CD40 gene polymorphism and its expression in children with Kawasaki disease from North India: a preliminary case–control study and meta-analysis
- 1Department of Pediatrics, All India Institute of Medical Sciences, Patna, India
- 2Allergy Immunology Unit, Department of Pediatrics, Advanced Pediatrics Centre, Post Graduate Institute of Medical Education and Research (PGIMER), Chandigarh, India
- 3Genetics and Metabolic Unit, Advanced Pediatrics Centre, Post Graduate Institute of Medical Education and Research, Chandigarh, India
Introduction: CD40 gene single-nucleotide polymorphisms (SNPs) have been associated with susceptibility and development of coronary artery abnormalities (CAAs) in children with Kawasaki disease (KD) in Japanese, Chinese, and Taiwanese populations. However, data on SNPs of the CD40 gene in patients with KD from the Indian subcontinent are not available. We studied the CD40 gene polymorphisms and its expression in children with KD from North India.
Methods: SNPs of the CD40 gene (rs4810485, rs1535045) were studied using Sanger sequencing. CD40 expression was studied by flow cytometry. Meta-analysis was carried out to assess the role of both SNPs of the CD40 gene in KD. GRADEpro GDT software (v.3.2) was used to assess the “certainty of evidence.”
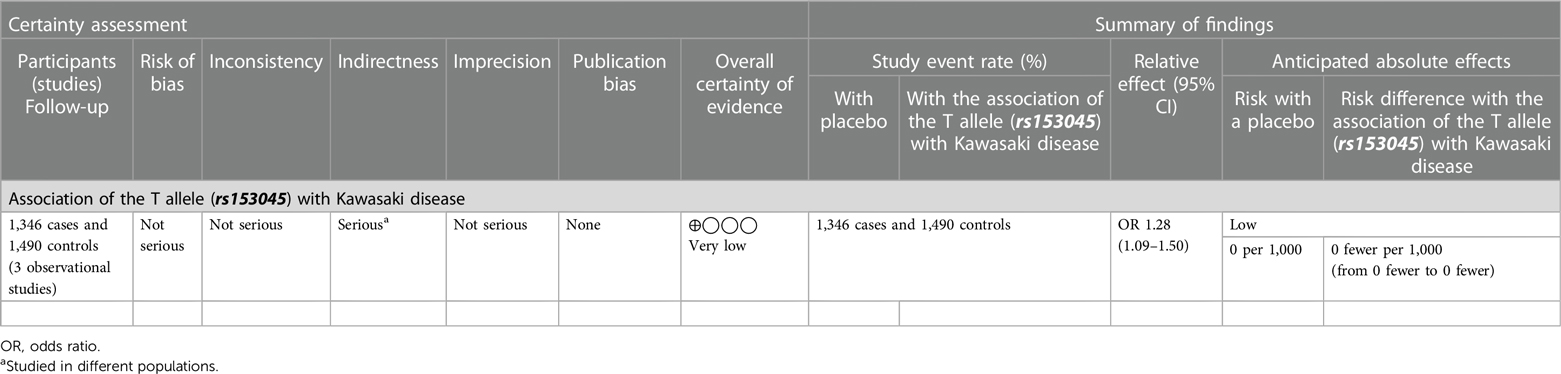
Results: Forty-one patients with KD and 41 age-, sex-matched febrile controls were enrolled. However, none of the alleles and genotypes of the CD40 gene were found to be associated with KD. CD40 expression was higher in KD and in KD with CAAs compared to controls, but it failed to reach statistical significance. In a meta-analysis, the T allele of rs153045 was found to be significantly associated with KD (OR = 1.28; 95% confidence interval (: 1.09–1.50; p = 0.002). The GRADE of evidence for this outcome, however, is of “ very low certainty.”
Conclusion: The present study found no association between SNPs (rs4810485 and rs153045) and susceptibility to KD. This could be a reflection of a modest sample size. CD40 expression was higher in KD and in KD with CAAs. In the meta-analysis, the T allele of rs153045 was significantly associated with KD. Our study confirms a significant genetic heterogeneity in KD among different ethnicities.
Introduction
Kawasaki disease (KD) is a medium-vessel vasculitis that predominantly affects young children (1, 2). The highest incidence of KD has been reported in Northeast Asian countries like Japan, South Korea, and Taiwan (3), ranging from 80 to 359 per 100,000 children below 5, and it continues to rise. Incidence data in the United States and Europe range from 15 to 20 per 100,000 children below 5. To date, no data on the nationwide incidence of KD has been gathered in India. Hospital-based studies in Chandigarh have shown that the incidence of KD is at least 5.35 per 100,000 children below 5 (4, 5). Despite more than 50 years of extensive research, the etiology of KD remains an enigma. Compared to other parts of the world, the high prevalence of KD in the Northeast Asian nations led to the speculation that the genetic endowment of an individual also plays a crucial role in KD evolution. Common genes implicated in KD are CD40, ACE, BLK, CASP3, FCGR2A, FGb, HLA-E, IL1A, IL6, ITPKC, LTA, MPO, PD1, SMAD3, CCL17, and TNF (6). However, genome-wide association studies (GWAS) found that three genes (viz., FCG2RA, BLK, and CD40) are consistently identified in patients with KD (7).
CD40 is a 48-kDA transmembrane protein expressed on the surface of platelets, neutrophils, monocytes, macrophages, and endothelial cells. Single-nucleotide polymorphisms (SNPs) of the CD40 gene have been reported to predispose to KD and increase the risk of coronary artery abnormalities (CAAs) in patients with KD in Japan, China, and Taiwan (8–10). However, there are no data on CD40 gene SNPs in patients with KD from the Indian subcontinent. Herein, we report the role of CD40 gene SNPs, viz., rs153045 and rs4810485, in Indian patients with KD. We have also assessed CD40 expression on B cells in children with KD.
Materials and methods
This case–control study was conducted at the Pediatric Rheumatology Clinic, Advanced Pediatrics Centre, Post Graduate Institute of Medical Education and Research, Chandigarh, India. The study protocol was approved by the Institute Thesis Committee and Institute Ethics Committee. The manuscript was approved by the Departmental Review Board.
The primary objective of the study was to assess the association of SNPs rs1535045 and rs4810485 with KD. The secondary objective was to ascertain CD40 expression on B lymphocytes in children with KD during the acute phase of the disease. We recruited 41 patients with KD, i.e., before administration of intravenous immunoglobulin (IVIg), admitted to the hospital over 1 year. We also enrolled an equal number of age-, sex-matched febrile controls from the inpatient department. Children with known chronic conditions, such as chronic kidney disease, celiac disease, nephrotic syndrome, congenital heart diseases, and neurological disorders, and children on corticosteroids or any other immunosuppressive therapy were excluded from the study.
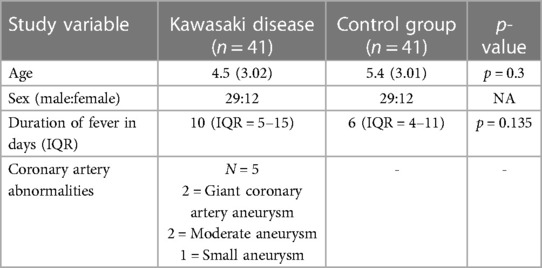
Diagnosis of KD was based on the 2017 American Heart Association treatment guideline (11). The demographic, clinical, echocardiographic, and treatment details of patients with KD were recorded on a predesigned proforma. 2D echocardiography (2DE) was carried out by a Philips EPIQ 7 ultrasound system. Coronary arteries are considered dilated when the Z score is between >2 and <2.5. Coronary artery Z scores of >2.5 are categorized as an aneurysm. The term CAAs encompasses both dilatations and aneurysms. IVIg resistance was considered when there was persistence of or reappearance of fever at least 36 h after completion of a full single IVIg infusion (2 g/kg).
Sample collection and DNA extraction
After obtaining written informed consent, 2 ml of peripheral venous blood was drawn by venipuncture in an ethylene diamine tetraacetate (EDTA) vial and a plain vial from each study subject and control under aseptic conditions. DNA was extracted using the DNA extraction kit and stored at −80°C until further analysis. Genomic DNA was isolated from 200 µl of EDTA blood samples using the QIAamp DNA Blood Mini Kit (Cat. No. 51106, Qiagen, Hilden, Germany).
DNA quality was determined using 1% agarose gel electrophoresis and staining with ethidium bromide. The purity of DNA was determined by measuring the optical density (OD) of samples at 260 and 280 nm using a TECAN Infinite M200 PRO with a Nanoquant plate [TECAN Group (Life Sciences and Diagnostics) AG, Switzerland], and DNA samples were stored at −80°C until further use.
Primer designing
The reference sequence (NG_007279.1) was obtained from the National Center for Biotechnology Information Search database (NCBI). Primers were designed by Primer BLAST. Designed oligos showed highly specific PCR products.
DNA sequencing
For genotyping of the SNPs of the CD40 gene, DNA sequencing was done by Sanger's chain termination method. The sequencing data were obtained from an automated sequencer (ABI PRISM 3100) and analyzed using FinchTV.
CD40 expression on B cells
The CD40 expression assay on B cells of KD patients and controls was carried out by flow cytometry. In total, 100 µl of whole blood was incubated with CD19 FITC (Beckman Coulter, Germany) alone and CD19 fluorescein isothiocyanate (FITC) with the CD40 phycoerythrin (PE) (Beckman Coulter, Germany) antibody for 15 min at room temperature in the dark. After incubation, the cells were lysed using 2 ml of NH4Cl solution for 15 min at room temperature in the dark. After lysis, the cells were centrifuged at 450g for 5 min; then, the supernatant was discarded. After the first spin, the cells were washed with 1× phosphate-buffered saline (PBS) and vortexed. After washing, cells were again centrifuged at 450g for 5 min. After discarding the supernatant, 300 μl of 1× PBS was added, and the cells were ready to acquire on the Navios Flow Cytometer (Beckman Coulter, Germany).
During acquisition, forward scatter (FSC) and side scatter (SSC) were adjusted to get clusters (i.e., lymphocytes, monocytes, and neutrophils) on a dot plot. Optimum CD40 expression was obtained in both tubes, i.e., CD19 alone (fluorescence minus one) and CD19 with CD40. On gating lymphocytes, CD19-positive cells labeled with PE were further gated on a separate dot plot. On gated CD19+ cells, CD40 expression was noted (labeled with FITC) on a histogram. The data were analyzed by Kaluza software.
Statistical analysis
The data were presented as a proportion for categorical variables and mean ± standard deviation for continuous variables. Categorical variables were compared between two groups by a χ2 or Fisher's exact test. Numerical variables were compared between two groups by Student's t-test or the Mann–Whitney U-test, depending upon the type of distribution. The magnitude of effect size was expressed as risk ratio or odds ratio with a 95% confidence interval (CI). A p-value of <0.05 was considered significant. Microsoft Excel version 2019 was used for data entry, and R software version 3.4 was used for data analysis. For the meta-analysis, Revman 5.4 version and Meta Genyo were used. We used GRADEpro GDT software (v 3.2) to assess the certainty of evidence.
Results
Genotype and allele frequencies of rs153405 in patients with KD and controls
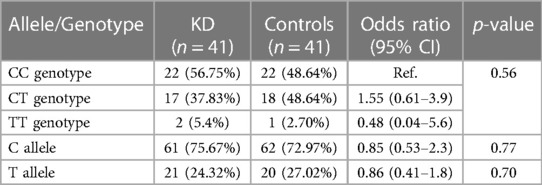
The baseline characteristics of children with KD (29 boys:12 girls) and the control group are depicted in Table 1. The mean age of children in the study group was 4.5 ± 3.01 years, whereas it was 5.4 ± 3.02 years in the control group. The study group had 22 complete and 19 incomplete KD cases. Genotypes were in the Hardy–Weinberg equilibrium. Of 41 genotypes, the frequency of the CC genotype was the highest (22; 53.65%), followed by CT (17; 41.46%) and TT (2; 4.87%). In the control group, the frequency of the CC genotype was the highest (22; 53.65%), followed by CT (18; 43.90%) and TT (1; 2%). Frequencies of the C and T alleles were 61 (74.40%) and 21 (25.60%) in the KD group 62 (75.60%) and 20 (24.39%) in the control group, respectively. The genotype and allele frequencies of both groups are summarized in Table 2. There was no significant difference in the frequency of SNP rs153045 between the study and control groups. Also, multivariate analysis revealed no genotype and allele association of rs153045 with KD (Table 3).
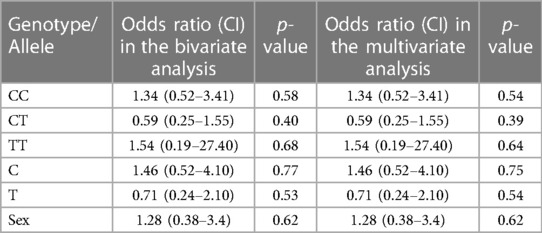
Table 3. Results of bivariate and multivariate logistic regression analyses of genotypes and alleles of SNP rs153045.
Meta-analysis of rs153045
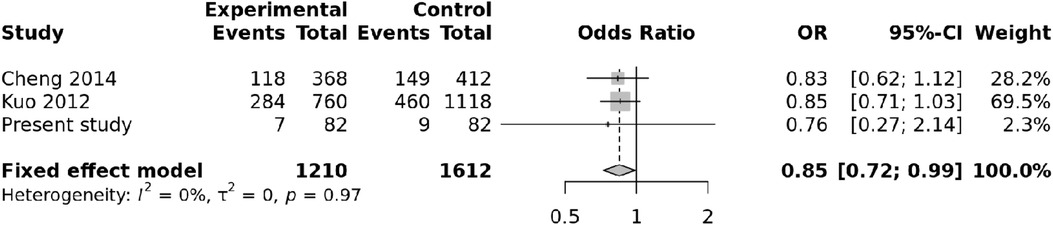
We also carried out a meta-analysis of the results of our study and two previous studies (9, 10). The study reported by Onouchi et al. was not included in the meta-analysis as polymorphisms reported in this study were different from those evaluated in the present study.
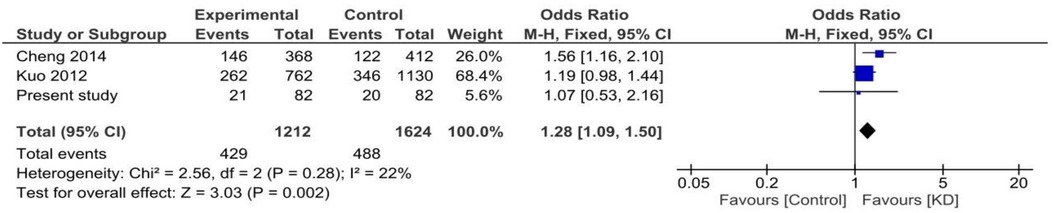
Both random and fixed-effect models were used. No association of genotypes of rs153045 with KD was found (p = 0.34), and there was no heterogeneity (I2 = 0%) (Figure 1). Similarly, no association was noted for the C allele. However, a meta-analysis found that the T allele was significantly associated with KD (p = 0.002) without significant heterogeneity (I2 = 22%) (Figure 2). There was no publication bias on the funnel plot (Egger's test; p = 0.94). However, this evidence was of “very low certainty” (Table 4).
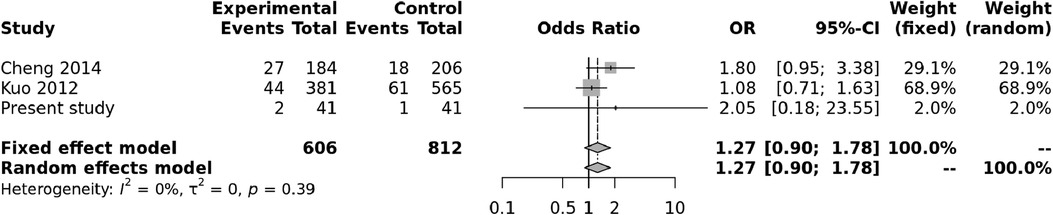
Figure 1. Forest plot showing the association of the rs153045 genotype with Kawasaki disease (recessive model).
Genotype and allele frequencies of rs4810485 in patients with KD and controls
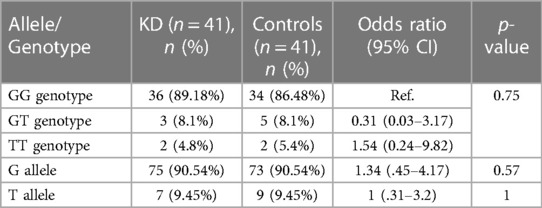
The genotype and allele frequency analyses of SNP rs4810485 are depicted in Table 5. Of 41 genotypes, the GG genotype was found in 36 (87.80%) patients, with three GT (7.3%) and two TT (4.8%) genotypes in the KD group. Frequencies of G and T alleles in the KD group were 75 (91.46%) and 7 (8.54%), respectively. Likewise, the control group had 34 GG (82.93%), five GT (12.19%), and two TT (4.8%) genotypes. The frequencies of G and T alleles were 73 (89.02%) and nine (10.98%), respectively. The difference in genotype and allele frequencies between KD and control groups was not statistically significant. The results of the multivariate analysis are presented in Table 6. There was no association of genotypes and alleles of rs4810485 with KD.
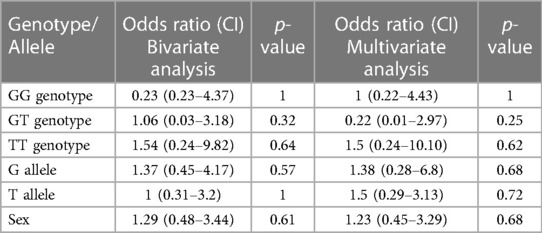
Table 6. Results of bivariate and multivariate logistic regression analyses of genotypes and alleles of SNP rs4810485.
Meta-analysis of rs4810485
We also carried out a meta-analysis of genotypes and alleles of rs4810485. No association was observed for genotypes of rs4810485 with KD (p = 0.86), and there was no heterogeneity (Figure 3). Pooled results and T alleles were also not associated with KD, p = 0.97, and there was no heterogeneity (Figure 4).
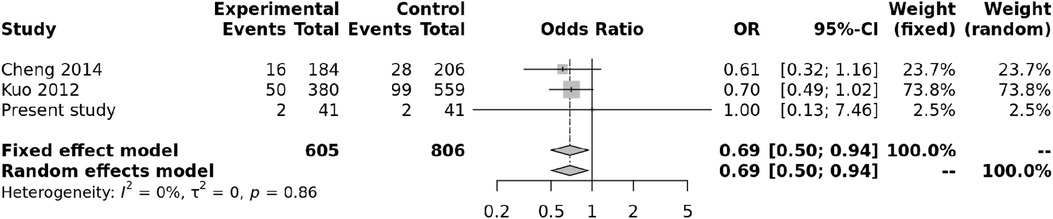
Figure 3. Forest plot showing the association of the rs4810485 genotype with Kawasaki disease (recessive model).
CD40 expression assay
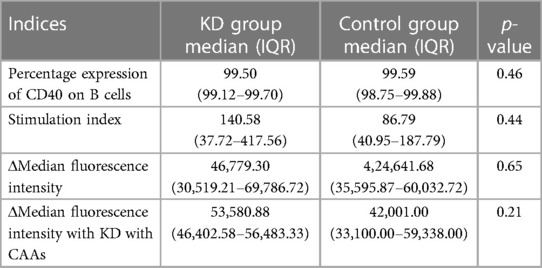
The CD40 expression assay on B (CD19+) cells was performed in patients with KD during the acute phase of the disease and in febrile controls. Flow cytometry parameters of both groups are depicted in Table 7. Although the median difference of the stained index (SI) and Δmedian fluorescence intensity (MFI) between the KD and control groups was not statistically significant, it was noted to be higher in the KD group. Similarly, compared to febrile controls, the ΔMFI was higher in patients with KD and CAAs; however, it failed to reach statistical significance (p = 0.21).
Discussion
KD is the leading cause of vasculitis worldwide, and coronary artery abnormalities may develop in up to 25% of untreated patients (12). A higher incidence of KD in Japan, Korea, and Taiwan suggests that genetic components play a role in the pathogenesis of KD (13, 14). Over the last few decades, extensive research has been carried out to identify the underlying genetic component that might predispose people to KD (15).
We evaluated the CD40 expression on B cells and SNPs of the CD40 gene (rs1535045 and rs4810485) in a single-center cohort of patients with KD from North India. There was no significant difference in the flow cytometry expression of CD40 on B cells and SNPs in the CD40 gene in patients with KD compared with controls. A meta-analysis of previously reported studies and the present study showed that the T allele of rs153045 polymorphism in the CD40 gene was significantly more common in patients with KD than controls.
The interaction of CD40–CD40ligand (hereafter CD40l) is believed to play a pivotal role in acute coronary syndrome (16–18). CD40–CD40l interaction leads to the proliferation of B cells, isotype switching, and release of proinflammatory cytokines (TNFα, IL10, and IL6) (19). CD40–CD0l signaling in non-hematopoietic cells also plays a crucial role in inflammation (20). However, its role in the evolution of KD (especially in KD with CAAs) is unclear.
In 2012, Onouchi et al. reported a strong association between CD40 gene polymorphisms (rs4813003) and KD development (8). Subsequently, two more studies have reported the association of different SNPs of the CD40 gene with KD (9, 10).
In the present study, the difference in genotypes of rs153045 (CC, CT, and TT) was not statistically significant between patients with KD and controls. The frequencies of C and T alleles in both groups were also not significantly different. CC was the dominant genotype, followed by CT and TT in both groups. Likewise, the C allele was the primary allele in both groups, followed by the T allele.
Genotype and allele constitution in the present study was similar to the Taiwanese population but differed from the Han Chinese population (9, 10). Cheng et al. have shown an association of rs153045 with KD (10). Furthermore, the authors illustrated an association of the T allele with KD susceptibility (9). Kuo et al. have also reported a similar association of the rs153045 T allele with KD. However, no association of the T allele with KD was observed in the present cohort (OR: 1.16; 95% CI: 0.54–2.4, p = 0.70). In the meta-analysis including these three studies, the association of the T allele with KD was significant.
A substantial difference in genotype distributions between KD patients and controls of SNP rs153045 has been documented by Cheng et al. from China. Similarly, Kuo et al. have reported an association of rs153045 with KD in the dominant model, although it was not significant in haplotype analysis. We observed no association of rs153045 genotypes with KD in our cohort. A meta-analysis on rs153045 genotypes also showed no significant association.
Regarding SNP rs4810485, GG genotypes and G alleles were the most common genotypes and alleles in both groups. The genotype and allele frequencies of SNP rs4810485 in our cohort differed from those in the Taiwanese and Chinese populations. The GT genotype has been reported as the most frequent genotype in both studies (9, 10). However, the G allele has been reported as the predominant allele in these studies. This finding is in accordance with our results.
In our study, the genotype and allele frequencies of rs4810485 between KD patients and controls (p = 0.54) were not statistically different. Our results were similar to the study by Cheng et al., where the authors also reported no association between rs4810485 and KD (9).
Although the risk of CAAs and KD with SNP rs4810485 has been reported by Kuo et al., this association was not evident in haplotype analysis (10). Further, the meta-analysis also showed no association of any genotypes and alleles of SNP rs4810485 with KD. The lack of association of genotypes of SNPs rs153045 and rs4810485 with KD in our cohort suggests that there is significant genetic heterogeneity in KD among different ethnicities. In addition, it is important to note that the phenotype of KD in India may be different from the rest of the world (21–23). This could be related to differences in genetic background.
To date, only two studies have explored the role of the CD40–CD40l pathway in KD (24, 25). We have previously demonstrated a significant elevation of CD40l expression in KD before the administration of intravenous immunoglobulin compared to healthy controls (24). However, it was not significantly different compared to the febrile controls. Furthermore, the soluble CD40l was not significantly different before IVIg administration. In a similar report, Wang et al. showed increased expression of CD40l on CD4 and CD8 T cells and platelets during the acute stage of KD compared to febrile controls (25). Furthermore, the CD40l expression was found to be significantly correlated with the occurrence of CAAs in KD.
To the best of our knowledge, no studies have demonstrated the CD40 expression in children with KD. We showed that CD40 expression on B cells was higher in KD and in KD with CAAs than in controls. However, this difference failed to reach a statistical significance. Our findings suggest that the interaction of CD40 and CD40l pathways may have a role in the inflammation and development of CAAs in children with KD.
The strength of our study was that genotypes were determined using Sanger sequencing, which is considered to be the gold standard. In addition, CD40 expression was also studied simultaneously. We have also carried out a meta-analysis of previously published studies to examine the relationship between different alleles and genotypes of the CD40 gene and the risk of KD. However, the sample size of our study was admittedly modest. This is understandable because the work pertains to the DM dissertation of the first author (PP); the work had to be concluded over a definite time period. Other lacunae of our study that functional validation and haplotype analysis could not be done. Hence, extrapolation of these findings to the community may be open to questions.
In conclusion, our study confirms genetic heterogeneity in KD among different populations. CD40 gene polymorphisms appear to be associated with (i) susceptibility to develop KD and (ii) KD with CAAs. Further studies with a larger sample size are required to validate these findings.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://api.medgag.com/cases/Data-CD40-and-Kawasaki-disease/
Ethics statement
The studies involving humans were approved by the Post Graduate Institute of Medical Education and Research. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent for participation in this study was provided by the participants’ legal guardians/next of kin. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
PP: Writing of initial draft of the manuscript, editing, and revision of manuscript at all stages of its production, acquisition of images, patient management, review of literature, and final approval. AJ: Inception of idea, evaluation, management, follow-up of the patient, and critical revision of the manuscript at all stages of production and final approval. RR: Writing of initial draft of the manuscript, editing and revision of the manuscript at all stages of production, patient management, and final approval. AK: Editing and revision of the manuscript, image acquisition, patient management, and final approval. PS: Editing and revision. DS: Editing and revision. AR: Editing and revision of the manuscript. RP: Editing and revision of the manuscript. SS: Editing and revision of the manuscript, patient management, overall supervision of clinical stage, and final approval. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Newberger JW, Son MBF. Kawasaki disease. In: Kliegman RM, Stanton BF, Schor NF, editors. Nelson text book of pediatrics. 20th ed. New York: Elsevier (2016). p. 1209–13.
2. Son MB, Sundel RP. Kawasaki disease. In: Petty RE, Laxer RM, Lindsay CB, Wedderburn LR, editors. Text book of pediatric rheumatology. 7th ed. Philadelphia, PA: Elsevier (2016). p. 467–83.
3. Singh S, Vignesh P, Burgner D. The epidemiology of Kawasaki disease: a global update. Arch Dis Child. (2015) 100(11):1084–8. doi: 10.1136/archdischild-2014-307536
4. Singh S, Aulakh R, Bhalla AK, Suri D, Manojkumar R, Narula N, et al. Is Kawasaki disease incidence rising in Chandigarh, North India? Arch Dis Child. (2011) 96(2):137–40. doi: 10.1136/adc.2010.194001
5. Singh S, Bhattad S. Kawasaki disease incidence at Chandigarh, North India, during 2009–2014. Rheumatol Int. (2016) 36(10):1391–7. doi: 10.1007/s00296-016-3543-y
6. Xie X, Shi X, Liu M. The roles of genetic factors in Kawasaki disease: a systematic review and meta-analysis of genetic association studies. PediatrCardiol. (2018) 39(2):207–25. doi: 10.1007/s00246-017-1760-0
7. Onouchi Y. The genetics of Kawasaki disease. Int J Rheum Dis. (2018) 21(1):26–30. doi: 10.1111/1756-185X.13218
8. Onouchi Y, Ozaki K, Burns JC, Shimizu C, Terai M, Hamada H, et al. A genome-wide association study identifies three new risk loci for Kawasaki disease. Nat Genet. (2012) 44(5):517–21. doi: 10.1038/ng.2220
9. Cheng S-C, Cheng Y-Y, Wu J-L. Association between gene polymorphism of CD40 gene and coronary artery lesion in Kawasaki disease. Zhongguo Dang Dai Er Ke Za Zhi. (2014) 16(10):1025–8. doi: 10.7499/j.issn.1008-8830.2014.10.013
10. Kuo H-C, Chao M-C, Hsu Y-W, Lin Y-C, Huang Y-H, Yu H-R, et al. CD40 gene polymorphisms associated with susceptibility and coronary artery lesions of Kawasaki disease in the Taiwanese population. Sci World J. (2012) 2012:520865. doi: 10.1100/2012/520865
11. McCrindle BW, Rowley AH, Newburger JW, Burns JC, Bolger AF, Gewitz M, et al. Diagnosis, treatment, and long-term management of Kawasaki disease: a scientific statement for health professionals from the American heart association. Circulation. (2017) 135(17):e927–99. doi: 10.1161/CIR.0000000000000484
12. Hedrich CM, Schnabel A, Hospach T. Kawasaki disease. Front Pediatr. (2018) 6:198. doi: 10.3389/fped.2018.00198
13. Nakamura Y, Yashiro M, Uehara R, Sadakane A, Tsuboi S, Aoyama Y, et al. Epidemiologic features of Kawasaki disease in Japan: results of the 2009–2010 nationwide survey. J Epidemiol. (2012) 22:216–21. doi: 10.2188/jea.JE20110126
14. Dergun M, Kao A, Hauger SB, Newburger JW, Burns JC. Familial occurrence of Kawasaki syndrome in North America. Arch Pediatr Adolesc Med. (2005) 159:876–81. doi: 10.1001/archpedi.159.9.876
15. Xie X, Shi X, Liu M. The roles of genetic factors in Kawasaki disease: a systematic review and meta-analysis of genetic association studies. Pediatr Cardiol. (2018) 39(2):207–25. doi: 10.1007/s00246-017-1760-0
16. Aukrust P, Muller F, Ueland T, et al. Enhanced levels of soluble and membrane-bound CD40 ligand in patients with unstable angina. Possible reflection of T lymphocyte and platelet involvement in the pathogenesis of acute coronary syndromes. Circulation. (1999) 100:614–20. doi: 10.1161/01.CIR.100.6.614
17. Garlichs CD, Eskafi S, Raaz D, et al. Patients with acute coronary syndromes express enhanced CD40 ligand/CD154 on platelets. Heart. (2001) 86:649–55. doi: 10.1136/heart.86.6.649
18. Lee Y, Lee WH, Lee SC, et al. CD40l activation in circulating platelets in patients with acute coronary syndrome. Cardiology. (1999) 92:11–16. doi: 10.1159/000006940
19. Khanna R, Cooper L, Kienzle N, Moss DJ, Burrows SR, Khanna KK. Engagement of CD40 antigen with soluble CD40 ligand up-regulates peptide transporter expression and restores endogenous processing function in Burkitt’s lymphoma cells. J Immunol. (1997) 159(12):5782–5. doi: 10.4049/jimmunol.159.12.5782
20. Schönbeck U, Libby P. The CD40/CD154 receptor/ligand dyad. Cell Mol Life Sci CMLS. (2001) 58(1):4–43. doi: 10.1007/PL00000776
21. Singh S, Bansal A, Gupta A, Kumar RM, Mittal BR. Kawasaki disease: a decade of experience from North India. Int Heart J. (2005) 46(4):679–89. doi: 10.1536/ihj.46.679
22. Singh S, Gupta MK, Bansal A, Kumar RM, Mittal BR. A comparison of the clinical profile of Kawasaki disease in children from Northern India above and below 5 years of age. Clin Exp Rheumatol. (2007) 25(4):654–7.17888228
23. Kushner HI, Macnee R, Burns JC. Impressions of Kawasaki syndrome in India. Indian Pediatr. (2006) 43(11):939–42.17151396
24. Jindal AK, Rawat A, Goel S, Shandilya J, Saikia B, Minz RW, et al. Expression of CD40 ligand on T cells and soluble CD40 ligand in children with Kawasaki disease: a single-center preliminary study from North India. J Clin Rheumatol. (2021) 27(5):194–200. doi: 10.1097/RHU.0000000000001283
Keywords: Kawasaki disease, CD40 gene, SNP rs153045, SNP rs4810485, CD40 expression
Citation: Patra PK, Jindal AK, Rikhi R, Kaur A, Srivastava P, Suri D, Rawat A, Pilania R and Singh S (2023) CD40 gene polymorphism and its expression in children with Kawasaki disease from North India: a preliminary case–control study and meta-analysis. Front. Pediatr. 11:1252024. doi: 10.3389/fped.2023.1252024
Received: 3 July 2023; Accepted: 31 August 2023;
Published: 21 September 2023.
Edited by:
Lovro Lamot, University of Zagreb, CroatiaReviewed by:
Xiaohui Li, Children's Hospital of Capital Institute of Pediatrics, ChinaDragana Lazarevic, University Clinical Center Nis, Serbia
© 2023 Patra, Jindal, Rikhi, Kaur, Srivastava, Suri, Rawat, Pilania and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pratap Kumar Patra patrapratap41@gmail.com
†ORCID Pratap Kumar Patra orcid.org/0000-0002-0338-7517
 Pratap Kumar Patra
Pratap Kumar Patra Ankur Kumar Jindal
Ankur Kumar Jindal Rashmi Rikhi
Rashmi Rikhi Anit Kaur
Anit Kaur Priyanka Srivastava
Priyanka Srivastava Deepti Suri
Deepti Suri Amit Rawat
Amit Rawat Rakesh Pilania
Rakesh Pilania Surjit Singh
Surjit Singh