- 1College of Plant Protection, Shandong Agricultural University, Tai’an, China
- 2Beijing Key Laboratory of Environment Friendly Management on Fruit Diseases and Pests in North China, Institute of Plant and Environment Protection, Beijing Academy of Agriculture and Forestry Sciences, Beijing, China
- 3Phytotoxicity Research Department, Central Agricultural Pesticide Laboratory, Agricultural Research Center, Giza, Egypt
The insect chemosensory system plays an important role in many aspects of insects’ behaviors necessary for their survival. Despite the complexity of this system, an increasing number of studies have begun to understand its structure and function in different insect species. Nonetheless, the chemosensory system in the orange spiny whitefly Aleurocanthus spiniferus, as one of the most destructive insect pests of citrus in tropical Asia, has not been investigated yet. In this study, the sensillum types, morphologies and distributions of the male and female antennae of A. spiniferus were characterized using scanning electron microscopy. In both sexes, six different sensilla types were observed: trichodea sensilla, chaetica sensilla, microtrichia sensilla, coeloconic sensilla, basiconic sensilla, and finger-like sensilla. Moreover, we identified a total of 48 chemosensory genes, including 5 odorant-binding proteins (OBPs), 12 chemosensory proteins (CSPs), 3 sensory neuron membrane proteins (SNMPs), 6 odorant receptors (ORs), 8 gustatory receptors (GRs), and 14 ionotropic receptors (IRs) using transcriptome data analysis. Tissue-specific transcriptome analysis of these genes showed predominantly expression in the head (including antennae), whereas CSPs were broadly expressed in both head (including the antennae) and body tissue of adult A. spiniferus. In addition, the expression profiling of selected chemosensory genes at different developmental stages was examined by quantitative real time-PCR which was mapped to the transcriptome. We found that the majority of these genes were highly expressed in adults, while AspiORco, AspiGR1, AspiGR2, and AspiIR4 genes were only detected in the pupal stage. Together, this study provides a basis for future chemosensory and genomic studies in A. spiniferus and closely related species. Furthermore, this study not only provides insights for further research on the molecular mechanisms of A. spiniferus-plant interactions but also provides extensive potential targets for pest control.
Introduction
In insects, the chemosensory system is extremely critical for detecting and discriminating specific chemical signals in the environment necessary for their survival and reproduction (Hallem et al., 2006; Knolhoff and Heckel, 2014; Kang et al., 2020). The insect peripheral chemosensory system comprises odorant receptors (ORs), gustatory receptors (GRs), ionotropic receptors (IRs), odorant binding proteins (OBPs), chemosensory proteins (CSPs), and sensory neuron membrane proteins (SNMPs) (Fleischer et al., 2018; Sun et al., 2020; Liu et al., 2021). These protein families have been identified from a large number of insect species, however, they still remain unidentified from several insect species.
Odorant binding proteins are small soluble olfactory proteins which are thought to be responsible for transporting hydrophobic odor molecules through the sensillum lymph to odorant receptors, which are housed on the dendritic membrane of olfactory sensory neurons (Wang et al., 2020; Tian et al., 2021). Previous studies have shown that OBPs are expressed selectively in different types of sensilla on the antenna, which are considered the minimum functional units for chemoreception. In general, OBPs show higher binding affinities with ligands in vitro. For instance, ApisOBP3 and SaveOBP7 showed a high binding affinity with aphid alarm pheromone, (E)-beta-farnesene, whereas, ApisOBP1, ApisOBP3, ApisOBP8, ApisOBP7, and SaveOBP7 showed a high binding affinity with plant volatiles (Qiao et al., 2009; Zhong et al., 2012).
Chemosensory proteins represent another class of small soluble proteins abundant in the lymph of chemosensilla (Pelosi et al., 2006). They are also broadly expressed in various organs, such as palps, proboscis, legs, wings, eyes, and pheromone glands (Hua et al., 2012, 2013; Gu et al., 2013; Liu et al., 2014; Zhu et al., 2016; Li et al., 2020). CSPs are different from OBPs in amino acid sequence and structure, but appear to be similar in functions, although better evidence is needed to clarify their role in olfaction (Calvello et al., 2005; Tian et al., 2021). The first CSP protein was discovered in the regenerating legs of the American cockroach, Periplaneta americana (Nomura et al., 1992). OS-D, a related CSP, was later cloned from Drosophila melanogaster antennae and is thought to be involved in pheromone binding (Mckenna et al., 1994). More insect CSP genes have recently been identified and characterized as a result of the completion of diverse insect genome sequences (Pelosi et al., 2017). Various numbers of CSP genes have been identified in different insect species. For instance, 4 CSPs were reported in D. melanogaster, six in Apis mellifera (Forêt et al., 2007), and 20 in Bombyx mori (Gong et al., 2007), 70 in Locusta migratoria (Picimbon et al., 2000), 43 in Aedes aegypti (Mei et al., 2018), three in Heliothis virescens (Picimbon et al., 2001) and 27 in Helicoverpa armigera (Agnihotri et al., 2021).
Odorant receptors were the first insect chemosensory receptor family which were identified using a bioinformatics screen of the D. melanogaster genome (Gao and Chess, 1999). The typical ORs are seven-transmembrane receptors with a reversed membrane topology. In general, ORs have a wide variety of odor affinities, and a single odorant molecule may bind to a number of olfactory receptors with variable affinities, which are dependent on physio-chemical features of molecules such as their molecular weights (Buck, 2004). Once the odorant interacts with the odorant receptor, it undergoes structural modifications and binds and activates the olfactory-type G protein on the inside of the olfactory receptor neuron. Activated olfactory receptors trigger nerve impulses that transmit information about the odor to the brain (Fleischer et al., 2018).
In insect gustatory organs, gustatory receptors GRs are a large gene family, which are implicated in host-seeking (Hallem et al., 2006; Agnihotri et al., 2016). Most of these GR proteins have the typical structure of seven transmembrane domains, were initially identified in the D. melanogaster genome based on a bioinformatic approach (Clyne et al., 2000). Further studies discovered that D. melanogaster has 68 gustatory receptor proteins, which are encoded by 60 gustatory receptor genes by alternative splicing (Dunipace et al., 2001; Scott et al., 2001; Robertson et al., 2003). The amino acid sequences of most gustatory receptor proteins are quite diverse, with just 8–12% sequence similarity. Some of this variance might help to increase the diversity of GRs’ responses to ligands (Robertson et al., 2003). GRs were classified as sugar receptors, CO2 receptors, GR43a-like receptors, bitter receptors, sex pheromone receptors, and unknown receptors based on the ligands to which they respond (Jones et al., 2007; Sato et al., 2011). With the development of insect genome sequencing, insect GR genes have been discovered in an increasing number of species: Anopheles gambiae has 52 Gr genes that encode 76 GR proteins (Hill et al., 2002), and A. aegypti has 79 GR genes that encode 114 GR proteins (Kent et al., 2008). Bombyx mori and Tribolium castaneum have 65 and 220 GR genes, respectively (Richards et al., 2008; Wanner and Robertson, 2008). Among all insect species investigated, H. armigera had the second-highest number of GR genes (197) (Xu et al., 2016).
Compared to other chemosensory gene families, SNMPs are a small family where only one or two members have been reported (SNMP1 and SNMP2). SNMP1 is found to be co-expressed with pheromone receptors in pheromone responsive neurons and seems to be an indicator of pheromone-responsive neurons (Jiang et al., 2016; Fleischer et al., 2018; Zhang et al., 2018). In contrast, SNMP2 is expressed in cells surrounding the neuron clusters supporting cells (Jiang et al., 2016). Recently, a novel SNMP gene, SNMP3 was found specifically expressed in the larval midgut of (B. mori), which assumed to be involved in the immune response to virus and bacterial infections (Zhang et al., 2018).
The orange spiny whitefly Aleurocanthus spiniferus is a serious insect pest of citrus, grapes and tea plants (Tang et al., 2015; Nugnes et al., 2020; Radonjić and Hrnčić, 2021). It also causes significant damage to more than 90 plant species from 38 families widely distributed throughout the world (Tang et al., 2015; Radonjić and Hrnčić, 2021). Due to the serious damage caused by this pest, it has been reported as quarantine pest in many countries (EPPO A2 list1). To date, there are limited studies on A. spiniferus that are mainly focused on population dynamics, insecticide selections, biological control and color plates (Mokrane et al., 2020; Nugnes et al., 2020; Tian et al., 2020). In this study, we investigated the structure, distribution, and abundance of the antennal sensilla in the adult male and female A. spiniferus by scanning electron microscopy. Transcriptome sequencing of A. spiniferus was performed to identify the candidate chemosensory genes. Moreover, tissue expression patterns of the putative chemosensory genes were assessed by quantitative real-time PCR (qPCR). These findings provide a basis for future chemosensory and genomic studies in A. spiniferus and closely related species.
Materials and Methods
Insect Materials
In this study, A. spiniferus were collected form tea cultivar ‘Huangjinya’ (Camellia sinensis) that were maintained in the greenhouse in Jinan, Shandong, China. Due to the low sex ratio of male, we are unable to get high quality and quantity of RNAs from male head tissues. Thus, we conducted the transcriptome analysis with the mixture of male and female head and bodies tissues. Heads with antennae (200 heads per replicate) and bodies only with thoraxes, legs, wings and abdomens (50 bodies per replicate) were dissected, collected in liquid nitrogen and then subjected to RNA extractions using RNAiso (Takara Bio, Tokyo, Japan) according to the manufacturer’s instructions. The RNA integrity was verified by 1% agarose gel electrophoresis and the quantity was assessed with a Nanodrop ND-2000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, United States).
Scanning Electron Microscopy
Approximately 50 female and male adults were used for the identification of antennal sensilla using scanning electron microscopy (SEM). Experiments were conducted followed the method previously descried by Zhang et al. (2015). Whole bodies of A. spiniferus were putted into 1.5 ml clean Eppendorf tubes and washed twice using 0.1 M phosphate-buffered saline (PBS, pH 7.2) each for 5 min. After the preliminary cleaning, all of these samples were transferred into ultrasonic bath for deep cleaning (250 W, 30 s). Cleaned samples were fixed in 2.5% glutaraldehyde at 4°C overnight. After the fixation, all samples were washed five times in PBS (0.1 M, pH 7.2) for 20 min each, and then incubated in osmium tetroxide for 15 h. Dehydration of all samples was conducted in ethanol series (45%, 55%, 75%, 85%, 95% for 30 min each, and 100% for 14 h). Then, all samples were transferred into new Eppendorf tubes with 0.5 ml 100% ethanol for 7 h. Dehydrated samples were rinsed in isoamyl acetate for 1 h each. Finally, all samples were dried, mounted on aluminum stubs and gold coated. Antennal sensilla were observed and recorded using ZEISS Ultra-55 Scanning Electron Microscope (Carl Zeiss Meditec, Oberkochen, Baden-Württemberg, Germany). Student’s t-test was used for the comparison of the difference between male and female (P < 0.05).
Transcriptome Sequences
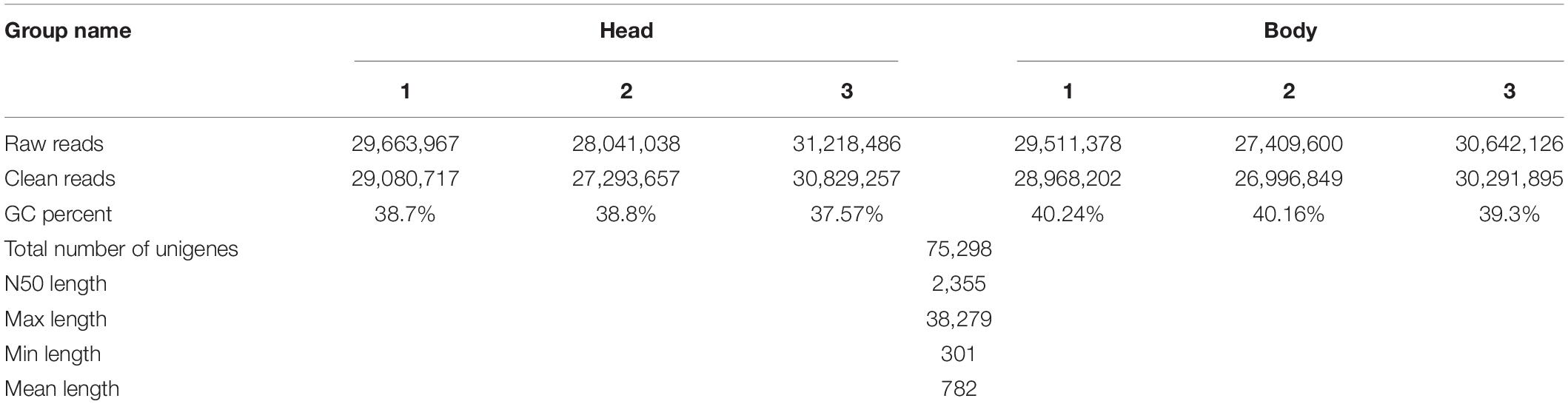
Three biological replicates of high quality and quantity RNAs from heads and bodies of A. spiniferus were subjected to cDNA library construction and sequencing on the Illumina, Inc. (San Diego, CA, United States) by Novogene Bioinformatics Technology Co., Ltd. (Beijing, China). Clean data (clean reads) were obtained by removing adapter-containing reads, higher N rate reads (N rates > 10%), and low-quality reads (50% bases with Q-score ≤ 5) from the raw data (raw reads) using in-house Perl scripts. Clean read assembly was carried out with the short-read assembly program Trinity with min_kmer_cov set to 2 by default and all other parameters set default. The annotation of unigenes was performed by NCBI BLASTx search against the Nr protein database, with an E-value threshold of 1 × 10––5. The blast results were then imported into the Blast2GO pipeline for GO annotation. The longest open reading frame ORF for each unigene was determined by the NCBI ORF Finder tool2. Differential expression analysis was performed using the DESeq2 R package (1.20.0). DESeq2 provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamini and Hochberg’s approach for controlling the false discovery rate. Genes with an adjusted P-value < 0.05 found by DESeq2 were assigned as differentially expressed. Expression levels were expressed in terms of FPKM values (fragments per kilobase per million reads), which was calculated by RSEM (RNA-Seq by Expectation-Maximization) with default parameters (Kang et al., 2017b). The sequences reported in this paper have been deposited in the GenBank SRA database (BioProject ID: PRJNA792195).
Verification of Candidate Chemosensory Genes in Aleurocanthus spiniferus
Genes annotated as chemosensory genes in A. spiniferus were further verified by BLASTp (E-value < 1 × 10–5 and Identity > 30%) in NCBI non-redundant protein sequences database with algorithm of PSI-BLAST. Furthermore, we also used the amino acid sequences of OBPs and CSPs of B. tabaci against our transcriptome database to avoid the omission of transcriptome annotation (Zeng et al., 2019). The signal peptide and conserved domains of OBPs and CSPs of A. spiniferus were predicted by SignalP 5.0 Server3 and SMART (simple modular architecture research tool4). Transmembrane domains in ORs, GRs and IRs were predicted by TMHMM - 2.05.
Sequence Alignment and Phylogenetic Analysis
Sequence alignment and phylogenetic analysis were conducted as described by Zeng et al. (2019). Amino acid sequences of candidate OBPs, CSPs, SNMPs, ORs, GRs, and IRs were aligned by ClustalW used gap opening penalty 10 and gap extension penalty 0.2. The alignments were further manually edited. Phylogenetic trees were subsequently constructed by the maximum likelihood method using MEGA X based on the model WAG and gamma distributed with bootstrap 1000 (Kumar et al., 2018). The trees were further edited using the ITOL tool (Letunic and Bork, 2019). All amino acid sequences used in this work are presented in Supplementary Table S1.
Expression Pattern Analysis of Chemosensory Genes by Quantitative Real-Time Polymerase Chain Reaction
RNAs of A. spiniferus from different tissues (heads and bodies) and developmental stages (second nymphs, third nymphs, puparia/fourth nymphs, female adults and male adults) were extracted by RNAiso (Takara Bio., Tokyo, Japan). The cDNA was synthesized from total RNA using FastQuant RT Kit (With gDNase) (Tiangen, Beijing, China) according to the standard manufacturer’s protocol. Gene-specific primers were designed by Primer Premier 6 (PREMIER Biosoft International, Palo Alto, CA, United States), which are listed in Supplementary Table S2. qPCR reaction was conducted in a total volume of 20 μL containing: 10 μL of 50× SYBR Premix Ex Taq, 0.8 μL of primer (10 mM), 0.8 μL of sample cDNA, and 7.6 μL sterilized ultra-pure grade H2O. The cycling conditions were as follows: 95°C for 30 s, followed by 40 cycles of 95°C for 5 s and 55°C for 30 s. Three technical and three biological replicates were used for each sample. Relative quantification was performed using the Comparative 2–ΔΔCT method (Livak and Schmittgen, 2001). Transcription levels of these chemosensory genes were normalized by reference gene RPS28 (Kang et al., 2017a; Kong et al., 2021). Heatmaps of chemosensory genes were constructed by pheatmap in R 4.0.4 as Liu et al. (2020) reported. Differences of selected chemosensory genes between male and female were subjected to Student’s t-test (P < 0.05), while one-way analysis of variance (ANOVA) followed by separation of means with the Fisher’s protected least significant difference (LSD) test (P < 0.05) was used for the difference among the different developmental stages.
Results
Morphology of Antennal Sensilla of Aleurocanthus spiniferus
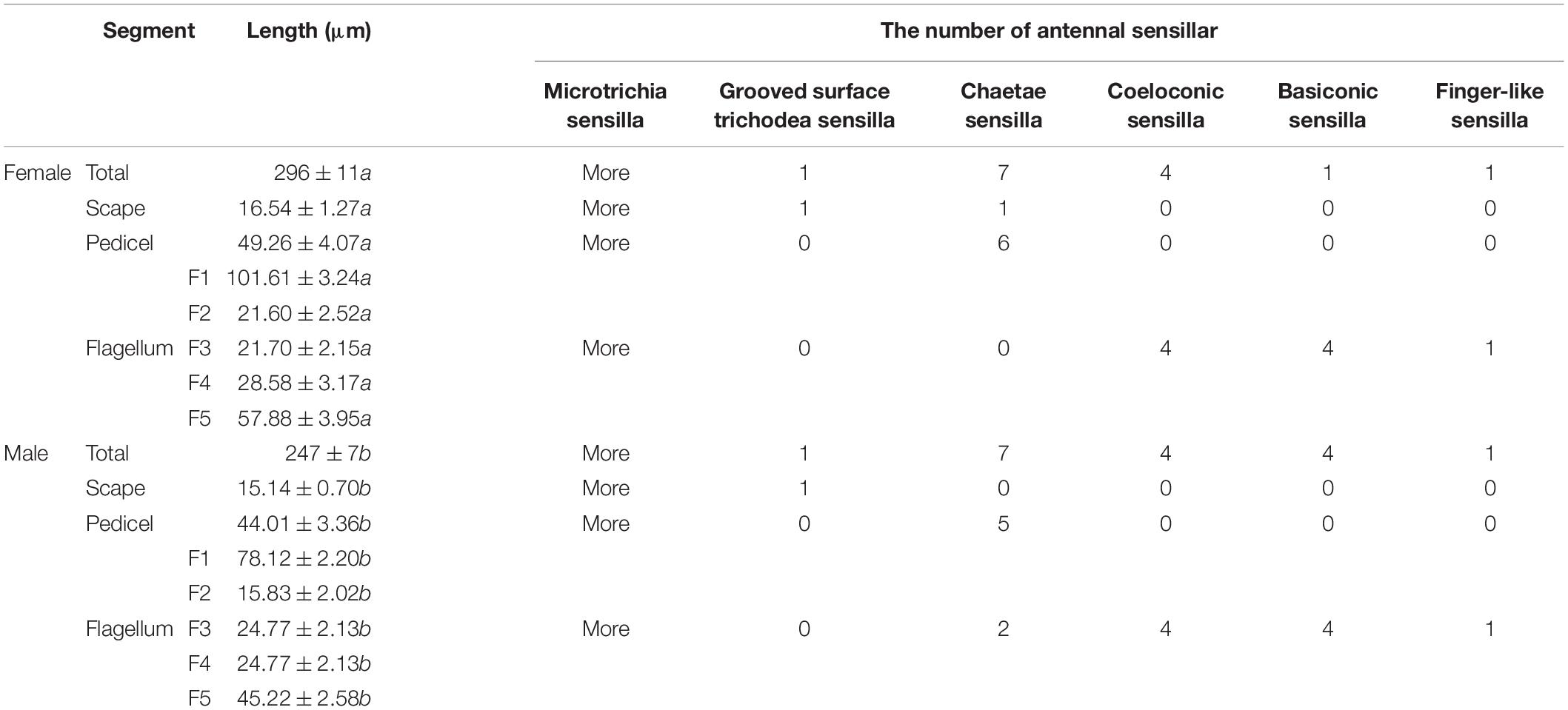
The length of female antennae was significantly longer than that of male (Table 1). Six different sensilla types were observed: trichodea sensilla, chaetica sensilla, microtrichia sensilla, coeloconic sensilla, basiconic sensilla, and finger-like sensilla. There was no difference of the distribution and structure of other sensilla between the two sexes (Figure 1A). Grooved surface trichodea sensilla were only found on the scape (Figure 1B and Table 1). Chaetae sensilla presented on the scape and pedicel female A. spiniferus, while it was found on the pedicel and flagellum (Figure 1C and Table 1). Finger-like sensilla was only found on the tips of the fifth flagellum of A. spiniferus (Figure 1D and Table 1). Basiconic sensilla looks like a sword and was found on the flagellar subsegment 5 (Figures 1E,F and Table 1). Coeloconic sensilla were surrounded by microtrichia sensilla, and microtrichia sensilla were the most abundant and widely distributed sensilla on the entire antennae of A. spiniferus (Figure 1G and Table 1).
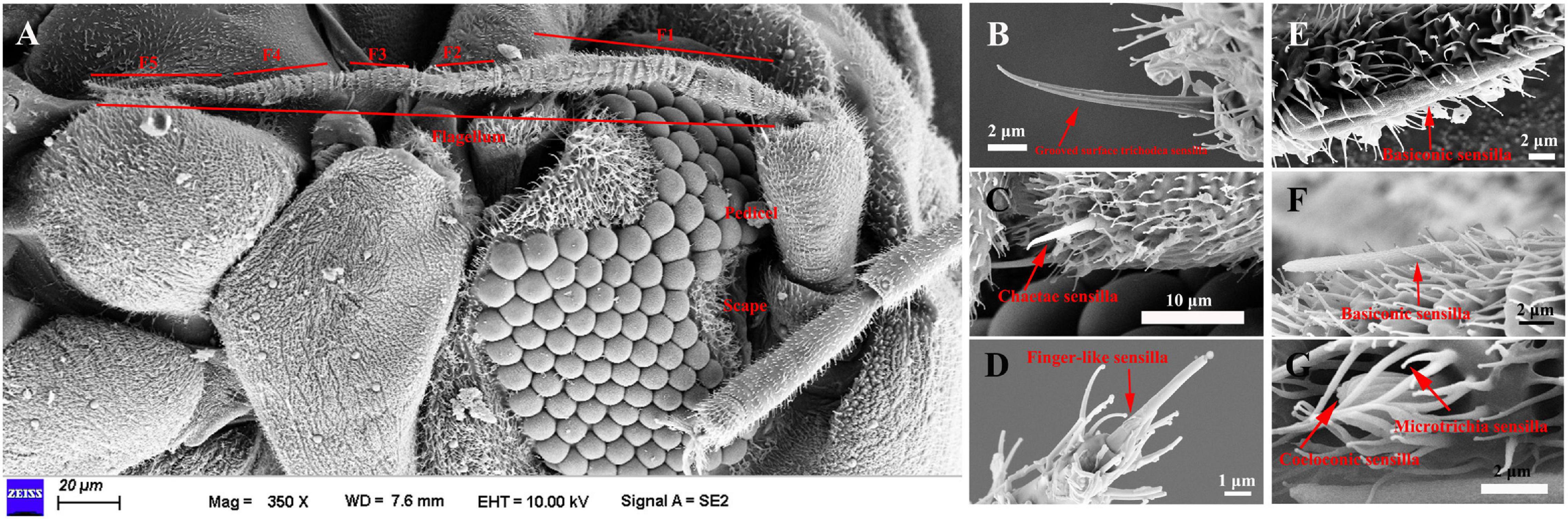
Figure 1. The types of sensilla present on A. spiniferus antennae. (A) Female antenna. (B) Grooved surface richodea sensilla (C) Chaetae sensilla. (D) Figure-like sensilla. (E) Basiconic sensilla. (F) Basiconic sensilla. (G) Coeloconic and microtrichia sensilla.
Transcriptome Analysis Data of Aleurocanthus spiniferus
The transcriptome data was presented in Table 2. The total number of unigenes was 75298. Max, Min, and mean length were 38279, 301, and 782 bp, respectively (Table 2). GC percent of sequences from bodies showed a little bit higher than that from heads (Table 2). Homology analyses results showed that the most similar sequences of 75.1% sequences were from B. tabaci (Supplementary Figure S1). Functional annotation was performed using NR, NT, KO, Swissprot, PFAM, GO, and KOG databases (Supplementary Table S3). Based on the GO categorization, differential expressed genes were enriched in protein metabolic process, hydrolase activity, cellular protein metabolic process, intracellular non-membrane-bounded organelle, non-membrane-bounded organelle and translation (Supplementary Figure S2).
Putative Chemosensory Genes in Aleurocanthus spiniferus
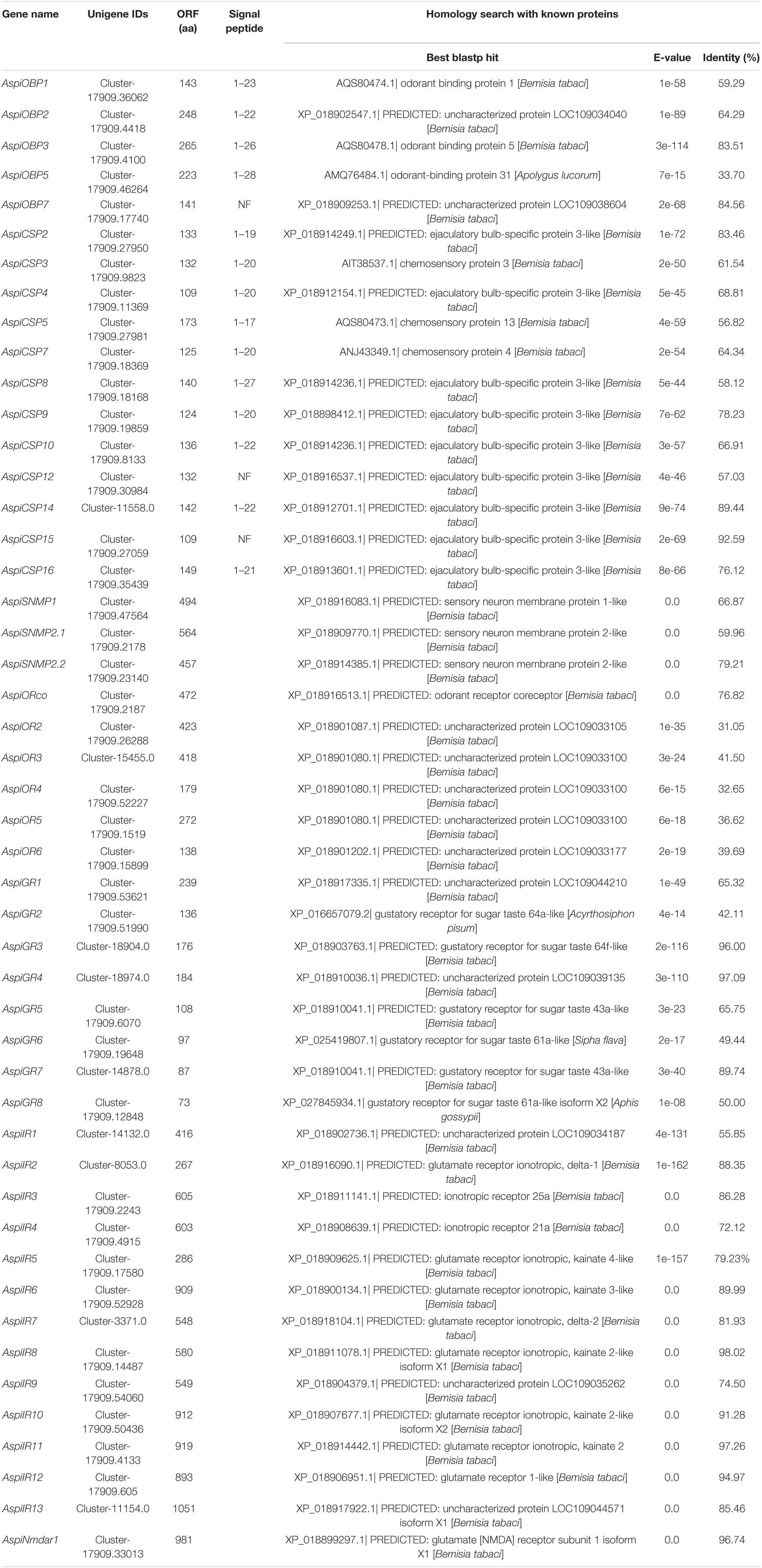
In this study, a total of five transcripts encoding candidate OBPs were identified in the transcriptome of A. spiniferus (Table 3). The number of putative OBPs was a little bit lower than that identified in the genome of B. tabaci (eight OBPs). All of the putative OBPs had full-length ORFs, and only AspiOBP7 without signal peptide (Table 3). A phylogenetic tree was constructed using the identified OBPs from whiteflies (A. spiniferus and B. tabaci), aphids (Acyrthosiphon pisum, Aphis glycines, Brevicoryne brassicae, Metopolophium dirhodum, Rhopalosiphum padi, Lipaphis erysimi, Aphis fabae, Aphis craccivora, Tuberolachnus salignus, Myzus persicae, Aphis gossypii, Drepanosiphum platanoidis, and Nasonovia ribisnigri), plant bugs (Apolygus lucorum and Adelphocoris lineolatus) and plant hoppers (Nilaparvata lugens and Sogatella furcifera) (Figure 2 and Supplementary Figure S3). In the phylogenetic tree, AspiOBP1, AspiOBP2, AspiOBP5, and AspiOBP7 were clustered with OBPs from B. tabaci, while AspiOBP3 was clustered with OBPs from aphids (Figure 2).
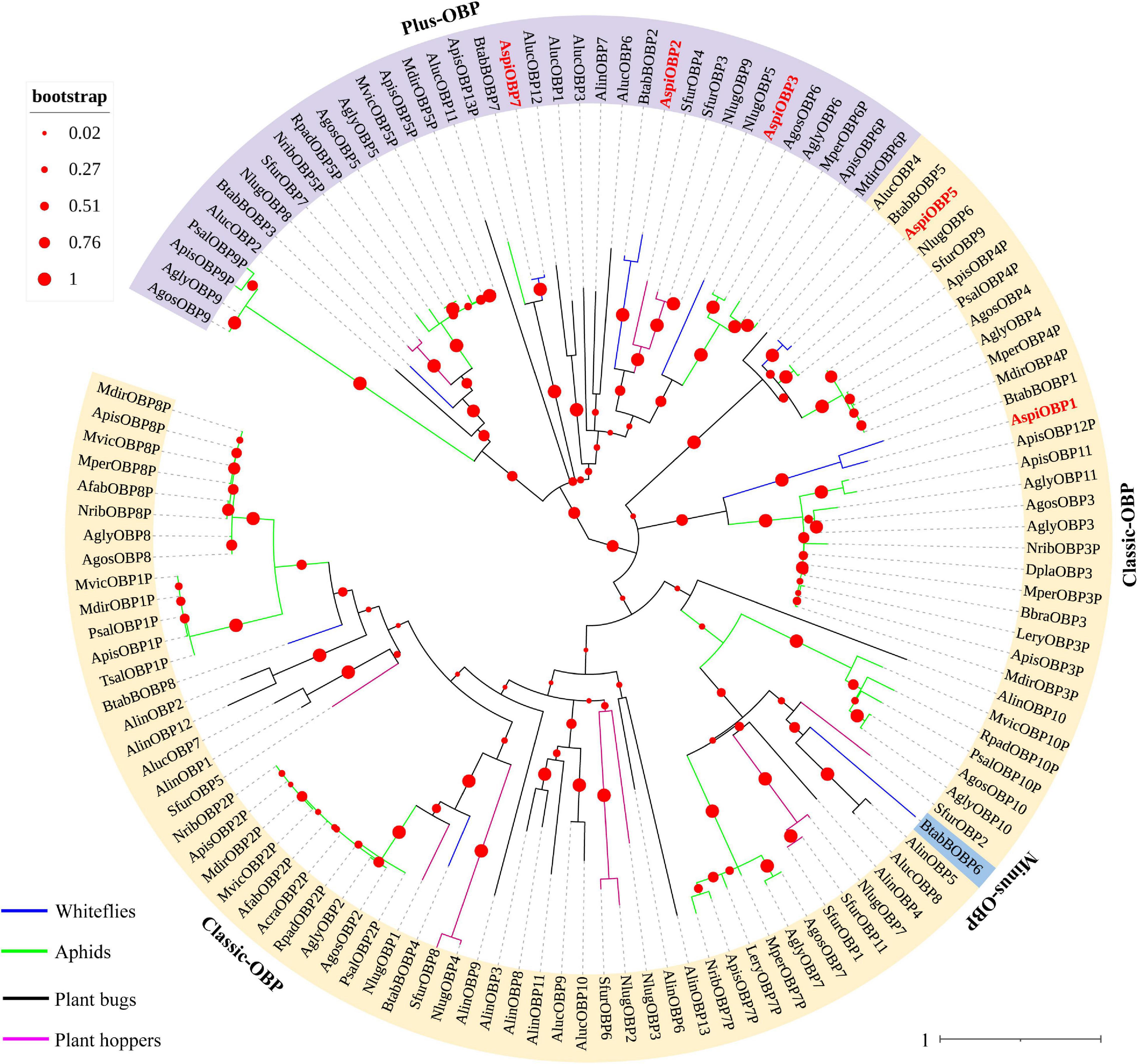
Figure 2. Phylogenetic analysis of putative odorant-binding proteins (OBPs) of A. spiniferus. The phylogenetic tree was built using OBP sequences from whitefly species (Btab, Bemisia tabaci; Aspi, A. spiniferus), aphid species (Apis, Acyrthosiphon pisum; Mper, Myzus persicae; Agos, Aphis gossypii; Psal, Pterocomma salicis; Agly, Aphis glycines; Mdir, Metopolophium dirhodum; Mvic, Megoura viciae; Bbra, Brevicoryne brassicae; Lery, Lipaphis erysimi; Afab, Aphis fabae; Acra, Aphis craccivora; Tsal, Tuberolachnus salignus; Dpla, Drepanosiphum platanoidis; Nrib, Nasonovia ribisnigri; Rpad, Rhopalosiphum padi), plant hoppers (Sfur, Sogatella furcifera; Nlug, Nilaparvata lugens), and plant bugs (Aluc, Apolygus lucorum; Alin, Adelphocoris lineolatus).
We identified 12 candidate CSPs in A. spiniferus (Table 3) and the number of putative CSPs identified was also lower than that in B. tabaci (19 CSPs). Of the 12 putative CSPs, all of them had full-length ORFs, and only AspiCSP12 and AspiCSP15 without signal peptide. To analyze the relationship between the CSPs in the different species, a phylogenetic tree was constructed and is presented in Figure 3, which includes the identified CSPs from whiteflies (A. spiniferus and B. tabaci), aphids (A. gossypii and M. persicae), plant bugs (A. lucorum and A. lineolatus) and plant hoppers (N. lugens and S. furcifera). In the phylogenetic tree, all of the identified CSPs were clustered with CSPs in B. tabaci (Figure 3).
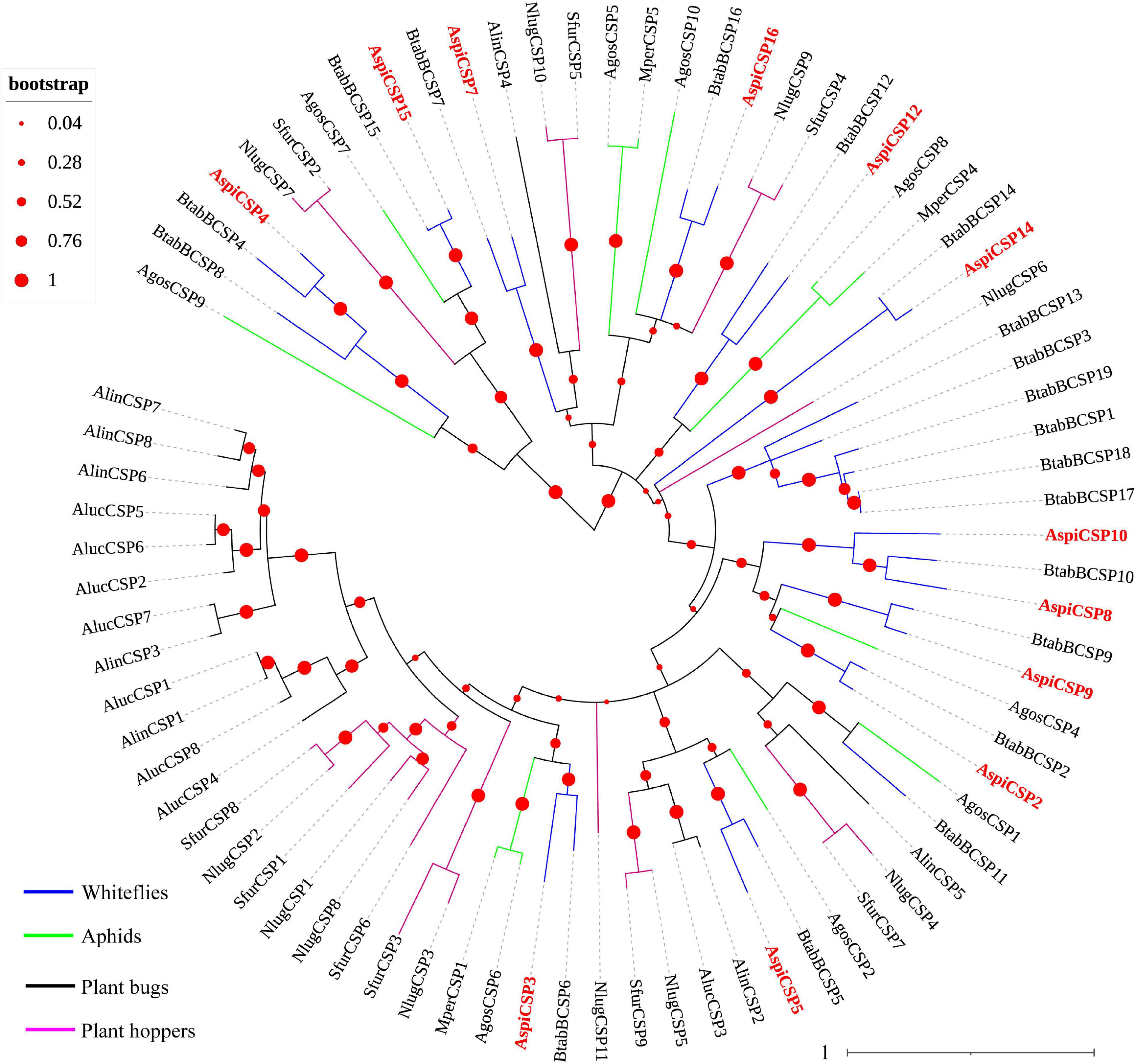
Figure 3. Phylogenetic analysis of putative chemosensory proteins (CSPs) of A. spiniferus. The phylogenetic tree was built using CSP sequences from whitefly species (Btab, B. tabaci; Aspi, A. spiniferus), aphid species (Apis, A. pisum; Agos, A. gossypii), plant hoppers (Sfur, S. furcifera; Nlug, N. lugens) and plant bugs (Aluc, A. lucorum; Alin, A. lineolatus).
Interestingly, there were three SNMPs identified in A. spiniferus that were significantly different from other Hemipteran insects (Table 3). The best hits by homology search in NCBI of these SNMPs were SNMPs from B. tabaci (Table 3). The phylogenetic tree showed that there were two distinct cluster SNMP1 (AspiSNMP1) and SNMP2 (AspiSNMP2.1 and AspiSNMP2.2; Figure 4).
We identified transcripts encoding six putative ORs (Table 3). Among these candidate ORs, AspiORco, AspiOR2, and AspiOR3 likely represented full-length genes, encoding proteins made up of more than 400 amino acids (Table 3). In the phylogenetic tree, AspiORco, AgosOrco1, RapdOrco1, and ApisOR1 were clustered in a specific subgroup called odorant co-receptor (Orco) with four transmembrane domains (Figure 5 and Supplementary Table S4). Rest of these identified ORs was clustered in a specific subgroup (Figure 5).
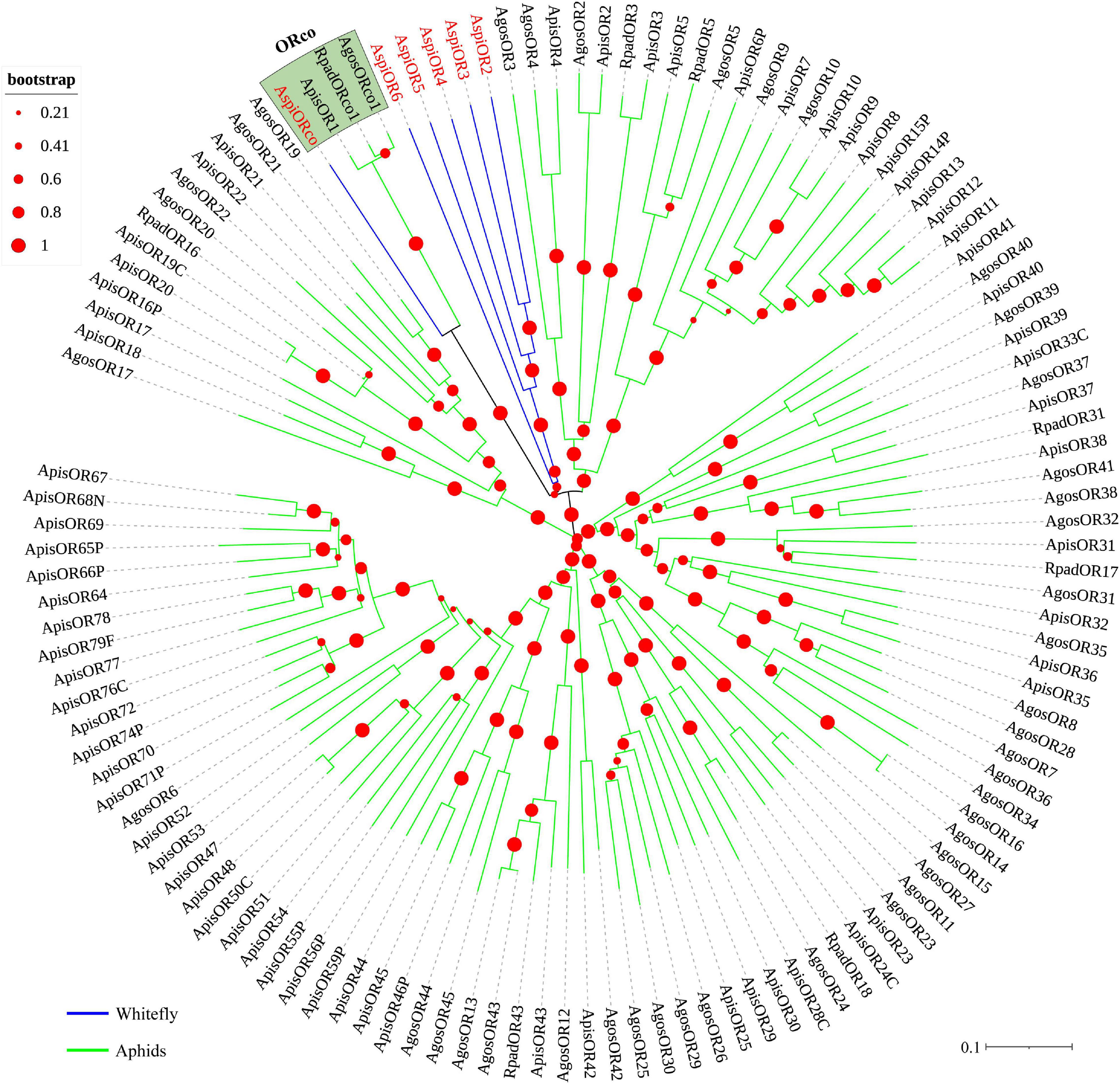
Figure 5. Phylogenetic analysis of putative odorant receptors (ORs) of A. spiniferus. The phylogenetic tree was built using OR sequences from whitefly specie (Aspi, A. spiniferus) and aphid species (Apis, A. pisum; Rpad, R. padi; Agos, A. gossypii).
For GRs, in this study, we identified eight candidate GRs from the transcriptome of A. spiniferus (Table 3). A phylogenetic tree was constructed with sequences from whitefly (A. spiniferus), aphids (A. pisum and R. padi) and fly (D. melanogaster). AspiGR14 was clustered with DmelGR63a and DmelGR21a as a CO2 receptor, while AspiGR3 were found in a clade with sugar receptors, which included GRs identified from D. melanogaster, A. pisum, and R. padi (Figure 6).
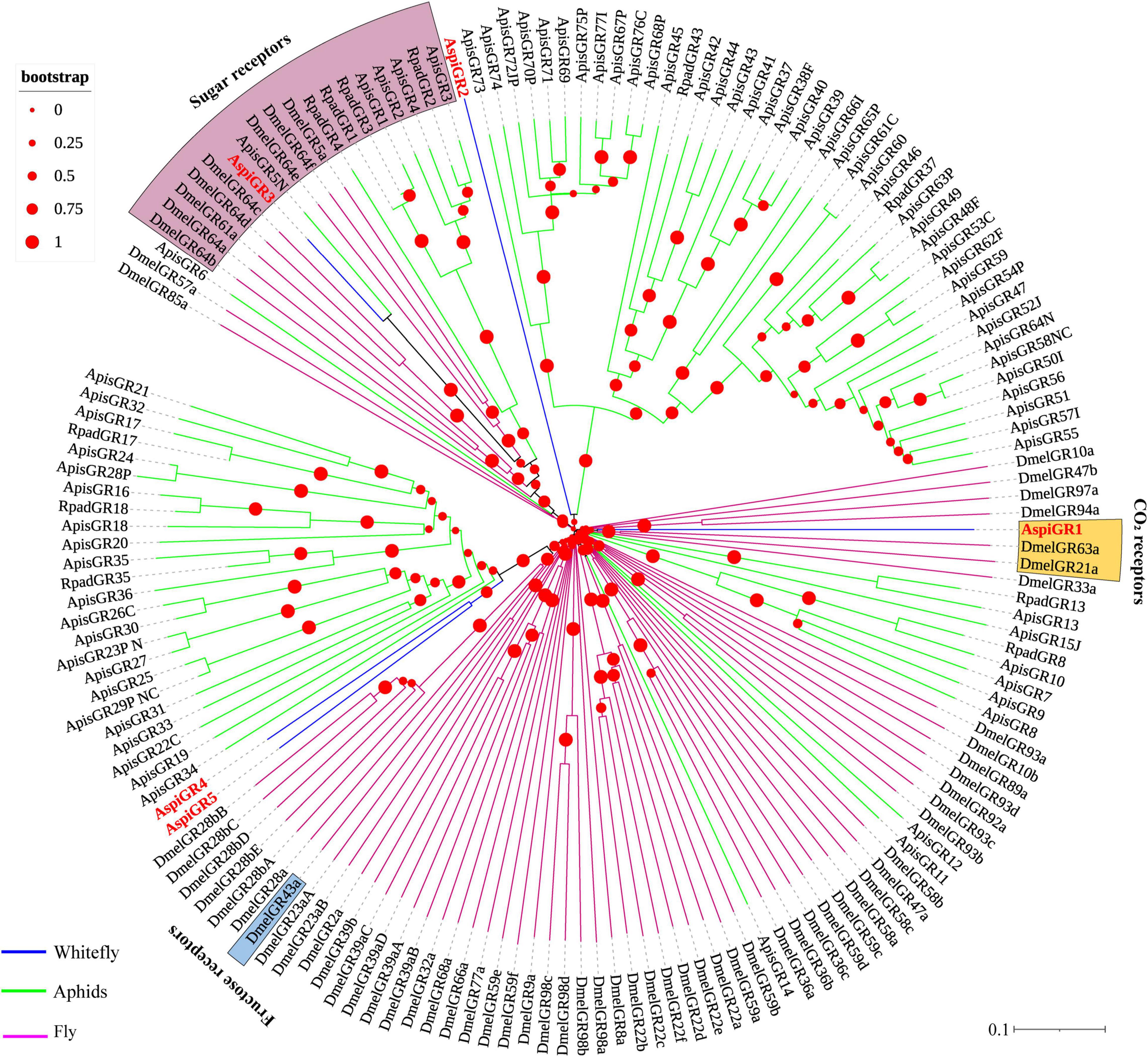
Figure 6. Phylogenetic analysis of putative gustatory receptors (GRs) of A. spiniferus. The phylogenetic tree was built using GR sequences from whitefly specie (Aspi, A. spiniferus), aphid species (Apis, A. pisum; Rpad, R. padi; Agos, A. gossypii) and fly (Drosophila melanogaster).
Fourteen putative IRs were identified from the transcriptome of A. spiniferus (Table 3). Among them, only AspiIR2, AspiIR5, and AspiIR8 were found to be a part of the full-length gene. The E-values for AspiIR3, AspiIR4, AspiIR6, AspiIR7, AspiIR8, AspiIR9, AspiIR10, AspiIR11, AspiIR12, AspiIR13, and AspiNmdar1 were zero as compared to the amino acid sequences of these genes in B. tabaci (Table 3). In the phylogenetic tree, almost all of these IRs were clustered in a known group, such as IR8a/IR25a (AspiIR3), IR21a (AspiIR4), IR40a (AspiIR5), IR75 (AspiIR9), IR76b (AspiIR7), IR93a (AspiIR1 and AspiIR2), and NMDA iGluRs (AspiNmdar1) (Figure 7).
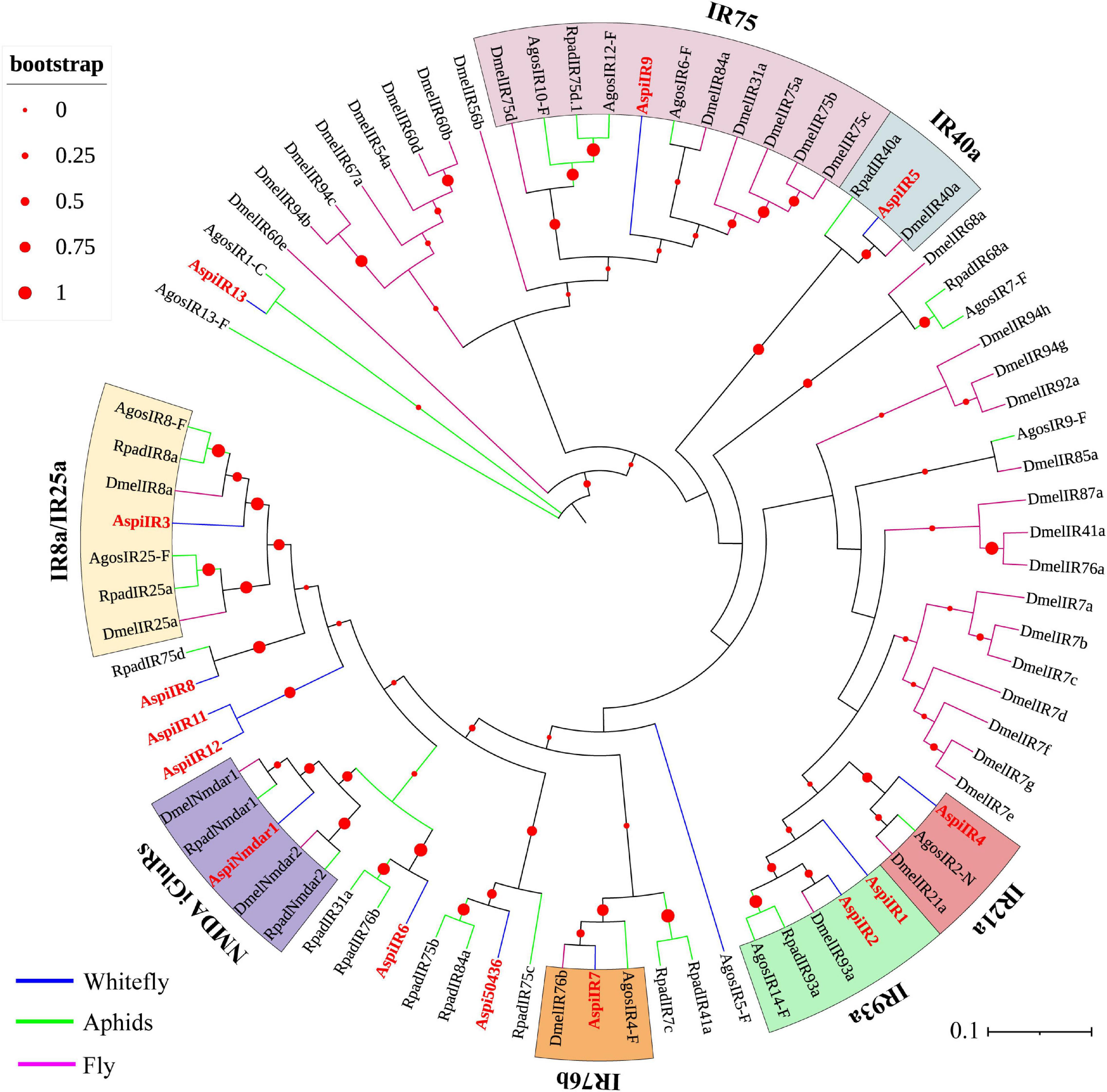
Figure 7. Phylogenetic analysis of putative ionotropic receptors (IRs) of A. spiniferus. The phylogenetic tree was built using IR sequences from whitefly specie (Aspi, A. spiniferus), aphid species (Apis, A. pisum; Rpad, R. padi; Agos, A. gossypii) and fly (Drosophila melanogaster).
Expression Profiles of Chemosensory Genes
Expression results of these selected chemosensory genes in different developmental stages showed that AspiOBP1 and AspiIR9 were more strongly expressed in nymphs than that in puparia and adults whereas expression of AspiOBP3 and AspiCSP12 in puparia and adults were significantly higher than that in nymphs (Figure 8). Surprisingly, AspiORco, AspiOR2, AspiGR1, AspiGR3, and AspiIR4 showed highest expression profiles in puparia among the developmental stages (Figure 8). On the contrary, AspiCSP10, AspiIR2, and AspiIR3 had the lowest expression in puparia. The expression of AspiOBP2 and AspiIR5 were significantly higher than that in nymphs and puparia (Figure 8). AspiGR6, AspiGR8, AspiIR8, and AspiIR13 presented a higher expression in nymphs and puparia than that in adults (Figure 8). The expression of AspiIR11 in second instar was significantly higher than other developmental stages (Figure 8).
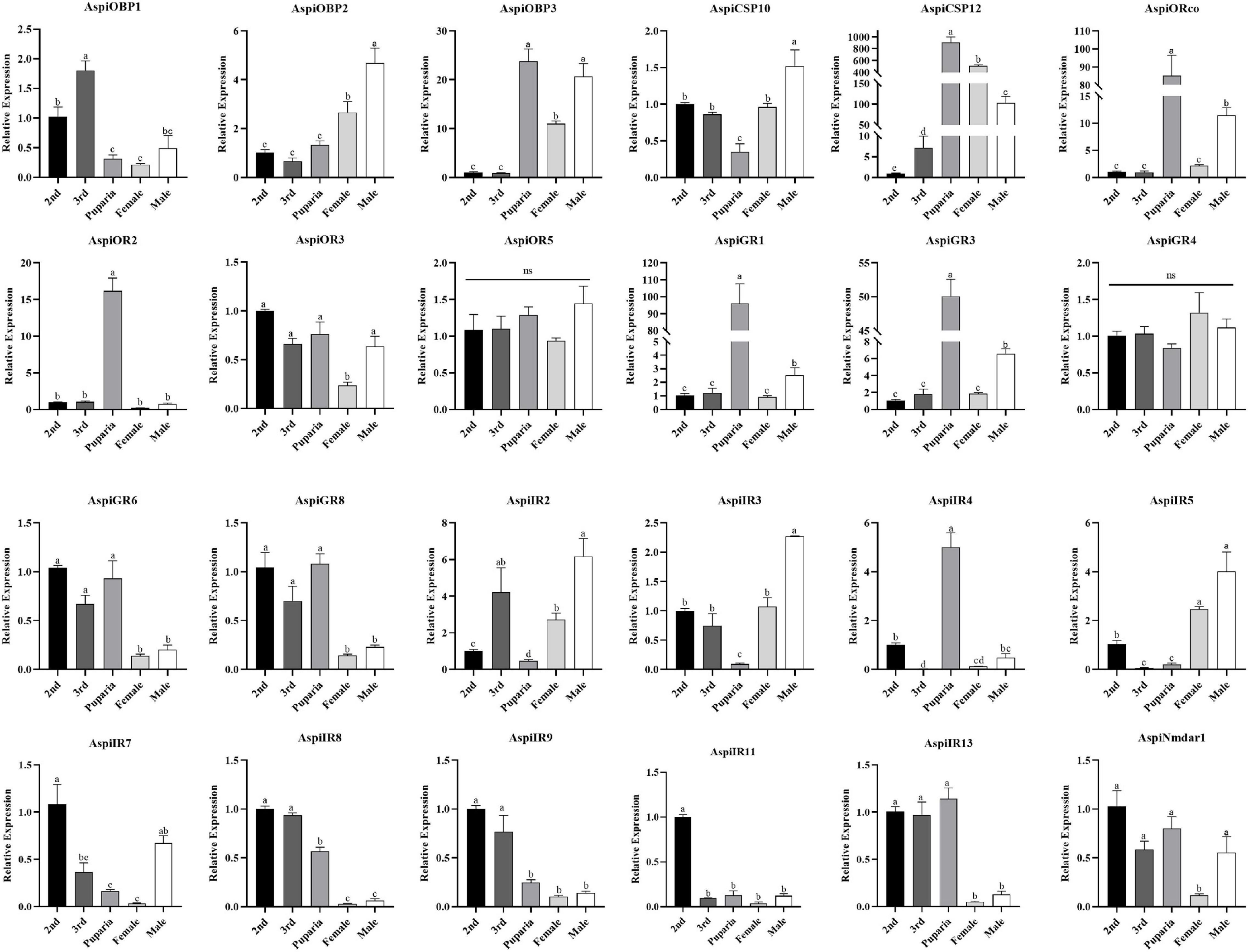
Figure 8. Quantitative real-time polymerase chain reaction (qPCR)-based expression profiling of selected chemosensory genes in different developmental stages of A. spiniferus.
Based on the transcriptome results, we found that all of OBPs and SNMPs, and major of ORs and IRs were more considerably expressed in head than in bodies (Figure 9A). Meanwhile, only five of 12 CSPs were predominately expressed in heads, and four of 12 CSPs highly expressed in bodies (Figure 9A). In addition, there were only two of eight GRs showed significant tissue-specific expression patterns (Figure 9A). qPCR validation of selected chemosensory genes showed that expressions of AspiOBP1, AspiOBP2, AspiOBP3, AspiCSP10, AspiORco, AspiOR2, AspiGR1, AspiGR6, AspiGR8, AspiNmdar1, AspiIR2, AspiIR3, AspiIR4, AspiIR7, AspiIR8, AspiIR9, AspiIR11, and AspiIR13 in heads were significantly higher than that in bodies, while AspiCSP12, AspiGR3, AspiGR4, and AspiIR5 were predominantly expressed in bodies (Figure 9B). There was no difference of the expression of ApisOR3 and AspiOR5 between heads and bodies (Figure 9B).
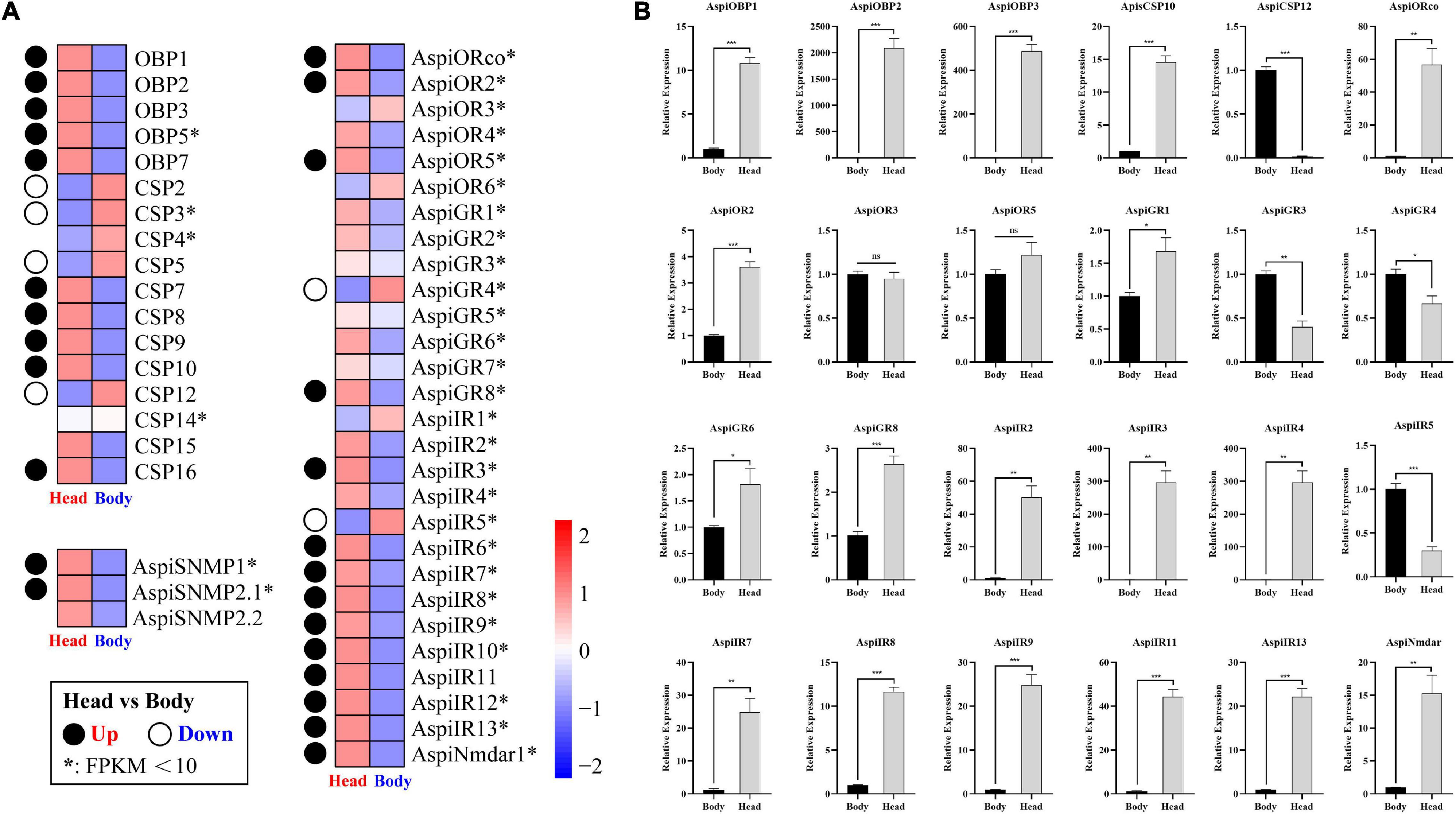
Figure 9. Expression profiles of chemosensory genes in different tissues. (A) Heatmap of chemosensory genes in the antennal transcriptome. Significance means an absolute value of log2Ratio ≥ 1 and FDR < 0.05. (B) Validation of selected chemosensory genes in different tissues by qPCR.
Discussion
Insects have a complex chemosensory system that accurately perceives external chemicals and plays a pivotal role in many insect life activities. Several studies have been conducted to understand the structure and function of the chemosensory system in different insect species, however, the chemosensory system in the orange spiny whitefly, A. spiniferus has not been investigated yet. The present study is the first report identifying the various types and distribution of the sensilla on the adult male and female antenna of A. spiniferus. Consistent with the results of two cryptic B. tabaci specie, length of male antenna was significantly longer than that of females, which was caused by the obviously smaller bodies of male A. spiniferus (Zhang et al., 2015). Furthermore, there was no differences in the composition and number of antennal sensilla between males and females. Contrary with that, in two cryptic B. tabaci specie, males had more chaetae sensilla (7) than females (5) (Zhang et al., 2015). Interestingly, in A. spiniferus, distribution of chaetae sensilla between males and females was different. In females, chaetae sensilla was observed in scape (1) and pedicel (6), while in males chaetae sensilla was found in pedicel (5) and flagellum (2). Differences of the distribution of chaetae sensilla might be involved in the different behaviors between males and females of A. spiniferus.
In this study, we systematically identified and chemosensory genes in A. spiniferus via transcriptomic analyses. A total of 48 candidate chemosensory genes including 5 OBPs, 12 CSPs, 3 SNMPs, 6 ORs, 8 GRs, and 14 IRs were predicted. The number of identified chemosensory receptors was close to B. tabaci that contains 9 OBPs, 18 CSPs, 7 ORs, and 17 GRs, but significantly lower than that in other hemipterans (A. pisum: 79 ORs, 77 GRs, 15 OBPs, and 1 SNMP; A. gossypii: 45 ORs, 14 IRs, 9 OBPs, 9 CSPs, and 1 SNMPs) (Chen et al., 2016; Wang et al., 2017; Xie et al., 2017; Zeng et al., 2019). However, the total number of OBPs and CSPs in whiteflies showed no contractions or expansion when compared with other hemipteran insects (Zeng et al., 2019). The reduction of numbers of ORs and GRs in whiteflies might result from their polyphagia and strong detoxification systems (Chen et al., 2016; Xie et al., 2017). Thereby, less ORs and GRs are enough for them to find their suitable host plants. In addition, as the number of IRs was similar with other insects, we speculated that IRs may work as ORs and GRs. Thus, the functional investigations about ORs, GRs and IRs are needed to figure out the reason of this phenomenon.
In this study, we found that all of these five OBPs were predominately expressed in the head. Expression of AspiOBP1, AspiOBP2, and AspiOBP3 across developmental stages showed that AspiOBP1 was more highly expressed in nymphs than that in pupae and adults whereas the expression of AspiOBP3 in pupae and adults was significantly higher than that in nymphs. AspiOBP2 was abundantly expressed in adults. Similarly, in Sitophilus zeamais, SzeaOBP1 showed highest expression at larval stage, while the expression of SzeaOBP28 at pupae and adult stage was significantly higher than that at larval stage (Zhang Y. et al., 2019). Furthermore, SzeaOBP1 showed broader binding affinity for plant volatile compounds than SzeaOBP28 (Zhang Y. et al., 2019). Silencing SzeaOBP1 reduced the preference of S. zeamais to its preferred volatiles (Zhang Y. et al., 2019). In B. tabaci, BtabOBP1, BtabOBP2, BtabOBP3, BtabOBP4, BtabOBP7, and BtabOBP8 were highly expressed in heads whereas BtabOBP5 predominately expressed in legs and wings. BtabOBP1, BtabOBP3, and BtabOBP4 have been demonstrated to bind oviposition repellent volatile, β-ionone and various volatiles to its specific chemosensory receptor (Li et al., 2019; Wang et al., 2020). Knockdown of BtabOBP3 in B. tabaci by RNAi resulted in a reverse olfactory behavior to β-ionone (Wang et al., 2020). Furthermore, silencing BtabOBP3 also reduced the preference of B. tabaci on ToCV-infected tomato plants and the ToCV transmission rate of B. tabaci (Shi et al., 2019). Besides the function of odorants perception, OBPs also have been found to been involved in other insect physiological process. For example, the adult A. lineolatus head predominately expressed AlinOBP14 showed a pronounced binding affinity for insect juvenile hormone III (Sun et al., 2019). In N. lugens, knockdown of the gene for NlugOBP3 not only reduced the response rate to seeding volatiles but also resulted in strikingly high nymph mortality (He et al., 2011).
Unlike the expression of OBPs is focused in the antennae or other olfactory sensilla in most insects, CSPs were found to be broadly expressed in various tissues including antennae, wings, legs and abdomen (Hua et al., 2013; Yang et al., 2014). For example, AlucCSP2 and AlucCSP3 of A. lucorum were specifically expressed in female wings, and showed high binding affinities with cotton secondary metabolites including gossypol, tannings, quercetin and rutin hydrate (Hua et al., 2013). In N. lugens, none of CSPs was predominately expressed in antennae (Yang et al., 2014). Leg highly expressed CSP3 and CSP8 of N. lugens strongly bound to plant volatiles (Waris et al., 2018). Injected with dsNlugCSP8 significantly reduced the attractive responses of N. lugens to nerolidol and hexanal (Waris et al., 2018). Furthermore, CSPs are also known to be involved in insecticide resistance. Overexpressing AgosCSP5 on Drosophila files showed higher resistance and survival in response to imidacloprid and cypermethrin than control flies (Li et al., 2021). In B. tabaci and Ophraella communa, BtabCSP11 and OcomCSP12 were strongly expressed in the female abdomen and ovary respectively, and both of them are involved in the reproduction (Ma et al., 2019; Zeng et al., 2020). Consistent with these results, in this work, we found AspiCSP12 was specifically expressed in female bodies. Additionally, AspiCSP12 also showed higher expression levels at puparia and adult stages than that in nymphs. However, AspiCSP10 showed lowest transcript abundance in puparia, and there was no difference of this gene between nymphs and adults. All of these results indicate that AspiCSP12 might have other physiological functions rather than just being involved in odorant perception.
Recently, more research has been focused on the function of three chemosensory receptor types: ORs, GRs, and IRs (Zhang R.B. et al., 2017; Yang et al., 2020, 2021). In this study, we identified six ORs in the A. spiniferus transcriptome. Among of these ORs, AspiORco, AspiOR2, and AspiOR5 were predominately expressed in head whereas other ORs showed broadly expressed in heads and bodies. Furthermore, the expression of ORco was significantly higher in male and puparia than that in other stages. Similar results were observed in aphids, and knockdown of SaveORco in S. avenae disrupted its response to plant volatiles and the aphid alarm pheromone, (E)-β-farnesene (Fan et al., 2015; Kang et al., 2018). Besides the ORco, in A. pisum, ApisOR5 is known as an essential receptor for its alarm pheromone E-β-farnesene, and ApisOR4 is involved in the recognition of plant volatiles (Zhang R.B. et al., 2017; Zhang R.B. et al., 2019). Silencing CquiOR114/117 in female Culex quinquefasciatus significantly impaired the blood feeding behavior (Wu et al., 2020). All of these results indicated that ORs especially for AspiORco, AspiOR2, and AspiOR5 in A. spiniferus might be involved in the plant volatiles perception.
As phloem-feeding insects, whiteflies can be affected and even killed by the phytochemicals in plant phloem sap, such as amino acids, sugars and other metabolites (Cui et al., 2017; Hasanuzzaman et al., 2018; Xia et al., 2021). To cope with this, whiteflies assess the suitability of a potential host plant and select the best plant as well as the best feeding region on the plant (Döring, 2014; Cui et al., 2017; Hu and Tsai, 2020). For example, the content of phenolic glycosides and amino acids in cottonwood leaves varies with their developmental stage and its host aphid Chaitophorous populicola was able to detect the difference and to track the preferred leaf stages to optimize its feeding (Gould et al., 2007). In M. persicae, the high glutamine concentration stimulated the feeding behavior (Cao et al., 2017). Overexpressing PrapGR28 from Pieris rapae in Drosophila files resulted in a strong preference to the food with sinigrin whereas the wild-type (WT) files showed avoidance (Yang et al., 2021). In B. mori, BmorGR66 mutant showed no significant feeding preference for both mulberry leaves and Mongolian oak leaves, while WT B. mori did not eat Mongolian oak leaves (Zhang Z.J. et al., 2019). Apart from the GRs, some IRs are also expressed in gustatory organs and are involved in gustation perception (Zhang et al., 2021). For example, IRs expressed in D. melanogaster leg sensilla also showed a response to food components such as sugar, salts, polyamines and bitter compounds (Ling et al., 2014; Hussain et al., 2016). In H. armigera, knockout of IR8a reduced the EAG responses and trend behavior to acetic acid (Zhang et al., 2021). Additionally, IR8a was found to be essential to detect human odors and water detection in Aedes aegypti (Raji et al., 2019a,b). Meanwhile, IR40a, IR93a and IR25a mediate the humidity preference in D. melanogaster (Enjin et al., 2016). Furthermore, IR25a and IR93a are also involved in the detection of temperature. Interestingly, Nmdars have been implicated in associative learning and memory in D. melanogaster and are essential factors for male offspring production in Diploptera punctata (Xia et al., 2005; Huang et al., 2015). Based on the A. spiniferus transcriptome, putative receptors for sugar in GRs, IR8a, IR40a, IR93a, and Nmdars were predicted according to the phylogenetic analyses. Almost all of GRs except AspiGR4 were widely expressed in heads and bodies whilst major of IRs exhibited higher expression in heads than that in bodies. Expressions of selected GRs and IRs showed that the highest expression of AspiGR1, AspiGR3, and AspiIR4 were at puparia. AspiIR5, which was clustered as IR40a, had higher expression in adults than that in nymphs and puparia. All of these results indicated that GRs and IRs in A. spiniferus might be involved in various biological processes and have critical roles in the survival.
Taken together, in this study, we systemically identified six types of sensilla on antennae of including grooved surface trichodea sensilla, chaetae sensilla, microtrichia sensilla, coeloconic sensilla, basiconic sensilla and finger-like sensilla via SEM and a total of 48 chemosensory genes in A. spiniferus including 5 OBPs, 12 CSPs, 3 SNMPs, 6 ORs, 8 GRs, and 14 IRs. Based on the transcriptome data, we developed a tissue-specific expression profile for each of the identified chemosensory genes in A. spiniferus, which might reveal an initial prediction of these genes’ function. Furthermore, we also analyzed the expression of 24 selected chemosensory genes across the developmental stages. In summary, this study not only provides strong background information and initial understanding on the chemosensory systems in host reception of this polyphagous insect but also provides extensive potential targets for pest control. In future, the further investigation about which gene is the key factor of plant perception and the suitable pest management target is needed to be done.
Data Availability Statement
The datasets presented in the study are deposited in the Genbank SRA database, accession number PRJNA792195. Our data has now been released in NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA792195).
Author Contributions
Z-WK designed the research, analyzed transcriptome data, constructed phylogenetic trees, and wrote the manuscript. Z-WK, Y-QG, and M-YL collected sample. Y-QG, Z-ZC, and C-YS performed SEM experiment. Y-QG, Z-ZC, and F-HL conducted qPCR and analyzed qPCR results. YD, H-PZ, Y-YX, and CQ edited the manuscript. Y-YX and Z-WK revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by the Modern Tea Industry Technology System of Shandong Province (SDAIT-19-04).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely thank all staff and students in the Laboratory of Insect Ecology and Physiology (Shandong Agricultural University). We also appreciate the constructive comments from the reviewers which greatly improved the quality of our paper.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2022.847895/full#supplementary-material
Supplementary Figure S1 | Homology analyses results. The BLASTx annotations of A. spiniferus antenna transcripts. (A) E-value distribution. (B) Similarity distribution. (C) Species distribution.
Supplementary Figure S2 | Functional annotation of A. spiniferus transcripts based on gene ontology (GO) categorization.
Supplementary Figure S3 | Alignment of OBPs (A) and CSPs (B) of A. spiniferus.
Supplementary Table S1 | Amino acid sequences of chemosensory genes from A. spiniferus and other insects that used for phylogenetic analyses.
Supplementary Table S2 | Primers used in this work.
Supplementary Table S3 | Summary of A. spiniferus transcriptome annotation.
Supplementary Table S4 | Transmembrane domains in ORs, GRs and IRs.
Footnotes
- ^ https://www.eppo.int/ACTIVITIES/plant_quarantine/A2_list
- ^ http://www.ncbi.nlm.nih.gov/gorf/gorf.html
- ^ http://www.cbs.dtu.dk/services/SignalP/
- ^ http://smart.emblheidelberg.de/
- ^ https://services.healthtech.dtu.dk/service.php?TMHMM-2.0
References
Agnihotri, A., Liu, N. Y., and Xu, W. (2021). Chemosensory proteins (CSPs) in the cotton bollworm Helicoverpa armigera. Insects 13:29. doi: 10.3390/insects13010029
Agnihotri, A. R., Roy, A. A., and Joshi, R. S. (2016). Gustatory receptors in Lepidoptera: chemosensation and beyond. Insect Mol. Biol. 25, 519–529. doi: 10.1111/imb.12246
Buck, L. B. (2004). Olfactory receptors and odor coding in mammals. Nutr. Rev. 62, S184–S188. doi: 10.1301/nr.2004.nov.S184-S188
Calvello, M., Brandazza, A., Navarrini, A., Dani, F. R., Turillazzi, S., Felicioli, A., et al. (2005). Expression of odorant-binding proteins and chemosensory proteins in some Hymenoptera. Insect Biochem. Mol. Biol. 35, 297–307. doi: 10.1016/j.ibmb.2005.01.002
Cao, H. H., Zhanga, Z. F., Wang, X. F., and Liu, T. X. (2017). Nutrition outweighs defense: myzus persicae (green peach aphid) prefers and performs better on young leaves of cabbage. BioRxiv [Preprint] doi: 10.1101/085159
Chen, W., Hasegawa, D. K., Kaur, N., Kliot, A., Pinheiro, P. V., Luan, J. B., et al. (2016). The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 14:110. doi: 10.1186/s12915-016-0321-y
Clyne, P. J., Warr, C. G., and Carlson, J. R. (2000). Candidate taste receptors in Drosophila. Science 287, 1830–1834. doi: 10.1126/science.287.5459.1830
Cui, H. Y., Guo, L. T., Wang, S. L., Xie, W., Jiao, X., Wu, Q. J., et al. (2017). The ability to manipulate plant glucosinolates and nutrients explains the better performance of Bemisia tabaci Middle East-Asia Minor 1 than Mediterranean on cabbage plants. Ecol. Evol. 7, 6141–6150. doi: 10.1002/ece3.2921
Döring, T. F. (2014). How aphids find their host plants, and how they don’t. Ann. Appl. Biol. 165, 3–26. doi: 10.1111/aab.12142
Dunipace, L., Meister, S., McNealy, C., and Amrein, H. (2001). Spatially restricted expression of candidate taste receptors in the Drosophila gustatory system. Curr. Biol. 11, 822–835. doi: 10.1016/S0960-9822(01)00258-5
Enjin, A., Zaharieva, E. E., Frank, D. D., Mansourian, S., Suh, G. S., Gallio, M., et al. (2016). Humidity sensing in Drosophila. Curr. Biol. 26, 1352–1358. doi: 10.1016/j.cub.2016.03.049
Fan, J., Zhang, Y., Francis, F., Cheng, D. F., Sun, J. R., and Chen, J. L. (2015). Orco mediates olfactory behaviors and winged morph differentiation induced by alarm pheromone in the grain aphid, Sitobion avenae. Insect Biochem. Mol. Biol. 64, 16–24. doi: 10.1016/j.ibmb.2015.07.006
Fleischer, J., Pregitzer, P., Breer, H., and Krieger, J. (2018). Access to the odor world: olfactory receptors and their role for signal transduction in insects. Cell Mol. Life Sci. 75, 485–508. doi: 10.1007/s00018-017-2627-5
Forêt, S., Wanner, K. W., and Maleszka, R. (2007). Chemosensory proteins in the honey bee: insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem. Mol. Biol. 37, 19–28. doi: 10.1016/j.ibmb.2006.09.009
Gao, Q., and Chess, A. (1999). Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics 60, 31–39. doi: 10.1006/geno.1999.5894
Gong, D. P., Zhang, H. J., Zhao, P., Lin, Y., Xia, Q. Y., and Xiang, Z. H. (2007). Identification and expression pattern of the chemosensory protein gene family in the silkworm, Bombyx mori. Insect Biochem. Mol. Biol. 37, 266–277. doi: 10.1016/j.ibmb.2006.11.012
Gould, G., Rifleman, P., Perez, A., and Coleman, J. S. (2007). Variation in Eastern cottonwood (Populus deltoides Bartr.) phloem sap content caused by leaf development may affect feeding site selection behavior of the aphid, Chaitophorous populicola Thomas (Homoptera: aphididae). Environ. Entomol. 36, 1212–1225.
Gu, S. H., Wu, K. M., Guo, Y. Y., Pickett, J. A., Field, L. M., Zhou, J. J., et al. (2013). Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics 14:636. doi: 10.1186/1471-2164-14-636
Hallem, E. A., Dahanukar, A., and Carlson, J. R. (2006). Insect odor and taste receptors. Annu. Rev. Entomol. 51, 113–135. doi: 10.1146/annurev.ento.51.051705.113646
Hasanuzzaman, A. T. M., Islam, M. N., Liu, F. H., Cao, H. H., and Liu, T. X. (2018). Leaf chemical compositions of different eggplant varieties affect performance of Bemisia tabaci (Hemiptera: aleyrodidae) nymphs and adults. J. Econ. Entomol. 111, 445–453. doi: 10.1093/jee/tox333
He, P., Zhang, J., Liu, N. Y., Zhang, Y. N., Yang, K., and Dong, S. L. (2011). Distinct expression profiles and different functions of odorant binding proteins in Nilaparvata lugens Stal. PLoS One 6:e28921. doi: 10.1371/journal.pone.0028921
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., et al. (2002). G protein coupled receptors in Anopheles gambiae. Science 298, 176–178. doi: 10.1126/science.1076196
Hu, F. Y., and Tsai, C. W. (2020). Nutritional relationship between Bemisia tabaci and its primary endosymbiont, Portiera aleyrodidarum, during host plant acclimation. Insects 11:498. doi: 10.3390/insects11080498
Hua, J. F., Zhang, S., Cui, J. J., Wang, D. J., Wang, C. Y., Luo, J. Y., et al. (2012). Identification and binding characterization of three odorant binding proteins and one chemosensory protein from Apolygus lucorum (Meyer-Dur). J. Chem. Ecol. 38, 1163–1170. doi: 10.1007/s10886-012-0178-7
Hua, J. F., Zhang, S., Cui, J. J., Wang, D. J., Wang, C. Y., Luo, J. Y., et al. (2013). Functional characterizations of one odorant binding protein and three chemosensory proteins from Apolygus lucorum (Meyer-Dur) (Hemiptera: miridae) legs. J. Insect Physiol. 59, 690–696. doi: 10.1016/j.jinsphys.2013.04.013
Huang, J., Hult, E. F., Marchal, E., and Tobe, S. S. (2015). Identification and characterization of the NMDA receptor and its role in regulating reproduction in the cockroach Diploptera punctata. J. Exp. Biol. 218, 983–990. doi: 10.1242/jeb.115154
Hussain, A., Zhang, M., Ucpunar, H. K., Svensson, T., Quillery, E., Gompel, N., et al. (2016). Ionotropic chemosensory receptors mediate the taste and smell of polyamines. PLoS Biol. 14:e1002454. doi: 10.1371/journal.pbio.1002454
Jiang, X., Pregitzer, P., Grosse-Wilde, E., Breer, H., and Krieger, J. (2016). Identification and characterization of two “Sensory Neuron Membrane Proteins” (SNMPs) of the desert locust, Schistocerca gregaria (Orthoptera: acrididae). J. Insect Sci. 16:33. doi: 10.1093/jisesa/iew015
Jones, W. D., Cayirlioglu, P., Kadow, I. G., and Vosshall, L. B. (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. doi: 10.1038/nature05466
Kang, Z. W., Liu, F. H., Pang, R. P., Yu, W. B., Tan, X. L., Zheng, Z. Q., et al. (2018). The identification and expression analysis of candidate chemosensory genes in the bird cherry-oat aphid Rhopalosiphum padi (L.). Bull. Entomol. Res. 108, 645–657. doi: 10.1017/S0007485317001171
Kang, Z. W., Tian, H. G., Liu, F. H., Liu, X., Jing, X. F., and Liu, T. X. (2017b). Identification and expression analysis of chemosensory receptor genes in an aphid endoparasitoid Aphidius gifuensis. Sci. Rep. 7:3939. doi: 10.1038/s41598-017-03988-z
Kang, Z. W., Liu, F. H., Tian, H. G., Zhang, M. Z., Guo, S. S., and Liu, T. X. (2017a). Evaluation of the reference genes for expression analysis using quantitative real-time polymerase chain reaction in the green peach aphid, Myzus persicae. Insect Sci. 24, 222–234. doi: 10.1111/1744-7917.12310
Kang, Z. W., Liu, F. H., Xu, Y. Y., Cheng, J. H., Lin, X. L., Jing, X. F., et al. (2020). Identification of candidate odorant-degrading enzyme genes in the antennal transcriptome of Aphidius gifuensis. Entomol. Res 51, 36–54. doi: 10.1111/1748-5967.12489
Kent, L. B., Walden, K. K. O., and Robertson, H. M. (2008). The gr family of candidate gustatory and olfactory receptors in the yellow-fever mosquito Aedes aegypti. Chem. Senses 33, 79–93. doi: 10.1093/chemse/bjm067
Knolhoff, L. M., and Heckel, D. G. (2014). Behavioral assays for studies of host plant choice and adaptation in herbivorous insects. Annu. Rev. Entomol. 59, 263–278. doi: 10.1146/annurev-ento-011613-161945
Kong, X., Li, Z. X., Gao, Y. Q., Liu, F. H., Chen, Z. Z., Tian, H. G., et al. (2021). Genome-wide identification of neuropeptides and their receptors in an aphid endoparasitoid wasp, Aphidius gifuensi. Insects 12:745. doi: 10.3390/insects12080745
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Letunic, I., and Bork, P. (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. doi: 10.1093/nar/gkz239
Li, F., Venthur, H., Wang, S., Homem, R. A., and Zhou, J. J. (2021). Evidence for the involvement of the chemosensory protein Agoscsp5 in resistance to insecticides in the cotton aphid, Aphis gossypii. Insects 12:335. doi: 10.3390/insects12040335
Li, F. Q., Li, D., Dewer, Y., Qu, C., Yang, Z., Tian, J., et al. (2019). Discrimination of oviposition deterrent volatile β-ionone by odorant-binding proteins 1 and 4 in the whitefly Bemisia tabaci. Biomolecules 9:563. doi: 10.3390/biom9100563
Li, Z. B., Zhang, Y. Y., An, X. K., Wang, Q., Khashaveh, A., Gu, S. H., et al. (2020). Identification of leg chemosensory genes and sensilla in the Apolygus lucorum. Front. Physiol. 11:276. doi: 10.3389/fphys.2020.00276
Ling, F., Dahanukar, A., Weiss, L. A., Kwon, J. Y., and Carlson, J. R. (2014). The molecular and cellular basis of taste coding in the legs of Drosophila. J. Neurosci. 34, 7148–7164. doi: 10.1523/JNEUROSCI.0649-14.2014
Liu, F. H., Wickham, J. D., Cao, Q. J., Lu, M., and Sun, J. H. (2020). An invasive beetle–fungus complex is maintained by fungal nutritional-compensation mediated by bacterial volatiles. ISME J. 14, 1–14. doi: 10.1038/s41396-020-00740-w
Liu, J. B., Liu, H., Yi, J. Q., Mao, Y. K., Li, J., Sun, D. L., et al. (2021). Transcriptome characterization and expression analysis of chemosensory genes in Chilo sacchariphagus (Lepidoptera Crambidae), a key pest of sugarcane. Front. Physiol. 12:636353. doi: 10.3389/fphys.2021.636353
Liu, Y. L., Guo, H., Huang, L. Q., Pelosi, P., and Wang, C. Z. (2014). Unique function of a chemosensory protein in the proboscis of two Helicoverpa species. J. Exp. Biol. 217, 1821–1826. doi: 10.1242/jeb.102020
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2–ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, C., Cui, S., Tian, Z., Zhang, Y., Chen, G., Gao, X., et al. (2019). OcomCSP12, a chemosensory protein expressed specifically by ovary, mediates reproduction in Ophraella communa (Coleoptera: chrysomelidae). Front. Physiol. 10:1290. doi: 10.3389/fphys.2019.01290
Mckenna, M. P., Hekmatscafe, D. S., Gaines, P., and Carlson, J. R. (1994). Putative Drosophila pheromone-binding proteins expressed in a subregion of the olfactory system. J. Biol. Chem. 269, 16340–16347. doi: 10.1016/S0021-9258(17)34013-9
Mei, T., Fu, W. B., Li, B., He, Z. B., and Chen, B. (2018). Comparative genomics of chemosensory protein genes (CSPs) in twenty-two mosquito species (Diptera: culicidae): identification, characterization, and evolution. PLoS One 13:e0190412. doi: 10.1371/journal.pone.0190412
Mokrane, S., Cavallo, G., Tortorici, F., Romero, E., Fereres, A., Djelouah, K., et al. (2020). Behavioral effects induced by organic insecticides can be exploited for a sustainable control of the orange spiny whitefly Aleurocanthus spiniferus. Sci. Rep. 10:15746. doi: 10.1038/s41598-020-72972-x
Nomura, A., Kawasaki, K., Kubo, T., and Natori, S. (1992). Purification and localization of p10, a novel protein that increases in nymphal regenerating Legs of Periplaneta americana (American Cockroach). Int. J. Dev. Biol. 36, 391–398.
Nugnes, F., Laudonia, S., Jesu, G., Jansen, M. G. M., Bernardo, U., and Porcelli, F. (2020). Aleurocanthus spiniferus (Hemiptera: aleyrodidae) in some european countries: diffusion, hosts, molecular characterization, and natural enemies. Insects 11:42. doi: 10.3390/insects11010042
Pelosi, P., Iovinella, I., Zhu, J., Wang, G. R., and Dani, F. R. (2017). Beyond chemoreception: diverse tasks of soluble olfactory proteins in insects. Biol. Rev. 93, 184–200. doi: 10.1111/brv.12339
Pelosi, P., Zhou, J. J., Ban, L. P., and Calvello, M. (2006). Soluble proteins in insect chemical communication. Cell Mol. Life Sci. 63, 1658–1676. doi: 10.1007/s00018-005-5607-0
Picimbon, J. F., Dietrich, K., Breer, H., and Krieger, J. (2000). Chemosensory proteins of Locusta migratoria (Orthoptera: acrididae). Insect Biochem. Mol. Biol. 30, 233–241. doi: 10.1016/S0965-1748(99)00121-6
Picimbon, J. F., Dietrich, K., Krieger, J., and Breer, H. (2001). Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem. Mol. Biol. 31, 1173–1181. doi: 10.1016/S0965-1748(01)00063-7
Qiao, H., Tuccori, E., He, X., Gazzano, A., Field, L., Zhou, J. J., et al. (2009). Discrimination of alarm pheromone (E)-β-farnesene by aphid odorant-binding proteins. Insect Biochem. Mol. Biol. 39, 414–419. doi: 10.1016/j.ibmb.2009.03.004
Radonjić, S., and Hrnčić, S. (2021). Spreading of Aleurocanthus spiniferus (Quaintance) (Hemiptera: aleyrodidae) in coastal area of Montenegro. Acta Hortic. 1308, 311–318. doi: 10.17660/ActaHortic.2021.1308.44
Raji, J. I., Gonzalez, S., and DeGennaro, M. (2019a). Aedes aegypti Ir8a mutant female mosquitoes show increased attraction to standing water. Commun. Integr. Biol. 12, 181–186. doi: 10.1080/19420889.2019.1681063
Raji, J. I., Melo, N., Castillo, J. S., Gonzalez, S., Saldana, V., Stensmyr, M. C., et al. (2019b). Aedes aegypti mosquitoes detect acidic volatiles found in human odor using the IR8a pathway. Curr. Biol. 29, 1253.e1257–1262.e1257. doi: 10.1016/j.cub.2019.02.045
Richards, S., Gibbs, R. A., Weinstock, G. M., Brown, S. J., Denell, R., Beeman, R. W., et al. (2008). The genome of the model beetle and pest Tribolium castaneum. Nature 452, 949–955. doi: 10.5167/uzh-2931
Robertson, H. M., Warr, C. G., and Carlson, J. R. (2003). Molecular evolution of the insect chemoreceptor gene superfamily in Drosophila melanogaster. Proc. Natl. Acad. Sci. U.S.A. 100, 14537–14542. doi: 10.1073/pnas.2335847100
Sato, K., Tanaka, K., and Touhara, K. (2011). Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. U.S.A. 108, 11680–11685. doi: 10.1073/pnas.1019622108
Scott, K., Brady, R., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C., et al. (2001). chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673. doi: 10.1016/S0092-8674(01)00263-X
Shi, X. B., Wang, X. Z., Zhang, D. Y., Zhang, Z. H., Zhang, Z., Cheng, J., et al. (2019). Silencing of odorant-binding protein gene OBP3 using RNA interference reduced virus transmission of tomato chlorosis virus. Int. J. Mol. Sci. 20:4969. doi: 10.3390/ijms20204969
Sun, D. D., Huang, Y., Qin, Z. J., Zhan, H. X., Zhang, J. P., Liu, Y., et al. (2020). Identification of candidate olfactory genes in the antennal transcriptome of the stink bug Halyomorpha halys. Front. Physiol. 11:876. doi: 10.3389/fphys.2020.00876
Sun, L., Li, Y., Zhang, Z., Guo, H., Xiao, Q., Wang, Q., et al. (2019). Expression patterns and ligand binding characterization of Plus-C odorant-binding protein 14 from Adelphocoris lineolatus (Goeze). Comp. Biochem. Physiol. B 227, 75–82. doi: 10.1016/j.cbpb.2018.10.001
Tang, X. T., Tao, H. H., and Du, Y. Z. (2015). Microsatellite-based analysis of the genetic structure and diversity of Aleurocanthus spiniferus (Hemiptera: aleyrodidae) from tea plants in China. Gene 560, 107–113. doi: 10.1016/j.gene.2015.01.050
Tian, J. H., Zhan, H. X., Dewer, Y., Zhang, B. Y., Qu, C., Luo, C., et al. (2021). Whitefly network analysis reveals gene modules involved in host plant selection, development and evolution. Front. Physiol. 12:656649. doi: 10.3389/fphys.2021.656649
Tian, Y. Y., Chen, Z. J., Huang, X. Q., Zhang, L. X., and Zhang, Z. Q. (2020). Evaluation of botanicals for management of piercing-sucking pests and the effect on beneficial arthropod populations in tea trees Camellia sinensis (L.) O. Kuntze (Theaceae). J. Insect Sci. 20:27. doi: 10.1093/jisesa/ieaa101
Wang, R., Hu, Y., Wei, P., Qu, C., and Luo, C. (2020). Molecular and functional characterization of one odorant-binding protein gene OBP3 in Bemisia tabaci (Hemiptera: aleyrodidae). J. Econ. Entomol. 113, 299–305. doi: 10.1093/jee/toz248
Wang, R., Li, F. Q., Zhang, W., Zhang, X. M., Qu, C., Tetreau, G., et al. (2017). Identification and expression profile analysis of odorant binding protein and chemosensory protein genes in Bemisia tabaci MED by head transcriptome. PLoS One 12:e0171739. doi: 10.1371/journal.pone.0171739
Wanner, K. W., and Robertson, H. M. (2008). The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629. doi: 10.1111/j.1365-2583.2008.00836.x
Waris, M. I., Younas, A., Ui Qamar, M. T., Hao, L., Ameen, A., Ali, S., et al. (2018). Silencing of chemosensory protein gene NlugCSP8 by RNAi induces declining behavioral responses of Nilaparvata lugens. Front. Physiol. 9:379. doi: 10.3389/fphys.2018.00379
Wu, Q., Li, C. X., Liu, Q. M., Guo, X. X., Shi, Q. M., Zhang, H. D., et al. (2020). RNA interference of odorant receptor CquiOR114/117 affects blood-feeding behavior in Culex quinquefasciatus. Acta Trop. 204:105343. doi: 10.1016/j.actatropica.2020.105343
Xia, J. X., Guo, Z. J., Yang, Z. Z., Han, H. L., Wang, S. L., Xu, H. F., et al. (2021). Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184, 1693.e1617–1705.e1617. doi: 10.1016/j.cell.2021.02.014
Xia, S., Miyashita, T., Fu, T. F., Lin, W. Y., Wu, C. L., Pyzocha, L., et al. (2005). NMDA receptors mediate olfactory learning and memory in Drosophila. Curr. Biol. 15, 603–615. doi: 10.1016/j.cub.2005.02.059
Xie, W., Chen, C. H., Yang, Z. Z., Guo, L. T., Yang, X., Wang, D., et al. (2017). Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. Gigascience 6, 1–7. doi: 10.1093/gigascience/gix018
Xu, W., Papanicolaou, A., Zhang, H. J., and Anderson, A. (2016). Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci. Rep. 6:23666. doi: 10.1038/srep23666
Yang, J., Guo, H., Jiang, N. J., Tang, R., Li, G. C., Huang, L. Q., et al. (2021). Identification of a gustatory receptor tuned to sinigrin in the cabbage butterfly Pieris rapae. PLoS Genet. 17:e1009527. doi: 10.1371/journal.pgen.1009527
Yang, K., Gong, X. L., Li, G. C., Huang, L. Q., Ning, C., and Wang, C. Z. (2020). A gustatory receptor tuned to the steroid plant hormone brassinolide in Plutella xylostella (Lepidoptera: plutellidae). eLife 9:e64114. doi: 10.7554/eLife.64114
Yang, K., He, P., and Dong, S. L. (2014). Different expression profiles suggest functional differentiation among chemosensory proteins in Nilaparvata lugens (Hemiptera: delphacidae). J. Insect Sci. 14:270. doi: 10.1093/jisesa/ieu132
Zeng, Y., Merchant, A., Wu, Q. J., Wang, S. L., Kong, L., Zhou, X. G., et al. (2020). A chemosensory protein BtabCSP11 mediates reproduction in Bemisia tabaci. Front. Physiol. 11:709. doi: 10.3389/fphys.2020.00709
Zeng, Y., Yang, Y. T., Wu, Q. J., Wang, S. L., Xie, W., and Zhang, Y. J. (2019). Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 26, 620–634. doi: 10.1111/1744-7917.12576
Zhang, H. J., Xu, W., Chen, Q. M., Sun, L. N., Anderson, A., Xia, Q. Y., et al. (2018). Functional characterization of sensory neuron membrane proteins (SNMPs). bioRxiv [Preprint] doi: 10.1101/262154
Zhang, R. B., Liu, Y., Yan, S. C., and Wang, G. R. (2019). Identification and functional characterization of an odorant receptor in pea aphid, Acyrthosiphon pisum. Insect Sci. 26, 58–67. doi: 10.1111/1744-7917.12510
Zhang, Y., Shen, C., Xia, D. S., Wang, J., and Tang, Q. F. (2019). Characterization of the expression and functions of two odorant-binding proteins of Sitophilus zeamais Motschulsky (Coleoptera: curculionoidea). Insects 10:409. doi: 10.3390/insects10110409
Zhang, Z. J., Zhang, S. S., Niu, B. L., Ji, D. F., Liu, X. J., Li, M. W., et al. (2019). A determining factor for insect feeding preference in the silkworm, Bombyx mori. PLoS Biol. 17:e3000162. doi: 10.1371/journal.pbio.3000162
Zhang, R. B., Wang, B., Grossi, G., Falabella, P., Liu, Y., Yan, S. C., et al. (2017). Molecular basis of alarm pheromone detection in aphids. Curr. Biol. 27, 55–61. doi: 10.1016/j.cub.2016.10.013
Zhang, X. M., Wang, S., Li, S., Luo, C., Li, Y. X., and Zhang, F. (2015). Comparison of the antennal sensilla ultrastructure of two cryptic species in Bemisia tabaci. PLoS One 10:e0121820. doi: 10.1371/journal.pone.0121820
Zhang, X. X., Yang, B., Sun, D. D., Guo, M. B., Zhang, J., and Wang, G. R. (2021). Ionotropic receptor 8a is involved in the attraction of Helicoverpa armigera to acetic acid. Insect Sci. [Online ahead of print] doi: 10.1111/1744-7917.12962
Zhong, T., Yin, J., Deng, S., Li, K., and Cao, Y. (2012). Fluorescence competition assay for the assessment of green leaf volatiles and trans-beta-farnesene bound to three odorant-binding proteins in the wheat aphid Sitobion avenae (Fabricius). J. Insect Physiol. 58, 771–781. doi: 10.1016/j.jinsphys.2012.01.011
Keywords: Aleurocanthus spiniferus, transcriptome, antennal sensilla, chemosensory genes, expression patterns
Citation: Gao Y-Q, Chen Z-Z, Liu M-Y, Song C-Y, Jia Z-F, Liu F-H, Qu C, Dewer Y, Zhao H-P, Xu Y-Y and Kang Z-W (2022) Characterization of Antennal Chemosensilla and Associated Chemosensory Genes in the Orange Spiny Whitefly, Aleurocanthus spiniferus (Quaintanca). Front. Physiol. 13:847895. doi: 10.3389/fphys.2022.847895
Received: 03 January 2022; Accepted: 20 January 2022;
Published: 28 February 2022.
Edited by:
Xin-Cheng Zhao, Henan Agricultural University, ChinaReviewed by:
Herbert Venthur, University of La Frontera, ChileZhao-Qun Li, Tea Research Institute (CAAS), China
Copyright © 2022 Gao, Chen, Liu, Song, Jia, Liu, Qu, Dewer, Zhao, Xu and Kang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yong-Yu Xu, xuyy@sdau.edu.cn; Zhi-Wei Kang, zwkang2010@126.com
†These authors have contributed equally to this work
 Yu-Qing Gao1†
Yu-Qing Gao1† Zhi-Fei Jia
Zhi-Fei Jia Fang-Hua Liu
Fang-Hua Liu Cheng Qu
Cheng Qu Hai-Peng Zhao
Hai-Peng Zhao Yong-Yu Xu
Yong-Yu Xu Zhi-Wei Kang
Zhi-Wei Kang