- 1College of Agriculture, Jilin Agricultural University, Changchun, China
- 2Crop Science Post-doctoral Station, Jilin Agricultural University, Changchun, China
Plants have evolved a series of sophisticated defense mechanisms to help them from harm. Ethylene Response Factor (ERF) plays pivotal roles in plant immune reactions, however, its underlying mechanism in maize with a defensive function to Exserohilum turcicum (E. turcicum) remains poorly understood. Here, we isolated and characterized a novel ERF transcription factor, designated ZmERF061, from maize. Phylogenetic analysis revealed that ZmERF061 is a member of B3 group in the ERF family. qRT-PCR assays showed that the expression of ZmERF061 is significantly induced by E. turcicum inoculation and hormone treatments with salicylic acid (SA) and methyl jasmonate (MeJA). ZmERF061 was proved to function as a nucleus-localized transcription activator and specifically bind to the GCC-box element. zmerf061 mutant lines resulted in enhanced susceptibility to E. turcicum via decreasing the expression of ZmPR10.1 and ZmPR10.2 and the activity of antioxidant defense system. zmerf061 mutant lines increased the expression of the SA signaling-related gene ZmPR1a and decreased the expression of the jasmonic acid (JA) signaling-related gene ZmLox1 after infection with E. turcicum. In addition, ZmERF061 could interact with ZmMPK6-1. These results suggested that ZmERF061 plays an important role in response to E. turcicum and may be useful in genetic engineering breeding.
Introduction
Northern corn leaf blight (NCLB) caused by Exserohilum turcicum (E. turcicum) is one of the most destructive fungal diseases of maize in the world (Leach et al., 1977; Galiano-Carneiro and Miedaner, 2017). Heavy infections of NCLB can result in yield losses of greater than 50% (Fajemisin and Hooker, 1974; Raymundo and Hooker, 1981; Perkins and Pedersen, 1987; Ding et al., 2015). Additionally, NCLB may cause a reduction of feeding value and increase the incidence of maize to stalk rot (Hooker et al., 1965; Fajemisin and Hooker, 1974). Host plant resistance is the most common strategy used to control NCLB through the deployment of qualitative and quantitative resistance. Several qualitative Ht genes such as Ht1, Ht2, Ht3, Htn1, HtM, HtP, HtNB, ht4, and rt have already been identified and mapped in maize (Ullstrup, 1963; Gevers, 1975; Hooker, 1977, 1981; Bentolila et al., 1991; Robbins and Warren, 1993; Simcox and Bennetzen, 1993; Carson, 1995; Ogliari et al., 2005; Hurni et al., 2015; Galiano-Carneiro and Miedaner, 2017). However, these Ht genes can quickly get ineffective and result in significant yield losses due to the emergence of new, virulent races (Welz and Geiger, 2000; McDonald and Linde, 2002). Quantitative resistance is considered to provide more durable disease and more useful in breeding process than qualitative resistance (St Clair, 2010). Therefore, it is vital to identify the important genes conferring quantitative resistance and elucidate their molecular mechanism for improving disease resistance of maize cultivars against E. turcicum.
Plants have evolved a series of sophisticated defense mechanisms to cope with the invading pathogens during their life span (Jones and Dangl, 2006; Dangl et al., 2013; Fu and Dong, 2013; Bigeard et al., 2015; Birkenbihl et al., 2017). In plant immunity system, there are two layers of immune responses, called pathogen/microbe-associated molecular pattern (PAMP or MAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI) (Cui et al., 2015; Couto and Zipfel, 2016; Zipfel and Oldroyd, 2017). During PTI and ETI responses, plants trigger a variety of immune reactions including the accumulation of reactive oxygen species (ROS), the activation of mitogen-activated protein kinase (MAPK) signaling pathway, and the expression of pathogenesis-related (PR) genes (Tsuda and Katagiri, 2010; Pieterse et al., 2012; Meng and Zhang, 2013; Baxter et al., 2014). In addition, ETI is often associated with programmed cell death (PCD), also called the hypersensitive response (HR), which occurs at the site of infection and prevents further invasion by the pathogen (Cui et al., 2015).
Transcription factors (TFs) play pivotal roles in plant immune reactions (Singh et al., 2002; Buscaill and Rivas, 2014; Huang et al., 2019). In recent studies, many TFs have been identified according to their conserved structural domain, such as APETALA2/Ethylene Response Factor (AP2/ERF), WRKY, NAC, and bZIP families (Eulgem and Somssich, 2007; Alves et al., 2013; Nuruzzaman et al., 2013; Huang et al., 2016). The AP2/ERF superfamily is a large plant-specific TF family and is defined by the conserved AP2/ERF domain that consists of 58 or 59 amino acids (Ohme-Takagi and Shinshi, 1995). The AP2/ERF superfamily is divided into the ERF family, AP2 family, and RAV family, based on the numbers and characteristics of the AP2/ERF domain (Licausi et al., 2013). The ERF family is further divided into two major subfamilies, DREBs and ERFs (Sakuma et al., 2002). ERF genes have been identified in many species, including Arabidopsis (Nakano et al., 2006), maize (Hao et al., 2020), rice (Nakano et al., 2006), wheat (Zhang et al., 2020), tomato (Yang et al., 2020), pepper (Jin et al., 2018), and soybean (Zhao et al., 2017). It has been well established that the ERF genes can specifically bind to the GCC-box element (AGCCGCC), which is present in the promoters of downstream defense-related genes (Ohme-Takagi and Shinshi, 1995; Fujimoto et al., 2000; Pré et al., 2008; Van der Does et al., 2013).
ERF genes as transcription activators or repressors are involved in modulating disease resistance reactions (Lu et al., 2013; Tian et al., 2015; Li et al., 2018). ERF transcription activators have been shown to positively regulate plant immune response against pathogens. For instance, overexpression of the transcription activators, AtERF1, Octadecanoid-Responsive Arabidopsis 59 (ORA59), AtERF5, AtERF6, AtERF15, and AtERF96 in Arabidopsis resulted in a significantly enhanced resistance against Botrytis cinerea (B. cinerea) through activating the expression of defense related genes, including PLANT DEFENSIN 1.2 (PDF1.2) (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003; Pré et al., 2008; Moffat et al., 2012; Catinot et al., 2015; Zhang et al., 2015). ZmERF105 is a transcription activator, and overexpression of ZmERF105 in maize enhanced resistance to E. turcicum, while the mutant of zmerf105 led to decreased resistance (Zang et al., 2020). Overexpression of TaPIE1, a transcription activator, exhibited significantly increased resistance to Rhizoctonia cerealis, while TaPIE1-underexpressing wheat exhibited the opposite trend (Zhu et al., 2014). In tomato, SlERF.A1, SlERF.B4, or SlERF.C3 functions as a transcription activator and has been found to positively regulate the plant resistance against B. cinerea (Ouyang et al., 2016). In contrast, several ERF transcription repressors that contain an ERF-associated Amphiphilic Repression (EAR) motif in their C-terminal regions negatively regulated the plant resistance to pathogens (Ohta et al., 2000). The transcription repressors AtERF4 and AtERF9 acted as negative regulators of resistance to Fusarium oxysporum and B. cinerea, respectively (McGrath et al., 2005; Maruyama et al., 2013).
ERF genes can coordinately integrate the salicylic acid (SA) and jasmonic acid (JA)/ethylene (ET) signaling pathways or antagonize them, to finely modulate the defense response to pathogens (Berrocal-Lobo and Molina, 2004; Zarei et al., 2011; Zhang et al., 2015, 2016; Wang et al., 2018). AtERF1, AtERF96, or ORA59 has been shown to positively regulate the Arabidopsis defense against B. cinerea through the JA/ET signaling pathway and negatively modulate immunity against Pseudomonas syringae pv. tomato (Pst) DC3000 through the SA signaling pathway (Berrocal-Lobo et al., 2002; Pré et al., 2008; Catinot et al., 2015). It was also found that AtERF15 is involved in resistance to Pst DC3000 and B. cinerea via the SA and JA/ET signaling pathways (Zhang et al., 2015). VqERF112, VqERF114, and VqERF072 acted as positive regulators of plant resistance against Pst DC3000 and B. cinerea through integrating the SA and JA/ET signaling pathways (Wang L. et al., 2020). AtERF11 positively regulated Arabidopsis resistance to Pst DC3000 by directly activating the transcription of AtBT4, which depends on the SA and ET signaling pathways (Zheng et al., 2019).
Recently, several ERF genes have shown to regulate the expression of their target genes through interaction with other proteins (Huang et al., 2016). ORA59 physically interacted with RELATED TO AP2.3 (RAP2.3) to increase the plant resistance against Pectobacterium carotovorum (Kim et al., 2018). GmERF5 and GmERF113 interacted with a BASIC HELIX-LOOP-HELIX TF (GmbHLH) to improve the soybean resistance against Phytophthora sojae (P. sojae) (Dong et al., 2015; Zhao et al., 2017). AtERF6 could interact with AtMPK6 and directly be phosphorylated by AtMPK6. The phosphorylation of AtERF6 increased its protein stability and thus constitutively activated defense genes (Meng et al., 2013; Wang et al., 2013).
A large number of ERF genes have been shown to regulate plant resistance against pathogens in many species, however, its underlying mechanism in maize with a defensive function to E. turcicum remains poorly understood. Previously, we identified a maize ERF gene whose expression was specifically induced by E. turcicum inoculation. Therefore, we isolated and characterized ZmERF061 from maize B73. ZmERF061 was proved to function as a nucleus-localized transcription activator and specifically bind to the GCC-box element. zmerf061 mutant lines resulted in decreased resistance to E. turcicum. In addition, ZmERF061 could interact with ZmMPK6-1. These results suggested that ZmERF061 plays an important role in response to E. turcicum and may be useful in genetic engineering breeding.
Materials and Methods
Plant Materials and Treatments
Exserohilum turcicum (mixed races), the seeds of the maize inbred lines Mo17 (resistant to E. turcicum), Huobai (resistant to E. turcicum), and B73 were obtained from Maize Breeding Team in Jilin Agricultural University, Changchun, China. The seedlings were grown in a glasshouse at 25°C under long-day (16 h light/8 h dark) and 70% relative humidity conditions. For hormone treatments, the seedlings of maize inbred line B73 were sprayed with 0.1 mM of methyl jasmonate (MeJA) and 0.5 mM of SA at the three-leaf stage. For E. turcicum inoculation, the seedlings of maize inbred line Mo17 and Huobai were inoculated with three drops of conidial suspensions at the six-leaf stage according to the method of Zang et al. (2020). The conidial suspensions were adjusted to 1 × 105 conidia ml–1. The leaves were sampled at 0, 2, 5, 10, and 24 h after hormone treatments and were collected at 0, 10, 24, and 72 h after E. turcicum inoculation, respectively. The leaves were frozen in liquid nitrogen and stored at -80°C for the subsequent quantitative real-time polymerase chain reaction (qRT-PCR) analysis. The special primers used for assays are listed in Supplementary Table S1.
qRT-PCR Analysis
Total RNA was extracted from maize leaves using TRIzol reagent (Invitrogen, China) according to the manufacturer’s instruction. Total RNA (1 μg) was used to reverse transcribe into complementary DNA (cDNA) with ReverTra Ace®, qPCR RT Kit (TOYOBO, Japan) following the manufacture’s instruction. qRT-PCR was performed using SYBR Mixture system (TOYOBO, Japan) on a QuantStudio 3 instrument (Thermo, United States). A maize Actin gene, ZmTub (GRMZM2G066191), was used as an internal control to normalize the data. The relative expression levels of genes were analyzed using the 2–Δ Δ CT method. The experimental data were determined using three independent biological repeats, and the significance analysis was performed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean.
Cloning and Bioinformatics Analysis of ZmERF061
The full-length coding sequence of ZmERF061 was isolated from the leaves of B73 by reverse transcription PCR (RT-PCR). The PCR product was cloned into the pMD18-T vector (TaKaRa, China), and the sequence was verified by sequencing (Sangon, China). ERF sequences from different species were downloaded from the NCBI database1, and the phylogenetic tree was built with MEGA 5.0 software using the neighbor joining (NJ) method. The amino acid sequence alignment was performed by DNAMAN software. The nucleic acid sequence and protein sequence of ZmERF061 were analyzed using ExPASy2 database.
Yeast Two-Hybrid Assay
The full-length coding sequence of ZmERF061 was inserted into pGBKT7 vector to generate the bait plasmid (pGBKT7-ZmERF061). The coding sequence of ZmMPK6-1 was cloned into pGADT7 vector to generate prey plasmid (pGADT7-ZmMPK6-1). The prey and bait plasmids were co-transformed into the yeast strain Y2H according to the manufacturer’s instructions (Clontech, United States). After selection on SD/-Trp/-Leu medium for 3 days at 30°C, the transformants were grown on SD/-Trp/-Leu/-His/-Ade medium containing X-α-Gal (20 μg ml–1). Yeast cells carrying the pGBKT7-p53 and pGADT7-SV40 plasmids were used as positive controls, and yeast cells harboring the pGBKT7-Lam and pGADT7-SV40 plasmids were used as negative controls.
Subcellular Localization and Bimolecular Fluorescence Complementation Assays
For subcellular localization of ZmERF061, fusion expression vector ZmERF061–green fluorescent protein (GFP) was constructed by inserting full-length coding sequence of ZmERF061 into the pCAMBIA1300 vector. For bimolecular fluorescence complementation (BiFC) assays, the full-length coding sequences of ZmERF061 and ZmMPK6-1 were fused into pUC-SPYCE and pUC-SPYNE vectors, respectively. The plasmids were transiently expressed in Nicotiana benthamiana (N. benthamiana) leaves by Agrobacterium-mediated method (Liu et al., 2010). The fluorescence signal in cells was photographed by a laser confocal microscope (Leica TCS SP2, Germany).
Yeast One-Hybrid Assay
Yeast one-hybrid assays were used to examine the binding of ZmERF061 to a GCC-box element and were performed according to the Matchmaker Gold Yeat One-Hybrid Library Screening System (Clontech, United States). The full-length coding sequence of ZmERF061 was cloned into the pGADT7 vector containing a GAL4 transcription activation domain, to generate the prey plasmid (pGADT7-ZmERF061). The synthesized DNA fragments harboring four tandem copies of the GCC-box (ATCCATAAGAGCCGCCACTAAAATAAGACCGATCAA) and mGCC (ATCCATAAGATCCTCCACTAAAATAAGACCGATC AA) were cloned into the pAbAi vector as bait plasmids (pAbAi-4 × GCC and pAbAi-4 × mGCC), respectively. The pGADT7-ZmERF061 plasmid was co-transformed with pAbAi-4 × GCC and pAbAi-4 × mGCC plasmids, into Y1H Gold yeast strain, respectively. The co-transformation yeasts were determined on SD/-Leu/-Ura medium supplemented with 200 ng ml–1 of AbA or 300 ng ml–1 of AbA and cultured at 30°C for 3 days. Positive (pGAD-rec-53 + pAbAi-p53) and negative (pGADT7 + pAbAi) controls were processed in the same manner.
Luciferase Activity Assay
The full-length coding sequence of ZmERF061 was cloned into the pGreenII 62-SK vector as effector, and four tandem copies of the GCC-box (ATCCATAAGAGCCGCCACTAAAATAAGACCGATCAA) were ligated into the pGREENII0800-LUC vector as reporter (4 × GCC-LUC). The effector and reporter plasmids were, respectively transferred into Agrobacterium tumefaciens GV3101 and co-transformed into N. benthamiana leaves by Agrobacterium-mediated method (Liu et al., 2010). The LUC activity was determined using commercial dual-LUC reaction reagents (Promega, United States) according to the previous report (Ma et al., 2018). Empty pGreenII 62-SK vector co-transformed with 4 × GCC-LUC was used as the negative control.
Pathogen Response Assays of zmerf061 Mutant Lines
Loss-of-function zmerf061 mutant lines (zmerf061 UFMu mutant, mu1014012) were obtained from the Maize Genetics Cooperation Stock Center. The homozygous mutant lines were obtained from self-fertilizing and identified by PCR. Then, two homozygous T4 zmerf061 mutant lines, named zmerf061-1 and zmerf061-2, were confirmed by qRT-PCR and used for further analyses. Artificial inoculation procedures were performed according to the methods described by Zang et al. (2020). The living ear leaves of zmerf061 mutant lines were infected with E. turcicum agar disks, and the detached leaves from inoculated plants were pictured at 5 days post-inoculation (dpi) with a Nikon D7000 camera for disease assays. The relative lesion area was evaluated using the Photoshop CS3 software according to Cui et al. (2009).
Detection of Enzyme Activities
For the enzyme activity assays, the fresh leaves (about 0.1 g) of W22 wild-type (WT) plants and zmerf061 mutant lines were harvested 24 h after inoculation with E. turcicum conidial suspension, and the plants that were treated with water served as control. The superoxide dismutase (SOD) and peroxidase (POD) activities were measured following the methods that described by Li et al. (2015).
Results
Cloning and Characterization of ZmERF061
ZmERF061 (GenBank Accession no. XM008670839), the ERF gene, was isolated from total RNA of maize by RT-PCR. Sequence analysis revealed that ZmERF061 contains a 1,071-bp open reading frame (ORF) encoding a polypeptide of 356 amino acids (aa) with predicted molecular mass of 37.783 kDa (pI 4.84). The results from searching the database3 indicated that ZmERF061 is located on chromosome 2 and does not have signal peptide. ZmERF061 contains a typical AP2/ERF domain, with conserved alanine (A) and aspartic acid (D) in it, suggesting that it belongs to the ERF family (Sakuma et al., 2002). The AP2/ERF domain contains conserved YRG and RAYD elements, which have been shown to play a vital role in GCC-box binding activity and protein interaction, respectively (Mazarel et al., 2002). ZmERF061 contains a conserved PXXSPXSP (X represents any amino acid) motif in the C-terminal region, which is believed to act as MPK phosphorylation sites (Meng et al., 2013). Additionally, ZmERF061 also possessed a nuclear targeting signal (NLS) sequence “AANKRKRQQL” (Figure 1). Blast search in NCBI revealed that ZmERF061 shares 67.75, 65.38, and 52.75% identity to SbERF104 (Protein ID: XP021319691), SiERF105 (Protein ID: XP004976417), and OsERF105 (Protein ID: XP015635116), respectively (Figure 2A). The phylogenetic tree analysis indicated that ZmERF061 belongs to B3 group (Sakuma et al., 2002). The prediction of the three-dimensional structure based on SWISS-MODEL database revealed that the ZmERF061 has a long C-terminal α-helix (α) surrounded by a three-stranded anti-parallel β-sheet (from β1 to β3) (Figure 2B).

Figure 1. Alignment of the ZmERF061 with other Ethylene Response Factor (ERF) proteins. The sequence alignment was performed using DNAMAN software. The AP2/ERF domain is indicated by a pink box. The one α-helix and three β-sheets are marked above the corresponding sequences. The YRG and RAYD elements are indicated with a black horizontal solid line. The conserved alanine and aspartic acid residues are marked by red triangles. NLS and PXXSPXSP motifs are marked with a black horizontal solid line. OsERF105 (XP015635116) is derived from Oryza sativa, SbERF104 (XP021319691) is derived from Sorghum bicolor, and SiERF105 (XP004976417) is derived from Setaria italica.
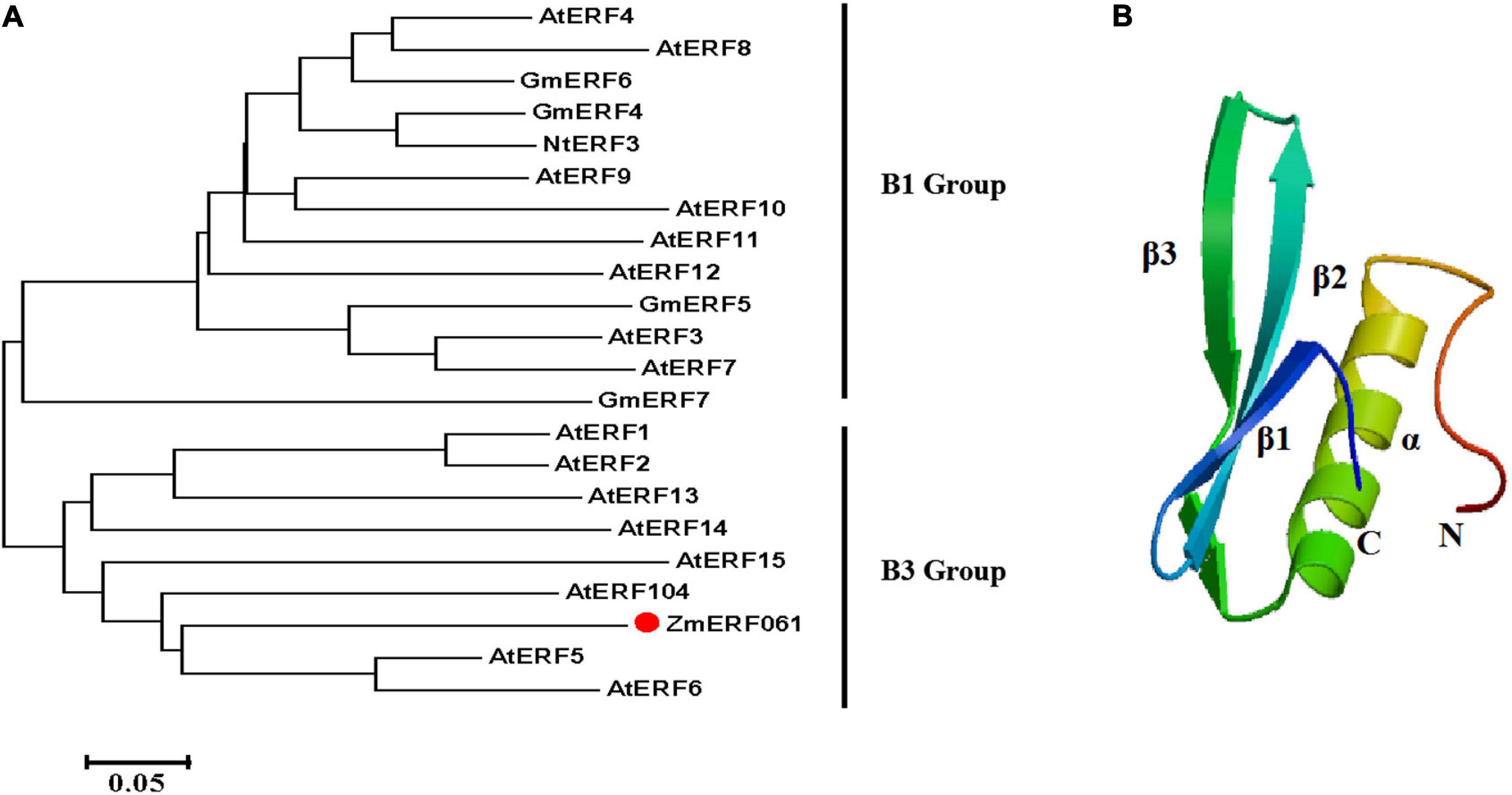
Figure 2. Phylogenetic analysis and three-dimensional structure of ZmERF061. (A) The phylogenetic tree was constructed with the MEGA 5.0 software using the neighbor joining method. ZmERF061 is indicated by the red dot. The accession numbers are as follows: AtERF1 (NP188965), AtERF2 (NP199533), AtERF3 (NP175479), AtERF4 (NP188139), AtERF5 (NP568679), AtERF6 (NP567529), AtERF7 (NP188666), AtERF8 (NP175725), AtERF9 (NP199234), AtERF10 (NP171876), AtERF11 (NP174159), AtERF12 (NP174158), AtERF13 (NP182011), AtERF14 (NP171932), AtERF15 (NP9850162), AtERF104 (NP_200968), GmERF4 (ACE76905), GmERF5 (AEX25891), GmERF6 (AEQ55267), GmERF7 (AEQ55266), and NtERF3 (BAJ72664). (B) Predicted three-dimensional structure of ZmERF061.
Expressions of ZmERF061 Responds to Pathogen Infection and Hormone Induction
To characterize the potential role of ZmERF061 in plant defense reaction, the expression profiles of ZmERF061 in both resistant maize inbred line Mo17 and susceptible maize inbred line Huobai following inoculation with E. turcicum were analyzed by qRT-PCR. The expression level of ZmERF061 in maize inbred Huobai was increased at 10 h but rapidly decreased at 24 h (0.69-fold) and 72 h (0.46-fold) compared with Huobai, however a significant upregulation of ZmERF061 expression is detected in the leaves from 10 to 72 h after E. turcicum in maize inbred line Mo17 (Figure 3A). These results indicated that ZmERF061 may play an important role in maize defense response to E. turcicum.
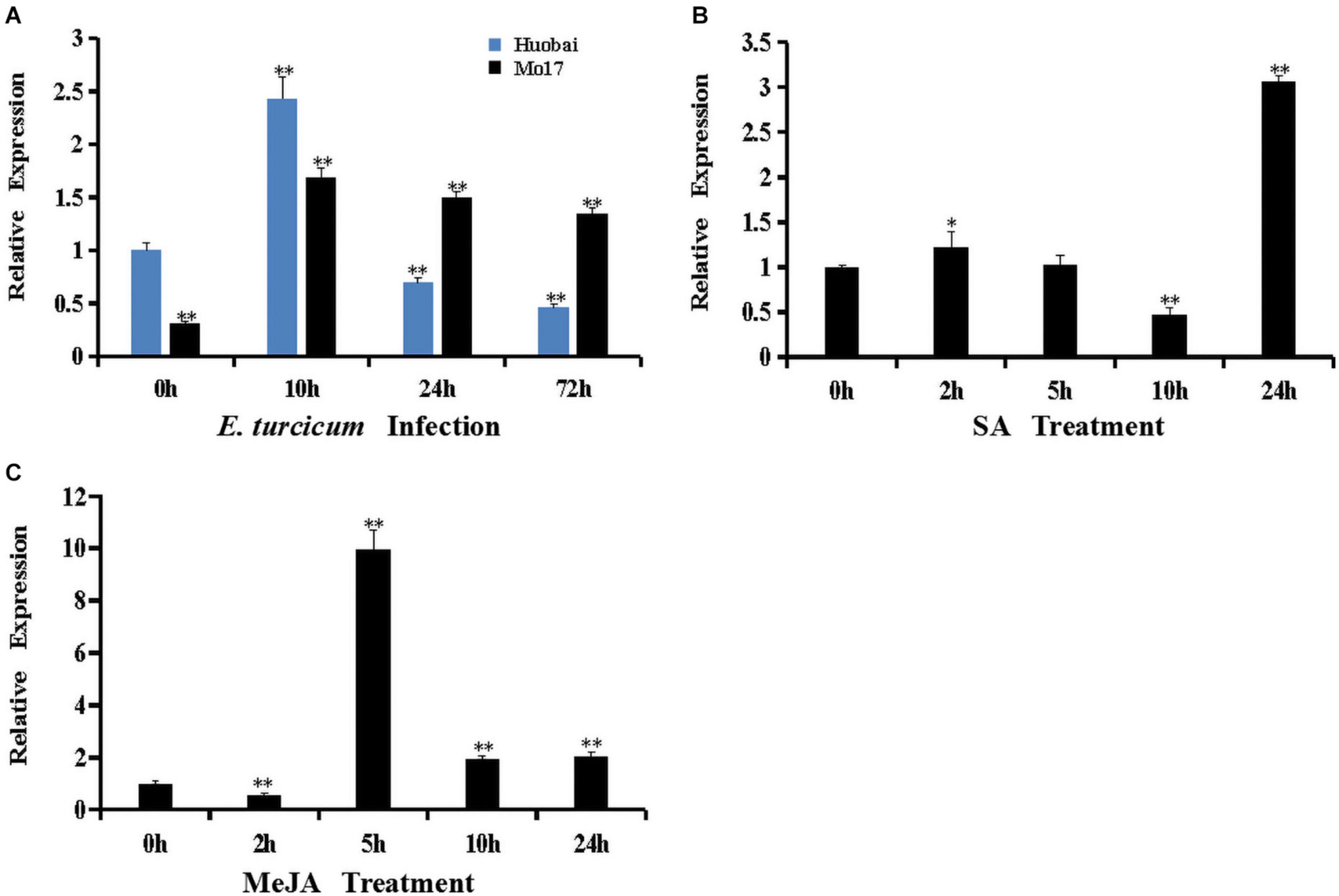
Figure 3. Expression of ZmERF061 was induced by Exserohilum turcicum inoculation and by hormone treatments with salicylic acid (SA) and methyl jasmonate (MeJA). (A) Expression of ZmERF061 in Mo17 and Huobai after inoculation with E. turcicum. The samples were collected at 0, 10, 24, and 72 h after E. turcicum infection. Relative expression levels were compared with Huobai at 0 h. (B) Expression of ZmERF061 in B73 after treatment with 0.5 mM of SA. (C) Expression of ZmERF061 in B73 after treatment with 0.1 mM of MeJA. The samples were collected at 0, 2, 5, 10, and 24 h after the initiation of treatments. Transcript levels were normalized to ZmTub (GRMZM2G066191). The relative expression levels of genes were analyzed using the 2– Δ Δ CT method. The experimental data were determined using three independent biological replicates, and the significance analysis was performed using Student’s t-test (*P < 0.05, **P < 0.01). Bars indicate standard error of the mean.
ERF genes are involved in a variety of defense signaling hormones, such as SA and JA. In our qRT-PCR assay, the transcript levels of ZmERF061 were analyzed in maize inbred line B73 after the application of SA and MeJA treatments. The expression of ZmERF061 was decreased at 2 h, then rapidly increased from 5 to 24 h, and peaked at 5 h by 9.96-fold after the application of MeJA treatment. In SA-treated plants, the expression level of ZmERF061 was lower at 10 h but peaked 3.06-fold higher at 24 h than that in the control. These results indicated that ZmERF061 responds to E. turcicum inoculation and is involved in the JA and SA signaling pathways (Figures 3B,C).
Subcellular Localization of ZmERF061
To elucidate the biological role of ZmERF061, the subcellular localization was analyzed in planta. The coding sequence of ZmERF061 was fused to GFP and was transiently expressed in leaf epidermal cells of N. benthamiana. The ZmERF061–GFP fusion protein was solely localized in nucleus, while the control GFP protein was located in both nucleus and cytoplasm. This result indicated that ZmERF061 is a nucleus-localized protein (Figure 4).
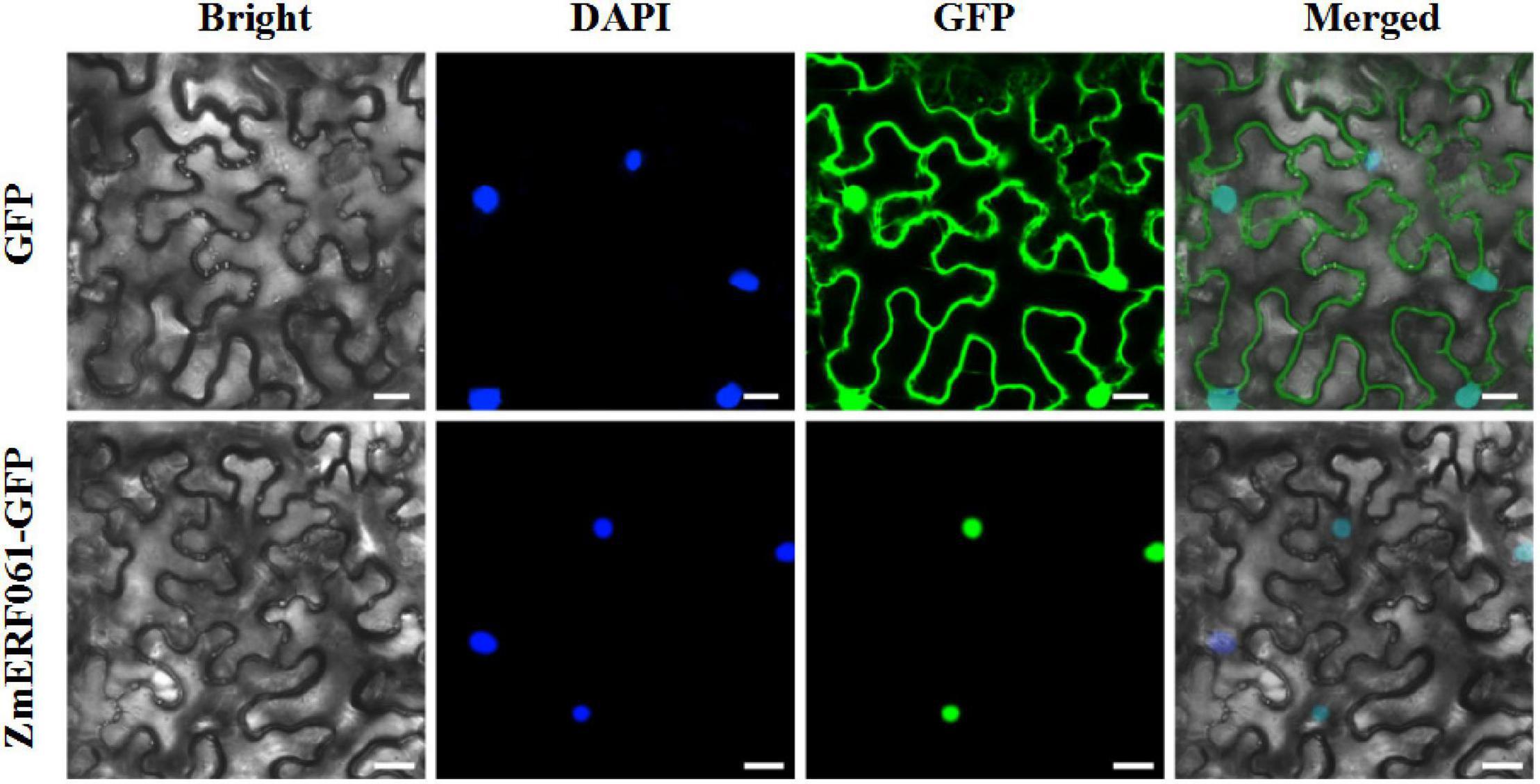
Figure 4. Subcellular localization of the ZmERF061 in Nicotiana benthamiana. Transient expression of the green fluorescent protein (GFP) (upper) and ZmERF061–GFP (bottom) in N. benthamiana. Fluorescence were observed using a laser confocal microscope. Bars = 25 μm.
ZmERF061 Binds to GCC-Box Element and Functions as a Transcriptional Activator
Previous studies have demonstrated that some ERF genes can bind to the GCC-box element (Guo et al., 2004; Franco-Zorrilla et al., 2014; Sun et al., 2016). A yeast one-hybrid assay was performed to investigate the binding characteristics of ZmERF061 to the GCC-box element. The yeast cells transfected with pGADT7-ZmERF061 and pAbAi-4 × GCC could grow on SD/-Leu/-Ura medium containing 200 ng ml–1 of AbA or 300 ng ml–1 of AbA. By contrast, yeast cells harboring the mutant bait cannot grow normally. These data suggested that ZmERF061 specifically binds to the GCC-box element (Figure 5A).
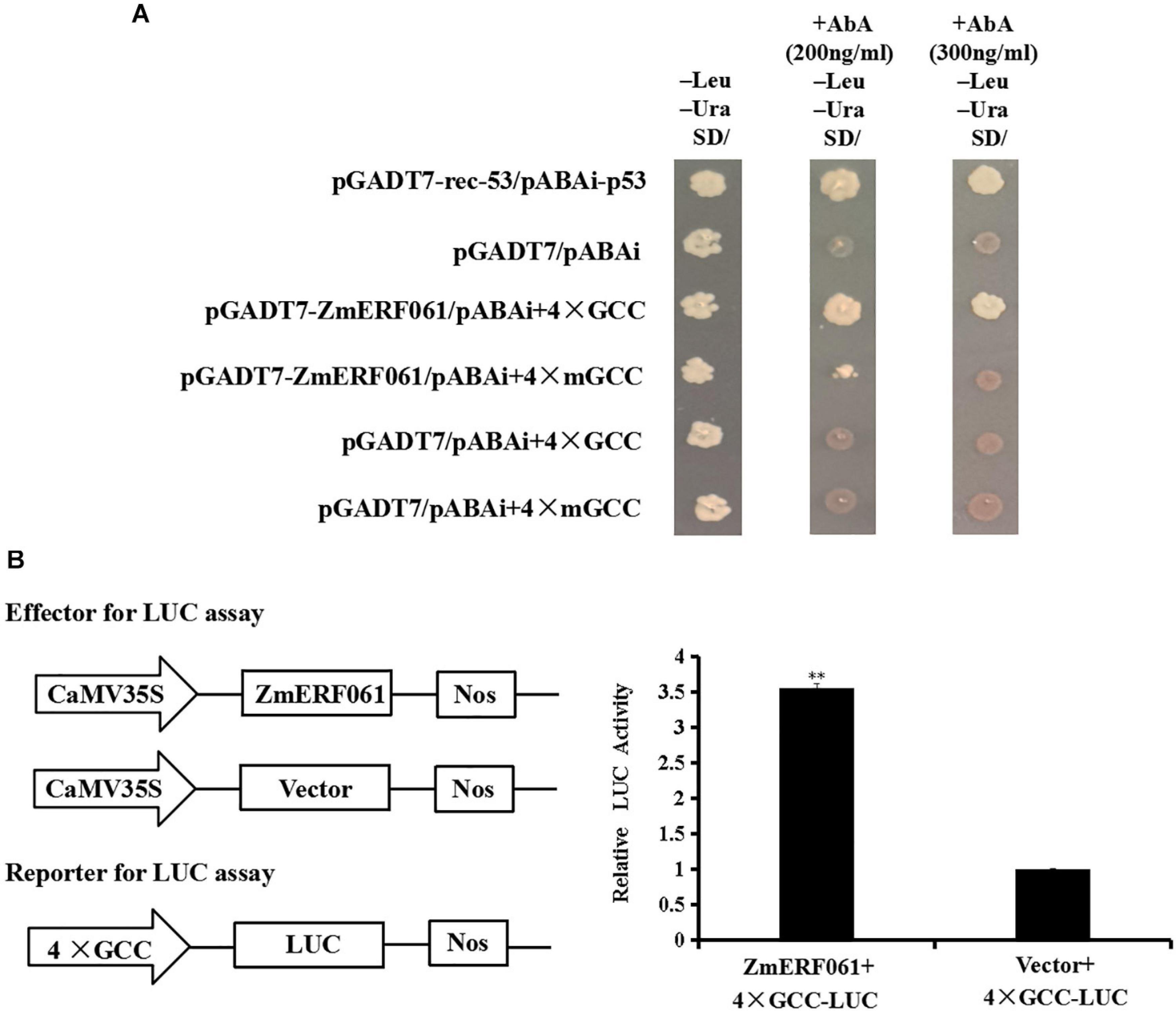
Figure 5. The binding activity of ZmERF061 to GCC-box element and transactivation activity. (A) The binding characteristics of ZmERF061 to GCC-box element. The full-length coding sequence of ZmERF061 was fused with the GAL4 transcription activation domain of vector pGADT7 to generate the prey plasmid (pGADT7-ZmERF061). Four tandem copies of the GCC-box and mGCC-box mutants were cloned into the pAbAi vector and used as bait. The yeast transformants were incubated for 3 days at 30°C on SD/-Leu/-Ura medium with 200 ng ml– 1 of AbA or 300 ng ml– 1 of AbA. Positive (pGAD-rec-53 + pAbAi-p53) and negative (pGADT7 + pAbAi) controls were processed in the same manner. (B) Transcription activity of ZmERF061. The full-length coding sequence of ZmERF061 was cloned into the pGreenII 62-SK vector as effector, and four tandem copies of the GCC-box were ligated into the pGREENII0800-LUC vector as reporter. Relative luciferase activity detected by transient co-transformation with reporter and effector into N. benthamiana. The experiment was performed using three independent biological replicates and analyzed using Student’s t-tests (**P < 0.01). Bars indicate standard error of the mean.
To determine whether ZmERF061 plays a role in transcription activation or inhibition, we performed a transient LUC assay. As shown in Figure 5B, the relative LUC activity of tobacco leaves transfected with the ZmERF061 effector and 4 × GCC-LUC reporter was approximately 3.55-fold higher than that of the control, indicating that ZmERF061 can activate the reporter gene transcription. These results demonstrated that ZmERF061 is able to bind to the GCC-box element and functions as a transcription activator.
zmerf061 Mutant Lines Decreased the Resistance to Exserohilum turcicum
To explore the role of ZmERF061 in mediating the maize resistance to E. turcicum, loss-of-function zmerf061 UFMu mutant lines were obtained from the Maize Genetics Cooperation Stock Center. Homozygous zmerf061 mutant lines were obtained from self-fertilizing and were characterized by PCR. Two homozygous T4 zmerf061 mutant lines, named zmerf061-1 and zmerf061-2, were confirmed by qRT-PCR. qRT-PCR analysis revealed that the expression levels of ZmERF061 have about 0.22-fold and 0.14-fold declines in zmerf061-1 and zmerf061-2 mutant lines, respectively (Figure 6B). WT plants and two independent T4 zmerf061 mutant lines were inoculated with E. turcicum to examine whether ZmERF061 is involved in pathogen resistance. The detached leaves from inoculated plants were pictured with a Nikon D7000 camera for disease assays. At 5 dpi, E. turcicum-caused lesions were significantly smaller on the leaves of the WT plants compared with the zmerf061 mutant lines, indicating that zmerf061 mutant lines decreased resistance to E. turcicum (Figures 6A,C). These results demonstrate that ZmERF061 positively regulates the maize resistance against E. turcicum.
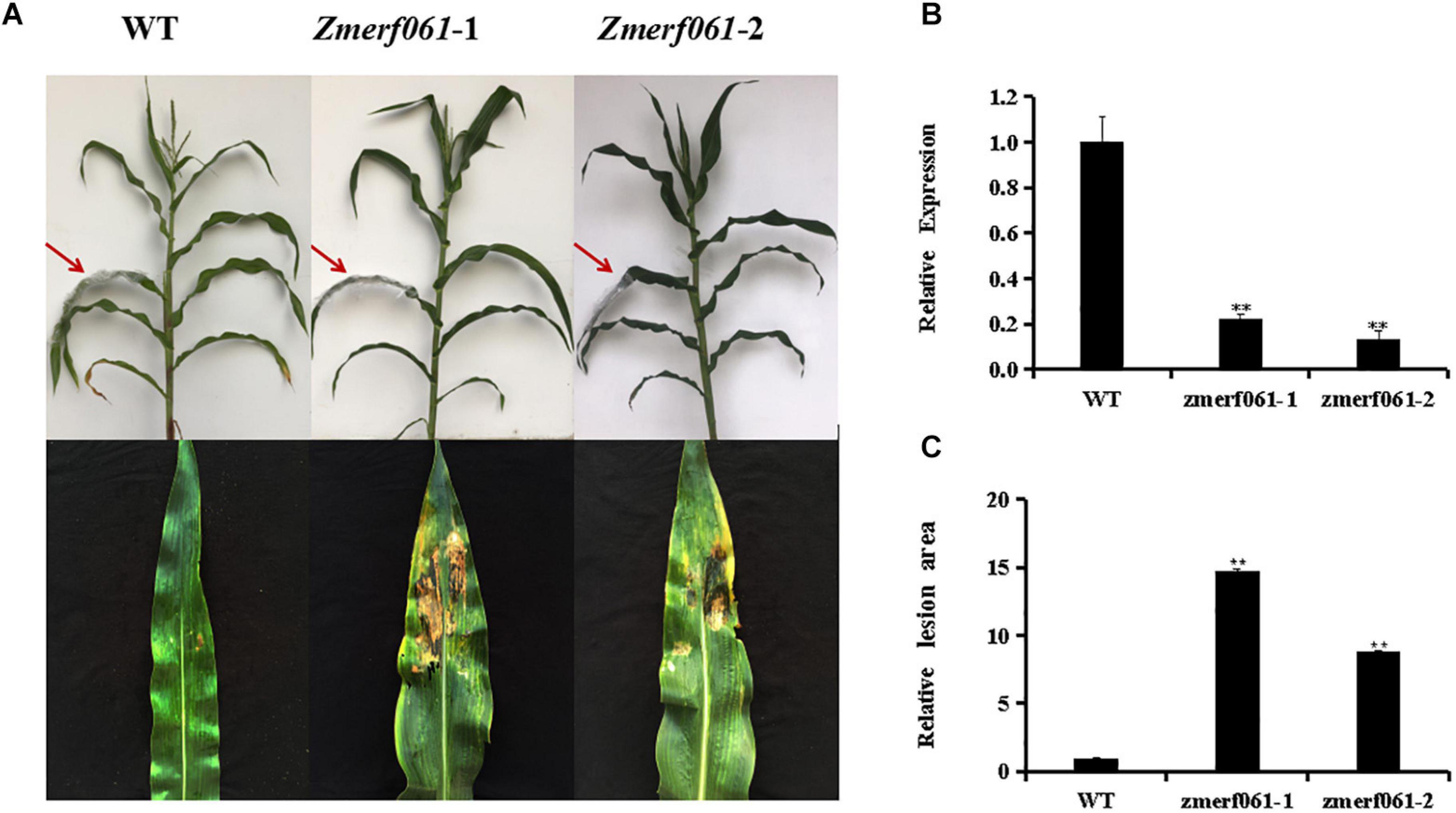
Figure 6. zmerf061 mutant lines decreased the resistance to Exserohilum turcicum. (A) Disease symptom on detached leaves at 5 days post inoculation (dpi). (B) Relative expression analysis of ZmERF061 in zmerf061 mutant lines. (C) The relative lesion areas of ZmERF061 mutant lines and wild-type (WT) leaves at 5 dpi. The experiment was performed using three independent biological replicates and analyzed using Student’s t-tests (**P < 0.01). Bars indicate standard error of the mean.
ZmERF061 Mutant Lines Attenuated Exserohilum turcicum-Induced Defense Response
To further investigate the physiological changes in zmerf061 mutant lines after infection with E. turcicum, we analyzed the activities of two important antioxidant enzymes, including SOD and POD. Under both the mock treatment and at 24 h after infection with E. turcicum, both SOD and POD activities were significantly decreased in zmerf061 mutant lines compared with WT plants (Figures 7A,B). These results indicate that ZmERF061 improves maize resistance against E. turcicum through affecting SOD and POD activities.
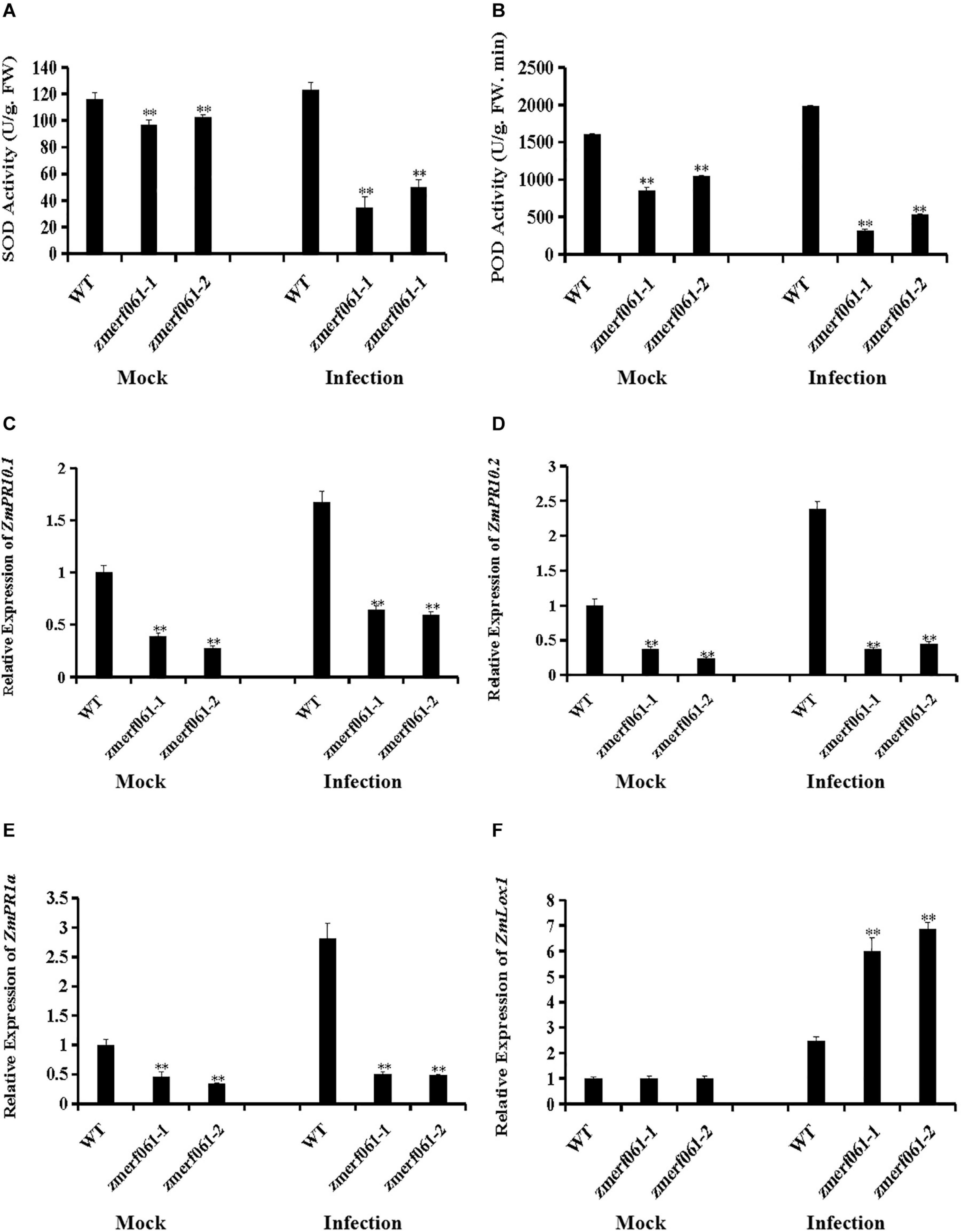
Figure 7. Altered antioxidant enzyme accumulation and defense gene expression in zmerf061 mutant lines after infection with Exserohilum turcicum. (A,B) The activities of superoxide dismutase (SOD) and peroxidase (POD) in zmerf061 mutant lines and wild-type (WT) plants at 24 h after E. turcicum inoculation. (C,D) Expression analysis of ZmPR10.1 and ZmPR10.2 in zmerf061 mutant lines and WT plants at 24 h after E. turcicum inoculation. (E,F) Expression analysis of salicylic acid (SA)-responsive gene ZmPR1a and jasmonic acid (JA)-responsive gene ZmLox1 in zmerf061 mutant lines and WT plants at 24 h after E. turcicum inoculation. The mock-treated WT sample was set to unity. Transcript levels of ZmPR10.1, ZmPR10.2, ZmPR1a, and ZmLox1 were normalized to ZmTub (GRMZM2G066191). The relative expression levels of genes were analyzed using the 2– Δ Δ CT method. The experiment was performed using three independent biological replicates and analyzed using Student’s t-tests (**P < 0.01). Bars indicate standard error of the mean.
To examine whether the increased susceptibility to E. turcicum in zmerf061 mutant lines was associated with transcription changes of defense-related genes, we measured the expression levels of two defense-related genes [i.e., ZmPR10.1 (GRMZM2G112488) and ZmPR10.2 (GRMZM2G112538)] in the WT plants and zmerf061 mutant lines after E. turcicum inoculation. Under both the mock treatment and at 24 h inoculation with E. turcicum, the expression levels of ZmPR10.1 and ZmPR10.2 were significantly lower in zmerf061 mutant lines than in the WT plants (Figures 7C,D). These results showed that ZmERF061 is involved in maize resistance to E. turcicum through regulating the expression of defense-related genes.
To explore whether ZmERF061 is required for SA-induced and JA-induced defense response, the expression levels of SA- and JA-responsive genes were analyzed by qRT-PCR after E. turcicum inoculation. Under both the mock treatment and at 24 h after infection with E. turcicum, the expression levels of SA-responsive gene, ZmPR1a (GRMZM2G465226), were significantly lower in zmerf061 mutant lines than in the WT plants (Figure 7E). By contrast, the expression levels of the JA signaling-related gene, ZmLox1 (GRMZM2G156861), were increased after E. turcicum inoculation in zmerf061 mutant lines compared with the WT plants (Figure 7F). These results demonstrated that ZmERF061 may regulate resistance against E. turcicum via the SA signaling pathway.
ZmERF061 Interacted With ZmMPK6-1
Recently, several B3 group ERF TFs have been shown to interact with MPK6 and involve in MAPK signaling cascade (Bethke et al., 2009; Wang et al., 2013). Amino acid sequence analysis has suggested that ZmERF061 contains putative MPK phosphorylation sites at its C terminus (Fujimoto et al., 2000). Thus, we anticipated that ZmERF061 might interact with ZmMPK6-1 (GRMZM2G002100), which shares high identity to AtMPK6 (At2g43790).
To test this hypothesis, yeast two-hybrid assays were performed. In yeast two-hybrid assay, the yeast cells co-transformed with pGBKT7-ZmERF061 and pGADT7-ZmMPK6-1 developed well on SD (-Trp/-Leu/-His/-Ade) medium containing X-a-Gal (20 μg ml–1), indicating that ZmERF061 can interact with ZmMPK6-1 (Figure 8A).
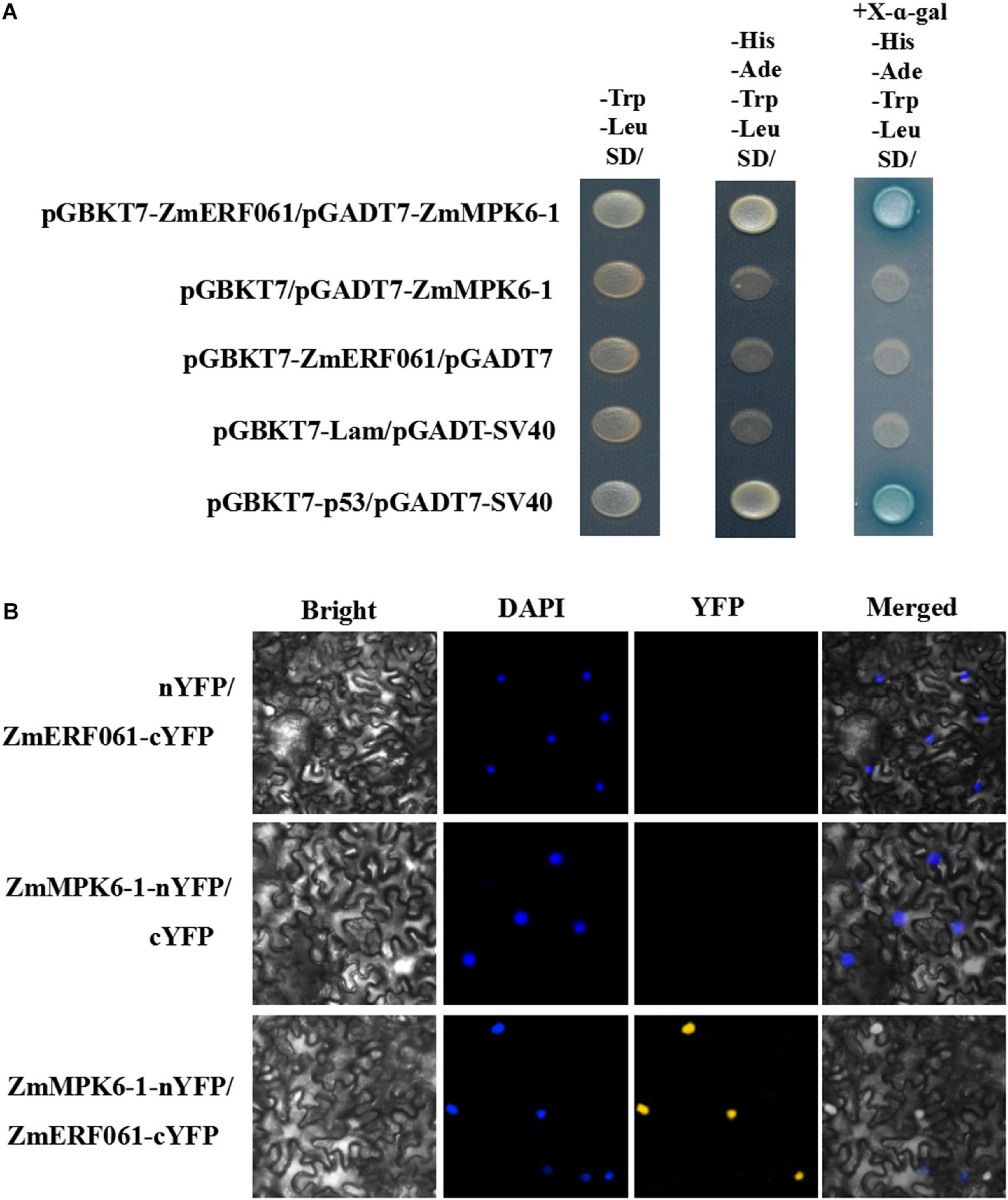
Figure 8. ZmERF061 physically interacts with ZmMPK6-1 in yeast cells and Nicotiana benthamiana. (A) The full-length ZmERF061 interacts with the ZmMPK6-1 in yeast cells. Transformed cells were spotted on the SD (-Leu/-Trp) medium, selective SD (-Leu/-Trp/-His/-Ade) medium, and selective SD (-Leu/-Trp/-His/-Ade) medium containing X-a-Gal (20 μg ml–1). Yeast Y2H gold cells carrying pGBKT7-p53 and pGADT7-SV40 served as a positive control, whereas co-expression of pGBKT7-Lam and pGADT7-SV40 was used as a negative control. (B) Bimolecular Fluorescence Complementation (BiFC) assay verifies the interaction between ZmERF061 and ZmMPK6-1 in N. benthamiana. ZmERF061-cYFP and ZmMPK6-1-nYFP were co-transfected into N. benthamiana. Fluorescence was observed using a laser confocal microscope. Bars = 25 μm.
To confirm the interaction between ZmERF061 and ZmMPK6-1, a BiFC assay was performed in leaf epidermal cells of N. benthamiana. As shown in Figure 8B, yellow fluorescence was displayed in leaf epidermal cells of N. benthamiana co-transformed with N-terminal yellow fluorescent protein (nYFP)-tagged ZmMPK6-1 and C-terminal YFP (cYFP)-tagged ZmERF061. These results indicated that ZmERF061 physically interacts with ZmMPK6-1.
Discussion
ERF TFs play critical roles in response to pathogen infection in plants. In maize, a total of 76 predicted ERF genes have been identified (Hao et al., 2020). In the present study, a novel ERF gene, ZmERF061, was firstly isolated and functionally characterized in maize. Sequence analysis showed that ZmERF061 is characterized as a member of B3 group in the ERF family (Figure 2A). Nucleus-localized ZmERF061 plays a positive role in plant resistance to E. turcicum (Figures 4, 6).
Recent investigations demonstrated that a number of ERF TFs as positive regulators involved in plant defense response against pathogens (Liang et al., 2008; Son et al., 2012; Lu et al., 2013). Constitutive overexpression of AtERF1, ORA59, AtERF5, AtERF6, or AtERF96 in Arabidopsis was shown to confer enhanced resistance to B. cinerea (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003; Pré et al., 2008; Moffat et al., 2012; Catinot et al., 2015). Arabidopsis plants overexpressing AtERF11 resulted in increased Pst DC3000 resistance, and loss-of-function of AtERF11 decreased Arabidopsis resistance to Pst DC3000 (Zheng et al., 2019). Overexpression of AtERF15, AcERF2, VqERF112, VqERF114, or VqERF072 in transgenic Arabidopsis showed improved resistance to Pst DC3000 and B. cinerea (Zhang et al., 2015; Sun et al., 2018; Wang L. et al., 2020). Transgenic Arabidopsis overexpression of MdERF11 led to enhanced resistance against Botryosphaeria dothidea (Wang J. H. et al., 2020). Silencing of SlERF.A1, SlERF.A3, SlERF.B4, or SlERF.C3 in tomato exhibited decreased resistance against B. cinerea (Ouyang et al., 2016). GmERF113 and GmERF5 positively regulated the soybean resistance to P. sojae (Dong et al., 2015; Zhao et al., 2017). Similar to these results, the expression levels of ZmERF061 were specifically induced by E. turcicum inoculation in maize inbred line Mo17 and Huobai (Figure 3A). zmerf061 mutant lines resulted in enhanced susceptibility against E. turcicum (Figure 6). The expression levels of defense-related genes (ZmPR10.1 and ZmPR10.2) were significantly compromised in zmerf061 mutant lines, compared with WT plans in response to E. turcicum infection (Figures 7C,D). These results demonstrated that ZmERF061 positively modulates immunity against E. turcicum in maize. In contrast, some ERF TFs also negatively regulate the resistance to pathogens. For example, OsERF922-overexpressing plants decreased the resistance to Magnaporthe oryzae, while RNAi-mediated suppression OsERF922 showed increased resistance (Liu et al., 2012). Knockout mutants of AtERF9 showed enhanced resistance to B. cinerea (Maruyama et al., 2013). Overexpression of AtERF014 decreased the resistance to B. cinerea (Zhang et al., 2016). AtERF19 negatively regulated Arabidopsis resistances to B. cinerea and Pst DC3000 (Huang et al., 2019).
Previous studies have shown that ERF genes function in plants’ immune response through modulating diverse hormone signaling molecules, such as SA, JA, and ET (Zarei et al., 2011; Zhang et al., 2016). Generally, ERF TFs regulate the Arabidopsis defense against necrotrophic pathogens through the JA/ET signaling pathway and negatively modulate immunity against (hemi)biotrophic pathogens through the SA signaling pathway (Berrocal-Lobo et al., 2002; Pré et al., 2008; Catinot et al., 2015). However, our present study demonstrated that relative expression levels of ZmERF061 were significantly induced by SA and the expression levels of SA-responsive defense gene (ZmPR1a) were decreased in zmerf061 mutant lines after infection with E. turcicum, indicating that ZmERF061 may be involved in E. turcicum resistance via the SA signaling pathway (Figures 3B, 7E). This is different from the functions of AtERF1, AtERF5, AtERF6, ORA59, or AtERF96, which have been reported to play positive roles in disease resistance against B. cinerea by promoting the JA/ET signaling pathway (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003; Pré et al., 2008; Moffat et al., 2012; Catinot et al., 2015). In addition, we demonstrated that the expression levels of JA-responsive defense gene (ZmLox1) were significantly induced in zmerf061 mutant lines after infection with E. turcicum, indicating that ZmERF061 is involved in modulating immune response through antagonizing the SA and JA/ET signaling pathways (Figure 7F). How ZmERF061 modulates the JA and SA signaling pathways to improve plant resistance is an interesting question for future studies.
To ensure survival and negate the adverse effects of ROS, plants have evolved a complete antioxidant defense system to remove extra ROS (Radwan et al., 2010; Hu et al., 2017). POD and SOD are key antioxidant enzymes to help scavenge extra ROS in plants, so that ROS are maintained at a low level to improve the plant resistance against pathogens (Mengiste, 2012). Overexpression of GmSnRK1.1 in soybean showed enhanced resistance to P. sojae through increasing the activity levels of SOD and POD, and GmSnRK1.1-RNAi plants exhibited opposite patterns (Wang et al., 2019). Our studies also showed that both SOD and POD activities were significantly lower in zmerf601 mutant lines after infection with E. turcicum than those in the WT plants (Figures 7A,B), suggesting that ZmERF061 may improve the resistance to pathogen in maize via regulating the plants’ antioxidant defense system.
Recently, some ERF genes have shown to be involved in plant defense reactions through interacting with other proteins (Meng et al., 2013; Dong et al., 2015). GmERF5 and GmERF113 interacted with GmbHLH to improve soybean resistance against P. sojae (Dong et al., 2015; Zhao et al., 2017). ORA59 could enhance Arabidopsis resistance against Pectobacterium carotovorum through interaction with RAP2.3 (Kim et al., 2018). In addition, several ERF genes can interact with MPK genes and are the substrates of pathogen-responsive MAPK signaling cascade (Popescu et al., 2009; Son et al., 2012; Cao et al., 2018). AtERF6 or AtERF104 could interact physically with AtMPK6 and be phosphorylated by AtMPK6 (Bethke et al., 2009; Meng et al., 2013; Wang et al., 2013). The upregulation of AtERF6 or AtERF104 in response to B. cinerea depends on AtMPK6 signaling cascade. Here, we found that ZmERF061 interacted physically with ZmMPK6-1 in yeast cells and N. benthamiana (Figure 8). We speculated that ZmERF061 may play an important role downstream of the ZmMPK6-1 signaling cascade in regulating maize defense against E. turcicum.
An increasing number of evidence indicated that ERF TFs can specifically bind to the GCC-box element to activate the expression of defense-related genes. AtERF1, ORA59, AtERF6, or AtERF96 could directly bind to the GCC-box element in the promoter of AtPDF1.2 to enable its transcription activity (Berrocal-Lobo et al., 2002; Lorenzo et al., 2003; Pré et al., 2008; Moffat et al., 2012; Catinot et al., 2015). GmERF3, TiERF1, or VaERF057 also conferred the ability to bind to the GCC-box element (Liang et al., 2008; Zhang et al., 2009; Sun et al., 2016). In this study, we found that ZmERF061 specifically binds to the GCC-box element by Y1H Gold yeast strain and in planta (Figure 5). These results suggested that ZmERF061 may directly activate the expression of downstream defense-related genes by interacting with the GCC-box element in their promoter regions. It will be helpful to characterize the direct target genes that are regulated by ZmERF061 during immune response against E. turcicum.
In conclusion, we isolated and characterized a novel ERF gene, ZmERF061, which was a nucleus-localized transcription activator and could specifically bind to the GCC-box element. The expression of ZmERF061 was induced by E. turcicum, SA, and MeJA. We also demonstrated that ZmERF061 plays a positive role in modulating plant resistance to E. turcicum through regulating the expression of downstream defense-related genes and antioxidant defense system. Moreover, we found that ZmERF061 can interact with ZmMPK6-1. These data will be important to elucidate the function and regulatory mechanisms of ZmERF061 in maize and provide a reference for breeding disease-resistant varieties.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/ Supplementary Material.
Author Contributions
LJ and WGY conceived and designed the experiments, contributed reagents, materials, and analysis tools. ZZ, ZW, and FZ performed the experiments and drafted the manuscript. WY, JC, and XR analyzed the data. All authors contributed to the article and approved the submitted version.
Funding
This research was supported by the Science and Technology Development Project of Jilin Province (20200402018NC), Doctor Foundation of Jilin Agricultural University (201801), and “13th 5 Year” Science and Technology Project of Education Department of Jilin Province (JJKH20180660KJ).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank the Maize Genetics Cooperation Stock Center for the maize mutants.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2021.630413/full#supplementary-material
Footnotes
- ^ http://www.ncbi.nlm.nih.gov/
- ^ https://www.expasy.org/
- ^ http://www.phytozome.net/maize; http://www.cbs.dtu.dk/serv-ices/signalp/
References
Alves, M. S., Dadalto, S. P., Gonçalves, A. B., De Souza, G. B., Barros, V. A., and Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Baxter, A., Mittler, A. R., and Suzuki, N. (2014). ROS as key players in plant stress signalling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Bentolila, S., Guitton, C., Bouvet, N., Sailland, A., Nykaza, S., and Freyssinet, G. (1991). Identification of an RFLP marker tightly linked to the Ht1 gene in maize. Theor. Appl. Genet. 82, 393–398. doi: 10.1007/BF00588588
Berrocal-Lobo, M., and Molina, A. (2004). Ethylene response factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum. Mol. Plant Microbe. Interact. 17, 763–770. doi: 10.1094/MPMI.2004.17.7.763
Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. doi: 10.1046/j.1365-313x.2002.01191.x
Bethke, G., Unthan, T., Uhrig, J. F., Pöschl, Y., Gust, A. A., Scheel, D., et al. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 8067–8072. doi: 10.1073/pnas.0810206106
Bigeard, J., Colcombet, J., and Hirt, H. (2015). Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 8, 521–539. doi: 10.1016/j.molp.2014.12.022
Birkenbihl, R. P., Liu, S., and Somssich, I. E. (2017). Transcriptional events defining plant immune responses. Curr. Opin. Plant Biol. 38, 1–9. doi: 10.1016/j.pbi.2017.04.004
Buscaill, P., and Rivas, S. (2014). Transcriptional control of plant defence responses. Curr. Opin. Plant Biol. 20, 35–46. doi: 10.1016/j.pbi.2014.04.004
Cao, F. Y., DeFalco, T. A., Moeder, W., Li, B., Gong, Y. C., Liu, X. M., et al. (2018). Arabidopsis ETHYLENE RESPONSE FACTOR 8 (ERF8) has dual functions in ABA signaling and immunity. BMC Plant Biol. 18:211. doi: 10.1186/s12870-018-1402-6
Carson, M. L. (1995). A new gene in maize conferring the “Chlorotic Halo” reaction to infection by Exserohilum turcicum. Plant Dis. 79, 717–720. doi: 10.1094/PD-79-0717
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2015). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate- and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Couto, D., and Zipfel, C. (2016). Regulation of pattern recognition receptor signalling in plants. Nat. Rev. Immun. 16, 537–552. doi: 10.1038/nri.2016.77
Cui, H. T., Tsuda, K., and Parker, J. E. (2015). Effector-triggered immunity: from pathogen perception to robust defense. Annu. Rev. Plant Biol. 66, 487–511. doi: 10.1146/annurev-arplant-050213-040012
Cui, H. W., Yang, Y. L., Li, J. T., Luo, W. F., Miao, A. M., Hu, Z. X., et al. (2009). A faster method for measuring relative lesion area on leaves based on software photoshop. J. Anhui Agric. Sci. 37, 10760–10762.
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 341, 746–751. doi: 10.1126/science.1236011
Ding, J. Q., Ali, F., Chen, G. S., Li, H. H., Mahuku, G., Yang, N., et al. (2015). Genome-wide association mapping reveals novel sources of resistance to northern corn leaf blight in maize. BMC Plant Biol. 15:206. doi: 10.1186/s12870-015-0589-z
Dong, L. D., Cheng, Y. X., Wu, J. J., Cheng, Q., Li, W. B., Fan, S. J., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007 04.020
Fajemisin, J. M., and Hooker, A. L. (1974). Predisposition for diplodia stalk rot in corn affected by three Helminthosporium leaf blights. Phytopathology 64, 1496–1499. doi: 10.1094/Phyto-64-1496
Franco-Zorrilla, J. M., Lopez-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., and Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
Fu, Z. Q., and Dong, X. N. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Galiano-Carneiro, A. L., and Miedaner, T. (2017). Genetics of resistance and pathogenicity in the maize/Setosphaeria turcica pathosystem and implications for breeding. Front. Plant Sci. 8:1490. doi: 10.3389/fpls.2017.01490
Gevers, H. O. (1975). A new major gene for resistance to Helminthosporium turcicum leaf blight in maize. Plant Dis. Rep. 59, 296–299.
Guo, Z. J., Chen, X. J., Wu, X. L., Ling, J. Q., and Xu, P. (2004). Overexpression of the AP2/EREBP transcription factor OPBP1 enhances disease resistance and salt tolerance in tobacco. Plant Mol. Biol. 55, 607–618. doi: 10.1007/s11103-004-1521-3
Hao, L. D., Shi, S. B., Guo, H. B., Li, M., Hu, P., Wei, Y. D., et al. (2020). Genome-wide identification and expression profiles of ERF subfamily transcription factors in Zea mays. Peer J. 8:e9551. doi: 10.7717/peerj.9551
Hooker, A. L. (1977). A second major gene locus in corn for chlorotic-lesion resistance to Helminthosporium turicum. Crop Sci. 17, 132–135. doi: 10.2135/cropsci1977.0011183x001700010035x
Hooker, A. L. (1981). Resistance to Helminthosporium turcicum from Tripsacum floridanum incorporated into corn. Maize Genet. Coop. Newsl. 55, 87–88.
Hooker, A. L., Nelson, R. R., and Hilu, H. M. (1965). Avirulence of Helminthosporium turcicum on monogeni resistant corn. Phytopathol. Notes 55, 462–463.
Hu, Y., Wu, Q. Y., Peng, Z., Sprague, S. A., Wang, W., Park, J., et al. (2017). Silencing of OsGRXS17 in rice improves drought stress tolerance by modulating ROS accumulation and stomatal closure. Sci. Rep. 7:15950. doi: 10.1038/s41598-017-16230-7
Huang, P. Y., Catinot, J., and Zimmerli, L. (2016). Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 67, 1231–1241. doi: 10.1093/jxb/erv518
Huang, P. Y., Zhang, J. S., Jiang, B. E., Chan, C., Yu, J. H., Lu, Y. P., et al. (2019). NINJA-associated ERF19 negatively regulates Arabidopsis pattern-triggered immunity. J. Exp. Bot. 70, 1033–1047. doi: 10.1093/jxb/ery414
Hurni, S., Scheuermann, D., Krattinger, S. G., Kessel, B., Wicker, T., Herren, G., et al. (2015). The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc. Natl. Acad. Sci. U.S.A. 112, 8780–8785. doi: 10.1073/pnas.1502522112
Jin, J. H., Wang, M., Zhang, H. X., Khan, A., Wei, A. M., Luo, D. X., et al. (2018). Genome-wide identification of the AP2/ERF transcription factor family in pepper (Capsicum annuum L.). Genome 61, 663–674. doi: 10.1139/gen-2018-0036
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kim, N. Y., Jang, Y. J., and Park, O. K. (2018). AP2/ERF family transcription factors ORA59 and RAP2.3 interact in the nucleus and function together in ethylene responses. Front. Plant Sci. 9:1675. doi: 10.3389/fpls.2018.01675
Leach, C. M., Fullerton, R. A., and Young, K. (1977). Northern leaf blight of maize in New Zealand: relationship of Drechslera turcia airspora to factors influencing sporulation, conidium development, and chlamydospore formation. Phytopathology 67, 629–636. doi: 10.1094/Phyto-67-629
Li, Z. J., Tian, Y. S., Xu, J., Fu, X. Y., Gao, J. J., Wang, B., et al. (2018). A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Bioch. 132, 683–695. doi: 10.1016/j.plaphy.2018.08.022
Li, Z. J., Zhu, B., Wang, B., Gao, J. J., Fu, X. Y., and Yao, Q. H. (2015). Stress responses to trichlorophenol in Arabidopsis and integrative analysis of alteration in transcriptional profiling from microarray. Gene 555, 159–168. doi: 10.1016/j.gene.2014.10.059
Liang, H. X., Lu, Y., Liu, H. X., Wang, F. D., Xin, Z. Y., and Zhang, Z. Y. (2008). A novel activator type ERF of Thinopyrum intermedium, TiERF1, positively regulates defence responses. J. Exp. Bot. 59, 3111–3120. doi: 10.1093/jxb/ern165
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, D. F., Chen, X. J., Liu, J. Q., Ye, J. C., and Guo, Z. J. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Liu, L. J., Zhang, Y. Y., Tang, S. Y., Zhao, Q. Z., Zhang, Z. H., Zhang, H. W., et al. (2010). An efficient system to detect protein ubiquitination by agroinfiltration in Nicotiana benthamiana. Plant J. 61, 893–903. doi: 10.1111/j.1365-313X.2009.04109.x
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Lu, X., Jiang, W. M., Zhang, L., Zhang, F., Zhang, F. Y., Shen, Q., et al. (2013). AaERF1 positively regulates the resistance to Botrytis cinerea in Artemisia annua. PLoS One 8:e57657. doi: 10.1371/journal.pone.0057657
Ma, Y. N., Xu, D. B., Li, L., Zhang, F., Fu, X. Q., Shen, Q., et al. (2018). Jasmonate promotes artemisinin biosynthesis by activating the TCP14-ORA complex in Artemisia annua. Sci. Adv. 4:eaas9357. doi: 10.1126/sciadv.aas9357
Maruyama, Y., Yamoto, N., Suzuki, Y., Chiba, Y., Yamazaki, K. I., Sato, T., et al. (2013). The Arabidopsis transcriptional repressor ERF9 participates in resistance against necrotrophic fungi. Plant Sci. 213, 79–87. doi: 10.1016/j.plantsci.2013.08.008
Mazarel, M., Puthoff, D. P., Hart, J. K., Rodermel, S. R., and Baum, T. J. (2002). Identification and characterization of a soybean ethylene responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Mol. Plant Microbe. Interact. 15, 577–586. doi: 10.1094/MPMI.2002.15.6.577
McDonald, A. B., and Linde, C. (2002). Pathogen population genetics, evolutionary potential and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. doi: 10.1146/annurev.phyto.40.120501.101443
McGrath, K. C., Dombrecht, B., Manners, J. M., Schenk, P. M., Edgar, C. I., Maclean, D. J., et al. (2005). Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via agenome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 139, 949–959. doi: 10.1104/pp.105.068544
Meng, X. Z., Xu, J., He, Y. X., Yang, K. Y., Mordorski, B., Liu, Y. D., et al. (2013). Phosphorylation of an ERF transcription factor by Arabidopsis MPK3/MPK6 regulates plant defense gene induction and fungal resistance. Plant Cell 25, 1126–1142. doi: 10.1105/tpc.112.109074
Meng, X. Z., and Zhang, S. Q. (2013). MAPK cascades in plant disease resistance signaling. Annu. Rev. Phytopathol. 51, 245–266. doi: 10.1146/annurev-phyto-082712-102314
Mengiste, T. (2012). Plant immunity to necrotrophs. Annu. Rev. Phytopathol. 50, 267–294. doi: 10.1146/annurev-phyto-081211-172955
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., and Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS One 7:e35995. doi: 10.1371/journal.pone.0035995
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Ogliari, J. B., Guimarães, M. A., Geraldi, I. O., and Camargo, L. E. A. (2005). New resistance genes in the Zea mays - Exserohilum turcicum pathosystem. Genet. Mol. Biol. 28, 435–439. doi: 10.1590/S1415-47572005000300017
Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. doi: 10.1105/tpc.7.2.173
Ohta, M., Ohme-Takagi, M., and Shinshi, H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22, 29–38. doi: 10.1046/j.1365-313x.2000.00709.x
Ouyang, Z. G., Liu, S. X., Huang, L. H., Hong, Y. B., Li, X. H., Huang, L., et al. (2016). Tomato SlERF.A1, SlERF.B4, SlERF.C3 and SlERF.A3, members of B3 group of ERF family, are required for resistance to Botrytis cinerea. Front. Plant Sci. 7:1964. doi: 10.3389/fpls.2016.01964
Perkins, J. M., and Pedersen, W. L. (1987). Disease development and yield losses associated with northern leaf blight on corn. Plant Dis. 71, 940–943. doi: 10.1094/PD-71-0940
Pieterse, C. M., Van der Does, D., Zamioudis, C., Leon-Reyes, A., and Van Wees, S. C. (2012). Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28, 489–521. doi: 10.1146/annurev-cellbio-092910-154055
Popescu, S. C., Popescu, G. V., Bachan, S., Zhang, Z. M., Gerstein, M., Snyder, M., et al. (2009). MAPK target networks in Arabidopsis thaliana revealed using functional protein microarrays. Genes Dev. 23, 80–92. doi: 10.1101/gad.1740009
Pré, M., Atallah, M., Champion, A., De Vos, M., Pieterse, C. M., and Memelink, J. (2008). The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 147, 1347–1357. doi: 10.1104/pp.108.117523
Radwan, D. E. M., Fayez, K. A., Mahmoud, S. Y., and Lu, G. Q. (2010). Modifications of antioxidant activity and protein composition of bean leaf due to bean yellow mosaic virus infection and salicylic acid treatments. Acta. Physiol. Plant 32, 891–904. doi: 10.1007/s11738-010-0477-y
Raymundo, A. D., and Hooker, A. L. (1981). Measuring the relationship between northern corn leaf blight and yield losses. Plant Dis. 65, 325–327. doi: 10.1094/pd-65-325
Robbins, W. A. Jr., and Warren, H. L. (1993). Inheritance of resistance to Exserohilum turcicum in ‘PI 209135’, ‘Mayorbela’ variety of maize. Maydica 38, 209–213.
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi- Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and coldinducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Simcox, K. D., and Bennetzen, J. L. (1993). Mapping the HtN resistance gene to the longarm of chromosome 8. Maize Genet. Coop. Newsl. 67, 118–119.
Singh, K. B., Foley, R. C., and Oñate-Sánchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. doi: 10.1016/s1369-5266(02)00289-3
Son, G. H., Wan, J. R., Kim, H. J., Nguyen, X. C., Chung, W. S., Hong, J. C., et al. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe. Interact. 25, 48–60. doi: 10.1094/MPMI-06-11-0165
St Clair, D. A. (2010). Quantitative disease resistance and quantitative resistance loci in breeding. Annu. Rev. Phytopathol. 48, 247–268. doi: 10.1146/annurev-phyto-080508-081904
Sun, X. H., Yu, G., Li, J. T., Liu, J. L., Wang, X. L., Zhu, G. L., et al. (2018). AcERF2, an ethylene-responsive factor of Atriplex canescens, positively modulates osmotic and disease resistance in Arabidopsis thaliana. Plant Sci. 274, 32–43. doi: 10.1016/j.plantsci.2018.05.004
Sun, X. M., Zhao, T. T., Gan, S. H., Ren, X. D., and Fang, L. C. (2016). Ethylene positively regulates cold tolerance in grapevine by modulating the expression of ethylene response factor 057. Sci. Rep. 6:24066. doi: 10.1038/srep24066
Tian, Z. D., He, Q., Wang, H. X., Liu, Y., Zhang, Y., Shao, F., et al. (2015). The potato ERF transcription factor StERF3 negatively regulates resistance to Phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 56, 992–1005. doi: 10.1093/pcp/pcv025
Tsuda, K., and Katagiri, F. (2010). Comparing signaling mechanisms engaged in pattern-triggered and effector-triggered immunity. Curr. Opin. Plant Biol. 13, 459–465. doi: 10.1016/j.pbi.2010.04.006
Ullstrup, A. J. (1963). Sources of resistance of monogenic and polygenic resistance to Helminthosporium turcicum in corn. Plant Dis. Rep. 47, 107–108.
Van der Does, D., Leon-Reyes, A., Koornneef, A., Van Verk, M. C., Rodenburg, N., Pauwels, L., et al. (2013). Salicylic acid suppresses jasmonic acid signaling downstream of SCFCOI1-JAZ by targeting GCC promoter motifs via transcription factor ORA59. Plant Cell 25, 744–761. doi: 10.1105/tpc.112.108548
Wang, J. H., Gu, K. D., Han, P. L., Yu, J. Q., Wang, C. K., and Zhang, Q. Y. (2020). Apple ethylene response factor MdERF11 confers resistance to fungal pathogen Botryosphaeria dothidea. Plant Sci. 291:110351. doi: 10.1016/j.plantsci.2019.110351
Wang, L., Liu, W. D., and Wang, Y. J. (2020). Heterologous expression of Chinese wild grapevine VqERFs in Arabidopsis thaliana enhance resistance to Pseudomonas syringae pv. tomato DC3000 and to Botrytis cinerea. Plant Sci. 293:110421. doi: 10.1016/j.plantsci.2020.110421
Wang, L., Wang, H. Y., He, S. Y., Meng, F. S., Zhang, C. Z., Fan, S. J., et al. (2019). GmSnRK1.1, a Sucrose Non-fermenting-1(SNF1)-related protein kinase, promotes soybean resistance to Phytophthora sojae. Front. Plant Sci. 10:996. doi: 10.3389/fpls.2019.00996
Wang, M. N., Zhu, Y. X., Han, R., Yin, W. C., Guo, C. L., Li, Z., et al. (2018). Expression of Vitis amurensis VaERF20 in Arabidopsis thaliana improves resistance to Botrytis cinerea and Pseudomonas syringae pv. tomato DC3000. Int. J. Mol. Sci. 19:696. doi: 10.3390/ijms19030696
Wang, P. C., Du, Y. Y., Zhao, X. L., Miao, Y. C., and Song, C. P. (2013). The MPK6-ERF6-ROS responsive cis-acting Element7/GCC box complex modulates oxidative gene transcription and the oxidative response in Arabidopsis. Plant Physiol. 161, 1392–1408. doi: 10.1104/pp.112.210724
Welz, H. G., and Geiger, H. H. (2000). Genes for resistance to northern corn leaf blight in diverse maize populations. Plant Breed. 119, 1–14. doi: 10.1046/j.1439-0523.2000.00462.x
Yang, H. H., Shen, F. Y., Wang, H. X., Zhao, T. T., Zhang, H., Jiang, J. B., et al. (2020). Functional analysis of the SlERF01 gene in disease resistance to S. lycopersici. BMC Plant Biol. 20:376. doi: 10.1186/s12870-020-02588-w
Zang, Z. Y., Lv, Y., Liu, S., Yang, W., Ci, J. B., Ren, X. J., et al. (2020). A novel ERF transcription factor, ZmERF105, positively regulates maize resistance to Exserohilum turcicum. Front. Plant Sci. 11:850. doi: 10.3389/fpls.2020.00850
Zarei, A., Körbes, A. P., Younessi, P., Montiel, G., Champion, A., and Memelink, J. (2011). Two GCC boxes and AP2/ERF-domain transcription factor ORA59 in jasmonate/ethylene-mediated activation of the PDF1.2 promoter in Arabidopsis. Plant Mol. Biol. 75, 321–331. doi: 10.1007/s11103-010-9728-y
Zhang, G. Y., Chen, M., Li, L. C., Xu, Z. S., Chen, X. P., Guo, J. M., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, H. J., Hong, Y. B., Huang, L., Li, D. Y., and Song, F. M. (2016). Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Sci. Rep. 6:30251. doi: 10.1038/srep30251
Zhang, H. J., Huang, L., Dai, Y., Liu, S. X., Hong, Y. B., Tian, L. M., et al. (2015). Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 6:686. doi: 10.3389/fpls.2015.00686
Zhang, L., Pan, L., Wu, J., Qiao, L. Y., Zhao, G. Y., Jia, J. Z., et al. (2020). Identification of a novel ERF gene, TaERF8, associated with plant height and yield in wheat. BMC Plant Biol. 20:263. doi: 10.1186/s12870-020-02473-6
Zhao, Y. L., Chang, X., Qi, D. Y., Dong, L. D., Wang, G. J., Fan, S. J., et al. (2017). A novel soybean ERF transcription factor, GmERF113, increases resistance to Phytophthora sojae infection in soybean. Front. Plant Sci. 8:299. doi: 10.3389/fpls.2017.00299
Zheng, X., Xing, J. H., Zhang, K., Pang, X., Zhao, Y. T., Wang, G. J., et al. (2019). Ethylene Response Factor ERF11 Activates BT4 Transcription to Regulate Immunity to Pseudomonas syringae. Plant Physiol. 180, 1132–1151. doi: 10.1104/pp.18.01209
Zhu, X. L., Qi, L., Liu, X., Cai, S. B., Xu, H. J., Huang, R. F., et al. (2014). The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164, 1499–1514. doi: 10.1104/pp.113.229575
Keywords: maize, ZmERF061, Exserohilum turcicum, transcription factor, defense response
Citation: Zang Z, Wang Z, Zhao F, Yang W, Ci J, Ren X, Jiang L and Yang W (2021) Maize Ethylene Response Factor ZmERF061 Is Required for Resistance to Exserohilum turcicum. Front. Plant Sci. 12:630413. doi: 10.3389/fpls.2021.630413
Received: 17 November 2020; Accepted: 14 January 2021;
Published: 09 March 2021.
Edited by:
Zuhua He, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Jin-Song Zhang, Institute of Genetics and Developmental Biology (CAS), ChinaHaitao Cui, Fujian Agriculture and Forestry University, China
Copyright © 2021 Zang, Wang, Zhao, Yang, Ci, Ren, Jiang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Liangyu Jiang, Liangyu0113@163.com; Weiguang Yang, Ywg789@126.com
†These authors have contributed equally to this work
 Zhenyuan Zang1†
Zhenyuan Zang1† Liangyu Jiang
Liangyu Jiang