- 1Unit of Pathology, Ospedale di Circolo, Azienda Socio Sanitaria Territoriale (ASST)-Sette Laghi, Varese, Italy
- 2Research Center for the Study of Hereditary and Familial Tumors, Department of Medicine and Surgery, University of Insubria, Varese, Italy
- 3Biosciences Laboratory, Istituto Ricerca e Cura a Carattere Scientifico (IRCCS) Istituto Romagnolo per lo Studio dei Tumori “Dino Amadori” – Istituto Romagnolo per lo Studio dei Tumori (IRST) S.r.l., Meldola, Italy
- 4Cogentech s.r.l. Società Benefit a Socio Unico, Milan, Italy
- 5Inter-Hospital Pathology Division, Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) MultiMedica, Milan, Italy
- 6Department of Medicine and Surgery, University of Insubria, Varese, Italy
Background: Lobular breast carcinoma (LBC) is considered an exceptionally rare disease in men, including only 1% of all male breast malignancies. The majority of LBCs have negative immunohistochemical staining for E-cadherin (CDH1) expression, and the loss of CDH1 function was traditionally implicated in the tumorigenesis of diffuse gastric cancer as well as LBC. It is well recognized that LBC in women could be involved in both hereditary breast and ovarian cancer (HBOC) and hereditary diffuse gastric cancer (HDGC) syndromes; however, there are no data present in literature about the involvement of male LBC in these inherited conditions.
Methods: BRCA1, BRCA2, and CDH1 genes were performed on DNA from peripheral blood using next-generation sequencing (NGS), Sanger sequencing, and multiplex ligation-dependent probe amplification analyses. BRCA2 and CDH1 somatic gene analyses were performed on breast tumoral DNA using the NGS sequencing approach.
Results and conclusions: Here, we describe two men affected by LBC, the carriers of a pathogenic variant of BRCA2 and CDH1 genes, respectively. Our data, including somatic and germline results, demonstrate a strong relationship between male LBC and HBOC/HDGC syndromes, excluding a sporadic origin of LBC in these two patients. Male LBC could represent a sentinel cancer for inherited syndrome identification, and early identification of cancer susceptibility could improve cancer prevention both for men and women in these families. The history of the LBC patient carrier of the CDH1 variant suggests to include male LBC genetic testing criteria and male breast surveillance in HDGC guidelines.
Introduction
Male breast cancer (MBC) is a rare entity, representing 1% of all breast, male and female, cancers. Invasive lobular breast carcinoma (LBC) is exceptionally rare in men, comprising only 1% of all male breast malignancies (1), probably because lobular development does not occur in the male breast. The origin of the lobular histological type of breast carcinoma in men remains largely unexplained and, due to its extremely uncommon occurrence, knowledge on the natural history of disease progression, clinical presentation, treatment management, and prognosis in men is, to date, very limited (2).
Invasive LBC is more likely to be estrogen and progesterone positive compared with invasive ductal carcinoma, and it is usually HER-2, p53, and EGFR negative. The majority of LBCs have negative immunohistochemical staining for E-cadherin expression, which is the main regulator of the lobular phenotype in breast cancer. E-cadherin dysfunction is responsible for the discohesive histomorphological characteristics of LBC. The loss of CDH1 function was traditionally implicated in the tumorigenesis of diffuse gastric cancer (DGC), and both LBC and DGC share the same histopathological characteristics, including individual or small clusters of discohesive cell growth pattern.
It is well known that a subset of MBCs is due to inherited conditions; in particular, BRCA1 and BRCA2 are the most involved genes. The genetic susceptibility linked to these genes contributes to the pathogenesis of many MBCs (3); however, the relationship between the MBC lobular histotype and cancer susceptibility is so far unknown.
A family history of breast and ovarian cancers is reported in approximately 15%–20% of MBCs, and approximately 10% of MBC patients carry BRCA2 variants, while few patients carry the pathogenic variants of BRCA1 (4).
Very recently, Rizzolo et al., in a large Italian study, identified MBC patients with the germline pathogenic variants of other genes including PTEN, TP53, PALB2, and CHEK2 (5), suggesting that MBC susceptibility could involve additional genes other than BRCA1/2.
Here, we describe two male patients affected by LBC, the carriers of BRCA2 and CDH1 germline pathogenic variants, respectively. These two cases suggest that male LBC could be associated with inherited cancer conditions.
Methods
Somatic Analysis
Immunohistochemistry (IHC) Analysis
Formalin-fixed, paraffin-embedded (FFPE) tumor sections were completely processed automatically on a VENTANA BenchMark ULTRA immunostainer using ultraView universal DAB detection Kit (Ventana Indianapolis, Indiana), as routinely. According to the suggested protocols, after antigen retrieval with a Cell Conditioning 1 solution, the sections were incubated with the following primary antibodies: anti-ER (clone SP1, Ventana), anti-PR (clone 1E2, Ventana), anti-HER2 (clone 4B5, Ventana), anti-p53 (clone DO7, Ventana), and anti-Ki67 (clone MIB1 1/100, DAKO).
DNA Extraction From Formalin-Fixed Paraffin-Embedded Tissue
Tumoral DNA was obtained after the macrodissection of FFPE samples with 80% of neoplastic cells, as previously described by Carnevali etal. (6).
Next-Generation Sequencing
For the analysis of BRCA2 somatic variants on patient 1, the NGS library was prepared with SOPHiA homologous recombination solution (SOPHiA GENETICS), starting from 80 ng of DNA extracted from FFPE tumor tissue, and was run on the Miseq System (Illumina). Results were analyzed with SOPHiA DDM platform v. 2.6.3 (SOPHiA GENETICS).
For the analysis of CDH1 somatic variants on patient 2, the NGS library was prepared with Nextera Flex for Enrichment (Illumina), starting from 100 ng of DNA extracted from FFPE tumor tissue, and enriched for a custom panel (Integrated DNA Technologies), including the CDH1 gene and other cancer predisposition genes. The enriched library was run on the Miseq System (Illumina), and results were analyzed with Miseq Reporter Software v. 2.6.2 (Illumina).
Methylation Analysis
CDH1 methylation was assessed, addressing six CpG sites (exact position: GrCh37/Hg19 chromosome 1668771035, 68771037, 68771045, 68771051, 68771059, 68771064, 68771073) in the gene promoter by pyrosequencing on bisulfite-converted DNA. About 200ng of DNA was bisulfite-treated by using EZ DNA Methylation Kit (Zymo Research, Irvine, CA, USA) and then amplified using TaKaRa EpiTaq™ HS reagents (Takara Bio Inc, Shiga, Japan). Primers used for PCR reactions were the following: Forward 5’-AGTAATTTTAGGTTAGAGGGTTA, Reverse 5’-Biotin- ACCACAACCAATCAACAAC while sequencing primer was 5’- ATTTTAGGTTAGAGGGTTAT. The cut-off value to call the presence of methylation was 10%.
Germline Analysis
DNA Extraction
Whole blood DNA was isolated through the MagCore® Super automatic workstation with the MagCore® Genomic DNA Whole Blood Kit (Diatech LabLine SRL, Jesi, Italy).
Sanger Sequencing
CDH1 coding exons (1-16) and flanking regions were simultaneously amplified at the annealing temperature of 60°C, with the AmpliTaq Gold kit (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Sequencing was performed on purified PCR products by using BigDye® Terminator v.3.1 Cycle Sequencing kit (Thermo Fisher Scientific, Inc.) and run on the 3500 Dx Genetic Analyzer (Life Technologies) after purification with Agencourt CleanSeq®-Beckman Coulter. Sequences were analyzed by Mutation Surveyor® Software v5.1.0 (SoftGenetics, LLC, State College, PA, USA). The same method was applied to confirm the BRCA2 mutation identified in case 2 by NGS. Primers are available on request.
Multiplex Ligation-Dependent Probe Amplification
The analysis of large deletions and duplications of CDH1, BRCA1, and BRCA2 was carried out on the DNA extracted from peripheral blood with the CDH1 SALSA MLPA KIT - P083 (C2), BRCA1 SALSA MLPA KIT - P002 (D1), BRCA2 SALSA MLPA KIT - P045 (C1) probemixes (MRC-Holland, Amsterdam, Netherlands), following manufacturer’s instructions. MLPA products were run on the 3730Xl DNA Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, Massachusetts, United States) with the Gene Mapper Module (Applied Biosystems; Thermo Fisher Scientific, Inc.). Results were analyzed through the Gene Marker Software v2.7.0 (SoftGenetics, LLC, State College, PA, USA).
Next-Generation Sequencing
Approximately 10–25 ng of dsDNA, according to Qubit dsDNA HS assay kits fluorometric quantification (Thermo Fisher Scientific Inc.), were amplified in multiplex reactions, spanning the entire coding region of the BRCA1 and BRCA2 genes, including flanking intronic sequences (+/-20bp). The NGS library was created using TruSeq Custom Amplicon v.1.2 (TSCA) technology (Illumina Inc., San Diego, CA, USA), and sequencing was performed on MiSeq (Illumina Inc.), using a 2 × 150 bp paired-end module. Data collection was done with the MiSeq Reporter (MSR) software v.2.6.2.3. The run quality was evaluated by Illumina Sequencing Analysis Viewer v.1.9.1, while the annotation of the vcf files was performed with a customized bioinformatics pipeline. Paired-end reads were mapped on the Human hg19 genome. A full coverage of the coding regions was obtained, with a minimum read depth of 50X and an average read depth of 3,500X/sample.
Variant Classification
The identified genetic variants were divided into five classes according to the International Agency for Research on Cancer (IARC) recommendations (7) and classified in accordance with the guidelines of the American College of Medical Genetics (ACMG) (8) and the most recent guidelines on CDH1 variant classification (9). The BRCA2 variant was also classified according to the ENIGMA Consortium guidelines (10), obtaining the same class of pathogenicity.
Results
Patient 1
We herein described the case of a 63-year-old man showing unilateral gynecomastia in the right breast and an underlying palpable nodule. Ultrasound examination showed an irregular right periareolar hypoechogenic nodular formation of 23 mm with suspected posterior extension. In the homolateral axillary site, a possible adenopathy of approximately 10 mm was visible. In December 2016, he underwent right mastectomy; sentinel lymph node biopsy intraoperative examination revealed metastatic cancer cells and, subsequently, complete axillary lymphadenectomy.
The final diagnosis, from the pathology examination, was LBC with a solid growth pattern, poorly differentiated. In addition, LBC showed perineoplastic vascular invasion; the nipple had focal infiltration and the areolar skin was affected by neoplastic infiltration up to the reticular dermis. The resection margins were free from neoplasia. Thirty axillary lymph nodes were examined, and none showed signs of metastasis.
The IHC analysis of ER, PgR, Her2/neu, MIB1, p53, and E-cadherin proteins on FFPE tissue sections demonstrated immunoreactivity for ER and PgR (90% and 75%, respectively) and the loss of p53 e E-cadherin expression (Figure 1). The proliferation index (MIB-1) was detected in 32% of neoplastic cells, and Her2/neu was evaluated as a 1+ score (ASCO guidelines 2018). TNM staging was pT1cN1a.
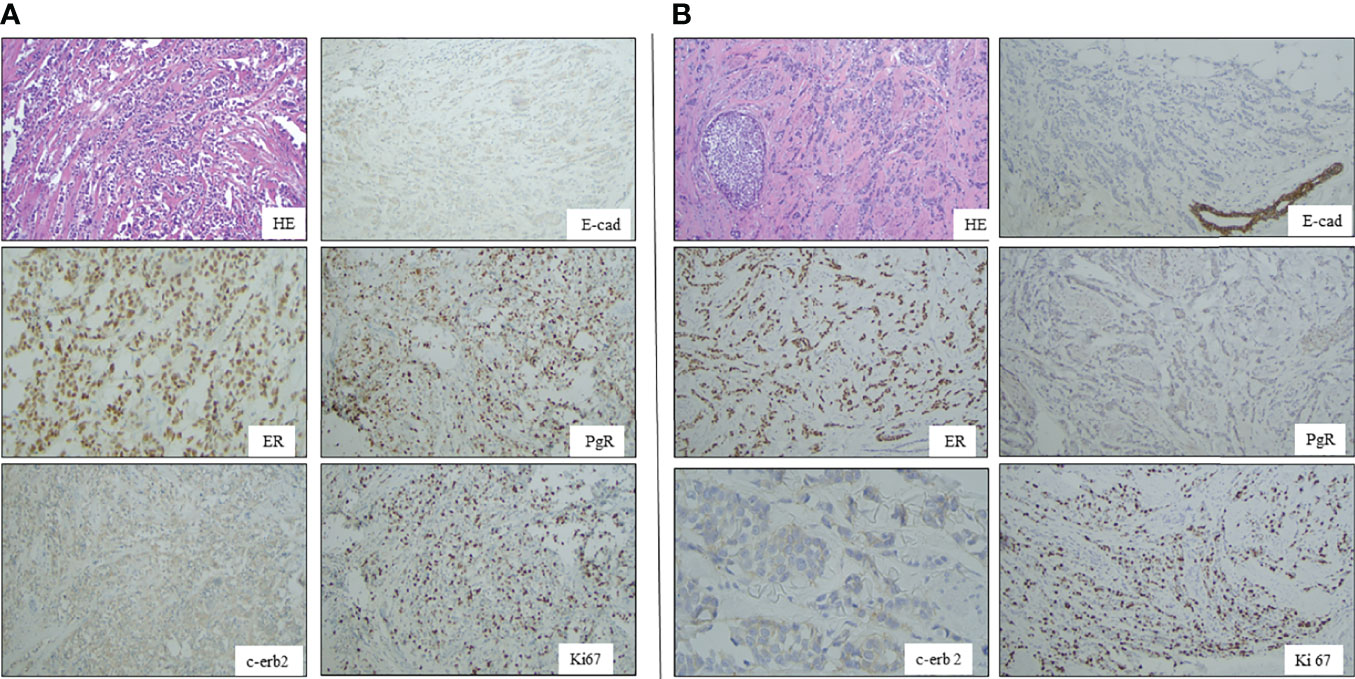
Figure 1 Panel A shows Hematoxilin-Eosin (HE) picture and immunohystochemical results of E-cadherin (B), estrogen receptor (D), progesterone receptor (C), HER2 and Ki67 expressions in breast cancer of case 1 Panel B: A shows Hematoxilin-Eosin (HE) picture and immunohystochemical results of E-cadherin (B), estrogen receptor (D), progesterone receptor (C), HER2 and Ki67 expressions in breast cancer of case 2.
After a multidisciplinary discussion, the patient underwent surgery and 4 cycles of adjuvant chemotherapy with anthracyclines, followed by 12 cycles q1 of paclitaxel. After a whole chemotherapy course, the patient underwent fractioned radiation therapy with a total dose of 50 and 66 Gy applied to the chest wall and to the surgical site, respectively. After radiotherapy, the patient began a 5-year treatment with tamoxifen; he is still alive in a follow-up after 5 years from diagnosis.
Family History and Germline Analysis
Case 1 (III.4 in a pedigree Figure 2A) was referred in 2016 to our Genetic Counseling Service, ASST Sette Laghi Varese, during his follow-up. As shown in Figure 2, the proband’s mother (II.2) died at age 85 after the diagnosis of bilateral breast cancer at ages 60 and 70, and the proband’s sister (III.2) developed breast cancer at age 60. There is no other report of malignancy in this family that, in agreement with FONCAM and NCCN guidelines (11, 12), was suspected of HBOC syndrome (ORPHA 145).
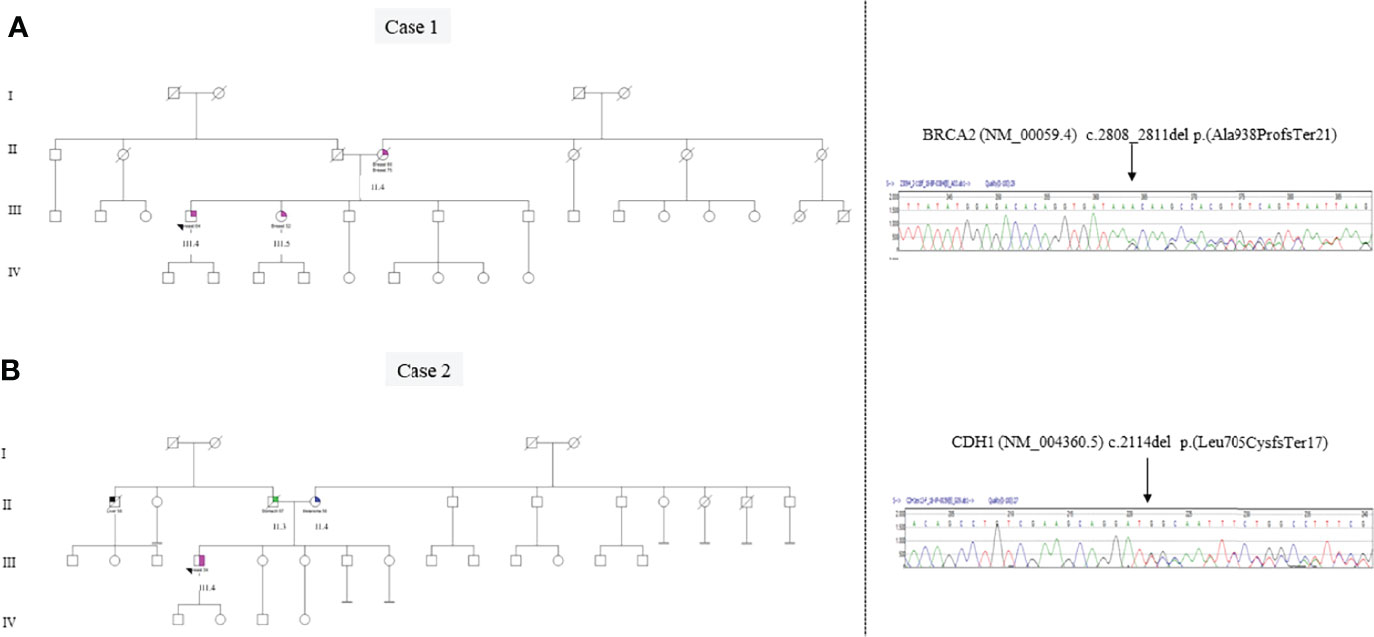
Figure 2 Panel A: Genetic pedigree of case 1 and Sanger sequencing electropherogram of the BRCA2 exon 11 variant c.2808_2811delACAA (p.Ala938ProfsTer21). The alignment to the reference sequence of BRCA2 NM_000059.3 was created by the Mutation Surveyor® Software (v5.1.0). Panel B: Genetic pedigree of case 2 and Sanger sequencing electropherogram of the CDH1 exon 13 variant c.2114delT (p.Leu705CysfsTer17). The alignment to the reference sequence of CDH1 NM_004360.3 was created by the Mutation Surveyor® Software (v5.1.0).
The genetic analysis of BRCA1 and BRCA2 genes in this patient revealed the germline pathogenic variant c.2808_2811delACAA p.(Ala938ProfsTer21) in heterozygosity (VAF ~0.5) in the BRCA2 gene (transcript NM_00059.4) using NGS and subsequently confirmed by Sanger sequencing.
The CDH1 gene was also analyzed using both Sanger Sequencing and MLPA approaches, and no variants were identified. The BRCA2 pathogenic variant results in a premature termination codon predicted to cause a truncated or absent BRCA2 protein due to nonsense-mediated decay, which is a commonly known mechanism for disease. It is seen in the general European population at a low overall frequency of 0.0018%, as reported in the Genome Aggregation Database (13). This variant is known to be one of the most common pathogenic variants in non-Ashkenazi Caucasians (14–16), and it has been reported in association with familial and early-onset breast and/or ovarian cancers (17–19) and also in individuals with prostate cancer (20, 21).
The BRCA2 pathogenic variant was also investigated in the patient’s two sons (IV.8, IV.9), who inherited the same BRCA2 pathogenic variant. No other members of this family decided to perform genetic testing.
Tumor Analysis
The molecular analysis of the BRCA2 gene on the tumoral DNA of case 1 revealed the presence of 8 variants, 7 of which were also present in the germline DNA, previously identified as germline variants from blood (Table 1). The software detected in the tumor tissue the BRCA2 variant c.865A>C p.(Asn289His) (rs766173) not present in the germline DNA with a variant allele frequency (VAF) of 0.013.
Three of the 7 BRCA2 variants, present both in the germline and tumoral DNA (rs206075, rs206076, rs169547), were in homozygosity (VAF~1.00). The other 4 variants (rs80359351, rs1801406, rs28897724, rs9534262) were in heterozygosity in the germline DNA (VAF~0.5) but respectively showed the VAFs of 0.75, 0.18, 0.23 and 0.76 in the tumoral DNA, indicating an allelic imbalance (AI) and suggesting the presence of a loss of heterozygosity (LOH).
Patient 2
In 2017, a 34-year-old Caucasian man had a diagnosis of right breast nodule in the peri-retroareolar site. Breast ultrasound showed a pseudo-nodular formation with poorly defined boundaries. The subsequent mammography showed a small parenchyma, while positron emission tomography (PET) analysis was negative. The patient underwent a right mastectomy with axillary dissection after the evaluation of a sentinel lymph node that was positive with 1 cm of the metastatic localization. The neoplasm had a maximum diameter of 1 cm, hard consistency, and irregular margins.
The histopathological analysis revealed a lobular invasive classical type (Indian file carcinoma) (Breast Tumours WHO Classification of Tumours, 5th Edition, Volume 2)), G2 infiltrating the perineural spaces, the skin derma of the areola, and nipple. No metastasis was observed among the 21 examined lymph nodes of the right axilla. The TNM staging of this tumor was pT1b pN1a G2.
Immunohistochemistry was done for ER, PgR, Her2/neu, MIB1, and p53 on FFPE sections, and positive immunoreactivity for ER and PgR (98% and 10%, respectively) was observed.
The proliferation index (MIB-1) revealed 40% immunoreactivity, and Her2/neu was evaluated as a 1+ score (ASCO guidelines 2013). Immunohistochemistry showed negativity for E-cadherin and p53 proteins (Figure 1).
The patient underwent 4 cycles of AC + 12 weekly paclitaxel chemotherapy cycles followed by long- term hormonotherapy with tamoxifen. Clinical follow-up was performed every year. During 4 years of follow-up, radiological follow-up through mammography or ultrasound, no signs of recurrence were observed.
Family History and Germline Analyses
Case 2 was referred to the Genetic Counseling Service of the Department of Pathology at Varese Hospital in 2018 after his LBC diagnosis.
As can be seen in the pedigree (Figure 2B), the patient’s father died at the age of 57 due to a DGC. His paternal uncle died at 58 years of age due to hepatocellular carcinoma without histological diagnosis, and the proband’s mother (II.4) was affected by melanoma at 50 years.
In case 2, the BRCA1 and BRCA2 genes were both analyzed by NGS, to identify single-nucleotide variations and small ins/del, and by MLPA to identify extended deletions/duplications non-detectable by sequencing. The CDH1 gene was also analyzed using both Sanger Sequencing and MLPA approaches. Germline analysis in the proband revealed the pathogenic variant c.2114delT p.(Leu705CysfsTer17) in the heterozygosity (VAF~0.5) CDH1 gene (transcript NM_004360.5), leading to the diagnosis of hereditary diffuse gastric cancer (HDGC) syndrome (ORPHA 26610) (Figure 2), while BRCA1/2 analysis did not reveal any variant. The pathogenic CDH1 variant results in a premature termination codon predicted to cause a truncated or absent E-cadherin protein due to nonsense-mediated decay. The identified pathogenic CDH1 variant is not present in population databases but has been already described in a DGC patient (22). Unfortunately, no other members of this family decided to perform CDH1 genetic testing for cancer prevention. Patient 2 was sent to a reference center for high-risk stomach surveillance for DGC, as suggested by international guidelines (Blair et al.).
Tumor Analysis
The analysis of somatic variants on case 2 revealed the presence of 7 variants in the CDH1 gene (Table 2). Two variants (rs3743674, rs1801552) were in homozygosity (VAF~1.00). The other 5 variants (rs16260, rs45625236, rs2276330, c.2114delT, rs1801026) showed VAFs of 0.17, 0.75, 0.76, 0.67, and 0.68, respectively, indicating an allelic imbalance (AI) and suggesting the presence of an LOH. The methylation analysis of the CDH1 promoter was negative, suggesting that the methylation mechanism is not involved in the carcinogenesis of this LBC.
Discussion
LBC is a distinct type of breast carcinoma characterized by its histopathological features of discohesive cells, representing 5%–15% of invasive breast carcinomas in women. The occurrence of LBC is exceptionally rare in men (16). In our Breast Unit, MBCs were systematically investigated for inherited conditions since 2008–2018 and 37 affected men were referred to Cancer Genetic Counselling.
Genetic analysis revealed an inherited condition involving MBC in 10 out of 37 (27%) cases, including 7 carriers of BRCA1/2 germline pathogenic variants (3 BRCA1 and 4 BRCA2) and 3 carriers of pathogenic variants of other genes including APC, MUTYH, and CDH1. Invasive ductal carcinomas were diagnosed in 35 out of 37 MBCs including sporadic and inherited cases. Only two MBC patients with a lobular histological type (2 out of 37) described here resulted to have an inherited cancer syndrome.
It is well recognized that LBC in women could be involved in both HBOC (ORPHA 145) and HDGC syndromes (ORPHA 26610) (23, 24); however, no data are present in literature about the involvement of male LBC in these inherited conditions.
Here, we described two men who are affected by LBC and carriers of BRCA2 and CDH1 pathogenic variants, respectively. Both of these patients are affected by two distinct inherited syndromes involving LBC, and, to our knowledge, this is the first report demonstrating the strong relationship between an inherited condition and LBC carcinogenesis in men.
Genetic analyses performed on the tumoral DNA of both patients suggested that, in case 1, male LBC arose from BRCA2 inactivation due to the presence of a germline pathogenic variant in the gene, followed by a possible somatic loss of the second allele (LOH) of the gene. Similarly, in case 2, LBC seems due to both the germline pathogenic variant of the CDH1 gene, followed by a possible loss of the second allele in tumoral DNA, which represents the second hit of CDH1 tumor suppressor gene inactivation process. Interestingly, this CDH1-mediated carcinogenesis correlated with a lobular morphological feature in an MBC.
The identification of inherited cancer syndromes through the genetic analysis of the two LBC male patients prompted us to offer a cascade genetic testing in both of these two families, in order to improve cancer prevention using dedicated breast and ovarian cancer surveillance protocol in the family members who are a carrier of the BRCA2 pathogenic variant and in breast and gastric cancer patients’ family members who are a carrier of the CDH1 pathogenic variant in agreement with the prevention guidelines of these syndromes (11, 12, 24). It is noteworthy that the endoscopic surveillance performed in patient 2 allowed to identify an early DGC.
In summary, our data including somatic and germline genetic results demonstrate a strong relationship between male LBC and BRCA2 and CDH1 genes, excluding a sporadic origin of LBC in these men.
Taking into account that male LBC is a very rare cancer, this condition could represent a sentinel cancer for inherited syndrome identification, and, furthermore, the early identification of inherited cancer syndromes could improve cancer prevention both for men and women in these families. In particular, the clinical and genetic history of the LBC patient carrier of the CDH1 pathogenic variant suggests us to include male LBC in the genetic testing criteria and male breast surveillance in local and HDGC guidelines (23).
Data Availability Statement
The data presented in the study are deposited in the Insubria Ethics Committee repository, approved in 2014 and revised in study number 147/2021. Further inquiries can be directed to the corresponding author/s.
Ethics Statement
The studies involving human participants were reviewed and approved by Comitato Etico dell’Insubria. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
IC: genetic counseling, data management, and manuscript writing. GT: somatic DNA analyses. VP: germline analysis. FS and DM: histological diagnosis and case revision. NS: methylation analyses. FR: clinical management. MT: design of the study, data management, and manuscript writing. All the authors of this manuscript directly participated in the planning, execution, and analysis of the study.
Conflict of Interest
Author VP was employed by Cogentech s.r.l. Società Benefit a Socio Unico.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We are grateful to Azzurra Cangiano for its dedication and psycho-oncological support to patients with high cancer risk.
References
2. Fentiman IS. The Biology of Male Breast Cancer. Breast (2018) 38:132–5. doi: 10.1016/j.breast.2018.01.001
3. Deb S, Lakhani SR, Ottini L, Fox SB. The Cancer Genetics and Pathology of Male Breast Cancer. Histopathology (2016) 68:110–8. doi: 10.1111/his.12862
4. Cardoso F, Bartlett JMS, Slaets L, Van Deurzen CHM, Van Leeuwen-Stok E, Porter P, et al. Characterization of Male Breast Cancer: Results of the EORTC 10085/TBCRC/BIG/NABCG International Male Breast Cancer Program. Ann Oncol (2018) 29:405–17. doi: 10.1093/annonc/mdx651
5. Mangone L, Ferrari F, Mancuso P, Carrozzi G, Michiara M, Falcini F, et al. Epidemiology and Biological Characteristics of Male Breast Cancer in Italy. Breast Cancer (2020) 27:724–31. doi: 10.1007/s12282-020-01068-1
6. Carnevali I, Riva C, Chiaravalli AM, Sahnane N, Di Lauro E, Viel A, et al. Inherited Cancer Syndromes in 220 Italian Ovarian Cancer Patients. Cancer Genet (2019) 237:55–62. doi: 10.1016/j.cancergen.2019.06.005
7. Plon SE, Eccles DM, Easton D, Foulkes WD, Genuardi M, Greenblatt MS, et al. Sequence Variant Classification and Reporting: Recommendations for Improving the Interpretation of Cancer Susceptibility Genetic Test Results. Hum Mutat (2008) 29:1282–91. doi: 10.1002/humu.20880
8. Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med (2015) 17:405–24. doi: 10.1038/gim.2015.30
9. Lee K, Krempely K, Roberts ME, Anderson MJ, Carneiro F, Chao E, et al. Specifications of the ACMG/AMP Variant Curation Guidelines for the Analysis of Germline CDH1 Sequence Variants. Hum Mutat (2015) 39:1553–68. doi: 10.1002/humu.23650
10. Consortium E. Evidence-Based Network for the Interpretation of Germline Mutant Allele. Available at: https://www.enigmaconsortium.org/.
11. Baratelli G, Bergonzi S, Bohm S, Bonanni B, Carcangiu ML, Di M, et al. Linee Guida F.O.N.Ca.M Sorveglianza E Trattamento Delle Donne Ad Alto Rischio Per Carcinoma Mammario Familiare. Attualità Senol (2008) 53:28–44.
12. Daly MB, Pal T, Berry MP, Buys SS, Dickson P, Domchek SM, et al. Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2021, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw (2021) 19:77–102. doi: 10.6004/jnccn.2021.0001
13. Gnomad. Available at: https://gnomad.broadinstitute.org.
14. Caputo S, Benboudjema L, Sinilnikova O, Rouleau E, Beroud C, Lidereau R. Description and Analysis of Genetic Variants in French Hereditary Breast and Ovarian Cancer Families Recorded in the UMD-BRCA1/BRCA2 Databases. Nucleic Acids Res (2012) 40:D992–1002. doi: 10.1093/nar/gkr1160
15. Hall MJ, Obeid EI, Schwartz SC, Mantia-Smaldone G, Forman AD, Daly MB. Genetic Testing for Hereditary Cancer Predisposition: BRCA1/2, Lynch Syndrome, and Beyond. Gynecol Oncol (2016) 140:565–74. doi: 10.1016/j.ygyno.2016.01.019
16. Infante M, Duran M, Acedo A, Sanchez-Tapia EM, Diez-Gomez B, Barroso A, et al. The Highly Prevalent BRCA2 Mutation C.2808_2811dedelacaa) Is Located in a Mutational Hotspot and has Multiple Origins. Carcinogenesis (2013) 34:2505–11. doi: 10.1093/carcin/bgt272
17. Walsh T, Casadei S, Lee MK, Pennil CC, Nord AS, Thornton AM, et al. Mutations in 12 Genes for Inherited Ovarian, Fallopian Tube, and Peritoneal Carcinoma Identified by Massively Parallel Sequencing. Proc Natl Acad Sci U.S.A. (2011) 108:18032–7. doi: 10.1073/pnas.1115052108
18. Zhang S, Royer R, Li S, Mclaughlin JR, Rosen B, Risch HA, et al. Frequencies of BRCA1 and BRCA2 Mutations Among 1,342 Unselected Patients With Invasive Ovarian Cancer. Gynecol Oncol (2011) 121:353–7. doi: 10.1016/j.ygyno.2011.01.020
19. De Juan I, Palanca S, Domenech A, Feliubadalo L, Segura A, Osorio A, et al. BRCA1 and BRCA2 Mutations in Males With Familial Breast and Ovarian Cancer Syndrome. Results of a Spanish Multicenter Study. Fam Cancer (2015) 14:505–13. doi: 10.1007/s10689-015-9814-z
20. Kote-Jarai Z, Leongamornlert D, Saunders E, Tymrakiewicz M, Castro E, Mahmud N, et al. BRCA2 Is a Moderate Penetrance Gene Contributing to Young-Onset Prostate Cancer: Implications for Genetic Testing in Prostate Cancer Patients. Br J Cancer (2011) 105:1230–4. doi: 10.1038/bjc.2011.383
21. Edwards SM, Evans DG, Hope Q, Norman AR, Barbachano Y, Bullock S, et al. Prostate Cancer in BRCA2 Germline Mutation Carriers is Associated With Poorer Prognosis. Br J Cancer (2010) 103:918–24. doi: 10.1038/sj.bjc.6605822
22. Tedaldi G, Pirini F, Tebaldi M, Zampiga V, Cangini I, Danesi R, et al. Multigene Panel Testing Increases the Number of Loci Associated With Gastric Cancer Predisposition. Cancers (Basel) (2019) 11(19):1340. doi: 10.3390/cancers11091340
23. Blair VR, Mcleod M, Carneiro F, Coit DG, D'addario JL, Van Dieren JM, et al. Hereditary Diffuse Gastric Cancer: Updated Clinical Practice Guidelines. Lancet Oncol (2020) 21:e386–97. doi: 10.1016/S1470-2045(20)30219-9
Keywords: male lobular breast cancer, inherited cancer syndromes, BRCA, CDH1, case report
Citation: Carnevali I, Tedaldi G, Pensotti V, Sahnane N, Micello D, Rovera F, Sessa F and Tibiletti MG (2022) Case Report: Male Lobular Breast Cancer in Hereditary Cancer Syndromes. Front. Oncol. 12:891426. doi: 10.3389/fonc.2022.891426
Received: 07 March 2022; Accepted: 20 April 2022;
Published: 24 May 2022.
Edited by:
Je-Keun Rhee, Soongsil University, South KoreaReviewed by:
Woochang Lee, University of Ulsan, South KoreaMiguel Angel Alcántara-Ortigoza, National Institute of Pediatrics, Mexico
Copyright © 2022 Carnevali, Tedaldi, Pensotti, Sahnane, Micello, Rovera, Sessa and Tibiletti. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ileana Carnevali, ileana.carnevali@asst-settelaghi.it
 Ileana Carnevali
Ileana Carnevali Gianluca Tedaldi
Gianluca Tedaldi Valeria Pensotti4
Valeria Pensotti4 Fausto Sessa
Fausto Sessa Maria Grazia Tibiletti
Maria Grazia Tibiletti
