- 1Key Laboratory of Soybean Biology of Chinese Ministry of Education, Key Laboratory of Soybean Biology and Breeding/Genetics of Chinese Agriculture Ministry, College of Agriculture, Northeast Agricultural University, Harbin, China
- 2Jilin Academy of Agricultural Sciences, Changchun, China
In some legume–rhizobium symbioses, host specificity is influenced by rhizobial type III effectors-nodulation outer proteins (Nops). However, the genes encoding host proteins that interact with Nops remain unknown. In this study, we aimed to identify candidate soybean genes associated with NopD, one of the type III effectors of Sinorhizobium fredii HH103. The results showed that the expression pattern of NopD was analyzed in rhizobia induced by genistein. We also found NopD can be induced by TtsI, and NopD as a toxic effector can induce tobacco leaf death. In 10 soybean germplasms, NopD played a positively effect on nodule number (NN) and nodule dry weight (NDW) in nine germplasms, but not in Kenjian28. Significant phenotype of NN and NDW were identified between Dongnong594 and Charleston, Suinong14 and ZYD00006, respectively. To map the quantitative trait locus (QTL) associated with NopD, a recombinant inbred line (RIL) population derived from the cross between Dongnong594 and Charleston, and chromosome segment substitution lines (CSSLs) derived from Suinong14 and ZYD00006 were used. Two overlapping conditional QTL associated with NopD on chromosome 19 were identified. Two candidate genes were identified in the confident region of QTL, we found that NopD could influence the expression of Glyma.19g068600 (FBD/LRR) and expression of Glyma.19g069200 (PP2C) after HH103 infection. Haplotype analysis showed that different types of Glyma.19g069200 haplotypes could cause significant nodule phenotypic differences, but Glyma.19g068600 (FBD/LRR) was not. These results suggest that NopD promotes S. fredii HH103 infection via directly or indirectly regulating Glyma.19g068600 and Glyma.19g069200 expression during the establishment of symbiosis between rhizobia and soybean plants.
Introduction
Soybean [Glycine max (L.) Merr.] is a widely grown commercial crop around the world and supplies a large amount of protein and oil for humans and animals (Xin et al., 2016). Nitrogen is an indispensable element for soybean growth and an important limiting factor in crop production. Nowadays, huge amounts of nitrogen fertilizers are applied to improve crop production, but nitrogen fertilizers can also cause negative effects, such as soil acidification, change of soil microbial diversity, soil compaction, and groundwater pollution (Vance, 2001). Biological nitrogen fixation (BNF) could sustainably supply large amounts of nitrogen for agricultural production and could reduce the application of synthetic nitrogen fertilizer (Ladha and Peoples, 2012). Legumes can recognize and accept various strains of Rhizobium to establish a symbiotic relationship; numerous different strains are present in different ecoregions (Zimmer et al., 2016). The recognition and acceptance of rhizobia by legumes are a complex process. Secretion of Nod factor from rhizobia induces the curling of root hair tips, rhizobial cells are wrapped by the curling tips, and then start to infect host cells. Rhizobia induce the development of infection threads in host cells by means of which rhizobia can be transported into the root cortical cells (Riely et al., 2013). When rhizobia symbiotically colonize soybean roots, the plants can fix atmospheric nitrogen (Riches et al., 2013). Sinorhizobium fredii strain HH103 can nodulate soybean efficiently, is a fast-growing rhizobia similar as model strain S. fredii NGR234. In recent years, the genome of S. fredii HH103 has been uncovered, and extensive analyses of its genome and transcriptome have paved a good foundation for gene functional characterization (Margaret et al., 2011; Weidner et al., 2012; Vinardell et al., 2015; López-Baena et al., 2016; Pérez-Montaño et al., 2016). Thus, it is an ideal strain for studying the molecular mechanisms symbiosis between soybean and rhizobium.
The establishment of an effective symbiotic interaction is a complex process that requires multiple signal exchanges between the legume and rhizobia (Miwa and Okazaki, 2017). Among those signals, type III effectors (T3E) play vital roles during the infection of the host with rhizobia. T3Es are secreted through the type 3 secretion system (T3SS) and are translocated into host cells. Within the host cells, the effectors change host signaling, including suppressing plant immunity systems and supplying a more favorable environment for rhizobial infection and multiplication. Similar to plant pathogen effectors, some rhizobial T3Es can also induce strong defense responses that suppress rhizobial infection after being recognized by host legume resistance proteins (Marie et al., 2003; Tampakaki, 2014; Staehelin and Krishnan, 2015; López-Baena et al., 2016). These findings support that T3Es from rhizobia could have either positive or negative influences on the establishment of symbioses. To date, 12 nodulation outer proteins (Nops), namely, NopA, NopAA (GunA), NopB, NopC, NopD, NopI, NopJ, NopL, NopM, NopP, NopT, and NopX, have been identified in S. fredii strain HH103 (López-Baena et al., 2016; Jiménez-Guerrero et al., 2019). Among these effectors, NopA and NopB are important components of the needle of the T3SS (Saad et al., 2005; Kim and Krishnan, 2014). NopAA (GunA) is a cellulase that is able to break down the soybean cell wall and so promote infection (Jiménez-Guerrero et al., 2019). NopAA increased GmPR1 expression at an early stage of symbiosis (Jiménez-Guerrero et al., 2019). NopC is secreted into soybean root cells, exerting a positive function during infection (Jiménez-Guerrero et al., 2015a). NopM, a NEL-domain E3 ubiquitin ligase, appears to induce target sumoylation and may dampen the flg22-induced burst in reactive oxygen species in tobacco (Xin et al., 2012). An HH103 T3SS mutant that failed to secrete T3Es altered the expression of GmPR1, suggesting that the T3E might be related to the defense response (Jiménez-Guerrero et al., 2015b). NopP is a substrate for plant kinases, and its secretion by strain USDA112 was associated with host effector-triggered immunity to regulate symbiotic incompatibility with Rj2 soybeans (Sugawara et al., 2018). Different effectors might have various functions during the establishment of symbiosis.
Rhizobial TtsI can regulate Nops expression during rhizobial infection. In a TtsI mutant, Nops expression was clearly suppressed (López Baena et al., 2008). NopD was first detected in culture supernatants of S. fredii strain HH103 induced by genistein (Rodrigues et al., 2007) and was regulated by TtsI (López-Baena et al., 2016). NopD showed homology to Blr1693, a putative outer protein of Bradyrhizobium japonicum. The C-terminal region of Blr1693 contains a domain with homology to the ubiquitin-like protease Ulp1 (Kaneko et al., 2002). XopD, one of the Xanthomonas campestris pv. vesicatoria T3E, belongs to the C48 cysteine peptidase family and encodes a ubiquitin-like protease 1 (Ulp1). Interestingly, bioinformatic analysis showed that the C-terminal region of S. fredii HH103 NopD shares sequence similarities with Blr1693 and XopD. XopD interacts with a small ubiquitin-like modifier (SUMO)-conjugated protein and removes the SUMO conjugate in plants during X. campestris pv. vesicatoria infection (Hotson et al., 2003), suggesting that NopD protease might similarly cleave SUMO modifications from SUMO-conjugated proteins. Besides desumoylation, XopD can play a role in host plant defense by interacting with the transcription factor MYB30 (Canonne et al., 2011). In tomato, XopD targeted the transcription factor SlERF4. This interaction influenced signaling in response to ethylene and promoted pathogen reproduction (Kim et al., 2013). S. fredii HH103 NopD is one of the Ulp1 proteins with similarities to XopD from Xanthomonas. NopD might influence the host cell signaling pathway in a similar way to XopD. However, no proteins that directly interact with NopD have yet been reported.
Numerous important traits in crops have been studied using quantitative trait locus (QTL) mapping to identify genes related to the target traits. Traits related to nodulation are controlled by various genes, and QTL mapping has been used to identify loci or genes associated with symbiosis (Hwang et al., 2014). Several loci related to nodulation have been mapped, such as rj1, rj2, rj3, rj4, rj5, rj6, rj7, and rj8 (Caldwell, 1966; Vest, 1970; Vest and Caldwell, 1972; Caetano-Anollés and Gresshoff, 1991; Vuong et al., 1996). Among these loci, rj2 and rj4 were recently cloned (Yang et al., 2010; Tang et al., 2016). The rj2 gene can associate with NopP to determine symbiotic specificity (Okazaki et al., 2013; Sugawara et al., 2018). Rj4 can regulate soybean compatibility and incompatibility with rhizobia, and interestingly, the Rj4 gene was found to associate with some T3Es of Bradyrhizobium elkanii to influence establishment of the symbiosis (Faruque et al., 2015). In recent studies in soybean, PP2C-related gene and RPK were detected by QTL mapping and were shown to interact with S. fredii HH103 NopL to regulate the infection of soybean root cells by rhizobia (Zhang et al., 2018). Via QTL mapping, S. fredii HH103 NopP was found to induce the expression of TLP and MAPK3 during rhizobium infection (Wang et al., 2018). The identification and study of T3Es and their interacting genes could enhance the understanding of the signal communication between host plants and rhizobia during the establishment of symbiosis.
In this study, we show that NopD can be secreted from S. fredii HH103 in the presence of genistein. The nodulation effect of NopD was analyzed on 10 soybean germplasms, including Charleston, Dongnong594, Suinong14, and ZYD00006. These varieties showed significant differences in nodulation phenotype after being inoculated with S. fredii HH103 (wild type), the NopD and TtsI mutants, respectively. The recombinant inbred line (RIL) population derived from Charleston × Dongnong594 was used to identify QTL loci related to NopD. The conditional QTL related to NopD were verified by the chromosome segment substitution lines (CSSLs). Finally, two genes Glyma.19g068600 (FBD/LRR) and Glyma.19g069200 (PP2C) located on the overlap region of QTL location were identified as candidate genes that can interact with NopD. The expression of both genes can be induced by NopD. However, the haplotype effect on nodule traits is different between 19g068600 and Glyma.19g069200.
Materials and Methods
Strains, Primers, and Plasmids in This Study
Bacterial strains S. fredii HH103, the derived mutants HH103ΩNopD, HH103ΩTtsI, and Escherichia coli DH5α were used are listed in Supplementary Table S2. Primers for gene cloning and qRT-PCR are listed in Supplementary Table S1. Plasmids used for mutant construction and studies of gene function are listed in Supplementary Table S2.
Construction of the HH103ΩNopD and HH103ΩTtsI
The construction of insertion mutants was performed as follows: a 1.4 kb fragment containing a 550 bp fragment upstream of the NopD ATG codon and an 850-bp fragment downstream of the ATG codon was cloned into pGWC, yielding plasmid pGWC-NopD1400. A SpeI restriction enzyme site was constructed close to the start codon of NopD using the Fast Mutagenesis System (Transgene Co., Beijing, China). Primers for site-directed mutagenesis are listed in Supplementary Table S1. A kanamycin Ω interposon was obtained from pEASY-Blunt with SpeI and then ligated into pGWC-NopD1400 SpeI site, yielding pGWC-NopD2400Ω. The 2,400 bp construct was then cloned into the suicide vector pJQ200SK (Quandt and Hynes, 1993) using XbaI and SmaI. The triparental mating was used to the transferred pJQ-NopD2400Ω from E. coli DH5α cells into S. fredii HH103 in the presence of pRK2013 helper plasmid (Figurski et al., 1979). Candidate mutant recombination colonies were obtained by screening for kanamycin resistance and growth on sucrose (5% w/v). Subsequently, positive mutants were screened by antibiotics and 5% sucrose. The candidate NopD and TtsI mutants were detected by PCR, qRT-PCR, and analysis of nodulation outer proteins. All the bacterial strains, primers, and plasmids used for mutant construction are shown in Supplementary Tables S1, S2.
RNA Isolation of Rhizobia and qRT-PCR Analyses of NopD
S. fredii strains HH103, HH103ΩNopD, and HH103ΩTtsI were incubated with shaking at 28°C in YM medium in the presence or absence of 3.7 μM genistein. Rhizobial RNA was extracted as described (Jiménez-Guerrero et al., 2015a), gDNA was treated by gDNA remover (Transgene Co., Beijing, China) to eliminate its effects on expression, and then RNA samples were synthesized into cDNA using TransScript® One-Step cDNA Synthesis SuperMix (Transgene Co., Beijing, China). qRT-PCR was performed with TransStart® Top Green qPCR SuperMix (Transgene Co., Beijing, China) in a Roche LightCycler 480 II System. The qRT-PCR program was as follows: denaturation at 94°C for 30 s, followed by 40 cycles of 94°C for 5 s, 60°C for 15 s, and 72°C for 10 s. The 16S rRNA gene was used as a reference gene to calibrate the transcript abundance values among different cDNA samples (Crespo-Rivas et al., 2007). The threshold cycle values were analyzed by the software in the Roche LightCycler 480 II. All sample harvests were performed with three biological replicates, and the individual values for each RNA sample were analyzed by three technical replications. The primers for expression analysis are listed in Supplementary Table S1.
Analysis of NopD in Nodulation Outer Proteins
The wild-type strain and two mutants were each cultured in 500 ml YM medium at 28°C until OD600 reached 0.6. The bacteria were cultured in the presence of 3.7 mM genistein for about 40 h at 28°C. After centrifugation of the cells at 8,000 g for 30 min (4°C), the supernatant was collected for purification of outer proteins. To eliminate contamination by bacteria and increase the protein concentration, the supernatant was filtered through MilliporeTM filter units (0.22 μM) (Millipore Co., Germany), then concentrated using MilliporeTM AmiconTM Ultra-15 (100 kDa) centrifugal filter units (Millipore Co., Germany). Proteins were precipitated in the presence of 10% w/v trichloroacetic acid for 20 h at 4°C then collected by centrifugation at 10,000 g for 20 min (4°C). After washing twice with cold 80% acetone (v/v), the precipitated proteins were resuspended in 8 M urea. Extracellular proteins from the different strains were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). For immunostaining, extracellular proteins were transferred onto nitrocellulose membranes, then the membranes were blocked with TBST pH 7.5 (per liter contains 3.03 g Tris, 8 g NaCl, and 1 ml Tween 20) containing 5% (w/v) skim milk, followed by incubation for 1 h with anti-NopD rabbit antiserum (1,000-fold dilution). Subsequently, the membranes were incubated with goat anti-rabbit immunoglobulin AP-conjugated secondary antibody (Abmart Co., China) for 1 h in accord with the supplier’s instructions, and reaction results were visualized using SuperSignal West Pico Chemiluminescent Substrate (Thermo Co., United States).
Agrobacterium tumefaciens-Mediated Transformation
Agrobacterium-mediated transformation by agroinfiltration was performed as follows: the NopD gene (GenBank: CEO91485.1) was cloned into the entry vector pGWC, and the entry clone was subsequently recombined into the destination vector pGWB17 using the Gateway® system (Invitrogen Co., United States). Plasmids pGWB17-NopD, the empty vector pGWB17-T, and pGWB17-HopQ1 were transformed into A. tumefaciens EHA105 by electroporation. Four-week-old Nicotiana benthamiana plants were used for transient expression: the A. tumefaciens culture was adjusted to OD600 0.5 using infiltration buffer (10 mM MgCl2, 10 mM MES-KOH pH 5.6, 150 μM acetosyringone). Top leaves were used for infiltrating, then at 0–4 days after infiltration, leaves were harvested for detection of cell death. Staining of N. benthamiana leaves was performed with trypan blue as described by Tennant (1964). Electrolyte leakage was used to evaluate cell death in leaf tissues by measuring ion conductivity (Mackey et al., 2002).
Nodulation Tests
For nodulation tests, the wild-type strain S. fredii HH103 and mutants HH103ΩNopD and HH103ΩTtsI were used. The soybean germplasms of different ecoregions and RIL population used are listed in Supplementary Table S3. Soybean seeds were sterilized with chlorine gas for 12–14 h, and then germinated into sterilized 300 ml plastic jar units containing nitrogen-free nutrient solution in the lower vessel. Each jar was kept in a greenhouse with 16 h/8 h light/dark at 26°C until plants grew to Vc stage, then plants were inoculated with 2 ml of 10 mM MgSO4 containing approximately 1 × 109 bacteria (Zhang et al., 2018). Thirty days after inoculation, all soybean roots were harvested for nodulation evaluation. The nodulation tests on each soybean germplasm were performed with five replicates and three independent experiments. The statistical significance of differences in phenotype was detected using t-tests.
The Conditional Quantitative Trait Locus Mapping of Nodulation-Related Traits
The experimental RIL population used in this study (n = 150) was derived from the cross of Charleston and Dongnong594 (Jiang et al., 2018). A high-density genetic map with 5,308 specific locus amplified fragment sequencing (SLAF-seq) markers had been constructed via this population (Qi et al., 2014). Recently, we also completed the genomic resequencing of CSSLs and their parents (unpublished). These genetic backgrounds supply a useful support to identify the candidate genes in interested QTL.
To detect the QTL underlying nodule-related traits, a composite interval mapping method was used with WinQTL Cartographer (Wang et al., 2018; Zhang et al., 2018). The detailed parameters followed the published method (Zhang et al., 2018). The nodule number (NN) and nodule dry weight (NDW) of RILs and their parents after inoculation with NopD mutant or the parental strain were used for QTL identification. The differences in phenotypic values were used to determine the location of conditional QTL. At the same time, the phenotype differences in RILs inoculated with HH103ΩTtsI or the parental strains were used to determine the conditional QTL’s location (Wang et al., 2018). The detailed method is as follows. When the operation was running, the control marker number was set to 5 and window size was 10 cM. The forward regression method and a walk speed of 0.5 cM were selected. The proportional and additive effects of variances interpreted by each specific QTL were obtained by composite interval mapping analysis. The log of the odds (LOD) score peaks higher than 3.0 (WinQTL Cartographer default threshold) was selected to indicate the existence of conditional QTL for the nodule traits inoculated with the two type strains, respectively. About the additive-effects signals, “+” indicates increasing allelic effects from “Dongnong 594” and “−” indicates decreasing allelic effects from “Charleston.” The 1,000 permutations of each genotypic marker against the phenotype in RIL population determined the experimental threshold levels for linkage. When the two values for a marker were greater than the critical value at p = 0.05, it indicated that the linkage was significant. CSSLs produced by the cross between soybean cultivar SN14 and wild soybean ZYD00006 (G. soja Sieb. & Zucc.) were used for verifying consensus QTL, according to the genetic map by Xin et al. (2016).
Annotation of Candidate Genes in Quantitative Trait Locus of NopD-Related
The “Williams 82. a2. v1” genome was the first published soybean genome and could provide valuable information for QTL mapping of soybean important traits (Brensha et al., 2012). Phytozome website1 and Soybase database2 integrated the soybean Williams 82 genome information and the latest information on the soybean genome uploaded by researchers, could provide necessary information for mapping. Our laboratory completed the construction of a high-density genetic map of the RIL population in the early stage (Qi et al., 2014). So the soybean genes in the major QTL intervals could be identified by combining the high-density genetic map information with two important database tools, and corresponding gene annotations were performed (Wang et al., 2018). Candidate genes involved in the plant immunity and signal conduction were selected from the annotation data, which were used for subsequent verification.
Verification of NopD Candidate Genes by qRT-PCR
qRT-PCR in various soybean germplasm materials was performed to verify candidate genes that potentially interacted with NopD. Root samples were harvested at 0, 12, 24, 36, 48, and 60 h post-inoculation with the wild-type strain HH103, NopD mutant, and TtsI mutant, respectively. The classic TRIzol reagent (Invitrogen Co., United States) was used for the extraction of total RNA from soybean roots, and total RNA was treated by gDNA remover (Transgene Co., Beijing, China) to eliminate its effects on expression, and then RNA samples were synthesized into cDNA by the TransScript® One-Step cDNA Synthesis SuperMix (Transgene Co., Beijing, China). qRT-PCR was performed as described above, and GmELF1b was used as the internal control to normalize the transcript amounts in different samples (Jian et al., 2008). The gene-specific primers for qRT-PCR are listed in Supplementary Table S1.
Haplotype Analysis of Candidate Genes Based on Chromosome Segment Substitution Line Population
Haplotype analysis of Glyma.19g068800 and Glyma.19g069200 was based on the resequencing data of CSSL populations in a subset of 142 soybean accessions. The genomic regions including the gene transcript sequence and approximately 2.0-kb promoter region of two genes upstream of the start codon were used for the haplotype analysis.
Results
NopD Can Be Secreted From S. fredii HH103 by Type 3 Secretion System
The NopD coding sequence was located in the plasmid pSfHH103d of S. fredii HH103. The promoter region contains a tts box (−257 bp), supporting the idea that NopD can be induced by TtsI (Figure 1A). By phylogenetic analysis, we can separate the NopD tree into two branches (I-Red and II-Green). Branch I comprises only three Sinorhizobium species. Branch II comprises Mesorhizobium and Bradyrhizobium species without Sinorhizobium species (Figure 1A).
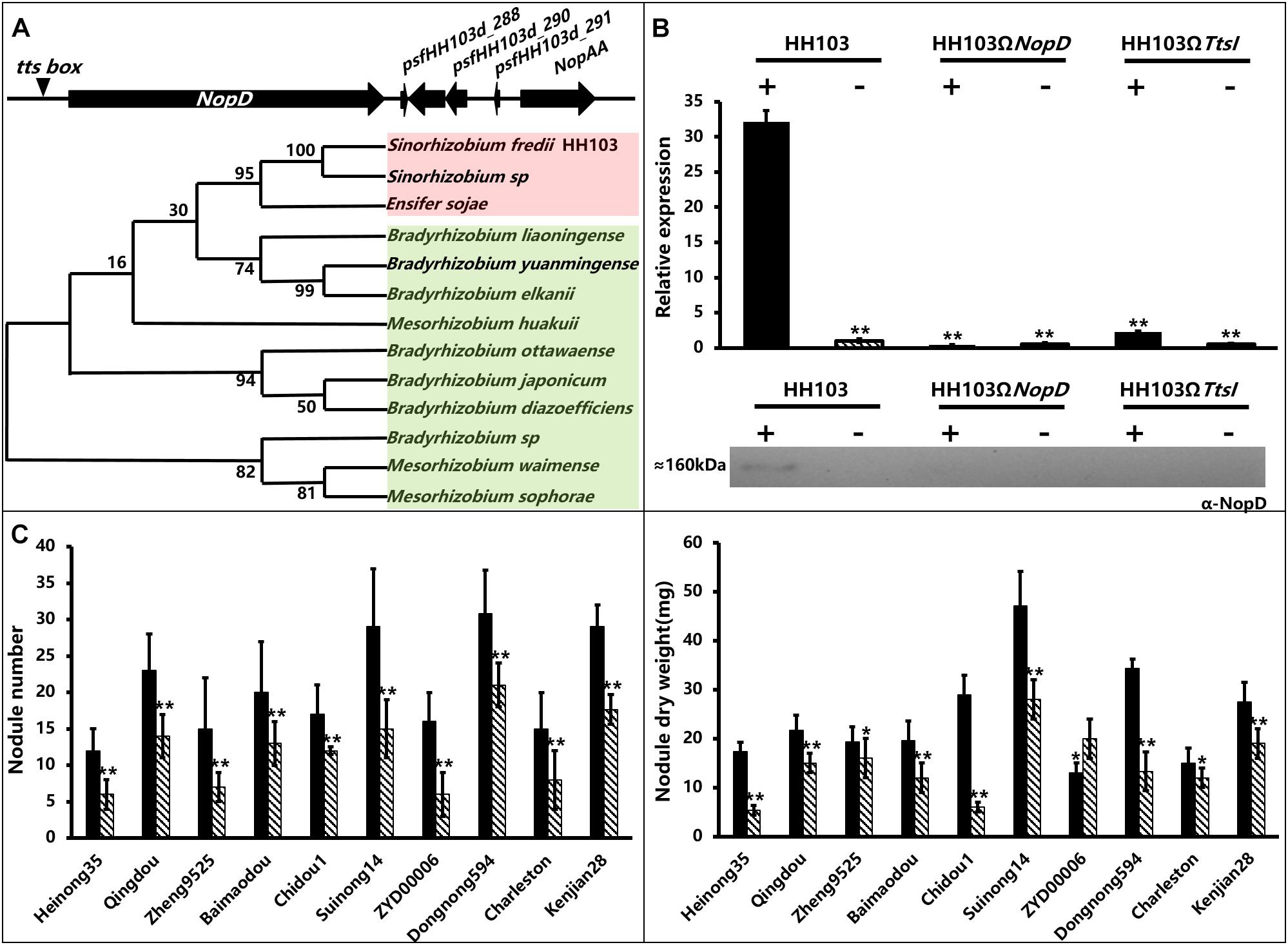
Figure 1. The information of NopD gene, detection of NopD protein induction by genistein and nodule traits of soybean plants inoculated with S. fredii wild-type HH103 or mutants HH103ΩNopD and HH103ΩTtsI. (A) Position of the NopD gene and phylogenetic tree of NopD from various rhizobial strains by neighbor-joining method using MEGA6.0 software. (B) Expression analysis of NopD in S. fredii wild-type strain HH103 and mutants HH103ΩNopD and HH103ΩTtsI with (+) or without (−) genistein (3.7 μM). Final expression was calculated relative to the expression of the wild-type strain HH103 in the absence of genistein. All sample harvests were performed with three biological replicates, and the individual values for each RNA sample were analyzed by three technical replications. The sample of the wild-type strain S. fredii HH103 in the absence of genistein was used as the control. Asterisks indicate significant differences at the level α = 1% (P < 0.01). Immunoblot analysis of NopD in extracellular protein extracts of the wild-type strain S. fredii HH103 and indicated mutant derivatives induced with or without genistein (3.7 μM). Immunoblots were performed with anti-NopD antibodies. (C) The analysis of phenotype was performed three times; significant differences were determined by t-tests; * indicates 0.01 ≤ P ≤ 0.05 and **indicates P ≤ 0.01. Soybean varieties (with origins): Heinong35 (Heilongjiang), Qingdou (Shanxi), Zheng9525 (Henan), Baimaodou (Zhejiang), Chidou1 (Inner Mongolia), Suinong14 (Heilongjiang), ZYD00006 (Heilongjiang), Charleston (America), Dongnong594 (Heilongjiang), Kenjian28 (Heilongjiang).
NopD’s expression was studied by qRT-PCR in strains HH103, HH103ΩNopD, and HH103ΩTtsI induced or not by genistein. The qRT-PCR results showed that genistein promoted the expression of NopD significantly in the wild strain (Figure 1B), but NopD expression was not detected in the NopD and TtsI mutant either with or without genistein. This result was consistent with the previous reports that NopD’s expression was downregulated in a flavonoid mutant, NodD1 mutant, and TtsI mutant (Pérez-Montaño et al., 2016). An antibody against NopD was used to detect the NopD protein in supernatants from HH103, HH103ΩNopD, and HH103ΩTtsI. Western blot results showed a band corresponding to NopD (about 160 kDa) in samples from S. fredii HH103 induced with genistein, but not in NopD or TtsI mutants (Figure 1B).
Nodulation Tests
To elucidate the role of NopD in establishing symbiosis, we collected 10 soybean varieties with differing genetic backgrounds from various ecoregions in China and United States. In most soybean germplasms, there were significant differences in NN and NDW after inoculation with the NopD mutant or the wild-type HH103 (Figure 1C). Only in Kenjian28, NopD had a negative effect on the NN and NDW, and there was no difference in NN of Charleston when inoculated with the wild strain or HH103ΩNopD. However, NopD had a positive effect on NN and NDW in eight of the 10 soybean germplasms, except for the NDW of ZYD00006 (Figure 1C). Thus, NopD exerting either positive or negative effects on the various soybean germplasms might be due to the different genetic backgrounds of germplasms. On the other hand, these results support NopD mainly played a positive effect on soybean nodulation.
Nodulation tests show that Dongnnong594 and Charleston, as the parents of RIL population, had a significant difference in NN and NDW when inoculated with wild strain and NopD mutant, the same situation also occurs in the CSSL populations with Suinong14 and ZYD00006 as their parents, so RIL and CSSL populations could be used to map important QTL interactions with NopD.
NopD Induces Leaf Death in Nicotiana benthamiana
Agrobacterium-mediated transient transformation was used to study NopD’s effects within tobacco leaves. pGWB17, a binary vector containing the cauliflower mosaic virus 35S promoter and the NopD coding sequence, was introduced into A. tumefaciens EHA105, which was then infiltrated into leaves of 4-week-old tobacco plants. Four days after infiltration, a clearly necrotic leaf zone (hypersensitive response with induced cell death) was observed in tissue transformed with NopD as well as in leaves expressing HopQ1 of Pseudomonas syringae, which was used as a positive control (Figure 2). These results indicated that NopD functions like an avirulence effector in tobacco. Trypan blue staining and electrolyte leakage were used to detect the effect of NopD expression in leaves (Figure 2).
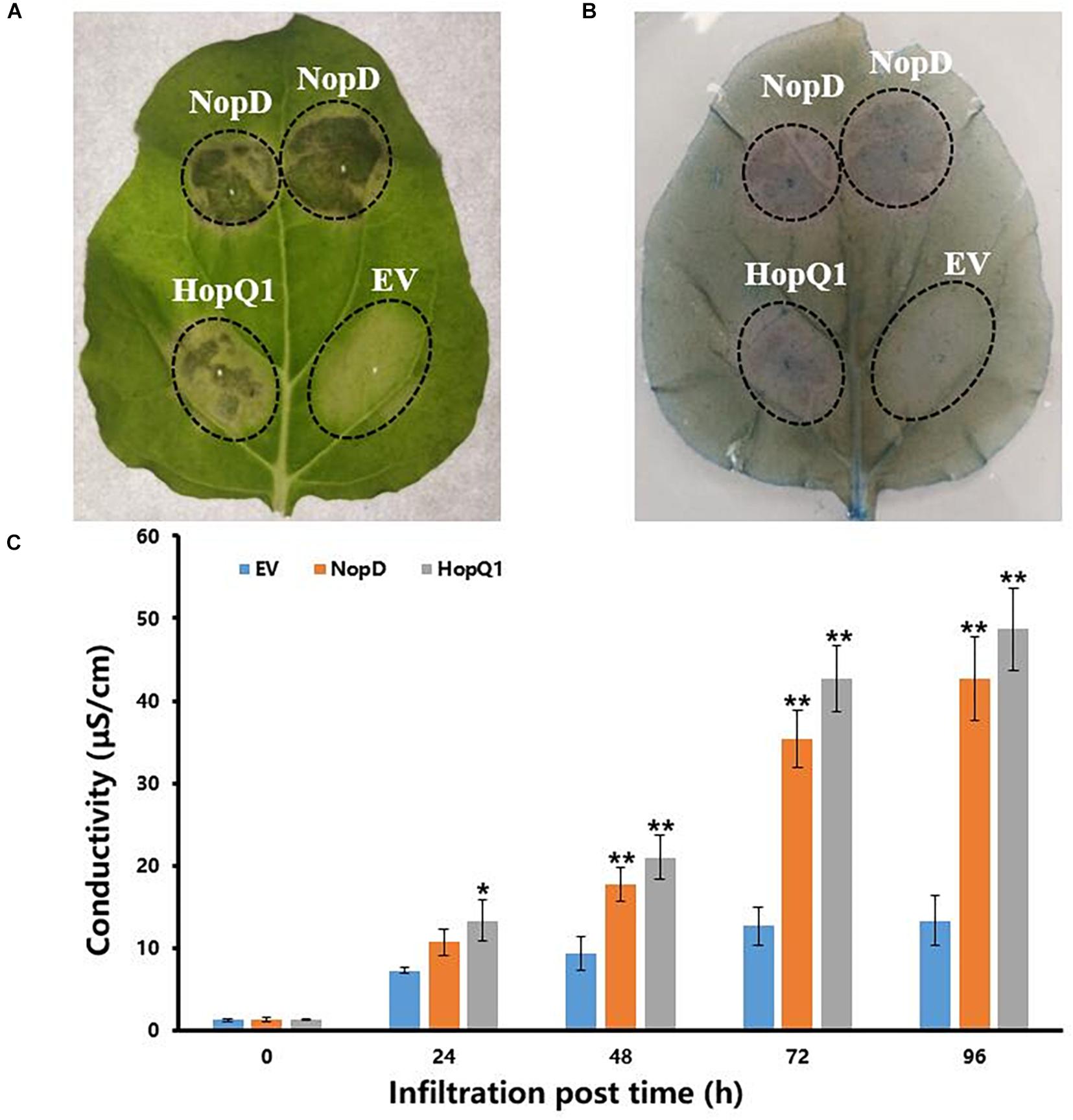
Figure 2. Effects of NopD on N. benthamina cells. Infiltrated zones are marked by circles (EV, infiltration zone with A. tumefaciens carrying the empty vector pGWB17-T). For comparison, A. tumefaciens carrying plasmid pGWB17-HopQ1-myc was also used for infiltration (expression of HopQ1 of P. syringae pv. tomato DC3000). (A) Leaves were photographed 4 days post infiltration. (B) Trypan blue-based cell death staining of a representative N. benthamiana leaf transiently expressing NopD and HopQ1 4 days post Agrobacterium infiltration. (C) Electrolyte leakage levels in leaves. “*” indicated the significant differences (p ≤ 0.05) and “**” indicated the significant differences (p ≤ 0.01).
Phenotype of Nodulation Analysis in Recombinant Inbred Lines
Nodulation tests on the soybean germplasms showed that there were significant differences in NN and NDW between Charleston and Dongnong594 when inoculated with the wild-type strain HH103 or NopD and TtsI mutants (Figure 1C and Table 1). The RIL population was derived from the cross between Charleston and Dongnong594. From the nodulation tests, the NopD mutant gave a higher average NN and NDW in the whole RIL population compared with inoculation with the wild-type strain or TtsI mutant. There were no significant differences in average NN or NDW of the whole RIL population after inoculation with the wild-type strain or the TtsI mutant. The RIL population has a more complex genetic background than the individual parents Charleston and Dongnong594, the genetic information between individuals were quite different, these individuals should have different responses to the wild strain, NopD mutant, or TtsI mutant, and this might have caused the observed differences compared with 10 soybean varieties in phenotype. So the nodule traits of the whole RIL population are not representative after inoculation with the wild strain, NopD mutant, and TtsI mutant. The more complex genetic background of the RIL population should facilitate the mining of QTL associated with the phenotype.
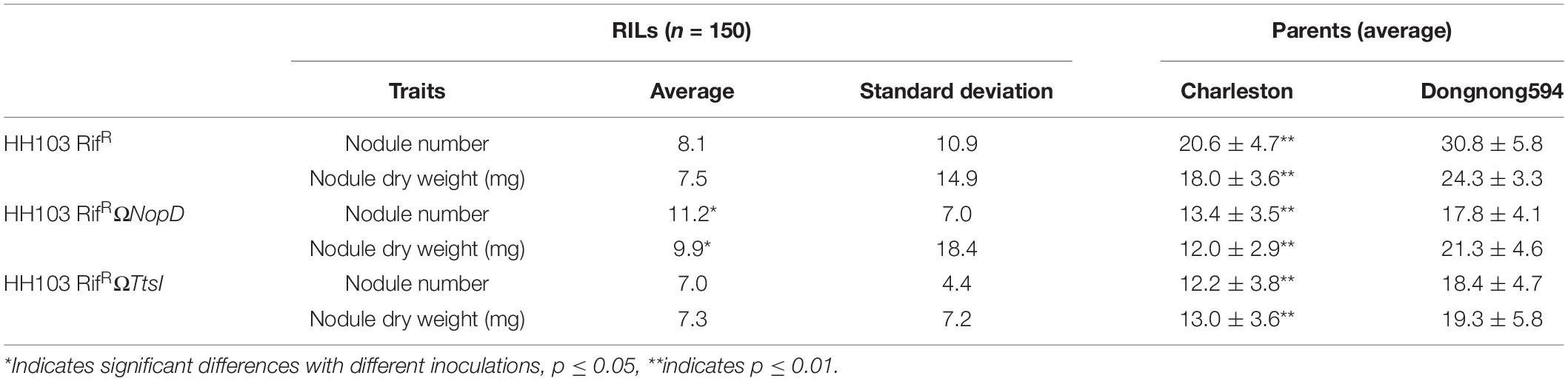
Table 1. Nodule traits of the recombinant inbred line (RIL) population inoculated with the wild-type strain and NopD and TtsI mutants.
Conditional Quantitative Trait Locus Mapping for Nodule Number and Nodule Dry Weight and Validation of Consensus Quantitative Trait Locus
Seven conditional QTL for NN and four conditional QTL for NDW were identified, after inoculation with S. fredii HH103 or the NopD and TtsI mutants (Table 2 and Figure 3). Seven conditional QTL associated with NN were located on chromosomes Gm02 (n = 2), Gm17 (n = 1), Gm18 (n = 1), and Gm19 (n = 3), and four conditional QTL for NDW were located on Gm08 (n = 1), Gm9 (n = 1), Gm11 (n = 1), and Gm19 (n = 1).
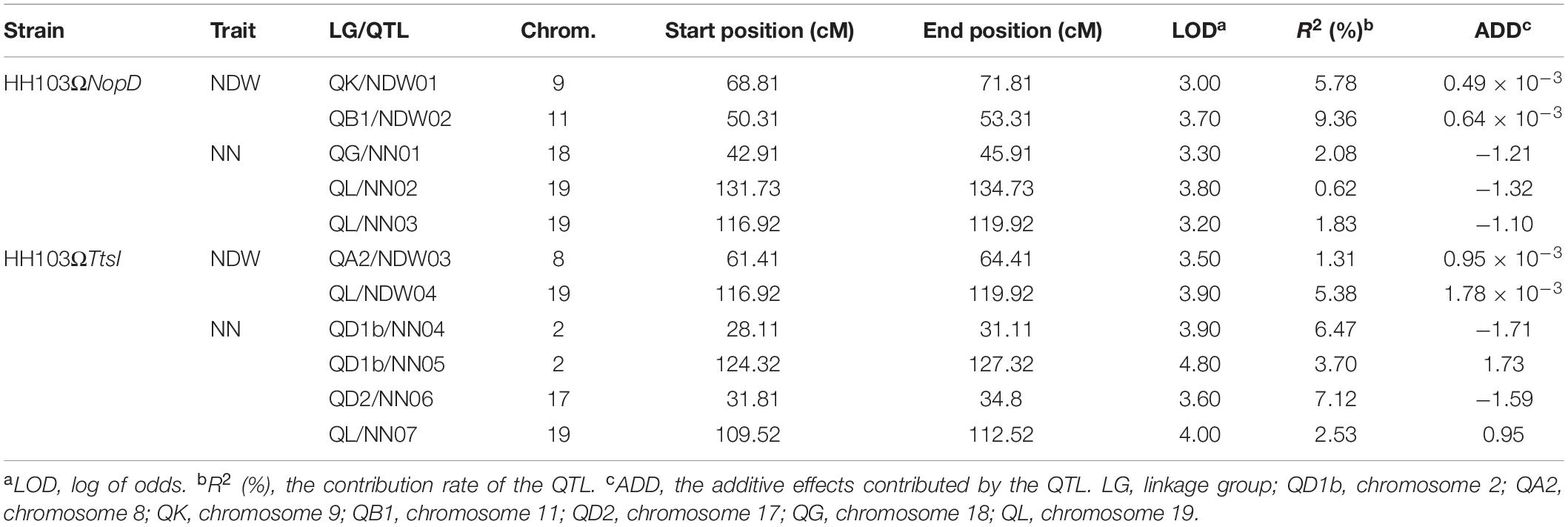
Table 2. Distribution of conditional quantitative trait locus (QTL) for nodule number (NN) and nodule dry weight (NDW) among linkage groups and chromosomes.
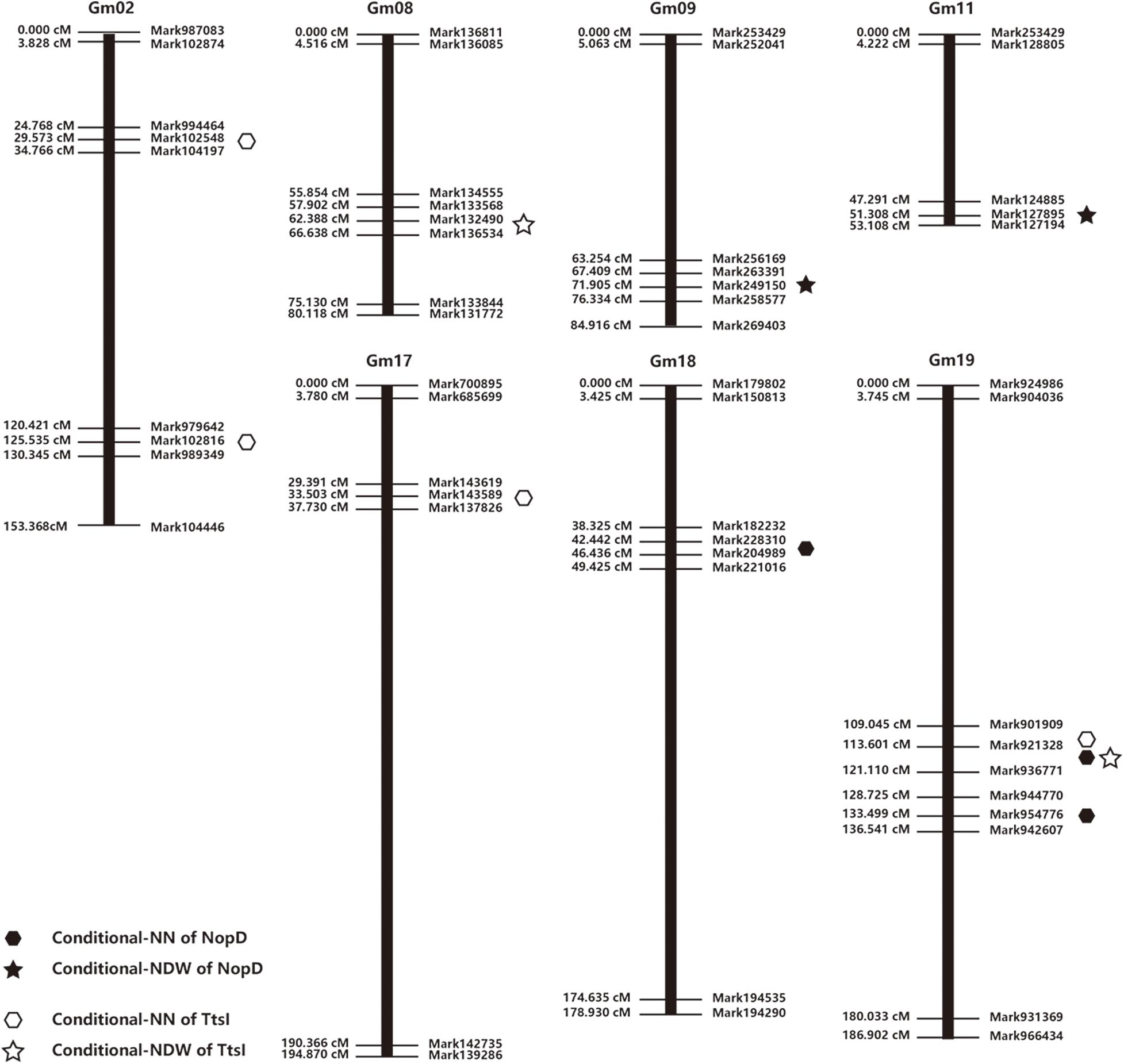
Figure 3. Distribution of conditional quantitative trait locus (QTL) for nodule number and nodule dry weight among linkage groups and chromosomes.
Overlapping conditional QTL (118.4 cM) for NN and NDW were identified on Chromosome 19, and we found two further conditional QTL (111.0 and 133.2 cM) close to these overlapping QTL. An earlier investigation of soybean revealed two QTL (99.7 and 108 cM) on Gm19 associated with nodule weight, and two QTL (97.5 and 108.2 cM) associated with nodule size (Hwang et al., 2014). This previously identified QTL support that in the region of QTL we identified is confident. So we selected this region as a candidate region that might contain gene response to NopD. To test our hypothesis, specific lines of CSSLs were identified and used for further work.
Nodulation tests showed significant differences in NN and NDW between Suinong14 and ZYD00006 after inoculation with wild-type strain HH103, NopD mutant, and TtsI mutant (Figure 1C). A CSSL population derived from the cross between Suinong14 and ZYD00006 was used to verify whether the overlapping QTL might be related to NN or NDW in other soybean populations (Supplementary Table S4). We selected six soybean lines from the CSSL, three of which (CSSL-600, CSSL-603, and CSSL-612) had higher NN and NDW than the parent Suinong14 after inoculation with NopD mutant and TtsI mutant, and another three of which (CSSL-519, CSSL-593, and CSSL-648) had lower NN and NDW than ZYD00006 after inoculation with NopD mutant and TtsI mutant. SSR markers located the conditional QTL on chromosome 19 when the RIL population were screened. Substituted chromosome fragments were also detected in related regions: we found an overlap region between the conditional QTL and the fragment of a CSSL (Sat_134–Satt398) on chromosome 19 (97.5–133.2 cM), these lines including CSSL-600, CSSL-603, and CSSL-612. No corresponding region was found in lines CSSL-519, CSSL-593, CSSL-648, or the parent ZYD00006 (Figure 4).
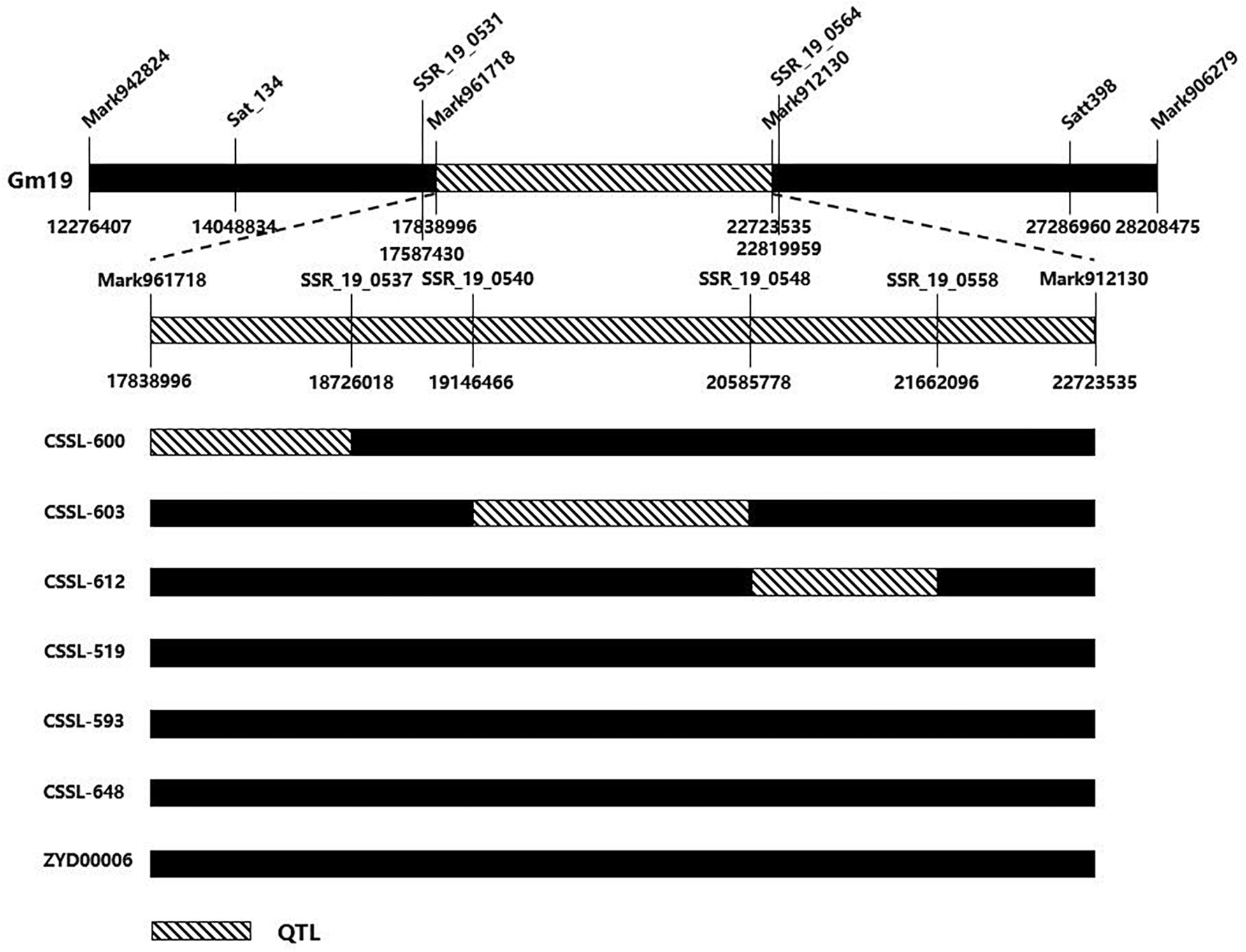
Figure 4. Validation of consensus quantitative trait locus (QTL) in chromosome segment substitution lines (CSSLs). The consensus QTL associated with nodulation phenotype on chromosome Gm19 (97.5–133.2 cM) had a corresponding partial region in the substituted wild soybean chromosomal segment on Gm19 in CSSL-600, CSSL-603, and CSSL-612. On the contrary, there was no corresponding region in CSSL-519, CSSL-593, CSSL-648, or ZYD00006.
Furthermore, our candidate QTL were close to those QTL in the previous reports and identified by the CSSL population. No QTL or genes interacting with NopD had been identified in previous reports, so it was interesting to determine whether the QTL identified in our work might interact with NopD.
Expression Analysis of Candidate Genes Associated With Nodulation Phenotype
In the confident QTL region, seven genes (Glyma.19g065800, Glyma.19g066800, Glyma.19g067200, Glyma.19g068300, Glyma.19g068600, Glyma.19g068800, and Glyma.19g069200) on Chromosome 19 associated with pathogen resistance, signal exchange, and symbiosis were selected for further analyses (Supplementary Table S5). To identify whether these genes have interaction with NopD, soybean Suinong14 was inoculated with the wild-type HH103, HH103ΩNopD, and HH103ΩTtsI. The expression pattern of these genes was detected by qRT-PCR (Figure 5). Among the seven genes, Glyma.19g067200 showed no expression in Suinong14 inoculated with any of the three strains because of no expression signal detected by qRT-PCR. At 12 h post-inoculation, the Glyma.19g065800 expression levels were significantly different comparing TtsI mutant with the wild strain and NopD mutant, at 24 and 60 h post-inoculation, the wild strain could induce the higher expression. It showed that NopD and TtsI mutant could induce a different expression pattern, indicating that Glyma.19g065800 had no obvious interaction with NopD. At 24 h post-inoculation with the wild-type strain HH103 and NopD mutant, the expression level of Glyma.19g066800 reached a maximum, but TtsI mutant did not have a similar trend. Glyma.19g068300 had the similar expression trend in soybean inoculated with three strains, and expression levels all changed around 1.0. At 12 h post-inoculation, the Glyma.19g068300 expression levels was significantly different inoculated with TtsI mutant compared to the wild strain and NopD mutant. These results showed that Glyma.19g068300 and Glyma.19g066800 had no obvious interaction with NopD similar with Glyma.19g065800. The expression of Glyma.19g068800 had a strange pattern; NopD mutant could induce the gene to reach two peaks at 24 and 60 h post-inoculation, its expression level was higher compared with inoculation with the wild strain and TtsI mutant which had a similar induction pattern, so we could not infer that Glyma.19g068800 could interact with NopD. Glyma.19g068600 and Glyma.19g069200 showed significantly different expression patterns in Suinong14 after inoculation with S. fredii HH103 and the two mutant strains. When Suinong14 was inoculated with S. fredii HH103, the expression patterns of Glyma.19g068600 and Glyma.19g069200 were similar. At 36 h post-inoculation with the wild-type strain HH103, the expression level of Glyma.19g068600 and Glyma.19g069200 reached a minimum, and then the expression level of these two genes increased. But in a clear difference from the wild-type strain, infection with the mutants HH103ΩNopD and HH103ΩTtsI showed no evident effects on the expression patterns of these two genes. At 36 h post-inoculation with HH103ΩNopD and HH103ΩTtsI, the expression level of Glyma.19g068600 was 6.7 and 5.2 times higher than expression induced by inoculation with HH103. Under the same conditions, the expression level of Glyma.19g069200 induced by HH103ΩNopD and HH103ΩTtsI was 10.7 and 12.0 times higher than that in plants induced by HH103, respectively. These qRT-PCR results supported the hypothesis that NopD interacted with Glyma.19g068600 and Glyma.19g069200 in the early stage of rhizobial infection.
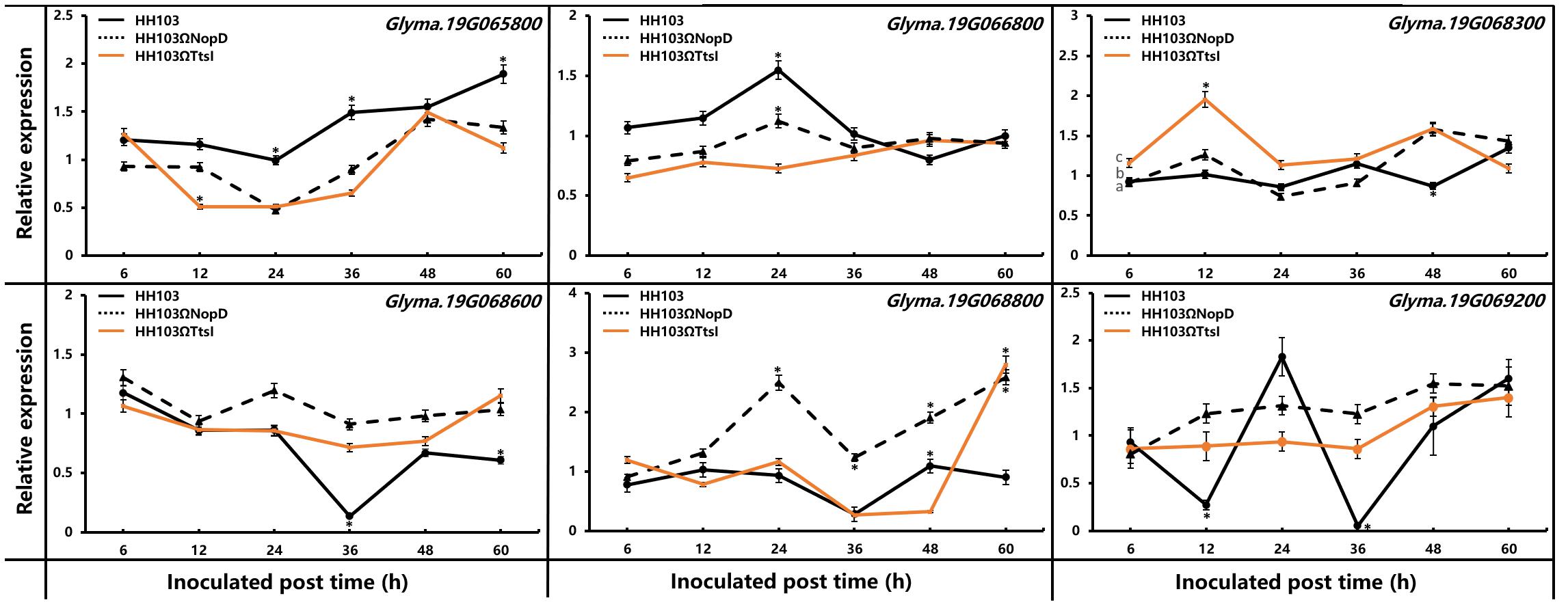
Figure 5. Relative expression of Glyma.19g065800, Glyma.19g066800, Glyma.19g068300, Glyma.19g068600, Glyma.19g068800, and Glyma.19g069200 was measured by qRT-PCR in RNA extracted from roots of soybean Suinong14 plants inoculated with S. fredii HH103 wild-type or mutants HH103ΩNopD and HH103ΩTtsI. Uninoculated Suinong14 plants were used as the control. The 2–ΔΔCt method was used to calculate the relative expression levels of candidate genes. Error bars indicate the mean ± standard error of three replications. Significant differences were determined by t-tests, and “*” indicated the significant differences (p ≤ 0.05) at the time point.
Haplotypes of Glyma.19g068600 and Glyma.19g069200 That Correlated With Nodulation Traits
Considering that Gm19g068600 and Gm19g069200 can be suppressed at a lower level in Suinong14 during the wild HH103 infecting (Figure 5) and nodule traits had the significant differences in parents of CSSL populations (Figure 1C). We analyzed the haplotypes of Gm19g068600 and Gm19g069200 in 142 accessions of CSSL populations. In total, 14 single-nucleotide polymorphisms (SNPs) and/or indels were found in the promoter and coding sequence of Gm19g068600 from the CSSL populations. According to 14 SNPs and/or indels, 142 soybean accessions were classified into eight haplotypes (Hap1–Hap8) (Figure 6A). Hap1, the largest group, includes 40 soybean accessions; Hap2 including 32 soybean accessions was the second largest group. In these two types, two SNPs are located in the exon, but these differences did not result in the amino acid change. Ten SNPs and indels located in the promoter sequence. The relative expression of gene was detected during some rhizobia strains infecting in Hap1 and Hap2, the relative expression showed that Gm19g068600 had the similar expression pattern in Hap1 accession and Hap2 accession after inoculated with the wild strain HH103, NopD mutant and TtsI mutant, respectively. The nodule traits of Hap1 and Hap2 accessions were further compared, the nodulation results show that the nodule traits of haplotypes do not have significant differences after inoculation with the wild HH103, NopD mutant, and TtsI mutant, respectively (Figure 6B). The relative expression and nodule trait analysis indicated that Gm19g068600 could be related to NopD, but haplotype difference could not influence expression pattern.
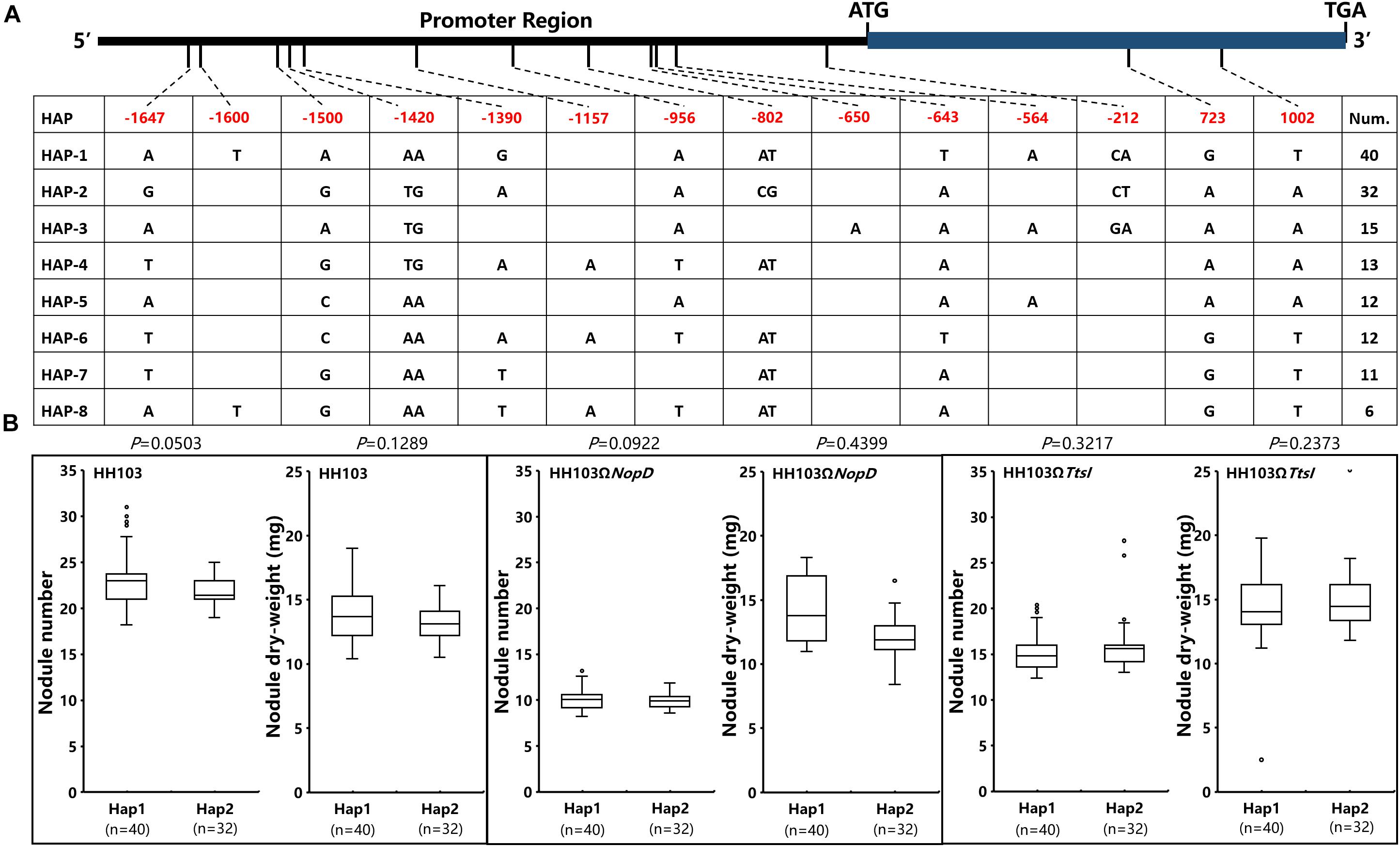
Figure 6. The analysis of Gm19g068600 haplotypes and nodule traits in two Hap accessions. (A) Gm19g068600 gene haplotypes (eight haplotypes) in chromosome segment substitution line (CSSL) populations (n = 142). The start codon site is set at “0” position. (B) Comparison and analysis of the nodule phenotype between haplotypes Hap1 (n = 40) and Hap2 (n = 32). n represents the number of the two haplotypes. Two-tailed t-test was used to detect the significance.
The haplotype analysis of Gm19g069200 shows that there were 15 SNPs and/or indels found in the promoter and coding sequence. Based on the 15 SNPs and/or indels, 142 soybean accessions were classified into seven haplotypes (Hap1–Hap7). Hap1, the largest group, includes 42 soybean accessions, and Hap2 was the second largest group, including 39 soybean accessions (Figure 7A). One SNP located in the exon, but the change did not result in the amino acid change, five SNPs or indels located in the promoter sequence. As it is known, the promoters always affects the function of genes by regulating gene expression, and the relative expression of Gm19g069200 was studied in one Hap1 accession and one Hap2 accession, respectively. These results show that the relative expression of Gm19g069200 had a significant difference in Hap1 and Hap2 soybean accession (Figure 8). Gm19g069200 could be regulated by NopD in HH103 via Hap1 accession, but not in Hap2 accession. The nodule traits of Hap1 and Hap2 accessions were further compared, and the nodulation results show that haplotypes have significantly different effects on nodule trait after inoculation with the wild strain, NopD mutant, and TtsI mutant. The NN and NDW of Hap1 soybean accessions were higher than the Hap2 soybean accessions, and the difference was significant (P < 0.01) (Figure 7B). Similar to the wild strain, NopD mutant and TtsI mutant could result in higher NN and NDW in Hap1 accessions than Hap2 accessions. NopD mutant and TtsI mutant could reduce NN and NDW compared with the wild strain in both Hap1 accessions and Hap2 accessions. These results suggested that Gm19g069200 can be associated with the rhizobia infection. Comparisons of nodule phenotypes of Hap1 accessions and Hap2 accessions inoculated with different strains show that the presence of NopD in the wild strain could cause a greater difference than the absence of NopD, indicating that NopD could be associated with Gm19g069200.
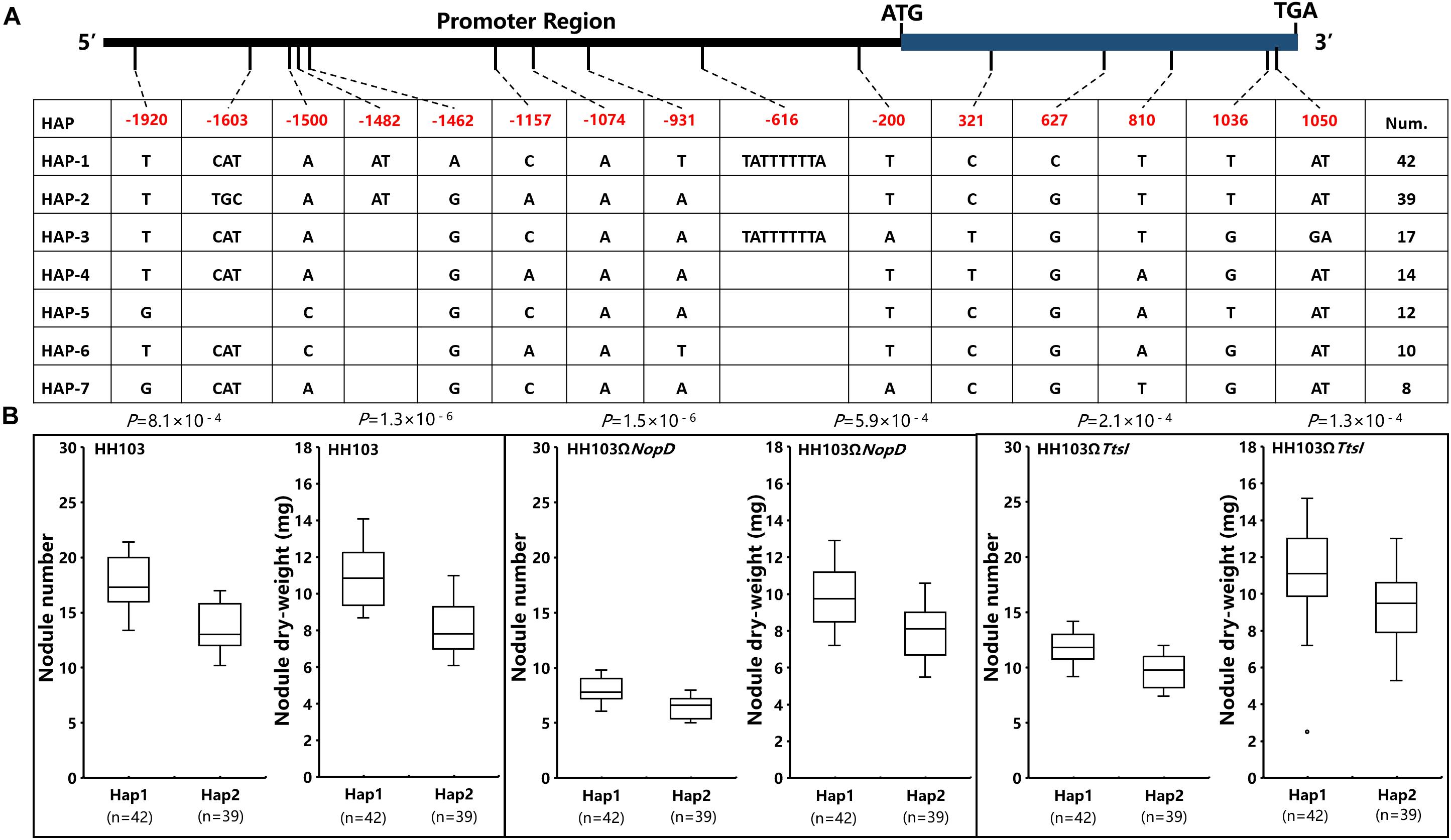
Figure 7. The analysis of Gm19g069200 haplotypes and nodule traits in two Hap accessions. (A) Gm19g069200 gene haplotypes (seven haplotypes) in chromosome segment substitution line (CSSL) populations (n = 142). The start codon site is set at “0” position. (B) Comparison and analysis of the nodule phenotype between haplotypes Hap1 (n = 42) and Hap2 (n = 39). n represents the number of the two haplotypes. Two-tailed t-test was used to detect the significance.
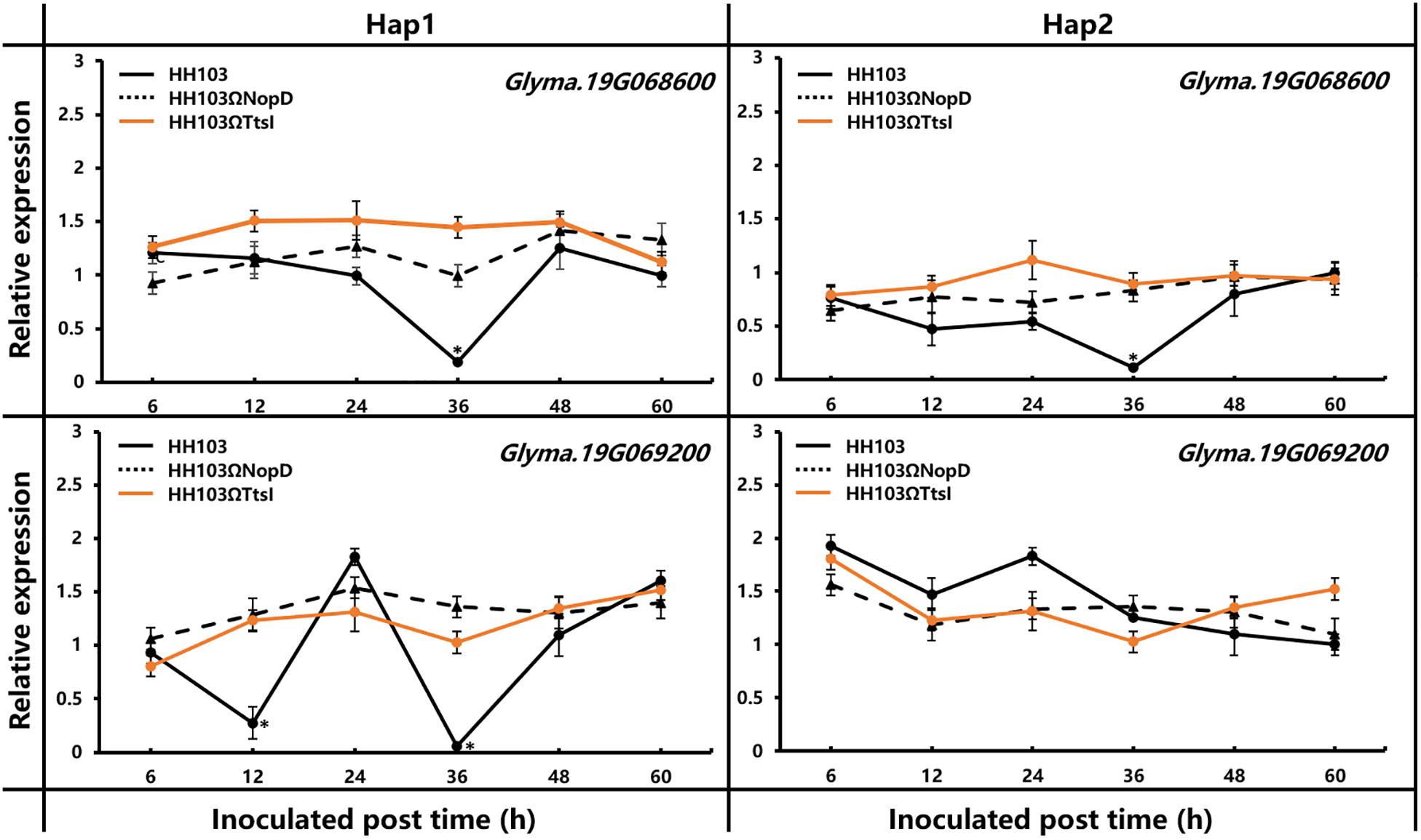
Figure 8. Relative expression of Glyma.19g066800 and Glyma.19g069200 was measured by qRT-PCR in RNA extracted from roots of Hap1 and Hap2 accession plants inoculated with S. fredii HH103 wild-type or mutants HH103ΩNopD and HH103ΩTtsI. Uninoculated soybean plants were used as the control. The 2–ΔΔCt method was used to calculate the relative expression levels of candidate genes. Error bars indicate the mean ± standard error of three replications. Significant differences were determined by t-tests, and ‘*’ indicated the significant differences (p ≤ 0.05) at the time point.
Discussion
The locus we identified in this study on soybean chromosome Gm08 overlaps with a previously identified QTL related to compatibility of soybean with Bradyrhizobium strains (Ramongolalaina et al., 2018). A QTL previously shown to be associated with nodule weight (Hwang et al., 2014) was found to be adjacent to the QTL on Gm18 in our study. In addition, a locus on Gm17 related to NopL, a T3SS effector of S. fredii HH103 (Zhang et al., 2018), overlaps with a locus adjacent to a QTL on Gm17 that we detected.
We identified two genes (Glyma.19g068600 and Glyma.19g069200) by qRT-PCR that had a similar expression pattern induced by the wild-type strain HH103 (showing a minimum of expression at 36 h after inoculation), but this pattern was not identified when soybean was inoculated with the mutant HH103ΩNopD or HH103ΩTtsI. This expression pattern suggests that Glyma.19g068600 and Glyma.19g069200 might be associated with the NopD-triggered signaling pathway activated after inoculation with rhizobia. Glyma.19g068600 encodes a protein belonging to the F-Box/LRR-repeat (FBD/LRR) protein family. FBD/LRR proteins have been identified in several plant species, for instance, in radish (Zhai et al., 2016), soybean (Chen et al., 2018), tomato (Du et al., 2014; Quintana-Camargo et al., 2015), and peanut (Agarwal et al., 2018). Some members of the FBD/LRR protein family have been found to be associated with pathogen resistance, such as tomato FBD/LRR3, where overexpression of FBD/LRR3 increased plant resistance to X. perforans (Du et al., 2014). One novel FBD/LRR gene was identified from a high-density genetic map, and expression of this gene was induced during pathogen resistance responses in peanut (Agarwal et al., 2018). Using proteome analysis, GmFBD/LRR was shown to have a different expression pattern in response to Phytophthora sojae infection. The FBD/LRR proteins contain an F-box domain and LRR-repeat domain. The F-box domain is usually associated with a transcription factor that participates in the defense response induced by jasmonic acid (Xu et al., 2009). The LRR-repeat domain has been identified in a functionally and evolutionarily diverse series of proteins. In these proteins, the LRR-repeat domains are essential to protein–protein interactions or signal transduction (Tornero et al., 1996). From the nodulation tests in this study, the NopD mutant decreased NN and NDW in most soybean germplasms, suggesting that NopD could play a positive role in rhizobial infection and nodule formation. Our qRT-PCR results confirmed that NopD can depress FBD/LRR expression during infection with the wild-type strain HH103. The haplotype analysis classified the haplotypes into eight types, and Hap1 and Hap2 were the major two types, but Hap1 and Hap2 could not change the gene expression pattern and nodule traits inoculated with the same rhizobia strain. The finding that FBD/LRR encodes an FBD/LRR protein was unexpected. Since some FBD/LRR proteins in other plants are known to be associated with resistance to pathogens, the interaction of NopD with an FBD/LRR protein is interesting, and so further research will be necessary to clarify the nature of this interaction.
Glyma.19g069200 encodes a protein phosphatase 2C (PP2C). The PP2C protein family is widely distributed in almost all plants, whether eukaryotic or prokaryotic plant. PP2C proteins interact with many signaling pathways. Overexpression of the rice PP2C gene OsBIPP2C1 in transgenic tobacco was found to suppress pathogen infection and enhance some abiotic tolerance (Hu et al., 2006). GmPP2C3a, a member of the soybean PP2C protein family, was identified as an antiviral protein that was able to suppress virus infection and spread (Seo et al., 2014). Earlier studies indicated that PP2Cs negatively regulate the mitogen-activated protein kinase (MAPK) pathways in yeast and Arabidopsis thaliana (Meskiene et al., 2003), as well as the MAPK pathway triggered by a bacterial flagellin (Cristina et al., 2010). The MAPK pathway might be involved in establishing the rhizobial symbiosis since MtTDY1, one of the MAPK pathway genes, was shown to be associated with nodule formation and to regulate the development of the root tip (Schoenbeck et al., 1999). In Lupinus albus, bradyrhizobia can activate MAPK genes SIMK and SAMK, and mutants of SIMK and SAMK inhibited bradyrhizobial infection (Fernandez-Pascual et al., 2006). In soybean, GMK1 has been identified as a MAPK homolog, and its expression was associated with infection by Bradyrhizobium japonicum USDA110 (Lee et al., 2008). T3SS is essential to rhizobial infection, and T3Es can also interact with MAPKs during establishment of symbiosis. The T3E NopP can induce MAPK3 expression at an early stage of rhizobial infection (Wang et al., 2018). In specific Lotus japonicus lines of a monogenic-recessive mutant carrying the symbiosis-associated locus, considerable changes to LjNPP2C1 were observed, suggesting that LjNPP2C1 may be functional during the early and late nodule development stages (Kapranov et al., 1999). NopL, another T3SS effector of HH103, was shown to suppress the expression of the PP2C-related protein Glyma.07g099700 during HH103 infection of soybean (Zhang et al., 2018). In this study, expression pattern and haplotype analysis of PP2C gene were studied; the result showed PPC2 to be depressed by the wild-type strain HH103 (which produces NopD) in Hap1 accession, but not in Hap2. This also explained that PP2C had a different expression regulation pattern because of the difference in promoter region. Nodule traits indicated that there was a significant difference between Hap1 and Hap2 inoculated with the wild strain HH103, NopD mutant, and TtsI mutant. These results suggesting that expression of PP2C could regulation nodule formation. It is interesting to detect the interaction mechanism between NopD and PP2C. However, further research is essential to fully clarify this interaction.
The T3Es have been shown to be secreted into legume cells via rhizobial T3SSs, in a similar way to many gram-negative pathogenic bacteria. In a previous study, the function of rhizobial T3Es in legume cells has been questioned (Büttner and Bonas, 2006). However, more recent research has confirmed that T3 effectors are essential to symbiosis (Schechter et al., 2010; Wenzel et al., 2010). NopD is a conserved T3Er in most rhizobial strains, and our nodulation tests showed that its secretion can positively influence the formation of nodules. The expression of NopD also induced tobacco leaf cell death, giving us evidence that NopD was directly or indirectly recognized by a non-host plant. In our study, NopD influenced the immunity pathway but had no effect on the symbiosis pathway (Figure 2). This is similar to XopD, one of the X. campestris pv. vesicatoria T3SS effectors, which belongs to the C48 cysteine peptidase family. XopD is able to suppress the host immunity and so promote infection by the pathogens. This suggested that NopD could influence the symbiosis between soybean and rhizobia by affecting signal transduction in the host immunity system. We propose that NopD inhibits FBD/LRR and PP2C expression, thereby promoting infection by the wild-type HH103.
In this work, FBD/LRR and PP2C were identified by QTL mapping and can be used to aid further detect of signaling communication induced by NopD. Further identification and clarification of the host genes involved in interactions with rhizobial effector molecules could enhance the understanding of symbiosis establishment.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation, to any qualified researcher.
Author Contributions
DX, QC, and CL conceived the study and designed and managed the experiments. ZQ, HJ, RZ, and XW provided soybean seeds. JinW, JieW, CM, JL, LC, and DY performed trials and collected data. JinW, QK, HZ, JieW, ZS, HLiu, ZZ, JZ, HLi, and QW completed statistical analyses of phenotypic data and wrote the manuscript. DX, QC, and JinW participated in correcting the manuscript. All authors contributed to writing the manuscript.
Funding
This work was supported by the Ministry of Science and Technology of the People’s Republic of China Project (Grant No. 2017YFE0111000), the Europe Horizon 2020 (EUCLEG, 727312), the National Natural Science Foundation of China (Grant Nos. 31771882, 31971899, 31400074, 31471516, 31271747, and 30971809), the Natural Science Foundation of Heilongjiang Province of China (Grant No. ZD201213), the Heilongjiang Postdoctoral Science Foundation (Grant No. LBH-Q16014), the Harbin Science Technology Project (Grant Nos. 2013RFQXJ005 and 2014RFXXJ012), and the Foundation for University Key Teachers from the Education Department of Heilongjiang Province in China (Grant No. 1254G011), University Project of Young Scientist (UNPYSCT-2015011).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Huw Tyson, Ph.D., from Liwen Bianji, Edanz Editing China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2020.00453/full#supplementary-material
Footnotes
References
Agarwal, G., Clevenger, J., Pandey, M. K., Wang, H., Shasidhar, Y., Chu, Y., et al. (2018). High-density genetic map using whole-genome resequencing for fine mapping and candidate gene discovery for disease resistance in peanut. Plant Biotechnol. J. 16, 1954–1967. doi: 10.1111/pbi.12930
Brensha, W., Kantartzi, S. K., Meksem, K., Grier, R. L. IV, Barakat, A., Lightfoot, D. A., et al. (2012). Genetic analysis of root and shoot traits in the ‘Essex’by ‘Forrest’recombinant inbred line (RIL) population of soybean [Glycine max (L.) Merr.]. J. Plant Genome Sci. 1, 1–9. doi: 10.5147/jpgs.2012.0051
Büttner, D., and Bonas, U. (2006). Who comes first? How plant pathogenic bacteria orchestrate type III secretion. Curr. Opin. Microbiol. 9, 193–200. doi: 10.1016/j.mib.2006.02.006
Caetano-Anollés, G., and Gresshoff, P. M. (1991). Plant genetic control of nodulation. Annu. Rev. Microbiol. 45, 345–382. doi: 10.1146/annurev.mi.45.100191.002021
Caldwell, B. E. (1966). Inheritance of a strain-specific ineffective nodulation in soybeans 1. Crop Sci. 6, 427–428.
Canonne, J., Marino, D., Jauneau, A., Pouzet, C., Brière, C., Roby, D., et al. (2011). The Xanthomonas type III effector XopD targets the Arabidopsis transcription factor MYB30 to suppress plant defense. Plant Cell 23, 3498– 3511.
Chen, Q. S., Yu, G. L., Zou, J. N., Jing, W. A. N. G., and Qiu, H. M. (2018). GmDRR1, a dirigent protein resistant to Phytophthora sojae in Glycine max (L.) Merr. J. Integr. Agric. 17, 1289–1298.
Crespo-Rivas, J. C., Margaret, I., Pérez-Montaño, F., López-Baena, F. J., Vinardell, J. M., Ollero, F. J., et al. (2007). A pyrF auxotrophic mutant of Sinorhizobium fredii HH103 impaired in its symbiotic interaction with soybean and other legumes. Int. Microbiol. 10, 169–176. doi: 10.2436/20.1501.01.24
Cristina, M. S., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant Biol. 61, 621–649. doi: 10.1073/pnas.252641899
Du, H., Li, W., Wang, Y., and Yang, W. (2014). Identification of genes differentially expressed between resistant and susceptible tomato lines during time-course interactions with Xanthomonas perforans race T3. PLoS One 9:e93476. doi: 10.1371/journal.pone.0093476
Faruque, O. M., Miwa, H., Yasuda, M., Fujii, Y., Kaneko, T., Sato, S., et al. (2015). Identification of Bradyrhizobium elkanii genes involved in incompatibility with soybean plants carrying the Rj4 allele. Appl. Environ. Microbiol. 81, 6710–6717. doi: 10.1128/AEM.01942-15
Fernandez-Pascual, M., Lucas, M. M., de Felipe, M. R., Boscá, L., Hirt, H., and Golvano, M. P. (2006). Involvement of mitogen-activated protein kinases in the symbiosis Bradyrhizobium-Lupinus. J. Exp. Bot. 57, 2735–2742. doi: 10.1093/jxb/erl038
Figurski, D. H., Meyer, R. J., and Helinski, D. R. (1979). Suppression of ColE1 replication properties by the Inc P-1 plasmid RK2 in hybrid plasmids constructed in vitro. J. Mol. Biol. 133, 295–318. doi: 10.1016/0022-2836(79)90395-4
Hotson, A., Chosed, R., Shu, H., Orth, K., and Mudgett, M. B. (2003). Xanthomonas type III effector XopD targets SUMO-conjugated proteins in planta. Mol. Microbiol. 50, 377–389. doi: 10.1046/j.1365-2958.2003.03730.x
Hu, X., Song, F., and Zheng, Z. (2006). Molecular characterization and expression analysis of a rice protein phosphatase 2C gene, OsBIPP2C1, and overexpression in transgenic tobacco conferred enhanced disease resistance and abiotic tolerance. Physiol. Plant. 127, 225–236. doi: 10.1111/j.1399-3054.2006.00671.x
Hwang, S., Ray, J. D., Cregan, P. B., King, C. A., Davies, M. K., and Purcell, L. C. (2014). Genetics and mapping of quantitative traits for nodule number, weight, and size in soybean (Glycine max L.[Merr.]). Euphytica 195, 419–434.
Jian, B., Liu, B., Bi, Y., Hou, W., Wu, C., and Han, T. (2008). Validation of internal control for gene expression study in soybean by quantitative real-time PCR. BMC Mol. Biol. 9:59. doi: 10.1186/1471-2199-9-59
Jiang, H., Ying, L. Y., Qin, H., Qi, H., Li, C., Wang, N., et al. (2018). Identification of major QTLs associated with first pod height and candidate gene mining in soybean. Front. Plant Sci. 9:1280. doi: 10.3389/fpls.2018.01280
Jiménez-Guerrero, I., Pérez-Montaño, F., Medina, C., Ollero, F. J., and López-Baena, F. J. (2015a). NopC is a Rhizobium-specific type 3 secretion system effector secreted by Sinorhizobium (Ensifer) fredii HH103. PLoS One 10:e0142866. doi: 10.1371/journal.pone.0142866
Jiménez-Guerrero, I., Pérez-Montaño, F., Monreal, J. A., Preston, G. M., Fones, H., Vioque, B., et al. (2015b). The Sinorhizobium (Ensifer) fredii HH103 type 3 secretion system suppresses early defense responses to effectively nodulate soybean. Mol. Plant Microbe 28, 790–799. doi: 10.1094/MPMI-01-15-0020-R
Jiménez-Guerrero, I., Pérez-Montaño, F., Zdyb, A., Beutler, M., Werner, G., Göttfert, M., et al. (2019). GunA of Sinorhizobium (Ensifer) fredii HH103 is a T3SS-secreted cellulase that differentially affects symbiosis with cowpea and soybean. Plant Soil 435, 15–26.
Kaneko, T., Nakamura, Y., Sato, S., Minamisawa, K., Uchiumi, T., Sasamoto, S., et al. (2002). Complete genomic sequence of nitrogen-fixing symbiotic bacterium Bradyrhizobium japonicum USDA110. DNA Res. 9, 189–197. doi: 10.1093/dnares/9.6.189
Kapranov, P., Jensen, T. J., Poulsen, C., De Bruijn, F. J., and Szczyglowski, K. (1999). A protein phosphatase 2C gene, LjNPP2C1, from Lotus japonicus induced during root nodule development. Proc. Natl. Acad. Sci. U.S.A. 96, 1738–1743. doi: 10.1073/pnas.96.4.1738
Kim, J. G., Stork, W., and Mudgett, M. B. (2013). Xanthomonas type III effector XopD desumoylates tomato transcription factor SlERF4 to suppress ethylene responses and promote pathogen growth. Cell Host Microbe 13, 143–154. doi: 10.1016/j.chom.2013.01.006
Kim, W. S., and Krishnan, H. B. (2014). A nopA deletion mutant of Sinorhizobium fredii USDA257, a soybean symbiont, is impaired in nodulation. Curr. Microbiol. 68, 239–246. doi: 10.1007/s00284-013-0469-4
Ladha, J. K., and Peoples, M. B. (eds) (2012). Management of Biological Nitrogen Fixation for the Development of More Productive and Sustainable Agricultural Systems: Extended Versions of Papers Presented at the Symposium on Biological Nitrogen Fixation for Sustainable Agriculture at the 15th Congress of Soil Science, Acapulco, Mexico, 1994, Vol. 65, (Dordrecht: Springer). doi: 10.1007/978-94-011-0053-3_1
Lee, H., Kim, J., Im, J. H., Kim, H. B., Oh, C. J., and An, C. S. (2008). Mitogen-activated protein kinase is involved in the symbiotic interaction between Bradyrhizobium japonicum USDA110 and soybean. J. Plant Biol. 51, 291–296. doi: 10.1007/BF03036129
López Baena, F. J., Vinardell González, J. M., Pérez Montaño, F. D. A., Crespo Rivas, J. C., Bellogín Izquierdo, R. A., Espuny Gómez, M. D. R., et al. (2008). Regulation and symbiotic significance of nodulation outer proteins secretion in Sinorhizobium fredii HH103. Microbiology 154, 1825–1836. doi: 10.1099/mic.0.2007/016337-0
López-Baena, F., Ruiz-Sainz, J., Rodríguez-Carvajal, M., and Vinardell, J. (2016). Bacterial molecular signals in the Sinorhizobium fredii-soybean symbiosis. Int. J. Mol. Sci. 17:E755. doi: 10.3390/ijms17050755
Mackey, D., Holt, B. F. III, Wiig, A., and Dangl, J. L. (2002). RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell 108, 743–754. doi: 10.1016/S0092-8674(02)00661-X
Margaret, I., Becker, A., Blom, J., Bonilla, I., Goesmann, A., Göttfert, M., et al. (2011). Symbiotic properties and first analyses of the genomic sequence of the fast growing model strain Sinorhizobium fredii HH103 nodulating soybean. J. Biotechnol. 155, 11–19. doi: 10.1016/j.jbiotec.2011.03.016
Marie, C., Deakin, W. J., Viprey, V., Kopciñska, J., Golinowski, W., Krishnan, H. B., et al. (2003). Characterization of Nops, nodulation outer proteins, secreted via the type III secretion system of NGR234. Mol. Plant Microbe 16, 743–751. doi: 10.1094/MPMI.2003.16.9.743
Meskiene, I., Baudouin, E., Schweighofer, A., Liwosz, A., Jonak, C., Rodriguez, P. L., et al. (2003). Stress-induced protein phosphatase 2C is a negative regulator of a mitogen-activated protein kinase. J. Biol. Chem. 278, 18945–18952. doi: 10.1074/jbc.M300878200
Miwa, H., and Okazaki, S. (2017). How effectors promote beneficial interactions. Curr. Opin. Plant Biol. 38, 148–154. doi: 10.1016/j.pbi.2017.05.011
Okazaki, S., Kaneko, T., Sato, S., and Saeki, K. (2013). Hijacking of leguminous nodulation signaling by the rhizobial type III secretion system. Proc. Natl. Acad. Sci. U.S.A. 110, 17131–17136. doi: 10.1073/pnas.1302360110
Pérez-Montaño, F., Jiménez-Guerrero, I., Acosta-Jurado, S., Navarro-Gómez, P., Ollero, F. J., Ruiz-Sainz, J. E., et al. (2016). A transcriptomic analysis of the effect of genistein on Sinorhizobium fredii HH103 reveals novel rhizobial genes putatively involved in symbiosis. Sci. Rep. 6:31592. doi: 10.1038/srep31592
Qi, Z., Huang, L., Zhu, R., Xin, D., Liu, C., Han, X., et al. (2014). A high-density genetic map for soybean based on specific length amplified fragment sequencing. PLoS One 9:e104871. doi: 10.1371/journal.pone.0104871
Quandt, J., and Hynes, M. F. (1993). Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene 127, 15–21. doi: 10.1016/0378-1119(93)90611-6
Quintana-Camargo, M., Méndez-Morán, L., Ramirez-Romero, R., Gurrola-Díaz, C. M., Carapia-Ruiz, V., Ibarra-Laclette, E., et al. (2015). Identification of genes differentially expressed in husk tomato (Physalis philadelphica) in response to whitefly (Trialeurodes vaporariorum) infestation. Acta Physiol. Plant. 37:29. doi: 10.1007/s11738-015-1777-z
Ramongolalaina, C., Teraishi, M., and Okumoto, Y. (2018). QTLs underlying the genetic interrelationship between efficient compatibility of Bradyrhizobium strains with soybean and genistein secretion by soybean roots. PLoS One 13:e0194671. doi: 10.1371/journal.pone.0194671
Riches, D., Porter, I. J., Oliver, D. P., Bramley, R. G. V., Rawnsley, B., Edwards, J., et al. (2013). Soil biological properties as indicators of soil quality in Australian viticulture. Aust. J. Grape Wine Res. 19, 311–323. doi: 10.1111/ajgw.12034
Riely, B. K., Larrainzar, E., Haney, C. H., Mun, J. H., Gil-Quintana, E., González, E. M., et al. (2013). Development of tools for the biochemical characterization of the symbiotic receptor-like kinase DMI2. Mol. Plant Microbe 26, 216–226. doi: 10.1094/MPMI-10-11-0276
Rodrigues, J. A., Lopez-Baena, F. J., Ollero, F. J., Vinardell, J. M., Espuny, M. D. R., Bellogín, R. A., et al. (2007). NopM and NopD are rhizobial nodulation outer proteins: identification using LC-MALDI and LC-ESI with a monolithic capillary column. J. Proteome Res. 6, 1029–1037. doi: 10.1021/pr060519f
Saad, M. M., Kobayashi, H., Marie, C., Brown, I. R., Mansfield, J. W., Broughton, W. J., et al. (2005). NopB, a type III secreted protein of Rhizobium sp. strain NGR234, is associated with pilus-like surface appendages. J. Bacteriol. 187, 1173–1181. doi: 10.1128/JB.187.3.1173-1181.2005
Schechter, L. M., Guenther, J., Olcay, E. A., Jang, S., and Krishnan, H. B. (2010). Translocation of NopP by Sinorhizobium fredii USDA257 into Vigna unguiculata root nodules. Appl. Environ. Microbiol. 76, 3758–3761. doi: 10.1128/AEM.03122-09
Schoenbeck, M. A., Samac, D. A., Fedorova, M., Gregerson, R. G., Gantt, J. S., and Vance, C. P. (1999). The alfalfa (Medicago sativa) TDY1 gene encodes a mitogen-activated protein kinase homolog. Mol. Plant Microbe 12, 882–893. doi: 10.1094/MPMI.1999.12.10.882
Seo, J. K., Kwon, S. J., Cho, W. K., Choi, H. S., and Kim, K. H. (2014). Type 2C protein phosphatase is a key regulator of antiviral extreme resistance limiting virus spread. Sci. Rep. 4:5905. doi: 10.1038/srep05905
Staehelin, C., and Krishnan, H. B. (2015). Nodulation outer proteins: double-edged swords of symbiotic rhizobia. Biochem. J. 470, 263–274. doi: 10.1042/BJ20150518
Sugawara, M., Takahashi, S., Umehara, Y., Iwano, H., Tsurumaru, H., Odake, H., et al. (2018). Variation in bradyrhizobial NopP effector determines symbiotic incompatibility with Rj2-soybeans via effector-triggered immunity. Nat. Commun. 9:3139. doi: 10.1038/s41467-018-05663-x
Tampakaki, A. P. (2014). Commonalities and differences of T3SSs in rhizobia and plant pathogenic bacteria. Front. Plant Sci. 5:114. doi: 10.3389/fpls.2014.00114
Tang, F., Yang, S., Liu, J., and Zhu, H. (2016). Rj4, a gene controlling nodulation specificity in soybeans, encodes a thaumatin-like protein but not the one previously reported. Plant Physiol. 170, 26–32. doi: 10.1104/pp.15.01661
Tennant, J. R. (1964). Evaluation of the trypan blue technique for determination of cell viability. Transplantation 2, 685–694. doi: 10.1097/00007890-196411000-00001
Tornero, P., Mayda, E., Gómez, M. D., Cañas, L., Conejero, V., and Vera, P. (1996). Characterization of LRP, a leucine-rich repeat (LRR) protein from tomato plants that is processed during pathogenesis. Plant J. 10, 315–330. doi: 10.1046/j.1365-313X.1996.10020315.x
Vance, C. P. (2001). Symbiotic nitrogen fixation and phosphorus acquisition, Plant nutrition in a world of declining renewable resources. Plant Physiol. 127, 390–397. doi: 10.1104/pp.010331
Vest, G. (1970). Rj3 A gene conditioning ineffective nodulation in soybean 1. Crop Sci. 10, 34–35. doi: 10.2135/cropsci1970.0011183X001000010013x
Vest, G., and Caldwell, B. E. (1972). Rj4-A gene conditioning ineffective nodulation in soybean1. Crop Sci. 12, 692–693. doi: 10.2135/cropsci1972.0011183X001200050042x
Vinardell, J. M., Acosta-Jurado, S., Zehner, S., Goettfert, M., Becker, A., Baena, I., et al. (2015). The Sinorhizobium fredii HH103 genome: a comparative analysis with S. fredii strains differing in their symbiotic behavior with soybean. Mol. Plant Microbe 28, 811–824. doi: 10.1094/MPMI-12-14-0397-FI
Vuong, T. D., Nickell, C. D., and Harper, J. E. (1996). Genetic and allelism analyses of hypernodulation soybean mutants from two genetic backgrounds. Crop Sci. 36, 1153–1158. doi: 10.2135/cropsci1996.0011183X003600050015x
Wang, J., Wang, J., Liu, C., Ma, C., Li, C., Zhang, Y., et al. (2018). Identification of soybean genes whose expression is affected by the Ensifer fredii HH103 effector protein NopP. Int. J. Mol. Sci. 19:3438. doi: 10.3390/ijms19113438
Weidner, S., Becker, A., Bonilla, I., Jaenicke, S., Lloret, J., Margaret, I., et al. (2012). Genome sequence of the soybean symbiont Sinorhizobium fredii HH103. J. Bacteriol. 194, 1617–1618.
Wenzel, M., Friedrich, L., Göttfert, M., and Zehner, S. (2010). The type III–secreted protein NopE1 affects symbiosis and exhibits a calcium-dependent autocleavage activity. Mol. Plant Microbe 23, 124–129. doi: 10.1094/MPMI-23-1-0124
Xin, D., Qi, Z., Jiang, H., Hu, Z., Zhu, R., Hu, J., et al. (2016). QTL location and epistatic effect analysis of 100-seed weight using wild soybean (Glycine soja Sieb. & Zucc.) chromosome segment substitution lines. PLoS One 11:e0149380. doi: 10.1371/journal.pone.0149380
Xin, D. W., Liao, S., Xie, Z. P., Hann, D. R., Steinle, L., Boller, T., et al. (2012). Functional analysis of NopM, a novel E3 ubiquitin ligase (NEL) domain effector of Rhizobium sp. strain NGR234. PLoS Pathog. 8:e1002707. doi: 10.1371/journal.ppat.1002707
Xu, G., Ma, H., Nei, M., and Kong, H. (2009). Evolution of F-box genes in plants: different modes of sequence divergence and their relationships with functional diversification. Proc. Natl. Acad. Sci. U.S.A. 106, 835–840. doi: 10.1073/pnas.0812043106
Yang, S., Tang, F., Gao, M., Krishnan, H. B., and Zhu, H. (2010). R gene-controlled host specificity in the legume-rhizobia symbiosis. Proc. Natl. Acad. Sci. U.S.A. 107, 18735–18740. doi: 10.1073/pnas.1011957107
Zhai, L., Xu, L., Wang, Y., Zhu, X., Feng, H., Li, C., et al. (2016). Transcriptional identification and characterization of differentially expressed genes associated with embryogenesis in radish (Raphanus sativus L.). Sci. Rep. 6:21652. doi: 10.1038/srep21652
Zhang, Y., Liu, X., Chen, L., Fu, Y., Li, C., Qi, Z., et al. (2018). Mining for genes encoding proteins associated with NopL of Sinorhizobium fredii HH103 using quantitative trait loci in soybean (Glycine max Merr.) recombinant inbred lines. Plant Soil 431, 245–255. doi: 10.1007/s11104-018-3745-z
Zimmer, S., Messmer, M., Haase, T., Piepho, H. P., Mindermann, A., Schulz, H., et al. (2016). Effects of soybean variety and Bradyrhizobium strains on yield, protein content and biological nitrogen fixation under cool growing conditions in Germany. Eur. J. Agron. 72, 38–46. doi: 10.1016/j.eja.2015.09.008
Keywords: symbiosis, T3SS effector, NopD, quantitative trait locus (QTL), haplotype, soybean
Citation: Wang J, Wang J, Ma C, Zhou Z, Yang D, Zheng J, Wang Q, Li H, Zhou H, Sun Z, Liu H, Li J, Chen L, Kang Q, Qi Z, Jiang H, Zhu R, Wu X, Liu C, Chen Q and Xin D (2020) QTL Mapping and Data Mining to Identify Genes Associated With the Sinorhizobium fredii HH103 T3SS Effector NopD in Soybean. Front. Plant Sci. 11:453. doi: 10.3389/fpls.2020.00453
Received: 13 December 2019; Accepted: 27 March 2020;
Published: 19 May 2020.
Edited by:
Deyue Yu, Nanjing Agricultural University, ChinaReviewed by:
Zhihui Shan, Oil Crops Research Institute, Chinese Academy of Agricultural Sciences, ChinaDan Zhang, Henan Agricultural University, China
Copyright © 2020 Wang, Wang, Ma, Zhou, Yang, Zheng, Wang, Li, Zhou, Sun, Liu, Li, Chen, Kang, Qi, Jiang, Zhu, Wu, Liu, Chen and Xin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyan Liu, cyliucn@126.com; Qingshan Chen, qshchen@126.com; Dawei Xin, xdawei@163.com
†These authors have contributed equally to this work
 Jinhui Wang1†
Jinhui Wang1† Zhaoming Qi
Zhaoming Qi Rongsheng Zhu
Rongsheng Zhu Xiaoxia Wu
Xiaoxia Wu Qingshan Chen
Qingshan Chen Dawei Xin
Dawei Xin