- 1Department of Gastroenterology, General Hospital of the Chinese People’s Liberation Army, Beijing, China
- 2Department of Health Technology and Informatics, Faculty of Health and Social Science, The Hong Kong Polytechnic University, Hunghom, Hong Kong SAR, China
Gastrointestinal cancers are a group of cancers occurred in gastrointestinal tissues with high morbidity and mortality rate. Although numerous studies were conducted on the investigation of gastrointestinal cancers, the real mechanisms haven’t been discovered, and no effective methods of prevention and treatment of gastrointestinal cancers have been developed. Autophagy, a vital catabolic process in organisms, have been proven to participate in various mechanisms and signaling pathways, thus producing a regulatory effect on various diseases. The role of autophagy in gastrointestinal cancers remains unclear due to its high complexity. In this review, firstly, the biological features of autophagy will be introduced. Secondly, the role of autophagy in three popular gastrointestinal cancers, namely esophageal cancer, gastric cancer, and colorectal cancer will be described and discussed by reviewing the related literature. We aimed to bring novel insights in exploring the real mechanisms for gastrointestinal cancers and developing effective and efficient therapeutic methods to treat gastrointestinal cancers.
Introduction
Gastrointestinal cancers are generally referred to as a group of cancers occurred in the gastrointestinal tissues, including the esophagus, stomach and colon/rectum (1). According to recent studies, it has been reported that each year, 4 million cases of gastrointestinal cancers are diagnosed in the world, and 2.7 million people die from gastrointestinal cancers annually (1–3). In a broad sense, gastrointestinal cancers are comprised of malignant tumors occurred in other digestive system tissues, including liver, small intestine, gallbladder, bile duct and pancreas (4, 5). Based on that knowledge, to keep it accurate and convincing, in our study, we focused on discussing the mechanisms of the three commonly acknowledged kinds of gastrointestinal cancers, including esophageal cancer, gastric cancer, and colorectal cancer. Although there are many different biological features of gastrointestinal cancers, several common risk factors have been discovered, including pro-tumoral genetic mutations, smoking, excessive alcohol intake, western diet, disturbance of gastrointestinal microbiota homeostasis and radioactive stimulation (6–9). In addition, the disturbance of normal gastrointestinal microenvironment is revealed to potentially cause gastrointestinal cancers, such as the pro-tumoral fibrosis and overwhelmingly local or overall inflammatory and immune responses (10–12). Besides those commonly acknowledged risk factors, some disorders have been proven to be closely related to the cause of gastrointestinal cancers. For instance, diabetes has been revealed to be associated with gastrointestinal cancer. Metformin, one of the most well-known anti-hyperglycemic drugs, was shown to reduce the incidence rate of gastrointestinal cancers in diabetic patients (4, 13). Based on that knowledge, the restoration of metabolic homeostasis and prevention of tumor-associated microenvironment formation might serve as potentially effective methods to treat gastrointestinal cancers.
Autophagy has been commonly recognized as a vital metabolic mechanism in organisms (14, 15). It is proven to be effective in degrading and recycling long-term and misfolded proteins, useless organelles under certain stressful conditions such as hypoxia, nutritional deficiency and inflammatory stimulation (16–18). So far, autophagy has been indicated to participate in numerous mechanisms and signaling pathways or cascades, thus regulating various kinds of diseases in many systems (19–22). For example, in digestive system, autophagy has been proven to contribute to the regulation of gastrointestinal microbiota (23–26). Although numerous studies reported and discussed the effect and mechanisms of autophagy in gastrointestinal cancers (27–34), the role of autophagy in gastrointestinal cancers is still not clear, and the real mechanisms remain unclarified. This situation makes it hard to use autophagy to prevent and treat gastrointestinal cancers.
In this paper, we will review related literature on the study of autophagy in three commonly diagnosed gastrointestinal cancers, including esophageal cancer, gastric cancer, and colorectal cancer. We will focus on the introduction and discussion of the controversial role of autophagy in such types of cancers, aiming to provide novel insight in the recognition of autophagy in gastrointestinal cancers, and seeking to develop an effective method to use autophagy to treat gastrointestinal cancers.
Part I: Biological characteristics of autophagy
The word “autophagy” was initially created in the 1960s by Dr. Christian de Duve, who was awarded the Nobel Prize in 1974 for the discovery of the lysosome as a new specialized membrane-bound organelle in animal cells (35). The word “autophagy” was derived from the Greek roots “auto” (self) and “phagy” (eat). It was referred to as the cellular metabolic processes in which cytoplasmic proteins and certain organelles were “eaten” by itself (to eat it self) (36–38). Since its initial discovery, the mechanisms of autophagy and its roles in cellular catabolic processes and diseases have been widely studied and recognized by researchers. In 2016, Dr. Yoshinori Ohsumi was awarded the Nobel Prize in Medicine or Physiology for his work of investigating the processes of cellular autophagy (39). So far, three types of classic autophagy have been divided based on the difference in mode of cargo delivery to the lysosomal lumen and physiological functions, including microautphagy, chaperone-mediated autophagy and macroautophagy (40–42). Microautophagy is recognized as a non-selective lysosomal process. Degrading proteins and organelles are engulfed via the invagination of the lysosomal/vacuolar membranes through microautphagy (43, 44). Chaperone-mediated autophagy is a form of selective autophagy which relies on the recognition of chaperons via targeted motif in the degrading proteins and lysosomal chaperons (45, 46). Macroautophagy is the most studied form of autophagy. Macroautophagy is referred to as a metabolic process with the functional unit of double-membraned autophagosomes, which are subsequently fused with lysosomes for further degradation and recycling (47, 48). Besides the classic classification of autophagy, some special forms of selective autophagy have been discovered and studied, including pexophagy, mitophagy, xenophagy and reticulophagy, etc. Those special forms of selective autophagy represent the special forms and functions of autophagy in certain organelles and conditions (49–51).
Autophagy-lysosomal system is recognized as one of the two classic protein degrading pathways along with ubiquitin-proteasome system (52, 53). The process of autophagy has been revealed to be mediated by more than 30 autophagy-related genes (Atgs), most of which are proven to be conserved in mammal cells (54, 55). According to our previous study, the induction of autophagy mainly follows two steps (56, 57) (illustrated in Figure 1). In the first step, under the challenge of stressful conditions such as starvation and hypoxia, cup-shaped phagophores with lipid bilayer membrane is formed to wrap around substrate materials. The formation of phagophores demands the formation of the Atg1 complex and Class III phosphatidylinositol 3-kinase (PI3K) complex, with Unc-51-like kinase (ULK1, aka Atg1 in yeast), FIP200, Atg13, Atg101 assembly for Atg1 complex and Beclin-1, Atg14, vacuolar protein sorting 15 (VPS15), and VSP34 for PI3K complex (58, 59). After the initiation, the bilayer membrane undergoes expansion, elongation and nucleation, which are sequestrated into double-membrane sphere-shaped autophagosomes. Such process is dependent on the formation of Atg16L1 complex, assembled by Atg5, Atg12 and Atg16L1 (60, 61). In addition, two ubiquitin-like proteins including Atg12 and Atg8 [LC3 (light-chain 3)] also participate the process of autophagosome formation. In the second step, autophagosomes dispose of “coat proteins (LC3-II)” on the surface of the membrane and fuse with lysosomes assisted by Atg3 and Atg7 to form the functional autolysosomes (62, 63). For the regulation of autophagy process, the Class I PI3K-mammalian target of rapamycin (mTOR) is shown as an inhibitory pathway of autophagy through the stimulation of mTOR complex 1 (mTORC1) (64). The Class III PI3K pathway is illustrated as an inductive pathway for autophagy with the formation of Class III PI3K-Beclin-1 complex (65, 66). So far, several kinds of autophagy inducers and inhibitors have been widely used in the modulation of autophagy level in both experimental studies and clinical practice. For instance, rapamycin is widely used to up-regulate the level of autophagy through the inhibition of mTORC1 activation (67, 68). For the inhibition of autophagy, the mechanism of 3-methyladenine (3-MA) for the inhibition of autophagy is through the inhibition of Class III PI3K complex formation (69). In addition, chloroquine is via the influence of the acidic environment of lysosomes and bafilomycin A1 is via the disturbance of the formation of autolysosomes (70–72).
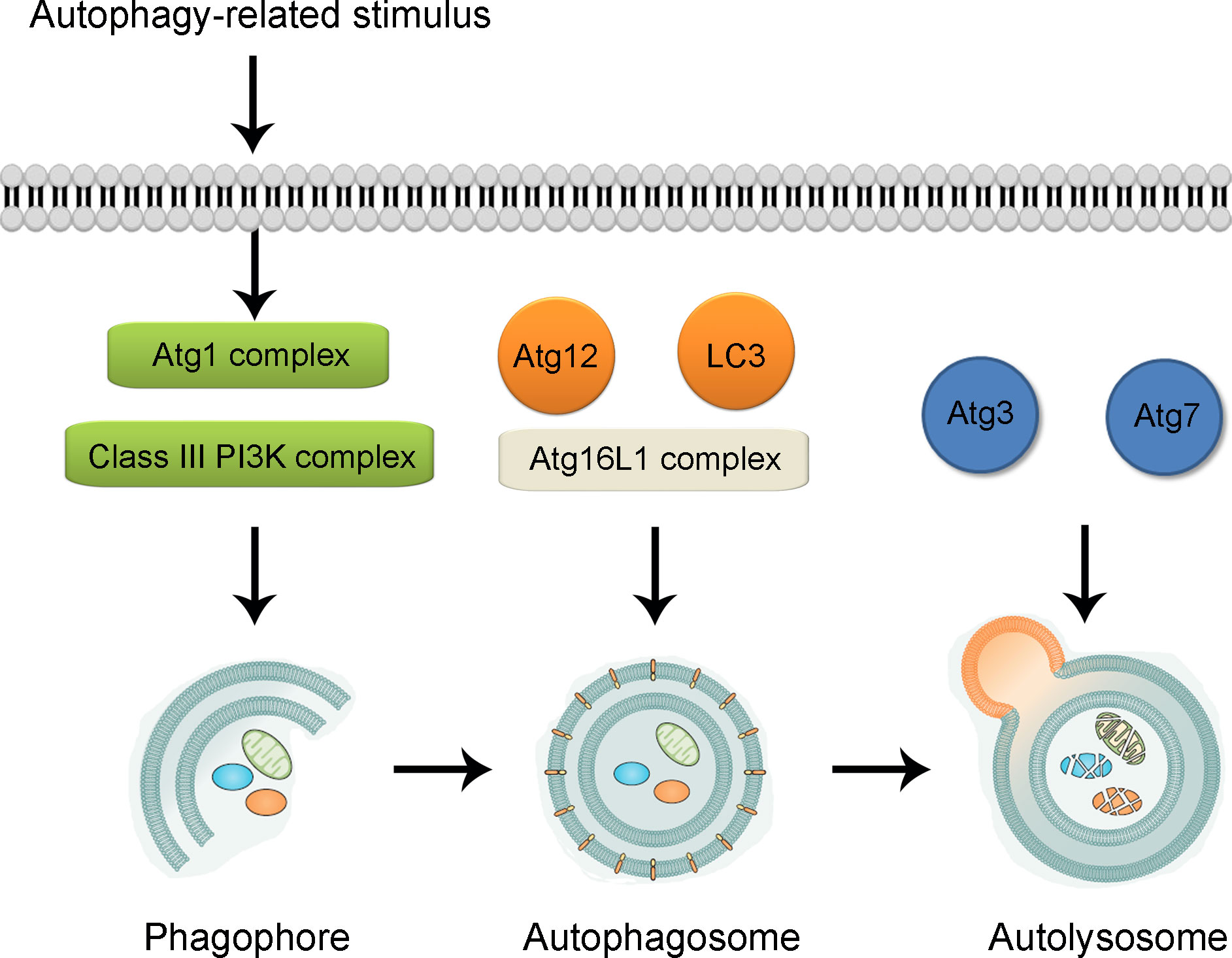
Figure 1 Illustration of biological process of autophagy. Under the challenge of autophagy-related stimulus, autophagy process is triggered through the formation of cup-shaped phagophore participated by Atg1 complex and Class III PI3K complex. With the formation of Atg16L1 complex and assistance of Atg12 and LC3, the bilayer membrane undergoes expansion, elongation and nucleation, which are sequestrated into double-membrane sphere-shaped autophagosomes. With the participation of Atg3 and Atg7, autophagosomes fuse with lysosomes to form the functional autolysosomes. Atg, autophagy-related gene. PI3K, phosphatidylinositol 3-kinase; LC3, light chain 3.
In the recent few decades, autophagy has been illustrated to participate or regulate numerous diseases in many systems, including atherosclerosis and hypertension (cardiovascular system) (73, 74), diabetes and obesity (metabolic system) (75, 76), ischemic stroke and multiple sclerosis (central nervous system) (77, 78), inflammatory bowel disease and gastritis (digestive system) (23, 79) as well as malignant tumors (80–82). In the treatment of cancers, the interventions to both induce and suppress autophagy have been shown as effective therapies, which indicates the complication of autophagy in the pathogenesis and progression of cancers (83). According to research literature, in the onset of cancers, autophagy can effectively prevent cancer cell formation via the correction of pro-tumoral genetic mutations and clearance of mutated cells (84, 85). Conversely, once cancer cells are formed, autophagy may promote cancer cell survival and growth through the autophagy-mediated cellular protection (80, 86). That knowledge reveals the difference between basal autophagy and stimulus-induced autophagy in cancers. The studies also indicate the different roles and effects of autophagy in the different stages of cancer progression.
Part II: The role of autophagy in gastrointestinal cancers
As discussed above, the role of autophagy is controversial in cancers. Such contradiction also exists in gastrointestinal cancers. In the following contents, the role of autophagy in several popular gastrointestinal cancers, including esophageal cancer, gastric cancer, and colorectal cancer, will be further discussed by reviewing previous related studies (illustrated in Figure 2).
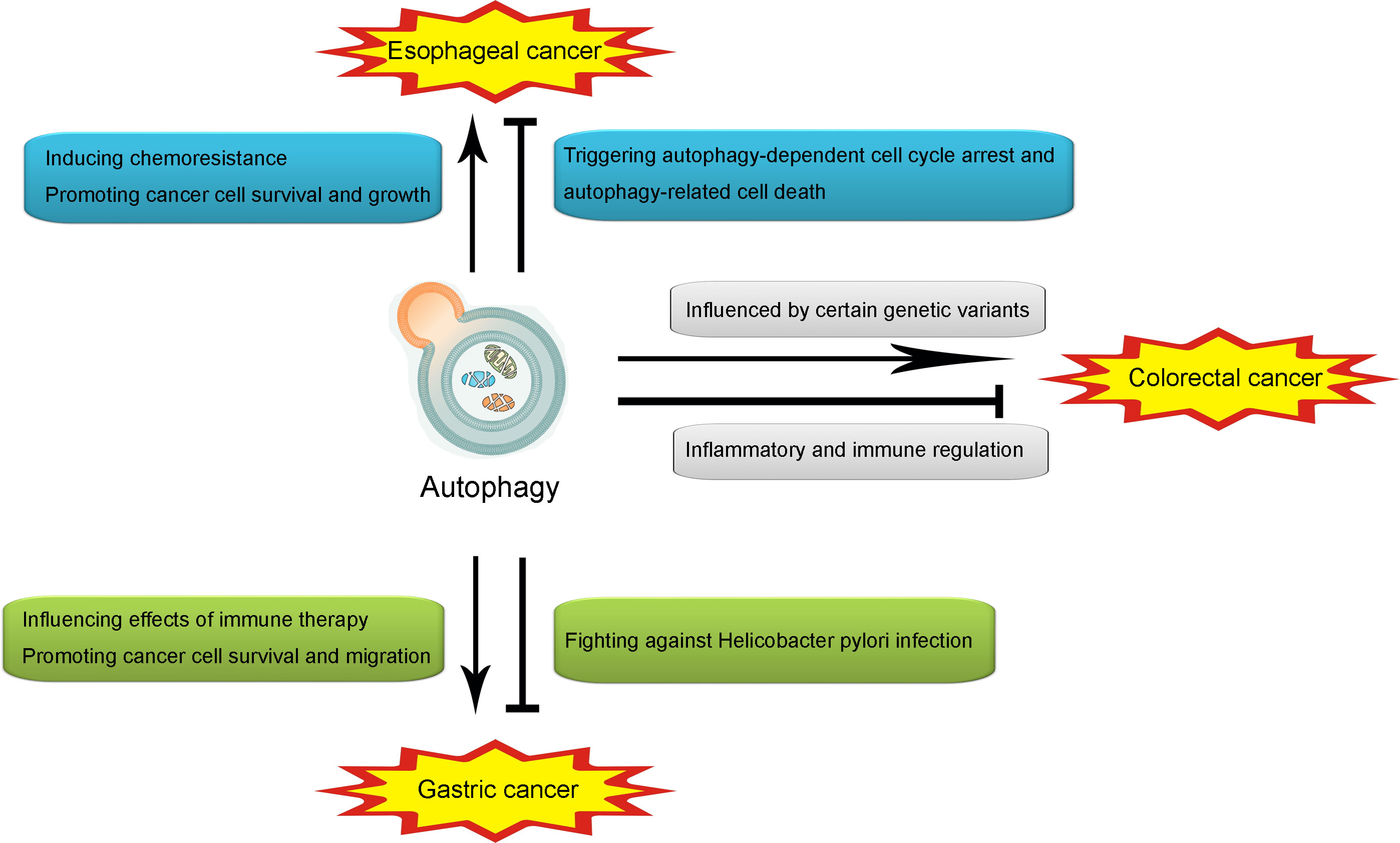
Figure 2 Illustration of the role of autophagy in esophageal cancer, gastric cancer and colorectal cancer. Autophagy may promote esophageal cancer through inducing chemoresistance and promoting cancer cell survival and growth. However, autophagy may also attenuate esophageal cancer through triggering autophagy-dependent cell cycle arrest and autophagy-related cell death. In gastric cancer, autophagy may promote the onset and development through influencing effects of immune therapy. However, autophagy may also attenuate it through fighting against Helicobacter pylori infection. In colorectal cancer, autophagy may promote the pathogenesis and progression through the influence of certain genetic variants in autophagy, while producing an attenuative effect through inflammatory and immune regulation.
Autophagy in esophageal cancer
Esophageal cancer is a type of malignant tumor that occurs in the esophagus, which is a long and hollow tubular organ ranging from throat to stomach (87, 88). According to recent statistics, although the incidence rates vary from different geographic locations, esophageal cancer is listed as one of the most common causes of cancer deaths all over the world (89, 90). The risk factors of esophageal cancer include Barrett’s esophagus, smoking, alcohol abuse, obesity, achalasia and frequent hot liquid intake, etc. (91, 92). The typical symptoms of esophageal cancer include dysphagia, chest pain, coughing and weight loss (92, 93). However, in some cases, due to its hidden onset, esophageal cancer is hard to be controlled and managed in the early stage. As a result, the research study of pathogenic mechanisms and specific screening markers for esophageal cancer is urgent.
So far, autophagy has been demonstrated to be closely associated with esophageal cancer. Four Atgs including DNAJB1, BNIP1, VAMP7 and TBK1 were shown to act as prognostic signature for the recognition of high- and low-risk groups among people (94). Another Atg signature including (VIM, UFM1, TSC2, SRC, MEFV, CTTN, CFTR and CDKN1A) were demonstrated to contribute to the improvement of the prediction of clinical outcomes in esophageal cancer patients (95). According to a research study, 22 autophagic long-chain noncoding ribonucleic acids were revealed to be highly correlated with the overall survival of esophageal adenocarcinoma patients, thus creating a novel prognostic model for esophageal adenocarcinoma (96).
For the role of autophagy in esophageal cancer, Fang et al. (97) reported that under the treatment of diketopyrrolopyrrole (DPP), autophagy was induced as a self-protective mechanism of esophageal cancer cells from DPP. Autophagy was also shown to mediate microRNA-193b-related chemoresistance on 5-fluorouracil (5-FU) treatment (98). Those findings indicated that the suppression of autophagy-mediated chemoresistance might be a potential strategy for adjuvant chemotherapy in esophageal cancer. In addition, microRNA-498 was shown to suppress esophageal cancer through the inhibition of autophagy and M2-like polarization of macrophages via mouse double minute 2 (MDM2)/activating transcription factor 3 (ATF3) signaling pathway (99). Jia et al. (100) reported that Phlorizin, an important member of the dihydrochalcone family derived from sweet tea, could suppress the progress of esophageal cancer through the promotion of cellular apoptosis and inhibit the level of autophagy in esophageal cancer cells. Those effects might lead to antagonizing the Janus kinase-2 (JAK2)/signal transducer and activator of transcription 3 (STAT3) signaling pathway. Furthermore, up-regulation of Beclin-1, a vital member of Atgs, can lead to a more aggressive esophageal squamous cell phenotype and a worse survival prognosis, thus indicating Beclin-1 to be a potential and promising prognostic biomarker and therapeutic target for patients with esophageal squamous cancer (101).
Conversely, some researches have reported the opposite effective of autophagy on esophageal cancer. According to a previous study, silencing autophagy via the inhibition of ATP6V1C1 has been shown to serve to enhance radiotherapy resistance in esophageal squamous cell carcinoma (102). In addition, the induction of autophagy via certain agents have been reported to trigger autophagy-dependent cell cycle arrest and autophagy-related cell death in cancer cells. For instance, dihydroartemisinin (DHA), the primary active derivative of artemisinin, produced an anti-tumor effect on esophageal cancer cells through the triggering of autophagy-dependent cell cycle arrest at the G2/M phase (103, 104). Therefore, further studies are demanded for the exploration of the role of autophagy in esophageal cancer.
Autophagy in gastric cancer
Gastric cancer, or stomach cancer, is amongst the most aggressive human malignant tumors all over the world, representing a heavy health burden (105, 106). An estimation of 90%-95% gastric cancers belong to adenocarcinomas. Besides, other types of gastric cancer can start in the stomach, including gastrointestinal stromal tumors (GISTs), neuroendocrine tumors and lymphomas, etc. (107, 108). So far, several risk factors have been commonly recognized, including western diet, obesity, age, gender and bacterial infection (109–111). Notably, Helicobacter pylori (H. pylori) infection is regarded as an independent risk factor, especially cancers in the lower (distal) part of the stomach (112). It is reported that gastric cancer patients are at higher infectious rate of H. pylori than people without gastric cancer (113). Like the situation of esophageal cancer, the prevention and management of gastric cancer in the early stage are difficult because of its latent symptoms in many cases.
In recent studies, the role of autophagy in gastric cancer has been widely investigated. According to many research studies, suppression of the level of autophagy might serve as an effective approach in the treatment of gastric cancer. For instance, the level of autophagy was shown to be associated with programmed cell death-1 (PD-1) with its ligand (PD-L1). Wang et al. (114) reported that inhibition of autophagy could enhance the expression of PD-L1, thus promoting the sensitivity to PD-L1-related immune therapy. In addition, UPR-induced autophagy activation was triggered by Sec62, a membrane protein of the endoplasmic reticulum that facilitated protein transport. Such pathway of autophagy induction was shown to contribute significantly to the metastasis of gastric cancer (115). In another study, Xu et al. (116) demonstrated that c-Jun N-terminal kinase (JNK)/extracellular signal-regulated kinase (ERK)-dependent autophagy was connected to gastric cancer cell survival, and the inhibition of JNK/ERK-dependent autophagy enhanced the Jaspine B derivative-induced gastric cancer cell death through the p62/Keap1/Nrf2 signaling pathway. Furthermore, oncogenic autophagy in gastric cancer cells were demonstrated to be controlled by mucolipin TRP cation channel 1 (MCOLN1), a lysosomal cation channel, via the mediation of zinc influx into the cytosol (117).
However, the situation of H. pylori infection has been shown to be different. It has been demonstrated that autophagy could protect against H. pylori infection (118). The H. pylori toxin vacuolating cytotoxin (VacA) and genetic deficiency of autophagy could promote H. pylori infection and thus contributing to the incidence of gastric cancer (118, 119). Sustained exposure to H. pylori was shown to inhibit autophagy process in gastric epithelial cells at least partly via the Nod1-nuclear factor (NF)-κB/mitogen-activated protein kinase (MAPK)-ERK/fork head box O 4 (FOXO4) signaling pathway (120). In addition, the induction of autophagy degrading functions by vitamin D3 could prevent gastric epithelial cells against H. pylori infection (121). Based on those findings, to fully take advantage of autophagy in the treatment of gastric cancer, the controversial roles of autophagy should be taken into thorough consideration, especially the effect of autophagy on H. pylori infection.
Autophagy in colorectal cancer
Colorectal cancer is a common malignant tumor of digestive tract, usually occurring at the junction of rectum and left half colon (122). It is considered to be the fourth most common malignant tumor, ranking third at 11% of all malignant tumors diagnosed all over the world (123, 124). The pathological types of colorectal cancer include adenocarcinoma, mucinous adenocarcinoma, squamous carcinoma and undifferentiated carcinoma, etc. The etiology of colorectal cancer is complicated and remains unclear. Currently, it is commonly recognized that the incidence of colorectal cancer is related to smoking, high-fat and low-cellulose diet, intestinal inflammation and other factors (125, 126). As for the occurrence and development mechanism of colorectal cancer, recent studies indicate that the inflammatory immune microenvironment of tumor cell growth plays an important role in such processes. Patients with chronic inflammatory bowel diseases, such as ulcerative colitis, have a significantly increased incidence of colorectal cancer, with enteritis-related colorectal cancer accounting for 6.7% of the total population during a 30-year follow-up period (127). The proportion of patients with ulcerative colitis who developed poor prognosis types of colorectal cancer such as signet ring cell carcinoma or myxoid carcinoma has increased significantly (128–130). Therefore, regulating the over-activation of inflammatory and immune reaction thus controlling the formation of tumor-associated inflammatory and immune microenvironment is vital for the prevention and treatment of colorectal cancer.
As shown in previous research and studies from us and other researchers, autophagy could function in the regulation of inflammatory and immune responses in many inflammation- and immune-related diseases, such as inflammatory bowel disease (IBD) (79, 131–135). Such inflammatory and immune regulating effects have also been revealed by numerous studies in colorectal cancer. For instance, autophagy was shown to couple the environmental signals and metabolic homeostasis to protect lineage and survival integrity of Treg cells, thus preventing tumor resistance and development of inflammatory disorders (136). The induction of mitophagy, a form of selective autophagy, could trigger the anti-tumor adaptive immunity during tumorigenesis (137). In addition, autophagy in intestinal epithelial cells was demonstrated to prevent tumorigenesis via the restoration of DNA damage and prevention of cell proliferation and inflammation, while deficiency in autophagy promoted tumor progression of colorectal cancer (138, 139). Induction of autophagy by certain small molecular agents were reported to protect against colitis-associated colorectal cancer via suppressing the NLR family pyrin domain containing 3 (NLRP3) inflammasome activation (140). Furthermore, as shown in our previous study, autophagy could attenuate the growth and metastasis of colorectal cancer through the modulation of neutrophil extracellular traps (NETs) and inflammasomes (141). However, another study revealed that the Thr300Ala variant in Atg16L1, one of the vital Atgs, was associated with improved overall survival in human colorectal cancer (142). Those findings indicated that certain mutation of Atgs might produce a therapeutic effect on colorectal cancer.
Conclusion
In this review, we have introduced and discussed the role of autophagy in three popular gastrointestinal cancers, including esophageal cancer, gastric cancer and colorectal cancer. As discussed, the effects of autophagy on the gastrointestinal cancers are controversial and complex. Although there have been many research studies conducted on this topic, the real mechanisms of autophagy in gastrointestinal cancers remain unclarified. To fully take advantage of autophagy in the treatment of gastrointestinal cancer, further studies are demanded on this topic.
Author’s contributions
B-ZS, N-LC and YY retrieved concerned literatures and wrote the manuscript. J-PL designed the figures. EL and HL revised the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Griffin-Sobel JP. Gastrointestinal cancers: Screening and early detection. Semin Oncol Nurs (2017) 33(2):165–71. doi: 10.1016/j.soncn.2017.02.004
2. Meyerhardt JA, Tepper JE, Venook AP. Special series: Advances in GI cancer. J Clin Oncol (2015) 33(16):1717–20. doi: 10.1200/JCO.2015.60.8661
3. Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer (2015) 136(5):E359–86. doi: 10.1002/ijc.29210
4. Khezri MR, Malekinejad H, Majidi-Zolbanin N, Ghasemnejad-Berenji M. Anticancer potential of metformin: focusing on gastrointestinal cancers. Cancer Chemother Pharmacol (2021) 87(5):587–98. doi: 10.1007/s00280-021-04256-8
5. Newsome RC, Yang Y, Jobin C. The microbiome, gastrointestinal cancer, and immunotherapy. J Gastroenterol Hepatol (2022) 37(2):263–72. doi: 10.1111/jgh.15742
6. Patel SG, Karlitz JJ, Yen T, Lieu CH, Boland CR. The rising tide of early-onset colorectal cancer: a comprehensive review of epidemiology, clinical features, biology, risk factors, prevention, and early detection. Lancet Gastroenterol Hepatol (2022) 7(3):262–74. doi: 10.1016/S2468-1253(21)00426-X
7. Moazzen S, Cortes-Ibanez FO, van der Vegt B, Alizadeh BZ, de Bock GH. Diet quality indices and gastrointestinal cancer risk: results from the lifelines study. Eur J Nutr (2022) 61(1):317–27. doi: 10.1007/s00394-021-02648-3
8. Lewandowska A, Rudzki G, Lewandowski T, Stryjkowska-Gora A, Rudzki S. Title: Risk factors for the diagnosis of colorectal cancer. Cancer Control (2022) 29:10732748211056692. doi: 10.1177/10732748211056692
9. Grady WM, Yu M, Markowitz SD. Epigenetic alterations in the gastrointestinal tract: Current and emerging use for biomarkers of cancer. Gastroenterology (2021) 160(3):690–709. doi: 10.1053/j.gastro.2020.09.058
10. Zhang B, Wu Q, Li B, Wang D, Wang L, Zhou YL. m(6)A regulator-mediated methylation modification patterns and tumor microenvironment infiltration characterization in gastric cancer. Mol Cancer (2020) 19(1):53. doi: 10.1186/s12943-020-01170-0
11. Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol (2021) 21(10):653–67. doi: 10.1038/s41577-021-00534-x
12. Lu Y, Li Y, Liu Q, Tian N, Du P, Zhu F, et al. MondoA-Thioredoxin-Interacting protein axis maintains regulatory T-cell identity and function in colorectal cancer microenvironment. Gastroenterology (2021) 161(2):575–91.e16. doi: 10.1053/j.gastro.2021.04.041
13. You JH, Song SO, Kang MJ, Cho YY, Kim SW, Suh SH, et al. Metformin and gastrointestinal cancer development in newly diagnosed type 2 diabetes: A population-based study in Korea. Clin Transl Gastroenterol (2020) 11(11):e00254. doi: 10.14309/ctg.0000000000000254
14. Kocak M, Ezazi Erdi S, Jorba G, Maestro I, Farres J, Kirkin V, et al. Targeting autophagy in disease: established and new strategies. Autophagy (2022) 18(3):473–95. doi: 10.1080/15548627.2021.1936359
15. Ma Q, Long S, Gan Z, Tettamanti G, Li K, Tian L. Transcriptional and post-transcriptional regulation of autophagy. Cells (2022) 11(3). doi: 10.3390/cells11030441
16. Dong Y, Wu Y, Zhao GL, Ye ZY, Xing CG, Yang XD. Inhibition of autophagy by 3-MA promotes hypoxia-induced apoptosis in human colorectal cancer cells. Eur Rev Med Pharmacol Sci (2019) 23(3):1047–54. doi: 10.26355/eurrev_201902_16992
17. Qureshi-Baig K, Kuhn D, Viry E, Pozdeev VI, Schmitz M, Rodriguez F, et al. Hypoxia-induced autophagy drives colorectal cancer initiation and progression by activating the PRKC/PKC-EZR (ezrin) pathway. Autophagy (2020) 16(8):1436–52. doi: 10.1080/15548627.2019.1687213
18. Tanida I, Ueno T, Kominami E. LC3 and autophagy. Methods Mol Biol (2008) 445:77–88. doi: 10.1007/978-1-59745-157-4_4
19. Klionsky DJ, Petroni G, Amaravadi RK, Baehrecke EH, Ballabio A, Boya P, et al. Autophagy in major human diseases. EMBO J (2021) 40(19):e108863. doi: 10.15252/embj.2021108863
20. Glick D, Barth S, Macleod KF. Autophagy: cellular and molecular mechanisms. J Pathol (2010) 221(1):3–12. doi: 10.1002/path.2697
21. Lamark T, Svenning S, Johansen T. Regulation of selective autophagy: the p62/SQSTM1 paradigm. Essays Biochem (2017) 61(6):609–24. doi: 10.1042/EBC20170035
22. Deretic V. Autophagy in inflammation, infection, and immunometabolism. Immunity (2021) 54(3):437–53. doi: 10.1016/j.immuni.2021.01.018
23. Larabi A, Barnich N, Nguyen HTT. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy (2020) 16(1):38–51. doi: 10.1080/15548627.2019.1635384
24. Wang T, Liu K, Wen L, Yang Y, Yin X, Liu K, et al. Autophagy and gastrointestinal diseases. Adv Exp Med Biol (2020) 1207:529–56. doi: 10.1007/978-981-15-4272-5_38
25. Lapaquette P, Bizeau JB, Acar N, Bringer MA. Reciprocal interactions between gut microbiota and autophagy. World J Gastroenterol (2021) 27(48):8283–301. doi: 10.3748/wjg.v27.i48.8283
26. Wang F, Sun N, Zeng H, Gao Y, Zhang N, Zhang W. Selenium deficiency leads to inflammation, autophagy, endoplasmic reticulum stress, apoptosis and contraction abnormalities via affecting intestinal flora in intestinal smooth muscle of mice. Front Immunol (2022) 13:947655. doi: 10.3389/fimmu.2022.947655
27. Mommersteeg MC, Simovic I, Yu B, van Nieuwenburg SAV, Bruno IMJ, Doukas M, et al. Autophagy mediates ER stress and inflammation in helicobacter pylori-related gastric cancer. Gut Microbes (2022) 14(1):2015238. doi: 10.1080/19490976.2021.2015238
28. Weng W, Goel A. Curcumin and colorectal cancer: An update and current perspective on this natural medicine. Semin Cancer Biol (2022) 80:73–86. doi: 10.1016/j.semcancer.2020.02.011
29. Yang R, Dong H, Jia S, Yang Z. Resveratrol as a modulatory of apoptosis and autophagy in cancer therapy. Clin Transl Oncol (2022) 24(7):1219–30. doi: 10.1007/s12094-021-02770-y
30. Han H, Yang C, Ma J, Zhang S, Zheng S, Ling R, et al. N(7)-methylguanosine tRNA modification promotes esophageal squamous cell carcinoma tumorigenesis via the RPTOR/ULK1/autophagy axis. Nat Commun (2022) 13(1):1478. doi: 10.1038/s41467-022-29125-7
31. Rahman MA, Ahmed KR, Rahman MH, Park MN, Kim B. Potential therapeutic action of autophagy in gastric cancer managements: Novel treatment strategies and pharmacological interventions. Front Pharmacol (2021) 12:813703. doi: 10.3389/fphar.2021.813703
32. Luo Y, Zheng S, Wu Q, Wu J, Zhou R, Wang C, et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy (2021) 17(12):4083–101. doi: 10.1080/15548627.2021.1901204
33. Wu Q, Ma J, Wei J, Meng W, Wang Y, Shi M. lncRNA SNHG11 promotes gastric cancer progression by activating the wnt/beta-catenin pathway and oncogenic autophagy. Mol Ther (2021) 29(3):1258–78. doi: 10.1016/j.ymthe.2020.10.011
34. Li GM, Li L, Li MQ, Chen X, Su Q, Deng ZJ, et al. DAPK3 inhibits gastric cancer progression via activation of ULK1-dependent autophagy. Cell Death Differ (2021) 28(3):952–67. doi: 10.1038/s41418-020-00627-5
35. Wen X, Klionsky DJ. At A glance: A history of autophagy and cancer. Semin Cancer Biol (2020) 66:3–11. doi: 10.1016/j.semcancer.2019.11.005
36. Parkhitko AA, Favorova OO, Henske EP. Autophagy: Mechanisms, regulation, and its role in tumorigenesis. Biochem (Mosc) (2013) 78(4):355–67. doi: 10.1134/S0006297913040044
37. Mamaev DV, Zvyagilskaya RA. Mitophagy in yeast. Biochem (Mosc) (2019) 84(Suppl 1):S225–32. doi: 10.1134/S000629791914013X
38. Tam SY, Wu VW, Law HK. Influence of autophagy on the efficacy of radiotherapy. Radiat Oncol (2017) 12(1):57. doi: 10.1186/s13014-017-0795-y
39. Van Noorden R, Ledford H. Medicine Nobel for research on how cells 'eat themselves'. Nature (2016) 538(7623):18–9. doi: 10.1038/nature.2016.20721
40. Cui H, Norrbacka S, Myohanen TT. Prolyl oligopeptidase acts as a link between chaperone-mediated autophagy and macroautophagy. Biochem Pharmacol (2022) 197:114899. doi: 10.1016/j.bcp.2021.114899
41. Liu J, Kuang F, Kroemer G, Klionsky DJ, Kang R, Tang D. Autophagy-dependent ferroptosis: Machinery and regulation. Cell Chem Biol (2020) 27(4):420–35. doi: 10.1016/j.chembiol.2020.02.005
42. Islam Khan MZ, Tam SY, Law HKW. Autophagy-modulating long non-coding RNAs (LncRNAs) and their molecular events in cancer. Front Genet (2018) 9:750. doi: 10.3389/fgene.2018.00750
43. Schuck S. Microautophagy - distinct molecular mechanisms handle cargoes of many sizes. J Cell Sci (2020) 133(17). doi: 10.1242/jcs.246322
44. Li J, Hochstrasser M. Microautophagy regulates proteasome homeostasis. Curr Genet (2020) 66(4):683–7. doi: 10.1007/s00294-020-01059-x
45. Dong S, Wang Q, Kao YR, Diaz A, Tasset I, Kaushik S, et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature (2021) 591(7848):117–23. doi: 10.1038/s41586-020-03129-z
46. Chen C, Wang D, Yu Y, Zhao T, Min N, Wu Y, et al. Legumain promotes tubular ferroptosis by facilitating chaperone-mediated autophagy of GPX4 in AKI. Cell Death Dis (2021) 12(1):65. doi: 10.1038/s41419-020-03362-4
47. Munz C. The macroautophagy machinery in MHC restricted antigen presentation. Front Immunol (2021) 12:628429. doi: 10.3389/fimmu.2021.628429
48. Nieto-Torres JL, Hansen M. Macroautophagy and aging: The impact of cellular recycling on health and longevity. Mol Aspects Med (2021) 82:101020. doi: 10.1016/j.mam.2021.101020
49. Deretic V, Kroemer G. Autophagy in metabolism and quality control: opposing, complementary or interlinked functions? Autophagy (2022) 18(2):283–92. doi: 10.1080/15548627.2021.1933742
50. Guan X, Iyaswamy A, Sreenivasmurthy SG, Su C, Zhu Z, Liu J, et al. Mechanistic insights into selective autophagy subtypes in alzheimer's disease. Int J Mol Sci (2022) 23(7). doi: 10.3390/ijms23073609
51. Li W, He P, Huang Y, Li YF, Lu J, Li M, et al. Selective autophagy of intracellular organelles: recent research advances. Theranostics (2021) 11(1):222–56. doi: 10.7150/thno.49860
52. Ji CH, Kwon YT. Crosstalk and interplay between the ubiquitin-proteasome system and autophagy. Mol Cells (2017) 40(7):441–9. doi: 10.14348/molcells.2017.0115
53. Sun-Wang JL, Yarritu-Gallego A, Ivanova S, Zorzano A. The ubiquitin-proteasome system and autophagy: self-digestion for metabolic health. Trends Endocrinol Metab (2021) 32(8):594–608. doi: 10.1016/j.tem.2021.04.015
54. Zhang J, Ding R, Wu T, Jia J, Cheng X. Autophagy-related genes and long noncoding RNAs signatures as predictive biomarkers for osteosarcoma survival. Front Cell Dev Biol (2021) 9:705291. doi: 10.3389/fcell.2021.705291
55. Hu LF. Epigenetic regulation of autophagy. Adv Exp Med Biol (2019) 1206:221–36. doi: 10.1007/978-981-15-0602-4_11
56. Shao BZ, Yao Y, Zhai JS, Zhu JH, Li JP, Wu K. The role of autophagy in inflammatory bowel disease. Front Physiol (2021) 12:621132. doi: 10.3389/fphys.2021.621132
57. Wang P, Shao BZ, Deng Z, Chen S, Yue Z, Miao CY. Autophagy in ischemic stroke. Prog Neurobiol (2018) 163-164:98–117. doi: 10.1016/j.pneurobio.2018.01.001
58. Fazeli G, Trinkwalder M, Irmisch L, Wehman AM. C. elegans midbodies are released, phagocytosed and undergo LC3-dependent degradation independent of macroautophagy. J Cell Sci (2016) 129(20):3721–31. doi: 10.1242/jcs.190223
59. Chang YY, Juhasz G, Goraksha-Hicks P, Arsham AM, Mallin DR, Muller LK, et al. Nutrient-dependent regulation of autophagy through the target of rapamycin pathway. Biochem Soc Trans (2009) 37(Pt 1):232–6. doi: 10.1042/BST0370232
60. Cadwell K, Liu JY, Brown SL, Miyoshi H, Loh J, Lennerz JK, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal paneth cells. Nature (2008) 456(7219):259–63. doi: 10.1038/nature07416
61. Boukhalfa A, Roccio F, Dupont N, Codogno P, Morel E. When the autophagy protein ATG16L1 met the ciliary protein IFT20. Autophagy (2021) 17(7):1791–3. doi: 10.1080/15548627.2021.1935004
62. Vujic N, Bradic I, Goeritzer M, Kuentzel KB, Rainer S, Kratky D, et al. ATG7 is dispensable for LC3-PE conjugation in thioglycolate-elicited mouse peritoneal macrophages. Autophagy (2021) 17(11):3402–7. doi: 10.1080/15548627.2021.1874132
63. Wan DF, Pan SS, Tong YS, Huang Y. Exercise preconditioning promotes autophagy to cooperate for cardioprotection by increasing LC3 lipidation-associated proteins. Front Physiol (2021) 12:599892. doi: 10.3389/fphys.2021.599892
64. Aoki M, Fujishita T. Oncogenic roles of the PI3K/AKT/mTOR axis. Curr Top Microbiol Immunol (2017) 407:153–89. doi: 10.1007/82_2017_6
65. Liu Y, Song A, Wu H, Sun Y, Dai M. Paeonol inhibits apoptosis of vascular smooth muscle cells via up-regulation of autophagy by activating class III PI3K/Beclin-1 signaling pathway. Life Sci (2021) 264:118714. doi: 10.1016/j.lfs.2020.118714
66. Shen JZ, Wu G, Guo S. Amino acids in autophagy: Regulation and function. Adv Exp Med Biol (2021) 1332:51–66. doi: 10.1007/978-3-030-74180-8_4
67. Chao X, Wang S, Fulte S, Ma X, Ahamed F, Cui W, et al. Hepatocytic p62 suppresses ductular reaction and tumorigenesis in mouse livers with mTORC1 activation and defective autophagy. J Hepatol (2022) 76(3):639–51. doi: 10.1016/j.jhep.2021.10.014
68. Yang H, Wen Y, Zhang M, Liu Q, Zhang H, Zhang J, et al. MTORC1 coordinates the autophagy and apoptosis signaling in articular chondrocytes in osteoarthritic temporomandibular joint. Autophagy (2020) 16(2):271–88. doi: 10.1080/15548627.2019.1606647
69. Du J, Xu Q, Zhao H, Jia X, Ba N, Peng F, et al. PI3K inhibitor 3-MA promotes the antiproliferative activity of esomeprazole in gastric cancer cells by downregulating EGFR via the PI3K/FOXO3a pathway. BioMed Pharmacother (2022) 148:112665. doi: 10.1016/j.biopha.2022.112665
70. Wang M, Zeng L, Su P, Ma L, Zhang M, Zhang YZ. Autophagy: A multifaceted player in the fate of sperm. Hum Reprod Update (2022) 28(2):200–31. doi: 10.1093/humupd/dmab043
71. Holm TM, Bian ZC, Manupati K, Guan JL. Inhibition of autophagy mitigates cell migration and invasion in thyroid cancer. Surgery (2022) 171(1):235–44. doi: 10.1016/j.surg.2021.08.024
72. Visser N, Lourens HJ, Huls G, Bremer E, Wiersma VR. Inhibition of autophagy does not re-sensitize acute myeloid leukemia cells resistant to cytarabine. Int J Mol Sci (2021) 22(5). doi: 10.3390/ijms22052337
73. Gatica D, Chiong M, Lavandero S, Klionsky DJ. The role of autophagy in cardiovascular pathology. Cardiovasc Res (2022) 118(4):934–50. doi: 10.1093/cvr/cvab158
74. Lin L, Zhang MX, Zhang L, Zhang D, Li C, Li YL. Autophagy, pyroptosis, and ferroptosis: New regulatory mechanisms for atherosclerosis. Front Cell Dev Biol (2021) 9:809955. doi: 10.3389/fcell.2021.809955
75. Sarparanta J, Garcia-Macia M, Singh R. Autophagy and mitochondria in obesity and type 2 diabetes. Curr Diabetes Rev (2017) 13(4):352–69. doi: 10.2174/1573399812666160217122530
76. Tao T, Xu H. Autophagy and obesity and diabetes. Adv Exp Med Biol (2020) 1207:445–61. doi: 10.1007/978-981-15-4272-5_32
77. Wnuk A, Kajta M. Steroid and xenobiotic receptor signalling in apoptosis and autophagy of the nervous system. Int J Mol Sci (2017) 18(11). doi: 10.3390/ijms18112394
78. Vezzani B, Carinci M, Patergnani S, Pasquin MP, Guarino A, Aziz N, et al. The dichotomous role of inflammation in the CNS: A mitochondrial point of view. Biomolecules (2020) 10(10). doi: 10.3390/biom10101437
79. Shao BZ, Wang SL, Fang J, Li ZS, Bai Y, Wu K. Alpha7 nicotinic acetylcholine receptor alleviates inflammatory bowel disease through induction of AMPK-mTOR-p70S6K-Mediated autophagy. Inflammation (2019) 42(5):1666–79. doi: 10.1007/s10753-019-01027-9
80. Li X, He S, Ma B. Autophagy and autophagy-related proteins in cancer. Mol Cancer (2020) 19(1):12. doi: 10.1186/s12943-020-1138-4
81. Rakesh R, PriyaDharshini LC, Sakthivel KM, Rasmi RR. Role and regulation of autophagy in cancer. Biochim Biophys Acta Mol Basis Dis (2022) 1868(7):166400. doi: 10.1016/j.bbadis.2022.166400
82. Fares HM, Lyu X, Xu X, Dong R, Ding M, Mi S, et al. Autophagy in cancer: The cornerstone during glutamine deprivation. Eur J Pharmacol (2022) 916:174723. doi: 10.1016/j.ejphar.2021.174723
83. Levy JMM, Towers CG, Thorburn A. Targeting autophagy in cancer. Nat Rev Cancer (2017) 17(9):528–42. doi: 10.1038/nrc.2017.53
84. Awi NJ, Yap HY, Armon S, Low JSH, Peh KB, Peh SC, et al. Association between autophagy and KRAS mutation with clinicopathological variables in colorectal cancer patients. Malays J Pathol (2021) 43(2):269–79.
85. Gamez-Garcia A, Bolinaga-Ayala I, Yoldi G, Espinosa-Gil S, Dieguez-Martinez N, Megias-Roda E, et al. ERK5 inhibition induces autophagy-mediated cancer cell death by activating ER stress. Front Cell Dev Biol (2021) 9:742049. doi: 10.3389/fcell.2021.742049
86. Li YJ, Lei YH, Yao N, Wang CR, Hu N, Ye WC, et al. Autophagy and multidrug resistance in cancer. Chin J Cancer (2017) 36(1):52. doi: 10.1186/s40880-017-0219-2
87. Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, et al. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): A randomised, open-label, phase 3 trial. Lancet (2021) 398(10294):27–40. doi: 10.1016/S0140-6736(21)00797-2
88. Lagergren J, Smyth E, Cunningham D, Lagergren P. Oesophageal cancer. Lancet (2017) 390(10110):2383–96. doi: 10.1016/S0140-6736(17)31462-9
89. Kang YK, Chen LT, Ryu MH, Oh DY, Oh SC, Chung HC, et al. Nivolumab plus chemotherapy versus placebo plus chemotherapy in patients with HER2-negative, untreated, unresectable advanced or recurrent gastric or gastro-oesophageal junction cancer (ATTRACTION-4): A randomised, multicentre, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol (2022) 23(2):234–47. doi: 10.1016/S1470-2045(21)00692-6
90. Xia C, Dong X, Li H, Cao M, Sun D, He S, et al. Cancer statistics in China and united states, 2022: profiles, trends, and determinants. Chin Med J (Engl) (2022) 135(5):584–90. doi: 10.1097/CM9.0000000000002108
91. Huang FL, Yu SJ. Esophageal cancer: Risk factors, genetic association, and treatment. Asian J Surg (2018) 41(3):210–5. doi: 10.1016/j.asjsur.2016.10.005
92. Uhlenhopp DJ, Then EO, Sunkara T, Gaduputi V. Epidemiology of esophageal cancer: update in global trends, etiology and risk factors. Clin J Gastroenterol (2020) 13(6):1010–21. doi: 10.1007/s12328-020-01237-x
93. Li S, Chen H, Man J, Zhang T, Yin X, He Q, et al. Changing trends in the disease burden of esophageal cancer in China from 1990 to 2017 and its predicted level in 25 years. Cancer Med (2021) 10(5):1889–99. doi: 10.1002/cam4.3775
94. Du H, Xie S, Guo W, Che J, Zhu L, Hang J, et al. Development and validation of an autophagy-related prognostic signature in esophageal cancer. Ann Transl Med (2021) 9(4):317. doi: 10.21037/atm-20-4541
95. Shi X, Li Y, Pan S, Liu X, Ke Y, Guo W, et al. Identification and validation of an autophagy-related gene signature for predicting prognosis in patients with esophageal squamous cell carcinoma. Sci Rep (2022) 12(1):1960. doi: 10.1038/s41598-022-05922-4
96. Wu L, Zheng Y, Ruan X, Wu D, Xu P, Liu J, et al. Long-chain noncoding ribonucleic acids affect the survival and prognosis of patients with esophageal adenocarcinoma through the autophagy pathway: construction of a prognostic model. Anticancer Drugs (2022) 33(1):e590–603. doi: 10.1097/CAD.0000000000001189
97. Fang F, Li Y, Chang L. Mechanism of autophagy regulating chemoresistance in esophageal cancer cells. Exp Mol Pathol (2020) 117:104564. doi: 10.1016/j.yexmp.2020.104564
98. Nyhan MJ, O'Donovan TR, Boersma AW, Wiemer EA. MiR-193b promotes autophagy and non-apoptotic cell death in oesophageal cancer cells. BMC Cancer (2016) 16:101. doi: 10.1186/s12885-016-2123-6
99. Li D, Yan M, Sun F, Song J, Hu X, Yu S, et al. miR-498 inhibits autophagy and M2-like polarization of tumor-associated macrophages in esophageal cancer via MDM2/ATF3. Epigenomics (2021) 13(13):1013–30. doi: 10.2217/epi-2020-0341
100. Jia Z, Xie Y, Wu H, Wang Z, Li A, Li Z, et al. Phlorizin from sweet tea inhibits the progress of esophageal cancer by antagonizing the JAK2/STAT3 signaling pathway. Oncol Rep (2021) 46(1). doi: 10.3892/or.2021.8088
101. Du H, Luo F, Shi M, Che J, Zhu L, Li H, et al. Beclin-1 is a promising prognostic biomarker in a specific esophageal squamous cell carcinoma population. Pathol Oncol Res (2021) 27:594724. doi: 10.3389/pore.2021.594724
102. Yao X, Chen H, Xu B, Lu J, Gu J, Chen F, et al. The ATPase subunit of ATP6V1C1 inhibits autophagy and enhances radiotherapy resistance in esophageal squamous cell carcinoma. Gene (2021) 768:145261. doi: 10.1016/j.gene.2020.145261
103. Ma Q, Liao H, Xu L, Li Q, Zou J, Sun R, et al. Autophagy-dependent cell cycle arrest in esophageal cancer cells exposed to dihydroartemisinin. Chin Med (2020) 15:37. doi: 10.1186/s13020-020-00318-w
104. Chen X, He LY, Lai S, He Y. Dihydroartemisinin inhibits the migration of esophageal cancer cells by inducing autophagy. Oncol Lett (2020) 20(4):94. doi: 10.3892/ol.2020.11955
105. Yeoh KG, Tan P. Mapping the genomic diaspora of gastric cancer. Nat Rev Cancer (2022) 22(2):71–84. doi: 10.1038/s41568-021-00412-7
106. Ajani JA, D'Amico TA, Bentrem DJ, Chao J, Cooke D, Corvera C, et al. Gastric cancer, version 2.2022, NCCN clinical practice guidelines in oncology. J Natl Compr Canc Netw (2022) 20(2):167–92. doi: 10.6004/jnccn.2022.0008
107. Cummings D, Wong J, Palm R, Hoffe S, Almhanna K, Vignesh S. Epidemiology, diagnosis, staging and multimodal therapy of esophageal and gastric tumors. Cancers (Basel) (2021) 13(3). doi: 10.3390/cancers13030582
108. Domenech-Ximenos B, Juanpere S, Serra I, Codina J, Maroto A. Duodenal tumors on cross-sectional imaging with emphasis on multidetector computed tomography: A pictorial review. Diagn Interv Radiol (2020) 26(3):193–9. doi: 10.5152/dir.2019.19241
109. Lee JWJ, Zhu F, Srivastava S, Tsao SK, Khor C, Ho KY, et al. Severity of gastric intestinal metaplasia predicts the risk of gastric cancer: A prospective multicentre cohort study (GCEP). Gut (2022) 71(5):854–63. doi: 10.1136/gutjnl-2021-324057
110. Yang Y, Jung GM. Metabolic syndrome as a risk factor for gastric cancer by gender. Iran J Public Health (2022) 51(1):216–8. doi: 10.18502/ijph.v51i1.8317
111. Song M, Camargo MC, Katki HA, Weinstein SJ, Mannisto S, Albanes D, et al. Association of antiparietal cell and anti-intrinsic factor antibodies with risk of gastric cancer. JAMA Oncol (2022) 8(2):268–74. doi: 10.1001/jamaoncol.2021.5395
112. Oster P, Vaillant L, Riva E, McMillan B, Begka C, Truntzer C, et al. Helicobacter pylori infection has a detrimental impact on the efficacy of cancer immunotherapies. Gut (2022) 71(3):457–66. doi: 10.1136/gutjnl-2020-323392
113. Shichijo S, Uedo N, Michida T. Detection of early gastric cancer after helicobacter pylori eradication. Digestion (2022) 103(1):54–61. doi: 10.1159/000519838
114. Wang X, Wu WKK, Gao J, Li Z, Dong B, Lin X, et al. Autophagy inhibition enhances PD-L1 expression in gastric cancer. J Exp Clin Cancer Res (2019) 38(1):140. doi: 10.1186/s13046-019-1148-5
115. Su S, Shi YT, Chu Y, Jiang MZ, Wu N, Xu B, et al. Sec62 promotes gastric cancer metastasis through mediating UPR-induced autophagy activation. Cell Mol Life Sci (2022) 79(2):133. doi: 10.1007/s00018-022-04143-2
116. Xu F, Xie Q, Li YW, Jing QQ, Liu XJ, Xu YC, et al. Suppression of JNK/ERK dependent autophagy enhances jaspine b derivative-induced gastric cancer cell death via attenuation of p62/Keap1/Nrf2 pathways. Toxicol Appl Pharmacol (2022) 438:115908. doi: 10.1016/j.taap.2022.115908
117. Qi J, Xing Y, Liu Y, Wang MM, Wei X, Sui Z, et al. MCOLN1/TRPML1 finely controls oncogenic autophagy in cancer by mediating zinc influx. Autophagy (2021) 17(12):4401–22. doi: 10.1080/15548627.2021.1917132
118. Raju D, Hussey S, Ang M, Terebiznik MR, Sibony M, Galindo-Mata E, et al. Vacuolating cytotoxin and variants in Atg16L1 that disrupt autophagy promote helicobacter pylori infection in humans. Gastroenterology (2012) 142(5):1160–71. doi: 10.1053/j.gastro.2012.01.043
119. Muhammad JS, Nanjo S, Ando T, Yamashita S, Maekita T, Ushijima T, et al. Autophagy impairment by helicobacter pylori-induced methylation silencing of MAP1LC3Av1 promotes gastric carcinogenesis. Int J Cancer (2017) 140(10):2272–83. doi: 10.1002/ijc.30657
120. He Y, Wang C, Zhang X, Lu X, Xing J, Lv J, et al. Sustained exposure to helicobacter pylori lysate inhibits apoptosis and autophagy of gastric epithelial cells. Front Oncol (2020) 10:581364. doi: 10.3389/fonc.2020.581364
121. Hu W, Zhang L, Li MX, Shen J, Liu XD, Xiao ZG, et al. Vitamin D3 activates the autolysosomal degradation function against helicobacter pylori through the PDIA3 receptor in gastric epithelial cells. Autophagy (2019) 15(4):707–25. doi: 10.1080/15548627.2018.1557835
122. Wang SL, Shao BZ, Zhao SB, Fang J, Gu L, Miao CY, et al. Impact of paneth cell autophagy on inflammatory bowel disease. Front Immunol (2018) 9:693. doi: 10.3389/fimmu.2018.00693
123. Zhang B, Wang HE, Bai YM, Tsai SJ, Su TP, Chen TJ, et al. Inflammatory bowel disease is associated with higher dementia risk: a nationwide longitudinal study. Gut (2021) 70(1):85–91. doi: 10.1136/gutjnl-2020-320789
124. Cox SR, Lindsay JO, Fromentin S, Stagg AJ, McCarthy NE, et al. Effects of low FODMAP diet on symptoms, fecal microbiome, and markers of inflammation in patients with quiescent inflammatory bowel disease in a randomized trial. Gastroenterology (2020) 158(1):176–88.e7. doi: 10.1053/j.gastro.2019.09.024
125. Yue T, Chen S, Zhu J, Guo S, Huang Z, Wang P, et al. The aging-related risk signature in colorectal cancer. Aging (Albany NY) (2021) 13(5):7330–49. doi: 10.18632/aging.202589
126. Burnett-Hartman AN, Lee JK, Demb J, Gupta S. An update on the epidemiology, molecular characterization, diagnosis, and screening strategies for early-onset colorectal cancer. Gastroenterology (2021) 160(4):1041–9. doi: 10.1053/j.gastro.2020.12.068
127. Yvellez OV, Rai V, Sossenheimer PH, Hart J, Turner JR, Weber C, et al. Cumulative histologic inflammation predicts colorectal neoplasia in ulcerative colitis: A validation study. Inflammation Bowel Dis (2021) 27(2):203–6. doi: 10.1093/ibd/izaa047
128. Potdar AA, Dube S, Naito T, Li K, Botwin G, Haritunians T, et al. Altered intestinal ACE2 levels are associated with inflammation, severe disease, and response to anti-cytokine therapy in inflammatory bowel disease. Gastroenterology (2021) 160(3):809–22.e7. doi: 10.1053/j.gastro.2020.10.041
129. Ke P, Shao BZ, Xu ZQ, Chen XW, Liu C. Intestinal autophagy and its pharmacological control in inflammatory bowel disease. Front Immunol (2016) 7:695. doi: 10.3389/fimmu.2016.00695
130. Guo R, Meng Q, Wang B, Li F. Anti-inflammatory effects of platycodin d on dextran sulfate sodium (DSS) induced colitis and e. coli lipopolysaccharide (LPS) induced inflammation. Int Immunopharmacol (2021) 94:107474. doi: 10.1016/j.intimp.2021.107474
131. Matsuzawa-Ishimoto Y, Hwang S, Cadwell K. Autophagy and inflammation. Annu Rev Immunol (2018) 36:73–101. doi: 10.1146/annurev-immunol-042617-053253
132. Reglero-Real N, Perez-Gutierrez L, Yoshimura A, Rolas L, Garrido-Mesa J, Barkaway A, et al. Autophagy modulates endothelial junctions to restrain neutrophil diapedesis during inflammation. Immunity (2021) 54(9):1989–2004.e9. doi: 10.1016/j.immuni.2021.07.012
133. Yao RQ, Ren C, Xia ZF, Yao YM. Organelle-specific autophagy in inflammatory diseases: A potential therapeutic target underlying the quality control of multiple organelles. Autophagy (2021) 17(2):385–401. doi: 10.1080/15548627.2020.1725377
134. Foerster EG, Mukherjee T, Cabral-Fernandes L, Rocha JDB, Girardin SE, Philpott DJ. How autophagy controls the intestinal epithelial barrier. Autophagy (2022) 18(1):86–103. doi: 10.1080/15548627.2021.1909406
135. Wang W, Liu Z, Yue W, Zhu L, Zhong H, Yang C, et al. Mucosa-colonizing microbiota correlate with host autophagy signaling in patients with inflammatory bowel disease. Front Microbiol (2022) 13:875238. doi: 10.3389/fmicb.2022.875238
136. Wei J, Long L, Yang K, Guy C, Shrestha S, Chen Z, et al. Autophagy enforces functional integrity of regulatory T cells by coupling environmental cues and metabolic homeostasis. Nat Immunol (2016) 17(3):277–85. doi: 10.1038/ni.3365
137. Ziegler PK, Bollrath J, Pallangyo CK, Matsutani T, Canli O, De Oliveira T, et al. Mitophagy in intestinal epithelial cells triggers adaptive immunity during tumorigenesis. Cell (2018) 174(1):88–101.e16. doi: 10.1016/j.cell.2018.05.028
138. Lucas C, Salesse L, Hoang MHT, Bonnet M, Sauvanet P, Larabi A, et al. Autophagy of intestinal epithelial cells inhibits colorectal carcinogenesis induced by colibactin-producing escherichia coli in Apc(Min/+) mice. Gastroenterology (2020) 158(5):1373–88. doi: 10.1053/j.gastro.2019.12.026
139. Liu X, Chen J, Long X, Lan J, Liu X, Zhou M, et al. RSL1D1 promotes the progression of colorectal cancer through RAN-mediated autophagy suppression. Cell Death Dis (2022) 13(1):43. doi: 10.1038/s41419-021-04492-z
140. Zhao Y, Guo Q, Zhao K, Zhou Y, Li W, Pan C, et al. Small molecule GL-V9 protects against colitis-associated colorectal cancer by limiting NLRP3 inflammasome through autophagy. Oncoimmunology (2017) 7(1):e1375640. doi: 10.1080/2162402X.2017.1375640
141. Shao BZ, Yao Y, Li JP, Chai NL, Linghu EQ. The role of neutrophil extracellular traps in cancer. Front Oncol (2021) 11:714357. doi: 10.3389/fonc.2021.714357
Keywords: autophagy, gastrointestinal cancer, esophageal cancer, gastric cancer, colorectal cancer
Citation: Shao B-Z, Chai N-L, Yao Y, Li J-P, Law HKW and Linghu E-Q (2022) Autophagy in gastrointestinal cancers. Front. Oncol. 12:975758. doi: 10.3389/fonc.2022.975758
Received: 22 June 2022; Accepted: 11 August 2022;
Published: 26 August 2022.
Edited by:
Kevin Ni, St George Hospital Cancer Care Centre, AustraliaReviewed by:
Sailong Zhang, Second Military Medical University, ChinaCopyright © 2022 Shao, Chai, Yao, Li, Law and Linghu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: En-Qiang Linghu, linghuenqiang@vip.sina.com; Helen Ka Wai Law, hthelen@polyu.edu.hk; Bo-Zong Shao, shaobozong@126.com
†These authors have contributed equally to this work
 Bo-Zong Shao
Bo-Zong Shao Ning-Li Chai1†
Ning-Li Chai1† Yi Yao
Yi Yao Helen Ka Wai Law
Helen Ka Wai Law En-Qiang Linghu
En-Qiang Linghu