Is Less Sensitive to BRs
The phenotype of osddm1b shown, such as erect and dark green leaves, dwarfism, and small grains, is typical BR response phenotypes in rice (Yamamuro et al., 2000; Hong et al., 2003). So, we hypothesize that OsDDM1b is involved in the BR response. We evaluated the sensitivity of osddm1b to 24-epibrassinolide (24-epiBL) using a lamina joint assay. As shown in the results, the second lamina of the wild type exhibited a 24-epiBL dosage-dependent inclination. By contrast, osddm1b did not show a blatant response (Figures 6A,B and Supplementary Figure 6A). The previous studies suggested that BR promoting coleoptile growth was an another indicator of a BR-responsive phenotype (Yamamuro et al., 2000; Li et al., 2002; Je et al., 2010). To analyze the response of coleoptiles to BR, we also performed a coleoptile elongation assay for BR sensitivity (Yamamuro et al., 2000). The wild-type and osddm1b seeds were germinated and grown under dark conditions in half MS medium containing different concentrations of 24-epiBL. The results showed that the relative elongation of coleoptile treatments was much weaker in osddm1b than in the wild type under BR treatment (Figure 6C), which suggests that osddm1b was less sensitive to BRs than the wild type.
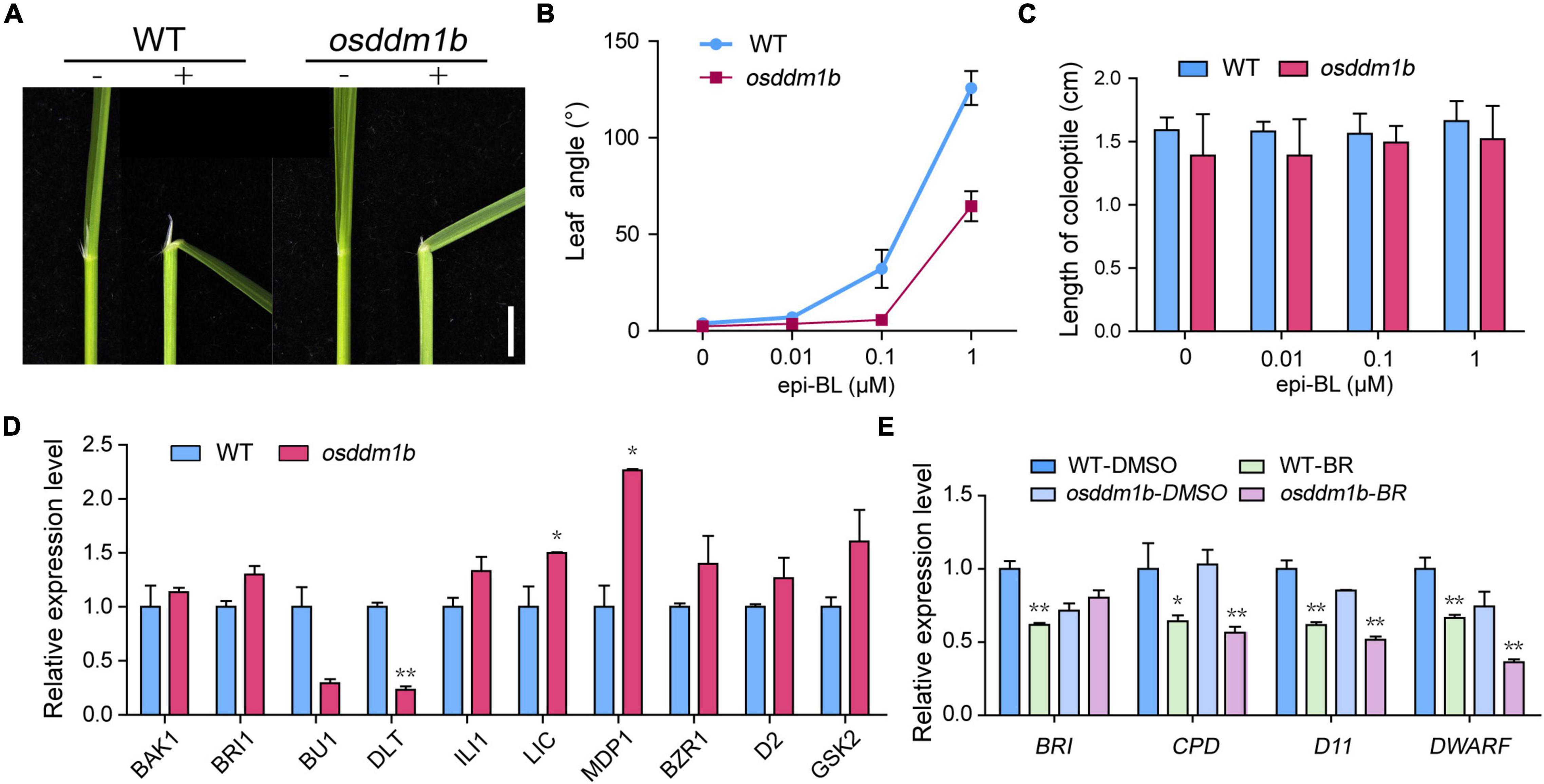
Figure 6. The OsDDM1b mutant is less sensitive to brassinolide (BL). Response to BR treatments, (A) Lamina joints of wild type (HY) and osddm1b. Bar = 2 mm. (B) Measurements of the lamina joint angle of wild type (HY) and osddm1b after various BR treatments. (C) Effect of BR on the coleoptile in wild-type (HY) and osddm1b seedling. (D) The expression level of BR signaling pathway-related genes and assayed using RNA from roots of 7-day-old seedlings cultured in half-strength MS medium. (E) Quantitative RT-PCR analysis of feedback-inhibited genes of BR biosynthesis. Data are given as means ± SD. **p < 0.01, *p < 0.05 compared with wild type (HY) using Student’s t-test.
OsDDM1b Influences BR Homeostasis and Signaling
According to the above results, loss-of-function of OsDDM1b decreased the response to BR treatments, which suggests that OsDDM1b may involve in BR signaling. Then, we analyzed the expression levels of BR-related genes, which include OsBAK1, OsBRI1, OsBU1, OsDLT, OsILI1, OsLIC, OsMDP1, OsBZR1, OsD2, and OsGSK2 in the wild type and osddm1b (Yamamuro et al., 2000; Hong et al., 2003; Ito et al., 2005; Duan et al., 2006; Bai et al., 2007; Koh et al., 2007; Wang et al., 2008; Tong et al., 2009, 2012; Zhang et al., 2009). In roots, the OsBAK1, OsBRI1, OsILI1, and OsBZR1 were slightly upregulated in osddm1b (Figure 6D). However, the expression level of OsBU1 and OsDLT, positive regulator of BR signaling, was significantly lower in osddm1b than in the wild type. The expression level of OsLIC, OsMDP1, and OsGSK2, as the negative regulator of BR, was also upregulated in osddm1b. OsD2, as a representative BR biosynthetic gene, was slightly upregulated in osddm1b. These results suggested that OsDDM1b influenced BR homeostasis and signaling.
It had been reported that the regulation of feedback-inhibited genes in BR biosynthesis usually defects mainly in BR signaling in mutants (Bai et al., 2007; Chen et al., 2013). To confirm this, we analyzed the expression of OsBRI1, OsCPD, OsD11, and OsDWARF (Yamamuro et al., 2000; Hong et al., 2002; Tanabe et al., 2005) genes in the wild type and osddm1b in the absence or presence of 24-epiBL. In osddm1b, BR’s inhibition of these feedback-inhibited genes was released, and they were all strongly suppressed in the mutant than wild type (Figure 6E). To find the essential answers of OsDDM1b in regulating BR biosynthesis, the amounts of endogenous BRs were measured in the mutants and wild type (Supplementary Figure 6B). The contents of endogenous BRs were increased by 36.01% in osddm1b compared with wild type, which indicated that OsDDM1b was required for the feedback inhibition of BR biosynthetic and signaling genes.
The Effect of Osddm1b on Grain Size-Related Genes
Grain size includes length, width, and thickness, as a quantitative trait locus (QTL), which is vital for grain yield and quality. Thus far, multiple QTLs have been cloned and studied, such as BG1, GL3.1, GL6, GL7, GLW7, GS2, GS5, GS9, GSN1, MKKK10, qLGY3, qTGW3, GW2, GW5, GW8, DEP1, RGB1, and XIAO, which contributed to rice yield (Song et al., 2007; Huang et al., 2009; Li et al., 2011; Jiang et al., 2012; Qi et al., 2012; Wang et al., 2012, 2015, 2019; Hu et al., 2015, 2018; Liu L. C. et al., 2015; Si et al., 2016; Liu et al., 2017, 2018; Guo et al., 2018; Xu et al., 2018; Ying et al., 2018; Zhao et al., 2018). Snf2 family genes, as the chromatin remodeling factors, regulate the transcription of the genes, and our data showed that loss-of-function of OsDDM1b resulted in abnormal grain size. To analyze the expression level of grain size-related genes in osddm1b, we detected several genes in inflorescence (length 8 cm) and seeds (1 DAF) in wild type and osddm1b using qRT-PCR. The results showed that GL3.1, GL7, GS2, GS5, GSN1, MKKK10, GW8, DEP1, RGB1, and XIAO were upregulated in osddm1b compared with wild type in inflorescence or seeds (Figure 7), and GL3.1, GL7, GS2, and DEP1 were significantly upregulated. It indicates that OsDDM1b negatively regulates their expression level, which affects the grain length (Figure 7A). The loss-of-function of GSN1, MKKK10, and XIAO caused small grain size, which was highly expressed in inflorescence and seeds in osddm1b, which suggests that OsDDM1b might negatively regulate the expression of these genes (Figures 7A,B). The expression of GS5 and GW2, which affects the milk-filling rate in rice, increased significantly in 1 DAF seeds of osddm1b. However, in-depth study is required for identifying that the loss-of-function of OsDDM1b influences milk-filling rate in rice.
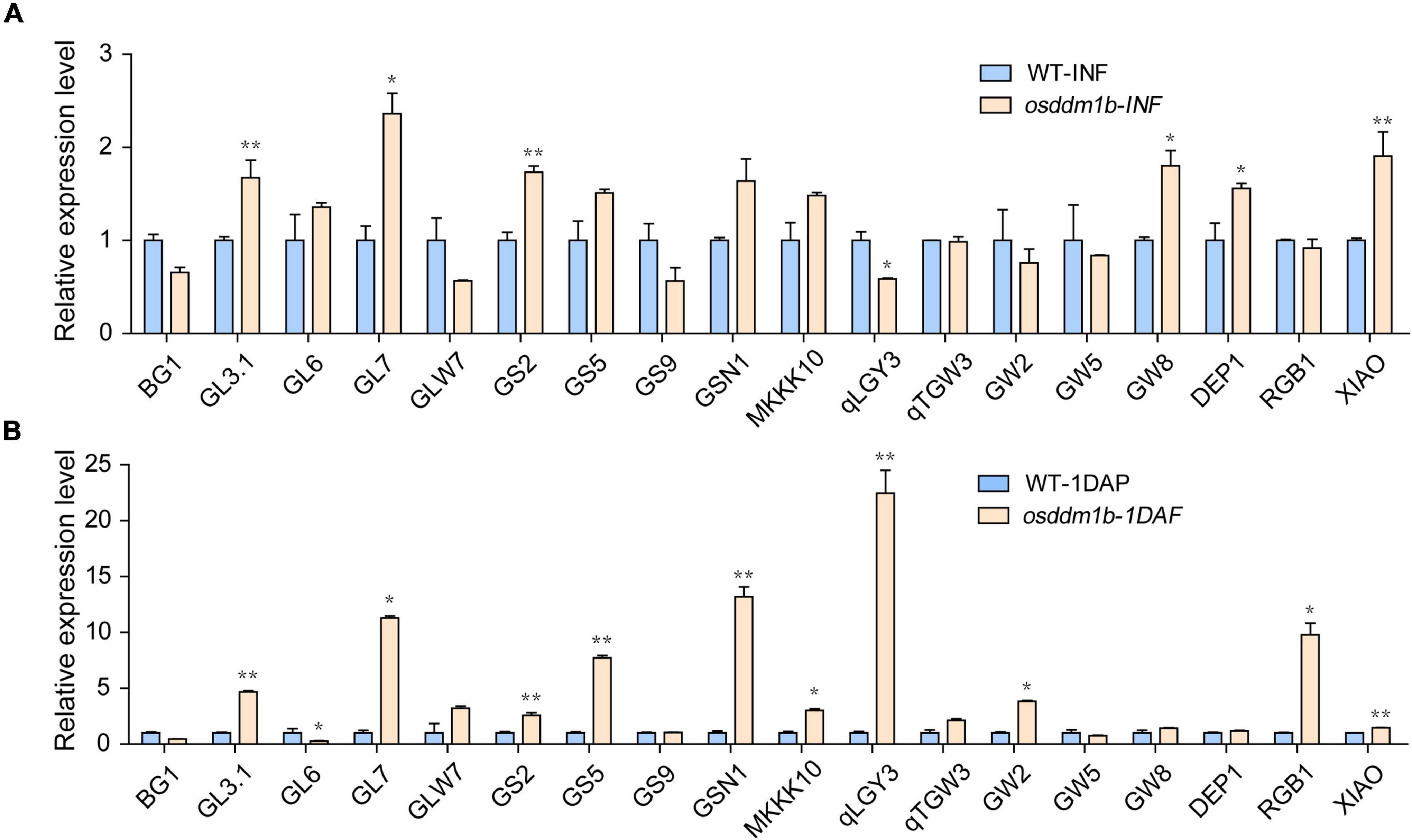
Figure 7. Comparative expression of the genes to determine grain size between wild-type (HY) and osddm1b plants. (A) The expression level of the genes for the determination of grain size by qRT-PCR using 8 cm panicle. (B) The relative expression of grain size determination genes in the seeds of one day after fertilization. Data are given as means ± SD. *p < 0.05, **p < 0.01 compared with wild type (HY) using Student’s t-test.
Discussion
Snf2 Gene Family in Rice
Snf2, as a sizeable gene family, has conserved domains (SNF2_N, DEAD and Helicase_C, HELICs) as in animal counterparts, which suggests that they may have a similar function to regulate the gene expression. There are 42 proteins in chromDB database that belong to Snf2 family in rice. The proteins of OsCHR723 and OsCHR744 (homologs of MOM1/CHR15 in Arabidopsis) do not have the Helicase_C domain, which is not the member of Snf2 family (Hu et al., 2013). Many genes exhibit specific expression patterns at the different developmental stages of various reproductive tissues (inflorescence, stamens, pistils, and seeds). In addition, some genes have been reported to regulate the abiotic or biotic stress responses, such as OsALT1 (alkaline tolerance 1, OsCHR706) (Guo et al., 2014) and OsBRHIS1 (Li et al., 2015). Here, we report the comparative expression profile of 40 Snf2 genes during the reproductive development to enrich the expression patterns and insights into the functions of this family. However, the responses of the Snf2 gene family under abiotic or biotic stresses have not been systematically analyzed.
The Function of DDM1 Is Conserved in Arabidopsis and Rice
DDM1 is a nucleosome remodeling ATPase and regulates the DNA damage that is critical influence for the chromatin structure in Arabidopsis. The RNAi mutation of DDM1 has a typical phenotype of demethylation (Shaked et al., 2006). The repeated sequence, even a single-copy sequence, is induced toward hypomethylation after repeated self-pollination, which activates the transposons (Kakutani et al., 1996). The mutation of DDM1 showed reduced fertility only after repeated self-pollination and produced several kinds of developmental abnormalities. DDM1 is conserved in regulating DNA methylation. For example, Lsh (lymphocyte-specific helicase), a homolog gene of DDM1, causes genomic DNA hypomethylation in the mouse (Dennis et al., 2001). The antisense OsDDM1 lines exhibit dwarf phenotypes, and some sublines were observed to be progressive loss of fertility even complete infertility. In addition, methylation is severely reduced in antisense OsDDM1 lines, and reduction is evident in later generations but not in the genetic transformation process. It is still unknown whether the demethylation reduction was mediated by the repression of the OsDDM1a/OsCHR746, OsDDM1b/OsCHR741, or both of them. Our data showed that these two homologous genes, OsDDM1a/OsCHR746 and OsDDM1b/OsCHR741, had similar expression patterns and slight differences during seed development, which indicates that the two genes might have diverse functions.
The Phenotype of Osddm1b on Regulating the Grain Size
The dwarfism phenotype was also observed in the T-DNA insertion mutation of OsDDM1b (Figures 2B,H,I and Supplementary Figures 2B,C). The male and female gametophyte formation was found to be normal as expected. However, the seed setting rate was partially decreased to 84.39%, which suggests that OsDDM1b might play a role in pollen tube growth or embryo development after fertilization (Supplementary Figure 2D–H). The dwarfism and partial fertility are similar to antisense OsDDM1 lines. The single mutation of OsDDM1a or OsDDM1b showed a normal phenotype during vegetative or reproductive stages, even in several generations (Tan et al., 2016). However, our data showed that osddm1b exhibited dwarfism and grain size variations. The T-DNA insertion in Tan’s mutation of OsDDM1b was in the third intron. However, in our experiment, the mutation was in the second exon (Figure 2A and Supplementary Figure 2A, see footnote 3), which suggests that this difference may be due to the position of T-DNA insertion. OsDDM1a and OsDDM1b, as the homologous genes, have a significantly similar genome and amino acid sequences. The primers of qRT-PCR were designed by aligning the CDS of two genes and finding the different bases among T-DNA insertion positions to improve primers’ specificity and obtain more accurate expression data (Figure 2A).
As an important agronomic trait, grain size is essential for crop genetics and breeding, which includes length, width, and thickness. Several genes have been reported to play the critical roles in regulating grain size. Cell proliferation and expansion are coordinately significant in the spikelet hull, which controls the final grain size. The expression levels of both OsDDM1 are more enriched in proliferating young tissues (Hu et al., 2013), which is consistent with the expression pattern of OsDDM1b by GUS staining (Figures 5B,C and Supplementary Figure 5). The results of the paraffin section and SEM suggested that OsDDM1b might promote latitudinal growth by increasing cell proliferation to contribute to the grain size (Figure 4 and Supplementary Figure 4). Many regulators are involved in controlling grain size, such as G protein signaling (DEP1, RGB1) (Huang et al., 2009; Zhang et al., 2021), the ubiquitin–proteasome pathway (GW2) (Song et al., 2007), the MAPK signaling pathway (GSN1, MKKK10, and MAPK6) (Liu S. Y. et al., 2015; Guo et al., 2018; Xu et al., 2018), phytohormone signaling (BG1, GL3.1, and GS5) (Li et al., 2011; Qi et al., 2012; Liu L. C. et al., 2015), transcriptional regulation (GL7, GLW7/OsSPL13, GS2/OsGRF4, GS9, GW8/OsSPL16, and qLGY3/OsMADS1) (Wang et al., 2012, 2015; Hu et al., 2015; Si et al., 2016; Liu et al., 2018; Zhao et al., 2018), and other regulators such as protein kinase (qTGW3/OsGSK5) (Hu et al., 2018). The creation of insertion mutations is a valuable tool for active transposons to analyze the function of endogenous genes (Hirochika, 2001). The abnormal grain size was caused by the defection of OsDDM1b, which affected the expression of grain size regulators.
There may be a balance mechanism in plants when the mutation of OsDDM1b displayed decreased grain length. In contrast, grain width and thickness were increased due to the compensation mechanism or regulated grain filling rate to balance the 1,000-grain weight with wild type. The higher expression levels of GS5 produced more comprehensive grains and increased grain filling rate. Then, GS5 was upregulated in the inflorescence (length 8 cm) and seeds (1 DAF) of osddm1b, which suggests that OsDDM1b might negatively regulate GS5 to affect the grain width. However, whether OsDDM1b can influence the grain filling rate is still unknown.
A Proposed Model for OsDDM1b Regulation on Grain Size
Above all, OsDDM1b (decrease in DNA methylation) contributes to grain size by regulating latitudinal cell proliferation and cell width to enhance the grain width and thickness, which decreases the grain length reversely. In addition, OsDDM1b, as a chromatin remodeling regulator, influences BR homeostasis and signaling pathway, the expression levels of BR, and grain size-related genes (Supplementary Figure 7). DNA methylation is involved in embryo and endosperm development and is required to determine the methylation pattern in osddm1b. The relationship between grain size and dynamic DNA methylation is necessary to be revealed with the future studies. This helps to understand the mechanism of epigenetic regulation in controlling grain size in rice.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
YQ and HZ conceived the initial screening and research plans and designed the experiments. MG, WZ, ZH, ZS, MY, CS, LL, AW, and JL performed most of the experiments. ZS, MY, CS, LL, and AW extracted the RNA. MM, MG, and WZ performed the qRT-PCR and analyzed the data. ZH and DT performed the paraffin section and SEM. YQ, HZ, and MG wrote the article with the contributions of all the authors. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Fujian Natural Science Foundation (2019J01425), the Science and Technology Program of Fujian Province (2019N5008), Scientific Research Foundation of Graduate School of Fujian Agriculture and Forestry University (324-1122yb069), College Students’ Innovative Entrepreneurial Training Plan Program (201510389081, 201610389062, and 201910389010), the National Natural Science Foundation of China (31970333), and a Guangxi Distinguished Experts Fellowship awarded to YQ.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.873993/full#supplementary-material
Supplementary Figure 1 | The relative expression level of Snf2 gene family in the wild-type (ZH11) seeds. (A) Highly expressed OsCHR714 in the S3 (7 DAF) stage, while OsCHR728 is just lowly expressed in the S5 (25 DAF) stage. (B) Preferentially expressed OsCHR720, 726, and 740 in the S4 (10 DAF) stage. (C) predominately expressed OsCHR717, 724, 743, and 746 in the S4 (10 DAF) or S5 (25 DAF) stage. S1, the seeds of 1 DAF; S2, the seeds of 3 DAF; S3, the seeds of 7 DAF; S4, the seeds of 10 DAF; S5, the seeds of 25 DAF.
Supplementary Figure 2 | Genotype and phenotype identification of OsDDM1b mutant. (A) The young seedlings of wild type (HY) and osddm1b in 10 days. Bar = 4 cm. (B) The internode length of wild type (HY) and osddm1b. Bar = 5 cm. (C,D) Viability of mature pollen grains in wild type (C), (HY) and osddm1b (D) as assessed by I2-KI staining. Bar = 200 μm. (E,F) Mature ovules in the wild type (E), (HY) and osddm1b (F). Bar = 25 μm. (G) The seed setting rate of wild type (HY) and osddm1b (n = 10). (H) The 1,000-grain weight of wild type (HY) and osddm1b (n = 27). (I–N) Comparation on grain length (I,J), width (K,M), and thickness (L,N) of wild type (HY) and osddm1b. (G–H) Bar = 5 mm. (I–L) Bar = 2 mm. Data are given as means ± SD. NSp > 0.05, NS: not significant.
Supplementary Figure 3 | Identification of T-DNA insertion site. A segment was amplified from osddm1b homozygous mutants DNA using the 2B-60109-Lp and Rb (2707) primers. The PCR fragments were sequenced and aligned with the genome of OsDDM1b and 2707 vector sequence by the SnapGene software, respectively.
Supplementary Figure 4 | Histological analysis of spikelet hulls and the effect of OsDDM1b on the expression of genes involved in the cell cycle. (A,B) Magnified views of the cross-section of wild type (HY) and osddm1b. The black and blue arrows indicate the rows of specialized cells with lower epidermis cells and rigid walls, respectively. Bar = 50 μm. (C) The expression levels of cell cycle-related genes in the 8-cm panicles. Data are given as means ± SD. **p < 0.01, *p < 0.05 compared with wild type (HY) using Student’s t-test.
Supplementary Figure 5 | The native OsDDM1b promoter drives GUS expression. (A–D) GUS signal was detected in young root tips (A), young leaf (B), mature leaf (C), stem node (D). Bar = 1 mm (A), Bar = 2 mm (B–D). (E–F) The stamen (E) and pistil (F) from 1–2, 2–3, 3–4, 4–5, 5–6, 6–7 cm, and mature spikelet, respectively. Bar = 500 μm.
Supplementary Figure 6 | The phenotype of wild type (HY) and osddm1b in response to BR treatment. (A) Lamina joint of the wild type (HY) and osddm1b in response to various concentrations of BR. Bar = 2 mm. (B) The contents of endogenous BR in the wild type (HY) and osddm1b. Data are given as means ± SD. **p < 0.01 compared with wild type (HY) using Student’s t-test.
Supplementary Figure 7 | A working model on OsDDM1b regulation on grain size. As an epigenetic regulator, OsDDM1b is involved in DNA methylation, which participates in multiple biological processes. For example, OsDDM1b affected the expression of key genes determining cell cycle, BR signal, and grain size. Abnormal cell proliferation can influence the character of spikelets resulting in grain size defection.
Footnotes
- ^ http://gsds.gao-lab.org/index.php
- ^ https://imagej.nih.gov/ij/
- ^ http://signal.salk.edu/cgi-bin/RiceGE
References
Andrew, F., Martin, D., Barton, G. J., and Tom, O. H. (2006). Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 34, 2887–2905. doi: 10.1093/nar/gkl295
Bai, M. Y., Zhang, L. Y., Gampala, S. S., Zhu, S. W., Song, W. Y., Chong, K., et al. (2007). Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc. Natl. Acad. Sci. U.S.A. 104, 13839–13844. doi: 10.1073/pnas.0706386104
Bargsten, J. W., Folta, A., Mlynarova, L., and Nap, J. P. (2013). Snf2 family gene distribution in higher plant genomes reveals DRD1 expansion and diversification in the tomato genome. PLoS One 8:e81147. doi: 10.1371/journal.pone.0081147
Brzezinka, K., Altmann, S., Czesnick, H., Nicolas, P., Gorka, M., Benke, E., et al. (2016). Arabidopsis FORGETTER1 mediates stress-induced chromatin memory through nucleosome remodeling. Elife 5:e17061. doi: 10.7554/eLife.17061
Cai, H. Y., Zhang, M., Chai, M. N., He, Q., Huang, X. Y., Zhao, L. H., et al. (2019). Epigenetic regulation of anthocyanin biosynthesis by an antagonistic interaction between H2A.Z and H3K4me3. New Phytol. 221, 295–308. doi: 10.1111/nph.15306
Cairns, B. R. (2005). Chromatin remodeling complexes: strength in diversity, precision through specialization. Curr. Opin. Genet. Dev. 15, 185–190. doi: 10.1016/j.gde.2005.01.003
Chen, L. S., Xiong, G. S., Cui, X., Yan, M. X., Xu, T., Qian, Q., et al. (2013). OsGRAS19 may be a novel component involved in the brassinosteroid signaling pathway in rice. Mol. Plant 6, 988–991. doi: 10.1093/mp/sst027
Cho, E. J., Choi, S. H., Kim, J. H., Kim, J. E., Lee, M. H., Chung, B. Y., et al. (2016). A mutation in plant-specific SWI2/SNF2-like chromatin-remodeling proteins, DRD1 and DDM1, delays leaf senescence in Arabidopsis thaliana. PLoS One 11:e0146826. doi: 10.1371/journal.pone.0146826
Choi, K., Park, C., Lee, J., Oh, M., Noh, B., and Lee, I. (2007). Arabidopsis homologs of components of the SWR1 complex regulate flowering and plant development. Development 134, 1931–1941. doi: 10.1242/dev.001891
Dennis, K., Tao, F., Geiman, T., Yan, Q., and Muegge, K. (2001). Lsh, a member of the SNF2 family, is required for genome-wide methylation. Genes Dev. 15, 2940–2944. doi: 10.1101/gad.929101
Duan, K., Li, L., Hu, P., Xu, S. P., Xu, Z. H., and Xue, H. W. (2006). A brassinolide-suppressed rice MADS-box transcription factor, OsMDP1, has a negative regulatory role in BR signaling. Plant J. 47, 519–531. doi: 10.1111/j.1365-313X.2006.02804.x
Efroni, I., Han, S. K., Kim, H. J., Wu, M. F., Steiner, E., Birnbaum, K. D., et al. (2013). Regulation of leaf maturation by chromatin-mediated modulation of cytokinin responses. Dev. Cell 24, 438–445. doi: 10.1016/j.devcel.2013.01.019
Farrona, S., Hurtado, L., Bowman, J. L., and Reyes, J. C. (2004). The Arabidopsis thaliana SNF2 homolog AtBRM controls shoot development and flowering. Development 131, 4965–4975. doi: 10.1242/dev.01363
Farrona, S., Hurtado, L., March-Diaz, R., Schmitz, R. J., Florencio, F. J., Turck, F., et al. (2011). Brahma is required for proper expression of the floral repressor FLC in Arabidopsis. PLoS One 6:e17997. doi: 10.1371/journal.pone.0017997
Folta, A., Bargsten, J. W., Bisseling, T., Nap, J. P., and Mlynarova, L. (2016). Compact tomato seedlings and plants upon overexpression of a tomato chromatin remodelling ATPase gene. Plant Biotechnol. J. 14, 581–591. doi: 10.1111/pbi.12400
Guo, M. X., Wang, R. C., Wang, J., Hua, K., Wang, Y. M., Liu, X. Q., et al. (2014). ALT1, a Snf2 family chromatin remodeling ATPase, negatively regulates alkaline tolerance through enhanced defense against oxidative stress in rice. PLoS One 9:e112515. doi: 10.1371/journal.pone.0112515
Guo, T., Chen, K., Dong, N. Q., Shi, C. L., Ye, W. W., Gao, J. P., et al. (2018). GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell 30, 871–888. doi: 10.1105/tpc.17.00959
Han, S. K., Sang, Y., Rodrigues, A., Wu, M. F., Rodriguez, P. L., Wagner, D., et al. (2012). The SWI2/SNF2 chromatin remodeling ATPase BRAHMA represses abscisic acid responses in the absence of the stress stimulus in Arabidopsis. Plant Cell 24, 4892–4906. doi: 10.1105/tpc.112.105114
Higo, H., Tahir, M., Takashima, K., Miura, A., Watanabe, K., Tagiri, A., et al. (2012). DDM1 (Decrease in DNA Methylation) genes in rice (Oryza sativa). Mol. Genet. Genom. 287, 785–792. doi: 10.1007/s00438-012-0717-5
Hirochika, H. (2001). Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant. Biol. 4, 118–122. doi: 10.1016/s1369-5266(00)00146-1
Hong, Z. Ueguchi-Tanaka, M. Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2002). Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 32, 495–508. doi: 10.1046/j.1365-313x.2002.01438.x
Hong, Z., Ueguchi-Tanaka, M., Shimizu-Sato, S., Inukai, Y., Fujioka, S., Shimada, Y., et al. (2003). A rice brassinosteroid-deficient mutant, ebisu dwarf (d2), is caused by a loss of function of a new member of cytochrome P450. Plant Cell 15, 2900–2910. doi: 10.1105/tpc.014712
Hu, J., Wang, Y. X., Fang, Y. X., Zeng, L. J., Xu, J., Yu, H. P., et al. (2015). A rare allele of GS2 enhances grain size and grain yield in rice. Mol. Plant 8, 1455–1465. doi: 10.1016/j.molp.2015.07.002
Hu, Y. F., Liu, D. N., Zhong, X. C., Zhang, C. J., Zhang, Q. F., and Zhou, D. X. (2012). CHD3 protein recognizes and regulates methylated histone H3 lysines 4 and 27 over a subset of targets in the rice genome. Proc. Natl. Acad. Sci. U.S.A. 109, 5773–5778. doi: 10.1073/pnas.1203148109
Hu, Y., Zhu, N., Wang, X., Yi, Q., Zhu, D., Lai, Y., et al. (2013). Analysis of rice Snf2 family proteins and their potential roles in epigenetic regulation. Plant Physiol. Biochem. 70, 33–42. doi: 10.1016/j.plaphy.2013.05.001
Hu, Z., Lu, S.-J., Wang, M.-J., He, H., Sun, L., Wang, H., et al. (2018). A novel QTL qTGW3 encodes the GSK3/SHAGGY-like kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol. Plant 11, 736–749. doi: 10.1016/j.molp.2018.03.005
Huang, X. Z., Qian, Q., Liu, Z. B., Sun, H. Y., He, S. Y., Luo, D., et al. (2009). Natural variation at the DEP1 locus enhances grain yield in rice. Nat. Genet. 41, 494–497. doi: 10.1038/ng.352
Hurtado, L., Farrona, S., and Reyes, J. C. (2006). The putative SWI/SNF complex subunit BRAHMA activates flower homeotic genes in Arabidopsis thaliana. Plant Mol. Biol. 62, 291–304. doi: 10.1007/s11103-006-9021-2
Ito, Y., Takaya, K., and Kurata, N. (2005). Expression of SERK family receptor-like protein kinase genes in rice. Biochim. Biophys. Acta Gene Struct. Expr. 1730, 253–258. doi: 10.1016/j.bbaexp.2005.06.007
Je, B. I., Hai, L. P., Park, S. J., Park, S. H., Kim, C. M., Xuan, Y. H., et al. (2010). RAV-Like1 maintains brassinosteroid homeostasis via the coordinated activation of BRI1 and biosynthetic genes in rice. Plant Cell 22, 1777–1791. doi: 10.1105/tpc.109.069575
Jeddeloh, J. A., Stokes, T. L., and Richards, E. J. (1999). Maintenance of genomic methylation requires a SWI2/SNF2-like protein. Nat Genet. 22, 94–97. doi: 10.1038/8803
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6, 3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x
Jiang, Y. H., Bao, L., Jeong, S. Y., Kim, S. K., Xu, C. G., Li, X. H., et al. (2012). XIAO is involved in the control of organ size by contributing to the regulation of signaling and homeostasis of brassinosteroids and cell cycling in rice. Plant J. 70, 398–408. doi: 10.1111/j.1365-313X.2011.04877.x
Kakimoto, T. J. P., and Physiology, C. (2011). The CKH2/PKL chromatin remodeling factor negatively regulates cytokinin responses in Arabidopsis Calli. Plant Cell Physiol. 52, 618–628. doi: 10.1093/pcp/pcr022
Kakutani, T., Jeddeloh, J. A., Flowers, S. K., Munakata, K., and Richards, E. J. (1996). Developmental abnormalities and epimutations associated with DNA hypomethylation mutations. Proc. Natl. Acad. Sci. U.S.A. 93, 12406–12411. doi: 10.1073/pnas.93.22.12406
Kanno, T., Mette, M. F., Kreil, D. P., Aufsatz, W., Matzke, M., and Matzke, A. J. M. (2004). Involvement of putative SNF2 chromatin remodeling protein DRD1 in RNA-directed DNA methylation. Curr. Biol. 14, 801–805. doi: 10.1016/j.cub.2004.04.037
Knizewski, L., Ginalski, K., and Jerzmanowski, A. (2008). Snf2 proteins in plants: gene silencing and beyond. Trends Plant Sci. 13, 557–565. doi: 10.1016/j.tplants.2008.08.004
Koh, S., Lee, S. C., Kim, M. K., Koh, J. H., Lee, S., An, G., et al. (2007). T-DNA tagged knockout mutation of rice OsGSK1, an orthologue of Arabidopsis BIN2, with enhanced tolerance to various abiotic stresses. Plant Mol. Biol. 65, 453–466. doi: 10.1007/s11103-007-9213-4
Kwon, C. S., Chen, C. B., and Wagner, D. (2005). WUSCHEL is a primary target for transcriptional regulation by SPLAYED in dynamic control of stem cell fate in Arabidopsis. Genes Dev. 19, 992–1003. doi: 10.1101/gad.1276305
Li, C. L., Gu, L. F., Gao, L., Chen, C., Wei, C. Q., Qiu, Q., et al. (2016). Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 48, 687–693. doi: 10.1038/ng.3555
Li, J., Wen, J. Q., Lease, K. A., Doke, J. T., Tax, F. E., and Walker, J. C. (2002). BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell 110, 213–222. doi: 10.1016/s0092-8674(02)00812-7
Li, X., Jiang, Y., Ji, Z., Liu, Y., and Zhang, Q. (2015). BRHIS1 suppresses rice innate immunity through binding to monoubiquitinated H2A and H2B variants. EMBO Rep. 16, 1192–1202. doi: 10.15252/embr.201440000
Li, Y. B., Fan, C. C., Xing, Y. Z., Jiang, Y. H., Luo, L. J., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Liu, J., Chen, J., Zheng, X., Wu, F., Lin, Q., Heng, Y., et al. (2017). GW5 acts in the brassinosteroid signalling pathway to regulate grain width and weight in rice. Nat. Plants 3:17043. doi: 10.1038/nplants.2017.43
Liu, L. C., Tong, H. N., Xiao, Y. H., Che, R. H., Xu, F., Hu, B., et al. (2015). Activation of big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 11102,2015.
Liu, Q., Han, R., Wu, K., Zhang, J., Ye, Y., Wang, S., et al. (2018). G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9
Liu, S. Y., Hua, L., Dong, S. J., Chen, H. Q., Zhu, X. D., Jiang, J. E., et al. (2015). OsMAPK6, a mitogen-activated protein kinase, influences rice grain size and biomass production. Plant J. 84, 672–681. doi: 10.1111/tpj.13025
Ma, X. D., Ma, J., Zhai, H. H., Xin, P. Y., Chu, J. F., Qiao, Y. L., et al. (2015). CHR729 Is a CHD3 protein that controls seedling development in rice. PLoS One 10:e0138934. doi: 10.1371/journal.pone.0138934
Mlynarova, L., Nap, J. P., and Bisseling, T. (2007). The SWI/SNF chromatin-remodeling gene AtCHR12 mediates temporary growth arrest in Arabidopsis thaliana upon perceiving environmental stress. Plant J. 51, 874–885. doi: 10.1111/j.1365-313X.2007.03185.x
Qi, P., Lin, Y. S., Song, X. J., Shen, J. B., Huang, W., Shan, J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Ren, D. Y., Cui, Y. J., Hu, H. T., Xu, Q. K., Rao, Y. C., Yu, X. Q., et al. (2019). AH2 encodes a MYB domain protein that determines hull fate and affects grain yield and quality in rice. Plant J. 100, 813–824. doi: 10.1111/tpj.14481
Ren, D. Y., Yu, H. P., Rao, Y. C., Xu, Q. K., Zhou, T. T., Hu, J., et al. (2018). “Two-floret spikelet’ as a novel resource has the potential to increase rice yield. Plant Biotechnol. J. 16, 351–353. doi: 10.1111/pbi.12849
Sang, Q., Pajoro, A., Sun, H., Song, B., Yang, X., Stolze, S. C., et al. (2020). Mutagenesis of a quintuple mutant impaired in environmental responses reveals roles for CHROMATIN REMODELING4 in the Arabidopsis floral transition. Plant Cell 32, 1479–1500. doi: 10.1105/tpc.19.00992
Shaked, H., Avivi-Ragolsky, N., and Levy, A. A. (2006). Involvement of the Arabidopsis SWI2/SNF2 chromatin remodeling gene family in DNA damage response and recombination. Genetics 173, 985–994. doi: 10.1534/genetics.105.051664
Si, L. Z., Chen, J. Y., Huang, X. H., Gong, H., Luo, J. H., Hou, Q. Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Song, X.-J., Huang, W., Shi, M., Zhu, M.-Z., and Lin, H.-X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Song, Z. T., Liu, J. X., and Han, J. J. (2021). Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 63, 438–450. doi: 10.1111/jipb.13064
Su, Z. X., Wang, L. L., Li, W. M., Zhao, L. H., Huang, X. Y., Azam, S. M., et al. (2017). Genome-Wide Identification of Auxin Response Factor (ARF) genes family and its tissue-specific prominent expression in pineapple (Ananas comosus). Trop. Plant Biol. 10, 86–96. doi: 10.1007/s12042-017-9187-6
Tan, F., Lu, Y., Jiang, W., Wu, T., Zhang, R., Zhao, Y., et al. (2018). DDM1 represses noncoding RNA expression and RNA-directed DNA methylation in heterochromatin. Plant Physiol. 177, 1187–1197. doi: 10.1104/pp.18.00352
Tan, F., Zhou, C., Zhou, Q. W., Zhou, S. L., Yang, W. J., Zhao, Y., et al. (2016). Analysis of chromatin regulators reveals specific features of rice DNA methylation pathways. Plant Physiol. 171, 2041–2054. doi: 10.1104/pp.16.00393
Tanabe, S., Ashikari, M., Fujioka, S., Takatsuto, S., Yoshida, S., Yano, M., et al. (2005). A novel cytochrome P450 is implicated in brassinosteroid biosynthesis via the characterization of a rice dwarf mutant, dwarf11, with reduced seed length. Plant Cell 17, 776–790. doi: 10.1105/tpc.104.024950
Tong, H. N., Jin, Y., Liu, W. B., Li, F., Fang, J., Yin, Y. H., et al. (2009). DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 58, 803–816. doi: 10.1111/j.1365-313X.2009.03825.x
Tong, H. N., Liu, L. C., Jin, Y., Du, L., Yin, Y. H., Qian, Q., et al. (2012). DWARF AND LOW-TILLERING acts as a direct downstream target of a GSK3/SHAGGY-like kinase to mediate brassinosteroid responses in rice. Plant Cell 24, 2562–2577. doi: 10.1105/tpc.112.097394
Vercruyssen, L., Verkest, A., Gonzalez, N., Heyndrickx, K. S., Eeckhout, D., Han, S.-K., et al. (2014). ANGUSTIFOLIA3 binds to SWI/SNF chromatin remodeling complexes to regulate transcription during Arabidopsis leaf development. Plant Cell 26, 210–229. doi: 10.1105/tpc.113.115907
Wang, A. H., Hou, Q. Q., Si, L. Z., Huang, X. H., Luo, J. H., Lu, D. F., et al. (2019). The PLATZ transcription Factor GL6 affects grain length and number in rice. Plant Physiol. 180, 2077–2090. doi: 10.1104/pp.18.01574
Wang, L., Xu, Y., Zhang, C., Ma, Q., Joo, S. H., Kim, S. K., et al. (2008). OsLIC, a novel CCCH-Type zinc finger protein with transcription activation, mediates rice architecture via brassinosteroids signaling. PLoS One 3:e3521. doi: 10.1371/journal.pone.0003521
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, Y. X., Xiong, G. S., Hu, J., Jiang, L., Yu, H., Xu, J., et al. (2015). Copy number variation at the GL7 locus contributes to grain size diversity in rice. Nat. Genet. 47, 944–948. doi: 10.1038/ng.3346
Xu, R., Duan, P. G., Yu, H. Y., Zhou, Z. K., Zhang, B. L., Wang, R. C., et al. (2018). Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol. Plant 11, 860–873. doi: 10.1016/j.molp.2018.04.004
Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S., Ashikari, M., et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell 12, 1591–1606. doi: 10.1105/tpc.12.9.1591
Ying, J.-Z., Ma, M., Bai, C., Huang, X.-H., Liu, J.-L., Fan, Y.-Y., et al. (2018). TGW3, a major QTL that negatively modulates grain length and weight in rice. Mol. Plant 11, 750–753. doi: 10.1016/j.molp.2018.03.007
Yu, X. Q., Xia, S. S., Xu, Q. K., Cui, Y. J., Gong, M., Zeng, D. L., et al. (2020). ABNORMAL FLOWER AND GRAIN 1 encodes OsMADS6 and determines palea identity and affects rice grain yield and quality. Sci. China Life Sci. 63, 228–238. doi: 10.1007/s11427-019-1593-0
Zeng, Y. X., Hu, C. Y., Lu, Y. G., Li, J. Q., and Liu, X. D. (2007). Diversity of abnormal embryo sacs in indica/japonica hybrids in rice demonstrated by confocal microscopy of ovaries. Plant Breed. 126, 574–580. doi: 10.1111/j.1439-0523.2007.01380.x
Zeng, Y. X., Hu, C. Y., Lu, Y. G., Li, J. Q., and Liu, X. D. (2009). Abnormalities occurring during female gametophyte development result in the diversity of abnormal embryo sacs and leads to abnormal fertilization in indica/japonica hybrids in rice. J. Integr. Plant Biol. 51, 3–12. doi: 10.1111/j.1744-7909.2008.00733.x
Zhang, D., Gao, S., Yang, P., Yang, J., Yang, S., and Wu, K. (2019). Identification and expression analysis of Snf2 family proteins in tomato (Solanum lycopersicum). Int. J. Genomics 2019:5080935. doi: 10.1155/2019/5080935
Zhang, D., Jing, Y., Jiang, Z., and Lin, R. (2014). The chromatin-remodeling Factor PICKLE integrates brassinosteroid and gibberellin signaling during skotomorphogenic growth in Arabidopsis. Plant Cell 26:2472. doi: 10.1105/tpc.113.121848
Zhang, D., Li, Y. H., Zhang, X. Y., Zha, P., and Lin, R. C. (2017). The SWI2/SNF2 chromatin-remodeling ATPase BRAHMA regulates chlorophyll biosynthesis in Arabidopsis. Mol. Plant 10, 155–167. doi: 10.1016/j.molp.2016.11.003
Zhang, D., Zhang, M., and Liang, J. (2021). RGB1 regulates grain development and starch accumulation through its effect on OsYUC11-mediated auxin biosynthesis in rice endosperm cells. Front. Plant Sci. 12:585174. doi: 10.3389/fpls.2021.585174
Zhang, H., Bishop, B., Ringenberg, W., Muir, W. M., and Ogas, J. (2012). The CHD3 Remodeler PICKLE associates with genes enriched for Trimethylation of Histone H3 Lysine 27. Plant Physiol. 159, 418–432. doi: 10.1104/pp.112.194878
Zhang, L. Y., Bai, M. Y., Wu, J. X., Zhu, J. Y., Wang, H., Zhang, Z. G., et al. (2009). Antagonistic HLH/bHLH transcription factors mediate brassinosteroid regulation of cell elongation and plant development in rice and Arabidopsis. Plant Cell 21, 3767–3780. doi: 10.1105/tpc.109.070441
Zhang, M., Liu, Y. H., Cai, H. Y., Guo, M. L., Chai, M. N., She, Z. Y., et al. (2020). The bZIP Transcription Factor GmbZIP15 negatively regulates salt- and drought-stress responses in soybean. Int. J. Mol. Sci. 21:7778. doi: 10.3390/ijms21207778
Zhao, D. S., Li, Q. F., Zhang, C. Q., Zhang, C., Yang, Q. Q., Pan, L. X., et al. (2018). GS9 acts as a transcriptional activator to regulate rice grain shape and appearance quality. Nat. Commun. 9:1240. doi: 10.1038/s41467-018-03616-y
Zhao, H. M., Guo, M. L., Yan, M. K., Cheng, H., Liu, Y. H., She, Z. Y., et al. (2020). Comparative expression profiling reveals genes involved in Megasporogenesis. Plant Physiology 182, 2006–2024. doi: 10.1104/pp.19.01254
Zhao, H. M., Maokai, Y., Cheng, H., Guo, M. L., Liu, Y. H., Wang, L. L., et al. (2021). Characterization of auxin transporter AUX, PIN and PILS gene families in pineapple and evaluation of expression profiles during reproductive development and under abiotic stresses. PeerJ 9:e11410. doi: 10.7717/peerj.11410
Keywords: rice, Snf2 family, OsDDM1b, grain size, cell proliferation, brassinosteroid
Citation: Guo M, Zhang W, Mohammadi MA, He Z, She Z, Yan M, Shi C, Lin L, Wang A, Liu J, Tian D, Zhao H and Qin Y (2022) OsDDM1b Controls Grain Size by Influencing Cell Cycling and Regulating Homeostasis and Signaling of Brassinosteroid in Rice. Front. Plant Sci. 13:873993. doi: 10.3389/fpls.2022.873993
Received: 11 February 2022; Accepted: 07 March 2022;
Published: 08 April 2022.
Edited by:
Zhaojun Ding, Shandong University, Qingdao, ChinaReviewed by:
Shengjun Li, Qingdao Institute of Bioenergy and Bioprocess Technology (CAS), ChinaShutang Tan, University of Science and Technology of China, China
Copyright © 2022 Guo, Zhang, Mohammadi, He, She, Yan, Shi, Lin, Wang, Liu, Tian, Zhao and Qin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Heming Zhao, zhaohm@ahstu.edu.cn; Yuan Qin, yuanqin001@foxmail.com
 Mingliang Guo
Mingliang Guo Wenchao Zhang
Wenchao Zhang Mohammad Aqa Mohammadi
Mohammad Aqa Mohammadi Zhimei He
Zhimei He Zeyuan She
Zeyuan She Maokai Yan
Maokai Yan Chao Shi5
Chao Shi5 Yuan Qin
Yuan Qin