- 1Guangxi Key Laboratory of Rice Genetics and Breeding, Rice Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, China
- 2State Key Laboratory of Crop Biology, Shandong Key Laboratory of Crop Biology, College of Agronomy, Shandong Agricultural University, Tai’an, China
- 3Beijing Key Laboratory of Crop Genetic Improvement, State Key Laboratory of Agrobiotechnology, College of Agronomy and Biotechnology, China Agricultural University, Beijing, China
- 4Department of Bioinformatics & Biotechnology, Government College University Faisalabad, Faisalabad, Pakistan
- 5Guangxi Lvhai Seed Co., Ltd., Nanning, China
- 6Guangxi Crop Genetic Improvement and Biotechnology Laboratory, Guangxi Academy of Agricultural Sciences, Nanning, China
Rice (Oryza sativa L.) is an important staple food crop for more than half of the world’s population. Enhancing the grain quality and yield of rice to meet growing demand remains a major challenge. Here, we show that OsMKK3 encode a MAP kinase kinase that controls grain size and chalkiness by affecting cell proliferation in spikelet hulls. We showed that OsSPL16, GS5, and GIF1 have a substantial effect on the OsMKK3-regulated grain size pathway. OsMKK3 has experienced strong directional selection in indica and japonica. Wild rice accessions contained four OsMKK3 haplotypes, suggesting that the OsMKK3 haplotypes present in cultivated rice likely originated from different wild rice accessions during rice domestication. OsMKK3-Hap1, gs3, and gw8 were polymerized to enhance the grain length. Polymerization of beneficial alleles, such as OsMKK3-Hap1, gs3, gw8, fgr, alk, chalk5, and wx, also improved the quality of hybrid rice. Overall, the results indicated that beneficial OsMKK3 alleles could be used for genomic-assisted breeding for rice cultivar improvement and be polymerized with other beneficial alleles.
Introduction
Rice (Oryza sativa L.) is an important staple food crop for over half of the world’s population. Grain yield is determined by three key factors: grain weight, number of grains per panicle, and number of panicles per plant. Grain size is an important breeding target that affects both yield and appearance quality and is determined by the length, width, and thickness of the grain. Studies of grain size can provide new insights that could be used to improve the yield and quality of rice and the rice domestication process (Shomura et al., 2008).
Many quantitative trait loci (QTLs) of grain length have been mapped, and some of these QTLs, such as GS3, OSLG3, qGL3, SMG1, and OsSPL13, have been cloned as well. GS3 encodes a protein of 232 amino acids with a putative PEBP-like domain, a transmembrane region, a putative TNFR/NGFR family cysteine-rich domain, and a VWFC module that is a negative regulator of grain size (Fan et al., 2006). GS3 is an evolutionarily important gene controlling grain size in rice by a C to A mutation in the second exon of GS3 (A allele) (Takano-Kai et al., 2009). GL3.1 encodes a protein phosphatase kelch (PPKL) family — Ser/Thr phosphatase that affects the phosphorylation of proteins in the spikelet and accelerates cell division. GL3.1 increases the grain length and results in higher yields (Qi et al., 2012). OSLG3 was found to encode an AP2 domain class transcription factor that positively regulates grain length and improves rice yield without affecting grain quality. OSLG3 alleles in indica and japonica evolved independently from distinct ancestors (Yu et al., 2017). OsSPL13 encodes a plant-specific transcription factor that regulates the cell size of the grain hull and enhances rice grain length and yield (Si et al., 2016). SMG1 is a mitogen−activated protein kinase kinase 4 that regulates grain size through its effect on the MAPK pathways and brassinosteroids (Duan et al., 2014). A study examining the function of OrMKK3 in Oryza officinalis Wall. ex Watt revealed that the plant height was lower, the grains were shorter, and the number of tillers was higher in plants overexpressing OrMKK3 than wild-type plants of the Nipponbare (Nip) cultivar (Pan et al., 2021). There is thus a need to study the genetic and molecular bases of grain size (Mao et al., 2010).
Rice breeders use natural variations in grain size to improve yield and quality, but only a few alleles of genes regulating grain shape have been widely used. Class A haplotypes of OsLG3 show a longer grain phenotype compared with class B haplotypes of OsLG3 in several cultivars (Yu et al., 2017). Hap-SLG of OsLG3b shows a longer grain phenotype than Hap-NIP and has much breeding potential for increasing grain length in indica (Yu et al., 2018). Application of the qgl3 allele can increase the grain yield through its positive effect on grain length, filling, and weight (Zhang et al., 2012). Xia et al. (2018) pyramided the GW7 allele from TFA and gs3 to develop new high-yielding indica hybrid rice varieties. Liu et al. (2018) combined the OsMADS1lgy3 allele with high yield-associated dep1-1 and gs3 alleles, which enhanced both rice yield and quality. More characterizations of gene–coding sequence–haplotype (gcHap) diversity would facilitate basic research and improvement of rice (Zhang et al., 2021).
Here, OsMKK3 was shown to be a positive regulator of grain length and height in rice. Characterization of natural variations in the OsMKK3 coding sequence (CDS) associated with grain length and chalkiness and favorable alleles could provide useful genetic resources for improving rice cultivars. By pyramiding OsMKK3, fgr, wx, alk, and gs3 alleles, Zhou et al. (2019) developed the new high-yielding indica hybrid rice variety Wantaiyou3158, which had higher yield and grain quality.
Materials and Methods
Plant Materials and Measurement of Grain and Yield Traits
Yexiang maintainer line (YXB) is an excellent quality hybrid rice parent that has been used to produce more than 30 varieties. The varieties have been planted widely in southern China. Rice was planted under natural field conditions at the Rice Research Institute of Guangxi Academy of Agricultural Sciences, Nanning, China in the summers of 2015–2021 (22.85°N, 108.26°E). The distance between plants within rows was 16.7 cm, and the distance between plants in separate rows was 20 cm. Field management, including irrigation, fertilizer application, and pest control, followed normal agricultural practices. Fully filled grains were used for measurements of grain width, length, and weight with a Wanshen SC-G automatic seed test system. All trait measurements were repeated at least 3 times. A total of 342 Guangxi common wild rice core germplasm accessions (Pan et al., 2018), a total of 419 Guangxi core germplasm landrace accessions (Yang et al., 2018), and 94 improved varieties (Supplementary Table 1) were used in this study.
Vector Construction and Rice Transformation
To generate the overexpression vector, the open reading frame of OsMKK3 was amplified from the cDNA of Nip (Supplementary Table 2) and cloned into the pMDC32 vector. sgRNA-Cas9 plant expression vectors were constructed as described previously (Mao et al., 2013).
Gene Genotyping by PARMS
The PARMS is a KASP-like SNP genotyping technique combined with ARMS, which is also referred to as allele-specific PCR (Newton et al., 1989; Heim and Meyer, 1990). The primer sequence information of SNP markers for rice was obtained from the rice 3K project (RFGB1). The primers were both designed by Primer Premier 5.0 (Supplementary Table 3). Genotyping tests were carried out with PARMS (Gentides, China). The PCR reactions were conducted in 384-well PCR plates for PARMS genotyping. The 5-μl PCR reaction system contained 2 × PARMS PCR reaction mix, each allele-specific primer (150 nM), locus-specific primer (400 nM), and 1.4 μl of alkaline lysis DNA template. 5-μl of mineral oil was added into each well of the PCR plate to prevent evaporation of the PCR mix. The thermal cycler program of PARMS was denaturation at 95°C for 15 min and 10 cycles of 95°C for 20 s, followed by annealing at 65°C for 1 min. The temperature was then decreased 0.8°C per cycle to the annealing temperature at 57°C, which was followed by 32 cycles of denaturation at 95°C for 20 s and annealing at 57°C for 1 min. The well plate was read using a TECAN Infinite M1000 plate reader; SNP calling and plots were conducted using an online software SNP Decoder2 combined with manual modification. Sequence analysis was performed following the method provided by Wuhan Gentides Biotech Co., Ltd.
DNA Extraction, RNA Extraction, Expression Analysis, and RNA-Seq
An EasyPure® Plant Genomic DNA Kit (Trans, China) was used for genomic DNA extraction. Panicles of YXB and YXB-Cr line of different lengths (3, 5, 10, 15, 20, and 25 cm) at the booting stage were flash-frozen in liquid nitrogen. Total RNA from the aforementioned tissues was extracted using a Takara MiniBEST Plant RNA Extraction Kit per the manufacturer’s instructions (Takara, Catalog no. 9769). Total RNA (2.0 μg) was used for cDNA synthesis with a PrimeScriptTM RT Reagent Kit with gDNA Eraser (TransScript® II One-Step RT-PCR SuperMix, Tran, Catalog no. AH411-02). The resulting cDNA samples were diluted five times and used as templates for PCR. We performed qRT-PCR using a TransStart® Green qPCR SuperMix and a CFX96 RealTime system (Bio-Rad, Hercules, CA, United States) following the manufacturer’s instructions. qRT-PCR was performed using 10-μL mixtures containing 5 μL of 2 × Green qPCR MasterMix, 1 μL of cDNA, 0.25 μL of each primer (10 μM), and 3.5 μL of ddH2O. Amplification steps were 95°C for 30 s, 40 cycles of 95°C for 5 s, and 60°C for 30 s, followed by 65°C for 5 s, 95°C for 15 s, 60°C for 30 s, and 95°C for 15 s. Each experiment was repeated at least three times. The qRT-PCR analysis was performed using the ΔΔCt method. Details on gene-specific primers used for real-time PCR are provided in Supplementary Table 1. qRT-PCR was performed in triplicate for each sample, and the Ubiquitin gene (LOC_Os03g13170) (Chen et al., 2021) was used as a control (∗P < 0.05, ∗∗P < 0.01; Student’s t-test).
RNA samples used for RNA-seq analysis were prepared from 10 and 20-cm panicles of YXB and YXB-cr line grown under normal field conditions with three biological replicates. RNA library sequencing was performed on an Illumina HiseqTM 2500/4000 platform by Gene Denovo Biotechnology Co., Ltd. (Shanghai, China). Sequence analysis was performed using the method provided by Majorbio. Additional detailed information is provided on the Majorbio website3.
Selection of Germplasm and Phylogenetic Analysis
The phylogenetic tree of rice core germplasm landrace accessions, Oryza rufipogon wild rice core germplasm accessions, and improved varieties was constructed based on SNPs from the rice 3K project (RFGB, see Text Footnote 1). A total of 419 rice core germplasm landrace accessions were from Guangxi province (Yang et al., 2018). A total of 351 O. rufipogon wild rice core germplasm accessions were from Guangxi province (Pan et al., 2018). A total of 96 improved varieties were conserved in the Rice Research Institute, Guangxi Academy of Agricultural Sciences. A neighbor-joining variety tree of rice varieties was constructed using MEGA 7.0. The number of bootstrap replicates was 1,000 (Kumar et al., 2016). The phylogenetic tree was visualized with the iTOL online tool (Letunic and Bork, 2019). An independent samples t-test was performed to identify differences in phenotypes between groups and control.
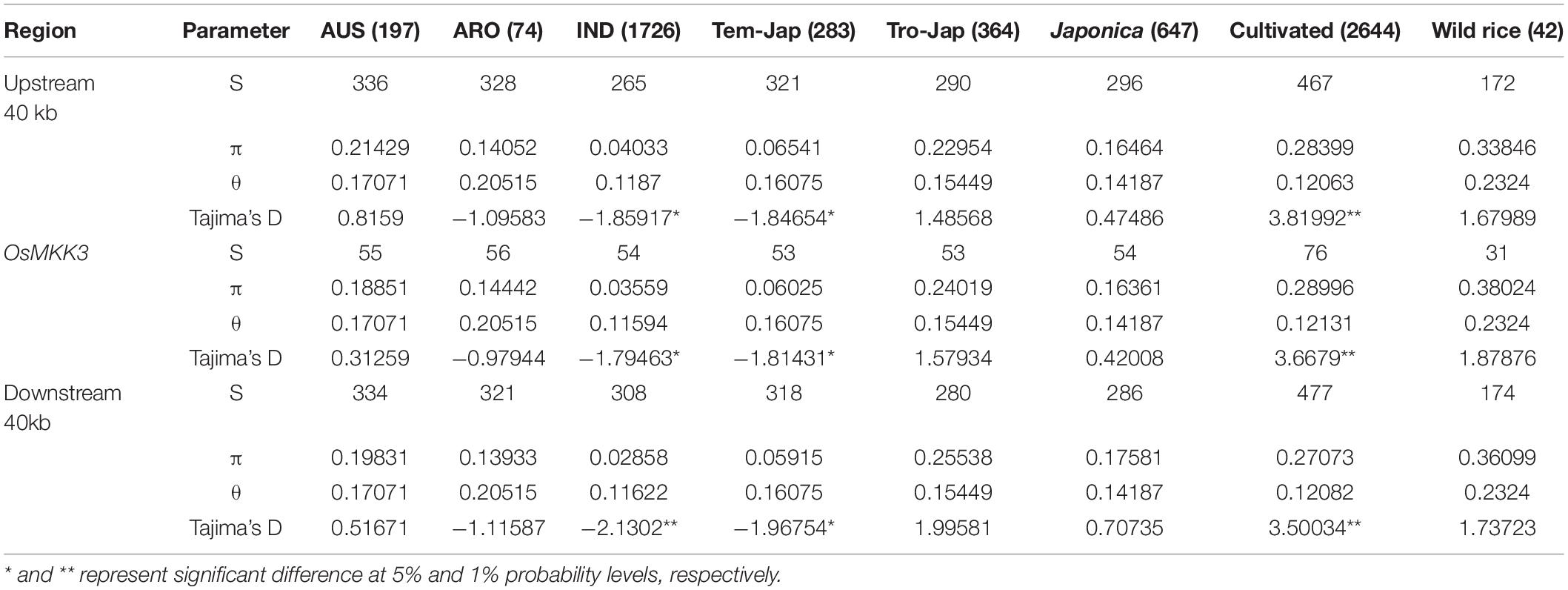
Nucleotide Diversity Analysis
The genomic sequences of 2,644 cultivated and 42 wild accessions were available from the 3K Rice Genome Project (3KRGP) (Alexandrov et al., 2015; Wang et al., 2018) and OryzaGenome4, respectively. The average nucleotide diversity (π and θ) and Tajima’s D for each subpopulation in OsMKK3 and flanking regions (40-kb) were calculated using DnaSP 5.10 (Librado and Rozas, 2009). The nucleotide diversity curves were acquired using a 40-bp window and 10-bp step length. Population differentiation statistics (FST) were calculated using VCFtools software (Danecek et al., 2011) with a 3,000-bp window and 300-bp step length.
Histological Analysis
To observe the morphology of starch granules in the grain of Nip, the transgenic line, YXB, and YXB-Cr line. Milled rice grains were transversely cut in the middle with a sharp knife. Samples were then cleaned, placed in an electron microscope fixator at room temperature for 2 h, and then transferred to 4°C for preservation. The samples were fixed, dehydrated, and dried with a critical point drier. Samples were attached to conductive carbon film double-sided tape and coated with gold under vacuum for 30 s. Milled rice grains for scanning electron microscopy were transversely cut in the middle with a sharp knife, attached to conductive carbon film double-sided tape, and coated with gold under vacuum for 30 s. The morphology of starch granules in the belly part of the endosperm was examined with a scanning electron microscope (Hitachi, SU8100, Wuhan Servicebio Technology) at an accelerating voltage of 12 kV. The analysis was based on at least three biological replications of mounted specimens. All procedures were carried out per the manufacturer’s protocol.
Results
OsMKK3 Regulates Grain Size, the Accumulation of Starch, and the Expression of Other Genes Involved in the Production of Rice
According to our previous study (Pan et al., 2021), OrMKK3 of O. officinalis Wall. ex Watt affects the morphology and grain size of rice. We performed a series of studies of OsMKK3 in rice to determine its function. Overexpression constructs containing the OsMKK3 CDS from Nip (N-OE) driven by the 35S promoter from tobacco cauliflower mosaic virus (CaMV35S) were separately introduced into Nip. N-OE lines showed significantly longer grains, higher grain length-width ratio, chalkiness degree, and greater higher plant height (Figures 1A–C). N-OE lines had a higher chalky grain rate than the no transgene line (NT). And N-OE lines showed few changes in grain width. In general, the expression of OsMKK3 regulated grain size, chalkiness, and height. In addition, we used a CRISPR-Cas9 system for targeted gene mutation of OsMKK3 in the Yexiang maintainer line (YXB) (Figure 1A). The target sequence (5′-AAATCTCAAGGGTGAGGCAAA-3′) was at sites +666–+667 within the fourth exon encoding the C-terminal of OsMKK3 (amino acid residues 222). These deletions lead to frameshifting mutations that result in differences in the C-terminal of OsMKK3 and incomplete peptides of OsMKK3; these changes result in a loss of OsMKK3 function. The grain of transgenic plants (Cr) was smaller than the wild-type YXB (Figures 1B,C). The Cr line showed a significant reduction in grain length, grain width, grain length-to-width ratio, and chalky rice rate (Figures 1B,C). This finding was consistent with the reduced production of rice and the change in chalkiness caused by the loss of OsMKK3 function. The results of the histological analysis indicated that the chaff of N-OE increased in size compared with that of NT, and the chaff of Cr became shorter compared with that of NT and YXB. Scanning electron microscopy images of transverse sections of N-OE grains indicated that this endosperm was filled with loosely packed, small, and spherical starch granules with large air spaces (Figure 1D), and the Cr endosperm consisted of densely packed, large, and irregularly shaped polyhedral starch granules (Figure 1E). These results suggest that OsMKK3 regulated the accumulation of starch and affected grain size. Furthermore, real-time quantitative PCR (qRT-PCR) analysis of GL3, GW2, GW8, SMG1, and GS3 was performed in the Cr line and YXB to study the development of spikes. These genes regulate grain size and cell cycle time (Figure 1F). The expression patterns of GL3 and PPKL1 were the same in YXB and the Cr line at 3, 5, 10, 15, 20, and 25 cm panicle length. The expression of GS3 was lower in YXB than in the Cr line. The expression of OsLG3 and SMG1 was higher in YXB than in the Cr line. These results indicate that the knockout of OsMKK3 affected GS3, OsLG3, and SMG1 expression. In short, OsMKK3 played an important role in cell development in rice, regulated genes involved in grain size and cell cycle time, and affected the grain size and accumulation of starch.
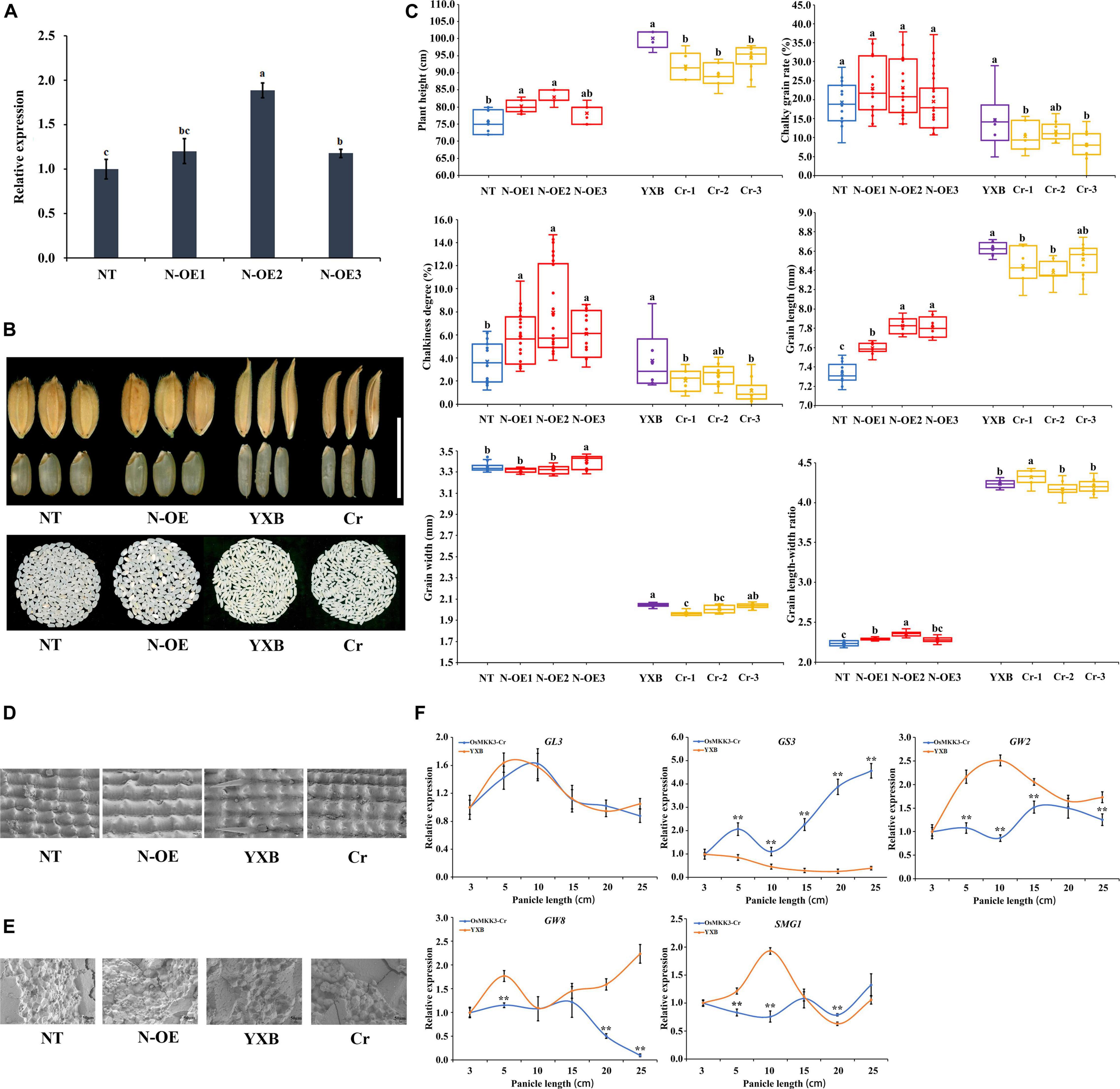
Figure 1. Identification of OsMKK3. (A) Relative expression of OsMKK3 in NT and transgenic plants. (B) Functional validation of the role of OsMKK3 in grain size and chalkiness by genetic transformation. (C) Comparison of the grain length, grain width, chalkiness rate, chalkiness score, and plant height between transgenic plants and NT. Data represent mean ± S.E.M. (n = 12). Student’s t-tests were used to generate P-values. (D) The epidermal cells of rice husk in transgenic plants and NT. Bar = 1 cm. (E) Scanning electron microscope images of chalkiness in transgenic plants and NT. Bar = 50 μm. (F) Relative expression of important genes in NT and transgenic plants. a, b, and c represent significant difference at 5% probability level. ** represents significant difference at 1% probability level.
OsMKK3 Regulates Genes in the Early Spike Development Stage to Control Grain Size and Chalkiness
To analyze the effects of OsMKK3 on the transcriptome of rice grain and spike, we studied 5- and 10-cm spikes in YXB and the Cr line. There were three biological replicates per condition for RNA-seq. Overall, more than 83.86 Gb of clean data were generated, and these were used for mapping onto the Os-Nipponbare-Reference-IRGSP-1.0 genome5. The results showed that the Q30 base percentage was above 93.03%. A total of 40,225 genes were expressed, 35,193 genes were identified as known genes, and 5,032 genes were identified as new genes. YXB_5cm was used as a control in the group YXB_5cm_vs_Cr line _5cm. The total differential expression analysis revealed 4,268 differentially expressed genes (DEGs), including 1,496 up-regulated DEGs and 2,772 down-regulated DEGs (Figure 2A). YXB_10cm was used as a control in the group YXB_10cm_vs_Cr line_10cm; the total number of DEGs was 830, including 485 up-regulated DEGs and 345 down-regulated DEGs (Figure 2B). DEGs in the 5-cm spike stage revealed that OsMKK3 affects the expression of several genes in early spike development to regulate traits such as grain size, height, and starch accumulation. To identify the common target genes of the OsMKK3 regulatory module, we next compared the genome-wide transcriptional profiles in the developing panicles, spikelet hulls, and starch accumulation of YXB and Cr line plants using weighted gene co-expression network analysis. The network was constructed from the filtered probes, and 18 co-expressed modules were identified. The module detection parameters were as follows: minimum module size 30, the module detection sensitivity deepSplit 2, and cut height for merging of modules 0.25 (meaning that modules whose eigengenes are correlated are merged). For example, the MEblack, MEpink, MEmagenta, MEgreen, MEsalmon, MEcyan, MEbrown, and MEred modules were related to up-regulated DEGs in the grain development stages, especially the MEsalmon module, which was strongly related to grain development; the red and brown modules were negatively related to the very early stage of grain development; and the MEtan, MEgrey60, and MEpurple modules were specifically related to down-regulated DEGs in the grain development stages (Figure 2C). In short, multiple modules were related to one or more grain development and starch accumulation stages associated with OsMKK3.
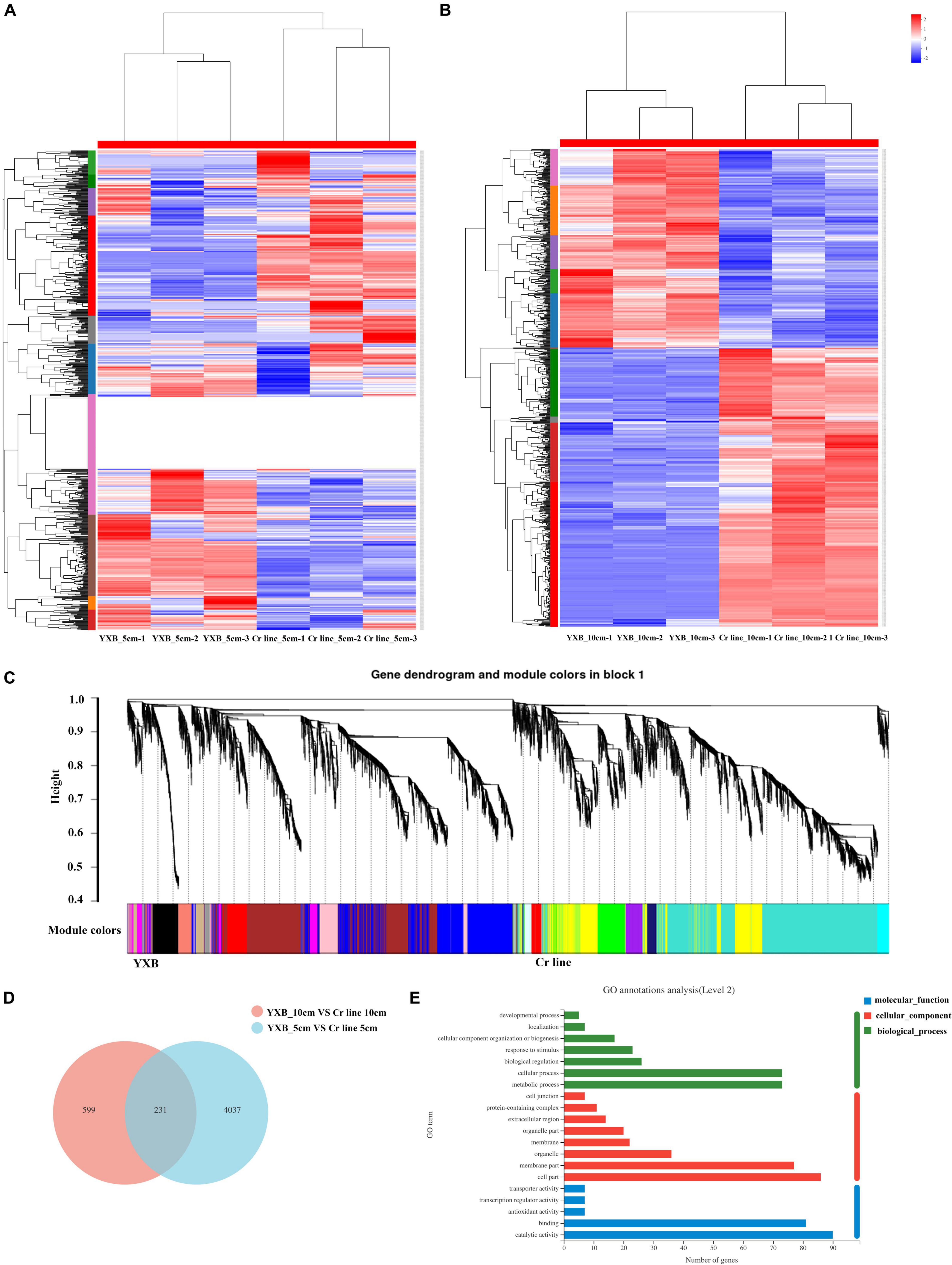
Figure 2. OsMKK3 regulates genes in the early spike development stage. (A) Hierarchical clustering of the DEGs for the YXB and the Cr line in the spike development stage. (B) Weighted gene co-expression network analysis of DEGs identified from YXB and the Cr line in the spike development stage. (C) Common DEGs expressed in YXB_5cm_vs_Cr line_5cm and YXB_10cm_vs_Cr line_10cm. (D) GO enrichment analysis of common DEGs. (E) GO term of common DEGs.
To identify the OsMKK3 pathways involved in spike development, we analyzed 231 common DEGs expressed in both YXB_5cm_vs_ Cr line_5cm and YXB_10cm_vs_cr line_10cm (Figure 2D). The results indicated that 90 DEGs were up-regulated in YXB_5cm_vs_ Cr line_5cm and YXB_10cm_vs_Cr line_10cm; 124 DEGs were down-regulated in YXB_5cm_vs_Cr line_5cm and YXB_10cm_vs_Cr line_10cm; 12 DEGs were down-regulated in YXB_5cm_vs_Cr line_5cm but up-regulated in YXB_10cm_vs_Cr line_10cm; and 5 DEGs were up-regulated in YXB_5cm_vs_Cr line_5cm but down-regulated in YXB_10cm_vs_Cr line_10cm. To investigate the biological functions of these DEGs, Gene Ontology (GO) enrichment analysis was performed with agriGO. DEGs of cellular process and metabolic process were more enriched compared with other GO terms within biological process. DEGs of cell part were more enriched compared with other GO terms within cellular component. DEGs of catalytic activity were most enriched in molecular function (Figure 2E). In the GO enrichment analysis, 137 DEGs were enriched in cellular component, 86 DEGs were enriched in cell part, and 76 DEGs were enriched in membrane part (Figure 2E). The results indicated that 231 DEGs may be regulated by OsMKK3 pathways. Most DEGs were enriched in cell development. To determine whether OsMKK3 affected genes controlling grain size and starch accumulation, we examined 22 genes that have been cloned and identified to be involved in grain size and starch accumulation in the spike development stage. In YXB_5cm_vs_cr line_5cm, there were three up-regulated genes: OsSPL16, GS5, and GIF1. Wx was up-regulated in YXB_5cm_vs_cr line_5cm. OsSPL16 was up-regulated in YXB_10cm_vs_Cr line_10cm. GS5, GIF1, and Wx were not detected in YXB_10cm_vs_Cr line_10cm. OsSPL16, GS5, GIF1, and Wx affect the OsMKK3-regulated grain size and chalkiness pathway; OsSPL16 in particular may be up-regulated in the OsMKK3 pathway. Overall, these results indicate that OsMKK3 is a key functional factor controlling grain size and chalkiness by regulating a series of genes.
OsMKK3 Has Undergone a Selective Sweep During the Domestication of Indica and Temperate Japonica
We performed a haplotype analysis of OsMKK3 in 3,110 cultivated varieties and 446 wild rice samples to investigate natural variation in OsMKK3 among rice germplasm accessions and identified six main haplotypes (Hap) for OsMKK3 (Figure 3A). Based on the phylogenetic analysis, the six haplotypes could be classified into two classes: haplotypes 1, 2, 3, and 6 in class A and haplotypes 4 and 5 in class B (Figure 3B). Phenotype analysis showed that cultivars with class A haplotypes had a longer grain phenotype compared with those with class B haplotypes. Hap1 is the main haplotype in both indica and japonica. We also calculated population differentiation statistics (FST) for OsMKK3 and its flanking regions between indica and japonica. FST in OsMKK3 was above the genome-wide threshold, indicating that there was genetic differentiation in OsMKK3 between indica and japonica subspecies (Figure 3C).
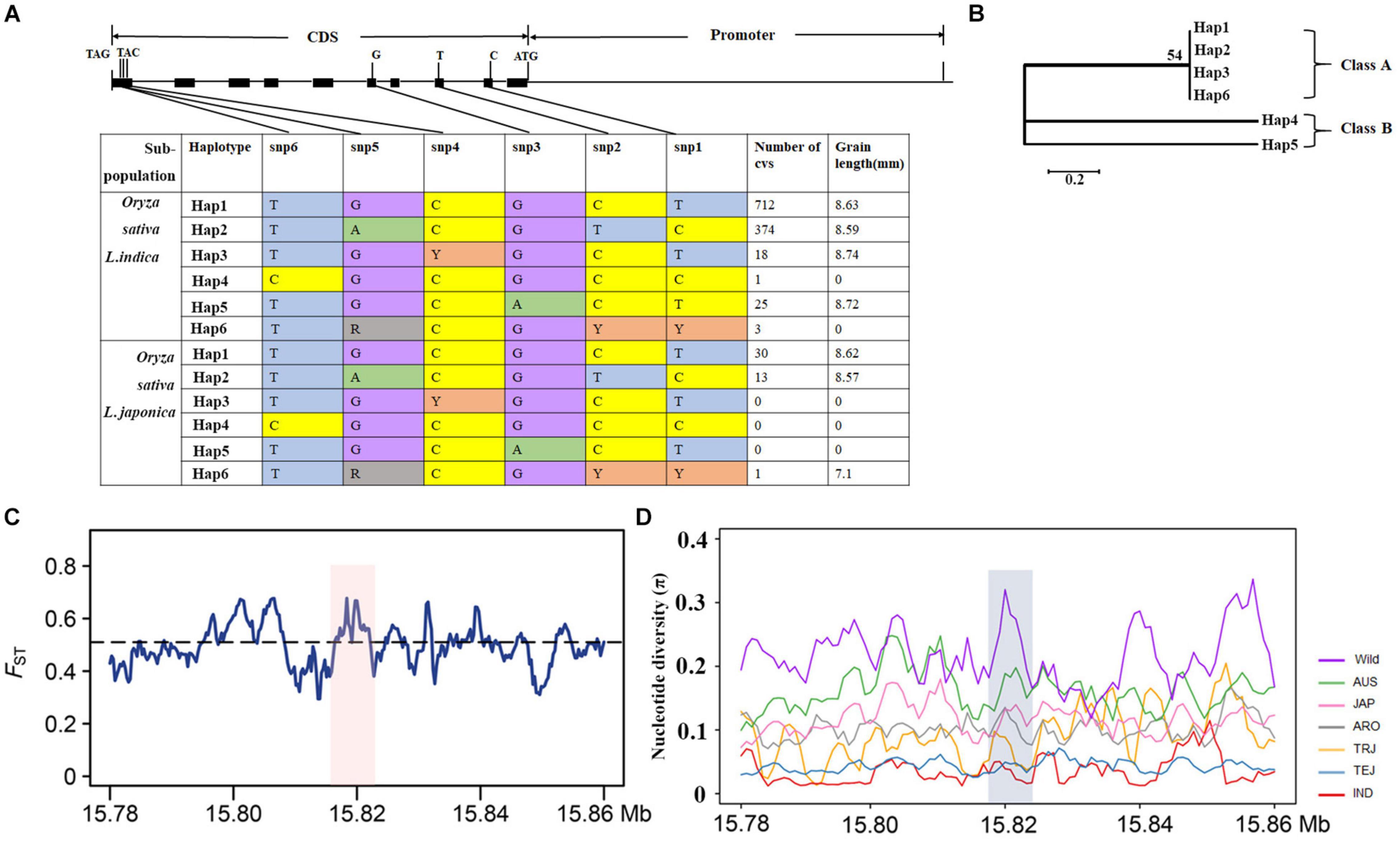
Figure 3. Haplotypes and genomic differentiation of OsMKK3. (A) Haplotype analysis of OsMKK3. Gene structure and natural variation between alleles from NIP. (B) Cladogram of six haplotypes. (C) Genetic differentiation of OsMKK3 and major associated loci. Blue and green lines represent the FST between indica and japonica. (D) Nucleotide diversity analysis in OsMKK3 and flanking regions (∼40 kb).
To reveal whether selection has acted on OsMKK3, 2,644 cultivated and 42 wild accessions were used to analyze the genetic diversity of OsMKK3 and its flanking regions. Compared with O. rufipogon, the nucleotide diversity of OsMKK3 was significantly lower in indica and temperate japonica. Tajima’s D-values for indica and temperate japonica were negative and statistically significant (Table 1), suggesting that OsMKK3 has experienced strong directional selection in these two subpopulations. Tajima’s D for cultivated varieties was positive and statistically significant, indicating that OsMKK3 had high polymorphism and might have experienced balancing selection. Given that directional selection might result in a selective sweep in the flanking region of selected genes, we examined the nucleotide diversity of 40-kb regions flanking OsMKK3 (Figure 3D). The average nucleotide diversity of OsMKK3-flanking regions in indica and temperate japonica was comparable to that of the OsMKK3 region but much lower than that in O. rufipogon populations. In addition, the Tajima’s D of OsMKK3-flanking regions in indica and temperate japonica was also negative and statistically significant. These findings indicated that OsMKK3 may have undergone a selective sweep during the domestication of indica and temperate japonica subspecies.
Selection Leads to Differences in the Main Haplotypes of Wild and Cultivated Rice
To characterize the geographic distribution of OsMKK3 haplotypes, we analyzed the geographic distribution of OsMKK3 in 1,703 cultivated varieties of the 3K project. Most japonica accessions were distributed in northern regions, whereas indica accessions were mostly distributed in southern regions. Cultivars distributed in China, Sri Lanka, Sierra Leone, Philippines, Malaysia, Madagascar, Korea, Indonesia, and India have more than four OsMKK3 haplotypes. OsMKK3-Hap1 is the main haplotype in 3K accessions (Figure 4A). A previous study indicated that Guangxi province, southern China is likely the region where cultivated rice was first developed (Huang et al., 2012). To characterize the distribution of OsMKK3 alleles in wild rice and cultivar accessions, we sequenced OsMKK3 in 342 Guangxi common wild rice core germplasm accessions, 419 Guangxi core germplasm landrace accessions, and 94 improved varieties (Supplementary Table 1). The results indicated that 278 wild accessions carried OsMKK3-Hap1, OsMKK3-Hap2, OsMKK3-Hap4, and OsMKK3-Hap6 at the OsMKK3 locus. A total of 64 wild accessions showed heterozygous haplotypes or haplotype deficiency at the OsMKK3 locus. One wild accession with Hap1 and one wild accession with Hap6 were from Laibin (Figure 4B). One wild accession with Hap2 was from Guigang (Figure 4B). Aside from these, all wild accessions had Hap4 of class B. One wild accession had Hap1, Hap2, and Hap6. These findings indicate that there is high diversity at the OsMKK3 locus. A total of 261 landraces in the putative zone of origin were analyzed. Among the landrace varieties, only one indica from Nanning had Hap4 (Figure 4B). A total of 6 indica and 3 japonica cultivars with Hap1, Hap2, and Hap3 and 5 wild rice accessions were from Guilin in the higher elevation zone (Figure 4B). In higher elevation zones such as Guilin, Liuzhou, Hechi, and Hezhou, japonica cultivars accounted for a large proportion of core germplasm accessions (Figure 4B). Hap1, Hap2, and Hap3 were present in improved varieties but not Hap4, Hap5, and Hap6. Almost all indica accessions and japonica accessions from Oryza rufipogon to O. sativa had OsMKK3-Hap. indica cultivars were in the lower elevation zone. OsMKK3 may have undergone selection from O. rufipogon to O. sativa; Hap4 was the most common in wild rice compared with Hap1, which was the most common in landrace varieties. Haplotype diversity was decreased in improved varieties.
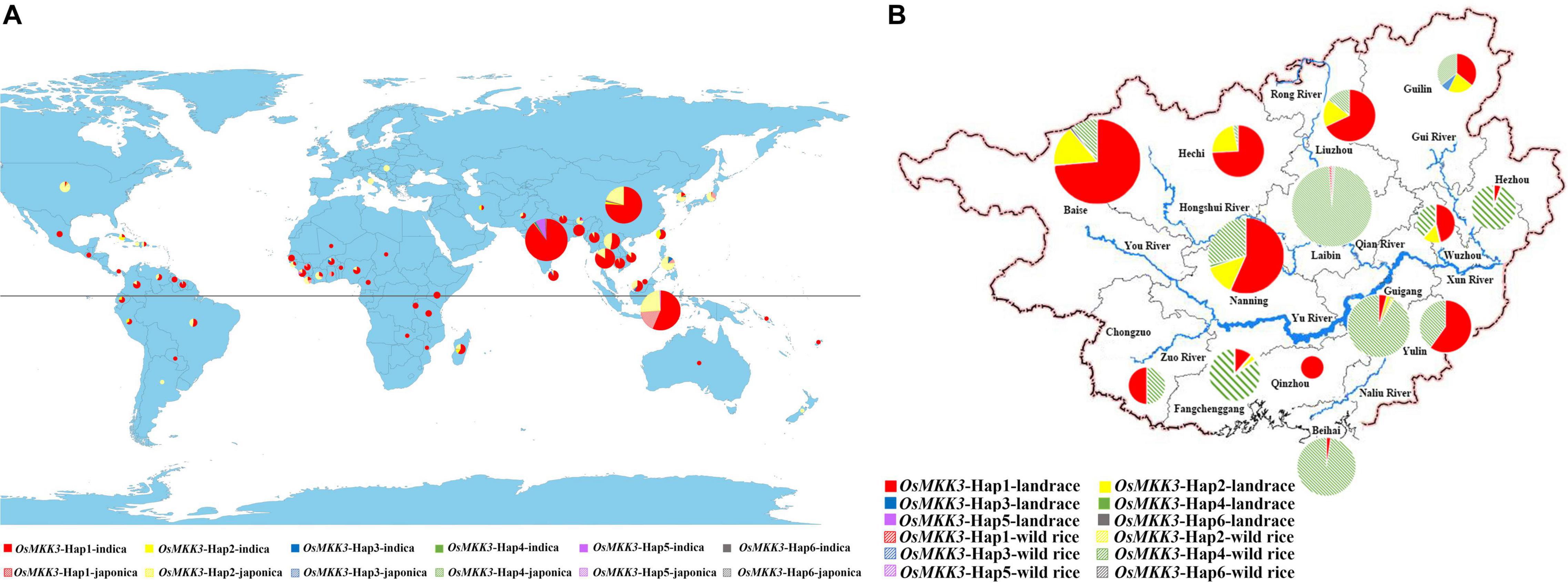
Figure 4. Distribution of the main OsMKK3 haplotypes in wild rice and cultivated rice in the world and Guangxi province. (A) Geographic origin of OsMKK3. Hap1–6 is represented by red, yellow, blue, green, purple, and gray, respectively. The indica and japonica cultivars are denoted by solid and dashed circles, respectively. (B) Geographic origin of OsMKK3 in Guangxi province. Hap1–6 is represented by red, yellow, blue, green, purple, and gray, respectively. The rice landraces and wild rice cultivars are denoted by solid and dashed circles, respectively.
OsMKK3-Hap1 and Hap2 Are Associated With Longer Grain Length in a GS3/gs3 Background in Indica and Japonica
To characterize the genetic interaction between OsMKK3 and GS3 in controlling grain length, we examined the haplotypes of OsMKK3 and GS3 in the rice 3K project, 342 Guangxi common wild rice core germplasm accessions, 419 Guangxi core germplasm landrace accessions, and 94 improved varieties. In the rice 3K project, grain length was longer with OsMKK3-Hap2 compared with other OsMKK3 haplotypes of indica in a GS3 background. Grain length was longer with OsMKK3-Hap1 and OsMKK3-Hap2 compared with other OsMKK3 haplotypes of indica in a gs3 background. Grain length was longer with OsMKK3-Hap1 compared with other OsMKK3 haplotypes of japonica in a GS3 background. Grain length was longer with OsMKK3-Hap2 compared with other OsMKK3 haplotypes of japonica in a gs3 background (Figure 5A). OsMKK3-Hap1 and OsMKK3-Hap2 thus appear to positively affect the grain length in indica and japonica in a GS3/gs3 background.
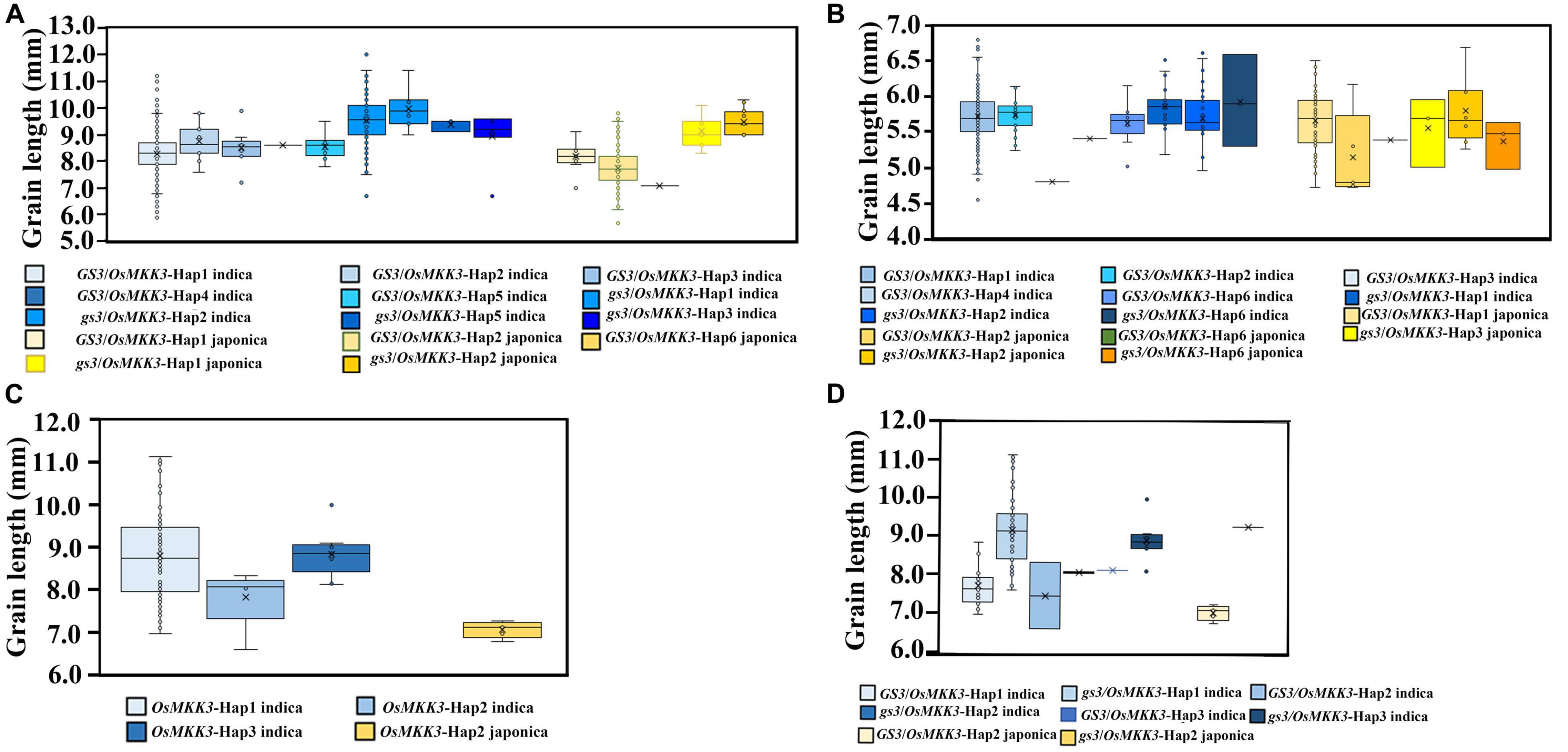
Figure 5. Genetic interactions between OsMKK3 and other grain length-related genes based on diverse germplasm accessions. Box plots and kernel density plots were generated for different groups. (A) Genetic interactions between OsMKK3 and GS3/gs3 in the rice 3K project. (B) Genetic interactions between OsMKK3 and GS3/gs3 in Guangxi core germplasm landrace accessions. (C) Effects of OsMKK3 in improved varieties. (D) Genetic interactions between OsMKK3 and GS3/gs3 in improved varieties.
To investigate the relationship between OsMKK3 and GS3 in Guangxi core germplasm landrace accessions, we examined their effects on grain length under different backgrounds. The results indicated that in Guangxi core germplasm landrace accessions, OsMKK3-Hap1 and OsMKK3-Hap2 positively affected the grain length of indica. OsMKK3-Hap1 had a stronger effect on the grain length of japonica (Figure 5B). In Guangxi core germplasm landrace accessions, the grain length of landraces was longer with OsMKK3-Hap1 and OsMKK3-Hap2 compared with other OsMKK3 haplotypes in a GS3 background of indica. Grain length of OsMKK3-Hap1 in a GS3 background significantly differed between japonica and indica. Furthermore, in the 94 improved varieties, grain length was longer with OsMKK3-Hap1 in indica (Figures 5C,D). These findings indicate that grain length is regulated by different OsMKK3 and GS3 haplotypes. To confirm the observed changes in OsMKK3 haplotype, we performed a phylogenetic analysis of OsMKK3 using Guangxi common wild rice core germplasm accessions, Guangxi core germplasm landrace accessions, and improved varieties. Phylogenetic analysis showed that grain length was increased in the improved varieties (Figure 6A). OsMKK3-Hap of breeding varieties was concentrated in class A (OsMKK3-Hap1, OsMKK3-Hap2, and OsMKK3-Hap3). Landraces and wild rice accessions were clearly distinguished in OsMKK3-Hap.
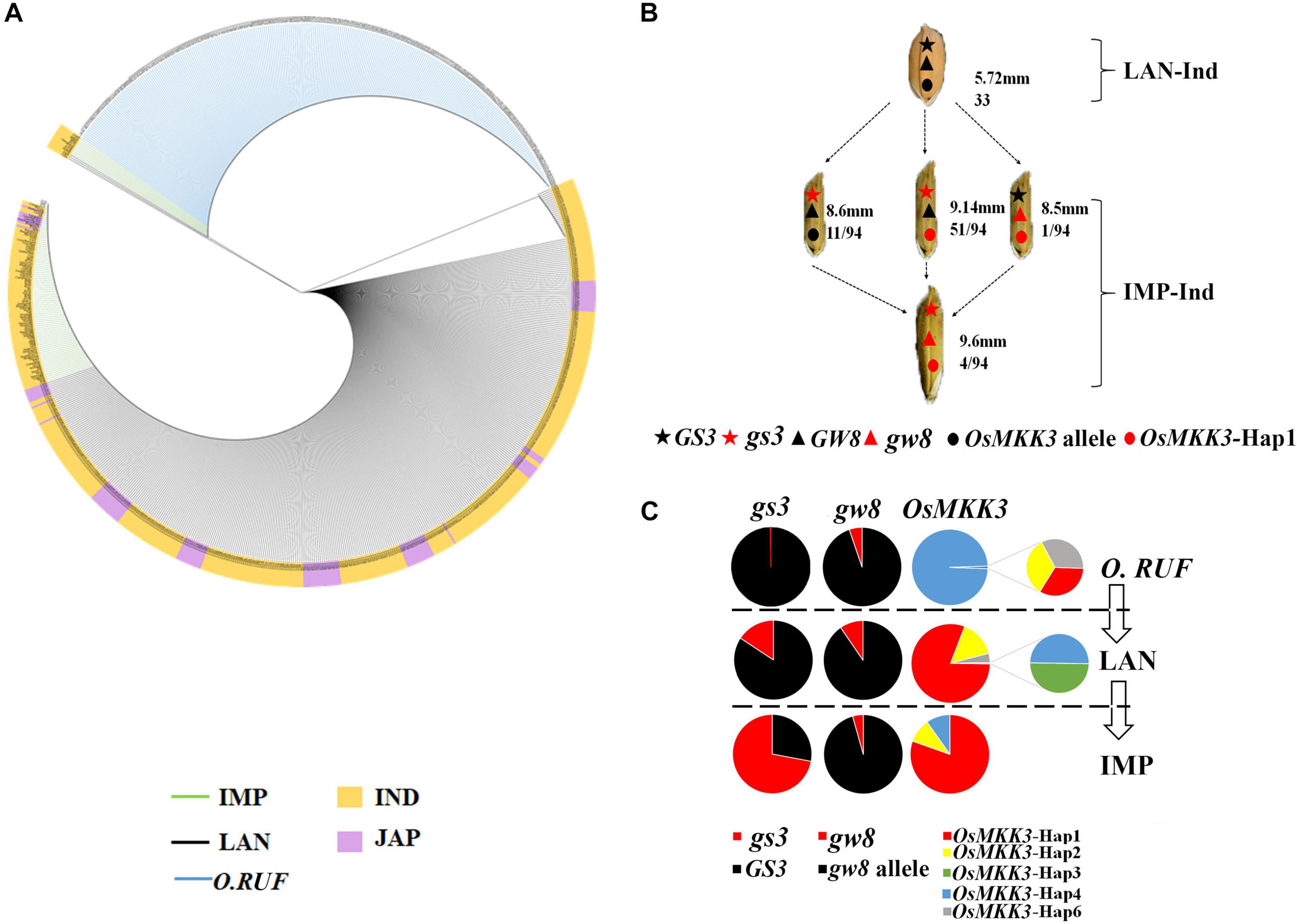
Figure 6. Breeding improvement of OsMKK3. (A) Phylogenetic analysis of haplotypes of OsMKK3 in wild rice accessions, Guangxi core germplasm landrace accessions, and improved varieties. The phylogenetic tree of varieties was constructed based on different functional SNPs in OsMKK3 CDS by MAGE 7.0. The blue line corresponds to wild rice, the black line corresponds to landraces, and the green line corresponds to improved varieties. (B) Improvement of the haplotype combination for three grain length-related genes, as in (A). Top numbers indicate average grain length; bottom numbers correspond to grain length-related genes, as in (A). Top numbers indicate average grain length; bottom numbers correspond to the accession number of a certain haplotype combination in the subgroup. (C) Spectra of allele frequencies comparing landrace and improved varieties at the causal polymorphisms of GS3, GW8, and OsMKK3 during modern breeding in the respective subgroups. LAN landraces, IMP improved varieties.
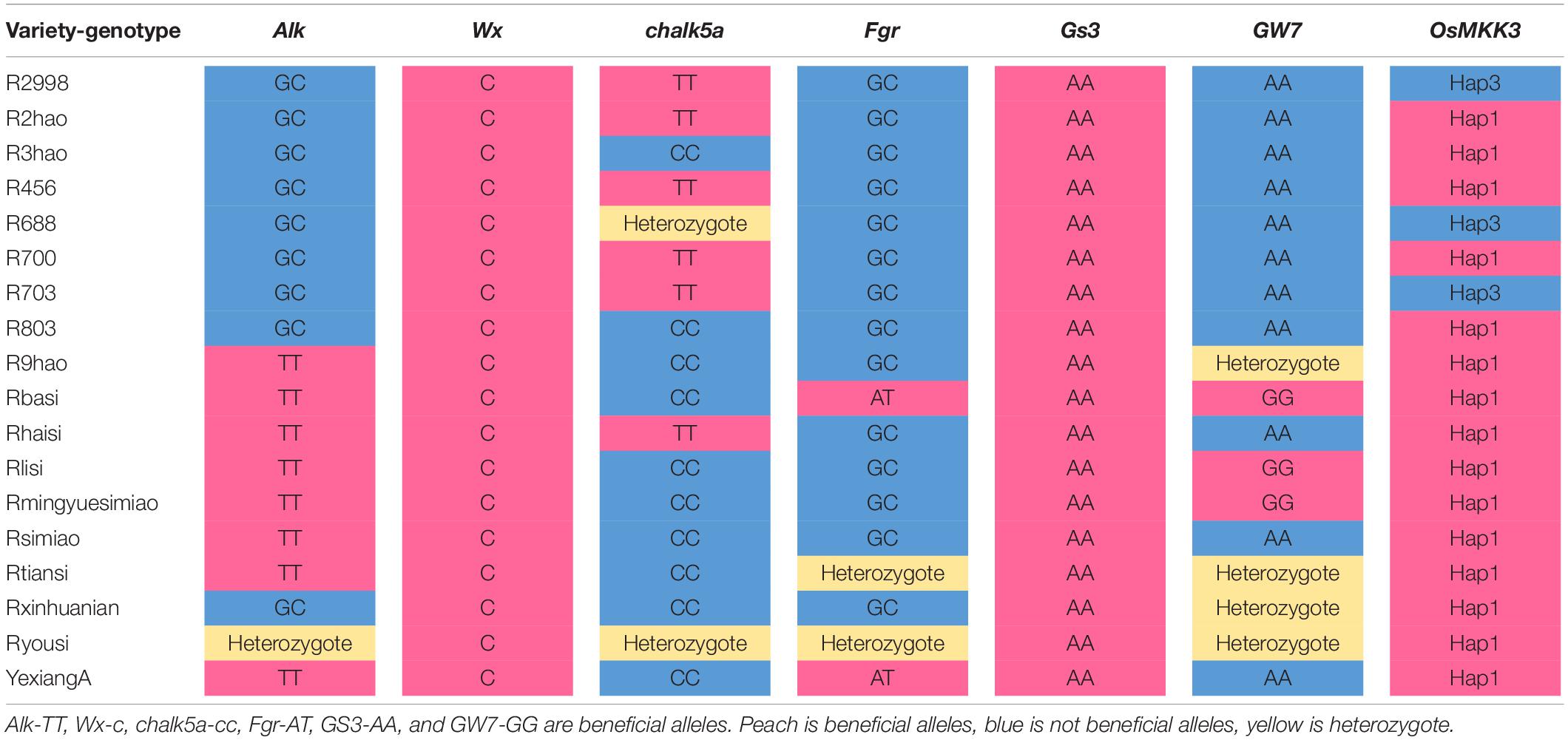
To determine whether OsMKK3 was involved in the control of grain size along with other genes, we analyzed the grain length of landraces using different gene haplotypes. Grain length was shorter with OsMKK3-Hap (not hap1), GS3, and GW8 than in groups with one more dominant genotype, such as OsMKK3-Hap1/gs3/GW8 and OsMKK3-Hap1/GS3/gw8. Grain length increased with OsMKK3-Hap1/gs3/gw8 (Figure 6B). These results indicated that the grain length in indica rice is improved by the presence of beneficial alleles of OsMKK3-Hap1, gs3, and gw8. gs3 is a beneficial allele that was only present in 0.31% of wild rice accessions, 15.71% of landrace accessions, and 72.04% of improved varieties. gw8 was also a beneficial allele that appeared in a few wild rice accessions, landrace accessions, and improved varieties. The OsMKK3 haplotypes were common in wild rice and included OsMKK3-Hap1, OsMKK3-Hap2, OsMKK3-Hap4, and OsMKK3-Hap6. OsMKK3-Hap4 was the most common haplotype in wild rice (Figure 6C). The haplotypes of OsMKK3 were abundant in landraces, and OsMKK3-Hap1 was the most common. OsMKK3-Hap1 was the most common haplotype in improved varieties, but OsMKK3-Hap2 and OsMKK3-Hap3 were also observed. Our analysis indicates that gs3 occurs widely in wild rice, landraces, and improved varieties. The beneficial allele Gw8 has not been widely used in improved varieties. OsMKK3-Hap1 is a beneficial allele that has been used in landraces and improved varieties.
The Aggregation of Many Beneficial Alleles Improves the Quality of Hybrid Rice
To determine the effect of aggregating many beneficial alleles on the quality of hybrid rice, we genotyped a series of hybrid parents of Alk, Wx, chalk5, fgr, gs3, gw7, and OsMKK3. Yexiang hybrid rice varieties were included in 16 combinations of hybrid rice (Supplementary Table 4). Yexiang hybrid rice has a ratio of length to width ranging from 2.6 to 4.1, chalkiness degree ranging from 0.1 to 7.5, gel consistency ranging from 53 to 84 mm, and amylose content ranging from 13 to 22.1%. Thus, Yexiang hybrid rice is high quality; it has a planted area of more than 1,088,666 ha (Supplementary Table 4)6. We determined the genotypes of some important genes in the 16 combinations of Yexiang hybrid rice. Sterile line Yexiang A had the beneficial alleles Alk, Wx, chalk5, fgr, gs3, and OsMKK3-hap1 (Table 2). The restorer lines R2hao, R456, and R700 had the beneficial alleles Wx and gs3. R3hao and R803 had the beneficial alleles Wx, chalk5, gs3, and OsMKK3-Hap1 (Table 2). Rlisi and Rmingyuesimiao had the beneficial alleles Alk, Wx, chalk5, gs3, gw7, and OsMKK3-Hap1 (Table 2). Rbasi had the greatest number of beneficial alleles (Alk, Wx, chalk5, fgr, gs3, gw7, and OsMKK3-Hap1) among the 16 restorer lines (Table 2). The 16 restorer lines contained the three beneficial alleles wx, gs3, and OsMKK3-Hap1/Hap3 (Table 2). The results showed that the quality of hybrid rice can be improved by aggregating more beneficial alleles of important genes. We identified the genotypes Alk, Wx, chalk5, fgr, gs3, gw7, and OsMKK3 in hybrid parents of the good quality hybrid rice variety Meiyou998 in the 2000s and the super hybrid rice variety Wantaiyou3158 in the 2010s to determine how polymerizing beneficial alleles could be used to breed super rice varieties. Meiyou998 had the following features: ratio of length to width 3.1, chalkiness degree 1.7, gel consistency 67 mm, and amylose content 21%. The sterile line MeiA had beneficial alleles of gw8, gs3, and OsMKK3-Hap1 in the 2000s. Minghui63 had the beneficial alleles gs3, wx, chalk5, and OsMKK3-Hap1 in the 1980s. Guanghui998 was bred from Minghui63 in the 1990s with the beneficial alleles gs3, alk, chalk5, and OsMKK3-Hap1 (Figure 7A). Thus, the hybrid rice variety Meiyou998 had the beneficial alleles gw8, alk, gs3, Wx, chalk5, and OsMKK3-Hap1. The results indicated that the quality of the hybrid rice variety Meiyou998 was regulated by gw8, alk, gs3, Wx, chalk5, and OsMKK3-Hap1. In addition, the genotypes of Alk, Wx, chalk5, fgr, gs3, gw7, and OsMKK3 were identified in the parents of Wantaiyou3158. The super hybrid rice variety Wantaiyou3158 had the following features: ratio of length to width 3.4, chalkiness degree 0.5, gel consistency 78 mm, and amylose content 13.3%. Gui99 had the beneficial alleles alk, gs3, Wx, and OsMKK3-Hap3 in the 1980s. In the 2000s, Gui582 was bred from Gui99 with alk, wxb, and OsMKK3-Hap1. In the 2010s, Gui3158 was bred from Gui582 with the beneficial alleles fgr, wx, alk, gs3, chalk, and OsMKK3-Hap1. The sterile line WantaiA had the beneficial alleles fgr, gs3, wx, chalk, and OsMKK3-Hap1 in the 2010s (Figure 7B). Thus, the hybrid rice variety Wantaiyou3158 had the beneficial alleles fgr, wx, alk, gs3, chalk, and OsMKK3-Hap1 in the 2010s. These results shed light on how the quality of hybrid rice has improved from the 1980s to now. The quality of hybrid rice was improved by a greater number of beneficial alleles when OsMKK3-Hap1 was aggregated in the sterile line and restorer line. The presence of beneficial alleles of important genes in homozygous form has greatly improved the quality of hybrid rice. OsMKK3-Hap1 had a particularly pronounced positive effect in improving the quality of hybrid rice.
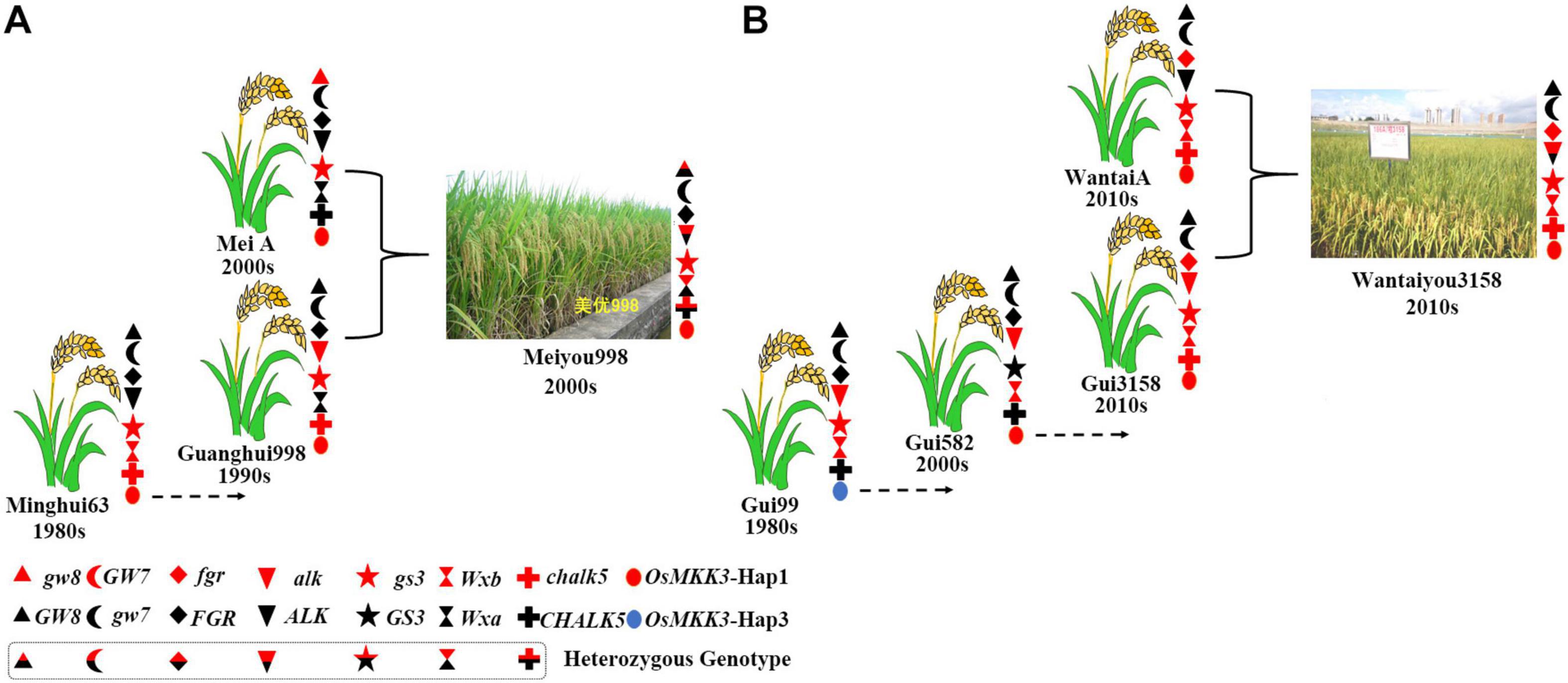
Figure 7. Aggregation of beneficial alleles in hybrid rice in different eras. (A) Aggregation of beneficial alleles in the hybrid rice variety Meiyou998 in the 2000s. (B) Aggregation of beneficial alleles in the hybrid rice variety Wantaiyou3158 in the 2010s.
Discussion
Exploring the genes involved in the production and quality of rice and employing them under an appropriate genetic background remains a major challenge. Grain size is one of the key factors associated with grain weight, which affects rice yield. Several genes for grain size such as GS3, GL3.1, GW6a, OsLG3, OsLG3b, gw8, GS5, SMG1, and OsSPL13 have been cloned from natural rice varieties (Fan et al., 2006; Yan et al., 2011; Wang et al., 2012; Li et al., 2014; Si et al., 2016; Xia et al., 2018; Yu et al., 2018). The genes involved in regulating the quality of rice such as chalk5 (Li et al., 2014), fgr (Bradbury et al., 2005), and wx (Zhou et al., 2016) have been used in breeding by molecular design. In this study, we showed that OsMKK3 is a positive regulator of grain size and chalkiness. It controls the grain length by enhancing cell proliferation in spikelet hulls. Genes with natural variations that regulate important agronomic traits have been used by breeders to improve crop yield and quality.
Identifying Pleiotropy Is Essential for Gene Mining and Utilization
There are two types of genes with genic pleiotropy: one is a gene with a single function but involved in multiple biological processes, such as fgr (Bradbury et al., 2005), wx (Zhou et al., 2016), and chalk5 (Li et al., 2014). Another is a gene with multiple functions that contributes to different traits (Chen et al., 2010), such as GS3 (Takano-Kai et al., 2009), Hd1 (Leng et al., 2020), and GS5 (Li et al., 2011). Additional difficulties associated with utilizing pleiotropic genes stem from the effect of adverse correlations between favorable traits and disadvantageous traits (Li et al., 2018). Identifying the function of new genes and obtaining information on new genes is essential for developing optimal utilization strategies. A loss-of-function mutation of GW8/OsSPL16 in Basmati rice is associated with the formation of a slender grain and better appearance quality (Wang et al., 2012). GL7/GW7 plays a role in grain size and chalkiness; the OsSPL16-GW7 module regulates cell division and rice endosperm (Wang et al., 2015). qSW5/GW5/GSE5 results in grain width diversity in rice and salt stress resistance through an association with the calmodulin protein OsCaM1−1 (Tian et al., 2019). Ghd8/DTH8 likely plays an important role in the signal network of photoperiodic flowering as a novel suppressor as well as in the regulation of plant height and yield potential (Wei et al., 2010). TGW6 encodes a novel protein with indole-3-acetic acid (IAA)-glucose hydrolase activity; control of the IAA supply limits the number of cells and grain length (Ishimaru et al., 2013). FLO2 plays an important role in regulating rice grain size and starch quality by affecting the accumulation of storage substances. Gong et al. (2017) used large populations of hybrid rice for genetic dissections of grain quality traits and recovered only very weak correlations between the grain length-to-width ratio and degree of chalkiness (Chen et al., 2020). Exploration of the genes involved in the regulation of rice yield and quality provides valuable information that could be used in future research. Identifying the function of genes involved in production and quality could also be used to improve the balance between rice yield and quality. In this study, we showed that OsMKK3 encodes a MAP Kinase Kinase 3 that regulates the grain length, chalky rice rate, and chalkiness degree. Overexpression of OsMKK3 in Nip plants increased the quality, grain length, and percentage of grain with chalkiness. The relationship between grain size and chalkiness is complex, which suggests that the genetic architectures of grain size and chalkiness differ. The discovery of new genes provided a genetic resource for rice yield and quality breeding. Based on the observed pleiotropy of genes, the relationships between genes should be an important future direction of breeding efforts.
Breeding Strategies Need to Be Tailored to Specific Breeding Objectives and Based on Mined Genes
Several hundred QTLs of rice yield and quality have been identified. Mining genes and identifying the haplotypes associated with rice yield and quality provide a foundation for molecular design breeding. Leng et al. (2020) detected 19 haplotypes in Hd1 and analyzed five major haplotypes in 123 major rice varieties, as well as the relationship between Hd1 alleles and yield-related traits. Backcrossing of the most preponderant allele Hap16 into the japonica variety Chunjiang06 improved yield without decreasing grain quality. Zhang et al. (2020) found that the null alleles were enriched in modern indica, and they introgressed the null sd1 allele into the elite japonica variety Daohuaxiang to improve the semi-dwarf line. Long-grain alleles of OsLG3b still have much breeding potential for increasing the grain length in indica (Yu et al., 2018). The long-grain allele of OsLG3 might have much potential value for improving the grain length in japonica breeding (Yu et al., 2017). GSE5 regulates grain size by affecting cell proliferation in spikelet hulls, which indicates that natural variation in the promoter region of GSE5 contributes to grain size diversity in rice (Duan et al., 2017). Molecular design breeding has been used to enhance the effects of haplotypes and rice breeding based on the mining of genes.
A previous study showed that rational design is a powerful strategy for addressing the challenges of future crop breeding, particularly the pyramiding of multiple complex traits (Zeng et al., 2017). Molecular design breeding models often differ in the balance that they achieve between yield and quality. Wang et al. (2015) developed new high-yielding indica hybrid rice varieties with improved yield and grain quality. Wang et al. (2012) converted a line with short and wide grains into one with slender grains by pyramiding the non-functional alleles gs3 and gw8 in line HJX74, which substantially improved grain quality. Zeng et al. (2017) pyramided genes that significantly contribute to grain quality and yield from Nip (NPB), Teqing, and LYP9 and developed new elite varieties. Wu et al. (2018) developed near-isogenic lines with a background of 9311, and 9311-OsMADS1lgy3 plants carrying haplotype 2 produced much slender and heavier grains than both 9311-OsMADS1NIP and 9311-OsMADS19311 plants. Li et al. (2020) pyramided the favorable haplotypes of five cloned rice grain quality genes, which resulted in a line with very low amylose content and high yield. Multiple patterns of gene aggregation and interaction require different practical strategies for molecular design breeding. The effect of gene aggregation in different backgrounds is an important research direction.
The increasing requirements of consumers underscore the need to develop both high-yield and superior quality rice (Zhang, 2007; Suwannaporn and Linnemann, 2008). Different breeding strategies have been developed for different breeding objectives. Breeding strategies should focus on yield-associated genes to breed high-yield rice. Gn1a and SCM2 control grain number (Ashikari et al., 2005; Ookawa et al., 2010), TAC1 and sd1 determine plant architecture (Yu et al., 2007), Hd1 increases the biomass of most indica varieties under short-day conditions (Yano et al., 2000), and Ghd7 and Ghd8 regulate grain yield, heading date, and plant height (Xue et al., 2008; Yan et al., 2011). Backbone parents with excellent properties should be used for breeding.
Breeding high-quality rice requires selecting quality genes, such as starch synthesis-related genes (SSRGs) (Tian et al., 2009), GS3, and qSW5, which control grain shape and grain weight (Fan et al., 2006; Shomura et al., 2008). Fgr is associated with the presence of 2-acetyl-1-pyrroline (Bradbury et al., 2005). Chalk5 encodes a vacuolar H+-translocating pyrophosphatase that affects grain chalkiness in rice (Li et al., 2014). Breeding high quality and environmentally adaptable rice requires balancing resistance genes and quality genes. In this study, we showed that OsMKK3 regulates grain size and chalkiness, but this gene still has much breeding potential for improving yield and quality when it is polymerized with other benefited alleles. The diversity of breeding strategies is likely to increase with additional research, which will potentially increase the difficulty of mining genes and characterizing their interactions and collocation patterns.
Conclusion
We cloned and identified OsMKK3, which encodes a MAP kinase kinase that controls grain size and chalkiness. OsMKK3 regulates many genes including OsSPL16, GS5, GIF1, and Wx at the early spike development stage. OsMKK3 has undergone a selective sweep during indica and temperate japonica domestication. Hap4 is the main haplotype in wild rice, and Hap1 is the main haplotype in cultivated rice. In hybrid rice with good yield and quality, OsMKK3-Hap1 was polymerized with beneficial alleles. Our findings confirmed that OsMKK3 enhances crop yield and quality.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: National Center for Biotechnology Information (NCBI) BioProject database under accession number PRJNA769799.
Author Contributions
GDe, DL, GDa, and LG designed and supervised the research. LC and YZ performed the experiments. HG, JL, HW, DQ, and CL analyzed the data. WZ and XY bred the rice variety. YL provided wild rice. YP wrote the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NSFC32060452, 32160447, 32060454, 32160645, and U20A2032), the key project of the Guangxi Natural Science Foundation (2018GXNSFDA281053 and 2019GXNSFBA245006), Natural Science Foundation of Shandong Province (ZR2020MC096), the project of the central government guides the development of local science and technology (GuikeZY20198015, GuikeZY19183020, and GuikeZY18057004), the project of the base and talent (Guike AD17129064, Guike AD20297093, GuiKe 2021JM50, GuiKe AD18281069, and 2018-15-Z06-KF16), the Base Business Project of Guangxi Academy of Agricultural Sciences (2018 QN21 and GuiNongKe 2020YM72), the Project of Guangxi Rice High-quality Breeding Research (Talent small highland for rice QN-08), and the China Postdoctoral Science Foundation (2021M693438).
Conflict of Interest
XY was employed by Guangxi Lvhai Seed Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.784037/full#supplementary-material
Abbreviations
Nip, Nipponbare; QTLs, many quantitative trait loci; PPKL, protein phosphatase kelch; gcHap, gene-coding sequence-haplotype; CaMV35S, tobacco cauliflower mosaic virus; N-OE, Overexpression constructs containing the OsMKK3 CDS from Nip; YXB, Yexiang maintainer line; NT, no transgene line; qRT-PCR, real-time quantitative PCR; Cr, transgenic plants by CRISPR-Cas9 system; DEGs, differentially expressed genes; Hap, haplotypes; MKK3, MAP kinase kinase 3.
Footnotes
- ^ http://www.rmbreeding.cn/Index/s
- ^ http://www.snpway.com/snpdecoder/
- ^ http://www.majorbio.com/
- ^ http://viewer.shigen.info/oryzagenome/
- ^ http://rice.plantbiology.msu.edu/index.shtml
- ^ https://www.ricedata.cn/variety/
References
Alexandrov, N., Tai, S., Wang, W., Mansueto, L., Palis, K., Fuentes, R. R., et al. (2015). SNP-Seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res. 43, D1023–D1027. doi: 10.1093/nar/gku1039
Ashikari, M., Sakakibara, H., Lin, S., Yamamoto, T., Takashi, T., Nishimura, A., et al. (2005). Cytokinin oxidase regulates rice grain production. Science 309, 741–745.
Bradbury, L. M. T., Fitzgerald, T. L., Henry, R. J., Jin, Q., and Waters, D. L. E. (2005). The gene for fragrance in rice. Plant Biotechnol. J. 3, 363–370.
Chen, L., Wang, Q., Tang, M., Zhang, X., Pan, Y., Yang, X., et al. (2021). QTL mapping and identification of candidate genes for heat tolerance at the flowering stage in rice. Front. Genet. 11:621871. doi: 10.3389/fgene.2020.621871
Chen, W., Cheng, Z., Liu, L., Wang, M., You, X., Wang, J., et al. (2010). Molecular basis of trait correlations. Trends Plant Sci. 15, 454–461.
Chen, Y., Zhu, A., Xue, P., Wen, X., Cao, Y., Wang, B., et al. (2020). Effects of GS3 and GL3.1 for grain size editing by CRISPR/Cas9 in rice. Rice Sci. 27, 405–413.
Danecek, P., Auton, A., Abecasis, G., Albers, C. A., Banks, E., DePristo, M. A., et al. (2011). The variant call format and VCFtools. Bioinform. 27, 2156–2158. doi: 10.1093/bioinformatics/btr330
Duan, P., Rao, Y., Zeng, D., Yang, Y., Xu, R., Zhang, B., et al. (2014). SMALL GRAIN 1, which encodes a mitogen-activated protein kinase kinase 4, influences grain size in rice. Plant J. 77, 547–557. doi: 10.1111/tpj.12405
Duan, P., Xu, J., Zeng, D., Zhang, B., Geng, M., Zhang, G., et al. (2017). Natural variation in the promoter of GSE5 contributes to grain size diversity in rice. Mol. Plant 10, 685–694. doi: 10.1016/j.molp.2017.03.009
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Gong, J., Miao, J., Zhao, Y., Zhao, Q., Feng, Q., Zhan, Q., et al. (2017). Dissecting the genetic basis of grain shape and chalkiness traits in hybrid rice using multiple collaborative populations. Mol. Plant 10, 1353–1356. doi: 10.1016/j.molp.2017.07.014
Heim, M., and Meyer, U. A. (1990). Genotyping of poor metabolisers of debrisoquine by allele-specific PCR amplification. Lancet 336, 529–532. doi: 10.1016/0140-6736(90)92086-w
Huang, X., Kurata, N., Wei, X., Wang, Z., Wang, A., Zhao, Q., et al. (2012). A map of rice genome variation reveals the origin of cultivated rice. Nature 490, 497–501. doi: 10.1038/nature11532
Ishimaru, K., Hirotsu, N., Madoka, Y., Murakami, N., Hara, N., Onodera, H., et al. (2013). Loss of function of the IAA-glucose hydrolase gene TGW6 enhances rice grain weight and increases yield. Nat. Genet. 45, 707–711. doi: 10.1038/ng.2612
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Leng, Y., Gao, Y., Chen, L., Yang, Y., Huang, L., Dai, L., et al. (2020). Using Heading date 1 preponderant alleles from indica cultivars to breed high-yield, high-quality japonica rice varieties for cultivation in south China. Plant Biotechnol. J. 18, 119–128. doi: 10.1111/pbi.13177
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, F., Xie, J., Zhu, X., Wang, X., Zhao, Y., Ma, X., et al. (2018). Genetic basis underlying correlations among growth duration and yield traits revealed by GWAS in rice (Oryza sativa L.). Front. Plant Sci. 9:650. doi: 10.3389/fpls.2018.00650
Li, L., Zheng, X., Zhang, X., Xu, K., Song, S., Su, J., et al. (2020). Combination of favorable gene haplotypes plays an important role in rice hybrid grain quality traits based on genome-wide association analysis. bioRxiv doi: 10.1101/2020.06.04.134023
Li, Y., Fan, C., Xing, Y., Jiang, Y., Luo, L., Sun, L., et al. (2011). Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat. Genet. 43, 1266–1269. doi: 10.1038/ng.977
Li, Y., Fan, C., Xing, Y., Yun, P., Luo, L., Yan, B., et al. (2014). Chalk5 encodes a vacuolar H(+)–translocating pyrophosphatase influencing grain chalkiness in rice. Nat. Genet. 46, 398–404. doi: 10.1038/ng.2923
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinform. 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, Q., Han, R., Wu, K., Zhang, J., Ye, Y., Wang, S., et al. (2018). G-protein βγ subunits determine grain size through interaction with MADS-domain transcription factors in rice. Nat. Commun. 9:852. doi: 10.1038/s41467-018-03047-9
Mao, H., Sun, S., Yao, J., Wang, C., Yu, S., Xu, C., et al. (2010). Linking differential domain functions of the GS3 protein to natural variation of grain size in rice. Proc. Natl. Acad. Sci. U.S.A. 107, 19579–19584. doi: 10.1073/pnas.1014419107
Mao, Y., Zhang, H., Xu, N., Zhang, B., Gou, F., and Zhu, J. K. (2013). Application of the CRISPR-Cas system for efficient genome engineering in plants. Mol. Plant 6, 2008–2011. doi: 10.1093/mp/sst121
Newton, C. R., Graham, A., Heptinstall, L. E., Powell, S. J., Summers, C., Kalsheker, N., et al. (1989). Analysis of any point mutation in DNA. The amplification refractory mutation system (ARMS). Nucleic Acids Res. 17, 2503–2516.
Ookawa, T., Hobo, T., Yano, M., Murata, K., Ando, T., Miura, H., et al. (2010). New approach for rice improvement using a pleiotropic QTL gene for lodging resistance and yield. Nat. Commun. 1:132. doi: 10.1038/ncomms1132
Pan, Y., Xu, Z., and Liang, Y. (2018). Genetic structure and core collection of common wild rice (Oryza rufipogon Griff.) in Guangxi. J. Plant Genet. 19, 498–509.
Pan, Y. H., Gao, L. J., Liang, Y. T., Zhao, Y., Liang, H. F., Chen, W. W., et al. (2021). OrMKK3 influences morphology and grain size in rice. J. Plant Biol. Jan 4, 1–14. doi: 10.1007/s12374-020-09290-2
Qi, P., Lin, Y. S., Son, X. J., Shen, J. B., Huang, W., Shan, J. X., et al. (2012). The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating Cyclin-T1;3. Cell Res. 22, 1666–1680. doi: 10.1038/cr.2012.151
Shomura, A., Izawa, T., Ebana, K., Ebitani, T., Kanegae, H., Konishi, S., et al. (2008). Deletion in a gene associated with grain size increased yields during rice domestication. Nat. Genet. 40, 1023–1028. doi: 10.1038/ng.169
Si, L., Chen, J., Huang, X., Gong, H., Luo, J., Hou, Q., et al. (2016). OsSPL13 controls grain size in cultivated rice. Nat. Genet. 48, 447–456. doi: 10.1038/ng.3518
Song, X. J., Kuroha, T., Ayano, M., Furuta, T., Nagai, K., Komeda, N., et al. (2015). Rare allele of a previously unidentified histone H4 acetyltransferase enhances grain weight, yield, and plant biomass in rice. Proc. Natl. Acad. Sci. U.S.A. 112, 76–81. doi: 10.1073/pnas.1421127112
Suwannaporn, P., and Linnemann, A. R. (2008). Rice-eating quality among consumers in different rice grain preference countries. J. Sens. Stud. 23, 1–13. doi: 10.1111/j.1745-459x.2007.00129.x
Takano-Kai, N., Jiang, H., Kubo, T., Sweeney, M., Matsumoto, T., et al. (2009). Evolutionary history of GS3, a gene conferring grain length in rice. Genet. 182, 1323–1334. doi: 10.1534/genetics.109.103002
Tian, P., Liu, J., Mou, C., Shi, C., Zhang, H., Zhao, Z., et al. (2019). GW5-Like, a homolog of GW5, negatively regulates grain width, weight and salt resistance in rice. J. Integr. Plant Biol. 61, 1171–1185. doi: 10.1111/jipb.12745
Tian, Z., Qian, Q., Liu, Q., Yan, M., Liu, X., Yan, C., et al. (2009). Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. U.S.A. 106, 21760–21765. doi: 10.1073/pnas.0912396106
Wang, S., Li, S., Liu, Q., Wu, K., Zhang, J., Wang, S., et al. (2015). The OsSPL16-GW7 regulatory module determines grain shape and simultaneously improves rice yield and grain quality. Nat. Genet. 47, 949–954. doi: 10.1038/ng.3352
Wang, S., Wu, K., Yuan, Q., Liu, X., Liu, Z., Lin, X., et al. (2012). Control of grain size, shape and quality by OsSPL16 in rice. Nat. Genet. 44, 950–954. doi: 10.1038/ng.2327
Wang, W., Mauleon, R., Hu, Z., Chebotarov, D., Tai, S., Wu, Z. L., et al. (2018). Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature 557, 43–49. doi: 10.1038/s41586-018-0063-9
Wei, X., Xu, J., Guo, H., Jiang, L., Chen, S., Yu, C., et al. (2010). DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 153, 1747–1758. doi: 10.1104/pp.110.156943
Wu, K., Xu, X., Zhong, N., Huang, H., Yu, J., Ye, Y., et al. (2018). The rational design of multiple molecular module-based assemblies for simultaneously improving rice yield and grain quality. J.Genet. Genomics 45, 337–341. doi: 10.1016/j.jgg.2018.03.007
Xia, D., Zhou, H., Liu, R., Dan, W., Li, P., Wu, B., et al. (2018). GL3.3, a novel QTL encoding a GSK3/SHAGGY-like Kinase, epistatically interacts with GS3 to produce extra-long grains in rice. Mol. Plant 11, 754–756. doi: 10.1016/j.molp.2018.03.006
Xue, W., Xing, Y., Weng, X., Zhao, Y., Tang, W., Wang, L., et al. (2008). Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40, 761–767. doi: 10.1038/ng.143
Yan, W. H., Wang, P., Chen, H. X., Zhou, H. J., Li, Q. P., Wang, C. R., et al. (2011). A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 4, 319–330. doi: 10.1093/mp/ssq070
Yang, X., Xia, X., Zeng, Y., Nong, B., Zhang, Z., Wu, Y., et al. (2018). Identification of candidate genes for gelatinization temperature, gel consistency and pericarp color by GWAS in rice based on SLAF-sequencing. PLoS One 13:e0196690. doi: 10.1371/journal.pone.0196690
Yano, M., Katayose, Y., Ashikari, M., Yamanouchi, U., Monna, L., Fuse, T., et al. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12, 2473–2484. doi: 10.1105/tpc.12.12.2473
Yu, B., Lin, Z., Li, H., Li, X., Li, J., Wang, Y., et al. (2007). TAC1, a major quantitative trait locus controlling tiller angle in rice. Plant J. 52, 891–898. doi: 10.1111/j.1365-313X.2007.03284.x
Yu, J., Miao, J., Zhang, Z., Xiong, H., Zhu, X., Sun, X., et al. (2018). Alternative splicing of OsLG3b controls grain length and yield in japonica rice. Plant Biotechnol. J. 16, 1667–1678. doi: 10.1111/pbi.12903
Yu, J., Xiong, H., Zhu, X., Zhang, H., Li, H., Miao, J., et al. (2017). OsLG3 contributing to rice grain length and yield was mined by Ho-LAMap. BMC Biol. 15:28. doi: 10.1186/s12915-017-0365-7
Zeng, D., Tian, Z., Rao, Y., Dong, G., Yang, Y., Huang, L., et al. (2017). Rational design of high-yield and superior-quality rice. Nat. Plants 3:17031. doi: 10.1038/nplants.2017.31
Zhang, F., Wang, C., Li, M., Cui, Y., Shi, Y., Wu, Z., et al. (2021). The landscape of gene–CDS–haplotype diversity in rice: properties, population organization, footprints of domestication and breeding, and implications for genetic improvement. Mol. Plant 14, 11–18. doi: 10.1016/j.molp.2021.02.003
Zhang, J., Wang, J., Wang, J., Zhao, Z., Guo, X., Lei, C., et al. (2019). Small Grain and Dwarf 2, encoding an HD-Zip II family transcription factor, regulates plant development by modulating gibberellin biosynthesis in rice. Plant Sci. 288, 110208. doi: 10.1016/j.plantsci.2019.110208
Zhang, L., Bian, Z., Ma, B., Li, X., Zou, Y., Xie, D., et al. (2020). Exploration and selection of elite Sd1 alleles for rice design breeding. Mol. Breed. 40:79.
Zhang, Q. (2007). Strategies for developing Green super rice. Proc. Natl. Acad. Sci. U.S.A. 104, 16402–16409. doi: 10.1073/pnas.0708013104
Zhang, X., Wang, J., Huang, J., Lan, H., Wang, C., Yin, C., et al. (2012). Rare allele of OsPPKL1 associated with grain length causes extra-large grain and a significant yield increase in rice. Proc. Natl. Acad. Sci. U.S.A. 109, 21534–21539. doi: 10.1073/pnas.1219776110
Zhou, H., Wang, L., Liu, G., Meng, X., Jing, Y., Shu, X., et al. (2016). Critical roles of soluble starch synthase SSIIIa and granule-bound starch synthase Waxy in synthesizing resistant starch in rice. Proc. Natl. Acad. Sci. U.S.A. 113, 12844–12849. doi: 10.1073/pnas.1615104113
Keywords: OsMKK3, grain size, rice domestication, haplotype, artificial selection, genetic interaction
Citation: Pan Y, Chen L, Zhao Y, Guo H, Li J, Rashid MAR, Lu C, Zhou W, Yang X, Liang Y, Wu H, Qing D, Gao L, Dai G, Li D and Deng G (2021) Natural Variation in OsMKK3 Contributes to Grain Size and Chalkiness in Rice. Front. Plant Sci. 12:784037. doi: 10.3389/fpls.2021.784037
Received: 27 September 2021; Accepted: 03 November 2021;
Published: 25 November 2021.
Edited by:
Shiv Kumar, International Center for Agricultural Research in the Dry Areas (ICARDA), MoroccoReviewed by:
Deyong Ren, China National Rice Research Institute, Chinese Academy of Agricultural Sciences (CAAS), ChinaWricha Tyagi, Central Agricultural University, India
Copyright © 2021 Pan, Chen, Zhao, Guo, Li, Rashid, Lu, Zhou, Yang, Liang, Wu, Qing, Gao, Dai, Li and Deng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lijun Gao, gaolijun@gxaas.net; Gaoxing Dai, dgx@gxaas.net; Danting Li, lidanting@gxaas.net; Guofu Deng, dengguofu163@163.com
†These authors have contributed equally to this work
 Yinghua Pan
Yinghua Pan Lei Chen
Lei Chen Yan Zhao
Yan Zhao Haifeng Guo3†
Haifeng Guo3† Muhammad Abdul Rehman Rashid
Muhammad Abdul Rehman Rashid