- 1Zhejiang Academy of Agricultural Sciences, Hangzhou, China
- 2State Key Laboratory of Silkworm Genome Biology, Southwest University, Chongqing, China
- 3Department of Biological Sciences, Texas Tech University, Lubbock, TX, USA
Ricinus communis displays a broad range of phenotypic diversity in size, with dwarf, common, and large-sized varieties. To better understand the differences in plant productivity between a high-stalk variety and a dwarf variety under normal growth conditions, we carried out a comparative proteomic study between Zhebi 100 (a high stalk variety) and Zhebi 26 (a dwarf variety) combined with agronomic and physiological analyses. Over 1000 proteins were detected, 38 of which differed significantly between the two varieties and were identified by mass spectrometry. Compared with Zhebi 100, we found that photosynthesis, energy, and protein biosynthesis related proteins decreased in abundance in Zhebi 26. The lower yield of the dwarf castor is likely related to its lower photosynthetic rate, therefore we hypothesize that the lower yield of the dwarf castor, in comparing to high stalk castor, could be increased by increasing planting density. Consequently, we demonstrated that at the higher planting density in Zhebi 26 (36,000 seedlings/hm2) can achieve a higher yield than that of Zhebi 100 (12,000 seedlings/hm2). Proteomic and physiological studies showed that for developing dwarf R. communis cultivar that is suitable for large scale-production (i.e., mechanical harvesting), it is imperative to identify the optimum planting density that will contribute to higher leaf area index, higher photosynthesis, and eventually higher productivity.
Introduction
The castor bean (Ricinus communis) is a tropical perennial shrub and field crop originated in Africa, but has now been introduced worldwide and is widely cultivated (Chan et al., 2010). It can be cross- and self-pollinated, and studies have revealed low levels of genetic variation among castor bean germplasm worldwide (Allan et al., 2008; Foster et al., 2010). Because of the nearly uniform ricinoleic acid content of castor oil and its unique fatty acid properties, castor beans are a valuable oilseed crop for the cosmetics industry, specialty lubricants, and biomedical and specialty chemical applications. R. communis has also been proposed as a potential source of biodiesel feedstock because of its high seed oil content (Da Silva et al., 2006), and the ease with which it can be cultivated in unfavorable crop-growing environments has contributed to its appeal as a crop in tropical developing nations. Furthermore, the castor plant is commonly cultivated in many counties for its leaves to feed the Eri silkworm (Attacus Cynthia ricini Boisduval), which provides Eri silk, a high-quality natural protein fiber.
R. communis is produced in about 30 countries for commercial purposes, among which India, China and Brazil account for >90% of the world's production (Severino et al., 2012). Mechanized castor production is possible and needs to become mandatory to sustain or increase global castor production. Because the three main countries producing castor are experiencing rapid economic and social development, the labor required for traditional castor production has become expensive (Severino et al., 2012). Currently, only limited areas of castor production are fully mechanized because of the lack of dwarf-internode and commercial cultivars (Baldanzi et al., 2003). Therefore, the main challenge in developing new cultivars is the adaptation of castor plants to mechanical harvesting. The development of appropriate new castor cultivars should be enhanced by improved knowledge of the genetics and molecular biology of the species. Mutation breeding has shown that irradiation of castor seeds and seedlings produces mutants with desirable characteristics including semi-dwarfs with higher yield potential and earlier maturity (Sujatha et al., 2008).
R. communis has a complex genetic background, which causes difficulties in genomic mutation and gene cloning. Transcriptome and gene expression analyses via measurement of mRNA levels have contributed greatly to characterizing mutants of rice and Arabidopsis. In contrast, there are obstacles to studying gene expression in R. communis. Moreover, levels of mRNAs, the key players in the cell, measured in qualitative terms do not always correlate well with phenotypes because of post-transcriptional regulation mechanisms. Therefore, proteome studies aimed at the complete set of genome-encoded proteins may complement the shortcoming of transcriptome approaches. Here, we present a study to identify the differences between a dwarf castor variety and a high stalk variety by using an integrated approach of proteomics, physiological analysis, and field experiments.
Dwarfism was an attractive phenotype of the “Green Revolution” and is still a desirable agronomic trait for crop cultivators as it is associated with high yield potential, improved lodging resistance, and higher fertility (Li et al., 2010). Since 1960s, new varieties of grain crops with short stems and substantially improved yields were developed (Conway and Toenniessen, 1999). Castor is a tropical plant that can be as short as two feet in height, or as tall as a moderate-sized tree. The development of improved varieties using molecular technologies will help ensure farmers across the world enjoy the economic potential of this crop (Auld et al., 2010). In this study, our main objective was to identify candidate genes and proteins that can be used to create a dwarf castor cultivar with high yield for agricultural applications and mechanical harvesting.
The specific aim of our research was to identify the differential protein changed in abundance comparing Zhebi 100 (a high stalk variety, 12,000 seedlings/hm2) and Zhebi 26 (a dwarf variety, 36,000 seedlings/hm2) in the highest yields, and find the relationship between castor yield and photosynthesis. Furthermore, we tested our hypothesis that increasing the effective photosynthetic area through increased planting density could make up for the low photosynthetic capacity of dwarf varieties.
Materials and Methods
Plant Materials
The experimental castor varieties, Zhebi 100 and Zhebi 26, were introduced from South Africa. In field experiments (Hangzhou, China; 120°12′ E, 30°16′ N, altitude 20–60 m), castor seeds were sown on April 15 during 2012–2015 with three replicates. The field experiment was based on complete randomized block design with three replicates at five different planting densities, i.e., 8000 seedlings/hm2, 12,000 seedlings/hm2, 24,000 seedlings/hm2, 36,000 seedlings/hm2, and 48,000 seedlings/hm2. Each replicate plot was 30 m2. For Zhebi 100, the plant distance was 100 cm, and the row spacing was 125, 83, 42, 28, and 21 cm, respectively. For Zhebi 26, the plant distance was 50 cm, and the row spacing was 250, 166, 84, 56, and 42 cm, respectively. Field management is carried out according to farmer's practice.
Ninety days after planting, six plants from each variety were randomly selected, and their morphological traits and photosynthetic parameters were measured. Mature leaves were taken from field R. communis plants in the experimental field. The samples were located at the same leaf position and orientation and were taken on the same day and hour during the active growth season (summer). After 90 days of growth, mature leaves and other plant tissues were selected for physiological measurements and protein extraction. Leaves were washed in situ with tap water, dried with filter paper, and then frozen in liquid nitrogen immediately. Samples were stored at −80°C until protein extractions were done.
Physiological Index Measurements
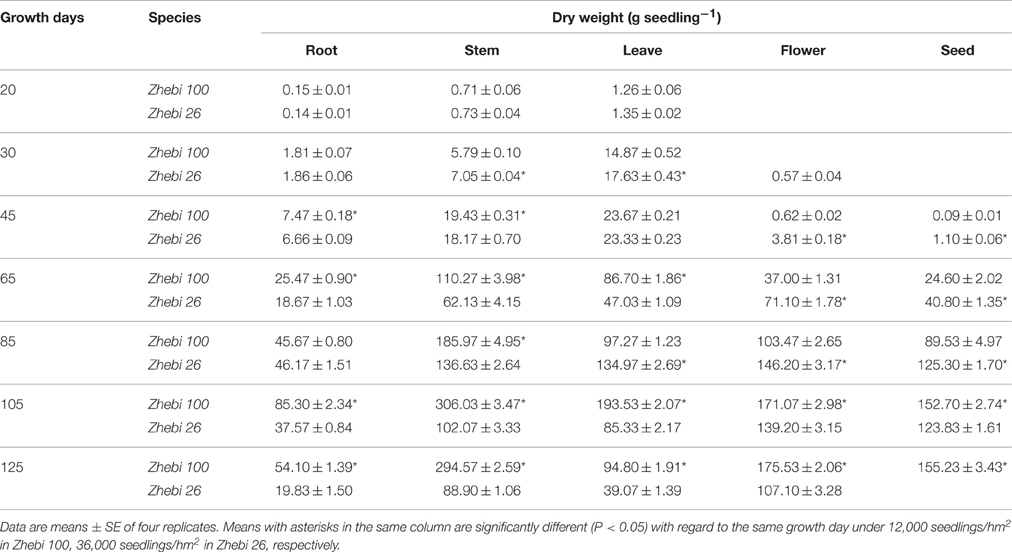
Seedling performance was assessed in terms of biomass, separated into roots, stems, leaves, flowers, and seeds under 12,000 seedlings/hm2 in Zhebi 100, 36,000 seedlings/hm2 in Zhebi 26, respectively. Plant materials were oven-dried (70°C) to constant weight. A mean value was obtained from 10 seedlings. Each treatment contained three biological replicates. We used the average of the four replicates of 10 seedlings as the mean value of seedling weight.
R. communis leaves (0.1 g of fresh weight) were prepared, and chlorophyll was extracted with ice-cold 80% v/v acetone. Absorption of the extract was measured at 663 and 646 nm with a spectrometer (Varian Cary 50 UV-VIS) and chlorophyll content was calculated with formulae proposed by Wellburn (1994). The formulae used were as follows:
Eight seedlings per castor variety were randomly selected for net photosynthetic rate (Pn) and chlorophyll fluorescence measurements. Pn was measured using a portable photosynthesis system (LI-6400, Li-Cor Inc., Lincoln, NE, USA), as described previously (Wei et al., 2015). The leaf area was measured using a leaf area meter of LI-3100 (Li-Cor Inc., Lincoln, Nebraska, USA). Leaf area index (LAI) was calculated as a ratio of the leaf area from a given land area to land area. According to the method of Chen et al. (2013), leaf chlorophyll fluorescence was measured using a pulse-amplitude-modulation fluorometer (PAM-2100, Heinz Walz, Effeltrich, Germany). The seedlings were exposed to sunlight for at least 15 min prior to Pn measurement. For each seedling, at least five measurements were made. Mean values were obtained from 8 replicates.
Protein Extraction
For Zhebi 100, the planting distance was 100 cm, and the row spacing was 83 cm, planting density is 12,000 seedlings/hm2. For Zhebi 26, the planting distance was 50 cm, and the row spacing was 56 cm, planting density is 36,000 seedlings/hm2. Leaves from these treatments were used for proteomics and Western blot analyses. Total R. communis leaf protein extraction was performed using the phenol extraction method (Hu et al., 2016), with slight modifications. Briefly, frozen R. communis leaves (1.0 g) were ground using a mortar and pestle in liquid nitrogen to a fine powder with an equal amount of polyvinyl polypyrrolidone. Next, the ground powder was homogenized in pre-cooled protein extraction buffer (20 mM Tris-HCl pH 7.5, 250 mM sucrose, 10 mM ethylene diamine tetraacetic acid, 1% Triton X-100, 1 mM 1,4-dithiothreitol and 1 mM phenylmethyl-sulfonyl fluoride) on ice. The pellets were then washed with ice-cold acetone for three times. The final washed protein pellets were dried by vacuum centrifugation to remove any remaining acetone and dissolved in lysis buffer (8 M urea, 2 M thiourea, 1% DTT, 4% CHAPS, 0.5% IPG buffer pH 4–7). The protein concentrations of the lysates were determined using a 2-D Quant Kit (GE Healthcare Amersham Bioscience, Little Chalfont, UK).
Two-Dimensional Electrophoresis and Data Analysis
Two-dimensional electrophoresis (2-DE) was conducted according to Hu et al. (2014c). The protein samples (500 μg) were loaded onto Immobiline DryStrips (18 cm long, pH 4–7, GE Healthcare) during the rehydration step at room temperature for 12 h. Isoelectric focusing (IEF) was performed using an Ettan IPGphor isoelectric focusing system (GE Healthcare) as follows: 300 V for 1 h, 500 V for 1 h, 1000 V for 1 h, a gradient to 8000 V for 4 h, and kept at 8000 V for a total of 80,000 volt-hours (Vh) at 20°C. After IEF, the focused strips were equilibrated in equilibration buffer as described by Hu et al. (2014a). For the second dimension electrophoresis, the proteins were separated on 15% SDS polyacrylamide gels. Subsequently, the gels were stained using Coomassie Brilliant Blue (CBB) R-250. The 2-DE gel images were acquired with an image scanner (Uniscan M3600, China) and analyzed using the PDQuest software (Version 8.01, Bio-Rad, Hercules, CA, USA). Protein spots with significant (>2-fold change) and reproducible changes were selected for MS analysis.
Identification of Proteins and Classification
Protein digestion and identification were performed according to Hu et al. (2014c). Protein spots were identified using a 4800 Plus MALDI-TOF/TOF Proteomics Analyzer (Applied Biosystems, USA). Tryptic peptide masses were searched according to the corresponding annotations in the National Center for Biotechnology Information non-redundant (NCBInr) database (release date: November 29, 2013), species restriction to R. communis and Viridiplantae (green plants) only when no proteins matched in R. communis using the MASCOT interface (Version 2.5, Matrix Science, London, UK). The following search parameters were used: no molecular weight restriction, one allowed missed trypsin cleavage, fixed modification of carbamidomethyl (C), variable modification of oxidation (Met), the peptide tolerance of 100 ppm, fragment mass tolerance of ± 0.4 Da and peptide charge of 1+. Protein identifications were validated with at least 3 peptides matched, keratin contamination was removed, and the MOWSE threshold was set over 60 (P < 0.05). The highest scoring peptide was then blast searched against the NCBI database and the protein was identified according to the alignment.
The functions of the identified proteins were searched against the UniProt (http://www.uniprot.org) and NCBI protein (http://www.ncbi.nlm.nih.gov) databases as described by Chen et al. (2014).
Western Blot Analysis
Western blot analysis was performed as described previously (Hu et al., 2016). The protein samples were separated by 12% w/v standard SDS-PAGE and then electroblotted onto polyvinylidene difluoride membranes. After transfer, the membranes were probed with the appropriate primary antibodies and anti-rabbit IgG horseradish peroxidase conjugated to alkaline phosphatase (Abcam, UK, 1:5000 dilution) to detect the primary antibodies. The primary antibodies for the RuBisCO large subunit (RuBisCO LSU; Agrisera, Sweden), ATP synthase (ATPase; Agrisera) and chloroplast Cu/Zn superoxide dismutase (Cu/Zn SOD; Agrisera) were diluted 1:5000, 1:2000, and 1:1000, respectively. β-actin (1:5000; Santa Cruz, CA, USA) was used as an internal control. The signals were detected using an enhanced chemiluminescence kit (TIAN-GEN, China) according to the manufacturer's instructions.
Statistical Analysis
Values in figures are expressed as means ± SE. The statistical significance of the data was analyzed using univariate analysis of variance (P < 0.05) (One-way ANOVA; SPSS for Windows, version 16.0).
Results
Yield Parameters in Two Castor Varieties
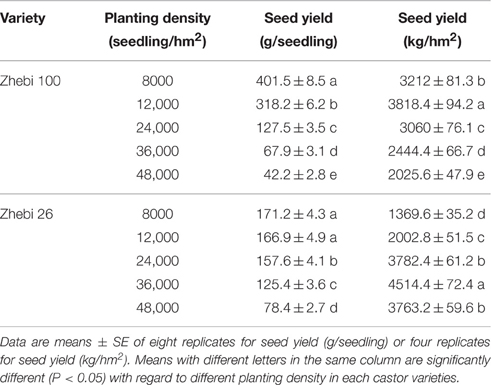
By reasonably increasing the planting densities of Zhebi 100 (12,000 seedlings/hm2) and Zhebi 26 (36,000 seedlings/hm2), we demonstrated that Zhebi 26 could achieve a higher yield than Zhebi 100 (Table 1).
Morphological and Physiological Differences between Ricinus communis Varieties
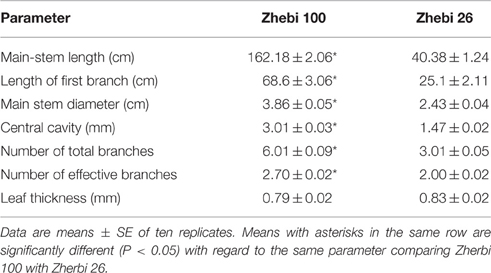
In the field experiment, we observed that the number of nodes and length per node of the main stem were higher in Zhebi 100 than in Zhebi 26 (Figure 1). Other parameters were also higher in Zhebi 100, including main stem length, main stem internode length, main stem diameter, central cavity, number of total branches, number of effective branches, and leaf thickness (Table 2).
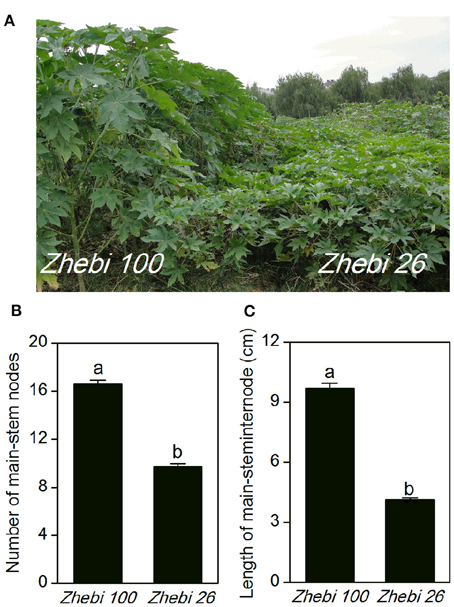
Figure 1. Morphological parameters in two castor varieties. (A) Phenotypes of the Zhebi 100 (left) and Zhebi 26 (right). (B) Number of main-stem nodes. (C) Length per node of main stem. Different letters above columns indicate significant difference at P < 0.05.
After 20–125 growth days, the dry weights of different organs during different developmental stages were measured in the two varieties (Table 3). The roots and aerial parts of Zhebi 100 showed higher biomass accumulation. Notably, compared with Zhebi 100, the flowering and maturing times of Zhebi 26 were earlier.
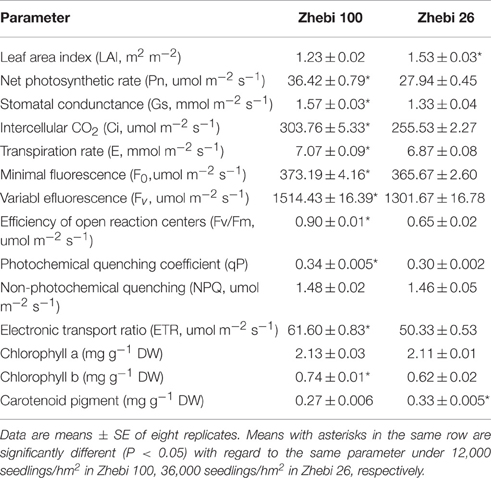
However, there was no significant difference in non-photochemical quenching (NPQ) or chlorophyll a between Zhebi 100 and Zhebi 26 (Table 4). But, almost all other physiological and photosynthetic parameters were obviously higher in Zhebi 100, including Pn, stomatal condunctance (Gs), intercellular CO2 (Ci), transpiration rate (E), minimal fluorescence (F0), variabl efluorescence (Fv), photochemical quenching coefficient (qP), efficiency of open reaction centers (Fv/Fm), electronic transport ratio (ETR), and chlorophyll b. Yet, compared with Zhebi 100, increased planting density contributed to higher LAI in Zhebi 26.
Identification and Functional Classification of Proteins Changed in Abundance Comparing Zherbi 100 and Zherbi 26
Because leaves are the major photosynthetic organs of broad-leaved crops, and leaf formation is a basic aspect of plant development and crop productivity. So we used leaf samples for experimental material. To better understand the differences in plant productivity between a high-stalk variety and dwarf variety, we carried out a comparative proteomic study comparing Zhebi 100 with Zhebi 26. A total of 38 protein spots were identified that were significantly different in abundances in the two-dimensional electrophoresis analysis of the two varieties (Figure 2, Table 5). These differential proteins belonged to a wide range of metabolic pathways. The data indicated that the identified proteins falling into eight functional categories according to their biological functions (Figure 3A), with the main groups being redox homeostasis and defense responses (23.7%), photosynthesis (21.0%), energy (18.4%), primary metabolic processes (7.9%), secondary metabolism, cell division and ion homeostasis (5.3%), and protein biosynthesis (2.6%).
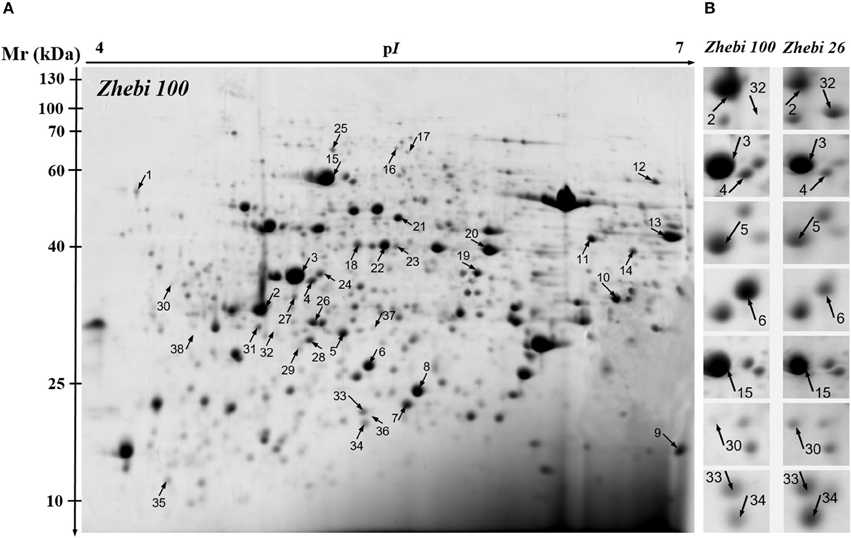
Figure 2. 2-DE analysis of proteins extracted from Ricinus communis leaves. The numbers assigned to the protein spots correspond to those listed in Table 5. (A) Representative CBB R250-stained 2D gel of total proteins from Zhebi 100. Arrows indicate 38 spots showing at least 2-fold changes (P < 0.05) analyzed by MALDI-TOF/TOF MS. (B) The enlarged window shows some differential protein spots changed in abundance between Zhebi 100 and Zhebi 26.
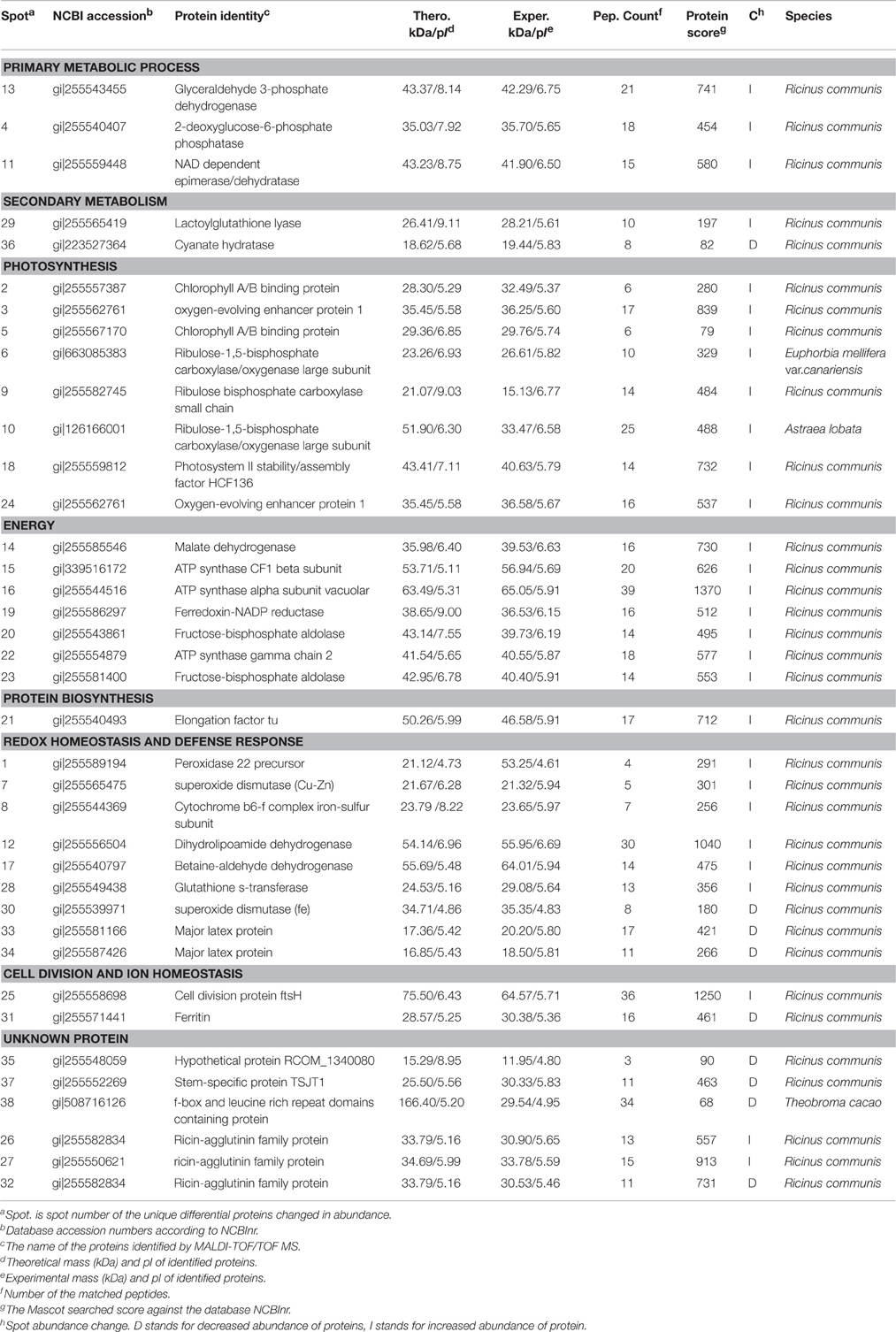
Table 5. Proteins identities using MALDI-TOF/TOF MS in Zherbi 100 compared with Zherbi 26 at the highest yield planting density.
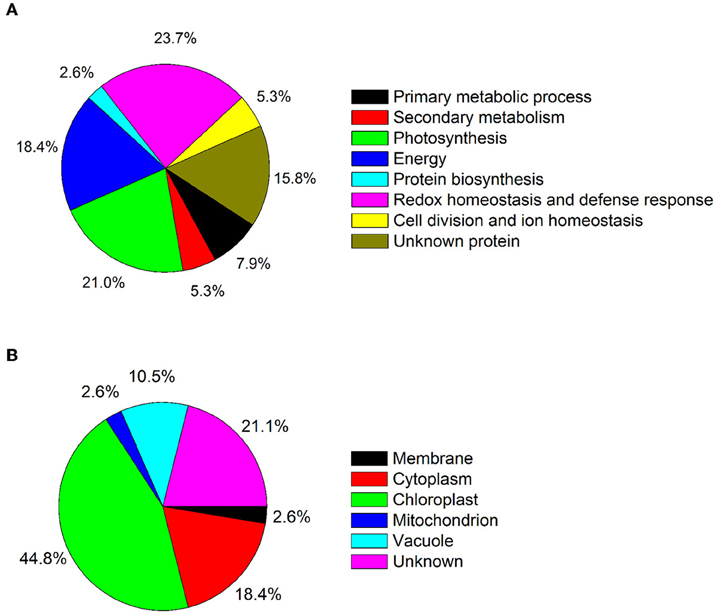
Figure 3. (A) Functional category distribution of 38 identified proteins. (B) Subcellular locations of the identified proteins.
In subcellular localization analysis, the majority of these proteins were located in the chloroplast (44.8%), followed by in the cytoplasm (18.4%), vacuole (10.5%), membrane (2.6%), and mitochondrion (2.6%) (Figure 3B).
Protein Abundance Analysis by Western Blot
The proteomic results revealed that the abundances of RuBisCO (spots 6, 10), ATP synthase (spots 15, 16, 22), and superoxide dismutase (Cu-Zn) (spot 7) were decreased in Zhebi 26 (Table 5). As shown in Figure 4, the protein abundance levels of RuBisCO LSU, ATPase and Cu/Zn SOD were indeed significantly decreased in Zhebi 26 according to Western blot analysis.
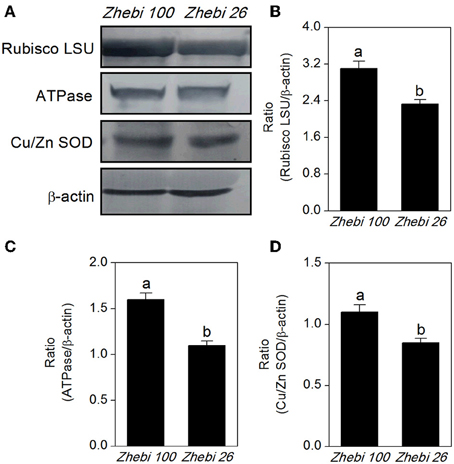
Figure 4. (A) The protein abundance levels of the ribulose-1,5-bisphosphate carboxylase large subunit (RuBisCO LSU), ascorbate peroxidase (APX) and superoxide dismutase (Cu-Zn) (Cu/Zn SOD) in Zhebi 100 and Zhebi 26 from western blotting. The relative abundance levels of RuBisCO LSU (B), ATPase (C), Cu/Zn SOD (D) were analyzed with the Quantity One software. β-actin was used as the internal control. Bars with different letters are significantly different from each other (P < 0.05).
Quality Assessment of Final Laboratory-Refined Oil in the Two Castor Varieties
In comparing with Zhebi 26, there does not appear to have major differences in total oil content in Zhebi 100 (Table 6). However, Zhebi 100 contains a significantly higher content of palm acid, whereas Zhebi 26 contains significantly higher amounts of ricinoleic acid, oleic acid, and linoleic acid.
Discussion
Metabolism Related Proteins
Leaf formation is a basic aspect of plant development and crop productivity because leaves are the major photosynthetic organs of broad-leaved crops. An understanding of the processes underlying the differences between R. communis varieties thus provides an insight into a basic biological process, modulation of which may have far-reaching significance for strategies to improve crops yield and the environment. Consistent with the growth and development indexes, Zhebi 100 showed higher biomass accumulation in its aerial parts and roots during different developmental stages (Table 3). Numerous early studies reported that glyceraldehyde 3-phosphate dehydrogenase is a key enzyme involved in glycolysis and cellular energy production associated with plant development (Giegé et al., 2003; Hu et al., 2014a). In this research, we found that glyceraldehyde-3-phosphate dehydrogenase C2 (spot 13) is increased expected in Zhebi 100.
A previous study reported that 2-deoxyglucose-6-phosphate phosphatase (spot 4) displays sugar-phosphatase activity, and plays an important role in plant carbon metabolism (Baskin et al., 2001). Furthermore, NAD dependent epimerase/dehydratase (spot 11) has racemase and epimerase activities, acting on carbohydrates and their derivatives, and takes part in pectin-related carbohydrate metabolic processes (Wang et al., 2008). In this study, the majority of protein spots (spots 4, 11) related to primary carbohydrate metabolism increased in abundance in Zhebi 100. This indicates that carbon metabolism is more active in Zhebi 100, providing more resources for tissue formation during development.
Photosynthesis, Energy Pathway and Protein Biosynthesis Related Proteins
Zhebi 100 leaves showed higher abundance of photosynthesis and energy pathway related proteins than Zhebi 26 (Table 5). This was supported by the observations that Zhebi 100 leaves have a higher total chlorophyll (a + b) content and net photosynthesis rate (Table 4). These results suggest that proteins involved in carbon assimilation, folding and assembly, and energy metabolism may work synchronously and show a correlation to the increased photosynthetic capacity in Zhebi 100 leaves. The key enzymes involved in these above processes are assembled into the RuBisCO complex and its activator RuBisCO activase (Portis and Parry, 2007); these enzymes showed higher accumulation levels in Zhebi 100 (spots 6, 9, 10; Table 5). Additionally, Western blot analysis showed that the RuBisCO large subunit was increased in abundance in Zhebi 100 leaves (Figure 4), Western blot result is consistent with the proteomic data for the selected protein at the highest yield planting density. These results support our speculation that a lower photosynthetic rate and primary carbon metabolism might be the reason for the slower plant growth and development in Zhebi 26. This was supported by Pn and chlorophyll fluorescence measurements; these photosynthetic indexes were markedly lower in Zhebi 26 as shown in Table 4. Moreover, similar patterns of decreased biomass accumulation of aerial parts were observed in Zhebi 26 compared with Zhebi 100 at the 85th day after planting (Table 3). Because the mature dates are different between Zhebi 26 and Zhebi 100: Zhebi 26's mature date is around the 85th day after planting (the growth of the aerial parts of Zhebi 26 stopped after the mature date), whereas Zhebi 100 continues to grow until its mature date around the 105th day. Elongation factor Tu (spot 21, Table 5), an essential component for protein synthesis (Hu et al., 2014c), also showed higher abundance in Zhebi 100 leaves. We speculate that Zhebi 100 has higher biosynthetic activity, and the participation of elongation factor Tu is required for the correct folding of the newly synthesized proteins.
In relation to energy production, malate dehydrogenase (spot 14, Table 5), which is implicated in NADPH photoactivation (Valledor et al., 2010), increased in abundance in Zhebi 100 leaves. Moreover, previous studies have indicated that sufficient ATP is necessary for plant growth and development (Jiang et al., 2007; Hu et al., 2014b). ATP is mainly produced by carbohydrate metabolism, such as glycolysis and the tricarboxylic acid cycle. Numerous papers have reported that several enzymes involved in glycolysis and the tricarboxylic acid cycle are associated with plant development (Palma et al., 2011; Cui et al., 2012). In this study, we identified seven proteins related to energy production pathways (Table 5). Among them, fructose-bisphosphate aldolase (spots 20, 23), an essential enzyme involved in the glycolytic pathway, showed enhanced abundance in Zhebi 100. We also found that the ATP synthase CF1 beta subunit (spot 15), ATP synthase vacuolar alpha subunit (spot 16) and ATP synthase gamma chain 2 (spot 22) had increased abundance in Zhebi 100. Ferredoxin-NADP reductase is a ubiquitous flavoenzyme that delivers NADPH or low potential one-electron donors (ferredoxin) to redox-based metabolisms in plastids and mitochondria (Ceccarelli et al., 2004). Donation of electrons by ferredoxin has been demonstrated in many other plastid enzymes, which may be essential for sufficient energy production in Zhebi 100.
Compared with Zhebi 26, Zhebi 100 displays a higher number of proteins increased in abundance (Table 5). These proteins are mainly related to chloroplast electron transfer chain, carbohydrate biosynthesis, and energy production, which may be the reason that Zhebi 100 possesses the higher photosynthesis and energy metabolic activity than Zhebi 26. In addition, we demonstrated that Zhebi 26 (36,000 seedlings/hm2) can achieve a higher yield than Zhebi 100 (12,000 seedlings/hm2) through increased planting density (Table 1). The reasonable explanation is that increased planting density contributes to higher LAI in Zhebi 26 compared with Zhebi 100 (Table 4). Higher LAI means higher light interception by plants, higher Pn, and consequently higher productivity. Therefore, Zhebi 26 can achieve a higher yield than Zhebi 100 through reasonably close planting (Figure 5).
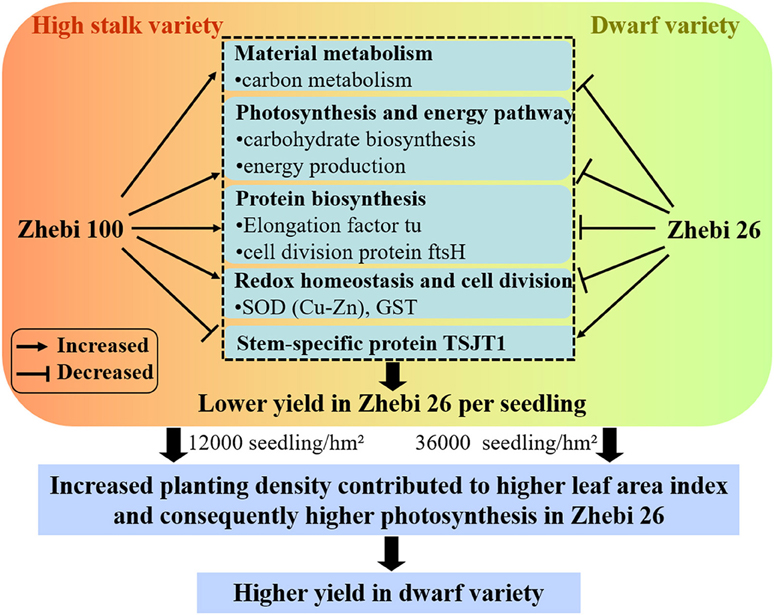
Figure 5. A schematic representation of different growth and development mechanisms in Zhebi 100 and Zhebi 26 via regulation of diverse biological processes.
Redox Homeostasis and Defense Response Related Proteins
Growing evidence indicates that, in redox homeostasis, reactive oxygen species (ROS) play a dual role in plant biology as both toxic byproducts of aerobic metabolism and key regulators of growth, development and defense pathways (Mittler et al., 2004; Yan et al., 2006). As the most important reaction in the cell elongation process, loosening of plant cell walls is associated with the production of ROS (Foreman et al., 2003; Kwon et al., 2007; Yang et al., 2008), which could damage plant cell if not removed efficiently, therefore increased antioxidation capacity is the hallmark of fast plant growth (Cui et al., 2012). However, plants can regulate ROS levels through complex mechanisms such as ROS scavenging with superoxide dismutase (Goossens et al., 2003) and glutathione S-transferase (GST) (Apel and Hirt, 2004; Hu et al., 2014b). In Zhebi 100, many antioxidant enzymes associated with redox homeostasis and antioxidation response were increased in abundance, including a peroxidase 22 precursor (spot 1), SOD (Cu-Zn) (spot 7), a cytochrome b6-f complex iron-sulfur subunit (spot 8), dihydrolipoamide dehydrogenase (spot 12), betaine-aldehyde dehydrogenase (spot 17) and GST (spot 28). Furthermore, Western blot analysis showed that the protein abundance of SOD (Cu-Zn) was increased in Zhebi 100 leaves (Figure 4), which was consistent with the proteomic data at the highest yield planting density. The findings presented above, together with previously published data, strongly suggest that ROS-mediated cell expansion may be an important mechanism regulating castor bean growth and development. On the other hand, enhanced abundance of these proteins may also imply that the anti-oxidative defense system and resistance to environmental stress is increased in Zhebi 100.
Proteins Related to Other Functions
Plant internode elongation is correlated with cell division and elongation (Cui et al., 2012). The increased abundance of the cell division protein ftsH (spot 25) is potentially important for the longer internodes in Zhebi 100 compared with Zhebi 26. In addition, little is known about the function of the stem-specific protein TSJT1 in plant growth and development. Notably, compared with Zhebi 26, the abundance of TSJT1 (spot 37) was decreased in Zhebi 100. As shown in Figure 1 and Table 2, Zhebi 26 exhibited a wide range of morphological phenotypes, such as dwarfism, compared with Zhebi 100. These results suggest that the intriguing stem-specific protein TSJT1 may act as a negative regulator in castor internode development. The specific function of this protein needs further study in the future.
In plants, ferritin is an essential regulator of iron homeostasis (Ravet et al., 2008). In our study, the abundance of ferritin (spot 31) was increased in Zhebi 26. Ferritin accumulation is induced by an excess of iron, as well as by abscisic acid, photoinhibition and ozone (Murgia et al., 2002). A secondary function of ferritin is protection from the oxidative stress of ROS (Ravet et al., 2008). In leaves, ferritin is an iron source at early stages of development for the synthesis of iron-containing proteins involved in photosynthesis (Briat and Lobréaux, 1997; Kobayashi and Nishizawa, 2012). The specific role of ferritin in redox homeostasis and photosynthesis response needs to further study in the future.
Author Contributions
WH and GS: Conceived and designed the research; WH, LC, XQ, HL, JW, and YB: Performed the research; NH, RH, and LS: Analyzed the data; WH and GS: Wrote the paper; and HZ: Revised this paper.
Funding
This study was supported by the State Key Laboratory Breeding Base for Zhejiang Sustainable Pest and Disease Control (2010DS700124-KF1405 and KF1612, 2015-cxzt-01), the State Key Laboratory of Silkworm Genome Biology (20120007), the National Natural Science Foundation of China (31571718, 31402140) and of Zhejiang province (LY15C020002), the International Cooperation, Innovation Programs of Zhejiang Academy of Agricultural Sciences (2014CX005), and the Shaoxing 330 Overseas Elites Program to GS.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01473
References
Allan, G., Williams, A., Rabinowicz, P. D., Chan, A. P., Ravel, J., and Keim, P. (2008). Worldwide genotyping of castor bean germplasm (Ricinus communis L.) using AFLPs and SSRs. Genet. Resour. Crop Evol. 55, 365–378. doi: 10.1007/s10722-007-9244-3
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Auld, D., Zanotto, M., McKeon, T., and Morris, J. (2010). “Castor,” in Oil Crops, eds J. Vollmann and I. Rajcan (New York, NY: Springer), 317–332.
Baldanzi, M., Fambrini, M., and Pugliesi, C. (2003). Redesign of the castorbean plant body plan for optimal combine harvesting. Ann. Appl. Biol. 142, 299–306. doi: 10.1111/j.1744-7348.2003.tb00254.x
Baskin, T. I., Remillong, E. L., and Wilson, J. E. (2001). The impact of mannose and other carbon sources on the elongation and diameter of the primary root of Arabidopsis thaliana. Funct. Plant Biol. 28, 481–488. doi: 10.1071/PP01047
Briat, J.-F., and Lobréaux, S. (1997). Iron transport and storage in plants. Trends. Plant Sci. 2, 187–193. doi: 10.1016/S1360-1385(97)85225-9
Ceccarelli, E. A., Arakaki, A. K., Cortez, N., and Carrillo, N. (2004). Functional plasticity and catalytic efficiency in plant and bacterial ferredoxin-NADP (H) reductases. Biochim. Biophys. Acta 1698, 155–165. doi: 10.1016/j.bbapap.2003.12.005
Chan, A. P., Crabtree, J., Zhao, Q., Lorenzi, H., Orvis, J., Puiu, D., et al. (2010). Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 28, 951–956. doi: 10.1038/nbt.1674
Chen, J., Hu, W.-J., Wang, C., Liu, T.-W., Chalifour, A., Shen, Z.-J., et al. (2014). Proteomic analysis reveals differences in tolerance to acid rain in two broad-leaf tree species, Liquidambar formosana and Schima superba. PLoS ONE 9:e102532. doi: 10.1371/journal.pone.0102532
Chen, J., Wang, W.-H., Liu, T.-W., Wu, F.-H., and Zheng, H.-L. (2013). Photosynthetic and antioxidant responses of Liquidambar formosana and Schima superba seedlings to sulfuric-rich and nitric-rich simulated acid rain. Plant Physiol. Biochem. 64, 41–51. doi: 10.1016/j.plaphy.2012.12.012
Conway, G., and Toenniessen, G. (1999). Feeding the world in the twenty-first century. Nature 402, C55–C58. doi: 10.1038/35011545
Cui, K., He, C. Y., Zhang, J. G., Duan, A. G., and Zeng, Y. F. (2012). Temporal and spatial profiling of internode elongation-associated protein expression in rapidly growing culms of bamboo. J. Proteome Res. 11, 2492–2507. doi: 10.1021/pr2011878
Da Silva, N. D. L., Maciel, M. R. W., Batistella, C. B., and Maciel Filho, R. (2006). Optimization of biodiesel production from castor oil. Appl. Biochem. Biotechnol. 130, 405–414. doi: 10.1385/ABAB:130:1:405
Foreman, J., Demidchik, V., Bothwell, J. H., Mylona, P., Miedema, H., Torres, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Foster, J. T., Allan, G. J., Chan, A. P., Rabinowicz, P. D., Ravel, J., Jackson, P. J., et al. (2010). Single nucleotide polymorphisms for assessing genetic diversity in castor bean (Ricinus communis). BMC Plant Biol. 10:13. doi: 10.1186/1471-2229-10-13
Giegé, P., Heazlewood, J. L., Roessner-Tunali, U., Millar, A. H., Fernie, A. R., Leaver, C. J., et al. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151. doi: 10.1105/tpc.012500
Goossens, A., Häkkinen, S. T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., et al. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. U.S.A. 100, 8595–8600. doi: 10.1073/pnas.1032967100
Hu, W.-J., Chen, J., Liu, T.-W., Liu, X., Wu, F.-H., Wang, W.-H., et al. (2014a). Comparative proteomic analysis on wild type and nitric oxide-overproducing mutant (nox1) of Arabidopsis thaliana. Nitric Oxide 36, 19–30. doi: 10.1016/j.niox.2013.10.008
Hu, W.-J., Chen, J., Liu, T.-W., Simon, M., Wang, W.-H., Wu, F.-H., et al. (2014b). Comparative proteomic analysis of differential responses of Pinus massoniana and Taxus wallichiana var. mairei to simulated acid rain. Int. J. Mol. Sci. 15, 4333–4355. doi: 10.3390/ijms15034333
Hu, W.-J., Chen, J., Liu, T.-W., Wu, Q., Wang, W.-H., Liu, X., et al. (2014c). Proteome and calcium-related gene expression in Pinus massoniana needles in response to acid rain under different calcium levels. Plant Soil 380, 285–303. doi: 10.1007/s11104-014-2086-9
Hu, W.-J., Wu, Q., Liu, X., Shen, Z.-J., Chen, J., Liu, T.-W., et al. (2016). Comparative proteomic analysis reveals the effects of exogenous calcium against acid rain stress in Liquidambar formosana Hance leaves. J. Proteome Res. 15, 216–228. doi: 10.1021/acs.jproteome.5b00771
Jiang, Y., Yang, B., Harris, N. S., and Deyholos, M. K. (2007). Comparative proteomic analysis of NaCl stress-responsive proteins in Arabidopsis roots. J. Exp. Bot. 58, 3591–3607. doi: 10.1093/jxb/erm207
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kwon, S. I., Lee, H., and An, C. S. (2007). Differential expression of three catalase genes in the small radish (Rhaphanus sativus L. var. sativus). Mol. Cells 24, 37–44.
Li, B., Xu, W., Xu, Y., Zhang, Y., Wang, T., Bai, Y., et al. (2010). Integrative study on proteomics, molecular physiology, and genetics reveals an accumulation of cyclophilin-like protein, TaCYP20-2, leading to an increase of Rht protein and dwarf in a novel GA-insensitive mutant (gaid) in wheat. J. Proteome Res. 9, 4242–4253. doi: 10.1021/pr100560v
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Murgia, I., Delledonne, M., and Soave, C. (2002). Nitric oxide mediates iron-induced ferritin accumulation in Arabidopsis. Plant J. 30, 521–528. doi: 10.1046/j.1365-313X.2002.01312.x
Palma, J. M., Corpas, F. J., and del Río, L. A. (2011). Proteomics as an approach to the understanding of the molecular physiology of fruit development and ripening. J. Proteomics 74, 1230–1243. doi: 10.1016/j.jprot.2011.04.010
Portis, A. R. Jr., and Parry, M. A. (2007). Discoveries in Rubisco (Ribulose 1,5-bisphosphate carboxylase/oxygenase): a historical perspective. Photosynth. Res. 94, 121–143. doi: 10.1007/s11120-007-9225-6
Ravet, K., Touraine, B., Boucherez, J., Briat, J. F., Gaymard, F., and Cellier, F. (2008). Ferritins control interaction between iron homeostasis and oxidative stress in Arabidopsis. Plant J. 57, 400–412. doi: 10.1111/j.1365-313X.2008.03698.x
Severino, L. S., Auld, D. L., Baldanzi, M., Cândido, M. J. D., Chen, G., Crosby, W., et al. (2012). A review on the challenges for increased production of castor. Agron. J. 104, 853–880. doi: 10.2134/agronj2011.0210
Sujatha, M., Reddy, T. P., and Mahasi, M. (2008). Role of biotechnological interventions in the improvement of castor (Ricinus communis L.) and Jatropha curcas L. Biotechnol. Adv. 26, 424–435. doi: 10.1016/j.biotechadv.2008.05.004
Valledor, L., Jorrín, J. V., Rodríguez, J. L., Lenz, C., Meijón, M., Rodríguez, R., et al. (2010). Combined proteomic and transcriptomic analysis identifies differentially expressed pathways associated to Pinus radiata needle maturation. J. Proteome Res. 9, 3954–3979. doi: 10.1021/pr1001669
Wang, C.-Y., Chiou, C.-Y., Wang, H.-L., Krishnamurthy, R., Venkatagiri, S., Tan, J., et al. (2008). Carbohydrate mobilization and gene regulatory profile in the pseudobulb of oncidium orchid during the flowering process. Planta 227, 1063–1077. doi: 10.1007/s00425-007-0681-1
Wei, J., Qiu, X., Chen, L., Hu, W., Hu, R., Chen, J., et al. (2015). The E3 ligase AtCHIP positively regulates Clp proteolytic subunit homeostasis. J. Exp. Bot. 66, 5809–5820. doi: 10.1093/jxb/erv286
Wellburn, R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents with spectrophotometers of different resolution. J. Plant Physiol. 144, 307–313. doi: 10.1016/S0176-1617(11)81192-2
Yan, S. P., Zhang, Q. Y., Tang, Z. C., Su, W. A., and Sun, W. N. (2006). Comparative proteomic analysis provides new insights into chilling stress responses in rice. Mol. Cell. Proteomics 5, 484–496. doi: 10.1074/mcp.M500251-MCP200
Keywords: Ricinus communis, castor bean productivity, agriculture, photosynthesis, plant proteomics
Citation: Hu W, Chen L, Qiu X, Lu H, Wei J, Bai Y, He N, Hu R, Sun L, Zhang H and Shen G (2016) Morphological, Physiological and Proteomic Analyses Provide Insights into the Improvement of Castor Bean Productivity of a Dwarf Variety in Comparing with a High-Stalk Variety. Front. Plant Sci. 7:1473. doi: 10.3389/fpls.2016.01473
Received: 05 July 2016; Accepted: 15 September 2016;
Published: 29 September 2016.
Edited by:
Setsuko Komatsu, National Institute of Crop Science, JapanReviewed by:
Mohammad-Zaman Nouri, Rice Research Institute of Iran, Deputy of Mazandaran, IranJose Tadeu Abreu Oliveira, Federal University of Ceará, Brazil
Copyright © 2016 Hu, Chen, Qiu, Lu, Wei, Bai, He, Hu, Sun, Zhang and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guoxin Shen, guoxin.shen@ttu.edu
 Wenjun Hu1
Wenjun Hu1 Rongbin Hu
Rongbin Hu Hong Zhang
Hong Zhang Guoxin Shen
Guoxin Shen