- 1College of Horticulture, Nanjing Agricultural University, Nanjing, China
- 2School of Life Sciences, Southern University of Science and Technology, Shenzhen, China
- 3Agricultural and Forestry Service Center, Suzhou, China
- 4Department of Plant Science, University of Manitoba, Winnipeg, MB, Canada
- 5Forestry and Pomology Research Institute, Shanghai Academy of Agricultural Sciences, Shanghai, China
TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors TEOSINTE BRANCHED1/CYCLOIDEA/PCF have been suggested to control the cell growth and proliferation in meristems and lateral organs. A total of 37 CsTCP genes were identified and divided into two classes, class I (PCF, group 1) and class II (CIN CYC/TB1, groups 2, and 3). The residues of TEOSINTE BRANCHED1/CYCLOIDEA/PCF of Camellia sinensis (Tea plant) (CsTCP) proteins between class I and class II were definitely different in the loop, helix I, and helix II regions; however, eighteen conserved tandem was found in bHLH. There are a large number of CsTCP homologous gene pairs in three groups. Additionally, most CsTCP proteins have obvious differences in motif composition. The results illuminated that CsTCP proteins in different groups are supposed to have complementary functions, whereas those in the same class seem to display function redundancies. There is no relationship between the number of CsTCP gene members and genome size, and the CsTCP gene family has only expanded since the divergence of monocots and eudicots. WGD/segmental duplication played a vital role in the expansion of the CsTCP gene family in tea plant, and the CsTCP gene family has expanded a lot. Most CsTCP genes of group 1 are more widely and non-specifically expressed, and the CsTCP genes of group 2 are mainly expressed in buds, flowers, and leaves. Most genes of group 1 and some genes of group 2 were up-/downregulated in varying degrees under different stress, CsTCP genes of group 3 basically do not respond to stress. TCP genes involved in abiotic stress response mostly belong to PCF group. Some CsTCP genes may have the same function as the homologous genes in Arabidopsis, but there is functional differentiation.
Introduction
As an important economical crop, the tea plant (Camellia sinensis) is widely planted in more than 52 countries across the world (Li et al., 2017b). The tea leaves are the main source of the most popular natural non-alcoholic beverages (Chen et al., 2007; Zhang et al., 2015). In tea plant, shoot branching greatly affects the overall plant architecture and other traits of plant, such as height, light harvesting efficiency, and leaf production, which influences the costs and benefits of agricultural production.
Branches are generated from axillary meristems of the axils of leaves, and then, the patterns of branching are conserved in angiosperms. There are few reports about branch development that plays a key role in the life of tea plant. Recently, Cao et al. (2020) revealed that zigzag-shaped shoot formation might be associated with the gravitropism response and polar auxin transport in tea plants (Cao et al., 2020). In addition, Yu et al. (2021) showed that the profiling of more than 40 developmental-related genes (CYC/GROWTH REGULATING FACTORs (GRFs), COTYLEDON 1 FACTORs (GIFs), CUP-SHAPED, PHAVOLUTA (PHV), and REVOLUTA (REV)) essentially proved their high expression levels in developing tea plant buds and leaves (Yu et al., 2021). However, the transcription factors that regulate the growth and development of shoot tips and the formation of tissues and organs in tea plant are rarely studied.
TEOSINTE BRANCHED1/CYCLOIDEA/PCF (TCP) transcription factors (TEOSINTE BRANCHED1/CYCLOIDEA/PCF) have been suggested to control the cell growth and proliferation in meristems and lateral organs (Martin-Trillo and Cubas, 2010). TCP domain was initially identified in four proteins encoded unrelated genes, from which the name “TCP” was derived: Teosinte branched1 (TB1) from maize (Zea mays), which participates in regulating apical dominance, inflorescence development, and some other processes of broad interest in maize developmental biology (Doebley et al., 1997); CYCLOIDEA (CYC) from snapdragon (Antirrhinum majus) (Luo et al., 1996) which regulates floral asymmetry, and the PROLIFERATING CELL FACTORS 1 and 2 (PCF1 and PCF2) from rice (Oryza sativa) which is involved in cell growth and proliferation in meristems and lateral organs (Kosugi and Ohashi, 1997).
TCP gene family is a transcription factor (TF), which contained a conserved non-canonical basic-helix-loop-helix (bHLH) domain with 59 conserved amino acid residues (Cubas et al., 1999). As plant-specific transcription factors, TCP genes are identified in basal land plant and freshwater algal genomes, such as in the Arabidopsis thaliana, poplar, rice, club-moss, and moss genomes, even in Rhodophyta and Prasinophyceae (Navaud et al., 2007). TCP proteins have been divided into two classes (Cubas, 2002), class I (PCF) and class II (CIN and CYC/TB1). Based on the widely existence of TCP genes in plants, TCP proteins are required for the multiple developmental pathways, especially in plant morphologies. Previous studies have reported their involvements in shoot branching (Martín-Trillo et al., 2011), controlling apical dominance (Doebley et al., 1997), and the formation of meristematic tissue (Faivre-Rampant et al., 2004). In addition, they also participated in the leaf and floral development (Luo et al., 1996; Palatnik et al., 2003; Aguilar-Martínez and Sinha, 2013), senescence (Huang and Irish, 2015), flavonoid biosynthesis (Li and Zachgo, 2013), plant immunity (Schommer et al., 2008; Lopez et al., 2015), and hormonal signaling including jasmonic acid (Schommer et al., 2008; Danisman et al., 2012), gibberellin (Daviere et al., 2014), and auxin (Li and Zachgo, 2013).
A total of 24 TCP proteins in Arabidopsis were divided into two classes. Class I (TCP-P, also known as PCF) have been indicated to function as the positive regulators of cell proliferation (Kosugi and Ohashi, 2002). For example, AtTCP14, AtTCP15 (Kieffer et al., 2011), AtTCP20 (Li et al., 2005; HERVé et al., 2009), AtTCP7, AtTCP8, AtTCP22, and AtTCP23 (Aguilar-Martínez and Sinha, 2013) proteins have been reported to play the important roles in processes of cell division and proliferation in leaf growth and development. Class II (TCP-C, also known as CIN and CYC/TB1), which duplicate from ancestral tb1-like gene, have been indicated to function as the regulators of branching signals within axillary buds and the morphogenesis of shoot lateral organs (Aguilar-MARTíNEZ et al., 2007; Finlayson, 2007; Koyama et al., 2007; Mitsuda et al., 2010). Thus, they will induce the plant branching and meristematic activity (Aguilar-MARTíNEZ et al., 2007; Koyama et al., 2007; Mitsuda et al., 2010). For instance, ectopic expression of AtTCP3 inhibits the formation of shoot meristem (Koyama et al., 2007). AtTCP5 is considered as an enhancer in axillary branch outgrowth (van Es et al., 2019), and AtTCP12 (BRANCHED2) and AtTCP18 (BRANCHED1) were involved in branching control (Aguilar-MARTíNEZ et al., 2007). Obviously, functional redundancy has been inferred in various members of the TCP groups (Cubas et al., 1999; Danisman et al., 2013), such as AtTCP14 and AtTCP15 (Kieffer et al., 2011; Steiner et al., 2012; van Es et al., 2019), which affect internode length and leaf shape and induce the branching and meristematic activity.
In Solanum lycopersicum, several genes play a key role in ripening, such as SlTCP12, SlTCP15, and SlTCP18 (Parapunova et al., 2014). In tobacco (Nicotiana tabacum), several TCP genes can affect the leaf development and growth such as NtTCP18 (Chen et al., 2016). There are 22 OsTCP genes in rice. The OsTCP proteins were divided into three groups, PCF, CIN, and CYC/TB1 groups (Yao et al., 2007). Overexpressing OsTB1 transgenic rice exhibited significant reduced lateral branch without the propagation of axillary buds being affected, which indicates that OsTB1 gene negatively regulates lateral branchings (Takeda et al., 2003). A total of 38 TCP genes were identified in Gossypium raimondii (Ma et al., 2014). Among them, the RNAi silenced GbTCP (GenBank accession no. DQ912941) transgenic line produced shorter fiber, a reduced lint percentage, and a lower fiber quality than the wild-type plants and overexpression of GbTCP in Arabidopsis enhanced root hair initiation and elongation. It obviously indicated that GbTCP regulated the fiber elongation and root hair development (Hao et al., 2012; Wang et al., 2013). A total of 52 TCP genes were identified in apple (Malus domestica) genome which were divided into three classes (classes 1, 2, and 3) (Xu et al., 2014).
In this study, 37 TCP proteins were identified in Camellia sinensis. The structural features, phylogenetic relations, and interaction networks of TEOSINTE BRANCHED1/CYCLOIDEA/PCF of Camellia sinensis (Tea plant) (CsTCP) proteins were analyzed. The expression profiles of 37 CsTCP genes in eight different tissues were surveyed to investigate their biological functions.
Materials and Methods
Genome-Wide Identification of TCP Genes in Tea Plant
The AtTCP protein sequence file was downloaded from the Arabidopsis Information Resource (TAIR)1 and put the AtTCP protein sequence on the Pfam protein analysis professional website2 search to obtain the hidden Markov model (HMM) profile of TCP domain (PF03634) (Eddy, 1998). The program HMMER 3.0 was used to search for CsTCP protein members in the tea plant protein sequence file (E-value < 1.0) that was downloaded from the Tea Plant Information Archive (TPIA)3 (Xia et al., 2019), and then, we acquired the protein sequence of CsTCP candidate genes. The conserved domains of candidate TCP proteins were identified one by one using the online websites of Pfam and SMART,4 and some sequences that did not contain TCP domains were removed.
Analyses of Phylogenetic Tree
The amino acid sequences of TCP proteins from Vitis vinifera, Arabidopsis thaliana, and Zea mays were obtained from the Plant Transcription Factor Databases.5 The amino acid sequences of all TCP proteins of Oryza sativa were derived from Rice Genome Annotation Project.6 The TCP proteins of Antirrh-inum majus were obtained from snapdragon genome database.7 All the TCP proteins in this study were aligned using MAFFT 7.0 (Katoh and Standley, 2013). A phylogenetic tree was constructed using the maximum likelihood estimate (ML) method by RAxML 8.0 software (Stamatakis, 2014).
Characteristics of TCP Proteins Analysis
The primary structure of TCP proteins was predicted using ProtParam tool.8 The Softberry Web Site9 was used to predict the subcellular localization of TCP proteins. The MEME (E < 1e-10) (Bailey et al., 2009)10 program was used to analyze protein structural motifs and set the maximum number of output motifs to 10. The DNAMAN 7 (Lynnon Corporation) was used to align the CsTCP domain sequences.
Gene Sequence Analysis for CsTCPs
Exon–intron structures of CsTCP genes were identified and visualized using TBtools (Chen et al., 2020a). Cis-element analysis of the 2,000 bp upstream sequences of each CsTCP gene at the five end of the cDNA was predicted using Plantcare program.11 Tandem duplications of TCP genes in the tea genome were identified by checking physical locations within a 200-kb adjacent region in individual chromosomes. The information for homologous gene pairs and syntenic relationships between tea plant and other species was analyzed using MCscan and using TBtools for visualization12 (Wang et al., 2012).
Mapping CsTCP on Chromosomes
CsTCP genes were mapped on chromosomes based on the whole-genome annotation from TPIA. The map was generated in the MapInspect software.
Plant Materials
About 1-year-old tea plant cultivars (C. sinensis cv. “longjing43”) were planted in an illuminating incubator at the Tea Science Research Institute, College of Horticulture, Nanjing Agriculture University, Jiangsu Province, China (32°03′ N, 118°46′E). The set of the incubator was controlled at 22°C temperature, 14/10 h (day/night).
Expression Pattern Analysis
About 2 weeks after raising seedlings in illuminating incubator, different development stages of tea leaves were sampled including a bud with first leaf (I), 2nd and 3rd leaves (II), natural leaves (4th, 5th, 6th leaves, III), and roots (IV), the epidermis and vascular tissue of unlignified stem (tender phloem (V), tender xylem (VI)) and lignified stem 9older phloem (VII), older xylem (VIII)] (Supplementary Figure 1). All the samples were frozen in liquid nitrogen and stored at –80°C for the following steps. The RNA of samples was extracted using RNA prep Pure Plant Kit (Polysaccharides and Polyphenolics-rich) from TIANGEN (Tiangen Biotech Co., Ltd., Beijing, China). The first-strand cDNA was synthesized using HiScript® II Q RT Super Mix (TaKaRa Biotech Co., Ltd., Dalian, China). The quantitative PCR primers were designed by Beacon designer 7.0 (Supplementary Table 1). Quantitative PCR was conducted in Switzerland Roche, Light Cycler® 480 II using SYBR GREEN dye (TaKaRa Biotech Co., Ltd., Dalian, China). The thermos cycle was set as follows: 95°C for 30 s; 40 cycles of 95°C for 10 s, and 60°C for 30 s. β-actin, as a reference gene of Camellia sinensis, was used as an internal control (Li et al., 2017a). Quantitative expression analysis in each sample was carried out with each of three biological and technique replicates. Relative gene expressions were analyzed using the 2-ΔΔCt method (Livak and Schmittgen, 2001).
Statistical Analysis
The experimental data were sorted and statistically analyzed using Excel 2019 (Microsoft Corp, Albuquerque, United States) software, significance analysis was performed using IBM SPSS Statistics 20.0 (IBM Corporation, New York, United States), all data analysis results were expressed as mean (n = 3) ± standard deviation (SD), and different lowercase letters indicate significant difference at p < 0.05 level. Graphs were made using GraphPad 8.0.1 (GraphPad Software, San Diego, United States) and TBtools v1.098.
Results
Identification and Sequence Analysis of TCP Gene Family in Tea Plant
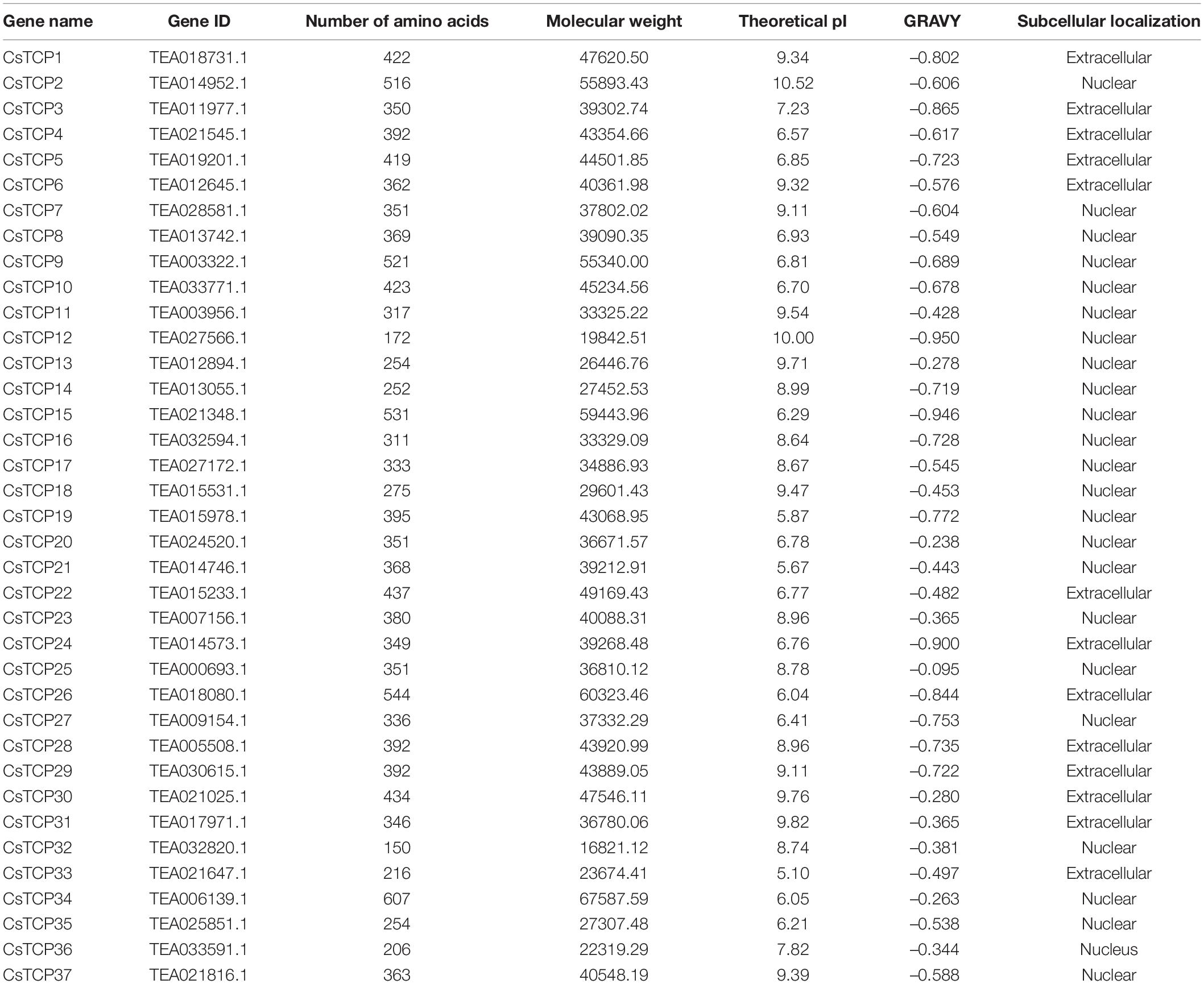
A total of 39 candidate CsTCP genes were obtained from the “Shuchazao” genome database TPIA (SCZ), and two of them were excluded: TEA005756.1 and TEA027571.1 due to the lack of TCP domain. Finally, 37 genes were renamed and included in this study (Table 1). The genome database of Camellia sinensis var. sinensis (CSS) cv. Huangdan (HD)(Wang et al., 2021) and Tieguanyin (TGY)(Zhang et al., 2021) with good assembly quality is worthy of reference. Therefore, we used the same method to identify the CsTCP gene family in the varieties of tea plant HD and TGY and constructed a phylogenetic tree to correspond to the CsTCP proteins. The maximum number of TCP proteins retrieved in SCZ was 37, 32 in HD, and 36 in TGY, all of which contained TCP-conserved domains (Supplementary Table 1). The annotation information of “Shuchazao” genome database is relatively perfect and widely used, and the number of TCP identified is the largest. Therefore, we analyze and discuss the TCP protein identified in SCZ.
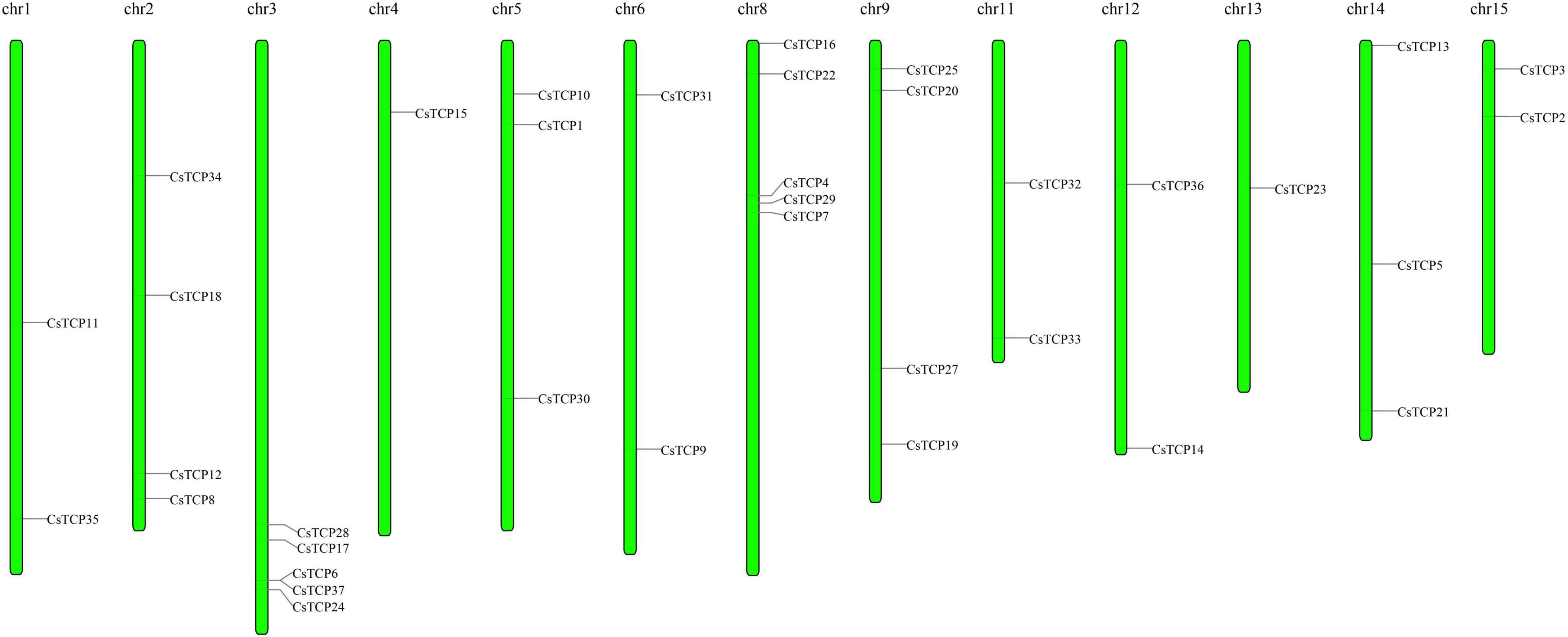
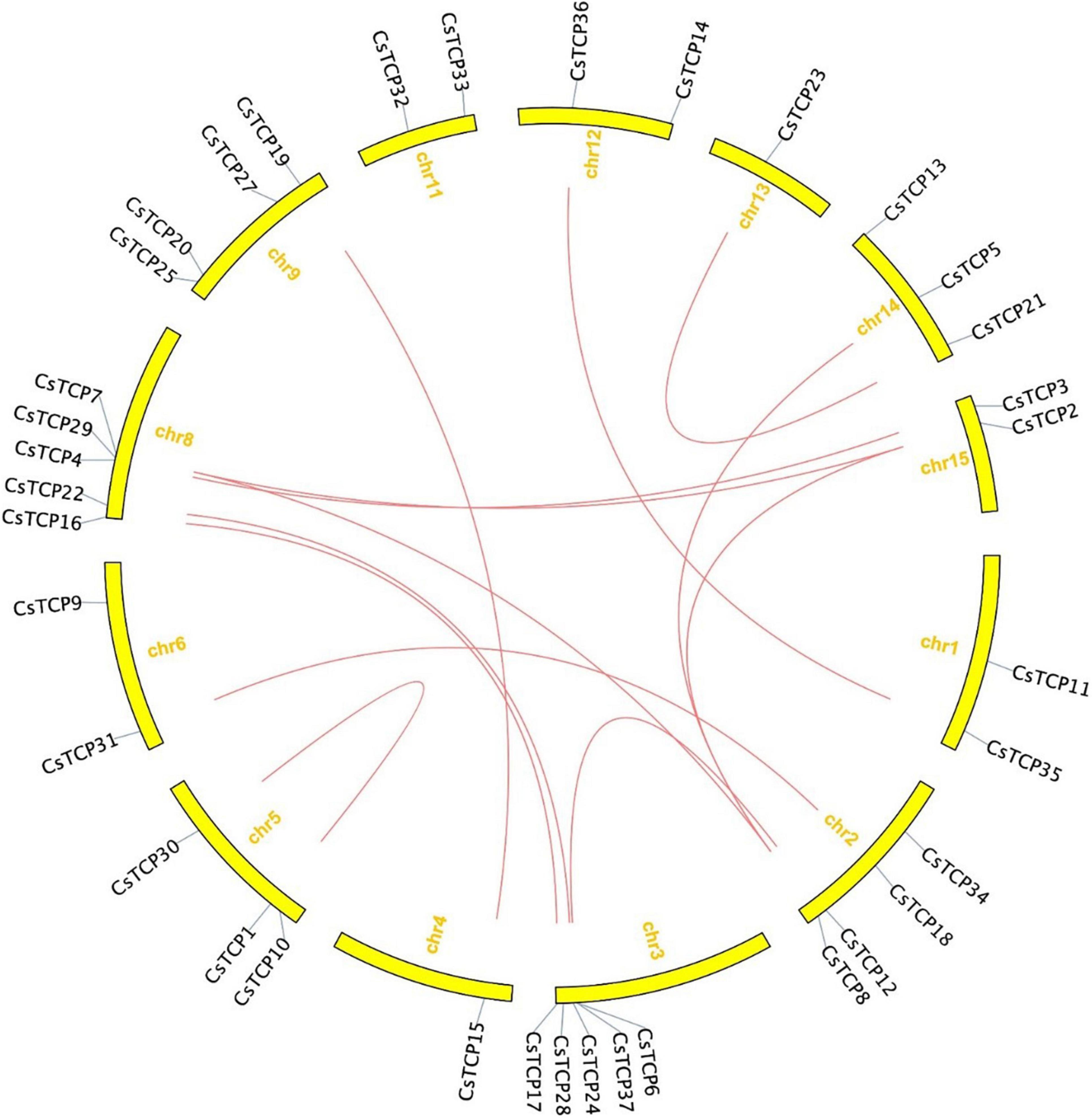
The length of amino acids in 37 CsTCP genes is ranged from 150 (CsTCP32) to 607 (CsTCP34). The pI of 56% of the CsTCP proteins was more than 7. In addition, the molecular weights of 37 CsTCP proteins are ranged from 16.8 kDa (CsTCP32) to 67.6 kDa (CsTCP34) (Table 1). A total of thirty-six CsTCP genes were mapped to 13 chromosomes (Chr) (Figure 1), and CsTCP26 was the only one that is not assembled on the chromosome. The distribution of CsTCP genes was uneven across all of the chromosomes from Chr 1–Chr 6, Chr 8, Chr 9, and Chr 11–Chr 15. Most TCP genes were found on Chr 3 (CsTCP6, CsTCP17, CsTCP24, CsTCP28, and CsTCP37) and Chr 8 (CsTCP4, CsTCP7, CsTCP16, CsTCP22, and CsTCP29). Chr 4 and Chr 13 had only one CsTCP gene (CsTCP5 and CsTCP23).
CsTCP Protein Sequence Analysis
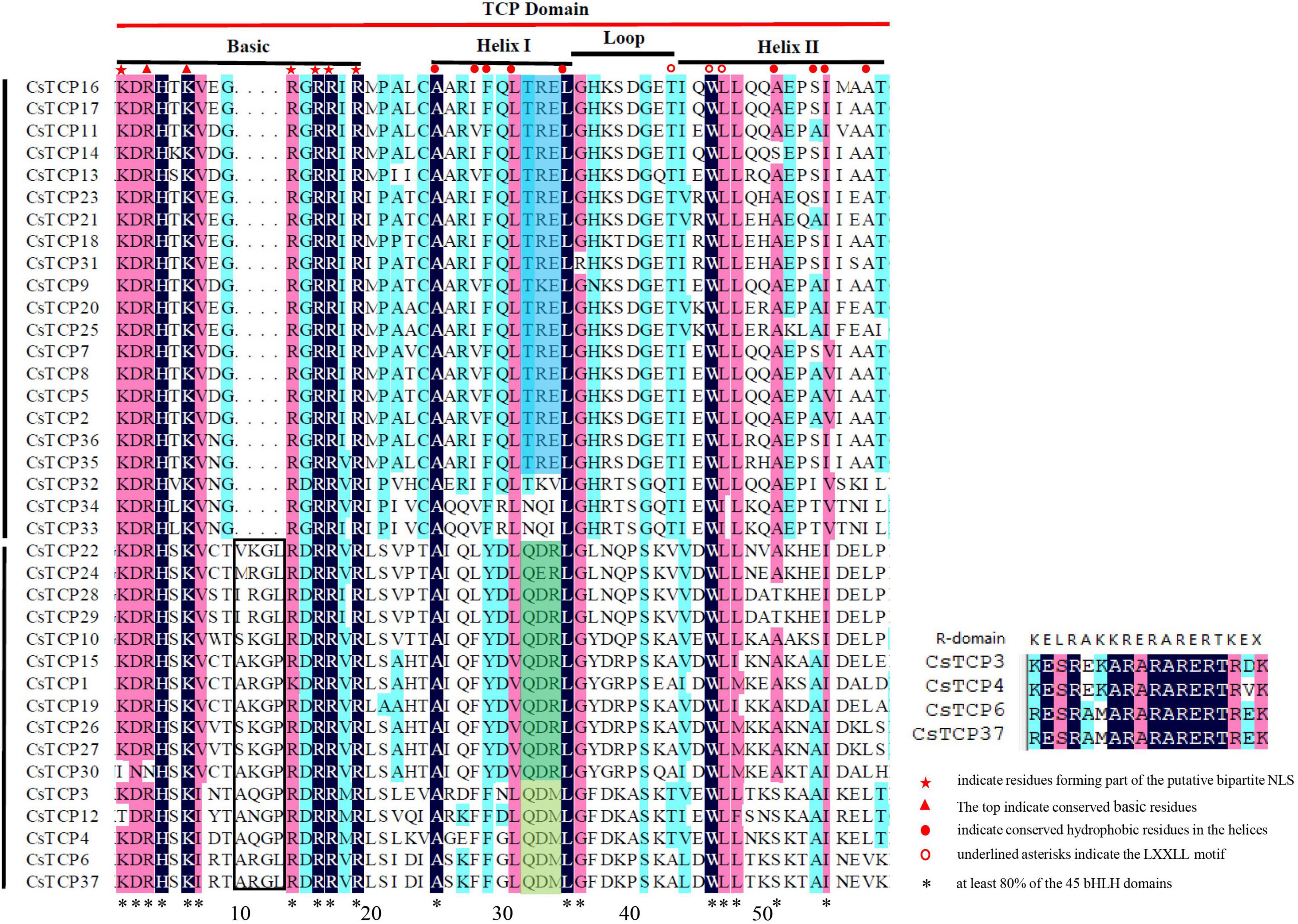
To clarify the sequence characteristics of CsTCP proteins, we performed multiple sequence alignment. In TCP domain with 59 residues, basic region is the most conservative, helix is less conservative, and loop region changes greatly (Yao et al., 2007). The basic-helix-loop-helix (bHLH) domain of CsTCP proteins is very similar to that of rice, Arabidopsis, and grape, indicating that the TCP domain is highly conserved among different species. We identified 18 residues that were identical in at least 80% of the 45 bHLH domains (Figure 2): 10 in the basic region (KDRHXKVXXRRRX R), seven hydrophobic residues in the two helices (21-A, 27-L, 31-L, 42-W, 43-L, 44-L, 51-I in our alignment), and a helix-breaking glycine (32-G in our alignment) in the loop between the helices (Figure 2). In addition to the bHLH domain, four CsTCP proteins shared an R domain comprising conserved polar residues (Figure 2).
Based on the characteristics of TCP domain, CsTCP proteins are divided into two class: class I and class II. Class I is PCF group (group 1) which contains 21 CsTCP proteins; class II contains two group, CIN and CYC/TB1. CIN group (group 2) has 11 CsTCP proteins and CYC/TB1 group (group 3) has 5 CsTCP proteins. Each group has unique sequence and structural characteristics. For example, in the basic region, PCF group has four amino acid residues less than CIN and CYC/TB1 group, which makes that the two types of TCP proteins have different but similar DNA-binding sites (class I is GGNCCAC and class II is GTGGNCCC). In terms of helix I and loop region residues, members of each group have obvious and unique sequence characteristics. For example, of the last three residues in the helix I region, members of group 1 were TRE, group 2 members were QDR, and group 3 members were QDM (Figure 2).
A total of thirty-seven CsTCP proteins were constructed into a phylogenetic tree (Figure 3A), and it was found that there were 15 pairs of homologous genes in the three groups, 9 pairs in the PCF group, CsTCP9/CsTCP11, CsTCP16/CsTCP17, CsTCP2/CsTCP7, CsTCP5/CsTCP8, CsTCP33/CsTCP34, CsTC P20/CsTCP25, CsTCP18/CsTCP31, CsTCP21/CsTCP23, and CsTCP35/CsTCP36; CIN group 4 pairs, CsTCP15/CsTCP19, CsTCP22/CsTCP24, CsTCP26/CsTCP27, CsTCP28/CsTCP29; CsTCP3/CsTCP4, and CsTCP6/CsTCP37. The homologous gene pairs showed similarities in gene structure and the motif composition of translated proteins. Among them, four pairs of homologous genes CsTCP20/CsTCP25, CsTCP21/CsTCP23, CsTCP28/CsTCP29, and CsTCP6/CsTCP37 had exactly the same gene structure and the motif compositions of the post-translational proteins (Figures 3B,C).
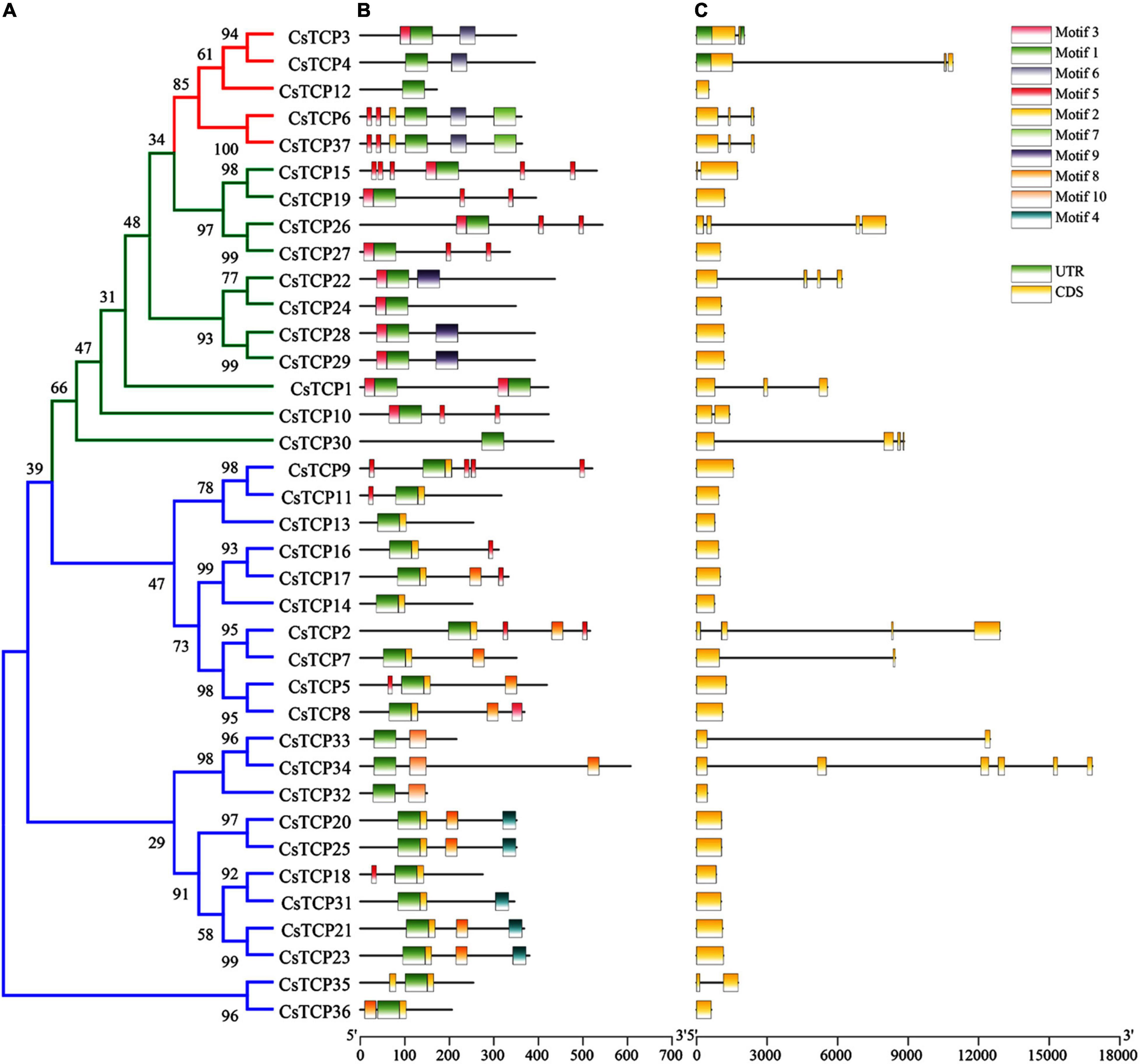
Figure 3. CsTCP protein and gene sequences analysis. (A) Phylogenetic tree (ML), the blue line represents the PCF group, the green line represents the CIN group, and the red line represents the CYC/TB1 group; (B) motif analysis; (C) gene structure analysis, horizontal lines indicate introns.
There are also large differences in the motif composition between the two class. A total of ten regular motifs were identified in CsTCP proteins (Figure 3B and Supplementary Figure 2). Motif 1 was found to distribute in TCP domain regions, so it exists in all CsTCP proteins. Motif 6 was distributed in R-domain regions. Most members of group 3 had an R domain (Figure 1), suggesting that they may have additional activity (Cubas et al., 1999). The proteins of PCF group mostly contain motif 2, and only CsTCP32, CsTCP33, and CsTCP34 do not contain motif 2, but they contain motif 10 which is not present in any other CsTCP proteins. A total of 11 CsTCP proteins contain motif 8 which is only present in the proteins of PCF groups. Except CsTCP30, all proteins of CIN group contain motif 3; the proteins of CYC/TB1 group contain motif 6, except CsTCP12. In addition, there are some interesting phenomena. For example, motif 5 exists in 14 TCP proteins, but most CsTCP proteins in class I contain only one motif 5, whereas CsTCP proteins in class II basically contain multiple motif 5. Some unique motifs exist only in certain CsTCPs, such as motif 4 exists only in CsTCP20/CsTCP25/CsTCP21/CsTCP23/CsTCP31, motif 9 exists only in CsTCP22/CsTCP28/CsTCP29, and motif 7 exists only in CsTCP6/CsTCP37.
The CsTCP gene structures show that the CsTCPs introns number is between 0 and 3, except for CsTCP34 which has 5 introns, and most of the CsTCPs (22/37) in tea plant have no intron (Figure 3C). Moreover, only CsTCP3 and CsTCP4 contain two and one UTR, respectively.
A total of five cis-acting elements were detected, which are participated in hormone response (229), stress response (53), and light response (405) and involved in growth and development regulation (28) and metabolism regulation (20) (Supplementary Figure 3). Among them, the proportion of light-responsive elements (55%) is the highest, followed by hormone-responsive elements (31%), stress-responsive elements (7%) and developmental and metabolic response elements (7%) are close, which is similar to PCF and CIN group, and CYC/TB1 group hormones (40%) accounted for more than other. Among hormone response elements, the ratio of abscisic acid (37%) and methyl jasmonate (33%) response elements was the highest, followed by auxin, gibberellin, and salicylic acid response elements.
Evolutionary Analysis of the CsTCP Gene Family
The study explored the evolution of the TCP gene family in tea plants by constructing phylogenetic trees among different species, comparing genomic information of TCP genes, and performing collinear analysis.
To study the evolution of CsTCP gene family, the phylogenetic tree was constructed by the TCP protein of Camellia sinensis, Zea mays, Oryza sativa, and Antirrhinum majus that three species first identified TCP genes, Arabidopsis thaliana that is the herbal model plant, and Vitis vinifera that is woody plant. All TCP proteins are divided into three groups: PCF, CIN, and CYC/TB1. Among them, there are 11 clades of TCP protein in monocots (rice and maize) and eudicots (snapdragon, Arabidopsis, grapevine, and tea plant) (Figure 4). In 4 clades, there are only TCP protein in tea and monocots. The TCP proteins of tea plant in 4 clades are CsTCP10, CsTCP14, CsTCP15, and CsTCP30, respectively. They may be new CsTCP proteins produced by the evolution in tea plant. In the phylogenetic tree, CsTCP proteins clustered together, mostly with TCP proteins of Arabidopsis and grapevine.
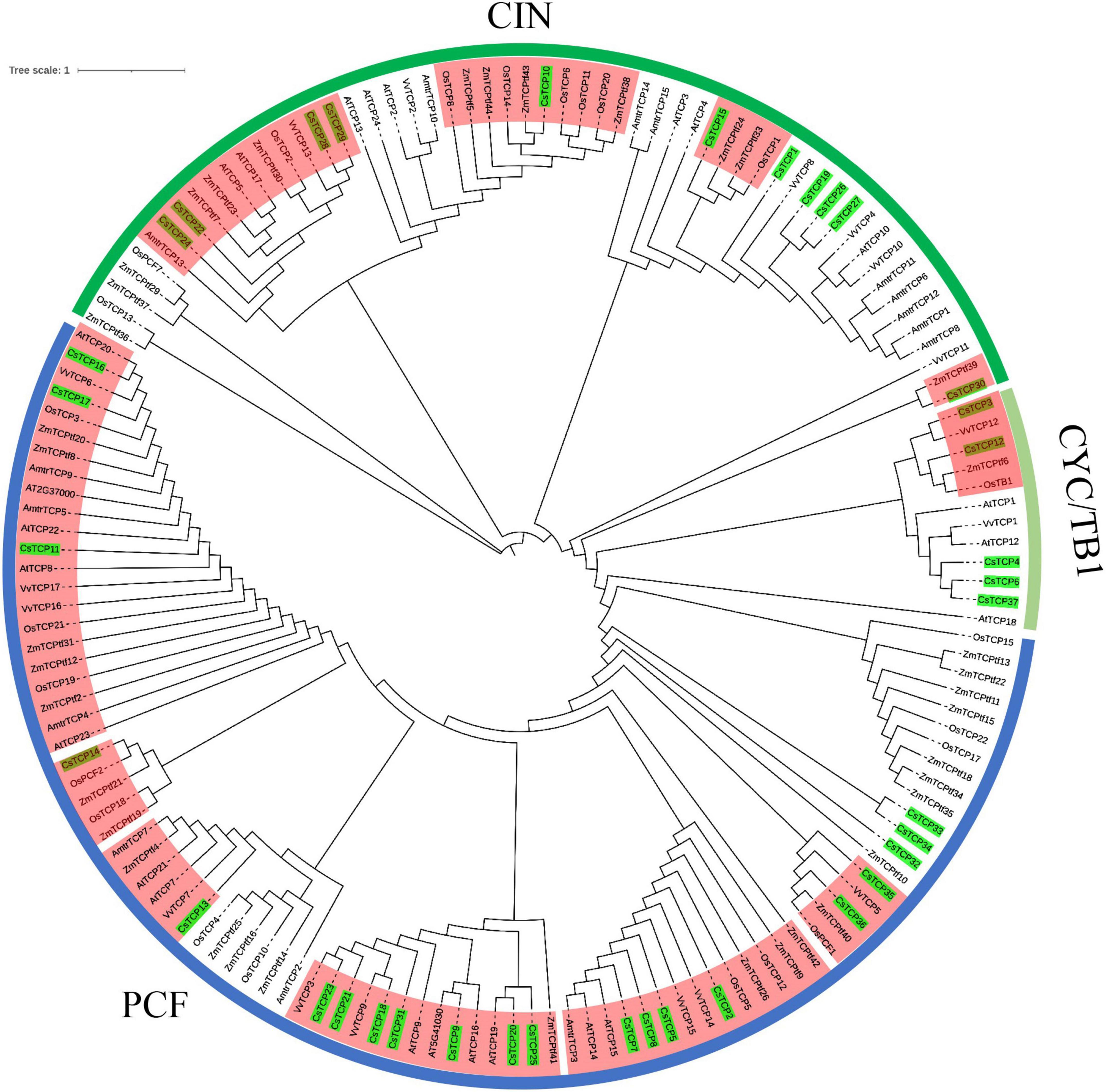
Figure 4. The phylogenetic analysis of TCP gene family among Camellia sinensis, Arabidopsis thaliana, Oryza sativa, Zea mays, Vitis vinifera, and Antirrhinum majus. The red shaded part is the clades containing both monocots and eudicots TCP proteins.
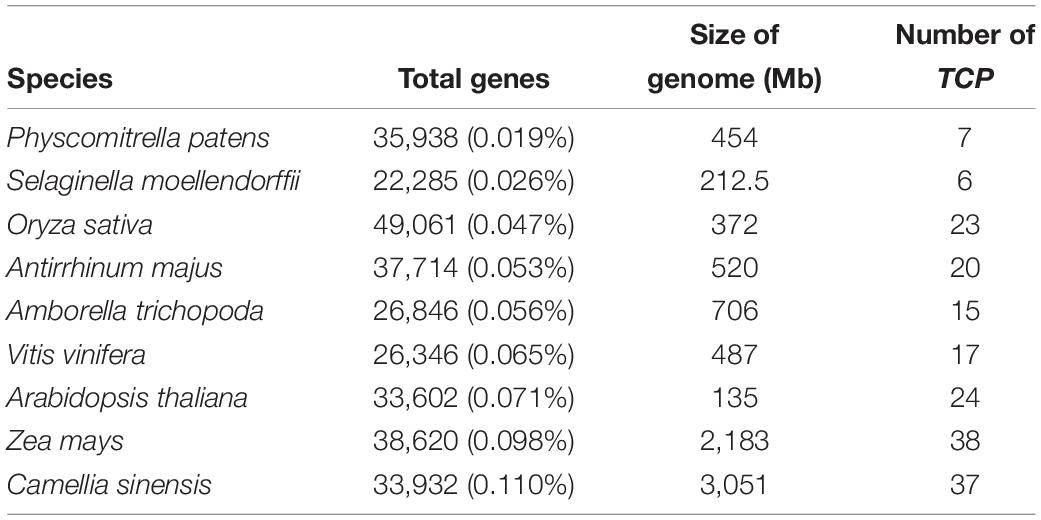
The number and proportion of TCP gene family in the nine species were compared and analyzed (Table 2). The number of TCP genes increases with the evolution of species from lower to higher. There are fewer TCP members in P. patens and S. moellendorffii than higher plants, which indicates that the TCP gene family has been expanded in higher plant. It is worth noting that the number of TCP genes in P. patens and S. moellendorffii is similar, and which in grapevine and snapdragon is also similar. But the genome size of moss is two times that of selaginella, the genome size of grape and snapdragon is similar. The number of CsTCP genes in tea is 1.5 times that of Arabidopsis, but the genome size of tea plant is 22 times that of Arabidopsis. It is found that the proportion of CsTCP gene members in the whole-genome in each species is not related to the genome size of species.
There are 13 syntenic pairs in tea plant, nine of which are homologous gene pairs, CsTCP2/CsTCP7, CsTCP5/CsTCP8, CsTCP16/CsTCP17, CsTCP18/CsTCP31, CsTCP21/CsTCP23, CsTCP35/CsTCP36, CsTCP15/CsTCP19, CsTCP22/CsTCP24, and CsTCP3/CsTCP4. Moreover, there are four syntenic pairs that are non-homologous gene pairs, CsTCP2/CsTCP8, CsTCP12/CsTCP37, and CsTCP1/CsTCP30 (Figure 5). Gene duplicated event analysis showed that the coordinates of transposed duplication and whole-genome duplication (WGD) events were detected in 35 CsTCP genes (Supplementary Table 2). CsTCP26 and CsTCP37 were not detected duplication events. The reason of former may be that it is not assembled on the chromosome.
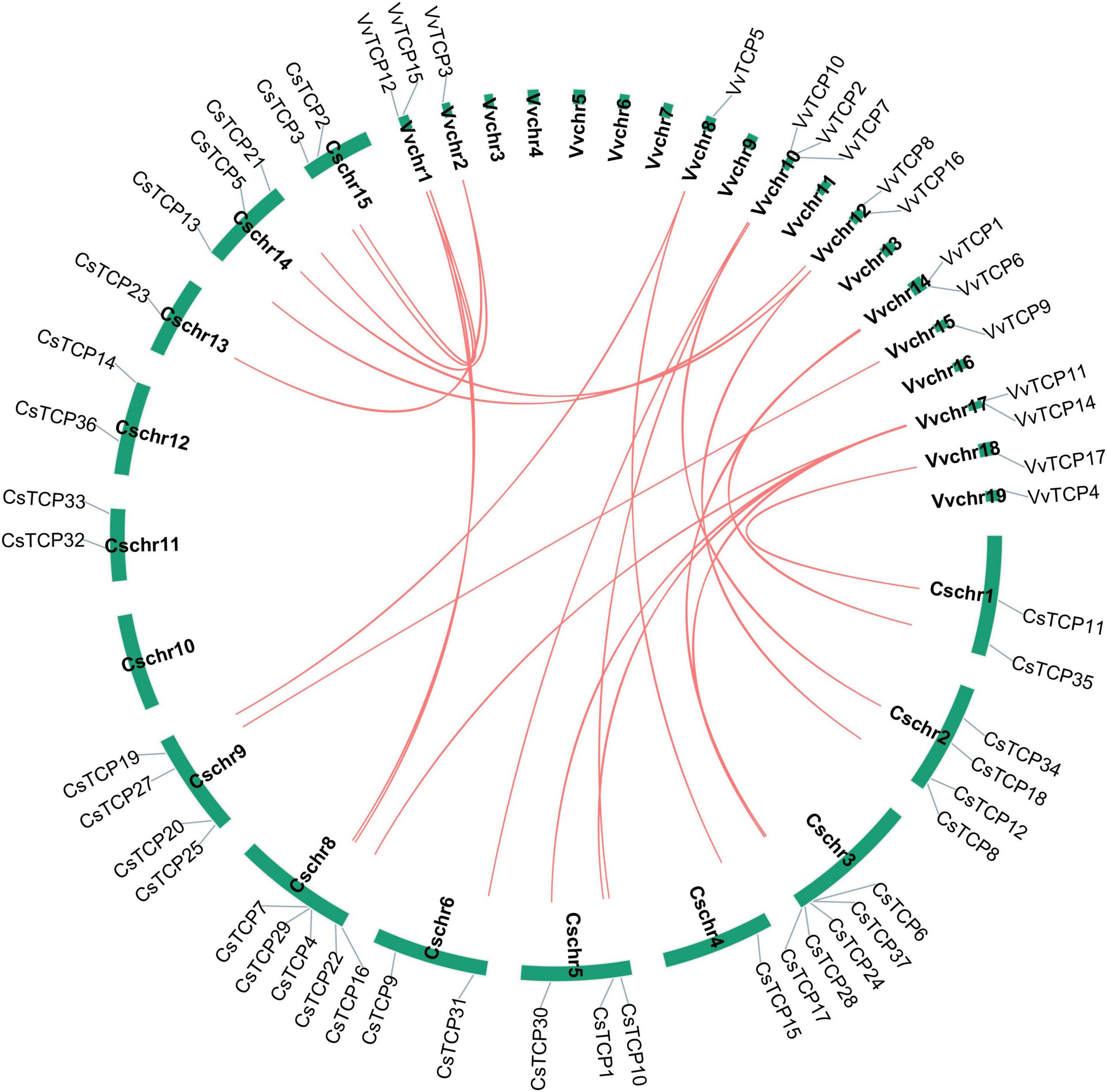
To further explore the origin and probable evolutionary mechanisms of the CsTCP gene family, we also investigated the syntenic blocks in tea plant and grapevine. A total of thirteen syntenic pairs were detected in tea plant, and 16 syntenic pairs were detected between tea plant and grapevine (Figure 6). The results showed that 16 VvTCP genes have syntenic counterpart in tea plants and VvTCP13 was excluded. Interestingly, the syntenic counterparts in tea plant of VvTCP2/VvTCP4/VvTCP8/VvTCP10 of CIN group belong to PCF group (except CsTCP10). In CYC/TB1 group, VvTCP1/CsTCP6, VvTCP1/CsTCP37, VvTCP11/CsTCP3, and VvTCP11/CsTCP4 are the syntenic pairs. CsTCP3, CsTCP4, CsTCP6, and CsTCP37 belong to CYC/TB1 group.
Expression Pattern Analysis of CsTCP Genes
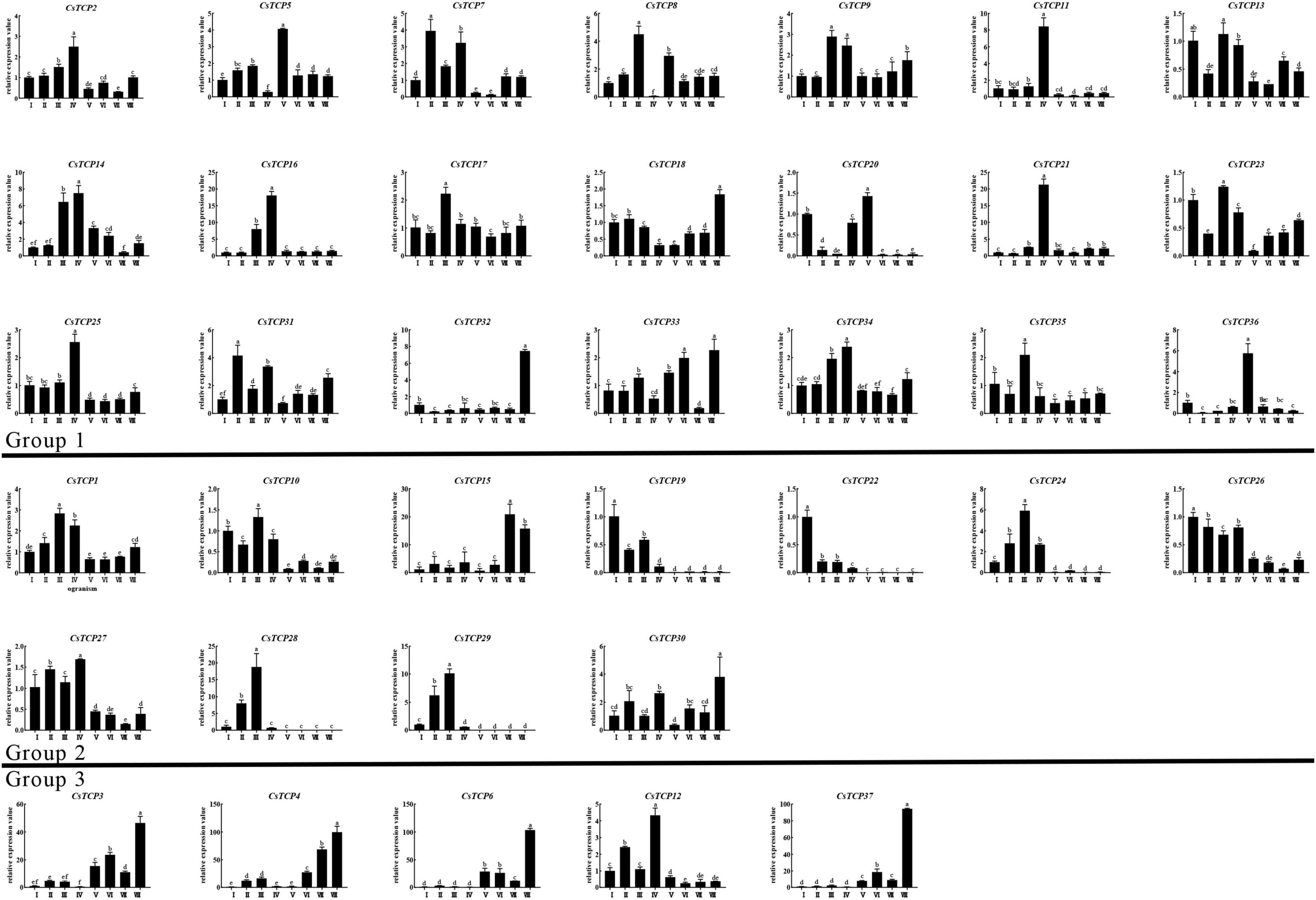
The expression patterns of CsTCP gene family in different tissues did not show significant differences among three groups (Figure 7). In root (IV), CsTCP1/CsTCP2/CsTCP7/CsTCP9/CsTCP11/CsTCP12/CsTCP14/CsTC P16/CsTCP21/CsTCP25/CsTCP27/CsTCP30/CsTCP31/CsTCP34 were highly expressed. The CsTCP gene family is expressed in leaves at different developmental stages. The expression levels of CsTCP19/CsTCP20/CsTCP22/CsTCP26/CsTCP32/CsTCP36 decreased gradually during leaf maturation, but the expression of CsTCP1/CsTCP2/CsTCP4/CsTCP5/CsTCP8/CsTCP9/CsTCP14/CsTCP16/CsTCP17/CsTCP21/CsTCP24/CsTCP28/CsTCP29/CsT CP31/CsTCP33/CsTCP34/CsTCP35 increased gradually.
The expression level of CsTCP genes in stems showed different characteristics in tender phloem (V), old phloem (VI), tender xylem (VII), and old xylem (VIII) (Figure 7). In phloem, the expression level of CsTCP5/CsTCP8/CsTCP14/CsTCP20/CsTCP33/CsTCP36 was higher; the expression level of CsTCP14 was high in old phloem (VI), and the rest was high in tender phloem. In xylem, the expression levels of CsTCP7/CsTCP9/CsTCP13/CsTCP18/CsTCP23/CsTCP25/CsTCP31/CsT CP32/CsTCP33/CsTCP15/CsTCP3/CsTCP4/CsTCP6/CsTCP30/CsTCP37 were high; among them, CsTCP15 is highly expressed in tender xylem (VII), and the rest is highly expressed in old xylem. Some CsTCP genes have special expression patterns and only show high-level expression in a single plant organ, such as CsTCP11/CsTCP21/CsTCP22/CsTCP28/CsTCP29/CsTCP32/CsT CP36/CsTCP15/CsTCP3/CsTCP4/CsTCP6/CsTCP37. Some genes of group 2, such as CsTCP15/CsTCP19/CsTCP10, and most genes of group 1, CsTCP9/CsTCP11/CsTCP13/CsT CP16/CsTCP17/CsTCP5/CsTCP8/CsTCP34/CsTCP20/CsTCP18/CsTCP21/CsTCP23, were downregulated in varying degrees under drought and salt stress (Supplementary Figure 4). In addition, CsTCP15/CsTCP19/CsTCP22 of group 2 and CsTCP9/CsTCP11/CsTCP16/CsTCP17/CsTCP23 of group 1 were upregulated under cold stress (Supplementary Figure 4). These stress responsive genes were also induced by MeJA, and the expression level had no significant correlation with the treatment time (Supplementary Figure 4).
Discussion
As an important economic crop in China, tea plant and its products have made significant contributions to Chinese agricultural industry. However, the molecular biological mechanisms of tea plant development have seldom been reported. TCP proteins play an important role in plant morphological evolution and development. TCP gene family has been identified in many plant species, such as Arabidopsis thaliana (Riechmann et al., 2000; Yao et al., 2007), Oryza sativa. L (Xiong et al., 2005), Lycopersicon esulentum Mill (Parapunova et al., 2014), and Gossypium raimondii (Ma et al., 2016). However, the identification of TCP gene family in tea plant is controversial and superficial. In this study, a variety of methods were used to identify the TCP gene family of tea plant, and the evolutionary process and functional characteristics of CsTCP proteins were analyzed.
Identification of TCP Gene Family in Camellia sinensis var. Sinensis Genome
In this study, 37 CsTCP proteins with TCP domain were identified, and three new CsTCP proteins CsTCP32, CsTCP33, and CsTCP34 were characterized compared to previous research, where 34 TCPs were found (Yu et al., 2021). Previous studies used the TCP protein of Arabidopsis and rice as queries for local BLAST searches against the TPIA. This method may lead to the elimination of some CsTCP proteins with TCP-conserved domain but low homology with the TCP protein of Arabidopsis and rice.
The residues of CsTCP proteins between class I and class II were definitely different in the loop, helix I, and helix II regions; however, a conserved tandem of tryptophan (W) and leucine (L) was found in helix II (Figure 1), which further indicates that CsTCP proteins may be functional redundancy. Many studies have shown that there is functional redundancy among TCP proteins in same group, for example, JAW-TCPs AtTCP7/AtTCP8/AtTCP22/AtTCP23 of CIN group (Aguilar-Martínez and Sinha, 2013) and AtTCP14/AtTCP15 of PCF group (Ferrero et al., 2021) in Arabidopsis. Thus, the mutation of single TCP gene will not cause plant phenotypic changes, such as AtTCP4/AtTCP10 (Koyama et al., 2017), BrrTCP2 (Du et al., 2017), and SlLA (Shleizer-Burko et al., 2011). Then, we speculate that the similar situation could happen in tea plant. Most TCP proteins have obvious differences in motif composition in tea plant, for example, motifs 2, 3, 5, 6, and 8 (Figure 3B). The special motif composition among different groups supports the functional differentiation of CsTCP protein. We conclude that CsTCP proteins in different groups are supposed to have complementary functions, whereas those in the same class could display the function redundancies, and the phylogenetic distribution of CsTCP proteins in the evolutionary tree among species is also supported this result (Figure 4).
TCP Gene Family in Camellia sinensis and Their Evolution
In this study, we found that there was no relationship between the number of CsTCP genes and the genome size (Table 2). Moreover, the number of TCP genes is increased with the evolution of species from lower to higher, and the TCP gene family has been expanded in higher plant (Martin-Trillo and Cubas, 2010). From the phylogenetic tree with six species, we observed that there were 11 well-supported clades in both tea plant and rice or maize genes (Figure 6), suggesting that the most recent common ancestor of eudicots and monocots had at least 11 TCP-conserved genes, because there are a few additional clades in only eudicots or monocots (rice and maize) genes, indicating that some TCP genes may lost. The number of TCP genes in the recent revolved plant species is probably over 11 (Yao et al., 2007). Therefore, the TCP gene family has only expanded since the divergence of monocots and eudicots in plant evolution history.
In plant genome, gene duplication and divergence are the essential steps for the gene family expansion and evolution of new function. To evaluate the effect of duplication on the CsTCP gene family, we first analyzed the duplicate events in CsTCP gene family. The results showed that 95% (35/37) CsTCP genes were duplicated from WGD/segmental event, and 30% (11/37) were also duplicated from transposed event (Supplementary Table 2). Transposed genes in tea plant are collinear with the genome ancestral plant species. The transposed genes were distributed in the 11 clades of the phylogenetic tree (Figure 4), suggesting that these genes are relatively conservative in the evolution. Moreover, thirteen syntenic pairs were detected in tea plant. The results demonstrated that WGD/segmental duplication played a vital role in the expansion of the CsTCP gene family.
To explore the evolution of CsTCP gene family, we analyzed their syntenic pairs in tea plant and between tea plant and grapevine. A total of nineteen CsTCP genes have counterparts in syntenic pairs (Figures 5, 6). A total of 11 of them belong to class I and eight belong to class II. The syntenic analysis between tea plant and grapevine showed that these genes located in corresponding syntenic blocks occurred before the divergence of tea plant and grapevine. In addition, previous study showed that, after core eudicot whole-genome triplication (WGT) with Vitis vinifera, C. sinensis has experienced additional WGD event (Chen et al., 2020b). Tea plant has experienced additional duplication event, resulting in a further increase of CsTCP gene numberin two classes, but the process of the event remains to be further studied.
Expression Profile Analysis of TCP Gene in Camellia sinensis
The CsTCP genes from group 3 are highly expressed in buds and stems (Figure 7 and Supplementary Figure 4), especially in the lignified stems (VII, VIII). The CsTCP genes from group 2 are mainly expressed in buds, flowers, and leaves (Figure 7 and Supplementary Figure 4). This is similar to the expression pattern in tomato. The expression levels in different organs vary widely between the tomato TCP genes, as well as between different organs for individual TCP genes (Parapunova et al., 2014). Most CsTCP genes of group 1 are more widely and non-specifically expressed in different tissues, as well as in tomato; the difference is that the CsTCP genes are expressed in all tissues, including buds, flowers, fruits, leaves, and stems (Supplementary Figure 4), whereas the SlTCP genes are mainly expressed in leaves, flowers, and fruits (Parapunova et al., 2014). It may be caused by the difference of the number of TCP genes in species. The CsTCP genes expression pattern in tissues are also similar to ZmTCP genes in maize that contains a large number of TCP genes (Ding et al., 2019).
CsTCP genes of group 3 basically did not show any expression difference in response to stress. This is similar to ZmTCP genes, and most of ZmTCP genes (13/19) of CYC/TB1 (group 3) in maize do not respond to stress induction (Ding et al., 2019). The expression levels of many CsTCP genes (16/32) changed under stress treatment, and these genes (12/16) mainly belong to PCF group. The result was also supported by the study of PCF-TCP genes in many species. In rice, most of the PCF group (group 1) genes participate in the stress response, OsPCF6 and OsTCP21 expression were largely induced by cold stress, and the downregulation of OsPCF6 and OsTCP21 resulted in enhanced tolerance to cold stress (Wang et al., 2014), and OsPCF5/OsPCF8 (Yang et al., 2013) and OsTCP19 (Mukhopadhyay and Tyagi, 2015) play the important roles in the stress response. In other plant species, TCP genes involved in abiotic stress response mostly belong to PCF group, such as in Phyllostachys edulis (Liu et al., 2020), Glycine max (Ling et al., 2020), Betula platyphylla (Li et al., 2020; Ren et al., 2021), and so on.
In the evolutionary tree, TCP family of tea plant and Arabidopsis can be divided into 9 clades (Supplementary Figure 5). The TCP genes of clades 8 and 9 belong to CIN group, and their expression levels are higher in leaves (Figure 7 and Supplementary Figure 4). CIN-TCPs have been found to play an important role in leaf development in Arabidopsis, including leaf primordium initiation (Alvarez et al., 2016), leaf expansion (Nath et al., 2003), leaf margin formation (Palatnik et al., 2003; Ori et al., 2007; Efroni et al., 2008), and leaf meristem differentiation (Navaud et al., 2007). In Arabidopsis, CIN-TCPs are divided into two clades, one is JAW-TCPs with miRNA319-binding site, which is regulated by miRNA319, and the other is TCP5-like clade without miRNA319-binding site. Clade 8 belongs to JAW-TCPs and clade 9 belongs to TCP5-like clade. Clade 7 contains AtBRC genes, such as BRC1 (AtTCP18) and BRC2 (AtTCP12). CsTCP3/CsTCP4 are specifically highly expressed in stems (Figure 7 and Supplementary Figure 4), and they fell into the same clade with AtBRC in the phylogenetic tree, so these two genes may be CsBRC1-like. CsTCP12 is specifically expressed in leaves (Figure 7 and Supplementary Figure 4) and may be the CsBRC2-like. In Arabidopsis, JAW-TCPs and TCP5-like are used as the enhancers for axillary branch growth, and Branched genes (AtTCP12 and AtTCP18) are used as the inhibitors to participate in plant branch development (Aguilar-MARTíNEZ et al., 2007; van Es et al., 2019). The function of TCP genes related to tea plant leafing, branching, and stress response in tea plant needs to be further studied.
In addition, we also found two interesting clades, clade 6 and clade 2 (Supplementary Figure 5). The number of CsTCP genes in most clades is large, but clade 6 contains only one CsTCP gene (CsTCP11) and three AtTCP genes (AtTCP8/AtTCP22/AtTCP23). A total of three AtTCP genes are involved in regulating leaf development, and there are redundancy functions among them (Danisman et al., 2013). CsTCP11 is highly expressed in lateral buds, fruits, and roots and induced by stress (Supplementary Figure 4), which seems to have different functions with the AtTCP genes in clade 6. Clade 2 contains only one AtTCP gene (AtTCP11) and five CsTCP genes (CsTCP32/CsTCP33/CsTCP34/CsTCP35/CsTCP36). The expression levels of these CsTCP genes in clade 2 are low under different tissues and stress treatments; however, their expression could be induced under MeJA treatment (Figure 7 and Supplementary Figure 4). The expression of CsTCP33 could not be detected under these treatments. CsTCP32/CsTCP33/CsTCP34 also did not exist in the other two tea varieties HD and TGY (Supplementary Table 1). It shows that these three genes either play a role in variety specificity or are non-functional genes.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://doi.org/10.6084/m9.figshare.19291454.v1.
Author Contributions
WF, YM, XZ, and XS designed the experiment. YM, XS, DZ, HQ, YW, and ZH performed the experiment. YM, XS, and LZ performed the search strategy and analyzed the data. YM and XS wrote the manuscript. ZZ, XZ, and WF paid for part of the study and provided revised suggestions. All authors read and approved the final manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31972460 and 31870680), the earmarked fund for China Agriculture Research System (CARS-19), the Key Research and Development Program of Jiangsu Province (BE2019379), the Jiangsu Agriculture Science and Technology Innovation Fund [CX(20)2004], the Innovation and Extension Projects of Forestry Science and Technology in Jiangsu Province [LYKJ-Changzhou(2020)03], the Changzhou Science and Technology Support Program (Agriculture CE20202003), and the Chuzhou Science and Technology Support Program (2020ZN009).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.840350/full#supplementary-material
Footnotes
- ^ https://www.Arabidopsis.org/
- ^ http://pfam.xfam.org/
- ^ http://tpia.teaplant.org/index.html
- ^ http://smart.embl.de/
- ^ http://planttfdb.cbi.pku.edu.cn/
- ^ http://rice.plantbiology.msu.edu/index.shtml
- ^ http://bioinfo.sibs.ac.cn/Am/index.php
- ^ https://web.expasy.org/protparam/
- ^ http://linux1.softberry.com/berry.phtml
- ^ https://meme-suite.org/meme/tools/meme
- ^ http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
- ^ https://github.com/tanghaibao/jcvi/wiki/MCscan-(Python-version)
References
Aguilar-Martínez, J. A., and Sinha, N. (2013). Analysis of the role of Arabidopsis class I TCP genes AtTCP7, AtTCP8, AtTCP22, and AtTCP23 in leaf development. Front Plant Sci. 4:406. doi: 10.3389/fpls.2013.00406
Aguilar-MARTíNEZ, J. A., Poza-CARRIóN, C., and Cubas, P. (2007). Arabidopsis BRANCHED1 acts as an integrator of branching signals within axillary buds. Plant Cell 19, 458–472. doi: 10.1105/tpc.106.048934
Alvarez, J. P., Furumizu, C., Efroni, I., Eshed, Y., and Bowman, J. L. (2016). Active suppression of a leaf meristem or chestrates determinate leaf growth. Elife 5:e15023.
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208.
Cao, H., Wang, F., Lin, H., Ye, Y., Zheng, Y., Li, J., et al. (2020). Transcriptome and metabolite analyses provide insights into zigzag-shaped stem formation in tea plants (Camellia sinensis). BMC Plant Biol. 20:98. doi: 10.1186/s12870-020-2311-z
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., Frank, M. H., et al. (2020a). TBtools: an Integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, J. D., Zheng, C., Ma, J. Q., Jiang, C. K., Ercisli, S., Yao, M. Z., et al. (2020b). The chromosome-scale genome reveals the evolution and diversification after the recent tetraploidization event in tea plant. Horticult. Res. 7:63. doi: 10.1038/s41438-020-0288-2
Chen, L., Chen, Y. Q., Ding, A. M., Chen, H., Xia, F., Wang, W. F., et al. (2016). Genome-wide analysis of TCP family in tobacco. Genet. Mol. Res. 15:15027728. doi: 10.4238/gmr.15027728
Chen, L., Zhou, Z.-X., and Yang, Y.-J. (2007). Genetic improvement and breeding of tea plant (Camellia sinensis) in China: from individual selection to hybridization and molecular breeding. Euphytica 154, 239–248.
Cubas, P. (2002). “Chapter 13-Role of TCP genes in the evolution of key morphological characters in angiosperms,” in Developmental Genetics and Plant Evolution, eds Q. C. B. Cronk, J. Hawkins, and R. M. Bateman (New York, NY: Taylor and Francis Ltd), 247–266.
Cubas, P., Lauter, N., Doebley, J., and Coen, E. (1999). The TCP domain: a motif found in proteins regulating plant growth and development. Plant J. 18, 215–222.
Danisman, S., van Dijk, A. D., Bimbo, A., van der Wal, F., Hennig, L., and de Folter, S. (2013). Analysis of functional redundancies within the Arabidopsis TCP transcription factor family. J. Exp. Bot. 64, 5673–5685. doi: 10.1093/jxb/ert337
Danisman, S., Van, D. E. R. W. A. L. F., Dhondt, S., Waites, R., de Folter, S., Bimbo, A., et al. (2012). Arabidopsis class I and class II TCP transcription factors regulate jasmonic acid metabolism and leaf development antagonistically. Plant Physiol. 159, 1511–1523. doi: 10.1104/pp.112.200303
Daviere, J. M., Wild, M., Regnault, T., Baumberger, N., Eisler, H., Genschik, P., et al. (2014). Class I TCP-DELLA interactions in inflorescence shoot apex determine plant height. Curr. Biol. 24, 1923–1928. doi: 10.1016/j.cub.2014.07.012
Ding, S., Cai, Z., Du, H., and Wang, H. (2019). Genome-Wide Analysis of TCP Family Genes in Zea mays L. Identified a Role for ZmTCP42 in drought tolerance. Int. J. Mol. Sci. 20:2762. doi: 10.3390/ijms20112762
Doebley, J., Stec, A., and Hubbard, L. (1997). The evolution of apical dominance in maize. Nature 386, 485–488.
Du, J., Hu, S., Yu, Q., Wang, C., Yang, Y., Sun, H., et al. (2017). Genome-Wide identification and characterization of BrrTCP transcription factorsin Brassica rapa ssp rapa. Front. Plant Sci. 8:1588. doi: 10.3389/fpls.2017.01588
Efroni, I., Blum, E., Goldshmidt, A., and Eshed, Y. A. (2008). Protracted and dynamic maturation schedule underlies Arabidopsis leaf development. Plant Cell 20, 2293–2306. doi: 10.1105/tpc.107.057521
Faivre-Rampant, O., Bryan, G. J., Roberts, A. G., Milbourne, D., Viola, R., Taylor, M. A., et al. (2004). Regulated expression of a novel TCP domain transcription factor indicates an involvement in the control of meristem activation processes in Solanum tuberosum. J. Exp. Bot. 55, 951–953. doi: 10.1093/jxb/erh082
Ferrero, L. V., Gastaldi, V., Ariel, F. D., Viola, I. L., and Gonzalez, D. H. (2021). Class I TCP proteins TCP14 and TCP15 are required for elongation and gene expression responses to auxin. Plant Mol. Biol. 105, 147–159. doi: 10.1007/s11103-020-01075-y
Finlayson, S. A. (2007). Arabidopsis teosinte Branched1-like 1 regulates axillary bud outgrowth and is homologous to monocot Teosinte Branched1. Plant Cell Physiol. 48, 667–677. doi: 10.1093/pcp/pcm044
Hao, J., Tu, L., Hu, H., Tan, J., Deng, F., Tang, W., et al. (2012). GbTCP, a cotton TCP transcription factor, confers fibre elongation and root hair development by a complex regulating system. J. Exp. Bot. 63, 6267–6281. doi: 10.1093/jxb/ers278
HERVé, C., Dabos, P., Bardet, C., Jauneau, A., Auriac, M. C., Ramboer, A., et al. (2009). In vivo interference with AtTCP20 function induces severe plant growth alterations and deregulates the expression of many genes important for development. Plant Physiol. 149, 1462–1477. doi: 10.1104/pp.108.126136
Huang, T., and Irish, V. F. (2015). Temporal control of plant organ growth by TCP transcription factors. Curr. Biol. 25, 1765–1770.
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kieffer, M., Master, V., Waites, R., and Davies, B. (2011). TCP14 and TCP15 affect internode length and leaf shape in Arabidopsis. Plant J. Cell Mol. Biol. 68, 147–158. doi: 10.1111/j.1365-313X.2011.04674.x
Kosugi, S., and Ohashi, Y. (1997). PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. Plant Cell 9, 1607–1619. doi: 10.1105/tpc.9.9.1607
Kosugi, S., and Ohashi, Y. (2002). DNA binding and dimerization specificity and potential targets for the TCP protein family. Plant J. 30, 337–348. doi: 10.1046/j.1365-313x.2002.01294.x
Koyama, T., Furutani, M., Tasaka, M., and Ohme-Takagi, M. (2007). TCP transcription factors control the morphology of shoot lateral organs via negative regulation of the expression of boundary-specific genes in Arabidopsis. Plant Cell 19, 473–484. doi: 10.1105/tpc.106.044792
Koyama, T., Sato, F., and Ohme-Takagi, M. (2017). Roles of miR319 and TCP transcription factors in leaf development. Plant Physiol. 175, 874–885.
Li, C., Potuschak, T., COLóN-Carmona, A., Gutiérrez, R. A., and Doerner, P. (2005). Arabidopsis TCP20 links regulation of growth and cell division control pathways. Proc. Natl. Acad. Sci. U.S.A. 102, 12978–12983. doi: 10.1073/pnas.0504039102
Li, H., Yuan, H., and Liu, F. (2020). BpTCP7 gene from Betula platyphylla regulates tolerance to salt and drought stress through multiple hormone pathways. Plant Cell Tissue Organ Cult. 141, 17–30.
Li, Q. H., Li, Y., Wu, X. Y., Zhou, L., Zhu, X., and Fang, W. (2017a). Metal transport protein 8 in Camellia sinensis confers superior manganese tolerance when expressed in yeast and Arabidopsis thaliana. Sci. Rep. 7:39915 doi: 10.1038/srep39915
Li, S., and Zachgo, S. (2013). TCP3 interacts with R2R3-MYB proteins, promotes flavonoid biosynthesis and negatively regulates the auxin response in Arabidopsis thaliana. Plant J. 76, 901–913. doi: 10.1111/tpj.12348
Li, W., Xiang, F., Zhong, M., Zhou, L., Liu, H., Li, S., et al. (2017b). Transcriptome and metabolite analysis identifies nitrogen utilization genes in tea plant (Camellia sinensis). Sci. Rep. 7:1693. doi: 10.1038/s41598-017-01949-0
Ling, L., Zhang, W., An, Y., Du, B., Wang, D., and Guo, C. (2020). Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genomics 20, 537–550. doi: 10.1007/s10142-020-00733-0
Liu, H., Gao, Y., Wu, M., Shi, Y., Wang, H., Wu, L., et al. (2020). TCP10, a TCP transcription factor in moso bamboo (Phyllostachys edulis), confers drought tolerance to transgenic plants. Environ. Exp. Bot. 172:104002.
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lopez, J. A., Sun, Y., Blair, P. B., and Mukhtar, M. S. (2015). TCP three-way handshake: linking developmental processes with plant immunity. Trends Plant Sci. 20, 238–245. doi: 10.1016/j.tplants.2015.01.005
Luo, D., Carpenter, R., Vincent, C., Copsey, L., and Coen, E. (1996). Origin of floral asymmetry in Antirrhinum. Nature 383, 794–799.
Ma, J., Wang, Q., Sun, R., Xie, F., Jones, D. C., and Zhang, B. (2014). Genome-wide identification and expression analysis of TCP transcription factors in Gossypium raimondii. Sci. Rep. 4:6645. doi: 10.1038/srep06645
Ma, X., Ma, J., Fan, D., Li, C., Jiang, Y., and Luo, K. (2016). Genome-wide identification of TCP Family transcription factors from populus euphratica and their involvement in leaf shape regulation. Sci. Rep. 6:32795. doi: 10.1038/srep32795
Martin-Trillo, M., and Cubas, P. (2010). TCP genes: a family snapshot ten years later. Trends Plant Sci. 15, 31–39. doi: 10.1016/j.tplants.2009.11.003
Martín-Trillo, M., Grandío, E. G., Serra, F., Marcel, F., Rodríguez-Buey, M. L., Schmitz, G., et al. (2011). Role of tomato BRANCHED1-like genes in the control of shoot branching. Plant J Cell Mol. Biol. 4, 701–714. doi: 10.1111/j.1365-313X.2011.04629.x
Mitsuda, N., Seki, M., Shinozaki, K., Ohme-Takagi, M., and Koyama, T. (2010). TCP transcription factors regulate the activities of ASYMMETRIC LEAVES1 and miR164, as well as the auxin response, during differentiation of leaves in Arabidopsis. Plant Cell 22, 3574–3588. doi: 10.1105/tpc.110.075598
Mukhopadhyay, P., and Tyagi, A. K. (2015). OsTCP19 influences developmental and abiotic stress signaling by modulating ABI4-mediated pathways. Sci. Rep. 5:9998.
Nath, U., Crawford, B. C., Carpenter, R., and Coen, E. (2003). Genetic control of surface curvature. Science 299, 1404–1407.
Navaud, O., Dabos, P., Carnus, E., Tremousaygue, D., and Hervé, C. (2007). TCP transcription factors predate the emergence of land plants. J. Mol. Evol. 65, 23–33. doi: 10.1007/s00239-006-0174-z
Ori, N., Cohen, A. R., Etzioni, A., Brand, A., Yanai, O., Shleizer, S., et al. (2007). Regulation of LANCEOLATE by miR319 is required for compound-leaf development in tomato. Nat. Genet. 39, 787–791.
Palatnik, J. F., Allen, E., and Wu, X. (2003). Control of leaf morphogenesis by microRNAs. Nature 425, 257–263.
Parapunova, V., Busscher, M., Busscher-Lange, J., Lammers, M., Karlova, R., Bovy, A. G., et al. (2014). Identification, cloning and characterization of the tomato TCP transcription factor family. BMC Plant Biol 14:157. doi: 10.1186/1471-2229-14-157
Ren, L., Li, F., Jiang, J., and Li, H. (2021). BpTCP3 transcription factor improves salt tolerance of betula platyphylla by reducing reactive oxygen species damage. Forests 12:1633.
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Schommer, C., Palatnik, J. F., Aggarwal, P., Chételat, A., Cubas, P., Farmer, E. E., et al. (2008). Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biol. 6:e230. doi: 10.1371/journal.pbio.0060230
Shleizer-Burko, S., Burko, Y., Ben-Herzel, O., and Ori, N. (2011). Dynamic growth program regulated by LANCEOLATE enables flexible leaf patterning. Development 138, 695–704. doi: 10.1242/dev.056770
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steiner, E., Yanai, O., Efroni, I., Ori, N., Eshed, Y., and Weiss, D. (2012). Class I TCPs modulate cytokinin-induced branching and meristematic activity in tomato. Plant Signal Behav. 7, 807–810. doi: 10.4161/psb.20606
Takeda, T., Suwa, Y., Suzuki, M., Kitano, H., Ueguchi-Tanaka, M., Ashikari, M., et al. (2003). The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 33, 513–520.
van Es, S. W., van der Auweraert, E. B., Silveira, S. R., Angenent, G. C., van Dijk, A. D. J., and Immink, R. G. H. (2019). Comprehensive phenotyping reveals interactions and functions of Arabidopsis thaliana TCP genes in yield determination. Plant J. 99, 316–328. doi: 10.1111/tpj.14326
Wang, M.-Y., Zhao, P.-M., Cheng, H.-Q., Han, L. B., Wu, X. M., Gao, P., et al. (2013). The cotton transcription factor TCP14 functions in auxin-mediated epidermal cell differentiation and elongation. Plant Physiol. 162, 1669–1680. doi: 10.1104/pp.113.215673
Wang, P., Yu, J., Jin, S., Chen, S., Yue, C., Wang, W., et al. (2021). Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Horticult. Res. 8:107. doi: 10.1038/s41438-021-00542-x
Wang, S. T., Sun, X. L., Hoshino, Y., Yu, Y., Jia, B., Sun, Z. W., et al. (2014). MicroRNA319 positively regulates cold tolerance by targeting OsPCF6 and OsTCP21 in rice (Oryza sativa L.). PLoS One 9:e91357. doi: 10.1371/journal.pone.0091357
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Xia, E.-H., Li, F.-D., Tong, W., Li, P. H., Wu, Q., Zhao, H. J., et al. (2019). Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol. J. 17, 1938–1953. doi: 10.1111/pbi.13111
Xiong, Y., Liu, T., Tian, C., Sun, S., Li, J., and Chen, M. (2005). Transcription factors in rice: a genome-wide comparative analysis between monocots and eudicots. Plant Mol. Biol. 59, 191–203. doi: 10.1007/s11103-005-6503-6
Xu, R., Sun, P., Jia, F., Lu, L., Li, Y., Zhang, S., et al. (2014). Genomewide analysis of TCP transcription factor gene family in Malus domestica. J. Genet. 93, 733–746. doi: 10.1007/s12041-014-0446-0
Yang, C., Li, D., Mao, D., Liu, X., Ji, C., Li, X., et al. (2013). Overexpression of microRNA319 impacts leaf morphogenesis and leads to enhanced cold tolerance in rice (Oryza sativa L.). Plant Cell Environ. 36, 2207–2218.
Yao, X., Ma, H., Wang, J., and Zhang, D. (2007). Genome-Wide comparative analysis and expression pattern of tcp gene families in Arabidopsis thaliana and Oryza sativa. J. Integr. Plant Biol. 49, 885–897.
Yu, S., Li, P., Zhao, X., Tan, M., Ahmad, M. Z., Xu, Y., et al. (2021). CsTCPs regulate shoot tip development and catechin biosynthesis in tea plant (Camellia sinensis). Horticult. Res. 8:104. doi: 10.1038/s41438-021-00538-7
Zhang, L., Ye, H., Weng, Y., and Chen, X. (2015). Comparison of tea quality and productive effectiveness for processing of longjing tea. J. Korean Tea Soc. 1, 95–97.
Keywords: genome-wide analysis, TCP gene family, evolution, expression pattern, Camellia sinensis
Citation: Shang X, Han Z, Zhang D, Wang Y, Qin H, Zou Z, Zhou L, Zhu X, Fang W and Ma Y (2022) Genome-Wide Analysis of the TCP Gene Family and Their Expression Pattern Analysis in Tea Plant (Camellia sinensis). Front. Plant Sci. 13:840350. doi: 10.3389/fpls.2022.840350
Received: 13 January 2022; Accepted: 13 May 2022;
Published: 01 July 2022.
Edited by:
Marco Landi, University of Pisa, ItalyReviewed by:
Liangsheng Zhang, Zhejiang University, ChinaNaixing Ye, Fujian Agriculture and Forestry University, China
Copyright © 2022 Shang, Han, Zhang, Wang, Qin, Zou, Zhou, Zhu, Fang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wanping Fang, fangwp@njau.edu.cn; Yuanchun Ma, myc@njau.edu.cn
 Xiaowen Shang
Xiaowen Shang Zhaolan Han1
Zhaolan Han1 Zhongwei Zou
Zhongwei Zou Lin Zhou
Lin Zhou Xujun Zhu
Xujun Zhu Wanping Fang
Wanping Fang