- 1Department of Crop Science, Faculty of Agriculture, Universiti Putra Malaysia, Selangor, Malaysia
- 2Department of Plant Breeding and Genetics, Faculty of Crop Production, Sindh Agriculture University Tandojam, Sindh, Pakistan
- 3Laboratory of Food Crops, Institute of Tropical Agriculture, Universiti Putra Malaysia, Selangor, Malaysia
- 4Department of Plant Protections, Faculty of Agriculture, Universiti Putra Malaysia, Selangor, Malaysia
- 5Agrotechnology and Bioscience Division, Malaysian Nuclear Agency, Selangor, Malaysia
- 6Department of Agronomy and Plant Breeding, Islamic Azad University of Yadegar-e-Imam Khomeini (RAH) Branch, Tehran, Iran
- 7Bangladesh Rice Research Institute, Gazipur, Bangladesh
Blast is the most common biotic stress leading to the reduction of rice yield in many rice-growing areas of the world, including Malaysia. Improvement of blast resistance of rice varieties cultivated in blast endemic areas is one of the most important objectives of rice breeding programs. In this study, the marker-assisted backcrossing strategy was applied to improve the blast resistance of the most popular Malaysian rice variety MR219 by introgressing blast resistance genes from the Pongsu Seribu 2 variety. Two blast resistance genes, Pi-b and Pi-kh, were pyramided into MR219. Foreground selection coupled with stringent phenotypic selection identified 15 plants homozygous for the Pi-b and Pi-kh genes, and background selection revealed more than 95% genome recovery of MR219 in advanced blast resistant lines. Phenotypic screening against blast disease indicated that advanced homozygous blast resistant lines were strongly resistant against pathotype P7.2 in the blast disease endemic areas. The morphological, yield, grain quality, and yield-contributing characteristics were significantly similar to those of MR219. The newly developed blast resistant improved lines will retain the high adoptability of MR219 by farmers. The present results will also play an important role in sustaining the rice production of Malaysia.
Introduction
The rice production system makes a vital contribution to the reduction of hunger and poverty. The fast growth of the world population demands an increase of 26% in rice production to fulfill the requirement (Khush, 2013). Rice production has widely increased after the green revolution, but the yield of superior varieties is still not increasing as farmers expect due to the influence of biotic and abiotic factors (Divya et al., 2014). The continuous supply of rice per demand of the consumer can only be achieved by maintaining a stable rice production, which is a challenge for rice breeders (Roychowdhury et al., 2012). Rice production can be managed by introducing new varieties possessing strong resistance against abiotic and biotic factors. Currently, DNA marker technology has immensely contributed to genetic improvement through the selection of desirable traits, such as disease resistance. Molecular markers are a valuable resource in marker-assisted backcross (MABC) breeding to monitor the disease resistance genes. Many rice cultivars resistant to biotic stress have been released and widely adopted by farmers with the application of marker-assisted selection (Xu and Crouch, 2008).
Blast is the one of the main diseases of rice crops causing crop loss in both temperate and tropical rice growing regions (Mackill and Bonman, 1992). The main agent causing this disease is the fungus Magnaporthe oryzae. Thus, blast resistance in rice plants has been one of the most important traits being pursued in breeding programs over several decades. Different breeding strategies have been adopted to achieve this serious challenge, such as the use of field resistance to blast disease and the introduction of resistance genes into the rice susceptible cultivar objective (Kushibuchi, 1997). As a result, several blast resistance rice varieties were introduced, but within a short period of time, they have become seriously blast susceptible because of the emergence of new pathotypes of blast fungus. This breakdown of blast resistance clearly indicates that the resistance cannot be widely achieved until true resistance genes are identified (Hittalmani et al., 2000). The continuous identification of resistance genes against blast can lead to genetic control over the new existing pathogens. To date, approximately 347 QTLs linked to blast resistance (Koide et al., 2009; Ballini et al., 2013) and more than 100 blast resistant genes have been identified from diversified rice germplasm (Divya et al., 2014). The identified blast R genes were found on all 12 rice chromosomes except 3, and most of them were in a cluster on chromosomes 6, 11, and 12 (Yang et al., 2013). Pi-b and Pi-kh have been used extensively in rice breeding programs in Japan, China, and Indonesia and are considered to be major blast resistance genes along with Pi-ta. Pi-b and Pi-kh are dominant major blast resistance genes conferring broad spectrum resistance to various isolates of the fungal pathogen M. oryzae (Wang et al., 1999; Sharma et al., 2005; Tanweer et al., 2015b). Pi-kh has been identified in many tropical Japonica varieties such as Tetep, and Pi-b in Indica varieties such as Thoku 11 9 (Conaway-Bormans et al., 2003). The dominant gene Pi-b, which confers high resistance to most Japanese blast races, has been mapped to the distal end of the long arm of chromosome 2 (Shinoda et al., 1971) and Pi-kh to the long arm of rice chromosome 11 (Sharma et al., 2005). The utilization of both of these blast resistance genes in marker assisted-selection breeding programs has been widely observed (Tanweer et al., 2015a).
Marker-assisted backcrossing has enormous potential to introduce the blast resistance genes into diverse rice cultivars (Collard and Mackill, 2008; Collard et al., 2008). Introgression of blast resistant genes into advanced improved rice lines is a cost-effective and environmentally friendly approach to combat yield losses (Wen and Gao, 2012). The main advantage of marker-assisted selection is the accuracy of selection of the true plant within the short breeding cycle to produce blast resistant rice varieties. Currently, the blast resistant breeding program has achieved greater success with the advent of marker-assisted selection (Ragimekula et al., 2013). Recently, blast resistance genes Piz5 and Pi54 have been introgressed into the genetic background of the PRR78 rice variety from donor parent C101A51 and Tetep, and blast resistant lines have been developed with the application of MABC breeding (Singh et al., 2012). The selection was based on foreground markers RM287 and RM206 by following repetitive backcrossing. The Pi1 leaf blast resistance gene has been introgressed into the D521 line derived from the donor line BL122 (Fu et al., 2012). With the application of MABC, 304 elite parental lines of hybrid rice have also been improved with bacterial blight and blast resistance genes (Zhou et al., 2003). Recently, IR64 cultivar submergence tolerant gene Sub1 has been introgressed into the OM1490 variety (Lang et al., 2011). The QTL Saltol derived from a salt tolerant variety also has been introgressed into popular cultivars of Vietnam (Huyen et al., 2012). These examples provide a great opportunity to develop blast resistant rice varieties through MABC breeding. In the present study, the MABC technique was applied to introgress blast resistant genes from the highly resistant rice variety Pongsu Seribu 2 to blast susceptible Malaysian rice cultivar MR219.
Materials and Methods
Developing Blast Resistant Lines
The crossing was performed between the parental lines of Pongsu Seribu 2 and MR219, and the F1 hybrid was produced (Supplementry Figure S1). After confirming the hybridity of the plants, true hybrid heterozygous plants were backcrossed with recurrent parent MR219, and BC1F1 generation seeds were produced. Foreground selection for the desired alleles and background selection for the recovery of the recurrent parent were performed. The plants with the desired allele and maximum recovery of recurrent parents were again backcrossed, and subsequently the BC2F1 generation was produced. The same steps were followed for the BC2F1 generation, and the best plants were selfed to produce the BC2F2 generation seed. The complete genome genotyping with SSR markers was performed, distributed over all 12 rice chromosomes. The true plants on the basis of genotype with the desired alleles were backcrossed in each generation. In every generation, the plant showing the heterozygous allele for Pongsu Seribu 2 was selected. At the final stage, 15 homozygous plants carrying target alleles along with a similar genome of MR219 in each chromosome were selected in the BC2F2 generation, and blast resistant lines were produced.
Microsatellite Analysis
Markers for Foreground Selection
The robust tightly linked marker RM208F 5′-tctgcaagccttgtctgatg-3′, RM208R 5′-taagtcgatcattgtgtggacc-3′ on chromosome 2 linked to the Pi-b gene (Wang et al., 1999) and RM206 5′-cccatgcgtttaactattct-3′, RM206R 5′-cgttccatcgatccgtatgg-3′ on chromosome 11 linked to the Pi-kh gene (Sharma et al., 2005) were used for selecting the target genes.
Markers for Background Selection
A total of 72 polymorphic markers were identified from 300 SSR markers with known chromosomal position covering all 12 rice chromosomes. SSR markers unlinked to the target gene covering the entire chromosome, including carrier chromosomes 2 and 11 that were polymorphic between the recurrent and donor parent, were used for the background selection to recover the recurrent parent genome. At least five polymorphic markers per chromosome were used to generate the data. The assessment of the recovery of the recurrent parent genome was based on the selection of SSR marker data that was carried out by using the software program Graphical Geno Types (GGTs) version 2.0.
DNA Extraction
Total genomic DNA was isolated from 21-days-old young fresh leaves of plants of each backcross generation using the CTAB method as mentioned by Doyle (1990) with minor changes.
PCR Amplification
For PCR amplification, the protocol described by McCouch et al. (2002) was adapted. The total volume of the PCR reaction was 15 μl, including 70 ng template DNA, 1.0 μl of forward primer, 1.0 μl of reverse primer, 7.4 μl master mix (premixed containing Taq DNA polymerase, dNTPs, and MgCl2) and 4.6 μl nuclease free water. PCR amplification was performed using the touch down PCR program using the following protocol: 94°C for 3 min followed by 10 cycles of 94°C for 30 s, 62°C for 1 min (decreasing 1°C per cycle), and 72°C for 30 s, and 30 cycles of 94°C for 30 s, 52°C for 1 min, 72°C for 2 min, and a final extension at 72°C for 10 min by rapid cooling to 4°C prior to analysis.
Gel Electrophoresis
The gel was prepared by mixing 3.0% metaphorTM agarose (Lonza) gel in 1× TBE buffer (0.05 M Tris, 0.05 M boric acid, 1 mM EDTA, pH 8.0). In total, 1 μl Midori green was also added for staining. The gel was run at 80 V for 80 min, and finally the amplified product was visualized in the Molecular imager® (GelDocTM XR, Bio-Rad Laboratories, Inc., USA).
Phenotypic Screening of Plants against Magnaporthe oryza Pathoype P7.2
The most virulent pathotype P7.2 of the M. oryzae isolate was provided by MARDI (Malaysian Agriculture and Research Development Institute). The plants of the donor parent, recurrent parent and BC2F2 generations were phenotypically screened in field conditions. The young plants of 21 days were inoculated by spraying spore suspension at a concentration of 1.5 × 105 conidia/ml, and 90% humidity was maintained by covering the plants with plastic bags to develop the disease. The inoculated plants were observed after 9 days of inoculation for blast disease lesions. The plants and blast lesion degrees (BLDs) were evaluated on the basis of 0-9 of the IRRI-SES scale (IRRI, 1996). The percentage of disease leaf area (%DLA) and blast lesion type (BLT) were scored as described by Correa-Victoria and Zeigler (1993). The percentage of DLA was calculated from 0 to 100%. For the BLT score, either 0 (highly resistant: no any symptoms), 1-2 (no sporulation, lesion 1–2 mm), 3 (little sporulation, round lesion), or 4 (heavy sporulation, spindle shaped lesion) were scored. For single-gene model analysis, if the plant showed lesion type 0-3, the plant was considered a resistant plant, and plants showing lesion type 4 or above were considered to be susceptible for the selected pathotype P7.2 in the selected BC2F2 population. For the two-gene model, blast resistance was classified as resistant (R) (1-2), moderately resistant (MR) (3), moderately susceptible (MS) (4-6) and susceptible (S) (7-9). The protocol of Singh et al. (2013) was followed with minor modification for plant disease reaction. The phenotypic segregation of plants for the two-gene model was calculated as 9:3:3:1 (R: MR: MS: S). A test for an effect of duplicate dominant gene action (epistasis) was analyzed by observing the resistant versus susceptible plants 15(R):1(S) ratio in the BC2F2 population. The plants showing a disease lesion score of 0-6 were resistant plants and 7-9 were susceptible plants.
Agronomic Performance of the Selected Best Lines of the BC2F2 Generation
The lines having a maximum recovery of the recurrent parent along with target genes and phenotypic similarity with the recurrent parent were used to observe the agro-morphological traits. Different parameters related to yield and yield contributing factors were recorded, such as days to 50% flowering, days to maturity, plant, panicles per plants, effective tillers per plants, panicle length, seed setting rate, full filled grain per plants seed setting rate, 1000-grain weight, yield per plant, grain length, grain width, flag leaf length and flag leaf width (Supplementry Table S1). These traits were recorded from all of the best selected lines of BC2F2 along with the recurrent parent.
Statistical Analysis
The BC2F2 population segregation data were analyzed using a chi-square test (χ2). An analysis for goodness of fit to the expected ratio of 3:1, 9:3:3:1, and 15:1 was calculated using the chi square formula χ2 = (O-E)2/E, where O represents observed value and E is the expected value. Analysis of a single marker was performed using SAS 9.3 software as mentioned by Divya et al. (2014). This analysis of single markers was fitted to the linear regression model: Y- bo+blx+e. The results were obtained as the estimate of R2 value and the F statistic for each marker. The R2 significance indicates the linkage of markers with the trait. The SSR genotyping data were analyzed using GGT software 2.0. The mean difference for the selected best lines of BC2F2 and the recurrent parent MR219 was analyzed using an independent t-test in the SAS 9.3 software.
Results
Marker-assisted Foreground Selection
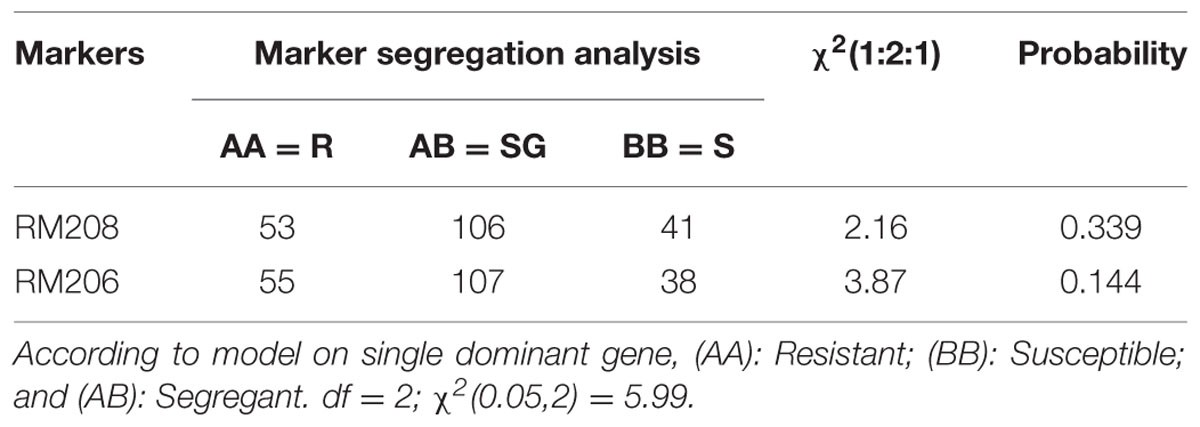
Crosses were made between the parental line of MR219 and Pongsu Seribu 2, and F1 seeds were produced. The best F1 plants were screened with foreground markers to identify the true F1 plants carrying the gene of interest in a heterozygous form. The six gene positive plants were backcrossed with the recurrent parent to generate the next generation BC1F1 seed. The BC1F1 plants were screened for the selection of a heterozygous allele at the putative Pi-b and Pi-kh locus with RM208 and RM206 markers along with phenotypic maximum similarity with the recurrent parent. The allele size in base pairs (bp) of both the parents MR219 and Pongsu Seribu 2 amplified by both markers has been given in Supplementry Table S2. The best plants of BC1F1 having an appearance similar to MR219 and carrying the target gene were again crossed with the recurrent parent and 320 plants of the BC2F1 generation were selected. Similarly, BC1F1 plants were screened to identify the plants in heterozygous form with maximum RPG recovery. Selfing was performed in the BC2F1 plants, and the BC2F2 generated seeds were grown and plants with similarity to MR219 along with homozygous resistant alleles using RM208 and RM206 were selected (Figure 1). The genotypic segregation of the BC2F2 population using linked markers RM208 and RM206 is shown in Table 1. Both of the markers represent a good fit to the expected marker segregation ratio (1:2:1) according to the expected Mendelian ratio. From this selection, 15 best improved blast resistant lines were evaluated and selected.
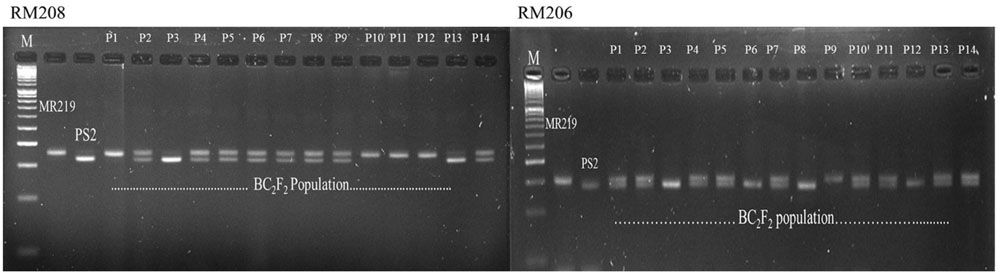
FIGURE 1. Genotyping of blast resistant improved homozygous lines using tightly linked marker RM208 and RM206.
Screening against Blast Disease in MR219 and Pongsu Seribu 2
Pongsu Seribu 2, the donor parent having Pi genes, expressed a strong spectrum of resistance against pathotype P7.2 with a score of 0-1 while the recurrent parent showed susceptibility with a score of 5-9 (Figure 2). The blast disease reaction is shown in Supplementry Figure S2.
Screening against Blast Disease in Improved Blast Resistant Lines of the BC2F2 Population
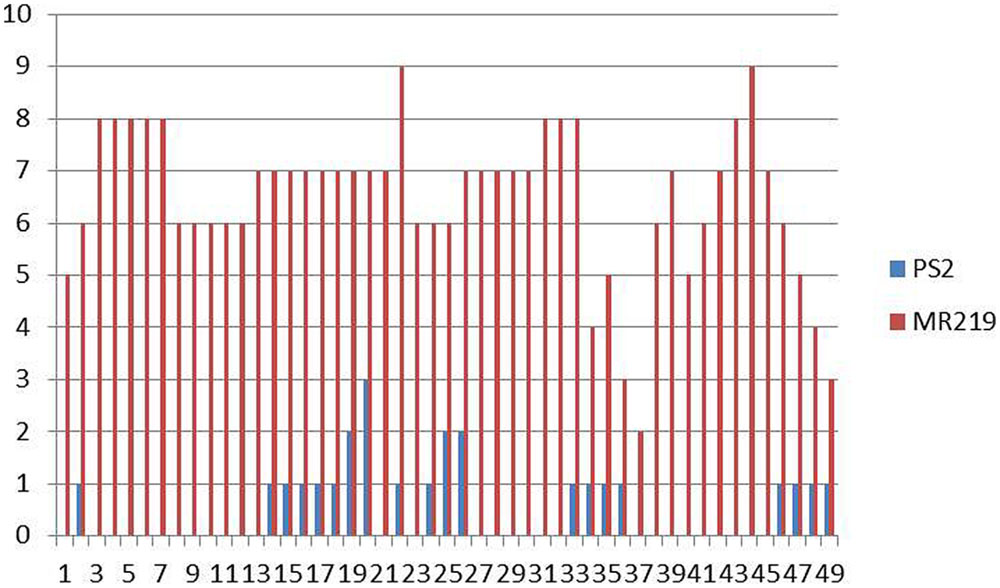
The advance 15 improved MR219 lines carrying blast resistance genes produced from both of these parents, 5-3-7-1, 5-3-7-4, 5-3-7-13, 5-3-7-19, 5-3-7-24, 5-3-7-29, 5-3-7-31, 5-3-7-36, 5-3-7-40, 5-3-7-69, 5-3-17-2, 5-3-17-4, 5-3-17-11, 5-3-17-19, 5-3-17-21, showed a great magnitude of resistance with a score of 0-1. The distribution of the BLD treated with M. oryzae pathotype P7.2 in the parental lines and advanced improved lines with introgressed blast resistant genes of the BC2F2 populations is summarized in Figure 3.
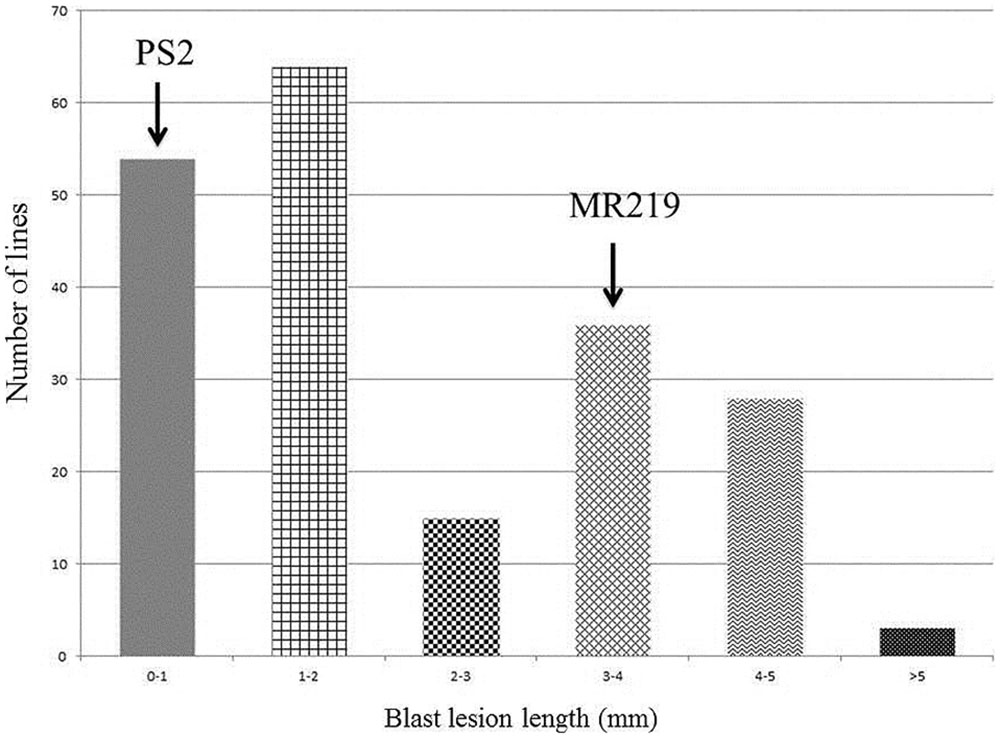
FIGURE 3. Calculation of the distribution of blast lesion length after inoculation with Magnaporthe oryzae pathotype P7.2 in parental lines along with improved introgressed blast resistant gene lines of the BC2F2 population (n = 200).
Assessment of the Phenotypic Segregation of Blast Resistant versus Susceptible Plants
A chi-square test was performed to evaluate the blast disease in the BC2F2 families. Different models, i.e., the single-gene model, two independent gene model and interaction of two different loci (Epistasis) were tested to assess which model population fit to the expected ratio. The number of expected resistant and susceptible plants for the phenotypic segregation ratio was not significantly different from the number of the observed resistant and susceptible plants and followed the expected Mendelian (3:1) ratio (Table 2).
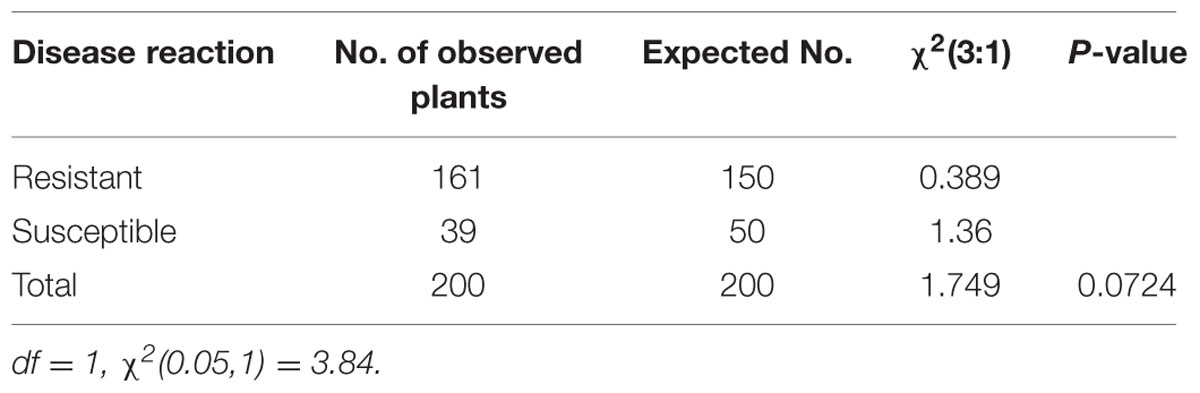
TABLE 2. Phenotypic segregation ratio of observed and expected number of resistant and susceptible plants in the BC2F2 population inoculated with highly virulent pathotype P7.2 of Magnaporthe oryzae.
The BC2F2 population did not follow the two-gene model. Phenotypic disease segregation of the BC2F2 population did not show a good fit to the expected 9:3:3:1 ratio (Table 3). The present results do not support the idea of the two-gene model, thus indicating that resistance to blast in the BC2F2 generation was not regulated by two different genes. Similarly, the chi-square value for an epistatic effect of the resistant versus susceptible plants does not segregate into 15:1 (Table 3) for the BC2F2 population, therefore the epistatic/two locus interaction was absent.
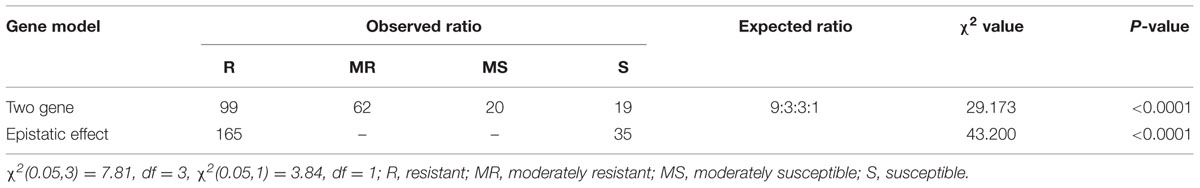
TABLE 3. Chi-square test for independent gene model (9:3:3:1) and epistatic effect (15:1) for blast resistance in BC2F2 population inoculated with pathotype P7.2 of Magnaporthe oryzae.
Marker-trait Association
Marker-trait association was analyzed by using SAS 9.3 software to identify the association among the resistance component, i.e., BLD, BLT and %DLA, with tightly linked polymorphic markers of corresponding blast resistance genes. Data of genotypic segregation of linked SSR markers obtained from the BC2F2 population were combined with phenotypic segregation data of the BC2F2 population for blast resistance traits. The data were interrogated to determine the significance level and linear model regression analysis for association between the marker genotypes and resistance component traits. The markers RM208 and RM206 showed significant association with BLD, percentage diseased leaf area (%DLA) and BLT with a simple linear regression (R2) value of more than 10 (Table 4).
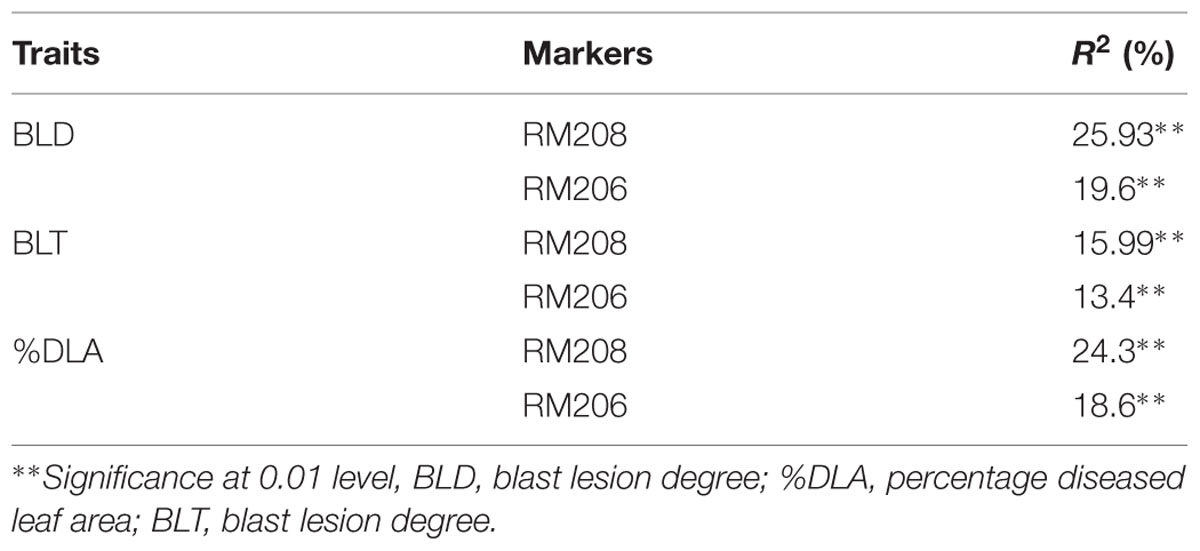
TABLE 4. Association between the marker and trait in the BC2F2 population analysed by regression analysis.
Variation and Correlation among Traits
The trait variations (means of the parents and BC2F2 population) for pathotype P7.2 are shown in Table 5. The average leaf blast disease severity score for recurrent parent MR219 was 6.63 for BLD, 3.43 for BLT and 61.43% for DLA. For the donor parent Pongsu Seribu 2 cultivar, the leaf blast disease severity score was 1.92 for BLD, 2.34 for BLT, and 12.4% for DLA. The parental cultivar showed a significantly different (P < 0.01) leaf blast resistance. In the BC2F2 population, the score for BLD, BLT, and DLA was 4.48, 2.5, and 47.18% with standard deviations of 2.24, 1.17, and 22.54, respectively. The selected best 15 lines from the BC2F2 population showed strong resistance against leaf blast for specific pathotype P7.2. The mean of disease severity of the selected improved lines for BLD, BLT, and DLA was 0.97, 0.88, and 3.96%. The disease severity showed a strong correlation among resistance components BLD, BLT, and %DLA in the BC2F2 families (Table 6).
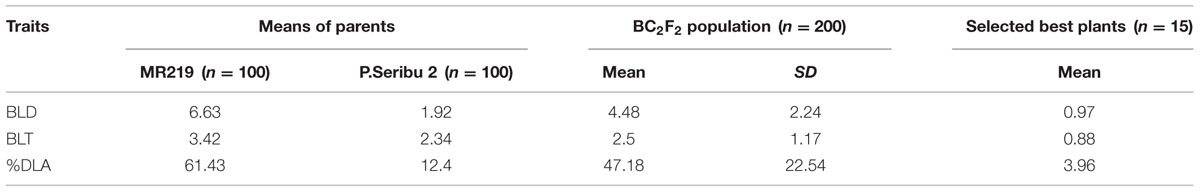
TABLE 5. Trait variation for selected pathotype P7.2 of Magnaporthe oryzae inoculated in BC2F2 population.
Recovery of the Recurrent Parent Genome in Selected Improved Homozygous Lines
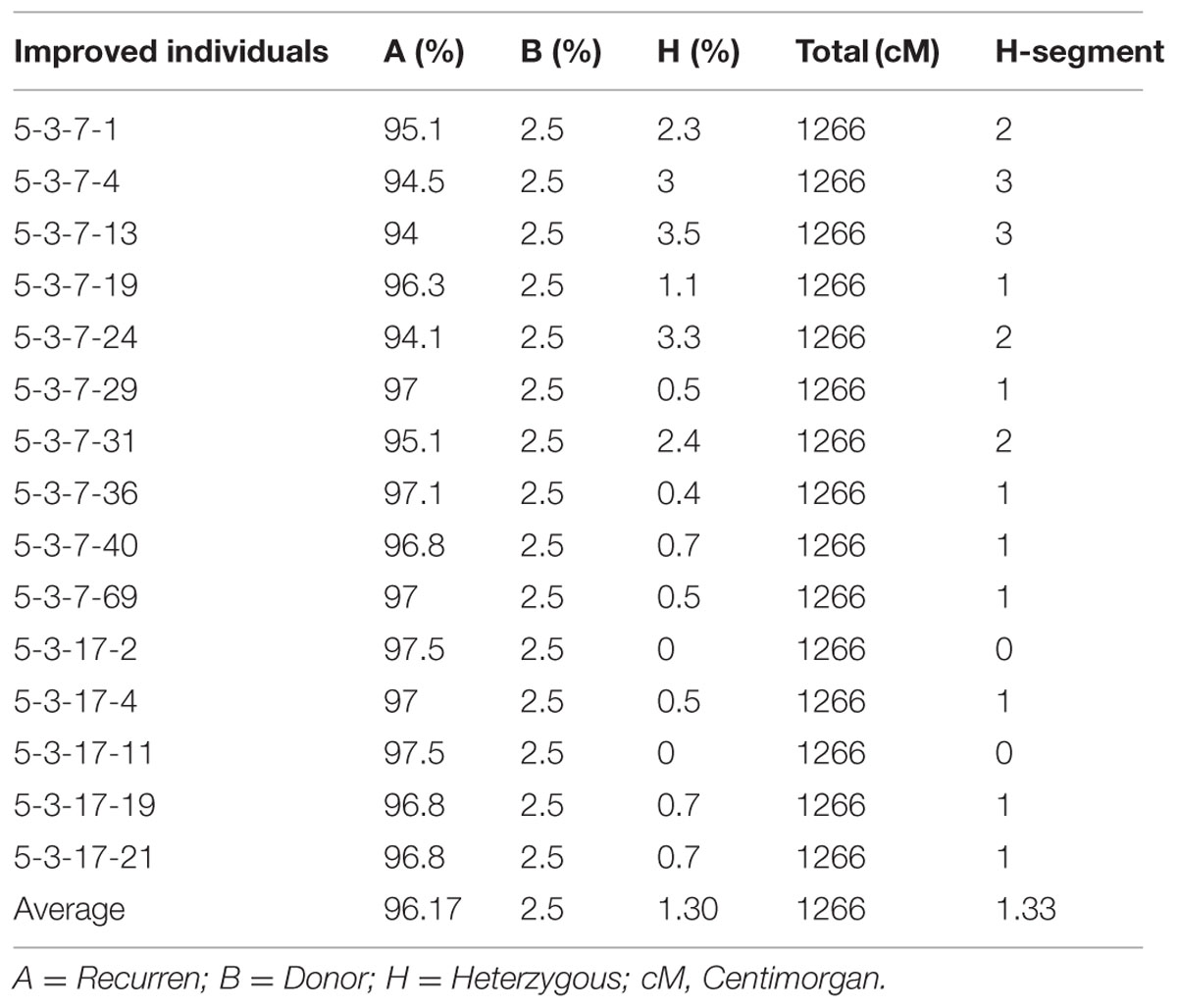
A total of 72 markers were used for background and the selection of improved blast resistant lines, and a genetic map was constructed covering 1266 cM with an average marker distance of 15.91 regions of the whole genome of Oryza sativa. A graphical representation of the carried chromosome 2 (putative Pi-b) and chromosome 6 (putative Pi-kh) of the selected improved blast resistance lines is shown in Figure 4. The minimum recovery of the recurrent parent genome in an improved lined was 94% and the maximum recovery in an improved line was 97.5% (Figure 5). Most of the residual segments from donor genome content were distributed on chromosomes 4, 9, and 10; however, other chromosomes were completely recovered. The percentage of chromosome segments derived from Pongsu Seribu 2 was 2.5% and remained constant in all of the advanced improved lines. The average proportions of the recurrent parent genome in all 15 improved lines were 96.17%, showing the maximum similarity observed at the phenotypic level with the recurrent parent (Table 7).
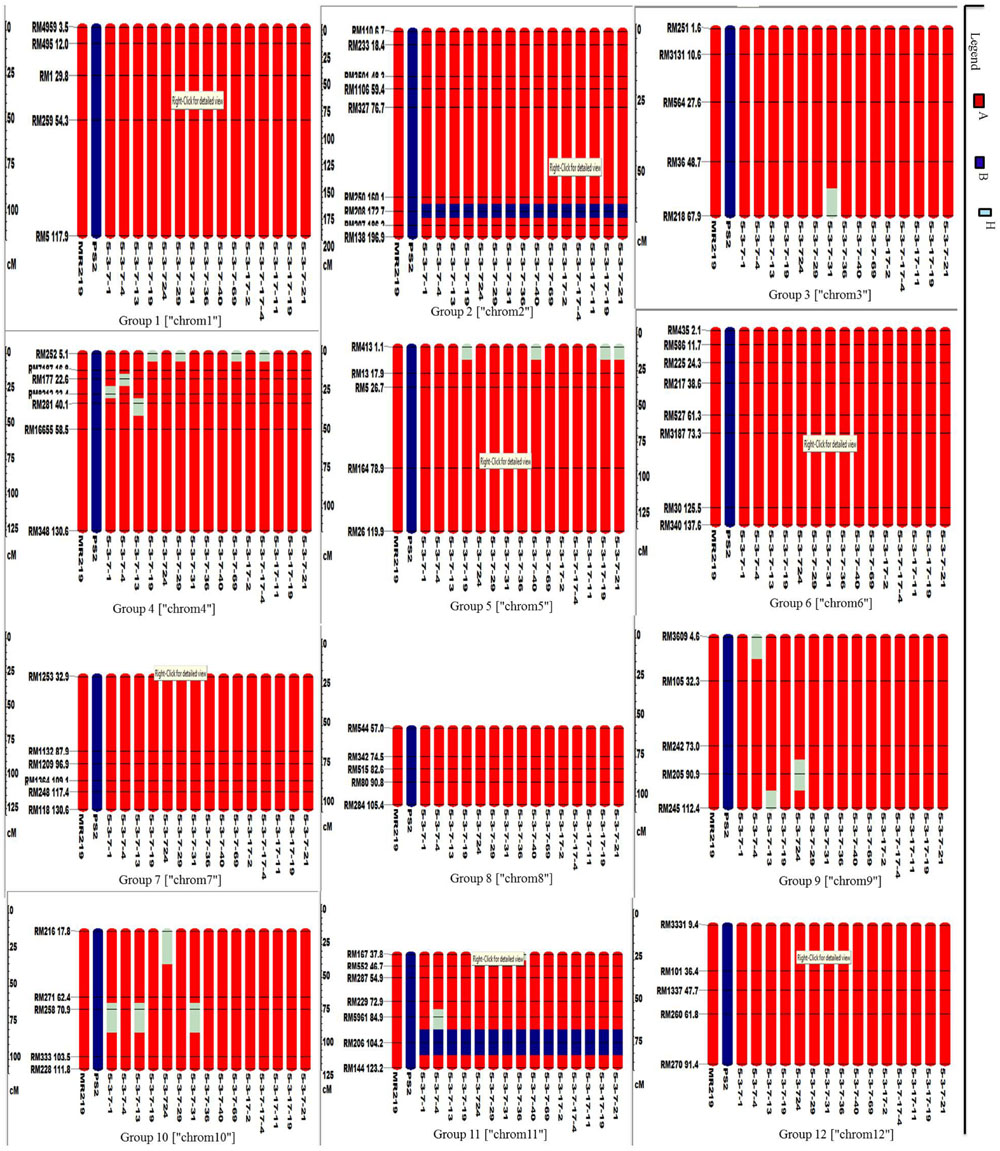
FIGURE 4. Graphical genotyping of selected 15 improved lines with introgressed Pi genes along with MR219 background developed in this study. The red color indicates homozygous regions for MR219, the blue color indicates homozygous regions for Pongsu Seribu 2 and the light green color indicates heterozygous regions.
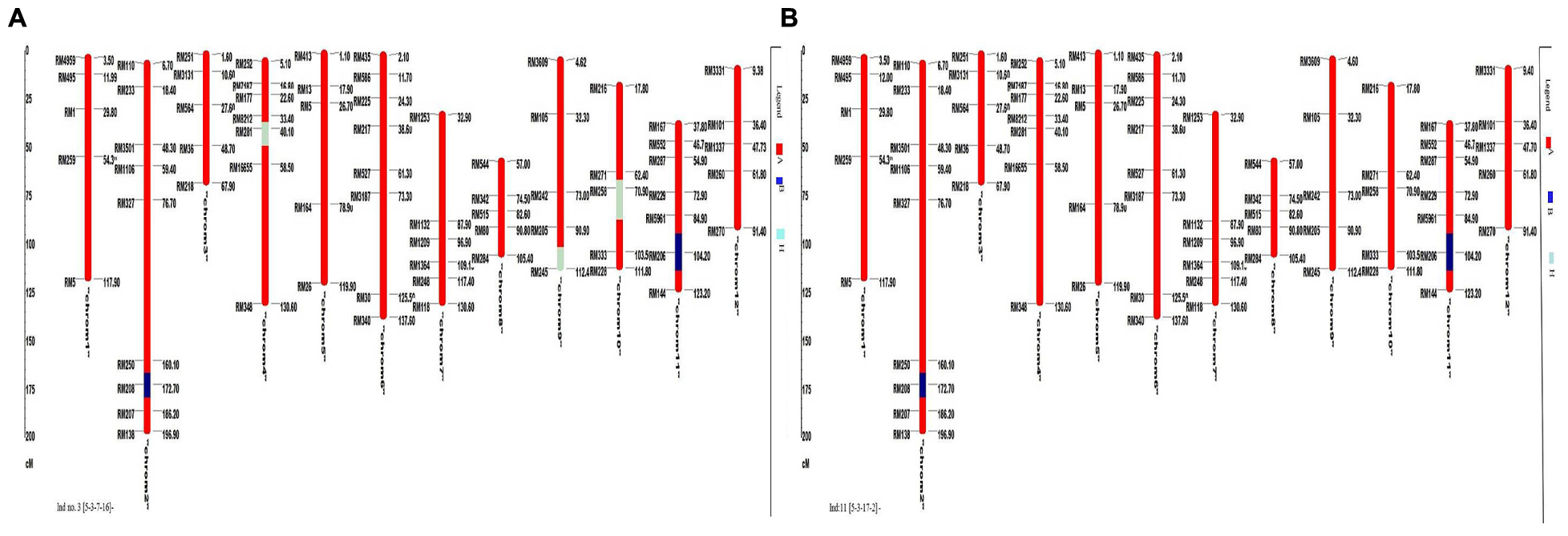
FIGURE 5. Graphical genotyping of the improved lines with the lowest (A) and highest (B) recovery among the best 15 improved lines. The red color indicates homozygous regions for MR219, the blue color indicates homozygous regions for Pongsu Seribu 2 and the light green color indicates heterozygous regions.
Comparison of Agro-morphological Performance of Improved Lines versus Recurrent Parent MR219
Agro-morphological traits of the advanced improved lines carrying additional blast resistant genes were measured and compared with the recurrent parent lines of MR219 (Table 8). There was minor variation in the days to flowering and total grain per panicles. However, other improved lines showed mostly similar performance and there was no significant differences found for other traits, such as plant height, days to maturity, total tiller/hill, effective tiller/hill, panicle length, no. of filled grains/panicle, seed setting rate, 1000 grain weight, yield/plant, grain length, grain width, flag leaf length, and flag leaf width.
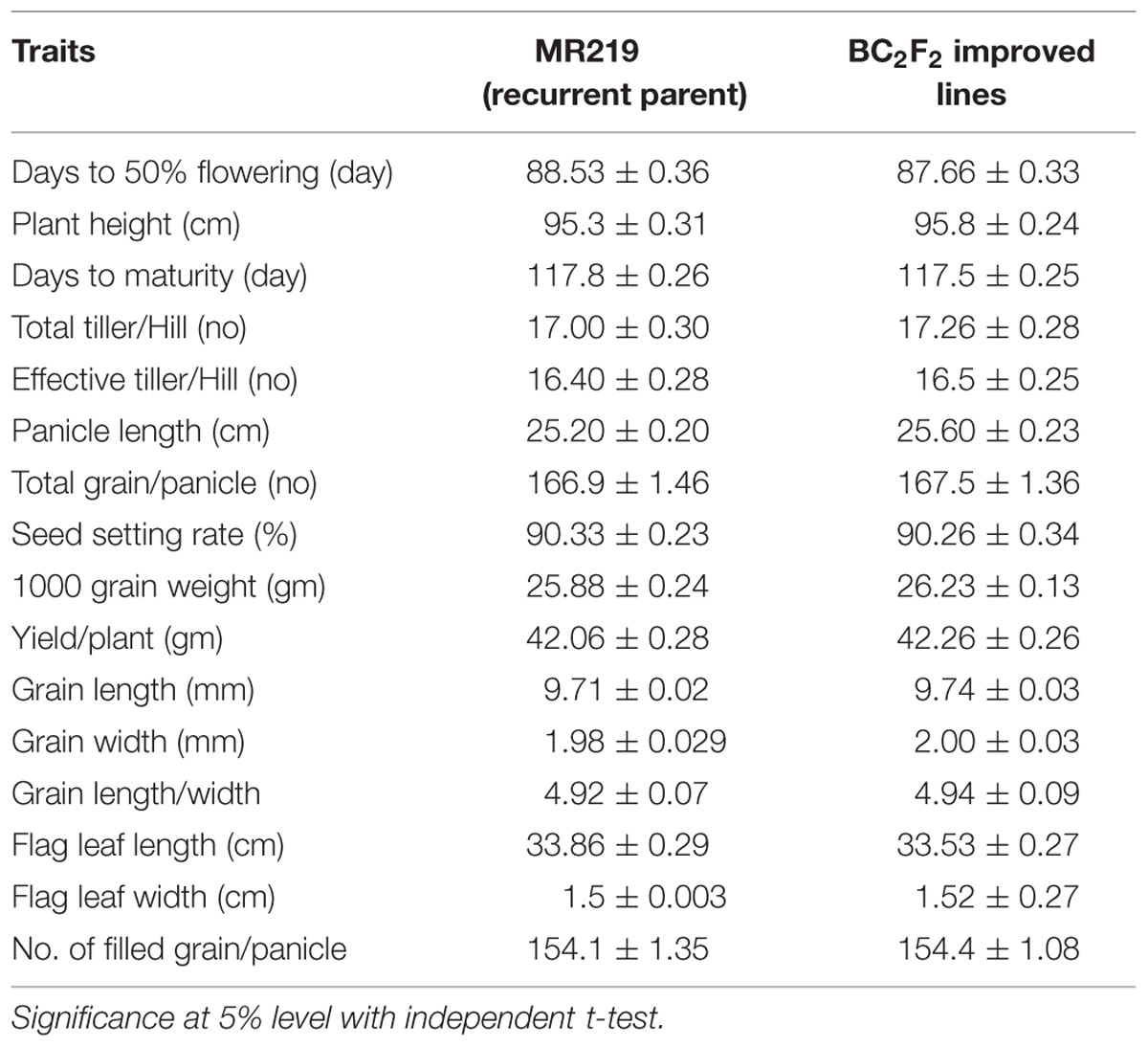
TABLE 8. Performance of major agronomic traits of BC2F2 improved lines carrying blast resistant (putative Pi-b and Pi-kh) gene.
Discussion
Rice production is always constrained by several biotic stresses, among which blast diseases impose both several yield and quality losses. These serious and most challenging issues could be overcome by utilizing resistance genes (Tabien et al., 2002). Pyramiding major resistance genes into elite rice cultivars with the application of conventional breeding were always hindered by environmental factors and the number of generations needed to achieve the goal. However, marker-assisted selection saves time and offers a very simple, efficient, and accurate method to improve the blast resistance of elite genotypes (Singh et al., 2012). The linkage drag can be minimized within a few generations, and the recurrent parent genotype can be stored easily with additional genes of interest (Joseph et al., 2004; Shanti et al., 2010). However, in the backcross breeding program, the choice of the recurrent parent plays a vital role (Ye and Smith, 2010). Blast resistance provided by identified genes is always race specific against the pathotype. Ultimately, the resistance of most of the varieties is lost due to variability of pathogenicity of the pathotype. The blast pathotypes of M. oryzae are able to change their virulence according to the environment. Therefore, the identification of more closely linked markers with blast resistance genes can help to introgress identified genes into improved cultivars through marker-assisted selection. The closely linked marker helps to monitor blast resistance genes for several generations (Jena and Mackill, 2008). Breeders have reduced the yield loss due to blast disease by introgressing the beneficial alleles from the wild rice genotype into elite high yielding rice cultivars (Brar and Khush, 1997). DNA marker technology has greatly facilitated the tagging of novel resistance genes from wild rice species and provides a straight forward way to identify and transfer the major genes from unadapted germplasm to adapted germplasm (Gu et al., 2013).
The present research was conducted to improve the blast resistance of the elite Malaysia rice variety MR219 through a MABC breeding approach along with phenotypic selection for agro-morphological traits. From previous studies, the IR64 rice variety was improved for blast resistance coupled with phenotypic selection for agro-morphological traits similar to our study (Sreewongchai et al., 2010). By using the MABC strategy, improved versions of the elite Basmati variety, Pusa Basmati 1, Pusa RH10 and KMR-3R were also released for bacterial blight resistance (Gopalakrishnan et al., 2008; Basavaraj et al., 2010; Hari et al., 2011). Narayanan et al. (2002) introgressed Piz-5 blast resistance gene into rice cultivar IR50 and improved the blast resistance. Singh et al. (2012) introgressed the blast resistance gene Pi-54 (previously known as Pi-kh) into Pusa Basmati 1 from donor parent Tetep. This is first report in Malaysia documenting the stacking of two major genes (Pi-kh and Pi-b) in elite rice cultivar MR219 through MABC breeding coupled with phenotypic selection for agro-morphological traits.
For PCR-based DNA markers used in the present study, RM208 tightly linked with the Pi-b gene (Wang et al., 1999; Roychowdhury et al., 2012) and RM206 tightly linked with the Pi-kh gene (Sharma et al., 2005; Singh et al., 2012; Hari et al., 2013). The RM208 marker presents on chromosome 2 and RM206 on chromosome 6 below the centromere. Both of these markers are highly polymorphic and can be detected very easily and therefore have great potential to serve as an important tool to introgress Pi-b and Pi-kh blast resistant genes into blast susceptible rice varieties. The importance and benefit of using tightly linked markers for gene pyramiding have been discussed earlier by Hittalmani et al. (2000) and Hayashi et al. (2006) for blast disease screening. However, the success of marker-assisted selection heavily depends upon the strong linkage between the marker and target gene. Thus, from the blast disease screening results, 15 best selected lines, 5-3-7-1, 5-3-7-4, 5-3-7-13, 5-3-7-19, 5-3-7-24, 5-3-7-29, 5-3-7-31, 5-3-7-36, 5-3-7-40, 5-3-7-69, 5-3-17-2, 5-3-17-4, 5-3-17-11, 5-3-17-19, 5-3-17-21, showed strong resistance against virulent pathotype P7.2 similar to the donor parent. Among the introgressed lines, BC2F2 with genes Pi-b and Pi-kh showed high resistance at both locations. The results of the phenotypic screening against blast disease reaction of the improved lines carrying the putative Pi-b and Pi-kh genes with a background of the recurrent parent MR219 conferred complete resistance to the highly virulent pathotype P7.2, indicating the strong bond between these markers with the trait.
Seventy-two polymorphic SSR markers between parental lines with at least five markers per chromosome were used for genetic background selection. Most of the recurrent parent segments were fully recovered in improved lines, but in some improved lines, some chromosomes were not recovered. Some heterozygous segments were found in some improved lines. The results are consistent with the finding of Tian et al. (2006) and Hirabayashi et al. (2010), who described that some regions of the BC2F2 generation may not be fully recovered if the marker-assisted selection is not performed until the BC3F2 generations. Some other biological factors may also be involved, such as gametophyte, heading date and hybrid sterility (Doi et al., 1997). Recurrent parent phenotype can be recovered in one or two backcrosses if more than one resistance gene is transferred from indica to japonica cultivars (Singh et al., 2001; Rajpurohit et al., 2011). Similarly, Shu (2009) transferred multiple resistance genes, such as Xa4, xa5 and Xa21, from the indicia cultivar to the japonica cultivar for BB resistance and mentioned that at least three backcrosses are required to recover the recurrent parent phenotype. In this study, a similar approach was adopted for foreground and background selection for higher recovery of the background genotype and introgression of target genes in the indica/indica cultivar in two backcrossed and one self-generation. This strategy is very effective in minimizing the cost and time required to recover the desirable recombinants to a considerable extent with target resistance genes in the indica/indica crosses.
The donor parent Pongsu Seribu 2 and the recurrent parent MR219 showed significantly different agro-morphological traits. However, in the blast resistant, improved lines of MR219, no apparent yield penalty was related with the presence of the blast resistance genes, and putative Pi-b and Pi-kh were observed. Therefore, the cultivation of our improved blast resistant lines would be of great advantage to reduce the yield losses in blast disease endemic areas. The introgression of blast resistance genes along with nearly complete recovery of the genome of the recurrent parents in improved advanced MR219 lines and yield is the greatest achievement of the current research. Yield and grain quality traits are multigenically encoded by loci that are distributed throughout the rice genome (Sundaram et al., 2008). In this study, a higher recovery of desirable improved plants of MR219 was obtained because of phenotypic-based selection for agro-morphological traits from the BC1F1 generation onward and screening of a quantifiable number of BC plants. The current strategies of phenotypic-based along with marker-based selection are consistent with results of Joseph et al. (2004) and Gopalakrishnan et al. (2008), who adopted the phenotypic-based selection for grain type and molecular-based for the target trait (i.e., bacterial blight and blast resistance). Improved blast resistant MR219 lines showed a similar agro-morphological performance in the field as a par recurrent parent MR219 with a minor acceptable difference. The mean value of blast resistance lines (carrying the Pi-b and Pi-kh genes) for all morphological characters were mostly similar with the recipient parent MR219, indicating that the performance of introgression lines is similar with MR219 for such traits. The present results strongly support that our phenotypic selection practice was efficient. These results are also consistent with the finding of Yoshimura et al. (1995) and Steele et al. (2006) who found that the offspring of resistant parental lines shows a similar or better level of resistance and has preferable quality and yield characteristics for further selection. Considering the agro-morphological traits, there was a significant difference for days of 50% flowering; some lines took the same time as MR219 and some lines were delayed in flowering. In MR219, flowering was significantly earlier under the proper irrigation (Rahim et al., 2012). The late flowering in some introgression lines was due to donor parent Pongsu Seribu 2, which takes more time compared to MR219. For the grain yield per plant, there was not any significant difference among the parental line and integration line. The present finding is similar to Sabu et al. (2006), who also did not find any significant difference in grain yield of parent lines and advanced backcrossed lines. The entire advanced breeding line (ABL) grain characteristics were similar to the recurrent parent (MR219). The number of panicles depends on the effective tillers number; if there are more effective tillers, there will be more panicles (Hossain et al., 2008). Biswas et al. (1998) also studied the genotypic difference of grain yield and reported that higher grain yield depended on the number of effective tillers per hill and number of grains per panicle. Shi et al. (2000) also reported that the exterior quality of the rice grain depends on the grain length and width. The proportion of the grain length to width in all backcross introgression lines and MR219 was a slender grain shape. According to Rafii et al. (2014), high grain length with low grain width could lead to a long shaped grain. Grain shape is controlled by triploid endosperm, cytoplasmic and maternal genes.
Until now, most of the breeders have introgressed a single major gene into blast susceptible varieties. The high level of instability in the pathogen genome could lead to break-down of the resistance based on the single gene (Hittalmani et al., 2000). The best way to sustain the resistance for long term is the incorporation of partial resistance or combining putative QTL or incorporating multiple genes that decrease the selection pressure on the pathogen, thus resistance remains for a long time (Bonman, 1992; Tabien et al., 2002; Lopez-Gerena, 2006). However, pyramiding major resistance genes into a single cultivar will be effective for a particular set of virulent pathotypes (Hittalmani et al., 2000). Ultimately, rice cultivar with durable resistance by accumulating major genes and QTL for partial resistance against M. oryzae is an ideal strategy to control blast disease.
Conclusion
The present study suggests that DNA markers for blast resistance (putative Pi-b and Pi-kh) genes are reliable for marker-assisted selection of blast resistance in rice breeding. The recovery of the recurrent parent along with the intogression of blast resistance enes with MABC breeding was much faster than that with conventional breeding. Fifteen improved blast resistance lines were produced from a backcross between the parental line MR219 and Pongsu Seribu 2. These improved blast resistant lines could be utilized as a source of genetic material for blast resistance with a high yielding background of MR219. The introgressed resistant genes Pi-b and Pi-kh are dominant blast resistance genes; therefore, the resistance in improved blast resistant lines will remain for long periods, thus enhancing the food security in Malaysia. These improved blast resistant lines have a practical breeding value without yield penalty by providing blast resistance against the highly virulent pathotype P7.2 that exists in Malaysia. Identifying the most resistant lines will lead to durable resistant rice varieties and serve as a source of genetic resistance in the rice germplasm, which will have a great impact on the rice yield sustainability and stability. To our knowledge, this is the first report on the successful introgression of blast resistant genes (Pi-b and Pi-kh) into the elite high yielding rice cultivar MR219 in Malaysia.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Long term Research Grant Scheme (LRGS), Food Security Project, Ministry of Higher Education, Malaysia, for the financial support to conduct research on rice breeding. The authors would also like to acknowledge Sindh Agriculture University Tandojam Sindh Pakistan for providing financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01002
References
Ballini, E., Nguyen, T. T., and Morel, J. B. (2013). Diversity and genetics of nitrogen-induced susceptibility to the blast fungus in rice and wheat. Rice 6, 32. doi: 10.1186/1939-8433-6-32
Basavaraj, S., Singh, V. K., Singh, A., Singh, A., Singh, A., Anand, D., et al. (2010). Marker-assisted improvement of bacterial blight resistance in parental lines of Pusa RH10, a superfine grain aromatic rice hybrid. Mol. Breeding 26, 293–305. doi: 10.1007/s11032-010-9407-3
Biswas, J. K., Hossain, M. A., Sarkar, B. C., Hassan, M., and Haque, M. Z. (1998). Yield performance of several rice varieties seeded directly as a late crop. Bangladesh J. Life Sci. 10, 47–52.
Bonman, J. (1992). “Durable resistance to rice blast disease-environmental influences,” in Breeding for Disease Resistance, eds R. Johnson and G. J. Jellis (Berlin: Springer), 115–123.
Brar, D. S., and Khush, G. S. (1997). Alien introgression in rice. Plant. Mol. Biol. 35–47. doi: 10.1023/A:1005825519998
Collard, B. C., and Mackill, D. J. (2008). Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philos. Trans. R. Soc. B Biol. Sci. 363, 557–572. doi: 10.1098/rstb.2007.2170
Collard, B. C., Vera Cruz, C. M., McNally, K. L., Virk, P. S., and Mackill, D. J. (2008). Rice molecular breeding laboratories in the genomics era: current status and future considerations. Int. J. Genomics 2008, 1.
Conaway-Bormans, C. A., Marchetti, M. A., Johnson, C. W., McClung, A. M., and Park, W. D. (2003). Molecular markers linked to the blast resistance gene Pi-z in rice for use in marker-assisted selection. Theor. Appl. Genet. 107, 1014–1020. doi: 10.1007/s00122-003-1338-5
Correa-Victoria, F. J., and Zeigler, R. S. (1993). Pathogenic variability in Pyricularia grisae at a rice blast hot spot breeding site in Eastern Colombia. Plant Dis. 77, 1029–1035. doi: 10.1094/PD-77-1029
Divya, B., Robin, S., Rabindran, R., Senthil, S., Raveendran, M., and Joel, A. J. (2014). Marker assisted backcross breeding approach to improve blast resistance in Indian rice (Oryza sativa) variety ADT43. Euphytica 200, 61–77. doi: 10.1007/s10681-014-1182-5
Doi, K., Iwata, N., and Yoshimura, A. (1997). The construction of chromosome introgression lines of African (Oryza glaberrima Steud.) in the background of Japonica (O. sativa L.). Rice Genet. Newslet. 1, 39–41.
Fu, C., Wu, T., Liu, W., Wang, F., Li, J., Zhu, X., et al. (2012). Genetic improvement of resistance to blast and bacterial blight of the elite maintainer line Rongfeng B in hybrid rice (Oryza sativa L.) by using marker-assisted selection. Af. J. Biotechnol. 11, 13104–13114.
Gopalakrishnan, S., Sharma, R. K., Rajkumar, K. A., Joseph, M., Singh, V. P., Singh, A. K., et al. (2008). Integrating marker assisted background analysis with foreground selection for identification of superior bacterial blight resistant recombinants in Basmati rice. Plant Breeding 127, 131–139. doi: 10.1111/j.1439-0523.2007.01458.x
Gu, J.-F., Qiu, M., and Yang, J.-C. (2013). Enhanced tolerance to drought in transgenic rice plants overexpressing C4 photosynthesis enzymes. Crop J. 1, 105–114. doi: 10.1016/j.cj.2013.10.002
Hari, Y., Srinivasarao, K., Viraktamath, B. C., Hariprasad, A. S., Laha, G. S., Ahmed, M. I., et al. (2011). Marker-assisted improvement of a stable restorer line, KMR-3R and its derived hybrid KRH2 for bacterial blight resistance and grain quality. Plant Breeding 130, 608–616.
Hari, Y., Srinivasarao, K., Viraktamath, B. C., Prasad, H., Arremsetty, S., Laha, G. S., et al. (2013). Marker-assisted introgression of bacterial blight and blast resistance into IR 58025B, an elite maintainer line of rice. Plant Breeding 132, 586–594. doi: 10.1111/pbr.12056
Hayashi, K., Yoshida, H., and Ashikawa, I. (2006). Development of PCR-based allele-specific and InDel marker sets for nine rice blast resistance genes. Theor. Appl. Genet. 113, 251–260. doi: 10.1007/s00122-006-0290-6
Hirabayashi, H., Sato, H., Nonoue, Y., Kuno-Takemoto, Y., Takeuchi, Y., Kato, H., et al. (2010). Development of introgression lines derived from Oryza rufipogon and O. glumaepatula in the genetic background of japonica cultivated rice (O. sativa L.) and evaluation of resistance to rice blast. Breeding Sci. 60, 604–612. doi: 10.1270/jsbbs.60.604
Hittalmani, S., Parco, A., Mew, T., Zeigler, R., and Huang, N. (2000). Fine mapping and DNA marker-assisted pyramiding of the three major genes for blast resistance in rice. Theor. Appl. Genet. 100, 1121–1128. doi: 10.1007/s001220051395
Hossain, M. B., Islam, M. O., and Hasanuzzaman, M. (2008). Influence of different nitrogen levels on the performance of four aromatic rice varieties. Int. J. Agri. Biol. 10, 693–696.
Huyen, L. T. N., Cuc, L. M., Ismail, A. M., and Ham, L. H. (2012). Introgression the salinity tolerance QTLs Saltol into AS996, the Elite rice variety of Vietnam. Am. J. Plant Sci. 3, 981–987. doi: 10.4236/ajps.2012.37116
IRRI (1996). Standard Evaluation System for Rice, 4th Edn. Manila: The International Network for Genetic Evaluation of Rice, Genetic Resources Center, International Rice Research Institute, 52.
Jena, K., and Mackill, D. (2008). Molecular markers and their use in marker-assisted selection in rice. Crop Sci. 48, 1266–1276. doi: 10.2135/cropsci2008.02.0082
Joseph, M., Gopalakrishnan, S., Sharma, R., Singh, V., Singh, A., Singh, N., et al. (2004). Combining bacterial blight resistance and Basmati quality characteristics by phenotypic and molecular marker-assisted selection in rice. Mol. Breeding 13, 377–387. doi: 10.1023/B:MOLB.0000034093.63593.4c
Khush, G. S. (2013). Strategies for increasing the yield potential of cereals: case of rice as an example. Plant Breeding 132, 433–436.
Koide, Y., Kobayashi, N., Xu, D., and Fukuta, Y. (2009). Resistance genes and selection DNA markers for blast disease in rice (Oryza sativa L.). Jpn. Agri. Res. Q. 43, 255–280. doi: 10.1016/j.crvi.2014.11.003
Kushibuchi, K. (1997). “Historical changes in rice cultivars,” in Science of the Rice Plant, Vol. 3, eds T. Matsuo, Y. Futsuhara, F. Kikuchi, and H. Yamaguchi (Tokyo: Food and Agriculture Policy Research Center), 837–875.
Lang, N. T., Tao, N., and Buu, B. C. (2011). Marker-assisted backcrossing (MAB) for rice submergence tolerance in Mekong delta. Omonrice 18, 11–21.
Lopez-Gerena, L. (2006). Mapping QTL Controlling Durable Resistance to Rice Blast in Cultivar Oryza llanos 5. Doctoral dissertation, Kanas State University, Manhattan, KS.
Mackill, D., and Bonman, J. (1992). Inheritance of blast resistance in near-isogenic lines of rice. Phytopathology 82, 746–749. doi: 10.1094/Phyto-82-746
McCouch, S. R., Teytelman, L., Xu, Y., Lobos, K. B., Clare, K., Walton, M., et al. (2002). Development and mapping of 2240 new SSR markers for rice (Oryza sativa L.). DNA Res. 9, 199–207. doi: 10.1093/dnares/9.6.199
Narayanan, N. N., Baisakh, N., Vera Cruz, N., Gnananmanickam, S. S., Datta, K., and Datta, S. K. (2002). Molecular breeding for the development of blast and bacterial blight resistance in rice cv. IR50. Crop Sci. 42, 2072–2079. doi: 10.2135/cropsci2002.2072
Rafii, M. Y., Zakiah, M. Z., Asfaliza, R., Haifaa, I., Latif, M. I., and Malek, M. A. (2014). Grain quality performance and heritability estimation in selected F1 rice genotypes. Sains Malaysiana. 43, 1–7.
Ragimekula, N., Varadarajula, N. N., Mallapuram, S. P., Gangimeni, G., Reddy, R. K., and Kondreddy, H. R. (2013). Marker assisted selection in disease resistance breeding. J. Plant Breed. Genet. 1, 90–109.
Rahim, H. A., Zarifth, S. K., Bhuiyan, M. A. R., Narimah, M. K., Wickneswari, R., Abdullah, M. Z., et al. (2012). “Evaluation and characterization of advanced rice mutant line of rice (Oryza sativa), MR219-4 and MR219-9 under drought condition,” in Proceedings of the Research and Development Seminar, 26–28 September 2012, Vol. 44, Bangi, 1–15.
Rajpurohit, D., Kumar, R., Kumar, M., Paul, P., Awasthi, A., Basha, P., et al. (2011). Pyramiding of two bacterial blight resistance and a semidwarfing gene in Type 3 Basmati using marker-assisted selection. Euphytica 178, 111–126. doi: 10.1007/s10681-010-0279-8
Roychowdhury, M., Jia, Y., Jia, M., Fjellstrom, R., and Cartwright, R. (2012). Identification of the rice blast resistance gene Pi-b in the national small grains collection. Phytopathology 102, 700–706. doi: 10.1094/PHYTO-02-12-0026-R
Sabu, K. K., Abdullah, M. Z., Lim, L. S., and Wickneswari, R. (2006). Development and evaluation of advanced backcross families of rice for agronomically important traits. Com. Biom. Crop Sci 1, 111–123.
Shanti, M., Shenoy, V., Devi, G. L., Kumar, V. M., Premalatha, P., Kumar, G. N., et al. (2010). Marker-assisted breeding for resistance to bacterial leaf blight in popular cultivar and parental lines of hybrid rice. J. Plant Pathol. 92, 495–501.
Sharma, T. R., Madhav, M. S., Singh, B. K., Shanker, P., Jana, T. K., Dalal, V., et al. (2005). High resolution mapping, cloning and molecular characterization of the Pi-kh gene of rice, which confers resistance to M. grisea. Mol. Genet. Genom. 274, 569–578. doi: 10.1007/s00438-005-0035-2
Shi, C., Zhu, J., Wu, J., and Fan, L. (2000). Genetic and genotype × environment interaction effects from embryo, endosperm, cytoplasm and maternal plant for rice grain shape traits of indica rice. Field Crop Res. 68, 191–198. doi: 10.1016/S0378-4290(00)00124-6
Shinoda, H., Toriyama, K., Yunoki, T., Ezuka, A., and Sakurai, Y. (1971). Studies on the varietal resistance of rice to blast. 6. Linkage relationship of blast resistance genes. Bull. Chugoku Agric. Exp. Stn. Ser. 20, 1–25. [In Japanese].
Shu, Q. Y. (2009). “Induced plant mutations in the genomics era,” in Proceedings of an International Joint FAO/IAEA Symposium (Vienna: FAO).
Singh, S., Sidhu, J. S., Huang, N., Vikal, Y., Li, Z., Brar, D. S., et al. (2001). Pyramiding three bacterial blight resistance genes (xa5, xa13 and Xa21) using marker-assisted selection into indica rice cultivar PR106. Theor. Appl. Genet. 102, 1011–1015. doi: 10.1007/s001220000495
Singh, V. K., Singh, A., Singh, S., Ellur, R. K., Choudhary, V., Sarkel, S., et al. (2012). Incorporation of blast resistance into “PRR78”, an elite Basmati rice restorer line, through marker assisted backcross breeding. Field Crop Res. 128, 8–16. doi: 10.1016/j.fcr.2011.12.003
Singh, V. K., Singh, A., Singh, S. P., Ellur, R. K., Singh, D., Krishnan, S. G., et al. (2013). Marker-assisted simultaneous but stepwise backcross breeding for pyramiding blast resistance genes Piz5 and Pi54 into an elite Basmati rice restorer line ‘PRR78’. Plant Breeding 132, 486–495.
Sreewongchai, T., Toojinda, T., Thanintorn, N., Kosawang, C., Vanavichit, A., Tharreau, D., et al. (2010). Development of elite indica rice lines with wide spectrum of resistance to Thai blast isolates by pyramiding multiple resistance QTLs. Plant Breeding 129, 176–180. doi: 10.1111/j.1439-0523.2009.01669.x
Steele, K. A., Price, A. H., Shashidhar, H. E., and Witcombe, J. R. (2006). Marker-assisted selection to introgress rice QTLs controlling root traits into an Indian upland rice variety. Theor. Appl. Genet. 112, 208–221. doi: 10.1007/s00122-005-0110-4
Sundaram, R. M., Vishnupriya, M. R., Biradar, S. K., Laha, G. S., Reddy, G. A., Rani, N. S., et al. (2008). Marker assisted introgression of bacterial blight resistance in Samba Mahsuri, an elite indica rice variety. Euphytica 160, 411–422. doi: 10.1007/s10681-007-9564-6
Tabien, R., Li, Z., Paterson, A., Marchetti, M., Stansel, J., and Pinson, S. (2002). Mapping QTLs for field resistance to the rice blast pathogen and evaluating their individual and combined utility in improved varieties. Theor. Appl. Genet. 105, 313–324. doi: 10.1007/s00122-002-0940-2
Tanweer, F. A., Rafii, M. Y., Sijam, K., Rahim, H. A., Ahmed, F., Ashkani, S., et al. (2015a). Identification of suitable segregating SSR markers for blast resistance in rice using inheritance and disease reaction analysis in backcross families. Austral. Plant Pathol. 44, 619–627. doi: 10.1007/s13313-015-0380-5ă
Tanweer, F. A., Rafii, M. Y., Sijam, K., Rahim, H. A., Ahmed, F., and Latif, M. A. (2015b). Cloning and characterization of two major blast resistance genes Pi-b and Pi-kh from Malaysian rice variety Pongsu Seribu 2. Plant Omics 8, 257–263.
Tian, F., Li, D. J., Fu, Q., Zhu, Z. F., Fu, Y. C., Wang, X. K., et al. (2006). Construction of introgression lines carrying wild rice (Oryza rufipogon Griff.) segments in cultivated rice (Oryza sativa L.) background and characterization of introgressed segments associated with yield-related traits. Theor. Appl. Genet. 112, 570–580. doi: 10.1007/s00122-005-0165-2
Wang, Z. X., Yano, M., Yamanouchi, U., Iwamoto, M., Monna, L., Hayasaka, H., et al. (1999). The Pi-b gene for rice blast resistance belongs to the nucleotide binding and leucine-rich repeat class of plant disease resistance genes. Plant J. 19, 55–64. doi: 10.1046/j.1365-313X.1999.00498.x
Wen, S., and Gao, B. (2012). Introgressing blast resistant gene Pi-9(t) into elite rice restorer luhui17 by marker-assisted selection. Rice Genom. Genet. 2, 31–36.
Xu, Y., and Crouch, J. H. (2008). Marker-assisted selection in plant breeding: from publications to practice. Crop Sci. 48, 391–407. doi: 10.1007/s00122-015-2499-8
Yang, D., Zhang, Y., Zhu, Z., Chen, T., Zhao, Q., Yao, S., et al. (2013). Substitutional mapping the cooked rice elongation by using chromosome segment substitution lines in rice. Mol. Plant Breed. 4, 107–115.
Ye, G., and Smith, K. F. (2010). Marker-for assisted gene pyramiding cultivar development. Plant Breed. Rev. 33, 219–256.
Yoshimura, S., Yoshimura, A., Iwata, N., McCouch, S., Abenes, M. L., Baraoidan, M. R., et al. (1995). Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol. Breeding 1, 375–387. doi: 10.1007/BF01248415
Keywords: rice, blast resistance, gene pyramiding, MABC, marker-assisted backcrossing, biotic stress
Citation: Tanweer FA, Rafii MY, Sijam K, Rahim HA, Ahmed F, Ashkani S and Latif MA (2015) Introgression of Blast Resistance Genes (Putative Pi-b and Pi-kh) into Elite Rice Cultivar MR219 through Marker-Assisted Selection. Front. Plant Sci. 6:1002. doi: 10.3389/fpls.2015.01002
Received: 18 May 2015; Accepted: 30 October 2015;
Published: 17 December 2015.
Edited by:
Janila Pasupuleti, International Crops Research Institute for the Semi-Arid Tropics, IndiaReviewed by:
Manish Kumar Pandey, International Crops Research Institute for the Semi-Arid Tropics, IndiaAre Ashok Kumar, International Crops Research Institute for the Semi-Arid Tropics, India
Copyright © 2015 Tanweer, Rafii, Sijam, Rahim, Ahmed, Ashkani and Latif. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohd Y. Rafii, mrafii@upm.edu.my
 Fatah A. Tanweer1,2
Fatah A. Tanweer1,2 Mohd Y. Rafii
Mohd Y. Rafii Fahim Ahmed
Fahim Ahmed Sadegh Ashkani
Sadegh Ashkani