Corrigendum: Research progress on microRNA-1258 in the development of human cancer
- Department of Thyroid and Breast Surgery, The Affiliated JiangNing Hospital of Nanjing Medical University, Nanjing, China
microRNAs (miRNAs) are small endogenous RNAs composed of 20-22 nucleotides that do not encode proteins, which regulate the expression of downstream genes by targeting the 3’ untranslated region of mRNA. Plentiful research has demonstrated that miRNAs participate in the initiation and development of diverse diseases and malignant tumors. miR-1258 exerts great influence on tumors, including tumor growth, distant metastasis, migration, invasion, chemosensitivity, cell glycolysis, apoptosis, and stemness. Interestingly, miR-1258 is a miRNA with explicit functions and has been investigated to act as a tumor suppressor in studies on various types of tumors. With accumulating research on miR-1258, it has been found to be used as a biomarker in the early diagnosis and prognosis prediction of tumor patients. In this review, we outline the development of miR-1258 research, describe its regulatory network, and discuss its roles in cancer. Additionally, we generalize the potential clinical applications of miR-1258. This review offers emerging perspectives and orientations for further comprehending the function of miR-1258 as a diagnostic and prognostic biomarker and potent therapeutic target in cancer.
Introduction
MicroRNAs (miRNAs) are non-coding RNAs composed of about 21-25 nucleotides, which are widely distributed from viruses to numerous cells (1). These miRNAs generally target one or more messenger RNAs (mRNAs), split them directly, or block their translation by binding to mRNAs, thereby blocking their protein production (2).
miRNAs are produced by endogenous transcription of primary transcripts, first cut in the nucleus by Drosha, generating stem-loop precursor miRNAs (about 70 nucleotides). Subsequently, Exportin5 transports precursor miRNA from the nucleus to the cytoplasm. Finally, mature miRNAs about 22 nucleotides in length are processed by Dicer (3).
Heretofore, most of the miRNAs found and reported are highly conserved among species, which are closely related to the significance of their functions (4). miRNAs play an essential role in the regulation of cell differentiation, tissue development, metabolism, and tumorigenesis (5, 6). Extensive studies have shown that miRNAs are dysregulated in tumors, and they widely participate in the whole process of tumor development as tumor suppressors or carcinogens (7–11). Moreover, miRNAs also act key roles in early diagnosis, treatment response predictors, and prognostic biomarkers of cancer (12–16).
miR-1258 is located in the first intron of its host gene ZNF385B (Zinc Finger Protein 385B) on chromosome 2q31.3 and plays a key regulatory role in intestinal barrier function, herpesvirus Lytic replication, bronchopulmonary dysplasia, brown adipose differentiation, and other diseases (17–20). miR-1258 is a miRNA with explicit functions and has been reported to act as a tumor suppressor in studies on diverse types of tumors. So far, there is no review on the research progress of miR-1258 in the development of human cancer. Hence, we first comprehensively and systematically reviewed the research progress of miR-1258 in the inhibitory role of human cancer and its detailed mechanism, to better translate the key role of miR-1258 into diagnostic and prognostic biomarkers and potential targets of cancer.
miR-1258 expression in human cancer
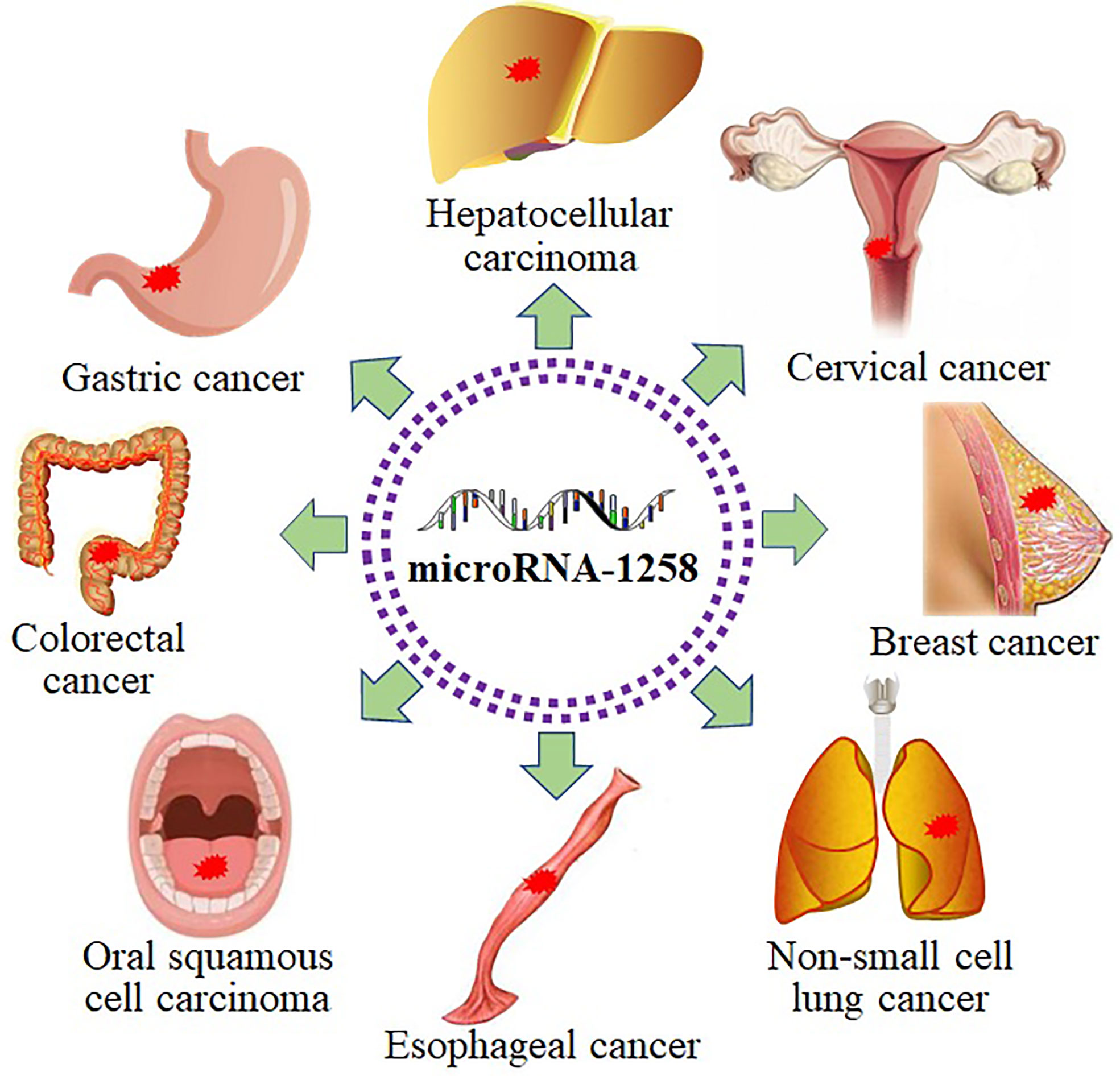
As shown in Table 1, miR-1258 was widely downregulated in the tumor tissues and cell lines of hepatocellular carcinoma (HCC) (21–25), gastric cancer (GC) (26), colorectal cancer (CRC) (27–29), oral squamous cell carcinoma (OSCC) (30), esophageal cancer (EC) (31), non-small cell lung cancer (NSCLC) (32–34), breast cancer (BC) (35–40), cervical cancer (CC) (41), myeloma (42), thyroid carcinoma (43), glioblastoma (44), and osteosarcoma (45) via the detection of qRT-PCR. In addition, the hypermethylation of the CpG island of the miR-1258 host gene was detected in ovarian cancer (OC), myeloma, prostate cancer, and BC via the methylation-specific PCR analysis (38, 42, 46–49). Since 2011, miR-1258 has been widely investigated and reported as a tumor suppressor.
The regulatory network of miR-1258 in cancer
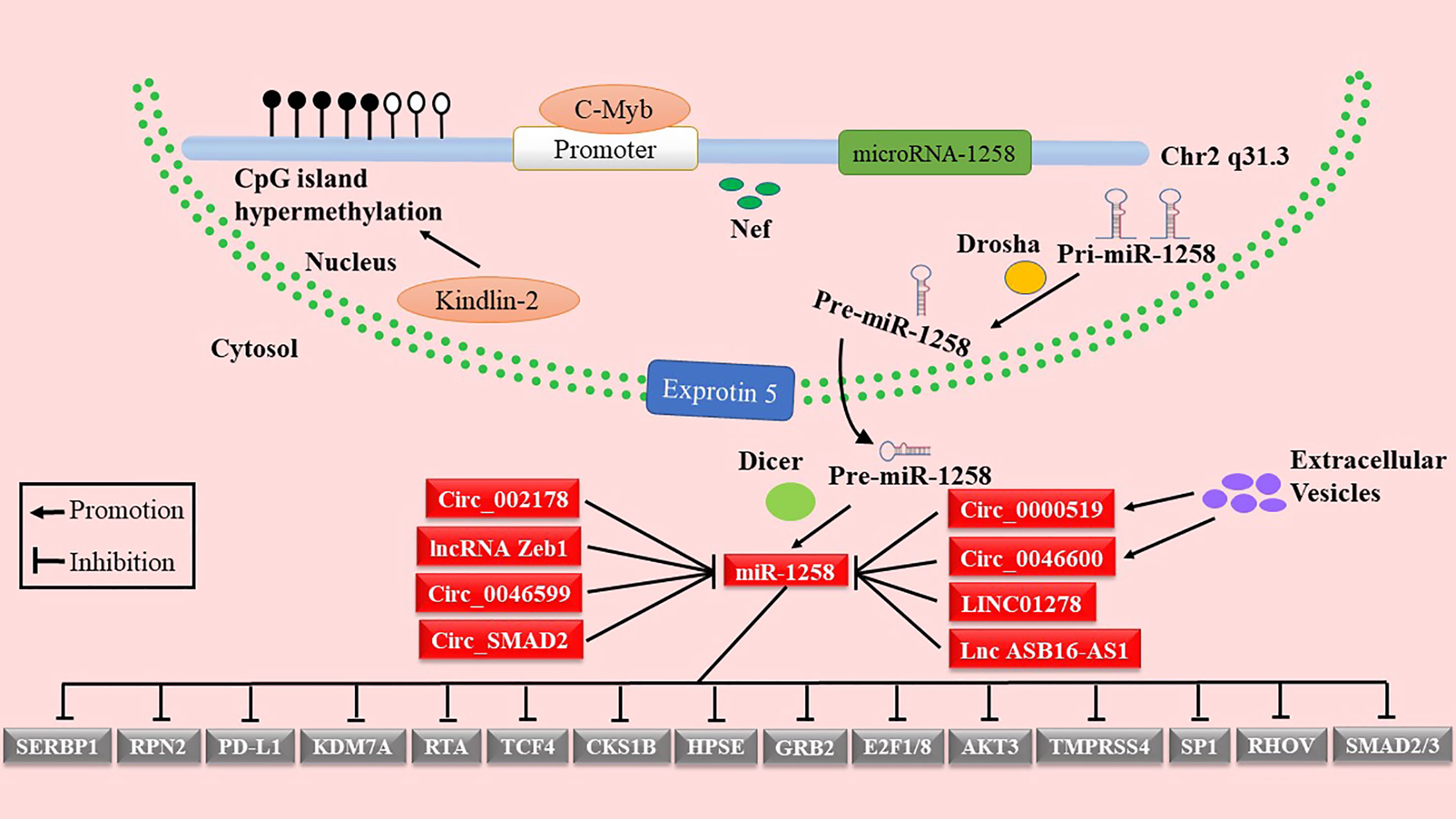
As shown in Figure 1 and Table 1, miR-1258 was regulated by Nef, c-Myb, and Kindlin-2 at the transcriptional stage. Yan et al. reported that negative factor (Nef), a secreted HIV-1 protein, elevated the expression of has-miR-1258 in primary effusion lymphoma cells (18). Kindlin-2 inhibited the transcription of miR-1258 by increasing methylation of the CpG island in the miR-1258 promoter (23). Moreover, c-Myb, a transcriptional factor, is directly bound to the promoter of has-miR-1258 to repress the transcription of miR-1258 (30). Furthermore, the hypermethylation of the CpG island of the miR-1258 promoter was demonstrated in the tissues of ovarian cancer, myeloma, prostate cancer, and BC, inhibiting the transcription of miR-1258 (38, 42, 46–49).
Accumulative studies indicated that the mature miRNAs were inactivated by lncRNAs or circRNAs through competing endogenous RNAs (ceRNAs) (50–52). miR-1258 was inactivated by LncRNA Zeb1 in the intestinal barrier (19). In addition, miR-1258 was targeted by circ_0046599, circ_0046600, and lncRNA LINC01278 in HCC (21, 22, 25). Besides, extracellular vesicles also acted key roles in the regulation of miR-1258 by transferring circ_0000519 as ceRNA in NSCLC (34). Zhang et al. noted that upregulated circ_SMAD2 suppressed the expression of miR-1258 through the ceRNAs mechanism in CRC (27). LncRNA ASB16-AS1, as sponge molecules, regulated the EC progression by absorbing miR-1258 (31). Li et al. reported that miR-1258 was directly regulated by circ_002178 via using luciferase reporter assay (35). Therefore, it is necessary to provide a theoretical basis for using miR-1258 as a potential therapeutic target by in-depth excavating of the regulatory mechanisms of miR-1258 in cancer.
The biological functions of miR-1258 in cancer
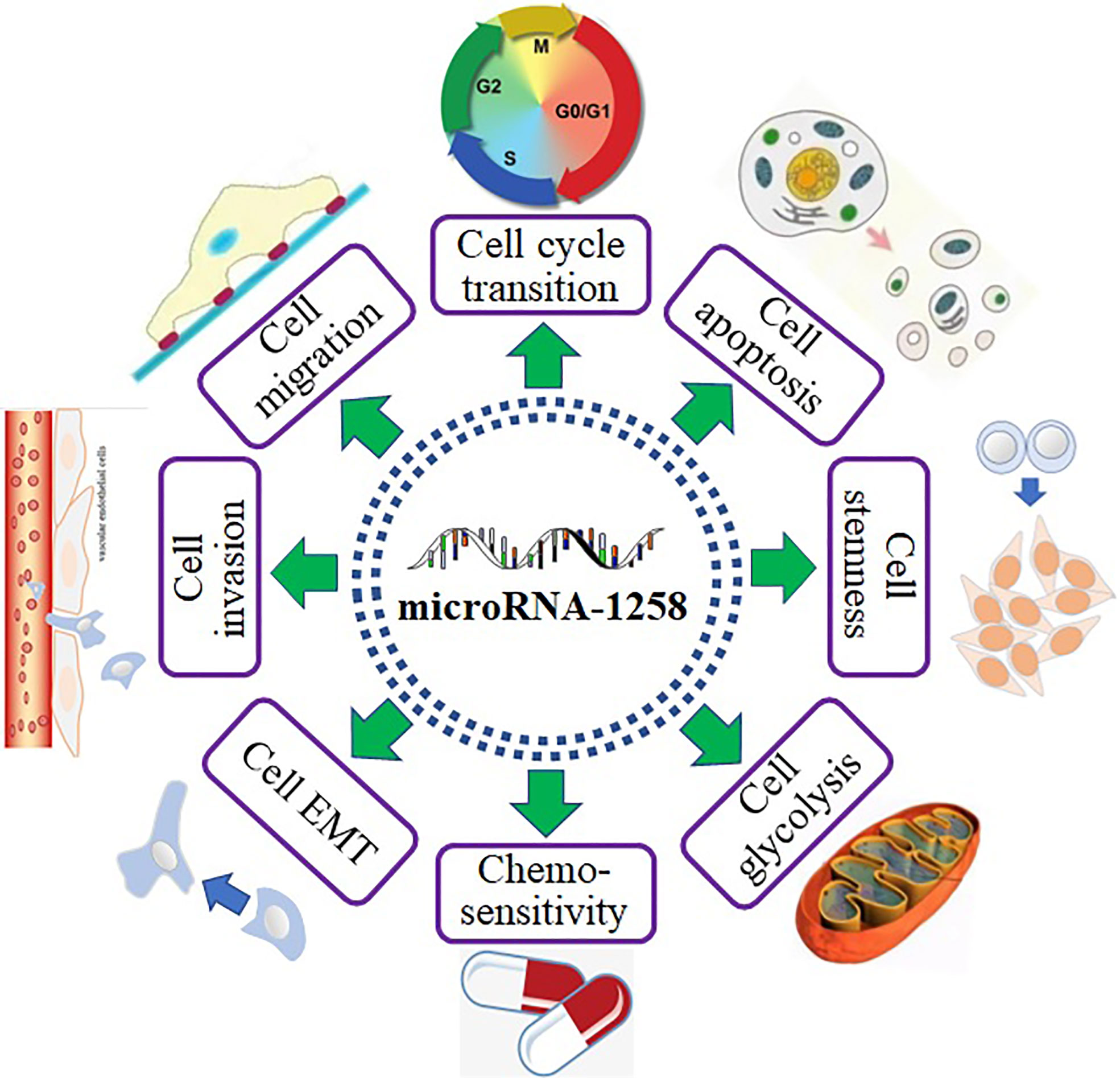
MiR-1258 profoundly inhibited tumor progression by binding to the mRNA of downstream genes (Figure 1). As shown in Figure 2, miR-1258 was involved in the biological processes of the cell cycle transition, cell apoptosis, cell stemness, cell migration and invasion, and EMT to restrain the progression of tumors. Also, miR-1258 repressed glycolysis metabolisms by targeting the mRNA of RPN2 to suppress the cell growth of HCC (21). The stemness of cancer cells plays a crucial role in the survival, proliferation, recurrence, and drug resistance of cancer (53). miR-1258 significantly suppressed the cell stemness and tumor progression of HCC by binding to the mRNA of cyclin-dependent kinase regulatory subunit 1B (CKS1B) (24). In the following parts, we comprehensively revealed the biological functions of miR-1258 as tumor suppressors.
Inhibition of the cancerogenic process in human cancer
MiR-1258 was extensively investigated in digestive system-related tumors as a tumor suppressor, including HCC (21–25), GC (26), CRC (27–29), OSCC (30), and EC (31). MiR-1258 inhibited cell viability and tumor progression by targeting key signaling pathway proteins and related transcription factors to function as a tumor suppressor in gastrointestinal cancer. Tumor cells are characterized by immortal and infinitely dividing cells (54). Overexpressed miR-1258 induced cell senescence and apoptosis and suppressed cell viability to mitigate tumor progression by binding to the targets in NSCLC (32–34). As a histone demethylase, KDM7A extensively affects the malignant biological behaviors of tumor cells by regulating cell cycle transition (55). The progression of BC was prohibited by miR-1258 through increasing cell apoptosis and stemness and decreasing cell viability and cell cycle transition via devitalizing key proteins, including KDM7A (35–38, 40). Furthermore, miR-1258 also functioned as a tumor suppressor in CC by restraining cell proliferation and enhancing cell apoptosis via targeting E2F1 (41). Besides, miR-1258 still repressing the tumor process in myeloma, thyroid carcinoma, glioblastoma, and osteosarcoma (42–45). Abovementioned facts indicated that miR-1258 was extensively involved in the development of human cancer (Figure 3).
Promotion of cell apoptosis and chemosensitivity
Currently, numerous small-molecule anti-cancer drugs are targeting molecules involved in cell apoptosis (56). Growth factor receptor binding protein 2 (GRB2) is a key adapter protein that activates the RAS/ERK signaling pathway, and its dysregulation can profoundly affect the process of cell apoptosis in cancer (57). miR-1258 inhibited the NSCLC progression via inducing cell apoptosis and senescence by directly targeting GRB2 (32). Tumor cells evade immune clearance by increasing the expression of PD-L1 on the surface and inhibiting T cell function via binding to PD-1 on the surface of T cells (58). Wang et al. reported that overexpressed miR-1258 inhibited cell proliferation and increased cell apoptosis by restraining the expression of PD-L1 in completely methylated myeloma cells (42). In addition, miR-1258 strengthened the cell apoptosis to repress cell proliferation by binding to mRNA of SERBP1, CKS1B, and E2F1 in HCC (22, 24) and BC (37). Furthermore, the sensitivity of chemotherapy was increased by enhancing cell apoptosis (59). Hu et al. indicated that upregulated miR-1258 greatly reinforced the sensitivity of HCC cells to chemotherapy drugs in vivo by restraining the expression of CKS1B (24).
Suppression of cell migration and invasion
The migratory and invasive ability of cells largely determines whether distant metastasis occurs in tumors, which is the main reason for the poor prognosis of patients with cancer (60). TGF-β (transforming growth factor-β)/Smad signaling pathway significantly regulates the biological behaviors of cell migration and invasion in cancer (61). Huang et al. found that miR-1258 inhibited the metastasis of cells by impairing the migratory and invasive ability of cells via targeting the mRNA of Smad2 and Smad3 in HCC (25). Besides, the translation of RPN2, TCF4, KDM7A, HPSE, TMPRSS4, and E2F1 was also suppressed by miR-1258 overexpression, according to recent research, which prevented cell invasion and migration in HCC (21, 23), CRC (27), BC (35, 40), and TC (43). It is impossible to disregard the roles of the matrix metalloproteinase (MMP) family in regulating cell migration and invasion (62). Qin et al. demonstrated that miR-1258 inhibited the transcription of MMP2 and PCNA through binding to the mRNA of E2F1 to depress the cell migration and invasion in glioblastoma (44). The abovementioned data indicated that miR-1228 regulated cell migration and invasion by mainly interacting with the TGF-β/Smad pathway and MMP family.
Inhibition of cell cycle transition
The cell cycle includes four consecutive phases of G0/G1, S, G2, and M, which is a set of organized and monitored events that are responsible for dividing cells into two daughter cells. The aberrant regulation of cell cycle transition has played a critical role in the growth and development of tumors (63). E2F family performs crucial functions in controlling cell cycle, maintaining genomic integrity, and coping with replication stress and DNA damage as transcriptional factors (64). miR-1258 arrested the cell cycle in G0/G1 phase by targeting E2F1 and E2F8 in CRC, BC, CC, and glioblastoma (28, 37, 41, 44). CKS1B is engaged in the transcription regulation of a series of genes involved in the cell cycle process, which is closely related to the abnormal cell proliferation of tumors (65). Overexpressed miR-1258 greatly inhibited cell cycle arrest in G0/G1 phase by directly repressing the expression of CKS1B in HCC (24). Additionally, miR-1258 inhibited the cell cycle transition by binding to mRNA of SERBP1, GRB2, and AKT3 in HCC (22), NSCLC (32), and osteosarcoma (45), respectively. These data proved that miR-1258, a potential therapeutic target, performed key roles in cell cycle arrest in cancer.
Role in EMT
Epithelial-mesenchymal transition (EMT) refers to the transformation of epithelial cells into invasive mesenchymal cells, which plays a crucial function in the invasion and metastasis of various types of cancer (66, 67). SP1, as a transcription factor, directly modulates the EMT and metastasis of cancer at transcriptional levels (68, 69). Overexpressed miR-1258 significantly repressed the EMT and metastasis in OSCC cells by targeting the mRNA of SP1 (30). In addition, Lin et al. demonstrated that miR-1258 suppressed the EMT and metastasis of HCC cells through targeting TCF4, a key member of the Wnt/β-catenin signaling pathway (23). Heparanase (HPSE) is a potent enzyme that fosters tumor growth, angiogenesis, and metastasis (70). Its dysregulation can produce a wide range of effects that significantly alter the microenvironment, stimulating cell growth and metastasis of tumors (71). Zhang et al. first revealed that upregulated miR-1258 inhibited breast cancer brain metastasis through targeting HPSE by using regulatory experiments, functional experiments, and clinical specimens’ validation (40). Moreover, overexpression of miR-1258 suppressed the cell metastasis through repressing the expression of RHOV in NSCLC both in vitro and in vivo (34). Collectively, miR-1258 can effectively regulate the EMT process and metastasis through targeting different genes at the post-transcriptional level in cancer.
Clinical application
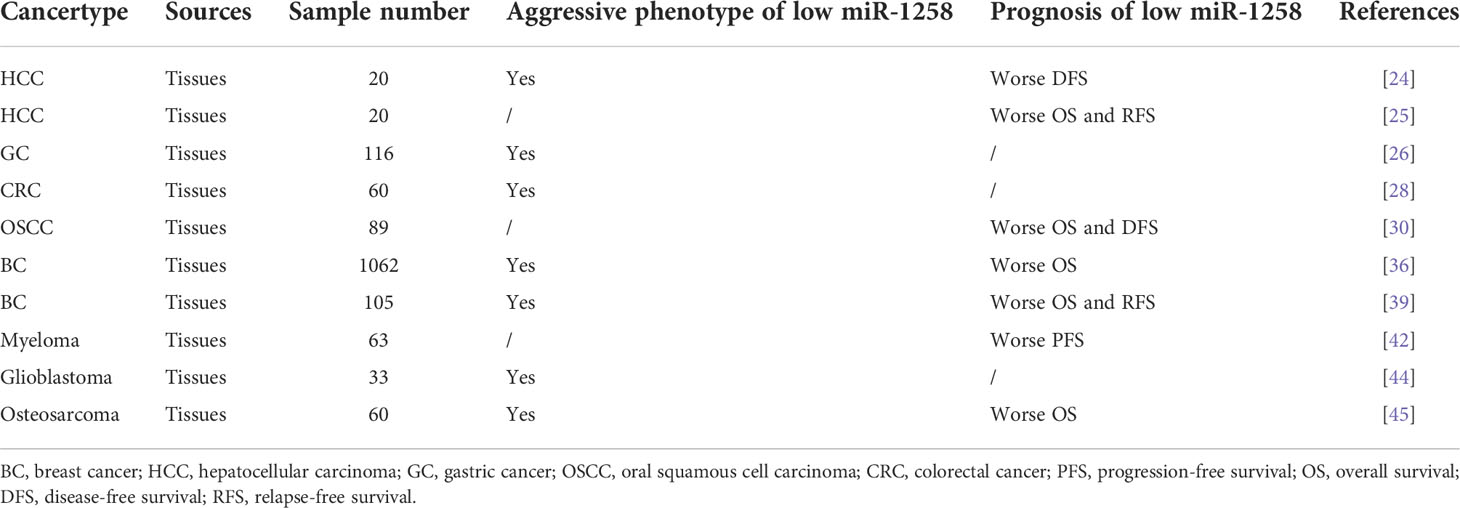
It has been reported that it is promising to manipulate these miRNAs for cancer treatment by combining effective applications of miRNA delivery systems, such as chemical modification of miRNAs, lipid-based miRNA delivery systems, and organic/inorganic composite nanoparticles (72). Besides, abundant studies have found that the differential profiles of miRNAs in circulation or tissues were closely correlated to the early diagnosis, clinical stage, response to therapy, and pathological characteristics of tumors (11). In addition, the abnormal expression of miRNAs can also be used to predict the long-term survival of tumor patients (73). The probability of distant metastasis and the clinical stage of OC can be predicted by the frequency of methylation in the promoter of the miR-1258 host gene. Metastatic patients had a twofold higher rate of miR-1258 methylation than non-metastatic OC patients did (46, 47). In addition, the methylation level of miR-1258 was positively correlated to the advanced clinical stage and pathological characteristics of OC, BC, and myeloma (38, 42, 48). Besides, the level of miR-1258 promoter methylation can accurately diagnose prostate cancer in clinical samples with 97.8% sensitivity and 100% specificity (49). Due to the important role of miR-1258 as a tumor suppressor, the level of its expression profoundly affected the prognosis and clinicopathological characteristics of tumor patients. As shown in Table 2, a lower level of miR-1258 meant an inferior progression-free survival (PFS) and a higher probability of recurrence in 63 patients of myeloma (42). Low expression of miR-1258 was not only associated with the advanced clinical stage but also meant worse overall survival (OS) and relapse-free survival (RFS) in BC patients (36, 39). In addition, the level of miR-1258 was negatively correlated with the probability of tumor recurrence and metastasis and poor disease-free survival (DFS), OS, and RFS in HCC (24, 25). Shi et al. demonstrated that miR-1258 was negatively correlated to advanced clinical stage and lymphatic vessel invasion by analyzing the postoperative pathological data of 116 GC patients (26). Furthermore, a lower expression of miR-1258 in ESCC patients meant a shorter OS and DFS (30). Qin et al. discovered that the level of miR-1258 expression was decreased with the elevation of pathological grade by analyzing the postoperative pathological results of 33 glioblastoma patients (44). In addition, patients with low miR-1258 were greatly related to bigger tumor size in CRC (28). The OS would be greatly shortened in osteosarcoma patients with a low level of miR-1258 expression. Meanwhile, decreased miR-1258 was strongly correlated to the malignant clinicopathological characteristics of patients with osteosarcoma (45). These results verified the significant role and prospective clinical relevance of miR-1258 as diagnostic and prognostic biomarkers in cancer.
Conclusions and prospects
At present, malignant tumors with high morbidity and mortality imposed a heavy burden on patients worldwide. Accumulative studies have been trying to reveal the etiology of tumor initiation and explore significant therapies. However, the mechanisms of tumorigenesis, recurrence, metastasis, and drug resistance remained unclear. Researchers reported that the expression of miR-1258 was considerably downregulated in tumor tissues and cell lines. To date, miR-1258 has been shown to act as a tumor suppressor in the development and progression of tumors, suppressing cell cycle transition, metastasis, stemness, migration, invasion, EMT, and glycolysis while boosting cell apoptosis and chemosensitivity. Furthermore, studies revealed that miR-1258 can be employed as a biomarker for early diagnosis and prognosis prediction in tumor patients.
In summary, with the deepening related research of miR-1258, the mechanism of miR-1258 in tumorigenesis and progression would be gradually disclosed. miR-1258 can be exploited as an indicator for early tumor diagnosis and prognosis as well as a potential target for tumor treatment, providing novel perspectives and orientations for precision therapy.
Author contributions
MQ and YC generated this topic. MQ and YX wrote the manuscript. MQ and GZ searched and collected all relevant literature. MQ and HY constructed the tables and figures. YC supervised and modified the manuscript. MQ and YX contributed equally to this study. All authors contributed to the article and approved the submitted version.
Funding
This research got funding from the Natural Science Foundation of Jiangsu Province (Grant No. BK20170142).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviation
BC, breast cancer; HCC, hepatocellular carcinoma; EC, esophageal cancer; NSCLC, non-small cell lung cancer; CC, cervical cancer; GC, gastric cancer; OSCC, oral squamous cell carcinoma; CRC, colorectal cancer; PFS, progression-free survival; OS, overall survival; DFS, disease-free survival; RFS, relapse-free survival.
References
1. Ambros V. microRNAs: tiny regulators with great potential. Cell (2001) 107(7):823–6. doi: 10.1016/S0092-8674(01)00616-X
2. Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell (2005) 123(4):631–40. doi: 10.1016/j.cell.2005.10.022
3. Lee Y, Ahn C, Han J, et al. The nuclear RNase III drosha initiates microRNA processing. Nature (2003) 425(6956):415–9. doi: 10.1038/nature01957
4. Ke XS, Liu CM, Liu DP, Liang CC. MicroRNAs: key participants in gene regulatory networks. Curr Opin Chem Biol (2003) 7(4):516–23. doi: 10.1016/S1367-5931(03)00075-9
5. Tsuchiya S, Okuno Y, Tsujimoto G. MicroRNA: biogenetic and functional mechanisms and involvements in cell differentiation and cancer. J Pharmacol Sci (2006) 101(4):267–70. doi: 10.1254/jphs.CPJ06013X
6. Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Curr Opin Cell Biol (2008) 20(2):214–21. doi: 10.1016/j.ceb.2008.01.006
7. Hu T, Shen H, Li J, Yang P, Gu Q, Fu Z. RFC2, a direct target of miR-744, modulates the cell cycle and promotes the proliferation of CRC cells. J Cell Physiol (2020) 235(11):8319–33. doi: 10.1002/jcp.29676
8. Qian W, Feng Y, Li J, et al. Construction of ceRNA networks reveals differences between distal and proximal colon cancers. Oncol Rep (2019) 41(5):3027–40. doi: 10.3892/or.2019.7083
9. Chen F, Chu L, Li J, et al. Hypoxia induced changes in miRNAs and their target mRNAs in extracellular vesicles of esophageal squamous cancer cells. Thorac cancer. (2020) 11(3):570–80. doi: 10.1111/1759-7714.13295
10. Ma M, Li J, Zhang Z, et al. The role and mechanism of microRNA-1224 in human cancer. Front Oncol (2022) 12:858892. doi: 10.3389/fonc.2022.858892
11. Li J, Sun J, Liu Z, et al. The roles of non-coding RNAs in radiotherapy of gastrointestinal carcinoma. Front Cell Dev Biol (2022) 10:862563. doi: 10.3389/fcell.2022.862563
12. Li J, Feng Y, Heng D, et al. Circulating non-coding RNA cluster predicted the tumorigenesis and development of colorectal carcinoma. Aging (2020) 12(22):23047–66. doi: 10.18632/aging.104055
13. Chen F, Xu B, Li J, et al. Hypoxic tumour cell-derived exosomal miR-340-5p promotes radioresistance of oesophageal squamous cell carcinoma via KLF10. J Exp Clin Cancer research: CR. (2021) 40(1):38. doi: 10.1186/s13046-021-01834-9
14. Zhang Y, Peng C, Li J, et al. Long non-coding RNA CCDC144NL-AS1 promotes cell proliferation by regulating the miR-363-3p/GALNT7 axis in colorectal cancer. J Cancer. (2022) 13(3):752–63. doi: 10.7150/jca.65885
15. Li J, Han X, Gu Y, et al. LncRNA MTX2-6 suppresses cell proliferation by acting as ceRNA of miR-574-5p to accumulate SMAD4 in esophageal squamous cell carcinoma. Front Cell Dev Biol (2021) 9:654746. doi: 10.3389/fcell.2021.654746
16. Zhang Z, Wang S, Ji D, et al. Construction of a ceRNA network reveals potential lncRNA biomarkers in rectal adenocarcinoma. Oncol Rep (2018) 39(5):2101–13. doi: 10.3892/or.2018.6296
17. Zhang Y, Kong X, Zhang J, Wang X. Functional analysis of bronchopulmonary dysplasia-related neuropeptides in preterm infants and miRNA-based diagnostic model construction. Comput Math Methods Med (2022) 2022:5682599. doi: 10.1155/2022/5682599
18. Yan Q, Ma X, Shen C, et al. Inhibition of kaposi’s sarcoma-associated herpesvirus lytic replication by HIV-1 nef and cellular microRNA hsa-miR-1258. J virology. (2014) 88(9):4987–5000. doi: 10.1128/JVI.00025-14
19. Yang X, Gao Y, Huang S, Su C, Wang J, Zheng N. Whole transcriptome-based ceRNA network analysis revealed ochratoxin a-induced compromised intestinal tight junction proteins through WNT/Ca(2+) signaling pathway. Ecotoxicology Environ safety. (2021) 224:112637. doi: 10.1016/j.ecoenv.2021.112637
20. Cao Y, Deng B, Zhang S, et al. Astragalus polysaccharide regulates brown adipogenic differentiation through miR-1258-5p-modulated cut-like homeobox 1 expression. Acta Biochim Biophys Sinica. (2021) 53(12):1713–22. doi: 10.1093/abbs/gmab151
21. Fang Q, Liu H, Zhou A, Zhou H, Zhang Z. Circ_0046599 promotes the development of hepatocellular carcinoma by regulating the miR-1258/RPN2 network. Cancer Manage Res (2020) 12:6849–60. doi: 10.2147/CMAR.S253510
22. Zhang D, Zhang Y, Zhang X, Zhai H, Sun X, Li Y. Circ_0046600 promotes hepatocellular carcinoma progression via up-regulating SERBP1 through sequestering miR-1258. Pathology Res practice. (2021) 228:153681. doi: 10.1016/j.prp.2021.153681
23. Lin W, Lin J, Li J, et al. Kindlin-2-miR-1258-TCF4 feedback loop promotes hepatocellular carcinoma invasion and metastasis. J gastroenterology. (2022) 57(5):372–86. doi: 10.1007/s00535-022-01866-8
24. Hu M, Wang M, Lu H, et al. Loss of miR-1258 contributes to carcinogenesis and progression of liver cancer through targeting CDC28 protein kinase regulatory subunit 1B. Oncotarget (2016) 7(28):43419–31. doi: 10.18632/oncotarget.9728
25. Huang WJ, Tian XP, Bi SX, et al. The β-catenin/TCF-4-LINC01278-miR-1258-Smad2/3 axis promotes hepatocellular carcinoma metastasis. Oncogene (2020) 39(23):4538–50. doi: 10.1038/s41388-020-1307-3
26. Shi J, Chen P, Sun J, et al. MicroRNA-1258: An invasion and metastasis regulator that targets heparanase in gastric cancer. Oncol letters. (2017) 13(5):3739–45. doi: 10.3892/ol.2017.5886
27. Zhang W, Wu G, Sun P, Zhu Y, Zhang H. circ_SMAD2 regulate colorectal cancer cells proliferation through targeting miR-1258/RPN2 signaling pathway. J Cancer. (2021) 12(6):1678–86. doi: 10.7150/jca.50888
28. Zhang Z, Li J, Huang Y, et al. Upregulated miR-1258 regulates cell cycle and inhibits cell proliferation by directly targeting E2F8 in CRC. Cell proliferation. (2018) 51(6):e12505. doi: 10.1111/cpr.12505
29. Hwang JS, Jeong EJ, Choi J, et al. MicroRNA-1258 inhibits the proliferation and migration of human colorectal cancer cells through suppressing CKS1B expression. Genes (2019) 10(11). doi: 10.3390/genes10110912
30. Zhang H, Jiang S, Guo L, Li X. MicroRNA-1258, regulated by c-myb, inhibits growth and epithelial-to-mesenchymal transition phenotype via targeting SP1 in oral squamous cell carcinoma. J Cell Mol Med (2019) 23(4):2813–21. doi: 10.1111/jcmm.14189
31. Jia Z, Wang PS, Yang Y, Zhu DY, Wang ZH, Wang W. [LncRNA ASB16-AS1 regulates the proliferation, migration and invasion of esophageal cancer cells by targeting miR-1258]. Zhonghua zhong liu za zhi [Chinese J oncology]. (2021) 43(7):762–8.
32. Jiang W, Wei K, Pan C, et al. MicroRNA-1258 suppresses tumour progression via GRB2/Ras/Erk pathway in non-small-cell lung cancer. Cell proliferation. (2018) 51(6):e12502. doi: 10.1111/cpr.12502
33. Liu H, Chen X, Gao W, Jiang G. The expression of heparanase and microRNA-1258 in human non-small cell lung cancer. Tumour biology: J Int Soc Oncodevelopmental Biol Med (2012) 33(5):1327–34. doi: 10.1007/s13277-012-0380-9
34. Wang R, Liu H, Dong M, Huang D, Yi J. Exosomal hsa_circ_0000519 modulates the NSCLC cell growth and metastasis via miR-1258/RHOV axis. Open Med (Warsaw Poland). (2022) 17(1):826–40. doi: 10.1515/med-2022-0428
35. Li W, Yang X, Shi C, Zhou Z. Hsa_circ_002178 promotes the growth and migration of breast cancer cells and maintains cancer stem-like cell properties through regulating miR-1258/KDM7A axis. Cell transplantation. (2020) 29:963689720960174. doi: 10.1177/0963689720960174
36. Sang M, Li A, Wang X, et al. Identification of three miRNAs signature as a prognostic biomarker in breast cancer using bioinformatics analysis. Trans Cancer Res (2020) 9(3):1884–93. doi: 10.21037/tcr.2020.02.21
37. Zhao X. miR-1258 regulates cell proliferation and cell cycle to inhibit the progression of breast cancer by targeting E2F1. BioMed Res Int (2020) 2020:1480819. doi: 10.1155/2020/1480819
38. Loginov VI, Burdennyy AM, Pronina IV, et al. [Novel miRNA genes hypermethylated in breast cancer]. Molekuliarnaia biologiia. (2016) 50(5):797–802. doi: 10.1134/S0026893316050101
39. Tang D, Zhang Q, Zhao S, et al. The expression and clinical significance of microRNA-1258 and heparanase in human breast cancer. Clin Biochem (2013) 46(10-11):926–32. doi: 10.1016/j.clinbiochem.2013.01.027
40. Zhang L, Sullivan PS, Goodman JC, Gunaratne PH, Marchetti D. MicroRNA-1258 suppresses breast cancer brain metastasis by targeting heparanase. Cancer Res (2011) 71(3):645–54. doi: 10.1158/0008-5472.CAN-10-1910
41. Peng X, Zhang Y, Gao J, Cai C. MiR-1258 promotes the apoptosis of cervical cancer cells by regulating the E2F1/P53 signaling pathway. Exp Mol pathology. (2020) 114:104368. doi: 10.1016/j.yexmp.2020.104368
42. Wang LQ, Kumar S, Calin GA, Li Z, Chim CS. Frequent methylation of the tumour suppressor miR-1258 targeting PDL1: implication in multiple myeloma-specific cytotoxicity and prognostification. Br J haematology. (2020) 190(2):249–61. doi: 10.1111/bjh.16517
43. Wang LJ, Cai HQ. miR-1258: a novel microRNA that controls TMPRSS4 expression is associated with malignant progression of papillary thyroid carcinoma. Endokrynologia Polska. (2020) 71(2):146–52. doi: 10.5603/EP.a2020.0009
44. Qin H, Gui Y, Ma R, et al. miR-1258 attenuates tumorigenesis through targeting E2F1 to inhibit PCNA and MMP2 transcription in glioblastoma. Front Oncol (2021) 11:671144. doi: 10.3389/fonc.2021.671144
45. Liu W, Zhou Z, Zhang Q, et al. Overexpression of miR-1258 inhibits cell proliferation by targeting AKT3 in osteosarcoma. Biochem Biophys Res Commun (2019) 510(3):479–86. doi: 10.1016/j.bbrc.2019.01.139
46. Braga EA, Loginov VI, Burdennyi AM, et al. Five hypermethylated MicroRNA genes as potential markers of ovarian cancer. Bull Exp Biol Med (2018) 164(3):351–5. doi: 10.1007/s10517-018-3988-y
47. Filippova EA, Loginov VI, Burdennyi AM, et al. Hypermethylated genes of MicroRNA in ovarian carcinoma: Metastasis prediction marker systems. Bull Exp Biol Med (2019) 167(1):79–83. doi: 10.1007/s10517-019-04465-5
48. Loginov VI, Burdennyy AM, Filippova EA, et al. [Hypermethylation of miR-107, miR-130b, miR-203a, miR-1258 genes associated with ovarian cancer development and metastasis]. Molekuliarnaia biologiia. (2018) 52(5):801–9. doi: 10.1134/S0026893318050102
49. Torres-Ferreira J, Ramalho-Carvalho J, Gomez A, et al. MiR-193b promoter methylation accurately detects prostate cancer in urine sediments and miR-34b/c or miR-129-2 promoter methylation define subsets of clinically aggressive tumors. Mol cancer. (2017) 16(1):26. doi: 10.1186/s12943-017-0604-0
50. Zhou R, Jia W, Gao X, et al. CircCDYL acts as a tumor suppressor in wilms’ tumor by targeting miR-145-5p. Front Cell Dev Biol (2021) 9:668947. doi: 10.3389/fcell.2021.668947
51. Qi X, Chen X, Zhao Y, Chen J, Niu B, Shen B. Prognostic roles of ceRNA network-based signatures in gastrointestinal cancers. Front Oncol (2022) 12:921194. doi: 10.3389/fonc.2022.921194
52. Guo L, Jia L, Luo L, et al. Critical roles of circular RNA in tumor metastasis via acting as a sponge of miRNA/isomiR. Int J Mol Sci (2022) 23(13). doi: 10.3390/ijms23137024
53. Boman BM, Wicha MS. Cancer stem cells: a step toward the cure. J Clin oncology: Off J Am Soc Clin Oncol (2008) 26(17):2795–9. doi: 10.1200/JCO.2008.17.7436
54. Ming D, Zhang S, Liu X, Xu C, Zhang X. Nondiploid cancer cells: Stress, tolerance and therapeutic inspirations. Biochim Biophys Acta Rev Cancer (2022), 188794.
55. Chaturvedi SS, Ramanan R, Waheed SO, Karabencheva-Christova TG, Christov CZ. Structure-function relationships in KDM7 histone demethylases. Adv Protein Chem Struct Biol (2019) 117:113–25. doi: 10.1016/bs.apcsb.2019.08.005
56. Wang J, Li D, Zhao B, Kim J, Sui G, Shi J. Small molecule compounds of natural origin target cellular receptors to inhibit cancer development and progression. Int J Mol Sci (2022) 23(5). doi: 10.3390/ijms23052672
57. Yablonski D. Bridging the gap: Modulatory roles of the Grb2-family adaptor, gads, in cellular and allergic immune responses. Front Immunol (2019) 10:1704. doi: 10.3389/fimmu.2019.01704
58. Hu X, Wang J, Chu M, Liu Y, Wang ZW, Zhu X. Emerging role of ubiquitination in the regulation of PD-1/PD-L1 in cancer immunotherapy. Mol therapy: J Am Soc Gene Ther (2021) 29(3):908–19. doi: 10.1016/j.ymthe.2020.12.032
59. Zeng Q, Ma X, Song Y, Chen Q, Jiao Q, Zhou L. Targeting regulated cell death in tumor nanomedicines. Theranostics (2022) 12(2):817–41. doi: 10.7150/thno.67932
60. Sleeman JP, Thiele W. Tumor metastasis and the lymphatic vasculature. Int J cancer. (2009) 125(12):2747–56. doi: 10.1002/ijc.24702
61. Dong H, Diao H, Zhao Y, et al. Overexpression of matrix metalloproteinase-9 in breast cancer cell lines remarkably increases the cell malignancy largely via activation of transforming growth factor beta/SMAD signalling. Cell proliferation. (2019) 52(5):e12633. doi: 10.1111/cpr.12633
62. Brinckerhoff CE, Matrisian LM. Matrix metalloproteinases: a tail of a frog that became a prince. Nat Rev Mol Cell Biol (2002) 3(3):207–14. doi: 10.1038/nrm763
63. Diaz-Moralli S, Tarrado-Castellarnau M, Miranda A, Cascante M. Targeting cell cycle regulation in cancer therapy. Pharmacol Ther (2013) 138(2):255–71. doi: 10.1016/j.pharmthera.2013.01.011
64. Tsantoulis PK, Gorgoulis VG. Involvement of E2F transcription factor family in cancer. Eur J Cancer (Oxford England: 1990). (2005) 41(16):2403–14. doi: 10.1016/j.ejca.2005.08.005
65. Shi W, Huang Q, Xie J, Wang H, Yu X, Zhou Y. CKS1B as drug resistance-inducing gene-a potential target to improve cancer therapy. Front Oncol (2020) 10:582451. doi: 10.3389/fonc.2020.582451
66. Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer metastasis Rev (2009) 28(1-2):15–33. doi: 10.1007/s10555-008-9169-0
67. Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol (2014) 15(3):178–96. doi: 10.1038/nrm3758
68. Li J, Peng W, Yang P, et al. MicroRNA-1224-5p inhibits metastasis and epithelial-mesenchymal transition in colorectal cancer by targeting SP1-mediated NF-κB signaling pathways. Front Oncol (2020) 10:294. doi: 10.3389/fonc.2020.00294
69. Liu Y, Song Y, Cao M, et al. A novel EHD1/CD44/Hippo/SP1 positive feedback loop potentiates stemness and metastasis in lung adenocarcinoma. Clin Trans Med (2022) 12(4):e836. doi: 10.1002/ctm2.836
70. Mayfosh AJ, Nguyen TK, Hulett MD. The heparanase regulatory network in health and disease. Int J Mol Sci (2021) 22(20). doi: 10.3390/ijms222011096
71. Kaur R, Deb PK, Diwan V, Saini B. Heparanase inhibitors in cancer progression: Recent advances. Curr Pharm design. (2021) 27(1):43–68. doi: 10.2174/1381612826666201113105250
72. Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug discovery. (2017) 16(3):203–22. doi: 10.1038/nrd.2016.246
Keywords: miR-1258, cancer, tumor suppressor, biological function, clinical application
Citation: Qian M, Xia Y, Zhang G, Yu H and Cui Y (2022) Research progress on microRNA-1258 in the development of human cancer. Front. Oncol. 12:1024234. doi: 10.3389/fonc.2022.1024234
Received: 21 August 2022; Accepted: 16 September 2022;
Published: 29 September 2022.
Edited by:
Jinhui Liu, Nanjing Medical University, ChinaReviewed by:
Youlong Huili, Affiliated Hospital of North China, University of Science and Technology, ChinaSrikanta Goswami, National Institute of Biomedical Genomics (NIBMG), India
Nur Damayanti, Purdue University Indianapolis, United States
Copyright © 2022 Qian, Xia, Zhang, Yu and Cui. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yiyao Cui, cyytt2000@sina.com
†These authors have contributed equally to this work
 Mengjia Qian
Mengjia Qian Yuke Xia
Yuke Xia Gong Zhang
Gong Zhang