- 1Department of Digestive Tumours, The Second Affiliated Hospital of Fujian Medical University, Quanzhou, China
- 2Centre of Neurological and Metabolic Research, The Second Affiliated Hospital of Fujian Medical University, Quanzhou, China
- 3Group of Neuroendocrinology, Garvan Institute of Medical Research, Sydney, NSW, Australia
Colorectal cancer (CRC) is the third most common malignancy in the world and one of the leading causes of cancer death; its incidence is still increasing in most countries. The early diagnostic accuracy of CRC is low, and the metastasis rate is high, resulting in a low survival rate of advanced patients. MicroRNAs (miRNAs) are a small class of noncoding RNAs that can inhibit mRNA translation and trigger mRNA degradation, and can affect a variety of cellular and molecular targets. Numerous studies have shown that miRNAs are related to tumour progression, immune system activity, anticancer drug resistance, and the tumour microenvironment. Dysregulation of miRNAs occurs in a variety of malignancies, including CRC. In this review, we summarize the recent research progress of miRNAs, their roles in tumour progression and metastasis, and their clinical value as potential biomarkers or therapeutic targets for CRC. Furthermore, we combined the roles of miRNAs in tumorigenesis and development with the therapeutic strategies of CRC patients, which will provide new ideas for the diagnosis and treatment of CRC.
Introduction
Colorectal cancer(CRC) is the third leading cause of cancer deaths in humans, with an overall incidence of approximately 5% and a 5-year survival rate of 40% to 60% (1). In 2018, there were approximately 1.8 million new CRC cases and 860,000 deaths. It is estimated that by 2040, the global CRC burden will increase by 72% to more than 3 million new cases, which will pose a serious threat to human health (2). Environmental and genetic factors play an important role in the pathogenesis of CRC (3). Dietary habits, smoking, low levels of physical activity, population ageing, and obesity are also factors that affect the pathogenesis of CRC (3). In recent years, although some progress has been made in the screening and treatment of CRC, the overall survival rate of patients with advanced stage disease remains low. Because the symptoms of CRC patients are not obvious in the early stage, and the prognosis is poor when it develops to the advanced stage, early detection and treatment are particularly important.
MicroRNAs are a group of single-stranded small noncoding RNAs of 21-23 nucleotides (nt) in length. They were first discovered and reported in 1993 (4), and an increasing number of studies have focused on the regulatory role of microRNAs since then. MicroRNAs play important roles in biological and pathological processes such as metabolism, apoptosis, differentiation, cell proliferation, cell cycle, invasion and metastasis, and are closely related to the occurrence and development of tumours. They regulate the expression of their target genes post transcriptionally and they may be involved in various physiological and pathological processes, including CRC metastasis, by affecting various factors in the human body (5).
Recent studies have shown that dysregulated microRNAs play an important role in the development and metastasis of CRC, and the abnormal expression of microRNAs may act as potential oncogenes or suppressors in the development of tumours. Disordered microRNAs may have carcinogenic or tumour suppressor functions, and can regulate some oncogenes and tumour suppressor genes. Similarly, they are also regulated by oncogenes and tumour suppressor genes (6). Studies have shown that alterations in the Wnt/β-catenin, EGFR, TGF β and TP53 signalling pathways can affect CRC survival, proliferation and metastasis, and specific miRNAs can lead to changes in these signalling pathways, thereby promoting or inhibiting tumorigenesis (7). The same microRNAs may act as a tumour promoter in one cancer and a tumour suppressor in another, so there is no need to study the role of the same microRNAs in different cancers. For example, miR-146a may have a carcinogenic effect in thyroid cancer and a tumour inhibitory effect in CRC (8).
As microRNAs could be used for the diagnosis and prognostic monitoring of CRC, their high tissue specificity and role in tumorigenesis make them novel biomarkers for diagnosing cancer and predicting patient outcomes (9). Meanwhile, due to the role of abnormal expression of microRNAs in tumour development and the therapeutic response, correcting miRNA deficiency or restoring miRNA function may be a new cancer treatment strategy.
In addition, the association of microRNAs with tumour angiogenesis, cell proliferation, metastasis, and apoptosis suggests that the related microRNAs may serve as potential targets for CRC therapy (10). This article reviews the roles of microRNAs in the occurrence, development and metastasis of CRC and provides new ideas for the diagnosis and treatment of CRC.
Colorectal cancer
CRC is one of the most common gastrointestinal malignancies, the incidence of CRC in young adults is rapidly increasing (11). Patient survival is closely related to tumour stage at diagnosis, with approximately 50% of patients dying from distant metastases (12). The diagnosis of CRC is generally based on the evaluation of symptoms or screening. However, because CRC has no obvious symptoms in the early stage, most tumours have already metastasized at the time of diagnosis.
The treatment of CRC includes primary tumour resection, radiotherapy, chemotherapy, targeted therapy, immunotherapy and so on. Despite advances in surgery and adjuvant therapy, cure rates and long-term survival have barely changed over the past few decades (13). Decreased chemotherapy sensitivity remains a major obstacle preventing effective treatment of advanced disease. The development of cancer resistance to chemotherapy also often leads to treatment failure. Although there are targeted therapies for CRC, there are still relatively few ways to improve survival (14). Therefore, we need to clarify the mechanism of tumour progression and find new therapeutic targets. CRC patients are still at risk of recurrence after surgical removal of the tumour. Routine surveillance of postoperative patients to detect recurrence during the early asymptomatic period is one of the ways to improve survival (15).
MicroRNAS
MicroRNAs are the most abundant small RNAs in animals and play a key role in the regulation of gene expression. They are involved in mRNA degradation by binding to the 3’-untranslated region (3’-UTR) and play important roles in cell differentiation, development, cell cycle regulation and apoptosis (6). It is estimated that microRNAs can regulate up to 30% of protein-coding genes in the human genome (16). Most microRNAs are detected in the cellular microenvironment, but circulating microRNAs or extracellular microRNAs can be detected in extracellular environments such as biological fluids. Circulating microRNAs exist as proteins or lipoprotein complexes in exosomes, microvesicles, apoptotic bodies, Argonaut protein complexes, and high-density lipoprotein complexes (17). These molecules are transported to recipient cells and regulate various physiological and pathological processes (18).
MicroRNAs are involved in the development and progression of cancer. Under specific conditions, microRNAs can act as both tumour promoters and tumour suppressors. Dysfunctional microRNAs can affect tumour progression, including maintaining proliferative signals, escaping growth inhibitors, resisting cell death, activating invasion and metastasis, and inducing angiogenesis (19). In recent years, an increasing number of studies have shown that microRNAs are not only potential biomarkers for CRC diagnosis and prognosis, but also potential therapeutic targets, and have broad application prospects in clinical diagnosis and treatment.
The role of microRNAs in tumour progression
Angiogenesis
Angiogenesis, the process of growing new blood vessels from venules of the existing capillaries, is an important step in tumour cell proliferation and metastasis (20). Studies have found that microRNAs can regulate all stages of angiogenesis (21). Approximately 33 different microRNA families have been reported to play a role in angiogenesis (22).
Zeng et al. (23) found that miR-25-3p secreted by CRC can be transferred to vascular endothelial cells through exosomes, destroy the integrity of the endothelial barrier, induce angiogenesis, and promote CRC metastasis. MTDH is a target gene of miR-375 in CRC. Han et al. (24) proved that the expression level of MTDH is negatively correlated with the expression of miR-375 in CRC. Inhibition of miR-375 expression in CRC can regulate cell proliferation and angiogenesis by increasing the expression of MTDH. Meanwhile, overexpression of miR-218 can significantly inhibit angiogenesis (25). In addition, miR-17~92 can inhibit CRC progression by inhibiting angio-genesis in tumours (26). Hu et al. (27) showed that exomiR-1229 has a positive effect on angiogenesis by activating the vascular endothelial growth factor (VEGF) pathway and may be a therapeutic target for inhibiting tumour angiogenesis. The recent findings of He et al. (28) revealed that miR-21-5p secreted by CRC cells is a key switch for cancer-induced angiogenesis and vascular permeability, and may also serve as a new target for cancer therapy. Moreover, hypoxia is closely related to angiogenesis. Targeting hypoxia-related microRNAs, such as miR-145, can inhibit CRC metastasis and may also help control tumour metastasis (29). In conclusion, the pathogenesis of cancer is related to the imbalance of angiogenesis, and miRNAs can regulate the related pathways of angiogenesis. Therefore, they are expected to become potential therapeutic targets for CRC.
Premetastatic niche formation
The primary tumour creates a favourable microenvironment for subsequent metastasis in the secondary organs and tissues, that is, the premetastatic niche. The premetastatic niche can increase angiogenesis and vascular permeability, thereby promoting metastasis (30). Therefore, analysis of the molecular and cellular components of the premetastatic niche in blood may contribute to the diagnosis and prognosis of cancer metastasis. The study by Shao et al. (31) showed that during the development of CRC, miR-21 secreted by primary CRC cells is phagocytosed by macrophages in the liver, thereby forming a premetastatic niche in the liver, and circulating CRC cells can settle there and survive. A recent study demonstrated that upregulated miR-135a-5p plays a key role in CRC liver metastasis by promoting the formation of a premetastatic niche through dual regulation of immunosuppression and cell adhesion (32). Furthermore, circulating tumour-derived exosomal miR-203 can promote distant metastasis by inducing host M2 macrophages to form a premetastatic niche (33). Exosomal miR-25-3p is also involved in the formation of the premetastatic niche and may serve as a blood-derived biomarker for CRC metastasis (23). These studies show that miRNAs can participate in the formation of the premetastatic niche and promote CRC metastasis. Quantitative blood detection of the level of relevant miRNAs in circulating exosomes may be helpful for the diagnosis of CRC metastasis and the preventive treatment of high-risk metastatic patients.
Cell proliferation and metastasis
Immortal proliferation of CRC cells is the basis of cancer development. MicroRNAs play an important role in the process of cell proliferation. Previous studies have shown that many microRNAs can affect the proliferation of CRC cells in different ways. For example, Huang et al. (34) found that upregulation of miR-17 could promote CRC proliferation. In contrast, miR-22 can inhibit the proliferation of CRC cells and slow the growth rate of tumours (35). In prostaglandin E2 (PGE2)-induced tumour cells, overexpression of miR-206 can reduce the proliferation of CRC cells (36), which may be a potential therapeutic target for PGE2-induced CRC cells. In addition, upregulation of miR-1258 and miR-500a-5p both inhibited tumour cell proliferation by blocking the cell cycle in G0/G1 (37, 38).
MicroRNAs can control multiple aspects of epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET) and support tumour progression and metastasis (39) (Figure 1). Exosomes from tumour cells can transfer miRNAs to normal cells, stimulating carcinogenesis and promoting metastasis (40). Exosomes promote EMT by targeting RASp21 protein activator 1 (RASA1) to deliver miR-NA-335-5p, thereby promoting CRC cell invasion and metastasis (41). In addition, miR-29b-3p can directly target progranulin (PGRN) to alter the downstream Wnt signalling pathway and promote EMT (42). The miR-496/RASSF6 axis can also promote EMT and CRC migration through Wnt signalling (43). The study by Wang et al. (44) showed that miR-25-3p, miR-130b-3p, and miR-425-5p can induce tumour cell proliferation and metastasis, and may be potential therapeutic targets for blocking CRC metastasis.
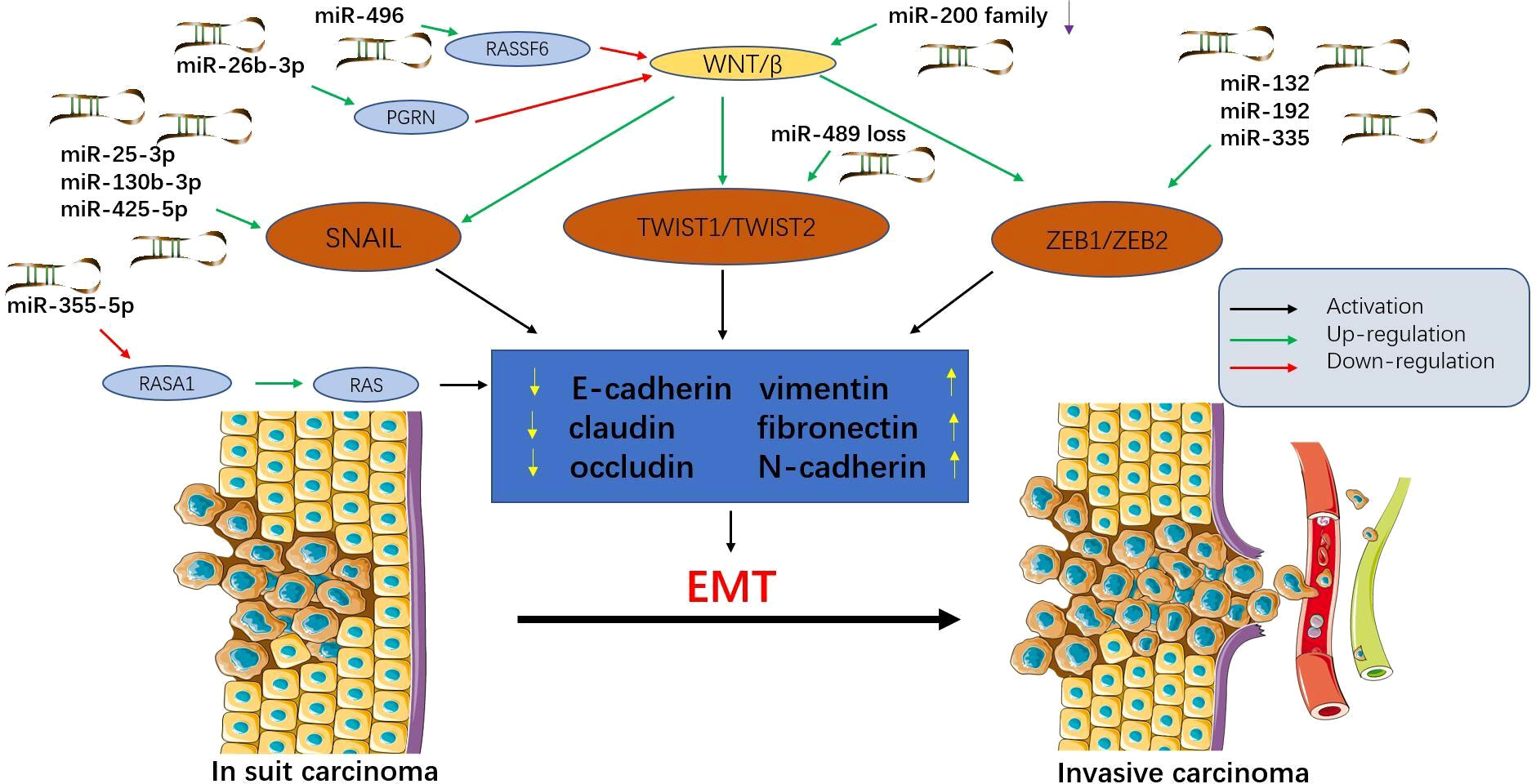
Figure 1 Epithelial-mesenchymal transition (EMT) is regulated by microRNAs in colorectal cancer (CRC). MicroRNAs affect multiple signalling pathways and participate in EMT by decreasing epithelial markers (e-cadherin, claudin, and occludin) and increasing interstitial markers (vimentin, fibronectin, and N-cadherin). During this process, epithelial cells acquire mesenchymal phenotypes, which play an important role in the progression and metastasis of CRC. Tumour cells that have undergone EMT can invade the local stroma and enter the vasculature, travel in the circulation, and finally establish a secondary tumour at a distant site. This figure summarizes some microRNAs involved in the EMT process in CRC.
In addition to cancer-promoting microRNAs, there are also cancer-suppressing microRNAs. Upregulation of miR-200c can inhibit EMT, thereby inhibiting tumour progression (45). Furthermore, the expression of miR-382-5p was significantly down-regulated in CRC tissues and cell lines. Upregulation of miR-382-5p expression can target NR2F2 and PD-L1, thereby inhibiting CRC cell proliferation and metastasis (46, 47). Przygodzka et al. (48) reported that miR-192 and miR-194 can inhibit snail-induced EMT and metastasis. Therefore, prevention of EMT may be a promising approach to block CRC metastasis. In a word, under normal physiological conditions, miRNAs can maintain the normal regulation of some cellular processes, and their abnormality will lead to abnormal growth and biosynthesis of cells, thus promoting or inhibiting the spread and metastasis of tumors.
Apoptosis
Apoptosis is a programmed death process that occurs during normal cell development and senescence. Chemotherapy forces cancer cells to undergo apoptosis by causing DNA damage or cell damage. Abnormal apoptosis is one of the pathogenic mechanisms of CRC and plays a role in the resistance to chemotherapeutic drugs and radiotherapy (49). MicroRNAs play an important role in tumour cell apoptosis and drug resistance. Activation of the caspase family of proteases is the main pathway for inducing apoptosis (50). MiR-433 can increase the expression of caspase-3 and caspase-9, thereby promoting apoptosis (51). Overexpression of miR-218 can also promote CRC cell apoptosis by increasing caspase-8 levels (52). In the Kyoto Encyclopedia of Genes and Genomes (KEGG) apoptosis pathway, miR-92a is associated with two apoptosis-related genes, CSF2RB and BCL2L1. Moreover, increased expression of miR-92a-3p in tumour tissue can improve patient survival time (53). Overexpression of miR-766 reduces CRC cell growth and induces apoptosis by inhibiting the MDM4/p53 pathway (54). MiR-27a-3p increases apoptosis through the ERK-MAPK pathway, while miR-422a induces apoptosis in CRC cells through the p38-MAPK pathway (55, 56). In contrast, mi-421 exerts an anti-apoptotic effect in CRC by downregulating caspase-3 (57). Therefore, the regulation of microRNAs will help to regulate the occurrence and development of CRC, promote cancer cell apoptosis, and alleviate drug resistance.
Immune system activity
Escape from immune system surveillance is an important link in tumorigenesis and development. Studies have shown that microRNAs may be involved in the immune escape process of CRC and are significantly associated with tumour survival. MicroRNAs may be involved in the differentiation of monocytes into M2 macrophages, which have been implicated in playing key roles in colon cancer (58, 59). Exosomes derived from M2 macrophages transfer miR-21-5p and miR-155-5p to CRC cells, promoting cell migration and invasion (60). The results of Ma et al. showed that M2 macrophage-derived exosomal miR-155-5p could promote immune escape by colon cancer, enhancing the progression of CRC (61). Studies have shown that miR-203-containing exosomes released by CRC cells can be internalized by monocytes, thereby promoting the expression of M2 markers (33). However, the PD-1/PD-L1 pathway, as an important immune checkpoint, is dysregulated in various human malignancies, including CRC, and is involved in tumorigenesis by inhibiting antitumor immune response. MiR-124 inhibits PD-L1 expression in CRC cells, which in turn promotes T-cell mediated anti-cancer responses (62). In conclusion, the interaction between miRNAs and immune checkpoints has great application prospects in the personalized treatment of CRC in the future.
Impact on the tumour microenvironment
Tumour growth and metastasis are highly dependent on the interaction between tumour and relevant microenvironment, and several miRNAs have been shown to play a key role in the interaction between tumour and tumour microenvironment (TME). In every step of tumour growth and metastasis, complex molecular interactions occur between cells in the tumour microenvironment, such as fibroblasts and immune-related cells (63). Cancer-associated fibroblasts (CAFs) affect tumour growth by regulating inflammation or direct cell-to-cell communication. Studies have shown that miRNA can alter chemokines secreted by fibroblasts to alter TME, thereby promoting migration and invasion (64). Tumour-derived microRNAs affect the matrix and immune cell components of the tumour microenvironment. In TME, miRNA is considered to be an important molecular mechanism for the interaction between tumour cells and immune cells. For example, miRNAs can control the production of chemokines or cytokines by tumour cells, which in turn affect the aggregation and expansion of immune cells (65). Tumour-associated macrophages (TAMs) are the key components of TME, and miRNAs play an important role in the regulation of TAMs on tumour progression. TAMs have been shown to be associated with a poor prognosis of CRC. TAMs can induce EMT in CRC cells by regulating the STAT3/miR-506-3p/FoxQ1 axis, thereby promoting metastasis (66). However, miR-195-5p could inhibit the polarization of M2-like TAMs, and patients with low miR-195-5p levels have significantly shorter overall survival times (67).
Mesenchymal stem cells (MSCs) are also an important part of the TME and play a key role in promoting tumour progression (68). In the TME, microRNAs generally have tumour-promoting effects and are an important direction for future cancer therapy. Although MSCs have some antitumor activity, microRNAs mediate immunosuppressive activity (69), which provides ideas for future cancer therapy. Intestinal microRNAs can influence the growth and composition of the intestinal microbiota (70). The pathogenesis of CRC is also associated with disorder in the microbiota, termed ecological disorder (71). Imbalances in microRNAs can affect the survival or gene expression of some beneficial bacteria in the microbiota. Dysfunctional microRNAs in tumour cells can be transmitted to stromal cells and immune cells, creating a more favourable microenvironment for tumour cells (72). Thus, microRNAs can modulate the microbiota, promoting the growth of beneficial bacteria and inhibiting the growth of cancer-causing bacteria. In short, the interaction between microRNAs and the TME may also be one of the entry points for antimetastatic treatment in the future.
The role of microRNAs in tumour tumourigenesis
MiRNAs also play an important role in the initiation of human cancer. MiRNAs are related to the pathogenesis of various types of human malignant tumours. In several types of cancer, the decreased expression of miR-34 and let-7 can trigger tumorigenesis, and the up-regulation of miR-34 and let-7 can lead to tumour growth inhibition (73). Moreover, there is ample evidence that miRNAs are closely related to the dysregulation of several key pathways in CRC. miR-31 is a potential driver of colon tumorigenesis by targeting EphB2 and EphA2 signalling pathways (74). Mamoori et al. (75) demonstrated that miR-21 expression was increased many times in colonic cancer stem cells compared to parental cells. Moreover, since the expression of miR-21 is increased, the expression level of PTEN in the colon bulb is decreased, and the Akt signalling pathway is activated, miR-21 is considered to play an important role in the tumorigenic regulation of colon cancer stem cells.
Inflammation also drives the steps of tumorigenesis. Jeffries et al. (76) found that miR-223 can regulate tumorigenesis at multiple levels, including by inhibiting the inflammatory tumour microenvironment and regulating the malignancy of cancer cells. And some studies have proved that the level of miR-223 can be used to predict the probability of CRC by sequencing circulating exosomal miRNAs (77). MiRNAs can be used as therapeutic targets and prediction means, and are potential tools for cancer management and treatment in the future. However, more research is needed before they can be applied to clinic.
Clinical applications
Early diagnosis
Increasing evidence suggests that miRNAs can serve as non-invasive biomarkers for CRC diagnosis and prognosis (Figure 2). They exist in the bloodstream in a highly stable form by binding to specific proteins or vesicles (78, 79). Karimi et al. (80) showed that miR-23a and miR-301 were upregulated in patients compared with healthy individuals, which can be used to distinguish CRC patients from normal subjects. Zhu et al. (81) found that miR-19a-3p, miR-21-5p and miR-425-5p were significantly upregulated in CRC patients compared with healthy individuals. Cheng et al. (82) found that the circulating abundance of exocrine miR-146a correlated with high levels of CD66 neutrophils. However, the proportion of tumour-infiltrating TCD8 cells decreased. MiR-146a is the main miRNA in the exosomes of CRC stem cells and can be used as a diagnostic biomarker. In addition, both miR-486-5p and miR-18b-5p have potential for use as non-invasive biomarkers for the early diagnosis of CRC (83, 84). Min et al. (85) found that miR-92b was differentially expressed in CRC patients and healthy individuals but could not be used to differentiate between CRC and adenoma. Even so, it has promise as a minimally invasive tool for the early diagnosis of CRC. In addition, miR-21, miR-155, and miR-221, which are expressed differently in colon and rectal cancers, can be used to distinguish colon and rectal cancer (86). In addition, the levels of miR-17-5p and miR-92a-3p isolated from serum exosomes were found to correlate with the pathological stage and grade of patients with CRC (87).
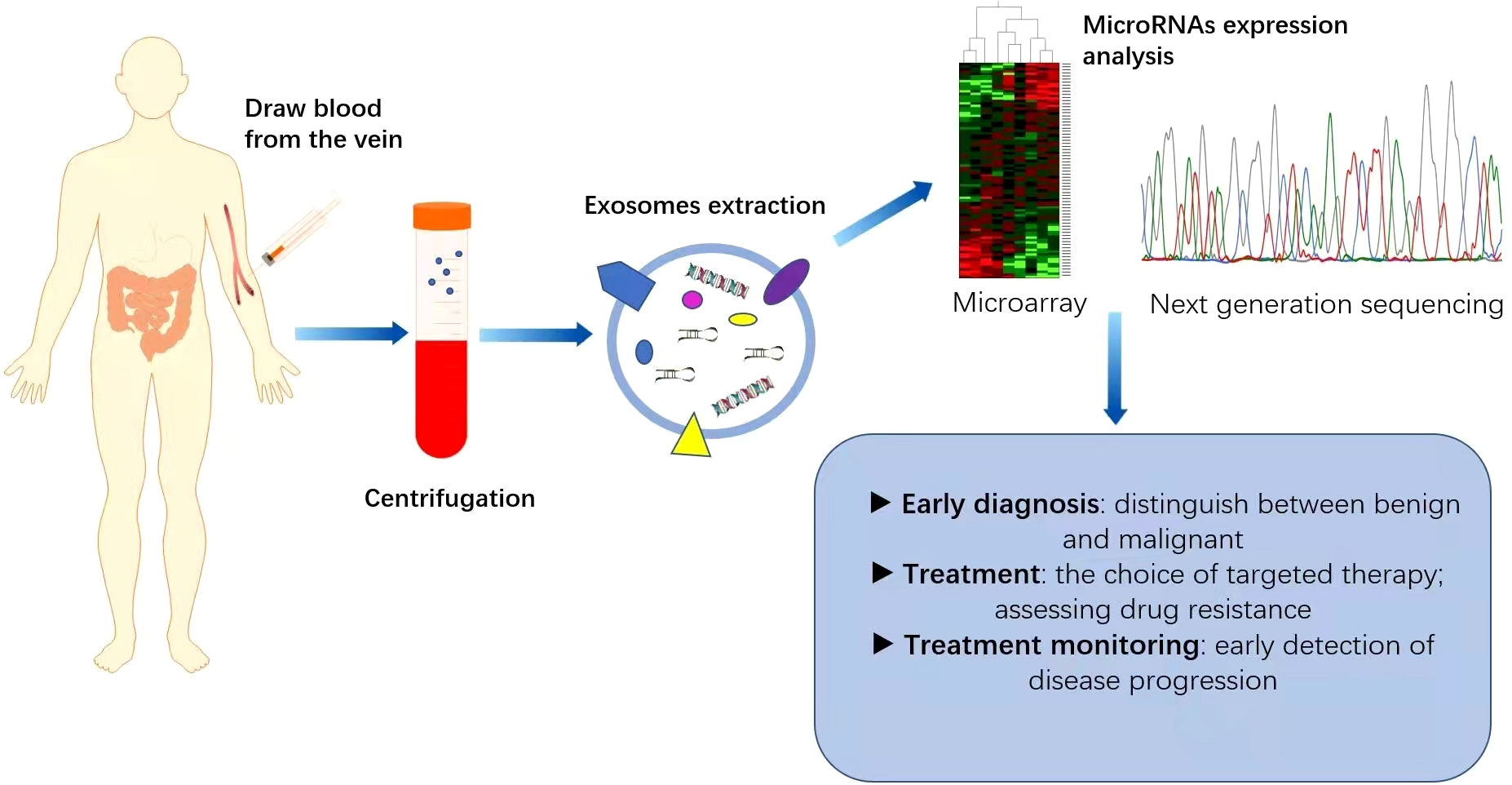
Figure 2 Application of miRNAs as biomarkers for colorectal cancer (CRC). MicroRNAs are generally expressed abnormally in CRC patients. Blood samples collected from CRC patients are used as a source of circulating exosomes and after their isolation, we can analyse the pattern of microRNA expression, which is helpful for the diagnosis and treatment of CRC. This can play an important role in the early diagnosis of cancer, identification of high-risk patients requiring intensive treatment, monitoring of drug efficacy and real-time monitoring of the effectiveness of treatment.
Numerous studies have shown that microRNAs in serum, exocrine and even faeces have the potential for early diagnosis. Decreased expression of miR-4478 and miR-1295-p in stool specimens is a noninvasive and effective diagnostic marker for CRC patients, which can be detected at an early stage of CRC, suggesting that it may be a promising CRC screening approach (88). Moody et al. (89) found that miR-20a in the faeces of CRC patients also serves as a potential prognostic biomarker. Furthermore, stool miR-135b-5p is not only a potential biomarker but also an ideal candidate intervention strategy for CRC patients (90). The establishment of appropriate miRNA biomarkers is very important for the early diagnosis of CRC. Of course, prospective studies with larger patient cohorts are needed to confirm the diagnostic value of these microRNAs. Further efforts are required before microRNAs in faeces can be used clinically.
Treatment options for patients with CRC require accurate assessment of TNM staging. Therefore, biomarkers that can accurately predict preoperative TNM staging will significantly improve the treatment efficiency of CRC. Bjørnetrø et al. (91) found that low levels of miR-486-5p and miR-181a-5p were associated with locally advanced dis-ease and lymph node metastasis, while high levels of miR-30d-5p were associated with metastatic progression. Orosz et al. (86) also evaluated the potential of several microRNAs to distinguish individual TNM stages. The results showed that the expression levels of miR-155, miR-34a, and miR-29a in the serum of TNMII, III, and IV patients were downregulated.
Treatment of CRC
Drug resistance
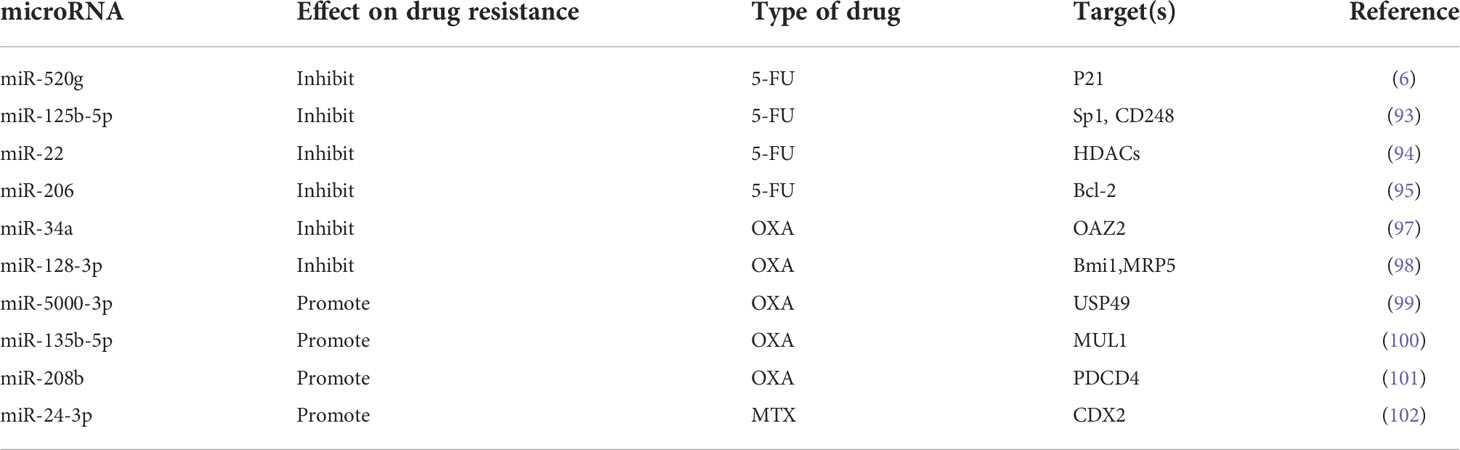
Although good progress has been made in the systemic treatment of tumours in recent years, in addition to surgery, chemotherapy is still the main treatment for CRC. The resistance of cancer cells to chemotherapy is a major factor leading to chemotherapy failure, often resulting in a poor prognosis. Many studies have shown that there is a certain relationship between tumour drug resistance and microRNA imbalance. Tumour drug resistance can occur through a variety of mechanisms, including apoptosis inhibition (92). Studies have shown that ectopic expression of miR-520 g resists 5-FU-induced apoptosis by inhibiting the expression of p21 (6). Decreased levels of miR-125b-5p have also been shown to contribute to tumour cell metastasis and 5-FU chemotherapy resistance (93). Similarly, miR-22 and miR-206 can also promote apoptosis induced by 5-FU (94, 95). Recent studies have shown that the tumour suppressor miR-27b-3p can increase the sensitivity of CRC cells to 5-FU (96). Oxaliplatin (OXA) resistance is also a major obstacle to the treatment of advanced CRC. Li et al. (97) reported that miR-34a was significantly downregulated in OXA-resistant patients, which could reduce OXA resistance by targeting OAZ2. In addition, studies have shown that miR-128-3p can enhance tumour sensitivity to chemotherapy and may become a promising OXA chemotherapy marker (98). In contrast, mir-5000-3p, mir-135b-5p, and mir-208b were associated with decreased sensitivity to OXA chemotherapy (99–101). Recent studies have shown that miR-24-3p can enhance the resistance of CRC cells to methotrexate (MTX) (102).
The hypothesis that drug resistance is the result of tumour-host interactions has been proposed, suggesting new strategies for overcoming the development of cancer chemotherapy resistance (103). Studies have shown that miR-21 and 5-FU combined with engineered exosomes can effectively reverse the drug resistance of 5-FU-resistant colon cancer cells and improve therapeutic efficiency (104). More efforts are needed to prevent cancer cells from developing resistance to chemotherapy and to try to resensitize cancer cells to chemotherapy drugs (Table 1).
There are two different types of CRC: “microsatellite stability” (MSS) and “microsatellite instability” (MSI). Cancers of MSS and MSI types promote tumorigenesis and progression through two distinct molecular pathways (105). Microsatellite stability-high (MSI-H) is caused by functional defects in the DNA mismatch repair (MMR) system. MSI-HCRC immune checkpoint molecules, such as PD-1 and PD-L1, have been shown to be resistant to the antitumor immune response (106). MicroRNAs can play a role in cancer-related immune responses by targeting immunosuppressive or immunostimulatory factors. It has been proven that miR-140-3p, miR-382-3p, miR-148a-3p, miR-93-5p, miR-200a-3p, miR-200c-3p, miR-138-5p and miR-15b-5p can regulate immune escape by inhibiting tumour PD-L1. They can also transform the immunosuppressive tumour microenvironment into a proinflammatory tumour microenvironment, enhancing the chemosensitivity of tumour cells (107). Therefore, it may be possible to alleviate the drug resistance of MSI-H CRC by regulating microRNAs.
Long noncoding RNAs (lncRNAs) are noncoding RNAs (ncRNAs) and microRNAs. Studies have shown that lncRNAs, as precursors of microRNAs, are also associated with drug resistance. For example, the lncRNA MIR100HG, a precursor of miR-100 and miR-125b, can lead to cetuximab resistance (108). The lncRNA-XIST/miR-125b-2-3p axis can also induce chemoresistance in CRC, but the specific mechanism by which it affects chemosensitivity has not been elucidated (109). The complex feedback loop between lncRNAs and microRNAs may provide new perspectives for the reversal of CRC drug resistance. In contrast, the lncRNA-XIST/miR-137 axis can enhance CRC glycolysis and chemotherapy resistance, providing a possible alternative to improve chemotherapy efficacy in CRC patients (110).
Therapeutic target and microRNA therapy
The aberrant expression of microRNAs plays an important role in the development of cancer and the response to anticancer drugs. Correcting microRNA defects or restoring microRNA function can be used as a new cancer treatment strategy. MicroRNAs have been proven to be therapeutic targets for CRC (111). For example, miR-135b has been shown to be upregulated in CRC and associated with tumour progression and a poor clinical prognosis. Therefore, tumour growth can be inhibited by reducing miR-135b. Studies have shown that blocking exocrine miR-25-3p in CRC can reduce the vascular permeability and metastasis of CRC, suggesting that miR-25-3p can be used as a therapeutic target for interfering with CRC metastasis (23).
With the development of high-throughput sequencing technology, the interaction of the gene expression network system comprised of messenger RNAs (mRNAs), miRNAs, lncRNAs and circular RNAs (circRNAs) in CRC progression has been discovered. It has been proven that lncRNA-miRNA cross-talk is a novel mechanism affecting CRC cell proliferation, invasion and metastasis (112). For example, lncRNA TUG1 can promote the growth and migration of CRC cells by secreting miR-145-5p, and the TUG1/miR-145-5p/TRPC6 pathway can serve as a target for CRC diagnosis and therapy (113). Liu et al. showed that the circIFT80/hsa-miR-370-3p/WNT7B signalling axis might also play a role in carcinogenesis (114). CircIFT80 inhibits the expression of hsa-miR-370-3p in CRC cell lines, thereby inhibiting apoptosis. Therefore, in addition to research on microRNAs, research on lncRNAs and circRNAs may also provide new ideas for the targeted therapy of CRC.
In recent years, the application of immune checkpoint inhibitors (ICIs), especially anti-PD-1 therapy, has greatly improved the efficiency of tumour treatment. However, the role of ICIs in CRC is generally limited to MSI-H tumours. The latest study by Liu et al. (115) found that miR-15b-5p downregulated the expression of PD-L1 at the protein level, inhibited tumorigenesis, and improved the sensitivity to anti-PD-1 therapy. Elevating the level of miR-15b-5p can improve the sensitivity of MSS CRC patients to ICI treatment. Blocking oncogenic microRNAs may adversely affect the physiological functions regulated by these microRNAs, thus requiring specific sites or cellular targets to avoid potential adverse effects. At the same time, extensive clinical trials are needed to evaluate the efficacy and safety of microRNAs as therapeutic targets in patients.
Despite advances in the application of immune checkpoint blockade therapy in malignancies, CRC patients usually only benefit if they have tumours with mismatch repair deletions or severe mutations in MSI-H (116). However, most tumours are MSS, so immunotherapy has a low response rate in treating CRC. Many studies have shown that microRNAs can modulate immune responses, and some of these microRNAs can inhibit the progression of CRC and are expected to be effective antineoplastic drugs. Since the disorder of microRNAs was first discovered in cancer, it has been studied extensively and uncovered new therapeutic possibilities. MiRNAs can regulate multiple signalling pathways of the immune system and have the advantage of multiple targets (117). Previous studies have shown that restoration of miR-34 expression can reduce the proliferative potential of CRC cells; thus, miR-34 can be used as a therapeutic drug (118). In addition, miR-34 can also increase tumour sensitivity to 5-Fu, thereby reversing drug resistance (119). Unfortunately, the therapeutic application of microRNAs is limited by technical barriers. MicroRNA molecules are unstable and are rapidly cleared from the blood, with only a small fraction absorbed by cells (120).
In some studies, exosomes have been used as transporters for microRNA drugs, and the lipid bilayer membrane of exosomes can protect exosomes from being degraded during blood circulation. Han et al. (121) used CBMSC-derived exosomes to infiltrate anti-miRNA-221 into solid tumours and significantly inhibited tumour growth. As a tumour suppressor microRNA, miR-124 can regulate several oncogenes and signalling pathways closely related to tumour growth and promote T-cell dependent immune responses. The study by Rezaei et al. (1) used CT-26-derived exosomes as a natural vehicle for miR-124-3p delivery, which elicited potent antitumor immune responses and reduced tumour growth. In the future, the response rate of immunotherapy may be significantly improved by increasing the technology of exosomes carrying microRNAs. However, the source of exosomes is limited and lacks targeting, there are still many challenges in future applications, and further research is needed. In addition, the efficacy and safety of microRNA therapy in patients need to be studied.
Prognosis of colon cancer
Predicting recurrence
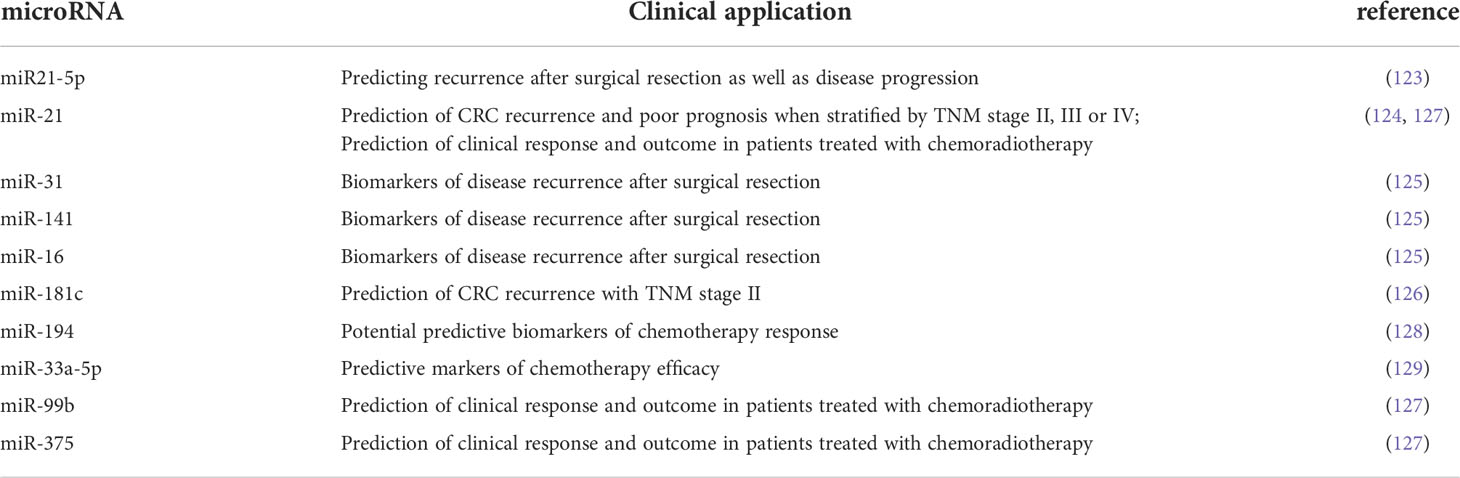
Approximately one-third of patients with CRC undergoing radical surgery will experience disease recurrence (13). Studies have shown that miRNAs can be used as biomarkers for predicting CRC recurrence, which is beneficial to the prognosis of CRC patients. The serum levels of exocrine miR-1229, miR-1224-5p, miR-223, let-7a, miR-150 and miR-21 in CRC patients were significantly increased, and then they decreased after resection (122). Plasma miR21-5p could be used to predict recurrence and disease progression after surgical resection (123). Studies have shown that serum exocrine miR-21 could be used to predict CRC recurrence and a poor outcome in TNM stage II, III, or IV (124). In addition, postoperative plasma miR-31, miR-141, and miR-16 have also been shown to be biomarkers of disease recurrence after surgical resection (125). In general, for patients with stage II CRC, surgical resection of the primary tumour is effective and may not require other treatment, but whether adjuvant chemotherapy should be used in patients with stage II CRC remains controversial (111). Yamazaki et al. (126) proposed that high expression of miR-181c plays a role in predicting recurrence of stage II CRC. Through the study of microRNAs, it is possible to assess which postoperative patients with stage II CRC may benefit from adjuvant therapy (Table 2).
Aberrant expression of microRNAs as biomarkers may contribute to individualized treatment of patients. A study by D’Angelo et al. (128) showed that miR-194 was a potential predictive biomarker of chemotherapy response. Meanwhile, other studies have found that miR-33a-5p, miR-21, miR-99b, and miR-375 can predict clinical response and outcomes in patients treated with radiotherapy and chemotherapy (127, 129). Yin et al. (130) established an in vitro tumour model called patient-derived tumour-like cell clusters (PTCs), which has been shown to be useful for assessing tumour sensitivity to drugs. By incorporating microRNAs as markers into this predictive model, real-time efficacy monitoring can be achieved to assess the benefit of chemotherapy or targeted therapy.
Metastasis of colon cancer
Approximately 50% of CRCs will metastasize in the advanced stage of malignant tumours, and distant metastasis is the main cause of death of CRC patients (Figure 3). Early detection and treatment of distant metastasis is of great significance to improve the long-term survival of CRC patients. MicroRNAs are significantly associated with tumour metastasis. Several microRNAs, including members of the miR-34 and miR-200 families, have been found to target the mRNAs of EMT transcription factors, such as ZEB1, ZEB2, and SNAIL (131). Downregulation of these microRNAs is associated with distant metastasis and advanced tumours. The liver is the most common metastatic site of CRC. The study by Hur et al. (132) showed that elevated serum miR-203 levels are closely associated with liver and systemic metastasis. Teng et al. (133) detected significantly elevated plasma miR-193a levels in CRC patients with liver metastasis. Lan et al. (60) found that miR-21-5p and miR-155-5p were transferred to CRC cells via exosomes and were key factors in promoting CRC metastasis. Preventing such messages may be a new strategy to suppress CRC metastasis. These microRNAs can be used as biomarkers to determine prognosis and predict distant metastasis. MiR-181a is significantly upregulated in CRC tissues of patients with liver metastases and promotes tumour cell growth and proliferation, which is closely associated with distant metastasis and poor survival (134). In contrast, miR-802 is negatively correlated with lymphatic and distant metastasis of CRC (135), and may be a regulatory target for suppressing metastasis.
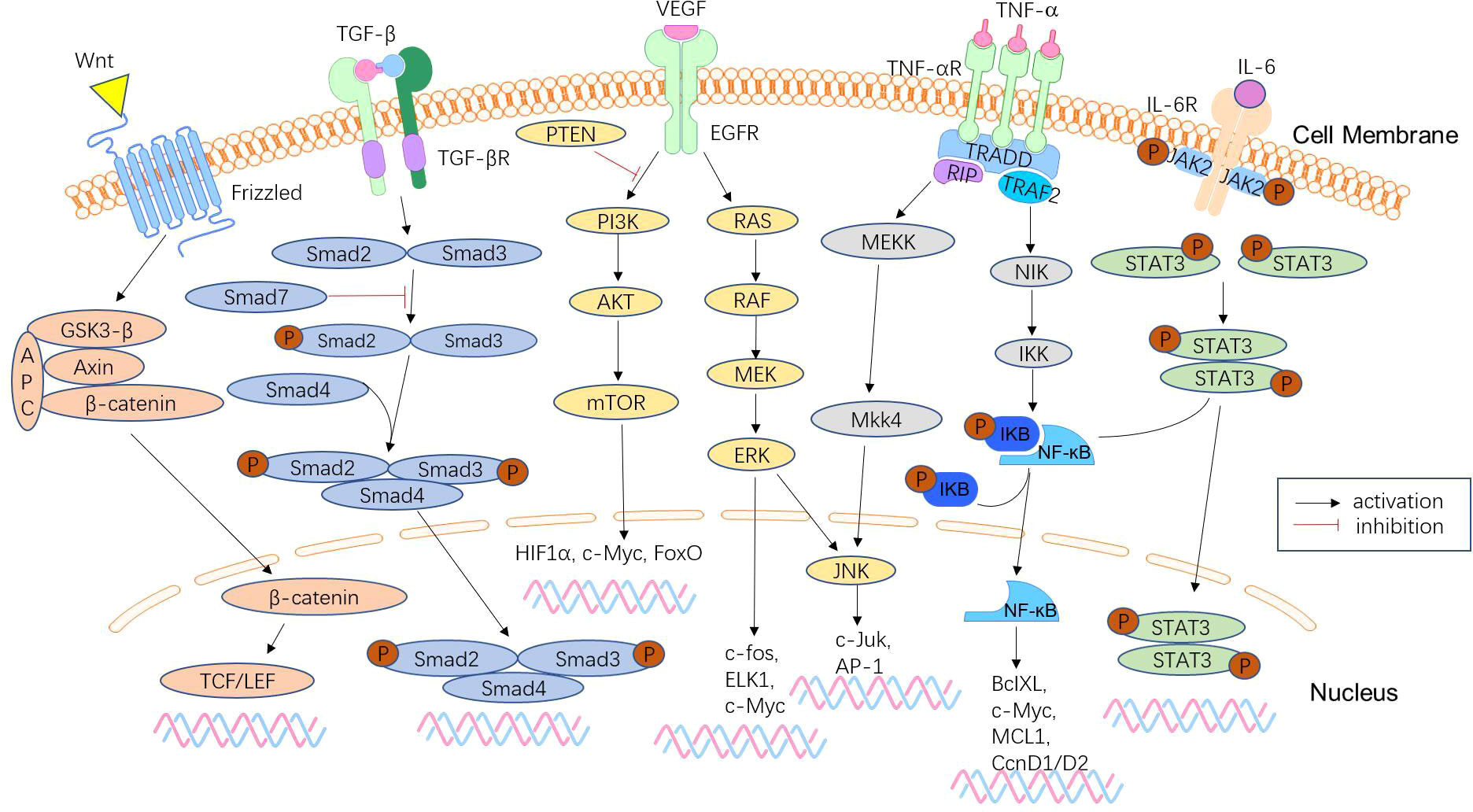
Figure 3 Signalling pathways involved in CRC metastasis. CRC metastasis is mediated by a complex network of signalling pathways, which include the Wnt/β-catenin signalling pathway, TGF-β/Smad pathway, phosphoinositide 3-kinase (PI3K)/phosphatase and tensin homologue (PTEN)/AKT pathway, KRAS-ERK signalling pathway, NF-κB signalling pathway, and JAK/STAT3 signalling pathway. These pathways lead to tumour anti-apoptosis, EMT, proliferation, and invasion.
Induction of muscular dystrophy
Cachexia is a complex metabolic and behavioural syndrome associated with underlying disease and is characterized by loss of skeletal muscle. Previous studies have found a significant correlation between skeletal muscle mass and circulating miR-21 expression in CRC patients, suggesting that assessment of serum miR-21 levels can be used to assess the risk of sarcopenia and cancer cachexia in patients with CRC (136). The results of Miao et al. (137) suggest that abundant microRNAs in tumour exosomes may induce muscle atrophy mainly by targeting Bcl-2-mediated apoptosis. In addition, the detection of serum miR-203 expression can be used to evaluate the risk of sarcopenia, and miR-203 may be a new therapeutic target for inhibiting sarcopenia in patients with CRC (138).
Conclusion and perspectives
MicroRNAs have a wide range of biological functions and are involved in many physiological and pathological processes, including cancer. An increasing number of studies have shown that microRNAs play an important role in the progression and metastasis of CRC. Specific microRNAs can be used to overcome diagnostic and therapeutic challenges of different types of tumours. The combination of novel microRNA markers with traditional biomarkers may help to improve the specificity and sensitivity of detection. Using microRNAs as new therapeutic targets to correct maladjusted microRNAs would be a promising approach for CRC therapy. In future studies, we should determine which biological fluids and assays are most suitable for CRC screening and which microRNA combinations have the best diagnostic performance. We should maximize the specificity of these microRNA biomarkers. At the same time, we should increase our understanding of the role of microRNAs in the molecular pathogenesis and treatment of cancer. This will facilitate the clinical application of microRNAs.
At present, some progress has been made in the study of microRNAs reversing drug resistance, but there are still few studies on immunotherapy resistance in MSI-H CRC. In addition, the biggest problem facing microRNA therapy is the choice of carrier. Nanoparticles or exosomes are used as carriers in the current studies. Both of these carriers have certain limitations, and more research is needed to overcome these difficulties and allow for their application in clinical practice. The roles and functions of individual microRNAs in CRC remain unclear and more research is needed. Investigating the effects of microRNAs on the occurrence, development and metastasis of CRC is of great significance for the diagnosis and treatment of CRC.
lncRNAs, circRNAs and microRNAs are all ncRNAs and have great potential in clinical applications. Accumulating evidence suggests that a complex regulatory net-work exists between lncRNAs, circRNAs, and microRNAs. They have great biological potential and may regulate CRC initiation, progression and metastasis. However, the exact mechanisms of how these interactions affect tumorigenesis and progression have not been fully revealed. Future analysis of different RNA molecules with potential crosstalk may provide new insights into the diagnosis and treatment of CRC, contributing to the improvement of biomarker prediction and the development of new treatments.
Author contributions
All authors contributed to the article and approved the submitted version.
Funding
We are supported by the National Natural Science Foundation of China (grant number 81970791), Science and Technology Bureau of Quanzhou (grant number 2020CT003), Fujian Province Scientific Foundation (grant number 2019J01168), and the Young and middle-aged backbone talent foundation of Fujian Provincial Commission of Health Construction (grant number 2020GGA058).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
CRC, colorectal cancer; 3’-UTR, 3’-untranslated regions; VEGF, vascular endothelial growth factor; PGE2, prostaglandin E2; EMT, epithelial-mesenchymal transition; MET, mesenchymal-to-epithelial transition; RASA1RASp21, protein activator 1; PGRN, progranulin; PD-1, programmed death-1; KEGGKyoto, Encyclopedia of Genes and Genomes; PD-L1, Programmed death ligand 1; MSCs, mesenchymal stem cells; TME, tumor microenvironment; TAMs, tumor-associated macrophages; 5-Fu, 5-Fluorouracil; OXA, Oxaliplatin; OAZ2O, rnithine Decarboxylase Antizyme 2; MTX, methotrexate; MSS, microsatellite stability; MSI, microsatellite instability; MSI-H, microsatellite instability-high; MMR, mismatch repair; lncRNAs, long non-coding RNAs; ncRNAs, non-coding RNAs; ICI, simmune checkpoint inhibitors; mRNAs, messenger RNAs; circRNAs, circleRNAs; PTCs, patient-derived tumor-like cell clusters.
References
1. Rezaei R, Baghaei K, Hashemi SM, Zali MR, Ghanbarian H, Amani D. Tumor-derived exosomes enriched by mirna-124 promote anti-tumor immune response in ct-26 tumor-bearing mice. Front Med (Lausanne) (2021) 8:619939. doi: 10.3389/fmed.2021.619939
2. Gunter SA MJ, Arnold M, Brenner H, Burn J, Casey G, Chan AT, et al. Meeting report from the joint iarc-nci international cancer seminar series: A focus on colorectal cancer. Ann Oncol (2019) 30(4):510–9. doi: 10.1093/annonc/mdz044/5307027
3. Thanikachalam K, Khan G. Colorectal cancer and nutrition. Nutrients (2019) 11(1):164. doi: 10.3390/nu11010164
4. Lee RC, Feinbaum RL, Ambros V. The c. elegans heterochronic gene Lin-4 encodes small rnas with antisense complementarity to &Ii-14. Cell (1993) 75 (5):843–54. doi: 10.1016/0092-8674(93)90529-y
5. Muhammad S, Kaur K, Huang R, Zhang Q, Kaur P, Yazdani HO, et al. Micrornas in colorectal cancer: Role in metastasis and clinical perspectives. World J Gastroenterol (2014) 20(45):17011–9. doi: 10.3748/wjg.v20.i45.17011
6. Wang H. Micrornas and apoptosis in colorectal cancer. Int J Mol Sci (2020) 21(15):5353. doi: 10.3390/ijms21155353
7. Balacescu O, Sur D, Cainap C, Visan S, Cruceriu D, Manzat-Saplacan R, et al. The impact of mirna in colorectal cancer progression and its liver metastases. Int J Mol Sci (2018) 19(12):3711. doi: 10.3390/ijms19123711
8. Iacona JR, Lutz CS. Mir-146a-5p: Expression, regulation, and functions in cancer. Wiley Interdiscip Rev RNA (2019) 10(4):e1533. doi: 10.1002/wrna.1533
9. Chen B, Xia Z, Deng YN, Yang Y, Zhang P, Zhu H, et al. Emerging microrna biomarkers for colorectal cancer diagnosis and prognosis. Open Biol (2019) 9(1):180212. doi: 10.1098/rsob.180212
10. Zhang N, Hu X, Du Y, Du J. The role of mirnas in colorectal cancer progression and chemoradiotherapy. BioMed Pharmacother (2021) 134:111099. doi: 10.1016/j.biopha.2020.111099
11. Stoffel EM, Murphy CC. Epidemiology and mechanisms of the increasing incidence of colon and rectal cancers in young adults. Gastroenterology (2020) 158(2):341–53. doi: 10.1053/j.gastro.2019.07.055
12. Corte H, Manceau G, Blons H, Laurent-Puig P. Microrna and colorectal cancer. Dig Liver Dis (2012) 44(3):195–200. doi: 10.1016/j.dld.2011.10.010
13. Boussios S, Ozturk MA, Moschetta M, Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, et al. The developing story of predictive biomarkers in colorectal cancer. J Pers Med (2019) 9(1):12. doi: 10.3390/jpm9010012
14. Chang YC, Chan MH, Li CH, Fang CY, Hsiao M, Chen CL. Exosomal components and modulators in colorectal cancer: Novel diagnosis and prognosis biomarkers. Biomedicines (2021) 9(8):931. doi: 10.3390/biomedicines9080931
15. van der Stok EP, Spaander MCW, Grunhagen DJ, Verhoef C, Kuipers EJ. Surveillance after curative treatment for colorectal cancer. Nat Rev Clin Oncol (2017) 14(5):297–315. doi: 10.1038/nrclinonc.2016.199
16. Felekkis K TE, Stefanou CH, Deltas C. Micrornas: A newly described class of encoded molecules that play a role in health and disease. HIPPOKRATIA (2010) 14 (4):236–40.
17. Sohel MH. Extracellular/Circulating micrornas: Release mechanisms, functions and challenges. Achievements Life Sci (2016) 10(2):175–86. doi: 10.1016/j.als.2016.11.007
18. Alves Dos Santos K, Clemente Dos Santos IC, Santos Silva C, Gomes Ribeiro H, de Farias Domingos I, Nogueira Silbiger V. Circulating exosomal mirnas as biomarkers for the diagnosis and prognosis of colorectal cancer. Int J Mol Sci (2020) 22(1):346. doi: 10.3390/ijms22010346
19. Peng Y, Croce CM. The role of micrornas in human cancer. Signal Transduct Target Ther (2016) 1:15004. doi: 10.1038/sigtrans.2015.4
20. Chang SH, Hla T. Gene regulation by rna binding proteins and micrornas in angiogenesis. Trends Mol Med (2011) 17(11):650–8. doi: 10.1016/j.molmed.2011.06.008
21. Tiwari A, Mukherjee B, Dixit M. Microrna key to angiogenesis regulation: Mirna biology and therapy. Curr Cancer Drug Targets (2018) 18(3):266–77. doi: 10.2174/1568009617666170630142725
22. Gallach S, Calabuig-Farinas S, Jantus-Lewintre E, Camps C. Micrornas: Promising new antiangiogenic targets in cancer. BioMed Res Int (2014) 2014:878450. doi: 10.1155/2014/878450
23. Zeng Z, Li Y, Pan Y, Lan X, Song F, Sun J, et al. Cancer-derived exosomal mir-25-3p promotes pre-metastatic niche formation by inducing vascular permeability and angiogenesis. Nat Commun (2018) 9(1):5395. doi: 10.1038/s41467-018-07810-w
24. Han SH, Mo JS, Park WC, Chae SC. Reduced microrna 375 in colorectal cancer upregulates metadherin-mediated signaling. World J Gastroenterol (2019) 25(44):6495–507. doi: 10.3748/wjg.v25.i44.6495
25. Lun W, Wu X, Deng Q, Zhi F. Mir-218 regulates epithelial-mesenchymal transition and angiogenesis in colorectal cancer Via targeting ctgf. Cancer Cell Int (2018) 18:83. doi: 10.1186/s12935-018-0575-2
26. Ma H, Pan JS, Jin LX, Wu J, Ren YD, Chen P, et al. Microrna-17~92 inhibits colorectal cancer progression by targeting angiogenesis. Cancer Lett (2016) 376(2):293–302. doi: 10.1016/j.canlet.2016.04.011
27. Hu HY, Yu CH, Zhang HH, Zhang SZ, Yu WY, Yang Y, et al. Exosomal mir-1229 derived from colorectal cancer cells promotes angiogenesis by targeting Hipk2. Int J Biol Macromol (2019) 132:470–7. doi: 10.1016/j.ijbiomac.2019.03.221
28. He Q, Ye A, Ye W, Liao X, Qin G, Xu Y, et al. Cancer-secreted exosomal mir-21-5p induces angiogenesis and vascular permeability by targeting Krit1. Cell Death Dis (2021) 12(6):576. doi: 10.1038/s41419-021-03803-8
29. Niu L, Yang W, Duan L, Wang X, Li Y, Xu C, et al. Biological implications and clinical potential of metastasis-related mirna in colorectal cancer. Mol Ther Nucleic Acids (2021) 23:42–54. doi: 10.1016/j.omtn.2020.10.030
30. Liu Y, Cao X. Characteristics and significance of the pre-metastatic niche. Cancer Cell (2016) 30(5):668–81. doi: 10.1016/j.ccell.2016.09.011
31. Shao Y, Chen T, Zheng X, Yang S, Xu K, Chen X, et al. Colorectal cancer-derived small extracellular vesicles establish an inflammatory premetastatic niche in liver metastasis. Carcinogenesis (2018) 39(11):1368–79. doi: 10.1093/carcin/bgy115
32. Sun H, Meng Q, Shi C, Yang H, Li X, Wu S, et al. Hypoxia-inducible exosomes facilitate liver-tropic premetastatic niche in colorectal cancer. Hepatology (2021) 74(5):2633–51. doi: 10.1002/hep.32009
33. Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, et al. Circulating exosomal microrna-203 is associated with metastasis possibly Via inducing tumor-associated macrophages in colorectal cancer. Oncotarget (2017). 8 (45):78598–613. doi: 10.18632/oncotarget.20009
34. Huang C, Liu J, Xu L, Hu W, Wang J, Wang M, et al. Microrna-17 promotes cell proliferation and migration in human colorectal cancer by downregulating Sik1. Cancer Manag Res (2019) 11:3521–34. doi: 10.2147/CMAR.S191087
35. Liu Y, Chen X, Cheng R, Yang F, Yu M, Wang C, et al. The Jun/Mir-22/Hur regulatory axis contributes to tumourigenesis in colorectal cancer. Mol Cancer (2018) 17(1):11. doi: 10.1186/s12943-017-0751-3
36. Park YR, Seo SY, Kim SL, Zhu SM, Chun S, Oh JM, et al. Mirna-206 suppresses Pge2-induced colorectal cancer cell proliferation, migration, and invasion by targetting Tm4sf1. Biosci Rep (2018) 38(5):BSR20180664. doi: 10.1042/BSR20180664
37. Tang W, Zhou W, Xiang L, Wu X, Zhang P, Wang J, et al. The P300/Yy1/Mir-500a-5p/Hdac2 signalling axis regulates cell proliferation in human colorectal cancer. Nat Commun (2019) 10(1):663. doi: 10.1038/s41467-018-08225-3
38. Zhang Z, Li J, Huang Y, Peng W, Qian W, Gu J, et al. Upregulated mir-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2f8 in crc. Cell Prolif (2018) 51(6):e12505. doi: 10.1111/cpr.12505
39. Ashrafizadeh M, Hushmandi K, Hashemi M, Akbari ME, Kubatka P, Raei M, et al. Role of Microrna/Epithelial-to-Mesenchymal transition axis in the metastasis of bladder cancer. Biomolecules (2020) 10(8):1159. doi: 10.3390/biom10081159
40. Ramesh Singh RP, Kounosuke W, Zhaohui L, Yin-Yuan M. Exosome-mediated transfer of mir-10b promotes cell invasion in breast cancer. Mol Cancer (2014) 13:256. doi: 10.1186/1476-4598-13-256
41. Sun X, Lin F, Sun W, Zhu W, Fang D, Luo L, et al. Exosome-transmitted mirna-335-5p promotes colorectal cancer invasion and metastasis by facilitating emt Via targeting Rasa1. Mol Ther Nucleic Acids (2021) 24:164–74. doi: 10.1016/j.omtn.2021.02.022
42. Ding D, Li C, Zhao T, Li D, Yang L, Zhang B. Lncrna H19/Mir-29b-3p/Pgrn axis promoted epithelial-mesenchymal transition of colorectal cancer cells by acting on wnt signaling. Mol Cells (2018) 41(5):423–35. doi: 10.14348/molcells.2018.2258
43. Wang H, Yan B, Zhang P, Liu S, Li Q, Yang J, et al. Mir-496 promotes migration and epithelial-mesenchymal transition by targeting Rassf6 in colorectal cancer. J Cell Physiol (2020) 235(2):1469–79. doi: 10.1002/jcp.29066
44. Wang D, Wang X, Si M, Yang J, Sun S, Wu H, et al. Exosome-encapsulated mirnas contribute to Cxcl12/Cxcr4-induced liver metastasis of colorectal cancer by enhancing M2 polarization of macrophages. Cancer Lett (2020) 474:36–52. doi: 10.1016/j.canlet.2020.01.005
45. O'Brien SJ, Carter JV, Burton JF, Oxford BG, Schmidt MN, Hallion JC, et al. The role of the mir-200 family in epithelial-mesenchymal transition in colorectal cancer: A systematic review. Int J Cancer (2018) 142(12):2501–11. doi: 10.1002/ijc.31282
46. Xie L, Pan Z. Circular rna Circ_0000467 regulates colorectal cancer development Via mir-382-5p/En2 axis. Bioengineered (2021) 12(1):886–97. doi: 10.1080/21655979.2021.1889130
47. Jin Y, Zhan X, Zhang B, Chen Y, Liu C, Yu L. Polydatin exerts an antitumor effect through regulating the mir-382/Pd-L1 axis in colorectal cancer. Cancer Biother Radiopharm (2020) 35(2):83–91. doi: 10.1089/cbr.2019.2999
48. Przygodzka P, Papiewska-Pajak I, Bogusz-Koziarska H, Sochacka E, Boncela J, Kowalska MA. Regulation of mirnas by snail during epithelial-to-Mesenchymal transition in Ht29 colon cancer cells. Sci Rep (2019) 9(1):2165. doi: 10.1038/s41598-019-39200-7
49. Watson AJ. Apoptosis and colorectal cancer. Gut (2004) 53(11):1701–9. doi: 10.1136/gut.2004.052704
50. Green DR, Llambi F. Cell death signaling. Cold Spring Harb Perspect Biol (2015) 7(12):a006080. doi: 10.1101/cshperspect.a006080
51. Li J, Mao X, Wang X, Miao G, Li J. Mir-433 reduces cell viability and promotes cell apoptosis by regulating Macc1 in colorectal cancer. Oncol Lett (2017) 13(1):81–8. doi: 10.3892/ol.2016.5445
52. Meng Q, Chen Y, Lian B, Shang Y, Yang H. Mir−218 promotes apoptosis of Sw1417 human colon cancer cells by targeting C−Flip. Oncol Rep (2018) 40 (2):916–22. doi: 10.3892/or.2018.6460
53. Slattery ML, Mullany LE, Sakoda LC, Wolff RK, Samowitz WS, Herrick JS. Dysregulated genes and mirnas in the apoptosis pathway in colorectal cancer patients. Apoptosis (2018) 23(3-4):237–50. doi: 10.1007/s10495-018-1451-1
54. Chen W, Cai G, Liao Z, Lin K, Li G, Li Y. Mirna-766 induces apoptosis of human colon cancer cells through the P53/Bax signaling pathway by Mdm4. Exp Ther Med (2019) 17(5):4100–8. doi: 10.3892/etm.2019.7436
55. Su C, Huang DP, Liu JW, Liu WY, Cao YO. Mir−27a−3p regulates proliferation and apoptosis of colon cancer cells by potentially targeting Btg1. Oncol Lett (2019) 18 (3):2825–34. doi: 10.3892/ol.2019.10629
56. Li P, Li Q, Zhang Y, Sun S, Liu S, Lu Z. Mir-422a targets Mapkk6 and regulates cell growth and apoptosis in colorectal cancer cells. BioMed Pharmacother (2018) 104:832–40. doi: 10.1016/j.biopha.2018.03.013
57. Zhou Y, Cheng X, Wan Y, Chen T, Zhou Q, Wang Z, et al. Microrna-421 inhibits apoptosis by downregulating caspase-3 in human colorectal cancer. Cancer Manag Res (2020) 12:7579–87. doi: 10.2147/CMAR.S255787
58. Grossman JG, Nywening TM, Belt BA, Panni RZ, Krasnick BA, DeNardo DG, et al. Recruitment of Ccr2(+) tumor associated macrophage to sites of liver metastasis confers a poor prognosis in human colorectal cancer. Oncoimmunology (2018) 7(9):e1470729. doi: 10.1080/2162402X.2018.1470729
59. Zhang LL, Zhang LF, Shi YB. Down-regulated paxillin suppresses cell proliferation and invasion by inhibiting M2 macrophage polarization in colon cancer. Biol Chem (2018) 399(11):1285–95. doi: 10.1515/hsz-2018-0002
60. Lan J, Sun L, Xu F, Liu L, Hu F, Song D, et al. M2 macrophage-derived exosomes promote cell migration and invasion in colon cancer. Cancer Res (2019) 79(1):146–58. doi: 10.1158/0008-5472.CAN-18-0014
61. Ma YS, Wu TM, Ling CC, Yu F, Zhang J, Cao PS, et al. M2 macrophage-derived exosomal microrna-155-5p promotes the immune escape of colon cancer by downregulating Zc3h12b. Mol Ther Oncolytics (2021) 20:484–98. doi: 10.1016/j.omto.2021.02.005
62. Roshani Asl E, Rasmi Y, Baradaran B. Microrna-124-3p suppresses pd-L1 expression and inhibits tumorigenesis of colorectal cancer cells Via modulating Stat3 signaling. J Cell Physiol (2021) 236(10):7071–87. doi: 10.1002/jcp.30378
63. Rupaimoole R, Calin GA, Lopez-Berestein G, Sood AK. Mirna deregulation in cancer cells and the tumor microenvironment. Cancer Discov (2016) 6(3):235–46. doi: 10.1158/2159-8290.CD-15-0893
64. Mitra AK, Zillhardt M, Hua Y, Tiwari P, Murmann AE, Peter ME, et al. Micrornas reprogram normal fibroblasts into cancer-associated fibroblasts in ovarian cancer. Cancer Discov (2012) 2(12):1100–8. doi: 10.1158/2159-8290.CD-12-0206
65. Xing Y, Ruan G, Ni H, Qin H, Chen S, Gu X, et al. Tumor immune microenvironment and its related mirnas in tumor progression. Front Immunol (2021) 12:624725. doi: 10.3389/fimmu.2021.624725
66. Lin X, Wang S, Sun M, Zhang C, Wei C, Yang C, et al. Mir-195-5p/Notch2-Mediated emt modulates il-4 secretion in colorectal cancer to affect M2-like Tam polarization. J Hematol Oncol (2019) 12(1):20. doi: 10.1186/s13045-019-0708-7
67. Wei C, Yang C, Wang S, Shi D, Zhang C, Lin X, et al. Crosstalk between cancer cells and tumor associated macrophages is required for mesenchymal circulating tumor cell-mediated colorectal cancer metastasis. Mol Cancer (2019) 18(1):64. doi: 10.1186/s12943-019-0976-4
68. Lazennec G, Jorgensen C. Concise review: Adult multipotent stromal cells and cancer: Risk or benefit? Stem Cells (2008) 26(6):1387–94. doi: 10.1634/stemcells.2007-1006
69. Whiteside TL. Exosome and mesenchymal stem cell cross-talk in the tumor microenvironment. Semin Immunol (2018) 35:69–79. doi: 10.1016/j.smim.2017.12.003
70. Yuan C, Burns MB, Subramanian S, Blekhman R. Interaction between host micrornas and the gut microbiota in colorectal cancer. mSystems (2018) 3(3):e00205–17. doi: 10.1128/mSystems.00205-17
71. Feng Q, Liang S, Jia H, Stadlmayr A, Tang L, Lan Z, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. Nat Commun (2015) 6:6528. doi: 10.1038/ncomms7528
72. Yuan C, Steer CJ, Subramanian S. Host(-)Microrna(-)Microbiota interactions in colorectal cancer. Genes (Basel) (2019) 10(4):270. doi: 10.3390/genes10040270
73. Khan AQ, Ahmed EI, Elareer NR, Junejo K, Steinhoff M, Uddin S. Role of mirna-regulated cancer stem cells in the pathogenesis of human malignancies. Cells (2019) 8(8):840. doi: 10.3390/cells8080840
74. De Robertis M, Mazza T, Fusilli C, Loiacono L, Poeta ML, Sanchez M, et al. Ephb2 stem-related and Epha2 progression-related mirna-based networks in progressive stages of crc evolution: Clinical significance and potential mirna drivers. Mol Cancer (2018) 17(1):169. doi: 10.1186/s12943-018-0912-z
75. Mamoori A, Gopalan V, Smith RA, Lam AK. Modulatory roles of micrornas in the regulation of different signalling pathways in Large bowel cancer stem cells. Biol Cell (2016) 108(3):51–64. doi: 10.1111/boc.201500062
76. Jeffries J, Zhou W, Hsu AY, Deng Q. Mirna-223 at the crossroads of inflammation and cancer. Cancer Lett (2019) 451:136–41. doi: 10.1016/j.canlet.2019.02.051
77. Ogata-Kawata H, Izumiya M, Kurioka D, Honma Y, Yamada Y, Furuta K, et al. Circulating exosomal micrornas as biomarkers of colon cancer. PLoS One (2014) 9(4):e92921. doi: 10.1371/journal.pone.0092921
78. Moridikia A, Mirzaei H, Sahebkar A, Salimian J. Micrornas: Potential candidates for diagnosis and treatment of colorectal cancer. J Cell Physiol (2018) 233(2):901–13. doi: 10.1002/jcp.25801
79. Mirzaei AS H, Jaafari MR, Goodarzi M, Mirzaei. Diagnostic HR. And therapeutic potential of exosomes in cancer: The beginning of a new tale? J Cell Physiol (2017) 232 (12):3250–60. doi: 10.1002/jcp.25739
80. Karimi N, Ali Hosseinpour Feizi M, Safaralizadeh R, Hashemzadeh S, Baradaran B, Shokouhi B, et al. Serum overexpression of mir-301a and mir-23a in patients with colorectal cancer. J Chin Med Assoc (2019) 82(3):215–20. doi: 10.1097/JCMA.0000000000000031
81. Mingxia Z, Zebo H, Danxia Z, Xin Z, Xia S, Lian-Wen Q, et al. A panel of microrna signature in serum for colorectal cancer diagnosis. Oncotarget (2017) 8 (10):17081–91. doi: 10.18632/oncotarget.15059
82. Cheng WC, Liao TT, Lin CC, Yuan LE, Lan HY, Lin HH, et al. Rab27b-activated secretion of stem-like tumor exosomes delivers the biomarker microrna-146a-5p, which promotes tumorigenesis and associates with an immunosuppressive tumor microenvironment in colorectal cancer. Int J Cancer (2019) 145(8):2209–24. doi: 10.1002/ijc.32338
83. Zhang H, Zhu M, Shan X, Zhou X, Wang T, Zhang J, et al. A panel of seven-mirna signature in plasma as potential biomarker for colorectal cancer diagnosis. Gene (2019) 687:246–54. doi: 10.1016/j.gene.2018.11.055
84. Liu X, Chen X, Zeng K, Xu M, He B, Pan Y, et al. DNA-Methylation-Mediated silencing of mir-486-5p promotes colorectal cancer proliferation and migration through activation of Plagl2/Igf2/Beta-catenin signal pathways. Cell Death Dis (2018) 9(10):1037. doi: 10.1038/s41419-018-1105-9
85. Min L, Chen L, Liu S, Yu Y, Guo Q, Li P, et al. Loss of circulating exosomal mir-92b is a novel biomarker of colorectal cancer at early stage. Int J Med Sci (2019) 16(9):1231–7. doi: 10.7150/ijms.34540
86. Orosz E, Kiss I, Gyongyi Z, Varjas T. Expression of circulating mir-155, mir-21, mir-221, mir-30a, mir-34a and mir-29a: Comparison of colonic and rectal cancer. In Vivo (2018) 32(6):1333–7. doi: 10.21873/invivo.11383
87. Susanne K-S, Maslova M, Pohl M, Eilert-Micus C, Schroers R, Schmiegel W, et al. Significance of liquid biopsy for monitoring and therapy decision of colorectal cancer. Trans Oncol (2018) 11 (2):213–20. doi: 10.1016/j.tranon.2017.12.010
88. Ghanbari R, Rezasoltani S, Hashemi J, Mohamadkhani A, Tahmasebifar A, Arefian E, et al. Expression analysis of previously verified fecal and plasma Dow-regulated micrornas (Mir-4478, 1295-3p, 142-3p and 26a-5p), in ffpe tissue samples of crc patients. Arch Iran Med (2017) 20(2):92–5. doi: 0172002/AIM.006
89. Moody L, Dvoretskiy S, An R, Mantha S, Pan YX. The efficacy of mir-20a as a diagnostic and prognostic biomarker for colorectal cancer: A systematic review and meta-analysis. Cancers (Basel) (2019) 11(8):1111. doi: 10.3390/cancers11081111
90. Li L, Wang A, Cai M, Tong M, Chen F, Huang L. Identification of stool mir-135b-5p as a non-invasive diaognostic biomarker in later tumor stage of colorectal cancer. Life Sci (2020) 260:118417. doi: 10.1016/j.lfs.2020.118417
91. Bjornetro T, Redalen KR, Meltzer S, Thusyanthan NS, Samiappan R, Jegerschold C, et al. An experimental strategy unveiling exosomal micrornas 486-5p, 181a-5p and 30d-5p from hypoxic tumour cells as circulating indicators of high-risk rectal cancer. J Extracell Vesicles (2019) 8(1):1567219. doi: 10.1080/20013078.2019.1567219
92. Mansoori B, Mohammadi A, Davudian S, Shirjang S, Baradaran B. The different mechanisms of cancer drug resistance: A brief review. Adv Pharm Bull (2017) 7(3):339–48. doi: 10.15171/apb.2017.041
93. Park GB, Jeong JY, Kim D. Modified tlr-mediated downregulation of mir-125b-5p enhances Cd248 (Endosialin)-induced metastasis and drug resistance in colorectal cancer cells. Mol Carcinog (2020) 59(2):154–67. doi: 10.1002/mc.23137
94. Hu Y, French SW, Chau T, Liu HX, Sheng L, Wei F, et al. Rarbeta acts as both an upstream regulator and downstream effector of mir-22, which epigenetically regulates Nur77 to induce apoptosis of colon cancer cells. FASEB J (2019) 33(2):2314–26. doi: 10.1096/fj.201801390R
95. Meng X, Fu R. Mir-206 regulates 5-fu resistance by targeting bcl-2 in colon cancer cells. Onco Targets Ther (2018) 11:1757–65. doi: 10.2147/OTT.S159093
96. Sun W, Li J, Zhou L, Han J, Liu R, Zhang H, et al. The c-Myc/Mir-27b-3p/Atg10 regulatory axis regulates chemoresistance in colorectal cancer. Theranostics (2020) 10(5):1981–96. doi: 10.7150/thno.37621
97. Li Y, Gong P, Hou JX, Huang W, Ma XP, Wang YL, et al. Mir-34a regulates multidrug resistance Via positively modulating Oaz2 signaling in colon cancer cells. J Immunol Res (2018) 2018:7498514. doi: 10.1155/2018/7498514
98. Liu T, Zhang X, Du L, Wang Y, Liu X, Tian H, et al. Exosome-transmitted mir-128-3p increase chemosensitivity of oxaliplatin-resistant colorectal cancer. Mol Cancer (2019) 18(1):43. doi: 10.1186/s12943-019-0981-7
99. Zhuang YY, Zhong W, Xia ZS, Lin SZ, Chan MC, Jiang K, et al. Mir-5000-3p confers oxaliplatin resistance by targeting ubiquitin-specific peptidase 49 in colorectal cancer. Cell Death Discovery (2021) 7(1):129. doi: 10.1038/s41420-021-00494-0
100. Wang H, Wang X, Zhang H, Deng T, Liu R, Liu Y, et al. The Hsf1/Mir-135b-5p axis induces protective autophagy to promote oxaliplatin resistance through the Mul1/Ulk1 pathway in colorectal cancer. Oncogene (2021) 40(28):4695–708. doi: 10.1038/s41388-021-01898-z
101. Ning T, Li J, He Y, Zhang H, Wang X, Deng T, et al. Exosomal mir-208b related with oxaliplatin resistance promotes treg expansion in colorectal cancer. Mol Ther (2021) 29(9):2723–36. doi: 10.1016/j.ymthe.2021.04.028
102. Zhang HW, Shi Y, Liu JB, Wang HM, Wang PY, Wu ZJ, et al. Cancer-associated fibroblast-derived exosomal microrna-24-3p enhances colon cancer cell resistance to mtx by down-regulating Cdx2/Heph axis. J Cell Mol Med (2021) 25(8):3699–713. doi: 10.1111/jcmm.15765
103. Alfarouk KO, Stock CM, Taylor S, Walsh M, Muddathir AK, Verduzco D, et al. Resistance to cancer chemotherapy: Failure in drug response from adme to p-gp. Cancer Cell Int (2015) 15:71. doi: 10.1186/s12935-015-0221-1
104. Liang G, Zhu Y, Ali DJ, Tian T, Xu H, Si K, et al. Engineered exosomes for targeted Co-delivery of mir-21 inhibitor and chemotherapeutics to reverse drug resistance in colon cancer. J Nanobiotechnol (2020) 18(1):10. doi: 10.1186/s12951-019-0563-2
105. Sugai T, Yoshida M, Eizuka M, Uesugii N, Habano W, Otsuka K, et al. Analysis of the DNA methylation level of cancer-related genes in colorectal cancer and the surrounding normal mucosa. Clin Epigenet (2017) 9:55. doi: 10.1186/s13148-017-0352-4
106. Ashizawa M, Okayama H, Ishigame T, Thar Min AK, Saito K, Ujiie D, et al. Mirna-148a-3p regulates immunosuppression in DNA mismatch repair-deficient colorectal cancer by targeting pd-L1. Mol Cancer Res (2019) 17(6):1403–13. doi: 10.1158/1541-7786.MCR-18-0831
107. Shadbad MA, Asadzadeh Z, Derakhshani A, Hosseinkhani N, Mokhtarzadeh A, Baghbanzadeh A, et al. A scoping review on the potentiality of pd-L1-Inhibiting micrornas in treating colorectal cancer: Toward single-cell sequencing-guided biocompatible-based delivery. BioMed Pharmacother (2021) 143:112213. doi: 10.1016/j.biopha.2021.112213
108. Yang S, Sun Z, Zhou Q, Wang W, Wang G, Song J, et al. Micrornas, long noncoding rnas, and circular rnas: Potential tumor biomarkers and targets for colorectal cancer. Cancer Manag Res (2018) 10:2249–57. doi: 10.2147/CMAR.S166308
109. Zeng ZL, Lu JH, Wang Y, Sheng H, Wang YN, Chen ZH, et al. The lncrna Xist/Mir-125b-2-3p axis modulates cell proliferation and chemotherapeutic sensitivity Via targeting Wee1 in colorectal cancer. Cancer Med (2021) 10(7):2423–41. doi: 10.1002/cam4.3777
110. Zheng H, Zhang M, Ke X, Deng X, Li D, Wang Q, et al. Lncrna Xist/Mir-137 axis strengthens chemo-resistance and glycolysis of colorectal cancer cells by hindering transformation from Pkm2 to Pkm1. Cancer Biomark (2021) 30(4):395–406. doi: 10.3233/CBM-201740
111. To KK, Tong CW, Wu M, Cho WC. Micrornas in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol (2018) 24(27):2949–73. doi: 10.3748/wjg.v24.i27.2949
112. Tang XJ, Wang W, Hann SS. Interactions among lncrnas, mirnas and mrna in colorectal cancer. Biochimie (2019) 163:58–72. doi: 10.1016/j.biochi.2019.05.010
113. Wang X, Bai X, Yan Z, Guo X, Zhang Y. The lncrna Tug1 promotes cell growth and migration Via the Tug1/Mir-145-5p/Trpc6 pathway in colorectal cancer. Biochem Cell Biol (2020) 99 (2):249–60. doi: 10.1139/bcb-2020-0017
114. Liu N, Jiang F, Chen Z. A preliminary study on the pathogenesis of colorectal cancer by constructing a hsa-Circrna-0067835-Mirna-Mrna regulatory network. Onco Targets Ther (2021) 14:4645–58. doi: 10.2147/OTT.S319300
115. Liu C, Liu R, Wang B, Lian J, Yao Y, Sun H, et al. Blocking il-17a enhances tumor response to anti-Pd-1 immunotherapy in microsatellite stable colorectal cancer. J Immunother Cancer (2021) 9(1):e001895. doi: 10.1136/jitc-2020-001895
116. Cagnoni AJ, Giribaldi ML, Blidner AG, Cutine AM, Gatto SG, Morales RM, et al. Galectin-1 fosters an immunosuppressive microenvironment in colorectal cancer by reprogramming Cd8(+) regulatory T cells. Proc Natl Acad Sci U.S.A. (2021) 118(21):e2102950118. doi: 10.1073/pnas.2102950118
117. Pottoo FH, Iqubal A, Iqubal MK, Salahuddin M, Rahman JU, AlHajri N, et al. Mirnas in the regulation of cancer immune response: Effect of mirnas on cancer immunotherapy. Cancers (Basel) (2021) 13(23):6145. doi: 10.3390/cancers13236145
118. Krajewska JB, Fichna J, Mosinska P. One step ahead: Mirna-34 in colon cancer-future diagnostic and therapeutic tool? Crit Rev Oncol Hematol (2018) 132:1–8. doi: 10.1016/j.critrevonc.2018.09.006
119. Akao Y, Noguchi S, Iio A, Kojima K, Takagi T, Naoe T. Dysregulation of microrna-34a expression causes drug-resistance to 5-fu in human colon cancer dld-1 cells. Cancer Lett (2011) 300(2):197–204. doi: 10.1016/j.canlet.2010.10.006
120. Di Martino MT, Campani V, Misso G, Gallo Cantafio ME, Gullà A, Foresta U, et al. In vivo activity of mir-34a mimics delivered by stable nucleic acid lipid particles (Snalps) against multiple myeloma. PLoS One (2014) 9(2):e90005. doi: 10.1371/journal.pone.0090005
121. Han S, Li G, Jia M, Zhao Y, He C, Huang M, et al. Delivery of anti-Mirna-221 for colorectal carcinoma therapy using modified cord blood mesenchymal stem cells-derived exosomes. Front Mol Biosci (2021) 8:743013. doi: 10.3389/fmolb.2021.743013
122. Ruiz-Lopez L, Blancas I, Garrido JM, Mut-Salud N, Moya-Jodar M, Osuna A, et al. The role of exosomes on colorectal cancer: A review. J Gastroenterol Hepatol (2018) 33(4):792–9. doi: 10.1111/jgh.14049
123. Fukada M, Matsuhashi N, Takahashi T, Sugito N, Heishima K, Yoshida K, et al. Postoperative changes in plasma Mir21-5p as a novel biomarker for colorectal cancer recurrence: A prospective study. Cancer Sci (2021) 112(10):4270–80. doi: 10.1111/cas.15065
124. Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y. Circulating exosomal microrna-21 as a biomarker in each tumor stage of colorectal cancer. Oncology (2017) 92(6):360–70. doi: 10.1159/000463387
125. Yuan Z, Baker K, Redman MW, Wang L, Adams SV, Yu M, et al. Dynamic plasma micrornas are biomarkers for prognosis and early detection of recurrence in colorectal cancer. Br J Cancer (2017) 117(8):1202–10. doi: 10.1038/bjc.2017.266
126. Yamazaki N KY, Taniguchi H, Kojima M, Kanemitsu Y, Saito N, Matsumura Y. High expression of mir-181c as a predictive marker of recurrence in stage ii colorectal cancer. Oncotarget (2017) 8 (4):6970–83. doi: 10.18632/oncotarget.14344
127. Campayo M, Navarro A, Benitez JC, Santasusagna S, Ferrer C, Monzo M, et al. Mir-21, mir-99b and mir-375 combination as predictive response signature for preoperative chemoradiotherapy in rectal cancer. PLoS One (2018) 13(11):e0206542. doi: 10.1371/journal.pone.0206542
128. D'Angelo E, Zanon C, Sensi F, Digito M, Rugge M, Fassan M, et al. Mir-194 as predictive biomarker of responsiveness to neoadjuvant chemoradiotherapy in patients with locally advanced rectal adenocarcinoma. J Clin Pathol (2018) 71(4):344–50. doi: 10.1136/jclinpath-2017-204690
129. Sasaki M, Ishikawa T, Ishiguro M, Okazaki S, Yamauchi S, Kikuchi A, et al. The effectiveness of plasma mir-33a-5p as a predictive biomarker for the efficacy of colorectal cancer chemotherapy. Oncol Lett (2021) 21(6):489. doi: 10.3892/ol.2021.12749
130. Yin S, Xi R, Wu A, Wang S, Li Y, Wang C, et al. Patient-derived tumor-like cell clusters for drug testing in cancer therapy. Sci Trans Med (2020) 12 (549):eaaz1723. doi: 10.1126/scitranslmed.aaz1723
131. Vu T, Datta PK. Regulation of emt in colorectal cancer: A culprit in metastasis. Cancers (Basel) (2017) 9(12):171. doi: 10.3390/cancers9120171
132. Hur K, Toiyama Y, Okugawa Y, Ide S, Imaoka H, Boland CR, et al. Circulating microrna-203 predicts prognosis and metastasis in human colorectal cancer. Gut (2017) 66(4):654–65. doi: 10.1136/gutjnl-2014-308737
133. Teng Y, Ren Y, Hu X, Mu J, Samykutty A, Zhuang X, et al. Mvp-mediated exosomal sorting of mir-193a promotes colon cancer progression. Nat Commun (2017) 8:14448. doi: 10.1038/ncomms14448
134. Wai Hon K, Zainal Abidin SA, Othman I, Naidu R. Insights into the role of micrornas in colorectal cancer (Crc) metabolism. Cancers (Basel) (2020) 12(9):2462. doi: 10.3390/cancers12092462
135. Wang X, Li D, Sun L, Shen G, Liu H, Guo H, et al. Regulation of the small gtpase ran by mir-802 modulates proliferation and metastasis in colorectal cancer cells. Br J Cancer (2020) 122(11):1695–706. doi: 10.1038/s41416-020-0809-7
136. Okugawa Y, Yao L, Toiyama Y, Yamamoto A, Shigemori T, Yin C, et al. Prognostic impact of sarcopenia and its correlation with circulating mir-21 in colorectal cancer patients. Oncol Rep (2018) 39(4):1555–64. doi: 10.3892/or.2018.6270
137. Miao C, Zhang W, Feng L, Gu X, Shen Q, Lu S, et al. Cancer-derived exosome mirnas induce skeletal muscle wasting by bcl-2-Mediated apoptosis in colon cancer cachexia. Mol Ther Nucleic Acids (2021) 24:923–38. doi: 10.1016/j.omtn.2021.04.015
Keywords: colon cancer, microRNAs, progression, metastasis, therapeutic strategies
Citation: Liang C, Yang J-B, Lin X-Y, Xie B-L, Xu Y-X, Lin S and Xu T-W (2022) Recent advances in the diagnostic and therapeutic roles of microRNAs in colorectal cancer progression and metastasis. Front. Oncol. 12:911856. doi: 10.3389/fonc.2022.911856
Received: 03 April 2022; Accepted: 26 September 2022;
Published: 13 October 2022.
Edited by:
Francesca Negri, University Hospital of Parma, ItalyReviewed by:
Letizia Gnetti MD, University Hospital of Parma, ItalyWen-Jian Meng, Sichuan University, China
Copyright © 2022 Liang, Yang, Lin, Xie, Xu, Lin and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tian-Wen Xu, xutianwen53@163.com; Shu Lin, shulin1956@126.com
 Chen Liang1
Chen Liang1 Shu Lin
Shu Lin Tian-Wen Xu
Tian-Wen Xu