- Donald Danforth Plant Science Center, St. Louis, MO, United States
Heterologous overexpression of Arabidopsis cellulase 1 (Atcel1) results in enhanced yield, early maturity, and increased biomass in dicotyledonous species like poplar and eucalyptus but has not been demonstrated in monocots. We produced transgenic Setaria viridis accession A10.1 plants overexpressing a monocotyledonous codon optimized (MCO) Atcel1. Agronomic characterization of the transgenic events showed that heterologous overexpression of MCOAtcel1 caused enhanced grain yield, shoot biomass, and accelerated maturation rate in the model grass species S. viridis under growth chamber conditions. The agronomic trait differences observed were consistent with previous reports in dicots but are here described in a monocot species and associated with increased seed yield. Overexpression of Atcel1 in S. viridis was shown to increase the number of panicles and seeds by 24–30%, enhance overall grain yield by up to 26%, and lead to a shoot dry biomass increase of 16–19%. Overexpression also reduced time to plant maturation and senescence by 12.5%. Our findings in S. viridis suggest that manipulation of Atcel1 has potential for developing early-maturing and higher-yielding monocotyledonous biomass crops suitable for climate-smart agriculture.
Introduction
By 2050 the human population is estimated to reach 9.15 billion, with food production having to increase by 60% to meet demand (Long et al., 2015). These goals must be met in the face of challenges presented by a changing climate. Achieving global food security therefore requires the development of staple crop cultivars with superior yield potential and stress adaptation. Wheat, rice, maize, pearl millet, and sorghum provide approximately 44% of the calories consumed per capita throughout the world (Reynolds et al., 2016). Securing higher yields through the enhancement of components such as grain yield, grain number, panicle number, and panicle length in these primary cereal crops is critical (Beche et al., 2014). Early maturity is a desirable trait as it reduces potential for exposure to post-anthesis biotic and abiotic stresses, thereby providing farmers with greater flexibility when faced with uncertain growing conditions (Vadez et al., 2012).
Heterologous overexpression of plant Endo-1, 4-β- glucanase (EGase) had been reported to result in enhanced yield, early maturity, and increased biomass in poplar and eucalyptus (Shani et al., 2004; Ledford, 2014). Plant EGases are included in the glycosyl hydrolase family 9 (GH9) and divided into three distinct classes (Urbanowicz et al., 2007). At least 25 EGases have been identified in Arabidopsis thaliana, categorized into membrane-anchored (class A), cell wall-targeted (class B), and carbohydrate binding (class C) sub-families (Urbanowicz et al., 2007). Arabidopsis cellulase 1 (Atcel1) belongs to GH9B subfamily and is thought to be involved in cell wall loosening that drives anisotropic cell enlargement; a fundamental process required for plant growth (Lipchinsky, 2013). Heterologous expression of Atcel1 in poplar and eucalyptus resulted in increased height, leaf area, biomass, dry weight, and accelerated maturation (Shani et al., 2004; Ledford, 2014). Similarly, expression of poplar cellulase PaPopCel1 promoted enhanced biomass and early maturity in Arabidopsis and sengon (Paraserianthes falcataria) (Park et al., 2003; Hartati et al., 2008). No such reports exist for overexpression of cel1 in the cereals. We performed studies in the monocot model Setaria viridis to investigate if heterologous overexpression of Atcel1 can promote similar enhancements in yield, biomass, and maturation rate in a monocotyledon.
Materials and Methods
Identification of Atcel1 (AT1G70710.1) Homologs in Setaria italica
Bioinformatics analysis1 of the S. italica genome was conducted using AT1G70710.1 amino acid sequence as a query to identify Atcel1 homologs. Additional phylogenetic analysis was performed using phylogeny.fr (Dereeper et al., 2008) to illustrate the parallel relationship between Atcel1 and its closest Setaria homolog.
Gene Cloning and Construction of Transformation Vectors
Sicel1 cDNA (GenBank ID Si016976m.g), the closest monocot homolog of AT1G70710.1 (Atcel1) was cloned from panicle tissue of 6-week old S. italica, Yugu-1 accession. Total RNA was isolated using TRIzol reagent (Ambion, Austin, United States) following manufacturer’s protocol and quantified using the Nanodrop2000 (Thermo Fisher Scientific, Austin, United States). One microgram of RNA was treated with DNase I (Ambion, Austin, United States) and reverse transcribed using the SuperScript III first strand cDNA synthesis kit (Invitrogen, Waltham, United States). Full-length cDNA (1509 bp) was PCR amplified by high fidelity TaKaRa (Clontech, Mountain View, United States) Taq polymerase, employing a two-step TaKaRa protocol (98°C for 3 min, 30 cycles of 98°C for 10 s and 68°C for 2 min, and 72°C for 10 min) using the specific primers Sicel1F (5′ATG CCG GCG GCG GTG CGG AG-3′) and Sicel1R (5′TCA GTC GCC GCC CGA CTC GG-3′). The 1509 bp full-length transcript was verified by Kpn1-HF restriction digestion (New England BioLabs, Ipswich, United States) and cloned into the pCR8/GW vector (Invitrogen, Waltham, United States), following the Gateway® cloning protocol. After sequence confirmation (Eurofins Scientific, St. Louis, United States) the pCR8/GW/Sicel1 clone was used as a template to recombine the gene insert into the GW® compatible pANIC10A overexpression vector (Mann et al., 2012), using LR clonase enzyme (Invitrogen, Waltham, United States). Similarly, a 1500 bp MCOAtcel1 coding sequence was designed and obtained from GenScript Inc., and transferred to the pANIC10A/MCOAtcel1 vector employing the GW® cloning strategy described above. The successful cloning of Sicel1 and MCOAtcel1 into pANIC10A vector was confirmed by sequencing (Eurofins Scientific, St. Louis, United States).
Production of Transgenic S. viridis Plants
The coding sequences of Sicel1 (1509 bp) and monocot codon optimized (MCO) Atcel1 (1482 bp) were fused to the constitutive maize ubiquitin (ZmUBI) promoter and cloned into binary vector pANIC10A (Mann et al., 2012). Sequence confirmed pANIC10A/MCOAtcel1 and pANIC10A/Sicel1 constructs plus the empty vector control (EVC) pANIC10A/EVC were transformed into Agrobacterium tumefaciens strain AGL1 and used to generate transgenic plants of S. viridis, accession A10.1 (Van Eck and Swartwood, 2015). Hygromycin resistant T0 plantlets were transferred to soil when at least 4 cm tall with well-developed roots system (Van Eck and Swartwood, 2015). Plantlets were washed in water to disassociate growth media from the roots, planted in 3-inch pots filled with Metro-Mix 360-soil mix (Hummert International, St. Louis, United States) and watered until saturated. A plastic dome was placed over the plantlets for 3 days after which plants were grown in a Conviron (Pembina, United States) growth chamber at 12 h day/12 h night photoperiod, 31°C day and 22°C night temperature with a relative humidity of 50–60%. Plants were watered daily and fertilized twice a week with 100 ppm of N. T0 plantlets were grown to maturity and selfed to obtain T1 seeds for further characterization. T1 generation S. viridis seeds were subjected to liquid smoke treatment to break dormancy (Sebastian et al., 2014) and propagated along with wild-type A10.1 seeds under the conditions described above.
Analysis of Transgenic Plants
T1 plants were genotyped 10–15 days after germination. Three hundred milligrams of leaf tissue obtained from the third fully expanded leaf was collected, placed in a 1.5 ml screw top tube and stored at −80°C. Frozen leaf tissue was crushed using TissueLyser II (QIAGEN, Germantown, United States) for 90 s at 30 Hz. One hundred milligrams of crushed leaf tissue was used for DNA isolation following the CTAB method (Fulton et al., 1995), and the remaining tissue powder saved for RNA extraction. Isolated DNA was re-suspended in 50 μl sterile Milli-Q water, quantified with a NanoDrop 2000 (Thermo Fisher Scientific, Austin, United States) and used for PCR analysis and Southern blot hybridization. RNA isolation from 200 mg of crushed leaf tissue was performed using the RNeasy Plant Mini Kit (QIAGEN, Germantown, United States), isolated RNA was quantified using the Nanodrop2000, and then treated with DNase I (Thermo Fisher Scientific, Austin, United States) following manufacturer’s protocol. One microgram of DNase I treated RNA was used to synthesize cDNA employing the SuperScript III first strand cDNA synthesis kit (Invitrogen, Waltham, United States) and stored at −20°C.
Setaria viridis plant lines transformed with pANIC10A/MCOAtcel1, pANIC10A/Sicel1, and pANIC10A/EVC were genotyped for presence of the hygromycin phosphotransferase (hpt) marker gene using primers hptF (5′-GAA CTC ACC GCG ACG TCT GTC GAG-3′) and hptR (5′-AAT GAC CGC TGT TAT GCG GCC AT-3′). Additionally, for plants transformed with pANIC10A/MCOAtcel1, presence of the MCOAtcel1 transgene was determined using primers MCOAtcel1F (5′-GAA GTG GGG CAC TGA CTA CC-3′) and MCOAtcel1R (5′- GTA TCC ATG TCC TCA GGC CG-3′). PCR amplification was performed in a 15 μl volume containing 1 μl of 1:5 diluted cDNA template, 1× Choice Taq Blue Master mix (Denville Scientific Incorporated, Holliston, United States), nuclease-free water, and 0.35 μM each of forward and reverse primers. The PCR conditions were: initial denaturation (94°C for 5 min), 35 cycles of amplification (denaturation 94°C for 45 s, annealing 55°C for 30 s, elongation 72°C for 30 s), and final elongation (72°C for 10 min). PCR products were resolved on 1.5% ethidium bromide stained agarose gels.
Southern Blot Analysis
Non-radioactive labeled DIG (Digoxigenin) Southern blot hybridization was used to determine T-DNA insert copy number in S. viridis plant lines (Brutnell et al., 2010). Ten micrograms of pooled DNA from hpt PCR positive T1 families/event for each construct was digested with Mfe1 restriction enzyme at 37°C for 4 h. Digested DNA was resolved on 1% agarose gel and transferred to a positively charged nitrocellulose membrane overnight, UV-crosslinked, and prepared for hybridization (pre-hybridization) using the DIG Easy Hyb (Roche, Branford, United States) solution. A probe was prepared complimentary to the hpt gene with DIG-labeled dNTPs and pANIC10A plasmid DNA as template, using forward primer 5′-TGGCAAACTGTGATGGACGA-3′ and reverse primer 5′- GGTTTCCACTATCGGCGAGT-3′. The membrane was hybridized overnight with the DIG-labeled hpt probe, washed with sequential low (2X SSC, 0.1% SDS) and high (0.5 X SSC, 0.1% SDS) stringency washes, and blocked with 1X blocking buffer (Roche, Branford, United States). The membrane was treated with anti-DIG AP Fab Fragments (Roche, Branford, United States) prepared in blocking buffer and washed three times with 1X washing buffer (1X maleic acid buffer, Tween 20). Detection was performed using the Tropix CDP-Star reagent (Applied Biosystems, Foster City, United States) for 5 min, undeveloped film exposed overnight and developed for imaging.
Identification of Single Copy T-DNA Integration Events in T2–T3 Generations
Both single and multiple T-DNA copy insertion lines identified by Southern blot hybridization were tracked through sexual generations using Mendelian segregation ratios to produce homozygous lines. Mendelian segregation ratios of T1 families were determined by PCR screening for presence of hpt and tested for goodness of fit for a 3:1 segregation by chi squared analysis. Progeny from transgenic lines possessing single T-DNA integrations were PCR tested for hpt marker segregation in the subsequent T2 generation to identify homozygous lines. Single copy homozygous families were advanced, genotyped, and phenotyped to the T3 generation.
Phenotypic Characterization of Transgenic Events
T1 families from three independent transgenic events plus EVC were selfed to obtain single T-DNA copy homozygous families, propagated, and subjected to phenotypic assessment up to the T3 generation. In each generation the S. viridis families were grown for 7 weeks and subjected to no watering for 1 week prior to harvesting. Plant growth parameters including total grain weight, grain number, panicle number, and primary panicle length were documented as a measure of agronomic performance at maturity. Plant development parameters including onset of flag leaf, inflorescence initiation, anthesis, seed setting, and senescence were documented in the T3 generation. For shoot dry matter content (DMC) determination, above ground parts of mature plants were harvested immediately after seed collection, placed in paper bags, and oven dried at 30°C for 1 week before assessing for DMC.
Data Analysis
Statistical differences in agronomic traits between the transgenic T3 families and controls were analyzed by a student t- test and the data presented as mean ± standard error of mean (SEM), with ∗p ≤ 0.05, ∗∗p ≤ 0.01 and ∗∗∗p ≤ 0.001.
Results
Identification of Atcel1 (AT1G70710.1) Homologs in Setaria italica
Phylogenetic analysis of endo-1, 4-β-glucanase gene families in S. italica using AtCEL1 amino acid sequence as a query, revealed two closely related genes, Si016976m.g and Si006281m.g, with pairwise scores of 71.34 and 70.93%, respectively (Figure 1). BLASTP analysis indicated that Si016976m.g (Score: 703.7) was more closely related to Atcel1 than Si006281m.g (Score: 688.3). Additional analysis using phylogeny.fr illustrated a parallel relationship between Atcel1 and its closest homolog (At1G23210.1), compared to Si016976m.g and Si006281m.g. Si016976m.g is less evolutionarily divergent than Si006281m.g, and more similar to Atcel1. Thus, Si016976m.g was henceforth referred to as Sicel1.
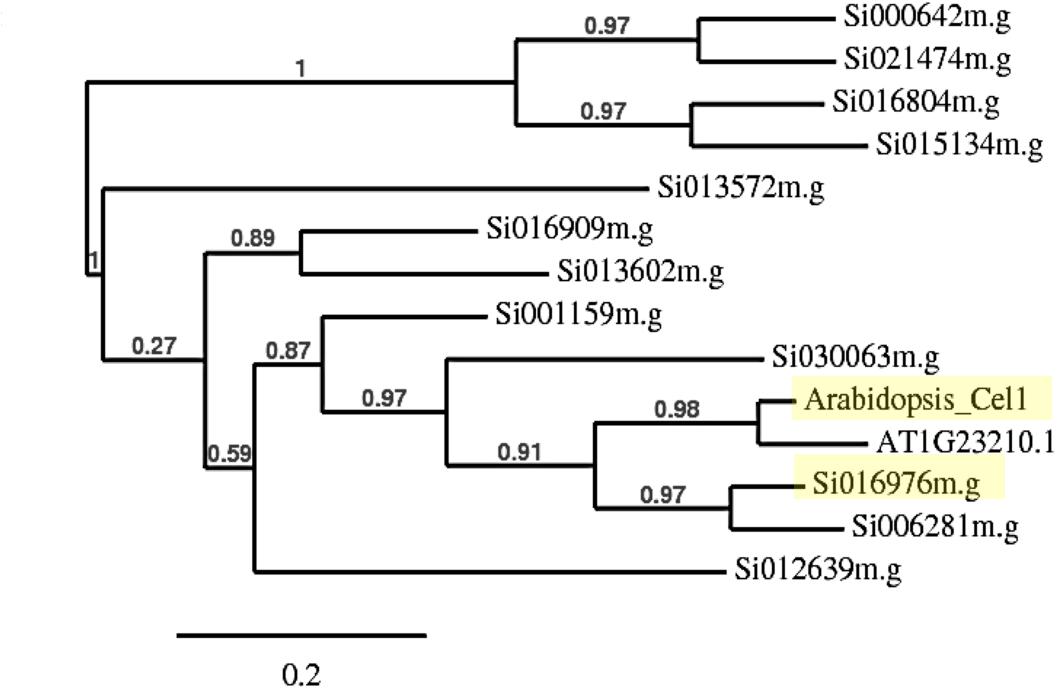
Figure 1. Neighbor-joining phylogenetic tree of Atcel1 homologs in S. italica. The relative branch lengths indicate that Si016976m.g is more closely related to Atcel1 (both highlighted in yellow) and therefore referred to as Sicel1.
Generation, Selection and Molecular Analysis of Transgenic S. viridis Events
Sequence-confirmed pANIC10A/MCOAtcel1 and pANIC10A/Sicel1 constructs plus the hpt selectable marker-only control (EVC) were used to generate transgenic plants of S. viridis, accession A10.1 (Van Eck and Swartwood, 2015). Ten, seven, and two independent transgenic events were recovered for pANIC10A/MCOAtcel1, pANIC10A/Sicel1, and EVC constructs, respectively. RT-PCR was performed, either with MCOAtcel1 specific primers (Figure 2A) or hpt (Supplementary Figure S1) primers, to confirm transgenic status of the plantlets. PCR-confirmed transgenic events were grown to maturity in soil and selfed. At each generation the S. viridis families were grown for 7 weeks, after which plant growth and development parameters were documented. None of the seven-pANIC10A/Sicel1 transgenic events recovered displayed discernable phenotypic differences in comparison to EVCs and wild type A10.1 controls at the T1 and T2 generations, and therefore were not studied further. In contrast, progeny from the three independent high MCOAtcel1 mRNA expressing events, 1287.4, 1258.7, and 1279.3 were characterized by enhanced height and vigor at both T1 and T2 generation (results not shown). Southern blot hybridization of T1 generation plants (Supplementary Figure S2) and PCR-based hpt marker segregation in the subsequent T3 generation (Figure 2B) were used to identify homozygous, single-copy transgenic lines in the T3 generation for further detailed trait analysis.
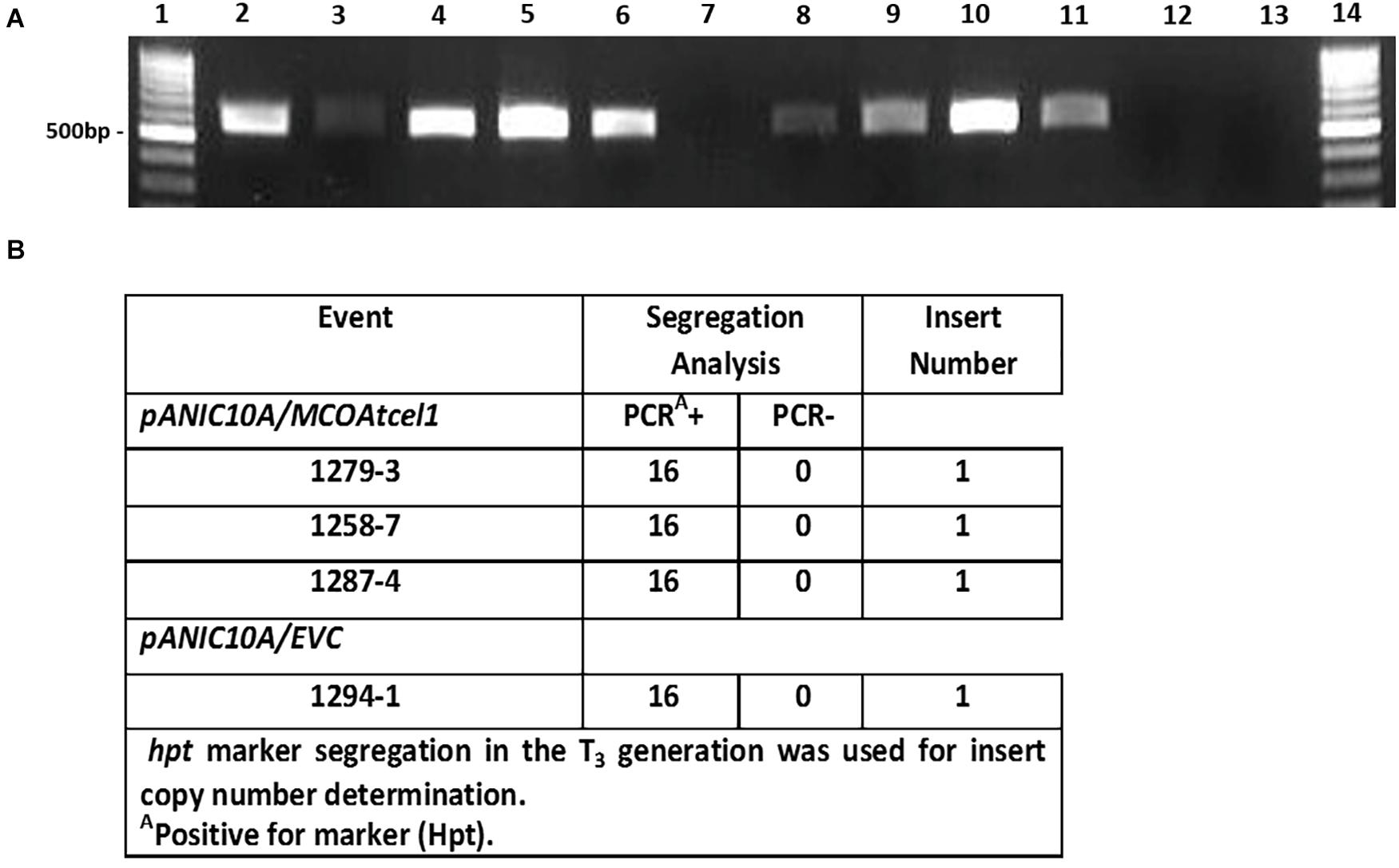
Figure 2. Molecular characterization of transgenic MCOAtcel1 events (A). Confirmation of MCOAtcel1 transgene expression in 10 independent transgenic S. viridis MCOAtcel1 T0 events (lane 2–11). The empty vector control T0 events tested negative for MCOAtcel1 transcript (lane. 12–13). Lane 1 and 14 1 kb marker DNA. (B) PCR-confirmation of single insert status of transgenic T3 families using hygromycin phosphotransferase (hpt) marker gene segregation.
Agronomic Trait Characterization of Transgenic MCOAtcel1 T3 Events
Sixteen individuals from the single-copy homozygous T3 MCOAtcel mRNA-expressing families from the three independent events 1287.4, 1258.7, and 1279.3 were grown to maturity and compared to the wild type A10.1 and EVC controls for primary panicle length, panicle number, grain number, total grain weight, and shoot dry matter content. Increased vigor and grain yield was observed in T3 plants from all three transgenic events. Grain yields increased significantly (p ≤ 0.001) by up to 26%, from an average of 1.19 ± 0.034 grams per plant in the controls to 1.62 ± 0.066 (26% gain), 1.63 ± 0.045 (26% gain), and 1.54 ± 0.045 (22% gain) per plant in the respective T3 families (Figure 3A). No change was seen in grain size, but assessment of grain yield components determined that total grain number increased (p ≤ 0.001) from an average of 818 ± 32 seeds per plant in controls to 1182 ± 52 (30% gain), 1108 ± 30 (26% gain), and 1179 ± 43 (30% gain) seeds per plant in the respective T3 families (Figure 3B).
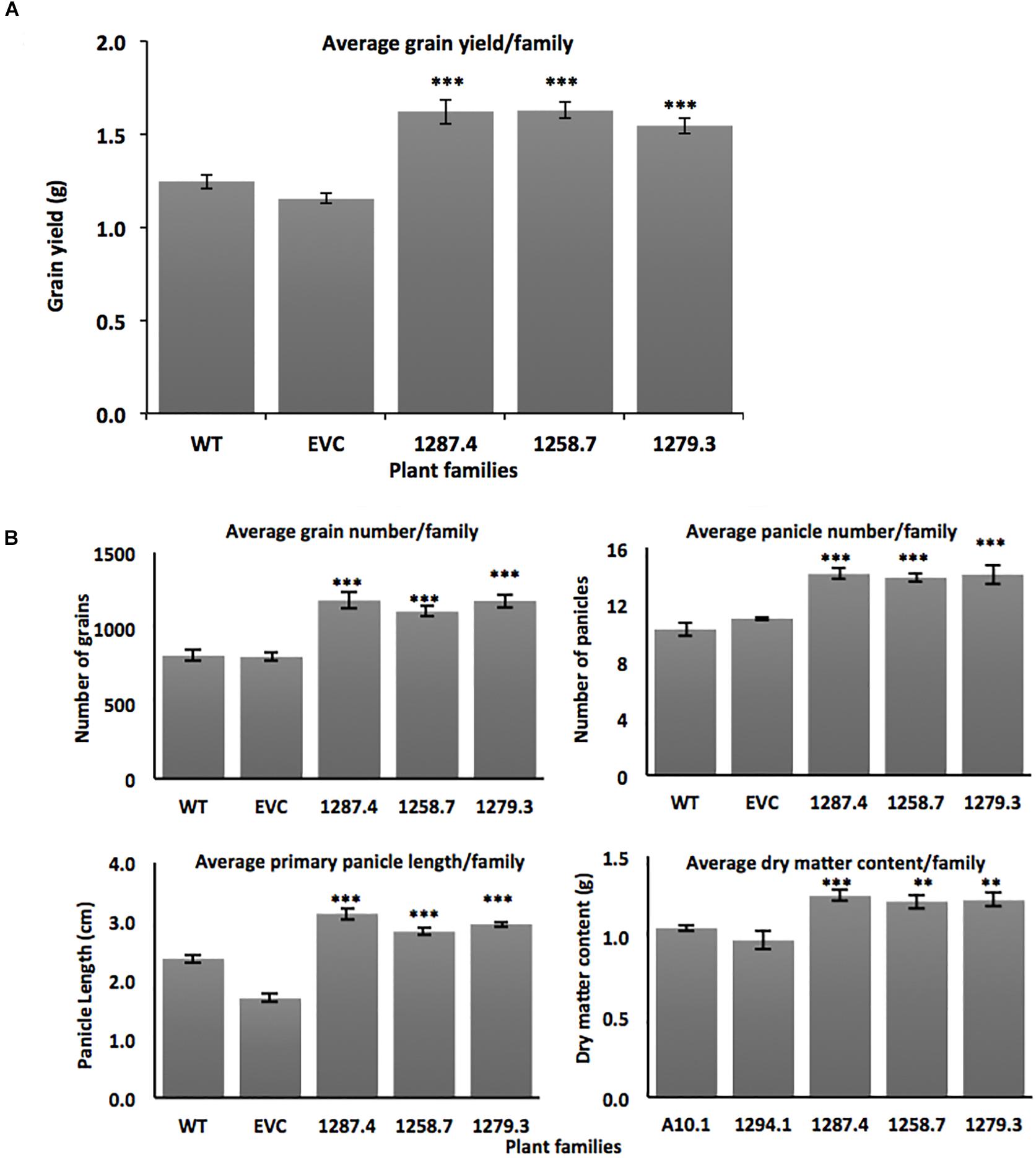
Figure 3. Agronomic trait characterization of transgenic MCOAtcel1 T3 events (A) Elevated grain yield in T3 S. viridis families transgenic for MCOAtcel1 in comparison to controls (n = 16 per family). (B) Elevated grain number, panicle number, primary panicle length, and vegetative dry matter content in T3 S. viridis families MCOAtcel1 in comparison to controls (n = 16 per family). Data are shown as mean ± SEM, with p ≤ 0.001(***)/p ≤ 0.01(**)/p ≤ 0.05(*). EVC (empty vector control) and WT (wild type A10.1).
We observed that yield increase was driven by an elevated number of seed-bearing panicles in the high MCOAtcel1 expressing T3 families. Seed-bearing panicle number increased significantly (p ≤ 0.001) from an average of 10 ± 0.32 per plant in the controls to 14 ± 0.34 (30% gain), 13 ± 0.31 (24% gain), and 14 ± 0.77 (30% gain) for panicles in the respective T3 families compared to controls (Figure 3B). Similarly, length of the primary panicle was significantly elevated (p ≤ 0.01) in the high MCOAtcel1 expresser T3 families, increasing from 2.1 cm ± 0.07 in controls to 3.1 ± 0.10 (32% gain), 2.8 ± 0.05 (25% gain), and 2.9 ± 0.04 (28% gain) cm per plant in the respective T3 families (Figure 3B). In addition to reproductive tissues, total vegetative shoot DMC, assessed after 7 days oven drying, was found to have increased significantly (p ≤ 0.01) in Atcel1 expressing plants. While controls averaged 1.01 g ± 0.037 per plant, the transgenic 1287.4, 1258.7, and 1279.3 families averaged 1.25 g ± 0.033 (19% gain), 1.21 g ± 0.042 (16% gain), and 1.22 g ± 0.041 (17% gain) per plant, respectively (Figure 3B).
Plants from T3 families were also characterized by a statistically significant pattern of accelerated developmental transitions. MCOAtcel1-expressing T3 families transitioned from the vegetative to reproductive phase earlier than control families, as characterized by early onset of flag leaf (transition phase), inflorescence initiation and anthesis (flowering), seed setting, and senescence. On average, the T3 families completed these transition events 7 days earlier than the controls (Figure 4A), representing a 12.5% reduction in life span of 56 to 49 days from germination to maturity (Figure 4B).
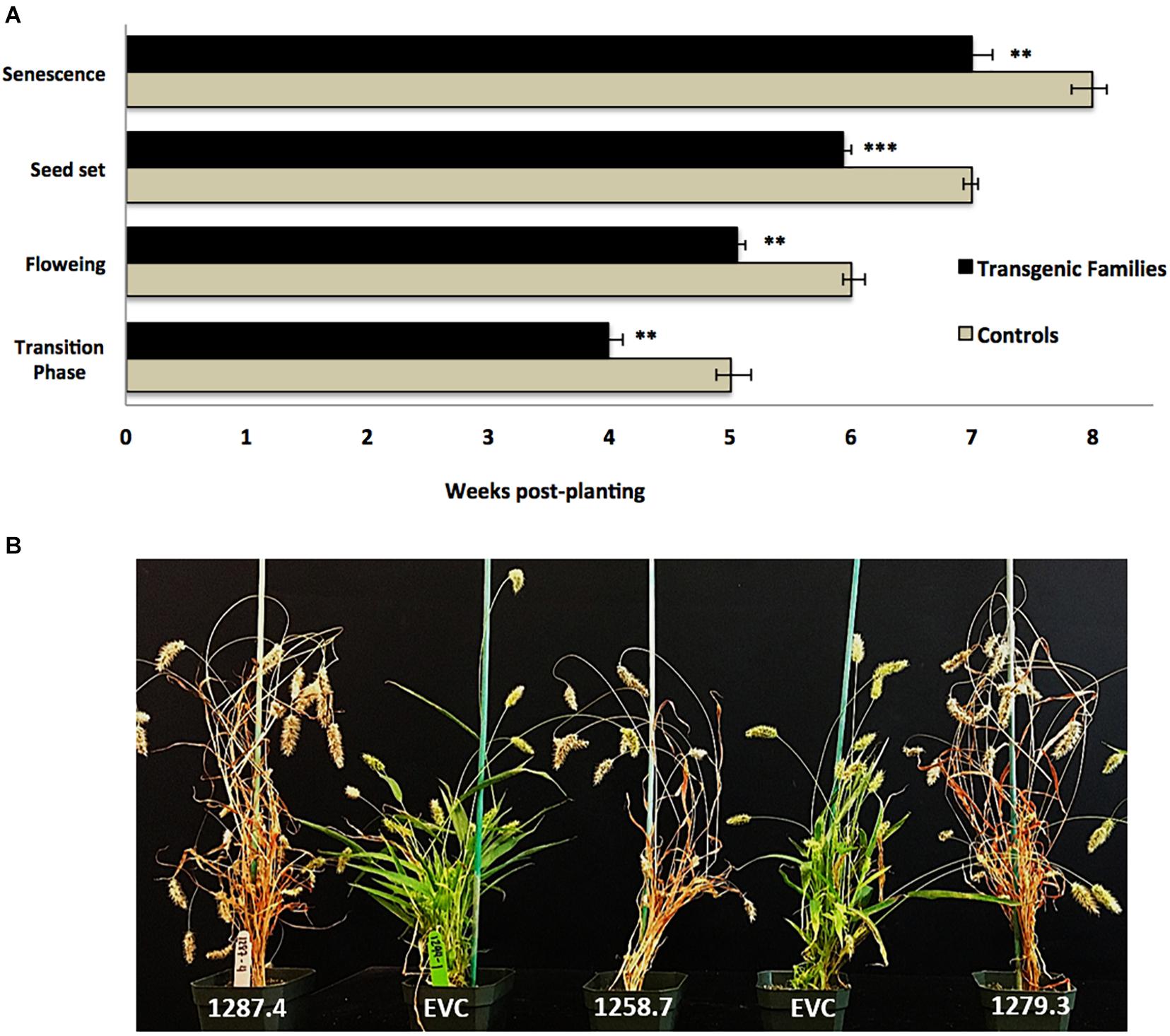
Figure 4. Developmental characterization of transgenic MCOAtcel1 T3 events (A) Accelerated developmental transitions in T3 S. viridis families transgenic for MCOAtcel1. Data are an average of all three transgenic families in comparison to controls. Data are shown as mean ± SEM, with p ≤ 0.001(***)/p ≤ 0.01(**)/p ≤ 0.05(*). (B) Early maturation of T3 S. viridis families transgenic for MCOAtcel1 in comparison to controls 7 weeks post-transplantation. On average plants of transgenic families completed their life cycle and reached senescence 7 days ahead of the empty vector (EVC), and wild-type A10.1 controls.
Discussion
Atcel1 belongs to the GH9B sub-family and is thought to be involved in cell wall loosening that drives anisotropic cell enlargement, a central process regulating the kinetics of plant growth and development (Buchanan et al., 2012; Lipchinsky, 2013). Endo-1,4-β-glucanases (glycosyl hydrolases) facilitate cell wall loosening by breaking down 1,4-β-glucosyl linkages and cell wall polysaccharides (Buchanan et al., 2012). The decreased cross-linking is proposed to increase cell wall plasticity which in turn leads to accelerated plant growth (Park et al., 2003). Phylogenetic analysis revealed two closely related genes, Si016976m.g and Si006281m.g in S. italica (Figure 1). Further analysis indicated Si016976m.g to be the closest homolog to Atcel and was designated as Sicel1.
Transgenic overexpression of Sicel1 did not confer a distinct phenotype in S. viridis. This was consistent with previous observation for overexpression of Atcel1 and PaPopCel1 in Arabidopsis and poplar, respectively (Park et al., 2003; Shani et al., 2004), and could be due to the homologous transgene-silencing (Matzke and Matzke, 1995). Conversely, transgenic S. viridis plants expressing the MCOAtcel1 under control of the robust, constitutive ZmUBI promoter displayed enhanced biomass, grain yield, and early maturity. MCOAtcel1 expressing plants were characterized by a 16–19% shoot biomass increment (Figure 3B), in a manner similar to the 20% enhancement in biomass production due to overexpression of Atcel1 in poplar (Shani et al., 2004). Enhanced biomass is a good indicator of high vigor and grain yield in cereal grasses (Capstaff and Miller, 2018). Indeed, plants from T3 families from all three independent MCOAtcel1 events studied displayed significantly increased grain weight, grain number, and panicle number (Figures 3A,B) compared to controls. Grain yield in cereals depends on component traits such as panicle length, panicle number, and grain weight (Reddy et al., 2013). Consistent with the above, we observed a 25–32%, 24–30%, and 26–30% increment in the component traits of primary panicle length, the total number of panicles, and total grain number, respectively, in the three MCOAtcel1 events (Figure 3B). The improvement of yield components resulted in up to a 26% increase in the grain yield (Figure 3A), confirming for the first time that increased seed yields can be achieved by heterologous up regulation of cel1.
Accelerated maturity due to overexpression of Atcel1 resulting in a 25% reduction in the time to harvest was reported in poplar and eucalyptus (Ledford, 2014). A similar response was observed in S. viridis. T3 families displayed accelerated growth and developmental trajectories, resulting in a 12.5% reduction in life span from germination to maturity (Figure 4A). Accelerated growth was statistically significant and likely due to early onset of juvenile-adult vegetative, adult vegetative-reproductive, and reproductive senescence transitions in the T3 families, in comparison to controls (Figures 4A,B).
Plant growth is driven by both cell division and cell expansion (Sun et al., 2017). Heterologous overexpression of Endo-1, 4-β- glucanases have previously been reported to result in expansion growth by trimming the disordered 1, 4-β- glucans (Park et al., 2003). Such expansion growth possibly resulted in the enhanced shoot biomass and accelerated growth observed in all three MCOAtcel1 overexpression events characterized in our study (Figures 3B, 4A). The enhanced shoot biomass in part could also be due to an increase in the leaf number and size that presumably results in higher grain yield due to increased capacity for photosynthesis, as reported previously (Weraduwage et al., 2015; Driever et al., 2017).
Accelerated maturity provides opportunities for reduced exposure to post-anthesis stress factors (Vadez et al., 2012) and enables development of crops for climate-smart agriculture. For example, early-maturing and high-yielding primary cereal crops could provide farmers with greater flexibility under increasingly uncertain climatic conditions to sustainably increase productivity and contribute to national food security. In developing country agricultural systems, such flexibility also helps mitigate poverty by enabling subsistence farmers to access off-farm revenue-generating employment (Kidane et al., 2006).
Heterologous overexpression of MCOAtcel1 caused enhanced grain yield, shoot biomass, and maturation rate in the model grass species S. viridis under growth chamber conditions. The agronomic trait differences observed were consistent with previous observations for dicots (Shani et al., 2004; Ledford, 2014), but are the first described in a monocot species. Importantly, increased seed yields are also achieved by overexpression of cel1. Our findings suggest that manipulation of Atcel1 has the potential for developing early-maturing, higher-yielding, monocot cereals and biomass crops that could be suitable for climate-smart agriculture. Studies should now be performed to evaluate if similar yield enhancements and accelerated maturation can be achieved in economically important cereal species under field conditions.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
BV directed the research, designed and conducted experiments, data analysis, and presentation, and wrote the manuscript. RP conducted experiments, collected data, and wrote manuscript. RW and AF conducted experiments. DB and SR conducted experiments and collected data. NT participated in data analysis and presentation and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge funding from the Donald Danforth Plant Science Center. The authors thank FuturaGene Ltd. for providing guidance in use of the cel1 technology, Kevin Lutke for generating Setaria transgenic events, and Rosana Segatto for help with harvesting Setaria families. RW and AF were supported by NSF-REU grant (NSF-DBI-REU-1156581).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.515078/full#supplementary-material
Supplementary Figure 1 | Molecular characterization of T0 Sicel1 and EVC events showing transgenic nature of the regenerated plants. PCR confirmation of hygromycin phosphotransferase (hpt) marker gene in 7 independent transgenic S. viridis Sicel1 (lane 3–9), and 2 empty vector controls (in duplicate, lane 13–16) T0 events. The pANIC10A vector and no template were used as positive (Lane 2, 10 and 11) and negative (Lane 12) controls, respectively, for the PCR. Lane 1, 1 kb marker DNA.
Supplementary Figure 2 | Confirmation of transgene integration. Southern blot analysis confirming T-DNA integration and copy number of the hpt marker gene in S. viridis plants of 1258.7, 1279.3, and 1287.4 T1 families and empty vector controls. The genomic DNA was digested with Mfe1 enzyme and probed with DIG labeled hpt probe.
Footnotes
References
Beche, E., Benin, G., da Silva, C. L., Munaro, L. B., and Marchese, J. A. (2014). Genetic gain in yield and changes associated with physiological traits in Brazilian wheat during the 20th century. Eur. J. Agron. 61, 49–59. doi: 10.1016/j.eja.2014.08.005
Brutnell, T. P., Wang, L., Swartwood, K., Goldschmidt, A., Jackson, D., Zhu, X.-G., et al. (2010). Setaria viridis: a model for C4 photosynthesis. Plant Cell 22, 2537–2544. doi: 10.1105/tpc.110.075309
Buchanan, M., Burton, R. A., Dhugga, K. S., Rafalski, A. J., Tingey, S. V., Shirley, N. J., et al. (2012). Endo-(1, 4)-β-Glucanase gene families in the grasses: temporal and spatial Co-transcription of orthologous genes1. BMC Plant Biol. 12:235. doi: 10.1186/1471-2229-12-235
Capstaff, N. M., and Miller, A. J. (2018). Improving the yield and nutritional quality of forage. Crops. 9, 1–18. doi: 10.1094/fg-2005-0512-01-rs
Dereeper, A., Guignon, V., Blanc, G., Audic, S., Buffet, S., Chevenet, F., et al. (2008). Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 1:36.
Driever, S. M., Simkin, A. J., Alotaibi, S., Fisk, S. J., Madgwick, P. J., Sparks, C. A., et al. (2017). Increased SBPase activity improves photosynthesis and grain yield in wheat grown in greenhouse conditions. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372:1730.
Fulton, T. M., Chunzoongse, J., and Tanksley, S. D. (1995). Microprep protocol for extraction of DNA from tomato and other herbaceous plants. Plant Mol. Biol. Rep. 3, 207–209. doi: 10.1007/bf02670897
Hartati, S., Sudarmonowati, E., Park, Y. W., Kaku, T., Kaida, R., Baba, K., et al. (2008). Over-expression of poplar cellulase accelerates growth and disturbs the closing movements of leaves in sengon. Plant Physiol. 147, 552–561. doi: 10.1104/pp.108.116970
Kidane, W., Maetz, M., and Dardel, P. (2006). Food Security and Agricultural Development in Sub-Saharan Africa. Rom: FAO.
Lipchinsky, A. (2013). How do expansins control plant growth? A model for cell wall loosening via defect migration in cellulose microfibrils. Acta Physiol. Plant. 35, 3277–3284. doi: 10.1007/s11738-013-1341-7
Long, S. P., Marshall-Colon, A., and Zhu, X.-G. (2015). Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 161:66.
Mann, D. G., Lafayette, P. R., Abercrombie, L. L., King, Z. R., Mazarei, M., Halter, M. C., et al. (2012). Gateway-compatible vectors for high-throughput gene functional analysis in switchgrass (Panicum virgatum L.) and other monocot species. Plant Biotechnol. J. 10, 226–236. doi: 10.1111/j.1467-7652.2011.00658.x
Matzke, M., and Matzke, A. (1995). How and why do plants inactivate homologous (trans) genes? Plant Physiol. 107, 679–685. doi: 10.1104/pp.107.3.679
Park, Y. W., Tominaga, R., Sugiyama, J., Furuta, Y., Tanimoto, E., Samejima, M., et al. (2003). Enhancement of growth by expression of poplar cellulase in Arabidopsis thaliana. Plant J. 33, 1099–1106. doi: 10.1046/j.1365-313x.2003.01696.x
Reddy, R. R., Madhusudhana, S., Murali, M. D. V., Chakravarthi, S. P., Mehtre, N., and Patil, J. V. (2013). Mapping QTL for grain yield and other agronomic traits in post-rainy sorghum [Sorghum bicolor (L.) Moench]. Theor. Appl. Genet. 126, 1921–1939. doi: 10.1007/s00122-013-2107-8
Reynolds, M. P., Quilligan, E., Aggarwal, P. K., et al. (2016). An integrated approach to maintaining cereal productivity under climate change. Glob. Food Sec. 8, 9–18.
Sebastian, J., Wong, M. K., Tang, E., and Dinneny, J. R. (2014). Methods to promote germination of dormant Setaria viridis seeds. PLoS One 9:e95109. doi: 10.1371/journal.pone.0095109
Shani, Z., Dekel, M., Tsabary, G., Goren, R., and Shoseyov, O. (2004). Growth enhancement of transgenic poplar plants by overexpression of Arabidopsis thaliana endo-1,4-β-glucanase (cel1). Mol Breed. 14, 321–330. doi: 10.1023/b:molb.0000049213.15952.8a
Sun, J., Cahill, T., Van Hautegem, K., Feys, C., Whipple, O., Novak, S., et al. (2017). Altered expression of maize Plastochron1 enhances biomass and seed yield by extending cell division duration. Nat. Commun. 8:14752.
Urbanowicz, B. R., Catalá, C., Irwin, D., Wilson, D. B., Ripoll, D. R., and Rose, J. K. C. (2007). A tomato endo-β-1,4-glucanase, SlCel9C1, represents a distinct subclass with a new family of carbohydrate binding modules (CBM49). J. Biol. Chem. 16, 12066–12074. doi: 10.1074/jbc.m607925200
Vadez, V., Hash, T., Bidinger, F. R., and Kholova, J. (2012). Phenotyping pearl millet for adaptation to drought. Front. Physiol. 3:386. doi: 10.3389/fphys.2012.00386
Van Eck, J., and Swartwood, K. (2015). Setaria viridis. Methods Mol. Biol. 1223, 57–67. doi: 10.1007/978-1-4939-1695-5_5
Keywords: Setaria viridis, enhanced grain yield, Arabidopsis cel1, shoot biomass, accelerated maturity, climate smart agriculture
Citation: Venkata BP, Polzin R, Wilkes R, Fearn A, Blumenthal D, Rohrbough S and Taylor NJ (2020) Heterologous Overexpression of Arabidopsis cel1 Enhances Grain Yield, Biomass and Early Maturity in Setaria viridis. Front. Plant Sci. 11:515078. doi: 10.3389/fpls.2020.515078
Received: 26 November 2019; Accepted: 02 October 2020;
Published: 10 November 2020.
Edited by:
James Lloyd, Stellenbosch University, South AfricaReviewed by:
Rosemary White, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaBradley Colin Campbell, The University of Queensland, Australia
Copyright © 2020 Venkata, Polzin, Wilkes, Fearn, Blumenthal, Rohrbough and Taylor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bala P. Venkata, bpvenkata@danforthcenter.org
 Bala P. Venkata
Bala P. Venkata Robert Polzin
Robert Polzin Nigel J. Taylor
Nigel J. Taylor