- 1National Clinical Research Center for Hematologic Diseases, Jiangsu Institute of Hematology, The First Affiliated Hospital of Soochow University, Suzhou, China
- 2Institute of Blood and Marrow Transplantation, Collaborative Innovation Center of Hematology, Soochow University, Suzhou, China
Chronic eosinophilic leukemia not otherwise specified (CEL-NOS) is classified as Myeloproliterative Neoplasms (MPN) and refers to chronic eosinophilic leukemia with some atypical recurrent genetic evidence(1). A rare fusion of ACSL6-ETV6 was previously identified in patients with the t(5;12) (q31; p13) karyotype(2). Here, we report a case of CEL-NOS with a translocation of t(5;12) (q31; p13) and identify IL3-ETV6 transcription, which has not been identified in hematologic diseases. In this patient, eosinophilia was observed. And compared with CEL-NOS patients without ETV6 fusion, a higher mRNA expression level of IL3 was found. After failing treatment with dasatinib, the patient was given hydroxyurea (HU). Subsequently his white blood cell (WBC) and eosinophils decreased significantly and remained in the normal range until publication. Due to the side effects, treatment with HU was replaced by PEG-interferon (PEG-IFN). What’s more, we summarized the case in our study and 21 patients with the karyotype of t(5; 12) (q31; p13) reported by other groups. It was found that most of them had similar clinical manifestations of eosinophilia and tyrosine kinase inhibitor (TKI) insensitivity. The ectopic mRNA expression of IL3 may be the main cause of eosinophilia, and HU and prednisone acetate (PAT), as well as IFN, were considered treatments for this group.
Introduction
Identification of specific chromosomal changes and recurrent gene translocations is crucial for the treatment and prognosis of patients with hematologic diseases, which is indispensable for the WHO classification of tumors of hematopoietic and lymphoid tissues. Recurrent IL3-IGH rearrangement [t(5;14)(q31.1;q32.1)] has been recognized as an entity of B-cell acute lymphoblastic leukemia, with distinct increased production of interleukin-3 (IL3) and subsequently characteristic reactive eosinophilia (1). It was supposed that ectopic expression of IL3 participated in the multistep process of leukemia (2). Additionally, hematologic malignancies with eosinophilia are often associated with rearrangements of genes such as PDGFRA (4q12), FILIP1-PDGFRB (5q31-33) and FGFR1 (8p11-12) (3). A fusion of ACSL6-ETV6 with chromosome translocation of t(5;12) (q31;p13) was first reported in 1999 (4). In the study by Yahata et al. (5), patients with this genetic abnormality often have accompanying eosinophilia. In our study, a novel IL3-ETV6 was identified in chronic eosinophilic leukemia not otherwise specified (CEL-NOS), and a significant increase in eosinophils and prominent overexpression of IL3 mRNA were found. Moreover, we found that 22 patients with t(5;12) (q31;p13) (including 1 patient in our study and 21 patients reported by other groups) had common clinical characteristics (4–20). It was suggested that eosinophilia or hematologic malignancies with eosinophilia may have more undifferentiated subtypes.
Case Report
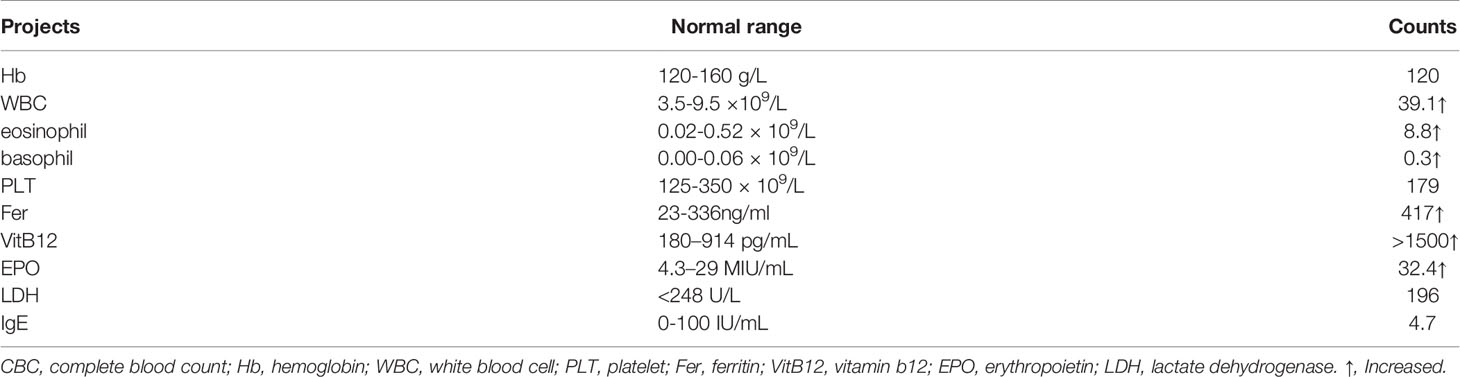
A 38-year-old man was admitted due to elevated levels of white blood cell (WBC) and eosinophils accompanied by splenomegaly on June 4, 2021. He had no history of radiation or drug exposure. The complete blood count and main biochemistry examinations were shown in Table 1. Bone marrow (BM) and peripheral blood (PB) smears showed a high ratio of eosinophils (26% in BM, 47% in PB) (Figures 1A, B), and the ratio of granulocytes: erythrocytes in BM was 13.3:1 (1.28-5.95:1). The percentage of myeloblasts in peripheral blood and bone marrow was 1%, respectively. Additionally, BM biopsy showed an increased number of myeloblasts dominated by eosinophils (>80%). The karyotype analysis revealed 46, XY, t(5;12) (q31; p13) (Figure 1C). BCORL1 (p.S803fs, VAF 19.26%), RUNX1 (p.S291fs, VAF36.05%), KMT2C (p.R894Q, VAF 5.69%), CCND1 (p.276_276del, VAF 5.11%), PTPN1 (1p.Y375C, VAF 27.38%), STAT5B (p.Q368fs, VAF 5.11%) mutations (Supplementary Table 1) were detected by next-generation sequencing covering 161 genes reportedly mutated in hematologic malignancies (Supplementary Table 2). Moreover, the nested polymerase chain reaction (PCR) for BCR-ABL1, FIP1L1-PDGFRα, ETV6-PDGFRα, and ETV6-PDGFRβ fusion genes, which are commonly detected in eosinophilia or hematologic malignancies with eosinophilia, were all negative. Supplementary medical history showed that the patient had skin itching and rash for more than 10 years.
Figure 1 (A) A significantly increased eosinophil ratio was shown on a bone marrow (BM) smear (Wright–Giemsa stained, x 100). (B) Significantly increased eosinophilia was observed on a peripheral blood (PB) smear (× 100), the arrows delineated eosinophils. (C) Karyotypic analysis of BM showed the translocation of t(5; 12) (q31; p13) (delineated by arrows). (D) RNA-sequencing analysis indicated the IL3-ETV6 fusion. (E) Circos plot displaying the interconnectivity between IL3 and ETV6. (F) Amplified IL3-ETV6 transcripts by RT–PCR. Control: water. The predicted product was 300bp, and the patient’s PCR product was indicated by red arrow. (G) Sanger sequencing of the PCR product (IL3-ETV6). (H) IL3 mRNA was obviously more highly expressed than in other eosinophilia patients with normal karyotypes. Control: CEL-NOS patients with normal karyotype, FPKM: fragments per kilobase of transcript sequence per millions base pairs sequenced. (I) Fluctuation of the patient’s peripheral white blood cell count (WBC, ×109/L) and eosinophil ratio (EOS, %) during treatment.
For further examination of the molecular abnormality, RNA sequencing using HiSeq (Illumina Inc, San Diego, CA, USA) was performed, which led to the identification of a novel fused mRNA, IL3-ETV6 (Figures 1D, E). In this translocation, exons 3 to 5 of IL3 on chromosome 5 were spliced, inverted and fused with exons 3 to 8 of ETV6 on chromosome 12. In addition, a transcript of GATA2-SOCS2 (exon 1 of GATA2 (5’UTR) fusing with exon 2 to 3 of SOCS2 (5’UTR)) was also detected (Supplementary Figure 1). Reverse transcription PCR (RT–PCR) was performed to confirm the IL3-ETV6 fusion by the following primers: forward (at IL3 intron 2-3), 5’AAATCA CAGAGACCCCAGC3’ and reverse (at ETV6 exon 3), 5’AAGG AGTTCATAGAGCACATCA3’, and a product of approximately 300 bp was observed as we predicted (Figure 1F). Sanger sequencing analysis of this product showed the detailed breakpoints and fusion segments (Figure 1G). The raw data can be downloaded in the Supplement 1. In addition, ectopic high-level expression of IL3 mRNA was also observed in patients with ACSL6-ETV6 (8, 11). Therefore, we reanalyzed the RNA-sequencing data. Compared with other CEL-NOS patients with normal karyotype, significantly higher (more than 700-fold change) fragments per kilobase of transcript sequence per million base pairs of IL3 mRNA in this patient was found (Figure 1H). Meanwhile, high mRNA expression levels of GATA2 and SOCS2 (more than 40-fold change and 30-fold change respectively) were also observed (Supplementary Figure 2).
The dynamic changes in WBC and eosinophil in this patient during treatment are shown (Figure 1I). Owing to the success of tyrosine kinase inhibitor (TKI) in CEL-NOS reported in previous cases, the patient was first treated with dasatinib 250 mg/day for 20 days, but the high WBC and eosinophil counts were maintained. Then, treatments with hydroxyurea (HU) 150 mg/day and prednisone acetate (PAT) 30 mg/day for 20 days were given, and the WBC count decreased from 38.7×109/L to 18×109/L, and eosinophil count decreased from 15.2×109/L to 2.29×109/L. However, due to the fever and gastrointestinal discomfort associated with HU, PEG-interferon (PEG-IFN) was used as an alternative. His WBC and eosinophil counts in peripheral blood soon returned to the normal range. The patient was then followed up for more than seven months, and the WBC and eosinophil counts remained stable by the time of this report.
Due to the rare incidence and limited description, we summarized the case in our study and 21 patients with the karyotype of t(5; 12) (q31; p13) reported by other groups from 1988 to 2021 (4–20) (Table 2). According to these reports, 19 cases were classified as MDS/MPD by the 2016 WHO criteria (21), 2 were AML and 1 was T-ALL. Most of these patients were male (17/22), and most of them had accompanying eosinophilia (normal range 0.4%-8%) (14/18). Unlike TKI treatment, which failed in these patients (4/4), HU with/without PAT or PEG-IFN treatment hydroxyurea or interferon treatment led to hematologic and cytogenetic remissions (12/16).
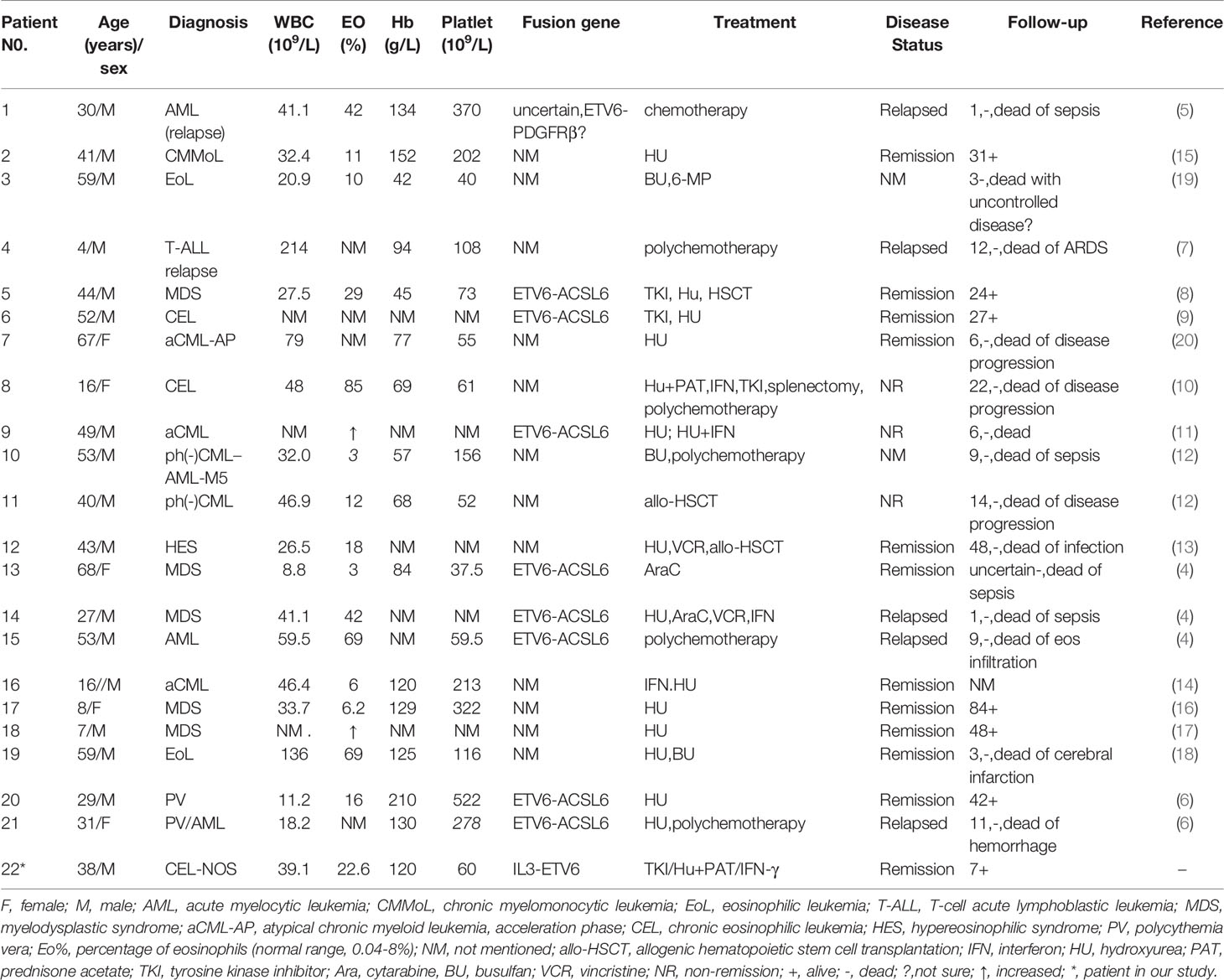
Table 2 Clinical features of 21 patients with t(5; 12) (q31; p13) with cases reported in the literature and the 1 patient in our center.
Discussion
CEL-NOS is a rare disease with eosinophilia and nonspecific clonal cytogenetic abnormalities. Patients diagnosed with CEL-NOS have the clinical manifestations of increased eosinophils and organ damage, with the risk of transformation into hematologic malignancies (3). However, the genetic abnormality of this disease has not been fully delineated, which will be essential for precise diagnosis and treatment.
Due to the rare incidence and limited description, we summarized all the reported cases with the karyotype of t(5; 12) (q31; p13). A fusion of ACSL6-ETV6 was previously identified in several patients with t(5;12) (q31; p13) (8, 9, 12, 14, 20). ACSL6, also named as ACS2, is considered as a suppressor gene in leukemia and involved in the metabolic process of leukemia cells (22). ETV6 on chromosome 12 plays a pivotal role in the regulation of myeloid hematopoiesis (23). It encodes a transcriptional repressor that plays a critical role in hematopoiesis and maintains HSCs. It is believed that the rearrangement of ETV6 and frequent loss of ETV6 expression could be genetic events that induce leukemia (24).
In our study, we did not observe ACSL6-ETV6 but a novel IL3-ETV6 transcript. 5q31-33 is a common fragile fragment that includes tissue-derived growth factor receptor B (PDGFRB), acyl-CoA synthetase long chain family member 6 (ACSL6), and interleukin 3 (IL3). In the chromatin 5q31, IL3 was adjacent to ACSL6 by 50KB. ACSL6-ETV6 was analyzed by FISH probes (a resolution at least 100kB-150KB) in previous reports (4, 6, 9, 11). Therefore, IL3 and ACSL6 could not be distinguished by FISH analysis, and we suspected that some patients expressing IL3-ETV6 were ignored due to technology limitations.
According to our description, the IL3 (exon 3-5) was reversely fused to ETV6 (exon 3-8), which resulted in the loss of the start codon for both genes. Out-of-frame fusions such as ACSL6-ETV6, IL3-IGH may be involved in pathogenesis was reported previously (1, 11). In all these cases, abnormally high mRNA expression of IL3 might be the molecular characteristic of t(5;12) (q31; p13). In our study, we found high expression level of IL3 mRNA by RNA sequencing. IL3 can promote proliferation and differentiation of eosinophils (11). Additionally, CD123, an IL3 receptor, was hypothesized to induce JAK-STAT-dependent cell survival and proliferation in an autocrine manner (25). IL3 rearrangement not only promotes the proliferation of eosinophils but also influences malignant blasts (1, 25). Thus, we speculated that high expression of IL3 may be a consequence of IL3-ETV6 fusion and may play a role in the pathogenesis. But it needs to be further confirmed.
In this case, exon 1 of GATA2 (5’UTR) fused with exon 2 of SOCS2 (5’UTR). We also found there were high mRNA expression levels of GATA2 and SOCS2. However, there were few literatures on the functions of GATA2 or SOCS2 in eosinophilia. We cannot find more evidence for the correlation of GATA2-SOCS2 with this disease. In summary, we considered that IL3-ETV6 is more likely to cause the progression of eosinophilia but not GATA2-SOCS2.
Similar clinical characteristics of eosinophilia and high mRNA expression levels of IL3 were found in IL3-ETV6 and ACSL6-ETV6 cases (8, 11). Thus, we believe that eosinophilia associated with IL3 high expression could be treated in a similar manner.
Although most patients with the karyotype of t(5; 12) (q31; p13) responded well to hydroxyurea and interferon, there were still risks of hematologic malignant progression and recurrence. Therefore, long-term follow-up should be conducted for this group. Some patients can achieve remission after chemotherapy or transplantation (13, 20). Allogeneic transplantation and intensive chemotherapy should be considered for patients with malignant transformation. In addition, JAK2 or CD123 might be a therapeutic target for some IL3 mRNA overexpressing and eosinophilia patients (25, 26). In study of Pellier et al. (16), an IL5 monoclonal antibody also effectively inhibited the proliferation of eosinophils. Recently, an adeno-associated virus (AAV) coding for an anti-eosinophil monoclonal antibody was shown to persistently suppress eosinophil numbers in blood, thus reducing eosinophil tissue invasion and organ dysfunction in a murine model of CEL-NOS (27). However, targeted therapy experience for patients with the karyotype of t(5; 12) (q31; p13), especially people with atypical recurrent molecular abnormalities such as IL3-ETV6, is still lacking.
In conclusion, we reported a novel IL3-ETV6 fusion in CEL-NOS with a high level of IL3 mRNA expression. RNA sequencing is valuable to identify some occult genetic abnormalities in eosinophilia and to help further understand the disease. By summarizing the previous literatures, the similar clinical characteristics and unfavorable outcomes of these patients with t(5; 12) (q31; p13) were found, which may indicate a novel subtype of hematologic malignancy.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA812744.
Ethics Statement
The studies involving human participants were reviewed and approved by the First Affiliated Hospital of Soochow University. The patients/participants provided their written informed consent to participate in this study. Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author Contributions
Contribution: TL and JA were the principal investigators. MW, CZ, and YZ performed most of the experiments. SC, QW and YX performed clinical analysis. CZ, TL, and JA wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the Excellent Youth Science Fund of Jiangsu Province (BK20211553), the Natural Science Foundation of China (81700139, 82070187, 81870120 and 82000157), the Key R&D Program of Jiangsu Province (BE2019655), the Translational Research Grant of NCRCH (2021ZKMB01) and the Natural Science Fund of Jiangsu Province (BK20170360, BK20190173).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2022.887945/full#supplementary-material
Supplement 1 | Raw sequencing data from patient’s RNA showed the IL3-ETV6 fusion.
References
1. Guenzel AJ, Smadbeck JB, Golden CL, Williamson CM, Benevides Demasi JC, Vasmatzis G, et al. Clinical Utility of Next Generation Sequencing to Detect IGH/IL3 Rearrangements [T(5;14)(Q31.1;Q32.1)] in B-Lymphoblastic Leukemia/Lymphoma. Ann Diagn Pathol (2021) 53:151761. doi: 10.1016/j.anndiagpath.2021.151761
2. Kobayashi K, Mizuta S, Yamane N, Ueno H, Yoshida K, Kato I, et al. Paraneoplastic Hypereosinophilic Syndrome Associated With IL3-IgH Positive Acute Lymphoblastic Leukemia. Pediatr Blood Cancer (2019) 66:e27449. doi: 10.1002/pbc.27449
3. Shomali W, Gotlib J. World Health Organization-Defined Eosinophilic Disorders: 2019 Update on Diagnosis, Risk Stratification, and Management. Am J Hematol (2019) 94:1149–67. doi: 10.1002/ajh.25617
4. Yagasaki F, Jinnai I, Yoshida S, Yokoyama Y, Matsuda A, Kusumoto S, et al. Fusion of TEL/ETV6 to a Novel ACS2 in Myelodysplastic Syndrome and Acute Myelogenous Leukemia With T(5;12)(Q31;P13). Genes Chromosomes Cancer (1999) 26:192–202. doi: 10.1002/(sici)1098-2264(199911)26:3<192:aid-gcc2<3.0.co;2-e
5. Yahata N, Ohyashiki K, Ohyashiki JH, Kimura Y, Miyazawa K, Kodama A, et al. Late Appearance of T(5;12)(Q31;P12) in Acute Myeloid Leukemia Associated With Eosinophilia. Cancer Genet Cytogenet (1998) 107:147–50. doi: 10.1016/s0165-4608(98)00102-2
6. Murati A, Adélaïde J, Gelsi-Boyer V, Etienne A, Rémy V, Fezoui H, et al. T(5;12)(Q23-31;P13) With ETV6-ACSL6 Gene Fusion in Polycythemia Vera. Leukemia (2006) 20:1175–8. doi: 10.1038/sj.leu.2404194
7. Kuru D, Yılmaz F, Berra SG, Canpolat C, Hacıhanefioğlu S, Tarkan Argüden Y, et al. A Childhood Acute Lymphoblastic Leukemia (ALL) Case With T(3;17)(Q23;P13),T(5;12)(Q31;P13),Inv(11)(P15q12). Turk J Haematol (2008) 25:152–4.
8. Katsura Y, Suzukawa K, Nanmoku T, Nemoto N, Machino T, Obara N, et al. Myelodysplastic Syndrome Accompanied by Basophilia and Eosinophilia With T(5;12)(Q31;P13). Cancer Genet Cytogenet (2007) 178:85–8. doi: 10.1016/j.cancergencyto.2007.05.020
9. Su RJ, Jonas BA, Welborn J, Gregg JP, Chen M. Chronic Eosinophilic Leukemia, NOS With T(5;12)(Q31;P13)/ETV6-ACSL6 Gene Fusion: A Novel Variant of Myeloid Proliferative Neoplasm With Eosinophilia. Hum Pathol (N Y) (2016) 5:6–9. doi: 10.1016/j.ehpc.2015.10.001
10. Luo RM, Wu SL, Tong CR, Qiu JY, Wu P, Lu DP. [FIP1L1/PDGFRalpha Fusion Gene-Negative Chronic Eosinophilic Leukemia With T(5;12)(Q31;P13): A Case Report and Review of Literatures]. Zhonghua Nei Ke Za Zhi (2008) 47:919–22. doi: 10.3321/j.issn:0578-1426.2008.11.013.
11. Cools J, Mentens N, Odero MD, Peeters P, Wlodarska I, Delforge M, et al. Evidence for Position Effects as a Variant ETV6-Mediated Leukemogenic Mechanism in Myeloid Leukemias With a T(4;12)(Q11-Q12;P13) or T(5;12)(Q31;P13). Blood (2002) 99:1776–84. doi: 10.1182/blood.v99.5.1776
12. Wessels JW, Fibbe WE, van der Keur D, Landegent JE, van der Plas DC, den Ottolander GJ, et al. T(5;12)(Q31;P12). A Clinical Entity With Features of Both Myeloid Leukemia and Chronic Myelomonocytic Leukemia. Cancer Genet Cytogenet (1993) 65:7–11. doi: 10.1016/0165-4608(93)90051-m
13. Juvonen E, Volin L, Koponen A, Ruutu T. Allogeneic Blood Stem Cell Transplantation Following non-Myeloablative Conditioning for Hypereosinophilic Syndrome. Bone Marrow Transplant (2002) 29:457–8. doi: 10.1038/sj.bmt.1703379
14. Berkowicz M, Rosner E, Rechavi G, Mamon Z, Neuman Y, Ben-Bassat I, et al. Atypical Chronic Myelomonocytic Leukemia With Eosinophilia and Translocation (5;12). A New Association. Cancer Genet Cytogenet (1991) 51:277–8. doi: 10.1016/0165-4608(91)90142-h
15. Srivastava A, Boswell HS, Heerema NA, Nahreini P, Lauer RC, Antony AC, et al. KRAS2 Oncogene Overexpression in Myelodysplastic Syndrome With Translocation 5;12. Cancer Genet Cytogenet (1988) 35:61–71. doi: 10.1016/0165-4608(88)90123-9
16. Pellier I, Le Moine PJ, Rialland X, François S, Baranger L, Blanchet O, et al. Myelodysplastic Syndrome With T(5;12)(Q31;P12-P13) and Eosinophilia: A Pediatric Case With Review of Literature. J Pediatr Hematol Oncol (1996) 18:285–8. doi: 10.1097/00043426-199608000-00010
17. Jani K, Kempski HM, Reeves BR. A Case of Myelodysplasia With Eosinophilia Having a Translocation T(5;12) (Q31;Q13) Restricted to Myeloid Cells But Not Involving Eosinophils. Br J Haematol (1994) 87:57–60. doi: 10.1111/j.1365-2141.1994.tb04870.x
18. Keene P, Mendelow B, Pinto MR, Bezwoda W, MacDougall L, Falkson G, et al. Abnormalities of Chromosome 12p13 and Malignant Proliferation of Eosinophils: A Nonrandom Association. Br J Haematol (1987) 67:25–31. doi: 10.1111/j.1365-2141.1987.tb02291.x
19. Yates P, Potter MN. Eosinophilic Leukaemia With an Abnormality of 5q31, the Site of the IL-5 Gene. Clin Lab Haematol (1991) 13:211–5. doi: 10.1111/j.1365-2257.1991.tb00271.x
20. Qian J, Wang QR, Liu J, Jiang SH, Ni XQ, Lin ZH, et al. Establishment and Characterization of a Rare Atypical Chronic Myeloid Leukemia Cell Line NT-1. Leuk Res (2014) 38:1111–6. doi: 10.1016/j.leukres.2014.06.008
21. Barbui T, Thiele J, Gisslinger H, Kvasnicka HM, Vannucchi AM, Guglielmelli P, et al. The 2016 WHO Classification and Diagnostic Criteria for Myeloproliferative Neoplasms: Document Summary and in-Depth Discussion. Blood Cancer J (2018) 8:15. doi: 10.1038/s41408-018-0054-y
22. Chen WC, Wang CY, Hung YH, Weng TY, Yen MC, Lai MD. Systematic Analysis of Gene Expression Alterations and Clinical Outcomes for Long-Chain Acyl-Coenzyme A Synthetase Family in Cancer. PloS One (2016) 11:e0155660. doi: 10.1371/journal.pone.0155660
23. Hock H, Shimamura A. ETV6 in Hematopoiesis and Leukemia Predisposition. Semin Hematol (2017) 54:98–104. doi: 10.1053/j.seminhematol.2017.04.005
24. Jakobczyk H, Jiang Y, Debaize L, Soubise B, Avner S, Sérandour AA, et al. ETV6-RUNX1 and RUNX1 Directly Regulate RAG1 Expression: One More Step in the Understanding of Childhood B-Cell Acute Lymphoblastic Leukemia Leukemogenesis. Leukemia (2022) 36:549–54. doi: 10.1038/s41375-021-01409-9
25. Fournier B, Balducci E, Duployez N, Clappier E, Cuccuini W, Arfeuille C, et al. B-ALL With T(5;14)(Q31;Q32); IGH-IL3 Rearrangement and Eosinophilia: A Comprehensive Analysis of a Peculiar IGH-Rearranged B-ALL. Front Oncol (2019) 9:1374. doi: 10.3389/fonc.2019.01374
26. Testa U, Pelosi E, Castelli G. CD123 as a Therapeutic Target in the Treatment of Hematological Malignancies. Cancers (Basel) (2019) 11:1358. doi: 10.3390/cancers11091358
Keywords: case report, fusion gene, t(5;12)(q31;p13), IL3-ETV6, CEL-NOS, eosinophilia
Citation: Zhao C, Wang M, Zhan Y, Xu Y, Chen S, Wang Q, An J and Liu T (2022) A Novel IL3-ETV6 Fusion in Chronic Eosinophilic Leukemia Not Otherwise Specified With t(5; 12) (q31; p13): A Case Report and Literature Review. Front. Oncol. 12:887945. doi: 10.3389/fonc.2022.887945
Received: 02 March 2022; Accepted: 26 April 2022;
Published: 07 June 2022.
Edited by:
Simona Soverini, University of Bologna, ItalyReviewed by:
Luisa Anelli, University of Bari Aldo Moro, ItalyLaura N. Eadie, South Australian Research and Development Institute, Australia
Copyright © 2022 Zhao, Wang, Zhan, Xu, Chen, Wang, An and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qinrong Wang, wangqr001@126.com; Jingnan An, ajn-yspa@hotmail.com; Tianhui Liu, liutianhui@suda.edu.cn; Yang Xu, xuyang1020@126.com
†These authors have contributed equally to this work
 Cenzhu Zhao
Cenzhu Zhao Man Wang1†
Man Wang1† Yang Xu
Yang Xu Suning Chen
Suning Chen Qinrong Wang
Qinrong Wang Tianhui Liu
Tianhui Liu