Corrigendum: Long non-coding RNA-TMPO-AS1 as ceRNA binding to let-7c-5p upregulates STRIP2 expression and predicts poor prognosis in lung adenocarcinoma
- 1Department of Thoracic Surgery, The Third Affiliated Hospital of Kunming Medical University, Kunming, China
- 2Laboratory of Animal Models and Human Disease Mechanisms of Chinese Academy of Sciences & Yunnan Province, Kunming Institute of Zoology, Kunming, China
- 3Department of Biology, Chemistry, Pharmacy, Free University of Berlin, Berlin, Germany
Background: Striatin-interacting protein 2 (STRIP2), also called Fam40b, has been reported to regulate tumor cell growth. But the role of STRIP2 in lung adenocarcinoma (LUAD) has not been discovered clearly. Thus, the aim of our study is to explore the function and underlying mechanism of STRIP2 in LUAD.
Methods: Expression of STRIP2 was determined using the Cancer Genome Atlas (TCGA), GTEx, Ualcan, and the Human Protein Altas databases. The Correlation of STRIP2 and survival was detected by PrognoScan and Kaplan–Meier plotter databases. Besides, the correlation between STRIP2 expression and tumor immune infiltration as well as immune checkpoints were analyzed by the ssGSEA method. The biological function of STRIP2 and its co-expression genes was determined by gene ontology (GO) and Genes and Genomes (KEGG), respectively. Finally, the expression level and biological function of STRIP2 in LUAD were determined by qPCR, CCK8, transwell, and wound healing assays.
Results: This manuscript revealed a significantly increased expression of mRNA and protein of STRIP2 in lung adenocarcinoma compared with the adjacent normal tissues. GEO and Kaplan–Meier plotter databases showed higher STRIP2 expression levels were correlated with poor prognosis survival of LUAD. Moreover, Cox regression analysis suggested that a higher STRIP2 level served as an independent risk factor in predicting deteriorative overall survival (OS) for LUAD patients. SsGSEA results showed STRIP2 expression level was positively correlated with infiltrating levels of Th2 cells in LUAD. Lastly, GO analysis indicated the biological processes were enriched in nuclear division and positive regulation of the cell cycle. KEGG signaling pathway analysis showed STRIP2 was correlated with the MAPK signaling pathway and the TNF signaling pathway. The GSEA database showed that STRIP2 was positively associated with the epithelial–mesenchymal transition, cell cycle, and TNF signaling pathway. The QRT-PCR assay showed that STRIP2 was upregulated in LUAD cell lines. Cell proliferation and migration were inhibited in LUAD by knockdown of STRIP2. Moreover, we confirmed that the TMPO-AS1/let-7c-5p/STRIP2 network regulates STRIP2 overexpression in LUAD and is associated with poor prognosis.
Conclusion: Our findings indicated that STRIP2 acted as a crucial oncogene in LUAD and was correlated with unfavorable survival and tumor infiltration inflation.
Introduction
Lung cancer is a worldwide epidemic malignancy among both women and men, and terribly causes globally cancer-related deaths (1). In China, the incidence and deaths of lung cancer have increased rapidly in recent years, revealing geographic and gender differences (2). Lung adenocarcinoma (LUAD) accounts for approximately more than 40% of the lung cancer incidence and poses a systemic threat due to more frequently occurring distant metastasis in LUAD (3). LUAD, with an overall survival of less than 5 years, is the most aggressive and rapid metastasis (4). For the past few years, immunotherapy has been highlighted as very effective in lung cancer therapy; however, it also has previously failed in lung cancer (5, 6). Therefore, the terrible therapeutic results of immunotherapy are the new obstacles for clinical application of immune checkpoint inhibitors in LUAD.
STRIP2 is a member of the striatin-interacting phosphatase and kinase (STRIPAK) complex that participates in regulating cell growth and migration (7). Increased STRIP2 has been observed in many cancers and is correlated with poor prognosis and unfavorable clinicopathological characteristics in gastric cancer and breast cancer cells (8, 9). High expression of STRIP2 implicates neoplastic growth, metastasis, chemoresistance, and shorter survival time in various human cancers (9). Increasing evidence has indicated that STRIP2 exerts its role as an oncogene by regulating various signaling pathways to regulate tumorigenesis and progression and counteract the effects of many chemotherapies. For instance, STRIP2 is involved in the P38-AKT-MMP-2 signaling pathway to regulate mouse aortic smooth muscle cell (MOVAS) proliferation and migration (10). There is little but strong evidence to demonstrate the connection between STRIP2 and immune cell infiltration in cancer. Despite STRIP2 serving as an oncogene in LUAD, no evidence demonstrates the association between STRIP2 and immune cell infiltration in LUAD.
Therefore, in this study, we detected the connection between STRIP2 and prognosis and immune infiltration in tumor samples of LUAD patients. Furthermore, GSEA and KEGG enrichment analyses were conducted to examine the potential signaling pathway of STRIP2 in LUAD progression. Finally, we performed a loss of function assay to determine the biological function of STRIP2 in LUAD. Our data highlight the crucial role of STRIP2 in tumor initiation, progression, clinical outcome, and immune infiltration in LUAD.
Materials and Methods
TCGA Datasets
We downloaded the RNA expression data and corresponding clinical information from the TCGA official website (https://portal.gdc.cancer.gov/). We used this data analysis to examine the correlation between STRIP2 expression and relevant clinical information, including pathological stage, andTNM stage.
UALCAN Database
UALCAN (http://ualcan.path.uab.edu/) is an online resource for the analysis of TCGA gene expression data (11). In this finding, we used UALCAN to examine the protein level of STRIP2 in LUAD.
The Human Protein Atlas (HPA)
HPA (https://proteinatlas.org/ contains information on normal tissue and tumor tissue protein levels of human gene expression profiles (12). In this study, we explored the protein expression of STRIP2 in lung cancer tissue.
The Clinical Proteomic Tumor Analysis Consortium (CPTAC) Common Data Analysis
The Clinical Proteomic Tumor Analysis Consortium (CPTAC) has produced large proteomics data sets from the mass spectrometric interrogation of tumor samples previously analyzed by The Cancer Genome Atlas (TCGA) program (13). In this study, we determined the expression of STRIP2 in lung cancer using the Clinical Proteomic Tumor Analysis Consortium (CPTAC) data.
AnnoLnc2 Database
AnnoLnc2 (http://annolnc.gao-lab.org/) is a one-stop portal to systematically annotate novel lncRNAs for humans and mice. lncRNAs with a comprehensive functional spectrum covering sequences, structure, expression, regulation, genetic association, and evolution (14). In this study, we used the AnnoLnc2 database to examine the subcellular localization and molecular coding potential of lncRNAs.
Gene Set Enrichment Analysis
In this study, we used the linkedomics database (http://www.linkedomics.org/login.php) to obtain the co-expression genes of STRIP2 in LUAD. We used the GSEA software and clusterProfiler package to perform KEGG enrichment analysis of the signaling pathway of STRIP2 in LUAD (15–17).
Cell Culture
The BEAS-2B cell line was purchased from the Cell Bank of theKunming Institute of Zoology and was cultured in BEGM media (Lonza, CC-3170). Lung cancer cell lines, namely, h1650, A549, SPC-A1, and H1975, were purchased from Cobioer, China with STR documents, and were cultured in RPMI-1640 medium (Corning) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin.
Cell Proliferation and Cell Migration Assay
The cell proliferation assay was performed as previously described (18). Indicated tumor cells were plated onto 12-well plates. The cell numbers were subsequently counted each day using an automatic cell analyzer countstar (Shanghai Ruiyu Biotech Co.). For the trans-well migration assay, 2 × 104 cells/well in 100 μl serum-free medium were plated in a 24-well plate chamber insert, and the lower chamber was filled with 10% FBS. After incubation for 24 h, cells were fixed with 4% PFA, washed and then stained with 0.5% crystal violet for further pictures to be captured.
Real-Time RT-PCR Assay
Using a real-time RT-PCR assay, cells were lysed by RNAiso Plus (Takara Bio, Beijing, China, Cat. 108-95-2). The primer used in this study is as follows: β-actin-F: AAGTGTGACGTGGACATCCGC, β-actin-R: CCGGACTCGTCATACTCCTGCT, STRIP2-F: AGGTGGTCAGTAGGGAACGG, and STRIP2-R: TGTAGCACATCGACCTCTGAA.
Statistical Analyses
The statistical analyses for Figures 1–4 were performed using R (V 3.6.3) and ROC curves to detect STRIP2 cutoff values using pROC packages. GraphPad Prism 7.0 was used for statistical analyses of the data regarding the biological function of STRIP2 (Figure 9). The significance of the data between two experimental groups was determined by Student’s t-test, and multiple group comparisons were analyzed by one-way ANOVA. P <0.05 (*), P <0.01 (**), and P <0.001 (***) were significant.
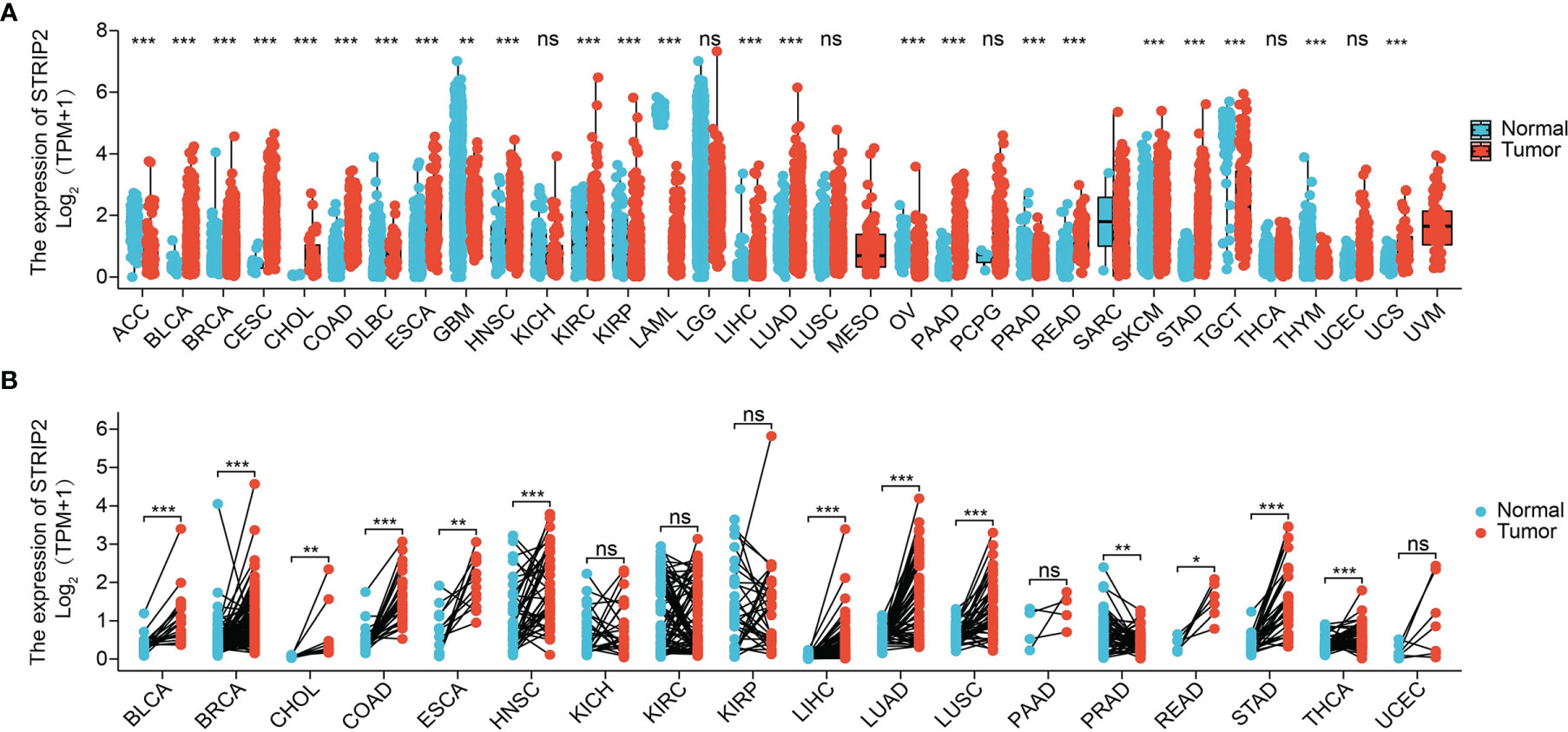
Figure 1 Expression pattern of STRIP2 from the perspective of pan-cancer. (A) STRIP2 was highly expressed in 24 of the 33 cancers compared with normal tissue. (B) The expression of STRIP2 in paired cancer tissues and adjacent normal tissues from the TCGA datasets. Cancer full name in Table 2. NS: P >0.05,*P <0.05, **P <0.01, ***P <0.001.
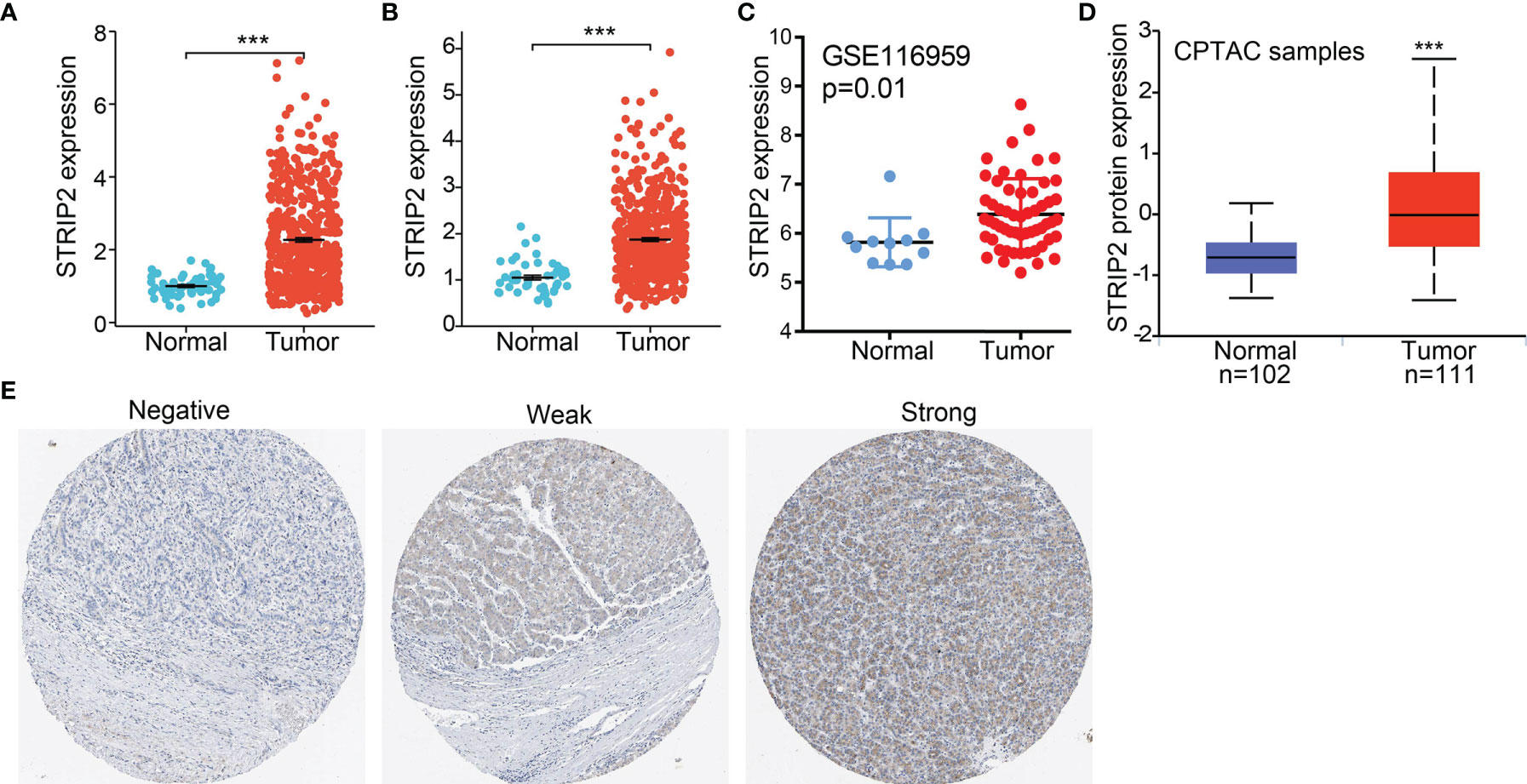
Figure 2 STRIP2 RNA and protein expression in LUAD. (A, B) STRIP2 mRNA expression levels in LUAD and LUSC patients and matched adjacent normal samples. (C) Validate the expression of STRIP2 in lung cancer. (D) STRIP2 protein expression level based on CPTAC. (E) STRIP2 protein levels based on Human Protein Atlas. ***P <0.001.
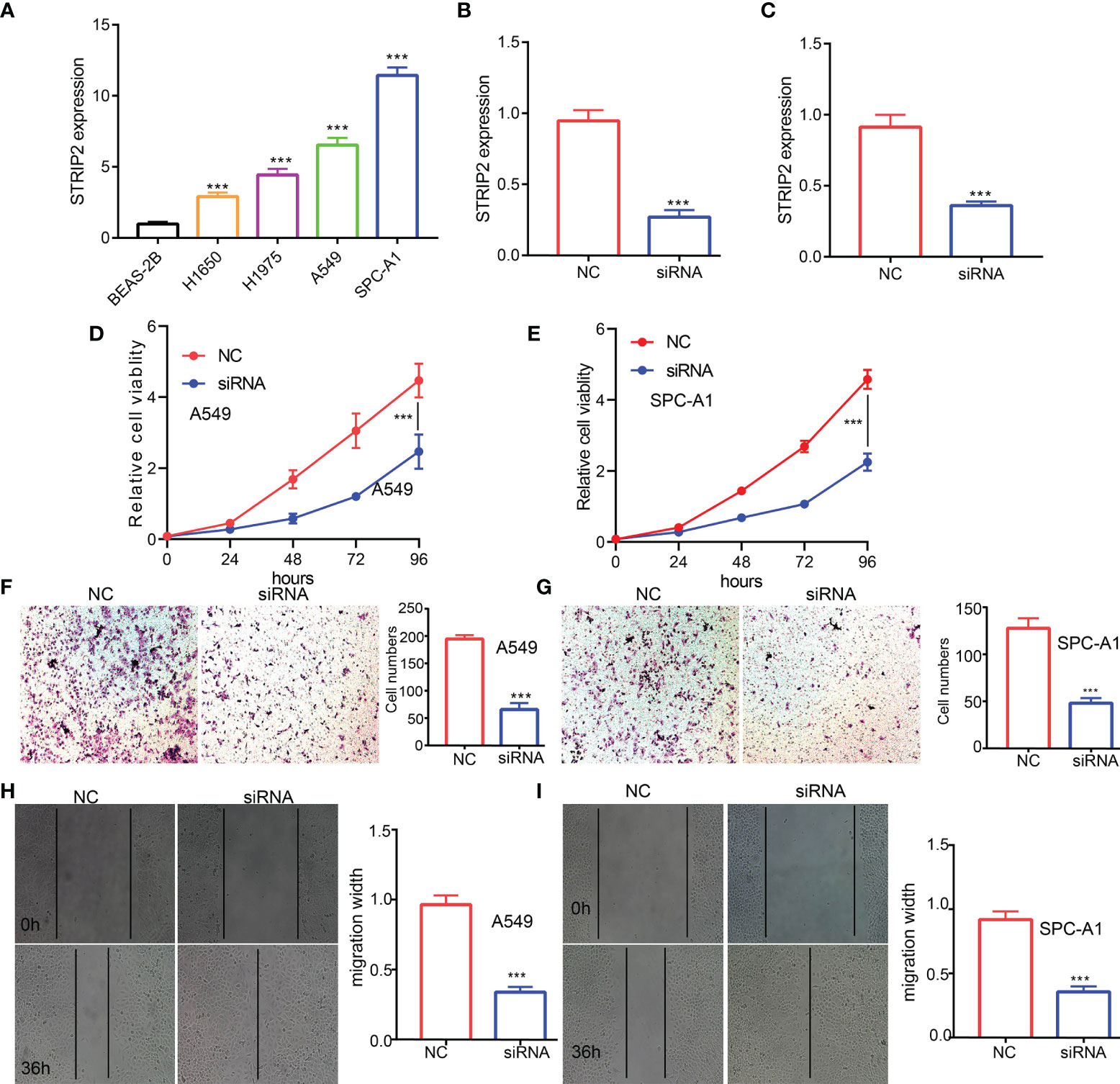
Figure 3 STRIP2 regulates LUAD cell proliferation and migration. (A) qPCR assay examines the expression level of STRIP2 in lung adenocarcinoma cancerous cell lines, namely, H1975, A549, and SPC-A1, compared to normal human bronchial epithelial cell line: BEAS-2B. (B, C) Establishment of STRIP2 knockdown cell lines in A549 and SPC-A1 verified by Real-time RT-PCR (D-I) Knockdown of STRIP2 significantly inhibits cell proliferation and migration in H1975 cells, as measured by CCK8, transwell, and Wound healing assays. NC, negative control; siRNA, STRIP2 siRNA. ***p <0.001.
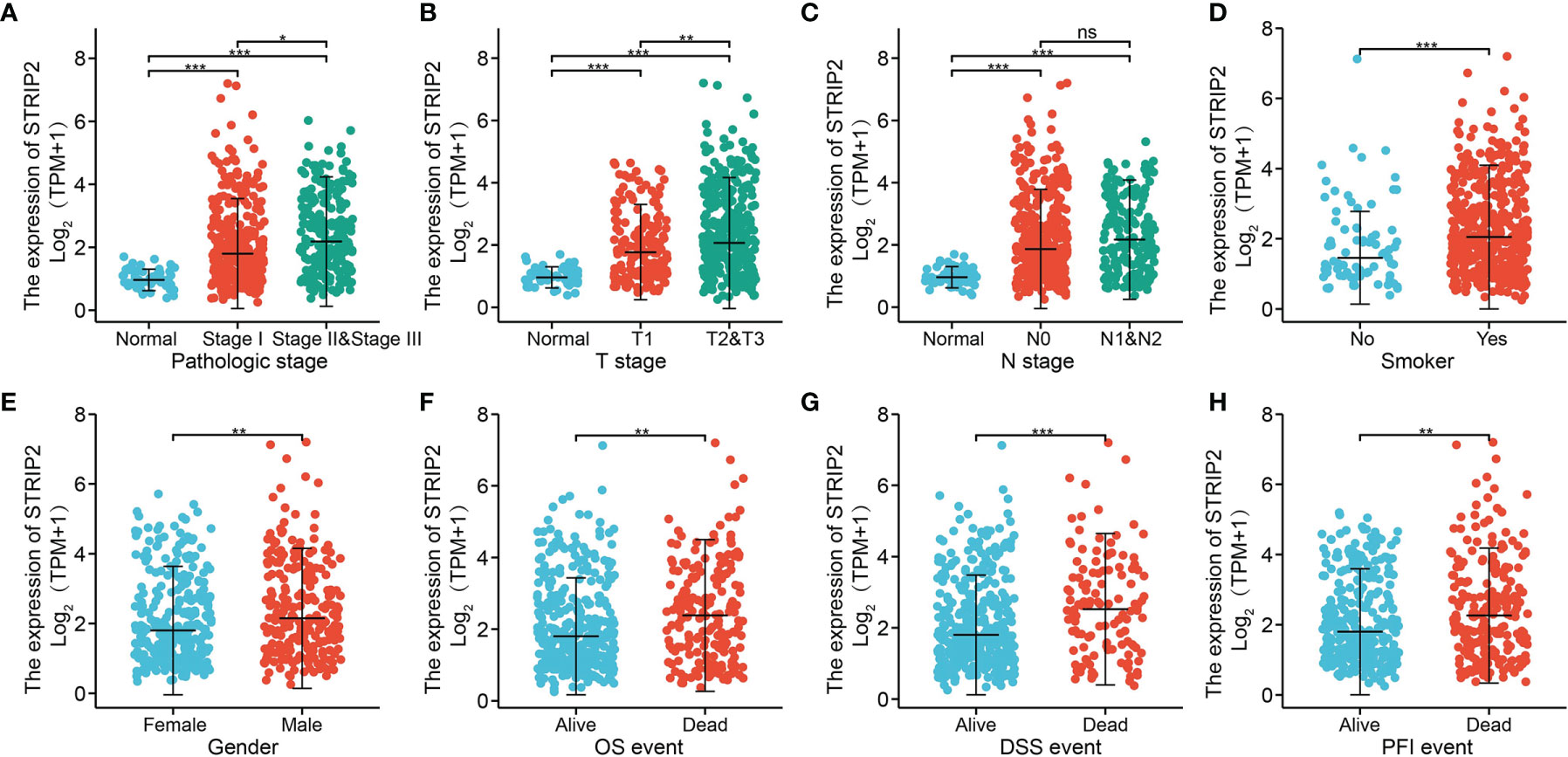
Figure 4 Clinical significance of STRIP2 in lung adenocarcinoma. Correlation between STRIP2 expression and clinical parameters, including, (A) pathological stage, (B–D) TNM stage, (E) gender, (F–H) OS, DSS, and PFS events. NS: P >0.05, *P <0.05, **P <0.01, ***P <0.001.
Results
Expression Level of STRIP2 in Human Cancers
STRIP2 expression was explored across tumor types in the TCGA database and GETx, followed by paired-difference analysis. Results show that STRIP2 was increased in 19 of the 33 cancers compared with normal tissue (Figure 1A). We also found that STRIP2 expression in paired cancer tissues and adjacent normal tissues in pan-cancer employed TCGA datasets. We found that STRIP2 levels were significantly higher in 12 of the 18 cancers compared with normal tissue (Figure 1B).
STRIP2 Was Upregulated in Lung Adenocarcinoma
To further determine STRIP2 mRNA and protein expression in LUAD, we analyzed STRIP2 expression data in TCGA and HPA. We found that STRIP2 was more highly expressed in LUAD and lung squamous cell carcinoma (LUSC) tumor tissues than in normal lung tissue (Figures 2A, B). Consistent with the results from the TCGA data, STRIP2 was significantly increased in lung tissue based on the Gene Expression Omnibus (GEO) dataset (Figure 2C). Moreover, we found that STRIP2 protein expression in LUAD was significantly higher than that in normal tissue (Figure 2D). As shown in Figure 2D, immunohistochemistry (IHC) results also confirmed that the upregulation of STRIP2 protein expression in lung cancer tissue compared to non-cancerous tissue (Figure 2E).
Depletion of STRIP2 Significantly Suppressed Proliferation and Migration of LUAD Cells
To examine the expression of STRIP2, we detected STRIP2 expression levels in LUAD cell lines using a qRT-PCR assay. Results confirmed that STRIP2 was significantly increased in lung cancer cell lines, especially in A549 and H1975 cells (Figure 3A). The qRT-PCR assay showed that the expression of STRIP2 mRNA was significantly decreased in H1975 cells after treatment with targeted siRNA (Figures 3B, C). The cholecystokinin octapeptide (CCK8), transwell, and wound healing assays demonstrated that STRIP2 depletion significantly inhibits the cell proliferation and cell migration ability of LUAD (Figures 3D–I). Collectively, these results demonstrate that STRIP2 was highly expressed in LUAD and significantly affected their proliferation and cell cycle.
Correlation Between STRIP2 Expression and Clinical Parameters
To examine the relationship between STRIP2 expression and clinical–pathological features in a LUAD sample. We found that STRIP2 expression was significantly associated with pathological stage, TN stage, smoking, gender, OS event, DSS event, and PFS event in patients with LUAD (Figures 4A–H). Moreover, logistics analysis results also show that upregulation of STRIP2 correlated with T stage (T2 & T3 & T4 vs. T1), N stage (N2 vs. N1), pathologic stage (Stage III & Stage IV vs. Stage I & Stage II), Gender (Male vs. Female), and smoker (Yes vs. No) (Table 1).
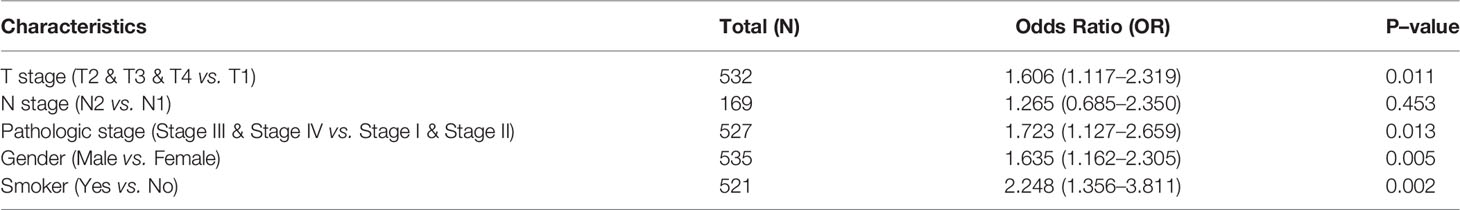
Table 1 Logistic regression analyzed the correlation between STRIP2 expression and clinical–pathological characteristics in LUAD.
STRIP2 Was Upregulated in LUAD and Predicts an Unfavorable Prognosis for LUAD Patients
Next, we analyzed the expression and clinical significance of STRIP2 in the TCGA database. We found that colorectal cancer (CRC) patients with higher STRIP2 levels had shorter overall survival, disease-specific survival, and progression survival (Figures 5A–C). We further examined the diagnostic value of STRIP2 in distinguishing LUAD samples from normal lung tissue. Receiver operating characteristic (ROC) curve analysis confirmed that the AUC value of STRIP2 was 0.828, with a 95% CI = 0.793–0.864 (Figure 5D). Then, we determined the prognostic values of STRIP2 in 4 independent GEO cohorts of lung cancer samples. Consistent with the results from the TCGA data, Kaplan–Meier analysis showed that patients with higher STRIP2 expression were associated with reduced overall survival time (Figures 6A–D). Collectively, these results suggest that STRIP2 was upregulated in lung cancer and that high expression levels of STRIP2 were associated with poor outcomes in lung cancer patients.

Figure 5 ROC and Kaplan–Meier curves of STRIP2. (A–C) Kaplan–Meier survival curves showed that lung adenocarcinoma patients with high STRIP2 expression exhibited poor overall survival, disease-specific survival, and progression-free survival of STRIP2 in LUAD determine by the TCGA-LUAD dataset. (D) ROC curves were used to determine the diagnostic value of STRIP2 in lung adenocarcinoma.
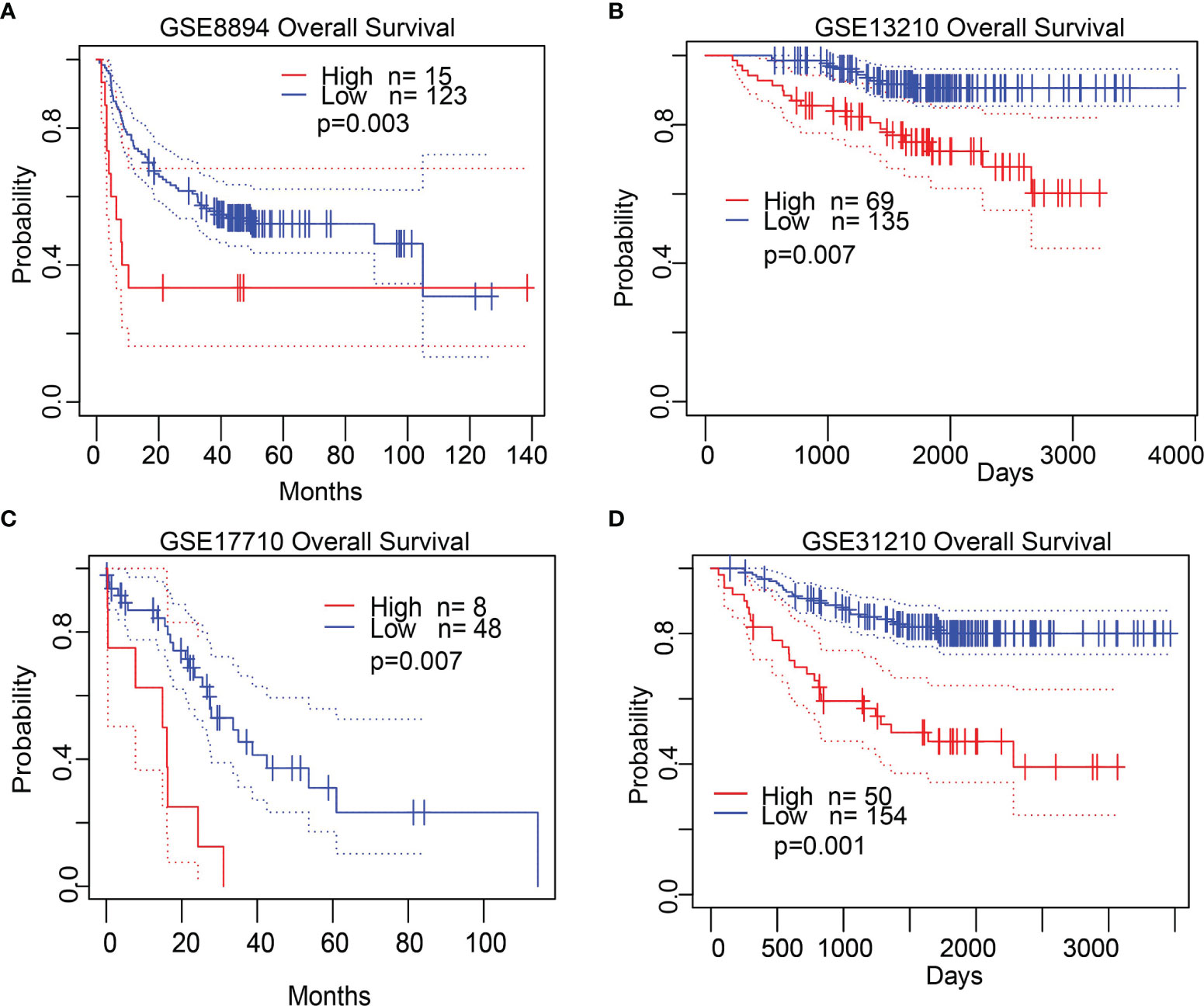
Figure 6 Validate the prognostic value of STRIP2 in lung cancer. (A–D) Kaplan–Meier survival curves showed that lung adenocarcinoma patients with high STRIP2 expression exhibited poor overall survival determine by the GEO dataset.
Validation of the Prognostic Value of STRIP2 Based on Various Subgroups
We further determined the prognostic values of STRIP2 in various clinical subgroups, namely, the pathological stage, tumor-node-metastasis (TNM) stage, gender, age, race, and smokers. Results suggest that upregulated STRIP2 levels are associated with poor clinical outcomes in patients with lung cancer (Figures 7A–C).
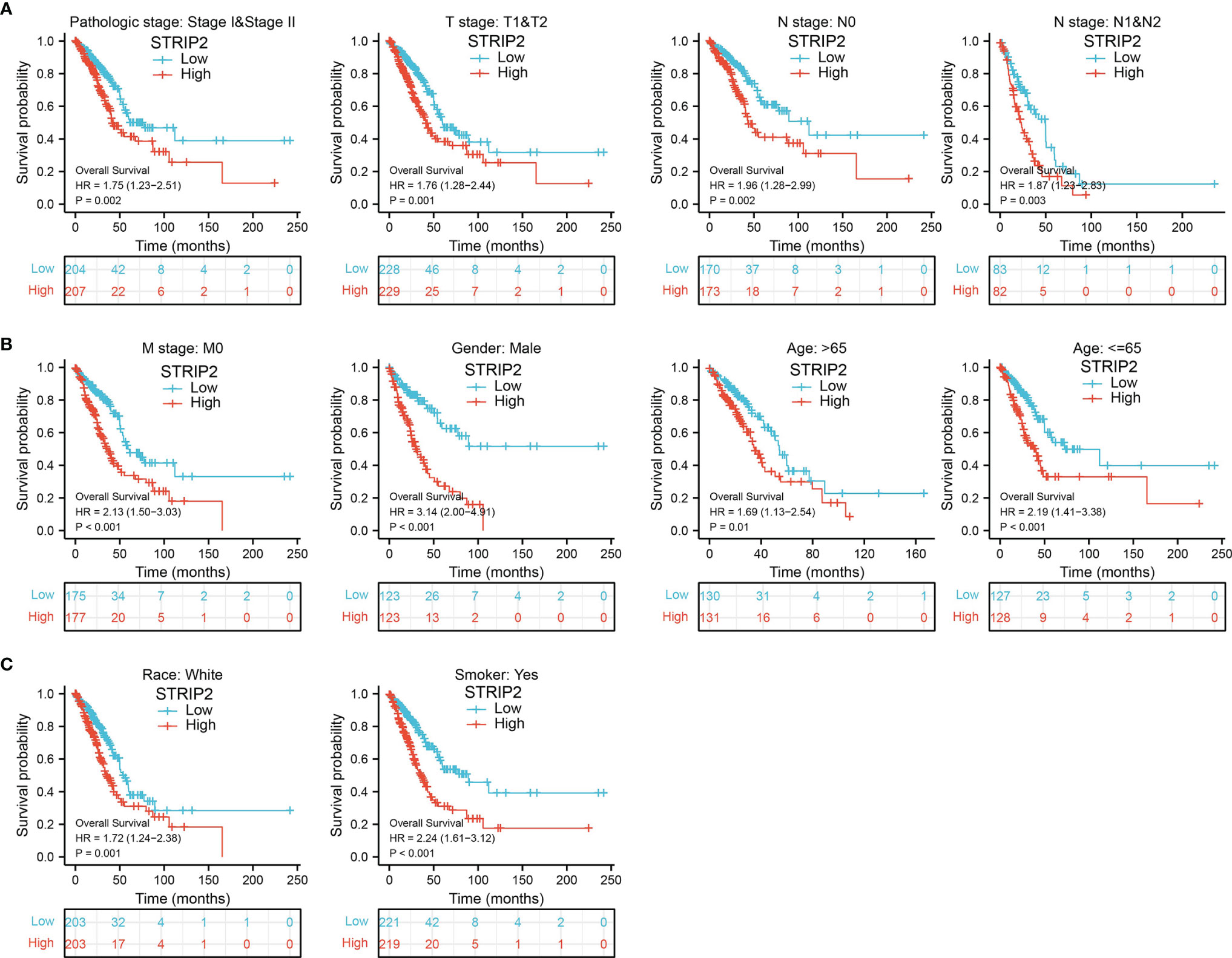
Figure 7 The overall survival of STRIP2 based on diverse subgroup. (A–C) Correlations between STRIP2 expression level and the overall survival in different clinical subgroups of LUAD in the TCGA database, including, stage I and II, T1 and T2, N0 and N1, M0, CR, Smoker, Gender, Age, and white.
Univariate and Multivariate Cox Regression Analyses of Different Parameters on Overall Survival
We performed Univariate Cox regression analysis in the TCGA-LAUD cohort to determine whether STRIP2 expression level might be a valuable prognostic biomarker. Univariate Cox regression analysis results show that high expression of STRIP2, pathologic stage, and TNM stage were associated with overall survival in LUAD patients (Figure 8A). Multivariate Cox regression analysis was performed to ascertain whether STRIP2 expression level could be an independent prognostic factor for patients with LUAD. We confirmed that increased STRIP2 expression was a significant independent prognostic factor in the TCGA-LAUD cohort that directly correlated with adverse clinical outcomes, along with pathological stage and N stage (Figure 8B).
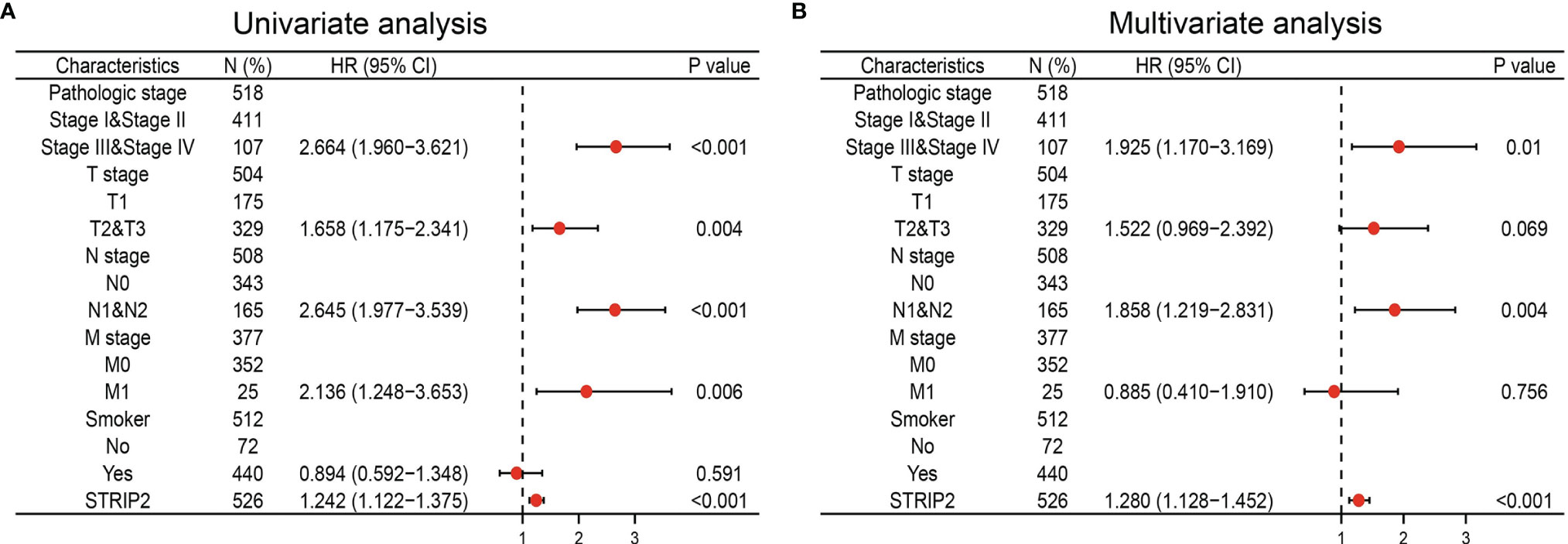
Figure 8 Forest plot of univariate and multivariate Cox regression analysis in LUAD. (A, B) The forest plot of univariate and multivariate Cox regression analysis in LUAD.
Construction and Validation of STRIP2 Based Nomogram
The results of the multivariate analysis confirmed that STRIP2 is an independent prognostic factor in LUAD. We then constructed a prediction model for overall survival, disease-free survival, and progression-free survival by integrating STRIP2 expression and pathological stage. We established a nomogram to integrate STRIP2 as a LUAD biomarker. Higher total points on the nomogram for overall survival, progression-free interval (PFI), and disease-specific survival, respectively, indicated a worse prognosis (Figures 9A–F).
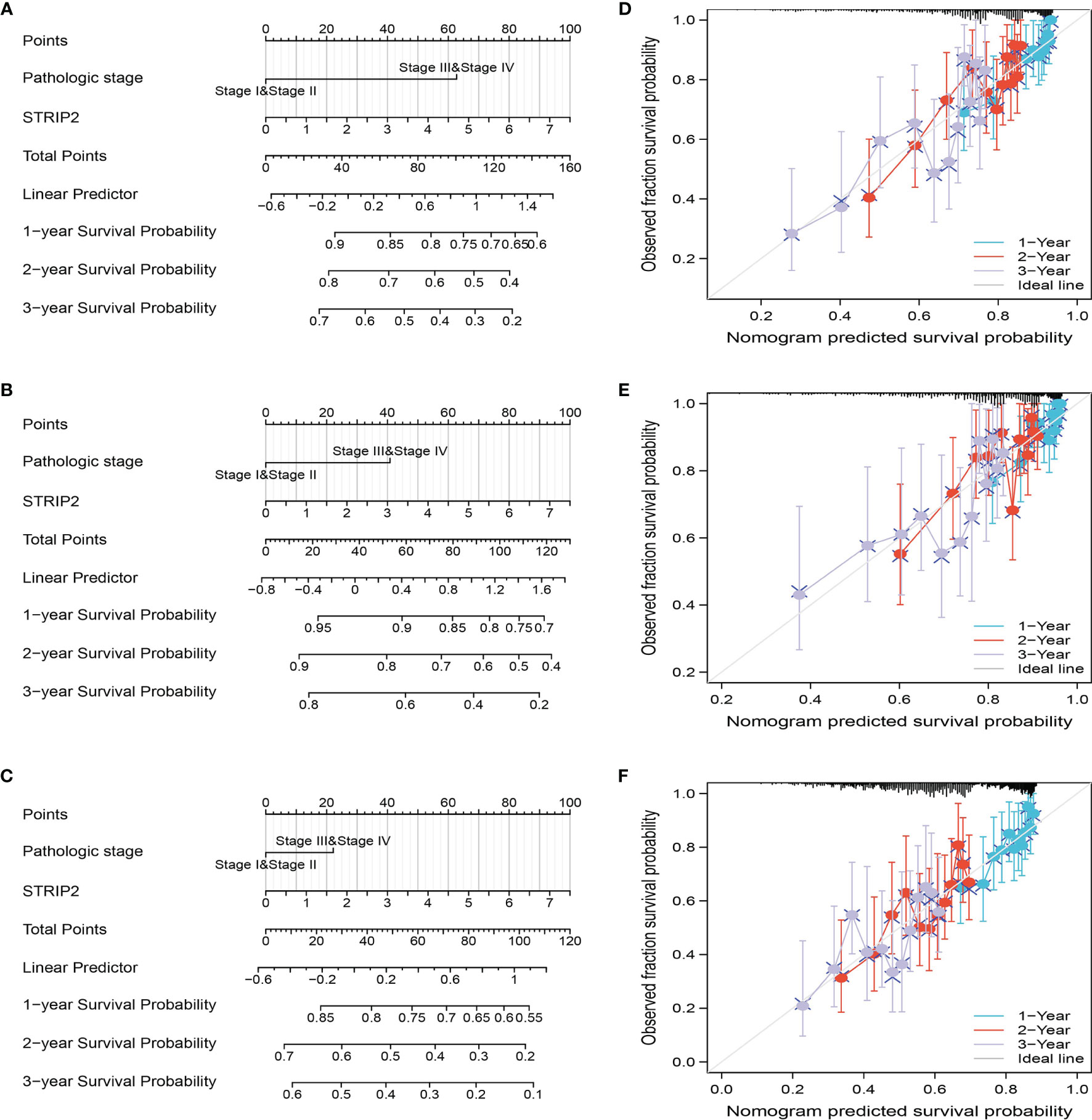
Figure 9 Construction and performance validation of the STRIP2 based nomogram for lung adenocarcinoma patients. (A–F) The calibration curve and Hosmer–Lemeshow test of nomograms in the TCGA-lung adenocarcinoma cohort for overall survival, disease-specific survival and progression-free survival.
KEGG and GSEA Enrichment Analysis
As shown in Figures 10A, B, the Linkedomic database is used to obtain the top 100 co-expressed genes by Pearson’s correlation analysis. In terms of biological processes, STRIP2 is mainly involved in organelle fission, nuclear division, positive regulation of the cell cycle, DNA replication, positive regulation of the cell cycle process, regulation of nuclear division, and positive regulation of the mitotic cell cycle (Figure 10C). KEGG enrichment analysis suggested that these genes participated in the MAPK signaling pathway, cell cycle, TNF signaling pathway, GnRH signaling pathway, and DNA replication (Figure 10D).
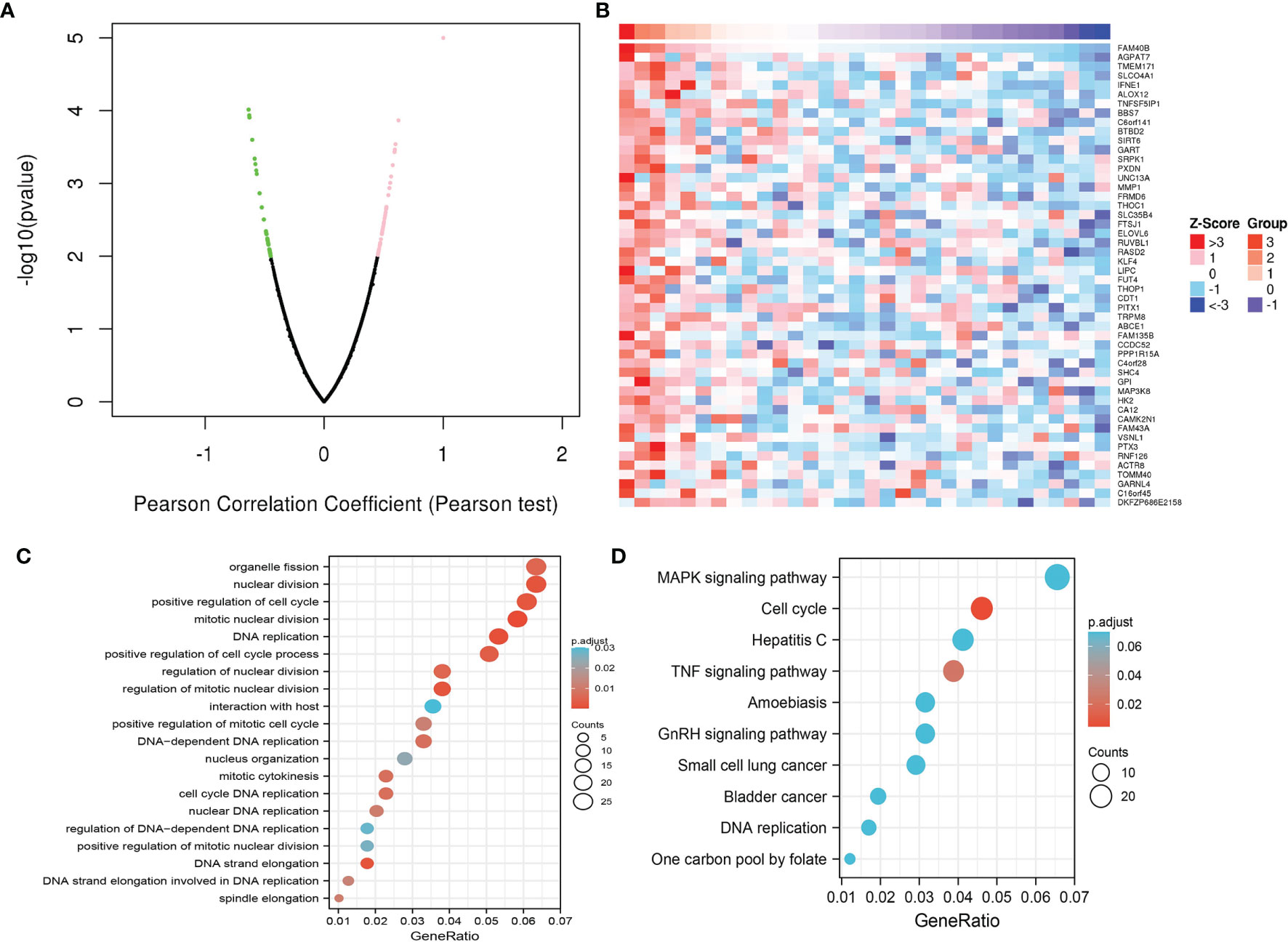
Figure 10 Functional enrichment analysis. (A, B) The correlation analysis of STRIP2 expression and its top 100 co-expressed gene network. (C, D) GO and KEGG enrichment analysis of co-expressed genes.
To explore the possible mechanism of STRIP2 in LUAD, GSEA analysis was performed on the differential genes. Gene set enrichment analysis (GSEA) also showed that pathways, namely, the MTOR signaling pathway, P53 signaling pathway, PI3K AKT MTOR signaling pathway, TNFA signaling pathway, DNA replication, apoptosis, IL2 STAT5 signaling pathway, MYC targets, hypoxia, GLYCOSIS, G2M Checkpoint, and EMT were significantly enriched in the high STRIP2 expression group (Figures 11A–C).
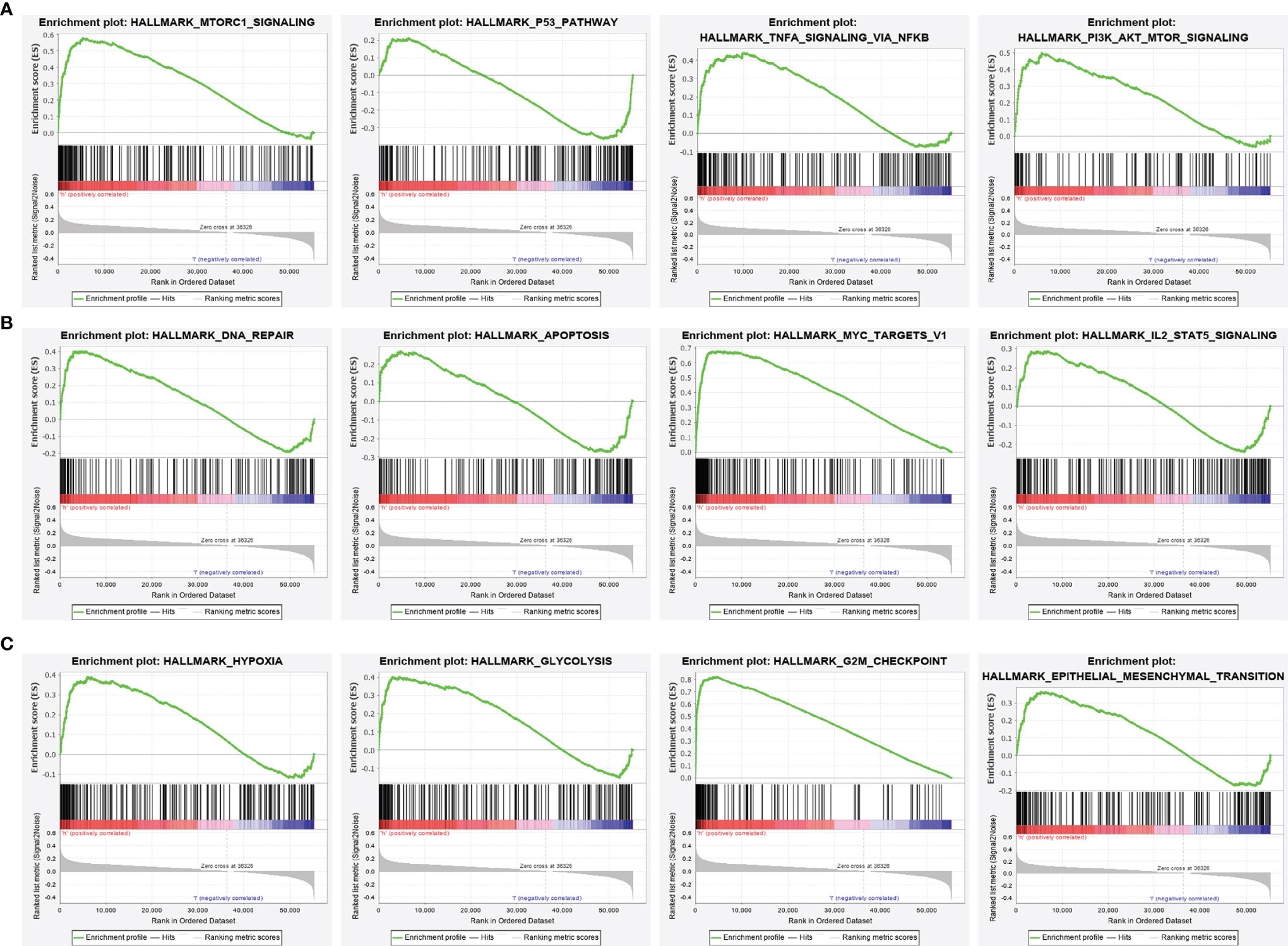
Figure 11 Identification of STRIP2 related signaling pathways in lung adenocarcinoma. (A–C) Identification of STRIP2 related signaling pathways by GSEA software.
Correlation Between STRIP2 Expression and Immune Infiltration
Given that the gene set enrichment analysis (GSEA) enrichment analysis above showed that STRIP2 may be correlated with the immune response regulation, we therefore examined the association between STRIP2 expression levels and immune cell infiltration, we used the ssGSEA algorithm to quantify the level of immune cell infiltration in the high- and low-expression groups of STRIP2. We determined that increased expression of STRIP2 was positively associated with the abundance of Th2 cells, NK CD56dim cells, Tgd, Tem, and neutrophils, and negatively associated with the abundance of macrophages, B cells, NK cells, DC, eosinophils, iDC, TFH, and mast cells in LUAD (Figures 12A–D).
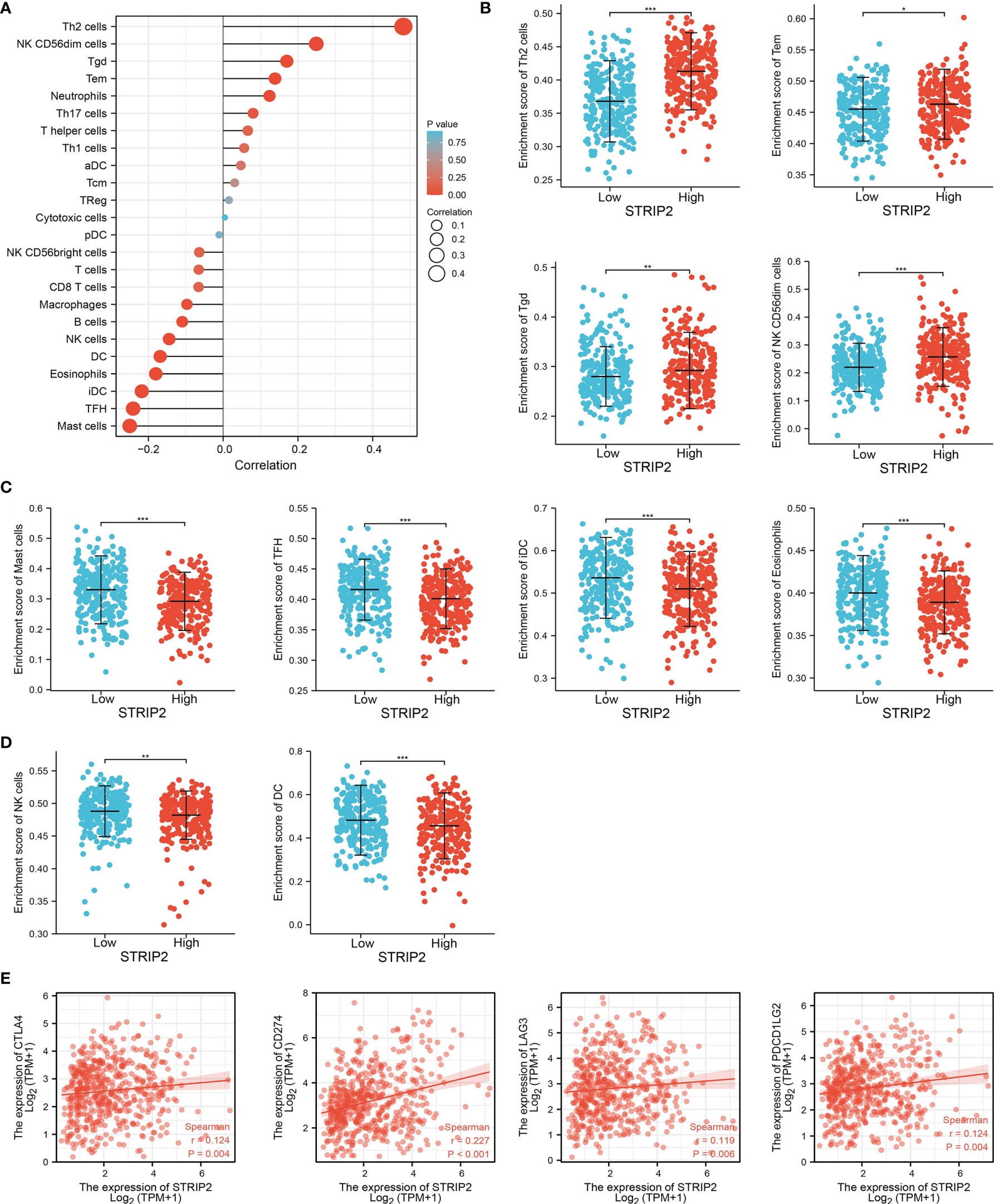
Figure 12 Correlation analysis of STRIP2 expression and infiltration levels of immune cells in LUAD. (A–D) The correlation between STRIP2 expression and the infiltration levels of 24 immune cells in LUAD by ssGSEA. (E) Correlation between STRIP2 and immune check points genes in LUAD. NS: P >0.05,*P <0.05, **P <0.01, ***P <0.001.
Given that immune checkpoints play a crucial role in tumor immunosuppression, we analyzed the correlation between STRIP2 expression and that of the immune checkpoint-related genes in LUAD using Pearson’s correlation analysis. The results confirmed that STRIP2 expression was significantly positively correlated with the expression of CD274, CTLA4, LAG3, and PDCD1LG2 in LUAD (Figure 12E). These results confirm that STRIP2 plays a crucial role in immune infiltration in LUAD.
Construction of the lncRNA–miRNA–mRNA Triple Regulatory Networks
To examine the lncRNA–miRNA–mRNA networks that modulate the expression of STRIP2 in LUAD, the Starbase database employed to obtain the potential miRNAs that bind with STRIP2, we obtained the 45 STRIP2-related miRNAs to construct a miRNA–mRNA co-expression network (Supplementary Table 1). In the miRNA–mRNA correlation analysis, we found a correlation between let-7c-5p expression and STRIP2 expression in LUAD (cor<−0.438, P-value <0.001; Figure 13A), and we found that let-7c-5p expression was significantly reduced in LUAD than in normal tissues and correlated with poor clinical outcomes (Figures 13B, C). ROC curve analysis of let-7c-5p showed an AUC value of 0.928 in lung cancer patients (Figure 13D). We also used the Starbase database to identify 62 potential target lncRNAs correlated with let-7c-5p in LUAD (Supplementary Table 2 and Figure 13E). Among them, TMPO-AS1, IER3-AS1, and AL360270.2 met the filtering conditions for correlation analysis with let-7c-5p (cor<−0.1, P-value <0.01), and only TMPO-AS1 was positively correlated with STRIP2 (cor >0.3, P-value <0.001) (Figures 13F, G). We also found that only TMPO-AS1 was significantly up-regulated in LUAD than in normal tissues, and its higher expression was associated with adverse clinical outcomes in patients with LUAD (Figures 13H–J). ROC curve analysis of TMPO-AS1 showed an AUC value of 0.898 in lung cancer patients (Figure 13K). Cellular localization analysis confirmed that TMPO-AS1 had the highest percentage in the cytoplasm and could not be translated into coding-proteins (Figures 13L, M). Since STRIP2 is upregulated in LUAD and associated with a poor prognosis, the TMPO-AS1/let-7c-5p/STRIP2 regulatory network could be used as a biomarker for poor prognosis and a new target for treating LUAD.
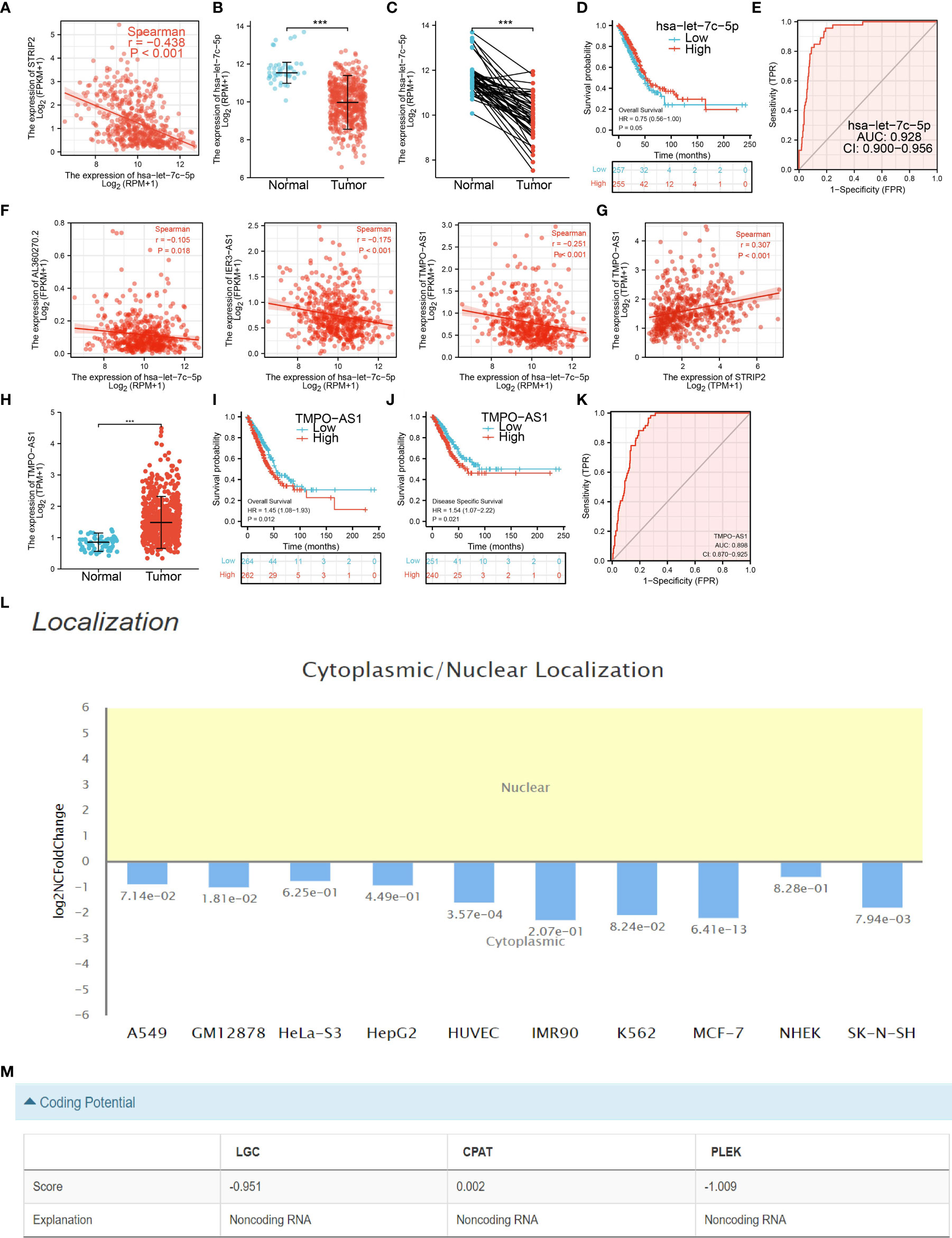
Figure 13 Analysis of the potential miRNAs and lncRNAs of STRIP2. (A) Correlations between STRIP2 expression and has-let-7c-5p in LUAD. (B–D) The expression level and prognostic value of has-let-7c-5p in LUAD. (E) ROC curve analyses and AUC values for has-let-7c-5p in lung cancer patients. (F) Correlations between has-let-7c-5p expression and lncRNAs (TMPO-AS1, IER3-AS1, and AL360270.2) in LUAD. (G) Correlations between STRIP2 expression and lncRNA-TMPO-AS1 in LUAD. (H–K) The expression levels, prognostic and diagnostic values of lncRNA-TMPO-AS1. (L, M) Subcellular localization and coding potential of lncRNA-TMPO-AS1 in LUAD, ***P <0.001.
Discussion
In this study, we found that STRIP2 expression was significantly higher in the LUAD than in the normal tissue at both transcriptional and protein levels. It was also upregulated in LUAD cell lines. The Kaplan–Meier curves and univariate analysis confirmed that STRIP2 expression is correlated with overall survival (OS), disease-specific survival (DSS), and progression-free survival (PFS) in the LUAD patients of the TCGA data. The GEO dataset also confirmed that patients with a high level of STRIP2 correlated with adverse clinical outcomes. Moreover, Cox regression analysis suggested that a higher STRIP2 level served as an independent risk factor in predicting deteriorative OS for LUAD patients. ROC curve analysis indicated that STRIP2 is a promising diagnostic biomarker for differentiating LUAD from normal tissues. We also established a nomogram to integrate STRIP2 as a LUAD biomarker; higher total points on the nomogram for overall survival, progression-free interval (PFS), and disease-specific survival, respectively, indicated a worse prognosis.
Previous studies have reported that STRIP2 is necessary for the onset of embryonic stem cell differentiation (19). Morpholino-mediated knockdown of STRIP2 results in severe abnormalities of the cardiovascular system (20). To further examine the functional role of STRIP2 in LUAD, we conducted an enrichment analysis between high- and low-expression groups based on STRIP2 mRNA expression. KEGG enrichment analysis suggested that these genes participated in the MAPK signaling pathway, cell cycle, TNF signaling pathway, GnRH signaling pathway, and DNA replication. Likewise, results of the GSEA analysis revealed that upregulated STRIP2 expression was associated with MTOR signaling pathway, P53 signaling pathway, PI3K AKT the MTOR signaling pathway, TNFA signaling pathway, DNA replication, apoptosis, IL2 STAT5 signaling pathway, MYC targets, hypoxia, GLYCOSIS, G2M Checkpoint, and EMT.
A previous study found that STRIP2 expression predicts prognosis in gastric cancer (8). In this finding, we found that STRIP2 was positively associated with the abundance of Th2 cells, NK CD56dim cells, Tgd, Tem, and neutrophils, and negatively associated with the abundance of macrophages, B cells, NK cells, DC, eosinophils, iDC, TFH, and mast cells in LUAD. Moreover, STRIP2 expression was significantly positively correlated with the expression of CD274, CTLA4, LAG3, and PDCD1LG2 in LUAD. These results confirm that STRIP2 plays a crucial role in immune infiltration in LUAD.
Given that GSEA enrichment results shown that STRIP2 may plays a central role in cell proliferation and EMT. We decide to examine the potential biological function of STRIP2 in LUAD. In vitro, we found that STRIP2 was increased in LUAD cells lines. Owing to A549 and SPCA1 has higher level of STRIP2. Therefore, we selected A549 and SPCA1 cells conducted assays. We found that depletion of STRIP2 in H1975 cells inhibited cell proliferation and migration. Based on above findings, we proposed that STRIP2 exerts an essential function in regulating pathologic progression of LUAD.
The crucial finding of this manuscript was to identify a prognosis-related ceRNA regulatory network (TMPO-AS1/let-7c-5p/STRIP2) in LUAD. In this ceRNA regulatory network, let-7c-5p was significantly and negatively correlated with STRIP2 expression, and TMPO-AS1 was significantly and negatively correlated with let-7c-5p expression, and significantly and positively correlated with STRIP2 expression. Furthermore, TMPO-AS1 and STRIP2 were significantly increased in LUAD tissues compared to normal tissues, and survival analysis results suggested that the higher expression group had a poorer prognosis compared to the low expression group, whereas let-7c-5p exhibited lower expression in LUAD tissues compared to normal tissues, and survival analysis revealed that the lower expression group had a poorer prognosis compared to the higher expression group. These results consistently indicate that TMPO-AS1/let-7c-5p/STRIP2 is a poor prognosis-associated ceRNA regulatory network in LUAD.
This study improves our understanding of the correlation between STRIP2 and LUAD, but some limitations still exist. First, although we explored the correlation between STRIP2 and immune infiltration in LUAD patients, there is a lack of experiments to validate the function of STRIP2 in the tumor microenvironment regulation of LUAD. Second, we found that knockdown of STRIP2 inhibits cell proliferation and cell migration of LUAD. However, the potential molecular mechanisms of STRIP2 in cancer progression need to be explored in further studies.
Conclusion
This finding describes, for the first time, the clinical relevance, immuno-oncology features and biological function of the TMPO-AS1/let-7c-5p/STRIP2 network, which upregulates STRIP2 expression in LUAD and is associated with adverse clinical outcomes. In summary, STRIP2 is a promising prognostic factor, and its future application may help determine the optimal treatment strategy for lung adenocarcinoma.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author Contributions
JW, YY, and LT designed this work and performed related assay. HZ and DZ analyzed the data. CL, LD, and XJ supervised and wrote the manuscript. All authors listed have made a substantial, direct, and intellectual contribution to the work and approved it for publication.
Funding
This work was supported by National Nature Science Foundation of China (82160508) and Yunnan Applied Basic Research Projects (YNWRMY-2019-067, 2019FE001) and Yunnan Province Specialized Training Grant for High-Level Healthcare Professionals (D-201614).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge support from the National Nature Science Foundation of China and the Department of Thoracic Surgery, The Third Affiliated Hospital of Kunming Medical University (Yunnan Tumor Hospital), Kunming, China.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.921200/full#supplementary-material
References
1. Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2019. CA Cancer J Clin (2019) 69(1):7–34. doi: 10.3322/caac.21551
2. Cao M, Chen W. Epidemiology of Lung Cancer in China. Thorac Cancer (2019) 10(1):3–7. doi: 10.1111/1759-7714.12916
3. Shi J, Hua X, Zhu B, Ravichandran S, Wang M, Nguyen C, et al. Somatic Genomics and Clinical Features of Lung Adenocarcinoma: A Retrospective Study. PloS Med (2016) 13(12):e1002162. doi: 10.1371/journal.pmed.1002162
4. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell Death-Based Treatment of Lung Adenocarcinoma. Cell Death Dis (2018) 9(2):117. doi: 10.1038/s41419-017-0063-y
5. Tsiouprou I, Zaharias A, Spyratos D. The Role of Immunotherapy in Extensive Stage Small-Cell Lung Cancer: A Review of the Literature. Can Respir J (2019) p:6860432. doi: 10.1155/2019/6860432
6. Steven A, Fisher SA, Robinson BW. Immunotherapy for Lung Cancer. Respirology (2016) 21(5):821–33. doi: 10.1111/resp.12789
7. Bai SW, Herrera-Abreu MT, Rohn JL, Racine V, Tajadura V, Suryavanshi N, et al. Identification and Characterization of a Set of Conserved and New Regulators of Cytoskeletal Organization, Cell Morphology and Migration. BMC Biol (2011) 9:54. doi: 10.1186/1741-7007-9-54
8. Wu J, Lu G, Zhou S, Jin Z, Fang F. MicroRNA-30c-2-3p Targets STRIP2 to Suppress Malignant Progression of Gastric Cancer Cells. J Biochem (2022) 171(4):451–7. doi: 10.1093/jb/mvac006
9. Madsen CD, Hooper S, Tozluogku M, Bruckbauer A, Fletcher G, Erler JT, et al. STRIPAK Components Determine Mode of Cancer Cell Migration and Metastasis. Nat Cell Biol (2015) 17(1):68–80. doi: 10.1038/ncb3083
10. Dai L, Zhou J, Li T, Qian Y, Jin L, Zhu C, et al. STRIP2 Silencing Inhibits Vascular Smooth Muscle Cell Proliferation and Migration via P38-AKT-MMP-2 Signaling Pathway. J Cell Physiol (2019) 234(12):22463–76. doi: 10.1002/jcp.28810
11. Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, et al. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia (2017) 19(8):649–58. doi: 10.1016/j.neo.2017.05.002
12. Navani S. Manual Evaluation of Tissue Microarrays in a High-Throughput Research Project: The Contribution of Indian Surgical Pathology to the Human Protein Atlas (HPA) Project. Proteomics (2016) 16(8):1266–70. doi: 10.1002/pmic.201500409
13. Rudnick PA, Markey SP, Roth J, Mirokhin Y, Yan X, Tchekhovskoi DV, et al. A Description of the Clinical Proteomic Tumor Analysis Consortium (CPTAC) Common Data Analysis Pipeline. J Proteome Res (2016) 15(3):1023–32. doi: 10.1021/acs.jproteome.5b01091
14. Ke L, Yang DC, Wang Y, Ding Y, Gao D. AnnoLnc2: The One-Stop Portal to Systematically Annotate Novel lncRNAs for Human and Mouse. Nucleic Acids Res (2020) 48(W1):W230–w238. doi: 10.1093/nar/gkaa368
15. Yu G, Wang LG, Han Y, He QY. Clusterprofiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics (2012) 16(5):284–7. doi: 10.1089/omi.2011.0118
16. Subramanian A, Tamayo AP, Mootha VK, Mukherjee S, Ebery BL, Gillette MA. Gene Set Enrichment Analysis: A Knowledge-Based Approach for Interpreting Genome-Wide Expression Profiles. Proc Natl Acad Sci U S A (2005) 102(43):15545–50. doi: 10.1073/pnas.0506580102
17. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: Analyzing Multi-Omics Data Within and Across 32 Cancer Types. Nucleic Acids Res (2018) 46(D1):D956–d963. doi: 10.1093/nar/gkx1090
18. Jiang X, Yuan Y, Tang L, Wang J, Liu Q, Zou X, et al. Comprehensive Pan-Cancer Analysis of the Prognostic and Immunological Roles of the METTL3/lncRNA-SNHG1/miRNA-140-3p/UBE2C Axis. Front Cell Dev Biol (2021) 9:765772. doi: 10.3389/fcell.2021.765772
19. Sabour D, Srinivasan SP, Rohani S, Wagh V, Gaspar JA, Panek D, et al. STRIP2 Is Indispensable for the Onset of Embryonic Stem Cell Differentiation. Mol Ther Methods Clin Dev (2017) 5:116–29. doi: 10.1016/j.omtm.2017.04.001
Keywords: striatin-interacting protein 2, prognosis biomarkers, tumor immune infiltration, lung adenocarcinoma, cell proliferation
Citation: Wang J, Yuan Y, Tang L, Zhai H, Zhang D, Duan L, Jiang X and Li C (2022) Long Non-Coding RNA-TMPO-AS1 as ceRNA Binding to let-7c-5p Upregulates STRIP2 Expression and Predicts Poor Prognosis in Lung Adenocarcinoma. Front. Oncol. 12:921200. doi: 10.3389/fonc.2022.921200
Received: 15 April 2022; Accepted: 09 May 2022;
Published: 14 June 2022.
Edited by:
Luciano Mutti, Temple University, United StatesReviewed by:
Jianyun Dong, First Affiliated Hospital of Southern University of Science and Technology, ChinaZhibin Yao, Fudan University Shanghai Cancer Center, China
Copyright © 2022 Wang, Yuan, Tang, Zhai, Zhang, Duan, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Li, chen.li.scholar@gmail.com; Xiulin Jiang, jiangxiulin@mail.kiz.ac.cn; Lincan Duan, duanmumuhuosan@163.com
†These authors have contributed equally to this work
 Juan Wang
Juan Wang Yixiao Yuan
Yixiao Yuan Lin Tang
Lin Tang Haoqing Zhai2
Haoqing Zhai2 Lincan Duan
Lincan Duan Xiulin Jiang
Xiulin Jiang