- Biotechnology Research Center, The University of Tokyo, Tokyo, Japan
The Brix domain is a conserved domain in several proteins involved in ribosome biogenesis in yeast and animals. In the Arabidopsis genome, six Brix domain-containing proteins are encoded; however, their molecular functions have not been fully characterized, as yet. Here we report the functional analysis of a Brix domain-containing protein, ARPF2, which is homologous to yeast Rpf2 that plays an essential role in ribosome biogenesis as a component of the 5S ribonucleoprotein particle. By phenotypic characterization of arpf2 mutants, histochemical GUS staining, and analysis using green fluorescence protein, we show that ARPF2 is an essential and ubiquitously expressed gene encoding a nucleolar protein. Co-immunoprecipitation and split-GFP-based bimolecular fluorescence complementation assays revealed that ARPF2 interacts with a protein named ARRS1, which is homologous to yeast Rrs1 that forms a complex with Rpf2 in yeast. Furthermore, the result of RNA immunoprecipitation assay indicated that ARPF2 interacts with 5S ribosomal RNA (rRNA) or the precursor of 5S rRNA, as well as with the internal transcribed spacer 2 in the precursors of 25S rRNA. Most intriguingly, we found that the overexpression of ARPF2 and ARRS1 leads to characteristic phenotypes, including short stem, abnormal leaf morphology, and long lifespan, in Arabidopsis. These results suggest that the function of Brix domain-containing ARPF2 protein in ribosome biogenesis is intimately associated with the growth and development in plants.
Introduction
Ribosome biogenesis is a complex process that requires numerous factors, including ribosomal proteins (RPs), ribosomal RNAs (rRNAs), and ribosome biogenesis factors (RBFs) (Henras et al., 2015; Weis et al., 2015a). In eukaryotes, ribosome biogenesis begins with the synthesis of 45S pre-rRNA, which functions as a scaffold for the assembly of RPs, RBFs, and small nucleolar RNAs to form the 90S pre-ribosomal particle in the nucleolus (Henras et al., 2015; Weis et al., 2015a). During the maturation steps, the 45S pre-rRNA in the 90S pre-ribosomal particle is processed into 18S, 5.8S, and 25S rRNA by the elimination of 5′- and 3′-external spacer regions [5′-external transcribed spacer (ETS) and 3′-ETS] and two internal spacer regions [internal transcribed spacer (ITS) 1 and ITS2], and the 90S pre-ribosomal particle is divided into the 40S pre-ribosome subunit, containing 18S rRNA, and the 60S pre-ribosome subunit, containing 5.8S and 25S rRNAs (Henras et al., 2015; Weis et al., 2015a). In the next step, the 5S ribonucleoprotein particle (RNP), comprising two RPs (Rpl5 and Rpl11 in Saccharomyces cerevisiae) and 5S rRNA, which is transcribed in the nucleoplasm, assembles into a 60S pre-ribosomal particle (Madru et al., 2015). Finally, the 40S and 60S pre-ribosomal subunits are exported into the cytoplasm and are re-assembled to form the functional mature ribosomes (Henras et al., 2015; Weis et al., 2015a).
The Brix domain (biogenesis of ribosomes in Xenopus domain) is a region of 150–180 amino acid residues in length, which is conserved in several proteins involved in ribosome biogenesis (Eisenhaber et al., 2001). For example, in S. cerevisiae, Brx1, Imp4, Ssf1/Ssf2, Rpf1, and Rpf2 contain this domain (Eisenhaber et al., 2001; Wehner and Baserga, 2002). Brx1 was the first Brix domain-containing protein that was identified; it was characterized as an rRNA-interacting protein involved in pre-rRNA processing in Xenopus laevis and S. cerevisiae (Kaser et al., 2001). Other Brix domain-containing proteins in S. cerevisiae, namely Imp4, Ssf1/2, Rpf1, and Rpf2, also bind to pre-rRNAs, and the depletion of Imp4, Ssf1, Rpf1, or Rpf2 results in defects in the processing of pre-rRNA (Lee and Baserga, 1999; Wehner and Baserga, 2002). Crystal analysis of the structures of Rpf2 and its homolog from Aspergillus nidulans revealed that Rpf2 tightly binds to a protein called Rrs1 through the N-terminal half of the Brix domain. Moreover, it was determined that the Rpf2–Rrs1 complex binds to 5S rRNA through the C-terminal half of the Brix domain of Rpf2 to form the pre-60S ribosomal subunit together with the 5S RNP (Asano et al., 2015; Kharde et al., 2015; Madru et al., 2015). Although Rpf2 alone can bind to 5S rRNA, Rpf2 was shown to more rigidly bind to 5S rRNA through forming the complex with Rrs1 (Madru et al., 2015). Furthermore, another Brix domain-containing protein, Imp4, was found to bind to U3 small nucleolar RNA and a U3 small nucleolar RNA-associated protein, Mpp10, through its Brix domain and it played a role in the nucleolar processing of pre-18S ribosomal RNA (Lee and Baserga, 1999; Granneman et al., 2003). Therefore, the Brix domain is supposed as a structural hub for interactions with both protein and RNA that are mediated by its N-terminal and C-terminal halves, respectively.
Brix domain-containing proteins likely play key roles in ribosome biogenesis in plants as well as in yeast and animals; however, plant Brix domain-containing proteins have not been well characterized as yet, although a role of Arabidopsis duplicated orthologs of yeast Brx1, BRX1-1 and BRX1-2, in pre-rRNA processing (Weis et al., 2015b) and lethality of the function-deficient mutants of the Arabidopsis homolog of yeast Ssf1/Ssf2, SNAIL1 (Hao et al., 2017), have been shown. In the current study, we characterized the Arabidopsis homolog of Rpf2, which binds to 5S RNP and is involved in ribosome biogenesis in S. cerevisiae (Asano et al., 2015; Kharde et al., 2015; Madru et al., 2015). To avoid confusion with RNA PROCESSING FACTOR2 (RPF2) that is required for 5′-end processing of mRNAs in the mitochondria of Arabidopsis (Jonietz et al., 2010), the Arabidopsis homolog of yeast Rpf2 is referred to as ARPF2 (Arabidopsis thalliana homolog of yeast Rpf2: At3g23620), hereafter. Our results indicate that ARPF2 is an essential gene that is ubiquitously expressed and encodes a nucleolar-localized protein. Moreover, it was also found that ARPF2 interacts with the Arabidopsis homolog of yeast Rrs1 (referred to as ARRS1, hereafter) and pre-rRNA in plant cells. These results suggest that ARPF2 and ARRS1 play roles similar to those of yeast Rpf2 and Rrs1 in the ribosome biogenesis. Most significantly, we found that the overexpression of ARPF2 and ARRS1 in Arabidopsis resulted in short height, abnormal leaf morphology, and an extended period of reproductive growth.
Materials and Methods
Phylogenetic Analysis
A phylogenetic tree of 23 Brix domain proteins from Homo sapiens, A. thaliana, Oryza sativa, and S. cerevisiae was created using MEGA7 (Kumar et al., 2016). The evolutionary history was inferred using the Neighbor-Joining method (Saitou and Nei, 1987). The evolutionary distances were computed using the Poisson correction method (Zuckerkandl and Pauling, 1965).
Plant Materials and Growth Conditions
Arabidopsis thaliana ecotype Col-0 was used as the wild type, and all T-DNA insertion lines and mutants used in this study were in the Col-0 background. The seeds of two T-DNA insertion lines, SALK_107828 for arpf2-1 and SAIL_314_A03 for arpf2-2, were provided by the Arabidopsis Biological Resource Center (Alonso et al., 2003) and the Syngenta Arabidopsis Insertion Library (Sessions et al., 2002), respectively. The seeds were sterilized and sown on agar plates containing half-strength Murashige and Skoog medium supplemented with 1% sucrose. After a 2-day cold treatment, the plates were placed at 23°C under diurnal light condition (16 h light/8 h dark, approximately 70 μmol m-2 s-1). After 2 weeks of growth, the seedlings were transferred onto peat containing nutrients (Sakatanotane Co., Yokohama, Japan) and further grown at 23°C under continuous light (∼70 μmol m-2 s-1).
Plasmid Construction
To generate plasmids for transient expression assay and for generation of transgenic plants, we obtained ARPF2, ARRS1, RPL4A and PRH75 cDNAs by RT-PCR using RNA from Col-0 seedlings. The sequences of primers used for RT-PCR are listed in Supplementary Table S1. The resulting PCR products were cloned into the pENTR/D-TOPO vector (Thermo Fisher Scientific Inc.). To construct the binary plasmids for expression of ARPF2 and ARRS1 fused to the N-terminus of GFP (ARPF2-GFP and ARRS1-GFP), ARPF2 and ARRS1 cDNAs were introduced into a Gateway binary vector, pGWB5, whereas RPL4A and PRH75 cDNAs were introduced into another Gateway binary vector, pGWB6, for the expression of RPL4A and PRH75 fused to the C-terminus of GFP (GFP-RPL4A and GFP-PRH75). The ARPF2 and ARRS1 cDNAs were also introduced into pGWB17 to produce expression vectors for ARPF2-MYC and ARRS1-MYC proteins. Additionally, ARPF2 cDNA was introduced into pB4GWnG to produce an expression vector for ARPF2-nGFP in which ARPF2 was fused to the N-terminal half of GFP (nGFP), whereas ARRS1 cDNA was introduced into pB4GWcG to produce an expression vector for ARRS1-cGFP in which ARRS1 was fused to the C-terminal half of GFP (cGFP). Furthermore, RPL4A cDNA was introduced into pB4nGGW and pB4cGGW to produce expression vectors for nGFP-RPL4A and cGFP-RPL4A in which RPL4A was fused to nGFP and cGFP, respectively. Plasmids, pGWB5/6/17, pB4GWnG, and pB4GWcG, were described earlier by Tanaka et al. (2012). To construct the pARPF2:GUS plasmid in which the ARPF2 promoter was located upstream of the GUS reporter gene, an approximately 1.5 kb DNA fragment containing the promoter and 5′-UTR regions of ARPF2 was amplified by PCR using genomic DNA of the wild type Col-0 plant and a set of PCR primers (Supplementary Table S1). The resulting product was initially cloned into the pENTR/D-TOPO vector, and subsequently transferred into pMDC164 (Curtis and Grossniklaus, 2003). All the PCR products and inserts were verified by DNA sequencing.
Generation of Transgenic Arabidopsis Plants
The wild type plants were transformed with the binary vectors for expression of ARPF2-GFP, ARPF2-MYC, ARRS1-MYC, APUM24-GFP (Maekawa et al., 2018), and GFP-PRH75, and pARPF2:GUS plasmid using the floral dip method (Clough and Bent, 1998) with Agrobacterium tumefaciens strain GV3101 (pMP90). The transgenic lines with T-DNA insertion(s) at a single locus were selected, and T3 progenies homozygous for the introduced gene were used for all the experiments. Two independent transgenic lines, ARPF2-GFPox#3 and ARPF2-MYCox#5, were subjected to phenotypic analysis of the plants overexpressing ARPF2.
Histochemical GUS Staining
GUS staining was performed in accordance with the standard protocol (Jefferson et al., 1987), with minor modifications. Various plant tissues were incubated in 80% acetone for 20 min at 4°C, and then washed with 50 mM sodium phosphate buffer (pH 7.0). Thereafter, the samples were submerged into the GUS staining buffer that contained 50 mM sodium phosphate buffer (pH 7.0), 100 mM of 5-bromo-4-chloro-3-indolyl β-D-glucuronide, 2 mM potassium ferricyanide, 2 mM potassium ferrocyanide, 10 mM EDTA (pH 8.0), and 0.1% Triton X-100. The staining reaction was carried out at 37°C in the dark for 4 h, and the samples were then washed with 70% ethanol to remove chlorophyll.
Subcellular Localization Analysis With Green Fluorescence Protein (GFP) and Split-GFP-Based Bimolecular Fluorescence Complementation (BiFC) Assays
The subcellular localization and split-GFP-based BiFC assays were performed by co-infiltrating Nicotiana benthamiana leaves (Kapila et al., 1997; D’Aoust et al., 2009) with derivatives of pGWB5, pB4GWnG, and pB4GWcG and the expression vector for FIB1-mCherry, as described previously (Maekawa et al., 2014). Two or three days after the infection, the GFP fluorescence was observed under a fluorescence microscope (BX51, Olympus Corp.) equipped with a cooled color digital camera (DP80, Olympus Corp.).
Co-immunoprecipitation Analysis
Co-immunoprecipitation analysis was performed by co-infiltrating N. benthamiana leaves (Kapila et al., 1997; D’Aoust et al., 2009) with the expression vectors for ARPF2-GFP and ARRS1-MYC. Three days after the infection, the infected leaves (∼100 mg) were frozen in liquid nitrogen and homogenized with zirconia balls in 400 μl of extraction buffer that contained 25 mM Tris-HCl (pH 7.5), 150 mM NaCl, 0.1% NP40, 10% glycerol, and protease inhibitor cocktail from Sigma-Aldrich Co. LLC (P9599 and 11697498001). The cell lysates were incubated with anti-GFP antibody (MBL Co., Ltd.) and protein A agarose beads (Sigma–Aldrich Co. LLC), and then the proteins interacting with the antibody were eluted with the SDS sample buffer (BioRad, Inc.) and resolved by SDS-PAGE. Following transfer to polyvinylidene difluoride membranes, the proteins were detected with anti-GFP (MBL Co., Ltd., 1:1,000) and anti-MYC (Merck Millipore, 1:1,000) antibodies. In a control experiment, ARRS1-MYC protein was transiently expressed alone or with ARPF2-GFP or GFP-RPL4A in N. benthamiana leaves. Immunoprecipitates obtained with anti-GFP antibody in the experiment were analyzed with anti-MYC and anti-GFP antibodies.
RNA Immunoprecipitation (RIP) Assay
The RIP assay was performed as described previously (Maekawa et al., 2018), except for the fact that immunoprecipitation was performed with an anti-GFP antibody (MBL Co., Ltd.). Quantitative PCR (qPCR) was performed using rRNA-related PCR primers (Supplementary Table S1), as described in “Expression Analysis”. The enrichment fold was calculated as the ratio of the RNA level in the eluate to that in the input sample.
Expression Analysis
Total RNA was prepared from seedlings using an ISOSPIN Plant RNA kit (Nippon Gene Co., Ltd., Japan) with a TURBO DNase kit (Thermo Fisher Scientific Inc.), and reverse transcription was performed with Prime Script RT Master Mix (Takara Bio Inc.). The PCR was performed with a StepOne Plus Real Time PCR System (Thermo Fisher Scientific Inc.) using a KAPA SYBR Fast Quantitative PCR kit (KAPA Biosystems, Inc.). The primers used are listed in Supplementary Table S1. The relative gene expression levels were calculated using the ΔΔCT method (Livak and Schmittgen, 2001) and normalized relative to the expression levels of UBQ10.
Quantification of Chlorophyll and Cell Death
Chlorophyll quantification was performed with a batch of three to five rosette leaves, as described by Porra et al. (1989). Cell death was quantified by electrolyte leakage. Leaf disks (4 mm diameter) of rosette leaves were soaked in milliQ water and electrolyte leakage was measured after 2 h of incubation at room temperature with a conductivity meter (Horiba Ltd., Japan). The total electrolytes were determined after boiling for 10 min, and the extent of cell death was assessed with relative electrolyte leakage rates.
Accession Numbers
The sequence data referred to in this manuscript can be found in the Arabidopsis Genome Initiative database under the following accession numbers: ARPF2 (At3g23620), ARRS1 (At2g37990), RPL4A (At3g09630), APUM24 (At3g16810), and PRH75 (At5g62190).
Results
Identification of the Arabidopsis Homolog of Yeast Rpf2
The Brix domain-containing proteins are divided into five subgroups in yeast (Eisenhaber et al., 2001; Wehner and Baserga, 2002). A phylogenetic analysis of Brix domain-containing proteins from several organisms (H. sapiens, A. thaliana, O. sativa, and S. cerevisiae) revealed that all the analyzed organisms possess at least one member of each subgroup, suggesting that they play distinct but conserved roles in each organism (Figure 1A). Although the Arabidopsis proteins, BRX1-1 and BRX1-2, redundantly function (Weis et al., 2015b; Maekawa et al., 2018), similarity between two rice proteins in the Brx1 subgroup (XP_015622643.1 and XP_015623195.1) was low. Thus, it is currently uncertain as to whether these rice proteins play similar roles in rice. All the Arabidopsis Brix domain-containing proteins possess the Brix domain in the central region but have no other characteristic domain, except for the coiled-coil domain in ARPF1 and SNAIL1 (Figure 1B). Alignment of amino acid sequences of Arabidopsis Brix domains indicated that the overall homology is not high and that only several amino acid residues are conserved in them (Supplementary Figure S1). This observation may suggest that each Brix domain mediates intereactions with distinct interaction partners.
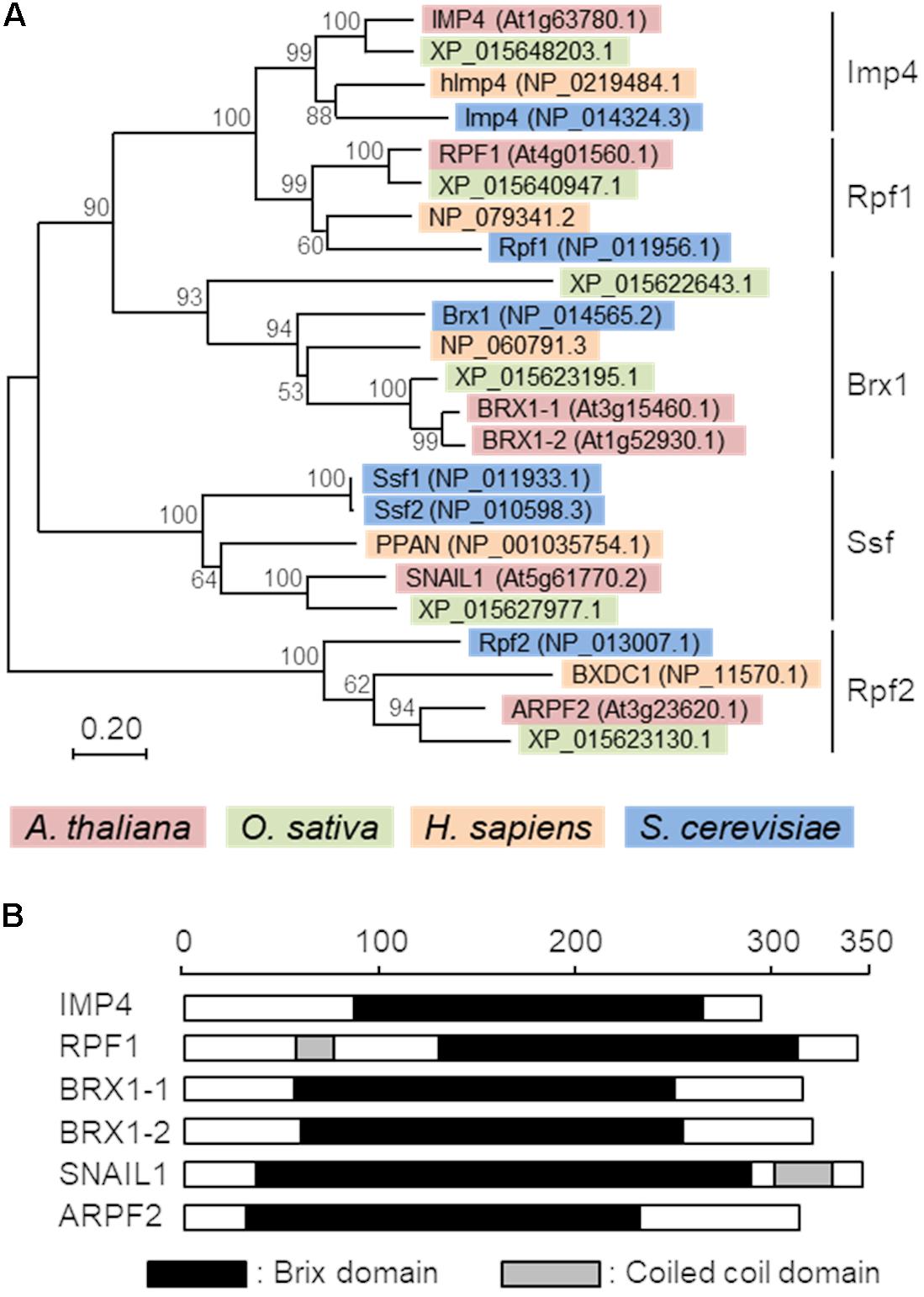
FIGURE 1. Phylogenetic tree and domain structures of Brix domain-containing proteins. (A) A phylogenic tree of Brix domain-containing proteins from Arabidopsis thaliana, Oryza sativa, Homo sapiens, and Saccharomyces cerevisiae. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown adjacent to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. (B) Schematic representation of the domain structures of six Arabidopsis Brix domain-containing proteins. Black and gray boxes indicate the Brix domains and coiled-coil domains, respectively. Protein architectures were deduced with SMART (Simple Modular Architecture Research Tool) (Letunic et al., 2015, http://smart.embl-heidelberg.de/). The scale indicates the number of amino acid residues.
Of the six Brix domain-containing proteins in Arabidopsis, ARPF2 is the closest to the yeast Rpf2 in terms of amino acid sequences, with 35% identities and 52% similarities with 6% gaps in their amino acid sequence (Supplementary Figure S2). The co-expression analysis of ARPF2 using the ATTEDII program (Obayashi et al., 2007) revealed that its expression pattern was similar to those of various ribosome-related genes including the other Brix domain-containing protein genes (Supplementary Figure S3), implying that Arabidopsis ARPF2 and yeast Rpf2 likely play a conserved role in ribosome biogenesis.
arpf2 Mutations Result in Aborted Embryos
To investigate the physiological role of ARPF2, we examined the phenotypes of two Arabidopsis T-DNA insertion lines in which the T-DNA insertions were located within the 1st intron (SALK_107828) and the 5’-UTR (SAIL_314_A03) of ARPF2. These mutant alleles were designated, arpf2-1 and arpf2-2 (Figures 2A,B). Although heterozygotes for arpf2-1 and arpf2-2 grew normally during the period for vegetative growth (Figure 2C), they developed aborted seeds (Figure 2D), suggesting that arpf2-1 and arpf2-2 are defective in gametogenesis and/or embryogenesis. Furthermore, we could not obtain homozygotes for the arpf2-1 or arpf2-2 allele. We, therefore, investigated the segregation ratio of the heterozygotes for arpf2-1 and arpf2-2 in the progeny after self-pollination. In both the cases, the transmission efficiency of the arpf2 mutation in the progeny population was much less than 100% (Table 1, χ2-test p < 0.001). Moreover, reciprocal crossing revealed that the transmission efficiency of the arpf2 mutation was also much less than 100% (χ2-test p < 0.001) in a progeny population produced by crossing both the wild type with an arpf2-1 heterozygote (Table 1). These results suggest that ARPF2 is an essential gene involved in both female and male gametogenesis as well as in embryogenesis.
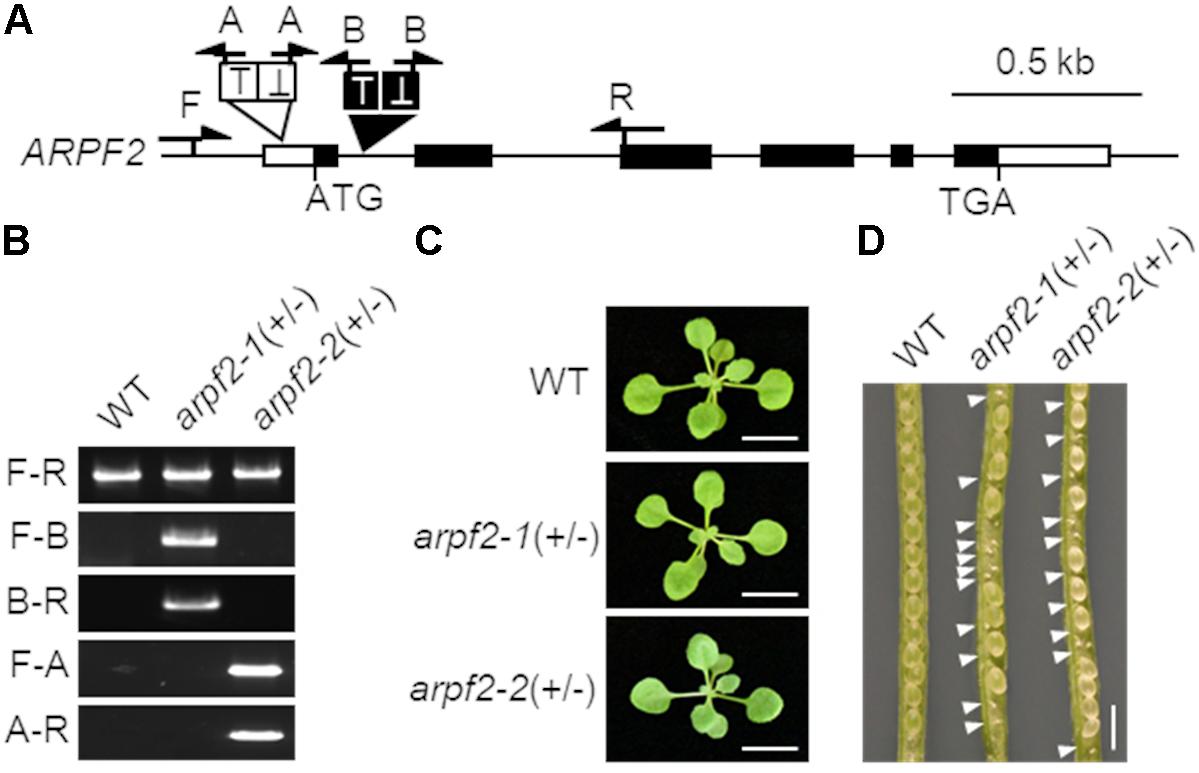
FIGURE 2. arpf2 mutations lead to embryonic lethality in Arabidopsis. (A) Schematic representation of the ARPF2 locus. White and black boxes indicate untranslated and coding regions in the exons of ARPF2, respectively. Black and white triangles indicate the positions of tandem T-DNA insertions in the arpf2-1 and arpf2-2 mutant alleles, respectively. Arrows indicate the positions of primers used for genotyping in (B). (B) PCR-based genotyping. Wild type (WT) and heterozygotes for the arpf2-1 or arpf2-2 allele [arpf2-1(+/–) and arpf2-2(+/–)] were analyzed using primers (F, R, A, and B) indicated in (A). Sets of primers F and R (F-R), primers F and A (F-A), primers A and R (A-R), primers F and B (F-B), and primers B and R (B-R) were applied. (C) Photographs of 2-week old plants grown on agar plates. Scale bars: 1 cm. (D) Photograph of seeds in fruits. White arrowheads indicate the aborted embryos. Scale bar: 1 mm.

TABLE 1. Segregation of rpf2-1 (+/-) and rpf2-2 (+/-) self-progeny and reciprocal crosses between rpf2-1 (+/-) and WT (+/+).
ARPF2 Is Expressed in a Variety of Tissues
To further characterize the ARPF2 gene, we examined the expression pattern of ARPF2 using the GUS reporter gene. An approximately 1.5 kb region upstream of the ARPF2 coding region was cloned and fused to the GUS reporter gene to make a construct, pARPF2:GUS, which was subsequently introduced into the wild type Arabidopsis (Col-0). Using T3 progenies of three independent transgenic lines harboring the pARPF2:GUS reporter construct, we checked the GUS expression in various tissues. In these three independent lines, the GUS expression patterns were similar. A typical expression pattern is shown in Figure 3. The ARPF2 promoter-driven GUS reporter gene was highly expressed in the shoot apexes of 2-day-old seedlings (Figure 3A) and almost ubiquitously expressed in the seedlings grown for 10 days, especially in the vascular bundles in shoots and roots, and in the primary and lateral root primordia (Figures 3B–G). In the inflorescence, GUS activity was detected in the pollen (Figures 3H,I). These results suggest that the ARPF2 promoter is probably active in a wide variety of tissues, particularly proliferating tissues, and therefore, the function of ARPF2 appears to be fundamental to most of the cells.
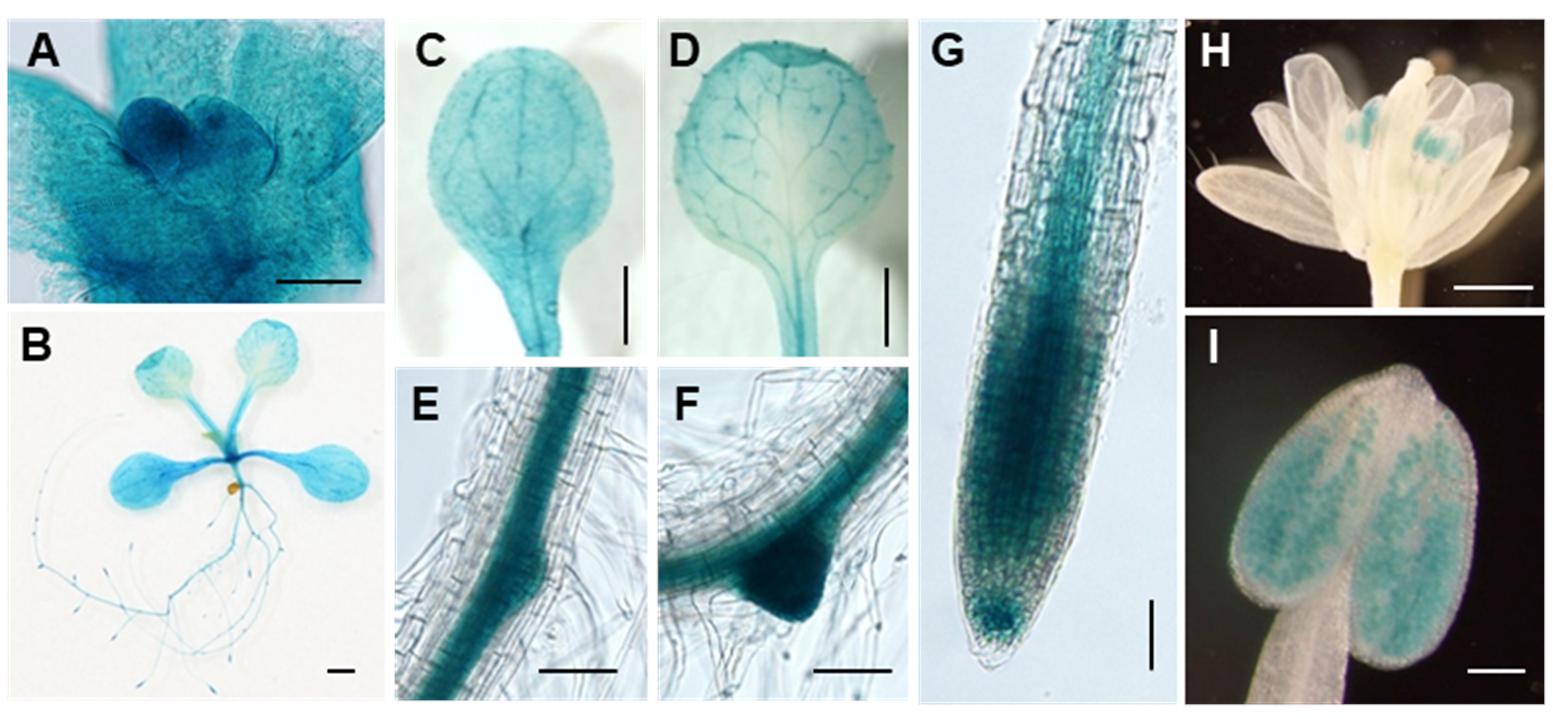
FIGURE 3. Tissue-specificity of ARPF2 expression. Histochemical GUS staining of transgenic plants in which GUS expression was driven with the ARPF2 promoter. Representatives of shoot apex of 2-day-old seedlings (A), whole seedling (B), cotyledon (C), true leaf (D), lateral root initiation (E,F), and primary root tip (G) of 10-day-old seedlings, flower (H), and pollen (I). Three independent lines were analyzed with similar results. Scare bars: 50 μm in (A), 1 mm in (B–D,H), 100 μm in (E–G,I).
APRF2 Interacts With ARRS1 in the Nucleolus
The cellular localization of ARPF2 was investigated by transiently expressing the ARPF2-GFP fusion protein in N. benthamiana leaves. Two days after the infection of Agrobacterium harboring the expression vector for ARPF2-GFP, the GFP fluorescence from ARPF2-GFP was detected only in the nucleolus. As shown in Figure 4A, the GFP fluorescence was perfectly merged with red fluorescence from mCherry fused to fibrillarin1 (FIB1), a nucleolus-localized marker protein (Barneche et al., 2000). The result indicates that like the yeast Rpf2 (Morita et al., 2002; Wehner and Baserga, 2002), ARPF2 is localized to the nucleolus.
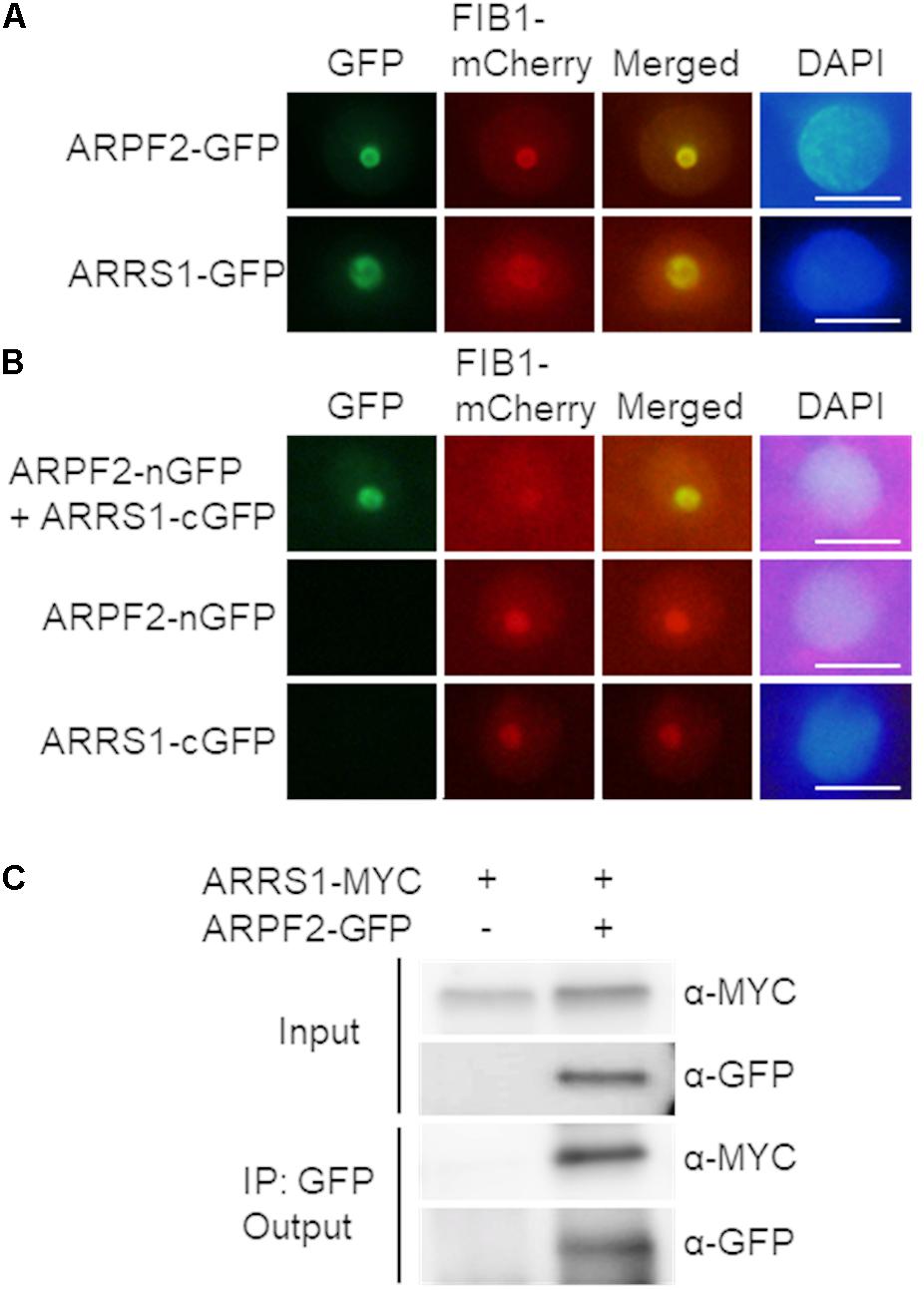
FIGURE 4. Interaction of ARPF2 with ARRS1 in the nucleolus. (A) Nucleolar localization of GFP fused to ARPF2 (ARPF2-GFP) and ARRS1 (ARRS1-GFP). Fluorescence from GFP and FIB1-mCherry (a nucleolar marker) was simultaneously monitored in Nicotiana benthamiana cells transiently expressing these proteins. Images for GFP were merged with those for fluorescence from FIB1-mCherry (Merged). DAPI staining indicates the nucleoplasm. Scale bars: 10 μm. (B) BiFC analyses for ARPF2 and ARRS1. ARPF2 fused to nGFP (ARPF2-nGFP) was co-expressed with cGFP-fused ARRS1 (ARRS1-cGFP) in N. benthamiana cells. The expression of cGFP and nGFP fused to the respective proteins alone served as negative controls. Scale bars: 10 μm. (C) Co-immunoprecipitation of ARPF2 and ARRS1. ARRS1-MYC was transiently expressed with or without ARPF2-GFP in N. benthamiana cells. The ARPF2-GFP protein in the crude cell extract (Input) was immunoprecipitated with an anti-GFP antibody, and the obtained precipitates (Output) were analyzed with anti-GFP and anti-MYC antibodies.
In yeast, Rpf2 directly binds to Rrs1, a protein essential for the maturation of 25S rRNA and the 60S ribosomal subunit assembly (Morita et al., 2002), with high affinity. Although yeast Rpf2 can bind to 5S rRNA directly, Rpf2 more rigidly binds to 5S rRNA through forming the complex with Rrs1 (Madru et al., 2015). Furthermore, the Rpf2–Rrs1 complex is shown to be an essential component for docking 5S RNP into the pre-60S ribosome (Gambe et al., 2009; Madru et al., 2015). To investigate the Rpf2–Rrs1 interaction in plant cells, we identified the Arabidopsis protein homologous to the yeast Rrs1. In the BLAST search with the amino acid sequence for yeast Rrs1, only one Arabidopsis protein that had not been characterized yet (At2g37990.1) was found to display a high homology to yeast Rrs1 (34% identity, 54% similarity) (Supplementary Figure S4). Therefore, this protein is presumably the homolog of yeast Rrs1. This protein is referred to as A. thaliana homolog of yeast Rrs1 (ARRS1), hereafter. Rrs1 is localized to the nucleolus in yeast (Gambe et al., 2009). The nucleolar localization of ARRS1 was consistently confirmed by transient expression of the ARRS1-GFP fusion protein in N. benthamiana leaves (Figure 4A). To test the interaction between ARPF2 and ARRS1 in vivo, we firstly performed BiFC analysis with nGFP and cGFP (Walter et al., 2004). We observed the GFP fluorescence in the nucleolus after transient co-expression of the ARPF2-nGFP fusion protein with ARRS1-cGFP in N. benthamiana leaves (Figure 4B). No GFP fluorescence was detected in cells expressing ARPF2-nGFP or ARRS1-cGFP alone (Figure 4B). Similarly, no GFP fluorescence was detected in a control experiment using RPL4A, a component of the ribosome large subunit (Barakat et al., 2001), further supporting a specific interaction between ARPF2 and ARRS1 (Supplementary Figure S5A). Furthermore, we also detected the ARPF2–ARRS1 interaction by co-immunoprecipitation analysis with lysates of cells transiently expressing both the GFP-tagged ARPF2 (ARPF2-GFP) and MYC-tagged ARRS1 (ARRS1-MYC) (Figure 4C). The immunoblot analysis of the immunoprecipitates obtained using anti-GFP and anti-MYC antibodies demonstrated that these proteins were co-immunoprecipitated. Furthermore, specificity of the interaction between ARPF2 and ARRS1 was also supported by the result of a control co-immunoprecipitation using RPL4A as a negative control (Supplementary Figure S5B). These results strongly suggest a specific interaction between ARRS1 and ARPF2 in the nucleolus.
ARPF2 Interacts With rRNAs in planta
We investigated whether ARPF2 got bound to pre-rRNA, as yeast Rpf2 was shown to interact with 5S rRNA directly and with 25S rRNA directly or indirectly (Asano et al., 2015; Kharde et al., 2015; Madru et al., 2015). For this purpose, we performed RIP assay in planta. We prepared cell lysates of the wild type and ARPF2-GFP expressing plants and performed immunoprecipitation with anti-GFP antibodies after cross-linking the proteins to the RNA using formaldehyde and sonication to fragment the RNAs. Then, the levels of rRNA co-immunoprecipitated with ARPF2-GFP were quantified through RT-qPCR with several pairs of primers that allowed the amplification of different regions of pre-rRNAs or mature rRNA (Figure 5A). Among the various fragments originating from the rRNA, the fragments that contained ITS2 and 5S rRNA (primer sets 4, 5, 6, and 9) were significantly enriched, depending on the expression of ARPF2-GFP (Figure 5B). The result suggests in vivo interactions of ARPF2 with 5S rRNA and precursors of 25S rRNA that contained ITS2.
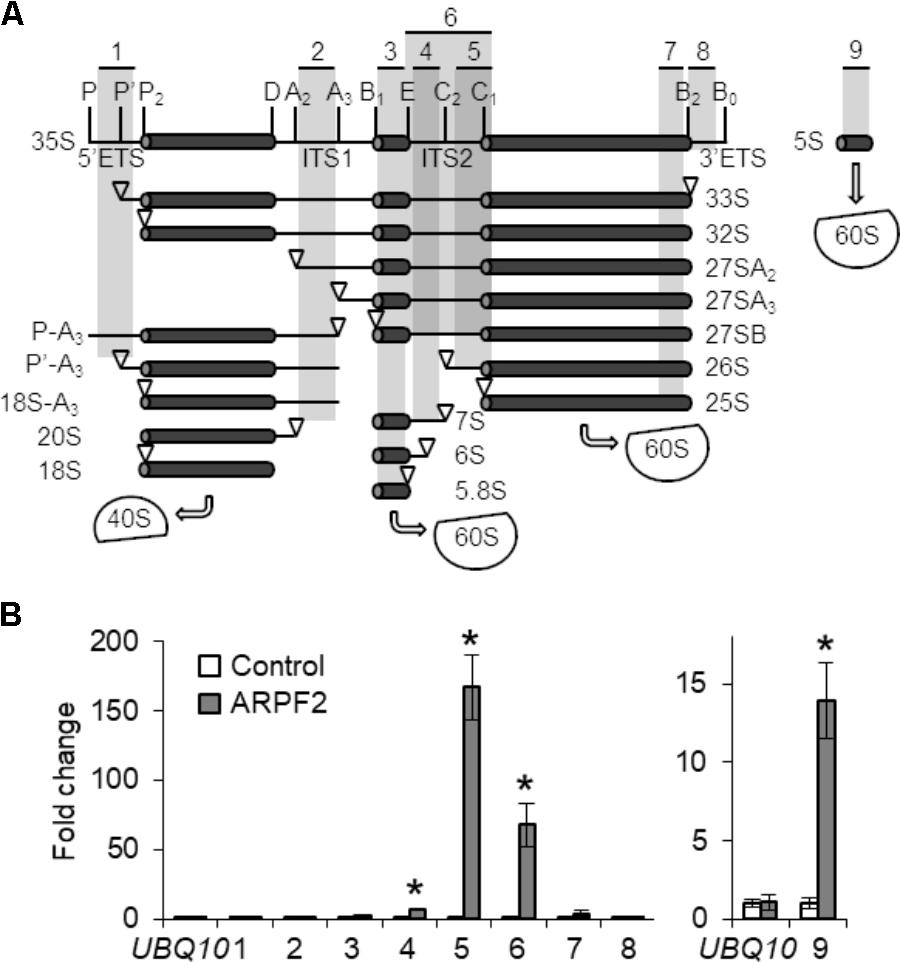
FIGURE 5. Binding of ARPF2 to pre-rRNA. (A) Scheme of the pre-rRNA processing pathway and processing intermediates in Arabidopsis. Vertical lines indicate the site of endo- or exonuclease processing steps. ETS and ITS are external and internal transcribed spacers, respectively. 40S and 60S are the 40S small subunit and the 60S large subunit, respectively. The positions of specific primers 1–9 used in (B) are indicated. (B) RT-qPCR analysis of RNA co-immunoprecipitated with wild type (Control) and ARPF2-GFP expressing transgenic (ARPF2) plants using anti-GFP antibodies. The enrichment of pre-rRNA or rRNA with specific regions [regions 1–9 in (A)] by co-immunoprecipitation was calculated with values obtained using the output sample versus those using the input sample. UBQ10 is shown as a non-binding control. Error bars represent SD of three biological replicates. Asterisks indicate statistically significant differences between values obtained with the wild type (Control) and ARPF2-GFP expressors (ARPF2) by Student’s t-test (p < 0.05).
ARPF2 and ARRS1 Overexpressors Show Shortened Stems, Abnormal Leaf Morphology, and Extension of Lifespan
To connect the function of ARPF2 with plant physiology, we generated transgenic Arabidopsis plants overexpressing ARPF2 or ARRS1. Phenotypic analysis was performed with two independent transgenic lines in which the overexpression of ARPF2 or ARRS1 was confirmed (Figure 6A). We did not find any phenotypic characteristics of these overexpressors in the early period for vegetative growth (Figure 6B), and the number of rosette leaves at bolting indicated that these overexpressors were comparable to the wild type in terms of the timing of flowering (Figure 6C). However, the 5-week-grown ARPF2 and ARRS1 overexpressors similarly displayed shorter stem than those of the wild type (Figures 6D,E), accompanied with the shortened lengths of internodes (Figures 6D,F). At this stage, the abnormality in the leaf shape became evident (Figures 6D,G). The height of the well-maturated overexpressors grown for 8-weeks were still shorter than those of the wild type plants (Figure 7A). In addition to this phenotypic characteristic, 8-week-old overexpressors were still in the growth phase, accompanied with living leaf cells containing a certain amount of chlorophyll, whereas the wild type plants were almost completely withered (Figures 7A–E). However, these overexpressors finally developed viable seeds. Because the overexpression of ARPF2 and ARRS1 had no effect on the timing of flowering (Figure 6C), it was unlikely that ARPF2 and ARRS1 overexpression caused a delay in growth and then extended the total length of life. Hence, these observations suggest that the overexpression of ARPF2 or ARRS1 greatly affected the growth in the reproduction phase and extended their lifespan.
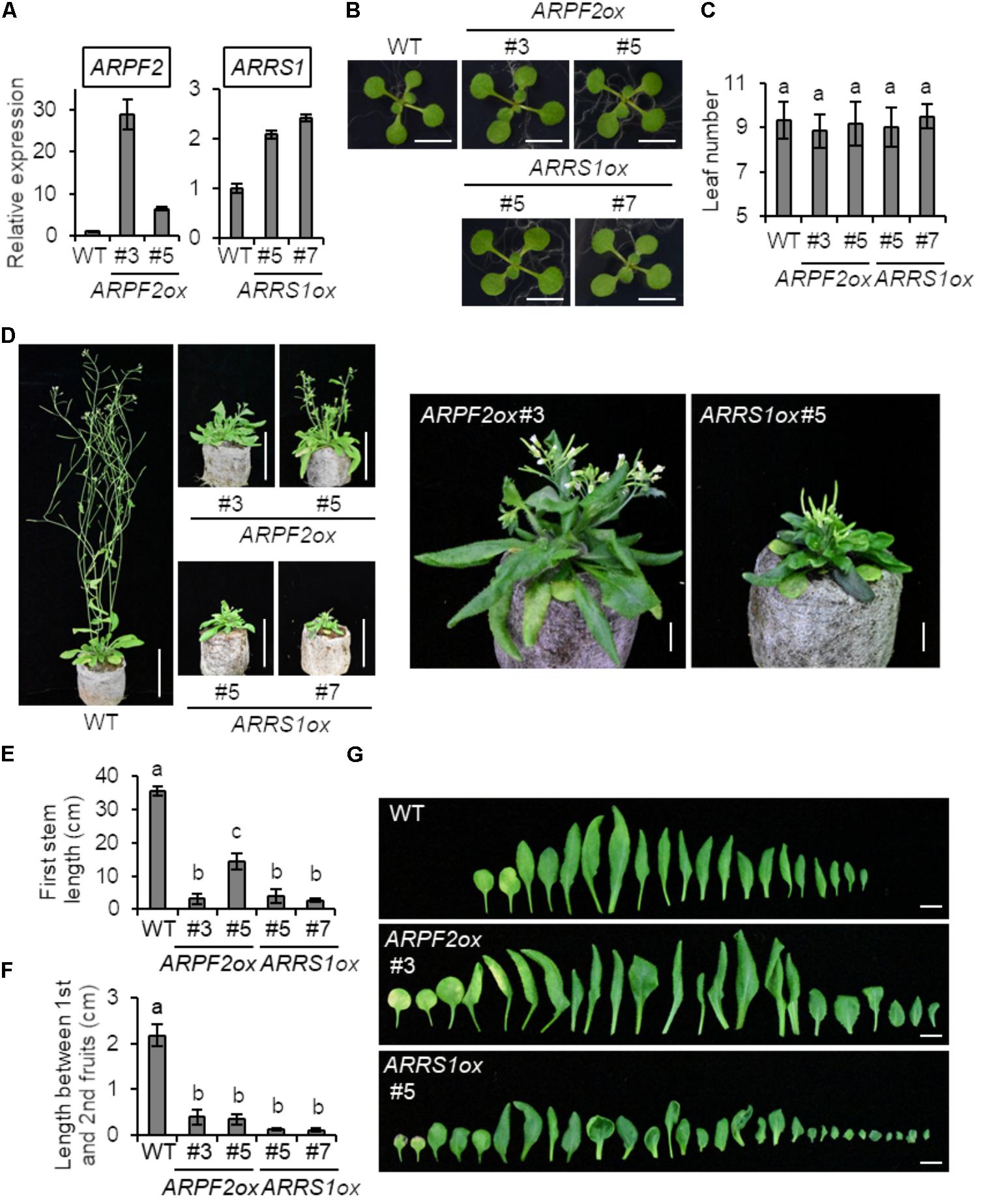
FIGURE 6. Phenotypic analysis of transgenic Arabidopsis overexpressing ARPF2 or ARRS1. (A) Levels of ARPF2 and ARRS1 transcripts in the wild type and ARPF2 or ARRS1 overexpressing plants. RNA was extracted from 10-day-old seedlings and technical triplicates were analyzed for quantification. UBQ10 was used as an internal control. The expression levels are relative to that in the wild type, and the values obtained with the wild type were set to 1. Error bars represent SD. (B) Photographs of 2-week-old seedlings grown on agar plates. Scale bars: 1 cm. (C) Numbers of rosette leaves at bolting. Error bars indicate SD (n = 6). (D) Photographs of the plants grown for 5-weeks. Scale bars: 5 cm. Enlarged view of ARPF2ox #3 and ARRS1ox #5 are also shown on the right side (scale bars: 1 cm). (E,F) Length of the first stem (E) and length between first and second fruits (F) of the plants grown for 5 weeks. Error bars indicate SD (n = 5). (G) Phenotype of rosette leaves of plants grown for 5 weeks. Scale bars: 1 cm. In (C,E,F) statistical significance was determined by ANOVA, followed by a Tukey–Kramer test under each experimental condition. Means that are significantly different from each other (p < 0.05) are indicated by different letters.
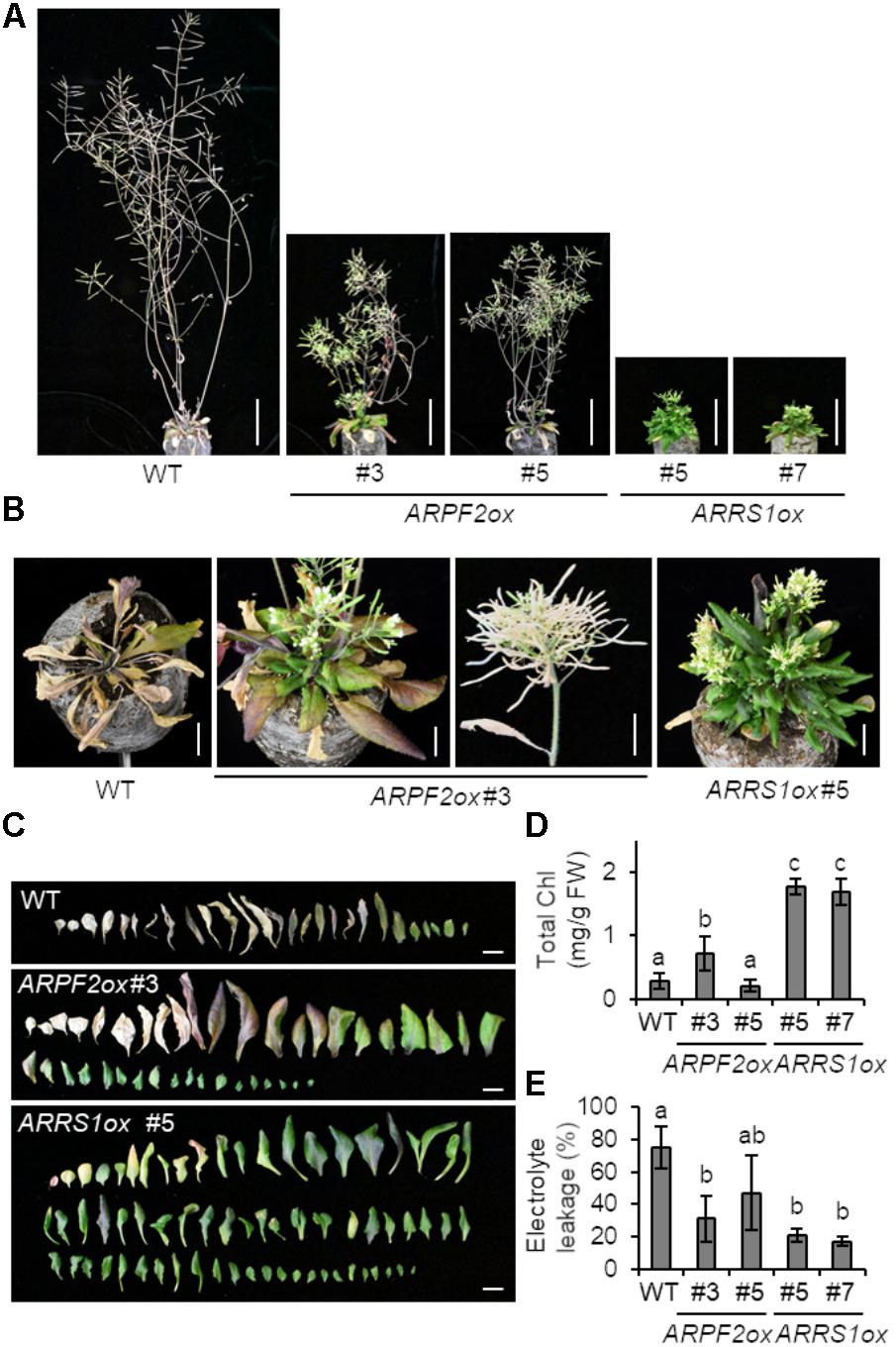
FIGURE 7. Extended lifespan of plants overexpressing ARPF2 or ARRS1. (A) Photographs of the plants grown for 8 weeks. Scale bars: 5 cm. (B) Enlarged view of (A) (scale bars: 1 cm). (C) Phenotype of rosette leaves of plants grown for 5-weeks. Scale bars: 1 cm. (D,E) Quantification of total chlorophyll (Chl) (D) and cell death (E) of the plants grown for 8-weeks. The extent of cell death was assessed as relative electrolyte leakage (percentage of total electrolytes). Error bars indicate SD (n = 5). In (D,E), the statistical significance was determined by ANOVA, followed by a Tukey–Kramer test. Means that are significantly different from each other (p < 0.05) are indicated by different letters.
To examine whether the overexpression of other ribosome biogenesis-related genes similarly induced the same phenotype, we produced transgenic Arabidopsis overexpressing APUM24 or PRH75 (Supplementary Figure S6). APUM24 encodes a pre-rRNA processing factor that plays an essential role in removing ITS2 through the interaction with ITS2 (Shanmugam et al., 2017; Maekawa et al., 2018), whereas PRH75/AtRH7 encodes a DEAD-box RNA helicase involved in rRNA processing (Nayak et al., 2013; Weis et al., 2015b; Huang et al., 2016). The growth of these overexpressors was comparable to that of the wild type (Supplementary Figure S6), indicating that the overexpression of ribosome biogenesis-related genes does not always induce phenotypes found in the ARPF2 and ARRS1 overexpressors, such as short stem, abnormal leaf morphology, and extended lifespan.
Discussion
Based on the findings in the current study, we conclude that ARPF2 is the Arabidopsis homolog of yeast Rpf2 and plays an essential role in ribosome biogenesis through its interactions with the Arabidopsis homolog of yeast Rrs1 (ARRS1), 5S rRNA, and 25S RNA precursors containing ITS2. We also suggest that the overexpression of ARPF2 or ARRS1 leads to unique phenotypic characteristics that are not produced by overexpressing other ribosome biogenesis-related genes.
The Brix domain-containing proteins can be divided into five subgroups that are probably responsible for distinct biological processes (Figure 1). Consistent with this idea, the double mutant of two Arabidopsis genes belonging to the Brx1 subgroup could not survive (Weis et al., 2015b). Furthermore, function-deficient mutants of a unique gene of the Ssf subgroup also showed lethality (Hao et al., 2017). Similarly, our finding of embryonic lethality in the arpf2 mutant (Figure 2 and Table 1), which was most likely caused by the ribosome biogenesis defects, also supports the idea that proteins in the distinct subfamily of the Brix domain protein family have essential but distinct roles.
By revealing the interactions of ARPF2 with ARRS1 in the nucleolus (Figure 4) and specific regions of rRNAs and/or their precursors in vivo (Figure 5), we also suggest that the function of ARPF2 is similar to those of yeast Rpf2 and its human homolog, BXDC1, in ribosome biogenesis. The function of the Rpf2–Rrs1 complex was proposed as a requirement of the 5S RNP (5S rRNA–Rpl5–Rpl11 complex) into the central protuberance of the pre-60S ribosome particles to correctly assemble the 5S RNP with the pre-60S ribosome particles in the nucleolus (Madru et al., 2015). In fact, depletion of Rpf2 led to a reduction in the rate of conversion of 27SB pre-rRNA to the mature 25S rRNA and in the impairment of nuclear export of pre-ribosomes (Wehner and Baserga, 2002; Zhang et al., 2007). Similarly, the complex of BXDC1 and hRRS1, human homolog of Rrs1, were proposed to mediate the assembly of the 5S RNP into the pre-ribosome particle, and depletion of BXDC1 caused processing defect of pre-rRNA in the large ribosomal subunit, mislocalization of Rpl5 and Rpl11 ribosomal proteins, and loss of nucleolar structure integrity (Donati et al., 2013; Sloan et al., 2013; Nicolas et al., 2016). Although we suggest that ARPF2 and ARRS1 constitute the 5S RNP in Arabidopsis, our results do not perfectly match with those of the published reports. The yeast Rpf2 has been shown to interact not only with 5S rRNA but also with 7S rRNA (Asano et al., 2015; Kharde et al., 2015; Madru et al., 2015). In another report, Rpf2 was also shown to bind to 35S, 27SA, and 27SB rRNAs in vivo (Wehner and Baserga, 2002; Zhang et al., 2007). The results of our RIP assay indicated the binding of ARPF2 to 5S rRNA and ITS2 that is present in the 25S RNA precursors (35S, 33S, 32S, 27SA, 27SAB, and 26S RNAs) but not in 25S RNA (Figure 5). Although our results do not rule out the binding of ARPF2 with 25S rRNA, the suggested regions recognized by ARPF2 and Rpf2 are subtly different. The exact identification of recognition sites of the Rpf2 subfamily proteins would be necessary to fully understand the Rpf2–Rrs1 complex-mediated process in ribosome biogenesis.
Very intriguingly, the overexpression of ARPF2 and ARRS1 did not induce any apparent phenotype in the early period for vegetative growth but caused drastic morphological changes, including short stem and abnormal leaf morphology, in the late period of vegetative growth and in the period for reproductive growth (Figures 6, 7). Furthermore, as the most interesting phenotypic characteristic, ARPF2- and ARRS1-overexpressors showed the extension of lifespan because of the extended period for reproductive growth (Figure 7). Overexpressing other ribosome-related factor genes (APUM24 and PRH75) did not give rise to such phenotypes (Supplementary Figure S6). Hence, these phenotypes appear to be due to the effects that are specifically linked to the overexpression of ARPF2 and ARRS1, although this possibility should be further examined in future. The molecular mechanism underlying these phenotypes is uncertain at this stage; however, we speculate that aberrant expression levels of ARPF2 and ARRS1 might generate incompleted forms of the ARPF2- and ARRS1-containing complexes that induce production of the unprocessed or miss-processed rRNA-containing ribonucleoprotein complexes and unexpected phenotypes. This highly speculative hypothesis should also be verified in future.
Although it is currently unknown whether the overexpression of the counterparts of ARPF2 or ARRS1 also induces a similar phenotype in animals or yeast, overexpressing the other rRNA processing factors, Dkc1 and Rrp5, has also been shown to extend the lifespan of mouse embryonic fibroblasts through an unknown mechanism (Nishimura et al., 2015). In the cells overexpressing Dkc1 or Rrp5, various cellular senescence-associated phenomena, including increases in the amount of nucleolar rRNA and the enlargement of nucleolus, occurred, accompanied with reduction in the amounts of p53, p21, and p16 that are key factors for cell senescence (Nishimura et al., 2015). Taken together with our findings, it is likely that the lifespan extension is selectively caused by the overexpression of particular rRNA processing factors in both animals and plants. Further analysis of the molecular mechanisms underlying this phenomenon would assist in fully understanding the ribosome biogenesis and might also provide a clue about the mechanism determining the lifespan of cells and living organisms.
Many ribosome-related genes have been characterized with loss-of-function mutants, thus far (reviewed in Horiguchi et al., 2012 and Weis et al., 2015a). However, it is worth pointing out that the findings in the current study highlight the potential usefulness of the plants overexpressing the ribosome-related genes for new valuable discoveries.
Author Contributions
SM and SY designed the research, analyzed the data and wrote the article. SM and YU performed the experiments.
Funding
This work was supported in part by JST, CREST (JPMJCR15O5) to SY, and JSPS KAKENHI to SY (Nos. 26221103, 18H03940), SM (No. 15J08368), and YU (No. 18J01554).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Junji Yamaguchi (Hokkaido University, Japan), Dr. Shoji Mano (National Institute of Basic Biology, Japan), and Dr. Tsuyoshi Nakagawa (Shimane University, Japan), the Arabidopsis Biological Resource Center and the Syngenta Arabidopsis Insertion Library for kindly providing the entry vector for FIB1 and the p35S:FIB1-mCherry vector, Gateway vectors for GFP/MYC-fused protein expression and BiFC assays, seeds of the SALK and SAIL T-DNA collection lines, respectively.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01177/full#supplementary-material
References
Alonso, J. M., Stepanova, A. N., Leisse, T. J., Kim, C. J., Chen, H., Shinn, P., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. doi: 10.1126/science.1086391
Asano, N., Kato, K., Nakamura, A., Komoda, K., Tanaka, I., and Yao, M. (2015). Structural and functional analysis of the Rpf2-Rrs1 complex in ribosome biogenesis. Nucleic Acids Res. 43, 4746–4757. doi: 10.1093/nar/gkv305
Barakat, A., Szick-Miranda, K., Chang, I. F., Guyot, R., Blanc, G., Cooke, R., et al. (2001). The organization of cytoplasmic ribosomal protein genes in the Arabidopsis genome. Plant Physiol. 127, 398–415. doi: 10.1104/pp.010265
Barneche, F., Steinmetz, F., and Echeverría, M. (2000). Fibrillarin genes encode both a conserved nucleolar protein and a novel small nucleolar RNA involved in ribosomal RNA methylation in Arabidopsis thaliana. J. Biol. Chem. 275, 27212–27220. doi: 10.1074/jbc.M002996200
Carroll, A. J., Heazlewood, J. L., Ito, J., and Millar, A. H. (2008). Analysis of the Arabidopsis cytosolic ribosome proteome provides detailed insights into its components and their post-translational modification. Mol. Cell. Proteomics 7, 347–69. doi: 10.1074/mcp.M700052-MCP200
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Curtis, M. D., and Grossniklaus, U. (2003). A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469. doi: 10.1104/pp.103.027979
D’Aoust, M. A., Lavoie, P. O., Belles-Isles, J., Bechtold, N., Martel, M., and Vézina, L. P. (2009). Transient expression of antibodies in plants using syringe agroinfiltration. Methods Mol. Biol. 483, 41–50. doi: 10.1007/978-1-59745-407-0_3
Donati, G., Peddigari, S., Mercer, C. A., and Thomas, G. (2013). 5S ribosomal RNA is an essential component of a nascent ribosomal precursor complex that regulates the Hdm2-p53 checkpoint. Cell Rep. 11, 87–98. doi: 10.1016/j.celrep.2013.05.045
Eisenhaber, F., Wechselberger, C., and Kreil, G. (2001). The Brix domain protein family – a key to the ribosomal biogenesis pathway? Trends Biochem. Sci. 26, 345–347. doi: 10.1016/S0968-0004(01)01851-5
Gambe, A. E., Matsunaga, S., Takata, H., Ono-Maniwa, R., Baba, A., Uchiyama, S., et al. (2009). A nucleolar protein RRS1 contributes to chromosome congression. FEBS Lett. 583, 1951–1956. doi: 10.1016/j.febslet.2009.05.033
Granneman, S., Gallagher, J. E., Vogelzangs, J., Horstman, W., van Venrooij, W. J., Baserga, S. J., et al. (2003). The human Imp3 and Imp4 proteins form a ternary complex with hMpp10, which only interacts with the U3 snoRNA in 60-80S ribonucleoprotein complexes. Nucleic Acids Res. 31, 1877–1887. doi: 10.1093/nar/gkg300
Hao, L., Wei, X., Zhu, J., Shi, J., Liu, J., Gu, H., et al. (2017). SNAIL1 is essential for female gametogenesis in Arabidopsis thaliana. J. Integr. Plant Biol. 59, 629–641. doi: 10.1111/jipb.12572
Henras, A. K., Plisson-Chastang, C., O’Donohue, M. F., Chakraborty, A., and Gleizes, P. E. (2015). An overview of pre-ribosomal RNA processing in eukaryotes. Wiley Interdiscip. Rev. RNA 6, 225–242. doi: 10.1002/wrna.1269
Horiguchi, G., Van Lijsebettens, M., Candela, H., Micol, J. L., and Tsukaya, H. (2012). Ribosomes and translation in plant developmental control. Plant Sci. 19, 24–34. doi: 10.1016/j.plantsci.2012.04.008
Huang, C. K., Shen, Y. L., Huang, L. F., Wu, S. J., Yeh, C. H., and Lu, C. A. (2016). The DEAD-box RNA helicase AtRH7/PRH75 participates in pre-rRNA processing, plant development and cold tolerance in Arabidopsis. Plant Cell Physiol. 57, 174–191. doi: 10.1093/pcp/pcv188
Jefferson, R. A., Kavanagh, T. A., and Bevan, M. W. (1987). GUS fusions: beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 20, 3901–3907.
Jonietz, C., Forner, J., Hölzle, A., Thuss, S., and Binder, S. (2010). RNA PROCESSING FACTOR2 is required for 5′ end processing of nad9 and cox3 mRNAs in mitochondria of Arabidopsis thaliana. Plant Cell 22, 443–453. doi: 10.1105/tpc.109.066944
Kapila, J., DeRycke, R., VanMontagu, M., and Angenon, G. (1997). An Agrobacterium-mediated transient gene expression system for intact leaves. Plant Sci. 122, 101–108. doi: 10.1016/S0168-9452(96)04541-4
Kaser, A., Bogengruber, E., Hallegger, M., Doppler, E., Lepperdinger, G., Jantsch, M., et al. (2001). Brix from Xenopus laevis and Brx1p from yeast define a new family of proteins involved in the biogenesis of large ribosomal subunits. Biol. Chem. 382, 1637–1647. doi: 10.1515/BC.2001.199
Kharde, S., Calviño, F. R., Gumiero, A., Wild, K., and Sinning, I. (2015). The structure of Rpf2-Rrs1 explains its role in ribosome biogenesis. Nucleic Acids Res. 43, 7083–7095. doi: 10.1093/nar/gkv640
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lee, S. J., and Baserga, S. J. (1999). Imp3p and Imp4p, two specific components of the U3 small nucleolar ribonucleoprotein that are essential for pre-18S rRNA processing. Mol. Cell. Biol. 19, 5441–5452. doi: 10.1128/MCB.19.8.5441
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, H. J., Liu, N. Y., Shi, D. Q., Liu, J., and Yang, W. C. (2010). YAO is a nucleolar WD40-repeat protein critical for embryogenesis and gametogenesis in Arabidopsis. BMC Plant Biol. 11:169. doi: 10.1186/1471-2229-10-169
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Madru, C., Lebaron, S., Blaud, M., Delbos, L., Pipoli, J., Pasmant, E., et al. (2015). Chaperoning 5S RNA assembly. Genes Dev. 29, 1432–1446. doi: 10.1101/gad.260349.115
Maekawa, S., Inada, N., Yasuda, S., Fukao, Y., Fujiwara, M., Sato, T., et al. (2014). The carbon/nitrogen regulator Arabidopsis toxicos En Levadura31 controls papilla formation in response to powdery mildew fungi penetration by interacting with SYNTAXIN OF PLANTS121 in Arabidopsis. Plant Physiol. 164, 879–887. doi: 10.1104/pp.113.230995
Maekawa, S., Ishida, T., and Yanagisawa, S. (2018). Reduced expression of APUM24, encoding a novel rRNA processing factor, induces sugar-dependent nucleolar stress and altered sugar responses in Arabidopsis thaliana. Plant Cell 30, 209–227. doi: 10.1105/tpc.17.00778
Matsumura, Y., Ohbayashi, I., Takahashi, H., Kojima, S., Ishibashi, N., Keta, S., et al. (2016). A genetic link between epigenetic repressor AS1-AS2 and a putative small subunit processome in leaf polarity establishment of Arabidopsis. Biol. Open 15, 942–954. doi: 10.1242/bio.019109
Morita, D., Miyoshi, K., Matsui, Y., Toh-E, A., Shinkawa, H., Miyakawa, T., et al. (2002). Rpf2p, an evolutionarily conserved protein, interacts with ribosomal protein L11 and is essential for the processing of 27 SB Pre-rRNA to 25 S rRNA and the 60 S ribosomal subunit assembly in Saccharomyces cerevisiae. J. Biol. Chem. 9, 28780–28786. doi: 10.1074/jbc.M203399200
Nayak, N. R., Putnam, A. A., Addepalli, B., Lowenson, J. D., Chen, T., Jankowsky, E., et al. (2013). An Arabidopsis ATP-dependent, dead-box rna helicase loses activity upon isoasp formation but is restored by protein isoaspartyl methyltransferase. Plant Cell 25, 2573–2586. doi: 10.1105/tpc.113.113456
Nicolas, E., Parisot, P., Pinto-Monteiro, C., de Walque, R., De Vleeschouwer, C., and Lafontaine, D. L. (2016). Involvement of human ribosomal proteins in nucleolar structure and p53-dependent nucleolar stress. Nat. Commun. 7:11390. doi: 10.1038/ncomms11390
Nishimura, K., Kumazawa, T., Kuroda, T., Katagiri, N., Tsuchiya, M., Goto, N., et al. (2015). Perturbation of ribosome biogenesis drives cells into senescence through 5S RNP-mediated p53 activation. Cell Rep. 10, 1310–1323. doi: 10.1016/j.celrep.2015.01.055
Obayashi, T., Kinoshita, K., Nakai, K., Shibaoka, M., Hayashi, S., Saeki, Y., et al. (2007). ATTED-II: a database of co-expressed genes and cis elements for identifying co-regulated gene groups in Arabidopsis. Nucleic Acids Res. 35, D863–D869. doi: 10.1093/nar/gkl783
Porra, R. J., Thompson, W. A., and Kriedemann, P. E. (1989). Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochem. Biophys. Acta 975, 384–394. doi: 10.1016/S0005-2728(89)80347-0
Saitou, N., and Nei, M. (1987). The neighbor-joining method: a new method for reconstructing phylogenyetic trees. Mol. Biol. Evol. 4, 406–425. doi: 10.1093/oxfordjournals.molbev.a040454
Sessions, A., Burke, E., Presting, G., Aux, G., McElver, J., Patton, D., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. doi: 10.1105/tpc.004630
Shanmugam, T., Abbasi, N., Kim, H. S., Kim, H. B., Park, N. I., Park, G. T., et al. (2017). An Arabidopsis divergent pumilio protein, APUM24, is essential for embryogenesis and required for faithful pre-rRNA processing. Plant J. 92, 1092–1105. doi: 10.1111/tpj.13745
Shinohara, N., Ohbayashi, I., and Sugiyama, M. (2014). Involvement of rRNA biosynthesis in the regulation of cuc1 gene expression and pre-meristematic cell mound formation during shoot regeneration. Front. Plant Sci. 28:159. doi: 10.3389/fpls.2014.00159
Sloan, K. E., Bohnsack, M. T., and Watkins, N. J. (2013). The 5S RNP couples p53 homeostasis to ribosome biogenesis and nucleolar stress. Cell Rep. 17, 237–247. doi: 10.1016/j.celrep.2013.08.049
Tanaka, Y., Kimura, T., Hikino, K., Goto, S., Nishimura, M., Mano, S., et al. (2012). “Gateway vectors for plant genetic engineering: overview of plant vectors, application for bimolecular fluorescence complementation (BiFC) and multigene construction,” in Genetic Engineering - Basics, New Applications and Responsibilities, ed. H. A. Barrera-Saldana (Croatia: InTech), 35–58. doi: 10.5772/32009
Walter, M., Chaban, C., Schütze, K., Batistic, O., Weckermann, K., Näke, C., et al. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40, 428–438. doi: 10.1111/j.1365-313X.2004.02219.x
Wehner, K. A., and Baserga, S. J. (2002). The sigma(70)-like motif: a eukaryotic RNA binding domain unique to a superfamily of proteins required for ribosome biogenesis. Mol. Cell 9, 329–339. doi: 10.1016/S1097-2765(02)00438-0
Weis, B. L., Kovacevic, J., Missbach, S., and Schleiff, E. (2015a). Plant-specific features of ribosome biogenesis. Trends Plant Sci. 20, 729–740. doi: 10.1016/j.tplants.2015.07.003
Weis, B. L., Palm, D., Missbach, S., Bohnsack, M. T., and Schleiff, E. (2015b). atBRX1-1 and atBRX1-2 are involved in an alternative rRNA processing pathway in Arabidopsis thaliana. RNA 21, 415–425. doi: 10.1261/rna.047563.114
Zhang, J., Harnpicharnchai, P., Jakovljevic, J., Tang, L., Guo, Y., Oeffinger, M., et al. (2007). Assembly factors Rpf2 and Rrs1 recruit 5S rRNA and ribosomal proteins rpL5 and rpL11 into nascent ribosomes. Genes Dev. 21, 2580–2592. doi: 10.1101/gad.1569307
Zhang, X. R., Qin, Z., Zhang, X., and Hu, Y. (2015). Arabidopsis SMALL ORGAN 4, a homolog of yeast NOP53, regulates cell proliferation rate during organ growth. J. Integr. Plant Biol. 57, 810–818. doi: 10.1111/jipb.12424
Keywords: Arabidopsis thaliana, Brix domain, lifespan, ribosome biogenesis, yeast Rpf2 homolog
Citation: Maekawa S, Ueda Y and Yanagisawa S (2018) Overexpression of a Brix Domain-Containing Ribosome Biogenesis Factor ARPF2 and its Interactor ARRS1 Causes Morphological Changes and Lifespan Extension in Arabidopsis thaliana. Front. Plant Sci. 9:1177. doi: 10.3389/fpls.2018.01177
Received: 28 April 2018; Accepted: 23 July 2018;
Published: 27 August 2018.
Edited by:
Diane C. Bassham, Iowa State University, United StatesReviewed by:
Hyun-Sook Pai, Yonsei University, South KoreaFrancisco Javier Medina, Centro de Investigaciones Biológicas (CIB), Spain
Copyright © 2018 Maekawa, Ueda and Yanagisawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuichi Yanagisawa, asyanagi@mail.ecc.u-tokyo.ac.jp
 Shugo Maekawa
Shugo Maekawa Yoshiaki Ueda
Yoshiaki Ueda Shuichi Yanagisawa
Shuichi Yanagisawa