- 1School of Biomedical Sciences at the Translational Research Institute (TRI), Queensland University of Technology (QUT), Brisbane, QLD, Australia
- 2Centre for Personalised Analysis of Cancers (CPAC), Brisbane, QLD, Australia
- 3Queensland Bladder Cancer Initiative (QBCI), Brisbane, QLD, Australia
- 4Australian Prostate Cancer Research Centre - Queensland (APCRC-Q), Brisbane, QLD, Australia
- 5Department of Urology, Princess Alexandra Hospital (PAH), Brisbane, QLD, Australia
- 6Department of Medical Oncology, Princess Alexandra Hospital (PAH), Brisbane, QLD, Australia
- 7School of Mechanical, Medical and Process Engineering, Queensland University of Technology (QUT), Brisbane, QLD, Australia
- 8Australian Research Council (ARC) Training Centre for Cell and Tissue Engineering, Queensland University of Technology (QUT), Brisbane, QLD, Australia
Precision medicine approaches that inform clinical management of individuals with cancer are progressively advancing. Patient-derived explants (PDEs) provide a patient-proximal ex vivo platform that can be used to assess sensitivity to standard of care (SOC) therapies and novel agents. PDEs have several advantages as a patient-proximal model compared to current preclinical models, as they maintain the phenotype and microenvironment of the individual tumor. However, the longevity of PDEs is not compatible with the timeframe required to incorporate candidate therapeutic options identified by whole exome sequencing (WES) of the patient’s tumor. This review investigates how PDE longevity varies across tumor streams and how this is influenced by tissue preparation. Improving longevity of PDEs will enable individualized therapeutics testing, and thus contribute to improving outcomes for people with cancer.
Introduction
Precision cancer medicine incorporates genetic sequencing results in the process of identifying optimal treatments for patients (1). Standard of care (SOC) therapies are typically prescribed based on anatomical origin and histological subtypes, however, this is not always the most effective treatment (1). Identifying genomic aberrations assists in understanding pathways driving tumor progression, which are often directly linked to the hallmarks of cancer (1–3). Understanding the functional significance of driver mutations in the context of a specific tumor is critical when moving towards clinical implementation of precision medicine techniques (1). Thus, there is a significant need for a patient-proximal ex vivo model reflecting the complex inter- and intra-tumor heterogeneity of the microenvironment (4, 5). An ex vivo model accurately reflecting patient responses to treatment will assist in identifying the druggable ‘Achilles’ heel’ of individual cancers. PDEs have been a focus of research for several types of solid tumor. However, the use of tissue explants is not limited to cancer research, with this culture system being used for research of several non-malignant human diseases including those of the liver (6) and inflammatory bowel disease (7).
Patient-Derived Explants and Cellular Viability
PDEs are physiologically representative of the patient’s disease (8, 9). They represent a significant advance in patient-derived models when compared to two-dimensional (2D) cell lines and complement other three-dimensional (3D) models such as tumoroids/organoids [extensively reviewed in (10, 11)]. Major benefits of PDEs over other patient-proximal models include response prediction to clinical therapies, where cultures can be generated rapidly post-surgery and used to gauge a patient’s response to various anti-cancer therapies, and a more accurate reflection of the intrinsic tumor microenvironment (TME) (12). A faithful recapitulation of inter- and intra-tumor heterogeneity within the TME is key for precision medicine and is often not replicated in other organotypic models (13). Longevity of the various components which comprise the TME (including immune cells, stromal cells, and tumor cells), ex vitro, is an important consideration when utilizing these models to investigate adjuvant and neoadjuvant treatments (14).
An intact TME allows for metabolically active tumor, stromal and immune cells, preserving their native cellular interactions (14). Consequently, PDEs may more closely reflect responses to SOC and molecularly determined therapeutics compared to other patient-derived models such as cell lines, tumoroids/organoids, or patient-derived xenografts (PDX). The importance of monitoring the presence and longevity of immune cells within PDEs harboring pro-immunogenic or immunosuppressive environments has become increasingly recognized with findings that tumor infiltrating immune cells (TIICs) are faithfully retained in PDEs (8, 15). In pancreatic PDEs, CD45+ TILs, including cytotoxic T lymphocytes (CTLs; CD3+, CD8+), regulatory T cells (Tregs; CD3+, FOXP3+) and macrophages (CD68+, CD163+, HLA-DR+) have been shown to be viable for 6 to 9 days in culture (8, 15, 16). Notably, long-term culture of ovarian cancer PDEs retained macrophages, Tregs, CTLs, and B cells (CD19+, CD20+) up to 21-days in agitation culture (17). Most studies, however, do not systematically describe the immune compartments and lack details regarding the longevity of other resident adaptive and innate/innate-like immune subsets, including natural killer cells, γδ T cells and myeloid-derived suppressor cells. The ability to quantify specific immune cell subtypes in PDEs is key to interpreting functional relevance and will ensure these platforms redefine preclinical testing of immunotherapies.
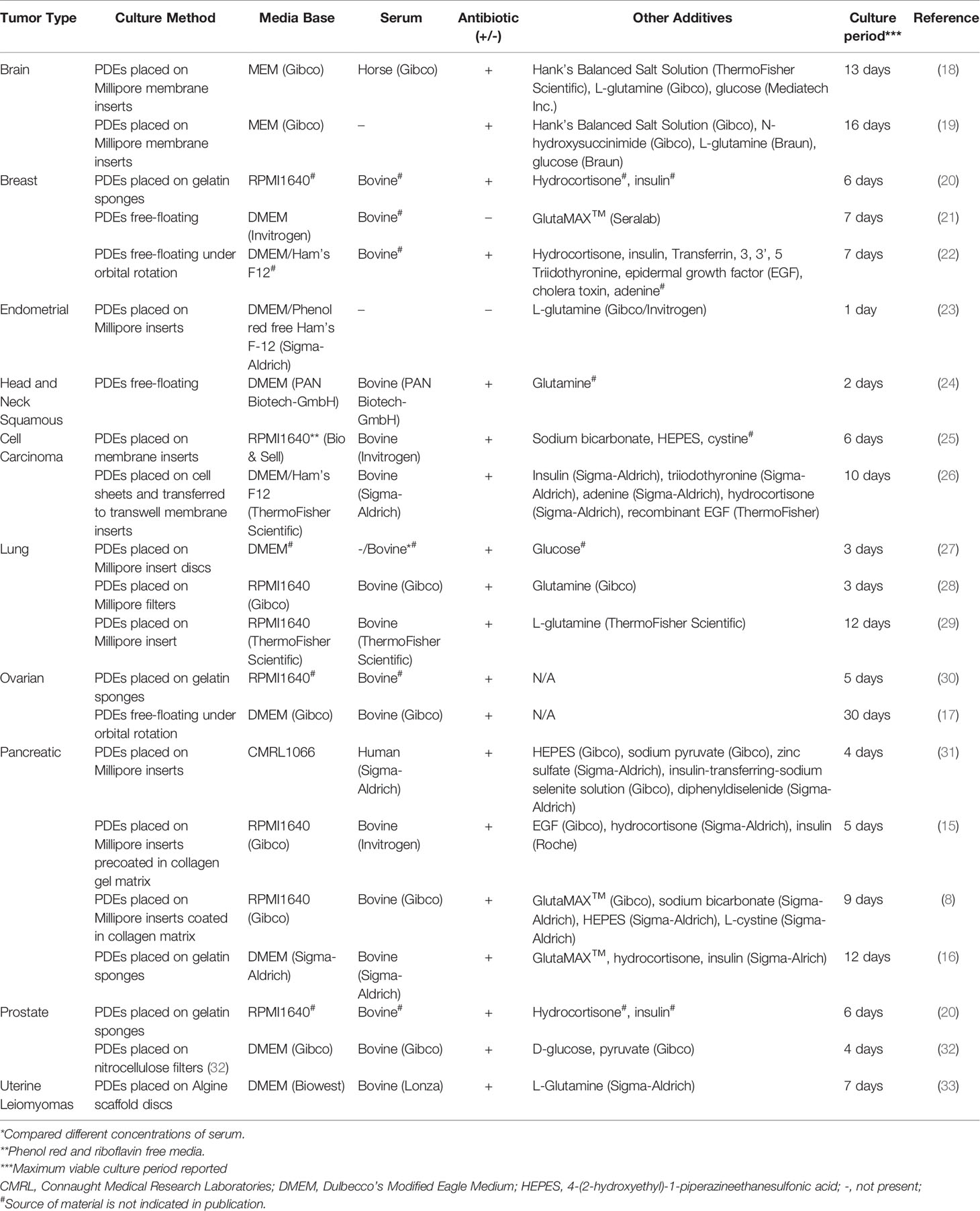
Important disadvantages of PDEs include the relative short-term viability of the cultures evident in some tumor types (Table 1) such as endometrial PDEs which were viable for 1 day (23). The lack of a functional vascular system has been suggested as the reason for this, as it impacts oxygen diffusion and waste removal (34–36). Indeed, Lee, You (26) observed the development of hypoxic cores in head and neck squamous cell carcinoma (HNSCC) PDEs, likely due to a lack of oxygen and nutrient diffusion through the explant. Despite the use of PDE culture systems dating back to the 1980s (37), these models have not been widely adopted into clinical practice due to this short-term viability. However, several groups, working across multiple tumor streams, have been investigating new approaches to improve and extend explant viability (summarized in Table 1) to accommodate whole exome sequencing (WES) turnaround times and thus enable functional testing of genomically identified drug targets.
PDEs can be generated by fragmentation or slicing of fresh tumor tissue. Slice thickness affects the length of time a PDE is viable in culture. A range of different slice thicknesses have been used (200 µm to 3 mm), although in most cases these do not appear to have been empirically determined. Naipal, Verkaik (22) investigated different tissue slice thickness in breast cancer but did not identify an optimal thickness suitable to all tumor types. Additionally, Parajuli and Doppler (36) assessed the effect of slice thickness on longevity using MMTV-neu mouse tumors, determining that a thickness of 160 µm maintained tissue architecture and culture viability most effectively for this tumor type.
Culturing Conditions
There is a lack of a consistent explant culturing methodology in the PDE literature. Table 1 summarizes the variety of culture techniques used across tumor types. Investigating universal principles and best practice approaches that could be applied across tumor types, including the incorporation of tumor-specific components would facilitate the integration of this precision medicine pipeline into clinical practice.
Tissue Preparation
A variety of tissue preparation strategies have been used to generate PDEs. The most common slicing methods are manual dissection with a scalpel and mechanized sectioning using either the vibratome (a vibrating blade microtome) or the McIlwain Tissue Chopper. Manual slicing is used to dissect excess fat or generate PDEs within a specific size range (30), however, it is becoming less common due to the lack of uniformity in explant thickness (16, 22). Two studies have directly compared the most common mechanized slicing technologies, the vibratome and Tissue Chopper, and found the Tissue Chopper more accurately and reproducibly produced slices of a specified thickness (19, 25). Furthermore, Holliday, Moss (21) were unable to slice breast tissue using a microtome due to the fatty nature of the tissue. By contrast, breast explants were successfully sliced using the Tissue Chopper resulting in improved tissue integrity when compared to a vibratome (22). Thus, where available, the McIlwain Tissue Chopper is currently the preferred approach for accurate slicing of tissues of varying densities.
Supplements
Supplements incorporated in PDE culture media are highly dependent on the tissue of origin (summarized in Table 1). When comparing the studies that used tissue specific supplements, there was no clear conclusion as to whether they improved longevity (22, 26, 31). However, Naipal, Verkaik (22) directly compared four types of media, two breast cancer-specific and two common culture media combinations, and found that the tissue specific media (listed in Table 1) was most effective. For HNSCC, Lee, You (26) reported that PDEs remained viable for 10 days, longer than the 2-6 days reported for other HNSCC PDE cultures in more basic media (25, 38). By contrast, in breast and pancreatic PDE studies the addition of tissue specific supplements did not improve viability compared to less complex culture media formulations (22, 31).
It is apparent that a combination of sodium bicarbonate, HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid) and cysteine has a positive effect on viability of PDE cultures across several tumor types. Gerlach, Merz (25) and Jiang, Seo (8) used these supplements in HNSCC and pancreatic cancer studies, respectively, where both had increased longevity compared to other studies. Sodium bicarbonate and HEPES are pH buffering reagents used in culture to control acidity (39). Cysteine is an essential amino acid important for protein synthesis and other metabolic functions, and helps promote proliferation in cancer cells (40). Zhang, van Weerden (41) found that a tissue-specific composition and media containing sodium bicarbonate helped improve viability compared to non-tissue-specific formulations when using PDX material. Given the fundamental roles of these supplements, they are likely to also be relevant to improving the longevity of explants for multiple tumor types, although this remains to be tested.
Culturing Techniques
Several culturing techniques have been adopted across tumor streams for PDE culture. The three most commonly used techniques are free-floating culture, grid or pore membrane supports, and gelatin sponge supports (42). Additionally, specialized platforms such as hydrogels and co-culture systems have been investigated in improving longevity of PDE cultures (26, 43, 44).
Free-floating culture of PDEs in the absence of rotation has been largely dismissed due to the overgrowth of cells resulting in monolayer growth (42), increase in apoptosis, and loss of tissue integrity (41). However, incorporating orbital shaking into this approach reduces these issues. Ovarian PDEs were still viable, proliferating, and metabolically active at the end of a 30-day orbital shaking, free-floating culture period, and no necrotic core was present indicating successful diffusion of nutrients (17). Similarly, Naipal, Verkaik (22) adopted this technique for breast cancer PDEs, suggesting it was most effective at nutrient diffusion and easy to replicate. By contrast, Davies, Dong (28) compared free-floating under rotation to a pore membrane in breast and prostate PDX cultures finding that the membrane culture was more successful at improving viability. Histological examination, and immunohistochemistry (IHC) for E-cadherin were performed in this study, where floating cultures showed the presence of more vacuoles, loss of E-cadherin, reduced proliferative capacity, and increased apoptosis compared to the membrane cultures, confirming that membrane cultures were able to maintain structural integrity over a longer culture period (28).
The use of a grid or pore membrane has been most adopted (19, 28). The most common membrane is the Millipore system due to its open access system allowing for easy transfer and access to culture media, enabling efficient gas exchange between the PDEs and culture media (25, 27, 31). Millipore inserts coated in collagen matrix derived from rat tail have also been used to culture pancreatic PDEs (8, 15). Lim, Chang (15) commented that the precoated membrane increased adherence of the tissue to the surface. However, there was no significant increase in viability using the precoated membrane compared to standard Millipore inserts.
The use of gelatin sponges that support the PDE at the air-liquid interface is a relatively new culture technique. Centenera, Raj (42) noted that sponges help the exchange of nutrients and waste by acting as a capillary surrogate in prostate PDE cultures. Centenera, Hickey (20) and Kokkinos, Sharbeen (16) also used sponges for PDE culturing, demonstrating a significant increase in viability compared to the other culture techniques, such as filter and membrane cultures in prostate and pancreatic PDE studies, respectively. The use of sponges aids in avoiding overgrowth of a cell monolayer from cells disseminating from the explant and helps maintain homeostasis through the exchange of nutrients and waste (20).
Hydrogels that mimic features of the extracellular matrix in tissue culture have become increasingly popular, due to progression from 2D to 3D culture platforms. A preliminary study by Hribar, Wheeler (43) investigated the feasibility of embedding PDEs in VersaGel®, a growth factor-free hydrogel. After three days tumor cells moved out of hydrogel-embedded explants, which was associated with remodeling of the extracellular matrix and mimicking tumor invasion. An advantage of hydrogels is the ability to retain secreted growth factors and to incorporate tissue-specific growth factors into the scaffold (45). These advantages allow for a highly customizable functionalized platform for PDE growth that can be used across tumor types.
Specialized experiments have been performed requiring unique culturing techniques that have not been widely adopted due to complexity and/or technical challenges. Given that angiogenesis is a hallmark of cancer and contributes to the complexity of the TME, this is a critical component missing in routine PDE cultures. Bazou, Maimon (44) used vasculature beds formed from co-culturing endothelial and smooth muscle cells to mimic angiogenetic formation in pancreatic cancer, resulting in PDE viability extending longer than most other culture systems at 3 weeks. Similarly, Lee, You (26) positioned HNSCC PDEs on a cell sheet comprised of matched patient epithelial and sub-epithelial cells to create a co-culture system. The results showed improved cell viability and longevity of PDEs when comparing the co-culture system to other non-matrix models and showed a reduction of viable cancer cells following SOC treatments. Despite the promise of these unique culture techniques, they are not readily available, difficult to establish, and expensive, making them a less attractive technique for widespread use.
Incubation Conditions
There is limited data addressing different cell culture incubation conditions, with most studies using 37°C, 5% CO2 and 21% oxygen. There is no published data addressing variation of temperature or CO2 concentration, however the effect of different oxygen levels on PDE viability has been examined. Misra, Moro (31) compared hyperoxic (41%), normoxic (21%) and hypoxic (less than 21%) oxygen levels concluding that hypoxic oxygen levels led to tissue degradation, but there was no significant difference in viability, proliferation, hypoxia, and metabolic activity between normoxic and hyperoxic conditions. Additionally, Davies, Dong (28) showed that lung and prostate PDX explants maintained in atmospheric oxygen (21%) had improved viability compared to tissues cultured under low oxygen conditions. Thus, atmospheric oxygen is likely to be most suitable for PDE incubation.
Genomic Sequencing for Personalized Therapeutic Options
The successful implementation of a precision medicine pipeline into clinical practice requires a robust patient-proximal model incorporating a timely analysis of each patient’s specific actionable genomic aberrations (typically identified through WES). Using the identification of genetic aberrations to tailor personalized patient care has become increasing popular, however, it is important to understand if this is able to be incorporated into healthcare systems and be utilized as a platform for the broader community (46). The molecular screening and therapeutics (MoST) trial is currently investigating the feasibility of incorporating molecular screening to assist in informing patient management (47). Previous studies have demonstrated that the combination of WES with functional testing of actionable mutations is feasible across multiple tumor types (48, 49). Incorporating WES outputs into a precision medicine pipeline resulted in enrolment of a patient, with metastatic lung adenocarcinoma and rapid progression on SOC therapy, in a phase 1 clinical trial of a cyclin-dependent kinase 4 (CDK4) inhibitor leading to an objective response (as per response evaluation criteria in solid tumors (RECIST) criteria) (49). This demonstrated how WES can assist in finding potential treatment alternatives for advanced cancers refractory to SOC therapies. Similarly, the Zero Childhood Cancer program have utilized whole genome sequencing (WGS) in combination with cell and PDX models, further demonstrating the feasibility of this platform (50, 51). Applying PDEs to screen drugs targeting actionable mutations may present a useful preclinical drug efficacy model.
A key problem associated with the broader application of a PDE platform is misalignment of the time required for comprehensive genomic analysis and PDE longevity. The turnaround time associated with receiving WES results is highly dependent on the technology used, and thus differs across studies, but is generally 3 to 8 weeks (48, 52). This is longer than the typical viable period of PDEs cultures, which has been reported to range from 1 to 30 days dependent on tumor type (Table 1). Preliminary studies have investigated cryopreservation as a method to improve longevity, where Ricciardelli, Lokman (30) demonstrated that cryopreserved tissue remained viable after re-culturing. This indicates that this method could be used for long-term storage of PDEs until sequencing results are returned to the treating clinicians. PDEs have been used to test a number of chemotherapies, targeted therapies, and immunotherapies targeting molecular mechanisms identified during the WES process (Table 2).
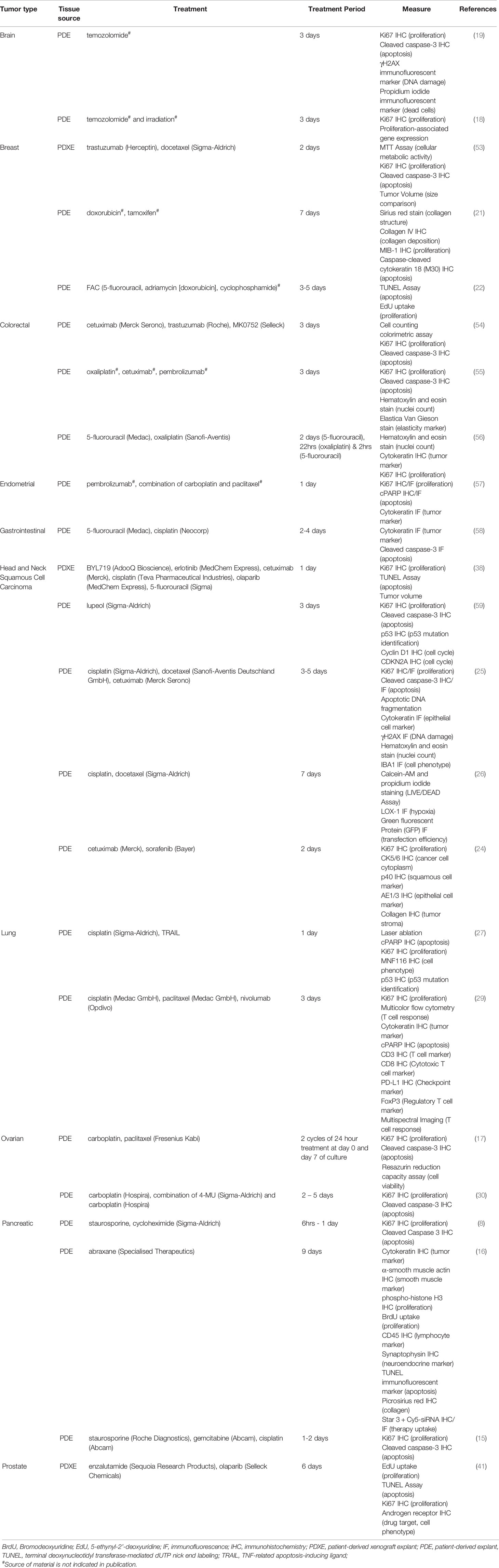
Table 2 Comparison of drug treatments in tumor explant cultures (PDEs and PDXEs) across tumor streams.
Chemotherapy
PDEs have been widely used for the evaluation of tumor response to chemotherapeutics, modeling clinical treatments that patients may receive as SOC (30, 56, 60). The possibility of resistance to chemotherapies means it is useful to test multiple regimens, where they are available, to determine the treatment(s) most likely to be effective for the individual patient. Merz, Gaunitz (19) treated glioblastoma PDEs derived from different tumors with temozolomide, a first line SOC agent, for 3 days, and found that individual tumors displayed different susceptibility to the treatment, consistent with clinical observations. Additionally, Ricciardelli, Lokman (30) treated PDEs with SOC chemotherapies such as carboplatin, which induced apoptosis in PDEs sensitive to chemotherapies but not in chemoresistant PDEs. This reinforces the relevance of adopting pipelines to probe patient-proximal models prior to clinical therapies.
Immunotherapy
Immunotherapies have increasingly become first-line for advanced cancers such as melanoma (61), colorectal (62) and lung (63, 64), due to outstanding tumor control in a subset of patients and better tolerability when compared to chemotherapy (65). While in its infancy, a growing number of tumor types, including endometrial (57), colorectal (54, 55) and head and neck (24, 38) cancer have adopted ex vivo treatment of PDEs to study CTL recruitment and immunotherapy efficacy. Seo, Jiang (66) showed that pancreatic ductal adenocarcinoma cell killing within the slice was mediated by CTL activity following 6-days of combined PD-1 and C-X-C chemokine receptor type 4 (CXCR4) blockade. Interestingly, anti-PD-1 (nivolumab) treatment of PD-L1+ melanoma PDEs increased the inter-cell distance between T cell subsets, demonstrating mechanisms whereby CTLs may avoid Treg-mediated immunosuppression in the TME following immunotherapy (67). As the numbers of studies incorporating PDEs with immunotherapy grows, future pipelines could be assessed in parallel with multiple mRNA and protein readouts to assess immunological response in patients eligible for anti-PD-1, anti-PD-L1 or anti-CTLA-4 treatment, as described in the Lombard Street Approach (68).
Targeted Therapy
Therapies focusing on exploiting specific molecular components to destroy proliferating cancer cells (69) are an important component of successful personalized therapeutics testing using PDEs. Although the concept of targeted therapies is more attractive than non-specific systemic treatments, side effects and resistance remain issues (70, 71). In that regard, targeted therapies are ideally informed by a WES pipeline to identify suitable and relevant candidates. Any molecular targets identified can be compared to FDA approved target therapies using ex vivo testing. Currently the feasibility of incorporating precision target therapies with functional testing of individual patient samples is poor due to the misalignment of WES turnaround times (48, 52, 72) and short-term viability associated with PDEs (Table 1). Nonetheless, targeted therapies such as erlotinib (EGFR inhibitor) and BYL719 (PI3K inhibitor) were shown to suppress the mitogen-activated protein kinase (MAPK) and protein kinase B (AKT) pathways respectively in PDEs (38). These signaling pathways are commonly upregulated during the tumorigenesis process, stimulating increased proliferation of cancer cells (73). PDE platform testing approaches can be used to determine whether such therapies might be useful for individual patients and avoid using treatments to which tumor cells are resistant and thus unnecessarily toxic.
The clinical predictive value of PDE culture as a patient-proximal testing platform needs further investigation. Brijwani, Jain (54) and Majumder, Baraneedharan (74) have used an ex vivo platform, based on PDE drug response, genomic and machine learning (CANscript), to predict patient responses in locally advanced/metastatic CRC and HNSCC. Their data reports that the sensitivity and specificity of this platform for clinical prediction exceeds 90% (54, 74).
Conclusion
PDEs provide a patient-proximal model that reflect intra- and inter-tumor heterogeneity. Most tumor types have limited viability timeframes with current PDE methodologies. To improve viability across tumor types, a combination of culturing techniques and media additives can be used to maintain a physiologically relevant microenvironment ex vivo. Importantly, PDEs have been proven to be an effective personalized testing platform, however the feasibility of incorporating WES in this pipeline is limited until either PDE longevity is extended or cryopreservation approaches can be used to store tissues until sequencing results are available.
Author Contributions
AT wrote the manuscript with contributions from all authors. All authors contributed to the manuscript and were involved in revisions and proof-reading. All authors approved the submitted version.
Funding
This research was supported by funding from a Princess Alexandra Research Foundation award (EW, IV, ET, PT), and the Medical Research Future Fund (MRFF) Rapid Applied Research Translation Program (Centre for Personalised Analysis of Cancers (CPAC; EW, ET, IV: Grant ID GA59729); LB acknowledges the support of grant 1159637 awarded through the 2018 Priority-driven Collaborative Cancer Research Scheme and co-funded by Cancer Australia and Leukemia Foundation of Australia; The Translational Research Institute receives support from the Australian Government.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Jackson SE, Chester JD. Personalised Cancer Medicine. Int J Cancer (2015) 137(2):262–6. doi: 10.1002/ijc.28940
2. Arnedos M, Vielh P, Soria J-C, Andre F. The Genetic Complexity of Common Cancers and the Promise of Personalized Medicine: Is There Any Hope? J Pathol (2014) 232(2):274–82. doi: 10.1002/path.4276
3. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
4. Bleijs M, van de Wetering M, Clevers H, Drost J. Xenograft and Organoid Model Systems in Cancer Research. EMBO J (2019) 38(15):e101654. doi: 10.15252/embj.2019101654
5. Senft D, Leiserson MDM, Ruppin E, Ronai ZA. Precision Oncology: The Road Ahead. Trends Mol Med (2017) 23(10):874–98. doi: 10.1016/j.molmed.2017.08.003
6. Palma E, Doornebal EJ, Chokshi S. Precision-Cut Liver Slices: A Versatile Tool to Advance Liver Research. Hepatol Int (2019) 13(1):51–7. doi: 10.1007/s12072-018-9913-7
7. Swanson KD, Theodorou E, Kokkotou E. Reproducing the Human Mucosal Environment Ex Vivo: Inflammatory Bowel Disease as a Paradigm. Curr Opin Gastroenterol (2018) 34(6):384–91. doi: 10.1097/MOG.0000000000000485
8. Jiang X, Seo YD, Chang JH, Coveler A, Nigjeh EN, Pan S, et al. Long-Lived Pancreatic Ductal Adenocarcinoma Slice Cultures Enable Precise Study of the Immune Microenvironment. OncoImmunology (2017) 6(7):e1333210. doi: 10.1080/2162402X.2017.1333210
9. Kenerson HL, Sullivan KM, Seo YD, Stadeli KM, Ussakli C, Yan X, et al. Tumor Slice Culture as a Biologic Surrogate of Human Cancer. Ann Trans Med (2020) 8(4):114. doi: 10.21037/atm.2019.12.88
10. van de Merbel AF, van der Horst G, van der Pluijm G. Patient-Derived Tumour Models for Personalized Therapeutics in Urological Cancers. Nat Rev Urol (2021) 18:33–45. doi: 10.1038/s41585-020-00389-2
11. Collins A, Miles GJ, Wood J, MacFarlane M, Pritchard C, Moss E. Patient-Derived Explants, Xenografts and Organoids: 3-Dimensional Patient-Relevant Pre-Clinical Models in Endometrial Cancer. Gynecol Oncol (2020) 156(1):251–9. doi: 10.1016/j.ygyno.2019.11.020
12. Powley IR, Patel M, Miles G, Pringle H, Howells L, Thomas A, et al. Patient-Derived Explants (PDEs) as a Powerful Preclinical Platform for Anti-Cancer Drug and Biomarker Discovery. Br J Cancer (2020) 122(6):735–44. doi: 10.1038/s41416-019-0672-6
13. Ayuso JM, Park K-Y, Virumbrales-Muñoz M, Beebe DJ. Toward Improved In Vitro Models of Human Cancer. APL Bioeng (2021) 5(1):010902. doi: 10.1063/5.0026857
14. Hynds RE, Frese KK, Pearce DR, Grönroos E, Dive C, Swanton C. Progress Towards Non-Small-Cell Lung Cancer Models That Represent Clinical Evolutionary Trajectories. Open Biol (2021) 11(1):200247. doi: 10.1098/rsob.200247
15. Lim CY, Chang JH, Lee WS, Lee KM, Yoon YC, Kim J, et al. Organotypic Slice Cultures of Pancreatic Ductal Adenocarcinoma Preserve the Tumor Microenvironment and Provide a Platform for Drug Response. Pancreatology (2018) 18(8):913–27. doi: 10.1016/j.pan.2018.09.009
16. Kokkinos J, Sharbeen G, Haghighi KS, Ignacio RMC, Kopecky C, Gonzales-Aloy E, et al. Ex Vivo Culture of Intact Human Patient Derived Pancreatic Tumour Tissue. Sci Rep (2021) 11. doi: 10.1038/s41598-021-81299-0
17. Abreu S, Silva F, Mendes R, Mendes TF, Teixeira M, Santo VE, et al. Patient-Derived Ovarian Cancer Explants: Preserved Viability and Histopathological Features in Long-Term Agitation-Based Cultures. Sci Rep (2020) 10. doi: 10.1038/s41598-020-76291-z
18. Haehnel S, Reiche K, Loeffler D, Horn A, Blumert C, Puppel S-H, et al. Deep Sequencing and Automated Histochemistry of Human Tissue Slice Cultures Improve Their Usability as Preclinical Model for Cancer Research. Sci Rep (2019) 9. doi: 10.1038/s41598-019-56509-5
19. Merz F, Gaunitz F, Dehghani F, Renner C, Meixensberger J, Gutenberg A, et al. Organotypic Slice Cultures of Human Glioblastoma Reveal Different Susceptibilities to Treatments. Neuro-Oncology (2013) 15(6):670–81. doi: 10.1093/neuonc/not003
20. Centenera MM, Hickey TE, Jindal S, Ryan NK, Ravindranathan P, Mohammed H, et al. A Patient-Derived Explant (PDE) Model of Hormone-Dependent Cancer. Mol Oncol (2018) 12(9):1608–22. doi: 10.1002/1878-0261.12354
21. Holliday DL, Moss MA, Pollock S, Lane S, Shaaban AM, Millican-Slater R, et al. The Practicalities of Using Tissue Slices as Preclinical Organotypic Breast Cancer Models. J Clin Pathol (2013) 66(3):253–5. doi: 10.1136/jclinpath-2012-201147
22. Naipal KAT, Verkaik NS, Sánchez H, van Deurzen CHM, den Bakker MA, Hoeijmakers JHJ, et al. Tumor Slice Culture System to Assess Drug Response of Primary Breast Cancer. BMC Cancer (2016) 16. doi: 10.1186/s12885-016-2119-2
23. Schäfer WR, Fischer L, Roth K, Jüllig AK, Stuckenschneider JE, Schwartz P, et al. Critical Evaluation of Human Endometrial Explants as an Ex Vivo Model System: A Molecular Approach. Mol Hum Reprod (2011) 17(4):255–65. doi: 10.1093/molehr/gaq095
24. Peria M, Donnadieu J, Racz C, Ikoli J-F, Galmiche A, Chauffert B, et al. Evaluation of Individual Sensitivity of Head and Neck Squamous Cell Carcinoma to Cetuximab by Short-Term Culture of Tumor Slices. Head Neck (2016) 38(S1):E911–E5. doi: 10.1002/hed.24126
25. Gerlach MM, Merz F, Wichmann G, Kubick C, Wittekind C, Lordick F, et al. Slice Cultures From Head and Neck Squamous Cell Carcinoma: A Novel Test System for Drug Susceptibility and Mechanisms of Resistance. Br J Cancer (2014) 110(2):479–88. doi: 10.1038/bjc.2013.700
26. Lee J, You JH, Shin D, Roh J-L. Ex Vivo Culture of Head and Neck Cancer Explants in Cell Sheet for Testing Chemotherapeutic Sensitivity. J Cancer Res Clin Oncol (2020) 146(10):2497–507. doi: 10.1007/s00432-020-03306-7
27. Karekla E, Liao W-J, Sharp B, Pugh J, Reid H, Quesne JL, et al. Ex Vivo Explant Cultures of Non-Small Cell Lung Carcinoma Enable Evaluation of Primary Tumor Responses to Anticancer Therapy. Cancer Res (2017) 77(8):2029–39. doi: 10.1158/0008-5472.CAN-16-1121
28. Davies EJ, Dong M, Gutekunst M, Närhi K, van Zoggel HJAA, Blom S, et al. Capturing Complex Tumour Biology In Vitro: Histological and Molecular Characterisation of Precision Cut Slices. Sci Rep (2015) 5. doi: 10.1038/srep17187
29. Junk D, Krämer S, Broschewitz J, Laura H, Massa C, Moulla Y, et al. Human Tissue Cultures of Lung Cancer Predict Patient Susceptibility to Immune-Checkpoint Inhibition. Cell Death Discov (2021) 7(1):264. doi: 10.1038/s41420-021-00651-5
30. Ricciardelli C, Lokman NA, Sabit I, Gunasegaran K, Bonner WM, Pyragius CE, et al. Novel Ex Vivo Ovarian Cancer Tissue Explant Assay for Prediction of Chemosensitivity and Response to Novel Therapeutics. Cancer Lett (2018) 421:51–8. doi: 10.1016/j.canlet.2018.02.006
31. Misra S, Moro CF, Del Chiaro M, Pouso S, Sebestyén A, Löhr M, et al. Ex Vivo Organotypic Culture System of Precision-Cut Slices of Human Pancreatic Ductal Adenocarcinoma. Sci Rep (2019) 9. doi: 10.1038/s41598-019-38603-w
32. van de Merbel AF, van der Horst G, van der Mark MH, van Uhm JIM, van Gennep EJ, Kloen P, et al. An Ex Vivo Tissue Culture Model for the Assessment of Individualized Drug Responses in Prostate and Bladder Cancer. Front Oncol (2018) 8:400. doi: 10.3389/fonc.2018.00400
33. Salas A, López J, Reyes R, Évora C, de Oca FM, Báez D, et al. Organotypic Culture as a Research and Preclinical Model to Study Uterine Leiomyomas. Sci Rep (2020) 10(1):5212. doi: 10.1038/s41598-020-62158-w
34. Köcher S, Beyer B, Lange T, Nordquist L, Volquardsen J, Burdak-Rothkamm S, et al. A Functional Ex Vivo Assay to Detect PARP1-EJ Repair and Radiosensitization by PARP-Inhibitor in Prostate Cancer. Int J Cancer (2019) 144(7):1685–96. doi: 10.1002/ijc.32018
35. Meijer TG, Naipal KAT, Jager A, van Gent DC. Ex Vivo Tumor Culture Systems for Functional Drug Testing and Therapy Response Prediction. Future Sci OA (2017) 3(2):FSO190. doi: 10.4155/fsoa-2017-0003
36. Parajuli N, Doppler W. Precision-Cut Slice Cultures of Tumors From MMTV-Neu Mice for the Study of the Ex Vivo Response to Cytokines and Cytotoxic Drugs. In Vitro Cell Dev Biol Anim (2009) 45(8):442–50. doi: 10.1007/s11626-009-9212-7
37. Freeman AE, Hoffman RM. In Vivo-Like Growth of Human Tumors In Vitro. Proc Natl Acad Sci USA (1986) 83(8):2694–8. doi: 10.1073/pnas.83.8.2694
38. Ghosh S, Prasad M, Kundu K, Cohen L, Yegodayev KM, Zorea J, et al. Tumor Tissue Explant Culture of Patient-Derived Xenograft as Potential Prioritization Tool for Targeted Therapy. Front Oncol (2019) 9(17). doi: 10.3389/fonc.2019.00017
39. Michl J, Park KC, Swietach P. Evidence-Based Guidelines for Controlling pH in Mammalian Live-Cell Culture Systems. Commun Biol (2019) 2:144. doi: 10.1038/s42003-019-0393-7
40. Combs JA, DeNicola GM. The Non-Essential Amino Acid Cysteine Becomes Essential for Tumor Proliferation and Survival. Cancers (2019) 11(5):678. doi: 10.3390/cancers11050678
41. Zhang W, van Weerden WM, de Ridder CMA, Erkens-Schulze S, Schönfeld E, Meijer TG, et al. Ex Vivo Treatment of Prostate Tumor Tissue Recapitulates In Vivo Therapy Response. Prostate (2019) 79(4):390–402. doi: 10.1002/pros.23745
42. Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex Vivo Culture of Human Prostate Tissue and Drug Development. Nat Rev Urol (2013) 10(8):483–7. doi: 10.1038/nrurol.2013.126
43. Hribar KC, Wheeler CJ, Bazarov A, Varshneya K, Yamada R, Buckley P, et al. A Simple Three-Dimensional Hydrogel Platform Enables Ex Vivo Cell Culture of Patient and PDX Tumors for Assaying Their Response to Clinically Relevant Therapies. Mol Cancer Ther (2019) 18(3):718–25. doi: 10.1158/1535-7163.MCT-18-0359
44. Bazou D, Maimon N, Gruionu G, Grahovac J, Seano G, Liu H, et al. Vascular Beds Maintain Pancreatic Tumor Explants for Ex Vivo Drug Screening. J Tissue Eng Regen Med (2018) 12(1):e318–e22. doi: 10.1002/term.2481
45. Freudenberg U, Hermann A, Welzel PB, Stirl K, Schwarz SC, Grimmer M, et al. A Star-PEG–heparin Hydrogel Platform to Aid Cell Replacement Therapies for Neurodegenerative Diseases. Biomaterials (2009) 30(28):5049–60. doi: 10.1016/j.biomaterials.2009.06.002
46. Vidgen ME, Williamson D, Cutler K, McCafferty C, Ward RL, McNeil K, et al. Queensland Genomics: An Adaptive Approach for Integrating Genomics Into a Public Healthcare System. NPJ Genomic Med (2021) 6(1):71. doi: 10.1038/s41525-021-00234-4
47. Thavaneswaran S, Sebastian L, Ballinger M, Best M, Hess D, Lee CK, et al. Cancer Molecular Screening and Therapeutics (MoST): A Framework for Multiple, Parallel Signal-Seeking Studies of Targeted Therapies for Rare and Neglected Cancers. Med J Aust (2018) 209(8):354–5. doi: 10.5694/mja18.00227
48. Réda M, Richard C, Bertaut A, Niogret J, Collot T, Fumet JD, et al. Implementation and Use of Whole Exome Sequencing for Metastatic Solid Cancer. EBioMedicine (2020) 51:102624. doi: 10.1016/j.ebiom.2019.102624
49. van Allen EM, Wagle N, Stojanov P, Perrin DL, Cibulskis K, Marlow S, et al. Whole-Exome Sequencing and Clinical Interpretation of Formalin-Fixed, Paraffin-Embedded Tumor Samples to Guide Precision Cancer Medicine. Nat Med (2014) 20(6):682–8. doi: 10.1038/nm.3559
50. Mould EV, Lau L, Arndt G, Barahona P, Cowley MJ, Ekert P, et al. Abstract 3111: Zero Childhood Cancer: A Comprehensive Precision Medicine Platform for Children With High-Risk Cancer. Cancer Res (2019) 79(13 Supplement):3111. doi: 10.1158/1538-7445.AM2019-3111
51. Rapport F, Smith J, O'Brien TA, Tyrrell VJ, Mould EVA, Long JC, et al. Development of an Implementation and Evaluation Strategy for the Australian 'Zero Childhood Cancer' (Zero) Program: A Study Protocol. BMJ Open (2020) 10(6):1–8. doi: 10.1136/bmjopen-2019-034522
52. Rennert H, Eng K, Zhang T, Tan A, Xiang J, Romanel A, et al. Development and Validation of a Whole-Exome Sequencing Test for Simultaneous Detection of Point Mutations, Indels and Copy-Number Alterations for Precision Cancer Care. NPJ Genomic Med (2016) 1. doi: 10.1038/npjgenmed.2016.19
53. Vesci L, Carollo V, Roscilli G, Aurisicchio L, Ferrara FF, Spagnoli L, et al. Trastuzumab and Docetaxel in a Preclinical Organotypic Breast Cancer Model Using Tissue Slices From Mammary Fat Pad: Translational Relevance Corrigendum in /or/35/1/602. Oncol Rep (2015) 34(3):1146–52. doi: 10.3892/or.2015.4074
54. Brijwani N, Jain M, Dhandapani M, Zahed F, Mukhopadhyay P, Biswas M, et al. Rationally Co-Targeting Divergent Pathways in KRAS Wild-Type Colorectal Cancers by CANscript Technology Reveals Tumor Dependence on Notch and Erbb2. Sci Rep (2017) 7. doi: 10.1038/s41598-017-01566-x
55. Martin SZ, Wagner DC, Hörner N, Horst D, Lang H, Tagscherer KE, et al. Ex Vivo Tissue Slice Culture System to Measure Drug-Response Rates of Hepatic Metastatic Colorectal Cancer. BMC Cancer (2019) 19. doi: 10.1186/s12885-019-6270-4
56. Sönnichsen R, Hennig L, Blaschke V, Winter K, Körfer J, Hähnel S, et al. Individual Susceptibility Analysis Using Patient-Derived Slice Cultures of Colorectal Carcinoma. Clin Colorectal Cancer (2018) 17(2):e189–e99. doi: 10.1016/j.clcc.2017.11.002
57. Collins A, Miles GJ, Powley IR, Hew R, Pringle JH, MacFarlane M, et al. Development of a Patient-Derived Explant Model for Prediction of Drug Responses in Endometrial Cancer. Gynecol Oncol (2021) 160(2):557–67. doi: 10.1016/j.ygyno.2020.11.033
58. Koerfer J, Kallendrusch S, Merz F, Wittekind C, Kubick C, Kassahun WT, et al. Organotypic Slice Cultures of Human Gastric and Esophagogastric Junction Cancer. Cancer Med (2016) 5(7):1444–53. doi: 10.1002/cam4.720
59. Bhattacharyya S, Sekar V, Majumder B, Mehrotra DG, Banerjee S, Bhowmick AK, et al. CDKN2A-P53 Mediated Antitumor Effect of Lupeol in Head and Neck Cancer. Cell Oncol (2017) 40(2):145–55. doi: 10.1007/s13402-016-0311-7
60. da Mata S, Franchi-Mendes T, Abreu S, Filipe B, Morgado S, Mesquita M, et al. Patient-Derived Explants of Colorectal Cancer: Histopathological and Molecular Analysis of Long-Term Cultures. Cancers (2021) 13(18). doi: 10.3390/cancers13184695
61. Larkin J, Chiarion-Sileni V, Gonzalez R, Grob J-J, Rutkowski P, Lao CD, et al. Five-Year Survival With Combined Nivolumab and Ipilimumab in Advanced Melanoma. N Engl J Med (2019) 381(16):1535–46. doi: 10.1056/NEJMoa1910836
62. Aparicio J, Esposito F, Serrano S, Falco E, Escudero P, Ruiz-Casado A, et al. Metastatic Colorectal Cancer. First Line Therapy for Unresectable Disease. J Clin Med (2020) 9(12):3889. doi: 10.3390/jcm9123889
63. Nasser NJ, Gorenberg M, Agbarya A. First Line Immunotherapy for Non-Small Cell Lung Cancer. Pharm (Basel) (2020) 13(11):373. doi: 10.3390/ph13110373
64. Socinski MA, Jotte RM, Cappuzzo F, Orlandi F, Stroyakovskiy D, Nogami N, et al. Atezolizumab for First-Line Treatment of Metastatic Nonsquamous NSCLC. N Engl J Med (2018) 378(24):2288–301. doi: 10.1056/NEJMoa1716948
65. Schirrmacher V. From Chemotherapy to Biological Therapy: A Review of Novel Concepts to Reduce the Side Effects of Systemic Cancer Treatment (Review). Int J Oncol (2019) 54(2):407–19. doi: 10.3892/ijo.2018.4661
66. Seo YD, Jiang X, Sullivan KM, Jalikis FG, Smythe KS, Abbasi A, et al. Mobilization of CD8(+) T Cells via CXCR4 Blockade Facilitates PD-1 Checkpoint Therapy in Human Pancreatic Cancer. Clin Cancer Res (2019) 25(13):3934–45. doi: 10.1158/1078-0432.CCR-19-0081
67. Boucherit N, Gorvel L, Olive D. 3D Tumor Models and Their Use for the Testing of Immunotherapies. Front Immunol (2020) 11:603640. doi: 10.3389/fimmu.2020.603640
68. Versluis JM, Thommen DS, Blank CU. Rationalizing the Pathway to Personalized Neoadjuvant Immunotherapy: The Lombard Street Approach. J Immunother Cancer (2020) 8(2):e001352. doi: 10.1136/jitc-2020-001352
69. Sledge GW. What Is Targeted Therapy? J Clin Oncol (2005) 23(8):1614–5. doi: 10.1200/JCO.2005.01.016
70. Bhullar KS, Lagarón NO, McGowan EM, Parmar I, Jha A, Hubbard BP, et al. Kinase-Targeted Cancer Therapies: Progress, Challenges and Future Directions. Mol Cancer (2018) 17:48. doi: 10.1186/s12943-018-0804-2
71. Pottier C, Fresnais M, Gilon M, Jérusalem G, Longuespée R, Sounni NE. Tyrosine Kinase Inhibitors in Cancer: Breakthrough and Challenges of Targeted Therapy. Cancers (2020) 12(3):731. doi: 10.3390/cancers12030731
72. Chung CCY, Leung GKC, Mak CCY, Fung JLF, Lee M, Pei SLC, et al. Rapid Whole-Exome Sequencing Facilitates Precision Medicine in Paediatric Rare Disease Patients and Reduces Healthcare Costs. Lancet Reg Health West Pac (2020) 1:100001. doi: 10.1016/j.lanwpc.2020.100001
73. Braicu C, Buse M, Busuioc C, Drula R, Gulei D, Raduly L, et al. A Comprehensive Review on MAPK: A Promising Therapeutic Target in Cancer. Cancers (2019) 11(10):1618. doi: 10.3390/cancers11101618
Keywords: precision medicine, patient-derived explants, whole exome sequencing, ex vivo, cancer
Citation: Templeton AR, Jeffery PL, Thomas PB, Perera MPJ, Ng G, Calabrese AR, Nicholls C, Mackenzie NJ, Wood J, Bray LJ, Vela I, Thompson EW and Williams ED (2021) Patient-Derived Explants as a Precision Medicine Patient-Proximal Testing Platform Informing Cancer Management. Front. Oncol. 11:767697. doi: 10.3389/fonc.2021.767697
Received: 31 August 2021; Accepted: 30 November 2021;
Published: 20 December 2021.
Edited by:
Chandra K. Singh, University of Wisconsin-Madison, United StatesReviewed by:
Arjun Singh, Memorial Sloan Kettering Cancer Center, United StatesSushmita Roy, University of Wisconsin - Madison, United States
Copyright © 2021 Templeton, Jeffery, Thomas, Perera, Ng, Calabrese, Nicholls, Mackenzie, Wood, Bray, Vela, Thompson and Williams. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elizabeth D. Williams, ed.williams@qut.edu.au
 Abby R. Templeton
Abby R. Templeton Penny L. Jeffery
Penny L. Jeffery Patrick B. Thomas
Patrick B. Thomas Mahasha P. J. Perera
Mahasha P. J. Perera Gary Ng
Gary Ng Alivia R. Calabrese
Alivia R. Calabrese Clarissa Nicholls
Clarissa Nicholls Nathan J. Mackenzie1,2,3,4
Nathan J. Mackenzie1,2,3,4 Jack Wood
Jack Wood Laura J. Bray
Laura J. Bray Erik W. Thompson
Erik W. Thompson Elizabeth D. Williams
Elizabeth D. Williams