- 1Department of Science and Education, Jinhua Guangfu Oncology Hospital, Jinhua, China
- 2Department of Gastroenterological Surgery, Jinhua Guangfu Oncology Hospital, Jinhua, China
- 3Department of Gastroenterology, Sir Run Run Shaw Hospital, Medical School of Zhejiang University, Hangzhou, China
- 4Central Laboratory, Affiliated Jinhua Hospital, Zhejiang University School of Medicine, Jinhua, China
The HOXC10 gene, a member of the HOX genes family, plays crucial roles in mammalian physiological processes, such as limb morphological development, limb regeneration, and lumbar motor neuron differentiation. HOXC10 is also associated with angiogenesis, fat metabolism, and sex regulation. Additional evidence suggests that HOXC10 dysregulation is closely associated with various tumors. HOXC10 is an important transcription factor that can activate several oncogenic pathways by regulating various target molecules such as ERK, AKT, p65, and epithelial mesenchymal transition-related genes. HOXC10 also induces drug resistance in cancers by promoting the DNA repair pathway. In this review, we summarize HOXC10 gene structure and expression as well as the role of HOXC10 in different human cancer processes. This review will provide insight into the status of HOXC10 research and help identify novel targets for cancer therapy.
Introduction
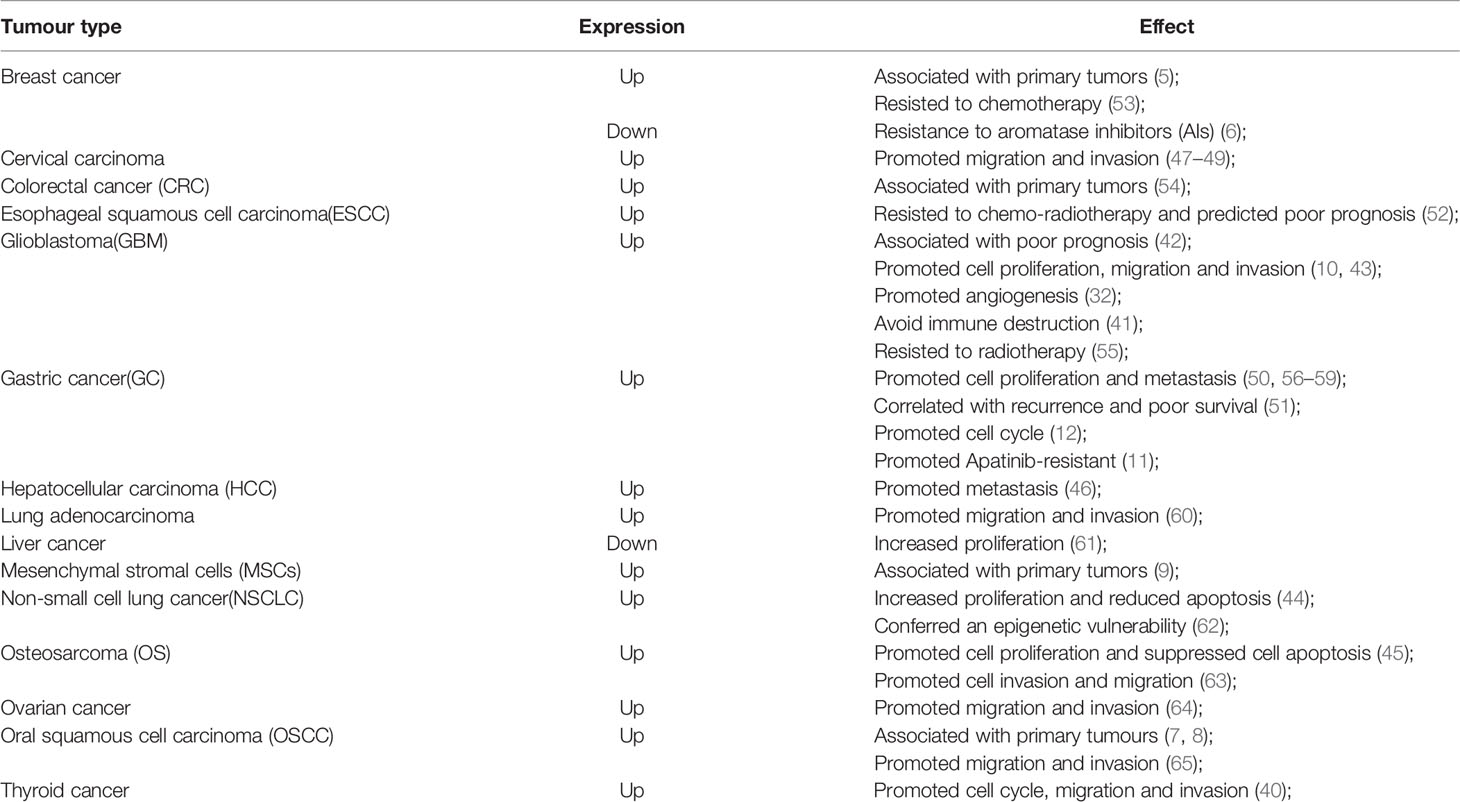
HOX genes, a highly conserved subgroup of the homologous box superfamily, play crucial roles in embryonic development (1). In mammals, HOX genes are divided into four clusters (HOXA, HOXB, HOXC, HOXD), which located on four different chromosomes (7p15, 17q21, 12q13, and 2q31) (2), with each cluster containing 9-11 members (3). To date, 39 HOX genes have been identified in mammals and are separated into 13 paralog groups according to the chromosomal position and sequence similarity in each cluster (4) (Figure 1A). The roles of HOX genes in embryonic development adhere strictly to three principles (1): 1) spatial collinearity (the HOX genes 3’ to 5’ position in a cluster is consistent with its expression along the anterior(A)-posterior(P) axis in animals), 2) posterior prevalence (HOX genes in the 5’ cluster will have a more dominant phenotype than those located in the 3’ cluster), and 3) temporal collinearity (the HOX genes expression sequences in each cluster corresponds to their position [3’ to 5’]) (1). HOX genes transcription usually occurs during the embryonic development and is lowly expressed in adult cells to participate in cell physiology (1, 14, 15). However, HOX genes re-expression occurs in different cancers and is associated with tumor initiation and progression (2, 16, 17). In recent decades, the roles of HOX genes in organogenesis and tumorigenesis have been studied in detail (1, 2, 18, 19). In 2014, Bhatlekar et al. (18) systematically summrized the HOX genes and their roles in human cancer development and concluded that specific HOX genes are expressed in cancers according to tissue type and tumor location. And HOXC family genes expression were upregulated in most solid tumors, including lung, colorectal and prostatic cancers (18). These authors also observed that of the 39 human HOX genes, only two of them (HOXC10, HOXC12) were not reported to be aberrantly expressed in a solid tumor (18). However, HOXC10, an important member of the HOXC family, was recently reported to be closely related to tumorigenesis. Thus, we have conducted a systematic review of the HOXC10 gene and its role in cancer.
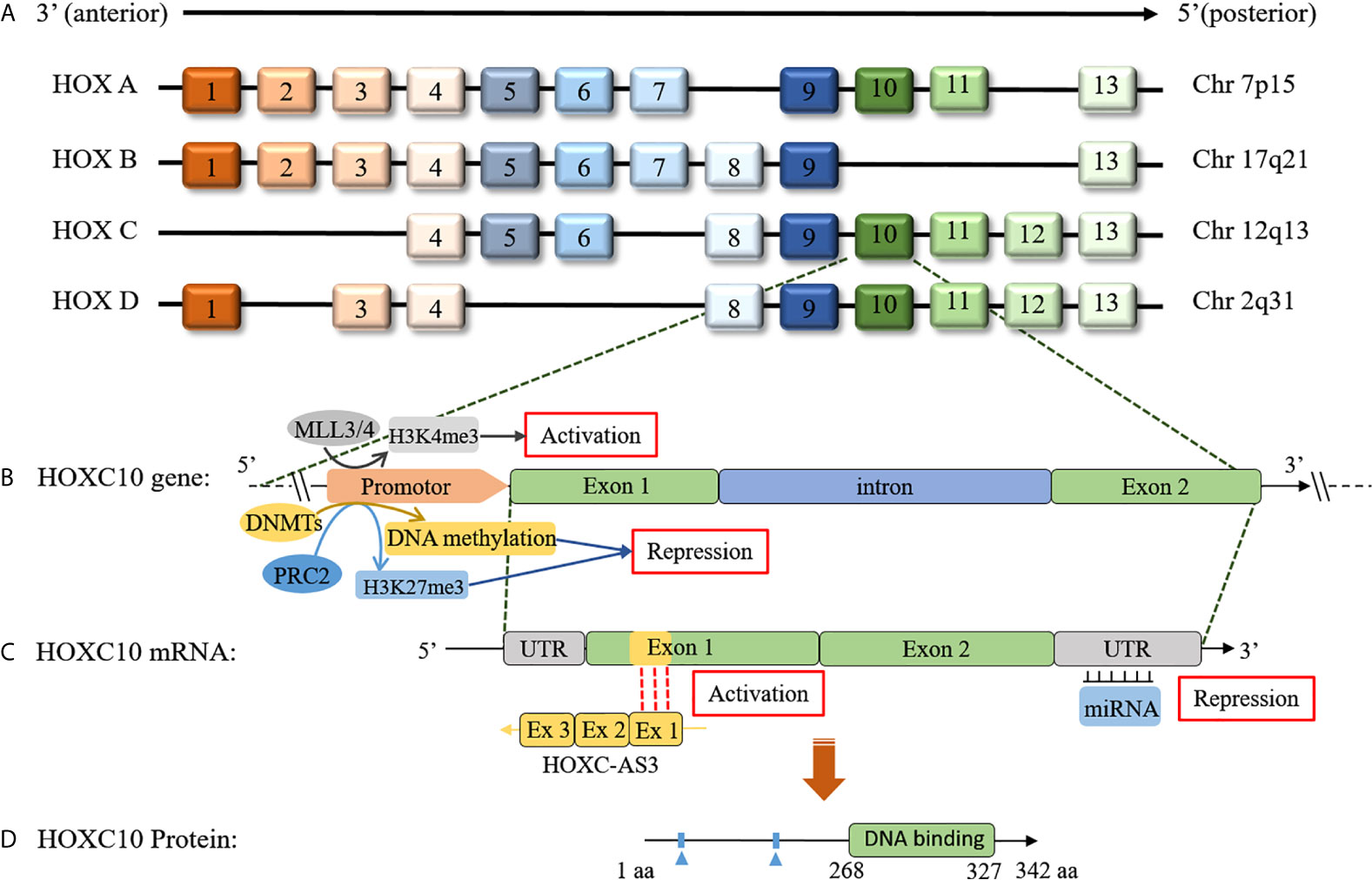
Figure 1 Schematic of the HOX clusters and the HOXC10 gene, mRNA, and protein. (A) In mammals, 39 members exist in HOX genes and are distributed in four clusters. The HOX genes are distributed adjacent to each other in the clusters, and each cluster contains 9-11 members. (B) The HOXC10 gene consists of an intron and two exons. HOXC10 transcription is upregulated by MLL3/4 by histone methylation (H3K4me3) (5). HOXC10 transcription is downregulated by DNMTs via DNA methylation (6) or by polycomb-repressive complex 2 (PRC2) via histone methylation (H3K27me3) (7, 8). (C) lncRNA HOXC-AS3, a natural antisense transcript from the first exon of the HOXC10 gene, can upregulate HOXC10 gene expression by interacting with exon 1 of HOXC10 (9). Additionally, miRNAs can post-transcriptionally suppress HOXC10 expression by base pairing with the 3’-UTR of HOXC10 mRNA (10–12). (D) HOXC10 is a conserved of transcription factor that promotes target genes expression by interacting with target genes through the DNA binding domains (13).
The HOXC10 gene, located on chromosome 12, which contains an intron and two exons in its gene sequence, encodes a protein with 342 amino acids (13) (Figures 1B–D). HOXC10, a highly conserved transcription factor, plays an important role in cellular identity and embryonic morphogenesis during development (20, 21). Considerable evidence has shown that HOXC10 is closely related to mammalian physiological processes. Earlier studies reported that HOXC10 was involved in regulating anterior/posterior pattern specification (22–24), limb regeneration (25–27), and lumbar motor neuron differentiation (28–31). HOXC10 is also associated with angiogenesis (32), fat metabolism (33–38), and sex regulation (39).
Similar to the HOX genes family expression patterns, HOXC10 maintains a low expression level to maintain normal physiological activities in most adult cells. However, HOXC10 appears to be re-expressed in various tumors. Here, we have shown the different HOXC10 expressions across 20 tumor samples and paired normal tissues with a dot plot via GEPIA2.0 (http://gepia2.cancer-pku.cn/) (Figure 2). Moreover, HOXC10 expression have been reported to be positively correlated with poor pathologic stage, and poor prognosis (40–43). High HOXC10 expression is significantly to enhance tumor proliferation (10, 43–45), invasiveness (46–49), recrudesce (50, 51) and drug resistance (11, 52, 53). Reports suggest that anomalous HOXC10 expression is strongly associated with the occurrence and progression of cancers (Table 1) and HOXC10 may be a potential prognostic factor and therapeutic target (42, 51, 56). HOXC10, as a highly conserved transcription factor, is also reported to cooperate with various target molecules, such as ERK (56), JNK (66), AKT (52, 60), VEGF-A (32), immunosuppression genes (41), caspase-3 (45), to drive tumorigenesis. These observations indicate that HOXC10 may be an important regulatory to drive tumorigenesis. In this review, we summarize the recent researches on the molecular mechanisms of HOXC10 in tumorigenesis, metastasis (migration and invasion, and epithelial mesencymal transition [EMT]), and drug resistance (Figure 3B). We also systematically summarize HOXC10 expression regulation mechanisms (Figure 3A). Further investigating the molecular mechanisms of HOXC10 overexpression and its role in tumorigenesis may give us new insights into oncogenesis and progression and enable designing new and more successful therapies for tumors.
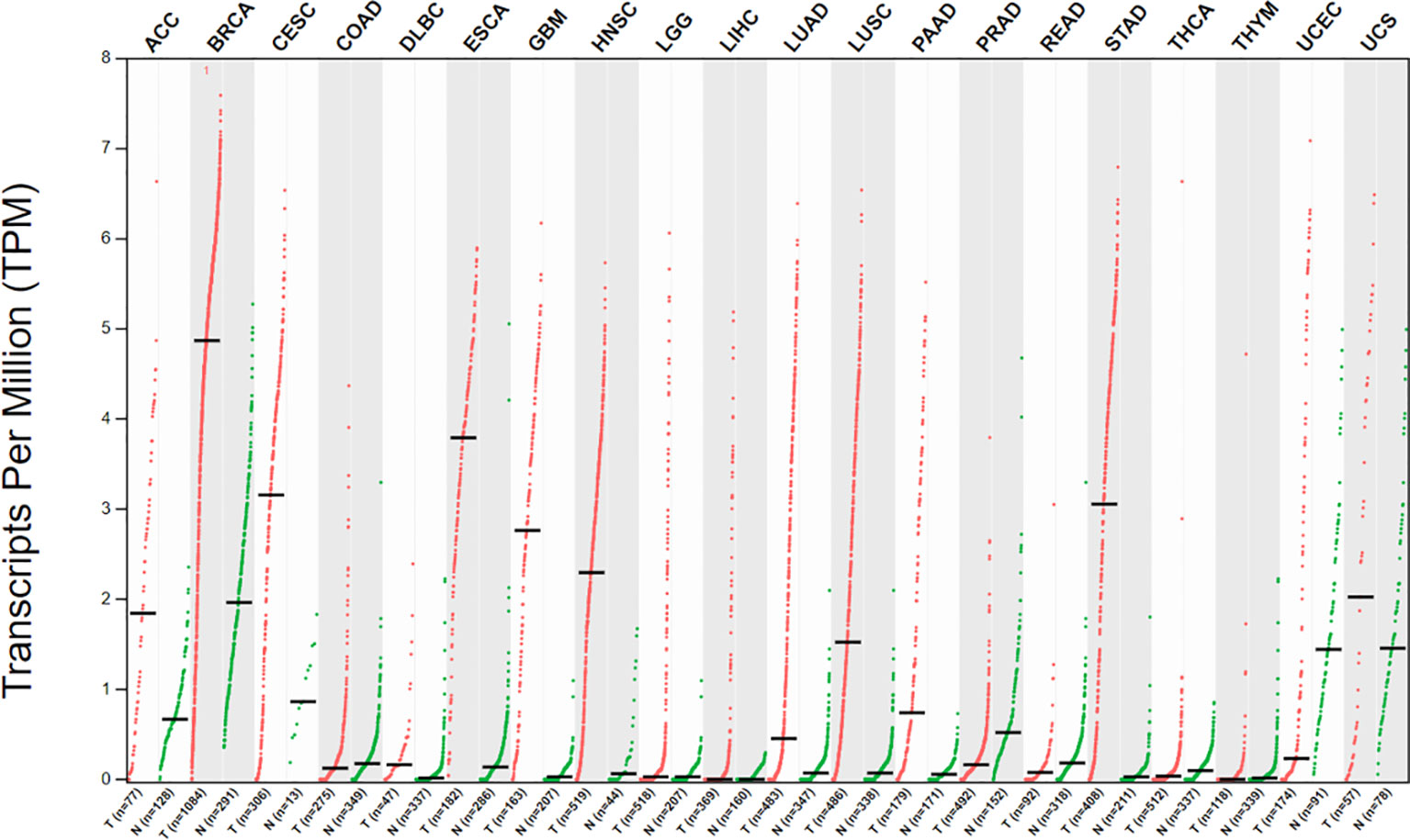
Figure 2 Dot plot showing different HOXC10 expression across 20 tumor samples and paired normal tissues. Each dots represents expression of samples. (X-axis: cancer type, Y-axis: log2[TPM + 1]) Tumors data are from the Cancer Genome Atlas (TCGA), and normal data are from the TCGA and GTEx database. Data were visualized using GEPIA2.0 (http://gepia2.cancer-pku.cn/).
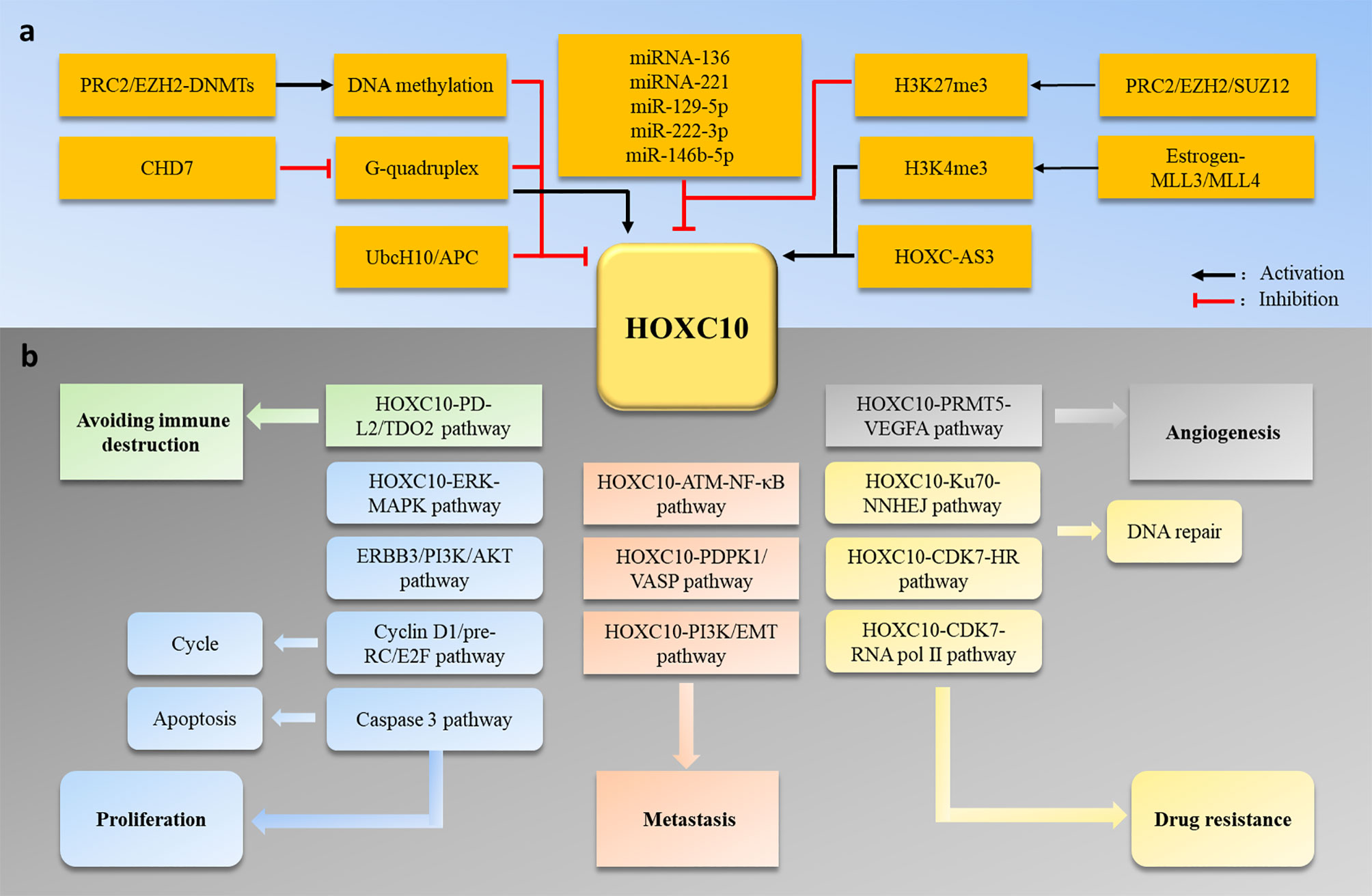
Figure 3 (A) HOXC10 expression is regulated by DNA methylation, histone methylation, miRNA, lncRNA, and the ubiquitin-degradation pathway. (B) Abnormal HOXC10 expression can induce tumor occurrence and development by promoting cell proliferation, metastasis, angiogenesis, drug resistance and avoidance of immune destruction.
HOXC10 Is Involved in Tumorigenesis, Metastasis, and Drug Resistance
Abnormal expression of HOXC10 has been reported in various tumors (Table 1). Anomalous HOXC10 expression is strongly associated with cancer occurrence and progression (40–43, 50, 51, 57). Several studies have revealed the HOXC10 molecular mechanisms that regulate tumor development. Dysregulated HOXC10 affects tumorigenesis in different ways, including cell proliferation (10, 43–45), the cell cycle (12), apoptosis (45), angiogenesis (32), invasion (46–49), drug resistance (52, 53, 55), and avoidance of immune destruction (41) (Figure 3). In this section, we systematically summarize the functions and potential molecular mechanisms of HOXC10 in tumor occurrence, metastasis and drug resistance.
Tumorigenesis
Dysregulation of HOXC10 expression is common in tumors and indicates that HOXC10 may contribute to tumor occurrence and development. Kim et al. used the TCGA data to compare the gene expressions in gastric cancer and normal tissues and found that HOXC10 expression was significantly promoted in gastric cancer (50). Miwa et al. used surgical specimens from gastric cancer patients with metastases and found that HOXC10 was the highest expressed gene in carcinoma compared with adjacent tissue (51). Yao et al. used tissue microarrays to test 73 gastric cancer patients and found that the HOXC10 expression level was strongly associated with tumor node metastasis (TNM) stage, lymph node metastasis, and distant metastasis (57). In gastric cancer cell lines, increasing HOXC10 expression significantly promoted cell proliferation and metastasis (50), and gastric cancer cells proliferation and invasion were inhibited via increased apoptosis after HOXC10 gene silencing (51). Guo et al. injected gastric cancer cells overexpressing HOXC10 into the intragastric walls of mice to obtain gastric cancer tumor-bearing mice and confirmed that HOXC10 overexpression increased the gastric cancer tumor volum in these mice (56). These studies indicated that HOXC10 induced gastric cancer occurrence and development by promoting gastric cancer cells proliferation, metastasis, and tumor growth. Cao et al. used bioinformatics to identify four survival-associated differentially expressed genes (OSMR, HOXC10, SCARA3, and SLC39A10) in glioblastomas and found that glioblastoma patients with abnormal HOXC10 expression had poor survival outcomes (42). Li et al. confirmed that HOXC10 was increased in glioblastomas compared with normal tissue, and HOXC10 expression was positively correlated with the high-grade of glioma (41). Moreover, HOXC10 knockdown inhibited the glioblastoma U87 cells proliferation, migration, and invasion (43). These results suggested that HOXC10 may be responsible for glioblastoma occurrence. Abnormal HOXC10 expression has been identified in other tumors, such as cervical cancer (49), breast cancer (5, 21, 43), non-small cell lung cancer (44), oral squamous cell carcinomas (7, 8) (Table 1). HOXC10 overexpression is also closely related to TNM stage, cell proliferation, metastasis, and tumor growth in these tumors. These studies indicated that HOXC10 may be a key regulator in inducing tumorigenesis and progression.
Next, we have systematically summarized the potential molecular mechanisms of HOXC10 in tumorigenesis. The mitogen-activated protein kinase (MAPK) signaling pathway, a clearest pathway in cancer biology, can induce carcinogenesis by activating the expression of proliferation-related genes and promoting cell overgrowth (67). In gastric cancer, HOXC10 promotes mRNA and protein expression of c-myc, c-jun, and p53, which are gene related to the MAPK signaling pathway (56). HOXC10 can also increase phosphorylation of c-Jun N-terminal kinase (JNK), extracellular signal-regulated kinase (ERK) and p38 without affecting their expressions (56). ERK, JNK, and p38 phosphorylation play an important roles in the MAPK signaling pathway (66). ERK1/2, a dominant component in the MAPK signaling pathway (68), is involved in regulating cell division. HOXC10 knockout significantly inhibited ERK phosphorylation and the tumor cell proliferation (37, 51). HOXC10 expression levels are positively correlated with FGFBP1 and SOX10 expression (51). FGFBP1 and SOX10 can also participate in regulating the MAPK signaling pathway (69, 70). These studies suggested HOXC10 expression levels were positively correlated with MAPK signaling pathway activation and that HOXC10 promoted cell proliferation and tumorigenesis by activating the MAPK signaling pathway. Conversely, Ma et al. (61) found that HOXC10 have negatively affected the MAPK signaling pathway, and MAPK signaling marker proteins increase significantly after HOXC10 knockdown in liver cells. This suggests that HOXC10 has diverse effects on the MAPK pathway in different tumors.
Aberrant activation of the phosphoinositide 3-kinase (PI3K/AKT) signaling pathway is the most frequent events in tumorigenesis and contributes to carcinogenic transformation by regulating cell proliferation, apoptosis, metastasis, and autophagy (71, 72). Elevated HOXC10 also accelerates cancer progression via the PI3K/AKT signaling pathway. HOXC10 can enhance PI3K phosphorylation (60) and promote the expression of pivotal genes in the PI3K/AKT pathway (43). Human Erb-b2 receptor tyrosine kinase 3 (ERBB3/HER3), an activator of the PI3K/AKT signaling pathway, can induce tumorigenesis and progression (73). Suo et al. found that HOXC10 upregulated ERBB3 transcription and activated the PI3K/AKT pathway by binding to the promoter of ERBB3 in esophageal squamous cell carcinoma cells (52). They further confirmed that ERBB3 silencing decreased PI3K and AKT phosphorylation upregulated by HOXC10 and significantly reduced esophageal squamous cell carcinoma cells proliferative capacity (52). These data demonstrated that HOXC10 promotes esophageal squamous cell carcinoma cell proliferation mainly via the ERBB3/PI3K/AKT axis.
HOXC10 can induce tumorigenesis by regulating angiogenesis and immunoregulation. Angiogenesis, the process of growth of new capillary blood vessels from existing capillaries, is important in tumor growth and metastasis (74). Tan et al. found that HOXC10 promoted angiogenesis in human glioma cells by upregulating of VEGF-A expression (32). Mechanistically, HOXC10 enriched H3R2me1, H3R2me2s and H3K4me3 on the VEGF-A promoter by interacting with PRMT5, which upregulated VEGF-A expression and angiogenesis (32). Avoiding immune destruction is a characteristic of tumors (75), and tumors can escape immune surveillance by regulating multiple immunosuppressive pathways (76). Li et al. found that HOXC10 expression was positively correlated with immunosuppression genes (CCL2, PD-L2, TGF-β2, TDO2) (41). Their further investigation revealed that HOXC10 could bind to the promoters of PD-L2 and TDO2 and promote their transcription (41). Previous studies have confirmed that PD-L2 (77), TDO2 (78), CCL2 (79), and TGF-β (80) can directly or indirectly inhibit T cell-mediated tumor clearance. Thus, we speculated that HOXC10 might help glioma cells escape immune surveillance by regulating the expression of immunosuppressive genes, leading to cancer occurrence and development.
HOXC10 can regulate the cell cycle and apoptosis in tumorigenesis. Guerra et al. revealed that HOXC10 overexpression promoted non-small cell lung cancer cells moving into the S phase, thus promoting cell proliferation (62). Their further research found that HOXC10 enhanced the expression of DNA replication genes (E2F family genes, pre-RC components) (62). The E2F gene family, a family of transcription factors, sits at the center of cell cycle gene expression and plays an important role in the cell cycle (81, 82). The pre-RC component can bind to DNA in G1 with well-defined steps and mark all potential starting points for replication (83). The DNA replication forks would stall and collapse with insufficient numbers of E2F and pre-RC components and cause DNA damage and cell death (84). These data indicate that HOXC10 might promote cell cycle progression by up-regulating the expression of E2F family genes and pre-RC components and lead to cell proliferation. Similarly, HOXC10 knockdown induced cell cycle blocking and inhibited thyroid cancer cell proliferation and invasion (40). In other studies, HOXC10 overexpression facilitated G1/S cell cycle transition by regulating the expression of cyclin D1 in gastric cancer cells (12). HOXC10 knockdown also promoted the expression and activity of caspase-3 and induced apoptosis (45). In conclusion, HOXC10 contributes to tumorigenesis by regulating the cell cycle and apoptosis.
Tumor Metastasis and Invasion
Metastasis is the main cause of high recurrence rates and low survival rates in cancer patients. Recent studies revealed that HOXC10 expression was strongly linked to tumor metastasis and invasion in various tumors. In studying cervical carcinoma, Zhai et al. used high-density oligonucleotide microarrays to compared gene expression in microdissected squamous epithelial samples from normal cervices, high-grade squamous intraepithelial lesions, and invasive squamous cell carcinomas, found HOXC10 have the highest expression in invasive squamous cell carcinomas (49). Additionally, invasive squamous cell carcinomas invasion was significantly decreased after HOXC10 knockdown (49). These data indicated that HOXC10 was a crucial mediator of invasion in cervical carcinoma. Miwaet al. used surgical specimens from gastric cancer patients with metastasis and found that HOXC10 was the highest expressed gene in the carcinoma tissues compared with adjacent tissues (51). Li et al. confirmed that HOXC10 silencing suppressed metastasis and invasion, whereas HOXC10 overexpression significantly enhanced metastasis in gastric cancer cell lines (58). These studies suggested that HOXC10 may be a novel biomarker of metastasis, invasion, and recurrence after radical resection of gastric cancer.
Further research revealed that the molecular mechanisms by which HOXC10 regulates tumor metastasis and invasion. Li et al. found that HOXC10 overexpression promoted metastasis and invasion by upregulating inflammatory cytokines in gastric cancer (58). In the tumor microenvironment, inflammatory factors, such as IL-6, TNF-α, and TGF-β, play an important roles in tumor occurrence, development, invasion and metastasis (85). Mechanistically, HOXC10 could activates the NF-κB pathway by binding to the p65 gene promotor and indirectly upregulating inflammatory cytokines (IL-6, TNF-α, TGF-β, EGF) in gastric cancer cells (58). Interestingly, inflammatory factor IL-1β can also promote HOXC10 expression via the JNK/c-Jun pathway and induce invasion and metastasis of hepatocellular carcinoma (46), indicating that HOXC10 can promote tumor metastasis by cooperating with inflammatory cytokines. Dang et al. (46) found that upregulated HOXC10 expression induced hepatocellular carcinoma metastasis by upregulating the expression of 3-phosphoinositide-dependent protein kinase 1 and vasodilator-stimulated phosphoprotein. Yao et al. found that HOXC10 promoted gastric cancer cell invasion and migration, and enhance the activity of ataxia telangiectasia-mutated gene (ATM) and NF-κB pathway (57). ATM, a member of the PI3/PI4 kinase family, plays an important role in DNA damage and repair (86). Activated ATM can be transferred to the cytoplasm and activate IkB kinase (IKK)-β (87). The NF-κB pathway also plays an intricate role in tumor metastasis (88, 89). Thus, HOXC10 induces gastric cancer cell invasion and migration through the ATM/NF-κB axis. However, further research is needed to confirm these results.
EMT, a biological process in which epithelial cells are endowed with mesenchymal cellular characteristics, can reduce cell-cell adhesion ability and enhance tumor cell migration and invasion (90, 91). Recent evidences revealed that HOXC10 regulated EMT to induce tumor invasion and metastasis. In human oral squamous cell carcinoma, Dai et al. (65) found that HOXC10 knockdown significantly decreased the expressions of N-cadherin, Vimentin, Snail, while E-cadherin expression was increased, and Wnt10B was markedly suppressed in shHOXC10-cell lines. Wnt10B, a secretory protein, can activate the Wnt/β-catenin pathway (92). Wnt10B overexpression promoted migration ability in oral squamous cell carcinoma cells, but this process was reversed after HOXC10 silencing (65). These data indicate that HOXC10 induces tumor metastasis via the WNT/EMT pathway in oral squamous cell carcinoma. HOXC10 was also shown to regulate the expression of EMT markers (MMP2/9, VCAM-1, vimentin and E-cadherin) in lung adenocarcinoma (60). Peng et al. confirmed that HOXC10 enhanced osteosarcoma cell metastasis by enhancing Slug transcription (64). Notably, Slug was the most important regulator of EMT in tumors (93) and was a surrogate marker of EMT in head and neck cancer (94). These studies confirmed that HOXC10 promotes tumor migration and invasion by activating EMT.
Drug Resistance
Drug resistance is a major reason for tumor therapy failure, and the underlying mechanisms must be explore to overcome it. Recent studies revealed that HOXC10 is closely related to the occurrence of drug resistance in various tumors. In ER-positive breast cancer, Pathiraja et al. found that HOXC10 promoters showed significant methylation enrichment in two breast cancer cell line models of aromatase inhibitors (AIs) resistance (6). Subsequent research demonstrated that silencing of HOXC10 by DNA methylation was a key process in AIs resistance (6). Sadik et al. found that ER-negative breast cancer with abnormal HOXC10 expression had shorter recurrence-free and overall survival after chemotherapy (53). Li et al. analyzed the gene expression profiles between radiotherapy patients and an untreated group and showed a significant different in the HOXC10 gene. HOXC10 overexpression also inhibited the efficacy of radiotherapy in gliomas (55). HOXC10 was also found to be involved in chemotherapy resistance in gastric cancer (11). HOXC10 knockdown can enhance the chemo-sensitivity of MGC-803/AP and AGS/AP cells.
DNA damage is a direct or indirect response to antitumor drug therapy, and tumors can induce the development of drug resistance by increasing DNA repair activity. HOXC10 was found to contribute to drug resistance in cancers by fine-tuning DNA repair. For double-strand breaks (DSB) repair, HOXC10 recruited homologous recombination (HR) repair proteins (RAD51, BRCA1) at the DNA damage sites. However, HOXC10 was undetectable at the I-Sce1 cleavage site, indicating that HOXC10 does not play a direct role in DSBs repair (53). Finally, Sadik et al. (53) confirmed that HOXC10 integrated HR functions by binding to and activating cyclin-dependent kinase, CDK7, which regulates transcription by phosphorylating the carboxy-terminal domain of RNA polymerase II. Non-homologous DNA end joining (NNHEJ) is another key pathway for repairing DSBs in eukaryotic cells (95). Suo et al. (52) found that HOXC10 directly bound to Ku70 and facilitated DNA damage repair by NHEJ in esophageal squamous cell carcinoma cells, thus conferring resistance to chemoradiotherapy in esophageal squamous cell carcinoma cells. These studies indicated that HOXC10 can induce tumor resistance to chemotherapy by enhancing DNA repair ability.
HOXC10 Expression Regulation
Our review has described the roles and mechanisms of HOXC10 in the different processes of human cancers. We also provided a comprehensive description of HOXC10 expression regulation. Specifically, HOXC10 expression is regulated by several epigenetic processes, including DNA (6, 50, 96) and histone (5, 97) methylation, posttranscriptional miRNA (10–12, 59, 61, 64, 98) and lncRNA (9, 99) modifications, and ubiquitin modifications (100).
DNA methylation causes changes in chromatin structure, DNA conformation, DNA stability and the methods by which DNA interacts with proteins to regulate gene expression (101). Studies have shown that HOXC10 expression is closely related to changes in DNA methylation, and DNA methylation generally functions as a repressive transcriptional signal. Lim et al. (96) found that HOXC10 levels increased after blocking DNA methylation with 5-azacytidine in adipocytes. Kim et al. (50) revealed that HOXC10 was significantly increased, and its CpG sites were hypomethylated in gastric cancer tissues compared with those of normal tissues. Bisulfite sequencing revealed that CpG sites in the first HOXC10 intronic region were hypomethylated in three gastric cancer tissues, and HOXC10 expression was increased in gastric cancer cell lines (AGS and SNU620) in response to 5-azacytidine treatment. In studying ER-positive breast cancer, Pathiraja et al. (6) found that the methylation of the HOXC10 promoter occurred in a CpG shore, and recruitment of EZH2 and H3K27me3 induced silencing of HOXC10 expression by increasing DNA methylation. A revious study confirmed that EZH2 serves as a recruitment platform for DNA methyltransferases (DNMTs), and EZH2 could interact with DNA methyltransferases (DNMTs) and activate DNMT activity (6). These results indicated that the interaction between EZH2 and DNMTs might promote HOXC10 DNA methylation, ultimately silencing HOXC10.
G-quadruplex (G4) refers to a four-stranded secondary structure formed by guanine-rich nucleic acid sequences through Hoogsteen hydrogen bonding in the DNA or RNA strand. Studies of G4 in humans and animals demonstrated that G4 is involved in a wide range of basic biological functions such as DNA replication, transcription, translation, and maintenance of telomeric structure (102). Zhang et al. (103) analyzed DNA sequences upstream of the HOXC10 transcription start site, verified the formation of G-quadruplex structures in the negative strand of the HOXC10 promoter and revealed that these structures could inhibit HOXC10 expression. These authors also confirmed that CHD7, a chromatin remodeling protein with DNA helicase activity, could associate with the HOXC10 promoter and likely unwind the G4 structures to enhance its gene expression (103). Conversely, Li et al. (44) found that a G4 formation in the HOXC10 promoter was required for elevated expression of HOXC10, and disruption of G4 formation could silence HOXC10 expression in non-small cell lung cancer cells. These studies indicated that G-quadruplex structure was closely correlated with HOXC10 expression, but the molecular mechanism was remains unclear.
Histone modification plays an important role in regulating gene expression in eukaryotes. Polycomb repressive complex 2 (PRC2), comprised of the H3K27 methylases EZH2, SUZ12 and EED, can catalyze mono-, di-, and trimethylation of lysine 27 on histone H3 (H3K27) (104). Previous studies revealed that HOX genes were canonical PRC2 targets (105) and HOXC10 was a direct PRC2 target, which was demonstrated using chromatin immunoprecipitation-X enrichment analysis and ENCODE datasets (97). Guerra et al. (62) reported that half of KRAS-mutant non-small cell lung cancer cells aberrantly expressed HOXC10, largely due to unappreciated defects in PRC2. Specifically, HOXC10 was more highly expressed in PRC2-low tumors. In addition, Marcinkiewicz et al. (7, 8) found that SUZ12, H3K27me3 and H3K9me3 were recruited in non-tumorigenic human OKF6-TERT1R compared with tumorigenic SCC-9 cells and concluded that altered PRC2 activity was associated with dysregulated HOXC10 expression in human oral squamous cell carcinoma. In breast cancer, HOXC10 overexpression has been widely reported. Ansari et al. (5) found that histone methylases MLL3 and MLL4 were bound estrogen-dependently to the ERE1 and ERE6 regions of the HOXC10 promoter and lead to enrichment of H3K4me3 and recruitment of RNA polymerase II, ultimately promoting HOXC10 gene expression. These studies suggested that histone methylation regulated altered HOXC10 expression in tumors.
MicroRNAs (miRNAs) are small noncoding RNAs that can degrade or suppress the translation of target mRNAs by base pairing with the 3’-untranslated region (3’UTR) (106). Several miRNAs, such as miR-129-5p (10–12), miR-222-3p (64), miR-146b-5p (98), miRNA-221 (61), and miRNA-136 (59), have been reported to play a key role in regulating HOXC10 expression. These miRNAs directly target the 3’UTR of HOXC10 to inhibit HOXC10 expression.
Antisense transcripts can regulate alternative splicing, transport and structural stability of the sense transcripts by forming double-stranded RNA structures with the sense transcript (107). Li et al. (9) identified that the natural antisense transcript, HOXCAS3, arises from intergenic regions of the HOXC10 gene. Their data showed that as a pair of protein-coding sense/non-coding antisense transcripts, HOXC-AS3 bound to HOXC10 thereby increasing its stability by reducing its decay. Additionally, enhancing the stability of HOXC10/HOXC-AS3 upregulated the HOXC10 expression. Similarly, Fu et al. (99) found that HOXC-AS3 might be involved in gastric adenocarcinoma by regulating the HOXC10 gene with a cis-effect.
Other researchers found that HOXC10 expression was also related to protein stability. Gabellini et al. (100) found that HOXC10 expression was reduced in the early G1 phase, abundant from the mid-G1 to G2 phases and undetectable in mitosis. Northern blot analysis showed that HOXC10 mRNA levels did not change, suggesting that HOXC10 levels may be regulated post-translationally. Further studies showed that HOXC10 could coimmunoprecipitate the APC subunit, CDC27, and protein degradation of HOXC10 was suppressed by expression of a dominant-negative form of UbcH10, an APC-associated ubiquitin-conjugating enzyme. These data implied that HOXC10 protein stability was regulated by the UbcH10/APC-mediated ubiquitination pathway.
Conclusion and Future Perspectives
In recent years, significant has been made progress in understanding the function of HOXC10 in various physiological and pathological process. Interestingly, the tissues that are developmentally regulated by HOXC10 during embryogenesis appear to be more likely to lead to malignancies. Choe et al. reported that HOXC10 and the other HOX10 paralogs were key to axial skeletal positioning and neural tissue development, and mutations in these genes could affect motor neuron patterning (28). Birth defects in humans caused by HOXC10 mutations appear to included skeletal and nervous system abnormalities (29, 30). In tumorigenesis, aberrant HOXC10 expression also appeared to contribute to the development of osteosarcomas (45, 63) and gliomas (10, 43). HOXC10 was also identified as the first truly “regeneration-specific” gene transcript (25). In axolotl, HOXC10 was not expressed during forelimb development, but was activated during forelimb regeneration (25, 27). HOXC10 may regulate tissue development by controlling cell proliferation and differentiation (20, 108). However, disruption the precise expression regulation of HOXC10 induced malformation in hindlimb (109) or tumorigenesis (18). Thus, HOXC10 plays dual roles in the process of development and carcinogenesis.
Accumulating research has showed that HOXC10 expression is dysregulated in various cancers and serves as an oncogenic driver in cancer processes (Table 1). In this review, we summarized the mechanism of HOXC10 in tumorigenesis and found that abnormal HOXC10 expression induced tumor occurrence by regulating cell proliferation, cycles, apoptosis, and angiogenesis and by avoiding immune destruction. Additionally, HOXC10 can regulate tumor invasion by regulating the NF-κB signal pathway, EMT and the expression of metastasis-related genes. Moreover, HOXC10 affects the drug treatment response and induce drug resistance in tumors (Figure 3B). These studies suggest that HOXC10 may be a potential prognostic factor and therapeutic target in cancer (42, 51, 56). Furthermore, we summarized that the aberrant HOXC10 expression in tumors is regulated by several epigenetic processes, including methylation of DNA and histone, posttranscriptional modifications of miRNA and lncRNA, and ubiquitin modifications (Figure 3A). Investigating the molecular mechanisms of abnormal HOXC10 expression in tumors is an essential step towards developing HOXC10 gene-targeted therapeutics and may help advance our understanding of cancer development and enable designing new therapeutic agents.
Although HOXC10 appears to play an important role in many cancers, its precise function remains unclear. To date, most of studies on HOXC10 expression and function are derived from retrospective analyses of patient tumors. These studies only hint at the mechanisms underlying the roles of these genes in oncogenesis, without adequate treatment information. Recently, Guerra et al. (62) reported that HOXC10 was overexpressed in half of KRAS-mutant non-small cell lung cancer cells, which led to more sensitivity to combined BET/MEK inhibitors in xenograft and patient-derived tumor xenograft (PDX) models. The efficacy of the combination depended on the inhibition of HOXC10 by BET inhibitors (62). The study indicated that HOXC10 may be a functional, predictive biomarker for BET inhibitor-based combinations in non-small cell lung cancer (62). Because HOXC10 can be easily detected using immunohistochemistry, thus it may provide a promising, clinically manageable biomarker for selecting patients. Furthermore, Miwa et al. (51) found that high levels of HOXC10 in gastric cancer tissues were significantly associated with worse prognosis, as well as hepatic and peritoneal recurrence. And the studies confirmed that HOXC10 can cooperate with inflammatory cytokines (58), ATM/NF-κB axis (57), and EMT (60, 64, 65) to promote tumor migration and invasion. High HOXC10 expression can also induce tumor drug resistance by increasing DNA repair activity (52, 53). These results suggest that the high levels of HOXC10 expression contributes to increase malignant phenotypes during cancer progression and may provide a valuable prognostic biomarker. Therefore, further insights into the molecular role of HOXC10 in tumors is urgent and may provide new insights regarding selective therapeutic targets that could be used to design new and better therapies. As studies of the HOXC10 gene in cancer progress, we expect HOXC10 to play a potential role in direct targeting and selection of targeted therapeutic approaches.
Author Contributions
All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Zhejiang Public Welfare Technology Research Program (LGF19H030018), Natural Science Foundation of Zhejiang province (LY21H160014). Jin Hua Science and Technology Plan Project (2018-3-3001C).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest
Glossary

References
1. Shah N, Sukumar S. The Hox Genes and Their Roles in Oncogenesis. Nat Rev Cancer (2010) 10(5):361–71. doi: 10.1038/nrc2826
2. Paco A, Aparecida de Bessa Garcia S, Leitao Castro J, Costa-Pinto AR, Freitas R. Roles of the HOX Proteins in Cancer Invasion and Metastasis. Cancers (2020) 13(1):10. doi: 10.3390/cancers13010010
3. Scott MP. Vertebrate Homeobox Gene Nomenclature. Cell (1992) 71(4):551–3. doi: 10.1016/0092-8674(92)90588-4
4. Contarelli S, Fedele V, Melisi D, Genes Family HOX. And Cancer: A Novel Role for Homeobox B9 in the Resistance to Anti-Angiogenic Therapies. Cancers (2020) 12(11):2399. doi: 10.3390/cancers12113299
5. Ansari KI, Hussain I, Kasiri S, Mandal SS. HOXC10 is Overexpressed in Breast Cancer and Transcriptionally Regulated by Estrogen Via Involvement of Histone Methylases MLL3 and MLL4. J Mol Endocrinol (2012) 48(1):61–75. doi: 10.1530/JME-11-0078
6. Pathiraja TN, Nayak SR, Xi Y, Jiang S, Garee JP, Edwards DP, et al. Epigenetic Reprogramming of HOXC10 in Endocrine-Resistant Breast Cancer. Sci Transl Med (2014) 6(229):229ra41. doi: 10.1126/scitranslmed.3008326
7. Marcinkiewicz KM, Gudas LJ. Altered Epigenetic Regulation of Homeobox Genes in Human Oral Squamous Cell Carcinoma Cells. Exp Cell Res (2014) 320(1):128–43. doi: 10.1016/j.yexcr.2013.09.011
8. Marcinkiewicz KM, Gudas LJ. Altered Histone Mark Deposition and DNA Methylation At Homeobox Genes in Human Oral Squamous Cell Carcinoma. J Cell Physiol (2014) 229(10):1405–16. doi: 10.1002/jcp.24577
9. Li B, Han H, Song S, Fan G, Xu H, Zhou W, et al. Hoxc10 Regulates Osteogenesis of Mesenchymal Stromal Cells Through Interaction With Its Natural Antisense Transcript Lnchoxc-AS3. Stem Cells (2019) 37(2):247–56. doi: 10.1002/stem.2925
10. Liu J, Cheng C, Jiao J, Huang W, Huang J, Sun J, et al. MircoRNA-129-5p Suppresses the Development of Glioma by Targeting HOXC10. Pathol Res Pract (2020) 216(4):152868. doi: 10.1016/j.prp.2020.152868
11. Yu J, Zhang X, Ma Y, Li Z, Tao R, Chen W, et al. Mir-129-5p Restrains Apatinib Resistance in Human Gastric Cancer Cells Via Downregulating Hoxc10. Cancer Biother Radiopharma (2021) 36(1):95–105. doi: 10.1089/cbr.2019.3107
12. He J, Ge Q, Lin Z, Shen W, Lin R, Wu J, et al. MiR-129-5p Induces Cell Cycle Arrest Through Modulating HOXC10/Cyclin D1 to Inhibit Gastric Cancer Progression. FASEB J Off Publ Fed Am Soc Exp Biol (2020) 34(6):8544–57. doi: 10.1096/fj.201903217R
13. de Stanchina E, Gabellini D, Norio P, Giacca M, Peverali FA, Riva S, et al. Selection of Homeotic Proteins for Binding to a Human DNA Replication Origin. J Mol Biol (2000) 299(3):667–80. doi: 10.1006/jmbi.2000.3782
14. Rux DR, Wellik DM. Hox Genes in the Adult Skeleton: Novel Functions Beyond Embryonic Development. Dev Dynamics Off Publ Am Assoc Anatomists (2017) 246(4):310–7. doi: 10.1002/dvdy.24482
15. Paco A, de Bessa Garcia SA, Freitas R. Methylation in HOX Clusters and Its Applications in Cancer Therapy. Cells (2020) 9(7):1613. doi: 10.3390/cells9071613
16. de Bessa Garcia SA, Araujo M, Pereira T, Mouta J, Freitas R. HOX Genes Function in Breast Cancer Development. Biochim Biophys Acta Rev Cancer (2020) 1873(2):188358. doi: 10.1016/j.bbcan.2020.188358
17. Li B, Huang Q, Wei GH. The Role of HOX Transcription Factors in Cancer Predisposition and Progression. Cancers (2019) 11(4):528. doi: 10.3390/cancers11040528
18. Bhatlekar S, Fields JZ, Boman BM. HOX Genes and Their Role in the Development of Human Cancers. J Mol Med (2014) 92(8):811–23. doi: 10.1007/s00109-014-1181-y
19. Errico MC, Jin K, Sukumar S, Care A. The Widening Sphere of Influence of HOXB7 in Solid Tumors. Cancer Res (2016) 76(10):2857–62. doi: 10.1158/0008-5472.CAN-15-3444
20. Akbas GE, Taylor HS. HOXC and HOXD Gene Expression in Human Endometrium: Lack of Redundancy With HOXA Paralogs. Biol Reprod (2004) 70(1):39–45. doi: 10.1095/biolreprod.102.014969
21. Abba MC, Sun H, Hawkins KA, Drake JA, Hu Y, Nunez MI, et al. Breast Cancer Molecular Signatures as Determined by SAGE: Correlation With Lymph Node Status. Mol Cancer Res MCR (2007) 5(9):881–90. doi: 10.1158/1541-7786.MCR-07-0055
22. Nelson CE, Morgan BA, Burke AC, Laufer E, DiMambro E, Murtaugh LC, et al. Analysis of Hox Gene Expression in the Chick Limb Bud. Development (1996) 122(5):1449–66. doi: 10.1242/dev.122.5.1449
23. Logan M, Tabin CJ. Role of Pitx1 Upstream of Tbx4 in Specification of Hindlimb Identity. Science (1999) 283(5408):1736–9. doi: 10.1126/science.283.5408.1736
24. Jain D, Nemec S, Luxey M, Gauthier Y, Bemmo A, Balsalobre A, et al. Regulatory Integration of Hox Factor Activity With T-box Factors in Limb Development. Development (2018) 145(6):dev159830. doi: 10.1242/dev.159830
25. Carlson MR, Komine Y, Bryant SV, Gardiner DM. Expression of Hoxb13 and Hoxc10 in Developing and Regenerating Axolotl Limbs and Tails. Dev Biol (2001) 229(2):396–406. doi: 10.1006/dbio.2000.0104
26. Christen B, Beck CW, Lombardo A, Slack JMW. Regeneration-Specific Expression Pattern of Three Posterior Hox Genes. Dev Dynamics (2003) 226(2):349–55. doi: 10.1002/dvdy.10231
27. Nicolas S, Papillon D, Perez Y, Caubit X, Le Parco Y. The Spatial Restrictions of 5’hoxc Genes Expression Are Maintained in Adult Newt Spinal Cord. Biol Cell (2003) 95(9):589–94. doi: 10.1016/j.biolcel.2003.09.004
28. Choe A, Phun HQ, Tieu DD, Hu YH, Carpenter EM. Expression Patterns of Hox10 Paralogous Genes During Lumbar Spinal Cord Development. Gene Expression Patterns GEP (2006) 6(7):730–7. doi: 10.1016/j.modgep.2005.12.004
29. Wu Y, Wang G, Scott SA, Capecchi MR. Hoxc10 and Hoxd10 Regulate Mouse Columnar, Divisional and Motor Pool Identity of Lumbar Motoneurons. Development (2008) 135(1):171–82. doi: 10.1242/dev.009225
30. Hostikka SL, Gong J, Carpenter EM. Axial and Appendicular Skeletal Transformations, Ligament Alterations, and Motor Neuron Loss in Hoxc10 Mutants. Int J Biol Sci (2009) 5(5):397–410. doi: 10.7150/ijbs.5.397
31. Bulajic M, Srivastava D, Dasen JS, Wichterle H, Mahony S, Mazzoni EO. Differential Abilities to Engage Inaccessible Chromatin Diversify Vertebrate Hox Binding Patterns. Development (2020) 147(22):dev194761. doi: 10.1242/dev.194761
32. Tan Z, Chen K, Wu W, Zhou Y, Zhu J, Wu G, et al. Overexpression of HOXC10 Promotes Angiogenesis in Human Glioma Via Interaction With PRMT5 and Upregulation of VEGFA Expression. Theranostics (2018) 8(18):5143–58. doi: 10.7150/thno.27310
33. Brune JE, Kern M, Kunath A, Flehmig G, Schon MR, Lohmann T, et al. Fat Depot-Specific Expression of HOXC9 and HOXC10 May Contribute to Adverse Fat Distribution and Related Metabolic Traits. Obesity (2016) 24(1):51–9. doi: 10.1002/oby.21317
34. Ferrannini G, Namwanje M, Fang B, Damle M, Li D, Liu Q, et al. Genetic Backgrounds Determine Brown Remodeling of White Fat in Rodents. Mol Metab (2016) 5(10):948–58. doi: 10.1016/j.molmet.2016.08.013
35. Ng Y, Tan SX, Chia SY, Tan HY, Gun SY, Sun L, et al. HOXC10 Suppresses Browning of White Adipose Tissues. Exp Mol Med (2017) 49(2):e292. doi: 10.1038/emm.2016.144
36. Kato H, Ario T, Kishida T, Tadano M, Osawa S, Maeda Y, et al. Homeobox A5 and C10 Genes Modulate Adaptation of Brown Adipose Tissue During Exercise Training in Juvenile Rats. Exp Physiol (2021) 106(2):463–74. doi: 10.1113/EP089114
37. Ma M, Wang C, Ao Y, He N, Hao F, Liang H, et al. HOXC10 Promotes Proliferation and Attenuates Lipid Accumulation of Sheep Bone Marrow Mesenchymal Stem Cells. Mol Cell Probes (2020) 49:101491. doi: 10.1016/j.mcp.2019.101491
38. Breitfeld J, Kehr S, Muller L, Stadler PF, Bottcher Y, Bluher M, et al. Developmentally Driven Changes in Adipogenesis in Different Fat Depots Are Related to Obesity. Front Endocrinol (2020) 11:138. doi: 10.3389/fendo.2020.00138
39. Yatsu R, Miyagawa S, Kohno S, Parrott BB, Yamaguchi K, Ogino Y, et al. RNA-Seq Analysis of the Gonadal Transcriptome During Alligator Mississippiensis Temperature-Dependent Sex Determination and Differentiation. BMC Genomics (2016) 17:77. doi: 10.1186/s12864-016-2396-9
40. Feng X, Li T, Liu Z, Shi Y, Peng Y. HOXC10 Up-Regulation Contributes to Human Thyroid Cancer and Indicates Poor Survival Outcome. Mol Biosyst (2015) 11(11):2946–54. doi: 10.1039/c5mb00253b
41. Li S, Zhang W, Wu C, Gao H, Yu J, Wang X, et al. HOXC10 Promotes Proliferation and Invasion and Induces Immunosuppressive Gene Expression in Glioma. FEBS J (2018) 285(12):2278–91. doi: 10.1111/febs.14476
42. Cao M, Cai J, Yuan Y, Shi Y, Wu H, Liu Q, et al. A Four-Gene Signature-Derived Risk Score for Glioblastoma: Prospects for Prognostic and Response Predictive Analyses. Cancer Biol Med (2019) 16(3):595–605. doi: 10.20892/j.issn.2095-3941.2018.0277
43. Guan Y, He Y, Lv S, Hou X, Li L, Song J. Overexpression of HOXC10 Promotes Glioblastoma Cell Progression to a Poor Prognosis Via the PI3K/AKT Signalling Pathway. J Drug Targeting (2019) 27(1):60–6. doi: 10.1080/1061186X.2018.1473408
44. Li M, Alsager JS, Wang Z, Cheng L, Shan B. Epigenetic Upregulation of HOXC10 in Non-Small Lung Cancer Cells. Aging (Albany NY) (2020) 12(17):16921–35. doi: 10.18632/aging.103597
45. Xie X, Xiao Y, Huang X. Homeobox C10 Knockdown Suppresses Cell Proliferation and Promotes Cell Apoptosis in Osteosarcoma Cells Through Regulating Caspase 3. OncoTargets Ther (2018) 11:473–82. doi: 10.2147/OTT.S143440
46. Dang Y, Chen J, Feng W, Qiao C, Han W, Nie Y, et al. Interleukin 1beta-Mediated HOXC10 Overexpression Promotes Hepatocellular Carcinoma Metastasis by Upregulating PDPK1 and VASP. Theranostics (2020) 10(8):3833–48. doi: 10.7150/thno.41712
47. Santin AD, Zhan F, Bignotti E, Siegel ER, Cane S, Bellone S, et al. Gene Expression Profiles of Primary HPV16- and HPV18-Infected Early Stage Cervical Cancers and Normal Cervical Epithelium: Identification of Novel Candidate Molecular Markers for Cervical Cancer Diagnosis and Therapy. Virology (2005) 331(2):269–91. doi: 10.1016/j.virol.2004.09.045
48. Hung YC, Ueda M, Terai Y, Kumagai K, Ueki K, Kanda K, et al. Homeobox Gene Expression and Mutation in Cervical Carcinoma Cells. Cancer Sci (2003) 94(5):437–41. doi: 10.1111/j.1349-7006.2003.tb01461.x
49. Zhai Y, Kuick R, Nan B, Ota I, Weiss SJ, Trimble CL, et al. Gene Expression Analysis of Preinvasive and Invasive Cervical Squamous Cell Carcinomas Identifies HOXC10 as a Key Mediator of Invasion. Cancer Res (2007) 67(21):10163–72. doi: 10.1158/0008-5472.CAN-07-2056
50. Kim J, Bae DH, Kim JH, Song KS, Kim YS, Kim SY. HOXC10 Overexpression Promotes Cell Proliferation and Migration in Gastric Cancer. Oncol Rep (2019) 42(1):202–12. doi: 10.3892/or.2019.7164
51. Miwa T, Kanda M, Umeda S, Tanaka H, Tanaka C, Kobayashi D, et al. Homeobox C10 Influences on the Malignant Phenotype of Gastric Cancer Cell Lines and its Elevated Expression Positively Correlates With Recurrence and Poor Survival. Ann Surg Oncol (2019) 26(5):1535–43. doi: 10.1245/s10434-019-07166-5
52. Suo D, Wang Z, Li L, Chen Q, Zeng T, Liu R, et al. HOXC10 Upregulation Confers Resistance to Chemoradiotherapy in ESCC Tumor Cells and Predicts Poor Prognosis. Oncogene (2020) 39(32):5441–54. doi: 10.1038/s41388-020-1375-4
53. Sadik H, Korangath P, Nguyen NK, Gyorffy B, Kumar R, Hedayati M, et al. Hoxc10 Expression Supports the Development of Chemotherapy Resistance by Fine Tuning DNA Repair in Breast Cancer Cells. Cancer Res (2016) 76(15):4443–56. doi: 10.1158/0008-5472.CAN-16-0774
54. Enteghami M, Ghorbani M, Zamani M, Galehdari H. HOXC10 is Significantly Overexpressed in Colorectal Cancer. Biomed Rep (2020) 13(3):18. doi: 10.3892/br.2020.1325
55. Li S, Shi J, Gao H, Yuan Y, Chen Q, Zhao Z, et al. Identification of a Gene Signature Associated With Radiotherapy and Prognosis in Gliomas. Oncotarget (2017) 8(51):88974–87. doi: 10.18632/oncotarget.21634
56. Guo C, Hou J, Ao S, Deng X, Lyu G. HOXC10 Up-Regulation Promotes Gastric Cancer Cell Proliferation and Metastasis Through MAPK Pathway. Chin J Cancer Res = Chung-kuo yen cheng yen chiu (2017) 29(6):572–80. doi: 10.21147/j.issn.1000-9604.2017.06.12
57. Yao S, He L, Zhang Y, Ye L, Lai Y, Huang L, et al. HOXC10 Promotes Gastric Cancer Cell Invasion and Migration Via Regulation of the NF-kappaB Pathway. Biochem Biophys Res Commun (2018) 501(3):628–35. doi: 10.1016/j.bbrc.2018.05.019
58. Li J, Tong G, Huang C, Luo Y, Wang S, Zhang Y, et al. HOXC10 Promotes Cell Migration, Invasion, and Tumor Growth in Gastric Carcinoma Cells Through Upregulating Proinflammatory Cytokines. J Cell Physiol (2020) 235(4):3579–91. doi: 10.1002/jcp.29246
59. Zheng J, Ge P, Liu X, Wei J, Wu G, Li X. MiR-136 Inhibits Gastric Cancer-Specific Peritoneal Metastasis by Targeting HOXC10. Tumour Biol J Int Soc Oncodevelop Biol Med (2017) 39(6):1010428317706207. doi: 10.1177/1010428317706207
60. Tang XL, Ding BX, Hua Y, Chen H, Wu T, Chen ZQ, et al. Hoxc10 Promotes the Metastasis of Human Lung Adenocarcinoma and Indicates Poor Survival Outcome. Front Physiol (2017) 8:557. doi: 10.3389/fphys.2017.00557
61. Ma K, Zhao C, Guo K, Fu Z, Che C, Dong B, et al. Low HOXC10 Expression in Liver Cancer Regulates Proliferation Via a Mechanism Involving miR-221 and the MAPK Signaling Pathway. Oncol Lett (2020) 20(5):127. doi: 10.3892/ol.2020.11988
62. Guerra SL, Maertens O, Kuzmickas R, De Raedt T, Adeyemi RO, Guild CJ, et al. A Deregulated Hox Gene Axis Confers an Epigenetic Vulnerability in KRAS-Mutant Lung Cancers. Cancer Cell (2020) 37(5):705–19.e6. doi: 10.1016/j.ccell.2020.03.004
63. Xiong W, Zhou Q, Liu G, Liu XS, Li XY. Homeodomain-Containing Gene 10 Inhibits Cell Apoptosis and Promotes Cell Invasion and Migration in Osteosarcoma Cell Lines. Tumour Biol J Int Soc Oncodevelop Biol Med (2017) 39(5):1010428317697566. doi: 10.1177/1010428317697566
64. Peng Y, Li Y, Li Y, Wu A, Fan L, Huang W, et al. HOXC10 Promotes Tumour Metastasis by Regulating the EMT-Related Gene Slug in Ovarian Cancer. Aging (Albany NY) (2020) 12(19):19375–98. doi: 10.18632/aging.103824
65. Dai BW, Yang ZM, Deng P, Chen YR, He ZJ, Yang X, et al. HOXC10 Promotes Migration and Invasion Via the WNT-EMT Signaling Pathway in Oral Squamous Cell Carcinoma. J Cancer (2019) 10(19):4540–51. doi: 10.7150/jca.30645
66. Johnson GL. Lapadat R. Mitogen-Activated Protein Kinase Pathways Mediated by ERK, JNK, and p38 Protein Kinases. Science (2002) 298(5600):1911–2. doi: 10.1126/science.1072682
67. Yuan J, Dong X, Yap J, Hu J. The MAPK and AMPK Signalings: Interplay and Implication in Targeted Cancer Therapy. J Hematol Oncol (2020) 13(1):113. doi: 10.1186/s13045-020-00949-4
68. Shin M, Franks CE, Hsu KL. Isoform-Selective Activity-Based Profiling of ERK Signaling. Chem Sci (2018) 9(9):2419–31. doi: 10.1039/c8sc00043c
69. Schulze D, Plohmann P, Hobel S, Aigner A. Anti-Tumor Effects of Fibroblast Growth Factor-Binding Protein (FGF-BP) Knockdown in Colon Carcinoma. Mol Cancer (2011) 10:144. doi: 10.1186/1476-4598-10-144
70. Cronin JC, Loftus SK, Baxter LL, Swatkoski S, Gucek M, Pavan WJ. Identification and Functional Analysis of SOX10 Phosphorylation Sites in Melanoma. PloS One (2018) 13(1):e0190834. doi: 10.1371/journal.pone.0190834
71. Mayer IA, Arteaga CL. The PI3K/AKT Pathway as a Target for Cancer Treatment. Annu Rev Med (2016) 67:11–28. doi: 10.1146/annurev-med-062913-051343
72. Hoxhaj G, Manning BD. The PI3K-AKT Network At the Interface of Oncogenic Signalling and Cancer Metabolism. Nat Rev Cancer (2020) 20(2):74–88. doi: 10.1038/s41568-019-0216-7
73. Soltoff SP, Carraway KL 3rd, Prigent SA, Gullick WG, Cantley LC. ErbB3 is Involved in Activation of Phosphatidylinositol 3-Kinase by Epidermal Growth Factor. Mol Cell Biol (1994) 14(6):3550–8. doi: 10.1128/mcb.14.6.3550
74. Li S, Xu HX, Wu CT, Wang WQ, Jin W, Gao HL, et al. Angiogenesis in Pancreatic Cancer: Current Research Status and Clinical Implications. Angiogenesis (2019) 22(1):15–36. doi: 10.1007/s10456-018-9645-2
75. Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell (2011) 144(5):646–74. doi: 10.1016/j.cell.2011.02.013
76. Swann JB, Smyth MJ. Immune Surveillance of Tumors. J Clin Invest (2007) 117(5):1137–46. doi: 10.1172/JCI31405
77. Latchman Y, Wood CR, Chernova T, Chaudhary D, Borde M, Chernova I, et al. Pd-L2 Is a Second Ligand for PD-1 and Inhibits T Cell Activation. Nat Immunol (2001) 2(3):261–8. doi: 10.1038/85330
78. Zhai L, Lauing KL, Chang AL, Dey M, Qian J, Cheng Y, et al. The Role of IDO in Brain Tumor Immunotherapy. J Neuro Oncol (2015) 123(3):395–403. doi: 10.1007/s11060-014-1687-8
79. Chang AL, Miska J, Wainwright DA, Dey M, Rivetta CV, Yu D, et al. Ccl2 Produced by the Glioma Microenvironment Is Essential for the Recruitment of Regulatory T Cells and Myeloid-Derived Suppressor Cells. Cancer Res (2016) 76(19):5671–82. doi: 10.1158/0008-5472.CAN-16-0144
80. Thomas DA, Massague J. TGF-Beta Directly Targets Cytotoxic T Cell Functions During Tumor Evasion of Immune Surveillance. Cancer Cell (2005) 8(5):369–80. doi: 10.1016/j.ccr.2005.10.012
81. Ren B, Cam H, Takahashi Y, Volkert T, Terragni J, Young RA, et al. E2F Integrates Cell Cycle Progression With DNA Repair, Replication, and G(2)/M Checkpoints. Genes Dev (2002) 16(2):245–56. doi: 10.1101/gad.949802
82. Emanuele MJ, Enrico TP, Mouery RD, Wasserman D, Nachum S, Tzur A. Complex Cartography: Regulation of E2F Transcription Factors by Cyclin F and Ubiquitin. Trends Cell Biol (2020) 30(8):640–52. doi: 10.1016/j.tcb.2020.05.002
83. Mechali M. Eukaryotic DNA Replication Origins: Many Choices for Appropriate Answers. Nat Rev Mol Cell Biol (2010) 11(10):728–38. doi: 10.1038/nrm2976
84. Zeman MK, Cimprich KA. Causes and Consequences of Replication Stress. Nat Cell Biol (2014) 16(1):2–9. doi: 10.1038/ncb2897
85. Ringelhan M, Pfister D, O’Connor T, Pikarsky E, Heikenwalder M. The Immunology of Hepatocellular Carcinoma. Nat Immunol (2018) 19(3):222–32. doi: 10.1038/s41590-018-0044-z
86. Miyamoto S. Nuclear Initiated NF-kappaB Signaling: NEMO and ATM Take Center Stage. Cell Res (2011) 21(1):116–30. doi: 10.1038/cr.2010.179
87. Sakamoto K, Hikiba Y, Nakagawa H, Hirata Y, Hayakawa Y, Kinoshita H, et al. Promotion of DNA Repair by Nuclear IKKbeta Phosphorylation of ATM in Response to Genotoxic Stimuli. Oncogene (2013) 32(14):1854–62. doi: 10.1038/onc.2012.192
88. Liu B, Sun L, Liu Q, Gong C, Yao Y, Lv X, et al. A Cytoplasmic NF-kappaB Interacting Long Noncoding RNA Blocks IkappaB Phosphorylation and Suppresses Breast Cancer Metastasis. Cancer Cell (2015) 27(3):370–81. doi: 10.1016/j.ccell.2015.02.004
89. Li Y, Lin Z, Chen B, Chen S, Jiang Z, Zhou T, et al. Ezrin/NF-Kb Activation Regulates Epithelial- Mesenchymal Transition Induced by EGF and Promotes Metastasis of Colorectal Cancer. Biomed Pharmacother (2017) 92:140–8. doi: 10.1016/j.biopha.2017.05.058
90. Suarez-Carmona M, Lesage J, Cataldo D, Gilles C. EMT and Inflammation: Inseparable Actors of Cancer Progression. Mol Oncol (2017) 11(7):805–23. doi: 10.1002/1878-0261.12095
91. Pastushenko I, Blanpain C. Emt Transition States During Tumor Progression and Metastasis. Trends Cell Biol (2019) 29(3):212–26. doi: 10.1016/j.tcb.2018.12.001
92. Wend P, Wend K, Krum SA, Miranda-Carboni GA. The Role of WNT10B in Physiology and Disease. Acta Physiol (2012) 204(1):34–51. doi: 10.1111/j.1748-1716.2011.02296.x
93. Recouvreux MV, Moldenhauer MR, Galenkamp KMO, Jung M, James B, Zhang Y, et al. Glutamine Depletion Regulates Slug to Promote EMT and Metastasis in Pancreatic Cancer. J Exp Med (2020) 217(9):e20200388. doi: 10.1084/jem.20200388
94. Steinbichler TB, Dudas J, Ingruber J, Glueckert R, Sprung S, Fleischer F, et al. Slug is A Surrogate Marker of Epithelial to Mesenchymal Transition (EMT) in Head and Neck Cancer. J Clin Med (2020) 9(7):2061. doi: 10.3390/jcm9072061
95. Lieber MR. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem (2010) 79:181–211. doi: 10.1146/annurev.biochem.052308.093131
96. Lim YC, Chia SY, Jin S, Han W, Ding C, Sun L. Dynamic DNA Methylation Landscape Defines Brown and White Cell Specificity During Adipogenesis. Mol Metab (2016) 5(10):1033–41. doi: 10.1016/j.molmet.2016.08.006
97. Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, et al. The Harmonizome: A Collection of Processed Datasets Gathered to Serve and Mine Knowledge About Genes and Proteins. Database J Biol Database Curation (2016) 2016:baw100. doi: 10.1093/database/baw100
98. Di W, Zhang W, Zhu B, Li X, Tang Q, Zhou Y. Colorectal Cancer Prompted Adipose Tissue Browning and Cancer Cachexia Through Transferring Exosomal Mir-146b-5p. J Cell Physiol (2020) 236(7):5399–10. doi: 10.1002/jcp.30245
99. Fu T, Ji X, Bu Z, Zhang J, Wu X, Zong X, et al. Identification of Key Long non-Coding RNAs in Gastric Adenocarcinoma. Cancer Biomarkers Sec A Dis Markers (2020) 27(4):541–53. doi: 10.3233/CBM-192389
100. Gabellini D, Colaluca IN, Vodermaier HC, Biamonti G, Giacca M, Falaschi A, et al. Early Mitotic Degradation of the Homeoprotein HOXC10 Is Potentially Linked to Cell Cycle Progression. EMBO J (2003) 22(14):3715–24. doi: 10.1093/emboj/cdg340
101. Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, et al. A DNA Methylation Reader Complex That Enhances Gene Transcription. Science (2018) 362(6419):1182–6. doi: 10.1126/science.aar7854
102. Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The Regulation and Functions of DNA and RNA G-Quadruplexes. Nat Rev Mol Cell Biol (2020) 21(8):459–74. doi: 10.1038/s41580-020-0236-x
103. Zhang X, Zhao B, Yan T, Hao A, Gao Y, Li D, et al. G-Quadruplex Structures At the Promoter of HOXC10 Regulate its Expression. Biochim Biophys Acta Gene Regul Mech (2018) 1861(11):1018–28. doi: 10.1016/j.bbagrm.2018.09.004
104. Laugesen A, Hojfeldt JW, Helin K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol Cell (2019) 74(1):8–18. doi: 10.1016/j.molcel.2019.03.011
105. Schuettengruber B, Bourbon HM, Di Croce L, Cavalli G. Genome Regulation by Polycomb and Trithorax: 70 Years and Counting. Cell (2017) 171(1):34–57. doi: 10.1016/j.cell.2017.08.002
106. Bartel DP. MicroRNAs: Genomics, Biogenesis, Mechanism, and Function. Cell (2004) 116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5
107. Faghihi MA, Wahlestedt C. Regulatory Roles of Natural Antisense Transcripts. Nat Rev Mol Cell Biol (2009) 10(9):637–43. doi: 10.1038/nrm2738
108. Duboule D. Vertebrate Hox Genes and Proliferation: An Alternative Pathway to Homeosis? Curr Opin Genet Dev (1995) 5(4):525–8. doi: 10.1016/0959-437x(95)90058-o
Keywords: HOXC10, tumorigenesis, metastasis, drug resistance, expression regulation
Citation: Fang J, Wang J, Yu L and Xu W (2021) Role of HOXC10 in Cancer. Front. Oncol. 11:684021. doi: 10.3389/fonc.2021.684021
Received: 22 March 2021; Accepted: 04 May 2021;
Published: 25 May 2021.
Edited by:
Shilpa S. Dhar, University of Texas MD Anderson Cancer Center, United StatesReviewed by:
Zhenjia Wang, University of Virginia, United StatesBruce Boman, Christiana Care Health Center, United States
Copyright © 2021 Fang, Wang, Yu and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenxia Xu, xuwenxia@zju.edu.cn; Liangliang Yu, ydyy30383@163.com
†These authors have contributed equally to this work and share first authorship
 Jinyong Fang
Jinyong Fang Jianjun Wang2†
Jianjun Wang2† Wenxia Xu
Wenxia Xu