- 1Department of Gastrointestinal Surgery, The First Affiliated Hospital of Wannan Medical College, Yijishan Hospital of Wannan Medical College, Wuhu, Anhui, China
- 2Key Laboratory of Non-coding RNA Transformation Research of Anhui Higher Education Institution, Wuhu, Anhui, China
- 3Non-coding RNA Research Center of Wannan Medical College, Yijishan Hospital, Wuhu, Anhui, China
- 4Department of Clinical Medicine, Clinical College of Anhui Medical University, Hefei, Anhui, China
Research on noncoding ribonucleic acids (ncRNAs) is mostly and broadly focused on microRNAs (miRNAs), cyclic RNAs (circRNAs), and long ncRNAs (lncRNAs), which have been confirmed to play important roles in tumor cell proliferation, invasion, and migration. Specifically, recent studies have shown that ncRNAs contribute to tumorigenesis and tumor development by mediating changes in enzymes related to lipid metabolism. The purpose of this review is to discuss the characterized ncRNAs involved in the lipid metabolism of tumors to highlight ncRNA-mediated lipid metabolism-related enzyme expression in malignant tumors and its importance to tumor development. In this review, we describe the types of ncRNA and the mechanism of tumor lipid metabolism and analyze the important role of ncRNA in tumor lipid metabolism and its future prospects from the perspectives of ncRNA biological function and lipid metabolic enzyme classification. However, several critical issues still need to be resolved. Because ncRNAs can affect tumor processes by regulating lipid metabolism enzymes, in the future, we can study the unique role of ncRNAs from four aspects: disease prevention, detection, diagnosis, and treatment. Therefore, in the future, the development of ncRNA-targeted therapy will become a hot direction and shoulder a major task in the medical field.
Introduction
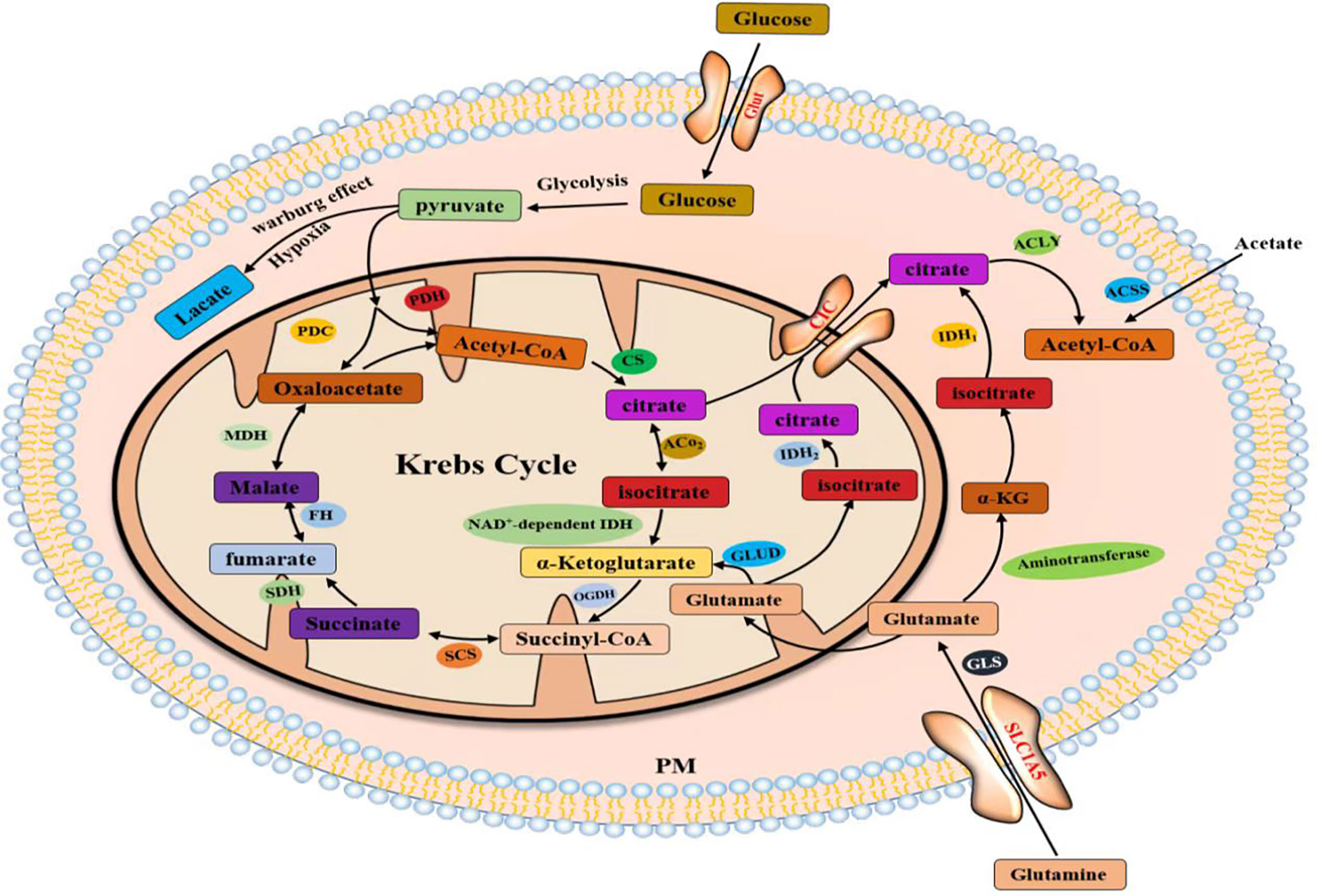
Lipids are hydrophobic molecules and include sterols, monoglycerides (MGs), diglycerides (DGs), triglycerides (TGs), phospholipids, and glycolipids. Lipids participate in the formation of membrane lipid bilayers and membrane-related protein structures. Phospholipids, glycolipids, and cholesterol are the main components of biofilms and significantly affect the fluidity of biofilms. Cholesterol is also a substrate for the synthesis of fat-soluble vitamins and steroid hormones (1). Lipids constitute the basic structure of cell membranes, and lipid anabolism is highly upregulated in human malignancies to meet the increased demand for membrane biosynthesis (2–4). Mammalian cells acquire lipids through two mechanisms, de novo synthesis and uptake from the environment. Acetyl-coA, as an important substrate for lipid synthesis and cholesterol synthesis, mainly comes from three sources: (1) glucose forms acetyl-CoA catalyzed by ATP citrate lyase (ACLY) through glycolysis and the Krebs cycle in mitochondria, (2) acetate is catalyzed in the cytoplasm by acetyl-CoA synthetase (ACSS) to produce acetyl-CoA, and (3) glutamate is produced by a series of reactions in the cytoplasm or mitochondria to citrate, which is catalyzed by ACLY to acetyl-CoA. Lipid uptake, storage, and fat formation are evident in many cancer types and contribute to rapid tumor growth. Lipid metabolism reprogramming is a newly discovered marker of malignant tumors (5–9).
With the continuous development of new research fields, researchers have realized that lipid metabolism is not just a self-regulation network. Technical developments in the human genome have revealed that noncoding RNAs are a new class of regulators in lipid metabolism (10, 11). Noncoding RNA is involved in tumor lipid metabolism through three ways: (1) as endogenous competing RNAs (ceRNAs) to regulate downstream mRNA stability, (2) miRNAs directly target mRNA of lipid-metabolizing enzymes, and (3) noncoding RNAs act as molecular decoys for RNA-binding proteins (RBPs). In this review, we have highlighted the regulating effects and mechanisms of noncoding RNAs on lipid metabolism and signaling, which may help advance the comprehension of the detailed modulatory networks of lipid metabolism and afford a better theoretical foundation for clinical diagnosis and treatment of metabolic diseases.
Noncoding RNAs in cancer
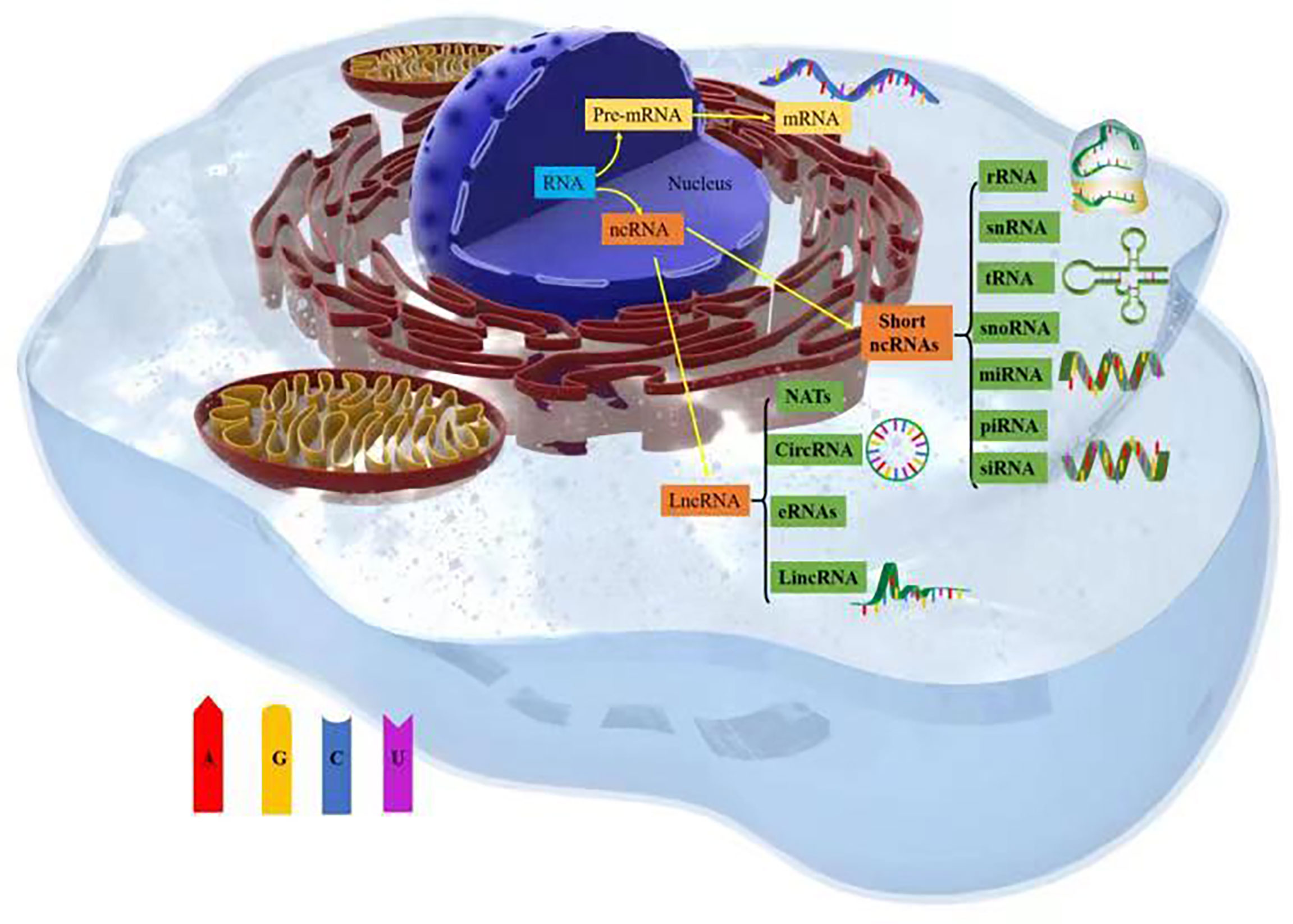
Thousands of unique noncoding ribonucleic acids (ncRNAs) are present within cells, and they mediate functional molecules that regulate cellular processes, including chromatin remodeling, transcription, posttranscriptional modification, and signal transduction (12). ncRNAs include a variety of RNA types, such as short ncRNAs (sncRNAs) and long ncRNAs (lncRNAs). sncRNAs include ribosomal RNAs (rRNAs), which are involved in mRNA translation; transfer RNAs (tRNAs); spliced small nuclear RNAs (snRNAs); small-interfering RNAs (siRNAs); small nucleolar RNAs (snoRNAs), which are involved in rRNA modification; and RNAs involved in targeted translation and inhibition of gene silencing (microRNAs [miRNAs]) and Piwi-interacting RNAs (piRNAs) (13).
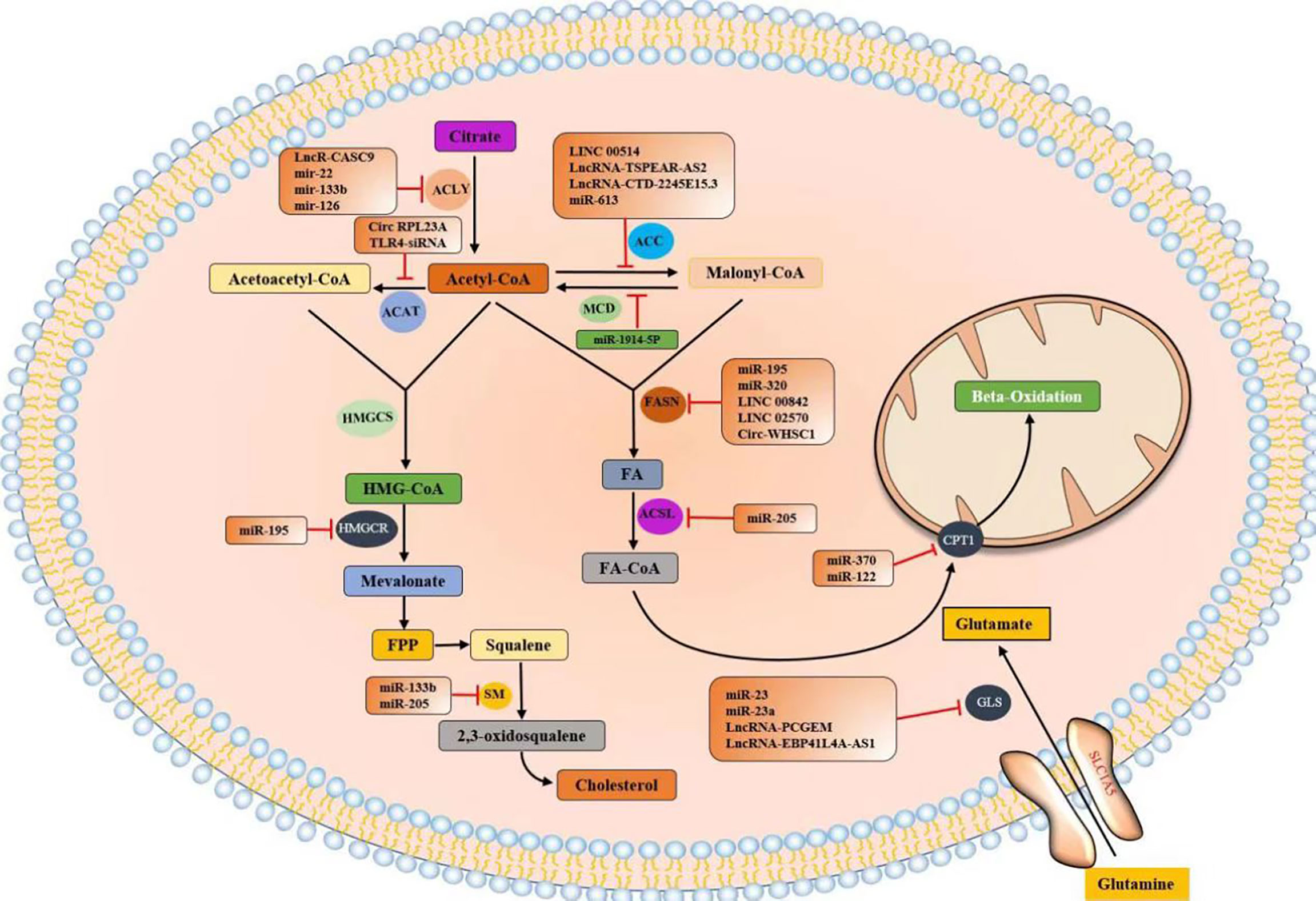
lncRNAs constitute a heterogeneous class of RNAs, including long intergenic ncRNAs (lincRNAs), circular RNAs (circRNAs), natural antisense transcripts (NATs), and enhancer RNAs (eRNAs) (14). These widespread ncRNAs are produced by the transcription of mammalian genomes and make up the majority of the transcribed genome, and only 1–2% of ncRNA transcripts are transcribed into proteins (15, 16). The roles played by miRNAs in cancer have been confirmed (17, 18), with thousands of papers reporting on the important roles that miRNAs play in cancer development and progression. For example, the degree to which miRNAs are conserved among species, expressed in different tissues and cell types, and participate in almost all biological processes (including cell cycle progression, growth, apoptosis, differentiation, and stress responses) and their ability to fine-tune gene expression by targeting multiple molecules have all been discussed (19). Notably, miR-130 and miR-494 can regulate cell survival and TNF-related apoptosis-induced ligand (TRAIL)–mediated therapy resistance in non-small-cell lung cancer (NSCLC) cell lines (20, 21). Therefore, ncRNAs play important roles in tumor cell metabolism (Figure 1).
Lipid metabolism in tumors
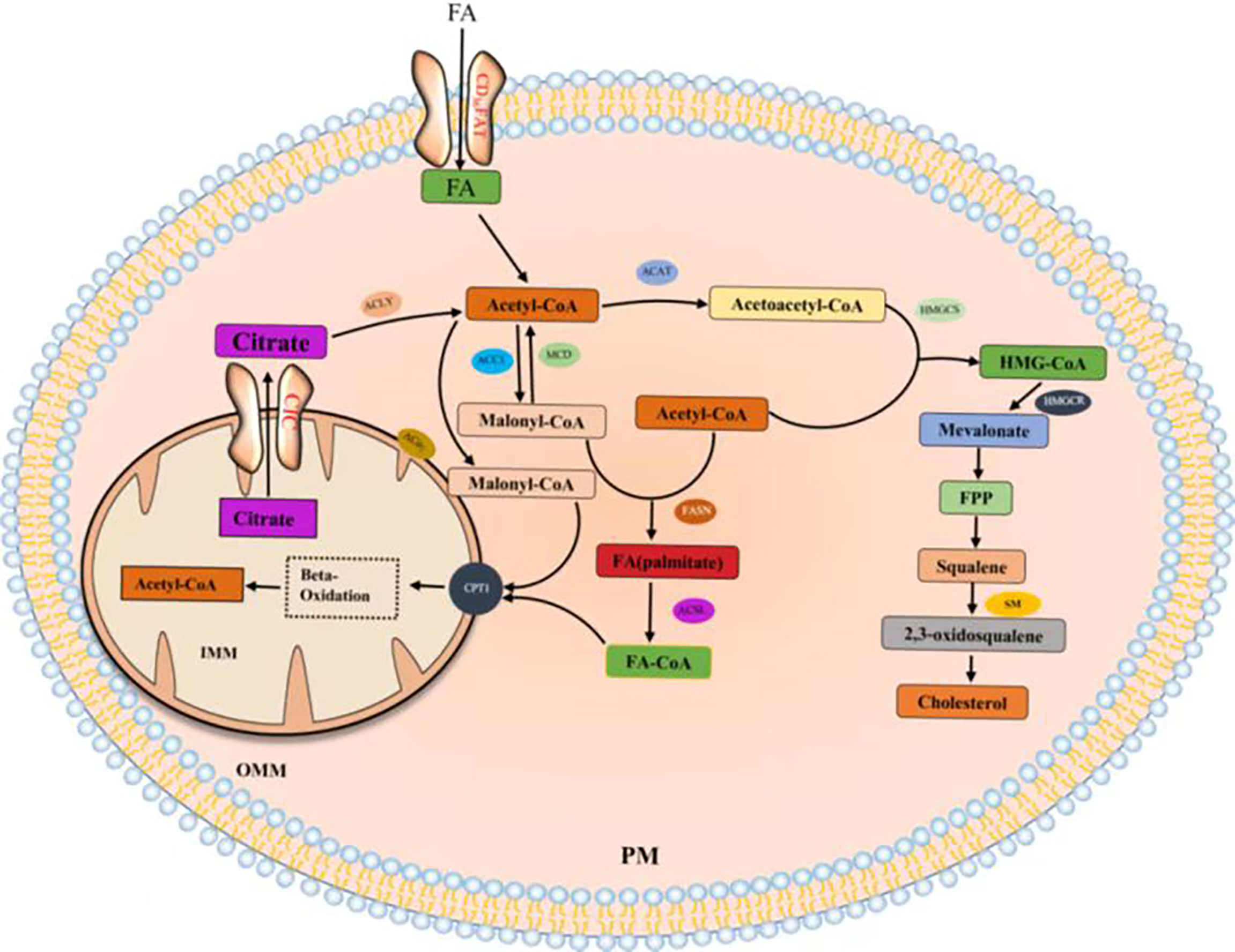
Citrate produced by glucose through glycolysis and Krebs cycle is mediated by a citrate carrier (CIC) into the cytoplasm and catalyzed by ATP-citrate lyase (ACLY) to generate acetyl-CoA (22, 23). Cytoplasmic acetyl-CoA can be extracted from acetic acid by acetyl-CoA synthase (ACSS) (24). Acetic acid (HAc) diffuses freely across cell membranes (25); in particular, under hypoxia, tumor cells capture acetate as an alternative carbon source (26–30). Citrate is obtained from glutamine, particularly from its carbon molecules, which contribute to citrate production (31, 32). Glutamine is transported into cells by a number of transporters, such as the extensively studied solute carrier family 1 neutral amino acid transporter member 5 (SLC1A5) (33). The catabolism of glutamine to glutamate is initiated by mitochondrial glutaminase (GLS). Glutamate is catalyzed by aminotransferase to produce α-ketoglutaric acid (α-kg) (34) and generate acetyl-CoA through a multistep enzymatic reaction or enter the mitochondrial tricarboxylic acid cycle to generate citrate, which is transported out of mitochondria by a CIC and catalyzed to produce acetyl-CoA through ACLY (Figure 2). Cytoplasmic acetyl-CoA is a substrate in fatty acid synthase (FAS). The interconversion of acetyl-CoA and malonyl-CoA in the cytoplasm is catalyzed by acetyl-CoA carboxylase (ACC) and malonyl-CoA decarboxylase (MCD) (35). ACC catalyzes the rate-limiting step in the fatty acid (FA) pathway. To enter a bioactive cell, an FA must be activated by acyl-CoA (FA-CoA) ligase (ACSL) to produce aliphatic FA-CoA (36, 37). FA-oxidation (FAO) is inhibited by the entry of FA-CoA into mitochondria, which is regulated by carnitine palmityl transferase 1 (CPT1) (37). Cholesterol synthesis begins with the condensation of two acetyl-CoA molecules mediated by cholesteryl transferase (ACAT) to form acetyl-CoA. Acetyl-acetyl-CoA is further condensed with a third acetyl-CoA molecule through 3-hydroxy-3-methylglutaryl-CoA synthase (HMGCS) to form 3-hydroxy-3-methylglutaryl-CoA (HMG-CoA). HMG-CoA reductase (HMGCR) is a glycoprotein located in the ER and catalyzes the rate-limiting step in cholesterol synthesis (38, 39). HMG-CoA is oxidized to 2,3-oxidized squalene by squalene monooxygenase (SQLE) through complex reactions. SQLE is the first oxidative step in cholesterol synthesis (39–41) (42–44) (Figure 3).
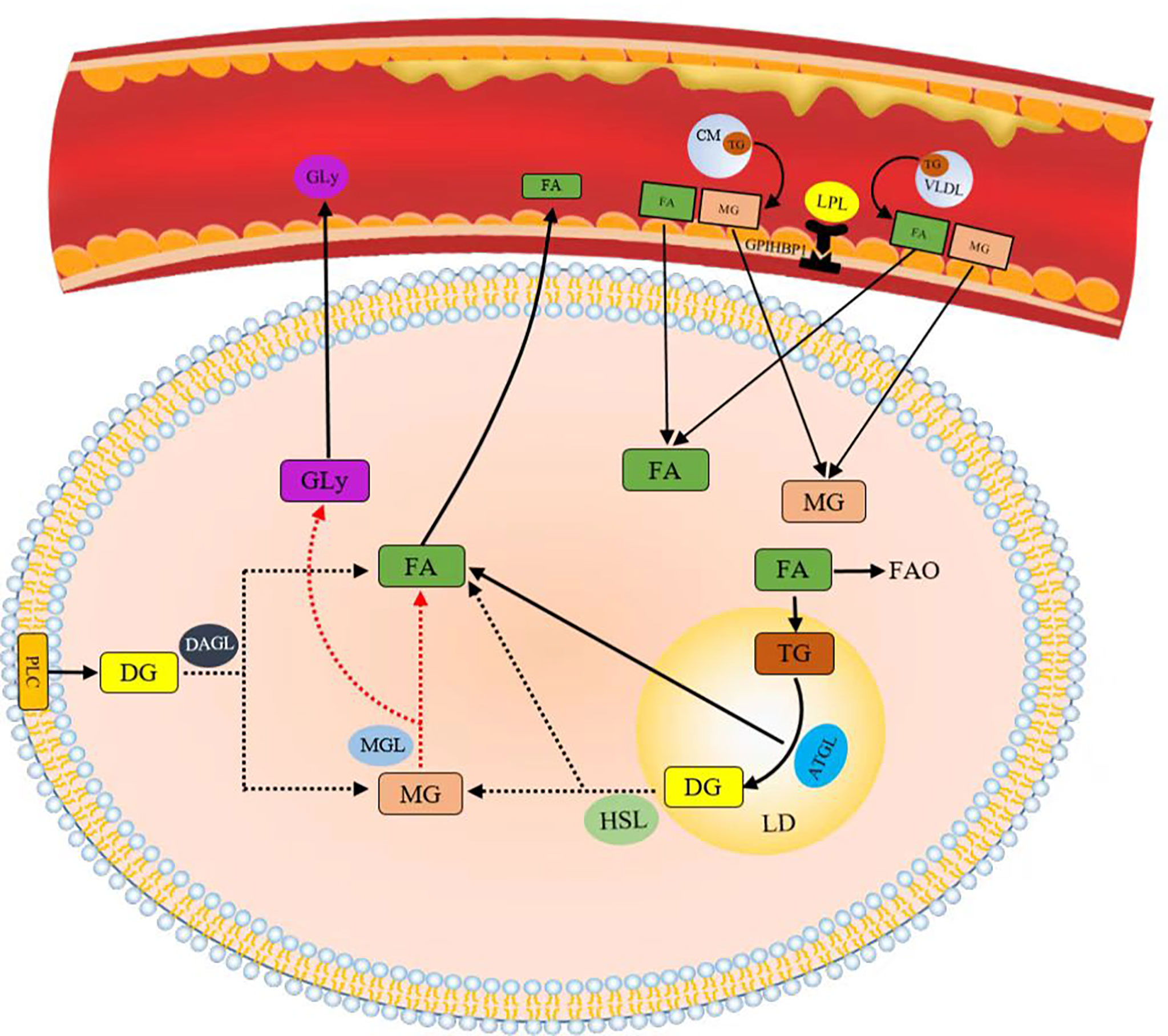
FA can be derived by intravascular and intracellular lipid decomposition. Lipoprotein lipase (LPL) is the rate-limiting component in plasma TG clearance and FA uptake by tissues (45). FA released by cyclic triTG hydrolysis can be absorbed by cells through CD36, a transmembrane channel protein with affinity for exogenous FA (46, 47). TG lipolysis is continuously performed by adipose triglyceride lipase (ATGL), hormone-sensitive lipase (HSL), and monoacylglycerol lipase (MGL) (48). ATGL plays an important catalytic role in the hydrolysis of TG to generate glycerol diesters (DGs) and FA in adipose and non-adipose tissues (49). HSL is the rate-limiting enzyme in the decomposition of DGs in adipose tissues (50). Phospholipase C (PLC) and diacylglycerol lipase (DGL) hydrolyze glycerol phospholipids to produce monoglycerol (MG) (Figure 4). Lipids constitute the basic structure of cell membranes. Phospholipids, glycolipids, and cholesterol are the main components of biofilms and significantly affect the fluidity of biofilms. At the cellular level, cholesterol in the cell membrane increases bilayer firmness and enhances its impermeability to water and ions. In addition to membrane formation, lipids are true second messengers that bind to specific sites to control channels and transporter gating. They also play a nonspecific role by changing the physical environment of channels and transporters, especially the protein–membrane interface (51). For example, phosphatidylinositol (4,5) diphosphate (PIP2) regulates K channels and changes ion channel permeability in sympathetic neurons (42). Lipid rafts are produced in the glycolipid-rich apical membrane of epithelial cells (43). They play an important role in post-Golgi transport, endocytosis, signal transduction, and many other membrane functions (44).
Noncoding RNAs play different biological roles
Noncoding RNAs in the cytoplasm can regulate mRNA stability and translation efficiency and interfere with posttranslational modification of proteins. LncRNAs and circRNAs mainly perform two functions. First, they interact with RNA, including regulating mRNA stability as an endogenous rival RNA (ceRNA), inducing miRNA degradation to regulate mRNA activity, forming double-stranded RNA with mRNA to regulate mRNA stability and inhibit target mRNA translation. Second, they interact with proteins, including acting as molecular decoys for RNA-binding proteins (RBPs) to participate in mRNA degradation, regulating posttranslational modifications by covering up posttranslational modification (PTM) sites or PTM enzyme binding sites, and acting as translation proteins and coding peptides.
For target transcripts, the biological function of miRNAs includes two aspects. One is that miRNA induces the cleavage and degradation of target mRNA through complete base pairing with the 3′ untranslated region (3′ UTR) of mRNA; the other is that miRNA cannot perfectly complement the target mRNA, and its effect is only to restrain the translation of mRNA. The consistent end result of these two aspects is that the protein expression products of target mRNA are reduced.
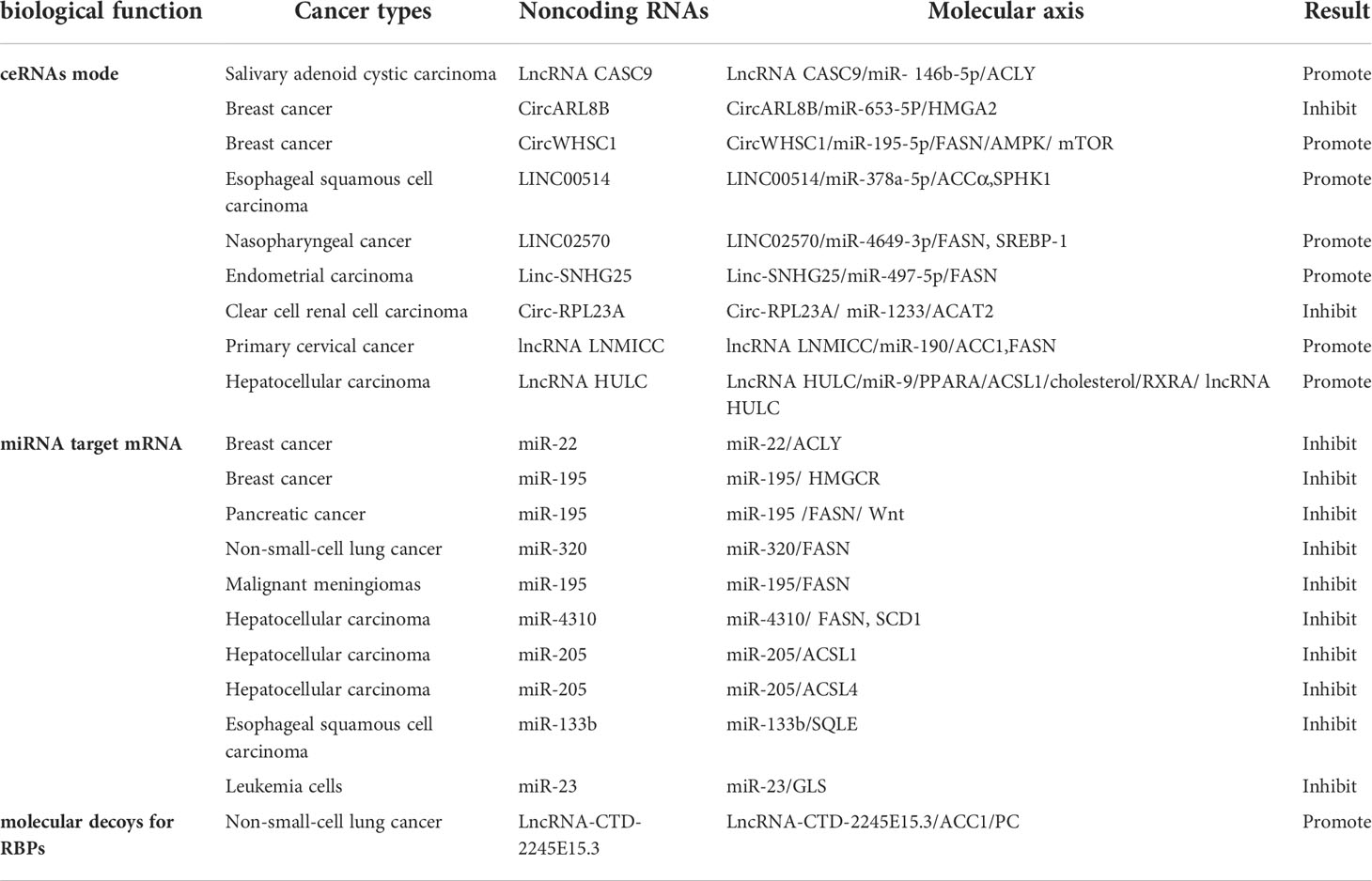
In this section, we review the role of noncoding RNAs in tumor lipid metabolism through different biological functions and promote or inhibit malignant tumor expression. We can clearly see the role of noncoding RNAs in different tumors with different pathways (Table 1).
As endogenous competing RNAs to regulate downstream mRNA stability
LncRNAs and circRNAs act as molecular sponges of miRNAs and adsorb miRNAs through base complementation pairing, resulting in the antagonism between mRNAs and lncRNAs or circRNAs. We have found that a considerable number of noncoding RNAs are involved in the progression of malignancy through this mode. Unexpectedly, the endogenous competitive RNA regulation mode was combined with a positive feedback loop to regulate the malignant progression of tumors.
In salivary adenoid cystic carcinoma (SACC) cells, studies have shown that lncRNA tumor susceptibility gene 9 (CASC9) promotes the malignant phenotype acquired by SACC cells by regulating miR-146b-5p/ACLY axis activation (52). The lncRNA CASC9 binds to miRNA-146b-5p and negatively regulates its expression, and miR-146b-5p directly targets the 3’ untranslated region (UTR) of ATP-citric acid lyase (ACLY) to degrade ACLY. Moreover, lncRNA CASC9 upregulates ACLY expression through competitive binding with miR-146b-5p.
In breast cancer (BC) studies, circARL8B inhibited tumor lipid metabolism and cell proliferation through the miR-653-5P/HMGA2 axis (53). Moreover, circWHSC1 acted as an oncogene to expedite BC evolution by modulating the miR-195-5p/FASN/AMPK/mTOR pathway (54). CircWHSC1 acts as a competitive endogenous RNA by sponging miR-195-5p, and miR-195-5p directly targets fatty acid synthase (FASN). miR-195-5p overexpression inhibits FASN expression and activates its downstream AMPK pathway.
In esophageal squamous cell carcinoma (ESCC), LINC00514 promotes ESCC cell proliferation and invasion through the absorption of miR-378a-5p by sponges as competitive endogenous RNA, leading to the upregulation of adipoformation-related proteins, including acetyl-coenzyme (Co)A carboxylase α, SPHK1, FAS, and stearoyl-CoA desaturase 1 (55).
In addition, LINC02570 was upregulated in patients with clinically advanced nasopharyngeal cancer (NPC) and promoted NPC progression by upregulating FASN and sterol regulatory element-binding protein-1 (SREBP-1) through the adsorption of miR-4649-3p (56). Targeting SREBP-1 Mediated Lipogenesis as Potential Strategies for Cancer (57).
Recently, studies have shown that linc-SNHG25 positively regulates FASN expression and promotes endometrial carcinoma (EC) malignant development by sponging miR-497-5p (58).
According to recent research, the overexpression of circ-RPL23A has been shown to inhibit cell cycle progression, proliferation, migration, and invasion. miR-1233 directly targets cholesteryl transferase 2 (ACAT2) and is a target of circ-RPL23A, which inhibits clear cell renal cell carcinoma (ccRCC) progression by competitively binding miR-1233 and thus upregulating ACAT2 expression (48).
In primary cervical cancer lymph node metastasis, LNMICC significant downregulation of acetyl-CoA carboxylase 1 (ACC1), FASN in the deregulation of FA metabolism, and miR-190 exert inhibitory effects on LNMICC expression by directly binding to LNMICC (59).
In hepatocellular carcinoma (HCC), lncRNA HULC forms a positive feedback loop to promote adipogenesis and enhance tumor proliferation through the HULC/miR-9/PPARA/ACSL1/cholesterol/RXRA/HULC pathway (60).
LncRNAs and circRNAs, as two kinds of ncRNAs, exert multitudinous biological functions and act as molecular sponges, relying on microRNA response elements (MREs) to competitively target microRNAs (miRNAs), thereby attenuating the degradation or inhibition of miRNAs to their own downstream protein-coding target genes and regulating the initiation and progression of neoplasms. Competing endogenous RNAs (ceRNAs) play an important role in the progression of tumor malignancy. Based on the theory of this model, we can design lncRNA-, circRNA-, or microRNA-targeted gene drugs to control the tumor process, and at the same time, the existence of tumors can be diagnosed by circulating tumor cell detection (Figure 5).
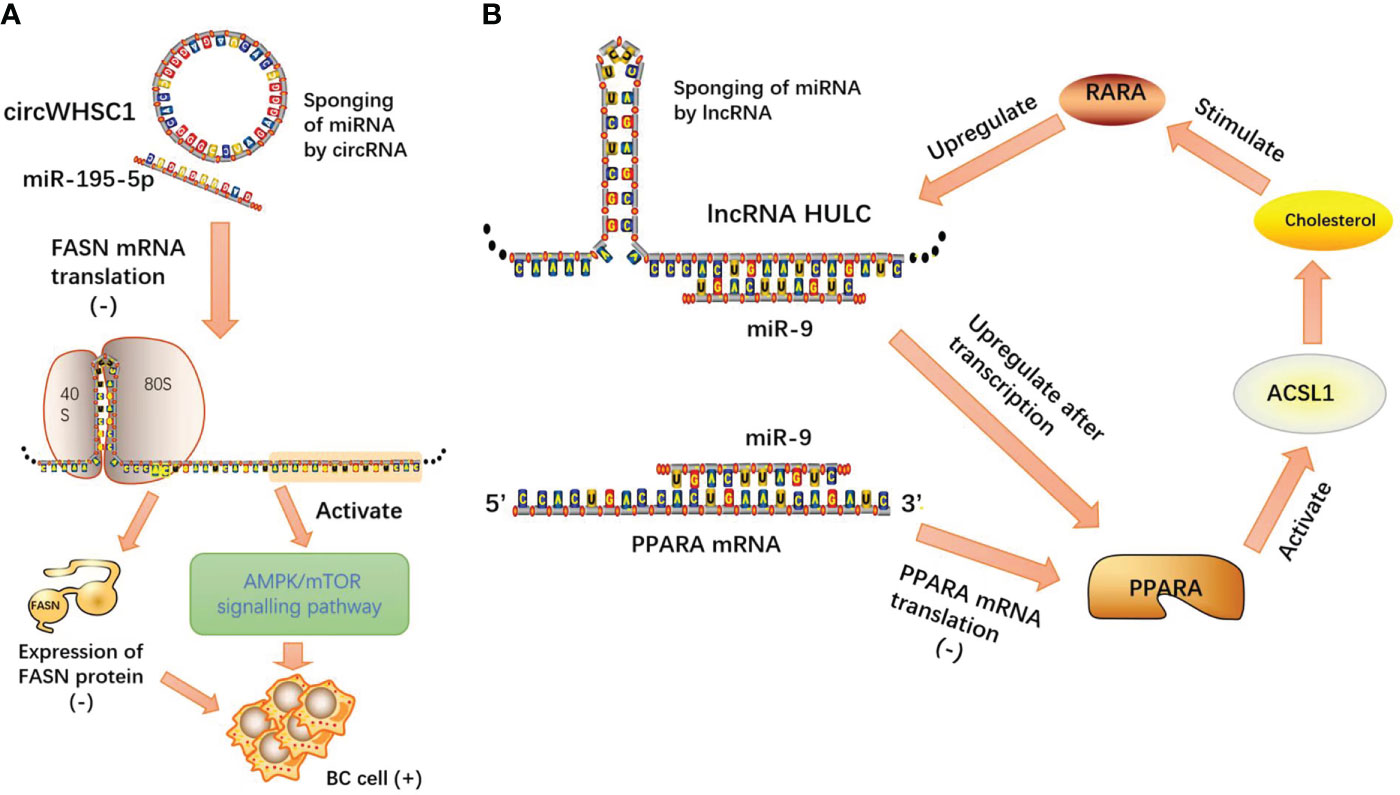
Figure 5 (A) CircWHSC1 acts as a competitive endogenous RNA by sponging miR-195-5p, and miR-195-5p directly targets FASN mRNA. miR-195-5p overexpression inhibits FASN expression and activates its downstream AMPK pathway; (B) lncRNA HULC forms a positive feedback loop to promote adipogenesis and enhance tumor proliferation through the HULC/miR-9/PPARA/ACSL1/cholesterol/RXRA/HULC pathway.
miRNAs directly target mRNA of lipid-metabolizing enzymes
The classical biological function of miRNA is to inhibit the translation of mRNA and to reduce the protein expression products of target mRNA by pairing with the base of mRNA. In addition, we unexpectedly found that miRNAs can target mRNAs of lipid metabolism enzymes through another miRNA.
In breast cancer research, miR-22 inhibits FA synthesis and elongation in tumor cells by targeting ACLY and FA elongase 6 (61). Moreover, HMGCR and FASN are direct targets of hsa-miR-195 in BC cells. hsa-miR-195 has been shown to target genes involved in de novo adipogenesis and to inhibit cell proliferation, migration, and invasion (62).
In pancreatic cancer-related studies, microRNA-195 directly targets the FAS enzyme and negatively regulates the expression of FAS, miRNA-195 overexpression inhibits the proliferation and invasion of pancreatic cancer cells, and miRNA-195 inhibits Wnt signaling in pancreatic cancer cells. The results showed that miRNA-195 inhibits pancreatic cancer cell proliferation and invasion by regulating the FASN/Wnt signaling pathway (63).
In lung cancer studies, it was found that miR-320 directly targets FASN to inhibit the growth, migration, and invasion of NSCLC cells and is significantly correlated with TNM classification and metastasis (64).
We found that FASN was significantly upregulated in malignant meningiomas. miR-195 directly targeted FASN. miR-195 has been demonstrated to play a tumor suppressive role in the occurrence and progression of malignant meningioma by targeting FASN (65).
By inhibiting FASN and stearyl CoA desaturase 1 (SCD1)–mediated lipid synthesis, miR-4310 inhibited HCC cell proliferation, migration, and invasion in vitro and inhibited HCC tumor growth and metastasis in vivo. These results indicate that miR-4310 plays an important role in HCC tumor growth and metastasis by targeting FASN and SCD1-mediated lipid synthesis pathways (66).
Studies have shown that miR-205 mediates the dysregulation of HCC lipid metabolism by targeting acyl-CoA long-chain family member 1 (ACSL1) mRNA, and ACSL1 is significantly affected by ncRNA-mediated lipid metabolism in HCC cells (67). In addition, miR-205 inhibited acyl-CoA long-chain family member 4 (ACSL4) expression at the mRNA and protein levels by targeting its 3’UTR. miR-205 expression promotes abnormal lipid metabolism in HCC by targeting ACSL4 (68).
In human ESCC cells and tissues, miR-133b expression was reported to be downregulated. SQLE is a direct downstream target gene of miR-133b. Exogenous miR-133b expression and SQLE knockdown reduced the rate of the epithelial–mesenchymal transition (EMT) in ESCC cells in vitro. These outcomes indicate that miR-133b–dependent SQLE potentially plays a key role in ESCC metastasis (69).
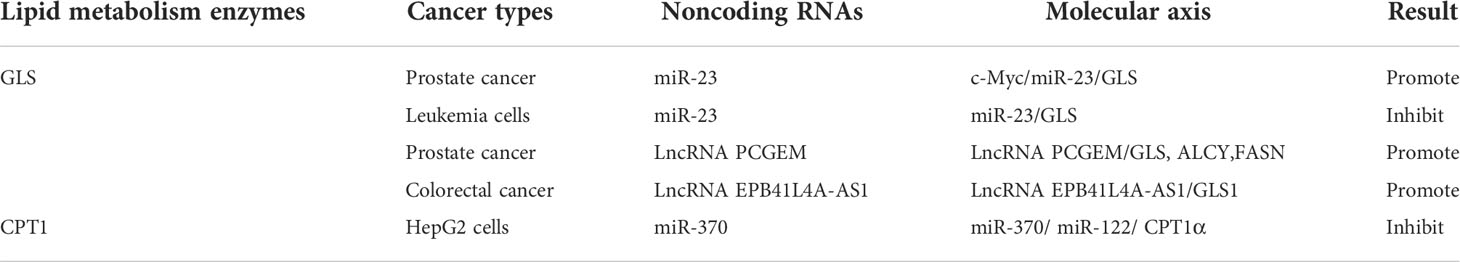
miR-23 targets GLS mRNA and inhibits GLS protein expression. The overexpression of miR-23a has been reported to impair glutamine metabolism and induce apoptosis in leukemia cells (70).
We consistently found that these miRNAs inhibit the enzyme function of lipid metabolism and reduce the production of metabolic enzymes through base complementary pairing with the mRNA of the enzyme. miRNA plays an important role in tumor lipid metabolism and inhibits malignant progression through its classical biological functions. Such intuitive effects make people have great interest in the research and development of miRNA mimics, especially the products harmless to human body after biological processing, which will provide a bright future for tumor treatment (Figure 6).
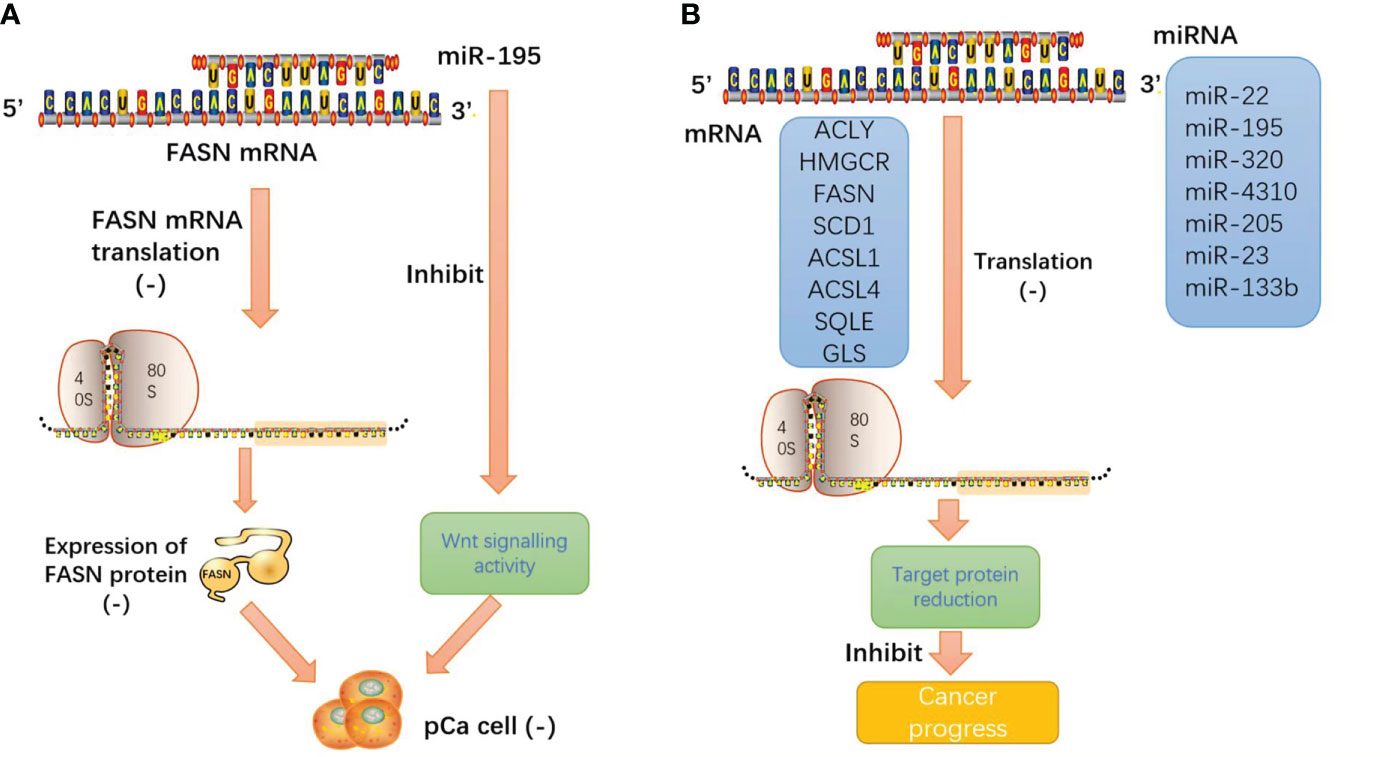
Figure 6 (A) miRNA-195 inhibits pancreatic cancer cell proliferation and invasion by regulating the FASN/Wnt signaling pathway; (B) miRNA induces the cleavage and degradation of target mRNA through base pairing with the 3′ untranslated region (3′ UTR) of mRNA. The result is that the protein expression products of target mRNA and the malignant behavior of tumors are inhibited.
Noncoding RNAs act as molecular decoys for RNA-binding proteins
Some specific RNA-binding domains of noncoding RNAs can be recognized and interact with RNA-binding proteins and are involved in various posttranscriptional regulatory processes, such as RNA shearing, transport, sequence editing, intracellular localization, and translation control, which play a role in the malignant biological behavior of tumors to a certain extent.
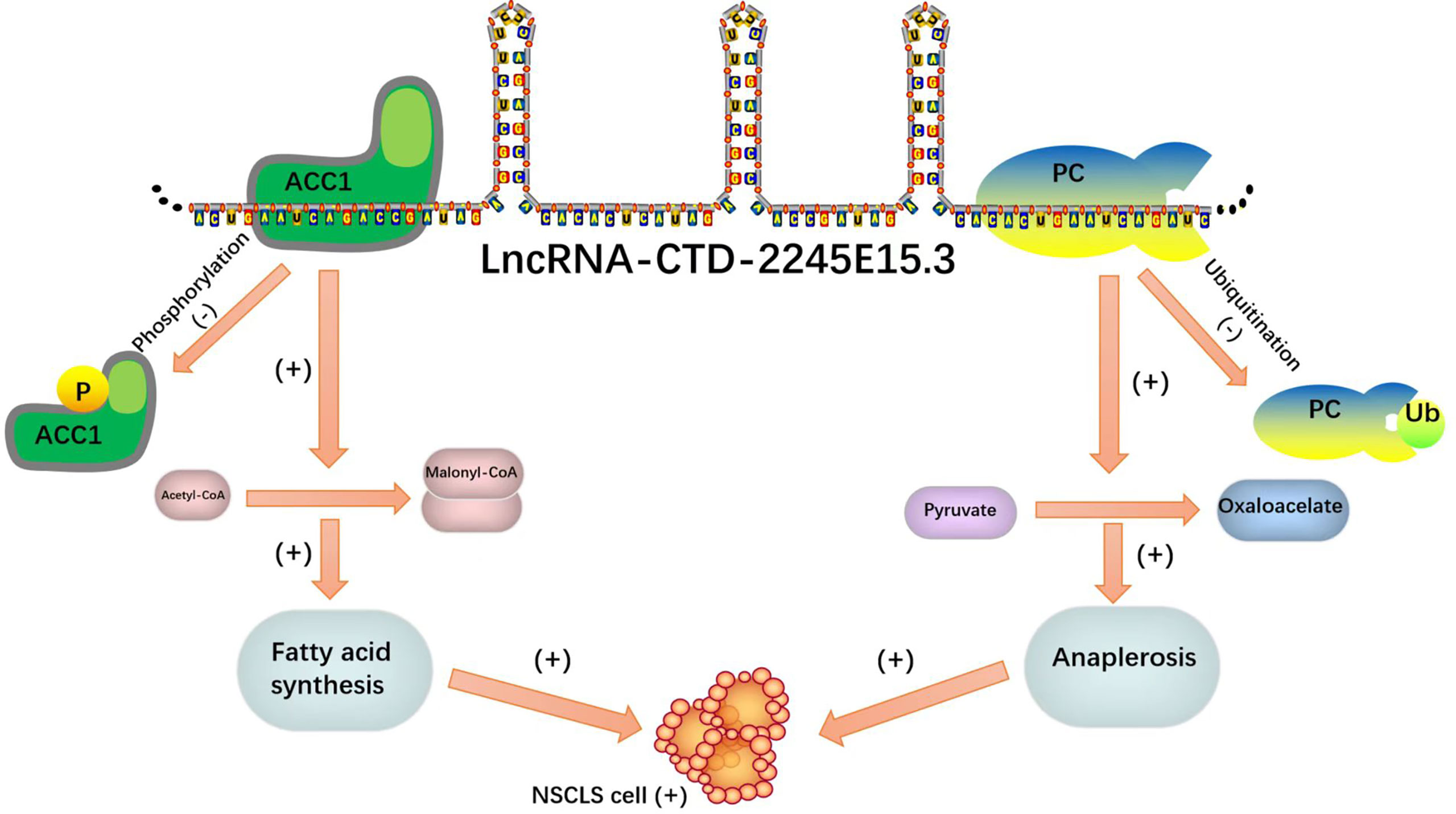
In recent studies on NSCLC, LncRNA-CTD-2245E15.3 exerts its carcinogenic function by binding ACC1 and pyruvate carboxylase (PC), which are key anabolic factors in biomolecule synthesis in rapidly proliferating tumor cells (71).
Lipid metabolism enzymes can bind to RNA binding proteins as ncRNA molecular decoys, inhibiting the ability of lipid metabolism enzymes to promote the progression of tumor malignancy. Such a binding mode can be a reasonable direction for tumor gene therapy. It is a good idea to design special probes to enhance the binding of ncRNA and RNA binding protein (RBP) for tumor targeted therapy (Figure 7).
Noncoding RNAs regulate the expression of lipid metabolism enzymes
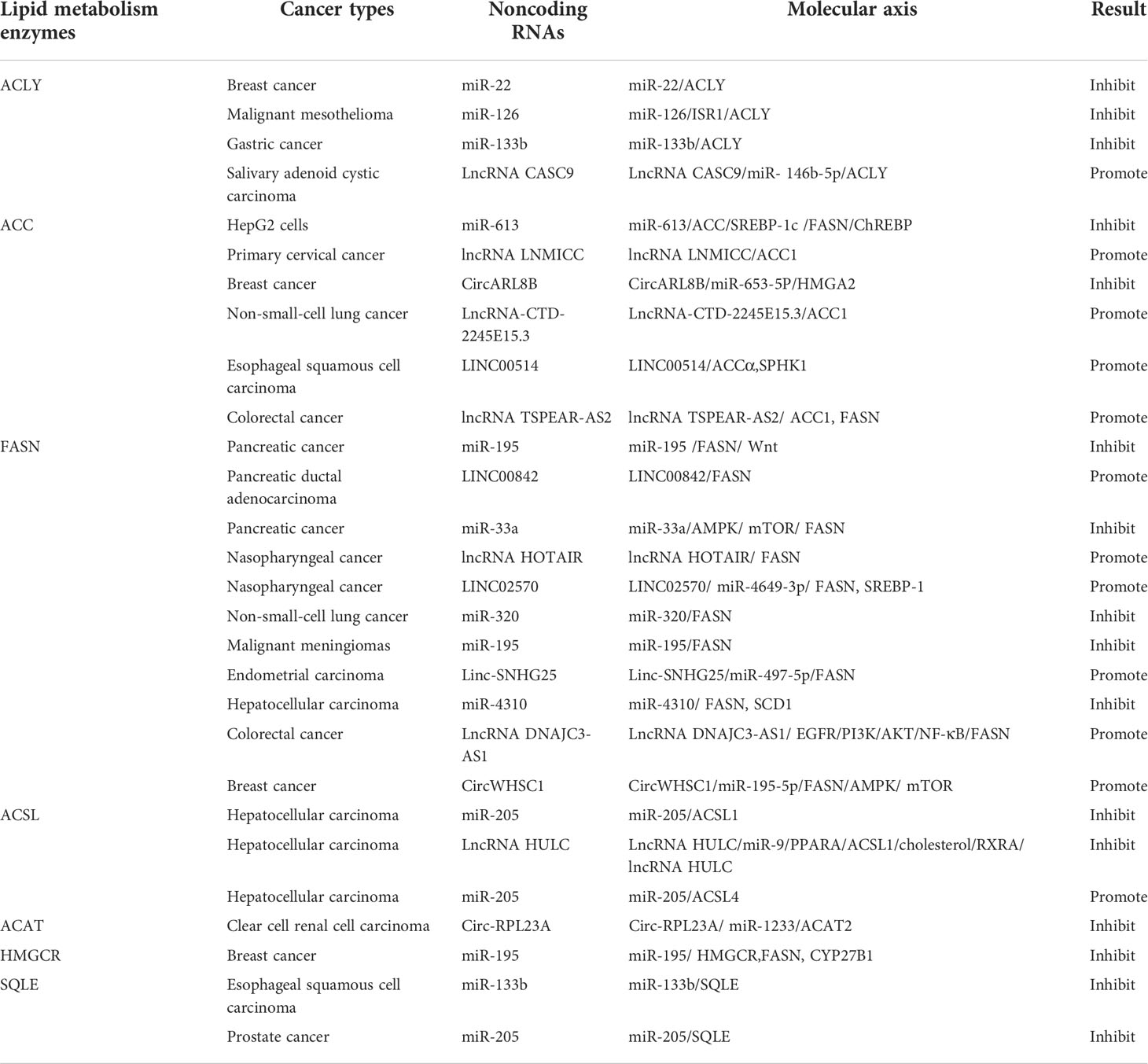
Tumor metabolic reprogramming mainly depends on the changes of metabolic enzymes, and noncoding RNA plays an important role in the regulation of tumor lipid metabolic enzymes. In order to have a more intuitive understanding of the regulation of various lipid metabolism enzymes, we used enzymes as clues to summarize the interference of various noncoding RNAs in tumor malignant biological behavior by regulating the expression level of tumor lipid metabolism enzymes and presented them in the form of tables and figures (Tables 2, 3 and Figure 8). We can clearly understand the leading role of noncoding RNA in tumor lipid metabolism, which provides a theoretical basis for noncoding RNA as a key molecule in targeted therapy and a solid theoretical basis for precision medicine in the future.
At present, the most studied lipid metabolism enzymes are ACLY, ACC, and FASN. ACLY is a cytoplasmic enzyme that catalyzes citric acid to produce acetyl-CoA and is an important component in the endogenous biosynthesis of FA and cholesterol (72). In the study of various tumors, including breast cancer, malignant mesothelioma, gastric cancer, salivary adenoid carcinoma, and noncoding RNA can affect the expression of lipid metabolism enzymes by directly binding to ACLY or by classical ceRNA mode, thus changing the malignant process of tumors. Relevant noncoding RNA mimics or inhibitors can be studied for molecular therapy of tumors.
ACC is a critical rate-limiting enzyme for FASN (73). In colorectal cancer (CRC) studies, in vitro experiments have shown that lncRNA TSPEAR-AS2 knockdown can reduce the TG content and ACC1 and FASN expressions in CRC cells. These outcomes suggest that lncRNA TSPEAR-AS2 can regulate FA metabolism in CRC (74). In the studies of primary cervical cancer, breast cancer, non-small-cell lung cancer (NSCLC), and esophageal carcinoma (ESCC), noncoding RNA can regulate ACC expression and affect tumor proliferation in various ways.
FASN is a key enzyme for the de novo synthesis of FA, plays an important role in lipid metabolism, and is associated with tumor-related signaling pathways (75). FASN plays a central role in lipid metabolism, so it has been extensively studied in pancreatic cancer, pancreatic ductal adenocarcinoma, NPC, and seven other tumors. Most of the research results show that noncoding RNA plays a role by binding miRNA or RBP or directly binding mRNA of lipid metabolism enzymes, and interferes with the biological behavior of tumor malignancy.
Future perspectives
Current research achievements and deficiencies
At present, we clearly understand the reactions involved in tumor lipid metabolism and the influence of ncRNAs on lipid metabolism-related enzymes, which change tumorigenesis and tumor development processes. These results provide us with a profound understanding of the influence of ncRNAs on tumor lipid metabolism and lay a solid foundation for the study of tumor lipid metabolism. Is this the end of research? Of course not. There are still unanswered questions.
First, ncRNAs are involved in the regulation of lipid metabolism in the cytoplasm, but their source is the nucleus. How do various ncRNAs cross the nuclear membrane into the cytoplasm? Current research has focused on the length of ncRNAs and related protein mediations (76, 77). Whether there are other factors, such as the sequence of ncRNA bases and the proportion of various bases, needs further investigation.
Second, everything in life has a life cycle; thus, are ncRNAs immortal? How are they regulated and degraded? The four currently known degradation pathways, namely microRNA, m6A, ribonuclease L (RNase L), and special structure-mediated degradation (78–81), remain to be further studied.
However, there are few studies on the regulation of several lipid metabolism enzymes by ncRNAs in the tumor metabolism process, such as MCD (82), ACSL (60, 67, 68), ACAT (48, 83), HMGCS, and HMGCR (62). These lipid metabolism enzymes will become research objects in the future.
Finally, some current studies have demonstrated that ncRNAs can directly act on the activities of lipid metabolism enzymes or their encoding gene regulatory enzymes. Whether there are other unknown signaling pathways that serve as a bridge between ncRNAs and lipid metabolism enzymes remains to be further investigated.
Future research directions for noncoding RNAs
Because ncRNAs can affect tumor processes by regulating lipid metabolism enzymes, in the future, we can study the unique role of ncRNAs from the four aspects of disease prevention, detection, diagnosis, and treatment.
In the future, as in the prevention of many diseases, ncRNA vaccines will be the first line of defense for various incurable diseases, including tumors. At present, the world is facing the challenge of a novel coronavirus. Scientists have developed relevant vaccines, such as the BNT162b2 mRNA COVID-19 vaccine (84) and the mRNA-1273 vaccine (85, 86), which have achieved good immune effects in clinical applications. A good tumor response rate was shown in clinical studies using mRNA vaccines in patients with advanced melanoma (87, 88). We believe that the advent of ncRNA vaccines will play an important role in tumor preventive immunity.
In terms of inspection and diagnosis, we will focus on the examination of patient body fluids, such as whole blood, plasma, serum, gastroenteric fluid, and urine, to detect and quantify the substances and related ncRNAs released by tumor cells to accurately diagnose diseases combined with other relevant information. Currently, circRNA and RNA splicing variants have been reported in lung cancer as cancer biomarkers and have achieved satisfactory results in evaluating therapeutic responses (89). Moreover, hsa_circ_002059 and hsa_circ_0000419 have been confirmed as potential novel and stable biomarkers for the diagnosis of gastric cancer (90, 91), and the role of hsa_circ_0004585 in CRC progression has been confirmed, indicating its potential as a biomarker for diagnosis (92). A recent survey showed that a total of 33 circulating miRNAs and six different panels of circulating miRNAs have been described for their diagnostic performance in EC diagnosis. A total of seven circulating miRNAs and one panel of circulating miRNAs have been associated with clinical and prognostic factors in EC (93). In a variety of tumor studies, significant progress has been made in the use of ncRNAs in extracellular fluid as cancer biomarkers (94).
Current tumor treatments include surgical excision, chemotherapy, and monoclonal antibody therapy. Based on the understanding of ncRNA, it has great potential as a gene-targeted therapy. At present, in lung cancer research, the effectiveness of a combination of mRNA vaccines and radiotherapy to eradicate solid tumors has been scientifically confirmed (95). An mRNA vaccine has also been reported to be safe and effective in inducing antigen-specific T-cell immunity in clinical studies of gastrointestinal tumors (96). For tumor resistance to chemotherapeutic drugs, there is a considerable amount of data to prove that noncoding RNA can enhance tumor sensitivity to chemotherapy. To date, a number of advanced miRNA detection methods with high specificity and sensitivity have been developed, such as chromatography-based methods and mass spectrometry-based nanomaterial-based methods. miRNA detection methods are going to focus on facilitating the development of noninvasive diagnosis and inhibiting cancer drug resistance (97). In a recent study, the combined use of microRNAs and long noncoding RNAs with chemotherapeutic compounds, as well as the induction of suicide genes, provided an innovative therapeutic approach for the management of glioblastoma (GBM) (98). It has been reported that miR-532–3p can directly target E26 oncogene homolog 1 (ETS1) and transglutaminase 2 (TGM2) to promote early apoptosis through the ETS1/TGM2 axis-mediated Wnt/β-catenin signaling pathway, and it activates the p53 signaling pathway and enhances the sensitivity to cisplatin and 5-FU in CRC treatment (99). Similarly, it was discovered that miR-365–3p inhibited the downstream expression of keratin 16 (KRT16) to enhance 5-FU–induced cytotoxicity through the c-Met/Src signaling pathway and curbed oral squamous cell carcinoma (OSCC) metastasis and cancer stemness (100). Therefore, ncRNAs not only can serve as targeted therapeutic sites but also can enhance the therapeutic effect of chemotherapy-resistant tumors and can also be combined with radiotherapy for the radical treatment of tumors. In the future, the development of ncRNA therapy will become a hot direction and shoulder a major task in the medical field.
Conclusions
In this paper, we systematically introduce the metabolism and function of lipids and their changes in tumorigenesis and explain the relationship between ncRNAs and tumor lipid metabolism, including how ncRNAs regulate tumor lipid metabolism and regulatory pathways. Finally, the progress of ncRNAs in disease prevention, detection, diagnosis, and treatment is discussed. Our goal is to take advantage of the characteristics and advantages of ncRNAs and use them in multiple stages of the disease. Current scientific and technological methods include gene knockout, molecular targeting, isothermal amplification-based methods, nanomaterial-based methods, chromatography-based methods, and mass spectrometry-based methods. We are very confident that ncRNAs can be incorporated into the diagnosis and treatment process to target rivals in the stage of disease detection and diagnosis, weaken rivals in the stage of prevention, and defeat rivals in the stage of treatment, providing a new platform for all-in-one tumor therapy.
Author contributions
YW, QL, SW and B-JW collected the related reports and drafted the manuscript. LX, X-XH and Y-BX revised the manuscript. YJ, HH, Q-SF, J-WW, QW, LQ, and T-TC participated in designing the review. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Research Project of Higher Education in Anhui Province (KJ2021A0857); the National Natural Science Foundation of China (81902515); Excellent Youth Talent Support Program (Key) of Anhui Province (gxyqzd2020029); Introducing Talents Natural Science Foundation of The First Affiliated Yijishan Hospital of Wannan Medical College (YR202006); the Natural Science Research Project of Higher Education in Anhui Province (KJ2019A0429); and the Natural Science Research Project of Higher Education in Anhui Province (KJ20190412).
Acknowledgments
We would like to acknowledge the reviewers for their helpful comments on this paper.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Luo J, Yang H, Song BL. Mechanisms and regulation of cholesterol homeostasis. Nat Rev Mol Cell Biol (2020) 21(4):225–45. doi: 10.1038/s41580-019-0190-7
2. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer (2007) 7(10):763–77. doi: 10.1038/nrc2222
3. Guo D, Bell EH, Chakravarti A. Lipid metabolism emerges as a promising target for malignant glioma therapy. CNS Oncol (2013) 2(3):289–99. doi: 10.2217/cns.13.20
4. Yoon S, Lee M-Y, Park SW, Moon J-S, Koh Y-K, Ahn Y-H, et al. Up-regulation of acetyl-CoA carboxylase alpha and fatty acid synthase by human epidermal growth factor receptor 2 at the translational level in breast cancer cells. J Biol Chem (2007) 282(36):26122–31. doi: 10.1074/jbc.M702854200
5. Abramson HN. The lipogenesis pathway as a cancer target. J Med Chem (2011) 54(16):5615–38. doi: 10.1021/jm2005805
6. Grossi-Paoletti E, Paoletti P, Fumagalli R. Lipids in brain tumors. J Neurosurg (1971) 34(3):454–5. doi: 10.3171/jns.1971.34.3.0454
7. Santos CR, Schulze A. Lipid metabolism in cancer. FEBS J (2012) 279(15):2610–23. doi: 10.1111/j.1742-4658.2012.08644.x
8. Röhrig F, Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer (2016) 16(11):732–49. doi: 10.1038/nrc.2016.89
9. Schulze A, Harris AL. How cancer metabolism is tuned for proliferation and vulnerable to disruption. Nature (2012) 491(7424):364–73. doi: 10.1038/nature11706
10. Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab (2019) 30(1):157–173.e7. doi: 10.1016/j.cmet.2019.05.009
11. Statello L, Guo CJ, Chen L-L, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol (2021) 22(2):96–118. doi: 10.1038/s41580-020-00315-9
12. Anastasiadou E, Jacob LS, Slack FJ. Non-coding RNA networks in cancer. Nat Rev Cancer (2018) 18(1):5–18. doi: 10.1038/nrc.2017.99
13. Noviello TMR, Ceccarelli F, Ceccarelli M, Cerulo L. Deep learning predicts short non-coding RNA functions from only raw sequence data. PloS Comput Biol (2020) 16(11):e1008415. doi: 10.1371/journal.pcbi.1008415
14. Boon RA, Jaé N, Holdt L, Dimmeler S. Long noncoding RNAs: From clinical genetics to therapeutic targets? J Am Coll Cardiol (2016) 67(10):1214–26. doi: 10.1016/j.jacc.2015.12.051
15. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al, et al. The transcriptional landscape of the mammalian genome. Science (2005) 309(5740):1559–63. doi: 10.1126/science.1112014
16. Djebali S, Davis CA, Merkel A, Dobin A, Lassmann T, Mortazavi A, et al. Landscape of transcription in human cells. Nature (2012) 489(7414):101–8. doi: 10.1038/nature11233
17. Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U.S.A. (2002) 99(24):15524–9. doi: 10.1073/pnas.242606799
18. Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, et al. Pre-b cell proliferation and lymphoblastic leukemia/high-grade lymphoma in e(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA (2006) 103(18):7024–9. doi: 10.1073/pnas.0602266103
19. Iorio MV, Croce CM. microRNA involvement in human cancer. Carcinogenesis (2012) 33(6):1126–33. doi: 10.1093/carcin/bgs140
20. Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene (2012) 31(5):634–42. doi: 10.1038/onc.2011.260
21. Romano G, Acunzo M, Garofalo M, Leva Di G, Cascione L, Zanca C, et al. MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci USA (2012) 109(41):16570–5. doi: 10.1073/pnas.1207917109
22. Srere PA. The citrate cleavage enzyme. i. distribution and purification. J Biol Chem (1959) 234:2544–7. doi: 10.1016/S0021-9258(18)69735-2
23. Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB, et al. ATP-citrate lyase links cellular metabolism to histone acetylation. Science (2009) 324(5930):1076–80. doi: 10.1126/science.1164097
24. Li X, Qian X, Lu Z. Local histone acetylation by ACSS2 promotes gene transcription for lysosomal biogenesis and autophagy. Autophagy (2017) 13(10):1790–1. doi: 10.1080/15548627.2017.1349581
25. Grime JM, Edwards MA, Rudd NC, Unwin PR. Quantitative visualization of passive transport across bilayer lipid membranes. Proc Natl Acad Sci USA (2008) 105(38):14277–82. doi: 10.1073/pnas.0803720105
26. Wellen KE, Thompson CB. A two-way street: reciprocal regulation of metabolism and signalling. Nat Rev Mol Cell Biol (2012) 13(4):270–6. doi: 10.1038/nrm3305
27. Comerford SA, Huang Z, Du X, Wang Y, Cai L, Witkiewicz AK, et al. Acetate dependence of tumors. Cell (2014) 159(7):1591–602. doi: 10.1016/j.cell.2014.11.020
28. Hosios AM, Vander Heiden MG. Acetate metabolism in cancer cells. Cancer Metab (2014) 2(1):27. doi: 10.1186/s40170-014-0027-y
29. Kamphorst JJ, Chung MK, Fan J, Rabinowitz JD, et al. Quantitative analysis of acetyl-CoA production in hypoxic cancer cells reveals substantial contribution from acetate. Cancer Metab (2014) 2:23. doi: 10.1186/2049-3002-2-23
30. Schug ZT, Peck B, Jones DT, Zhang Q, Grosskurth S, Alam IS, et al. Acetyl-CoA synthetase 2 promotes acetate utilization and maintains cancer cell growth under metabolic stress. Cancer Cell (2015) 27(1):57–71. doi: 10.1016/j.ccell.2014.12.002
31. Li X, Jiang Y, Meisenhelder J, Yang W, Hawke DH, Zheng Y, et al. Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol Cell (2016) 61(5):705–19. doi: 10.1016/j.molcel.2016.02.009
32. Metallo CM, Gameiro PA, Bell EL, Mattaini KR, Yang J, Hiller K, et al. Reductive glutamine metabolism by IDH1 mediates lipogenesis under hypoxia. Nature (2011) 481(7381):380–4. doi: 10.1038/nature10602
33. Bhutia YD, Babu E, Ramachandran S, Ganapathy V. Amino acid transporters in cancer and their relevance to "glutamine addiction": novel targets for the design of a new class of anticancer drugs. Cancer Res (2015) 75(9):1782–8. doi: 10.1158/0008-5472.CAN-14-3745
34. Moreadith RW, Lehninger AL. The pathways of glutamate and glutamine oxidation by tumor cell mitochondria. role of mitochondrial NAD(P)+-dependent malic enzyme. J Biol Chem (1984) 259(10):6215–21.
35. Brownsey RW, Boone AN, Elliott JE, Kulpa JE, Lee WM. Regulation of acetyl-CoA carboxylase. Biochem Soc Trans (2006) 34(Pt 2):223–7. doi: 10.1042/BST0340223
36. Steensels S, Ersoy BA. Fatty acid activation in thermogenic adipose tissue. Biochim Biophys Acta Mol Cell Biol Lipids (2019) 1864(1):79–90. doi: 10.1016/j.bbalip.2018.05.008
37. Cooper DE, Young PA, Klett EL, Coleman RA. Physiological consequences of compartmentalized acyl-CoA metabolism. J Biol Chem (2015) 290(33):20023–31. doi: 10.1074/jbc.R115.663260
38. Hegardt FG. Mitochondrial 3-hydroxy-3-methylglutaryl-CoA synthase: a control enzyme in ketogenesis. Biochem J (1999) 338(Pt 3):569–82. doi: 10.1042/bj3380569
39. Sharpe LJ, Brown AJ. Controlling cholesterol synthesis beyond 3-hydroxy-3-methylglutaryl-CoA reductase (HMGCR). J Biol Chem (2013) 288(26):18707–15. doi: 10.1074/jbc.R113.479808
40. Yoshioka H, Coates HW, Chua NK, Hashimoto Y, Brown AJ, Ohgane K, et al. A key mammalian cholesterol synthesis enzyme, squalene monooxygenase, is allosterically stabilized by its substrate. Proc Natl Acad Sci USA (2020) 117(13):7150–8. doi: 10.1073/pnas.1915923117
41. Alphonse PA, Jones PJ. Revisiting human cholesterol synthesis and absorption: The reciprocity paradigm and its key regulators. Lipids (2016) 51(5):519–36. doi: 10.1007/s11745-015-4096-7
42. Kruse M, Vivas O, Traynor-Kaplan A, Hille B. Dynamics of phosphoinositide-dependent signaling in sympathetic neurons. J Neurosci (2016) 36(4):1386–400. doi: 10.1523/JNEUROSCI.3535-15.2016
43. Simons K, van Meer G. Lipid sorting in epithelial cells. Biochemistry (1988) 27(17):6197–202. doi: 10.1021/bi00417a001
44. Simons K, Ikonen E. Functional rafts in cell membranes. Nature (1997) 387(6633):569–72. doi: 10.1038/42408
45. Beigneux AP, Allan CM, Sandoval NP, Cho GW, Heizer PJ, Jung RS, et al. Lipoprotein lipase is active as a monomer. Proc Natl Acad Sci USA (2019) 116(13):6319–28. doi: 10.1073/pnas.1900983116
46. Pepino MY, Kuda O, Samovski D, Abumrad NA. Structure-function of CD36 and importance of fatty acid signal transduction in fat metabolism. Annu Rev Nutr (2014) 34:281–303. doi: 10.1146/annurev-nutr-071812-161220
47. Su X, Abumrad NA. Cellular fatty acid uptake: a pathway under construction. Trends Endocrinol Metab (2009) 20(2):72–7. doi: 10.1016/j.tem.2008.11.001
48. Cheng L, Cao H, Xu J, Xu M, He W, Zhang W, et al. Circ_RPL23A acts as a miR-1233 sponge to suppress the progression of clear cell renal cell carcinoma by promoting ACAT2. J Bioenerg Biomembr (2021) 53(4):415–28. doi: 10.1007/s10863-021-09901-8
49. Kulminskaya N, Oberer M. Protein-protein interactions regulate the activity of adipose triglyceride lipase in intracellular lipolysis. Biochimie (2020) 169:62–8. doi: 10.1016/j.biochi.2019.08.004
50. Zimmermann R, Strauss JG, Haemmerle G, Schoiswohl G, Birner-Gruenberger R, Riederer M, et al. Fat mobilization in adipose tissue is promoted by adipose triglyceride lipase. Science (2004) 306(5700):1383–6. doi: 10.1126/science.1100747
51. Hilgemann DW, Dai G, Collins A, Lariccia V, Magi S, Deisl C, et al. Lipid signaling to membrane proteins: From second messengers to membrane domains and adapter-free endocytosis. J Gen Physiol (2018) 150(2):211–24. doi: 10.1085/jgp.201711875
52. Xie H, Tang J, Lu L, Li B, Wang M. CASC9 plays a role in salivary adenoid cystic carcinoma in vitro by upregulation of ACLY. Oral Dis (2022) 28(2):352–63. doi: 10.1111/odi.13759
53. Wu H, Xu J, Gong G, Zhang Y, Wu S. CircARL8B contributes to the development of breast cancer via regulating miR-653-5p/HMGA2 axis. Biochem Genet (2021) 59(6):1648–65. doi: 10.1007/s10528-021-10082-7
54. Chen Q, Yang Z, Ding H, Li H, Wang W, Pan Z, et al. CircWHSC1 promotes breast cancer progression by regulating the FASN/AMPK/mTOR axis through sponging miR-195-5p. Front Oncol (2021) 11:649242. doi: 10.3389/fonc.2021.649242
55. Wang X, Liu H, Zhang Q, Zhang X, Qin Y, Zhu G, et al. LINC00514 promotes lipogenesis and tumor progression in esophageal squamous cell carcinoma by sponging miR−378a−5p to enhance SPHK1 expression. Int J Oncol (2021) 59(5):86. doi: 10.3892/ijo.2021.5266
56. Liu F, Wei J, Hao Y, Lan J, Li W, Weng J, et al. Long intergenic non-protein coding RNA 02570 promotes nasopharyngeal carcinoma progression by adsorbing microRNA miR-4649-3p thereby upregulating both sterol regulatory element binding protein 1, and fatty acid synthase. Bioengineered (2021) 12(1):7119–30. doi: 10.1080/21655979.2021.1979317
57. Zhao Q, Lin X, Wang G. Targeting SREBP-1-Mediated lipogenesis as potential strategies for cancer. Front Oncol (2022) 12:952371. doi: 10.3389/fonc.2022.952371
58. He Y, Xu S, Qi Y, Tian J, Xu F. Long noncoding RNA SNHG25 promotes the malignancy of endometrial cancer by sponging microRNA-497-5p and increasing FASN expression. J Ovarian Res (2021) 14(1):163. doi: 10.1186/s13048-021-00906-w
59. Shang C, Wang W, Liao Y, Chen Y, Liu T, Du Q, et al. LNMICC promotes nodal metastasis of cervical cancer by reprogramming fatty acid metabolism. Cancer Res (2018) 78(4):877–90. doi: 10.1158/0008-5472.CAN-17-2356
60. Cui M, Xiao Z, Wang Y, Zheng M, Song T, Cai X, et al. Long noncoding RNA HULC modulates abnormal lipid metabolism in hepatoma cells through an miR-9-mediated RXRA signaling pathway. Cancer Res (2015) 75(5):846–57. doi: 10.1158/0008-5472.CAN-14-1192
61. Koufaris C, Valbuena GN, Pomyen Y, Tredwell GD, Nevedomskaya E, Lau C-He, et al. Systematic integration of molecular profiles identifies miR-22 as a regulator of lipid and folate metabolism in breast cancer cells. Oncogene (2016) 35(21):2766–76. doi: 10.1038/onc.2015.333
62. Singh R, Yadav V, Kumar S, Saini N. MicroRNA-195 inhibits proliferation, invasion and metastasis in breast cancer cells by targeting FASN, HMGCR, ACACA and CYP27B1. Sci Rep (2015) 5:17454. doi: 10.1038/srep17454
63. Xu Z, Li C, Qu H, Li H, Gu Q, Xu J, et al. MicroRNA-195 inhibits the proliferation and invasion of pancreatic cancer cells by targeting the fatty acid synthase/Wnt signaling pathway. Tumour Biol (2017) 39(6):1010428317711324. doi: 10.1177/1010428317711324
64. Lei T, Zhu Y, Jiang C, Wang Y, Fu J, Fan Z, et al. MicroRNA-320 was downregulated in non-small cell lung cancer and inhibited cell proliferation, migration and invasion by targeting fatty acid synthase. Mol Med Rep (2016) 14(2):1255–62. doi: 10.3892/mmr.2016.5370
65. Song LR, Li D, Weng JC, Li CB, Wang L, Wu Z, et al. MicroRNA-195 functions as a tumor suppressor by directly targeting fatty acid synthase in malignant meningioma. World Neurosurg (2020) 136:e355–64. doi: 10.1016/j.wneu.2019.12.182
66. Li H, Chen Z, Zhang Y, Yuan P, Liu J, Ding L, et al. MiR-4310 regulates hepatocellular carcinoma growth and metastasis through lipid synthesis. Cancer Lett (2021) 519:161–71. doi: 10.1016/j.canlet.2021.07.029
67. Cui M, Wang Y, Sun B, Xiao Z, Ye L, Zhang X, et al. MiR-205 modulates abnormal lipid metabolism of hepatoma cells via targeting acyl-CoA synthetase long-chain family member 1 (ACSL1) mRNA. Biochem Biophys Res Commun (2014) 444(2):270–5. doi: 10.1016/j.bbrc.2014.01.051
68. Cui M, Xiao Z, Sun B, Wang Y, Zheng M, Ye L, et al. Involvement of cholesterol in hepatitis b virus X protein-induced abnormal lipid metabolism of hepatoma cells via up-regulating miR-205-targeted ACSL4. Biochem Biophys Res Commun (2014) 445(3):651–5. doi: 10.1016/j.bbrc.2014.02.068
69. Qin Y, Zhang Y, Tang Q, Jin L, Chen Y. SQLE induces epithelial-to-mesenchymal transition by regulating of miR-133b in esophageal squamous cell carcinoma. Acta Biochim Biophys Sin (Shanghai) (2017) 49(2):138–48. doi: 10.1093/abbs/gmw127
70. Rathore MG, Saumet A, Rossi J-F, de Bettignies C, Tempé D, Lecellier CH, et al. The NF-κB member p65 controls glutamine metabolism through miR-23a. Int J Biochem Cell Biol (2012) 44(9):1448–56. doi: 10.1016/j.biocel.2012.05.011
71. Wang C, Meng X, Zhou Y, Yu J, Li Q, Liao Z, et al. Long noncoding RNA CTD-2245E15.3 promotes anabolic enzymes ACC1 and PC to support non-small cell lung cancer growth. Cancer Res (2021) 81(13):3509–24. doi: 10.1158/0008-5472.CAN-19-3806
72. Zaidi N, Swinnen JV, Smans K. ATP-citrate lyase: a key player in cancer metabolism. Cancer Res (2012) 72(15):3709–14. doi: 10.1158/0008-5472.CAN-11-4112
73. Wang C, Ma J, Zhang N, Yang Q, Jin Y, Wang Y, et al. The acetyl-CoA carboxylase enzyme: a target for cancer therapy? Expert Rev Anticancer Ther (2015) 15(6):667–76. doi: 10.1586/14737140.2015.1038246
74. Peng Y, Xu C, Wen J, Zhang Y, Wang M, Liu X, et al. Fatty acid metabolism-related lncRNAs are potential biomarkers for predicting the overall survival of patients with colorectal cancer. Front Oncol (2021) 11:704038. doi: 10.3389/fonc.2021.704038
75. Zhang J, Song Y, Shi Q, Fu L. Research progress on FASN and MGLL in the regulation of abnormal lipid metabolism and the relationship between tumor invasion and metastasis. Front Med (2021) 15(5):649–56. doi: 10.1007/s11684-021-0830-0
76. Wan Y, Hopper AK. Size matters: conserved proteins function in length-dependent nuclear export of circular RNAs. Genes Dev (2018) 32(9-10):600–1. doi: 10.1101/gad.316216.118
77. Li Z, Kearse MG, Huang C. The nuclear export of circular RNAs is primarily defined by their length. RNA Biol (2019) 16(1):1–4. doi: 10.1080/15476286.2018.1557498
78. Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature (2013) 495(7441):333–8. doi: 10.1038/nature11928
79. Park OH, Ha H, Lee Y, Boo SH, Kwon DH, Song HK, et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol Cell (2019) 74(3):494–507.e8. doi: 10.1016/j.molcel.2019.02.034
80. Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell (2019) 177(4):865–880.e21. doi: 10.1016/j.cell.2019.03.046
81. Fischer JW, Busa VF, Shao Y, Leung AKL. Structure-mediated RNA decay by UPF1 and G3BP1. Mol Cell (2020) 78(1):70–84.e6. doi: 10.1016/j.molcel.2020.01.021
82. de Oliveira da Silva B, Alberici LC, Ramos LF, Silva CM, da Silveira MB, Dechant CR, et al. Altered global microRNA expression in hepatic stellate cells LX-2 by angiotensin-(1-7) and miRNA-1914-5p identification as regulator of pro-fibrogenic elements and lipid metabolism. Int J Biochem Cell Biol (2018) 98:137–55. doi: 10.1016/j.biocel.2018.02.018
83. Ye K, Wu Y, Sun Y, Lin J, Xu J, et al. TLR4 siRNA inhibits proliferation and invasion in colorectal cancer cells by downregulating ACAT1 expression. Life Sci (2016) 155:133–9. doi: 10.1016/j.lfs.2016.05.012
84. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med (2020) 383(27):2603–15. doi: 10.1056/NEJMoa2034577
85. Jackson LA, Anderson EJ, Rouphael NG, Roberts PC, Makhene M, Coler RN, et al. An mRNA vaccine against SARS-CoV-2 - preliminary report. N Engl J Med (2020) 383(20):1920–31. doi: 10.1056/NEJMoa2022483
86. Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, et al. Evaluation of the mRNA-1273 vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med (2020) 383(16):1544–55. doi: 10.1056/NEJMoa2024671
87. Wilgenhof S, Corthals J, Heirman C, van Baren N, Lucas S, Kvistborg P, et al. Phase II study of autologous monocyte-derived mRNA electroporated dendritic cells (TriMixDC-MEL) plus ipilimumab in patients with pretreated advanced melanoma. J Clin Oncol (2016) 34(12):1330–8. doi: 10.1200/JCO.2015.63.4121
88. Wilgenhof S, Van Nuffel AMT, Benteyn D, Corthals J, Aerts C, Heirman C, et al. A phase IB study on intravenous synthetic mRNA electroporated dendritic cell immunotherapy in pretreated advanced melanoma patients. Ann Oncol (2013) 24(10):2686–93. doi: 10.1093/annonc/mdt245
89. de Fraipont F, Gazzeri S, Cho WC, Eymin B. Circular RNAs and RNA splice variants as biomarkers for prognosis and therapeutic response in the liquid biopsies of lung cancer patients. Front Genet (2019) 10:390. doi: 10.3389/fgene.2019.00390
90. Li P, Chen S, Chen H, Mo X, Li T, Shao Y, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta (2015) 444:132–6. doi: 10.1016/j.cca.2015.02.018
91. Tao X, Shao Y, Lu R, Ye Q, Xiao B, Ye G, et al. Clinical significance of hsa_circ_0000419 in gastric cancer screening and prognosis estimation. Pathol Res Pract (2020) 216(1):152763. doi: 10.1016/j.prp.2019.152763
92. Tian J, Xi X, Wang J, Yu J, Huang Q, Ma R, et al. CircRNA hsa_circ_0004585 as a potential biomarker for colorectal cancer. Cancer Manag Res (2019) 11:5413–23. doi: 10.2147/CMAR.S199436
93. Bloomfield J, Sabbah M, Castela M, Mehats C, Uzan C, Canlorbe G, et al. Clinical value and molecular function of circulating MicroRNAs in endometrial cancer regulation: A systematic review. Cells (2022) 11(11):1836. doi: 10.3390/cells11111836
94. Pardini B, Sabo AA, Birolo G, Calin GA. Noncoding RNAs in extracellular fluids as cancer biomarkers: The new frontier of liquid biopsies. Cancers (Basel) (2019) 11(8):1170. doi: 10.3390/cancers11081170
95. Fotin-Mleczek M, Zanzinger K, Heidenreich R, Lorenz C, Kowalczyk A, Kallen KJ, et al. mRNA-based vaccines synergize with radiation therapy to eradicate established tumors. Radiat Oncol (2014) 9:180. doi: 10.1186/1748-717X-9-180
96. Cafri G, Gartner JJ, Zaks T, Hopson K, Levin N, Paria BC, et al. mRNA vaccine-induced neoantigen-specific T cell immunity in patients with gastrointestinal cancer. J Clin Invest (2020) 130(11):5976–88. doi: 10.1172/JCI134915
97. Hu XY, Song Z, Yang ZW, Li JJ, Liu J, Wang HS, et al. Cancer drug resistance related microRNAs: recent advances in detection methods. Analyst (2022) 147(12):2615–32. doi: 10.1039/D2AN00171C
98. Ghaemi S, Fekrirad Z, Zamani N, Rahmani R, Arefian E, et al. Non-coding RNAs enhance the apoptosis efficacy of therapeutic agents used for the treatment of glioblastoma multiform. J Drug Target (2022) 30(6):589–602. doi: 10.1080/1061186X.2022.2047191
99. Gu C, Cai J, Xu Z, Zhou S, Ye L, Yan Q, et al. MiR-532-3p suppresses colorectal cancer progression by disrupting the ETS1/TGM2 axis-mediated wnt/β-catenin signaling. Cell Death Dis (2019) 10(10):739. doi: 10.1038/s41419-019-1962-x
100. Huang WC, Jang TH, Tung SL, Yen TC, Chan SH, Wang LH, et al. A novel miR-365-3p/EHF/keratin 16 axis promotes oral squamous cell carcinoma metastasis, cancer stemness and drug resistance via enhancing β5-integrin/c-met signaling pathway. J Exp Clin Cancer Res (2019) 38(1):8. doi: 10.1186/s13046-019-1091-5
Keywords: noncoding RNA, lipid metabolism, tumor, diagnosis, treatment
Citation: Wang Y, Li Q, Wang S, Wang B-j, Jin Y, Hu H, Fu Q-s, Wang J-w, Wu Q, Qian L, Cao T-t, Xia Y-b, Huang X-x and Xu L (2022) The role of noncoding RNAs in cancer lipid metabolism. Front. Oncol. 12:1026257. doi: 10.3389/fonc.2022.1026257
Received: 23 August 2022; Accepted: 20 October 2022;
Published: 14 November 2022.
Edited by:
Thomas W. Grunt, Medical University of Vienna, AustriaReviewed by:
Thasni Karedath, Qatar Biomedical Research Institute, QatarChunyan Sun, Huazhong University of Science and Technology, China
Copyright © 2022 Wang, Li, Wang, Wang, Jin, Hu, Fu, Wang, Wu, Qian, Cao, Xia, Huang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Li Xu, xulixuli1973@163.com; Xiao-xu Huang, xiaoxuhuang1984@163.com
†These authors have contributed equally to this work
 Ye Wang1,2,3†
Ye Wang1,2,3† Qian Li
Qian Li Yan Jin
Yan Jin Long Qian
Long Qian Li Xu
Li Xu