Role of GDNF, GFRα1 and GFAP in a Bifidobacterium-Intervention Induced Mouse Model of Intestinal Neuronal Dysplasia
- 1Department of Pediatric Surgery, Qilu Hospital, Cheeloo College of Medicine, Shandong University, Jinan, China
- 2Department of Pediatric Surgery, Dezhou People's Hospital, Dezhou, China
Objective: To investigate the effects of glial cell-derived neurotrophic factor (GDNF), GDNF family receptor alpha 1 (GFRα1), and glial fibrillary acidic protein (GFAP) on colonic motility in a mouse model of intestinal neuronal dysplasia by intervention with Bifidobacterium and to explore the influence of Bifidobacterium on enteric glial cells (EGCs).
Methods: Western blotting and qRT-PCR were employed to detect the expression of GFRα1 and GFAP in colonic tissues of mice with or without Tlx2 mutations, and ELISA was used to detect the expression of GDNF in serum. IHC was used to detect the appearance of the ganglion cells. Subsequently, Tlx2 homozygous mutant (Tlx2−/−) mice were treated with Bifidobacterium. Colonic motility was measured before and after intervention by measuring the glass bead expelling time. The variations in abdominal circumference and GDNF, GFRα1, and GFAP expression were measured. In addition, 16SrRNA gene sequencing was performed to detect the abundance of the intestinal microbiota.
Results: The mRNA and protein expression of GFRα1 and GFAP was decreased in the colonic tissues of Tlx2−/− mice and GDNF expression was decreased in serum compared with Tlx2+/− and WT mice. After confirming the colonization of Bifidobacterium by 16S rRNA gene sequencing, the expelling time and abdominal distension were ameliorated, and the expression of GFAP, GDNF, and GFRα1 was increased.
Conclusions: The expression of GDNF, GFRα1, and GFAP is associated with colonic motility. The altered expression of EGC-related factors suggested that Bifidobacterium may be involved in the EGC activation process. The amelioration of IND symptoms after intervention with Bifidobacterium prompted the elicitation of adjuvant therapy.
Introduction
Intestinal neuronal dysplasia (IND) is a disorder of the enteric nervous system (ENS) associated with intestinal dysmotility (1). Similar to Hirschsprung's disease (HSCR), patients with IND show severe prolonged defecation time, constipation, abdominal distension, and even intestinal obstruction. However, the pathological features of IND are distinct, and include: hyperplasia of the submucosal nerve plexus, immaturity of ganglia, and hypertrophy of the nerve trunks in clinical cases (2, 3). Since the first description by Meier-Ruge in 1971 (4), the definition of IND is still a subject of controversy. Although the histopathological diagnostic criteria for IND are continuously updated, its etiology and pathogenesis have not been elucidated. Glial cell line-derived neurotrophic factor (GDNF), secreted by enteric neuroglial cells (EGCs), are regarded as one of the most important neurotrophic pathways in the formation of the ENS (5). GDNF binds to GDNF family receptor alpha 1 (GFRα1), causing phosphorylation of the tyrosine kinase receptor RE-arranged during transfection (RET). Signals through the activation of downstream signaling pathways direct the development of the ENS, including neuroblast migration and axonal outgrowth (6, 7).
Gut microbiome can affect the function and number of enteric neurons (8–10), and the early gut microbiota plays an important role in the development of the ENS after birth (11, 12). Several studies have shown that abnormalities in the myenteric plexus and a decrease of intestinal motility can be observed in early postnatal germ-free mice (11). While there are hundreds of species of bacteria in the human intestine, bifidobacteria is a key microbial player in the infant gut microbiota and has a beneficial effect on gut microbiota composition, leading to reduced necrotizing enterocolitis (NEC) incidence (13, 14). Furthermore, bifidobacterium can affect the expression of inflammatory factors in EGCs to inhibit intestinal inflammation and upregulate the expression of enteric glial cell–derived nerve growth factor (NGF) and neurotrophin 3 (NT-3) (15). These findings prompted us to determine whether colonic dysmotility of IND can be improved by the probiotic components of intestinal flora through activation with EGCs.
To verify this hypothesis, Tlx2 homozygous mutant (Tlx2−/−) mice were chosen as the model. T-Cell Leukemia Homeobox Protein 2 (Tlx2), a member of orphan homeobox-containing transcription factor family, is also known as HOX11L1, Ncx and Enx (16). Tlx2 has been proved to be crucial to the development of ENS. Tlx2−/− mice show colonic dysmotility with hyperplasia of intestinal ganglion and immature enteric neurons after birth, which has been confirmed to be consistent with IND (17, 18). Glial fibrillary acidic protein (GFAP), a type III intermediate filament (IF) protein that is highly expressed in mature and activated EGCs (19–21), was chosen as a marker of EGCs. This study aimed to explore the effects of GDNF, GFRα1, and GFAP on colonic motility and the influence of Bifidobacterium animalis on EGCs, and to further clarify the potential pathogenesis of IND.
Materials and Methods
Animal and Sample Preparation
The study was approved by the Ethics Committee of Qilu Hospital of Shandong University (IACUC Issue No. Dull-2020-013). The mice were treated according to the animal use guidelines of the Animal Care and Use Committee (ACUC) of Qilu Hospital, Shandong University. Genechem (Shanghai, China) used CRISPR/Cas9 gene-targeting technology to knock out 173 bp of the second exon nucleotide sequence of the C57BL/6 mouse Tlx2 gene, resulting in Tlx2+/− mice. Tlx2+/− mice were interbred to obtain Tlx2−/− mice. The mice were reared under specific pathogen-free conditions. At the age of 3 weeks, different genotypes were screened out through genotype identification (WT Tlx2+/− and Tlx2−/−). Colonic motilities and abdominal circumference were measured 1 d before or 1 d after the intervention. Feces were collected 1 d after the intervention and before colonic motility measurement. Blood samples and segments of the distal colon were harvested after anesthesia, and were stored at −80°C.
Genotyping
The genotype of mice was identified by Southern blotting. Then the mice were divided into three groups (group WT Tlx2+/− and Tlx2−/−, nine mice for each group) according to their genotypes for experiments before probiotic intervention. Genomic DNA was isolated from mouse tails using a Mouse Tail DNA Extraction kit (CWBio, Beijing, China). The primer sequences for Tlx2 are shown in Table 1. Subsequently, the genomic DNA was amplified by PCR and separated by 1.2% agarose gel electrophoresis. Images were acquired using the BIO-RAD gel image acquisition system (Bio-Rad, Hercules, CA, USA).
Measure of Mouse Colonic Motility
Colonic motility was measured 1 d before or 1 d after the intervention. A small glass bead with a diameter of 2.5 mm was inserted slowly into the colon of the mice to a distance of 2 cm with a smooth glass rod (2.5 mm diameter). After checking that the glass bead was completely pushed in, the mouse was placed on a clean surgical drape, and the expulsion was recorded.
Bifidobacterium animalis Intervention
Bifidobacterium animalis (AS1.1852, biobw) was cultured in Bifidobacterium nutrient liquid medium (Haibo, Qingdao, China) in an anaerobic incubator at 37°C, and Spectroscopy Photometric Microplate Reader (ThermoFisher, MA, US) was used to monitor the density of the bacterial colony (OD 600). The bacterial colony density was adjusted to 1 × 109 CFU/ml before intervention. Tlx2−/− mice with a suitable weight at the age of 3–4 weeks were divided into three groups (eight mice for each group). Bifidobacterium animalis group (group BB) was given live Bifidobacterium liquid according to the weight of each mouse by retention-enema at 0.2 ml/20 g (0.2 × 109 CFU/20 g) daily for 3 weeks. The solution should be prepared before each intervention everyday and used in time after dilution to maintain the probiotic activity. The Bifidobacterium nutrient medium group (group NM) was administered the same dose of nutrient medium, and the normal saline/blank control group (group NS) was administered the same dose of normal saline.
Western Blotting
Western blotting was used to analyze the relative expression levels of GFRα1 and GFAP proteins in the colonic tissue. The Minute™ Total Rapid Protein Ext Kit (Invent, MN, USA) was used to isolate total protein from the full-thickness colonic tissue. Equal amounts of protein were separated on a 10% SDS-PAGE gel and then transferred to a PVDF membrane. The PVDF membrane was incubated with primary antibodies at 4°C overnight after 5% BSA blocking. The membrane was then washed with 1 × TBST and incubated with secondary antibody for 1 h at room temperature. ECL kit (Millipore, MA, USA) was used for chemiluminescence, and the gray values were calculated. Table 2 shows the details of the antibodies.
Quantitative Real-Time PCR
The qPCR assay was employed to investigate the relative expression of GDNF, GFRα1, and GFAP in colon tissue at the mRNA level. Total mRNA was isolated using TRIzol reagent (Ambion, USA). After concentration measurement, mRNA was reverse transcribed to cDNA using an RT-PCR kit (Toyota, Japan). The qPCR reactions were performed with SYBR Green PCR Master Mix (Toyota, Japan) on a Roche 480 real-time fluorescent PCR instrument. Primers were selected from the Beacon Designer software (Premier Biosoft, Palo Alto, California, USA). The detailed information is shown in Table 1. The 2−ΔΔCt values for each group were calculated for analysis. Colon sample of each mouse was repeated three times, and a melting curve was used to confirm specificity.
Immunohistochemistry Staining
Immunohistochemistry was used to detect the appearance of ganglion and glial cells. The colon tissue was fixed in 4% paraformaldehyde for 24 h and then embedded in 5-μm paraffin. After dewaxing and antigen retrieval, the slices were blocked with goat serum and incubated with the primary antibodies (1:500) overnight at 4°C, then incubated with secondary antibodies (1:200) for 1 h at 37°C. All antibody incubation and washing steps were performed in PBS at pH 7.4. DAB (ZSbio, Beijing, China) staining was monitored under a microscope. Finally, the sections were counterstained with hematoxylin and coverslipped. The integrated optical density (IOD) of positive staining within five random fields from paraffin sections of each mouse was measured at 20 × magnification using Image-Pro Plus 6.0 image analysis software (Media Cybernetics, Bethesda, MD, USA). The average value was used for statistics.
ELISA
An ELISA kit (Boster, Wuhan, China) was used to detect the level of GDNF in serum according to the manufacturer's instructions. Serum was added to a 96-well plate (100 μl per well) for detection. The OD values were measured at 450 nm after the reaction, and the concentrations were calculated.
16S rRNA Gene Sequencing
Total bacterial DNA was extracted from fecal samples using the E.Z.N.A. Stool DNA Kit (Omega, Inc., USA). The V3-V4 region of the bacterial 16S rRNA gene was amplified using the universal primers 338F (5-ACTCCTACGGGAGGCAGCAG-3) and 806R (5-GGACTACHVGGGTWTCTAAT-3). PCR products were purified with AMPure XT magnetic beads (Beckman Coulter Genomics, Danvers, MA, USA), and quantified using Qubit (Invitrogen, California, USA). The amplicon library was prepared for sequencing, and its size and quantity were assessed using an Agilent 2100 Bioanalyzer (Agilent, CA, USA) and the Library Quantification Kit for Illumina (Kapa Biosciences, Woburn, MA, USA). The amplicon library was paired-end sequenced using the Illumina MiSeq platform at a commercial company (LC-Bio Technology Co., Ltd, Hang Zhou, China).
Statistics and Analysis
All statistical analyses were performed using GraphPad Prism®8 software, and the results are presented as the mean ± SD. T-test was used for comparisons between two independent groups, and one-way analysis of variance (ANOVA) and Tukey's test were used for comparisons between multiple groups; P < 0.05 was considered statistically significant.
Results
Tlx2−/− Mice Showed Decreases in the Expression of GDNF, GFRα1 and GFAP
The allele of the wild-type was 568 bp, whereas the mutant was 395 bp. T1x2 −/− mice obtained by interbreeding were genotyped by Southern blot analysis (Figure 1A). To confirm whether the expressions of GDNF, GFRα1, and GFAP were different among WT, Tlx2+/−, and Tlx2−/− mice, the colonic samples and sera of these mice were used for analysis. The protein and mRNA expression of GFRα1 and GFAP in colonic tissues of Tlx2−/− mice were lower than those in Tlx2+/− and WT mice (P < 0.05) (Figures 1B–E,I). Compared to Tlx2+/− mice and WT mice, the protein expression of GDNF in serum and its mRNA expression in colonic samples of Tlx2−/− mice was also decreased (P < 0.05) (Figures 1F,G). Meanwhile, IHC staining showed that GFAP was mainly expressed in the EGCs of the colonic myenteric plexus (Figure 1J). The IOD of positive staining indicated that GFAP was significantly downregulated in colonic tissues of Tlx2−/− mice compared to other tissues (Figure 1H).
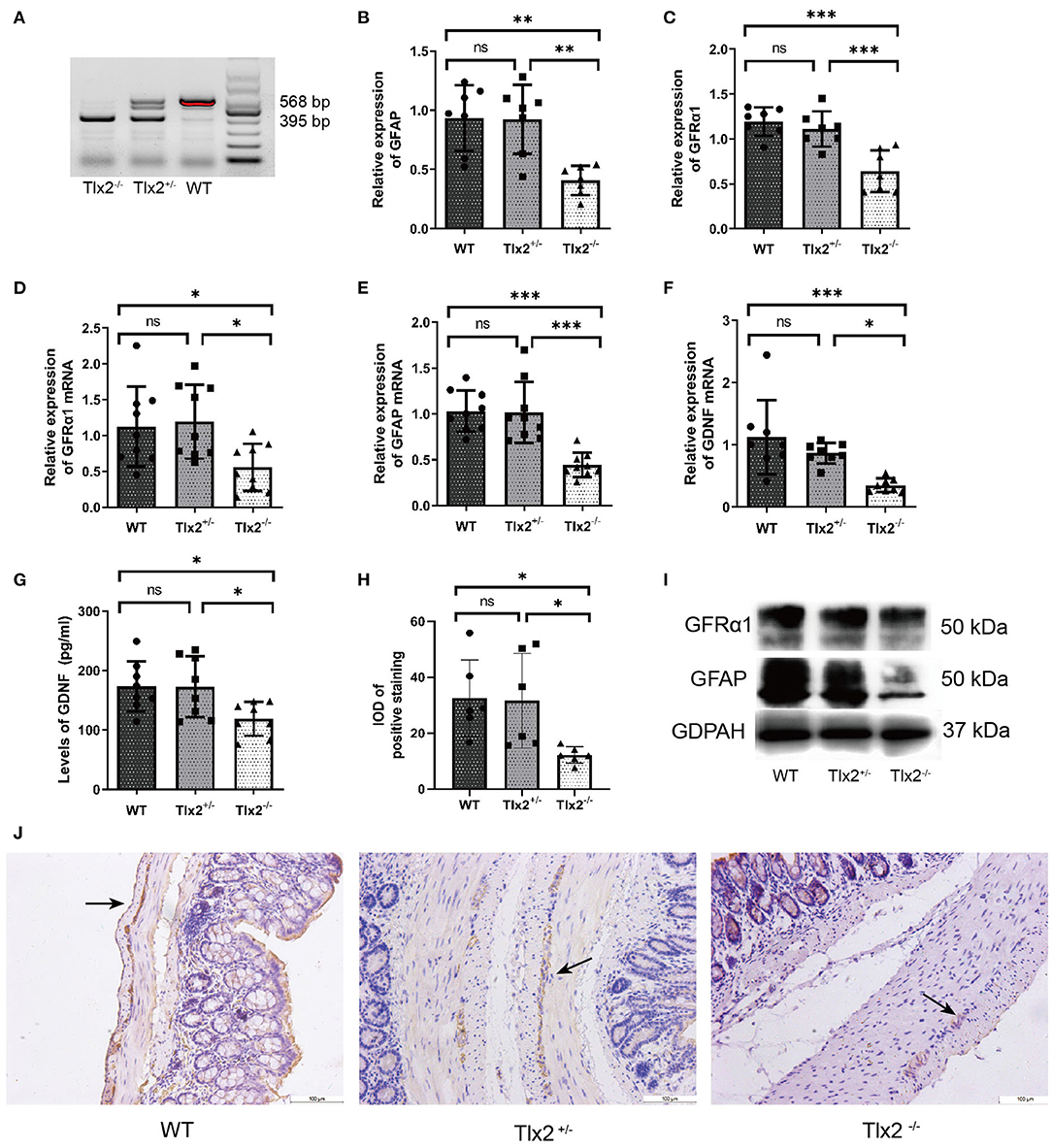
Figure 1. Example genotyping and decreased expressions of GDNF, GFRα1 and GFAP in Tlx2−/− mice. (A) Genotyping; the wild-type allele is 568 bp, and the mutant allele is 395 bp. (B,C) Western blot indicated that the protein expressions of GFAP and GFRα1 in colonic tissues of Tlx2−/− mice were decreased (n = 7 for each group). (D–F) The mRNA expressions of GDNF, GFRα and GFAP were decreased in colon tissues of Tlx2−/− mice (n = 9 for each group). (G) The expression level of GDNF in serum of Tlx2−/− mice was lower than that in Tlx2+/− mice and WT mice (n = 8 for each group). (H) IOD of positive staining indicated that GFAP was significantly downregulated in the colonic tissues of Tlx2−/− mice (n = 6 for each group). (I) Representative western blot analysis and detail bands of other mice were attached in Supplementary Figure 1. (J) GFAP was mainly expressed in EGCs of the colonic myenteric plexus (ns P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; GAPDH, glyceraldehyde-3-phosphate; Scale bar: 100 μm).
The Expelling Time of Tlx2−/− Mice Was Shortened and the Abdominal Circumference Was Smaller After Bifidobacterium Intervention
To confirm that the Bifidobacterium intervention was effective, 16S rRNA gene sequencing was performed. The relative abundances of intestinal flora were presented in a hierarchical clustering heatmap and Bifidobacterium in group BB was much higher than that in groups NM and NS (Figures 2A,B). The time of bead expulsion in different groups of Tlx2−/− mice before and after enema was recorded (Figure 2F). The expelling time of group BB after enema was shorter than that before the intervention (P < 0.05). However, nutrient medium and normal saline did not affect colonic motility (Figure 2C). The abdominal circumference of mice in group BB after intervention was significantly smaller than that of the other two groups (P < 0.05) (Figures 2D,F) and the differences in abdominal circumference before and after Bifidobacterium intervention in the BB group were smaller than those in the other groups (p < 0.05; Figure 2E). The abdominal distension in the BB group was relieved (Figure 2G). The gross anatomy of the intestines revealed that the fecal retention of mice in group BB was mild, and the dilatation of the cecum, distal ileum, and proximal colon was relieved compared with other groups (Figure 2G).
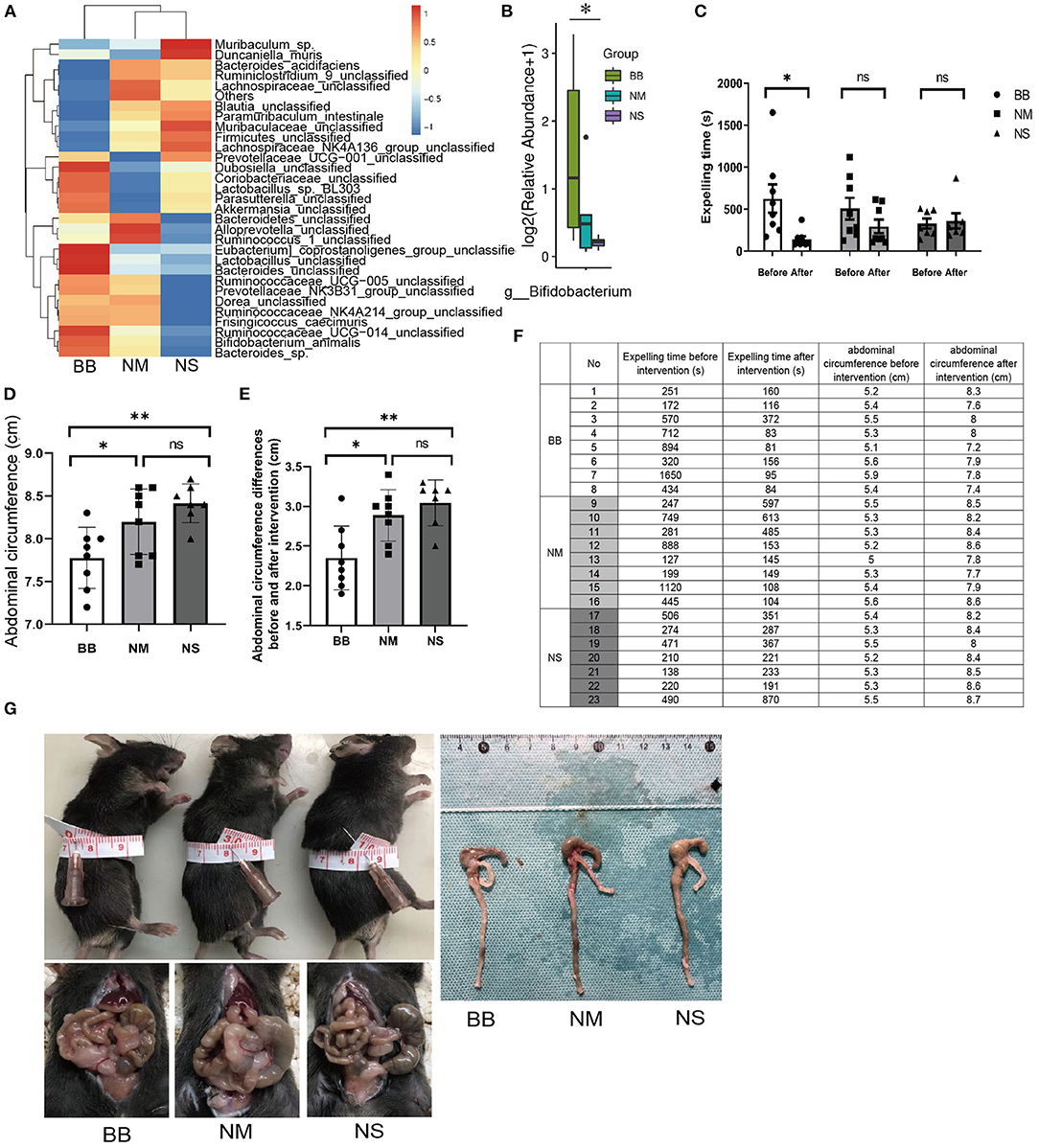
Figure 2. Measurement of the colonic motility and abdominal circumference of mice in groups BB, NM and NS. (A) Hierarchical clustering heatmap of the top 30 bacterial species of each group. (B) The relative abundance of Bifidobacterium among groups BB, NM and NS. (C,F) T After Bifidobacterium intervention, the expelling time in BB group was shortened compared to groups NM and NS. (D,F,G) The abdominal circumference after Bifidobacterium intervention among the three groups. (E,F) Differences in abdominal circumference before and after Bifidobacterium intervention in group BB was smaller than the other groups. (ns P > 0.05; *P < 0.05; **P < 0.01).
The Expression of GDNF, GFRα1 and GFAP Was Increased in Tlx2−/− Mice After Intervention
The protein expression of GFAP and GFRα1 was significantly increased in the BB group (Figures 3A–C), which was consistent with the qRT-PCR results (Figures 3D,F). ELISA was used to detect the expression level of GDNF in the serum, confirming the upregulated expression of GDNF in the BB group (Figure 3G). Similarly, the qRT-PCR assay revealed that mRNA expression of GDNF increased in the colonic tissues of mice in the BB group (Figure 3E), and there was no statistically significant difference between the other two groups. The location and expression of GFAP are shown by IHC (Figures 3H,I). As can be seen, the stain-positive cells of GFAP presented tan granules. The IOD of positive staining in group BB was higher than that in groups NM and NS, which was consistent with the results of WB and PCR.
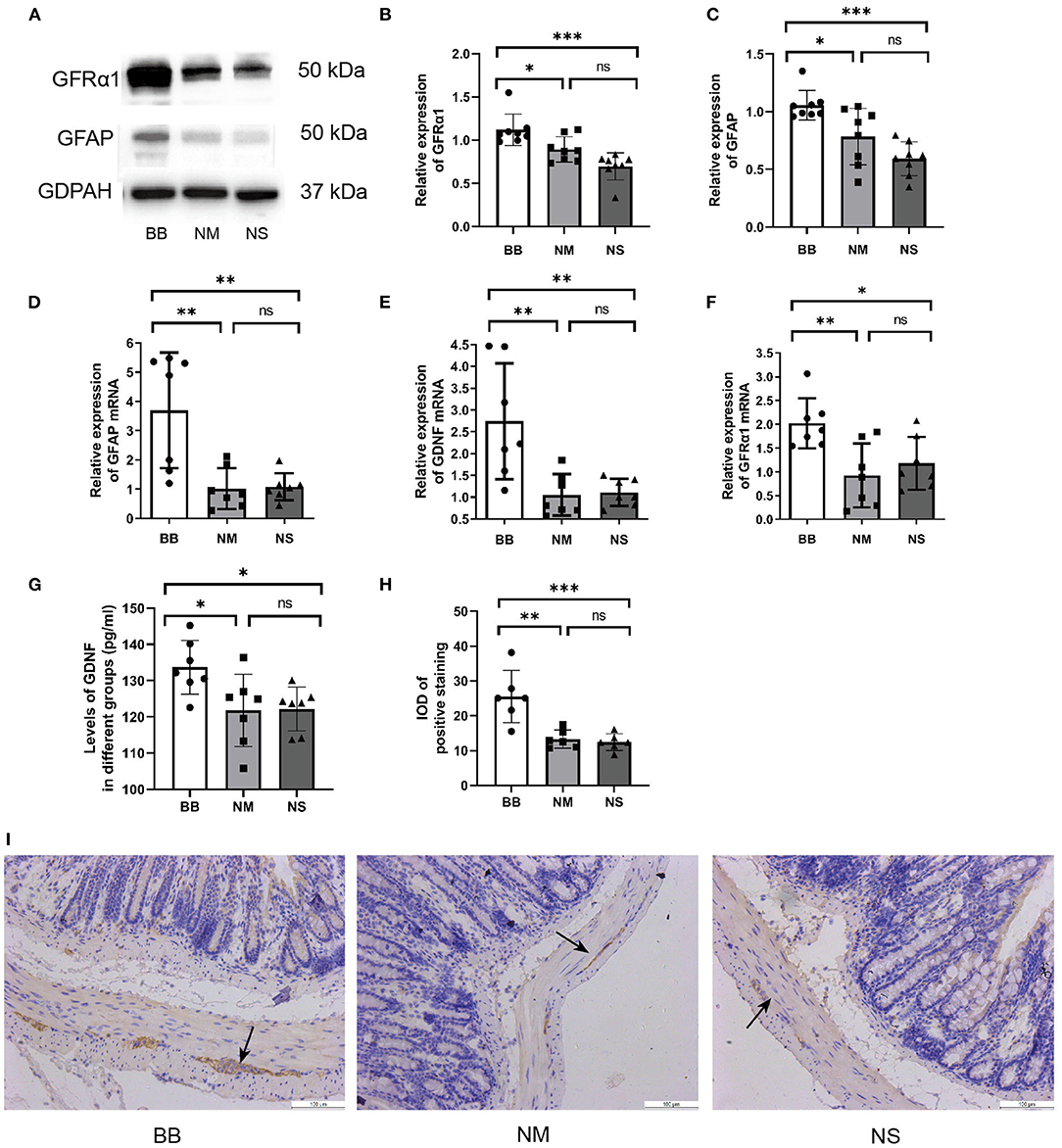
Figure 3. Expression of GDNF, GFRα1 and GFAP was increased in group BB. (A–C) The protein expression of GFAP and GFRα in the colonic tissues of group BB was higher than those in the other two groups (n = 8 for each group), detail bands of other mice can be found at Supplementary Figure 1. (D–F) The mRNA expressions of GDNF, GFRα and GFAP in colon tissues of group BB were increased (n = 7 for each group). (G) The serum levels of GDNF in group BB increased compared to those in groups NM and NS after intervention (n = 7 for each group). (H) The IOD of positive staining in group BB was increased compared to that of the NM and NS groups (n = 6 for each group). (I) Representative results of IHC staining of GFAP in different groups under 20 × microscope (ns P > 0.05; *P < 0.05; **P < 0.01; ***P < 0.001; GAPDH, glyceraldehyde-3-phosphate; Scale bar: 100 μm).
Discussion
The pathogenesis of IND is unresolved; however, several mechanisms have been proposed, including secondary infections, inflammation, and developmental failures (22–24). It has been confirmed that the seriously impaired colonic motility of IND is related to abnormal innervation between ENS and intestinal smooth muscle cells (25, 26). Tlx2−/− mice, a verified model of IND, usually show hyperplasia of the intestinal ganglion and persistence of immature enteric neurons after birth (18, 27). In our previous studies, Tlx2−/− mice showed obvious abdominal distension and impaired colonic motility, and 26% (33/127) of the mice died 8 weeks after birth (25), which may be related to histopathological changes in the colonic tissues of Tlx2−/− mice.
The main origin of the ENS is neural crest cells (28). EGCs and enteric neurons, vital components of the ENS, show interactive influences during the maturation and function of the ENS (28), and GDNF and GFRα1 play significant roles in this process (6, 7, 29). In this study, the expression of GDNF, GFRα1, and GFAP in Tlx2−/− mice was lower than that in Tlx2+/− and WT mice, confirming that the three may participate in the pathogenesis of IND. Alternatively, we observed higher expression of GDNF, GFRα1, and GFAP in mice after intervention with Bifidobacterium. In addition, the expelling times in group BB were significantly shortened. All of these results indicate possible relationship between GDNF, GFRα1, and GFAP and the relief of colonic motility dysfunction, which might explain the colonic motility dysfunction resulting from the immaturity of ganglion cells.
The gut microbiota affects the activation of the immune system by modulating antigen-specific adaptive immune responses, resulting in physiological functions (8, 30). Bacteria metabolites regulate the intestinal microenvironment by modulating the release of inflammatory factors (31). Bifidobacterium, the main probiotic component of the gut microbiota in humans, can reduce inflammatory responses by inhibiting the NF-κB and external cell signaling pathways (32). It has been confirmed that EGCs can be regulated by Bifidobacterium through inhibition of inflammation and up-regulation of nerve growth factors in vitro (15). In our study, the Bifidobacterium intervention in IND model mice caused the colonization of Bifidobacterium and increased expression of GDNF, GFRα1, and GFAP in colonic tissues. The variations in the expression of GDNF, GFRα1, and GFAP suggested that Bifidobacterium may participate in EGC activation and nutritional factor upregulation, but the specific mechanism of interaction between Bifidobacterium and EGC should be further researched.
IND and HSCR show similar clinical features, including constipation, abdominal distension, and even intestinal obstruction. In this study, after intervention by Bifidobacterium, the expelling time of Tlx2−/− mice was shortened and abdominal distension was reduced. The recovery of the impaired colonic motility confirmed the positive influence of Bifidobacterium on colonic motility, which is consistent with our clinical experience. These results are also consistent with recent findings that fecal microbiota transplantation can improve intestinal function (33, 34).
As shown above, the expression of GDNF, GFRα1, and GFAP decreased in Tlx2−/− mice and increased after Bifidobacterium intervention, while the IND-related symptoms were relieved after Bifidobacterium intervention. We can conclude that the expression of GDNF, GFRα1, and GFAP is associated with colonic motility dysfunction and the pathogenesis of IND. The altered expression of EGC-related genes suggested that Bifidobacterium may be involved in the activation process of EGCs. The amelioration of IND symptoms after intervention with Bifidobacterium prompted the elicitation of adjuvant therapy.
Data Availability Statement
The datasets presented in this article are not readily available because the dataset also forms part of an ongoing study. Requests to access the datasets should be directed to 591043945@qq.com.
Ethics Statement
The animal study was reviewed and approved by the Ethics Committee of Qilu Hospital of Shandong University.
Author Contributions
WL and TZ: design of the work, acquisition, analysis, and interpretation of data for the work. JT, XY, CR, ZC, and PH: feeding of laboratory animal and collection of specimens. QZ and AL: drafting the work or revising it critically for important intellectual content. All authors contributed to the article and approved the submitted version.
Funding
This work was funded by the National Natural Science Foundation of China (Projects Nos. 81873846 and 82071682).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to express our gratitude to: Key Laboratory of Cardiovascular Remodeling and Function Research of Ministry of Education. Laboratory of Basic Medical Sciences. Institute of Stomatology of Shandong University.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fped.2021.795678/full#supplementary-material
Supplementary Figure 1. (A) Western blot indicated that the protein expressions of GFAP and GFRα1 in colonic tissues of Tlx2−/− mice were decreased. (B) The protein expression of GFAP and GFRα in the colonic tissues of group BB was higher than those in the other two groups.
References
1. Kapur RP, Reyes-Mugica M. Intestinal neuronal dysplasia type B: an updated review of a problematic diagnosis. Arch Pathol Lab Med. (2019) 143:235–43. doi: 10.5858/arpa.2017-0524-RA
2. Yamataka A, Hatano M, Kobayashi H, Wang K, Miyahara K, Sueyoshi N, et al. Intestinal neuronal dysplasia-like pathology in Ncx/Hox11L 1 gene-deficient mice. J Pediatr Surg. (2001) 36:1293–6. doi: 10.1053/jpsu.2001.25797
3. Terra SA, de Arruda Lourencao PL., M GS, H AM, Rodrigues MAM. A critical appraisal of the morphological criteria for diagnosing intestinal neuronal dysplasia type B. Mod Pathol. (2017) 30:978–85. doi: 10.1038/modpathol.2017.4
4. Meier-Ruge W. [Casuistic of colon disorder with symptoms of Hirschsprung's disease (author's transl)]. Verh Dtsch Ges Pathol. (1971) 55:506–10.
5. Jing S, Wen D, Yu Y, Holst PL, Luo Y, Fang M, et al. GDNF-induced activation of the ret protein tyrosine kinase is mediated by GDNFR-alpha, a novel receptor for GDNF. Cell. (1996) 85:1113–24. doi: 10.1016/S0092-8674(00)81311-2
6. Ibanez CF, Andressoo JO. Biology of GDNF and its receptors - relevance for disorders of the central nervous system. Neurobiol Dis. (2017) 97(Pt B):80–9. doi: 10.1016/j.nbd.2016.01.021
7. Blennerhassett MG, Lourenssen SR. Obligatory activation of SRC and JNK by GDNF for survival and axonal outgrowth of postnatal intestinal neurons. Cell Mol Neurobiol. (2021). doi: 10.1007/s10571-021-01048-9. [Epub ahead of print].
8. Muller PA, Matheis F, Schneeberger M, Kerner Z, Jove V, Mucida D. Microbiota-modulated CART(+) enteric neurons autonomously regulate blood glucose. Science. (2020) 370:314–21. doi: 10.1126/science.abd6176
9. Tuganbaev T, Honda K. Non-zero-sum microbiome immune system interactions. Eur J Immunol. (2021) 51:2120–36. doi: 10.1002/eji.202049065
10. Kabouridis PS, Lasrado R, McCallum S, Chng SH, Snippert HJ, Clevers H, et al. The gut microbiota keeps enteric glial cells on the move; prospective roles of the gut epithelium and immune system. Gut Microbes. (2015) 6:398–403. doi: 10.1080/19490976.2015.1109767
11. Collins J, Borojevic R, Verdu EF, Huizinga JD, Ratcliffe EM. Intestinal microbiota influence the early postnatal development of the enteric nervous system. Neurogastroenterol Motil. (2014) 26:98–107. doi: 10.1111/nmo.12236
12. Kabouridis PS, Pachnis V. Emerging roles of gut microbiota and the immune system in the development of the enteric nervous system. J Clin Invest. (2015) 125:956–64. doi: 10.1172/JCI76308
13. Mohan R, Koebnick C, Schildt J, Schmidt S, Mueller M, Possner M, et al. Effects of Bifidobacterium lactis Bb12 supplementation on intestinal microbiota of preterm infants: a double-blind, placebo-controlled, randomized study. J Clin Microbiol. (2006) 44:4025–31. doi: 10.1128/JCM.00767-06
14. Hui Y, Smith B, Mortensen MS, Krych L, Sorensen SJ, Greisen G, et al. The effect of early probiotic exposure on the preterm infant gut microbiome development. Gut Microbes. (2021) 13:1951113. doi: 10.1080/19490976.2021.1951113
15. Yang PC, Li XJ, Yang YH, Qian W, Li SY, Yan CH, et al. The Influence of Bifidobacterium bifidum and Bacteroides fragilis on enteric glial cell-derived neurotrophic factors and inflammasome. Inflammation. (2020) 43:2166–77. doi: 10.1007/s10753-020-01284-z
16. Borghini S, Di Duca M, Santamaria G, Vargiolu M, Bachetti T, Cargnin F, et al. Transcriptional regulation of TLX2 and impaired intestinal innervation: possible role of the PHOX2A and PHOX2B genes. Eur J Hum Genet. (2007) 15:848–55. doi: 10.1038/sj.ejhg.5201852
17. Hatano M, Aoki T, Dezawa M, Yusa S, Iitsuka Y, Koseki H, et al. A novel pathogenesis of megacolon in Ncx/Hox11L 1 deficient mice. J Clin Invest. (1997) 100:795–801. doi: 10.1172/JCI119593
18. Kato Y, Miyahara K, Hatano M, Hasegawa Y, Seki T, Frykman PK, et al. Immature enteric neurons in Ncx/Hox11L. 1 deficient intestinal neuronal dysplasia model mice. Pediatr Surg Int. (2009) 25:961–5. doi: 10.1007/s00383-009-2451-6
19. Grundmann D, Loris E, Maas-Omlor S, Huang W, Scheller A, Kirchhoff F, et al. Enteric glia: S100, GFAP, and beyond. Anat Rec (Hoboken). (2019) 302:1333–44. doi: 10.1002/ar.24128
20. Cossais F, Leuschner S, Barrenschee M, Lange C, Ebsen M, Vogel I, et al. Persistent increased enteric glial expression of S100beta is associated with low-grade inflammation in patients with diverticular disease. J Clin Gastroenterol. (2019) 53:449–56. doi: 10.1097/MCG.0000000000001011
21. Turco F, Sarnelli G, Cirillo C, Palumbo I, De Giorgi F, D'Alessandro A, et al. Enteroglial-derived S100B protein integrates bacteria-induced Toll-like receptor signalling in human enteric glial cells. Gut. (2014) 63:105–15. doi: 10.1136/gutjnl-2012-302090
22. Sacher P, Briner J, Hanimann B. Is neuronal intestinal dysplasia (NID) a primary disease or a secondary phenomenon? Eur J Pediatr Surg. (1993) 3:228–30. doi: 10.1055/s-2008-1063549
23. Goldstein AM, Cox NJ. Complex simplicity and Hirschsprung's disease. N Engl J Med. (2019) 380:1478–9. doi: 10.1056/NEJMe1902827
24. Puri P. Intestinal neuronal dysplasia. Semin Pediatr Surg. (2003) 12:259–64. doi: 10.1053/j.sempedsurg.2003.08.007
25. Wang D, Gao N, Zhou T, Zhang Q, Wang J, Li A. Effect of Neuroligin1 and Neurexin1 on the colonic motility in a mouse model of neuronal intestinal dysplasia. Gastroenterol Res Pract. (2020) 2020:9818652. doi: 10.1155/2020/9818652
26. Wang J, Du H, Mou YR, Niu JY, Zhang WT, Yang HC, et al. Abundance and significance of neuroligin-1 and glutamate in Hirschsprung's disease. World J Gastroenterol. (2015) 21:7172–80. doi: 10.3748/wjg.v21.i23.7172
27. Shirasawa S, Yunker AM, Roth KA, Brown GA, Horning S, Korsmeyer SJ. Enx (Hox11L1)-deficient mice develop myenteric neuronal hyperplasia and megacolon. Nat Med. (1997) 3:646–50. doi: 10.1038/nm0697-646
28. Niesler B, Kuerten S, Demir IE, Schafer KH. Disorders of the enteric nervous system - a holistic view. Nat Rev Gastroenterol Hepatol. (2021) 18:393–410. doi: 10.1038/s41575-020-00385-2
29. Giuffre M, Moretti R, Campisciano G, da Silveira ABM, Monda VM, Comar M, et al. You talking to me? says the Enteric Nervous System (ENS) to the microbe how intestinal microbes interact with the ENS. J Clin Med. (2020) 9:3705. doi: 10.3390/jcm9113705
30. Honda K, Littman DR. The microbiota in adaptive immune homeostasis and disease. Nature. (2016) 535:75–84. doi: 10.1038/nature18848
31. Levy M, Thaiss CA, Zeevi D, Dohnalova L, Zilberman-Schapira G, Mahdi JA, et al. Microbiota-modulated metabolites shape the intestinal microenvironment by regulating NLRP6 inflammasome signaling. Cell. (2015) 163:1428–43. doi: 10.1016/j.cell.2015.10.048
32. O'Mahony L, McCarthy J, Kelly P, Hurley G, Luo F, Chen K, et al. Lactobacillus and bifidobacterium in irritable bowel syndrome: symptom responses and relationship to cytokine profiles. Gastroenterology. (2005) 128:541–51. doi: 10.1053/j.gastro.2004.11.050
33. Tian Y, Zuo L, Guo Q, Li J, Hu Z, Zhao K, et al. Potential role of fecal microbiota in patients with constipation. Therap Adv Gastroenterol. (2020) 13:1756284820968423. doi: 10.1177/1756284820968423
Keywords: intestinal neuronal dysplasia (IND), gut microbiota, Bifidobacterium, homozygous mutant mice of Tlx2, GDNF
Citation: Liu W, Zhou T, Tian J, Yu X, Ren C, Cao Z, Hou P, Zhang Q and Li A (2022) Role of GDNF, GFRα1 and GFAP in a Bifidobacterium-Intervention Induced Mouse Model of Intestinal Neuronal Dysplasia. Front. Pediatr. 9:795678. doi: 10.3389/fped.2021.795678
Received: 15 October 2021; Accepted: 20 December 2021;
Published: 14 January 2022.
Edited by:
Weibing Tang, Nanjing Medical University, ChinaReviewed by:
Yuying Liu, University of Texas Health Science Center at Houston, United StatesSven Flemming, University Hospital of Wuerzburg, Germany
Copyright © 2022 Liu, Zhou, Tian, Yu, Ren, Cao, Hou, Zhang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qiangye Zhang, zhangqiangye@qiluhospital.com; Aiwu Li, liaiwuxie@aliyun.com
†These authors have contributed equally to this work
 Wei Liu
Wei Liu Tingting Zhou
Tingting Zhou Jinqiu Tian1
Jinqiu Tian1