- 1Biosystematics Group, Experimental Plant Sciences, Wageningen University and Research, Wageningen, Netherlands
- 2Department of Horticulture, Institute of Plant Breeding, Genetics and Genomics, University of Georgia, Tifton Campus, Tifton, GA, United States
Parthenogenesis is the spontaneous development of an embryo from an unfertilized egg cell. It naturally occurs in a variety of plant and animal species. In plants, parthenogenesis usually is found in combination with apomeiosis (the omission of meiosis) and pseudogamous or autonomous (with or without central cell fertilization) endosperm formation, together known as apomixis (clonal seed production). The initiation of embryogenesis in vivo and in vitro has high potential in plant breeding methods, particularly for the instant production of homozygous lines from haploid gametes [doubled haploids (DHs)], the maintenance of vigorous F1-hybrids through clonal seed production after combining it with apomeiosis, reverse breeding approaches, and for linking diploid and polyploid gene pools. Because of this large interest, efforts to identify gene(s) for parthenogenesis from natural apomicts have been undertaken by using map-based cloning strategies and comparative gene expression studies. In addition, engineering parthenogenesis in sexual model species has been investigated via mutagenesis and gain-of-function strategies. These efforts have started to pay off, particularly by the isolation of the PsASGR-BabyBoom-Like from apomictic Pennisetum, a gene proven to be transferable to and functional in sexual pearl millet, rice, and maize. This review aims to summarize the current knowledge on parthenogenesis, the possible gene candidates also outside the grasses, and the use of these genes in plant breeding protocols. It shows that parthenogenesis is able to inherit and function independently from apomeiosis and endosperm formation, is expressed and active in the egg cell, and can induce embryogenesis in polyploid, diploid as well as haploid egg cells in plants. It also shows the importance of genes involved in the suppression of transcription and modifications thereof at one hand, and in embryogenesis for which transcription is allowed or artificially overexpressed on the other, in parthenogenetic reproduction. Finally, it emphasizes the importance of functional endosperm to allow for successful embryo growth and viable seed production.
Introduction
Parthenogenesis and Spontaneous Embryo Development
Parthenogenesis is the spontaneous development of an embryo from an unfertilized egg cell: parthenos = virgin, genesis = creation. It naturally occurs in a variety of plant and animal species, particularly in lower plants such as mosses and algae and species-rich invertebrate groups such as insects, nematodes, and crustaceans, but also in c. 10% of the fern and 1% of the flowering plant species, and as rare examples in vertebrates (Bell, 1982; Suomalainen et al., 1987; Asker and Jerling, 1992; Schön et al., 2009; Hand and Koltunow, 2014; Grusz, 2016). In plants, the egg cell develops within a female gametophyte, which is a multicellular organism that arises from a megaspore, a product of female meiosis. The female gametophyte is thus haploid (1n) and alternates with the diploid (2n) sporophytic generation after fertilization of the egg cell with a haploid male sperm cell (e.g., Niklas and Kutschera, 2010; Schmidt et al., 2015; Bowman et al., 2016). In more primitive plants, the mosses and ferns, female gametophytes are relatively large, free-living organisms and egg cells develop in special regions, the archegonia. In higher plants, the female gametophyte (also embryo sac) is highly reduced. Circa 70% of the angiosperm species produce female gametophytes of the Polygonum-type, which consists of seven cells only: two gametes, the egg cell and central cell, and five accessory cells, the two synergids and three antipodals (Maheshwari, 1950). Animals lack an intermediate organism similar to the female gametophyte. Here, the egg cell is a direct product of meiosis and, as such, similar to the megaspore.
Parthenogenesis usually occurs in combination with a mechanism that keeps or restores the diploid chromosome number, since haploid offspring are usually less fit or non-viable in nature. Depending on the mechanism involved, true or partial clones of the mother are produced. In angiosperms, one of two types of apomeiosis (apo = without) occur: apospory, in which the gametophyte develops directly from a sporophytic cell of the ovule, or diplospory, in which meiosis is omitted, restituted, or preceded by endoreplication in the megaspore mother cell (Nogler, 1984; Asker and Jerling, 1992). In both cases, true clones of the mother plant are formed given that in diplospory, chromosome restitution happens before crossing-over has initiated, and after endoreplication, copy- rather than sister-chromosome pairing occurs. However, there are reported exceptions of recombination in diplosporous apomicts, e.g., in dandelion (Malecka, 1965). Apomeiosis can also be facultative, in which part of the offspring is produced by sexual means. This is found in diplosporous species, e.g., Erigeron (Noyes, 2005), but particularly also in pseudogamous aposporous species in which ovule-derived embryo sacs develop next to the reduced embryo sac and autonomous versus the sexually derived embryo are competing for resources, e.g., Paspalum (Ortiz et al., 2013). Similar mechanisms of apomeiosis exist in parthenogenetic animals, although here more often meiosis still occurs, involving either haploid offspring or restoration of diploidy through various mechanisms (Avise, 2008).
Successful embryo development depends on a third factor, the nutrition of the embryo. In angiosperms, the embryo is nourished by the endosperm, a tissue that in sexual individuals arises via fertilization of the central cell. The process of double fertilization in which the egg cell and central cell each are fertilized by one of two clonal sperm cells is unique to flowering plants (see for a review, e.g., Dresselhaus et al., 2016). In most apomictic species, endosperm development is pseudogamous, requiring fertilization of the central cell, whereas in a minority of apomictic species, the endosperm develops autonomously. In both cases, the usual maternal (m) versus paternal (p) genome ratio of 2m : 1p in the endosperm might be altered, which can severely affect seed development in many plant species (Scott et al., 1998; Autran et al., 2005; Kradolfer et al., 2013). Apomicts evolved different adaptations to overcome this requirement, e.g., in pseudogamous panicoid grasses only four nuclei comprise the aposporous embryo sac with predominantly unreduced, uni-nucleate central cells fertilized by a reduced sperm (Ozias-Akins, 2006). In animals, embryo nutrition is provided by the mother in one of many ways, without the need for a second fertilization event. In some parthenogenetic animal species, however, embryo development needs activation by a sperm without the fusion of gametes, known as gynogenesis or sperm-dependent parthenogenesis. Parthenogenesis and apomeiosis together, combined with either pseudogamous or autonomous endosperm formation, is defined as apomixis (sensu stricto) or agamspermy, the clonal seed formation (reviewed by, e.g., Pupilli and Barcaccia, 2012; Hand and Koltunow, 2014; Conner and Ozias-Akins, 2017).
Parthenogenesis is one form of apogamy and is sometimes also used in a wider sense, including spontaneous embryo development from (a) gametophytic cell(s) other than the egg cell, which is particularly common in lower plants (Asker and Jerling, 1992). Apogamy sensu lato includes, in addition, the spontaneous development of an embryo from a sporophytic cell, known as somatic embryogenesis. This process lacks the development of an embryo sac, endosperm, and seed coat. A classic example of somatic embryogenesis is the spontaneous embryo formation at leaf margins in Kalanchoë spp. (Garcês et al., 2007). A particular form of it is adventitious embryony or polyembryony in which the embryo(s) develop(s) from a sporophytic cell of the ovule (Nogler, 1984). Another special form is in vitro embryogenesis in which embryos develop ex-planta usually from microspores (pollen) or, less frequently, female gametophytic cells (gametophytic embryogenesis), or protoplasts, leaves, hypocotyls, or other plant tissues (sporophytic embryogenesis), often indirectly via the formation of a callus (Horstman et al., 2017). The reprogramming to progress into embryogenesis occurs under the influence of external stimuli such as hormones, heat stress, or overexpression of particular TFs (Hand et al., 2016; Ikeuchi et al., 2016). Although successful in a range of species, many species or particular genotypes can be (very) recalcitrant to in vitro embryogenesis and unable to produce embryos with any of the known stimuli (Ochatt et al., 2010; Soliìs-Ramos et al., 2012). Identifying a gene that is able to induce parthenogenesis particularly in these recalcitrant species and genotypes would be very valuable as a tool in plant breeding.
The different forms of embryogenesis are summarized in Figure 1. In this review, we focus on parthenogenesis in the strict sense, concerning the spontaneous development of an embryo from an unfertilized egg cell, and in flowering plants. Nevertheless, the induction of embryo development from any other cell or tissue may include commonalities with this process in the plant egg cell and, where overlapping, this will be considered in addition.
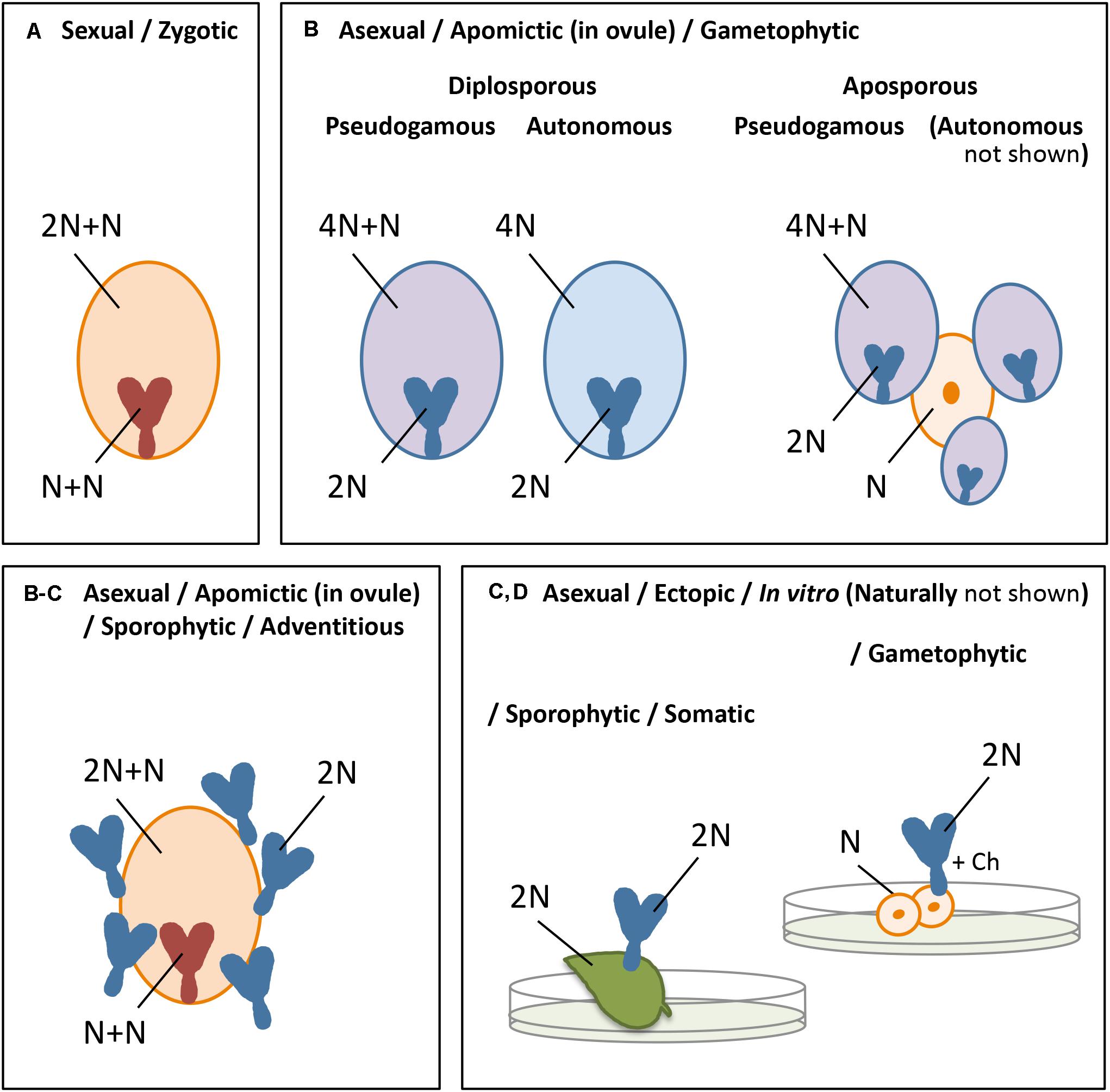
Figure 1. Summary of the different forms of embryogenesis in plants, showing the embryo and endosperm originated from a mature embryo sac (A,B) or the embryo ectopically (C,D) after sexual (A) or asexual (B–D) reproduction, with orange indicating the sexual process, blue the asexual or apomictic process, pink apomictic reproduction with fertilization of the central cell, and N = chromosome set after reduction division: (A) zygotic embryogenesis, involving chromosome reduction (N) and gamete fusion (N+N for the embryo, 2N+N for the endosperm), (B) apomictic embryogenesis, occurring in the ovule, either gametophytic apomixis in which an embryo sac arises from an unreduced megaspore (diplospory) or sporophytic cell of the ovule, usually adjacent to a sexually derived spore or developing embryo sac (apospory), and parthenogenetic (spontaneous) embryo development and autonomous (spontaneous) or pseudogamous (after fertilization of the central cell) endosperm formation, or sporophytic apomixis in which the embryo arises directly from a sporophytic cell of the ovule, often as polyembryony and alongside a sexually derived embryo and endosperm (C) somatic/sporophytic embryogenesis, involving ectopic embryo development from sporophytic cells, and (D) gametophytic embryogenesis, idem from a gametophytic cell. The latter two (C,D) omit the formation of an embryo sac, endosperm, and a seed coat, and occur naturally, for example, from leaf margins or ovular cells (C), gametophytic tissue in lower plants or, e.g., a synergid (D), but are particularly known from in vitro embryogenesis in which embryos are formed in culture, after external induction, particularly from protoplasts, leaf, the hypocotyl or other plant tissues (C), or microspores (D).
Historical Aspects of Parthenogenesis
The use of “Parthenogenesis” to denote asexual reproduction in plants followed its use in insects, as by Owen (1849): “On parthenogenesis, or the successive procreating individuals from a single ovum” (1849) and von Siebold (1856): “Wahre Parthenogenesis bei Schmetterlingen und Biene, ein Beitrag zur Fortpflanzungsgeschichte der Thiere” (1856). In fact, apomixis rather than parthenogenesis is meant here, also including apomeiosis and endosperm development. The recognition that some plant species are capable of asexual reproduction came only two decades after the universality of “the law of sexual reproduction” was established, realizing that this also holds for plants (c. 1830) (Nogler, 2007). Many discussions and numerous investigations in the two prior centuries preceded recognition (reviewed by Bergsma, 1857). Most of these studies included dioecious plant species, particularly Cannabis, Mercurialis, Spinacia, Curcurbits, Silenes, and Ricinus, and some monoecious species with separate male and female flowers, e.g., maize. However, due to the state of knowledge at that time and limited technical possibilities, all studies violated to a certain degree one or both of two essential conditions: (1) complete isolation of the plants and (2) exact observations (Bergsma, 1857). Regarding (1), some studies were done even before the discovery of the pollen or knowledge of its function (e.g., Camerarius, 1694), and (2), particularly the occasional formation of male organs in female flowers remained unrecognized. A relationship between asexual seed production and either annual plants or monoecious species has also been suggested (among the early investigators: Spallanzani, L., c. 1770–1785, Bernhardi, J., c. 1834–1839, Lecoq, H., c. 1858–1867, and Naudin, C., c. 1861–1867; see Bergsma, 1857). Ultimately, only one of these reports of asexual seed production was confirmed and is now considered as its first proof, namely, that Coelebogyne ilicifolia (presently: Alchornea ilicifolia; Euphorbiaceae) produces perfect seeds without any apparent action of pollen (Smith, 1841). After better fixing and staining methods became available, this example of asexual seed was revealed to result from polyembryony rather than parthenogenesis (Strasburger, 1877). It took another two decades to confirm that true parthenogenesis indeed exists in angiosperms, verified on the basis of careful observations in Antennaria alpina (Juel, 1898). Apomeiosis in this species involves the omission of meiosis in the megaspore mother cell, followed by two mitotic divisions resulting in four unreduced spores, now known as the Antennaria-type of diplospory. Subsequently, parthenogenesis was proven to occur in other species, including Taraxacum and Hieracium (Murbeck, 1904), and involve additional modes of apomeiosis (Juel, 1906; Rosenberg, 1906; reviewed by Nogler, 2007). According to our current knowledge, ∼400 species from different plant families are able to produce seeds without fertilization, and apomixis evolved numerous times in plants (Carman, 1997; van Dijk and Vijverberg, 2005). This suggests that parthenogenesis likely also relies on more than one genetic mechanism.
Egg Cell Arrest and the Trigger for Embryogenesis
In parthenogenetic reproduction egg cell arrest, as is found in sexual reproduction prior to fertilization, is absent or strongly reduced. Cytological investigations indicate a short period of egg cell arrest at least in some apomictic species, followed by precocious (before anthesis) embryo development (Nogler, 1984), e.g., in dandelion (van Baarlen et al., 2002) and the wild relative of wheat, Tripsacum dactyloides (Grimanelli et al., 2003). In sexual plants at the end of female gametophyte patterning (see for a review Tekleyohans et al., 2017), the mature egg cell is characterized by highly condensed repressive chromatin and a relatively quiescent transcriptional state (Garcia-Aguilar et al., 2010; Pillot et al., 2010). This is hypothesized to be necessary for attaining totipotency in the zygote and early embryo (Baroux and Grossniklaus, 2015). After fertilization and karyogamy, structural changes in the chromatin are necessary to enable access to the DNA for transcription and the replication machinery. Whether the egg cell in apomicts also undergoes a (brief) period of chromatin repression and transcriptional silencing before embryogenesis initiates is yet unknown, but likely if indeed needed to obtain totipotency. Parthenogenesis may then involve factors that are responsible for the spontaneous de-repression of chromatin and activation of transcription. Alternatively, the egg cells in apomicts bypass a chromatin repressive and transcriptionally silent state and need reprogramming. In this context, it is interesting to know the chromatin state in initial cells of embryogenesis other than egg cells, e.g., in somatic embryogenesis, to search for parallels. In any case, factors that are involved in chromatin-remodeling and transcriptional regulation are candidates to play a role in the parthenogenetic pathway.
In egg cell arrest and embryo development, a role for signals from the surrounding tissue is indicated, particularly from the companion cells, the central cell in the mature gametophyte and endosperm in the developing seeds (Grossniklaus, 2011). In sexual plant reproduction, the central cell also arrests until fertilization, but shows chromatin that is depleted from repressive marks and displays a more active transcriptional competence (Baroux and Grossniklaus, 2015). This allows for the expression of maternal alleles and TEs, the latter thought to serve the production of 24 nucleotide siRNAs to reinforce silencing of TEs in the egg cell (Ibarra et al., 2012; Roche et al., 2016; Martinez and Köhler, 2017). The former, the expression of maternal alleles, may contribute to the differential expression of maternally and paternally inherited alleles in the early endosperm after fertilization, as is also the results of imprinted genes (Wang and Köhler, 2017). In many species, the endosperm is, therefore, sensitive to a maternal to paternal dosage, and deviations from this lead to endosperm failure and embryo arrest (Scott et al., 1998; Kradolfer et al., 2013). Silencing of TEs in the egg cell by small RNAs from the surrounding nucellar tissue is also reported. A study in Arabidopsis shows reactivation of TEs in the egg cells of plants that are mutant for ARGONAUT9 (AGO9), a small RNA binding protein of the RNA Induced Silencing Complex (RISC) (Olmedo-Monfil et al., 2010). Another study shows higher overall transcription levels in early embryos of parthenogenetic Tripsacum x maize hybrids as compared to embryos of sexual maize, supporting reduced silencing under parthenogenetic conditions (Garcia-Aguilar et al., 2010). Taken together, findings suggest that: (1) dedicated (TE-)silencing pathways, involving companion cells and surrounding ovular tissue, result in dynamic patterns of transcriptional suppression in the egg cell, and (2) the m : p balance in the endosperm is important for proper functioning of the endosperm, which in turn is essential for embryo survival. They imply that changes in the accompanying cells and, as a result, in the communication to the egg cell, for example, changes in genes involved in DNA (de-)methylation or small RNA pathways, may have evolved in parthenogenesis.
In the sexual model species Arabidopsis, central cell arrest requires control by the PRC2, an evolutionarily conserved complex that is involved in the suppression of development via the regulation of epigenetic modulation (reviewed by Mozgova and Hennig, 2015; Wang and Köhler, 2017). The PRC2 maintains the repressive state of its target genes by preserving the tri-methylation of the N-terminal tail of histone H3 on lysine 27 (H3K27me3), a mark of transcriptional silencing. Different PRC2s exist, with the one involved in seed development containing the fertilization independent seed (FIS)-class proteins: MEDEA (MEA) (Grossniklaus et al., 1998), FIS2 (Luo et al., 1999), FERTILIZATION-INDEPENDENT ENDOSPERM (FIE) (Ohad et al., 1999), and MULTICOPY SUPPRESSOR OF IRA1 (MSI1) (Köhler et al., 2003a). Mutations in one of the FIS-class genes result in autonomous endosperm formation, showing diploid nuclei and development until cellularization (Chaudhury et al., 1997). Mutations in MSI1 result in spontaneous embryo development in addition, although, with early embryo abortion up to the c. 20-cell stage (Guitton and Berger, 2005). These non-viable, haploid embryos express molecular markers and polarity similar to the diploid wild-type embryos produced by fertilization. Mutants of the FIS-class genes mea and fis2 also rarely show embryo-like structures (Chaudhury et al., 1997). Since the penetrance of FIS-mutants on autonomous endosperm development is highest for msi1 (Köhler et al., 2003a), possibly the egg cell is able to undergo spontaneous development also in other FIS-mutants, but with lower penetrance (Chaudhury et al., 1997). Later studies showed that the functional requirement of the FIS-PRC2 could be bypassed by increasing the maternal genome dosage in the endosperm (Kradolfer et al., 2013), and that the FIS-PRC2 functions in the repression of maternal alleles of paternally expressed imprinted genes (reviewed in Wang and Köhler, 2017). The authors proposed that the FIS-PRC2 evolved concomitantly with sexual endosperm and the angiosperms. This is particularly interesting in the context of apomixis, in which the ability to reproduce sexually is lost or modified and the maternal genome dosage in the endosperm is usually increased in pseudogamous apomicts and unique in autonomous apomicts. Apomictic species may thus have become independent from the FIS-PRC2, either because it has a (relatively) modified expression or they evolved changed requirements for it. Unraveling this changes in more detail may give clues for parthenogenetic reproduction.
Despite the great discoveries discussed above, the precise molecular mechanism(s) by which the egg cell achieves its competence and is activated for embryogenesis is still unknown. In animals, early embryogenesis mainly depends on maternal genetic information deposited in the egg cell before fertilization (Tadros and Lipshitz, 2009; Eckersley-Maslin et al., 2018). During the MZT and ZGA, maternal transcripts are degraded and zygotic ones synthesized. In flowering plants, the large cytoplasm of the egg cell also allows for the deposition of maternally derived molecules. Single cell type transcriptome analyses confirmed that the egg cell of the dicot Arabidopsis (Wüst et al., 2010) and monocots rice (Anderson et al., 2013) and maize (Chen et al., 2017) is stocked with RNAs, proteins and other molecules that support embryogenesis upon activation. Circa 30–40% of the total number of genes are expressed in the egg cell, a percentage not notably lower than in other (gametic) cells. Several evidences suggest that embryogenesis in plants also mainly relies on maternal transcripts (Autran et al., 2011), although paternal contribution soon after fertilization is also reported (Del Toro-De León et al., 2014; Anderson et al., 2017), and hypothesized to trigger embryogenesis of fertilized egg cells (Khanday et al., 2018). If particularly or solely maternal transcripts are involved in the initiation of embryogenesis and MZT, parthenogenetic embryo development might be similar to that in sexual reproduction. However, if paternal factors are involved in addition, alternatives for their need should have been evolved in parthenogenesis, e.g., by activation of usually silent maternal transcripts. Transcripts over-represented in the egg cells of Arabidopsis include TF-families, particularly those of type I MADS domain, RWP-RK domain, and reproductive meristem (Wüst et al., 2010). In addition, PIWI/ARGONAUTE/ZWILLE (PAZ) domain encoding genes are upregulated, supporting a role for epigenetic regulation through small RNA pathways, and the AUXIN RESPONSE FACTOR 17 (ARF17) is enriched, suggesting the involvement of auxin. An interesting recent finding that highlights the importance of auxin in embryogenesis regulation is the identification of an auxin-response network that suppresses embryo development from the suspensor in Arabidopsis (Radoeva et al., 2018). In rice and maize egg cells, TFs are also over-represented, as are genes involved in transcriptional regulation and nucleic acid binding (Anderson et al., 2013; Chen et al., 2017). A comparative transcriptome analysis between egg cells and zygotes in maize shows ZGA to involve c. 10% of the genome (Chen et al., 2017). Particularly genes that encode transcriptional regulators are activated in ZGA and chromatin assembly is modified, while the egg cell becomes primed to activate the translational machinery. In summary, data show that a range of molecules known to play a role in development are stored in the egg cell and ready for use in embryogenesis. They suggest that only a trigger is needed to release the repressive state and activate transcription and translation in order to initiate this.
In vertebrate egg cells, different evidence suggests that the key trigger for egg cell activation is a rise in intercellular Ca2+, initiated by the fertilizing sperm and responsible for all further downstream reactions (Horner and Wolfner, 2008; Machaty, 2016). An increase of internal Ca2+ is also detected in zygotes of maize (Digonnet et al., 1997; Antoine et al., 2001) and wheat (Pónya et al., 2014) in in vitro fertilization experiments after the fusion of the gametes. Subsequently, cell wall material is formed, likely representing a block to polyspermy. A role for Ca2+ in cell–cell communication during plant fertilization was suggested by detecting a Ca2+ maximum at pollen tube rupture (Dresselhaus and Franklin-Tong, 2013). A short Ca2+ transient in both the egg and central cell was associated with pollen tube burst and sperm cell arrival, while a second extended Ca2+ transient solely in the egg cell was correlated with successful fertilization (Denninger et al., 2014). Although rising upon the fusion of gametes, a Ca2+ rise alone apparently is insufficient to trigger parthenogenesis in plants. In some parthenogenetic organisms of other kingdoms, such as insects, stimuli imparted to the egg cell during ovulation or egg-laying, or non-sperm-based signals, e.g., a change in ionic strength or pH, can trigger egg cell activation (Horner and Wolfner, 2008). In vitro protocols for DH-production make use of external stimuli such as a change in ion concentration or other abiotic stress factors to induce embryogenesis in micro- and megaspores (Germanà, 2006; Belogradova et al., 2009; Islam and Tuteja, 2012; Hand et al., 2016). However, these external stimuli are not generally applicable, and despite being successful in some species and genotypes others can be completely recalcitrant to such triggers. Nevertheless, they suggest that the breaking of egg cell arrest and/or release of the repressive chromatin and transcriptional silent state may (also) involve a change in (internal) physiology, particularly involving Ca2+. Searching for factors that underlie such changes likely aid in defining the molecular basis of parthenogenesis.
In summary, data show that egg cells in sexually reproducing species undergo a period of arrest that goes together with condensed, repressive chromatin and silenced transcription, and an egg cell that is stocked with molecules ready for use in embryo development. The molecular mechanism(s) or trigger by which the egg cell is activated and embryogenesis initiates is yet unknown, but results suggest the involvement of factors that release the chromatin repressive and transcriptionally silent state, e.g., genes involved in (de-)methylation, small RNAs, and hormones or a change in physiology. Particularly the inactivation or modification of the FIS-PRC2 may play a role in these changes. Parthenogenetic egg cells lack arrest or arrest for only a (very) short period, and it is unknown whether this implies that chromatin repression and transcriptional silencing are also omitted. Since a quiescent state is hypothesized to be necessary to attain totipotency in the zygote, probably this state occurs also in parthenogenetic eggs, but only for a (very) short period. In any case, factors that are involved in chromatin-remodeling or transcriptional regulation are likely candidates in the parthenogenesis developmental pathway.
Genetic Control of Parthenogenesis in Apomicts and Its Independency From Apomeiosis and Polyploidy
Parthenogenesis in angiosperms has long been thought to be a process that was initiated by apomeiosis and unknown to occur independently from it. Also the genetic control of parthenogenesis and apomeiosis was long assumed to rely on a single (master) gene or one locus with tightly linked genes (Mogie, 1992). However, some of the former genetic models (Richards, 1970; Nogler, 1984; Asker and Jerling, 1992) and more recent genetic mapping and other studies (reviewed by Vijverberg and van Dijk, 2007; Hand and Koltunow, 2014), showed that parthenogenesis is able to segregate from apomeiosis. Among the first evidence for this came from artificial crosses in the common dandelion (Taraxacum officinale), using diploid sexual female x triploid apomictic male crosses, resulting in small amounts of diploid, triploid, and tetraploid hybrid offspring, corresponding to fertilization of the haploid egg cell with a haploid, diploid, or triploid sperm, respectively (Tas and van Dijk, 1999; van Dijk et al., 1999). All tetraploid and some triploid hybrids showed spontaneous seed formation, thus displaying all apomixis elements. The other triploid hybrids produced (near-) diploid (type-A; three plants), triploid (type-B; four plants), or tetraploid (type-C; two plants) offspring after pollination with haploid pollen. Additional cytological investigations (van Baarlen et al., 2002) and molecular marker studies (van Dijk et al., 2003) showed that type-B hybrids were diplosporous, parthenogenetic, and lacked endosperm autonomy, while type-C hybrids were diplosporous, with autonomous endosperm formation, but lacking parthenogenesis. Apart from demonstrating that apomeiosis, parthenogenesis, and also endosperm autonomy, were inherited independently from each other, these results established that parthenogenesis functions independently from endosperm autonomy and vice versa in dandelion. Some offspring of the type-B triploids originated from parthenogenetic development (2n + 0 embryos), whereas the remainder resulted from fertilization of the egg cell (2n + n) suggesting incomplete, ∼2/3rd penetrance of parthenogenesis in these hybrids (van Dijk et al., 1999). Apparently, the usual precocious development of the embryo, as occurs in full apomicts, was disturbed, possibly as a result of the separation of parthenogenesis from apomeiosis or from modifiers or enhancers. All type-A triploids and the diploid hybrids gave rise to diploid offspring only after pollination with haploid pollen, suggesting that they were true sexuals. The absence of apomeiosis in diploid hybrids was later confirmed by the absence of a linked microsatellite marker in progeny from a similar cross (van Dijk et al., 2009). It was suggested to be the result of a genetic load associated to long-term asexual reproduction that becomes apparent and lethal in haploid gametes. This would then possibly also hold for parthenogenesis. Although presumed, the absence of parthenogenesis in the type-A triploids and diploid hybrids and the independent acting of parthenogenesis from apomeiosis were not explicitly demonstrated.
Shortly afterward, the separate inheritance of apomeiosis and parthenogenesis was confirmed in another diplosporous Asteraceae, Erigeron annuus (Noyes, 2000; Noyes and Rieseberg, 2000). In a follow-up, Noyes et al. (2007) re-confirmed the existence of one locus for diplospory (D) and a separate locus for parthenogenesis and endosperm autonomy (F), and showed that spontaneous development (F) occurred in the presence as well as absence of D, although, with early embryo abortion in the meiotic context. Parthenogenesis could thus act as an embryogenesis inducer in the absence of apomeiosis, whereas a possible role in embryo growth needed verification. All offspring investigated in these studies were triploid, implying imbalanced meiosis, resulting in aberrant chromosome numbers in most of the egg cells. Apparently, parthenogenesis can act in an aneuploid context, although with early embryo arrest. More recently, Noyes and Wagner (2014) showed that in autonomously produced di-haploid offspring from a tetraploid synthetic apomict (genotype: Dddd/Ffff) with incomplete, c. 50% penetrance of diplospory, D segregated 1:1 and parthenogenesis (F) was present in all (genotypes: Dd/Ff and dd/Ff). This confirmed that parthenogenesis is able to act and give rise to viable offspring independently from apomeiosis, at least, in a di-haploid context. Due to the absence of the genotypes Dd/ff and dd/ff among the offspring obtained without pollination, one could infer that the functioning of F is based on its presence and expression in the female gametophyte rather than on signals from the surrounding sporophytic maternal tissue. The study also mentioned rare, spontaneous development of seeds by some of the di-haploid parthenogenetic plants, that germinated normally and grew-out into haploid plants up to flowering (Noyes and Wagner, 2014). This showed that parthenogenesis is able to function in a haploid context, although, with (very) low efficiency. The low haploid survival rate is likely a result of recessive-lethal selection against the parthenogenesis locus, as was suggested in one of the previous studies in Erigeron for the absence of apomixis elements from diploid offspring (Noyes and Rieseberg, 2000).
Currently, the separate inheritance of apomeiosis and parthenogenesis is confirmed for most apomictic model species, including aposporous and diplosporous apomicts and monocots as well as dicots, as is implied by, e.g., a cytological study in Poa pratensis that shows two aposporous, non-parthenogenetic individuals (Albertini et al., 2001); flow cytometric analysis in Hypericum perforatum, indicating parthenogenetic development in 10% of the reduced, di-haploid offspring in the presence of pseudogamous endosperm formation (Barcaccia et al., 2006); idem in the guinea grass Panicum maximum, resolving eight different reproductive pathways of seed development (Kaushal et al., 2008); a gamma deletion mapping study in Hieracium caespitosum, showing one locus for Loss-Of-Apospory (LOA) and one for Loss-Of-Parthenogenesis (LOP) (Catanach et al., 2006); comparative mapping studies in Pennisetum squamulatum versus Cenchrus ciliarus, resolving one aposporous recombinant that lacks parthenogenesis (Conner et al., 2013); and backcrossing experiments in Allium, resulting in diplosporous individuals with and without parthenogenetic development (Yamashita et al., 2012). The independent inheritance of endosperm autonomy from parthenogenesis is also supported by other studies, e.g., in Hieracium, the factor AutE was found to function independently from apospory and parthenogenesis (Ogawa et al., 2013). Mapping studies support the dominant monogenic/-locus inheritance of both, apomeiosis and parthenogenesis. The usual co-segregation of the apomixis elements apparently is the result of their location in complex, repeat, and transposon-rich, non-recombining and in some species hemizygous genomic regions, particularly illustrated by the >50 Mbp long ASGR in P. squamulatum (Akiyama et al., 2004) and the Apomixis Controlling Locus in Paspalum simplex (Labombarda et al., 2002). This co-segregation likely evolved as a prerequisite for each of the apomixis elements alone to survive, since their separate occurrence will be untenable in the long-term due to their creation of plant lines with accumulating increasing or decreasing ploidy levels (Asker and Jerling, 1992).
Recently, the long-term studies in natural apomicts paid off by resolving the BABY BOOM-Like (BBML) gene in P. squamulatum as a candidate gene for parthenogenesis (Conner et al., 2015; next paragraph). Transgenes of PsASGR-BBML were able to induce parthenogenesis in the tetraploid sexual relative P. glaucum, supporting their function in di-haploid eggs. PsASGR-BBMLpromoter-GUS analysis provided evidence for the expression of parthenogenesis in the egg cells of P. glaucum, where GUS expression was observed from 1 day before anthesis and in the post-fertilization developing embryos. These observations confirmed the function of a parthenogenesis gene on the basis of its presence and expression in the egg cell, rather than the companion central cell and/or surrounding sporophytic tissue. In a more recent study, it was shown that PsASGR-BBML transgenes were also able to induce parthenogenesis in haploid eggs of sexual diploid rice and maize (Conner et al., 2017). This clearly demonstrated that parthenogenesis is fully functional also in the haploid context, and supports that the usual absence of parthenogenesis from haploidy is likely a result of recessive-lethal selection against associated flanking genomic regions.
In summary, studies in natural apomicts show that parthenogenesis usually co-segregates with apomeiosis, but is able to segregate and function independently from it. Parthenogenesis can also segregate and act independently from autonomous endosperm formation. Most studies indicate monogenic/-locus, dominant inheritance of parthenogenesis. It functions normally in di-haploid egg cells, and is able to induce embryogenesis in aneuploid eggs, although, with early embryo abortion. Parthenogenesis is virtually absent from reduced, haploid plant egg cells, but rare observations of its functioning in haploid eggs have been reported for di-haploid parthenogenetic Erigeron (see above) and a diploid apomictic Hieracium plant (Bicknell, 1997). The isolation of the parthenogenesis inducing PsASGR-BBML gene, and the demonstration of its function as a transgene in haploid eggs from Pennisetum, rice, and maize, confirms that parthenogenesis is able to act independently from polyploidy. It suggests that the absence of the trait from haploidy is likely explained by a genetic load in linked genomic regions. Finally, the results support the gametic presence and expression of parthenogenesis rather than non-cell-autonomous signaling from companion cells or surrounding, sporophytic tissue.
Candidate Genes for Parthenogenesis
The PsASGR-BabyBoom-Like Gene
A BABY BOOM (BBM)-Like gene was discovered in the natural apomictic grass P. squamulatum while skim-sequencing bacterial artificial chromosome clones that were linked to the ASGR (Conner et al., 2008). BBM genes were originally identified in Brassica napus (Boutilier et al., 2002) and shown to induce somatic embryogenesis in B. napus and Arabidopsis upon ectopic expression. They are part of a large gene family characterized by the APETALA 2/ETHYLENE RESPONSE FACTOR (AP2/ERF) DNA-binding domain (Jofuku et al., 1994; Riechmann and Meyerowitz, 1998). This TF-family of almost 150 members in Arabidopsis (Sakuma et al., 2002) and 157 members in rice (Nakano et al., 2006) is divided into groups of genes containing either one or two AP2 domains. The one-domain ERF-like genes typically are involved in biotic or abiotic stress response whereas the two-domain AP2-like genes function in growth and development (Floyd and Bowman, 2007). BBM genes along with AINTEGUMENTA (ANT) and PLETHORA (PLT) genes belong to the AINTEGUMENTA-Like (AIL) subclade within the eudicotANT (euANT) class of AP2/ERF DNA-binding domain genes, all of which function during embryogenesis (Horstman et al., 2014). PsASGR-BBML protein sequence is most similar to other BBML genes from related apomictic species: Cenchrus ciliaris and Pennisetum spp., but also to BBM genes in Setaria italica (foxtail millet) and Oryza sativa (rice), and yet more distantly related to BBM genes in Zea mays and Sorghum bicolor (El Ouakfaoui et al., 2010; Conner et al., 2015). It is surprising that the rice BBM1 is more closely related to PsASGR-BBML than to any BBM copy in maize and sorghum, given that rice and the panicoid grasses that include Pennisetum, sorghum, and maize, diverged from one another around 60 million years ago.
None of the AIL genes in Arabidopsis is expressed in pre-fertilization gametic cells during sexual reproduction (Horstman et al., 2014). On the other hand, after transformation to sexual P. glaucum (pearl millet), the PsASGR-BBML gene was shown to be expressed in egg cells prior to fertilization and to be sufficient for the initiation of embryos in the absence of fertilization (Conner et al., 2015). Since tetraploid pearl millet was the transgenic background, parthenogenetic development of the reduced eggs gave rise to diploid progeny. A second cycle of parthenogenesis resulted in true haploids, which expectedly were sterile. Sterile haploids also were derived through parthenogenesis of reduced egg cells in rice and maize (Conner et al., 2017). Among these three transgenic cereals, it was demonstrated that both the native PsASGR-BBML promoter and an egg-cell-specific Arabidopsis promoter (DD45; Steffen et al., 2007) provided the appropriate temporal regulation to enable fertilization-independent embryo formation. Mature haploid seed formation was irregular possibly as a result of asynchronous embryo-endosperm development. This first demonstration of parthenogenesis gene function opens the door for synthesizing apomixis in cereal crops once the capacity to produce unreduced gametes at high frequency is installed.
Interestingly, it was recently found that a wild-type rice BBM1 (Os-BBM1) transgene under an Arabidopsis egg-cell-specific promoter (DD45) was also able to initiate embryogenesis in rice egg cells without fertilization (Khanday et al., 2018). This supported the close relationship of PsASGR-BBML with Os-BBM1 and the functionality of the associated AP2-like domain in parthenogenesis rather than an evolved novel capability in functional domains. Most interestingly, it was shown that Os-BBM1 lacks expression in the egg cells of rice, but is expressed in sperm cells, whereas only male BBM1-transcripts are expressed in early zygotes. This suggests the requirement of fertilization in embryogenesis for the transmission of male-genomic factors that are maternally silenced. It would imply that in parthenogenesis, an essential, normally maternally imprinted gene, may have become maternally expressed. This is supported by another interesting recent study that shows maternal expression of the normally imprinted gene PHERES1 (PHE1; Köhler et al., 2003b) in apomictic Boechera (Kirioukhova et al., 2018), as is further discussed in Section “The FIS-PRC2 and RETINOBLASTOMA RELATED1”. It nicely brings together the many studies that indicate the involvement of diverse repression mechanisms in egg cell arrest and the associated factors that putatively play a role in the release of this repression (see section “Egg Cell Arrest and the Trigger for Embryogenesis” above).
The “Salmon System” in Wheat
Investigations in the context of parthenogenesis were done in the “Salmon system” of wheat (Triticum aestivum) already some decades ago. This system was developed for use in haploid production after it was recognized that transfer of the nucleus of the sexual cultivar “Salmon” to cytoplasm of the grass genus Aegilops resulted in lines that were capable of autonomous embryo development (Tsunewaki and Mukai, 1990). In the “Salmon” line, the short arm of chromosome 1B of wheat has been replaced by the short arm of chromosome 1R of rye. Tsunewaki and Mukai (1990) concluded that besides a cytoplasmic Restorer of fertility (Rfv1) factor, two nuclear genes were involved in spontaneous embryo development: the inducer gene Parthenogenesis gain (Ptg) that is under sporophytic control, and the suppressor gene Suppressor of parthenogenesis (Spg) that is under gametophytic control. These two genes were concomitantly exchanged with the chromosome 1 arm. Two other researchers came to the same conclusion in a 1B/1R-translocation system in durum wheat (Hsam and Zeller, 1993). To improve the system for in vivo investigation of parthenogenesis, three isogenic homozygous lines were produced, the male fertile sexual line Ae. aestivum-Salmon (aS), and the male sterile parthenogenetic lines Ae. caudata-Salmon (cS) and Ae. kotschyi-Salmon (kS) (Matzk et al., 1995). Comparative protein analysis from ovary extracts of these three lines resolved one protein that was uniquely expressed in the two parthenogenetic lines from 3 days before and during anthesis. This protein, P115.1, was characterized as a α-tubulin polypeptide. Tubulin α-chains are the major constituent of microtubules and function in GTP-binding and, regarding their function, could possibly also be a result of parthenogenesis rather than its cause. Further studies on isolated egg cells from the three isogenic lines and a common wheat line indicated that parthenogenetic development is independent from ovary-derived signals (Kumlehn et al., 2001). This encouraged the researches to focus on the egg cells and construct cDNA libraries from them. Analysis of these libraries delivered a number of egg cell specific candidates among which were RWP-RK domain (RKD)-containing TFs. These were subject to later studies in Arabidospis (Köszegi et al., 2011; Tedeschi et al., 2017) and Marchantia polymorpha (Rövekamp et al., 2016; Koi et al., 2016) (see next paragraph). Some eggs of the parthenogenetic lines showed a second nucleolus, a characteristic of zygotes isolated from sexual lines (Naumova and Matzk, 1998; Kumlehn et al., 2001). Together, the results showed that parthenogenesis apparently is an inherent property of the egg cell and not the surrounding tissue and is able to establish zygotic competence in the absence of fertilization.
RWP-RK Domain (RKD)-Containing Transcription Factor
Encouraged by their identification in wheat egg cell cDNA libraries (Kumlehn et al., 2001; former paragraph) RKD-TF homologs were searched for and investigated in Arabidopsis thaliana, which resolved a total of five AtRKDs (Köszegi et al., 2011). At least two of them were preferentially expressed in the egg cell and, interestingly, their ectopic expression induced cell proliferation and activated an egg cell-like transcriptome. Members of the RWP-RK domain family contain the MINUS DOMINANCE (MID) factor, which is in the distantly related green algae species Chlamydomonas reinhardtii required for gamete differentiation (Ferris and Goodenough, 1997). Since the RKD-TFs of Arabidopsis are highly redundant and the genes are conserved over the plant kingdom, the single copy homologous RKD-TF in Marchantia polymorpha was used for functional analysis (Koi et al., 2016; Rövekamp et al., 2016). MpRKD showed wide expression in M. polymorpha, but preferentially high in antheridia, developing egg cells, and sperm precursor cells. Lines with downregulated expression showed large cells at the base of the archegonium, indicating that egg cell specification occurs on the bases of anatomy and position, however, in the absence of specific molecular markers (Rövekamp et al., 2016). These cells underwent cell divisions instead of entering the quiescent egg cell stage, suggesting a role of MpRKD in establishing and/or maintaining the quiescent state of the egg cell prior to fertilization. MpRKD mutants lacked effects on the overall morphology of reproductive organs, but showed striking defects in egg and sperm cell differentiation (Koi et al., 2016). Together, these results indicate that RKD-TFs are evolutionary conserved regulators of germ cell differentiation in land plants and particularly act in the gametophyte-to-sporophyte transition by preventing the egg cell from entering mitosis in the absence of fertilization, i.e., by suppressing parthenogenesis.
The FIS-PRC2 and RETINOBLASTOMA RELATED1
Until the recent findings of PsASGR-BBML and RKD-TFs, a candidate for parthenogenesis in plants mentioned in literature was the FIS-PRC2 gene MSI1 (Köhler et al., 2003a; Guitton and Berger, 2005; section “Egg Cell Arrest and the Trigger for Embryogenesis”). It was isolated via a mutant screen in the sexual model A. thaliana and searched for because MSI-like homologs in yeast (MSI1) and mammals (RbAp46/48) were found to be involved in chromatin metabolism (Hennig et al., 2003). At that time, it became apparent that the modulation of chromatin structure played an important role in the regulatory decisions and gene expression during development, also in plants. As discussed above, the FIS-PRC2 is involved in gene suppression during seed development, particularly affecting the endosperm, whereas MSI1, and to a lesser extent also other FIS-class genes, affects embryo initiation in addition (Chaudhury et al., 1997). Other studies showed a role for the FIS-PRC2 in balancing the maternal versus paternal gene dosage, by showing plants with an increased maternal dosage resembling FIS-mutant phenotypes (Kradolfer et al., 2013). The results indicate that a release of gene suppression alone is insufficient to obtain viable seeds, but that this, particularly or solely, is a result of failure of the endosperm and maybe not embryo. Investigations on the role of the endosperm showed that, indeed, endosperm cellularization impacts embryo growth, and FIS-mutant embryos could be rescued on appropriate medium in vitro (Hehenberger et al., 2012). Also in other modes of in vivo induced parthenogenesis, such as via pollination with irradiated pollen or triploid inducer lines that results in haploid embryo development in some species (Germanà, 2006), embryos need to be rescued and cultivated in vitro due to failure of the endosperm. This indicates that release of the repressive state in the egg cell can be sufficient for the initiation of embryo development, however, finding clues for restoration of endosperm development is also necessary for successful parthenogenetic seed development.
The FIS-class genes FIS2 and MEA are imprinted genes. They are silenced throughout the life cycle of the plant, but become active in the female gametophyte, especially in the central cell, and remain expressed and active in the endosperm after fertilization, whereas the paternal alleles remain silent (Wang and Köhler, 2017). MEA is involved in the control of embryo growth in sexual species by repressing the maternal allele expression of the TF PHE1 (Köhler et al., 2003b). Thus, PHE1 is also imprinted, but expressed from the paternal allele only. A recent study asked the question what would happen with embryo growth in autonomous apomicts, where paternal alleles are absent (Kirioukhova et al., 2018). It was hypothesized that the silencing of maternal alleles might have become reduced or relieved during the evolution of apomixis, allowing maternally imprinted genes to be expressed from the maternal allele. This was tested for PHE in sexual versus asexual Boechera, a close relative of A. thaliana. In apomictic Boechera, the maternal PHE-like allele indeed was expressed, indicating a reversion of the imprinting status of this gene. In addition, a heavily methylated 3′MR was deleted from the PHE-alleles in apomicts, allowing increase of their expression. The authors proposed a model in which parthenogenesis in Boechera evolved via changes in epigenetic regulation of imprinted genes based on changes in DNA methylation (see Figure 3 in Kirioukhova et al., 2018). This shows parallelisms to an artificially induced case of parthenogenesis in mice through the loss of distal DNA methylation, resulting in maternal activation of the paternally expressed Insulin-like growth factor 2 (Igf2) gene (Kono et al., 2004). Thus, a modified role in transcriptional regulation of maternal alleles is indicated and interesting to further investigate in the context of parthenogenesis.
FIS-mutant phenotypes are resembled also by phenotypes of RETINOBLASTOMA RELATED1 (RBR1) mutants (Ebel et al., 2004; Johnston et al., 2010). This gene is related to the tumor suppressor gene RB in mammals, which has a role in inhibiting cell cycle progression. RBR1 in plants functions in cell cycle control during gametogenesis, with mutants showing supernumerary nuclei at the micropylar end and impaired cellularization (Johnston et al., 2008, 2010). Polar nuclei do not fuse in rbr gametophytes and cell-type-specific markers usually lack expression. RBR1 represses the G1/S-phase transition through inhibiting E2F transcription, and this, in turn, involves RBR1-phosphorylation that influences the RBR1-E2F interaction (Boniotti and Gutierrez, 2001; Kuwabara and Gruissem, 2014). Cyclin-dependent kinase (CDK) A combined with cyclin (CYC) regulatory subunit D (CDKA/CYCD; serine-threonine protein kinase) is involved in this phosphorylation. In interaction with MSI1, RBR1 also plays a role in the downregulation of METHYLTRANSFERASE1 (MET1) (Jullien et al., 2008). Reduction of MET1 in the central cell is essential for the activation of FIS2 and thus for the FIS-PRC2. Indeed, FIS2 expression is reduced in rbr-gametophytes (Johnston et al., 2010). Thus, RBR1 and genes associated to its functioning and/or to cell cycle progression are additional candidates to be involved in the suppression of spontaneous embryo and endosperm development and, as such, to have a role in parthenogenesis.
Genes Involved in the Induction of Ectopic Embryogenesis
A number of other genes, mostly TFs, have been reported to be involved in the induction of embryogenesis ectopically and/or in vitro after overexpression (excellent reviews by Radoeva and Weijers, 2014; Horstman et al., 2017). They are mentioned here for completeness, but we refer to the other reviews for their listing and details, since a specific role in parthenogenesis is yet undetermined. Whether they are part of one or a few larger networks also needs further elucidation. Among them are AP2-TF family genes, including BBM (AIL2/PLT4) (see also section “The PsASGR-BabyBoom-Like gene”, above) most other AIL-genes, and WOUND INDUCED DEDIFFERENTIATION 1 (WIND1) (Horstman et al., 2017) that support embryogenesis. In addition, most genes of the “LAFL”-network, namely, LEAFY COTYLEDON 1 (LEC1), LEC1-Like (L1L), and LEC2, another member of the RWP-RK domain-containing family, RKD4 (see also section “RWP-RK Domain (RKD)-Containing Transcription Factor”), and the homeodomain TF WUSCHEL (WUS). There are also genes that function more indirectly by increasing the capacity for embryogenesis, such as AGAMOUS-Like 15 (AGL15) and SOMATIC EMBYOGENESIS RECEPTOR KINASE 1 (SERK1). Last, some genes are known to be involved in the suppression of embryogenesis, including the chromatin-helicase-DNA binding gene (CHD3/4)-Like chromatin remodeling factor encoded by PICKLE (PKL), genes of the PRC1 and -2 (see sections “Egg Cell Arrest and the Trigger for Embryogenesis” and “The FIS-PRC2 and RETINOBLASTOMA RELATED1”), and the HIGH-LEVEL EXPRESSION OF SUGAR-INDUCIBLE GENES VAL1 and VAL2. Yet knowing the putative role of parental expression of Os-BBM1 in embryogenesis, it is relevant to investigate this also for the other genes mentioned.
In summary, a PsASGR-BBML gene has been isolated and verified as the first parthenogenesis gene by demonstrating its functionality in related, sexual relative grasses such as pearl millet and rice, but not yet in eudicots. Other candidates and studies support that the suppression of spontaneous embryo and endosperm development in sexual reproduction is under tight epigenetic control and release of this control allows for the initiation of spontaneous embryo and endosperm development. This is shown to involve FIS-PRC2 genes and genes associated to it and to cell cycle control. Although initiated, mutants of these genes show early embryo arrest and endosperm development up to cellularization, indicating that a release of transcriptional suppression alone is not enough to obtain viable seeds. Functional endosperm is important in addition, either because of probable roles in the regulation of embryogenesis, but especially also to nourish the embryo. Restoring endosperm development is, therefore, necessary for successful seed development via parthenogenesis. Alternatively, the haploid embryos can be cultivated in vitro after embryo rescue, as is also done in some of the other haploid induction methods currently used in DH-production. Interesting recent results show that Os-BBM1 is paternally expressed, maternally silenced, and hypothesized to induce embryogenesis in rice egg cells after fertilization. Other recent results support a role for evolved changes in apomicts in this context, by showing the normally paternally expressed PHE-Like genes to be maternally expressed in apomictic Boechera. The results converge upon the importance of genes involved in the suppression of transcription and modifications thereof in apomicts at one hand and genes involved in the developmental process for which either transcription is allowed or artificially overexpressed on the other in parthenogenetic reproduction. In Table 1 and Figure 2 this convergence is summarized.
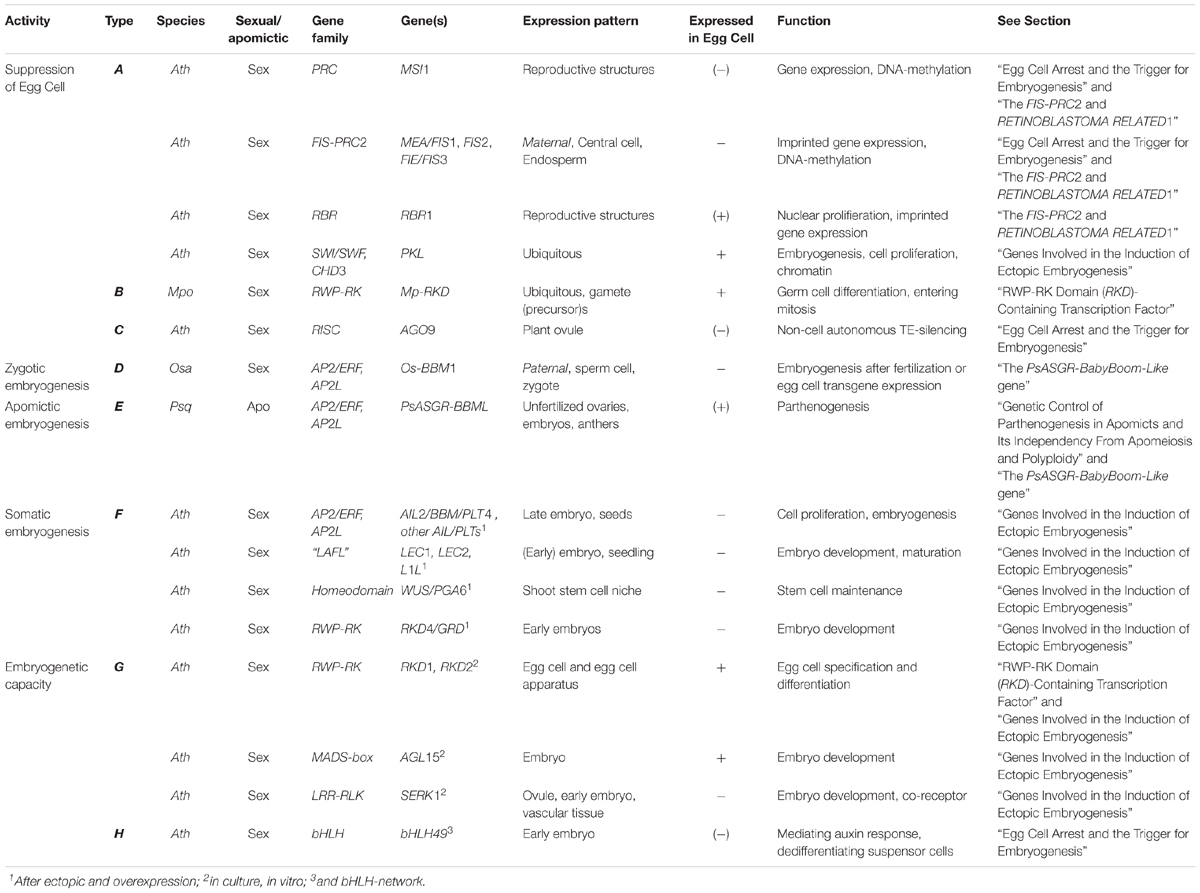
Table 1. Genes important in either the suppression or activation of embryogenesis in vivo or in vitro in plants and potentially relevant for parthenogenesis.
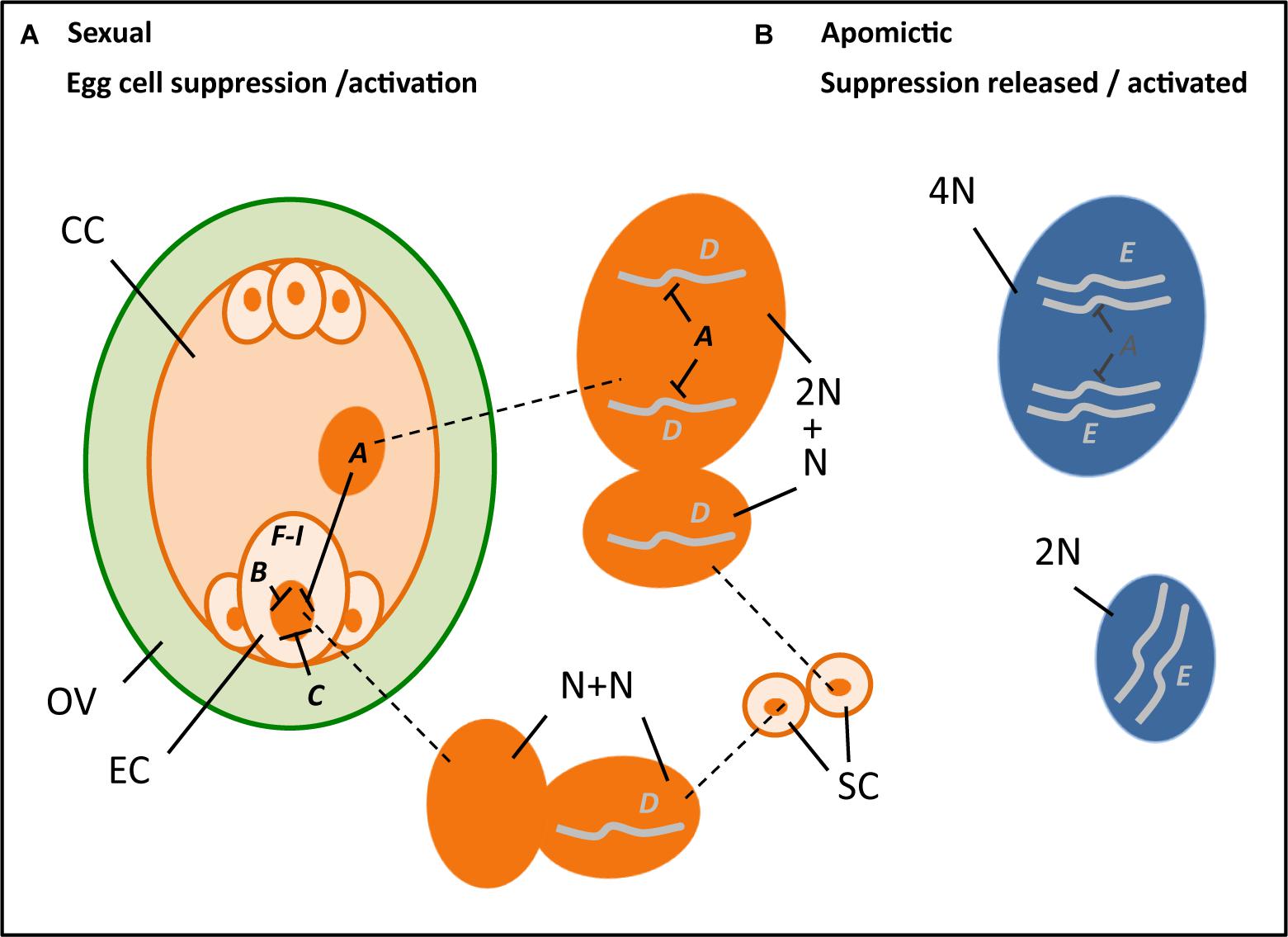
Figure 2. Hypothetical cartoon summarizing the potential processes and genes involved in parthenogenesis (A–I, with ---| denoting suppression) (see also Table 1), with left an ovule (OV, green) that encloses a reduced embryo sac (orange), including an egg cell (EC), central cell (CC), two synergids (next to the EC), and three antipodal cells, and outside it two sperm cells (SC, orange), in the middle the enlarged nuclei (dark orange) of the central cell (top, 2N; N = chromosome set after reduction division), sperm cells (middle, each N) and egg cell (bottom, N) at karyogamy, and right the unreduced nuclei (dark blue) of an apomictic central cell (top, 4N) and egg cell (bottom, 2N). A, B, and C denote genes involved in the suppression of transcription in the egg cell, particularly present in the central cell (A) notably the FIS-PRC2, egg cell (B) possibly RKD-TF, and the surrounding ovular tissue (C) potentially AGO9. D and E denote genes that support embryogenesis either after fertilization and transferring a paternally expressed, maternally silenced gene (D) as hypothesized for Os-BBM1, or via natural expression in an apomict (E) as is found for PsASGR-BBML and possibly involves reduced or absence of suppression. F–I denote genes and other stimuli that are found to be involved in embryogenesis either after ectopic overexpression to induce somatic embryogenesis (F), for example, LEC2, increase embryogenesis capacity (G), e.g., SERK1, being involved in auxin response (H) as recently found for bHLH49, or changes in physiological conditions and abiotic stress factors (I) such as Ca2+ and heat stress.
The Use of Parthenogenesis in Plant Breeding
The ultimate goal of identifying a gene for parthenogenesis is to apply it in protocols for breeding line production in order to induce either gametophytic or sporophytic embryogenesis in a variety of cell types and plant species. This is of particular interest in the context of DHs production, a method that is widely used for the instant production of homozygous lines via haploid induction technology followed by chromosome doubling methods (reviewed by, e.g., Germanà, 2006, 2011; Belogradova et al., 2009; Islam and Tuteja, 2012; Hand et al., 2016; Ikeuchi et al., 2016; Horstman et al., 2017). Current methods include, among others, in vivo induction of spontaneous haploid embryo formation at (very) low frequencies via wide crosses, crosses with triploids, or by using irradiated pollen, usually followed by embryo rescue and in vitro embryo cultivation, and in vitro induction of embryogenesis in response to different (a)biotic stress factors. These methods, however, need time-intensive and species-specific protocol development and lack application in a number of important crops. Although successful in particular species or genotypes, others can be completely recalcitrant to produce DHs. Understanding the genetic basis of egg cell activation and the initiation of embryogenesis will largely contribute to the production of a wide variety of DHs.
Most interesting is the combination of parthenogenesis with various forms of modified meiosis. Particularly the combination with apomeiosis, the omission of first meiosis, resulting in unreduced gametes that maintain all or most of the genetic and epigenetic variation of the mother plant, is interesting in order to produce clonal seeds. This enables the maintenance of vigorous F1-hybrids that are usually produced via extensive crossing and careful selection procedures involving five or more growing seasons, however, segregate in the subsequent generation. Being able to re-grow valuable F1-hybrids over more than one generation has high potential for food security and the increasing demand on food. Proof-of-principle for synthetic clonal reproduction was obtained by Marimuthu et al. (2011) using either the DYAD mutant [Ravi et al., 2008; one allele of SWITCH1 (SWI1): Mercier et al., 2001; Agashe et al., 2002] or the “turning MEIOSIS into MITOSIS” (MiMe) variant (d’Erfurth et al., 2009) to obtain unreduced gametes in combination with the CENTROMERE-SPECIFIC HISTON 3 (CENH3) mutant (Ravi et al., 2011) to fertilize the central cell without genomic contributing to the embryo. This method is now awaiting improvements to produce unreduced gametes at high frequency as well as identify or produce CENH3-Like variants in crops. Another interesting modified meiosis variant is the omission of second meiosis to produce (near-)homozygous gametes, using mutants of OMISSION OF SECOND DIVISION 1 (OSD1) (d’Erfurth et al., 2009). This is particularly of interest in the (near-)Reverse Breeding approach in which (near-)homozygous parental lines and chromosome substitution lines are produced in one generation (Dirks et al., 2009). Reverse Breeding relies on the suppression of recombination during the first meiosis and omission of chromatid separation in the second. All these protocols need the induction of embryo development from the gametes produced and are awaiting capacity for in vivo or in vitro embryo induction. A third very useful application of parthenogenesis in plant breeding is the potential to link diploid with polyploid gene pools, in alternation with apomeiosis. This makes it, for instance, much easier to cross-in interesting characters of diploid wild relatives to the usual tetraploid crop varieties in potato. All three aims contribute to the control of plant reproduction and breeding and are highly relevant in order to optimize crop development and increase plant productivity.
Conclusion and Future Perspective
This review provides a summary of current knowledge on parthenogenesis in plants obtained from studies in natural apomicts and mutants in sexual model species. Several lines of evidence from natural apomicts support that parthenogenesis inherits and functions independently from apomeiosis and endosperm formation, and that it is a monogenic and dominant trait. Results also show that parthenogenesis is expressed and active in the egg cell and independent from signals from the surrounding sporophytic tissue. Parthenogenesis functions normally in reduced di-haploid egg cells, resulting in viable embryos that grow into di-haploid plants. It also works in true haploid eggs, but with (much) lower frequency in producing viable plants and with the haploid offspring usually being infertile, and in aneuploid eggs, although with early embryo abortion. The reason that parthenogensis is usually absent from haploids in nature is indicated to be the result of a linked deleterious genetic load. The relationship of parthenogenesis to the central cell and endosperm is more complicated. The central cell plays a role in egg cell suppression in sexual reproduction (see Figure 2), and functional endosperm is needed for successful embryo development. It is not entirely clear if the parthenogenetic egg cell also undergoes a (short period) of quiescence in which the chromatin is repressed and transcription is silenced. These processes might be necessary for obtaining totipotency in the zygote, but has to be confirmed. Studies in sexual model systems, particularly Arabidopsis, support that the release of egg cell repression and transcriptional silencing results in the initiation of embryo development. Genes that are involved in this, particularly genes of the FIS-PRC2 or related to cell cycle control, e.g., RBR1, and the recently uncovered RKD-TFs, may therefore have changed or become ineffective in apomicts. A possible mechanism for this could be a change in the target gene sequence by which the silencing is reduced, e.g., by a deletion of a region involved in heavy methylation as was found in PHE-alleles in apomictic Boechera (Kirioukhova et al., 2018). Similarly, the functioning of PsASGR-BBML in apomicts, and the maternal silencing of the related Os-BBM1 in sexual rice, may hint to a reduction of maternal silencing of BBM-Like in apomictic grasses. The release of gene suppression particularly in the FIS-PRC2 mutants also affects the central cell, leading to spontaneous endosperm development up to cellularization. Embryos arrest at an early stage in these mutants, and this may also involve failure of the endosperm, since they can be rescued by in vitro cultivation. Embryos obtained with PsASGR-BBML also need either embryo rescue or fertilization of the central cell in order to allow endosperm development and embryo growth progression. The sexual endosperm is maternally to paternally genome dosage-dependent in most species (2m : 1p), whereas in apomicts, this dosage is usually highly disturbed without any obvious effect. Apparently, apomicts have evolved several mechanisms to overcome these requirements. One study supports that the function of the FIS-PRC2, and thus transcriptional silencing, can be suppressed by an increase of the maternal dosage in the endosperm, leading to a FIS-mutant-like phenotype. Whether this mimics the situation in the central cell of apomicts has to be resolved (see Figure 2). Altogether, the results show that parthenogenesis involves changes in epigenetic regulation needed to allow genes that are essential in embryo induction to be expressed from the maternal allele. They also show that parthenogenesis functions independently from endosperm development, but that the absence or failure of the endosperm impacts successful embryo and plant development. Insight into the mechanisms that have developed in apomicts to overcome the failure of the endosperm, and the development of methods to restore endosperm production, for example, through artificial crosses to fertilize the central cell, is needed and is the next challenge in successful seed production via parthenogenesis.
In the near future, establishing the function of the emerging parthenogenesis candidates in a wider sense is one of the main priorities as is the identification and/or engineering of parthenogenesis in non-grass species. A second priority is to transfer the knowledge to crop species to make it useful in plant breeding and in in vitro embryogenesis protocols. Third is to improve the experimental separation between the functioning of the egg cell and embryo growth progression on one hand and the dependence of the central cell and endosperm on the other in research on parthenogenesis, and to further unravel the mechanisms that underlie spontaneous endosperm formation. Fourth, to combine parthenogenesis with the different forms of “omission of meiosis” in order to use it as a tool in plant breeding, e.g., for clonal seed production/apomixis. Also interesting is to further unravel other mechanisms that have evolved in apomicts, such as the possible differences in egg cell quiescence as well as the ways in which the endosperm overcomes the maternal to paternal dosage requirements.
Author Contributions
KV designed and wrote the manuscript. PO-A wrote some parts of the manuscript. PO-A and MS gave valuable review comments.
Funding
The review is written as part of the Dutch Scientific Organization (NWO), Applied and Engineering Sciences (STW/TTW) fellowship 13700: PARtool: The molecular basis of plant embryo development without fertilization (Parthenogenesis), and its use as a tool in breeding line production, to KV and MS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
ASGR, apospory-specific genomic region; DHs, doubled haploids; FIS, fertilization-independent seed; MZT, maternal-to-zygote transition; PRC2, polycomb repressive complex 2; TEs, transposable elements; TF, transcription factor; ZGA, zygotic genome activation.
References
Agashe, B., Prasad, C. K., and Siddiqi, I. (2002). Identification and analysis of DYAD: a gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129, 3935–3943.
Akiyama, Y., Conner, J. A., Goel, S., Morishige, D. T., Mullet, J. E., Hanna, W. W., et al. (2004). High-resolution physical mapping in Pennisetum squamulatum reveals extensive chromosomal heteromorphism of the genomic region associated with apomixis. Plant Physiol. 134, 1733–1741. doi: 10.1104/pp.103.033969
Albertini, E. A., Porceddu, F., Ferranti, L., Reale, G., Barcaccia, B., Romano, B., et al. (2001). Apospory and parthenogenesis may be uncoupled in Poa pratensis: a cytological investigation. Sex. Plant Reprod. 14, 213–217. doi: 10.1007/s00497-001-0116-2
Anderson, S. N., Johnson, C. S., Chesnut, J., Jones, D. S., Khanday, I., Woodhouse, M., et al. (2017). The zygotic transition is initiated in unicellular plant zygotes with asymmetric activation of parental genomes. Dev. Cell 43, 349–358. doi: 10.1016/j.devcel.2017.10.005
Anderson, S. N., Johnson, C. S., Jones, D. S., Conrad, L. J., Gou, X., Russell, S. D., et al. (2013). Transcriptomes of isolated Oryza sativa gametes characterized by deep sequencing: evidence for distinct sex-dependent chromatin and epigenetic states before fertilization. Plant J. 76, 729–741. doi: 10.1111/tpj.12336
Antoine, A. F., Faure, J. E., Dumas, C., and Feijo, J. A. (2001). Differential contribution of cytoplasmic Ca2+ and Ca2+-influx to gamete fusion and egg activation in maize. Nat. Cell Biol. 3, 1120–1123. doi: 10.1038/ncb1201-1120
Autran, D., Baroux, C., Raissig, M. T., Lenormand, T., Wittig, M., Grob, S., et al. (2011). Maternal epigenetic pathways control parental contributions to Arabidopsis early embryogenesis. Cell 145, 707–719. doi: 10.1016/j.cell.2011.04.014
Autran, D., Huanca-Mamani, W., and Vielle-Calzada, J. P. (2005). Genomic imprinting in plants: the epigenetic version of an Oedipus complex. Curr. Opin. Plant Biol. 8, 19–25. doi: 10.1016/j.pbi.2004.11.011
Avise, J. C. (2008). Clonality: The Genetics, Ecology and Evolution of Sexual Abstinence in Vertebrate Animals. Oxford: Oxford University Press. doi: 10.1093/acprof:oso/9780195369670.001.0001
Barcaccia, G., Arzenton, F., Sharbel, T. F., Varotto, S., Parrini, P., and Lucchin, M. (2006). Genetic diversity and reproductive biology in ecotypes of the facultative apomict Hypericum perforatum L. Heredity 96, 322–334. doi: 10.1038/sj.hdy.6800808
Baroux, C., and Grossniklaus, U. (2015). The maternal-to-Zygotic transition in flowering plants: evidence, mechanisms, and plasticity. Curr. Top. Dev. Biol. 113, 351–371. doi: 10.1016/bs.ctdb.2015.06.005
Bell, G. (1982). The Masterpiece of Nature: the Evolution and Genetics of Sexuality. Berkeley CA: University of California Press.
Belogradova, K., Lewicka, I., Heberle-Bors, E., and Touraev, A. (2009). “An overview on tobacco doubled haploid,” in Advances in Haploid Production in Higher Plants, eds A. Touraev, B. P. Forster, and S. M. Jain (New York, NY: Springer), 75–85. doi: 10.1007/978-1-4020-8854-4_5
Bicknell, R. A. (1997). Isolation of a diploid, apomictic plant of Hieracium aurantiacum. Sex. Plant Reprod. 10, 168–172. doi: 10.1007/s004970050084
Boniotti, M. B., and Gutierrez, C. (2001). A cell-cycle-regulated kinase activity phosphorylates plant retinoblastoma protein and contains, in Arabidopsis, a CDKA/cyclin D complex. Plant J. 28, 341–350. doi: 10.1046/j.1365-313X.2001.01160.x
Boutilier, K., Offringa, R., Sharma, V. K., Kieft, H., Ouellet, T., Zhang, L., et al. (2002). Ecotopic expression of BABY BOOM triggers a conversion from vegetative to embryonic growth. Plant Cell 14, 1737–1749. doi: 10.1105/tpc.001941
Bowman, J. L., Sakakibara, K., Furumizu, C., and Dierschke, T. (2016). Evolution in the cycles of life. Annu. Rev. Genet. 50, 133–154. doi: 10.1146/annurev-genet-120215-035227
Camerarius, R. J. (1694). Ueber das Geschlecht der Pflanzen. (De sexu Plantarum Epistola.). Leigpzig: W. Engelmann.
Carman, J. G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 61, 51–94. doi: 10.1111/j.1095-8312.1997.tb01778.x
Catanach, A. S., Erasmuson, S. K., Podivinsky, E., Jordan, B. R., and Bicknell, R. (2006). Deletion mapping of genetic regions associated with apomixis in Hieracium. Proc. Natl. Acad. Sci. U.S.A. 103, 18650–18655. doi: 10.1073/pnas.0605588103
Chaudhury, A. M., Ming, L., Miller, C., Craig, S., Dennis, E. S., and Peacock, W. J. (1997). Fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 94, 4223–4228. doi: 10.1073/pnas.94.8.4223
Chen, J., Strieder, N., Krohn, N. G., Cyprys, P., Sprunck, S., Engelmann, J. C., et al. (2017). Zygotic genome activation occurs shortly after fertilization in maize. Plant Cell 29, 2106–2125. doi: 10.1105/tpc.17.00099
Conner, J., Podio, M., and Ozias-Akins, P. (2017). Haploid embryo production in rice and maize induced by PsASGR-BBML transgenes. Plant Reprod. 30, 41–52. doi: 10.1007/s00497-017-0298-x
Conner, J. A., Goel, S., Gunawan, G., Cordonnier-Pratt, M. M., Johnson, V. E., Liang, C., et al. (2008). Sequence analysis of bacterial artificial chromosome clones from the apospory-specific genomic region of Pennisetum and Cenchrus. Plant Physiol. 147, 1396–1411. doi: 10.1104/pp.108.119081
Conner, J. A., Gunawan, G., and Ozias-Akins, P. (2013). Recombination within the apospory specific genomic region leads to the uncoupling of apomixis components in Cenchrus ciliaris. Planta 238, 51–63. doi: 10.1007/s00425-013-1873-5
Conner, J. A., Mookkan, M., Huo, H., Chae, K., and Ozias-Akins, P. (2015). A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. U.S.A. 112, 11205–11210. doi: 10.1073/pnas.1505856112
Conner, J. A., and Ozias-Akins, P. (2017). Apomixis: engineering the ability to harness hybrid vigor in crop clants. Methods Mol. Biol. 1669, 17–34. doi: 10.1007/978-1-4939-7286-9_2
Del Toro-De León, G., García-Aguilar, M., and Gillmor, C. S. (2014). Non-equivalent contributions of maternal and paternal genomes to early plant embryogenesis. Nature 514, 624–627. doi: 10.1038/nature13620
Denninger, P., Bleckmann, A., Lausser, A., Vogler, F., Ott, T., Ehrhardt, D. W., et al. (2014). Male-female communication triggers calcium signatures during fertilization in Arabidopsis. Nat. Commun. 5:4645. doi: 10.1038/ncomms5645
d’Erfurth, I., Jolivet, S., Froger, N., Catrice, O., Novatchkova, M., and Mercier, R. (2009). Turning meiosis into mitosis. PLoS Biol. 7:e1000124. doi: 10.1371/journal.pbio.1000124
Digonnet, C., Aldon, D., Leduc, N., Dumas, C., and Rougier, M. (1997). First evidence of a calcium transient in flowering plants at fertilization. Development 124, 2867–2874.
Dirks, R., van Dun, K., de Snoo, C. B., van den Berg, M., Lelivelt, C. L. C., Voermans, W., et al. (2009). Reverse breeding: a novel breeding approach based on engineered meiosis. Plant Biotechnol. J. 7, 837–845. doi: 10.1111/j.1467-7652.2009.00450.x
Dresselhaus, T., and Franklin-Tong, N. (2013). Male-female crosstalk during pollen germination, tube growth and guidance, and double fertilization. Mol. Plant 6, 1018–1036. doi: 10.1093/mp/sst061
Dresselhaus, T., Sprunck, S., and Wessel, G. M. (2016). Fertilization mechanisms in flowering plants. Curr. Biol. 26, R125–R139. doi: 10.1016/j.cub.2015.12.032
Ebel, C., Mariconti, L., and Gruissem, W. (2004). Plant retinoblastoma homologues control nuclear proliferation in the female gametophyte. Nature 429, 776–780. doi: 10.1038/nature02637
Eckersley-Maslin, M. A., Alda-Catalinas, C., and Reik, W. (2018). Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat. Rev. Mol. Cell. Biol. 19, 436–450. doi: 10.1038/s41580-018-0008-z
El Ouakfaoui, S., Schnell, J., Abdeen, A., Colville, A., Labbé, H., Han, S., et al. (2010). Control of somatic embryogenesis and embryo development by AP2 transcription factors. Plant Mol. Biol. 7, 313–326. doi: 10.1007/s11103-010-9674-8
Ferris, P. J., and Goodenough, U. W. (1997). Mating type in Chlamydomonas is specified by mid, the minus-dominance gene. Genetics 146, 859–869.
Floyd, S. K., and Bowman, J. L. (2007). The ancestral developmental tool kit of land plants. Int. J. Plant Sci. 168, 1–35. doi: 10.1086/509079
Garcês, H. M., Champagne, C. E., Townsley, B. T., Park, S., Malhó, R., Pedroso, M. C., et al. (2007). Evolution of asexual reproduction in leaves of the genus Kalanchoë. Proc. Natl. Acad. Sci. U.S.A. 104, 15578–15583. doi: 10.1073/pnas.0704105104
Garcia-Aguilar, M., Michaud, C., Leblanc, O., and Grimanelli, D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22, 3249–3267. doi: 10.1105/tpc.109.072181
Germanà, M. A. (2006). Doubled haploid production in fruit crops. Plant Cell Tiss. Org. 86, 131–146. doi: 10.1007/s11240-006-9088-0
Germanà, M. A. (2011). Gametic embryogenesis and haploid technology as valuable support to plant breeding. Plant Cell Rep. 30, 839–857. doi: 10.1007/s00299-011-1061-7
Grimanelli, D., García, M., Kaszas, E., Perotti, E., and Leblanc, O. (2003). Heterochronic expression of sexual reproductive programs during apomictic development in Tripsacum. Genetics 165, 1521–1531.
Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M. A., and Gagliano, W. B. (1998). Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280, 446–450. doi: 10.1126/science.280.5362.446
Grossniklaus, U. (2011). Plant germline development: a tale of cross-talk, signaling, and cellular interactions. Sex. Plant Reprod. 24, 91–95. doi: 10.1007/s00497-011-0170-3
Grusz, A. L. (2016). A current perspective on apomixis in ferns. J. Syst. Evol. 54, 656–665. doi: 10.1111/jse.12228
Guitton, A. E., and Berger, F. (2005). Loss of function of MULTICOPY SUPPRESSOR OF IRA 1 produces nonviable parthenogenetic embryos in Arabidopsis. Curr. Biol. 15, 750–754. doi: 10.1016/j.cub.2005.02.066
Hand, M. L., de Vries, S., and Koltunow, A. M. (2016). A comparison of in vitro and in vivo asexual embryogenesis. Methods Mol. Biol. 1359, 3–23. doi: 10.1007/978-1-4939-3061-6_1
Hand, M. L., and Koltunow, A. M. (2014). The genetic control of apomixis: asexual seed formation. Genetics 97, 441–450. doi: 10.1534/genetics.114.163105
Hehenberger, E., Kradolfer, D., and Köhler, C. (2012). Endosperm cellularization defines an important developmental transition for embryo development. Development 139, 2031–2039. doi: 10.1242/dev.077057
Hennig, L., Taranto, P., Walser, M., Schönrock, N., and Gruissem, W. (2003). Arabidopsis MSI1 is required for epigenetic maintenance of reproductive development. Development 130, 2555–2565. doi: 10.1242/dev.00470
Horner, V. L., and Wolfner, M. F. (2008). Transitioning from egg to embryo: triggers and mechanisms of egg activation. Dev. Dyn. 237, 527–544. doi: 10.1002/dvdy.21454
Horstman, A., Bemer, M., and Boutilier, K. (2017). A transcriptional view on somatic embryogenesis. Regeneration 4, 201–216. doi: 10.1002/reg2.91
Horstman, A., Willemsen, V., Boutilier, K., and Heidstra, R. (2014). AINTEGUMENTA-Like proteins: hubs in a plethora of networks. Trends Plant Sci. 19, 146–157. doi: 10.1016/j.tplants.2013.10.010
Hsam, S. L. K., and Zeller, F. J. (1993). Haploid production in durum wheat by the interaction of Aegilops kotschyi cytoplasm and 1BL/1RS chromosomal interchange. Theor. Appl. Genet. 86, 951–954. doi: 10.1007/BF00211046
Ibarra, C. A., Feng, X., Schoft, V. K., Hsieh, T. F., Uzawa, R., Rodrigues, J. A., et al. (2012). Active DNA demethylation in plant companion cells reinforces transposon methylation in gametes. Science 337, 1360–1364. doi: 10.1126/science.1224839
Ikeuchi, M., Ogawa, Y., Iwase, A., and Sugimoto, K. (2016). Plant regeneration: cellular origins and molecular mechanisms. Development 143, 1442–1451. doi: 10.1242/dev.134668
Islam, S. M., and Tuteja, N. (2012). Enhancement of androgenesis by abiotic stress and other pretreatments in major crop species. Plant Sci. 182, 134–144. doi: 10.1016/j.plantsci.2011.10.001
Jofuku, K. D., den Boer, B. G. W., van Montague, M., and Okamuro, J. K. (1994). Control of Arabidopsis flower and seed development by the homeotic gene APETELA2. Plant Cell 6, 1211–1225. doi: 10.1105/tpc.6.9.1211
Johnston, A. J., Kirioukhova, O., Barrell, P. J., Rutten, T., Moore, J. M., Baskar, R., et al. (2010). Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet. 6:e1000988. doi: 10.1371/journal.pgen.1000988
Johnston, A. J., Matveeva, E., Kirioukhova, O., Grossniklaus, U., and Gruissem, W. (2008). A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. PLoS Genet. 6:e1000988. doi: 10.1371/journal.pgen.1000988
Juel, H. O. (1898). Parthenogenesis bei Antennaria alpina (L.) R. Br. Botanischer Centralblatt. 74, 369–372.
Juel, H. O. (1906). Die tetradenteilung bei Taraxacum und anderen cichorieen. K. Sven. Vetenskapsakad. Handl. 39, 1–21.
Jullien, P. E., Mosquna, A., Ingouff, M., Sakata, T., Ohad, N., and Berger, F. (2008). Retinoblastoma and its binding partner MSI1 control imprinting in Arabidopsis. PLoS Biol. 12:e194. doi: 10.1371/journal.pbio.0060194
Kaushal, P., Malaviya, D. R., Roy, A. K., Pathak, S., Agrawal, A., Khare, A., et al. (2008). Reproductive pathways of seed development in apomictic guinea grass (Panicum maximum Jacq.) reveal uncoupling of apomixis components. Euphytica 164, 81–92. doi: 10.1007/s10681-008-9650-4
Khanday, I., Skinner, D., Yang, B., Mercier, R., and Sundaresan, V. (2018). A male-expressed rice embryogenic trigger redirected for asexual propagation through seeds. Nature 565, 91–95. doi: 10.1038/s41586-018-0785-8
Kirioukhova, O., Shah, J. N., Larsen, D. S., Tayyab, M., Mueller, N. E., Govind, G., et al. (2018). Aberrant imprinting may underlie evolution of parthenogenesis. Sci. Rep. 8:10626. doi: 10.1038/s41598-018-27863-7
Köhler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U., and Gruissem, W. (2003a). Arabidopsis MSI1 is a component of the MEA/FIE Polycomb group complex and required for seed development. EMBO J. 22, 4804–4814.
Köhler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003b). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553.
Koi, S., Hisanaga, T., Sato, K., Shimamura, M., Yamato, K. T., Ishizaki, K., et al. (2016). An evolutionarily conserved plant RKD factor controls germ cell differentiation. Curr. Biol. 26, 1–7. doi: 10.1016/j.cub.2016.05.013
Kono, T., Obata, Y., Wu, Q., Niwa, K., Ono, Y., Yamamoto, Y., et al. (2004). Birth of parthenogenetic mice that can develop to adulthood. Nature 428, 860–864. doi: 10.1038/nature02402
Köszegi, D., Johnston, A. J., Rutten, T., Altschmied, L., Kumlehn, J., Wüst, S. E., et al. (2011). Members of the RKD transcription factor family induce an egg cell-like gene expression program. Plant J. 67, 280–291. doi: 10.1111/j.1365-313X.2011.04592.x
Kradolfer, D., Hennig, L., and Köhler, C. (2013). Increased maternal genome dosage bypasses the requirement of the FIS polycomb repressive complex 2 in Arabidopsis seed development. PLoS Genet. 9:e1003163. doi: 10.1371/journal.pgen.1003163
Kumlehn, J., Kirik, V., Czihal, A., Altschmied, L., Matzk, F., Lörz, H., et al. (2001). Parthenogenetic egg cells of wheat: cellular and molecular studies. Sex. Plant Reprod. 14, 239–243. doi: 10.1007/s00497-001-0115-3
Kuwabara, A., and Gruissem, W. (2014). Arabidopsis retinoblastoma-related and Polycomb group proteins: cooperation during plant cell differentiation and development. J. Exp. Bot. 65, 2667–2676. doi: 10.1093/jxb/eru069
Labombarda, P., Busti, A., Caceres, M. E., Pupilli, F., and Arcioni, S. (2002). An AFLP marker tightly linked to apomixis reveals hemizygosity in a portion of the apomixis-controlling locus in Paspalum simplex. Genome 45, 513–519. doi: 10.1139/g02-014
Luo, M., Bilodeau, P., Koltunow, A., Dennis, E., Peacock, W., and Chaudhury, A. M. (1999). Genes controlling fertilization-independent seed development in Arabidopsis thaliana. Proc. Natl. Acad.Sci. U.S.A. 96, 296–301. doi: 10.1073/pnas.96.1.296
Machaty, Z. (2016). Signal transduction in mammalian oocytes during fertilization. Cell Tissue Res. 363, 169–183. doi: 10.1007/s00441-015-2291-8
Maheshwari, P. (1950). An Introduction to the Embryology of Angiosperms. New York, NY: McGraw-Hill. doi: 10.5962/bhl.title.5681
Malecka, J. (1965). Embryological studies in Taraxacum palustre. Acta Biol. Cracov. Bot. 8, 223–235.
Marimuthu, M. P., Jolivet, S., Ravi, M., Pereira, L., Davda, J. N., Cromer, L., et al. (2011). Synthetic clonal reproduction through seeds. Science 331:876. doi: 10.1126/science.1199682
Martinez, G., and Köhler, C. (2017). Role of small RNAs in epigenetic reprogramming during plant sexual reproduction. Curr. Opin. Plant. Biol. 36, 22–28. doi: 10.1016/j.pbi.2016.12.006
Matzk, F., Meyer, H. M., Bäumlein, H., Balzer, H. J., and Schubert, I. (1995). A novel approach to the analysis of the initiation of embryo development in Gramineae. Sex. Plant Reprod. 8, 266–272. doi: 10.1007/BF00229382
Mercier, R., Vezon, D., Bullier, E., Motamayor, J. C., Sellier, A., Lefèvre, F., et al. (2001). SWITCH1 (SWI1): a novel protein required for the establishment of sister chromatid cohesion and for bivalent formation at meiosis. Genes Dev. 15, 1859–1871. doi: 10.1101/gad.203201
Mozgova, I., and Hennig, L. (2015). The polycomb group protein regulatory network. Annu. Rev. Plant Biol. 66, 269–296. doi: 10.1146/annurev-arplant-043014-115627
Murbeck, S. (1904). Parthenogenese bei den Gattungen Taraxacum und Hieracium. Bot. Notiser. 6, 285–296.
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432. doi: 10.1104/pp.105.073783
Naumova, T. N., and Matzk, F. (1998). Differences in the initiation of the zygotic and parthenogenetic pathway in the Salmon lines of wheat: ultrastructural studies. Sex. Plant Reprod. 11, 121–130. doi: 10.1007/s004970050129
Niklas, K. J., and Kutschera, U. (2010). The evolution of the land plant life cycle. New Phytol. 185, 27–41. doi: 10.1111/j.1469-8137.2009.03054.x
Nogler, G. A. (1984). “Gametophytic apomixis,” in Embryology of Angiosperms, ed. B. M. Johri (Berlin: Springer-Verlag), 475–518. doi: 10.1007/978-3-642-69302-1_10
Nogler, G. A. (2007). “The discovery of parthenogenesis: a long journey to the truth,” in Apomixis: Evolution, Mechanisms and Perspectives, Vol. 147, eds E. Hörandl, U. Grossniklaus, P. J. van Dijk, and T. F. Sharbel (Ruggell: Gantner Verlag), 25–35.
Noyes, R. D. (2000). Biogeographical and evolutionary insights on Erigeron and allies (Asteraceae) from ITS sequence data. Plant Syst. Evol. 220, 93–114. doi: 10.1007/BF00985373
Noyes, R. D. (2005). Inheritance of apomeiosis (diplospory) in fleabanes Erigeron (Asteraceae). Heredity 94, 193–198. doi: 10.1038/sj.hdy.6800597
Noyes, R. D., Baker, R., and Mai, B. (2007). Mendelian segregation for two-factor apomixis in Erigeron annuus (Asteraceae). Heredity 98, 92–98. doi: 10.1038/sj.hdy.6800907
Noyes, R. D., and Rieseberg, L. H. (2000). Two independent loci control agamospermy (apomixis) in the triploid flowering plant Erigeron annuus. Genetics 155, 379–390.
Noyes, R. D., and Wagner, J. D. (2014). Dihaploidy yields diploid apomicts and parthenogens in Erigeron (Asteraceae). Am. J. Bot. 101, 865–874. doi: 10.3732/ajb.1400008
Ochatt, S. J., Atif, R. M., Patat-Ochatt, E. M., Jacas, L., and Conreux, C. (2010). Competence versus recalcitrance for in vitro regeneration. Not. Bot. Horti Agrobot. 38, 102–108.
Ogawa, D., Johnson, S. D., Henderson, S. T., and Koltunow, A. M. (2013). Genetic separation of autonomous endosperm formation (AutE) from two other components of apomixis in Hieracium. Plant Reprod. 26, 113–123. doi: 10.1007/s00497-013-0214-y
Ohad, N., Yadegari, R., Margossian, L., Hannon, M., Michaeli, D., Harada, J., et al. (1999). Mutations in FIE, a WD polycomb group gene, allow endosperm development without fertilization. Plant Cell 11, 407–416. doi: 10.1105/tpc.11.3.407
Olmedo-Monfil, V., Duraìn-Figueroa, N., Arteaga-Vaìzquez, M., Demesa-Areìvalo, E., Autran, D., Grimanelli, D., et al. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632. doi: 10.1038/nature08828
Ortiz, J. P., Quarin, C. L., Pessino, S. C., Acuña, C., Martínez, E. J., Espinoza, F., et al. (2013). Harnessing apomictic reproduction in grasses: what we have learned from Paspalum. Ann. Bot. 112, 767–787. doi: 10.1093/aob/mct152
Owen, R. (1849). On Parthenogenesis, or the Successive Production of Procreating Individuals From a Single Ovum. London: J. van Voorst.
Ozias-Akins, P. (2006). Apomixis: developmental characteristics and genetics. Crit. Rev. Plant Sci. 25, 199–214. doi: 10.1080/07352680600563926
Pillot, M., Baroux, C., Vazquez, M. A., Autran, D., Leblanc, O., Vielle-Calzada, J. P., et al. (2010). Embryo and endosperm inherit distinct chromatin and transcriptional states from the female gametes in Arabidopsis. Plant Cell 22, 307–320. doi: 10.1105/tpc.109.071647
Pónya, Z., Corsi, I., Hoffmann, R., Kovács, M., Dobosy, A., Kovács, A. Z., et al. (2014). When isolated at full receptivity, in vitro fertilized wheat (Triticum aestivum, L.) egg cells reveal [Ca2+ ] cyt oscillation of intracellular origin. Int. J. Mol. Sci. 15, 23766–23791. doi: 10.3390/ijms151223766
Pupilli, F., and Barcaccia, G. (2012). Cloning plants by seeds: Inheritance models and candidate genes to increase fundamental knowledge for engineering apomixis in sexual crops. J. Biotechnol. 159, 291–311. doi: 10.1016/j.jbiotec.2011.08.028
Radoeva, T., Lokerse, A. S., Llavata-Peris, C. I., Wendrich, J., Xiang, D., Liao, C. Y., et al. (2018). A robust auxin response network controls embryo and suspensor development through a bHLH transcriptional module. Plant Cell doi: 10.1105/tpc.18.00518 [Epub ahead of print].
Radoeva, T., and Weijers, D. (2014). A roadmap to embryo identity in plants. Trends Plant Sci. 19, 709–716. doi: 10.1016/j.tplants.2014.06.009
Ravi, M., Marimuthu, M. P., and Siddiqi, I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451, 1121–1124. doi: 10.1038/nature06557
Ravi, M., Shibata, F., Ramahi, J. S., Nagaki, K., Chen, C., Murata, M., et al. (2011). Meiosis-specific loading of the centromere-specific histone CENH3 in Arabidopsis thaliana. PLoS Genet. 7:e1002121. doi: 10.1371/journal.pgen.1002121
Richards, A. J. (1970). Eutriploid facultative agamospermy in Taraxacum. New Phytol. 69, 761–774. doi: 10.1111/j.1469-8137.1970.tb02461.x
Riechmann, J. L., and Meyerowitz, E. M. (1998). The AP2/EREBP family of plant transcription factors. Biol. Chem. 379, 633–646.
Roche, B., Arcangioli, B., and Martienssen, R. A. (2016). RNA interference is essential for cellular quiescence. Science 354:aah5651. doi: 10.1126/science.aah5651
Rosenberg, O. (1906). Über die embryobildung in der Gattung Hieracium. Ber. Deut. Bot. Ges. 24, 157–161.
Rövekamp, M., Bowman, J. L., and Grossniklaus, U. (2016). Marchantia MpRKD regulates the gametophyte-sporophyte transition by keeping egg cells quiescent in the absence of fertilization. Curr. Biol. 26, 1–8. doi: 10.1016/j.cub.2016.05.028
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Schmidt, A., Schmid, M. W., and Grossniklaus, U. (2015). Plant germline formation: common concepts and developmental flexibility in sexual and asexual reproduction. Development 142, 229–241. doi: 10.1242/dev.102103
Schön, I., Martens, K., and van Dijk, P. J. (eds) (2009). Lost Sex: The Evolutionary Biology of Parthenogenesis. Dordrecht: Springer. doi: 10.1007/978-90-481-2770-2
Scott, R. J., Spielman, M., Bailey, J., and Dickinson, H. G. (1998). Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125, 3329–3341.
Smith, J. (1841). Notice of a plant which produces perfect seeds without any apparent action of pollen. Trans. Linn. Soc. Lond. 18, 509–512. doi: 10.1111/j.1095-8339.1838.tb00200.x
Soliìs-Ramos, L. Y., Andrade-Torres, A., Sáenz Carbonell, L. A., Oropeza Saliìn, C. M., and Castanþo de la Serna, E. (2012). Somatic Embryogenesis in Recalcitrant Plants in Embryologenesis. Available at: www.intechopen.com
Steffen, J. G., Kang, I. H., Macfarlane, J., and Drews, G. N. (2007). Identification of genes expressed in the Arabidopsis female gametophyte. Plant J. 51, 281–292. doi: 10.1111/j.1365-313X.2007.03137.x
Strasburger, E. (1877). Über befruchtung und zelltheilung. Zeitschr. Naturwissenschaften 11, 435–536.
Suomalainen, E., Saura, A., and Lokki, J. (1987). Cytology and Evolution in Parthenogenesis. Boca Raton FL: CRC Press.
Tadros, W., and Lipshitz, H. D. (2009). The maternal-to-zygotic transition: a play in two acts. Development 136, 3033–3042. doi: 10.1242/dev.033183
Tas, I. C., and van Dijk, P. J. (1999). Crosses between sexual and apomictic dandelions (Taraxacum). I. The inheritance of apomixis. Heredity 83, 707–714. doi: 10.1046/j.1365-2540.1999.00619.x
Tedeschi, F., Rizzo, P., Rutten, T., Altschmied, L., and Bäumlein, H. (2017). RWP-RK domain-containing transcription factors control cell differentiation during female gametophyte development in Arabidopsis. New Phytol. 213, 1909–1924. doi: 10.1111/nph.14293
Tekleyohans, D. G., Nakel, T., and Groß-Hardt, R. (2017). Patterning the female gametophyte of flowering plants. Plant Physiol. 173, 122–129. doi: 10.1104/pp.16.01472
Tsunewaki, K., and Mukai, Y. (1990). “Wheat haploids through the Salmon method,” in Biotechnology Agriculture Forestry, Vol. 13, ed. Y. P. S. Bajaj (Berlin: Springer), 460–478. doi: 10.1007/978-3-662-10933-5_25
van Baarlen, P., De Jong, J. H., and van Dijk, P. J. (2002). Comparative cyto-embryological investigations of sexual and apomictic dandelions (Taraxacum) and their apomictic hybrids. Sex. Plant Reprod. 15, 31–38. doi: 10.1007/s00497-002-0132-x
van Dijk, P. J., de Jong, J. H., Vijverberg, K., and Biere, A. (2009). “An apomixis-gene’s view on dandelions,” in Lost Sex: The Evolutionary Biology of Parthenogenesis, eds I. Schön, K. Martens, and P. J. van Dijk (Dordrecht: Springer), 475–493.
van Dijk, P. J., Tas, I. C., Falque, M., and Bakx-Schotman, T. (1999). Crosses between sexual and apomictic dandelions (Taraxacum). II. The breakdown of apomixis. Heredity 83, 715–721. doi: 10.1046/j.1365-2540.1999.00620.x
van Dijk, P. J., van Baarlen, P., and de Jong, J. H. (2003). The occurrence of phenotypically complementary apomixis-recombinants in crosses between sexual and apomictic dandelions (Taraxacum officinale). Sex. Plant Reprod. 16, 71–76. doi: 10.1007/s00497-003-0177-5
van Dijk, P. J., and Vijverberg, K. (2005). “The significance of apomixis in the evolution of the angiosperms: a reappraisal,” in Plant Dpecies-Level Dystematics. New Perspectives on Pattern and Process, Vol. 143, eds F. T. Bakker, L. W. Chatrou, B. Gravendeel, and P. B. Pelzer (Ruggell: Gantner Verlag), 101–116.
Vijverberg, K., and van Dijk, P. J. (2007). “Genetic linkage mapping of apomixis loci,” in Apomixis: Evolution, Mechanisms and Perspectives, Vol. 147, eds E. Hörandl, U. Grossniklaus, P. J. van Dijk, and T. F. Sharbel (Ruggell: Gantner Verlag), 137–158.
von Siebold, C. T. E. (1856). Wahre Parthenogenesis bei Schmetterlingen und Biene, ein Beitrag Zur Fortpflanzungsgeschichte der Thiere. Leigpzig: W. Engelmann.
Wang, G., and Köhler, C. (2017). Epigenetic processes in flowering plant reproduction. J. Exp. Bot. 68, 797–807. doi: 10.1093/jxb/erw486
Wüst, S. E., Vijverberg, K., Schmidt, A., Weiss, M., Gheyselinck, J., Lohr, M., et al. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20, 506–512. doi: 10.1016/j.cub.2010.01.051
Yamashita, K., Nakazawa, Y., Namai, K., Amagai, M., Tsukazaki, H., Wako, T., et al. (2012). Modes of inheritance of two apomixis components, diplospory and parthenogenesis, in Chinese chive (Allium ramosum) revealed by analysis of the segregating population generated by back-crossing between amphimictic and apomictic diploids. Breed. Sci. 62, 160–169. doi: 10.1270/jsbbs.62.160
Keywords: apomixis, embryogenesis, embryo induction, PsASGR-BabyBoom-Like (PsASGR-BBML), doubled haploids, parthenogenesis, Pennisetum, Taraxacum
Citation: Vijverberg K, Ozias-Akins P and Schranz ME (2019) Identifying and Engineering Genes for Parthenogenesis in Plants. Front. Plant Sci. 10:128. doi: 10.3389/fpls.2019.00128
Received: 15 November 2018; Accepted: 24 January 2019;
Published: 19 February 2019.
Edited by:
Emidio Albertini, University of Perugia, ItalyReviewed by:
Stefan de Folter, Centro de Investigación y de Estudios Avanzados (CINVESTAV), MexicoMasaru Ohme-Takagi, Saitama University, Japan
Copyright © 2019 Vijverberg, Ozias-Akins and Schranz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kitty Vijverberg, kitty.vijverberg@wur.nl
 Kitty Vijverberg
Kitty Vijverberg Peggy Ozias-Akins
Peggy Ozias-Akins M. Eric Schranz
M. Eric Schranz