- 1State Key Laboratory of Crop Biology, Shandong Agricultural University, Tai’an, China
- 2College of Forestry, Shandong Agricultural University, Tai’an, China
Environmental stresses are major constraints that limit the leaf productivity and quality of mulberry. LncRNAs have emerged as important regulators in response to biotic and abiotic stresses in plants. However, the functions and mechanisms of most lncRNAs remain largely unknown. A novel lncRNA designated as MuLnc1 was found to be cleaved by mul-miR3954 and produce secondary siRNAs in a 21 nt phase in mulberry. It was demonstrated that one of the siRNAs produced, si161579, can silence the expression of the calmodulin-like protein gene CML27 of mulberry (MuCML27). When MuCML27 was heterologously expressed in Arabidopsis, the transgenic plants exhibited enhanced resistance to Botrytis cinerea and Pseudomonas syringae pv tomato DC3000. In addition, the transgenic MuCML27-overexpressing Arabidopsis plants are more tolerant to salt and drought stresses. Furthermore, the network of mul-miR3954-MuLnc1-siRNAs-mRNAs was modeled to elucidate the interaction between lncRNAs and sRNAs with mRNAs. All of these, taken together, suggest that MuLnc1 was associated with environmental stress in mulberry and may be considered as a potential genetic improvement target gene of mulberry. The information provided may shed light on the complicated gene expression regulatory mechanisms in mulberry stress responses.
Introduction
Mulberry (Morus spp), an important food plant of the silkworms (Bombyx mori), is widely cultivated throughout the world in subtropical and temperate regions and is subjected to various environmental stresses during its entire life cycle (Ji et al., 2010). These stresses represent the leading constraints to leaf quality and productivity of mulberry, and feeding these stressed leaves affects larval development and cocoon characters of silkworm (Philip et al., 1994). Thus, it is fundamental to develop mulberry varieties with increased tolerance to environmental stresses to improve mulberry characteristics. Plant has developed complex signal transduction pathways to adapt to environmental stresses, and it is important to elucidate the expression networks of the genes involved in the signal transduction pathways to develop strategies to enhance the crop tolerance to environmental stresses (Xiong et al., 2002). However, the genetic control of tolerance to biotic and abiotic stresses is very complex, and the plant’s response to these stresses is accompanied by the activation of many genes associated with the perception and transduction of stress signals and by modulation of expression of defense-related genes (Atkinson and Urwin, 2012).
Previous research on molecular mechanisms underlying plant stress tolerance is largely focused on the functions of protein-coding genes (Wang J. et al., 2015). Recently, however, a large number of non-coding RNAs (ncRNAs) have been identified and found to play important roles in various biological processes including plant responses to environmental stresses (Ghildiyal and Zamore, 2009; Zhu and Wang, 2012; Joshi et al., 2016; Zhao et al., 2016). The ncRNAs comprise a class of RNAs such as miRNAs, siRNAs, ta-siRNAs and long non-coding RNAs (lncRNAs) (Axtell, 2013; Hirose et al., 2014; Bazin et al., 2017). Plant miRNAs are a class of small ncRNAs (20–24 nucleotides) which negatively regulate their targets via either mRNA degradation or translational repression (Pasquinelli, 2012). Besides guiding cleavage of target mRNA, it has been demonstrated that some miRNA-mediated cleavages of transcripts are capable of triggering the production of secondary siRNAs called trans-acting siRNAs (ta-siRNAs). ta-siRNAs can regulate the expression of their target genes to which they have limited sequence similarity in a way similar to miRNA (Yoshikawa et al., 2005; Manavella et al., 2012; Zhao et al., 2015). In contrast to miRNAs and siRNAs, lncRNAs are mRNA-like transcripts longer than 200 nucleotides that do not encode proteins (Quinn and Chang, 2016). In the last few years, lncRNAs have been considered as important regulators implicated in modulating transcription and as well as regulation of gene expression at post-transcriptional level, such as modulation of RNA stability and translation (Ponting et al., 2009; Wang and Chang, 2011; Heo et al., 2013; Wang T. et al., 2015; Chen et al., 2016). There were evidences showed that some lncRNAs serve as precursors of miRNAs and siRNAs which can perform their regulatory functions (Hirsch et al., 2006; Ma et al., 2014). Moreover, some lncRNAs harbor potential miRNA regulatory elements and are potentially targets of miRNAs or act as endogenous target mimics for miRNAs and participate in the miRNA regulatory network (Jalali et al., 2013; Wu et al., 2013; Wang J. et al., 2015). Therefore, ncRNAs can act as regulatory molecules involved in diverse biological processes and responses to biotic or abiotic stresses in plants (Mercer et al., 2009; Zhang et al., 2014; Wang T. et al., 2015; Chung et al., 2016; Joshi et al., 2016), and there are well-orchestrated regulatory interaction networks between various ncRNAs (Nacher and Araki, 2010; Collins, 2011; Jalali et al., 2013). However, in most cases, the interplay between the regulatory pathways of different ncRNAs remains largely unknown. As far as we know, there was only one report about the lncRNAs in mulberry so far (Song et al., 2016). Therefore, identification and characterization of stress-responsive ncRNAs and defining their regulatory networks will improve our understanding of the mechanisms underlying mulberry responses to environmental stresses.
In this study, based on the ncRNAs and transcriptome information attained, we report a novel lncRNA, MuLnc1, which was targeted and cleaved by mul-miR3954 and generated secondary siRNAs in a 21 nt phase in mulberry. One of the siRNAs produced, si161579, can direct the cleavage of the calmodulin-like protein gene CML27 (MuCML27) transcript of mulberry. Furthermore, we modeled a network of interactions between miRNAs, lncRNAs, siRNAs and protein-coding mRNAs. It may provide a basis to further study of the complicated gene expression regulatory mechanisms in mulberry stress responses and to explore the genes and ncRNAs which have a great potential application value in mulberry biotechnology in future.
Materials and Methods
Plant Materials
One-year-old mulberry cutting seedlings of Husang 32 (M. multicaulis) were grown in a growth chamber at 26°C with 12 h of light and 90% humidity. Arabidopsis thaliana (Col-0) plants were incubated under standard greenhouse conditions (22°C, humidity 90% and 12 h of light).
MiRNA Target Prediction and lncRNA Identification
The sixth leaves of five plants were sampled and mixed together, and the total RNA from the leaves sampled for transcriptome was isolated using TRIzol® reagent (Invitrogen, Carlsbad, CA, United States). The cDNA library was constructed and sequenced according the method described by Xie et al. (2013). The sequences obtained by high-throughput sequencing were chosen to predict the mul-miR3954 targets using psRNATarget1. The predicted miRNA targets were validated using the degradome sequencing data of the leaves of Husang 32 with the software package, CleaveLand 3.0, as previously described (Addo-Quaye et al., 2008). The Coding Potential Calculator (CPC2), CNCl and Pfam were used to predict transcripts with coding potential, and the transcripts with CPC score<-1 and CNCl score <0 were discarded. The remaining transcripts with length >200 nt and open reading frames (ORFs) <80 bp were considered as lncRNAs. The secondary siRNAs were identified based on the sequences from small RNA library sequencing data of the leaves of Husang 32 according to the method described before (Gai et al., 2014).
qRT-PCR Analysis
The TRIzol® reagent (Invitrogen, Carlsbad, CA, United States) was used to extract total RNAs of the leaf samples, and the RNAs extracted were digested with DNase I before being used. qRT-PCR was performed on the Rotor-Gene 3000A instrument (Corbett Research, Sydney, Australia) with the SYBR Premix Ex TaqTM kit (Takara, Shanghai, China) for mRNA and the PrimeScriptTM miRNA qPCR Starter Kit Ver.2.0 (Takara, Shanghai, China) for miRNAs according to the manufacturer’s protocol. The actin and U6 genes were chosen as reference genes for normalization of mRNA and miRNA, respectively. The comparative cycle threshold (Ct) method (Livak and Schmittgen, 2001) was used to revaluate the relative gene expression levels. All samples were assayed in triplicate. The primes used for qRT-PCR are given in the Supplementary Table S1.
RNA and Protein Gel Blotting
Total RNAs extracted were firstly separated on (1.2% w/v) formaldehyde denatured agarose gel and then were transferred onto nylon Hybond N membrane. The blots were hybridized with the probes labeled by digoxigenin (DIG) using the PCR DIG Probe Synthesis Kit (Roche, Germany). Prehybridization, hybridization with probe, washing, and detection steps were performed according to the protocol described by Liu et al. (2008). For the mul-miR3954, si165369, si47311, si161579 and si24218 northern blot analyses, sRNAs were extracted with miRCURYTM RNA Isolation Kit (Takara, Shanghai, China). Specific digoxigenin-labeled RNA probes complementary to the mul-miR3954 and each siRNA were synthesized by Roche Ltd (Roche, Shanghai, China). Prehybridization, hybridization, washing, and detection were performed as previously described method (Yang et al., 2008). The probes used are given in the Supplementary Table S2.
The prepared proteins were mixed with 5 × SDS–PAGE sample buffer and heated for 3 min at 95°C and then were subjected to SDS/PAGE on 12% (w/v) polyacrylamide gels. After electrophoresis, proteins were transferred electrophoretically onto nitrocellulose membranes for protein gel blot analysis. Polyclonal anti-MuCML27 antibodies were generated by immunizing rabbits with purified MuCML27 protein, and the protein was detected by western blots using horseradish peroxidase (HRP)-conjugated polyclonal anti-MuCML27. Western blot analysis was conducted according to the procedure described by Poppenberger et al. (2011).
Construction of si161579 Expression Vector
To study the function of si161579, the vector OE-si161579 over-expressing si161579 was constructed as described by Wang et al. (2010) with modifications. Firstly, a genomic fragment of the ath-miR165 backbone of 176 base pairs was amplified using primers miR165F and miR165R and cloned into pBI121. The mature ath-miR165-producing DNA region was substituted by si161579 using overlapping PCR with the combination of primers si161579 I, si161579 II, si161579 III, si161579 IV, miR165F and miR165R as described before (Schwab et al., 2006). The sequences of the primers used for the vector construction are listed in Supplementary Table S3.
Production of Transgenic Plant Lines
The mature mul-miR3954-producing DNA region was cloned from mulberry DNA, and the RNA isolated from leaves of mulberry was used to synthesize cDNA which was used as template to clone the MuCML27 and MuLnc1 genes, respectively. Primers (Supplementary Table S3) used to amplify the genes were designed based on the gene sequences (Supplementary Table S4). The genes cloned were then ligated individually into the binary vector pBI121 under the control of the CaMV 35S promoter. Then the vector was introduced into Agrobacterium tumefaciens strain GV3101 and transformed into WT Arabidopsis plants by floral dipping. Transformants were selected and confirmed by qRT-PCR and northern blot. The T3 generations from independent ectopic expression transgenic lines were randomly chosen for further functional studies.
Detection of Resistance Against Pathogens
The 4-week-old Arabidopsis plants were used for resistance analysis. Inoculation with Pst DC3000 was conducted by spraying the bacterial suspensions (105 CFU mL-1) of Pseudomonas syringae pv tomato DC3000 (Pst DC3000) onto the rosette leaves. Bacterial growth within the leaf was monitored by a serial-dilution assay (Katagiri et al., 2002). Detached rosette leaves were inoculated with 2-mm-diameter mycelium plugs taken from the actively growing colonies of Botrytis cinerea grown on PDA medium and placed in a covered Petri dish to retain moisture and incubated at 22°C. Each treatment was conducted independently at least three times.
Drought and Salt Stress
For drought treatments, Arabidopsis seedlings were grown in soil under regular watering regime for 4 weeks. Then the seedling plants were irrigated with 60 gL-1 PEG-6000 to simulate drought stress. For salt tolerance assay, 4-week-old Arabidopsis seedlings were irrigated with 1/8 concentrated MS salt solution supplemented with 250 mM NaCl. All the experiments were performed three times and three replicates for each treatment were used.
Malondialdehyde and Proline Measurements
Fresh leaf material was boiled in 3% sulfosalicylic acid for 10 min with constant shaking. After cooling and filtering, the filtrate was used for proline content assay following the method described by Shan et al. (2007). For malondialdehyde (MDA) concentration measurements, fresh leaf material was homogenized using 5% (w/v) trichloroacetic acid and the homogenate was centrifuged at 12,000 × g for 15 min. The supernatant was mixed with an equal volume of thiobarbituric acid (0.5% in 20% [w/v] trichloroacetic acid) and then the mixture was boiled for 25 min at 100°C, followed by centrifugation for 5 min at 7,500 × g. The supernatant was used for MDA concentration measurement according to the procedure as described previously (Peever and Higgins, 1989). These experiments were repeated at least three times.
Results
MuLnc1 Transcript as a Target of mul-miR3954 in Mulberry
Based on the transcriptome information attained, one lncRNA, MuLnc1, was identified. Interestingly, MuLnc1 was predicted to be a target of mul-miR3954 (Figure 1A), and this was further experimentally verified by mRNA degradome sequencing (Figure 1B). To explore whether MuLnc1 functioned as a target transcript of mul-miR3954, the correlation between the expression levels of MuLnc1 and mul-miR3954 in mulberry tissues were examined by qRT-PCR. The results showed that MuLnc1 was expressed at relatively high level in the ripe fruits, but its transcript level was very low in the roots, phloem and leaves (Figure 2A), whereas the expression level of mul-miR3954 was very low in the ripe fruits, but its transcript level was relatively high in the roots, phloem and leaves. This suggested that MuLnc1 and mul-miR3954 expressions were highly tissue-specific, and there was a very good negative correlation between their expression levels (Figure 2B), and this is in accord with the function of miRNA in guiding target mRNA cleavage.
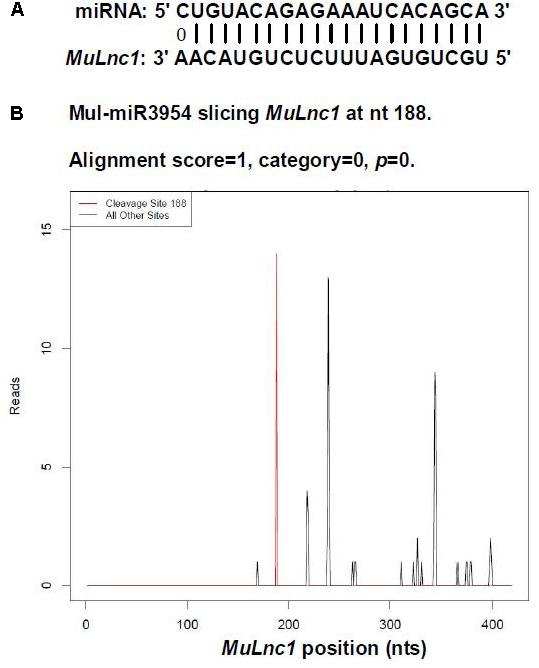
FIGURE 1. Identification of MuLnc1 as a target of mul-miR3954 in mulberry. (A) Base-pairing interaction between mul-miR3954 and MuLnc1. (B) Validation of the MuLnc1 degradation by degradome sequencing.
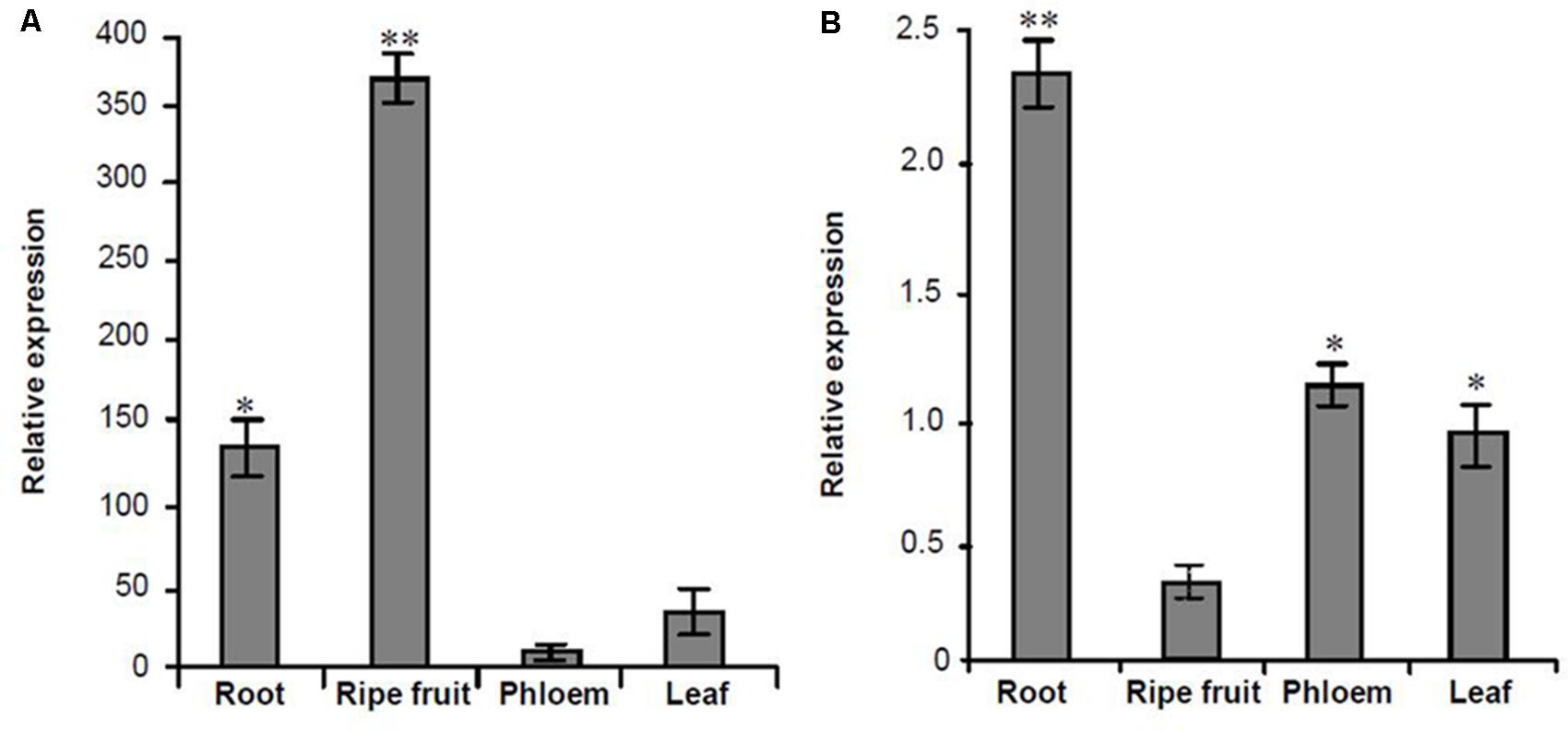
FIGURE 2. Quantitative analysis the expression levels of MuLnc1 and mul-miR3954 in different tissues of mulberry using qRT-PCR. (A) MuLnc1 abundance analysis by qRT-PCR. The relative gene expression was evaluated using comparative Ct method taking actin as the reference gene. (B) Mul-miR3954 abundance analysis by qRT-PCR. The relative abundance was evaluated using comparative Ct method taking U6 as the reference. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. Asterisks indicate significant difference based on Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
Secondary siRNAs Triggered by mul-miR3954-Mediated Cleavage of MuLnc1
Interestingly, some siRNAs in our small RNA library sequencing data were found to match to the MuLnc1 transcript downstream of mul-miR3954 binding site (Figure 3A), and northern blot results confirmed the existence of these siRNAs (Figure 3B). Thus, MuLnc1 transcript might be cleaved by mul-miR3954 and generate the secondary siRNAs. Since the efficient genetic transformation system has not been established in mulberry trees, and the MuLnc1 and mul-miR3954 do not exist in Arabidopsis, to further explore whether the MuLnc1 is targeted by mul-miR3954, the MuLnc1 and MUL-MIR3954 genes were cloned and introduced into Arabidopsis plants, respectively. Genome PCR analysis results showed that both the MuLnc1 and MUL-MIR3954 genes were integrated into the Arabidopsis genome (Figures 4A,B), and qRT-PCR and northern blot analysis results indicated that the MuLnc1 transcript and mature mul-miR3954 were successfully expressed in the transgenic Arabidopsis (Figures 4C–F). The hybrids between MuLnc1 and MUL-MIR3954 transgenic Arabidopsis were produced and used to analyze the cleavage of MuLnc1 transcript by mul-miR3954. qRT-PCR and northern blot assays showed that there was no difference in the expression levels of mul-miR3954 between the hybrid and MUL-MIR3954 transgenic plants (Figure 5A). However, the expression levels of MuLnc1 were significantly lower in the hybrid plants than those in the MuLnc1 transgenic plants (Figure 5B). This suggested that mul-miR3954 can target and cleave MuLnc1 transcript in the hybrid plants. Moreover, four siRNAs, si47311, si161579 and si24218, generated from MuLnc1 transcript were detected in the hybrid plants, but not detected in the MuLnc1 and MUL-MIR3954 transgenic plants (Figures 5C–E). Therefore, mul-miR3954 can cleave MuLnc1 transcript and trigger the production of secondary siRNAs from MuLnc1 transcript in vivo. Moreover, we found that these siRNAs generated from the downstream of the mul-miR3954 binding sites were in a 21-nt “phased” pattern which is a characteristic feature of ta-siRNA. Thus, these siRNAs were potential ta-siRNAs.
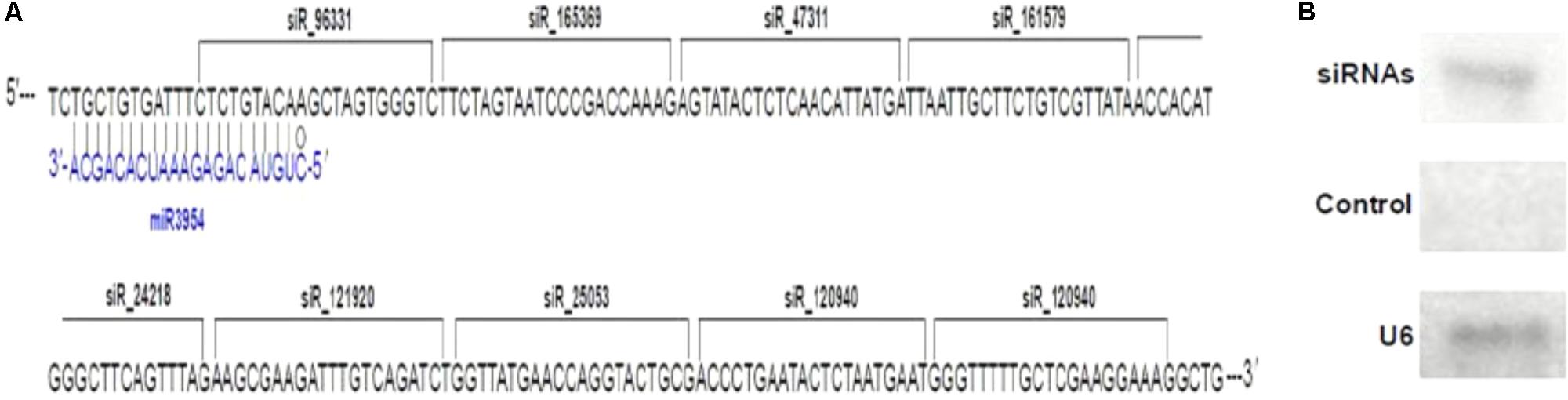
FIGURE 3. Production of secondary siRNAs from MuLnc1 transcript. (A) The siRNAs matching to the MuLnc1 transcript downstream of mul-miR3954 binding site. (B) Confirmation of the secondary siRNAs generated from MuLnc1 using northern blot analysis. sRNAs extracted were used for northern blot analysis. RNA oligonucleotides complementary to the specific sequence of MuLnc1 where the siRNAs were generated were used as probes, and RNA oligonucleotides complementary to the specific sequence of the 5′end of MuLnc1 where no siRNA generated were used as control probes.
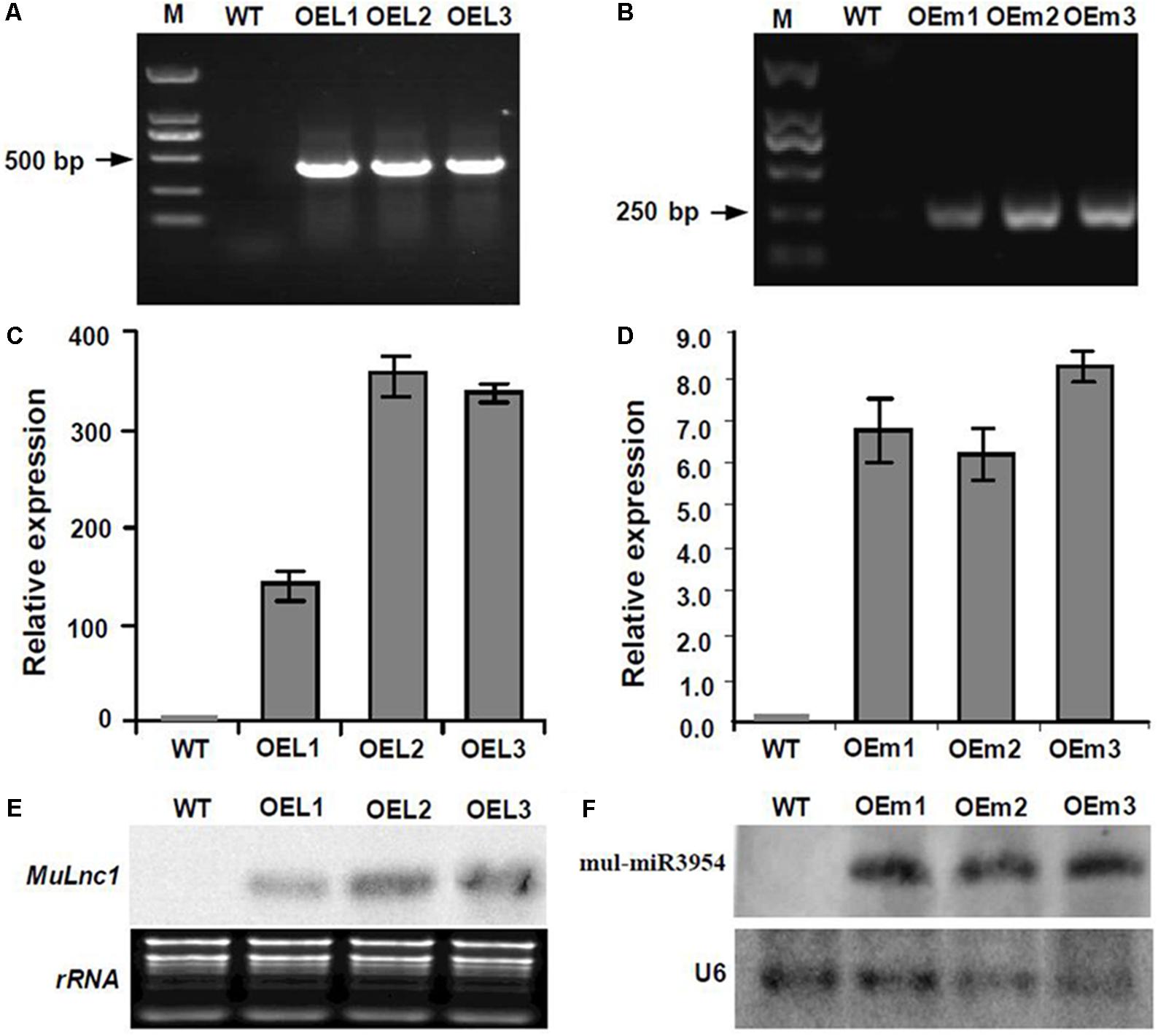
FIGURE 4. Analysis of the expression of Mulnc1 and mul-miR3954 in the MuLnc1 and MUL-MIR3954 transgenic plants. (A) PCR analysis showing that the MuLnc1 gene was integrated into the Arabidopsis genome (B) PCR analysis showing that the MUL-MIR3954 gene was integrated into the Arabidopsis genome. (C) Analysis of the expressions of MuLnc1 by qRT-PCR. The actin gene was amplified as a reference gene. (D) Analysis of the expressions of mul-miR3954 by qRT-PCR. The U6 gene was amplified as a reference gene. The relative gene expressions were evaluated using comparative Ct method. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. (E,F) Analysis of the expressions of MuLnc1 (E) and mul-miR3954 (F) by northern blot. WT: wild type Arabidopsis. OEL1-3: MuLnc1 transgenic lines. OEm1-3: MUL-MIR3954 transgenic lines.
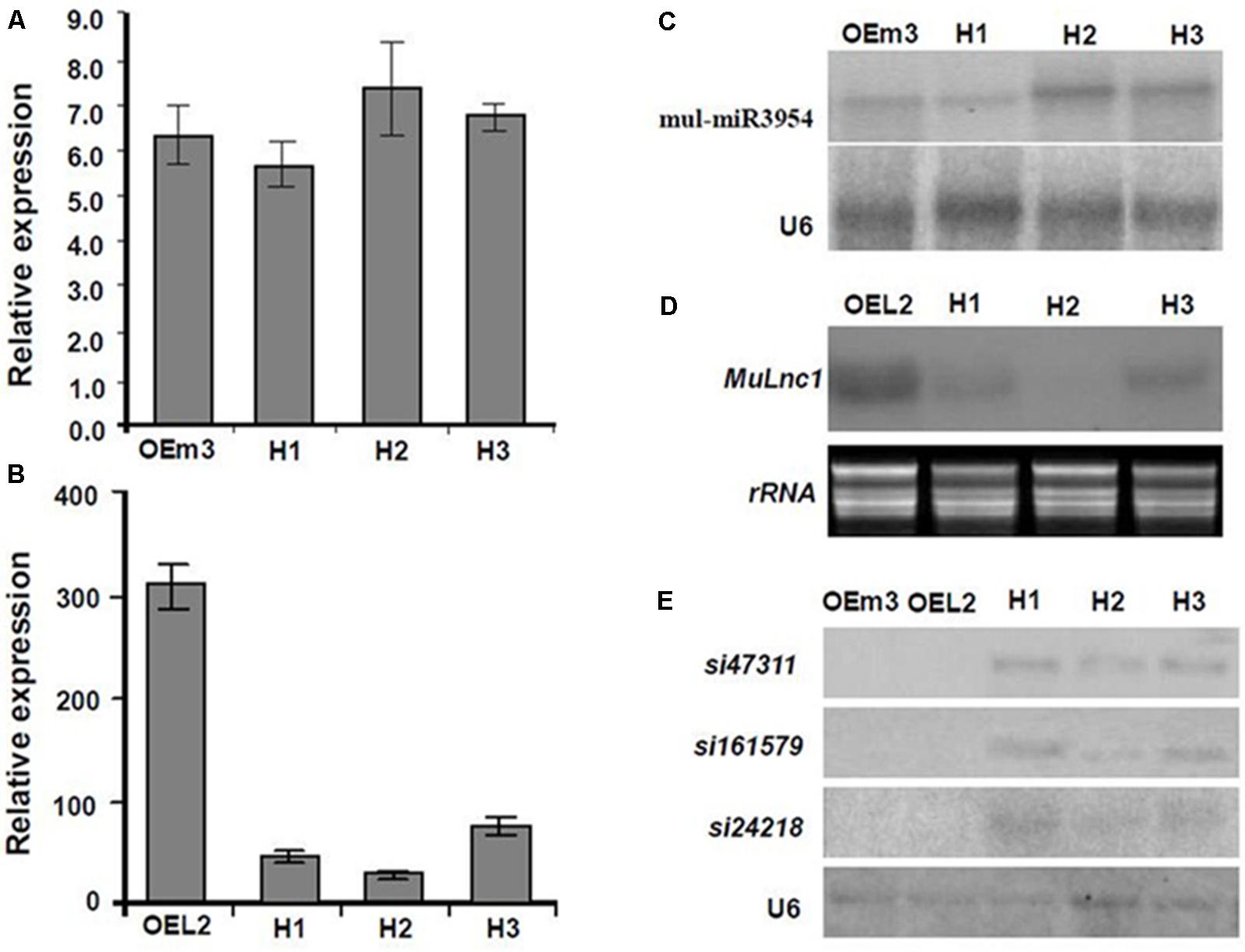
FIGURE 5. Analysis of the expression of Mulnc1, mul-miR3954, si47311, si161579, and si24218 in the MuLnc1 and MUL-MIR3954 transgenic plants and their hybrid plants. (A,B) Analysis of the expressions of mul-miR3954 (A) and MuLnc1 (B) by qRT-PCR. (C–E) Northern blot analysis of the expressions of mul-miR3954 (C), MuLnc1 (D), si47311, si161579, and si24218 (E). The relative gene expressions were evaluated using comparative Ct method, and the U6 and actin genes were amplified as reference genes for mul-miR3954 and MuLnc1 normalization, respectively. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. OEm3, the MUL-MIR3954 transgenic line. OEL2, the MuLnc1 transgenic line. H1-3, hybrid plants of OEL2 and OEm3 transgenic plants.
Silencing of MuCML27 Gene Guided by si161579
Analysis of the mRNA degradome data indicated that the siRNA, si161579, generated from MuLnc1 transcript can target the mulberry calmodulin-like protein 27 gene (MuCML27) (Supplementary Figure S1). To examine whether the si161579 is functional in vivo, the expression level of MuCML27 in different tissues of mulberry was determined. The results showed that the expression level of MuCML27 was relatively high in the ripe fruits, but it was low in the roots, phloem and leaves (Figure 6). This suggested that there was a negative correlation between MuCML27 and siRNA-triggering mul-miR3954, and the si161579 generated from MuLnc1 transcript is functional and able to down regulate its target. To further verify the cleavage of MuCML27 by si161579, the artificial vector OE-si161579 over-expressing si161579 was constructed first and then was transformed into Arabidopsis thaliana. Analyses of qRT-PCR and northern blot showed that the si161579 can be expressed in the transgenic Arabidopsis thaliana plants (OEs) successfully (Figures 7A,B). Moreover, the MuCML27 gene was also cloned and transformed into Arabidopsis thaliana. qRT-PCR, northern blot and western blot analyses showed that the transgenic plants (OEc) expressed the MuCML27 gene successfully (Figures 7C–E). Then the OEsiR-ARF vector constructed was transformed into the OEc plants, and the transgenic lines (OEsc) obtained were used to explore the si161579–mediated MuCML27 gene silencing. qRT-PCR and northern blot analyses showed that the si161579 was successfully expressed in the OEsc transgenic Arabidopsis plants (Figures 7F–G). Interestingly, the mRNA and protein abundances of MuCML27 in the OEsc plants were significant lower than those in the OEc plants (Figures 7H–J). These results indicated that the si161579 can silence and down-regulated the expression of MuCML27 gene in the OEsc plants, and the silencing may occur at the level of transcription.
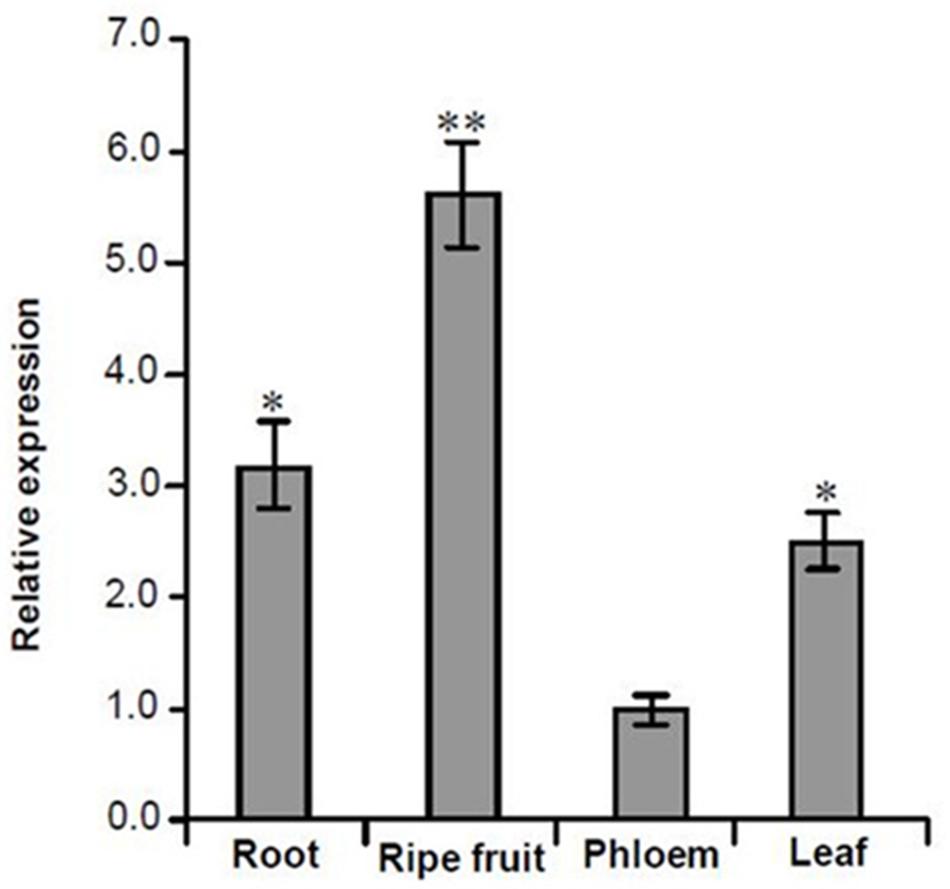
FIGURE 6. Quantitative analysis the expression levels of MuCML27 in different tissues of mulberry using qRT-PCR. The relative gene expression was evaluated using comparative Ct method taking actin as the reference gene. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. Asterisks indicate significant difference based on Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
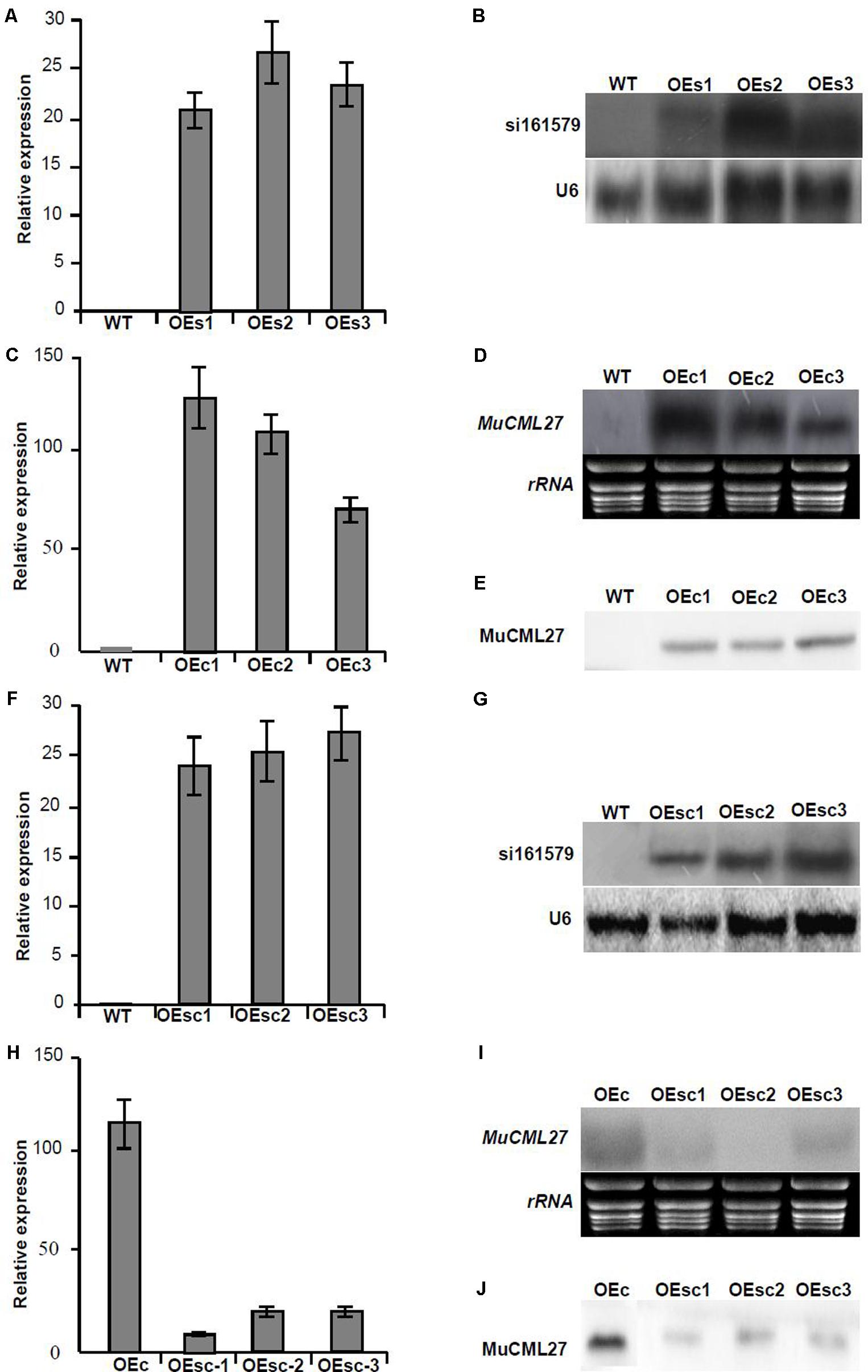
FIGURE 7. Analysis of the expression of si161579 and MuCML27. (A,B) Analysis of the expressions of si161579 in the OEs plants by qRT-PCR (A) and northern blot (B). (C–E) Analysis of the expressions of MuCML27 in the OEc plants by qRT-PCR (C), northern blot (D) and western blot (E). (F,G) Analysis of the expressions of si161579 in OEsc plants by qRT-PCR (F) and northern blot (G). (H–J) Analysis of the expressions of MuCML27 in the OEsc plants by qRT-PCR (H), northern blot (I) and western blot (J). The relative gene expressions were evaluated using comparative Ct method, and the U6 and actin genes were amplified as reference genes for si161579 and mRNA normalization, respectively. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. WT: wild type Arabidopsis. OEs1-3, si161579 transgenic lines. OEc1-3, MuCML27 transgenic lines. OEsc1-3, Co-expression MuCML27 and si161579 transgenic lines.
Silencing of MuCML27 Guided by si161579 Involved in Plant Response to Biotic Stress
Recent evidences suggest that several members of calmodulin-like protein (CMLs) play important roles in both abiotic and biotic responses including plant-microbe interactions (Perochon et al., 2011; Zeng et al., 2015). To find out whether the MuCML27 gene is involved in plant responses to pathogens, the OEc and OEsc plants were inoculated with B. cinerea and Pst DC3000, respectively. Rosette leaves were detached from 4-week-old Arabidopsis plants and inoculated with B. Cinerea, and development of disease symptoms was analyzed 4 days after inoculation (DAI). The results showed that expanding disease yellow lesions were yielded in the leaves of wild type and OEsc plants, and the symptoms in the leaves of wild type plants were the same as those in the OEsc plants. However, the disease spot size in the inoculated leaves of OEc plants was smaller than that in the inoculated leaves of wild type and OEsc plant (Figure 8A). When the plants were inoculated with Pst DC3000, there were severe disease symptoms showing gray-brown lesion with chlorosis in the wild type and OEsc Arabidopsis plants. However, the disease symptoms were not evident in the leaves of OEc Arabidopsis plants (Figures 8B,C), although mild chlorosis or necrosis was occasionally observed. To determine whether the depressive symptom development reflected the restricted growth of Pst DC3000 inside the leaves, the bacterial growth in the inoculated leaves was monitored. Arabidopsis leaf samples were taken at 36 h after inoculation, and the strain Pst DC3000 reaches the concentrations of 105 colony-forming units (CFU) cm-2 of leaf in the inoculated leaves of wild type Arabidopsis. Growth of the strain in the inoculated leaves of OEsc plants was essentially the same as that in the inoculated leaves of wild type Arabidopsis. Whereas growth of Pst DC3000 strains was extremely inhibited in the inoculated leaves of OEc plants (Figure 8D). Therefore, these results indicated that overexpressing MuCML27 can also enhance plant resistance both to Pst DC3000 and B. cinerea, and si161579 indeed silences the MuCML27 gene and inhibits the functions of MuCML27 in plant response to biotic stress.
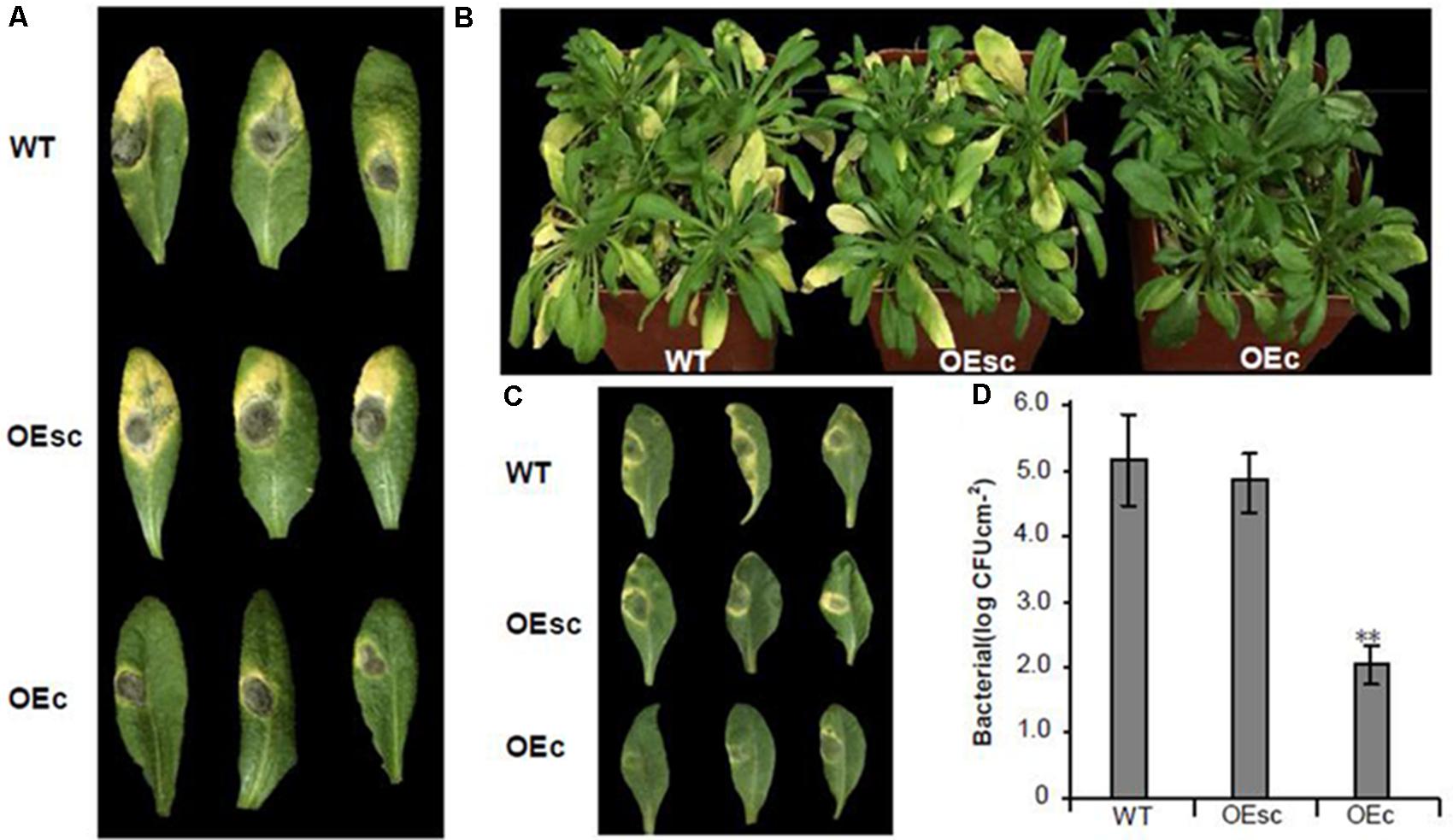
FIGURE 8. Resistance test in Arabidopsis against B. cinerea and Pst DC3000. (A) Symptoms observed 4 DAI on the leaves of 4-week-old Arabidopsis inoculated with 5-mm-diameter plugs of B. cinerea. (B) Disease symptoms in Arabidopsis plants sprayed with 106 CFU mL-1 suspension of Pst DC3000. (C) Detached leaves of 4-week-old Arabidopsis plants were infiltrated with 106 CFU mL-1 suspension of Pst DC 3000. Photographs were taken 3 DAI. (D) Bacterial populations in the inoculated leaves 36 h after inoculation with Pst DC3000. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. Double stars above the columns indicate significant differences according to Student’s t-test (P < 0.05). WT: wild type Arabidopsis. OEs, si161579 transgenic plants. OEc, MuCML27 transgenic plants. OEsc, Co-expression MuCML27 and si161579 transgenic plants.
Silencing of MuCML27 Guided by si161579 Involved in Plant Abiotic Stress Response
To explore whether silencing of MuCML27 mediated by si161579 is associated with plant tolerance to abiotic stress, we compared the drought and salt tolerance of the OEc, OEsc and wild-type Arabidopsis. When the plants were exposed to drought stress, the leaves of wild-type and OEsc plants became severe rolling and wilting at 12 days after withholding water. However, the leaves of OEc plants showed less rolling and wilting symptoms as compared with the wild-type and OEsc ones during the drought stress process (Figure 9). When the plants were exposed to salt stress, the leaves of wild-type and OEsc plants became wilted and curled after treatment with 250 mM NaCl for 2 weeks and the plants had a severe retardation in their growth. Whereas the growth inhibition level in the OEc plants was relatively less (Figure 9). In addition, the proline and malondialdehyde (MDA) contents in the leaves of wild type and OEc, OEsc plants under salt and drought stresses were also measured. The results showed that under salt and drought treatments, the proline level was higher but the MDA content was significantly lower in the OEc plants compared to that in the OEsc and wild type plants (Figure 10). These demonstrated that MuCML27 transgenic plants exhibit increased both drought and salt tolerance, and silencing of MuCML27 guided by si161579 inhibited the functions of MuCML27 in plant response to drought and salt stresses.

FIGURE 9. Silencing of MuCML27 guided by si161579 involved in plant response to abiotic stress. Drought stress was imposed on 4-week-old wild type, OEsc, and OEc Arabidopsis plants grown in soil and watered with 60 gL-1 PEG-6000. The photographs were taken 12 d after the drought treatments showing the differences in the reactions of the plants to the drought stress. Salt stress was imposed on 4-week-old wild type, OEsc, and OEc Arabidopsis plants by irrigating with 1/8 concentrated MS solution supplemented with 250 mM NaCl. Photographs of representative seedlings were taken 2 weeks after stress. WT: wild type Arabidopsis. OEs, si161579 transgenic plants. OEc, MuCML27 transgenic plants. OEsc, Co-expression MuCML27 and si161579 transgenic plants.
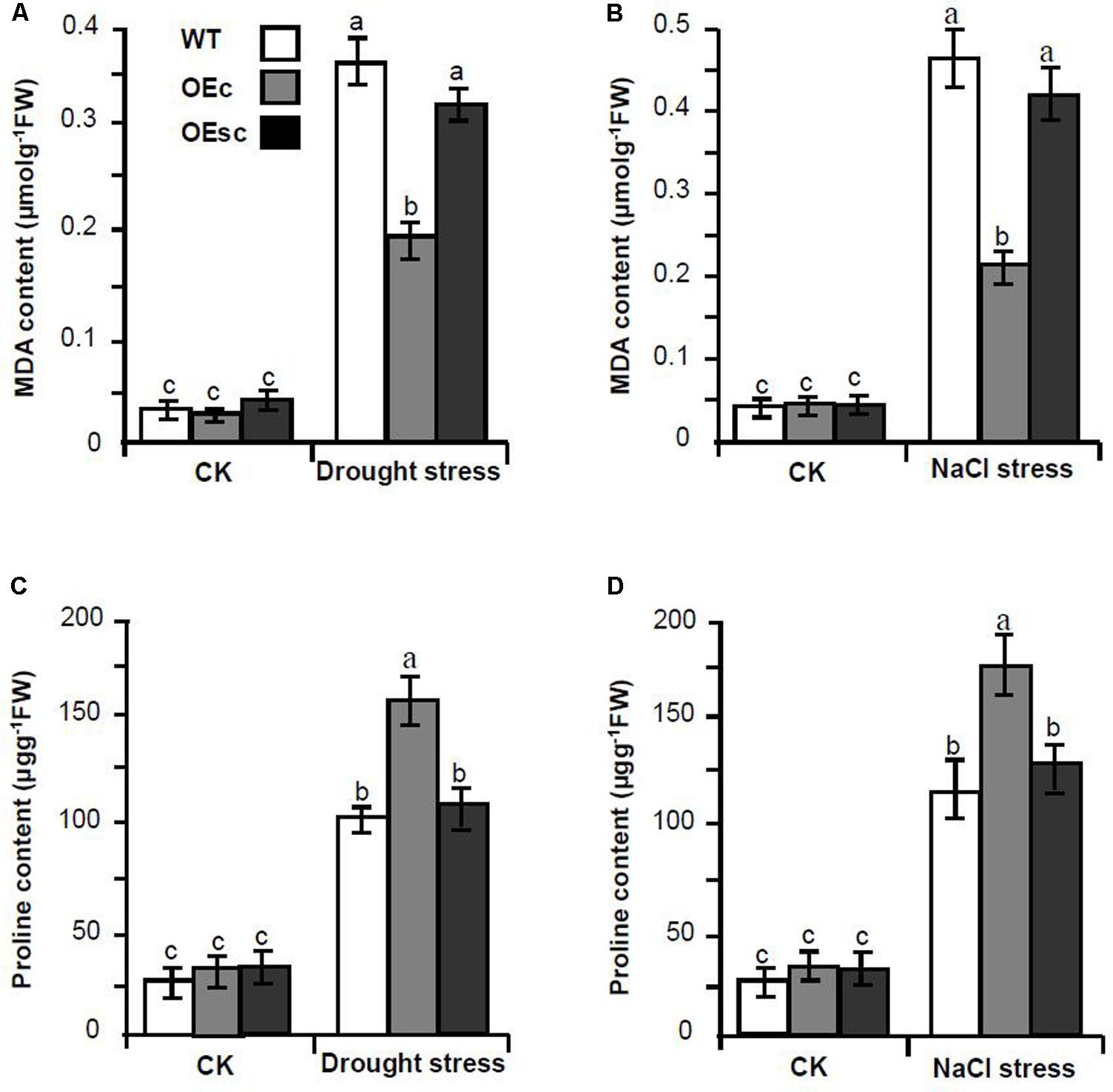
FIGURE 10. Comparison of MDA levels and proline amount between wild type and transgenic Arabidopsis plants under salt or drought stress treatments. (A,B) Comparison of MDA levels between WT, OEc and OEsc plants under drought (A) and salt (B) stress treatments. (C,D) Comparison of proline amount between OEc, OEsc, and WT plants under drought (C) and salt (D) stress treatments. Drought stress was imposed on 4-week-old wild type and transgenic Arabidopsis plants growing in vermiculite watered with 60 gL-1 PEG-6000. The figures were taken 12 d after the drought treatments. Salt stress was imposed on 4-week-old wild type and transgenic Arabidopsis plants by irrigating with 1/8 concentrated MS solution supplemented with 250 mM NaCl. The figures were taken two weeks after stress. CK, plant grown under normal conditions without salt or drought stress treatments. Assays were performed three times, each time with three replicates. Values are given as the mean ± SD of three experiments in each group. Different letters above the columns indicate significant differences according to Student’s t-test (P < 0.05). WT: wild type Arabidopsis. OEs, si161579 transgenic plants. OEc, MuCML27 transgenic plants. OEsc, Co-expression MuCML27 and si161579 transgenic plants.
Discussion
Although recent studies have discovered thousands of lncRNAs, only a small number of lncRNAs have been sufficiently described, and the functions and molecular mechanisms of most lncRNAs still remain elusive (Franco-Zorrilla et al., 2007; Swiezewski et al., 2009; Heo and Sung, 2011; Ding et al., 2012; Wang et al., 2014; Wang T. et al., 2015; Chen et al., 2016). In this study, a novel lncRNA, MuLnc1, was identified in mulberry. Blast searches of MuLnc1 against the Long Non-coding RNA Database v2.0 yielded none homolog, indicating it was lack of evolutionary conservation.
Plant lncRNAs may bind to specific miRNAs and function as ceRNAs (known as “target mimicry”) to protect the miRNA targets (Wang J. et al., 2015). In this study, MuLnc1 was predicted to be one of the targets of mul-miR3954, and the negative correlation expression pattern between MuLnc1 and mul-miR3954 and the degradome sequencing results supported the predicted results. It was reported that miR3954 was also involved in cotton responses to salt and drought stresses and was associated with drought response in tomato (Yang et al., 2013; Candar-Cakir et al., 2016). Therefore, MuLnc1-mul-miR3954 crosstalk might be an important regulator of gene expression in the response to environmental stress and many other biological processes in mulberry.
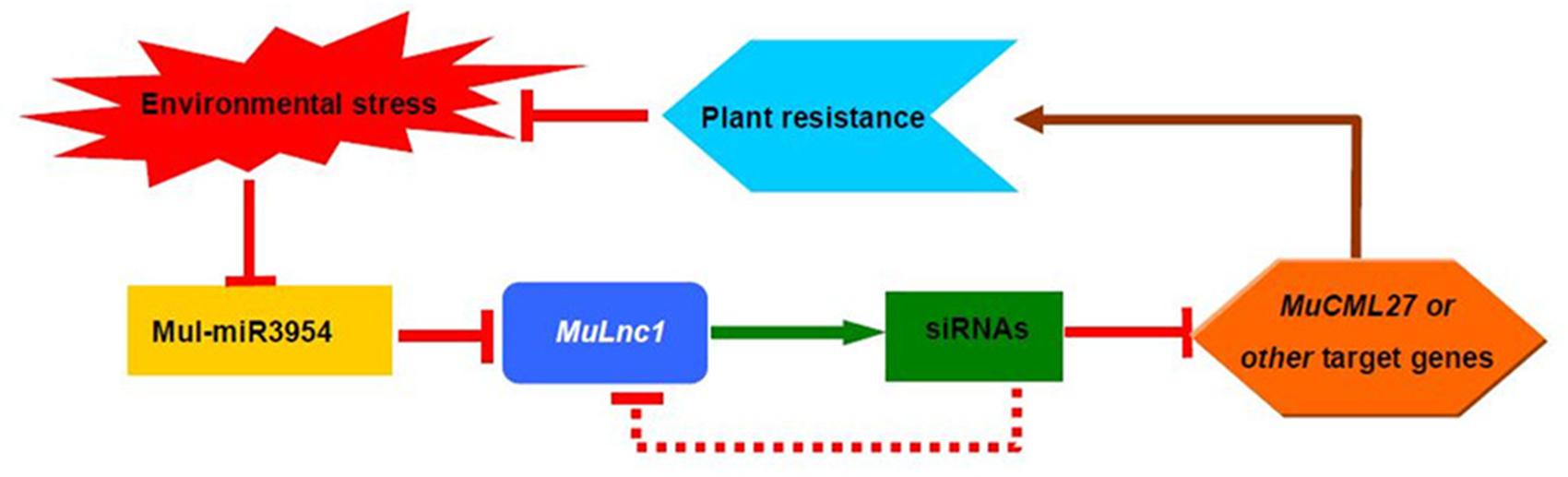
Previous studies showed that lncRNAs may function as cis- and trans-regulators of gene activity in many different ways (Hirsch et al., 2006; Liu et al., 2015), and some lncRNAs are processed to shorter ncRNAs such as miRNAs and siRNAs for functioning (Reinhart et al., 2002; Ben Amor et al., 2009; Jalali et al., 2012; Ma et al., 2014). Analysis of the sRNAs libraries of mulberry resulted in the identification of numerous 21-nt siRNAs that were mapped to both strands of the MuLnc1 downstream of mul-miR3954 binding site (Figure 3A), suggesting that MuLnc1 may function as siRNA precursors. It was reported that 22-nucleotide miRNAs are sufficient to trigger secondary siRNA biogenesis in plants (Chen et al., 2010; Cuperus et al., 2010). Further research confirmed that the asymmetrically positioned bulged bases in the miRNA : miRNA∗ duplex, regardless of miRNA or miRNA∗ length, determine the production of plant secondary siRNAs and are sufficient for the initiation of transitivity (Manavella et al., 2012; Liang et al., 2013). There were asymmetrically positioned bulged bases in the miRNA: miRNA∗ duplex of mul-miR3954 (Supplementary Figure S2). So mul-miR3954 may trigger the production of secondary siRNAs which were in a 21-nt “phased” pattern and were potential ta-siRNAs (Figure 3). Therefore, these potential ta-siRNAs may regulate the expression of MuLnc1 transcript in cis; moreover, they may function in trans to target other genes. Our data showed that one of the siRNAs, si161579, can target and silence the expression of the MuCML27 gene belonging to the CML family whose members are important Ca2+ sensors and play significant roles in mediating plant stress tolerance (White and Broadley, 2003; Chinpongpanich et al., 2011, 2012; Vadassery et al., 2012). Our results showed that the MuCML27 transgenic plants exhibited increased resistance to biotic and abiotic stresses. Besides the si161579, the siRNAs originated from the MuLnc1 were predicted to target many other protein-coding genes with diverse functions. Therefore, as the precursor of these siRNAs, the MuLnc1 gene may be involved in different physiological processes, and there was a complex regulatory network of mul-miR3954-MuLnc1-si161579-MuCML27 involved in the response to environmental stress in mulberry. Based on the information obtained, the complex regulatory network of mul-miR3954-MuLnc1-siRNAs-MuCML27 involved in mulberry response to environmental stress is outlined in Figure 11.
In addition, it was reported that lncRNAs are widely involved in regulation of gene expression including transcription, mRNA splicing and translation, and can affect the stability of RNA-protein complexes and regulate the activities of proteins (Zhu and Wang, 2012; Kornienko et al., 2013; Chekanova, 2015). Since the complete genome information of mulberry is not available, this hinders predicting the target genes and the interaction protein of MuLnc1. So more research is needed to understand the functions of MuLnc1, and MuLnc1 can be used as a target for genetic improvement of mulberry in the future.
Conclusion
The novel lncRNA, MuLnc1, was found to be cleaved by mul-miR3954 and produce secondary siRNAs in a 21-nt “phased” pattern in mulberry. One of the siRNAs produced, si161579, can silence the expression of the MuCML27 gene, and the transgenic Arabidopsis plants ectopically expressing MuCML27 gene showed more resistant to biotic and abiotic stresses. Therefore, there was a complex regulatory network of mul-miR3954-MuLnc1-si161579-MuCML27 involved in mulberry response to environmental stresses. All of these, taken together, suggest that MuLnc1 is associated with environmental stress in mulberry, and has potential applications in mulberry genetic improvement in the future.
Notes
Raw data of transcriptome, sRNA datasets and degradome can be obtained from SRA database with the accession number SRP139984, SRP139984, and SRP140041, respectively.
Author Contributions
Y-PG and X-LJ designed the research. S-SY generated and characterized Arabidopsis transgenics. Y-NZ performed quantitative RT-PCR. H-NZ and H-LZ designed and performed northern and western blot assays. X-LJ wrote the manuscript with input from all authors. All authors read and approved the final manuscript.
Funding
This work was funded by the National Natural Science Foundation of China (Nos. 30972366, 31070573, and 31100478), Natural Science Foundation of Shandong Province (Nos. ZR2015CM008 and ZR2015CM019), and Modern Agricultural Technology System of Shandong Province (No. SDAIT-18-04).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.00669/full#supplementary-material
FIGURE S1 | Identification of MuCML27 as a target of si161579 in mulberry by degradome sequencing.
FIGURE S2 | Predicted secondary structure of mul-miR3954. Red indicates miRNA guide strand and black arrows mark asymmetric bulges.
TABLE S1 | The sequences of northern blot probes.
TABLE S2 | The primers used for qRT-PCR.
TABLE S3 | The primers used for gene cloning and vector construction.
TABLE S4 | Sequences of Mul-MIR3954, MuLnc1, and MuCML27 genes.
Footnotes
References
Addo-Quaye, C., Eshoo, T. W., Bartel, D. P., and Axtell, M. J. (2008). Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 18, 758–762. doi: 10.1016/j.cub.2008.04.042
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Axtell, M. J. (2013). ShortStack: comprehensive annotation and quantification of small RNA genes. RNA 19, 740–751. doi: 10.1261/rna.035279.112
Bazin, J., Baerenfaller, K., Gosai, S. J., Gregory, B. D., Crespi, M., and Bailey-Serres, J. (2017). Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc. Natl. Acad. Sci. U.S.A. 114, E10018–E10027. doi: 10.1073/pnas.1708433114
Ben Amor, B., Wirth, S., Merchan, F., Laporte, P., d’Aubenton-Carafa, Y., Hirsch, J., et al. (2009). Novel long non-protein coding RNAs involved in Arabidopsis differentiation and stress responses. Genome Res. 19, 57–69. doi: 10.1101/gr.080275.108
Candar-Cakir, B., Arican, E., and Zhang, B. (2016). Small RNA and degradome deep sequencing reveals drought-and tissue-specific microRNAs and their important roles in drought-sensitive and drought-tolerant tomato genotypes. Plant Biotechnol. J. 14, 1727–1746. doi: 10.1111/pbi.12533
Chekanova, J. A. (2015). Long non-coding RNAs and their functions in plants. Curr. Opin. Plant Biol. 27, 207–216. doi: 10.1016/j.pbi.2015.08.003
Chen, H. M., Chen, L. T., Patel, K., Li, Y. H., Baulcombe, D. C., and Wu, S. H. (2010). 22-nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U.S.A. 107, 15269–15274. doi: 10.1073/pnas.1001738107
Chen, M., Wang, C., Bao, H., Chen, H., and Wang, Y. (2016). Genome-wide identification and characterization of novel lncRNAs in Populus under nitrogen deficiency. Mol. Genet. Genomics 291, 1663–1680. doi: 10.1007/s00438-016-1210-3
Chinpongpanich, A., Limruengroj, K., Phean-O-Pas, S., Limpaseni, T., and Buaboocha, T. (2012). Expression analysis of calmodulin and calmodulin-like genes from rice, Oryza sativa L. BMC Res. Notes 5:625. doi: 10.1186/1756-0500-5-625
Chinpongpanich, A., Wutipraditkul, N., Thairat, S., and Buaboocha, T. (2011). Biophysical characterization of calmodulin and calmodulin-like proteins from rice, Oryza sativa L. Acta Biochim. Biophys. Sin. 43, 867–876. doi: 10.1093/abbs/gmr081
Chung, P. J., Jung, H., Jeong, D. H., Ha, S. H., Choi, Y. D., and Kim, J. K. (2016). Transcriptome profiling of drought responsive noncoding RNAs and their target genes in rice. BMC Genomics 17:563. doi: 10.1186/s12864-016-2997-3
Collins, L. J. (2011). The RNA infrastructure: an introduction to ncRNA networks. Adv. Exp. Med. Biol. 722, 1–19. doi: 10.1007/978-1-4614-0332-6_1
Cuperus, J. T., Carbonell, A., Fahlgren, N., Garcia-Ruiz, H., Burke, R. T., Takeda, A., et al. (2010). Unique functionality of 22 nt miRNAs in triggering RDR6-dependent siRNA biogenesis from target transcripts in Arabidopsis. Nat. Struct. Mol. Biol. 17, 997–1003. doi: 10.1038/nsmb.1866
Ding, J., Lu, Q., Ouyang, Y., Mao, H., Zhang, P., Yao, J., et al. (2012). A long noncoding RNA regulates photoperiod-sensitive male sterility, an essential component of hybrid rice. Proc. Natl. Acad. Sci. U.S.A. 109, 2654–2659. doi: 10.1073/pnas.1121374109
Franco-Zorrilla, J. M., Valli, A., Todesco, M., Mateos, I., Puga, M. I., Rubio-Somoza, I., et al. (2007). Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet. 39, 1033–1037. doi: 10.1038/ng2079
Gai, Y. P., Li, Y. Q., Guo, F. Y., Yuan, C. Z., Mo, Y. Y., Zhang, H. L., et al. (2014). Analysis of Phytoplasma-responsive sRNAs provide insight into the pathogenic mechanisms of mulberry yellow dwarf disease. Sci. Rep. 4:5378. doi: 10.1038/srep05378
Ghildiyal, M., and Zamore, P. D. (2009). Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 10, 94–108. doi: 10.1038/nrg2504
Heo, J. B., Lee, Y. S., and Sung, S. (2013). Epigenetic regulation by long noncoding RNAs in plants. Chromosome Res. 21, 685–693. doi: 10.1007/s10577-013-9392-6
Heo, J. B., and Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hirose, T., Mishima, Y., and Tomari, Y. (2014). Elements and machinery of non-coding RNAs: toward their taxonomy. EMBO Rep. 15, 489–507. doi: 10.1002/embr.201338390
Hirsch, J., Lefort, V., Vankersschaver, M., Boualem, A., Lucas, A., Thermes, C., et al. (2006). Characterization of 43 non-protein-coding mRNA genes in Arabidopsis, including the MIR162a-derived transcripts. Plant Physiol. 140, 1192–1204. doi: 10.1104/pp.105.073817
Jalali, S., Bhartiya, D., Lalwani, M. K., Sivasubbu, S., and Scaria, V. (2013). Systematic transcriptome wide analysis of lncRNA-miRNA interactions. PLoS One 8:e53823. doi: 10.1371/journal.pone.0053823
Jalali, S., Jayaraj, G. G., and Scaria, V. (2012). Integrative transcriptome analysis suggest processing of a subset of long non-coding RNAs to small RNAs. Biol. Direct 7:25. doi: 10.1186/1745-6150-7-25
Ji, X., Lu, G., Gai, Y., Gao, H., Lu, B., Kong, L., et al. (2010). Colonization of Morus alba L. by the plant growth-promoting and antagonistic bacterium Burkholderia cepacia strain Lu10-1. BMC Microbiol. 10:243. doi: 10.1186/1471-2180-10-243
Joshi, R. K., Megha, S., Basu, U., Rahman, M. H., and Kav, N. N. V. (2016). Genome wide identification and functional prediction of long non-coding RNAs responsive to Sclerotinia sclerotiorum infection in Brassica napus. PLoS One 11:e0158784. doi: 10.1371/journal.pone.0158784
Katagiri, F., Thilmony, R., and He, S. Y. (2002). The Arabidopsis thaliana-Pseudomonas syringae interaction. Arabidopsis Book 1:e0039. doi: 10.1199/tab.0039
Kornienko, A. E., Guenzl, P. M., Barlow, D. P., and Pauler, F. M. (2013). Gene regulation by the act of long non-coding RNA transcription. BMC Biol. 11:59. doi: 10.1186/1741-7007-11-59
Liang, G., Li, Y., He, H., Wang, F., and Yu, D. (2013). Identification of miRNAs and miRNA-mediated regulatory pathways in Carica papaya. Planta 238, 739–752. doi: 10.1007/s00425-013-1929-6
Liu, J. H., Inoue, H., and Moriguchi, T. (2008). Salt stress-mediated changes in free polyamine titers and expression of genes responsible for polyamine biosynthesis of apple in vitro shoots. Environ. Exp. Bot. 62, 28–35. doi: 10.1016/j.envexpbot.2007.07.002
Liu, X., Hao, L., Li, D., Zhu, L., and Hu, S. (2015). Long non-coding RNAs and their biological roles in plants. Genomics Proteomics Bioinformatics 13, 137–147. doi: 10.1016/j.gpb.2015.02.003
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative RT-PCR and the 2-ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, X., Shao, C., Jin, Y., Wang, H., and Meng, Y. (2014). Long non-coding RNAs: a novel endogenous source for the generation of Dicer-like 1-dependent small RNAs in Arabidopsis thaliana. RNA Biol. 11, 373–390. doi: 10.4161/rna.28725
Manavella, P. A., Koenig, D., and Weigel, D. (2012). Plant secondary siRNA production determined by microRNA-duplex structure. Proc. Natl. Acad. Sci. U.S.A. 109, 2461–2466. doi: 10.1073/pnas.1200169109
Mercer, T. R., Dinger, M. E., and Mattick, J. S. (2009). Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 10, 155–159.
Nacher, J. C., and Araki, N. (2010). Structural characterization and modeling of ncRNA-protein interactions. Biosystems 101, 10–19. doi: 10.1016/j.biosystems.2010.02.005
Pasquinelli, A. E. (2012). MicroRNAs and their targets: recognition, regulation and an emerging reciprocal relationship. Nat. Rev. Genet. 13, 271–282. doi: 10.1038/nrg3162
Peever, T. L., and Higgins, V. J. (1989). Electrolyte leakage, lipoxygenase, and lipid peroxidation induced in tomato leaf tissue by specific and nonspecific elicitors from Cladosporium fulvum. Plant Physiol. 90, 867–875. doi: 10.1104/pp.90.3.867
Perochon, A., Aldon, D., Galaud, J. P., and Ranty, B. (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie 93, 2048–2053. doi: 10.1016/j.biochi.2011.07.012
Philip, T., Gupta, V. P., Govindaiah Bajpai, A. K., and Datta, R. K. (1994). Diseases of mulberry in India-research priorities and management strategies. Int. J. Trop. Plant Dis. 12, 1–21.
Ponting, C. P., Oliver, P. L., and Reik, W. (2009). Evolution and functions of long noncoding RNAs. Cell 136, 629–641. doi: 10.1016/j.cell.2009.02.006
Poppenberger, B., Rozhon, W., Khan, M., Husar, S., Adam, G., Luschnig, C., et al. (2011). CESTA, a positive regulator of brassinosteroid biosynthesis. EMBO J. 30, 1149–1161. doi: 10.1038/emboj.2011.35
Quinn, J. J., and Chang, H. Y. (2016). Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 17, 47–62. doi: 10.1038/nrg.2015.10
Reinhart, B. J., Weinstein, E. G., Rhoades, M. W., Bartel, B., and Bartel, D. P. (2002). MicroRNAs in plants. Genes Dev. 16, 1616–1626. doi: 10.1101/gad.1004402
Schwab, R., Ossowski, S., Riester, M., Warthmann, N., and Weigel, D. (2006). Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18, 1121–1133. doi: 10.1105/tpc.105.039834
Shan, D. P., Huang, J. G., Yang, Y. T., Guo, Y. H., Wu, C. A., Yang, G. D., et al. (2007). Cotton GhDREB1 increases plant tolerance to low temperature and is negatively regulated by gibberellic acid. New Phytol. 176, 70–81. doi: 10.1111/j.1469-8137.2007.02160.x
Song, X., Sun, L., Luo, H., Ma, Q., Zhao, Y., and Pei, D. (2016). Genome-wide identification and characterization of long non-coding RNAs from mulberry (Morus notabilis) RNA-seq Data. Genes 7:E11. doi: 10.3390/genes7030011
Swiezewski, S., Liu, F., Magusin, A., and Dean, C. (2009). Cold-induced silencing by long antisense transcripts of an Arabidopsis polycomb target. Nature 462, 799–802. doi: 10.1038/nature08618
Vadassery, J., Scholz, S. S., and Mithöfer, A. (2012). Multiple calmodulin-like proteins in Arabidopsis are induced by insect-derived (Spodoptera littoralis) oral secretion. Plant Signal. Behav. 7, 1277–1280. doi: 10.4161/psb.21664
Wang, J., Gao, X., Li, L., Shi, X., Zhang, J., and Shi, Z. (2010). Overexpression of Osta-siR2141 caused abnormal polarity establishment and retarded growth in rice. J. Exp. Bot. 61, 1885–1895. doi: 10.1093/jxb/erp378
Wang, J., Yu, W., Yang, Y., Li, X., Chen, T., Liu, T., et al. (2015). Genome-wide analysis of tomato long non-coding RNAs and identification as endogenous target mimic for microRNA in response to TYLCV infection. Sci. Rep. 5:16946. doi: 10.1038/srep16946
Wang, T., Liu, M., Zhao, M., Chen, R., and Zhang, W. (2015). Identification and characterization of long non-coding RNAs involved in osmotic and salt stress in Medicago truncatula using genome-wide high-throughput sequencing. BMC Plant Biol. 15:131. doi: 10.1186/s12870-015-0530-5
Wang, K. C., and Chang, H. Y. (2011). Molecular mechanisms of long noncoding RNAs. Mol. Cell 43, 904–914. doi: 10.1016/j.molcel.2011.08.018
Wang, Y., Fan, X., Lin, F., He, G., Terzaghi, W., Zhu, D., et al. (2014). Arabidopsis noncoding RNA mediates control of photomorphogenesis by red light. Proc. Natl. Acad. Sci. U.S.A. 111, 10359–10364. doi: 10.1073/pnas.1409457111
White, P. J., and Broadley, M. R. (2003). Calcium in plants. Ann. Bot. 92, 487–511. doi: 10.1093/aob/mcg164
Wu, H. J., Wang, Z. M., Wang, M., and Wang, X. J. (2013). Widespread long noncoding RNAs as endogenous target mimics for microRNAs in plants. Plant Physiol. 161, 1875–1884. doi: 10.1104/pp.113.215962
Xie, M., Huang, Y., Zhang, Y., Wang, X., Yang, H., Yu, O., et al. (2013). Transcriptome profiling of fruit development and maturation in Chinese white pear (Pyrus bretschneideri Rehd). BMC Genomics 14:823. doi: 10.1186/1471-2164-14-823
Xiong, L., Schumaker, K. S., and Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14(Suppl.), S165–S183. doi: 10.1105/tpc.000596
Yang, W. Q., Lai, Y., Li, M. N., Xu, W. Y., and Xue, Y. B. (2008). A novel C2-domain phospholipid-binding protein, OsPBP1, is required for pollen fertility in rice. Mol. Plant 1, 770–785. doi: 10.1093/mp/ssn035
Yang, X., Wang, L., Yuan, D., Lindsey, K., and Zhang, X. (2013). Small RNA and degradome sequencing reveal complex miRNA regulation during cotton somatic embryogenesis. J. Exp. Bot. 64, 1521–1536. doi: 10.1093/jxb/ert013
Yoshikawa, M., Peragine, A., Park, M. Y., and Poethig, R. S. (2005). A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 19, 2164–2175. doi: 10.1101/gad.1352605
Zeng, H., Xu, L., Singh, A., Wang, H., Du, L., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Zhang, W., Han, Z., Guo, Q., Liu, Y., Zheng, Y., Wu, F., et al. (2014). Identification of maize long non-coding RNAs responsive to drought stress. PLoS One 9:e98958. doi: 10.1371/journal.pone.0098958
Zhao, J., He, Q., Chen, G., Wang, L., and Jin, B. (2016). Regulation of non-coding RNAs in heat stress responses of plants. Front. Plant Sci. 7:1213. doi: 10.3389/fpls.2016.01213
Zhao, M., San León, D., Mesel, F., García, J. A., and Simón-Mateo, C. (2015). Assorted processing of synthetic trans-acting siRNAs and its activity in antiviral resistance. PLoS One 10:e0132281. doi: 10.1371/journal.pone.0132281
Keywords: mulberry (Morus spp), lncRNAs, miR3954, siRNA, calmodulin-like protein gene CML27, environmental stress
Citation: Gai Y-P, Yuan S-S, Zhao Y-N, Zhao H-N, Zhang H-L and Ji X-L (2018) A Novel LncRNA, MuLnc1, Associated With Environmental Stress in Mulberry (Morus multicaulis). Front. Plant Sci. 9:669. doi: 10.3389/fpls.2018.00669
Received: 03 January 2018; Accepted: 02 May 2018;
Published: 29 May 2018.
Edited by:
Dirk Albert Balmer, Syngenta, SwitzerlandReviewed by:
Vitantonio Pantaleo, Istituto per la Protezione Sostenibile delle Piante, ItalyKashmir Singh, Panjab University, India
Copyright © 2018 Gai, Yuan, Zhao, Zhao, Zhang and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xian-Ling Ji, xlji@sdau.edu.cn
†These authors have contributed equally to this work.
 Ying-Ping Gai
Ying-Ping Gai Shuo-Shuo Yuan2†
Shuo-Shuo Yuan2† Xian-Ling Ji
Xian-Ling Ji